Introduction
The H3K4me3 histone mark is frequently found at the
promoters of genes that are undergoing active transcription
(1,2). In mammals, to methylate H3K4 there
are six methyltransferases that can function as components of
complexes (3), whilst there are
six demethylases that can remove these methyl groups from this
mark. Specifically, two of these demethylases, namely
lysine-specific demethylase (LSD or KDM)1/KDM1A and LSD2/KDM1B,
belong to the flavin adenine dinucleotide-dependent homologues of
the amine oxidase family, which can remove methyl groups from
dimethylated and monomethylated H3K4 (4). By contrast, four KDM5 proteins,
KDM5A, KDM5B, KDM5C and KDM5D, are members of the Jumonji (Jmj)C
class of Fe (II)- and 2-oxoglutarate-dependent proteins, which can
demethylate H3K4me3 and H3K4me2 through the JmjC domain (5). Interacting with repressor complexes,
the KDM1 and KDM5 demethylases can repress gene transcription and
are overexpressed in a wide range of cancers, including renal cell
carcinoma and head and neck cancers (4,5).
KDM5B (also known as JARID1B or PLU-1) was first identified as
being upregulated in breast (6,7)
and prostate cancer (8). These
two types of cancer have been the focus of previous studies in
oncogenesis (9,10).
The KDM5 family of demethylases are unique among
those that use an Fe2+- and 2-oxoglutarate-dependent
mechanism for removing methyl groups (5,6).
The catalytic core of KDM5 is separated into the JmjN and JmjC
domains by sequences, which include the A-T rich interaction domain
(ARID or BRIGHT domain) and the plant homeodomain 1 (PHD1)
(5,6). The DNA-binding ARID domains in KDM5A
and B (Fig. 1A) can bind to
GC-rich sequences (11,12), whilst the PHD1 domain interacts
with unmethylated H3K4 (13).
However, in both KDM5A and B proteins, the JmjN and JmjC domains
lie adjacent to each other and interact closely (14-16).
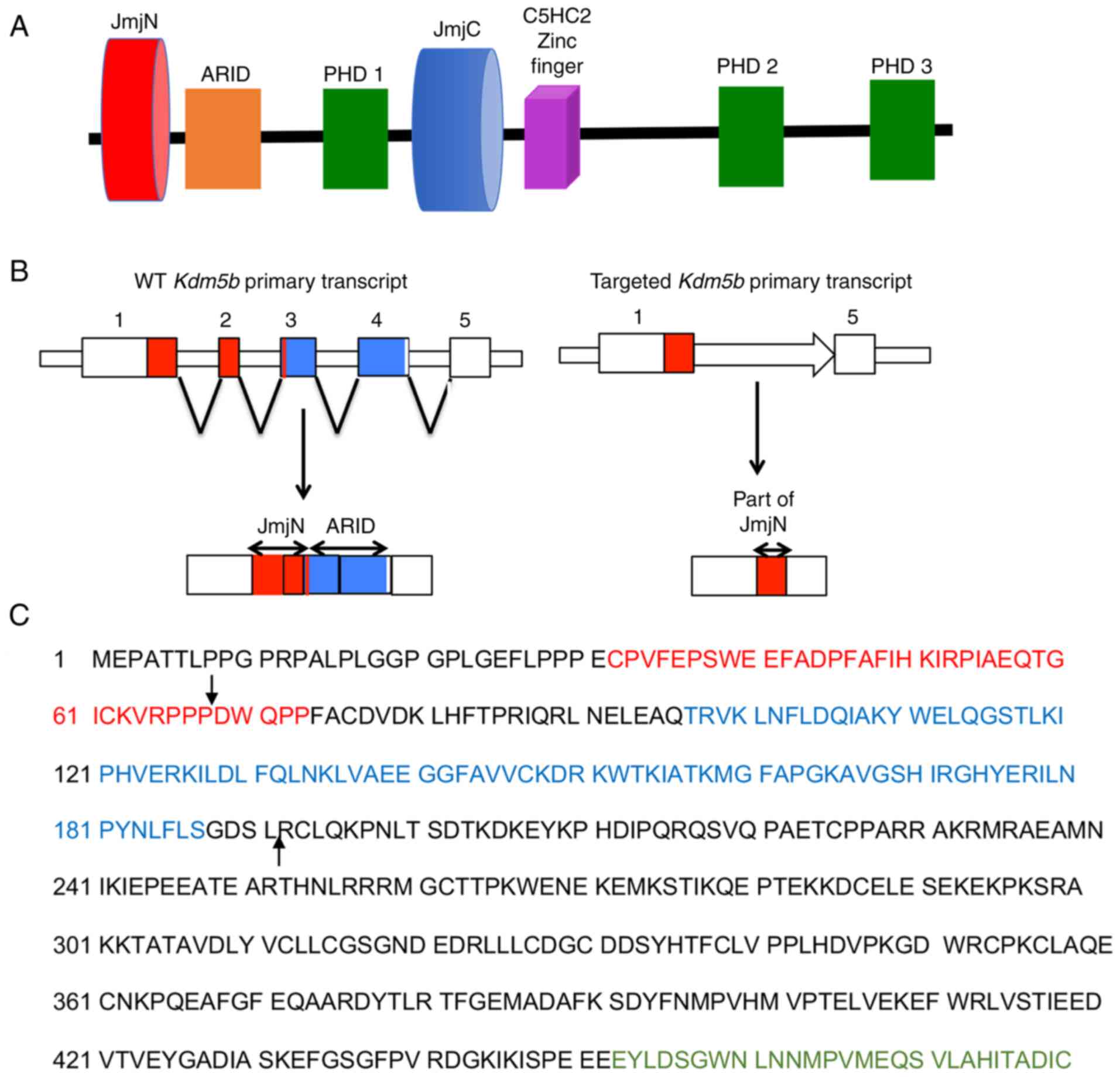 | Figure 1Kdm5B RNA is spliced from exon
1 to exon 5 in the mouse ΔARID-Kdm5B construct. (A) Linear
domain structure of mouse KDM5B. (B) Schematic representation
depicting the mRNA splicing event of the WT-Kdm5b primary
transcript that results in the production of the ΔARID-Kdm5B
transcript. Left, red areas in exons 1, 2 and 3 represent the JmjN
domain; blue areas in exons 3 and 4 represent the ARID domain.
