Introduction
The accurate early diagnosis of breast cancer (BC)
is critical for the management of the disease; however, there are
currently no established non-invasive epigenetic biomarkers for BC
diagnosis or screening. BC is the most frequent type of cancer
(14%), after lung cancer (14.6%) (1), and has been reported to be the leading
cause of cancer-associated death among females worldwide (2). BC accounts for ~40% of all types of
cancer that develop after the age of 40 years, while 6.6% of cases
arise prior to the age of 40 years (3). The prognosis of BC is associated with
early diagnosis, which is detected using mammography (4), while the CA15-3 and CA27-29 markers
are used for monitoring the recovery of the disease and the
effectiveness of treatment (5,6).
Male BC has a low frequency and accounts for 1% of
diagnoses annually (7). The
incidence rate of male BC prior to the age of 50 years is 0.2 per
100,000 individuals, which increases to 6.3 per 100,000 individuals
after the age of 65 years. This means that the risk of BC
developing in males, as well as in females, increases with age
(8).
Most cases of BC are considered sporadic in nature,
as they are mainly associated with environmental factors (9); ~20% are familial, of which 5–10% are
represented by cases of hereditary breast and ovarian cancer, due
to mutations that are transmitted in an autosomal dominant manner.
In total, ~25% of cases are due to germline mutations in major
susceptibility genes, such as breast cancer (BRCA)1 and 2, while
1–3% of mutations occur in susceptibility genes, such as checkpoint
kinase 2, ATM serine/threonine kinase (ATM), phosphatase and tensin
homolog, tumor protein p53 (TP53) and partner and localizer of
BRCA2 (10,11).
BC is highly heterogeneous at both the histological
and molecular levels. In fact, in the beginning, it was studied as
a complex multi-step process based on genetic alterations with the
subsequent activation of cellular proto-oncogenes and/or
inactivation of tumor suppressor genes. The interplay between
genetic and environmental risk factors is guided by precise
epigenetic programs, mainly by DNA methylation changes, leading to
a dysregulation of key molecular pathways involved in breast
carcinogenesis (12). Differences
in DNA methylation profiles between patients with BC and healthy
controls may aid in clarifying the molecular basis of BC
development and, potentially, provide useful prognostic and/or
diagnostic biomarkers (13). For
instance, global hypomethylation and hypermethylation of CpG
islands are molecular events that occur early in patients with BC,
suggesting potential regions of sensitivity to disease onset
(14). In addition, most DNA
methylation changes were reported during the progression from
healthy breast tissue to ductal carcinoma in situ (DCIS),
while the epigenetic changes from DCIS to invasive BC were minimal.
Therefore, DNA methylation changes are involved in early
carcinogenesis and may represent a marker for early diagnosis
(15).
Recently, it has been observed that DNA methylation
profiles may be mapped, not only in tumor-specific tissue and
peripheral blood biospecimens, but also by extracting circulating
tumor (ct)DNA from plasma using liquid-based assays, allowing the
evaluation of both primary lesion and metastases (16). Detection of ctDNA may be used both
as an early screening for cancer and for the diagnosis of minimal
residual disease. Furthermore, it may be useful to monitor the
response to pharmacological treatments, for a personalized
therapeutic strategy. This method is based on the possibility of
isolating ctDNA from the blood, thus overcoming the challenges
associated with tissue biopsy, due to tumor localization and/or the
small size of the sample recovered. ctDNA detection is easy to
perform and represents a source of valuable information on the
biology of tumors and a minimally invasive test used to evaluate
epigenetic alterations (2).
The aim of the present review was to update the
pathogenic DNA methylation signatures emerging from studies
performed on tissue samples, peripheral blood and ctDNA using
liquid-based assays, from both female and male patients with BC,
including triple-negative BC (TNBC). The potential role of these
signatures as novel prognostic and diagnostic biomarkers, as well
as alternative drug targets in patients with therapy resistance, is
also discussed.
Focus on basic mechanisms of DNA
methylation
DNA methylation changes are widely influenced by
environmental events, such as aging, lack of physical activity,
stress, depression, high alcohol consumption and air pollution, as
well as biological processes, such as inactivation of the X
chromosome, genomic imprinting, reprogramming of the genome during
differentiation, development and survival, and genetic/molecular
alterations (17,18). All of these events, individually or
in combination, are sensitive to the development of neoplasms and
influence its course (19). The DNA
methylation process consists of the addition of a methyl group to
the pyrimidine ring of cytosine in the CpG dinucleotides, which is
mediated by DNA methyltransferase (DNMT) enzymes. In general,
methylated DNA mediates transcriptional repression of
tissue-specific genes. By contrast, hypomethylation favors an
increase in gene expression, leading to the activation of specific
genes. When the promoter region is methylated, gene expression is
reduced, as the proteins within the chromodomain are able to bind
to the methylated groups and prevent the recruitment of activator
proteins. The promoters and CpG islands, corresponding to actively
expressed genes, are generally hypomethylated in disease as
compared with those in healthy subjects (20); the same may occur in patients with
cancer, in whom there may be a focal hypermethylation of the
promoter due to the presence of CpG islands in the first exonic
region of almost half of the genes that cause genomic instability.
Several tumors develop due to methylation of numerous CpG islands
within the genome or a secondary event to somatic genetic mutations
in regulatory genes (21). DNA
methylation is regulated in a tissue-specific manner, but it is
also largely influenced by the genomic context (22). As a general paradigm, DNA
hypermethylation at regulatory regions, such as promoters and CpG
islands, may repress the transcription of genes, thereby acting as
a tumor suppressor. However, a positive association between DNA
methylation levels, at both the intragenic (introns and exons) and
intergenic regions (enhancers), and gene expression features is
gaining relevance due to its impact on cancer risk. On the other
hand, the different effects of DNA methylation in the promoter vs.
the gene remain to be further clarified (23).
DNA methylation changes and their potential
clinical utility in patients with BC
Certain tumor suppressor genes are frequently
hypermethylated in BC tissues and are detected in the early disease
stage (24). DNA methylation
changes between healthy and malignant breast tissue may be
considered as both prognostic and diagnostic biomarkers in BC.
Analysis of tissue biopsy
Gene-specific DNA methylation changes
as prognostic biomarkers
Several gene-specific DNA methylation changes have
been identified, suggesting that epigenetic alterations may have
prognostic value in BC. In Table I,
major results regarding the potential clinical value of DNA
methylation in tissue biopsy are reported.
 | Table I.Gene-specific methylation in tissue
from female and malea
patients with breast cancer. |
Table I.
Gene-specific methylation in tissue
from female and malea
patients with breast cancer.
| Author (year) | Epigenetic
alteration | Gene | Potential clinical
utility | Ref. |
|---|
| Shargh et al
(2014) |
Hypermethylation | E-cadherin | Prognostic
biomarker | (25) |
| Avraham et
al (2014) |
Hypermethylation | ALX4, FEV, HOXA11,
LYL1, NEUROG1, PAX, MGMT, SOX10, SREBF1, TP73, TRIM29 | Prognostic
biomarkers | (26) |
| de Almeida et
al (2019) |
Hypermethylation | WT1, BCL9, SMYD3,
ZNF154, ZNF177, HOXD9, ITIH5, TMEM132C, TDRD10, RNF220, RIMBP2,
PRAC2, EFCAB1, ANKRD53 | Prognostic
biomarkers | (27) |
| Salta et al
(2018) |
Hypermethylation | APC, BRCA1, FOXA1,
PSAT1, CCND2, RASSF1A, SCGB3A1 | Prognostic
biomarkers | (2) |
| Yang et al
(2019) |
Hypermethylation | IL15RA | Prognostic
biomarker | (28) |
| Mao et al
(2015) |
Hypomethylation | CRY2 | Prognostic
biomarker | (29) |
| Sasidharan Nair
et al (2018) |
Hypomethylation | PD-1, CTLA-4,
TIM-3, LAG-3 | Prognostic
biomarkers | (30) |
| Cui et al
(2020) |
Hypomethylation | KPNA2 | Prognostic
biomarker | (31) |
Among these, there is E-cadherin, which is involved
in cell-cell adhesion via its association with catenins. The
silencing of the E-cadherin gene by genetic or epigenetic changes
leads to tumorigenesis. The methylation profile of the E-cadherin
gene promoter was mapped in 50 BC tissues as compared with that in
50 normal breast samples. In agreement with previous studies, the
results indicated hypermethylation of the E-cadherin promoter in
94% of tissues, with an association with an aggressive tumor
phenotype in infiltrating BC (25).
