Introduction
Diffuse large B-cell lymphoma (DLBCL) is a type of
lymphoid malignancy that accounts for 25-35% of non-Hodgkin's
lymphoma (NHL) and 37% of B-cell tumors (1). Germinal center B-cell-like (GCB) and
activated B-cell-like (ABC) are the two main molecular subtypes of
DLBCL (2). DLBCL occurs in lymph
node or extranodal structures and is pathologically characterized
by a diffuse growth pattern of large B-cell proliferation that
replaces these sites (3). Due to
its lack of obvious symptoms in the early stages and similar
symptoms to other diseases, it is often characterized by evident
heterogeneity and complexity, which is also one of the main reasons
for the poor prognosis and the low 5-year survival rate (30-80%) of
patients (4,5). Therefore, the exploration of novel
therapeutic targets for DLBCL is of utmost importance.
Long non-coding RNAs (lncRNAs) refer to RNA
molecules of >200 nucleotides in length that do not encode
proteins. There is evidence to indicate that lncRNAs play an
important role in malignant B-cells and serve as potential markers
for the diagnosis and progression of DLBCL; thus, they may play
carcinogenic or tumor suppressive functions in the DLBCL process.
For example, lncRNA MALAT1 (6),
lncRNA FIRRE (7) and lncRNA NEAT1
(8) have been shown to promote
tumorigenesis, and conversely, lncRNA SMAD5-AS1 (9), lncRNA PANDA (10) and lncRNA RP11-468E2.5 (11) have been shown to inhibit tumor
function. Previous studies have indicated that lncRNA growth arrest
specific 5 (GAS5) is abnormally expressed in colorectal cancer,
gastric cancer, glioma and other types of cancer, and functions as
a tumor suppressor gene (12-15). Moreover, GAS5 has been found to be
associated with cell survival and with the progression of B-cell
lymphoma (16). Previous studies
have also discovered that GAS5 is abnormally expressed in patients
with DLBCL, indicating its involvement in the pathogenesis of DLBCL
(17,18). Hence, the identification of the
molecular function and the biological targets of GAS5 may prove to
be helpful for attenuating the progression of DLBCL.
MicroRNAs (miRNAs or miRs) are small single-stranded
RNAs that play an important regulatory role by regulating target
gene transcription. miR-18a-5p has been demonstrated to play an
oncogenic role in lung, nasopharyngeal, prostate, colorectal and
breast cancers, and is widely involved in cell proliferation,
apoptosis and other phenotypes (19-23). The upregulated expression of
miR-18a-5p was previously predicted using a database of
differentially expressed miRNAs in human cancers v2.0 (dbDEMC 2.0)
in lymphoma and its high expression level in DLBCL tissue was show
to potentially play a carcinogenic role (24). Moreover, a previous study
confirmed that the expression of miR-18a-5p was also elevated in
B-cell lymphoma samples (25). In
addition, GAS5 has been shown to target and regulate the expression
of miR-18a-5p in cancers cells (26). However, the molecular function of
miR-18a-5p in DLBCL remains unclear.
The transcription factor, Runt-related transcription
factor 1 (RUNX1), also known as acute myeloid leukemia 1 (AML1), is
involved in regulating the development of hematopoietic stem cells
and is closely related to the occurrence and development of
hematological malignancies (27).
Furthermore, a previous study demonstrated that RUNX1 specifically
regulated the transcriptional activity of its target gene Ebf1, and
participated in B-lymphocyte development (28). Of note, a previous study by the
authors demonstrated that miR-18a-5p targeted the regulation of
RUNX1 and then participated in the permeability of the blood-tumor
barrier (29). However, whether
the interaction among GAS5, miR-18a-5p and RUNX1 affects the
biological behavior of DLBCL cells and plays a carcinogenic role
remains to be further investigated.
The present study aimed to determine the expression
levels of GAS5, miR-18a-5p and RUNX1 in DLBCL cell lines, and to
further investigate the potential molecular mechanisms among them
in DLBCL. The findings presented herein may provide novel
therapeutic targets for DLBCL.
Materials and methods
Cell line culture and transfection
The ABC DLBCL cell line (OCI-Ly3, BNCC338435), GCB
DLBCL cell line (TMD8, BNCC340121) (9) and the human B-lymphocyte (GM12878,
BNCC341257) were all purchased from the American Type Culture
Collection (ATCC). The Burkitt's lymphoma cell line, Raji, was
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences (TCHu 44). The cells were cultured in
the Iscove's modified Dulbecco's medium (IMDM) supplemented with
20% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml
streptomycin in 5% CO2 at 37°C. The cells were digested
by trypsin and spread on a 24-well plate, and further cultured in
IMDM (Gibco; Thermo Fisher Scientific, Inc.) in an incubator at
37°C for 2-3 days. The 2 µg lentiviral vector containing the
GAS5 sequence (Lv-GAS5) was purchased from Shanghai GenePharma Co.,
Ltd. and transfected into the OCI-Ly3/TMD8 cells with 10
µg/µl polybrene [40804ES76, 1:1,000; Yeasen
Biotechnology (Shanghai) Co., Ltd.] to construct
GAS5-overexpressing cells. In addition, cells transfected with the
20 nmol/l empty vector (GenePharma) were considered as Lv. Other
transfection targets, including a total of 25 pmol/l miR-18a-5p
mimics/inhibitor, 100 nmol/l si-RUNX1/RUNX1 overexpression plasmid
(pcDNA3.1) (Sangon Biotech Co., Ltd.) and their negative controls
(NC) were transfected into the cells using Lipofectamine
3000® transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) at room temperature for 48 h, respectively. The
cells were collected for subsequent experiments at 48 h following
transfection. Cells without transfection were considered as
controls. The sequences of the transfection targets are presented
in Table I.
 | Table ISequences of the transfection targets
in the present study. |
Table I
Sequences of the transfection targets
in the present study.
| Gene | Sequences (5′ to
3′) |
|---|
| miR-18a-5p
mimics |
UAAGGUGCAUCUAGUGCAGAUAG |
| NC mimics |
UUCUCCGAACGUGUCACGUTT |
| miR-18a-5p
inhibitor |
CCCUAUCUGCACUAGAUGCACCU |
| NC inhibitor |
CAGUACUUUUGUGUAGUACAA |
| si-RUNX1 |
ACGAATCACACTGAATGCAAACC |
| si-NC |
TGCTTAGTGTGACTTACGTTTGG |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® reagent (500 µl) (Sangon
Biotech Co., Ltd.) was used to extract the total RNA, and 500 ng
RNA were used to synthesize cDNA for reverse transcription using
the 5X Prime Script RT Master Mix (Takara Bio, Inc.). The cDNA (1
µl) was used as the template for RT-qPCR using SYBR qPCR
Master Mix (Takara Bio, Inc.) according to the manufacturer's
protocol. The PCR reaction conditions were 95°C for 5 min, 95°C for
30 cycles, 60°C for 30 sec, 72°C for 30 sec, and finally 72°C for 7
min. The expression levels were normalized to the
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels and
analyzed using the 2−ΔΔCq (30) method. The primer sequences used
are presented in Table II.
 | Table IIPrimer sequences used in RT-qPCR in
the present study. |
Table II
Primer sequences used in RT-qPCR in
the present study.
| Primers | Sequences (5′ to
3′) |
|---|
|
lncRNA-GAS5-Forward |
GCAAGCCTAACTCAAGCCATTG |
|
lncRNA-GAS5-Reverse |
CTTGCTCCACACAGTGTAGTC |
| RUNX1-Forward |
CCTCAGGTTTGTCGGTCGAA |
| RUNX1-Reverse |
CTTGCGGTGGGTTTGTGAAG |
| BAX-Forward |
CATGGGCTGGACATTGGACT |
| BAX-Reverse |
CAAAGTAGGAGAGGAGGCCG |
| miR-18a-5p-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACctatct |
|
miR-18a-5p-Forward |
CGTTATAAGGTGCATCTAGTGC |
|
miR-18a-5p-Reverse |
GTGCAGGGTCCGAGGT |
| U6-RT |
AACGCTTCACGAATTTGCGT |
| U6-Forward |
CTCGCTTCGGCAGCACA |
| U6-Reverse |
AACGCTTCACGAATTTGCGT |
| GAPDH-Forward |
GTTCGTCATGGGTGTGAACC |
| GAPDH-Reverse |
CATCCACAGTCTTCTGGGTG |
RNA fluorescence in situ hybridization
(FISH)
The location of GAS5 in two cell lines (OCI-Ly3 and
TMD8) was identified using the Ribo™ FISH kit (Guangzhou RiboBio
Co., Ltd.) according to the manufacturer's instructions. Briefly,
6×104 cells/well were fixed with 4% paraformaldehyde for
10 min at room temperature, followed by permeabilization with 0.5%
Triton, and washing three times with phosphate-buffered saline
(PBS) after discarding the permeabilization solution. Each well was
then supplemented with a pre-hybridization solution (Reagent A,
derived from the RiboBio FISH kit) and blocked with blocking
solution (Reagent C, derived from the RiboBio FISH kit) at 37°C for
30 min. Cy3-labeled lncRNA FISH probes (included with the kit) were
then synthesized and used to identify GAS5. The probe mix (2.5
µl and 20 µM) was added to 100 µl
hybridization solution (Reagent B, derived from the RiboBio FISH
kit), and then mixed and added to the cells, followed by
hybridization overnight at 37°C. Furthermore, the cells were washed
three times with hybridization washing solution, and then the
nuclei were stained with DAPI (Reagent D, derived from the RiboBio
FISH kit) for 10 min at room temperature, and fixed on a glass
slide for fluorescence detection. The aforementioned process was
carried out in a dark environment. Focus was placed on five fields
of view, and the cells were observed under a TCS SP5II confocal
microscope (Leica Microsystems GmbH).
