|
1
|
Orkin SH and Zon LI: Hematopoiesis: An
evolving paradigm for stem cell biology. Cell. 132:631–644. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miyoshi H, Shimizu K, Kozu T, Maseki N,
Kaneko Y and Ohki M: t(8;21) breakpoints on chromosome 21 in acute
myeloid leukemia are clustered within a limited region of a single
gene, AML1. Proc Natl Acad Sci USA. 88:10431–10434. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Look AT: Oncogenic transcription factors
in the human acute leukemias. Science. 278:1059–1064. 1997.
View Article : Google Scholar
|
|
4
|
Cancer Genome Atlas Research Network; Ley
TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley
K, Triche TJ Jr, Laird PW, et al: Genomic and epigenomic landscapes
of adult de novo acute myeloid leukemia. N Engl J Med.
368:2059–2074. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miyoshi H, Kozu T, Shimizu K, Enomoto K,
Maseki N, Kaneko Y, Kamada N and Ohki M: The t(8;21) translocation
in acute myeloid leukemia results in production of an AML1-MTG8
fusion transcript. EMBO J. 12:2715–2721. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Frank R, Zhang J, Uchida H, Meyers S,
Hiebert SW and Nimer SD: The AML1/ETO fusion protein blocks
transactivation of the GM-CSF promoter by AML1B. Oncogene.
11:2667–2674. 1995.PubMed/NCBI
|
|
7
|
Meyers S, Lenny N and Hiebert SW: The
t(8;21) fusion protein interferes with AML-1B-dependent
transcriptional activation. Mol Cell Biol. 15:1974–1982. 1995.
View Article : Google Scholar
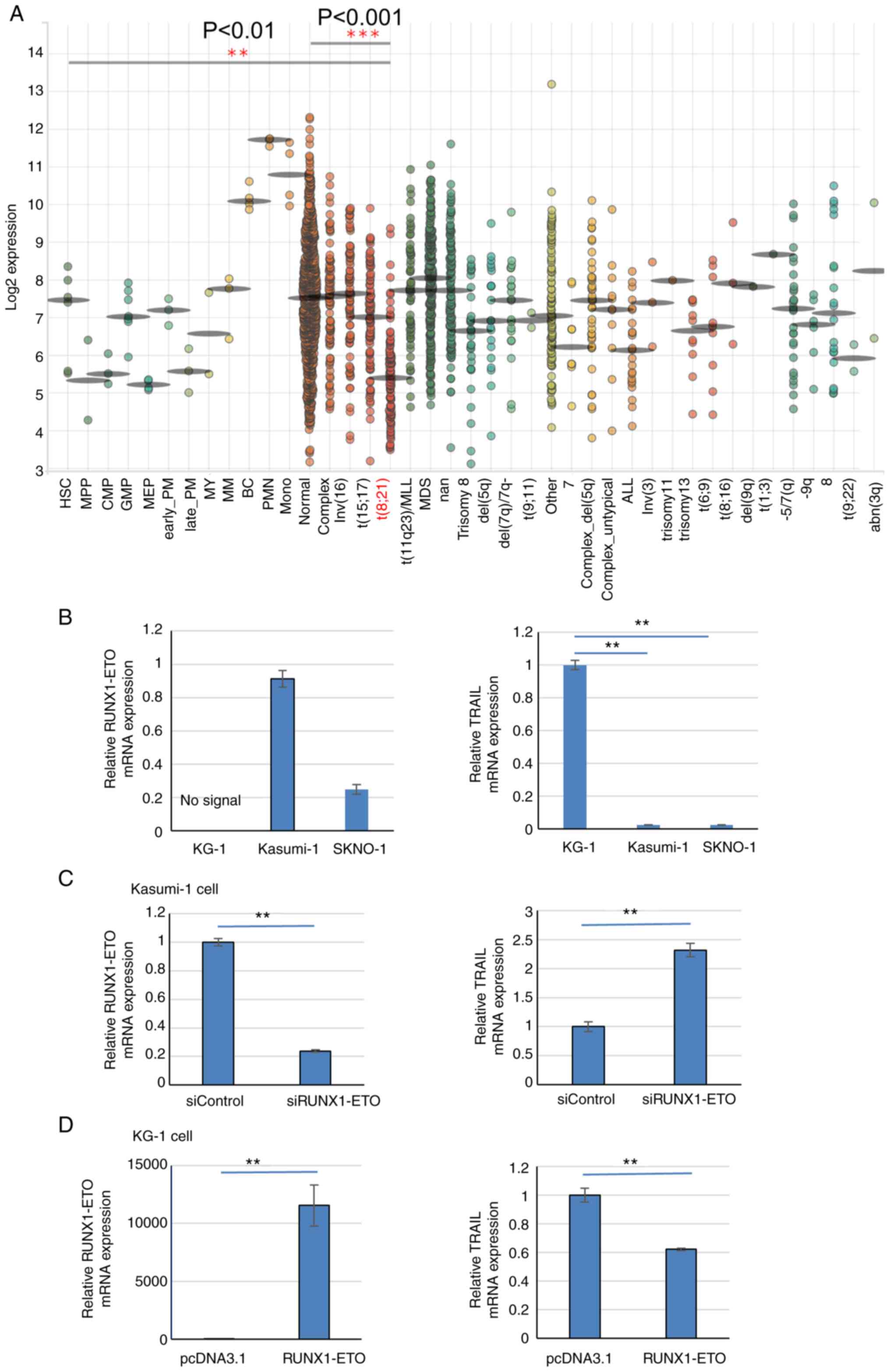
|
|
8
|
Osato M, Asou N, Abdalla E, Hoshino K,
Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K
and Ito Y: Biallelic and heterozygous point mutations in the runt
domain of the AML1/PEBP2alphaB gene associated with myeloblastic
leukemias. Blood. 93:1817–1824. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song WJ, Sullivan MG, Legare RD, Hutchings
S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, et
al: Haploinsufficiency of CBFA2 causes familial thrombocytopenia
with propensity to develop acute myelogenous leukaemia. Nat Genet.
23:166–175. 1999. View
Article : Google Scholar
|
|
10
|
Mizutani S, Yoshida T, Zhao X, Nimer SD,
