Introduction
Esophageal cancer (ESCA), as one of the most lethal
malignant tumors of the digestive tract, exhibits rapid
proliferative ability and early metastatic potential, and its
incidence is gradually increasing worldwide (1,2).
ESCA presents as two primary histological types: i) Esophageal
squamous cell carcinoma (ESCC) and; ii) esophageal adenocarcinoma
(3). ESCC, the predominant
histological type, accounts for >80% of all cases of ESCA
(4), particularly in the Henan
province of China (5,6). Despite significant progress in the
molecular targeted therapy and other strategies of management, the
5-year survival rate of patients remains low (15-25%) (7,8).
Therefore, there remains an urgent need to further understand the
pathogenesis of ESCC and to identify early biomarkers for the
diagnosis and therapy of ESCC, which will provide novel
opportunities for the treatment of patients with ESCC.
Long non-coding RNAs (lncRNAs) are a class of
transcripts >200 nucleotides in length without protein coding
potential (9). Increasing
evidence has demonstrated that lncRNAs are closely implicated in
numerous tumor phenotypes through a variety of molecular
mechanisms, including transcriptional regulation, epigenetic
modification, microRNA (miRNA/miR) sponging and RNA decay (10-12). With the rapid development of
sequencing and molecular biology tech- nologies, multiple
ESCC-related lncRNAs have been identified, including lncRNA H19
(13), cancer susceptibility 9
(14,15) and fragile X mental retardation
1-antisense RNA 1 (16), which
have been widely implicated in the development and progression of
ESCC via various mechanisms. Current data suggest that lncRNAs may
constitute novel, promising biomarkers and therapeutic targets for
patients with ESCC.
Long intergenic non-protein coding RNA 514
(LINC00514) is localized at chromosome 16p13.3, and it is 3,221-bp
long (17). LINC00514 has been
reported to be closely associated with the development and
progression of tumors, including osteosarcoma (18), papillary thyroid cancer (19) and neuroendocrine prostate cancer
(20). Sphingosine kinase 1
(SPHK1) is localized at chromosome 17q25.1, and it has been shown
to participate in lipogenesis and tumor progression in a variety of
tumors (21-23). However, to the best of our
knowledge, the roles and molecular mechanisms of LINC00514 in ESCC
remain to be investigated.
The present study investigated the expression and
correlation of LINC00514 and SPHK1 in ESCC tissues and cells, and
determined the clinical value of LINC000514 and SPHK1 in
Tumor-Node-Metastasis (TNM) stage, lymph node metastasis and
prognosis of patients with ESCC. Functionally, the roles of
LINC00514 in cell proliferation, invasion and lipogenesis were
explored. Mechanistically, the present study demonstrated the
competing endogenous (ceRNA)-mediated mechanism of LINC00514,
through its ability to absorb miR-378a-5p to promote SPHK1
expression and lipogenesis in ESCC cells, further promoting the
proliferation and invasion of these cells. Taken together, the
present data highlighted the role of LINC00514 in ESCC
proliferation, invasion and lipogenesis, and identified a novel
lipogenesis-related pathway based on LINC00514 and
LINC00514/miR-378a-5p/SHPK1 signaling in ESCC cells. Therefore, the
LINC00514/miR-378a-5p/SHPK1 signaling axis identified in the
current study may be a novel and promising therapeutic target for
patients with ESCC.
Materials and methods
Tissue samples
A total of 85 ESCC and corresponding normal tissues
were obtained from The First Affiliated Hospital of Zhengzhou
University (Zhengzhou, China).
The age range of all the patients was 41-85 years,
and the median age was 64 years old. The clinicopathological
features of the patients with ESCC enrolled in the present study
were as follows: i) Sex: Male, 55 cases vs. female, 30 cases; ii)
age: <60 years old, 38 cases vs. ≥60 years old, 47 cases; iii)
smoking history: Smoker, 41 cases vs. non-smoker, 44 cases; iv)
alcohol consumption: Drinker, 49 cases vs. non-drinker, 36 cases;
v) tumor diameter, <4 cm, 51 cases vs. ≥4 cm, 34 cases;
differentiation degree: High/moderate differentiation, 52 cases vs.
poor differentiation, 33 cases; vi) TNM stage (24): I-II, 55 cases vs. III-IV, 30
cases; and vii) metastasis status: Lymph node metastasis, 28 cases
vs. absence of lymph node metastasis, 57 cases.
Tumor tissue samples were confirmed as ESCC using
hematoxylin and eosin staining by experienced pathologists at The
First Affiliated Hospital of Zhengzhou University as routine.
Informed consent for the use of all samples in the present study
was obtained from each patient, and the study was approved by the
Research and Ethics Committee of The First Affiliated Hospital of
Zhengzhou University.
Public database analysis
starBase version 3.0 online soft- ware based on The
Cancer Genome Atlas (TCGA) database was used to investigate the
expression levels of LINC00514, SPHK1 and miR-378a-5p in patients
with ESCA and healthy individuals (starbase.sysu.edu.cn/panCancer.php), and the P-value
of differential expression was directly obtained from the website.
Gene Expression Profiling Interactive Analysis (GEPIA) was employed
to determine SPHK1 expression. The Gene Expression Omnibus (GEO)
dataset, GSE111011, was used to assess the expression of SPHK1 in 7
cases of ESCC and paired normal samples, whereas the GSE43732 was
used to analyze the expression of miR-378a-5p in 119 cases of ESCC
and paired normal samples. LncBase Predicted v.2
(carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted)
(25) was used to predict the
binding site of LINC00514 and miR-378a-5p. TargetScan (targetscan.org/vert_71/) and miRDB (mirdb.org/) were employed to predict the down- stream
target genes of miR-378a-5p.
Cell lines and culture
Human ESCC cell lines (Eca109, KYSE150, KYSE30,
KYSE450 and KYSE70) and normal esophageal epithelial Het-1A cells
were obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences, and were maintained in RPMI-1640
medium supplemented with 10% FBS (Gibco; Invitrogen; Thermo Fisher
Scientific, Inc.) in a humidified incubator with 5% CO2.
All cells were routinely tested and verified as negative for
mycoplasma.
Cell transfection
LINC00514 small interfering RNA (siRNA)s 1, 2 and 3
were designed and chemically synthesized by Guangzhou RiboBio Co.,
Ltd. miR-378-5p mimic, negative control (NC) mimic, miR-378a-5p
inhibitor and NC inhibitor were purchased from Shanghai GenePharma
Co., Ltd. SPHK1 siRNA was purchased from Santa Cruz Biotechnology,
Inc. pcDNA3.1 and pcDNA3.1-LINC00514/SPHK1 were constructed by
TsingKe Biological Technology.
The aforementioned constructs were transfected into
KYSE150 and KYSE30 cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
Cell Counting Kit-8 (CCK-8) assay
The proliferation of ESCC KYSE150 and KYSE30 cells
was assayed in triplicate according to the manufacturer's protocol.
Briefly, KYSE150 and KYSE30 cells (~2×103 cells/well)
were seeded into 96-well plates. At the time of measurement, CCK-8
reagent (Beyotime Institute of Biotechnology) was added to the
wells, and the absorbance value at 450 nm was determined using a
microplate reader (Thermo Fisher Scientific, Inc.) to evaluate the
proliferative ability of KYSE150 and KYSE30 cells.
Transwell assay
Cell invasion was determined using Transwell
chambers coated with Matrigel (BD Biosciences). Briefly, the
transfected KYSE150 and KYSE30 cells (1×105 cells) were
seeded in the upper layer of the chamber, whereas the bottom
chamber contained 20% FBS. A total of 48 h after seeding, the cells
that had invaded were fixed using methanol for 30 min at room
temperature, and then stained with 0.1% of crystal violet for 15
min at room temperature. Finally, the number of invasive cells was
determined using a light microscope (Leica Microsystems, Inc.)
(magnification, ×200).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. For mRNA analysis, RT-qPCR was
performed using a Quant One Step qRT-PCR kit (SYBR Green; cat. no.
FP303; Tiangen Biotech Co., Ltd.) on an ABI 7500 series PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) using the
following specific primers: LINC00514 forward 5'-CAT CCA GAT TTG
GGC CCC TT-3' and reverse, 5'-CAT GCC TGA CCA CGA ATC CT-3'
(product length, 231 bp); SPHK1 forward, 5'-CAG TGG TCG GTT GCG
GAC-3' and reverse, 5'-GCA CGT GAC TCC GGA AGA G-3' (product
length, 150 bp); and GAPDH forward, 5'-TCA TCA TCT CTG CCC CCT
CT-3' and reverse 5'-GAT GGC ATG GAC TGT GGT CA-3' (product length,
188 bp). The thermocycling conditions were: 95°C 30 sec; followed
by 30 cycles of 95°C for 5 sec, 50°C for 30 sec and 72°C for 30
sec. The results were normalized to GAPDH using the
2−∆∆Cq method (26).
For miR-378a-5p analysis, total RNA was reverse
transcribed using a miRcute Plus miRNA First-Strand cDNA kit
according to the manufacturer's protocol (Tiangen Biotech Co.,
Ltd.). qPCR amplification for miR-378a-5p was performed using the
miRcute Plus miRNA qPCR kit (SYBR Green) (Tiangen Biotech Co.,
Ltd.) and the following specific forward primers along with the
reverse primers from the kit: miR-378a-5p forward, 5'-CTC CTG ACT
CCA GGT CCT G TG T-3' and U6 forward, 5'-CTC GCT TCG GCA GCA CA-3'.
The thermocycling conditions were: 95°C for 30 sec; followed by 40
cycles of 95°C for 5 sec and 60°C for 30 sec. The results were
normalized to GAPDH using the 2−∆∆Cq method.
Subcellular fractionation
A Cell Nucleus and Cytoplasm RNA Isolation kit
(Shaanxi Yuan Beibei Biological Technology Co., Ltd.) was used to
extract the nuclear and cytoplasmic RNA according to the
manufacturer's instructions. Briefly, KYSE150 and KYSE30 cells
(3×107 cells) were harvested by centrifugation at 2,000
× g for 10 min at room temperature, and washed with ice-cold PBS.
Cells were then resuspended in 300 µl PBS and centrifuged at
3,000 × g for 10 min at 4°C. The supernatant was obtained as the
cytoplasmic fraction. The precipitate was collected and resuspended
in 300 µl PBS and then centrifuged at 3,000 × g for 10 min at 4°C.
The precipitate was collected as the nuclear fraction.
Total RNA was isolated using TRIzol®.
Finally, the cytoplasmic and nuclear RNA was reverse transcribed to
cDNA using a PrimeScript™ RT kit with gDNA Eraser (cat. no. RR047A;
Takara Bio, Inc.) according to the manufacturer's protocol, were
further investigated using RT-qPCR utilizing LINC00514, U6 and
GAPDH-specific primers as described above. GAPDH and U6 were used
as cytoplasmic and nuclear controls, respectively.
Fluorescence in situ hybridization (FISH)
assay
LINC00514 probe (NR_033861.1, 5'-AAC GGA CCG GGA ACC
CAG CCA GGT CGG GGC CGA AGG GGC TGG GGT GGC TGGGGGAG A-3') was
synthesized and labeled using cyanine 3 (Cy3) by Shanghai
GenePharma Co., Ltd. FISH assays were performed using an RNA FISH
kit (Shanghai GenePharma, Co., Ltd.) according to manufacturer's
instructions. Briefly, KYSE150 and KYSE30 cells were grown in
24-well plates with glass cover slips for 24 h. After
immobilization using 4% paraformaldehyde for 15 min at room
temperature and permeabilization using Triton X-100 for 15 min at
room temperature, KYSE150 and KYSE30 cells were hybridized with 20
µM Cy3-labeled LINC00514 probe, and DAPI was used to stain the
nuclei of KYSE150 and KYSE30 cells for 10 min in dark at room
temperature. Images were observed using a fluorescence microscope
(magnification, ×200).