Avraham et al (26) performed a study on specific DNA
methylation between healthy breast tissue and normal tissues, and
in tumor breast tissue and other neoplastic tissues, such as colon,
lung and endometrial cancer. The methylation profile of certain
genes was observed to be altered in neoplastic breast tissue,
including the ALX homeobox 4 (ALX4), FEV transcription factor, ETS
family member (FEV), homeobox A11 (HOXA11), LYL1 basic
helix-loop-helix family member (LYL1), neurogenin 1 (NEUROG1),
paired box 9 (PAX9), O-6-methylguanine-DNA methyltransferase
(MGMT), SRY-box transcription factor 10 (SOX10), sterol regulatory
element binding transcription factor 1 (SREBF1), tumor protein P73
(TP73) and tripartite motif 29 (TRIM29) genes. Specifically, in
healthy tissues, ALX4, FEV, HOXA11, LYL1, NEUROG1, PAX9, MGMT,
SOX10, SREBF1 and TP73 exhibited promoter hypomethylation, while in
neoplastic tissues, including the mammary gland, promoter
hypermethylation and reduced expression levels were present. In
addition, TRIM29 promoter hypomethylation in normal breast tissue
and hypermethylation in other healthy tissues was observed. In the
neoplastic breast tissue, there was hypermethylation of the
promoter with reduced gene expression, while hypomethylation was
observed in the other neoplastic tissues. This suggested that
epigenetic alterations may be associated with tissue-specific
susceptibility and may be involved in cancer progression (26).
De Almeida et al (27) analyzed DNA methylation and gene
expression profiles in BC tissue and matched normal tissue. In
addition to WT1 transcription factor (WT1), zinc finger protein
(ZNF)154, BCL9 transcription coactivator, homeobox D9 (HOXD9), SET
and MYND domain containing 3 (SMYD3), inter-α-trypsin inhibitor
heavy chain 5 (ITIH5) and ZNF177, methylation was found in seven
other genes, namely ring finger protein (RNF), transmembrane
protein 132C (TMEM132C), tudor domain containing (TDRD10), EF-hand
calcium binding domain 1 (EFCAB1), RIMS binding protein 2 (RIMBP2),
ankyrin repeat domain 53 (ANKRD53) and PRAC2 small nuclear protein
(PRAC2), also known as C17orf93, of which no association with
cancer has been previously observed. These authors reported
hypermethylation in breast tissue as compared with that in healthy
tissue. Furthermore, the RNF220, TMEM132C, TDRD10, EFCAB1, RIMBP2,
ANKRD53 and PRAC2 genes had promoter hypermethylation with
subsequent reduction in gene expression as compared with that in
healthy tissue. To evaluate the prognostic ability of the CpG
sites, survival curves were generated. Of the seven genes, only
PRAC2, TDR10 and TMEM132C had significant prognostic value. In
fact, they exhibited a different regulation of gene expression
between tumor and healthy breast tissue, as well as an association
with poor prognosis. In particular, the gene expression level of
PRAC2 was upregulated in tumor tissue, while there was a decrease
in TMEM132C and TDR10, despite promoter hypermethylation (27).
Furthermore, the prognostic performance of
promoter-related DNA methylation was evaluated in seven genes,
including adenomatous polyposis coli (APC), BRCA1, forkhead box A1
(FOXA1), phosphoserine aminotransferase 1 (PSAT1), cyclin D2
(CCND2), ras association domain family member 1 (RASSF1A) and
secretoglobin family 3A member 1 (SCGB3A1), which were
hypermethylated in 137 breast tissues as compared to normal breast
tissue. Disease-specific survival curves and disease-free survival
curves were drawn for methylation status, which revealed that
higher DNA methylation levels were associated with shorter
disease-specific survival (2).
Another study suggested that IL-15 receptor α
(IL15RA) hypermethylation may be associated with the development
and progression of BC by regulating the expression levels of other
genes. By comparing the methylation data of 316 breast tumor
tissues with 21 healthy breast tissues, it was observed that
hypermethylation of IL15RA led to upregulation of the proline rich
11 (PRR11), nucleolar and spindle-associated protein 1 (NUSAP1) and
homeobox C11 (HOXC11) genes and a reduced regulation of the SH3 and
cysteine rich domain 2 (STAC2) genes. Using Gene Ontology and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, it
was determined that PRR11, NUSAP1, STAC2 and HOXC11 methylation was
associated with the ‘cell adhesion-related molecular pathway’ and,
therefore, BC progression (28).
The involvement of DNA methylation in regulating the
cryptochrome circadian regulator 2 (CRY2) gene, which has a key
role in breast tumorigenesis, was observed, suggesting a strong
association with BC progression. The methylation of the CRY2 gene
was evaluated in 1,881 patients with BC and was observed to be
hypomethylated, with the downregulation of gene expression in BC
tissues, as compared with that in healthy tissue. This reduction in
CRY2 regulation was due to estrogen receptor (ER) negativity,
resulting in a higher tumor grade and shorter survival time for
patients with BC (29).
The expression levels of different immune checkpoint
genes, including T cell immunoglobulin and mucin domain-containing
protein 3 (TIM-3), programmed cell death protein-1 (PD-1),
lymphocyte activating 3 (LAG-3) and cytotoxic T-lymphocyte
associated protein 4 (CTLA-4), were analyzed in 8 breast tumor
tissues and 8 healthy breast tissues. Upregulation was associated
with hypomethylation of the promoter in breast tumor tissues as
compared with that in healthy breast tissues, suggesting that these
modifications may be utilized as prognostic biomarkers in BC
(30).
Methylation of the karyopherin α-2 (KPNA2) gene was
also analyzed in 33 male and female BC tissues, as compared with
that in 20 healthy tissues. The KPNA2 protein is a nuclear import
factor that is involved in nucleocytoplasmic transport, cell
proliferation, migration and invasion in tumors. The association
between KPNA2 expression and prognosis in BC was evaluated using
overall survival, relapse-free survival, distant metastasis-free
survival and post-progression survival time curves in patients with
BC. KPNA2 may serve as a potential indicator of poor prognosis, as
promoter hypomethylation of the gene was reported in patients with
a lower survival rate (31,32).
The WT1 gene and promoter methylation was analyzed
in a panel of normal breast epithelial and BC tissues. Contrary to
previous reports, WT1 was indicated to be hypermethylated and
expressed in 32% of BC tissue, but not in normal breast epithelium,
suggesting that WT1 may not have a tumor suppressor role in BC
(33).
In both male and female BC, promoter
hypermethylation in tumor tissue was frequently determined to be
high in the BRCA2, mutS homolog 6, WT1, PAX5 and 6, cadherin 13,
GATA binding protein 5, killin, P53 regulated DNA replication
inhibitor, thrombospondin 1, glutathione S-transferase pi 1
(GSTP1), MGMT, TP53, TP73, estrogen receptor 1 (ESR1), CD44 antigen
(CD44), cell adhesion molecule 1, retinoic acid receptor β (RARB),
PYD and CARD domain containing, Von Hippel-Lindau tumor suppressor
(VHL), cyclin dependent kinase inhibitor 2A (CDKN2A), ATM,
checkpoint with forkhead and ring finger domains (CHFR), RB
transcriptional corepressor 1 (RB1), BRCA1 and serine/threonine
kinase 11 (STK11) genes. Compared with that in females, promoter
methylation of the VHL, ESR1, CDKN2A, CD44, CHFR, BRCA2, RB1 and
STK11 genes was lower in males. This appears to be due to the
presence of higher levels of estrogen in females compared with
those in males. This suggests that there are differences in male
and female breast carcinogenesis with respect to promoter
methylation (34).
Analysis of DNA extracted from peripheral
blood
Global DNA methylation changes as risk
biomarkers
Several studies (35–41)
have evaluated global DNA methylation levels in blood using various
methods and BC samples and controls, and concluded that a high
level of methylation was associated with poor survival time, while
lower methylation was associated with improved prognosis (35). The strategies included measuring the
percentage of methylated DNA using a luminometric methylation assay
(LUMA), measuring the methylation of repetitive DNA elements
(LINE-1, Alu or Sat2) using pyrosequencing, the concentration of
5-methyldeoxycytosine (5-mdC) using liquid chromatography-mass
spectrometry and the MethyLight assay, as surrogates of global DNA
methylation levels.
Global hypomethylation has been suggested to cause
genomic instability and lead to an increase in cancer risk. A total
of three studies using the LUMA assay were performed between 2012
and 2014 (36–38), which reported different results.
Specifically, Delgado-Cruzata et al (36) determined similar global DNA
methylation levels between BC cases and controls, while Xu et
al (37) observed low global
DNA methylation levels in BC cases, and Kuchiba et al
(38) observed high global DNA
methylation in patients with BC. In addition, Delgado-Cruzata et
al (36) measured the
methylation of repetitive DNA elements and detected low levels in
patients with BC compared with those in the controls.