Taniwaki M and Okuda T: Loss of RUNX1/AML1 arginine-methylation
impairs peripheral T cell homeostasis. Br J Haematol. 170:859–873.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matsumura T, Nakamura-Ishizu A, Muddineni
SSNA, Tan DQ, Wang CQ, Tokunaga K, Tirado-Magallanes R, Sian S,
Benoukraf T, Okuda T, et al: Hematopoietic stem cells acquire
survival advantage by loss of RUNX1 methylation identified in
familial leukemia. Blood. 136:1919–1932. 2020. View Article : Google Scholar
|
|
12
|
DiFilippo EC, Coltro G, Carr RM,
Mangaonkar AA, Binder M, Khan SP, Rodriguez V, Gangat N, Wolanskyj
A, Pruthi RK, et al: Spectrum of abnormalities and clonal
transformation in germline RUNX1 familial platelet disorder and a
genomic comparative analysis with somatic RUNX1 mutations in
MDS/MPN overlap neoplasms. Leukemia. 34:2519–2524. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okuda T, van Deursen J, Hiebert SW,
Grosveld G and Downing JR: AML1, the target of multiple chromosomal
translocations in human leukemia, is essential for normal fetal
liver hematopoiesis. Cell. 84:321–330. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Q, Stacy T, Binder M, Marin-Padilla
M, Sharpe AH and Speck NA: Disruption of the Cbfa2 gene causes
necrosis and hemorrhaging in the central nervous system and blocks
definitive hematopoiesis. Proc Natl Acad Sci USA. 93:3444–3449.
1996. View Article : Google Scholar
|
|
15
|
North T, Gu TL, Stacy T, Wang Q, Howard L,
Binder M, Marín-Padilla M and Speck NA: Cbfa2 is required for the
formation of intra-aortic hematopoietic clusters. Development.
126:2563–2575. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Taniuchi I, Osato M, Egawa T, Sunshine MJ,
Bae SC, Komori T, Ito Y and Littman DR: Differential requirements
for Runx proteins in CD4 repression and epigenetic silencing during
T lymphocyte development. Cell. 111:621–633. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ichikawa M, Asai T, Saito T, Seo S,
Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M and
Hirai H: AML-1 is required for megakaryocytic maturation and
lymphocytic differentiation, but not for maintenance of
hematopoietic stem cells in adult hematopoiesis. Nat Med.