Dual luciferase reporter assay
Dual luciferase reporter assays were performed to
determine the interaction of miR-378a-5p with LINC00514 or SPHK1 in
KYSE150 and KYSE30 cells. Recombinant vectors,
pmirGLO-LINC00514-wild type (WT) and pmirGLO-LINC00514-mutant
(MUT), as well as pmirGLO-SPHK1-WT and pmirGLO-SPHK1-MUT (all from
TsingKe Biological Technology) along with miR-378a-5p mimic and NC
mimic were co-transfected into KYSE150 and KYSE30 cells using
Lipofectamine® 2000. Luciferase activity was determined
using the Dual-Luciferase Reporter assay system (Promega
Corporation) 48 h after transfection according to the
manufacturer's protocol.
RNA immunoprecipitation (RIP) assay
RIP assay was performed in KYSE150 and KYSE30 cells
using an RNA-Binding Protein Immunoprecipitation kit
(MilliporeSigma) according to the manufacturer's protocol. Briefly,
RIP lysates were prepared from KYSE150 and KYSE30 cells transfected
with miR-378a-5p mimic or NC mimic using a Magna RIP™ RNA-binding
Protein Immunoprecipitation kit (cat. no. 17-700; MilliporeSigma)
by centrifugation at 1,000 × g for 5 min at 4°C. Subsequently, 100
µl lysate was subjected to IP using 5 µl normal mouse IgG or
5 µl anti-argonaute (Ago)2 antibody in the kit. The presence
of LINC00514 or SPHK1 and miR-378a-5p enriched on beads was
determined using RT-qPCR with the corresponding specific
primers.
Western blotting
Total protein was extracted from ESCC cells using
RIPA lysis buffer (Beijing Solarbio Science & Technology Co.,
Ltd.), and the protein concentration was determined using a
Bradford assay. The proteins (100 µg/lane) were loaded on
10% SDS-gels, resolved using SDS-PAGE and then transferred to PVDF
membranes (MilliporeSigma). After blocking with skimmed milk for 2
h at room temperature, the membranes were incubated with primary
antibodies against SPHK1 (cat. no. ab109522; 1:1,000), fatty acid
synthetase (FASN) (cat. no. ab128870; 1:10,000), acetyl-CoA
carboxylase α (ACACA) (cat. no. ab109368; 1:1,000), stearoyl-CoA
desaturase 1 (SCD1) (cat. no. ab236868; 1:1,000) and β-actin (cat.
no. ab115777; 1:200) (Abcam) overnight at room temperature.
Subsequently, a secondary horseradish peroxidase-conjugated
AffiniPure goat anti-rabbit IgG (AmyJet Scientific Inc.; cat. no.
111-035-003; 1:20,000) was added to the PVDF membranes. Finally,
enhanced chemiluminescence reagent (Beyotime Institute of
Biotechnology) was used to visualize the signals, and the
quantification of the blots was analyzed using ImageJ version 1.8.0
(National Institutes of Health).
Statistical analysis
GraphPad Prism version 8.0 (GraphPad Software, Inc.)
was used to analyze the experimental data, which are presented as
the mean ± standard deviation. The association between LINC00514,
SPHK1, miR-378a-5p and the clinicopathological features were
investigated using a χ2 test. Survival analysis was
performed using a log-rank test. Spearman's rank correlation
analysis was used to analyze the non-parametric data, and Pearson's
correlation coefficient analysis was used to examine the parametric
data. For the matched samples, the data was analyzed using a
Wilcoxon signed rank test, and for non-matched samples, the data
was compared using a Mann-Whitney U test. Comparisons between two
groups were determined using a Student's t-test, whereas
comparisons between ≥3 groups were analyzed using a one-way ANOVA
with a Dunnett's or Tukey's post hoc test to assess the difference
between two groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Upregulated expression of LINC00514 in
ESCC tissues is predictive of a poor prognosis
To explore the expression pattern of LINC00514 in
ESCA tissues, starBase and RT-qPCR assays were used to determine
the expression of LINC00514 in ESCA tissues. Data from starBase
revealed that ESCA tissues exhibited higher expression levels of
LINC00514 compared with those of normal samples (P<0.05;
Fig. 1A). Furthermore, RT-qPCR
analysis showed that the expression of LINC00514 in the 85 ESCC
tissues was also higher compared with that of the paired normal
samples (P<0.0001; Fig. 1B).
Notably, patients with ESCC and high LINC00514 expression exhibited
a lower survival rate than those with low LINC00514 expression
(Fig. 1C).
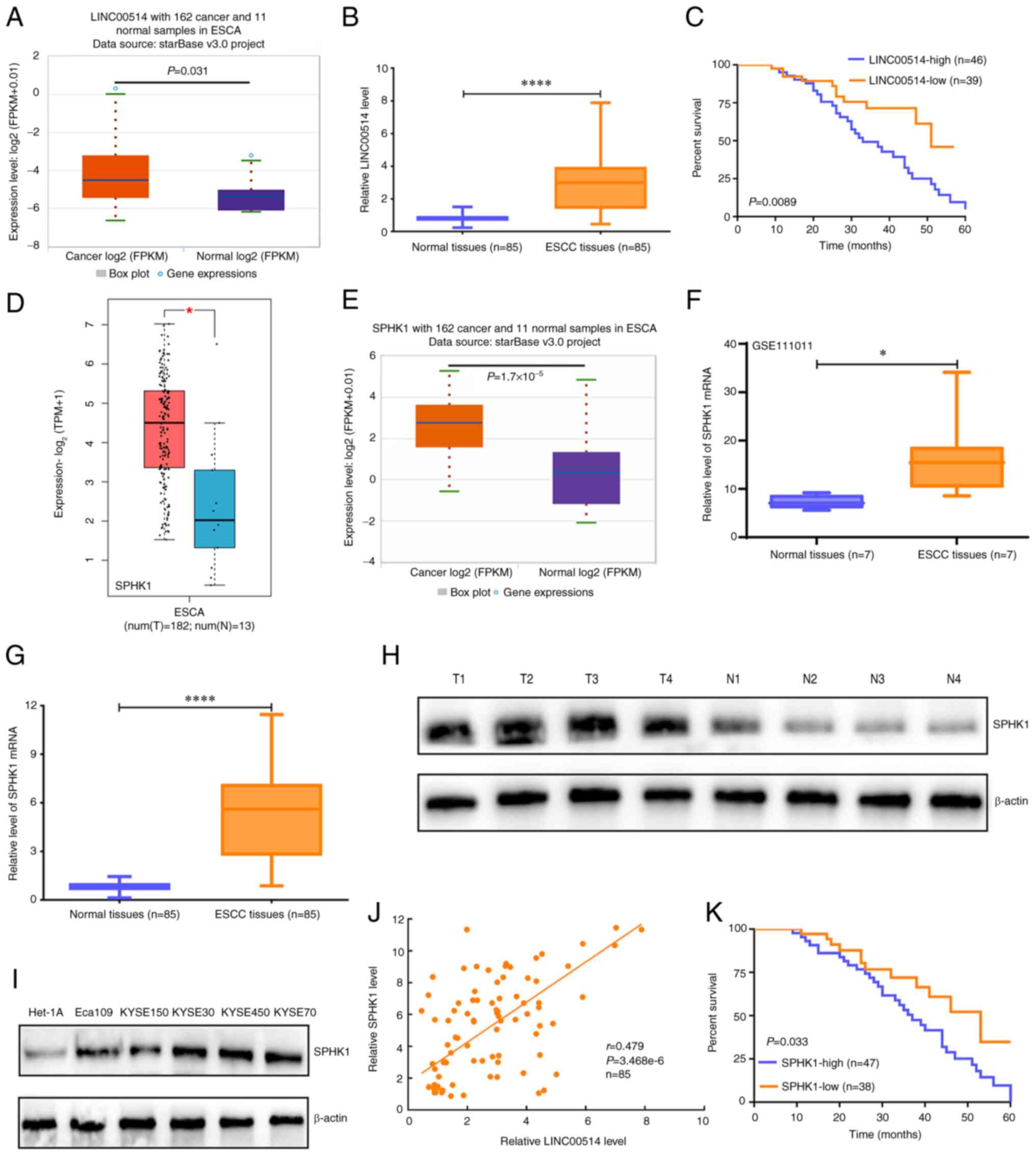 | Figure 1Expression and prognostic value of
LINC00514 and SPHK1 in ESCC. (A) starBase was used to investigate
the expression of LINC00514 in 162 esophageal cancer samples and 11
normal samples. (B) Reverse transcription-quantitative PCR was used
to evaluate the expression levels of LINC00514 in 85 ESCC and
paired normal samples using a Wilcoxon signed rank test.
****P<0.0001. (C) A log-rank test was used to analyze
the effects of LINC00514 expression on the survival rate of
patients with ESCC. (D) Gene Expression Profiling Interactive
Analysis online software analysis of SPHK1 level in 182 ESCA and 13
normal samples. (E) starBase analysis was performed to determine
the expression of SPHK1 in 162 ESCA and 11 normal samples. (F) The
GSE111011 Gene Expression Omnibus dataset was used to detect the
levels of SPHK1 in seven ESCC and paired normal samples using
paired a Student's t-test. *P<0.05. (G) Reverse
transcription-quantitative PCR assay was used to determine the
SPHK1 levels in 85 ESCC and paired normal samples, and the data was
analyzed using a Wilcoxon signed rank test. (H) SPHK1 protein
expression in four randomly selected paired tissues. β-actin was
used as the loading control. (I) SPHK1 protein expression in ESCC
cell lines (Eca109, KYSE150, KYSE30, KYSE450 and KYSE70) and the
normal esophageal epithelial cell line Het-1A. β-actin was used as
a loading control. (J) Spearman's rank correlation analysis of the
correlation between LINC00514 and SPHK1 expression in the 85 ESCC
and paired normal samples. (K) Log-rank analysis of the effect of
SPHK1 expression on the survival rate of patients with ESCC.
****P<0.0001. LINC00514, long intergenic
nonprotein-coding RNA 00514; ESCC, esophageal squamous cell
carcinoma; SPHK1, sphingosine kinase 1; ESCA, esophageal cancer; T,
tumor; N, normal. |
In addition, it was found that LINC00514 was
frequently upregulated in 13 other tumor types and downregulated in
two other tumor types, including kidney chromophobe (KICH) and lung
squamous cell carcinoma (Fig.
S1). These data suggest that LINC00514 is closely associated
with tumor development and progression in ESCC, and it may be a
novel prognostic predictor for patients with ESCC.
Upregulated expression of SPHK1 in ESCC
tissues
To analyze the expression pattern of SPHK1 in ESCA
tissues, data from TCGA and GEO, as well as from the RT-qPCR
analysis were used to determine the expression of SPHK1 in ESCA
tissues. GEPIA online software showed that SPHK1 expression in ESCA
tissues was significantly higher than that in normal tissues
(P<0.05; Fig. 1D), which was
consistent with the data from starBase online software and the GEO
dataset, GSE111011 (Fig. 1E and
F).
Further analysis of the 85 cases of ESCC and
corresponding normal tissues demonstrated that SPHK1 expression in
ESCC tissues was markedly higher than that in paired normal tissues
(P<0.0001; Fig. 1G), which was
further validated in four randomly selected ESCC and paired normal
tissues (Fig. 1H) as well as in a
number of ESCC cell lines (Fig.
1I).
To further elucidate the correlations between
LINC00514 and SPHK1, Pearson's correlation analysis was used, which
revealed that LINC00514 expression exhibited a positive correlation
with SPHK1 expression in the 85 ESCC tissues (Fig. 1J). Importantly, patients with ESCC
who had a high SPHK1 expression level exhibited reduced survival
rates compared with those of patients with ESCC who had low SPHK1
expression (Fig. 1K). In
addition, SPHK1 expression levels were higher in 13 other tumor
types compared with those in the respective paired normal tissues,
whereas it was only downregulated in KICH (Fig. S2). These findings suggest that
SPHK1 may function as an oncogene in different tumor types,
particularly in ESCC, and may be a novel prognostic predictor for
patients with ESCC.