Cell viability and proliferation
assay
Cell viability was assessed using the
3-(4,5-dimethylthiazol-2-yl)-5-(3-carbox
ymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) kit
(ab197010; Abcam). Briefly, the cells were seeded in a 96-well
plate at 5.0×103/well, and 20 µl MTS solution
were then added and incubated in an incubator at 37°C for 3 h. The
absorbance at 450 nm was detected using a microplate analyzer
(Tecan Group, Ltd.). Cell proliferation was measured using a
5-ethynyl-2′-deoxyuridine (EdU) kit (C0085S; Beyotime Institute of
Biotechnology) following the manufacturer's instructions. Firstly,
2X EdU working solution (20 µM) were added to the 6-well
plate and incubated for 4 h at 37°C. Following centrifugation at
1,000 × g for 5 min at room temperature, 1 ml of PBS was added to
resuspend the cells and cell suspension drops were added to the
slide and spread evenly, and further placed in an oven at 50°C for
1 h and fixed with 4% paraformaldehyde (P0099; Beyotime Institute
of Biotechnology) at room temperature for 1 min. Subsequently, the
cells were treated with washing solution (P0106; Beyotime Institute
of Biotechnology) and permeability solution (P0097, Beyotime
Institute of Biotechnology); the click reaction solution (derived
from the EdU kit) was then added to evenly cover the cell surface
followed by incubated at room temperature for 30 min in the dark
environment. Finally, 1 ml 1X Hoechst 33342 solution (C1025,
Beyotime Institute of Biotechnology) was used to incubate the cells
for 30 min at room temperature and avoid lighting for 10 min. After
washing, the fluorescence detection was performed.
Cell cycle assay
The cells were collected by centrifugation at 1,000
× g for 5 min at 4°C. Subsequently, cells were fixed with 1 ml
pre-cooled 70% ethanol overnight at 4°C. Propidium iodide (PI)
staining solution (0.5 ml) was then added to each tube of cell
samples for resuspension, followed by incubation at 37°C in a dark
environment for 30 min. Finally, the red fluorescence and light
scattering at the excitation wavelength of 488 nm were detected
using a flow cytometer (FACScan; BD Biosciences). The data were
analyzed using FlowJo 10 software.
Cell apoptosis assay
The number of apoptotic cells was detected using
flow cytometry. Firstly, the cells were washed twice with
pre-cooled PBS and centrifuged at 300 × g for 5 min at 4°C.
Following PBS absorption, 100 µl 1X binding buffer (Nanjing
KeyGen Biotech Co., Ltd.) were added to resuspend the cells.
Subsequently, 5 µl Annexin V-FITC and 10 µl PI
staining solution (Beyotime Institute of Biotechnology) were added
and mixed gently, while being protected from light and reacted for
10-15 min at room temperature. Finally, 400 µl 1X binding
buffer were added, mixed and placed on ice, and subsequently
detected using flow cytometry (FACScan; BD Biosciences) within 1 h.
The data were analyzed using FlowJo 10 software.
Luciferase reporter assay
The wild-type and mutant-type 3′UTR sequences of
RUNX1 were inserted into the pmirGLO dual luciferase reporter
vector (E1330; Promega Corporation) to construct wild-type (WT) and
mutated-type (MUT) luciferase reporter plasmids. The luciferase
plasmid (200 ng) was then combined with 60 nM miR-18a-5p
mimics/inhibitor and co-transfected into OCI-Ly3 cells.
Lipofectamine 2000® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for transfection. Following
transfection, the cells were washed with pre-cooled PBS, and then
lysed using a dual luciferase reporter gene detection kit (E1960;
Promega Corporation). Subsequently, 30 µl Firefly luciferase
detection reagent were added to the lysis solution, and the
relative light unit (RLU) was measured using a multifunctional
microplate reader (Fluoroskan ascent FL, Thermo Fisher Scientific,
Inc.). Renilla luciferase detection solution (30 µl)
was then added to the lysate for RLU determination. Finally, the
relative luciferase activity was calculated (E1960; Promega
Corporation).
Western blot analysis
The cells were washed with pre-cooled PBS and the
culture plate was then placed on ice. Subsequently, 10 µl
phenylmethanesulfonyl fluoride (PMSF) were added followed by lysis
on ice for 30 min, centrifugation at 15,000 × g for 5-10 min at 4°C
and storage at -20°C. The protein extracts were separated through
10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis
(SDS-PAGE) and then transferred onto a polyvinylidene fluoride
(PVDF) membrane. The membrane was then rinsed with Tris-buffered
saline with Tween-20 (TBST) for 5 min and transferred to a 5%
skimmed milk powder blocking solution, and then sealed on a shaker
at room temperature for 2 h. Primary antibodies against RUNX1
(DF6785, 1:1,000; Affinity Biosciences), BAX (AF0120, 1:1,000;
Affinity Biosciences) and GAPDH (P04406, 1:1,000; Affinity
Biosciences) were added to the membrane and incubated overnight at
4°C. The diluent of the secondary antibody goat anti-rabbit IgG
H&L (HRP) (S0001, 1:5,000; Affinity Biosciences) was then added
to the membrane and incubated for 1 h at room temperature. After
the incubation was completed, TBST was used for rinsing three times
for 5 min at room temperature each time. Enhanced chemiluminescence
reagent (Amersham Biosciences) and ImageJ software (version 1.48;
National Institutes of Health) were used to detect protein bands
and to analyze the optical density. GAPDH was used as a loading
control.
RNA antisense purification (RAP) and RNA
pull-down assay
The binding effect of GAS5 and miR-18a-5p was
detected using the RAP kit (Bes5103; BersinBio) and RNA pull down
kit (Thermo Fisher Scientific, Inc.). The biotin-labeled GAS5 probe
(Guangzhou RiboBio Co., Ltd.) was combined with the connection
region of GAS5, and the oligonucleotide probe (Guangzhou RiboBio
Co., Ltd.) was used as a control. The probe was added to the lysed
cells in proportion, and incubated with a vertical mixer for 5 h at
room temperature. Subsequently, the RNA enriched on the magnetic
beads (Thermo Fisher Scientific, Inc.) was washed multiple times
with RNA elution buffer, and the miRNA bound in the complex was
then extracted using TRIzol reagent (R0016, Beyotime Institute of
Biotechnology) and quantitatively analyzed using RT-qPCR.
Subsequently, 50 nmol/l biotin-labeled miR-18a-5p was transfected
into the OCI-ly3 cells for 48 h, and the cells were then lysed with
0.1% NP-40 (P0013F; Beyotime Institute of Biotechnology),
centrifugation at 12,000 × g for 5 min at 4°C. incubated for 1 h.
The combination of 500 µg streptavidin magnetic beads
(HY-K0208, MedChemExpress) and 200 pmol biotin-labeled miR-18a-5p
(Guangzhou RiboBio Co., Ltd.) were added to the RNA and mixed
gently at room temperature, incubated for 30 min at room
temperature. Biotin-scramble was used as a negative control for
biotin labeled miR-18a-5p, and the experimental operation was
consistent with the above. Elution buffer (Thermo Fisher
Scientific) was added to collect the RNA complex pulled down, and
this was then analyzed using RT-qPCR as described above.
Chromatin immunoprecipitation (ChIP)
The protein-gene interactions were identified using
ChIP assay with the Simple Chip Enzymatic Chromatin IP kit (9002S;
Cell Signaling Technology, Inc.). The cells were treated with
formaldehyde and incubated at 37°C for 15 min, and cross-linking
was terminated using glycine. The cells were washed and centrifuged
at 1,000 × g for 5 min at room temperature, and 2 ml cell lysis
buffer were then added to resuspend cells. Subsequently, the cells
were placed on ice for incubation for 15 min for lysis, and were
finally sonicated to share DNA to an average size of 500 bp. The
chromatin solution was cleaned with protein A-agarose beads (Cell
Signaling Technology, Inc) and incubated overnight with normal IgG
(ab172730, 5 µg; Abcam) or anti-RUNX1 antibody (ab272456, 5
µg; Abcam) at 4°C. After the incubation was complete,
protein A-agarose beads were added and inverted for 2 h at 4°C.
Protein G-agarose beads were used to capture the chromatin-immune
complex, and the immune complex was eluted with an elution buffer
containing 1% SDS and 0.1 M NaHCO3 (Cell Signaling
Technology, Inc). NaCl and proteinase K were then mixed and used to
reverse cross-linking at 65°C for 1 h. The interaction of RUNX1 and
the binding region was analyzed using RT-qPCR. The primers
sequences were as follows: R1 forward, CCTGTAATCCCAGCACTTTG and
reverse, CTCGGCTTACTGCAACCTCTG; and R2 forward,
CAAGGTCACGATCTCAGCTC and reverse, CTGTAATCCCAGCACTTTGGG.
Bioinformatics prediction
The targeted miRNA of lncRNA GAS5 was predicted
using online tools, including starBase (http://starbase.sysu.edu.cn/), DIANA (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php)
and NPInter (http://bigdata.ibp.ac.cn/npinter4/browse/). The
potential mRNA targets of miR-18a-5p was predicted using online
tools, including TargetScan (http://www.targetscan.org/vert_72/), PicTar
(https://pictar.mdc-berlin.de/), TarBase
(http://www.microrna.gr/tarbase) and
microT_CDS (http://www.microrna.gr/microT-CDS). The transcription
factor binding sites were predicted by JASPAR (http://jaspar.genereg.net/) software.
Statistical analysis
GraphPad Prism 8.0 (GraphPad Software, Inc.) was
used for statistical analysis. These data are presented as the mean
± standard deviation (SD). One-way ANOVA (followed by Tukey's test)
and an unpaired t-test were used for the determination of
significant differences between groups. The experiments were
carried out independently three times under the same conditions.
P<0.05 was considered to indicate a statistically significant
difference.