10:299–304. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Growney JD, Shigematsu H, Li Z, Lee BH,
Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR,
et al: Loss of Runx1 perturbs adult hematopoiesis and is associated
with a myeloproliferative phenotype. Blood. 106:494–504. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Bruijn M and Dzierzak E: Runx
transcription factors in the development and function of the
definitive hematopoietic system. Blood. 129:2061–2069. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang S, Wang Q, Crute BE, Melnikova IN,
Keller SR and Speck NA: Cloning and characterization of subunits of
the T-cell receptor and murine leukemia virus enhancer core-binding
factor. Mol Cell Biol. 13:3324–3339. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ogawa E, Maruyama M, Kagoshima H, Inuzuka
M, Lu J, Satake M, Shigesada K and Ito Y: PEBP2/PEA2 represents a
family of transcription factors homologous to the products of the
Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci
USA. 90:6859–6863. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Meyers S, Downing JR and Hiebert SW:
Identification of AML-1 and the (8;21) translocation protein
(AML-1/ETO) as sequence-specific DNA-binding proteins: The runt
homology domain is required for DNA binding and protein-protein
interactions. Mol Cell Biol. 13:6336–6345. 1993.PubMed/NCBI
|
|
23
|
Kanno T, Kanno Y, Chen LF, Ogawa E, Kim WY
and Ito Y: Intrinsic transcriptional activation-inhibition domains
of the polyomavirus enhancer binding protein 2/core binding factor
alpha subunit revealed in the presence of the beta subunit. Mol
Cell Biol. 18:2444–2454. 1998. View Article : Google Scholar
|
|
24
|
Kitabayashi I, Yokoyama A, Shimizu K and
Ohki M: Interaction and functional cooperation of the
leukemia-associated factors AML1 and p300 in myeloid cell
differentiation. EMBO J. 17:2994–3004. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Otto F, Lübbert M and Stock M: Upstream
and downstream targets of RUNX proteins. J Cell Biochem. 89:9–18.
2003. View Article : Google Scholar
|
|
26
|
Rossetti S and Sacchi N: RUNX1: A microRNA
hub in normal and malignant hematopoiesis. Int J Mol Sci.
14:1566–1588. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Imperato MR, Cauchy P, Obier N and Bonifer
C: The RUNX1-PU.1 axis in the control of hematopoiesis. Int J
Hematol. 101:319–329. 2015. View Article : Google Scholar
|
|
28
|
Wargnier A, Legros-Maida S, Bosselut R,
Bourge JF, Lafaurie C, Ghysdael CJ, Sasportes M and Paul P:
Identification of human granzyme B promoter regulatory elements
interacting with activated T-cell-specific proteins: implication of
Ikaros and CBF binding sites in promoter activation. Proc Natl Acad
Sci USA. 92:6930–6934. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Elagib KE, Racke FK, Mogass M, Khetawat R,
Delehanty LL and Goldfarb AN: RUNX1 and GATA-1 coexpression and
cooperation in megakaryocytic differentiation. Blood.
101:4333–4341. 2003. View Article : Google Scholar
|
|
30
|
Verma AK, Wheeler DL, Aziz MH and
Manoharan H: Protein kinase cepsilon and development of squamous
cell carcinoma, the nonmelanoma human skin cancer. Mol Carcinog.
45:381–388. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wiley SR, Schooley K, Smolak PJ, Din WS,
Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C and Smith
CA: Identification and characterization of a new member of the TNF
family that induces apoptosis. Immunity. 3:673–682. 1995.