Right, the mRNA splicing event of the engineered Kdm5b
primary transcript resulting in the splicing together of exons 1
and 5, which deletes the ARID domain and part of the JmjN domain.
(C) WT murine KDM5B protein sequence showing the JmjN domain (red,
residues 32-73), the ARID domain (blue, 97-187), and the beginning
of the JmjC domain (green, 453-619). Arrows indicate the beginning
and end of the spliced region observed in the ΔARID-KDM5. Jmj,
Jumonji; ARID, A-T rich interaction domain; PHD1, plant homeodomain
1; KDM5, lysine-specific demethylase 5B; WT, wild-type. |
Previous studies using the expression of mutated
constructs found that deletions of either the ARID, JmjN or JmjC
domains led to the loss of histone demethylase function (7,8).
However, linking the JmjN and JmjC domains of KDM5B, which results
in the deletion of the ARID and PHD1 domains, did not decrease
demethylase activity in vitro (17). In addition, a shortened construct
of KDM5B, which contains deletion of the PHD1 domain and most of
the ARID domain (amino acids 102-369 deleted), retains its
demethylase activity (14).
However, although apparently dispensable in terms of demethylase
activity, the PHD1 domain can influence the re-modelling of the
catalytic core by binding to H3K4me0 (15,18).
Human and mouse KDM5B share a 95% sequence homology
at protein level (6). To
investigate the function of KDM5B, a number of knockout (KO)
genotypes of mouse have been developed. The Kdm5b KO C57BL/6
mice developed by Albert et al (19), which targeted exon 6, resulted in
neonatal lethality due to respiratory failure resulting from
neurological abnormalities. Another homozygous Kdm5b KO mice
developed by Catchpole et al (20) generated on the same C57BL/6
background, which targeted exon 1, caused early embryonic
lethality. In addition, a Kdm5b KO mice developed by Zou
et al (21), with a
deletion between exon 2 and exon 3, remain viable and fertile on
the C57BL/6 background. However, when bred on the FVB/N background,
both males and females show increased rates of mortality and
females showed reduced fertility and abnormal mammary gland
development. It is possible that targeting different domains in
KDM5B can explain different phenotypes (21).
A Kdm5b transgenic mouse model was previously
developed (20), where a splicing
event led to the removal of exons 2, 3 and 4 via the splicing of
exon 1 to 5. This mouse genotype is known as the ΔARID mouse, which
is viable and fertile, with the only phenotype being a transient
delay in mammary gland development (20). In this mouse, in-frame splicing of
the primary transcript from exons 1-5 resulted in the deletion of
the entire ARID domain. However, this splicing event also resulted
in the truncation of the JmjN domain, with amino acids Asp69,
Trp70, Gln71, Pro72 and Pro73 deleted from the carboxyl end.
However, all other domains, including the PHD1 and JmjC domains,
remain intact (Fig. 1B).
Whilst the JmjN domain is clearly required for
demethylase activity, neither the deletion nor the mutation studies
aforementioned examined whether the partial deletion of the JmjN
domain together with the ARID domain affects demethylase activity.
Since the ΔARID mice with this deletion remain viable and fertile
(20), it is important to
establish whether the KDM5B protein they express has demethylase
activity. In the present study this question was addressed by
developing a plasmid construct encoding ΔARID-KDM5B and assaying
demethylase activity in Cos-1 cells transfected with this
construct. In silico homology modelling followed by
molecular dynamics (MD) simulations was then applied to document
any changes in the domains of ΔARID-KDM5B, with the aim of
providing supportive evidence for the loss of demethylase
activity.
Materials and methods
Construction of the Kdm5b-ΔARID construct
in the pcDNA3.1 plasmid
Mouse WT-Kdm5b cDNA was originally isolated
from a mouse mammary cell line 410.4 (obtained from the originator,
Dr Gloria Heppner, Michigan Cancer Foundation, Detroit, USA) by
Madsen et al (22). To
create the KDM5B-ΔARID construct, as previously seen in the
ΔARID mice (20), the
KDM5B sequence corresponding to the splicing of exon 1 to
exon 5 was designed by the authors and synthesised by GenScript for
ligation into the pcDNA3.1 plasmid (cat. no. V87020; Thermo Fisher
Scientific, Inc.), which was originally carrying the WT-KDM5B
sequence but was cut out using NotI and Bsu36I.
Fig. 1C shows the effect of this
splicing event on the protein sequence.
Immunofluorescence staining
Mouse WT-Kdm5b, mouse ΔARID-Kdm5b cDNA
and a human ΔJmjC construct were all encoded in the pcDNA3.1
plasmid (Sigma-Aldrich; Merck KGaA). They were transfected into
Cos-1 cells using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. In total, 0.5 µg plasmids were used per
transfection. Cos-1 (cat. no. CRL-1650; American Type Culture
Collection) were routinely cultured in DMEM (Thermo Fisher
Scientific, Inc.) with 10% foetal calf serum (Thermo Fisher
Scientific, Inc.), at 37°C and 5% CO2, The plasmid
containing KDM5B with a deletion in the JmjC domain, derived from
the WT-Kdm5b cDNA, was provided by Dr Degui Chen, State Key
Laboratory of Molecular Biology, Shanghai Institutes for Biological
Sciences (Shanghai, China) (8).