Other studies, performed using different methods,
have evaluated the methylation level of LINE-1 repeats and the
results provided a consensus opinion, namely that there were no
differences between BC cases and healthy controls. Cho et al
(39) observed global
hypomethylation in patients with BC using the 5-mdC assay. This
diversity of response depends not only on the method used, but also
on the timing with which the sampling was performed and the time
taken to perform the analysis. In fact, various factors may
influence the results. For instance, if the sampling was performed
while the subjects were undergoing therapeutic treatment, this may
easily alter the result. In addition, the presence of the disease
may lead to a different distribution of the cells in the
bloodstream (39). Other studies
have been performed comparing global DNA methylation levels and the
risk of cancer, with inconsistent results (40,41);
certain studies concluded that there was no significant association
between global DNA methylation levels and BC risk, while others
revealed a positive association between methylation level and BC
risk. This suggests that the association between global DNA
methylation and BC risk remains to be further clarified (40).
In a previous study, the association between global
DNA methylation and physical activity or global DNA methylation and
BC risk was evaluated. A higher level of global methylation in
patients performing physical activity over longer periods of time
was observed. In addition, lower cancer risk was determined in
patients with a higher level of global methylation. Furthermore,
where possible, global DNA methylation levels were evaluated from a
peripheral blood sample 3 years prior to the onset of cancer. It
was observed that higher levels of methylation reduced the risk of
developing the neoplasm. Therefore, this study suggested that
long-term physical activity was associated with global DNA
methylation and that the latter was associated with a decreased
risk of BC (41).
Gene-specific DNA methylation as risk
biomarkers
Various studies have evaluated the methylation
levels of tumor suppressor genes associated with BC development by
analyzing leukocyte DNA from patients with BC and in controls. The
evaluation of DNA methylation in leukocytes is of particular
importance, as peripheral blood is easier to obtain at multiple
time-points than tissues. Furthermore, it allows the identification
of early risk markers, even in healthy individuals, and allows the
easy evaluation of differences in methylation levels between
healthy individuals and patients with disease. In Table II, major results regarding the
potential clinical value of DNA methylation in peripheral blood are
reported.
 | Table II.Gene-specific methylation in
peripheral blood from female patients with breast cancer. |
Table II.
Gene-specific methylation in
peripheral blood from female patients with breast cancer.
| Author (year) | Epigenetic
alteration | Gene | Potential clinical
utility | Ref. |
|---|
| Cho et al
(2015) |
Hypermethylation | BRCA1 | Risk biomarker | (39) |
| Brennan et
al (2012) |
Hypermethylation | ATM | Risk biomarker | (42) |
| Widschwendter et
al (2008) |
Hypermethylation | HYAL2, NUP155,
ZNF217, PTGS, TITF1, NEUROD1 SFRP1 | Risk
Biomarkers | (43) |
| Shirkavand et
al (2018) |
Hypermethylation | DOK7 | Prognostic
biomarkers | (44) |
|
|
Hypomethylation | VIM, CXCR4 |
|
|
| Zmetakova et
al (2013) |
Hypermethylation | ESR1, TIMP3 | Prognostic
biomarkers | (45) |
The role of gene-specific methylation in peripheral
blood, as a marker of BC risk, is uncertain. Certain studies have
evaluated whether promoter hypermethylation of tumor suppressor
genes, which are frequently methylated in BC, may be used as a
biomarker for BC risk. BRCA1 and ATM promoter methylation was
analyzed in 1,021 BC samples and 1,036 controls, and in 640 BC
samples and 741 controls, respectively. Hypermethylation was
observed in patients with BC, indicating that DNA methylation
levels in BRCA1 and ATM may serve as a marker of BC risk and, as
peripheral blood is a comparatively more accessible biospecimen,
this may be valuable in epigenome-wide association studies
(39,42). In a case-control study including
1,083 patients with BC, it was investigated whether DNA methylation
was associated with BC risk. The risk of cancer onset was indicated
to be increased when the methylation levels of the hyaluronidase 2
(HYAL2), nucleoporin 155 (NUP155), ZNF217, post-transcriptional
gene silencing (PTGS), thyroid transcription factor-1 (TITF1),
neuronal differentiation 1 (NEUROD1) and secreted frizzled related
protein 1 (SFRP1) genes were low (43). To date, only a small number of
studies have evaluated gene-specific DNA methylation as risk
biomarkers. Further studies are required to determine whether DNA
methylation may be a new tool to predict the risk of BC.
Gene-specific DNA methylation changes
as prognostic biomarkers
Shirkavand et al (44) investigated the DNA methylation
status of the docking protein 7 (DOK7), VIM, C-X-C chemokine
receptor type 4 (CXCR4) and SAM pointed domain-containing ETS
transcription factor genes in 60 patients with BC and 40 controls.
The percentage of promoter methylation changes in the patients with
BC and normal specimens were analyzed using a gel analyzer software
(GenAnalyzer 2010). The results suggested that the VIM and CXCR4
genes, the latter of which is a chemokine receptor involved in
cancer progression, were hypomethylated in patients with BC as
compared with that in the healthy individuals. Hypermethylation of
the DOK7 gene in BC vs. controls was present. It was indicated that
hypermethylation of the DOK7 gene in patients with BC may be used
as a biomarker for cancer diagnosis, and that VIM and CXCR4
hypomethylation may be used as a biomarker for BC prognosis
(44). To determine whether the DNA
methylation level in peripheral blood may be used as a prognostic
biomarker, DNA methylation levels in the ESR1 and TIMP
metallopeptidase inhibitor 1 (TIMP3) genes were analyzed in
patients with BC and controls. Different results have been
obtained: Zmetakova et al (45) determined promoter hypermethylation
in patients with BC compared with that in the controls, while
Widschwendter et al (43)
did not observe any significant changes in the methylation levels
between the controls and patients with BC.
Analysis of ctDNA and circulating
tumor cells (CTCs) using liquid biopsy
Epigenetic analysis may be performed on circulating
material, such as CTCs and ctDNA using liquid biopsy, starting from
a simple and non-invasive blood sample. It is an advantageous
procedure, easy to perform with high specificity and sensitivity in
minimal residual disease quantification, and is able to monitor the
response to therapy and/or any resistance (46). CTCs are rare cells with variable
morphology and may be obtained from primary BC or from free
metastasis in the bloodstream, which invade other tissues or organs
and cause injury. It is also possible for CTC groups called
‘clusters’ to be present, resulting in more aggressive metastases
(47). Numerous studies have
analyzed genes with specific methylation using liquid biopsy. In
Table III, major results about
the potential clinical value of DNA methylation in liquid biopsy
were reported. DNA methylation patterns may be used as a biomarker
for the risk and survival prognosis of patients with cancer.
 | Table III.Gene-specific methylation in liquid
biopsies from female patients with breast cancer. |
Table III.
Gene-specific methylation in liquid
biopsies from female patients with breast cancer.
| Author (year) | Epigenetic
alterations | Gene | Potential clinical
utility | Ref. |
|---|
| Liu et al
(2015) |
Hypermethylation | BRCA1, FHIT | Risk biomarker | (48) |
| Ahmed et al
(2010) |
Hypermethylation | DAPK | Risk biomarker | (49) |
| Kloten et al
(2013) |
Hypermethylation | RASSF1A | Risk biomarker | (50) |
| Swellam et
al (2015) |
Hypermethylation | APC, RARB | Risk biomarker | (51) |
| Bao-Caamano et
al (2020) |
Hypermethylation | RASSF1A, GSTP1 | Prognostic
biomarkers | (52) |
| Chimonidou et
al (2017) |
Hypermethylation | CST6, SOX17,
BRMS1 | Prognostic
biomarkers | (53) |
| Zurita et al
(2010) |
Hypermethylation | SFN | Prognostic
biomarker | (55) |
| Radpour et
al (2011) |
Hypermethylation | GSTP1, TIMB3 | Prognostic
biomarkers | (56) |
| Chimonidou et
al (2013) |
Hypermethylation | CST6 | Prognostic
biomarker | (57) |
Gene-specific DNA methylation changes
as risk biomarkers
The development of cancer has been indicated to be
associated with epigenetic changes. A previous study estimated the
ctDNA methylation levels of fragile histidine triad diadenosine
triphosphatase (FHIT) and BRCA1 promoters in 30 patients with BC
and 30 healthy individuals. Statistical analysis was used to
analyze the differences in the methylation levels between the
groups and the results suggested that the methylation levels of the
BRCA1 and FHIT promoters were higher in the patients with BC
compared with those in the healthy individuals. The FHIT
methylation level was also indicated to be associated with BC and
may be useful for BC diagnosis (48).
Death-associated protein kinase 1 (DAPK) and RASSF1A
methylation was evaluated in 26 patients with BC, 16 patients with
benign breast disease and 12 age-matched healthy controls. APC and
RARB methylation was also analyzed in 121 patients with BC, 79
patients with benign breast disease and 66 healthy volunteers.
Statistical analysis was performed to analyze the positivity rates
of the investigated genes. DAPK, RASSF1A, APC and RARB genes had
higher hypermethylation frequencies in patients with BC compared
with those in the controls (49–51),
suggesting that they may be valuable biomarkers for BC detection.