Association between LINC00514 and SPHK1
expression levels with the clinicopathological features in patients
with ESCC
To unveil the possible biological functions of
LINC00514 and SPHK1 in ESCC, the present study further investigated
the associations between the expression levels of LINC00514 and
SPHK1 and the clinicopathological features. It was found that the
expression of LINC00514 and SPHK1 was closely associated with TNM
stage and lymph node metastasis in patients with ESCC, but was not
associated with patients' sex, age, smoking history, alcohol
consumption, tumor diameter or degree of differentiation (Tables I and II). These findings suggest that
LINC00514 and SPHK1 may participate in the progression and
metastasis of ESCC.
 | Table IAssociations between LINC00514
expression and the clinicopathological features of the patients
with ESCC. |
Table I
Associations between LINC00514
expression and the clinicopathological features of the patients
with ESCC.
| Features | n | LINC00514
expression
| χ2 | P-value |
|---|
| + | − |
|---|
| Sex | | | | 2.172 | 0.141 |
| Male | 55 | 33 | 22 | | |
| Female | 30 | 13 | 17 | | |
| Age, years | | | | 1.261 | 0.262 |
| <60 | 38 | 18 | 20 | | |
| ≥60 | 47 | 28 | 19 | | |
| Smoking | | | | 1.500 | 0.221 |
| Yes | 41 | 25 | 16 | | |
| No | 44 | 21 | 23 | | |
| Drinking | | | | 1.196 | 0.274 |
| Yes | 49 | 29 | 20 | | |
| No | 36 | 17 | 19 | | |
| Tumor diameter,
cm | | | | 1.335 | 0.248 |
| <4 | 51 | 25 | 26 | | |
| ≥4 | 34 | 21 | 13 | | |
| Differentiation
degree | | | | 1.968 | 0.161 |
| High/moderate | 52 | 25 | 27 | | |
| Poor | 33 | 21 | 12 | | |
|
Tumor-Node-Metastasis stage | | | | 4.710 | 0.030a |
| I-II | 55 | 25 | 30 | | |
| III-IV | 30 | 21 | 9 | | |
| Lymph node
metastasis | | | | 5.039 | 0.025a |
| Yes | 28 | 20 | 8 | | |
| No | 57 | 26 | 31 | | |
 | Table IIAssociations of SPHK1 expression with
the clinicopathological features of patients with ESCC. |
Table II
Associations of SPHK1 expression with
the clinicopathological features of patients with ESCC.
| Features | n | SPHK1 expression
| χ2 | P-value |
|---|
| + | − |
|---|
| Sex | | | | 0.526 | 0.468 |
| Male | 55 | 32 | 23 | | |
| Female | 30 | 15 | 15 | | |
| Age, years | | | | 1.746 | 0.186 |
| <60 | 38 | 18 | 20 | | |
| ≥60 | 47 | 29 | 18 | | |
| Smoking | | | | 0.337 | 0.562 |
| Yes | 41 | 24 | 17 | | |
| No | 44 | 23 | 21 | | |
| Drinking | | | | 1.646 | 0.200 |
| Yes | 49 | 30 | 19 | | |
| No | 36 | 17 | 19 | | |
| Tumor diameter,
cm | | | | 2.031 | 0.154 |
| <4 | 51 | 25 | 26 | | |
| ≥4 | 34 | 22 | 12 | | |
| Differentiation
degree | | | | 0.616 | 0.433 |
| High/moderate | 52 | 27 | 25 | | |
| Poor | 33 | 20 | 13 | | |
|
Tumor-Node-Metastases stage | | | | 8.567 | 0.003a |
| I-II | 55 | 24 | 31 | | |
| III-IV | 30 | 23 | 7 | | |
| Lymph node
metastasis | | | | 6.559 | 0.010a |
| Yes | 28 | 21 | 7 | | |
| No | 57 | 26 | 31 | | |
LINC00514 promotes the proliferation and
invasion of ESCC cells by regulating lipogenesis-related
proteins
In order to reveal the biological functions of
LINC00514 in ESCC, RT-qPCR was used to detect the expression of
LINC00514 in a number of ESCC cell lines. It was found that all
ESCC cell lines exhibited higher levels of LINC00514 expression
compared with the Het-1A cells, a normal esophageal epithelial cell
line (all P<0.0001; Fig. 2A),
which was consistent with the data derived from the ESCC
tissues.
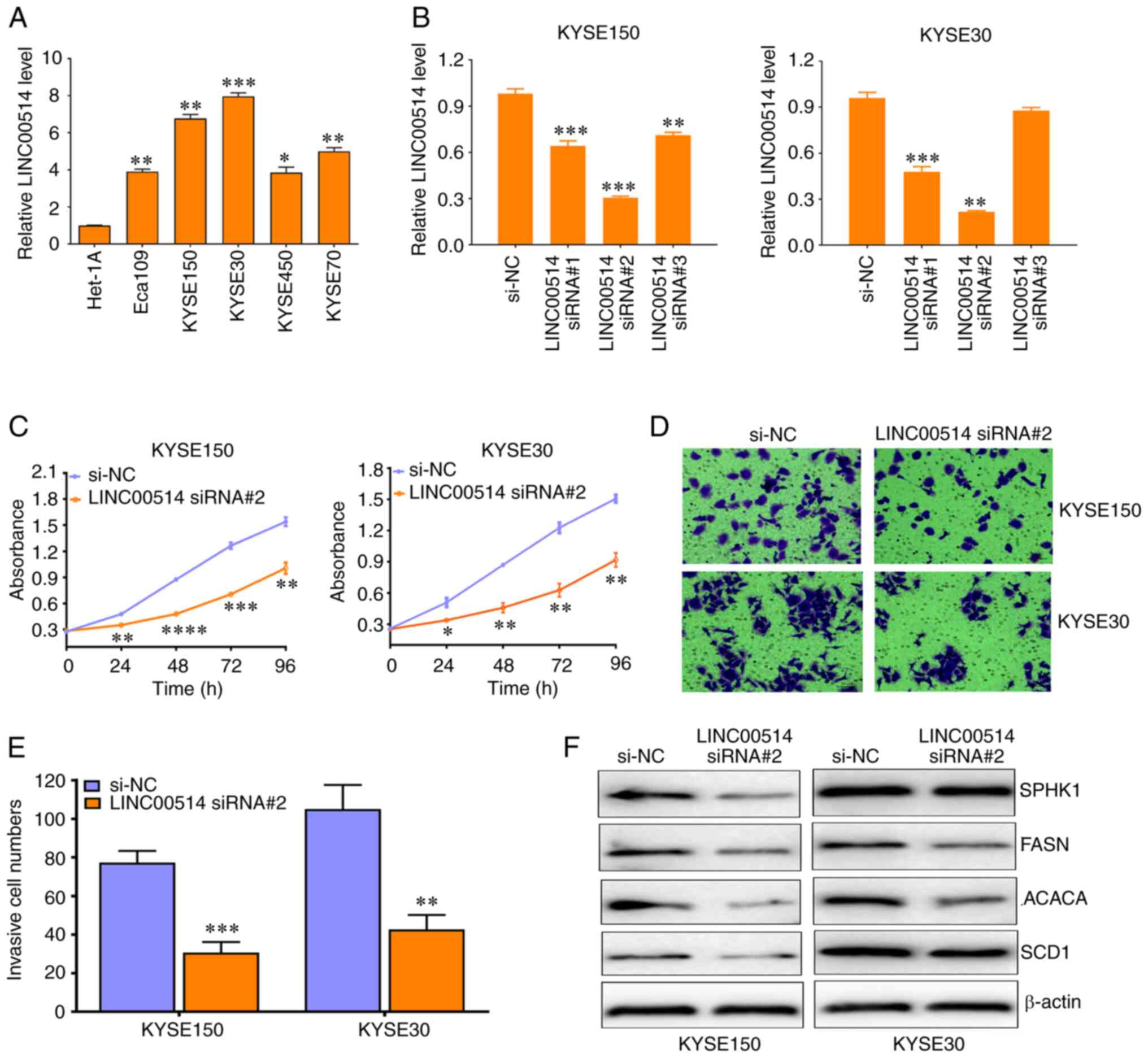 | Figure 2LINC00514 Knockdown suppresses the
proliferation, invasion and expression of lipogenesis-related
proteins in ESCC cells. (A) Reverse transcription-quantitative PCR
assay of LINC00514 expression in various ESCC cell lines, including
Eca109, KYSE150, KYSE30, KYSE450 and KYSE70 as well as in the
normal esophageal epithelial Het-1A cell line. Data were compared
using an ANOVA followed by a post hoc Dunnett's test.
*P<0.05, **P<0.01,
***P<0.001 vs. Het-1A. (B) Three siRNAs specific for
LINC00514 significantly downregulated the expression of LINC00514
in KYSE150 and KYSE30 cells. Data were compared using an ANOVA
followed by a post hoc Dunnett's test. (C-E) LINC00514 siRNA2
suppressed the proliferation and invasion of KYSE150 and KYSE30
cells. (F) LINC00514 siRNA2 reduced the expression of
lipogenesis-related proteins (SPHK1, FASN, ACACA and SCD1) in
KYSE150 and KYSE30 cells. *P<0.05,
**P<0.01, ***P<0.001,
****P<0.0001 vs. si-NC group. LINC00514, long
intergenic nonprotein-coding RNA 00514; ESCC, esophageal squamous
cell carcinoma; SPHK1, sphingosine kinase 1; FASN, fatty acid
synthetase; ACACA, acetyl-CoA carboxylase α; SCD1, stearoyl-CoA
desaturase 1; siRNA, small interfering RNA. |
In addition, three siRNAs against LINC00514 markedly
downregulated the expression of LINC00514 in KYSE150 and KYSE30
cells, which harbored the highest levels of LINC00514, and
LINC00514 siRNA2 displayed the most effective interference efficacy
in these cell lines (Fig. 2B).
Functionally, knockdown of LINC00514 significantly suppressed the
proliferation of KYSE150 and KYSE30 cells after 24, 48, 72 and 96 h
(P<0.05; Fig. 2C), as well as
their invasive ability after 48 h (Fig. 2D and E). In addition, knockdown of
LINC00514 led to the downregulation of expression of
lipogenesis-related proteins, including SPHK1, FASN, ACACA and SCD1
(Fig. 2F). By contrast,
pcDNA3.1-LINC00514 significantly increased the expression of
LINC00514 in KYSE150 and KYSE30 cells (Fig. 3A), significantly increased cell
proliferation and invasion in KYSE150 and KYSE30 cells, and led to
an increase in the protein expression levels of SPHK1, FASN, ACACA
and SCD1 (Fig. 3B-E). These
findings suggest that LINC00514 may function as an oncogene by
affecting lipogenesis in ESCC cells.