In total, 24 h after transfection the cells were fixed in 4%
paraformaldehyde for 15 min at room temperature, permeabilised with
Triton X-100 for 10 min at room temperature, washed and
sequentially stained with an antibody to KDM5B at a dilution of
1:700 for 2 h at room temperature. This antibody was an in-house
rabbit antiserum that was raised to the C-terminal domain of human
KDM5B corresponding to amino acid residues 1283-1473 (expressed and
produced in E coli M15). This antibody shows specificity for
KDM5B and reacts with both human and mouse KDM5B (23). To identify H3K4me3 staining, a
mouse monoclonal antibody (cat. no. ab1012; dilution, 1:100; Abcam)
was used, with incubation for 2 h at room temperature. Binding of
the primary antibodies was visualised using Alexa Fluor®
546 donkey anti-rabbit IgG (cat. no. A10040) and Alexa
Fluor® 488 goat anti-mouse IgG (cat. no. A-10680;
Molecular Probes; Thermo Fisher Scientific, Inc.) secondary
antibodies, both diluted to 1:500 and incubation was for 1 h at
room temperature. Cells were then mounted in mounting medium
containing DAPI (cat. no. ab104139; Abcam) and visualized under a
Zeiss AX10 immunofluorescence microscope (magnification, ×630).
Histone demethylation analysis
Mouse WT-Kdm5b and mouse ΔARID-Kdm5b
cDNA both encoded in the pcDNA3.1 plasmid and the vector control
(pcDNA3.1; Sigma-Aldrich; Merck KGaA), were transfected into 293T
cells using Polyethylenimine (cat. no. 23966-2; Polysciences, Inc.)
with 10 µg plasmid used per transfection. 293T cells (ATCC;
cat. no. CRL-3216) were routinely cultured in DMEM with 10% foetal
calf serum, at 37°C and 5% CO2. After 48 h cells were
suspended in a hypotonic cell lysis buffer (5 mM HEPES, 50 mM KCl,
10 mM MgSO4 .7H2O, 0.05% NP-40, 3
mM DTT, 1 mM PMSF and a cocktail of protease inhibitors), incubated
at room temperature for 1 min and centrifuged for 5 min at 1,000 ×
g at 4°C. The cell pellet was then resuspended and washed three
times in ice-cold RSB washing buffer (10 mM NaCl, 10 mM Tris-HCl pH
8.0 and 3 mM MgCl2). Subsequently, 0.4% trypan blue
staining was used to check for complete nuclear extraction as
viewed under a light microscope. After nuclear extraction, intact
nuclei were positive for trypan blue staining under the microscope
and are smaller in size compared with those in control 293T cells.
Intact cells with whole nuclei were stained negative.
Finally, nuclear proteins were extracted using RIPA
buffer (50 mM Tris HCl, 150 mM NaCl, 1.0% (v/v) NP-40, 0.5% (w/v)
sodium deoxycholate, 1.0 mM EDTA, 0.1% (w/v) SDS and 0.01% (w/v)
sodium azide, pH 7.4) with protease inhibitors. The nuclear
extracts were incubated on ice for 10 min before being cleared by
centrifugation at 100,000 × g for 10 min at 4°C. Protein
concentration was measured using Bradford Assay (Bio-Rad
Laboratories, Inc.). The demethylation activity was measured using
the histone H3(K4) demethylase activity quantification assay kit
from Abcam (cat. no. ab113455) using 5 µg each extract.
Statistical analysis
A two-tailed student's t-test was used to analyse
the difference between the WT-KDM5B and ΔARID-KDM5B results from
the demethylase assay. Data are presented as the mean ± standard
deviation. P≤0.05 was considered to indicate a statistically
significant difference. The results presented represent three
biological replicates.
Homology modeling
The Swiss-Model webserver (https://swissmodel.expasy.org) (24,25) was used for the homology modelling
of the mouse WT-KDM5B structural model using the FASTA-formatted
target protein sequence with the UniProt entry number 'Q80Y84'
(https://www.uniprot.org/uniprot/Q80Y84). The crystal
structure of human WT-KDM5A with the PDB ID 5K4L was used as the
template (16). The template and
KDM5B shared a sequence identity of 64.43%. The 3D structure of the
KDM5A in the PDB format was without any gaps and all of the
segments were solved. The mutant ΔARID-KDM5B (missing amino acid
residues 69-191; Fig. 1C) was
generated by manipulating the primary file in a text editor. All
systems were minimised and equilibrated using the AMBER version 16
software (https://ambermd.org/) (26) before performing the MD
simulations. Evaluation of the homology model was performed by
calculating the Z-score using AMBER 16. The Z-score is an
estimation of the comparability of the model to the
experimentally-derived structures with similar sizes of the target
protein. The Z-score for the ΔARID-KDM5B model was 0.68±0.05.
Z-scores ~0.0 would indicate a native-like structure, whilst
Z-scores <-4.0 would indicate a low-quality model. Therefore,
the Z score of the present model lied within the range of scores
calculated for proteins of similar size with experimentally
determined structures, indicating a good overall quality of the
built model.
MD simulations
Topology and coordinate files for the WT-KDM5B and
ΔARID-KDM5B systems were generated in TIP3P water using AMBER16
(https://ambermd.org/) (26) with the 'tLEaP module' of AMBER 16
(26). The systems were minimised
in two stages using the AMBER 16 package program. In the first
step, 1,000 steps of minimisation with restraint on solvent were
performed, which this was followed by 2,500 steps of minimisation
without restraint. MD was performed in three stages using the AMBER
program. First, 500-psec heating steps were accomplished from
0-300K with restraint on solvents. Subsequently, 500 ps
equilibration steps in constant temperature was run before 100 ns
sampling or production steps (NPT) finally completed the
simulation.
Periodic boundary conditions were applied during the
simulations. The NPT runs used the Langevin algorithm (https://ambermd.org/) (26), whilst the pressure was controlled
using the isotropic position scaling protocol in the AMBER
barostat. The Particle Mesh Ewald method was employed with a
cut-off radius of 12 Å for electrostatic and van der Waals
interactions for proteins (27).