View Article : Google Scholar
|
|
32
|
LeBlanc HN and Ashkenazi A: Apo2L/TRAIL
and its death and decoy receptors. Cell Death Differ. 10:66–75.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jurisic V, Bumbasirevic V, Konjevic G,
Djuricic B and Spuzic I: TNF-alpha induces changes in LDH isotype
profile following triggering of apoptosis in PBL of non-Hodgkin's
lymphomas. Ann Hematol. 83:84–91. 2004. View Article : Google Scholar
|
|
34
|
Jurisic V, Srdic-Rajic T, Konjevic G,
Bogdanovic G and Colic M: TNF-α induced apoptosis is accompanied
with rapid CD30 and slower CD45 shedding from K-562 cells. J Membr
Biol. 239:115–122. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stuckey DW and Shah K: TRAIL on trial:
Preclinical advances in cancer therapy. Trends Mol Med. 19:685–694.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Twomey JD, Kim SR, Zhao L, Bozza WP and
Zhang B: Spatial dynamics of TRAIL death receptors in cancer cells.
Drug Resist Updat. 19:13–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakata S, Yoshida T, Horinaka M, Shiraishi
T, Wakada M and Sakai T: Histone deacetylase inhibitors upregulate
death receptor 5/TRAIL-R2 and sensitize apoptosis induced by
TRAIL/APO2-L in human malignant tumor cells. Oncogene.
23:6261–6271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Farooqi AA, Naqvi SK, Perk AA, Yanar O,
Tabassum S, Ahmad MS, Mansoor Q, Ashry MS, Ismail M, Naoum GE and
Arafat WO: Natural agents-mediated targeting of histone
deacetylases. Arch Immunol Ther Exp (Warsz). 66:31–44. 2018.
View Article : Google Scholar
|
|
39
|
Zhou M, Yuan M, Zhang M, Lei C, Aras O,
Zhang X and An F: Combining histone deacetylase inhibitors (HDACis)
with other therapies for cancer therapy. Eur J Med Chem.
226:1138252021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Falschlehner C, Schaefer U and Walczak H:
Following TRAIL's path in the immune system. Immunology.
127:145–154. 2009. View Article : Google Scholar :
|
|
41
|
Zerafa N, Westwood JA, Cretney E, Mitchell
S, Waring P, Iezzi M and Smyth MJ: TRAIL deficiency accelerates
hematological malignancies. J Immunol. 175:5586–5590. 2005.
View Article : Google Scholar
|
|
42
|
Cretney E, Takeda K, Yagita H, Glaccum M,
Peschon JJ and Smyth MJ: Increased susceptibility to tumor
initiation and metastasis in TNF-related apoptosis-inducing
ligand-deficient mice. J Immunol. 168:1356–1361. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Finnberg N, Klein-Szanto AJ and El-Deiry
WS: TRAIL-R deficiency in mice promotes susceptibility to chronic
inflammation and tumorigenesis. J Clin Invest. 118:111–123. 2008.
View Article : Google Scholar
|
|
44
|
Wang Q, Ji Y, Wang X and Evers BM:
Isolation and molecular characterization of the 5′-upstream region
of the human TRAIL gene. Biochem Biophys Res Commun. 276:466–471.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Allen JE and El-Deiry WS: Regulation of
the human TRAIL gene. Cancer Biol Ther. 13:1143–1151. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Azahri NS and Kavurma MM: Transcriptional
regulation of tumour necrosis factor-related apoptosis-inducing
ligand. Cell Mol Life Sci. 70:3617–3629. 2013. View Article : Google Scholar
|
|
47
|
Nebbioso A, Carafa V, Conte M, Tambaro FP,
Abbondanza C, Martens J, Nees M, Benedetti R, Pallavicini I,
Minucci S, et al: c-Myc modulation and acetylation is a key HDAC
inhibitor target in cancer. Clin Cancer Res. 23:2542–2555. 2017.
View Article : Google Scholar
|
|
48
|
Barbetti V, Tusa I, Cipolleschi MG, Rovida
E and Sbarba PD: AML1/ETO sensitizes via TRAIL acute myeloid
leukemia cells to the pro-apoptotic effects of hypoxia. Cell Death
Dis. 4:e5362013. View Article : Google Scholar :
|
|
49
|
Okuda T, Takeda K, Fujita Y, Nishimura M,
Yagyu S, Yoshida M, Akira S, Downing JR and Abe T: Biological
characteristics of the leukemia-associated transcriptional factor
AML1 disclosed by hematopoietic rescue of AML1-deficient embryonic
stem cells by using a knock-in strategy. Mol Cell Biol. 20:319–328.