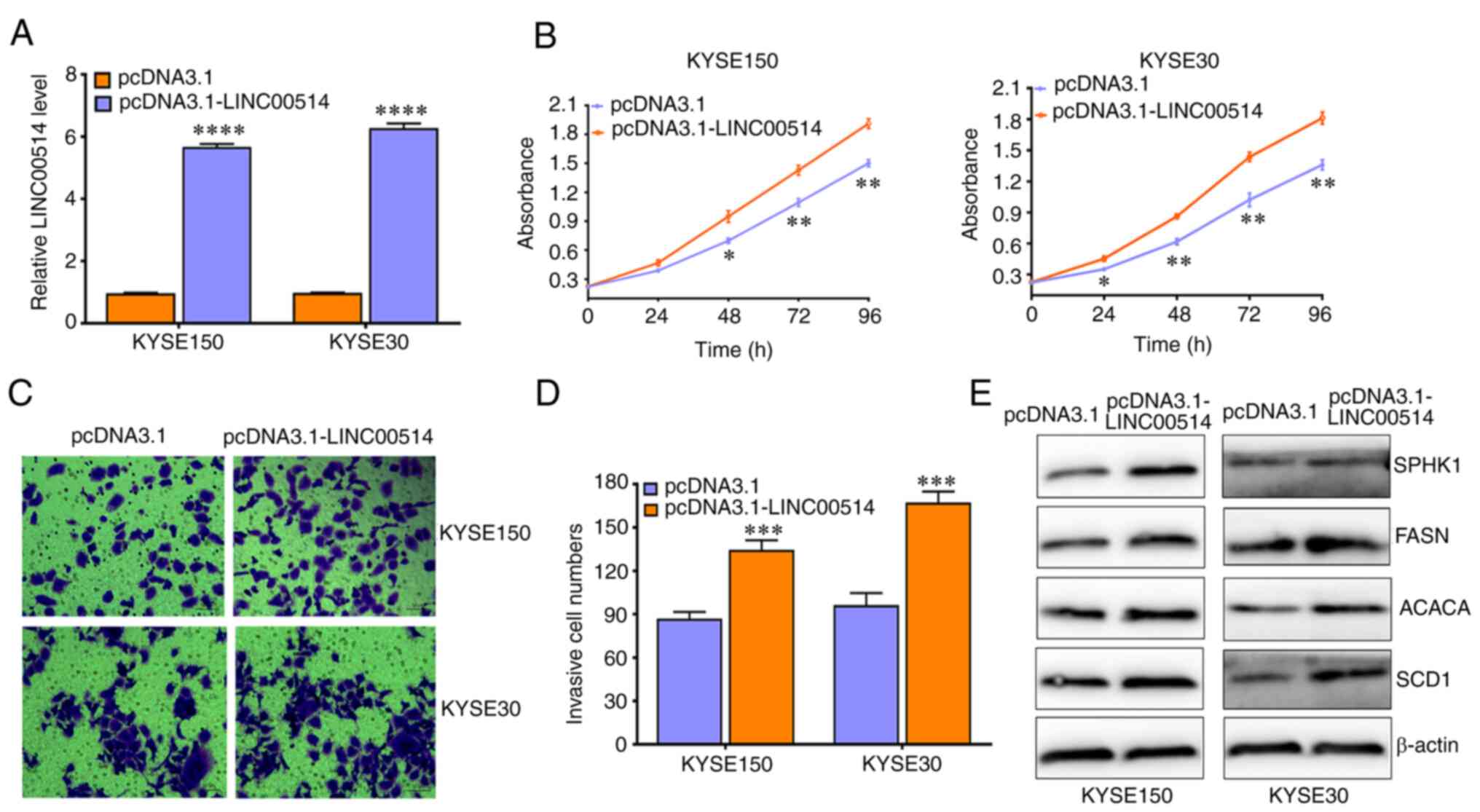 | Figure 3LINC00514 promotes the proliferation,
invasion and expression of lipogenesis-related proteins in ESCC
cells. (A) pcDNA3.1-LINC00514 transfection significantly
upregulated the expression of LINC00514 in KYSE150 and KYSE30
cells. (B-D) LINC00514 overexpression promoted the proliferation
and invasion of KYSE150 and KYSE30 cells. (E) LINC00514
overexpression promoted the expression of lipogenesis-related
proteins (SPHK1, FASN, ACACA and SCD1) in KYSE150 and KYSE30
cells.*P<0.05, **P<0.01,
***P<0.001, ****P<0.0001. LINC00514,
long intergenic nonprotein-coding RNA 00514; ESCC, esophageal
squamous cell carcinoma; SPHK1, sphingosine kinase 1; FASN, fatty
acid synthetase; ACACA, acetyl-CoA carboxylase α; SCD1,
stearoyl-CoA desaturase 1; siRNA, small interfering RNA. |
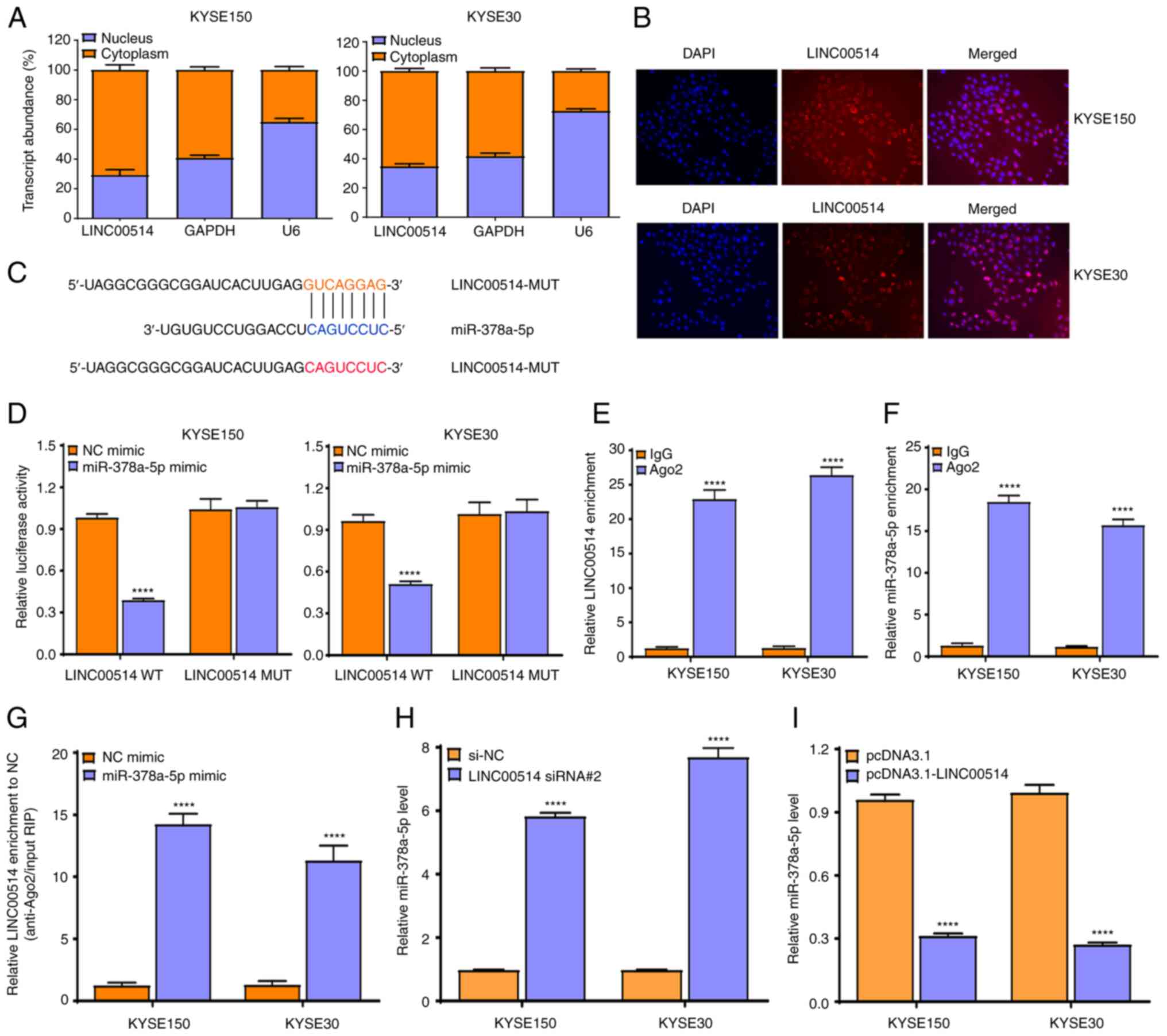
LINC00514 affects miR-378a-5p expression
by acting as a ceRNA
To further explore the possible molecular mechanisms
of LINC00514 in ESCC cells, a localization assay was performed
using RT-qPCR. It was found that LINC00514 was primarily localized
in the cytoplasm of KYSE150 and KYSE30 cells (Fig. 4A), which was validated using a
FISH assay (Fig. 4B), suggesting
that LINC00514 functions via a ceRNA-mediated mechanism in ESCC
cells.
Subsequently, LncBase Predicted v.2 which is
integrated into DIANA Tools was used to predict the possible miRNAs
that bound to LINC00514, and it was found that LINC00514 harbored
binding sites for miR-378a-5p (Fig.
4C).
To further confirm the interaction of LINC00514 with
miR-378a-5p, a dual-luciferase reporter assay was conducted to
investigate the binding of LINC00514 with miR-378a-5p. It was found
that the luciferase intensity was markedly reduced by
co-transfecting miR-378a-5p mimic and pmirGLO-LINC00514-WT vector
into KYSE150 and KYSE30 cells, but not by co-transfecting
miR-378a-5p mimic and pmirGLO-LINC00514-MUT vector (which lacks the
miR-378-5p binding site) (Fig.
4D).
To further validate this result, an Ago2-RIP assay
was performed, which revealed that endogenous LINC00514 and
miR-378a-5p were preferentially enriched in Ago2-RIP, compared with
the findings in control IgG-RIP (Fig.
4E and F). Importantly, LINC00514 enrichment was much higher in
the miR-378a-5p mimic group than that in the NC mimic group
(P<0.0001; Fig. 4G),
suggesting that LINC00514 and miR-378a-5p appeared in the same
RNA-induced silencing complex.
To verify the effects of LINC00514 knockdown or
over- expression on the expression of miR-378a-5p in ESCC cells,
RT-qPCR was performed, and it was found that LINC00514 depletion
notably enhanced the expression of miR-378a-5p (P<0.0001;
Fig. 4H), whereas LINC00514
overexpression markedly suppressed the expression of miR-378a-5p
(P<0.0001; Fig. 4I). These
findings suggest that LINC00514 directly affects the expression of
miR-378a-5p by acting as a ceRNA in ESCC cells.
Associations between miR-378a-5p
expression and clinicopathological features of patients with
ESCC
Considering the interaction of LINC00514 with
miR-378a-5p in ESCC cells, whether miR-378a-5p participated in the
development and progression of ESCC was next investigated.
miR-378a-5p was closely associated with TNM stage and lymph node
metastasis in patients with ESCC (P<0.01), but was not
associated with patients' sex, age, smoking habits, drinking
habits, tumor diameter or degree of differentiation (Table III). These findings suggest that
miR-378a-5p may participate in the progression and metastasis of
ESCC.
 | Table IIIAssociations of miR-378a-5p
expression with clinicopathological features of patients with
ESCC. |
Table III
Associations of miR-378a-5p
expression with clinicopathological features of patients with
ESCC.