Results
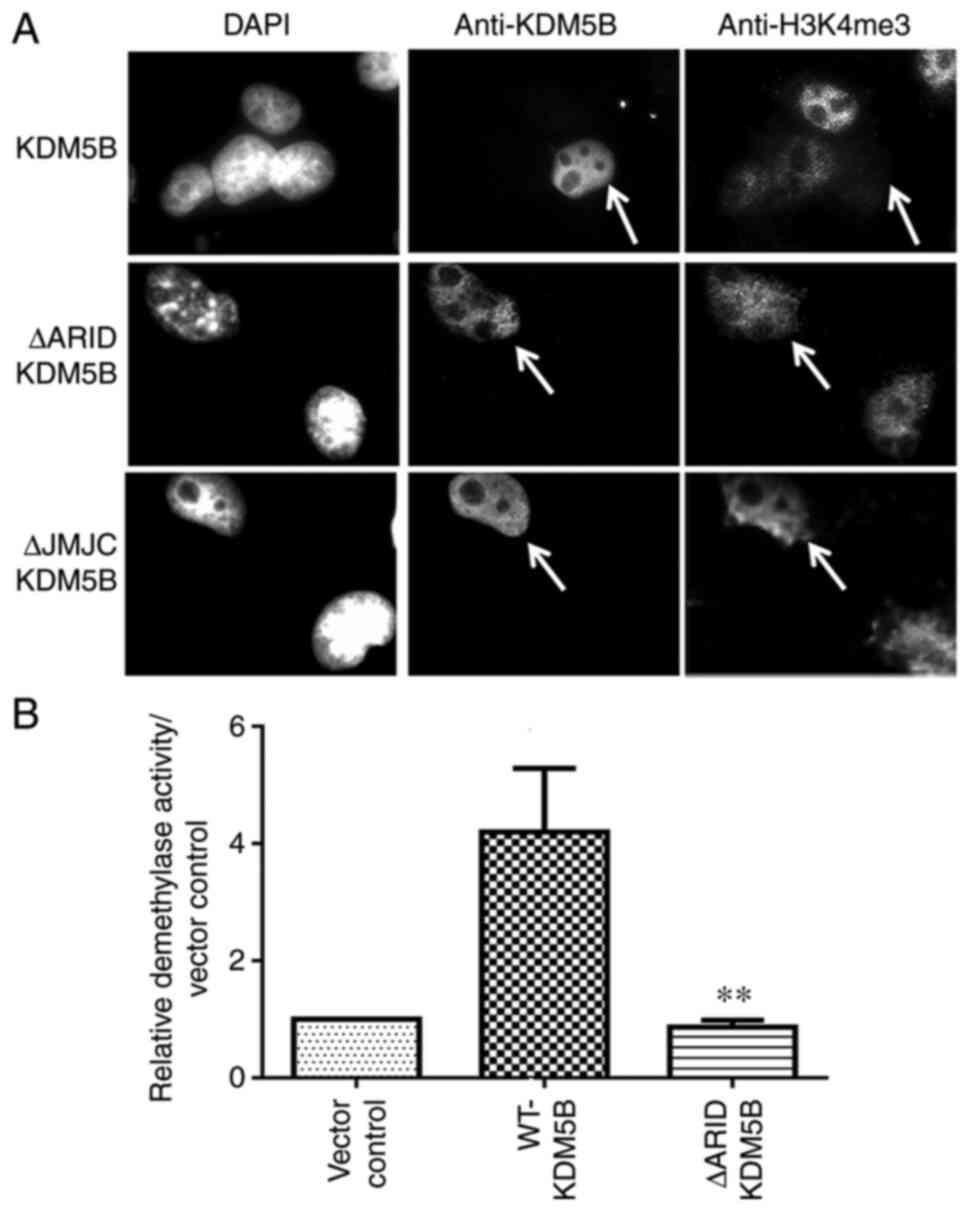
Mouse ΔARID-KDM5B does not have
demethylase activity
The deletion that occurred in the ΔARID-KDM5B by
splicing out exons 2-4 resulted in the expression of a smaller
transcript and protein with the ARID domain (residues 96-188) and
part of the JmjN domain deleted (residues 69-73; Fig. 1B). cDNAs encoding the
WT-Kdm5b and the ΔARID-Kdm5b sequence were
transfected into Cos-1 cells, which do not endogenously express
KDM5B (23). The cells were
co-stained with antibodies for H3K4me3 and KDM5B. A human
KDM5B construct with the JmjC domain deleted, which was
known to lack demethylase activity (7,8),
was also transfected into Cos-1 cells. Cells expressing the
ΔARID-KDM5B protein showed no clear reduction in the level of
H3K4me3, which was also observed in cells transfected with the
catalytically-dead JmjC mutant construct (Fig. 2A). However, H3K4me3 was markedly
downregulated in cells expressing WT-KDM5B. These data indicate
that the deletion that occurred in murine ΔARID-KDM5B results in
the loss of demethylase activity.
To further confirm these findings, in vitro
demethylase assays were performed on 293T cells transfected with
WT-Kdm5b, ΔARID-Kdm5b or vector control construct.
The nuclear proteins were extracted and subjected to an ELISA-based
demethylase activity assay. As shown in Fig. 2B, there was a significant loss of
demethylase activity in the ΔARID-KDM5B protein compared with that
in the wild-type protein, with the ΔARID-Kdm5b nuclear
extract yielding similar results to the vector control. Together
with the results from immunofluorescence staining, these data
indicate that the loss of five amino acids from the JmjN, in
addition to the ARID domain, results in the loss of enzymatic
activity of KDM5B.
MD simulation reveals conformational
changes between wild type KDM5B and DARID-KDM5B
Molecular modelling of WT-KDM5B and ΔARID-KDM5B
after 100 ns MD simulation was performed (Fig. 3A and B). As with the structural
models, the JmjN and JmjC domains are in close proximity to each
other in WT-KDM5B, whereas this association appeared to be
disturbed in ΔARID-KDM5B (Fig. 3A and
B). Further comparison of these two models obtained from the
last frame of 100 nsec MD simulation revealed that the JmjC domain
appeared more compact in the ΔARID-KDM5B model compared with that
of WT-KDM5B (Fig. 3A and B). This
observation was further evidenced by comparing the atomic
positional fluctuations of the Cα atoms in WT-KDM5B compared with
those in ΔARID-KDM5B during the MD simulation (Fig. 3C). Although there were differences
in the root mean square fluctuations within the truncated JmjN
domain of ΔARID-KDM5B, these positional fluctuations were
particularly pronounced in the JmjC domain compared with WT-KDM5B
(Fig. 3C and D). This suggests
that deletion of the 69-DWQPP-73 sequence and the ARID domain
resulted in conformational changes in the JmjC domain (residues
453-619) that increased the flexibility of this domain (Fig. 3D). This observation was further
supported by the time dependence of root mean square deviation
(RMSD) for the backbone atoms relative to the starting structure
during 50 ns MD simulations in both WT-KDM5B and ΔARID-KDM5B
(Fig. 3E). The RMSD curves show
that both simulations have reached equilibrium after ~30 nsec,
which was indicated by the relatively stable RMSD values from 30
nsec onwards until the end of the simulations (Fig. 3E). However, the ΔARID-KDM5B had a
much higher RMSD value of of 8-10 Å, compared with 4-5 Å for
WT-KDM5B (Fig. 3E), meaning that
ΔARID-KDM5B initially had a higher degree of instability but with
time reached a stable state. This suggests greater fluctuations and
altered dynamics in the protein structure of ΔARID-KDM5B.