2000. View Article : Google Scholar
|
|
50
|
Fukushima-Nakase Y, Naoe Y, Taniuchi I,
Hosoi H, Sugimoto T and Okuda T: Shared and distinct roles mediated
through C-terminal subdomains of acute myeloid
leukemia/Runt-related transcription factor molecules in murine
development. Blood. 105:4298–4307. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ito Y: Oncogenic potential of the RUNX
gene family: 'Overview'. Oncogene. 23:4198–4208. 2004. View Article : Google Scholar
|
|
52
|
Kanno Y, Kanno T, Sakakura C, Bae SC and
Ito Y: Cytoplasmic sequestration of the polyomavirus enhancer
binding protein 2 (PEBP2)/core binding factor alpha (CBFalpha)
subunit by the leukemia-related PEBP2/CBFbeta-SMMHC fusion protein
inhibits PEBP2/CBF-mediated transactivation. Mol Cell Biol.
18:4252–4261. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Goyama S, Yamaguchi Y, Imai Y, Kawazu M,
Nakagawa M, Asai T, Kumano K, Mitani K, Ogawa S, Chiba S, et al:
The transcriptionally active form of AML1 is required for
hematopoietic rescue of the AML1-deficient embryonic para-aortic
splanchno-pleural (P-Sp) region. Blood. 104:3558–3564. 2004.
View Article : Google Scholar
|
|
54
|
Liu H and Naismith JH: An efficient
one-step site-directed deletion, insertion, single and
multiple-site plasmid mutagenesis protocol. BMC Biotechnol.
8:912008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Asou H, Tashiro S, Hamamoto K, Otsuji A,
Kita K and Kamada N: Establishment of a human acute myeloid
leukemia cell line (Kasumi-1) with 8;21 chromosome translocation.
Blood. 77:2031–2036. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
57
|
Horinaka M, Yoshida T, Tomosugi M, Yasuda
S, Sowa Y and Sakai T: Myeloid zinc finger 1 mediates sulindac
sulfide-induced upregulation of death receptor 5 of human colon
cancer cells. Sci Rep. 4:60002014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kagoshima H, Akamatsu Y, Ito Y and
Shigesada K: Functional dissection of the alpha and beta subunits
of transcription factor PEBP2 and the redox susceptibility of its
DNA binding activity. J Biol Chem. 271:33074–33082. 1996.
View Article : Google Scholar
|
|
59
|
Bagger FO, Sasivarevic D, Sohi SH, Laursen
LG, Pundhir S, Sønderby CK, Winther O, Rapin N and Porse BT:
BloodSpot: A database of gene expression profiles and
transcriptional programs for healthy and malignant haematopoiesis.
Nucleic Acids Res. 44(D1): D917–D924. 2016. View Article : Google Scholar :
|
|
60
|
Rapin N, Bagger FO, Jendholm J,
Mora-Jensen H, Krogh A, Kohlmann A, Thiede C, Borregaard N,
Bullinger L, Winther O, et al: Comparing cancer vs normal gene
expression profiles identifies new disease entities and common
transcriptional programs in AML patients. Blood. 123:894–904. 2014.