| Features | n | LINC00514
expression
| χ2 | P-value |
|---|
| + | − |
|---|
| Sex | | | | 0.353 | 0.552 |
| Male | 55 | 22 | 33 | | |
| Female | 30 | 14 | 16 | | |
| Age, years | | | | 0.708 | 0.400 |
| <60 | 38 | 18 | 20 | | |
| ≥60 | 47 | 18 | 29 | | |
| Smoking | | | | 1.079 | 0.299 |
| Yes | 41 | 15 | 26 | | |
| No | 44 | 21 | 23 | | |
| Drinking | | | | 0.112 | 0.738 |
| Yes | 49 | 20 | 29 | | |
| No | 36 | 16 | 20 | | |
| Tumor diameter,
cm | | | | 2.321 | 0.128 |
| <4 | 51 | 25 | 26 | | |
| ≥4 | 34 | 11 | 23 | | |
| Differentiation
degree | | | | 0.193 | 0.660 |
| High/moderate | 52 | 23 | 29 | | |
| Poor | 33 | 13 | 20 | | |
|
Tumor-Node-Metastasis stage | | | | 6.869 | 0.009a |
| I-II | 55 | 29 | 26 | | |
| III-IV | 30 | 7 | 23 | | |
| Lymph node
metastasis | | | | 7.488 | 0.006a |
| Yes | 28 | 6 | 22 | | |
| No | 57 | 30 | 27 | | |
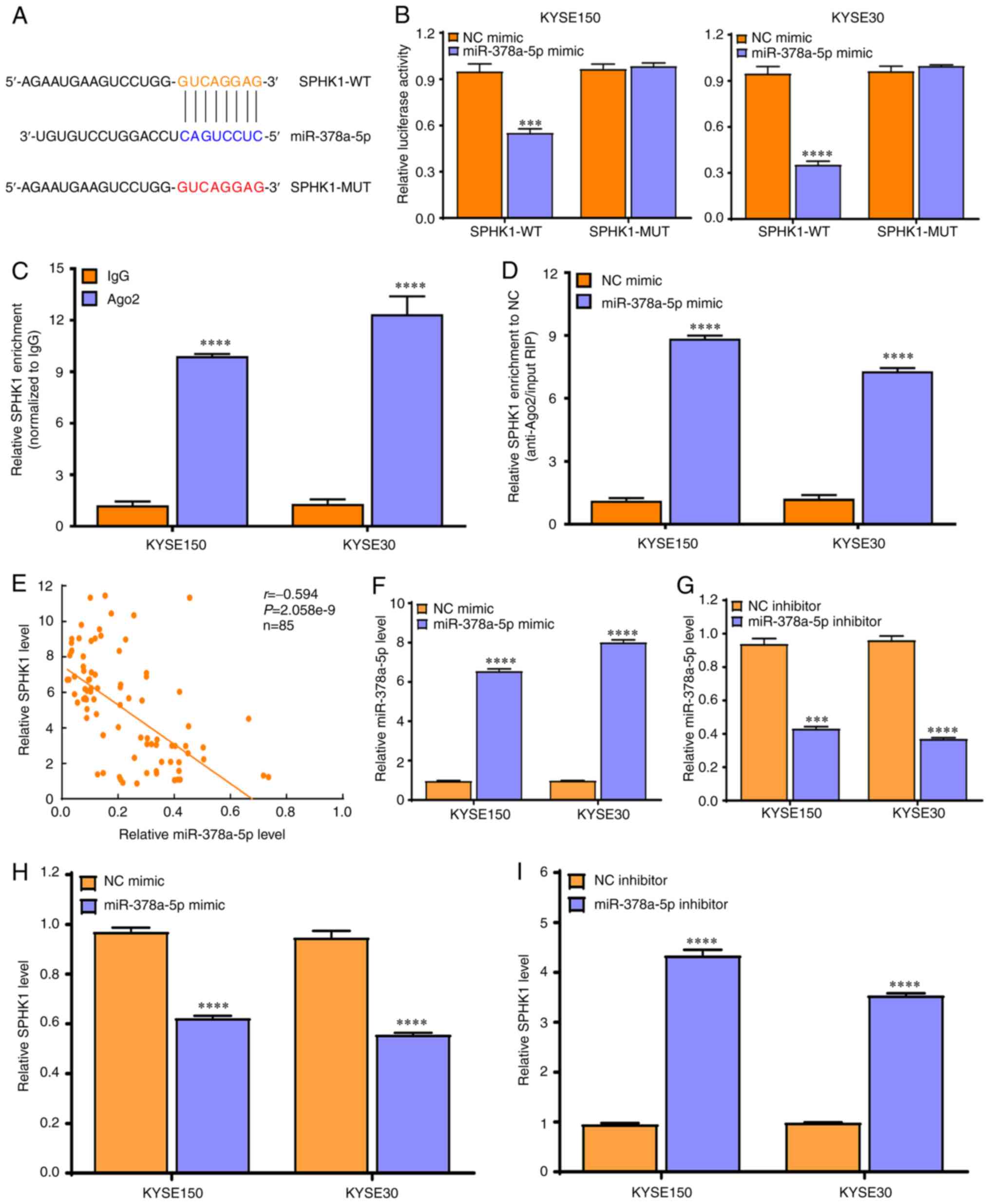
SPHK1 is the direct target of miR-378a-5p
in ESCC cells
To further investigate the downstream molecular
mechanisms of miR-378a-5p in ESCC cells, the expression of
miR-378a-5p in ESCC tissues was analyzed. The data derived from
TCGA and the GEO datasets revealed that miR-378a-5p expression was
markedly reduced in ESCA (Fig.
5A) and ESCC tissues (Fig.
5B), which was validated by RT-qPCR in 85 cases of ESCC and
paired normal tissues (Fig. 5C).
In addition, the miR-378a-5p levels in a number of ESCC cell lines
(Eca109, KYSE150, KYSE30, KYSE450 and KYSE70) was significantly
lower than that in the normal esophageal epithelial cell line
Het-1A (all P<0.0001; Fig.
5D).
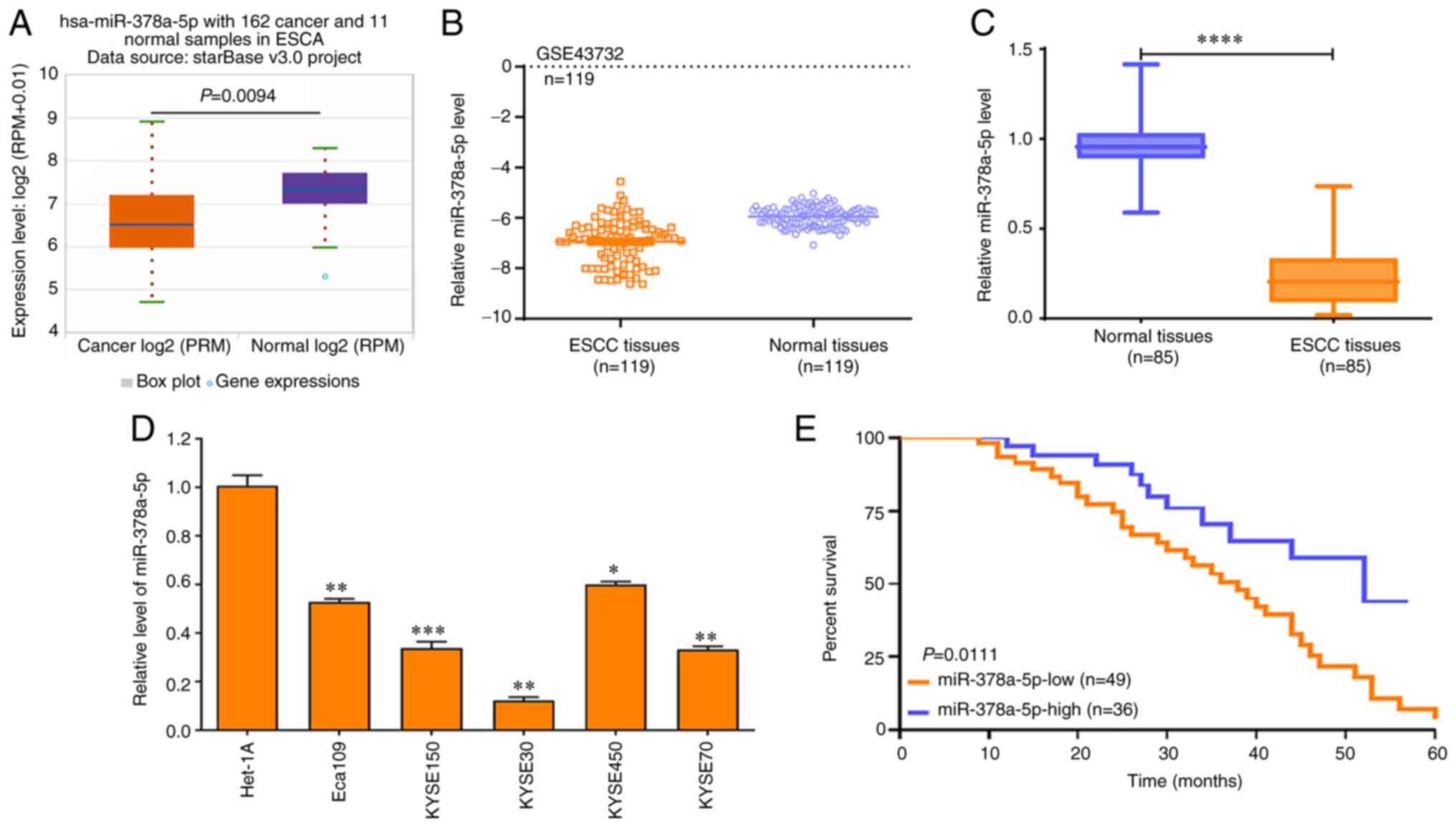 | Figure 5miR-378a-5p expression is low in ESCC
tissues. (A) starBase was used to determine the expression of
miR-378a-5p in 162 esophageal cancer and 11 normal samples. (B) The
GSE43732 Gene Expression Omnibus dataset was used to assess the
levels of miR-378a-5p in 119 ESCC and paired normal samples. Data
were compared using a paired Student's t-test. (C) RT-qPCR analysis
was used to determine the miR-378a-5p levels in 85 ESCC and paired
normal samples. Data were analyzed using a Wilcoxon signed rank.
(D) RT-qPCR was used to determine the miR-378a-5p levels in various
ESCC cell lines (Eca109, KYSE150, KYSE30, KYSE450 and KYSE70) and a
normal esophageal epithelial cell line (Het-1A), and the data were
compared using a one way ANOVA followed by a Dunnett's post hoc
test. (E) A log-rank test was used to analyze the effects of
miR-378a-5p expression on the survival rate of patients with ESCC.
*P<0.05, **P<0.01,
***P<0.001, ****P<0.0001. ESCC,
esophageal squamous cell carcinoma; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR. |
Notably, patients with ESCC and high miR-378a-5p
levels exhibited improved survival rates compared with patients
with ESCC and low miR-378a-5p levels (Fig. 5E). Bioinformatics analysis showed
that miR-378a-5p exhibited lower levels in another 11 different
tumor types compared with those in the corresponding paired normal
tissues (Fig. S3). These data
suggest that miR-378a-5p may function as a tumor suppressor in
multiple tumor types.
TargetScan and miRDB online software were used to
predict the possible downstream target genes of miR-378a-5p, and it
was found that SPHK1 may be the potential target of miR-378a-5p
(Fig. 6A). To validate this
predicted result, a dual-luciferase reporter assay was performed,
which showed that luciferase activity was markedly reduced in
KYSE150 and KYSE30 cells co-transfected with miR-378a-5p mimic and
pmirGLO-SPHK1-WT vector, whereas this was not observed with the
pmirGLO-SPHK1-MUT vector, which lacks the miR-378-5p binding site
(Fig. 6B).
To further validate this result, an Ago2-RIP assay
was performed, which revealed that SPHK1 was markedly enriched in
the Ago2-RIP compared with that observed in the control IgG-RIP
(Fig. 6C). Notably, SPHK1
enrichment was much higher in the miR-378a-5p mimic group than that
in the NC mimic group (P<0.0001; Fig. 6D).
Further evaluation demonstrated that miR-378a-5p
expression exhibited a negative correlation with SHPK1 expression
in 85 ESCC tissues compared with their paired normal tissues
(Fig. 6E). In addition,
miR-378a-5p mimic significantly promoted the expression of
miR-378a-5p in KYSE150 and KYSE30 cells (Fig. 6F), whereas miR-378a-5p inhibitor
markedly suppressed the expression of miR-378a-5p in KYSE150 and
KYSE30 cells (Fig. 6G).
Finally, it was found that the miR-378a-5p mimic
significantly downregulated the expression of SPHK1 in KYSE150 and
KYSE30 cells (Fig. 6H), whereas
miR-378a-5p inhibitor increased the expression of SPHK1 in KYSE150
and KYSE30 cells (Fig. 6I). These
data confirm that SPHK1 is a direct molecular target of miR-378a-5p
in ESCC cells.
Roles of the LINC00514/miR-378a-5p/SPHK1
signaling axis in the proliferation and invasion of ESCC cells
To clarify the underlying roles and related
molecular mechanisms of the LINC00514/miR-378a-5p/SPHK1 signaling
axis in the proliferation and invasion of ESCC cells, KYSE150 and
KYSE30 cells were transfected with miR-378a-5p mimic or inhibitor
combined with pcDNA3.1-LINC00514, or pcDNA3.1-SPHK1 combined with
LINC00514 siRNA2 or SPHK1 siRNA.
Next, CCK-8 and Transwell assays were used to
examine the proliferation and invasion of the transfected cells.
The results showed that miR-378a-5p mimic markedly suppressed the
proliferation and invasion of KYSE150 and KYSE30 cells, which was
partly reversed by pcDNA3.1-LINC00514 and pcDNA3.1-SPHK1 (Fig. 7A-C).