Results
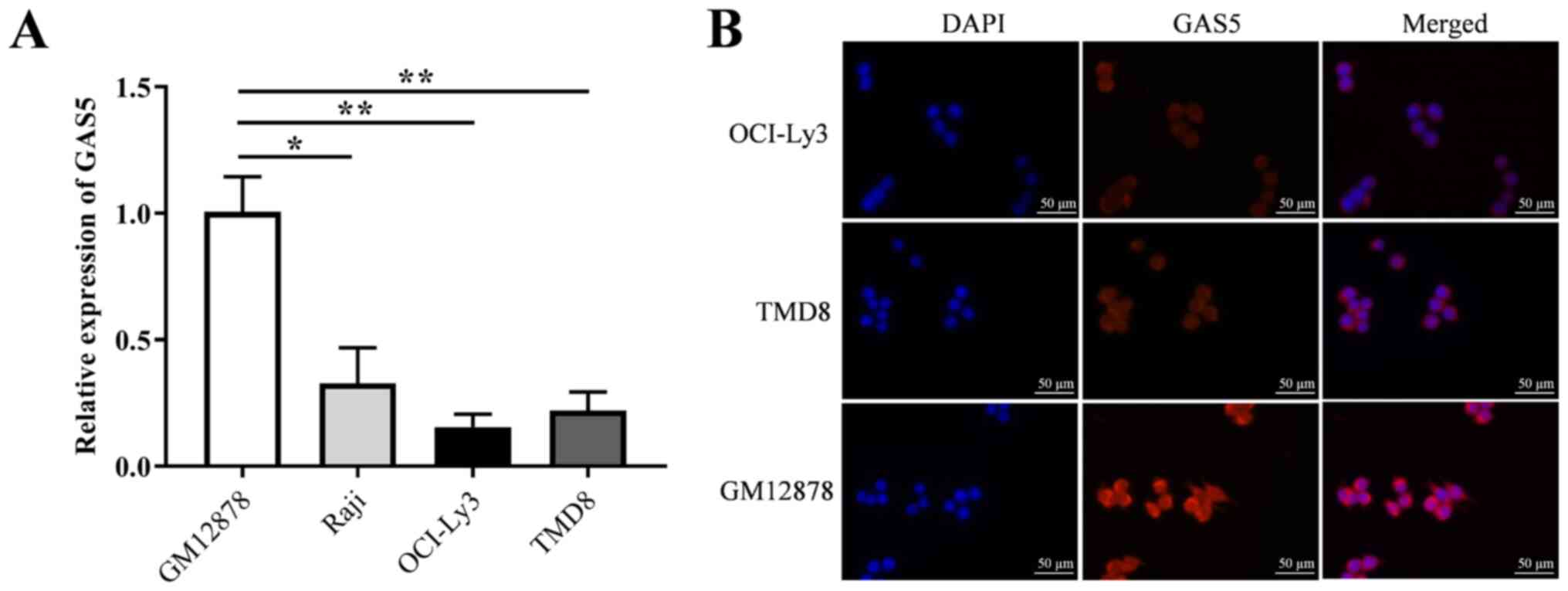
GAS5 expression is downregulated in DLBCL
cell lines
The expression of GAS5 in three DLBCL cell lines
(Raji, OCI-ly3 and TMD8) and a human normal lymphocyte cell line
(GM12878) was detected using RT-qPCR. The results revealed that the
expression level of GAS5 in the DLBCL cells was significantly lower
than that in the GM12878 cell lines (Fig. 1A). In addition, the expression of
GAS5 in the OCI-Ly3 and TMD8 cells was lower than that in the Raji
cells; thus, the subcellular localization of GAS5 was observed in
the OCI-Ly3, TMD8 and GM12878 cells. The corresponding results of
fluorescence detection revealed that the expression level of GAS5
in the OCI-Ly3 and TMD8 cells was markedly lower than that in the
GM12878 cells, and GAS5 was mainly located in the cytoplasm
(Fig. 1B).
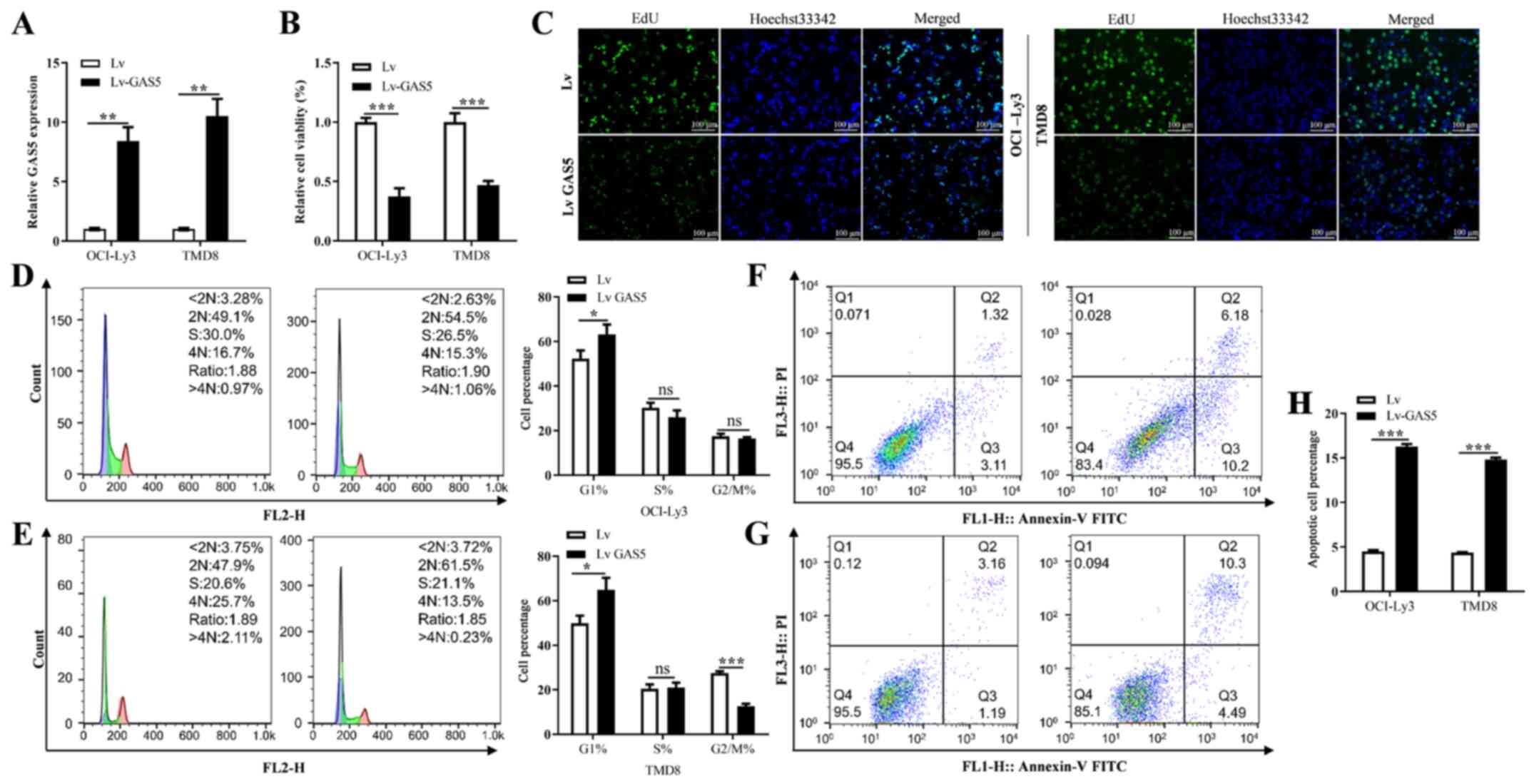
Overexpression of GAS5 regulates the
proliferation, cell cycle progression and apoptosis of DLBCL
cells
To evaluate the function of GAS5 in the
proliferation, cell cycle progression and apoptosis of DLBCL cells,
firstly, lentiviral vector containing the GAS5-encoding sequence
was transfected into the OCI-Ly3 and TMD8 cells. The results
revealed transfection with the GAS5 vector significantly promoted
its expression level when compared with the empty vector group
(Fig. 2A). The overexpression of
GAS5 significantly decreased the viability and proliferation of the
OCI-Ly3 and TMD8 cells when compared with the empty vector group
(Fig. 2B and C). In addition,
GAS5 overexpression markedly suppressed the cell cycle progression
of the DLBCL cells, leading to G1 cycle arrest (Fig. 2D and E). Furthermore, the
overexpression of GAS5 significantly increased the proportion of
apoptotic cells when compared with the empty vector group (Fig. 2F, G and H).