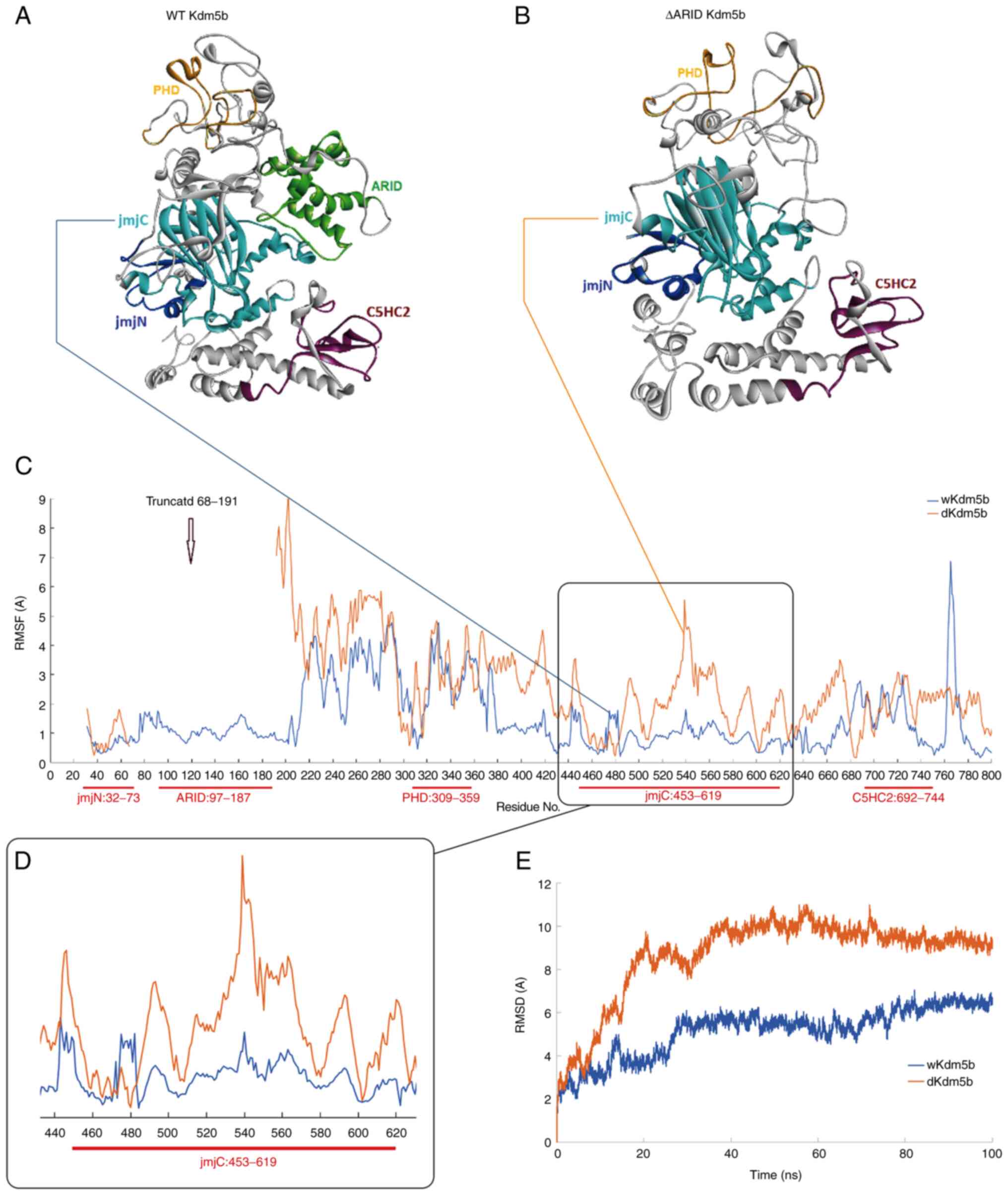 | Figure 3ΔARID-KDM5B shows changes in the
conformation of the JmjC domain. Molecular models of (A) WT-KDM5B
and (B) ΔARID-KDM5B obtained from the last frame of the 100 nsec MD
simulation. (C) Atomic positional fluctuations, in Å, of Cα atoms
in the WT-KDM5B, represented by the blue line, compared with those
in the ΔARID-KDM5B, represented by the brown line. (D) The residues
in the JmjC domain of the ΔARID-KDM5B show notably increased
degrees of flexibility compared with that in WT-KDM5B. (E) Time
dependence of root mean square deviation (RMSD), also in Å, for the
backbone atoms relative to the starting structure during 100 nsec
MD simulations of both WT-KDM5B (blue) and ΔARID-KDM5B (brown).
Jmj, Jumonji; ARID, A-T rich interaction domain; KDM5,
lysine-specific demethylase 5B; RMSF, root mean square
fluctuations; RMSD, root mean square deviation; WT, wild-type; MD,
molecular dynamics. |
Discussion
Both JmjN and JmjC are required for the core
demethylase enzymatic activity (7,8).