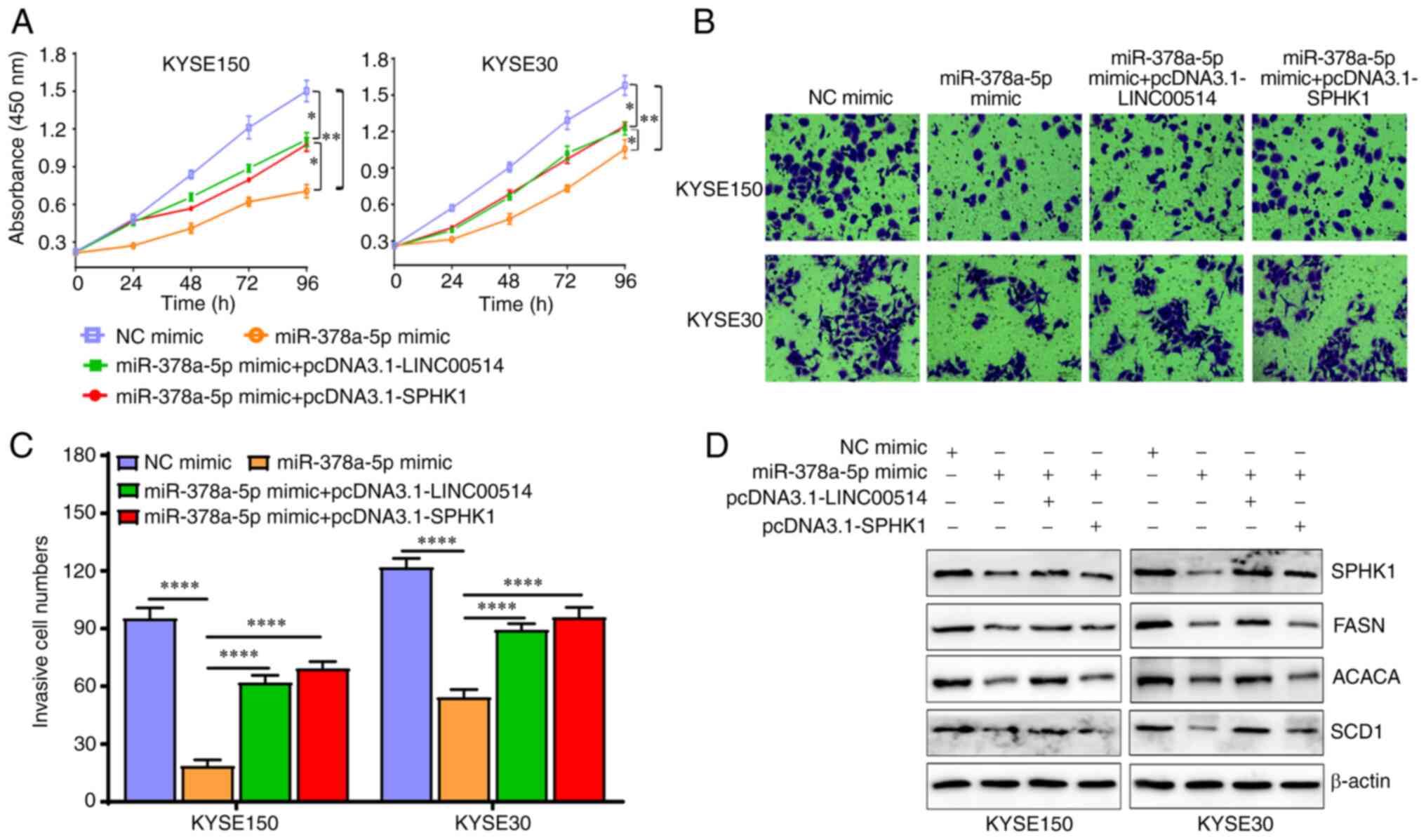 | Figure 7miR-378a-5p exerts biological
functions that are dependent on the levels of LINC00514 and SPHK1
in ESCC cells. (A) ESCC cell proliferation was assessed using a
CCK-8 assay in the NC mimic, miR-378a-5p mimic, miR-378a-5p mimic +
pcDNA3.1-LINC00514 and pcDNA3.1-SPHK1 groups. (B) Cell invasion
ability was assessed using a Transwell chamber assay. (C)
Quantification of the number of cells that had invaded in each
group. Data was compared using a one-way ANOVA followed by a
posy-hoc Tukey's post hoc test. (D) Western blotting was used to
analyze the expression of SPHK1, FASN, ACACA and SCD1 proteins in
various groups. *P<0.05, **P<0.01,
****P<0.0001. SPHK1, sphingosine kinase 1; ESCC,
esophageal squamous cell carcinoma; miR, microRNA; LINC00514, long
intergenic nonprotein-coding RNA 00514; siRNA, small interfering
RNA; NC, negative control; FASN, fatty acid synthetase; ACACA,
acetyl-CoA carboxylase α; SCD1, stearoyl-CoA desaturase 1. |
Importantly, miR-378a-5p mimic downregulated the
expression of lipogenesis-related proteins, including SPHK1, FASN,
ACACA and SCD1, which was also partly reversed by
pcDNA3.1-LINC00514 and pcDNA3.1-SPHK1 (Fig. 7D). Conversely, miR-378a-5p
inhibitor markedly promoted the proliferation and invasion of
KYSE150 and KYSE30 cells, whereas LINC00514 siRNA2 and SPHK1 siRNA
significantly reduced the promotive efficacy of miR-378a-5p
inhibitor on the proliferation and invasion of KYSE150 and KYSE30
cells (Fig. 8A-C). miR-378a-5p
inhibitor induced an upregulation in the protein expression levels
of SPHK1, FASN, ACACA and SCD1, which was also in part reversed by
LINC00514 siRNA2 and SPHK1 siRNA (Fig. 8D).
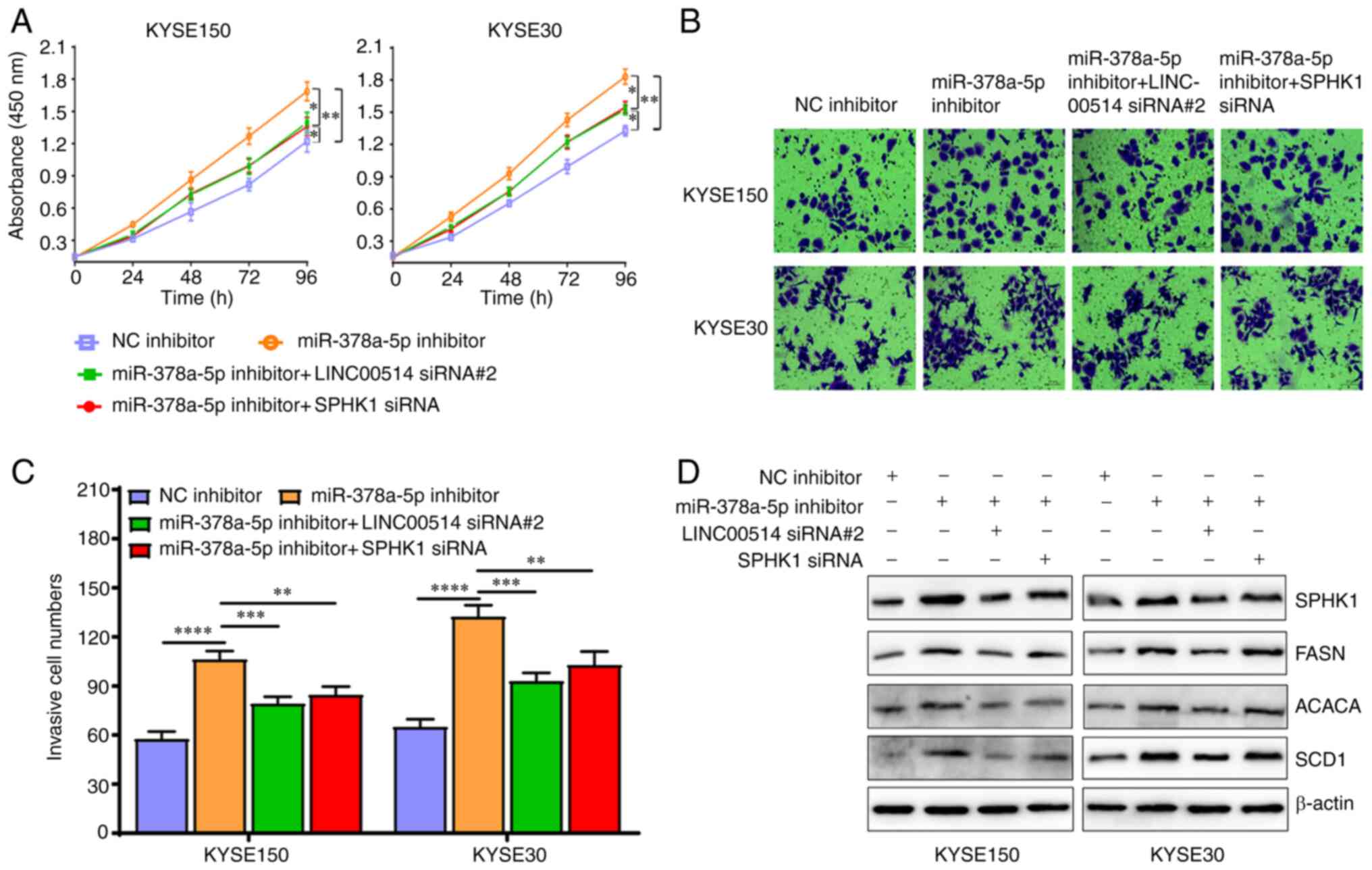 | Figure 8Effects of miR-378a-5p inhiation are
partly reversed by the downregulation of LINC00514 and SPHK1 in
ESCC cells. (A) ESCC cell proliferation was assessed using a CCK-8
assay in the NC inhibitor, miR-378a-5p inhibitor, miR-378a-5p
inhibitor + LINC00514 siRNA2 and SPHK1 siRNA groups. (B) Cell
invasive ability was detected using a Transwell chamber assay in
the various groups. (C) Quantification of the number of cells that
had invaded in the various groups. Data was compared using a
one-way ANOVA with a post hoc Tukey's test. (D) Western blot
analysis of the expression of SPHK1, FASN, ACACA and SCD1
expression in the various groups. *P<0.05,
**P<0.01, ***P<0.001,
****P<0.0001. SPHK1, sphingosine kinase 1; ESCC,
esophageal squamous cell carcinoma; miR, microRNA; LINC00514, long
intergenic nonprotein-coding RNA 00514; siRNA, small interfering
RNA; NC, negative control; FASN, fatty acid synthetase; ACACA,
acetyl-CoA carboxylase α; SCD1, stearoyl-CoA desaturase 1. |
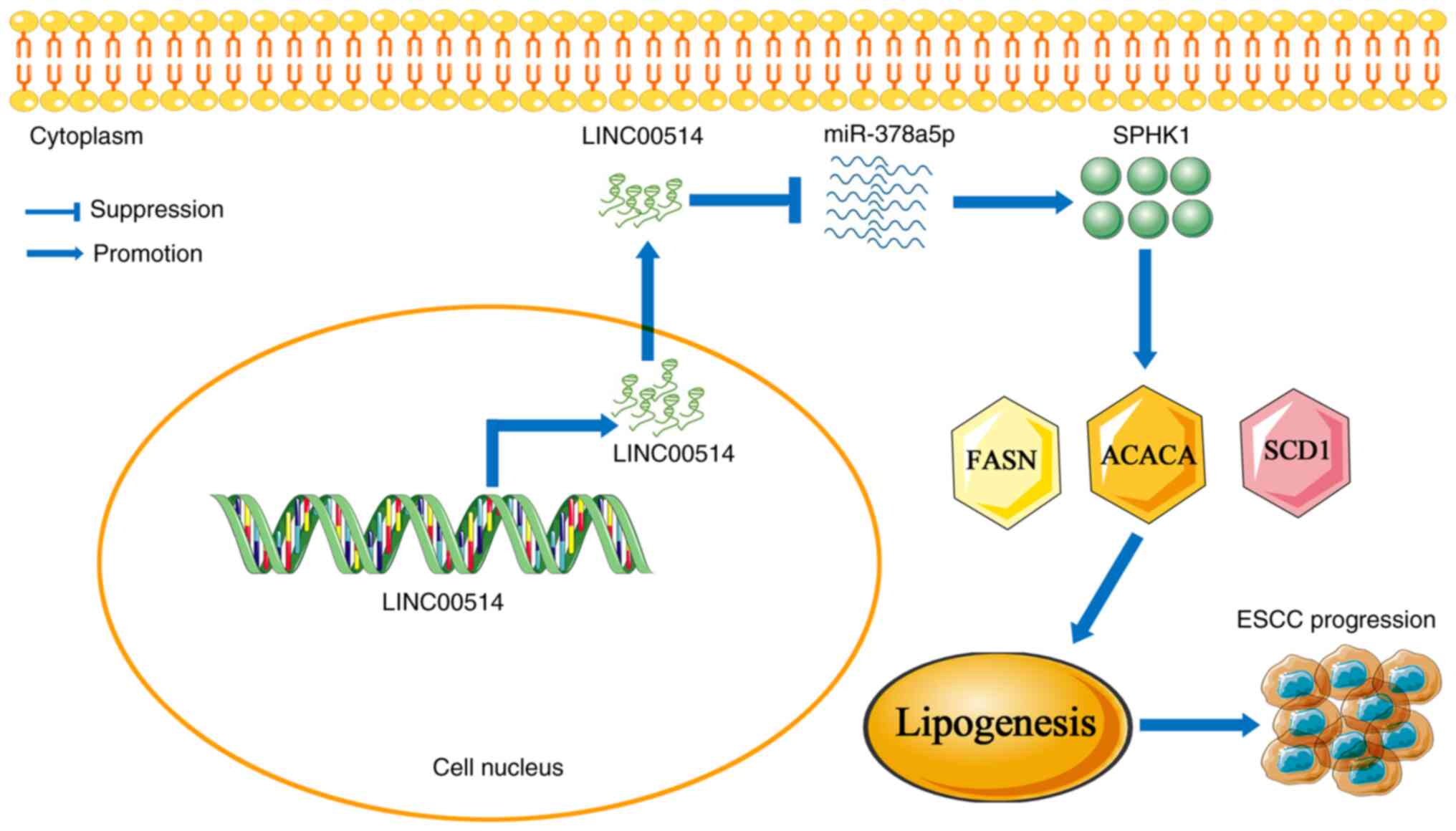
Overall, these data suggest that the
LINC00514/miR-378a-5p/SPHK1 signaling axis may play a pivotal role
in ESCC lipogenesis, and manipulating this signaling axis may be a
novel therapeutic strategy for patients with ESCC (Fig. 9).