Further studies confirming these results and the markers may be
useful in a clinical setting in the diagnosis and management of
BC.
Gene-specific DNA methylation changes
as prognostic biomarkers
In a recent study on CTCs, promoter hypermethylation
of certain tumor suppressor genes, including RASSF1A, CCND2, GSTP1,
HIC ZBTB transcriptional repressor 1 (HIC1), RARβ and DAPK, was
analyzed. In particular, the methylation of the RASSF1A gene was
associated with BC progression and metastases, while GSTP1
methylation was associated with chemotherapy treatment response and
patient survival time (52).
Chimonidou et al (53)
evaluated whether the DNA methylation status in CTCs and ctDNA was
comparable and whether it reflected the status of the primary
tumor. The study compared the methylation status in both CTCs and
ctDNA in three genes, SOX17, cystatin E/M (CST6) and BRMS1
transcriptional repressor and anoikis regulator (BRMS1) in 153
patients with BC and healthy individuals. CST6, a tumor suppressor
gene, has been associated with the inhibition of proliferation,
migration, invasion and bone metastases in BC. SOX17 has been
associated with the regulation of the Wnt/β-catenin signaling
pathway. BRMS1 is involved in chromatin remodeling. A correlation
between CTCs and ctDNA for the SOX17 promoter methylation was both
observed in patients with early and metastatic BC, but not for CST6
and BRMS1. DNA methylation analysis of SOX17 may thus be of
prognostic value (53).
In a study using different methylated specific CTC
clusters, hypomethylation in binding sites in the octamer-binding
transcription factor 4 (OCT4), nanog homeobox (NANOG), SOX2 and
SIN3 transcription regulator family member A (SIN3A) genes, which
have been associated with transcription, proliferation and
stability, was observed and associated with poor prognosis. In the
same study, in an experiment on single cells, application of
reagents that led to cluster dissociation caused a change in DNA
methylation with hypermethylation in the binding sites of OCT4,
SOX2, NANOG and SIN3A and the consequent reduction of gene
expression. This may be due to increased intracellular calcium and
the loss of cell-to-cell junction after treatment of CTC clusters
with dissociating compounds (54).
For certain genes, the degree of methylation in the
CTC regions matched that observed in the neoplastic tissue, BC cell
lines and peripheral blood, suggesting a more aggressive neoplasm.
In particular, promoter hypermethylation of the RASSF1A, GSTP1 and
APC genes was reported in tissue and BC cell lines, in the CCND2
and RARβ genes in tissue, and in the HIC1, DAPK1 and TIMP3 genes in
BC cell lines. Furthermore, BRCA1 and ESR1 promoter
hypermethylation in tissue, BC cell lines and peripheral blood was
also observed (2,32,35,34,39,45).
Other studies using ctDNA have confirmed an
association between high methylation levels in certain tumor
suppressor genes and poor prognosis in BC. It was reported that the
stratifin (SFN), GSTP1, CST6 and TIMP3 genes were always
hypermethylated in BC samples (55–57).
DNA methylation changes in TNBC
An estimated 10–20% of BC cases are classified as
TNBC. This is the most aggressive type of BC compared with the
other subtypes, due to its clinicopathological characteristics,
including early onset, relapse and higher frequency of developing
lung, liver and central nervous system metastases. Patients with
metastatic TNBC have unfavorable prognosis, as their cells do not
express the ER, progesterone receptor and HER2. The absence of
these receptors still makes it difficult to formulate a targeted
therapy with a consequently higher mortality rate (58). Germinal BRCA1 and BRCA2 mutations
occur in 19.5% of TBNC, which may vary according to family history
and ethnicity (59). Epigenetic
alterations in TNBC are more frequent than in other subtypes of BC
(60).
Gene-specific DNA methylation changes
as prognostic biomarkers
A study including 119 BC samples and 118 healthy
samples analyzed the methylation profiles of genes whose
methylation pattern identifies the TNBC subtype. Out of these,
seven were indicated to be differentially methylated and associated
with clinical conditions. Overall survival analysis of the selected
methylated genes in BC was performed and the family with sequence
similarity 150, member B, maturase K, interferon-induced protein
35, Wnt family member 10A and SKI family transcriptional
corepressor 1 genes were determined to be hypomethylated, with gene
upregulation, and were associated with favorable patient survival.
In addition, actin binding LIM protein 1 and carnitine
palmitoyltransferase 1A had low gene expression and were associated
with poor patient outcome. According to a KEGG analysis, the ‘cell
movement’, ‘cell proliferation’ and ‘cell differentiation
processes’ pathways were associated with these genes, supporting
the roles of these seven differentially methylated genes as
potential markers for TNBC prognosis (61).
A recent study also analyzed several hypomethylated
genes in 50 patients with TNBC and 24 healthy controls. In the TNBC
tissues, promoter hypomethylation in ADAM metallopeptidase domain
12 (ADAM12), tetraspanin 9 (TSPAN9) and Von Willebrand factor C and
EGF domains (VWCE) genes was observed. The VWCE gene promoted
cancer development and progression. The TSPAN9 gene was associated
with cell development, tumor proliferation and invasion, while
ADAM12 was associated with proteolytic process, apoptosis, cell
cycle and cell adhesion. ADAM12 hypomethylation was associated with
a more unfavorable prognosis than the other two genes. Furthermore,
ADAM12 knockdown decreased TNBC cell proliferation and migration,
suggesting that it may be a potential therapeutic target (62).
The role of BRCA1 gene methylation in 239 TNBC cases
was analyzed to investigate the association between clinical data
and BRCA1 gene methylation. Of note, BRCA1 DNA methylation was
observed in 57.3% of cases. Multivariate analyses further indicated
that BRCA1 promoter methylation was an independent predictor of
overall survival and disease-free survival. In addition, the BRCA1
promoter was also associated with a significant decrease in overall
survival time, suggesting BRCA1 promoter methylation may serve as a
biomarker for TNBC prognosis (63).
Furthermore, it was demonstrated that Tet
methylcytosine dioxygenase 1 (TET1) was specifically overexpressed
in patients with TNBC and associated with a shorter overall
survival time. This revealed a previously uncharacterized role of
TET1 as an oncogene, with hypomethylation and the activation of
oncogenic signaling pathways. Several previous studies in BC have
reported TET1 to be a tumor suppressor; thus, there is evidence
suggesting that TET1 may function as both an oncogene and a tumor
suppressor, depending on the cellular context (64).
Another study reported that the expression of the
ganglioside GD3 (GD3) gene was markedly higher in patients with
ER-negative BC compared with that in patients with ER-positive BC
and also highly expressed in TNBC compared to other types of BC.
This increase in expression was associated with the hypomethylation
of the ST8 α-N-acetyl-neuraminide α-2,8-sialyltransferase 1 gene.
Elevated expression of GD3 in human BC cells increased their
proliferation, migration, invasion and colony formation ability,
suggesting that GD3 may be a potential prognostic biomarker in TNBC
(65). In Table IV, the main results regarding the
potential clinical value of DNA methylation in TNBC are
presented.
 | Table IV.Gene-specific methylation in
triple-negative breast cancer. |
Table IV.
Gene-specific methylation in
triple-negative breast cancer.