Although separated by other sequences, in the 3-D protein structure
these domains lie adjacent to each other (14-18). The present study revealed that the
ΔARID-KDM5B protein, where the ARID domain and five amino acids
from the carboxyl end of the JmjN domain are deleted, has lost its
demethylase activity. While previous mutational experiments show
that deletion of the JmjN domain results in loss of enzyme activity
(7,8), the downstream effects of the partial
deletion the JmjN domain as previously seen in the ΔARID mouse has
not been previously investigated. The splice variant of KDM5B
expressed in the ΔARID mouse is the only form of KDM5B RNA
expressed and produced by using a targeting vector designed to
remove exons 2-4 (20). Compared
with the embryonic lethal KDM5B KO strains where several strains
were identified, only one strain of ΔARID mouse could be
identified, indicating that the splicing of exon 1 to exon 5 is a
rare event (20). Since the last
15 bases in exon 2 translate into the final five amino acid
residues (Asp69, Trp70, Gln71, Pro72 and Pro73) of the JmjN domain,
these amino acids were deleted from the expressed protein (20). These five amino acids are 100%
conserved between the mouse and human KDM5 families (6) and tend to pack closely with the JmjC
domain (14-16), suggesting that the deletion of
these amino acids would negatively affect catalytic function.
Previous studies by Horton et al (17) and Johansson et al (14) strongly support the concept that
deletion of the ARID and PHD1 domains from KDM5B would not abolish
demethylase activity per se. However, the PHD1 domain can be
crucial for the recruitment of KDM5A or KDM5B to H3K4me0 (13-15). The PHD1 domain is not deleted in
ΔARID-KDM5B.
Modelling and subsequent MD simulation of the
protein confirms that the JmjN and JmjC regions of the protein are
juxtaposed in WT-KDM5B (14,17). In addition, the residues in the
truncated JmjN domain of ΔARID-KDM5B showed increased degrees of
fluctuations compared with WT-KDM5B, suggesting a change in
conformation. Changes in the atomic positional fluctuations of the
JmjC domain were also observed during the MD simulation, which
provides in silico findings that support the experimental
observation that ΔARID-KDM5B has no demethylase activity. However,
deletion of the ΔARID domain and five amino acids in the JmjN
domain did not result in differences in the conformation of other
domains flanking the JmjC domain, highlighting the importance of
the JmjN and JmjC domains in the catalytic activity of KDM5B.
Indeed, the fact that the PHD1 domain did not show any significant
fluctuations suggests that this demethylase-null KDM5B protein
retains its function in the PHD1 domain, including its recruitment
to H3K4me0 (13). RMSD analysis
in the present study suggests that despite the greater
fluctuations, ΔARID-KDM5B is a stable protein, since both the
WT-KDM5B and ΔARID-KDM5B systems reached equilibrium after 30 nsec
and remained stable for the remaining 70 nsec of the
simulation.
To the best of our knowledge, ΔARID mice express an
experimentally-induced variant of KDM5B (20) that has not been reported to be
observed in humans. However, other variants of KDM5B have been
reported in patients with intellectual disability (ID) (28). One such variant showing the loss
of exon 4, which encodes a part of the ARID domain and leads to an
in-frame change, has been identified in identical twins with ID
(28). Since sufficient amounts
of variant protein could not be isolated, its effect on demethylase
activity could not be assayed. Nevertheless, this finding from a
patient with ID emphasises the importance of checking brain
function in mice expressing ΔARID-KDM5B. Indeed, KDM5B is highly
expressed in embryonic mouse brain (22), and in the adult brain KDM5B
negatively regulates the neurogenesis of neural stem cells
(29). In addition, in
Drosophila, flies lacking the KDM5 demethylase activity
(LID) remain viable and fertile but show behavioural defects
(30).
KDM5B was identified as being upregulated in breast
and prostate cancer (6-8) and it has been widely studied in
these cancers. A KDM5B variant (RBP2-H1), which can be found at
lower levels in some normal tissues, such as testis (23), has also been found to be expressed
at higher levels in the majority of melanomas (31). This isoform contains additional
amino acids (aa238-274) corresponding to exon 6 (31), which is normally absent in the
dominant form of KDM5B. In breast cancer, KDM5B is expressed most
highly in the estrogen receptor-positive subgroup and is classified
as a luminal lineage driving oncogene (32). However, it was also found to be
upregulated in other cancers, including bladder, lung, gastric and
liver (9,33-35). Evidence that KDM5B can drive
cancer cell proliferation comes from previous observations that its
levels of expression correlated with poor prognosis and that
knocking down its expression resulted in the inhibition of cell
proliferation in some cancer cell lines, including colorectal and
hepatocellular carcinoma lines (36-38). These data have led to the search
for small-molecule weight inhibitors of KDM5B for potential
clinical applications (14,16,39-42). The inhibitors that are being
developed, with a view to targeting the KDM5 proteins for cancer
therapy, are primarily focused on the inhibition of demethylase
function (14,16,39-42).
In the present study, the importance of the ARID
domain and the five amino acids of the C-terminus of the JmjN
domain on the demethylase function were not compared. This should
be addressed in a future study. However, results from the present
study show that the ΔARID-KDM5B is catalytically inactive for the
demethylation of methylated H3K4, yet mice expressing this mutant
remain healthy and fertile (20).
By contrast, Kdm5b-knockout is either embryonic lethal
(20) or results in neonatal
lethality (19). This indicates
that although the KDM5B protein serves crucial functions in
development, its demethylase activity is dispensable. It is
therefore important to evaluate if these functions and the domains
responsible for their activity are involved in oncogenesis. The
ΔARID mouse model could provide a model for investigating this and
for addressing other questions, including whether the
demethylase-independent effect on mitochondrial function seen in
the Drosophila KDM5 analogue LID, is also seen in mammalian
KDM5B (30).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SJ and KMR performed the homology modelling and
molecular dynamics stimulation. KMR contributed designing and
interpreting the simulation modelling experiments. SC made the
constructs and performed the transfection studies, JC and CWES
performed the demethylase assays, JB and JTP conceived and designed
the study, and interpreted the data. SC, JB and JTP confirmed the
authenticity of the data presented in Figs. 1 and 2. JC and CWES confirm the authenticity
of the data presented in Fig. 2,
SJ and KMR confirm the authenticity of the data presented in
Fig. 3. All authors read and
approved the final version of this manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Bernstein BE, Kamal M, Lindblad-Toh K,
Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK,
Kulbokas EJ III, Gingeras TR, et al: Genomic maps and comparative
analysis of histone modifications in human and mouse. Cell.