Discussion
An increasing number of studies have revealed that
numerous lncRNAs are widely implicated in tumor initiation,
development, progression and metastasis (21,27,28). lncRNAs have been regarded as
excellent candidates for biomarkers and therapeutic targets in a
large number of tumor types (29-33). Multiple lncRNAs exhibited
differential expression in numerous different human tumors, and
were confirmed to harbor diagnostic and prognostic potential
(34-37). LINC00514 was reported to be
involved in the development and progression of neuroendocrine
prostate cancer. Mi et al (38) found that the expression of
LINC00514 was markedly upregulated in osteosarcoma tissues and
cells, and high levels of LINC00514 were positively associated with
advanced tumor stages, distant metastasis and reduced overall
survival of patients. Another study on osteosarcoma confirmed that
LINC00514 was upregulated in osteosarcoma tissues and cells, and an
increased LINC00514 level was associated with tumor size, TNM stage
and distant metastasis. Furthermore, patients with osteosarcoma and
high LINC00514 levels had a shorter overall survival rate (18).
Despite the fact that the role of LINC00514 has been
investigated in various tumor types, its clinical value and
possible biological functions remain to be identified in ESCC, to
the best of our knowledge. The current study found that LINC00514
exhibited high expression levels in ESCC tissues and cell lines,
and this high expression was closely associated with TNM stage,
lymph node metastasis and reduced survival rates in patients with
ESCC. These data highlighted the important clinical value of
LINC00514 in ESCC.
Several studies demonstrated that LINC00514 was
implicated in the regulation of cell proliferation and metastasis
of different tumors. Yu et al (18) verified that LINC00514 knockdown
suppressed cell proliferation in vitro and in vivo,
as well as colony formation, migration and invasion in
osteosarcoma. LINC00514 functioned as a ceRNA by directly absorbing
miR-708-5p, and consequently promoting cell proliferation (38). In addition, LINC00514
downregulation suppressed the proliferation, migration and invasion
of papillary thyroid cancer cells, which was achieved by sponging
miR-204-3p to increase the expression of cell division cycle 23
(19). The present study found
that LINC00514 knockdown markedly inhibited the proliferation and
invasion of ESCC cells, whereas LINC00514 overexpression promoted
the proliferation and invasion of ESCC cells. Further research
confirmed that LINC00514 knockdown notably reduced the expression
of lipogenesis-related proteins (SPHK1, FASN, ACACA and SCD1) in
ESCC cells, and the opposite results were obtained when LINC00514
was upregulated. These data suggest that LINC00514 may participate
in the regulation of cell proliferation, invasion and lipogenesis
in ESCC cells. However, whether lipogenesis-related proteins
participate in the proliferation and invasion of ESCC remains to be
deter- mined, and will form the basis of future experiments into
the possible biological functions mediated by LINC00514 in ESCC
cells.
Based on the close association between the functions
of lncRNAs and their subcellular localization (39-41), a subcellular localization assay
was performed using RT-qPCR in the present study. It was found that
LINC00514 was primarily localized in the cytoplasm of ESCC cells,
suggesting that LINC00514 functions as a ceRNA in ESCC. It was also
found that LINC00514 directly regulated the miR-378a-5p level in
ESCC cells. miRNAs function as negative regulators of downstream
target genes by binding to their 3'-untranslated region (42). miR-378a-3p and miR-378a-5p belong
to the two mature strands of miR-378a localized at chromosome 5q32,
which was previously known as miR-378 (43). miR-378a has been confirmed to be
implicated in the metabolism of lipids and xenobiotics, as well as
in lipid storage, the glycolytic pathway and in mitochondrial
function (44,45). Pan et al (46) found that miR-378a-5p was reduced
in renal tissues and renal cell carcinoma cells, and patients with
a high miR-378a-5p level exhibited longer overall survival rates
than those of patients with low miR-378a-5p levels. In addition,
previous reports have demonstrated that miR-378a-5p is implicated
in the regulation of metabolism and angiogenesis (43,47). The present study found low
expression of miR-378a-5p in ESCC tissues, and this was closely
associated with TNM stage and lymph node metastasis, suggesting
that the LINC00514/miR-378a-5p signaling axis may be an important
therapeutic target for patients with ESCC.
Lipogenesis contributes to membrane synthesis,
provides a source of energy for tumor cells and promotes oncogenic
signaling (48); thus,
reprogramming lipid metabolism may be a potential therapeutic
target in cancer treatment (49).
Lipogenesis has been reported to be closely associated with tumor
development, progression and metastasis (50-52). SPHK1 is an oncogenic enzyme that
phosphorylates sphingosine to produce sphingosine-1-phosphate, and
plays a pivotal role in multiple cellular processes (53,54). Increasing evidence has
demonstrated that SPHK1 is upregulated in gastric carcinoma
(55), colorectal cancer
(56) colon cancer (57) and ESCC (58). In the present study, SPHK1 was
confirmed as a direct molecular target of miR-378a-5p, and SPHK1
was closely associated with TNM stage, lymph node metastasis and
poor prognosis of patients with ESCC.
To further clarify the role of the
LINC00514/miR-378a-5p/SPHK1 signaling axis in ESCC cell
proliferation and invasion, CCK-8 and Transwell chamber assays were
employed to investigate the function of this axis. The current data
revealed that miR-378a-5p mimic significantly suppressed cell
proliferation and invasion, and the expression of
lipogenesis-related proteins in ESCC cells, which was partly
reversed by over-expression of LINC00514 and SPHK1. However, the
opposite results were obtained with miR-378a-5p alone or combined
with LINC00514 and SPHK1 siRNAs. These results suggest that the
LINC00514/miR-378a-5p/SPHK1 signaling axis may be closely
associated with ESCC development and progression, and may be a
novel and promising therapeutic target for patients with ESCC.
Thus, in future studies, the function of the
LINC00514/miR-378a-5p/SPHK1 signaling axis will be further assessed
in vivo, to lay the foundation for targeting of this
signaling axis as a potential therapeutic option in the treamtent
of patients with ESCC.
In conclusion, the current data demonstrated that
LINC00514 and SPHK1 expression levels were upregulated in ESCC
tissues and cells, and this high expression of LINC00514 and SPHK1
was correlated with TNM stage, lymph node metastasis and poor
prognosis in patients with ESCC. LINC00514 knockdown inhibited cell
proliferation and invasion, and reduced the expression of
lipogenesis-related proteins, whereas LINC00514 overexpression
accelerated the proliferation and invasion of ESCC cells, and
promoted the expression of lipogenesis-related proteins.
Mechanistically, LINC00514 functioned as a ceRNA to sponge
miR-378a-5p, thereby indirectly upregulating SPHK1 expression,
which further promoted the expression of the lipogenesis-related
proteins FASN, ACACA and SCD1, and thus promoting ESCC progression.
The current data may provide novel evidence for the use of
LINC00514/miR-378a-5p/SPHK1 signaling axis-based targeted therapy
in patients with ESCC.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
RTF, XW and HTL conceived and designed the present
study. XW, HTL and QZ performed the experiments. XYZ, YQ, GZZ and
JHD analyzed and interpretated the data. FW and XXY contributed to
analysis of the data. RTF wrote the original manuscript. FW and XXY
reviewed and revised the manuscript. All authors read and approved
the final manuscript. XW and HTL confirm the authenticity of all
the raw data.
Ethics approval and consent to
participate
The present study was approved by the Research and
Ethics Committee of The First Affiliated Hospital of Zhengzhou
University. Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
We would like to thank Professor Shenglei Li and
Associate Professor Yue Xu, Department of Pathology of the First
Affiliated Hospital of Zhengzhou University, for their assistance
in identification of the ESCC tissues and normal tissues.
References
|
1
|
Zhang Y: Epidemiology of esophageal
cancer. World J Gastroenterol. 19:5598–5606. 2013. View Article : Google Scholar
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
3
|
Stoecklein NH, Hosch SB, Bezler M, Stern
F, Hartmann CH, Vay C, Siegmund A, Scheunemann P, Schurr P, Knoefel
WT, et al: Direct genetic analysis of single disseminated cancer
cells for prediction of outcome and therapy selection in esophageal
cancer. Cancer Cell. 13:441–453. 2008. View Article : Google Scholar
|
|
4
|
Arnold M, Soerjomataram I, Ferlay J and
Forman D: Global incidence of oesophageal cancer by histological
subtype in 2012. Gut. 64:381–387. 2015. View Article : Google Scholar
|
|
5
|
Yang H, Liu H, Chen Y, Zhu C, Fang W, Yu
Z, Mao W, Xiang J, Han Y, Chen Z, et al: Neoadjuvant
chemoradiotherapy followed by surgery versus surgery alone for
locally advanced squamous cell carcinoma of the esophagus
(NEOCRTEC5010): A phase III multicenter, randomized, open-label
clinical trial. J Clin Oncol. 36:2796–2803. 2018. View Article : Google Scholar
|
|
6
|
Pickens A and Orringer MB: Geographical
distribution and racial disparity in esophageal cancer. Ann Thorac
Surg. 76(Suppl): S1367–S1369. 2003. View Article : Google Scholar
|
|
7
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar
|
|
8
|
Ohashi S, Miyamoto S, Kikuchi O, Goto T,
Amanuma Y and Muto M: Recent advances from basic and clinical
studies of esophageal squamous cell carcinoma. Gastroenterology.