View Article : Google Scholar
|
|
61
|
Kohlmann A, Kipps TJ, Rassenti LZ, Downing
JR, Shurtleff SA, Mills KI, Gilkes AF, Hofmann WK, Basso G,
Dell'orto MC, et al: An international standardization programme
towards the application of gene expression profiling in routine
leukaemia diagnostics: The microarray innovations in LEukemia study
prephase. Br J Haematol. 142:802–807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Haferlach T, Kohlmann A, Wieczorek L,
Basso G, Kronnie GT, Béné MC, De Vos J, Hernández JM, Hofmann WK,
Mills KI, et al: Clinical utility of microarray-based gene
expression profiling in the diagnosis and subclassification of
leukemia: Report from the international microarray innovations in
leukemia study group. J Clin Oncol. 28:2529–2537. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Klein HU, Ruckert C, Kohlmann A, Bullinger
L, Thiede C, Haferlach T and Dugas M: Quantitative comparison of
micro-array experiments with published leukemia related gene
expression signatures. BMC Bioinformatics. 10:4222009. View Article : Google Scholar
|
|
64
|
Warnat-Herresthal S, Perrakis K, Taschler
B, Becker M, Baßler K, Beyer M, Günther P, Schulte-Schrepping J,
Seep L, Klee K, et al: scalable prediction of acute myeloid
leukemia using high-dimensional machine learning and blood
transcriptomics. iScience. 23:1007802020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wouters BJ, Löwenberg B,
Erpelinck-Verschueren CA, van Putten WL, Valk PJ and Delwel R:
Double CEBPA mutations, but not single CEBPA mutations, define a
subgroup of acute myeloid leukemia with a distinctive gene
expression profile that is uniquely associated with a favorable
outcome. Blood. 113:3088–3091. 2009. View Article : Google Scholar :
|
|
66
|
Taskesen E, Bullinger L, Corbacioglu A,
Sanders MA, Erpelinck CA, Wouters BJ, van der Poel-van de
Luytgaarde SC, Damm F, Krauter J, Ganser A, et al: Prognostic
impact, concurrent genetic mutations, and gene expression features
of AML with CEBPA mutations in a cohort of 1182 cytogenetically
normal AML patients: Further evidence for CEBPA double mutant AML
as a distinctive disease entity. Blood. 117:2469–2475. 2011.
View Article : Google Scholar
|
|
67
|
Taskesen E, Babaei S, Reinders MM and de
Ridder J: Integration of gene expression and DNA-methylation
profiles improves molecular subtype classification in acute myeloid
leukemia. BMC Bioinformatics. 16(Suppl 4): S52015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yang L, Wang L, Zhao CH, Zhu XJ, Hou Y,
Jun P and Hou M: Contributions of TRAIL-mediated megakaryocyte
apoptosis to impaired megakaryocyte and platelet production in
immune thrombocytopenia. Blood. 116:4307–4316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Melloni E, Secchiero P, Celeghini C,
Campioni D, Grill V, Guidotti L and Zauli G: Functional expression
of TRAIL and TRAIL-R2 during human megakaryocytic development. J
Cell Physiol. 204:975–982. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Crist SA, Elzey BD, Ludwig AT, Griffith
TS, Staack JB, Lentz SR and Ratliff TL: Expression of TNF-related
apoptosis-inducing ligand (TRAIL) in megakaryocytes and platelets.
Exp Hematol. 32:1073–1081. 2004. View Article : Google Scholar
|
|
71
|
Lam K, Muselman A, Du R, Harada Y, Scholl
AG, Yan M, Matsuura S, Weng S, Harada H and Zhang DE: Hmga2 is a
direct target gene of RUNX1 and regulates expansion of myeloid
progenitors in mice. Blood. 124:2203–2212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Rini D and Calabi F: Identification and
comparative analysis of a second runx3 promoter. Gene. 273:13–22.
2001. View Article : Google Scholar
|
|
73
|
Ashkenazi A, Pai RC, Fong S, Leung S,
Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert
A, et al: Safety and antitumor activity of recombinant soluble Apo2
ligand. J Clin Invest. 104:155–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yoshida T, Shiraishi T, Nakata S, Horinaka
M, Wakada M, Mizutani Y, Miki T and Sakai T: Proteasome inhibitor
MG132 induces death receptor 5 through CCAAT/enhancer-binding
protein homologous protein. Cancer Res. 65:5662–5667. 2005.
View Article : Google Scholar
|
|
75
|
Yoshida T, Maoka T, Das SK, Kanazawa K,
Horinaka M, Wakada M, Satomi Y, Nishino H and Sakai T:
Halocynthiaxanthin and peridinin sensitize colon cancer cell lines
to tumor necrosis factor-related apoptosis-inducing ligand. Mol
Cancer Res. 5:615–625. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Jurisić V, Spuzić I and Konjević G: A
comparison of the NK cell cytotoxicity with effects of TNF-alpha
against K-562 cells, determined by LDH release assay. Cancer Lett.
138:67–72. 1999. View Article : Google Scholar
|
|
77
|
Jurisic V, Bogdanovic G, Kojic V, Jakimov
D and Srdic T: Effect of TNF-alpha on Raji cells at different
cellular levels estimated by various methods. Ann Hematol.
85:86–94. 2006. View Article : Google Scholar
|