120:169–181. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cruz C, Della Rosa M, Krueger C, Gao Q,
Horkai D, King M, Field L and Houseley J: Tri-methylation of
histone H3 lysine 4 facilitates gene expression in ageing cells.
Elife. 7:e340812018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roa CR and Dou Y: Hijacked in cancer: The
KMT2 (MLL) family of methyltransferases. Nat Rev Cancer.
15:334–346. 2015. View
Article : Google Scholar
|
|
4
|
Maiques-Diaz A and Somervaille TC: LSD1:
Biologic roles and therapeutic targeting. Epigenomics. 8:1103–1016.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harmeyer KM, Facompre ND, Herlyn M and
Basu D: JARID1 histone demethylases: Emerging targets in cancer.
Trends Cancer. 3:713–725. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu PJ, Sundquist K, Baeckstrom D, Poulsom
R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P,
Freemont P and Taylor-Papadimitriou J: A novel gene (PLU-1)
containing highly conserved putative DNA/chromatin binding motifs
is specifically up-regulated in breast cancer. J Biol Chem.
274:15633–15645. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamane K, Tateishi K, Klose RJ, Fang J,
Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P
and Zhang Y: PLU-1 is an H3K4 demethylase involved in
transcriptional repression and breast cancer cell proliferation.
Mol Cell. 25:801–812. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z,
Ma Y, Yu Y, Lin H, Chen AP and Chen CD: JARID1B is a histone H3
lysine 4 demethylase up-regulated in prostate cancer. Proc Natl
Acad Sci USA. 104:19226–19231. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xhabija B and Kidder BL: KDM5B is a master
regulator of the H3K4-methylome in stem cells, development and
cancer. Semin Cancer Biol. 57:79–85. 2019. View Article : Google Scholar :
|
|
10
|
Jose A, Shenoy GG, Sunil Rodrigues G,
Kumar NAN, Munisamy M, Thomas L, Kolesar J, Rai G, Rao PPN and Rao
M: Histone demethylase KDM5B as a therapeutic target for cancer
therapy. Cancers (Basel). 12:21212020. View Article : Google Scholar
|
|
11
|
Scibetta AG, Santangelo S, Coleman J, Hall
D, Chaplin T, Copier J, Catchpole S, Burchell J and
Taylor-Papadimitriou J: Functional analysis of the transcription
repressor PLU-1/JARID1B. Mol Cell Biol. 27:7220–7235. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tu S, Teng YC, Yuan C, Wu YT, Chan MY,
Cheng AN, Lin PH, Juan LJ and Tsai MD: The ARID domain of H3K4
demethylase RBP2 binds to a DNA CCGCCC motif. Nat Struct Mol Bio.
15:419–421. 2008. View Article : Google Scholar
|
|
13
|
Klein BJ, Piao L, Xi Y, Rincon-Arano H,
Rothbart SB, Peng D, Wen H, Larson C, Zhang X, Zheng X, et al: The
histone-H3K4-specific demethylase KDM5B binds to its substrate and
product through distinct PHD fingers. Cell Rep. 6:315–335. 2014.
View Article : Google Scholar
|
|
14
|
Johansson C, Velupillai S, Tumber A,
Szykowska A, Hookway ES, Nowak RP, Strain-Damerell C, Gileadi C,
Philpott M, Burgess-Brown N, et al: Structural analysis of human
KDM5B guides histone demethylase inhibitor development. Nat Chem
Biol. 12:539–545. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Longbotham JE, Chio CM, Dharmarajan V,
Trnka MJ, Torres IO, Goswami D, Ruiz K, Burlingame AL, Griffin PR
and Fujimori DG: Histone H3 binding to the PHD1 domain of histone
demethylase KDM5A enables active site remodeling. Nat Commun.
10:942019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vinogradova M, Gehling VS, Gustafson A,
Arora S, Tindell CA, Wilson C, Williamson KE, Guler GD, Gangurde P,
Manieri W, et al: An inhibitor of KDM5 demethylases reduces
survival of drug-tolerant cancer cells. Nat Chem Biol. 12:531–538.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Horton JR, Engstrom A, Zoeller EL, Liu X,
Shanks JR, Zhang X, Johns MA, Vertino PM, Fu H and Cheng X:
Characterization of a linked jumonji domain of the KDM5/JARID1
family of histone H3 lysine 4 demethylases. J Biol Chem.
291:2631–2646. 2016. View Article : Google Scholar :
|
|
18
|
Torres IO, Kuchenbecker KM, Nnadi CI,
Fletterick RJ, Kelly MJ and Fujimori DG: Histone demethylase KDM5A
is regulated by its reader domain through a positive-feedback
mechanism. Nat Commun. 6:62042015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Albert M, Schmitz SU, Kooistra SM,
Malatesta M, Morales Torres C, Rekling JC, Johansen JV, Abarrategui
I and Helin K: The histone demethylase Jarid1b ensures faithful
mouse development by protecting developmental genes from aberrant
H3K4me3. PLoS Genet. 9:e10034612013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Catchpole S, Spencer-Dene B, Hall D,
Santangelo S, Rosewell I, Guenatri M, Beatson R, Scibetta AG,
Burchell JM and Taylor-Papadimitriou J: PLU-1/JARID1B/KDM5B is
required for embryonic survival and contributes to cell
proliferation in the mammary gland and in ER+ breast
cancer cells. Int J Oncol. 38:1267–1277. 2011.PubMed/NCBI
|
|
21
|
Zou MR, Cao J, Liu Z, Huh SJ, Polyak K and
Yan Q: Histone demethylase jumonji AT-rich interactive domain 1B
(JARID1B) controls mammary gland development by regulating key
developmental and lineage specification genes. J Biol Chem.