149:1700–1715. 2015. View Article : Google Scholar
|
|
9
|
Morris KV and Mattick JS: The rise of
regulatory RNA. Nat Rev Genet. 15:423–437. 2014. View Article : Google Scholar
|
|
10
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar
|
|
11
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar
|
|
12
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar
|
|
13
|
Chen MJ, Deng J, Chen C, Hu W, Yuan YC and
Xia ZK: lncRNA H19 promotes epithelial mesenchymal transition and
metastasis of esophageal cancer via STAT3/EZH2 axis. Int J Biochem
Cell Biol. 113:27–36. 2019. View Article : Google Scholar
|
|
14
|
Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang
K, Guan X, Yang K and Bai Y: lncRNA CASC9 promotes esophageal
squamous cell carcinoma metastasis through upregulating LAMC2
expression by interacting with the CREB-binding protein. Cell Death
Differ. 25:1980–1995. 2018. View Article : Google Scholar
|
|
15
|
Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X,
Meng H, Guan X, Yang K and Bai Y: Up-regulation of lncRNA CASC9
promotes esophageal squamous cell carcinoma growth by negatively
regulating PDCD4 expression through EZH2. Mol Cancer. 16:1502017.
View Article : Google Scholar
|
|
16
|
Li W, Zhang L, Guo B, Deng J, Wu S, Li F,
Wang Y, Lu J and Zhou Y: Exosomal FMR1-AS1 facilitates maintaining
cancer stem-like cell dynamic equilibrium via TLR7/NFκB/c-Myc
signaling in female esophageal carcinoma. Mol Cancer. 18:222019.
View Article : Google Scholar
|
|
17
|
Tao S, Chen Q, Lin C and Dong H: Linc00514
promotes breast cancer metastasis and M2 polarization of
tumor-associated macrophages via Jagged1-mediated notch signaling
pathway. J Exp Clin Cancer Res. 39:1912020. View Article : Google Scholar
|
|
18
|
Yu D, Xu X, Li S and Zhang K: LINC00514
drives osteosarcoma progression through sponging microRNA-708 and
consequently increases URGCP expression. Aging. 12:6793–6807. 2020.
View Article : Google Scholar
|
|
19
|
Li X, Zhong W, Xu Y, Yu B and Liu H:
Silencing of lncRNA LINC00514 inhibits the malignant behaviors of
papillary thyroid cancer through miR-204-3p/CDC23 axis. Biochem
Biophys Res Commun. 508:1145–1148. 2019. View Article : Google Scholar
|
|
20
|
Ramnarine VR, Alshalalfa M, Mo F, Nabavi
N, Erho N, Takhar M, Shukin R, Brahmbhatt S, Gawronski A, Kobelev
M, et al: The long noncoding RNA landscape of neuroendocrine
prostate cancer and its clinical implications. Gigascience.
7:giy0502018. View Article : Google Scholar
|
|
21
|
Jafari N, Drury J, Morris AJ, Onono FO,
Stevens PD, Gao T, Liu J, Wang C, Lee EY, Weiss HL, et al: De novo
fatty acid synthesis-driven sphingolipid metabolism promotes
metastatic potential of colorectal cancer. Mol Cancer Res.
17:140–152. 2019. View Article : Google Scholar
|
|
22
|
Jairajpuri DS, Mohammad T, Adhikari K,
Gupta P, Hasan GM, Alajmi MF, Rehman MT, Hussain A and Hassan MI:
Identification of sphingosine kinase-1 inhibitors from bioactive
natural products targeting cancer therapy. ACS Omega.
5:14720–14729. 2020. View Article : Google Scholar
|
|
23
|
Imbert C, Montfort A, Fraisse M,
Marcheteau E, Gilhodes J, Martin E, Bertrand F, Marcellin M,
Burlet-Schiltz O, Peredo AG, et al: Resistance of melanoma to
immune checkpoint inhibitors is overcome by targeting the
sphingosine kinase-1. Nat Commun. 11:4372020. View Article : Google Scholar
|
|
24
|
Arends MJ, Fukayama M, Klimstra DS, Lam
AKY, Nagtegaal ID, Odze RD, Paradis V, Park YN, Rugge M,
Salto-Tellez M, et al: TNM staging of tumours of the oesophagus.
WHO classification of tumours of the digestive system. The WHO
Classification of Tumours Editorial Board IARC Press; Lyon: pp.
252019
|
|
25
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S,
Zeng Z, He C, Liu ML, Huang K, Zhong JX, et al: Hypoxia-induced
lncRNA-BX111 promotes metastasis and progression of pancreatic
cancer through regulating ZEB1 transcription. Oncogene.
37:5811–5828. 2018. View Article : Google Scholar
|
|
28
|
Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang
Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN, et al: Long noncoding RNA
MALAT1 suppresses breast cancer metastasis. Nat Genet.
50:1705–1715. 2018. View Article : Google Scholar
|
|
29
|
Renganathan A and Felley-Bosco E: Long
noncoding RNAs in cancer and therapeutic potential. Adv Exp Med
Biol. 1008:199–222. 2017. View Article : Google Scholar
|
|
30
|
Arun G, Diermeier SD and Spector DL:
Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol
Med. 24:257–277. 2018. View Article : Google Scholar
|
|
31
|
Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G and
Zhu YS: MALAT1: A potential biomarker in cancer. Cancer Manag Res.
10:6757–6768. 2018. View Article : Google Scholar
|
|
32
|
Qiu L, Tang Q, Li G and Chen K: Long
non-coding RNAs as biomarkers and therapeutic targets: Recent
insights into hepatocellular carcinoma. Life Sci. 191:273–282.
2017. View Article : Google Scholar
|
|
33
|
Cossu AM, Mosca L, Zappavigna S, Misso G,
Bocchetti M, De Micco F, Quagliuolo L, Porcelli M, Caraglia M and
Boccellino M: Long non-coding RNAs as important biomarkers in
laryngeal cancer and other head and neck tumours. Int J Mol Sci.
20:34442019. View Article : Google Scholar
|
|
34
|
Peng Z, Liu C and Wu M: New insights into
long noncoding RNAs and their roles in glioma. Mol Cancer.
17:612018. View Article : Google Scholar
|
|
35
|
Chen Y, Bi F, An Y and Yang Q:
Identification of pathological grade and prognosis-associated
lncRNA for ovarian cancer. J Cell Biochem. 120:14444–14454. 2019.
View Article : Google Scholar
|
|
36
|
Chao Y and Zhou D: lncRNA-D16366 is a
potential biomarker for diagnosis and prognosis of hepatocellular
carcinoma. Med Sci Monit. 25:6581–6586. 2019. View Article : Google Scholar
|
|
37
|
Yang J, Li C, Mudd A and Gu X: lncRNA PVT1
predicts prognosis and regulates tumor growth in prostate cancer.
Biosci Biotechnol Biochem. 81:2301–2306. 2017. View Article : Google Scholar
|
|
38
|
Mi LD, Sun CX, He SW and Du GY:
SP1-Induced upregulation of lncRNA LINC00514 promotes tumor
proliferation and metastasis in osteosarcoma by regulating miR-708.
Cancer Manag Res. 12:3311–3322. 2020. View Article : Google Scholar
|
|
39
|
Chaumeil J, Le Baccon P, Wutz A and Heard
E: A novel role for Xist RNA in the formation of a repressive
nuclear compartment into which genes are recruited when silenced.
Genes Dev. 20:2223–2237. 2006. View Article : Google Scholar
|
|
40
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar
|
|
41
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar
|
|
42
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
43
|
Krist B, Florczyk U,
Pietraszek-Gremplewicz K, Jozkowicz A and Dulak J: The role of
miR-378a in metabolism, angiogenesis, and muscle biology. Int J
Endocrinol. 2015:2817562015. View Article : Google Scholar
|
|
44
|
Carrer M, Liu N, Grueter CE, Williams AH,
Frisard MI, Hulver MW, Bassel-Duby R and Olson EN: Control of
mitochondrial metabolism and systemic energy homeostasis by
microRNAs 378 and 378*. Proc Natl Acad Sci USA.
109:15330–15335. 2012. View Article : Google Scholar
|
|
45
|
Eichner LJ, Perry MC, Dufour CR, Bertos N,
Park M, St-Pierre J and Giguère V: miR-378(*) mediates metabolic
shift in breast cancer cells via the PGC-1beta/ERRgamma
transcriptional pathway. Cell Metab. 12:352–361. 2010. View Article : Google Scholar
|
|
46
|
Pan X, Zhao L, Quan J, Liu K, Lai Y, Li Z,
Zhang Z, Xu J, Xu W, Guan X, et al: miR-378a-5p acts as a tumor
suppressor in renal cell carcinoma and is associated with the good
prognosis of patients. Am J Transl Res. 11:2207–2218. 2019.
|
|
47
|
Cui Z, Liu QL, Sun SQ, Jiao K, Liu DR,
Zhou XC and Huang L: miR-378a-5p inhibits angiogenesis of oral
squamous cell carcinoma by targeting KLK4. Neoplasma. 67:85–92.
2020. View Article : Google Scholar
|
|
48
|
Zadra G, Photopoulos C and Loda M: The fat
side of prostate cancer. Biochim Biophys Acta. 1831:1518–1532.
2013. View Article : Google Scholar
|
|
49
|
Cheng C, Geng F, Cheng X and Guo D: Lipid
metabolism reprogramming and its potential targets in cancer.
Cancer Commun (Lond). 38:272018. View Article : Google Scholar
|
|
50
|
Ishay-Ronen D, Diepenbruck M, Kalathur
RKR, Sugiyama N, Tiede S, Ivanek R, Bantug G, Morini MF, Wang J,
Hess C and Christofori G: Gain Fat-lose metastasis: Converting
invasive breast cancer cells into adipocytes inhibits cancer
metastasis. Cancer Cell. 35:17–32.e6. 2019. View Article : Google Scholar
|
|
51
|
Porporato PE, Payen VL, Baselet B and
Sonveaux P: Metabolic changes associated with tumor metastasis,
part 2: Mitochondria, lipid and amino acid metabolism. Cell Mol
Life Sci. 73:1349–1363. 2016. View Article : Google Scholar
|
|
52
|
Tousignant KD, Rockstroh A, Taherian Fard
A, Lehman ML, Wang C, McPherson SJ, Philp LK, Bartonicek N, Dinger
ME, Nelson CC and Sadowski MC: Lipid uptake is an androgen-enhanced
lipid supply pathway associated with prostate cancer disease
progression and bone metastasis. Mol Cancer Res. 17:1166–1179.
2019. View Article : Google Scholar
|
|
53
|
Li W, Yu CP, Xia JT, Zhang L, Weng GX,
Zheng HQ, Kong QL, Hu LJ, Zeng MS, Zeng YX, et al: Sphingosine
kinase 1 is associated with gastric cancer progression and poor
survival of patients. Clin Cancer Res. 15:1393–1399. 2009.
View Article : Google Scholar
|
|
54
|
Facchinetti MM, Gandini NA, Fermento ME,
Sterin-Speziale NB, Ji Y, Patel V, Gutkind JS, Rivadulla MG and
Curino AC: The expression of sphingosine kinase-1 in head and neck
carcinoma. Cells Tissues Organs. 192:314–324. 2010. View Article : Google Scholar
|
|
55
|
Wang Z, Qu H, Gong W and Liu A:
Up-regulation and tumor-promoting role of SPHK1 were attenuated by
miR-330-3p in gastric cancer. IUBMB Life. 70:1164–1176. 2018.
View Article : Google Scholar
|
|
56
|
Bae GE, Do SI, Kim K, Park JH, Cho S and
Kim HS: Increased sphingosine kinase 1 expression predicts distant
metastasis and poor outcome in patients with colorectal cancer.
Anticancer Res. 39:663–670. 2019. View Article : Google Scholar
|
|
57
|
Liu SQ, Su YJ, Qin MB, Mao YB, Huang JA
and Tang GD: Sphingosine kinase 1 promotes tumor progression and
confers malignancy phenotypes of colon cancer by regulating the
focal adhesion kinase pathway and adhesion molecules. Int J Oncol.
42:617–626. 2013. View Article : Google Scholar
|
|
58
|
Nemoto M, Ichikawa H, Nagahashi M, Hanyu
T, Ishikawa T, Kano Y, Muneoka Y and Wakai T: Phospho-Sphingosine
Kinase 1 expression in lymphatic spread of esophageal squamous cell
carcinoma. J Surg Res. 234:123–131. 2019. View Article : Google Scholar
|