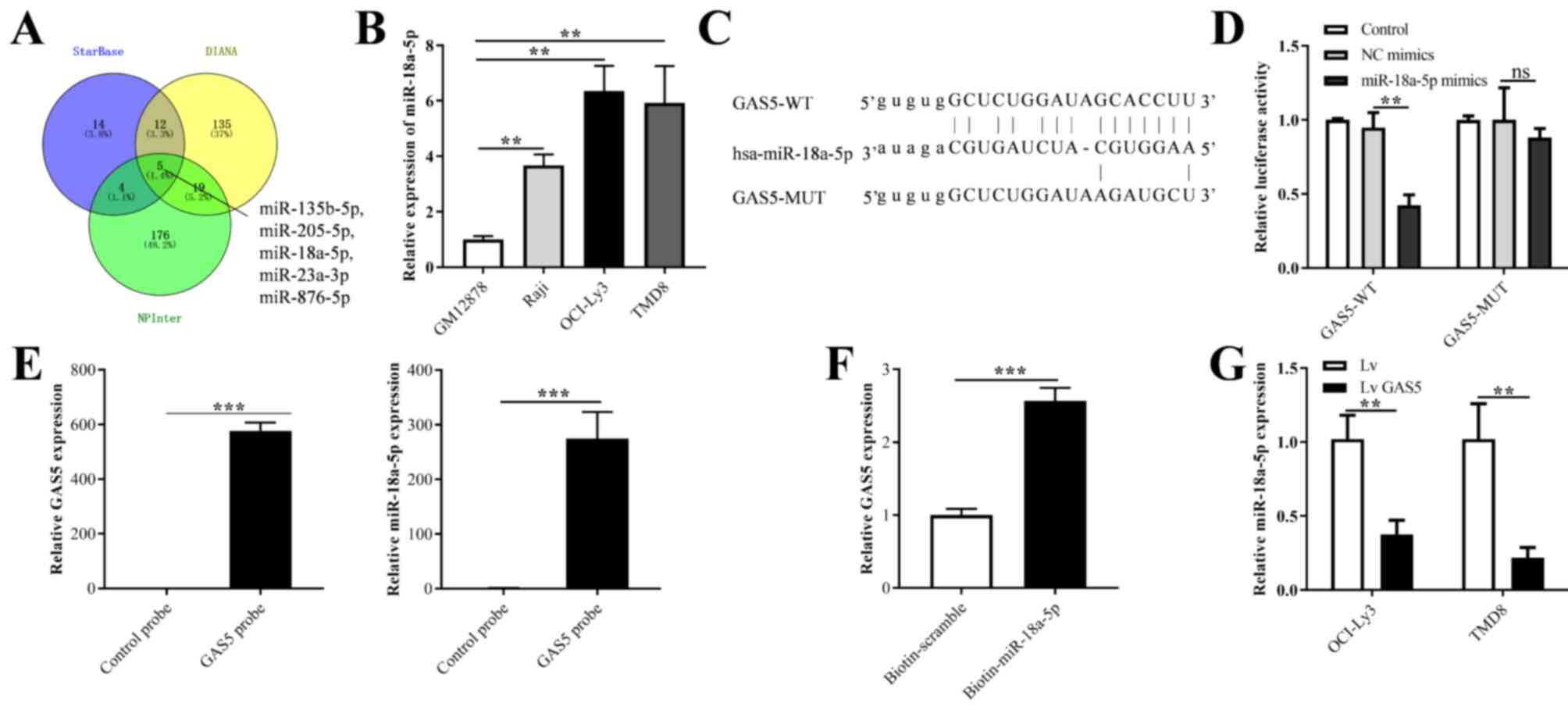
GAS5 functions as a molecular sponge for
miR-18a-5p
Three databases (starBase, DIANA and NPInter) were
used to predict the target miRNAs of GAS5 (Table SI). As illustrated in Fig. 3A, five common miRNAs, including
miR-135b-5p, miR-205-5p, miR-18a-5p, miR-23a-3p and miR-876-5p were
predicted. Based on previous studies mentioned in the
'Introduction' (24,25) and the database prediction results,
miR-18a-5p was shown to be highly expressed in B-cell lymphoma,
indicating that miR-18a-5p may be involved in the progression of
DLBCL. Therefore, miR-18a-5p was selected for the follow-up
mechanistic analysis. As was expected, the expression level of
miR-18a-5p in the three DLBCL cell lines (Raji, OCI-ly3 and TMD8)
was significantly higher than that in the GM12878 cell line
(Fig. 3B). In addition, the
binding site between GAS5 and miR-18a-5p was predicted, and the
binding sequence of CGUGGAA was targeted and recognized by GAS5
(Fig. 3C). Moreover, the OCI-Ly3
cell line was used for mechanistic analysis, and therefore, the
luciferase activity of the miR-18a-5p mimics and GAS5-WT
co-transfection group was significantly decreased in the OCI-Ly3
cells when compared with that in the GAS5-MUT group. Additionally,
no significant changes in luciferase activity were observed in the
control and NC mimic groups (Fig.
3D). The results of RT-qPCR revealed that the expression level
of GAS5 increased with the combination of the biotin-labeled GAS5
probe and GAS5, indicating the specific binding between the probe
and GAS5. In addition, RT-qPCR revealed that the probe specifically
bound by GAS5 'pulled down' a greater amount of miR-18a-5p in the
total RNA of OCI-Ly3 cells than in the control group (Fig. 3E). Furthermore, the RNA pulled
down by biotin-labeled miR-18a-5p was enriched by magnetic beads by
RNA Pull-down. The corresponding RT-qPCR results confirmed that
miR-18a-5p could specifically bind to GAS5 compared with the
biotin-scramble (Fig. 3F).
Furthermore, the overexpression of GAS5 decreased the relative
expression level of miR-18a-5p in the OCI-Ly3 and TMD8 cells when
compared with the control group (Fig.
3G).
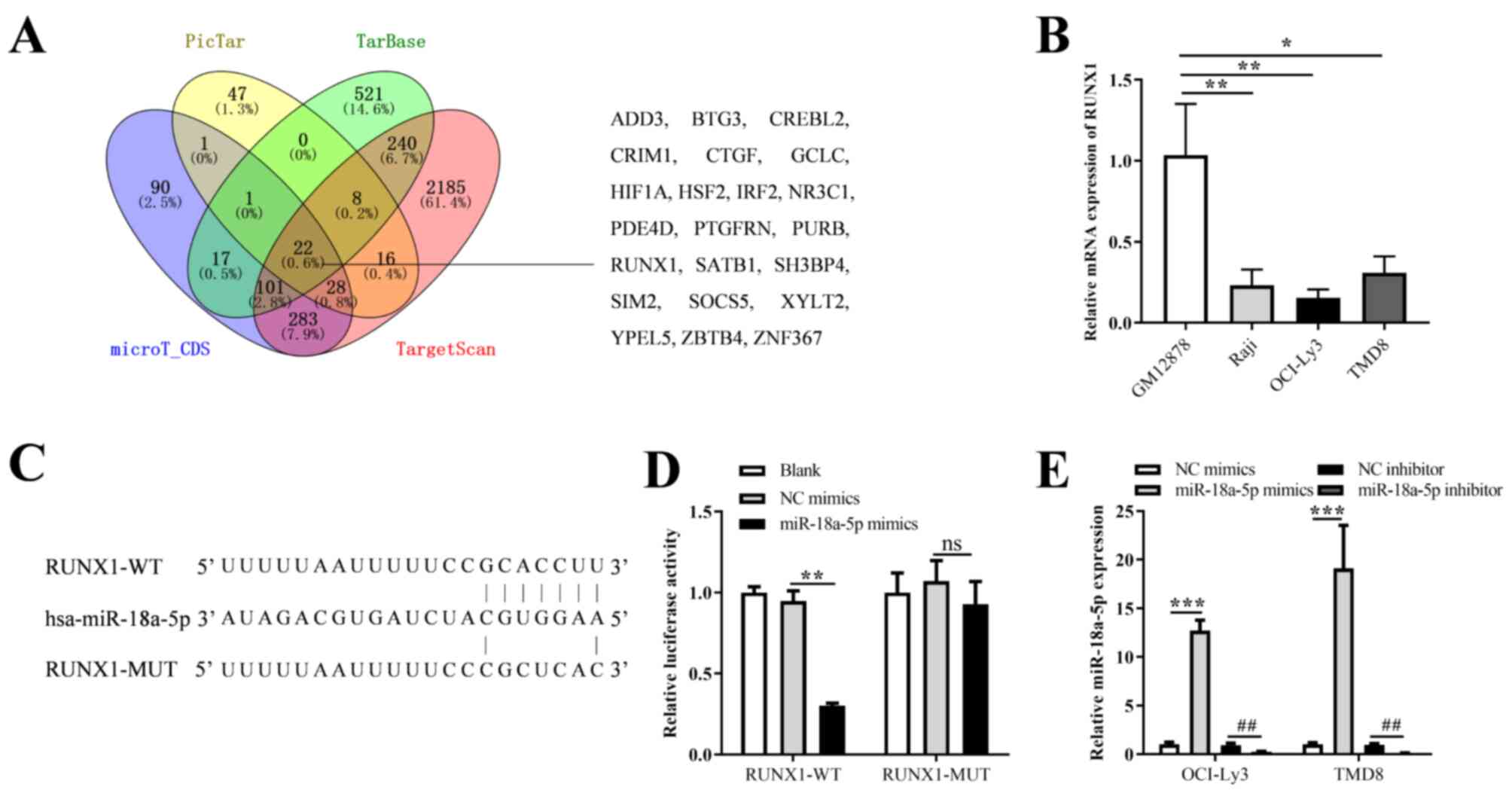
GAS5 functions as a ceRNA by sponging
miR-18a-5p
Four databases (TargetScan, PicTar, TarBase and
microT_CDS) were used to predict the target genes of miR-18a-5p
(Table SII). As illustrated in
Fig. 4A, 22 common genes (ADD3,
BTG3, CREBL2, CRIM1, CTGF, GCLC, HIF1A, HSF2, IRF2, NR3C1, PDE4D,
PTGFRN, PURB, RUNX1, SATB1, SH3BP4, SIM2, SOCS5, XYLT2, YPEL5,
ZBTB4 and ZNF367) were predicted. Based on the previous studies
mentioned in the 'Introduction' (27-29) and the database predictions
results, RUNX1 was selected for the follow-up mechanistic analysis.