| Author (year) | Epigenetic
alteration | Genes | Potential clinical
utility | Ref. |
|---|
| Chen et al
(2019) |
Hypomethylation | MATK, IFI35,
FAM150B, SKOR1, WNT10A, ABLIM1, CPT1A | Prognostic
biomarkers | (61) |
| Mendaza et
al (2020) |
Hypomethylation | ADAM12, TSPAN9,
VWCE | Prognostic
biomarkers | (62) |
| Zhu et al
(2015) |
Hypermethylation | BRCA1 | Prognostic
biomarker | (63) |
DNA methylation changes may induce drug
resistance in patients with BC
Gene-specific DNA methylation in patients with BC
represents a valuable tool in clinical practice, which contributes
not only to the early diagnosis of the disease, but also to risk
stratification and therapeutic treatment (66). It is well-known that activation of
the BRCA1 gene leads to cellular damage repair, which is highly
compromised when epigenetic alterations occur with subsequent
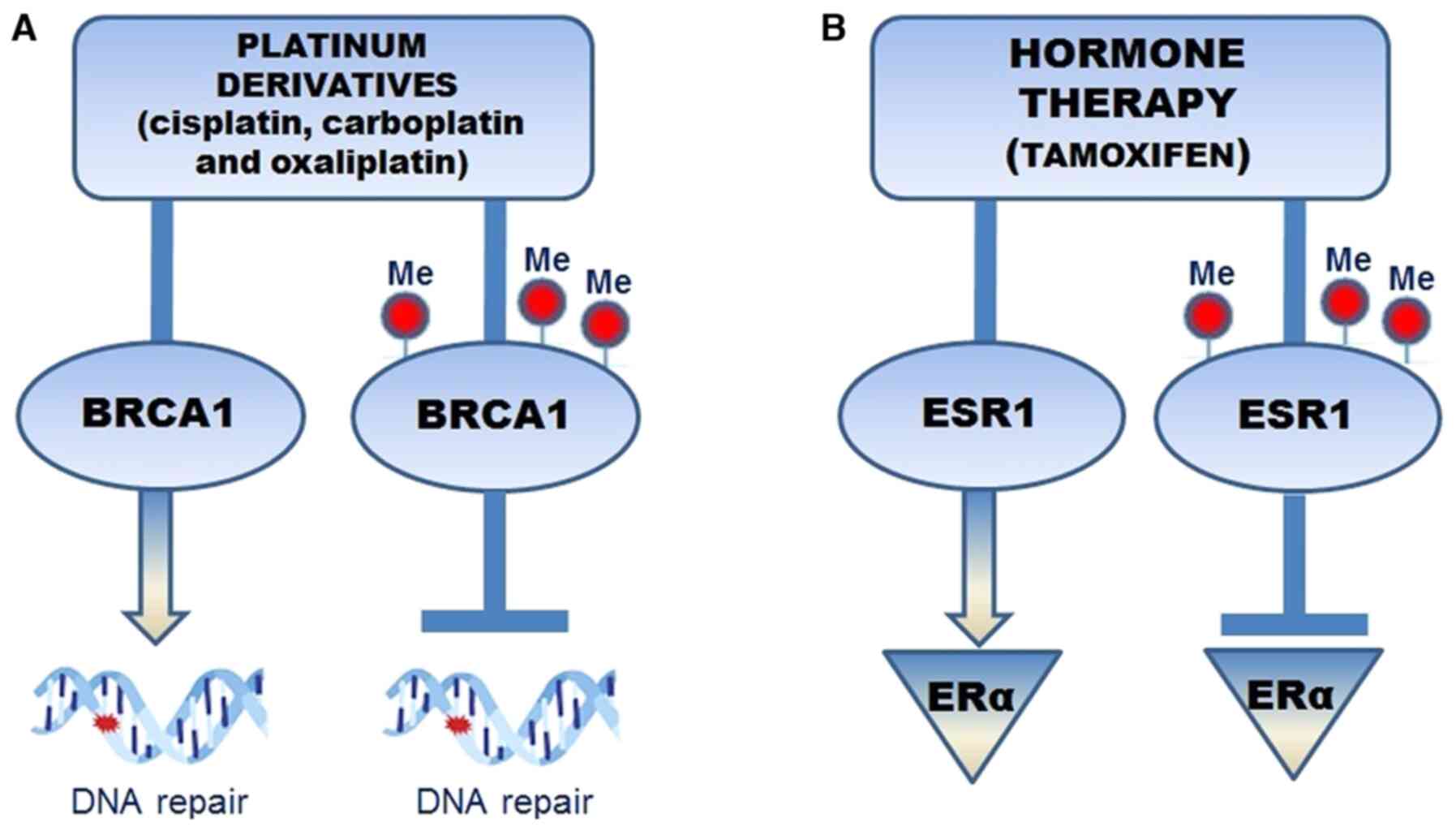
promoter hypermethylation and reduction in gene activity (Fig. 1A). In the treatment of patients with
BC using chemotherapeutic agents, such as platinum and its
derivatives (cisplatin, carboplatin and oxaliplatin), the ability
of BRCA1 to repair the DNA cross-links is inhibited. Therefore,
BRCA1 hypermethylation may become a predictive element for
therapeutic treatments (67).
Epigenetic alterations may disrupt the balance between ERα
coactivators and corepressors and have been associated with poor
prognosis and endocrine therapy resistance (68).
ESR1 methylation, which codes for ER-α, appears to
represent a predictive biomarker for the endocrine treatment
efficacy in breast tumors that do not respond to hormone therapy.
It has been observed that tamoxifen, an anti-estrogen drug,
administered to patients with BC, inhibited ER-α dimerization and
activation, preventing relapse. ERS1 methylation determined the
absence of ER expression and was associated with hormone therapy
resistance (69) (Fig. 1B). Furthermore, one study performed
on human BC cell lines (SKBr3 and AU565) evaluated the epigenetic
biomarkers associated with trastuzumab resistance in patients with
HER2-positive BC. It was indicated that hypermethylation of the
transforming growth factor β-induced (TGFBI) promoter led to
epigenetic silencing of the gene and trastuzumab resistance
(70).
The GeparSixto trial evaluated the effect of MGMT
promoter methylation on the response and survival time of patients
with TNBC treated with carboplatin therapy. A total of 210 TNBC
tumors were divided into two therapy groups, namely those with and
without carboplatin. No statistically significant difference in
therapeutic response was observed (71). DNA methylation changes among two BC
cell lines, MCF-7 and MDA-MB-231 (TNBC), were evaluated after
administration of doxorubicin and paclitaxel. In the treated MCF-7
cells, promoter methylation changes were observed, with changes in
cyclin A1 and prostaglandin-endoperoxide synthase 2 gene
expression. In the MDA-MB-231 cells, ESR1 hypomethylation was
observed to not increase gene expression. In cells treated with
doxorubicin only, GSTP1 and MGMT hypomethylation was observed with
an increase in gene expression levels. Furthermore, in MDA-MB-231
cells treated with doxorubicin and paclitaxel, a synergistic effect
on MMP9 gene expression was observed, which was different from that
in the cells treated with doxorubicin or paclitaxel alone. The
molecular changes observed suggested that doxorubicin or paclitaxel
administration does not always produce a synergistic effect and
further studies are required to consider them as prognostic and
therapeutic response markers (72).
Epi-drugs in combination therapy for BC
treatment
Recently, attention has focused on the epi-drugs
used to overcome epigenetic alterations and hormonal therapy
resistance, such as DNMT inhibitors (DNMTIs). The DNMTIs decitabine
(DAC) and 5-azacytidine (AZA) are agents involved in DNA
demethylation (73).
Several studies combining the efficacy of DNMTIs,
histone deacetylases inhibitors (HDACIs) and vorinostat, have been
performed in BC. A synergistic effect of both epigenetic drugs with
anticancer therapy and of the combination of the epi-drugs
themselves was determined (74,75).
Initially, DAC and AZA had a synergistic effect in combination with
HDACIs in preclinical and clinical studies in a different cancer
type (76); however, no synergistic
effect between AZA and entinostat was observed in a phase II
clinical trial in patients with hormone-refractory BC (77). The combination of HDACIs and DNMTIs
in BC cell lines resulted in re-expression of ER. In a preclinical
study, BC cell lines with resistance to tamoxifen were generated,
leading to promoter hypermethylation of E-cadherin with decreased
expression. After AZA administration, E-cadherin demethylation was
observed, along with re-established sensitivity to tamoxifen
(78).
A further experiment suggested that administration
of DNMTIs with poly (ADP-ribose) polymerase (PARP)-inhibitors
enhanced the cytotoxic effect of PARP inhibition in TNBC cell lines
(79). In addition, it was
suggested that DNMTIs may cause homologous recombination deficiency
in BRCA wild-type TNBC cells, similar to BRCA-mutant cancer cells
(80).