289:17620–17633. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Madsen B, Spencer-Dene B, Poulsom R, Hall
D, Lu PJ, Scott K, Shaw AT, Burchell JM, Freemont P and
Taylor-Papadimitriou J: Characterisation and developmental
expression of mouse Plu-1, a homologue of a human nuclear protein
(PLU-1) which is specifically up-regulated in breast cancer. Mech
Dev. 119(Suppl 1): S239–S246. 2002. View Article : Google Scholar
|
|
23
|
Barrett A, Madsen B, Copier J, Lu PJ,
Cooper L, Scibetta AG, Burchell J and Taylor-Papadimitriou J: PLU-1
nuclear protein, which is upregulated in breast cancer, shows
restricted expression in normal human adult tissues: A new
cancer/testis antigen? Int J Cancer. 101:581–586. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arnold K, Bordoli L, Kopp J and Schwede T:
The SWISS-MODEL workspace: A web-based environment for protein
structure homology modelling. Bioinformatics. 22:195–201. 2006.
View Article : Google Scholar
|
|
25
|
Schwede T, Kopp J, Guex N and Peitsch M:
Swiss-Model: An automated protein homology-modeling server. Nucleic
Acids Res. 31:3381–3385. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Case DA, Cheatham TE III, Darden T, Gohlke
H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B and Woods
RJ: The Amber biomolecular simulation programs. J Comput Chem.
26:1668–1688. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Darden T, York D and Pedersen L: Particle
mesh Ewald: An N log (N) method for Ewald sums in large systems. J
Chem Phys. 98:10089–10092. 1993. View Article : Google Scholar
|
|
28
|
Lebrun N, Mehler-Jacob C, Poirier K,
Zordan C, Lacombe D, Carion N, Billuart P and Bienvenu T: Novel
KDM5B splice variants identified in patients with developmental
disorders: Functional consequences. Gene. 679:305–313. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou Q, Obana EA, Radomski KL, Sukumar G,
Wynder C, Dalgard CL and Doughty ML: Inhibition of the histone
demethylase Kdm5b promotes neurogenesis and derepresses Reln
(reelin) in neural stem cells from the adult subventricular zone of
mice. Mol Biol Cell. 27:627–639. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zamurrad S, Hatch HAM, Drelon C,
Belalcazar HM and Secombe J: A Drosophila model of intellectual
disability caused by mutations in the histone demethylase KDM5.
Cell Rep. 22:2359–2369. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kuźbicki Ł, Lange D, Stanek-Widera A and
Chwirot BW: Prognostic significance of RBP2-H1 variant of JARID1B
in melanoma. BMC Cancer. 17:8542017. View Article : Google Scholar
|
|
32
|
Yamamoto S, Wu Z, Russnes HG, Takagi S,
Peluffo G, Vaske C, Zhao X, Moen Vollan HK, Maruyama R, Ekram MB,
et al: JARID1B is a luminal lineage-driving oncogene in breast
cancer. Cancer Cell. 25:762–777. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hayami S, Yoshimatsu M, Veerakumarasivam
A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue
H, et al: Overexpression of the JmjC histone demethylase KDM5B in
human carcinogenesis: Involvement in the proliferation of cancer
cells through the E2F/RB pathway. Mol Cancer. 9:592010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shen X, Zhuang Z, Zhang Y, Chen Z, Shen L,
Pu W, Chen L and Xu Z: JARID1B modulates lung cancer cell
proliferation and invasion by regulating p53 expression. Tumour
Biol. 36:7133–7142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tang B, Qi G, Tang F, Yuan S, Wang Z,
Liang X, Li B, Yu S, Liu J, Huang Q, et al: JARID1B promotes
metastasis and epithelial-mesenchymal transition via PTEN/AKT
signaling in hepatocellular carcinoma cells. Oncotarget.
6:12723–12739. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yan G, Li S, Yue M, Li C and Kang Z:
Lysine demethylase 5B suppresses CC chemokine ligand 14 to promote
progression of colorectal cancer through the Wnt/β-catenin pathway.
Life Sci. 264:1187262021. View Article : Google Scholar
|
|
37
|
Shigekawa Y, Hayami S, Ueno M, Miyamoto A,
Suzaki N, Kawai M, Hirono S, Okada KI, Hamamoto R and Yamaue H:
Overexpression of KDM5B/JARID1B is associated with poor prognosis
in hepatocellular carcinoma. Oncotarget. 9:34320–34335. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Montano MM, Yeh IJ, Chen Y, Hernandez C,
Kiselar JG, de la Fuente M, Lawes AM, Nieman MT, Kiser PD,
Jacobberger J, et al: Inhibition of the histone demethylase, KDM5B,
directly induces re-expression of tumor suppressor protein HEXIM1
in cancer cells. Breast Cancer Res. 21:1382019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sayegh J, Cao J, Zou MR, Morales A, Blair
LP, Norcia M, Hoyer D, Tackett AJ, Merkel JS and Yan Q:
Identification of small molecule inhibitors of Jumonji AT-rich
interactive domain 1B (JARID1B) histone demethylase by a sensitive
high throughput screen. J Biol Chem. 288:9408–9417. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tumber A, Nuzzi A, Hookway ES, Hatch SB,
Velupillai S, Johansson C, Kawamura A, Savitsky P, Yapp C,
Szykowska A, et al: Potent and selective KDM5 inhibitor stops
cellular demethylation of H3K4me3 at transcription start sites and
proliferation of MM1S myeloma cells. Cell Chem Biol. 24:371–380.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Højfeldt JW, Agger K and Helin K: Histone
lysine demethylases as targets for anticancer therapy. Nat Rev Drug
Discov. 12:917–930. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kristensen LH, Nielsen AL, Helgstrand C,
Lees M, Cloos P, Kastrup JS, Helin K, Olsen L and Gajhede M:
Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals
strong substrate recognition in vitro and identifies
2,4-pyridine-dicarboxylic acid as an in vitro and in cell
inhibitor. FEBS J. 279:1905–1914. 2012. View Article : Google Scholar : PubMed/NCBI
|