As was expected, the expression level of RUNX1 was significantly
lower in the three DLBCL cell lines (Raji, OCI-ly3 and TMD8),
whereas it was higher in the GM12878 cells (Fig. 4B). RUNX1 was identified as a novel
target of miR-18a-5p, and the binding site of GCACCUU was then
predicted (Fig. 4C). In addition,
the results of luciferase reporter assay revealed a significant
reduction in the luciferase activity following co-transfection with
the miR-18a-5p mimic and RUNX1-WT when compared to co-transfection
with the control or NC mimics and RUNX1-WT groups, while no
significant difference was observed in the RUNX1-MUT transfection
groups (Fig. 4D). To further
confirm the interaction between miR-18a-5p and RUNX1, miR-18a-5p
mimics and inhibitor were transfected into the OCI-ly3 and TMD8
cells. The results revealed that miR-18a-5p expression was
increased and suppressed following transfection with miR-18a-5p
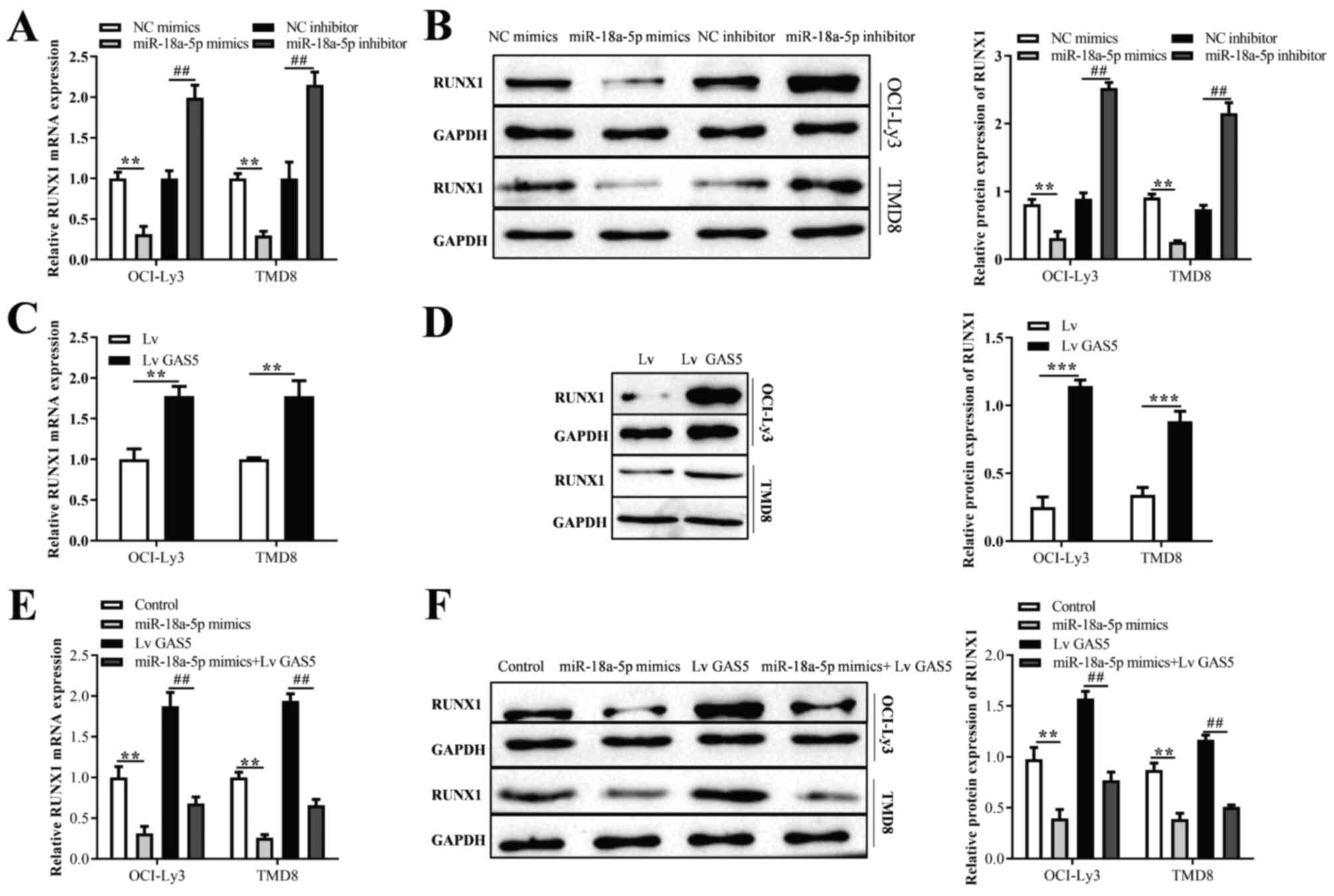
mimics and inhibitor, respectively (Fig. 4E). Moreover, it was found that the
overexpression or inhibition of miR-18a-5p significantly decreased
or increased the mRNA and protein expression of RUNX1, respectively
(Fig. 5A and B). In addition, the
results of RT-qPCR and western blot analysis revealed that the
overexpression of GAS5 enhanced the mRNA and protein expression of
RUNX1 (Fig. 5C and D).
Furthermore, compared with the OCI-ly3 and TMD8 cells transfected
with miR-18a-5p mimics or GAS5 overexpression vector alone, those
co-transfected with miR-18a-5p mimics and GAS5 overexpression
vector exhibited an increased or decreased RUNX1 expression,
respectively (Fig. 5E and F).
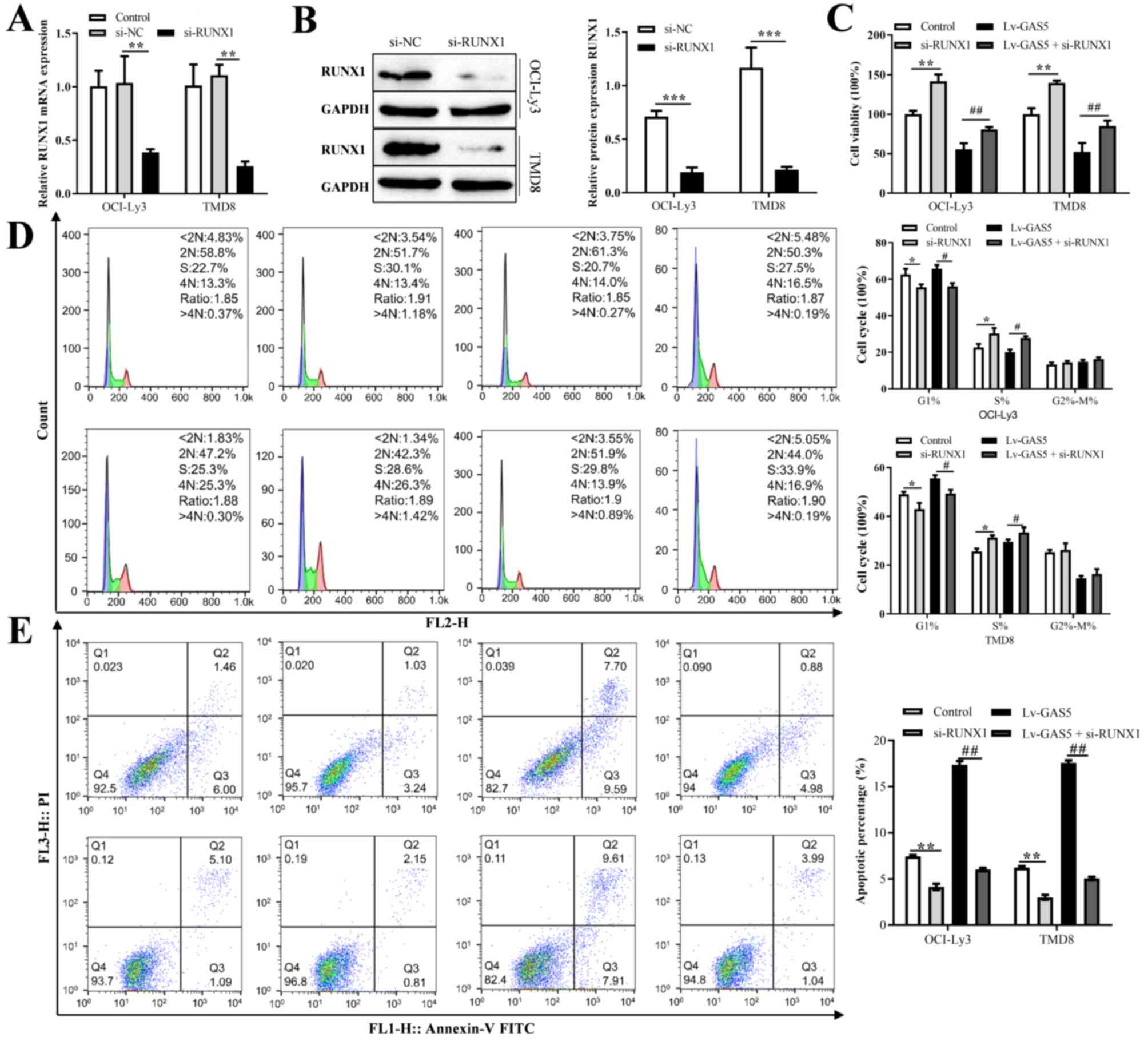
Knockdown of RUNX1 reverses the
GAS5-induced inhibition of the proliferation of DLBCL cells
To explore whether RUNX1 was involved in the
proliferation, cell cycle progression and apoptosis of DLBCL cells
regulated by GAS5, the expression level of RUNX1 was examined
following the knockdown of RUNX1. The results revealed that the
expression of RUNX1 was significantly decreased compared with the
control group (Fig. 6A).
Furthermore, the knockdown of RUNX1 decreased its protein
expression level (Fig. 6B). The
results of rescue experiments then revealed that co-transfection of
the OCI-LY3 and TMD8 cells with GAS5 overexpression vector and
si-RUNX1 reversed the inhibitory effects of GAS5 overexpression on
the proliferation of OCI-LY3 and TMD8 cells; the cell cycle arrest
in the G1 phase and the promotion of cell apoptosis (Fig. 6C-E).