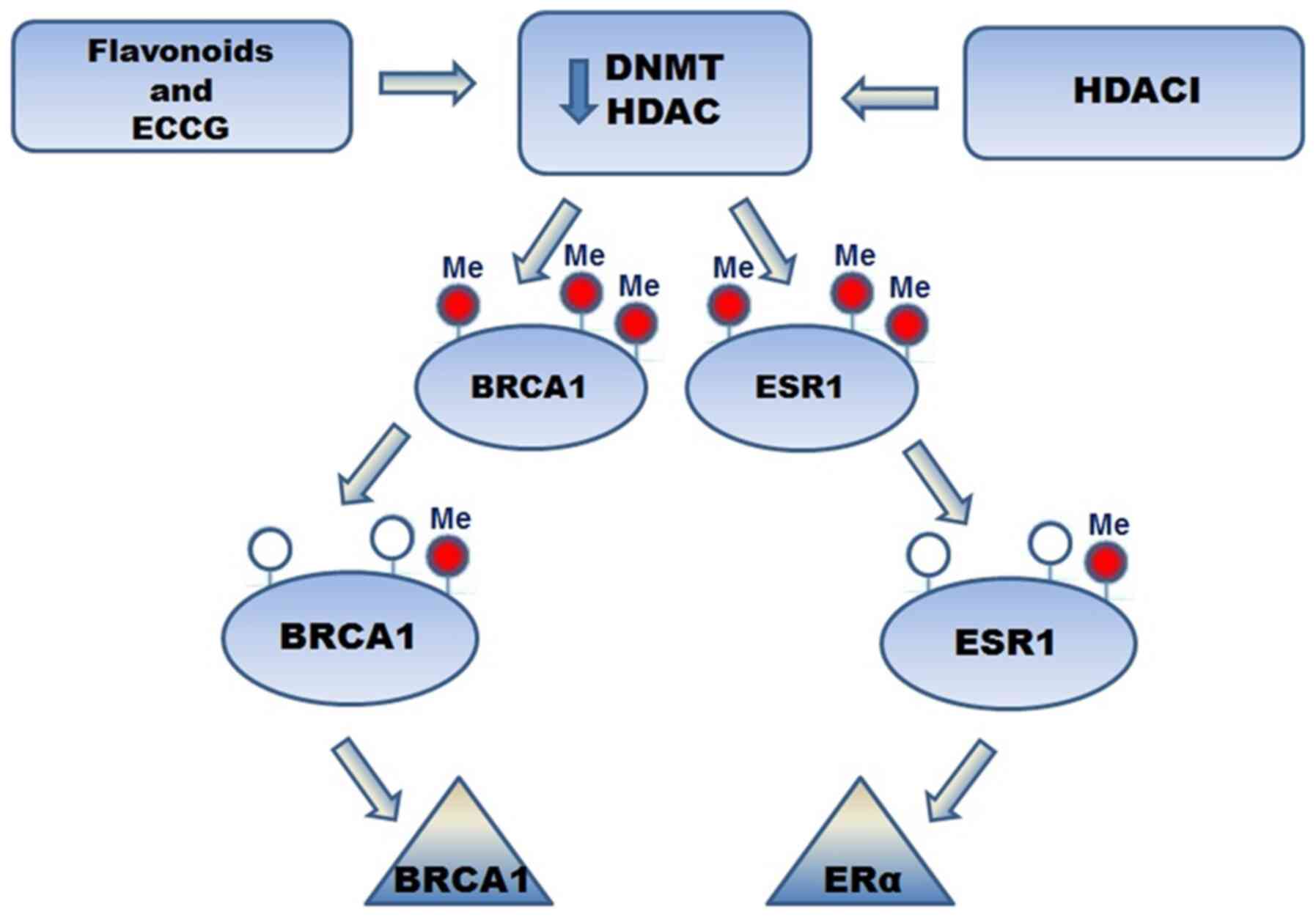
Several studies have demonstrated that isoflavone
intake leads to neoplasm reduction in BC.
Epigallocatechin-3-gallate (EGCG) is one of most studied flavonoids
in BC. It acts as an inhibitor of DNMT and HDAC. It also induced
the expression of tumor suppressor genes, leading to a reduction in
the development of metastasis and cancer progression. Treatment
with EGCG and HDACIs led to re-expression of ERα in ER-negative BC.
Furthermore, EGCG induced ER re-expression in ER-negative BC and
has decreased DNMT activity in BC cell lines. Regarding the effect
of EGCG and flavonoids on BRCA1 and ESR-1 promoter methylation, a
significant decrease in BRCA1 and ESR-1 promoter methylation was
observed, which led to increased expression levels in ER-negative
BC (81) (Fig. 2). In another study, the possibility
of converting aggressive TNBC cells into a less aggressive
phenotype using epigenetic drugs was analyzed (82). Guadecitabine (DNMTI) and entinostat
(HDACI) had antitumor effects in patient-derived xenograft mouse
models (82).
Despite these encouraging results, to date, the use
of epi-drugs has marked limitations due to adverse reactions,
including a high degree of cytotoxicity (13,83).
This has not stopped their use; however, further investigation is
required to determine the correct dose between epi-drugs and/or
anticancer treatments and to open new possibilities to increase the
number of targets and personalized therapies, with a considerable
reduction of side effects.
Conclusion
Numerous studies have reported epigenetic
alterations, such as DNA methylation, which led to changes in
expression of oncogenes and tumor suppressor genes in patients with
BC (84). Studies performed on
tissues and whole blood have detected several hypo- and
hypermethylated genes in both male and female patients with BC.
Detection of gene-specific methylation using liquid
biopsy may facilitate early cancer diagnosis and assist with
monitoring pharmacological treatment in order to obtain a
personalized and targeted therapy. The sensitivity to treatment
using chemotherapy, hormone and immunotherapy may be altered by
gene-specific DNA methylation. In recent studies, attention has
focused on epigenetic drugs. In particular, the association of the
demethylation agents DNMTI and HDACI, administered alone and/or in
combination with anticancer therapies, has led to remarkable
results, particularly in patients with BC and resistant to
anticancer treatment. Furthermore, studies on epigenetic
alterations represent a valid tool for the search for prognostic
biomarkers and for improving therapeutic treatments for patients
with cancer.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
Authors' contributions
MTV and CN conceived the study. GDE, GB and GC
performed the literature search/selection drafted the manuscript
and prepared the figures. MTV, GF, GFN and CN revised and edited
the manuscript. All authors read and approved the final manuscript.
Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Malvezzi M, Carioli G, Bertuccio P,
Boffetta P, Levi F, La Vecchia C and Negri E: European cancer
mortality predictions for the year 2017, with focus on lung cancer.
Ann Oncol. 28:1117–1123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Salta S, P Nunes S, Fontes-Sousa M, Lopes
P, Freitas M, Caldas M, Antunes L, Castro F, Antunes P, Palma de
Sousa S, et al: A DNA methylation-based test for breast cancer
detection in circulating cell-free DNA. J Clin Med. 7:4202018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oltra SS, Peña-Chilet M, Flower K,
Martinez MT, Alonso E, Burgues O, Lluch A, Flanagan JM and Ribas G:
Acceleration in the DNA methylation age in breast cancer tumours
from very young women. Sci Rep. 9:149912019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schiano C, Soricelli A, De Nigris F and
Napoli C: New challenges in integrated diagnosis by imaging and
osteo-immunology in bone lesions. Expert Rev Clin Immunol.
15:289–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeong S, Park MJ, Song W and Kim HS:
Current immunoassay methods and their applications to clinically
used biomarkers of breast cancer. Clin Biochem. 78:43–57. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schiano C, Franzese M, Pane K, Garbino N,
Soricelli A, Salvatore M, de Nigris F and Napoli C: Hybrid
18F-FDG-PET/MRI measurement of standardized uptake value coupled
with yin yang 1 signature in metastatic breast cancer. a
preliminary study. Cancers (Basel). 11:14442019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gucalp A, Traina TA, Eisner JR, Parker JS,
Selitsky SR, Park BH, Elias AD, Baskin-Bey ES and Cardoso F: Male
breast cancer: A disease distinct from female breast cancer. Breast
Cancer Res Treat. 173:37–48. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khan NAJ and Tirona M: An updated review
of epidemiology, risk factors, and management of male breast
cancer. Med Oncol. 38:392021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Romagnolo DF, Daniels KD, Grunwald JT,
Ramos SA, Propper CR and Selmin OI: Epigenetics of breast cancer:
Modifying role of environmental and bioactive food compounds. Mol
Nutr Food Res. 60:1310–1329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vietri MT, Molinari AM, Caliendo G, De
Paola ML, Giovanna D, Gambardella AL, Petronella P and Cioffi M:
Double heterozygosity in the BRCA1 and BRCA2 genes in Italian
family. Clin Chem Lab Med. 51:2319–2324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vietri MT, Caliendo G, Casamassimi A,
Cioffi M, De Paola ML, Napoli C and Molinari AM: A novel PALB2
truncating mutation in an Italian family with male breast cancer.
Oncol Rep. 33:1243–1247. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pasculli B, Barbano R and Parrella P:
Epigenetics of breast cancer: Biology and clinical implication in
the era of precision medicine. Semin Cancer Biol. 51:22–35. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sher G, Salman NA, Khan AQ, Prabhu KS,
Raza A, Kulinski M, Dermime S, Haris M, Junejo K and Uddin S:
Epigenetic and breast cancer therapy: promising diagnostic and
therapeutic applications. Semin Cancer Biol. Aug 25–2020.(Epub
ahead of print). doi: org/10.1016/j.semcancer.2020.08.009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bhat SA, Majid S, Wani HA and Rashid S:
Diagnostic utility of epigenetics in breast cancer - A review.
Cancer Treat Res Commun. 19:1001252019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park HL: Epigenetic biomarkers for
environmental exposures and personalized breast cancer prevention.
Int J Environ Res Public Health. 17:11812020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stewart CM and Tsui DWY: Circulating
cell-free DNA for non-invasive cancer management. Cancer Genet.
228-229:169–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Colao A, de Nigris F, Modica R and Napoli
C: Clinical epigenetics of neuroendocrine tumors: The road ahead.