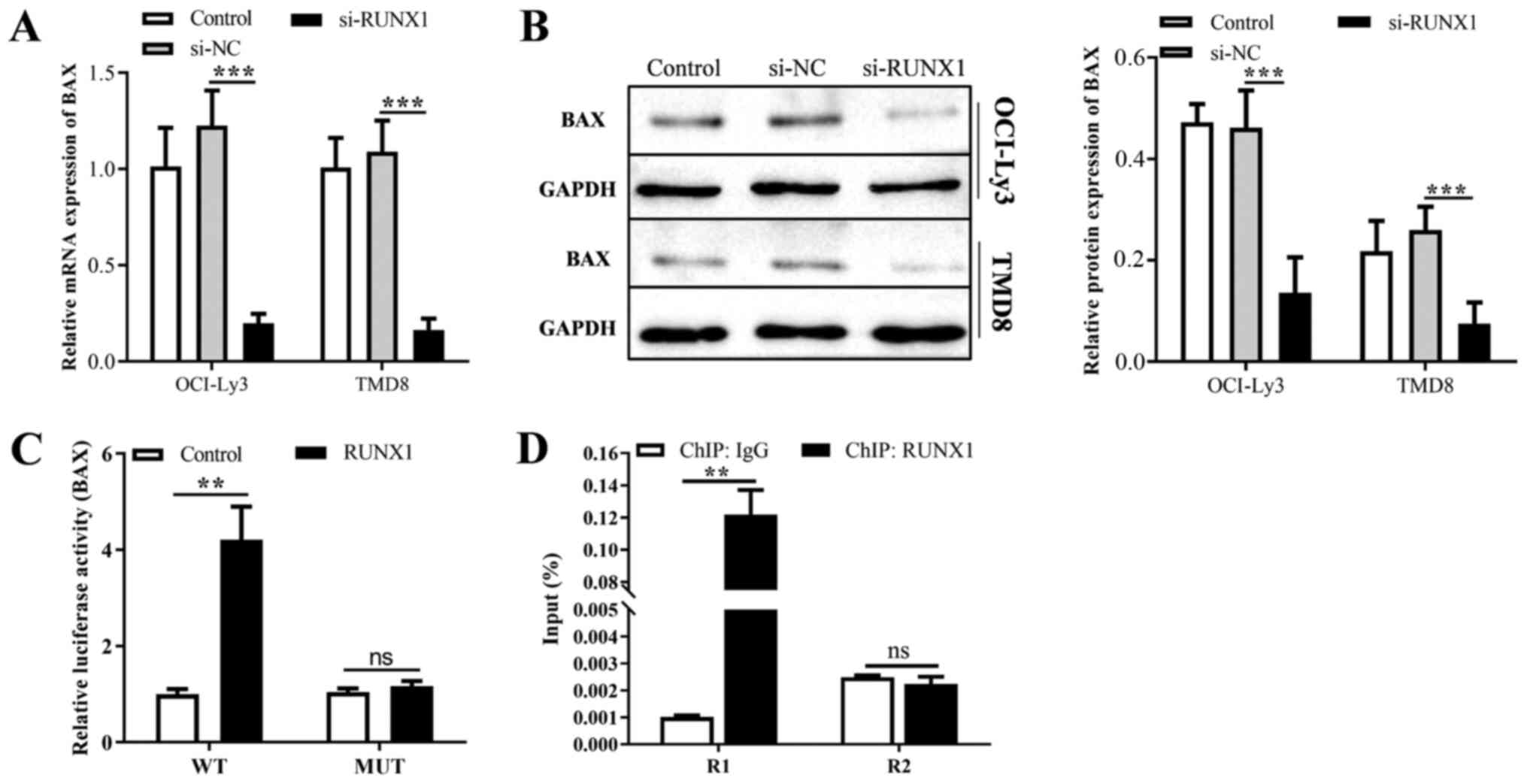
RUNX1 promotes the expression of BAX and
binds to its promoter
JASPAR database query results revealed that a
potential binding site ACTTGAGGT of RUNX1 was found within the
sequence 2000 upstream of the promoter region of BAX. The results
of RT-qPCR and western blot analysis revealed that the knockdown of
RUNX1 decreased the mRNA and protein expression of BAX (Fig. 7A and B). Moreover, luciferase
activity was used to evaluate the binding of RUNX1 to the BAX
promoter. As was expected, RUNX1-WT increased the activity of the
BAX promoter (Fig. 7C).
Furthermore, immunoprecipitation assay of the protein-DNA complexes
was performed, and the results of ChIP-qPCR assay confirmed that
RUNX1 protein enriched the binding site and enhanced the
interaction within the BAX promoter in OCI-Ly3 cells, but not
binding to the control region (Fig.
7D). In summary, lncRNA GAS5 promotes the cell cycle arrest,
suppresses the proliferation and promotes the apoptosis of OCI-Ly3
and TMD8 cells by sponging miR-18a-5p and regulating the expression
of RUNX1 (Fig. 8).
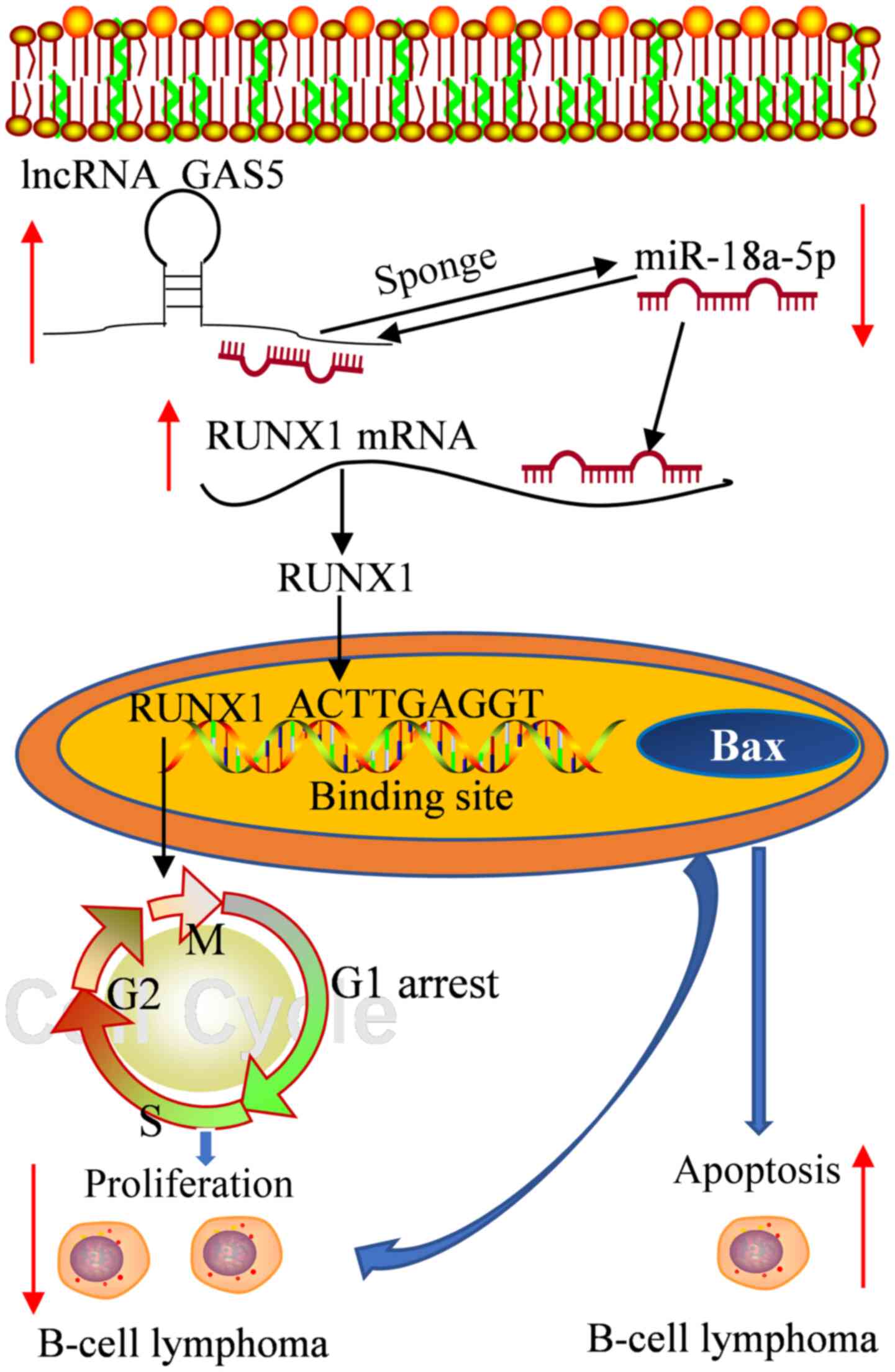 | Figure 8Schematic diagram of the mechanisms
through which lncRNA GAS5/miR-18a-5p/RUNX1 mediates DLBCL cell
proliferation and apoptosis. lncRNA GAS5 attenuates the expression
of miR-18a-5p, whereas it promotes that of RUNX1; miR-18a-5p
attenuates the expression of RUNX1 and interferes with the
expression of RUNX1 by GAS5. On the whole, lncRNA GAS5 promotes the
expression of RUNX1 by sponging miR-18a-5p, which in turn promotes
the cell cycle G1 phase arrest, suppresses proliferation and
promotes the apoptosis of OCI-Ly3 and TMD8 cells. In addition,
RUNX1 binds to the promoter region of BAX, and thus may play a role
in the apoptosis and proliferation of DLBCL cells. lncRNA, long
non-coding RNA; GAS5, growth arrest specific 5; RUNX1, Runt-related
transcription factor 1; DLBCL, diffuse large B-cell lymphoma. |
Discussion
The present study first established the interaction
among GAS5, miR-18a-5p and RUNX1 in DLBCL, and concluded that GAS5
inhibited the proliferation and G1 cycle progression, whereas it
promoted the apoptosis of DLBCL cells by functioning as a ceRNA to
sponge miR-18a-5p and modulate RUNX1 expression. These findings may
provide potential novel therapeutic targets for the treatment of
DLBCL (Fig. 7).
The abnormal expression of lncRNAs has been
identified as a main factor involved in the progression of DLBCL.
The association between lncRNAs and cell proliferation, and the
apoptosis of DLBCL has been previously demonstrated; for example,
lncRNA SNHG16 (31), lncRNA OR3A4
(32), lncRNA AFAP1-AS1 (33) have been shown to play a role in
B-cell lymphoma. A previous study demonstrated that a low GAS5
level was involved in the development of DLBCL and was associated
with a poor prognosis; moreover, the results of protein-protein
interaction network (PPI) network analysis revealed that GAS5
negatively regulated the cell cycle, apoptosis, differentiation,
autophagy and other cell functions (18). This finding is in agreement with
findings of previous studies demonstrating that GAS5 plays the role
of a tumor suppressor gene by inhibiting the proliferation and
promoting the apoptosis of tumor cells in glioma and ovarian
cancers (20,34,35). Indeed, the overexpression of GAS5
has been shown to reduce the expression of the anti-apoptotic
protein, Bcl-2, to promote the apoptosis of bladder cancer and
cervical cancer cells (36).
However, to date, to the best of our knowledge, there is no direct
evidence to verify the association between GAS5, and the
proliferation and apoptosis of DLBCL cells. In the present study,
it was also found that GAS5 was expressed at low levels in DLBCL
cells, indicating the role of GAS5 in this disease. Functionally,
GAS5 reduced the proliferation of DLBCL cells and enhanced cell
apoptosis. In addition, the results revealed that GAS5 induced G1
cell cycle arrest, which was also similar with the findings of a
previous study (37). However,
another study demonstrated that GAS5 was highly expressed in DLBCL,
whereas its expression was low in follicular lymphoma (FL) blood
samples by analyzing a cohort of previously published datasets from
the Gene Expression Omnibus (GEO; accession no. GSE53820), and the
lncRNA-mRNA co-expression network analysis results revealed that
GAS5 was associated with cell anti-apoptosis, cell cycle and other
functions (38). This may be due
to the fact that only six DLBCL blood samples were included in that
study. Moreover, differential analysis and multiple tests on all
genes in the DLBCL and FL samples were performed using the online
analysis tool, GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/); the results
revealed that the expression level of GAS5 in DLBCL and FL was no
longer significant (the adjusted P-value was 0.53) (Table SIII).
Mechanistically, the findings of the present study
validated that GAS5 functioned as a miRNA sponge in DLBCL. As
previously reported, GAS5 may participate in the specific DLBCL
process as a key regulator of the ceRNA network (38). The interaction between lncRNAs and
miRNAs involves the regulation of target mRNAs, and lncRNAs may
sponge and reduce miRNA expression to compete for the regulatory
effects of miRNAs on target gene mRNAs, also known as the ceRNA
mechanism. Previous studies have revealed that GAS5 competes with
miRNAs to regulate mRNA expression as a ceRNA (20,39). Notably, the present study
predicted that miR-18a-5p was the potential target gene of by GAS5
through three online prediction database and validated that
miR-18a-5p was directly targeted by GAS5 in OCI-Ly3 cells by using
luciferase reporter and RNA pull-down assays. Furthermore, the
present study demonstrated that miR-18a-5p expression was markedly
upregulated in DLBCL cell lines, and exhibited a strong affinity
with GAS5, indicating that GAS5 may function in DLBCL mainly
through miR-18a-5p. Similarly, it has been proven in previous
studies that miR-18a-5p plays a key carcinogenic role in several
tumors and is negatively regulated by GAS5 in glioma (20) and prostate cancer cells (26). In prostate cancer, α-solanine is
involved in the regulation of tumor inhibition of miR-18a-5p;
however, those studies did not include RUNX1.
Of note, the present study demonstrated that RUNX1
was expressed at low levels in DLBCL and was targeted by GAS5 and
miR-18a-5p, suggesting an involvement of RUNX1 in DLBCL cells.
Functionally, GAS5 decreased the proliferation of DLBCL cells, and
induced G1 phase arrest and cell apoptosis. However, in rescue
experiments, co-transfection with GAS5 overexpression and si-RUNX1
reversed the effects of GAS5 on the proliferation, cell cycle
progression and apoptosis of DLBCL cells. Similar results have also
been observed in other tumors. For example, RUNX1 has been shown to
markedly inhibit the lncRNA NEF-induced proliferation of gastric
cancer cells (40) and the lncRNA
CASC2-induced proliferation of malignant melanoma cells (41). However, some studies have
indicated that RUNX1 can promote tumor development; for example,
RUNX1 has been shown to reverse the anti-proliferative and
pro-apoptotic effects on glioma cells following the knockdown of
lncRNA HCP5 (42), or to enhance
the lncRNA RNCR3-induced progression of colorectal cancer (43). In addition, previous studies have
also revealed that RUNX1 regulates G1 to S cell cycle progression
in adult hematopoietic stem cells (44), and the knockdown of RUNX1 has been
shown to lead to a decrease in cell proliferation and to G1 cell
cycle arrest in epithelial ovarian carcinoma (45); these findings were similar to
those of the present study. The aforementioned results suggest that
GAS5 and RUNX1 may be potential therapeutic targets for DLBCL.
Furthermore, RUNX1 is a transcription factor that is
well known in the development of cancers for its dual role in the
transcription of specific genes. For example, RUNX1 binds to the
promoter region of the glioma oncogene astrocyte elevated gene-1
(AEG-1), enhances the activity of the AEG-1, and then induces the
proliferation of glioma cells (42). Of note, the results of RT-qPCR,
and luciferase activity and ChIP assays in the present study proved
that RUNX1 increased the activity of BAX and enhanced BAX
expression. It has been reported that BAX is a key regulatory
protein related to apoptosis and the proliferation of DLBCL cells
(46). A previous study also
identified that RUNX1 enhanced the transcriptional activity of BAX,
and si-RUNX1 was shown to play an anti-apoptotic role in human
colon carcinoma (47). Based on
these studies, RUNX1 may be a key factor in the BAX-mediated cell
proliferation and apoptosis of DLBCL, which may further confirm the
tumor suppressor effect of RUNX1 in DLBCL in the present study.
In conclusion, the present study provides evidence
that lncRNA GAS5 plays an anti-tumor and anti-proliferative role in
DLBCL cells. In addition, the potential mechanism identified was
that it inhibited the proliferation and G1 cycle progression, and
promoted the apoptosis of DLBCL cells by functioning as a ceRNA to
regulate RUNX1. These findings provide potential novel therapeutic
targets for DLBCL.
Supplementary Data
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YM and YJ performed the experiments and collected
the data, and confirmed the authenticity of all the raw data. YM
was a major contributor to the writing of the manuscript. XC and MQ
were responsible for data analysis and visualization. WZ and YW
conceived and designed the study, and they were major contributors
in critically revising the manuscript. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Offner F, Samoilova O, Osmanov E, Eom HS,
Topp MS, Raposo J, Pavlov V, Ricci D, Chaturvedi S, Zhu E, et al:
Frontline rituximab, cyclophosphamide, doxorubicin, and prednisone
with bortezomib (VR-CAP) or vincristine (R-CHOP) for non-GCB DLBCL.
Blood. 126:1893–1901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alizadeh AA, Eisen MB, Davis RE, Ma C,
Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al:
Distinct types of diffuse large B-cell lymphoma identified by gene
expression profiling. Nature. 403:503–511. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee WJ, Won KH, Won CH, Chang SE, Choi JH,
Moon KC, Park CS, Huh J, Suh C and Lee MW: Secondary cutaneous
diffuse large B-cell lymphoma has a higher international prognostic
index score and worse prognosis than diffuse large B-cell lymphoma,
leg type. Acta Derm Venereol. 96:245–250. 2016. View Article : Google Scholar
|
|
4
|
Castillo JJ, Winer ES and Olszewski AJ:
Sites of extranodal involvement are prognostic in patients with
diffuse large B-cell lymphoma in the rituximab era: An analysis of
the Surveillance, Epidemiology and End Results database. Am J
Hematol. 89:310–314. 2014. View Article : Google Scholar
|
|
5
|
Li S, Young KH and Medeiros LJ: Diffuse
large B-cell lymphoma. Pathology. 50:74–87. 2018. View Article : Google Scholar
|
|
6
|
Wang QM, Lian GY, Song Y, Huang YF and
Gong Y: LncRNA MALAT1 promotes tumorigenesis and immune escape of
diffuse large B cell lymphoma by sponging miR-195. Life Sci.
231:1163352019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi X, Cui Z, Liu X, Wu S, Wu Y, Fang F
and Zhao H: LncRNA FIRRE is activated by MYC and promotes the
development of diffuse large B-cell lymphoma via Wnt/β-catenin
signaling pathway. Biochem Biophys Res Commun. 510:594–600. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qian CS, Li LJ, Huang HW, Yang HF and Wu
DP: MYC-regulated lncRNA NEAT1 promotes B cell proliferation and
lymphomagenesis via the miR-34b5p-GLI1 pathway in diffuse large
B-cell lymphoma. Cancer Cell Int. 20:872020. View Article : Google Scholar
|
|
9
|
Zhao CC, Jiao Y, Zhang YY, Ning J, Zhang
YR, Xu J, Wei W and Kang-Sheng G: Lnc SMAD5-AS1 as ceRNA inhibit
proliferation of diffuse large B cell lymphoma via Wnt/β-catenin
pathway by sponging miR-135b-5p to elevate expression of APC. Cell
Death Dis. 10:2522019. View Article : Google Scholar
|
|
10
|
Wang Y, Zhang M, Xu H, Wang Y, Li Z, Chang
Y, Wang X, Fu X, Zhou Z, Yang S, et al: Discovery and validation of
the tumor-suppressive function of long noncoding RNA PANDA in human
diffuse large B-cell lymphoma through the inactivation of MAPK/ERK
signaling pathway. Oncotarget. 8:72182–72196. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang L, Zhao XH, Mao YL, Wang JF, Zheng
HJ and You QS: Long non-coding RNA RP11-468E2.5 curtails colorectal
cancer cell proliferation and stimulates apoptosis via the JAK/STAT
signaling pathway by targeting STAT5 and STAT6. J Exp Clin Cancer
Res. 38:4652019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Wang Y, Zhang CG, Xiao H-J, Xiao HJ,
Hu J-M, Hou JM and He JD: Effect of long non-coding RNA Gas5 on
proliferation, migration, invasion and apoptosis of colorectal
cancer HT-29 cell line. Cancer Cell Int. 18:42018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Y, Yin L, Chen C, Zhang X and Wang S:
Long non-coding RNA GAS5 inhibits migration and invasion in gastric
cancer via interacting with p53 protein. Dig Liver Dis. 52:331–338.