Front Endocrinol (Lausanne). 11:6043412020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schiano C, Casamassimi A, Rienzo M, de
Nigris F, Sommese L and Napoli C: Involvement of mediator complex
in malignancy. Biochim Biophys Acta. 1845:66–83. 2014.PubMed/NCBI
|
|
19
|
Sarno F, Benincasa G, List M, Barabasi AL,
Baumbach J, Ciardiello F, Filetti S, Glass K, Loscalzo J, Marchese
C, et al International Network Medicine Consortium, : Clinical
epigenetics settings for cancer and cardiovascular diseases:
Real-life applications of network medicine at the bedside. Clin
Epigenetics. 13:662021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benincasa G, Franzese M, Schiano C,
Marfella R, Miceli M, Infante T, Sardu C, Zanfardino M, Affinito O,
Mansueto G, et al: DNA methylation profiling of
CD04+/CD08+ T cells reveals pathogenic
mechanisms in increasing hyperglycemia: PIRAMIDE pilot study. Ann
Med Surg (Lond). 60:218–226. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rodgers KM, Udesky JO, Rudel RA and Brody
JG: Environmental chemicals and breast cancer: An updated review of
epidemiological literature informed by biological mechanisms.
Environ Res. 160:152–182. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Klutstein M, Nejman D, Greenfield R and
Cedar H: DNA methylation in cancer and aging. Cancer Res.
76:3446–3450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murtha M and Esteller M: Extraordinary
cancer epigenomics: Thinking outside the classical coding and
promoter box. Trends Cancer. 2:572–584. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brooks J, Cairns P and Zeleniuch-Jacquotte
A: Promoter methylation and the detection of breast cancer. Cancer
Causes Control. 20:1539–1550. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shargh SA, Sakizli M, Khalaj V, Movafagh
A, Yazdi H, Hagigatjou E, Sayad A, Mansouri N, Mortazavi-Tabatabaei
SA and Khorram Khorshid HR: Downregulation of E-cadherin expression
in breast cancer by promoter hypermethylation and its relation with
progression and prognosis of tumor. Med Oncol. 31:2502014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Avraham A, Cho SS, Uhlmann R, Polak ML,
Sandbank J, Karni T, Pappo I, Halperin R, Vaknin Z, Sella A, et al:
Tissue specific DNA methylation in normal human breast epithelium
and in breast cancer. PLoS One. 9:e918052014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
de Almeida BP, Apolónio JD, Binnie A and
Castelo-Branco P: Roadmap of DNA methylation in breast cancer
identifies novel prognostic biomarkers. BMC Cancer. 19:2192019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang H, Zhou L, Chen J, Su J, Shen W, Liu
B, Zhou J, Yu S and Qian J: A four-gene signature for prognosis in
breast cancer patients with hypermethylated IL15RA. Oncol Lett.
17:4245–4254. 2019.PubMed/NCBI
|
|
29
|
Mao Y, Fu A, Hoffman AE, Jacobs DI, Jin M,
Chen K and Zhu Y: The circadian gene CRY2 is associated with breast
cancer aggressiveness possibly via epigenomic modifications. Tumour
Biol. 36:3533–3539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sasidharan Nair V, El Salhat H, Taha RZ,
John A, Ali BR and Elkord E: DNA methylation and repressive H3K9
and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4,
TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast
cancer. Clin Epigenetics. 10:782018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cui X, Jing X, Wu X, Xu J, Liu Z, Huo K
and Wang H: Analyses of DNA methylation involved in the activation
of nuclear karyopherin alpha 2 leading to identify the progression
and prognostic significance across human breast cancer. Cancer
Manag Res. 12:6665–6677. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shukla S, Penta D, Mondal P and Meeran SM:
Epigenetics of breast cancer: Clinical status of epi-drugs and
phytochemicals. Adv Exp Med Biol. 1152:293–310. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Loeb DM, Evron E, Patel CB, Sharma PM,
Niranjan B, Buluwela L, Weitzman SA, Korz D and Sukumar S: Wilms'
tumor suppressor gene (WT1) is expressed in primary breast tumors
despite tumor-specific promoter methylation. Cancer Res.
61:921–925. 2001.PubMed/NCBI
|
|
34
|
Vermeulen MA, van Deurzen CHM, Doebar SC,
de Leng WW, Martens JW, van Diest PJ and Moelans CB: Promoter
hypermethylation in ductal carcinoma in situ of the male breast.
Endocr Relat Cancer. 26:575–584. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tao C, Luo R, Song J, Zhang W and Ran L: A
seven-DNA methylation signature as a novel prognostic biomarker in
breast cancer. J Cell Biochem. 121:2385–2393. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Delgado-Cruzata L, Wu HC, Perrin M, Liao
Y, Kappil MA, Ferris JS, Flom JD, Yazici H, Santella RM and Terry
MB: Global DNA methylation levels in white blood cell DNA from
sisters discordant for breast cancer from the New York site of the
Breast Cancer Family Registry. Epigenetics. 7:868–874. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu Z, Bolick SC, DeRoo LA, Weinberg CR,
Sandler DP and Taylo JA: Epigenome-wide association study of breast
cancer using prospectively collected sister study samples. J Natl
Cancer Inst. 105:694–700. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kuchiba A, Iwasaki M, Ono H, Kasuga Y,
Yokoyama S, Onuma H, Nishimura H, Kusama R, Tsugane S and Yoshida
T: Global methylation levels in peripheral blood leukocyte DNA by
LUMA and breast cancer: A case-control study in Japanese women. Br
J Cancer. 110:2765–2771. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cho YH, McCullough LE, Gammon MD, Wu HC,
Zhang YJ, Wang Q, Xu X, Teitelbaum SL, Neugut AI, Chen J, et al:
Promoter hypermethylation in white blood cell DNA and breast cancer
risk. J Cancer. 6:819–824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tang Q, Cheng J, Cao X, Surowy H and
Burwinkel B: Blood-based DNA methylation as biomarker for breast
cancer: A systematic review. Clin Epigenetics. 8:1152016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Boyne DJ, O'Sullivan DE, Olij BF, King WD,
Friedenreich CM and Brenner DR: Physical activity, global DNA
methylation, and breast cancer risk: A systematic literature review
and meta-analysis. Cancer Epidemiol Biomarkers Prev. 27:1320–1331.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Brennan K, Garcia-Closas M, Orr N,
Fletcher O, Jones M, Ashworth A, Swerdlow A, Thorne H, Riboli E,
Vineis P, et al KConFab Investigators, : Intragenic ATM methylation
in peripheral blood DNA as a biomarker of breast cancer risk.
Cancer Res. 72:2304–2313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Widschwendter M, Apostolidou S, Raum E,
Rothenbacher D, Fiegl H, Menon U, Stegmaier C, Jacobs IJ and
Brenner H: Epigenotyping in peripheral blood cell DNA and breast
cancer risk: A proof of principle study. PLoS One. 3:e26562008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shirkavand A, Boroujeni ZN and Aleyasin
SA: Examination of methylation changes of VIM, CXCR4, DOK7, and
SPDEF genes in peripheral blood DNA in breast cancer patients.
Indian J Cancer. 55:366–371. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zmetakova I, Danihel L, Smolkova B, Mego
M, Kajabova V, Krivulcik T, Rusnak I, Rychly B, Danis D, Repiska V,
et al: Evaluation of protein expression and DNA methylation
profiles detected by pyrosequencing in invasive breast cancer.
Neoplasma. 60:635–646. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Benincasa G, Mansueto G and Napoli C:
Fluid-based assays and precision medicine of cardiovascular
diseases: The ‘hope’ for Pandora's box? J Clin Pathol. 72:785–799.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Aceto N, Bardia A, Miyamoto DT, Donaldson
MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al:
Circulating tumor cell clusters are oligoclonal precursors of
breast cancer metastasis. Cell. 158:1110–1122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu L, Sun L, Li C, Li X, Zhang Y, Yu Y
and Xia W: Quantitative detection of methylation of FHIT and BRCA1
promoters in the serum of ductal breast cancer patients. Biomed
Mater Eng. 26 (Suppl 1):S2217–S2222. 2015.PubMed/NCBI
|
|
49
|
Ahmed IA, Pusch CM, Hamed T, Rashad H,
Idris A, El-Fadle AA and Blin N: Epigenetic alterations by
methylation of RASSF1A and DAPK1 promoter sequences in mammary
carcinoma detected in extracellular tumor DNA. Cancer Genet
Cytogenet. 199:96–100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kloten V, Becker B, Winner K, Schrauder
MG, Fasching PA, Anzeneder T, Veeck J, Hartmann A, Knüchel R and
Dahl E: Promoter hypermethylation of the tumor-suppressor genes
ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast
cancer screening. Breast Cancer Res. 15:R42013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Swellam M, Abdelmaksoud MDE, Sayed Mahmoud
M, Ramadan A, Abdel-Moneem W and Hefny MM: Aberrant methylation of
APC and RARβ2 genes in breast cancer patients. IUBMB Life.