2020. View Article : Google Scholar
|
|
14
|
Huang W, Shi Y, Han B, Wang Q, Zhang B, Qi
C and Liu F: LncRNA GAS5-AS1 inhibits glioma proliferation,
migration, and invasion via miR-106b-5p/TUSC2 axis. Hum Cell.
33:416–426. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng S, Li M, Miao K and Xu H: lncRNA
GAS5-promoted apoptosis in triple-negative breast cancer by
targeting miR-378a-5p/SUFU signaling. J Cell Biochem.
121:2225–2235. 2020. View Article : Google Scholar
|
|
16
|
Nakamura Y, Takahashi N, Kakegawa E,
Yoshida K, Ito Y, Kayano H, Niitsu N, Jinnai I and Bessho M: The
GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a
result of t(1;3)(q25;q27) in a patient with B-cell lymphoma. Cancer
Genet Cytogenet. 182:144–149. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dousti F, Shahrisa A, Ansari H, Hajjari M,
Tahmasebi Birgani Y, Mohammadiasl J and Tahmasebi Birgani M: Long
non-coding RNAs expression levels in diffuse large B-cell lymphoma:
An in silico analysis. Pathol Res Pract. 214:1462–1466. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Senousy MA, El-Abd AM, Abdel-Malek RR and
Rizk SM: Circulating long non-coding RNAs HOTAIR, Linc-p21, GAS5
and XIST expression profiles in diffuse large B-cell lymphoma:
Association with R-CHOP responsiveness. Sci Rep. 11:20952021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luengo-Gil G, García-Martínez E,
Chaves-Benito A, Conesa-Zamora P, Navarro-Manzano E,
González-Billalabeitia E, García-Garre E, Martínez-Carrasco A,
Vicente V and Ayala de la Peña F: Clinical and biological impact of
miR-18a expression in breast cancer after neoadjuvant chemotherapy.
Cell Oncol (Dordr). 42:627–644. 2019. View Article : Google Scholar
|
|
20
|
Liu Q, Yu W, Zhu S, Cheng K, Xu H, Lv Y,
Long X, Ma L, Huang J, Sun S, et al: Long noncoding RNA GAS5
regulates the proliferation, migration, and invasion of glioma
cells by negatively regulating miR-18a-5p. J Cell Physiol.
234:757–768. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang C, Zhang X, Wang HM, Liu XM, Zhang
XJ, Zheng B, Qian GR and Ma ZL: MicroRNA-18a-5p functions as an
oncogene by directly targeting IRF2 in lung cancer. Cell Death Dis.
8:e27642017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miao WJ, Yuan DJ, Zhang GZ, Liu Q, Ma HM
and Jin QQ: lncRNA CASC2/miR 18a 5p axis regulates the malignant
potential of nasopharyngeal carcinoma by targeting RBBP8. Oncol
Rep. 41:1797–1806. 2019.
|
|
23
|
Liang B, Zhou C, Cui S, Lu H, Xu R, Xue D,
Zou S and He X: Upregulation of miR-18a-5p promotes the
proliferation of prostate cancer via inhibiting the expression of
SLC40A1. Pathol Res Pract. 224:1534482021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lim EL, Trinh DL, Scott DW, Chu A,
Krzywinski M, Zhao Y, Robertson AG, Mungall AJ, Schein J, Boyle M,
et al: Comprehensive miRNA sequence analysis reveals survival
differences in diffuse large B-cell lymphoma patients. Genome Biol.
16:182015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ, et al: A microRNA polycistron as a potential human
oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang J, Hao T, Sun J, Wei P and Zhang H:
Long noncoding RNA GAS5 modulates α-Solanine-induced
radiosensitivity by negatively regulating miR-18a in human prostate
cancer cells. Biomed Pharmacother. 112:1086562019. View Article : Google Scholar
|
|
27
|
Sood R, Kamikubo Y and Liu P: Role of
RUNX1 in hematological malignancies. Blood. 129:2070–2082. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seo W, Ikawa T, Kawamoto H and Taniuchi I:
Runx1-Cbfβ facilitates early B lymphocyte development by regulating
expression of Ebf1. J Exp Med. 209:1255–1262. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miao YS, Zhao YY, Zhao LN, Wang P, Liu YH,
Ma J and Xue YX: MiR-18a increased the permeability of BTB via
RUNX1 mediated down-regulation of ZO-1, occludin and claudin-5.
Cell Signal. 27:156–167. 2015. View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Zhu Q, Li Y, Guo Y, Hu L, Xiao Z, Liu X,
Wang J, Xu Q and Tong X: Long non-coding RNA SNHG16 promotes
proliferation and inhibits apoptosis of diffuse large B-cell
lymphoma cells by targeting miR-497-5p/PIM1 axis. J Cell Mol Med.
23:7395–7405. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng H, Zhao B and Wang Y: FOXM1-induced
upregulation of lncRNA OR3A4 promotes the progression of diffuse
large B-cell lymphoma via Wnt/β-catenin signaling pathway. Exp Mol
Pathol. 115:1044512020. View Article : Google Scholar
|
|
33
|
Gao H, Sun Y, Chen J, Jin H and Yang W:
Long non-coding RNA AFAP1-AS1 promotes cell growth and inhibits
apoptosis by binding to specific proteins in germinal center
B-cell-like diffuse large B-cell lymphoma. Am J Transl Res.
12:8225–8246. 2020.
|
|
34
|
Li J, Yang C, Li Y, Chen A, Li L and You
Z: LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome
formation. Biosci Rep. 38:382018.
|
|
35
|
Li G, Cai Y, Wang C, Huang M and Chen J:
LncRNA GAS5 regulates the proliferation, migration, invasion and
apoptosis of brain glioma cells through targeting GSTM3 expression.
The effect of LncRNA GAS5 on glioma cells. J Neurooncol.
143:525–536. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao W, Shan B, He D, Cheng Y, Li B, Zhang
C and Duan C: Recent progress in characterizing long noncoding RNAs
in cancer drug resistance. J Cancer. 10:6693–6702. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang Y, Shen Z, Yan Y, Wang B, Zhang J,
Shen C, Li T, Ye C, Gao Z, Peng G, et al: Long non-coding RNA GAS5
inhibits cell proliferation, induces G0/G1 arrest and apoptosis,
and functions as a prognostic marker in colorectal cancer. Oncol
Lett. 13:3151–3158. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tian L, He Y, Zhang H, Wu Z, Li D and
Zheng C: Comprehensive analysis of differentially expressed
profiles of lncRNAs and mRNAs reveals ceRNA networks in the
transformation of diffuse large B-cell lymphoma. Oncol Lett.
16:882–890. 2018.PubMed/NCBI
|
|
39
|
Gu J, Wang Y, Wang X, Zhou D, Wang X, Zhou
M and He Z: Effect of the LncRNA GAS5-MiR-23a-ATG3 axis in
regulating autophagy in patients with breast cancer. Cell Physiol
Biochem. 48:194–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang X, Jiang X, Zhou L, Wang Z, Huang H
and Wang M: LncRNA NEF is involved the regulation of gastric
carcinoma cell proliferation by targeting RUNX1. Mol Med Rep.
19:2051–2056. 2019.PubMed/NCBI
|
|
41
|
Zhang Y, Qian W, Feng F, Cao Q, Li Y, Hou
Y, Zhang L and Fan J: Upregulated lncRNA CASC2 may inhibit
malignant melanoma development through regulating miR-18a-5p/RUNX1.
Oncol Res. 27:371–377. 2019. View Article : Google Scholar
|
|
42
|
Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H,
Xi Z, Li Z and Liu Y: Role of HCP5-miR-139-RUNX1 feedback loop in
regulating malignant behavior of glioma cells. Mol Ther.
24:1806–1822. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu G, Wang H, Yuan D, Yao J, Meng L, Li K,
Zhang Y, Dang C and Zhu K: RUNX1-activated upregulation of lncRNA
RNCR3 promotes cell proliferation, invasion, and suppresses
apoptosis in colorectal cancer via miR-1301-3p/AKT1 axis in vitro
and in vivo. Clin Transl Oncol. 22:1762–1777. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Friedman AD: Cell cycle and developmental
control of hematopoiesis by Runx1. J Cell Physiol. 219:520–524.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Keita M, Bachvarova M, Morin C, Plante M,
Gregoire J, Renaud MC, Sebastianelli A, Trinh XB and Bachvarov D:
The RUNX1 transcription factor is expressed in serous epithelial
ovarian carcinoma and contributes to cell proliferation, migration
and invasion. Cell Cycle. 12:972–986. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Asciolla JJ, Renault TT and Chipuk JE:
Examining BCL-2 family function with large unilamellar vesicles. J
Vis Exp. 68:42912012.
|
|
47
|
Wu D, Ozaki T, Yoshihara Y, Kubo N and
Nakagawara A: Runt-related transcription factor 1 (RUNX1)
stimulates tumor suppressor p53 protein in response to DNA damage
through complex formation and acetylation. J Biol Chem.
288:1353–1364. 2013. View Article : Google Scholar :
|