67:61–68. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bao-Caamano A, Rodriguez-Casanova A and
Diaz-Lagares A: Epigenetics of circulating tumor cells in breast
cancer. Adv Exp Med Biol. 1220:117–134. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chimonidou M, Strati A, Malamos N, Kouneli
S, Georgoulias V and Lianidou E: Direct comparison study of DNA
methylation markers in EpCAM-positive circulating tumour cells,
corresponding circulating tumour DNA, and paired primary tumours in
breast cancer. Oncotarget. 8:72054–72068. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dart A: Methylated clusters. Nat Rev
Cancer. 19:1252019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zurita M, Lara PC, del Moral R, Torres B,
Linares-Fernández JL, Arrabal SR, Martínez-Galán J, Oliver FJ and
Ruiz de Almodóvar JM: Hypermethylated 14-3-3-sigma and ESR1 gene
promoters in serum as candidate biomarkers for the diagnosis and
treatment efficacy of breast cancer metastasis. BMC Cancer.
10:2172010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Radpour R, Barekati Z, Kohler C, Lv Q,
Bürki N, Diesch C, Bitzer J, Zheng H, Schmid S and Zhong XY:
Hypermethylation of tumor suppressor genes involved in critical
regulatory pathways for developing a blood-based test in breast
cancer. PLoS One. 6:e160802011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chimonidou M, Tzitzira A, Strati A,
Sotiropoulou G, Sfikas C, Malamos N, Georgoulias V and Lianidou E:
CST6 promoter methylation in circulating cell-free DNA of breast
cancer patients. Clin Biochem. 46:235–240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pareja F and Reis-Filho JS:
Triple-negative breast cancers - a panoply of cancer types. Nat Rev
Clin Oncol. 15:347–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Okuma HS and Yonemori K: BRCA gene
mutations and poly(ADP-Ribose) polymerase inhibitors in
triple-negative breast cancer. Adv Exp Med Biol. 1026:271–286.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fackler MJ, Cho S, Cope L, Gabrielson E,
Visvanathan K, Wilsbach K, Meir-Levi D, Lynch CF, Marks J, Geradts
J, et al: DNA methylation markers predict recurrence-free interval
in triple-negative breast cancer. NPJ Breast Cancer. 6:32020.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chen X, Zhang J and Dai X: DNA methylation
profiles capturing breast cancer heterogeneity. BMC Genomics.
20:8232019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mendaza S, Ulazia-Garmendia A,
Monreal-Santesteban I, Córdoba A, Azúa YR, Aguiar B, Beloqui R,
Armendáriz P, Arriola M, Martín-Sánchez E, et al: ADAM12 is a
potential therapeutic target regulated by hypomethylation in
triple-negative breast cancer. Int J Mol Sci. 21:9032020.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhu X, Shan L, Wang F, Wang J, Wang F,
Shen G, Liu X, Wang B, Yuan Y, Ying J, et al: Hypermethylation of
BRCA1 gene: Implication for prognostic biomarker and therapeutic
target in sporadic primary triple-negative breast cancer. Breast
Cancer Res Treat. 150:479–486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Good CR, Panjarian S, Kelly AD, Madzo J,
Patel B, Jelinek J and Issa JJ: TET1-mediated hypomethylation
activates oncogenic signaling in triple-negative breast cancer.
Cancer Res. 78:4126–4137. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li W, Zheng X, Ren L, Fu W, Liu J, Xv J,
Liu S, Wang J and Du G: Epigenetic hypomethylation and upregulation
of GD3s in triple negative breast cancer. Ann Transl Med.
7:7232019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
van Hoesel AQ, Sato Y, Elashoff DA, Turner
RR, Giuliano AE, Shamonki JM, Kuppen PJ, van de Velde CJ and Hoon
DS: Assessment of DNA methylation status in early stages of breast
cancer development. Br J Cancer. 108:2033–2038. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Laham-Karam N, Pinto GP, Poso A and
Kokkonen P: Transcription and translation inhibitors in Cancer
treatment. Front Chem. 8:2762020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Garcia-Martinez L, Zhang Y, Nakata Y, Chan
HL and Morey L: Epigenetic mechanisms in breast cancer therapy and
resistance. Nat Commun. 12:17862021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Martínez-Galán J, Torres-Torres B, Núñez
MI, López-Peñalver J, Del Moral R, Ruiz De Almodóvar JM, Menjón S,
Concha A, Chamorro C, Ríos S, et al: ESR1 gene promoter region
methylation in free circulating DNA and its correlation with
estrogen receptor protein expression in tumor tissue in breast
cancer patients. BMC Cancer. 14:592014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Palomeras S, Diaz-Lagares Á, Viñas G,
Setien F, Ferreira HJ, Oliveras G, Crujeiras AB, Hernández A, Lum
DH, Welm AL, et al: Epigenetic silencing of TGFBI confers
resistance to trastuzumab in human breast cancer. Breast Cancer
Res. 21:792019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Jank P, Gehlhaar C, Bianca L, Caterina F,
Andreas S, Karn T, Marmé F, Sinn HP, van Mackelenbergh M, Sinn B,
et al: MGMT promoter methylation in triple negative breast cancer
of the GeparSixto trial. PLoS One. 15:e02380212020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hamadneh L, Abu-Irmaileh B, Al-Majawleh M,
Bustanji Y, Jarrar Y and Al-Qirim T: Doxorubicin-paclitaxel
sequential treatment: Insights of DNA methylation and gene
expression changes of luminal A and triple negative breast cancer
cell lines. Mol Cell Biochem. 476:3647–3654. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
de Nigris F, Ruosi C and Napoli C:
Clinical efficiency of epigenetic drugs therapy in bone
malignancies. Bone. 143:1156052021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Buocikova V, Rios-Mondragon I, Pilalis E,
Chatziioannou A, Miklikova S, Mego M, Pajuste K, Rucins M, Yamani
NE, Longhin EM, et al: Epigenetics in breast cancer therapy-new
strategies and future nanomedicine perspectives. Cancers (Basel).
12:36222020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Schröder R, Illert AL, Erbes T, Flotho C,
Lübbert M and Duque-Afonso J: The epigenetics of breast cancer -
Opportunities for diagnostics, risk stratification and therapy.
Epigenetics. 23:1–13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Griffiths EA and Gore SD: DNA
methyltransferase and histone deacetylase inhibitors in the
treatment of myelodysplastic syndromes. Semin Hematol. 45:23–30.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Connolly RM, Li H, Jankowitz RC, Zhang Z,
Rudek MA, Jeter SC, Slater SA, Powers P, Wolff AC, Fetting JH, et
al: Combination epigenetic therapy in advanced breast cancer with
5-azacitidine and entinostat: A phase ii national cancer
institute/stand up to cancer study. Clin Cancer Res. 23:2691–2701.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang Q, Gun M and Hong XY: Induced
tamoxifen resistance is mediated by increased methylation of
e-cadherin in estrogen receptor-expressing breast cancer cells. Sci
Rep. 9:141402019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Muvarak NE, Chowdhury K, Xia L, Robert C,
Choi EY, Cai Y, Bellani M, Zou Y, Singh ZN, Duong VH, et al:
Enhancing the cytotoxic effects of PARP inhibitors with DNA demethy
lating agents - a potential therapy for cancer. Cancer Cell.
30:637–650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
McLaughlin LJ, Stojanovic L, Kogan AA,
Rutherford JL, Choi EY, Yen RC, Xia L, Zou Y, Lapidus RG, Baylin
SB, et al: Pharmacologic induction of innate immune signaling
directly drives homologous recombination deficiency. Proc Natl Acad
Sci USA. 117:17785–17795. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Selvakumar P, Badgeley A, Murphy P, Anwar
H, Sharma U, Lawrence K and Lakshmikuttyamma A: Flavonoids and
other polyphenols act as epigenetic modifiers in breast cancer.
Nutrients. 12:7612020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Su Y, Hopfinger NR, Nguyen TD, Pogash TJ,
Santucci-Pereira J and Russo J: Epigenetic reprogramming of
epithelial mesenchymal transition in triple negative breast cancer
cells with DNA methyltransferase and histone deacetylase
inhibitors. J Exp Clin Cancer Res. 37:3142018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Scognamiglio G, De Chiara A, Parafioriti
A, Armiraglio E, Fazioli F, Gallo M, Aversa L, Camerlingo R,
Cacciatore F, Colella G, et al: Patient-derived organoids as a
potential model to predict response to PD-1/PD-L1 checkpoint
inhibitors. Br J Cancer. 121:979–982. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Terracciano D, Terreri S, de Nigris F,
Costa V, Calin GA and Cimmino A: The role of a new class of long
noncoding RNAs transcribed from ultraconserved regions in cancer.
Biochim Biophys Acta Rev Cancer. 1868:449–455. 2017. View Article : Google Scholar : PubMed/NCBI
|