Introduction
RNA modifications have emerged as a crucial process
in post-transcriptional gene regulation and are essential for RNA
biogenesis and functions. As the most prevalent chemical
modification of eukaryotic RNA, N6 methyladenosine (m6A)
modification of mRNA exerts a notable influence on mRNA stability,
splicing, localization, transport and translation efficiency
(1–4). m6A modifications also occur
on non-coding RNA, including ribosomal, transfer and small nuclear
RNA (5). Ribosomal RNA (rRNA) is
highly abundant in the total RNA of cells, and rRNA modifications
play an important role in regulating ribosome structure and
function (6). In humans, the
m6A modifications present in rRNA are located at two
specific sites: 1832 on 18S rRNA (m6A1832)
and 4220 on 28S rRNA (m6A4220) (7). The 28S rRNA
m6A4220 is catalyzed by rRNA
N6-adenosine-methyltransferase ZCCHC4, which is essential for
translation in ribosomes, cell proliferation and tumorigenesis
(8–10).
Mammalian rRNA N6-adenosine-methyltransferase METTL5
(METTL5), a member of the conserved methyltransferase-like protein
(METTL) family, was recently identified to methylate A1832 in 18S
rRNA (11). The 18S rRNA
m6A1832 modification is located in a critical
position in the decoding center, therefore suggesting its potential
importance in translation regulation (10). Indeed, a recent study indicated that
METTL5 promoted translation initiation, and the METTL5-mediated
m6A1832 modification may encourage the
decoding center to interact with mRNA undergoing active translation
(11). However, knowledge
concerning the biological functions of METTL5 is limited at
present. Several studies have revealed that METTL5 has an essential
role in the pluripotency and differentiation of mouse embryonic
stem cells, brain development and neural function in multiple
species (12–15). However, the other biological
functions of METTL5 are largely undefined, particularly in cancer.
Therefore, in the present study, the potential oncogenic activity
and the underlying mechanisms of METTL5 in pancreatic cancer were
evaluated.
c-Myc, which is located at chromosome 8q24, is one
of the three transcriptional activators of the Myc family (c-, N-
and L-Myc). c-Myc lies at a critical stage of a number of
growth-promoting signal transduction pathways, including ERK, PI3K,
AKT, MAPK and Wnt (16), and
therefore is involved in a variety of biological activities,
including ribosomal and mitochondrial biosynthesis, the cell cycle,
cell survival, proliferation, apoptosis, cell competition,
metabolic reprogramming and tumor progression (17–20).
Aberrant expression of c-Myc has been reported to be involved in
70% of human cancers, including lung, colon, breast, prostate,
colorectal and bladder cancer (20,21).
Elevated expression of c-Myc has also been identified to be a key
driver of pancreatic ductal adenocarcinoma (PDAC) through
modulating multiple oncogenic pathways, such as enhancing the
epithelial-mesenchymal transition and TGF-β signaling pathway
(22). A variety of factors lead to
the aberrant activation of c-Myc in cancer (23). The transcription of c-Myc is
mediated by transcription factors, such as NF-κB and nuclear factor
of activated T cells cytoplasmic 1, via the regulation of its
promoter activity. Meanwhile, the aberrant downregulation of
microRNAs (miRNAs/miRs), including Let7a, miR-145, miR-34a,
miR-375, miR-494 and miR-148a, has also been reported to lead to
the overactivation of c-Myc and further drive the progression of
PDAC (24). In addition,
post-translational modifications, such as acetylation and
phosphorylation, have been shown to enhance the protein stability
of c-Myc (25). Recent studies
showed that the mRNA m6A methyltransferase METTL3
enhanced c-Myc expression by increasing mRNA m6A levels
in prostate and lung cancer (19,26),
suggesting that c-Myc mRNA modification plays a key role in
c-Myc-centered oncogenic pathway regulation. Accordingly,
intervention of c-Myc expression through epigenetic pathways may
provide a potential therapeutic strategy for pancreatic cancer.
In the present study, the potential oncogenic
activity and the underlying mechanisms of METTL5 in pancreatic
cancer were investigated. It was demonstrated that METTL5 enhanced
c-Myc translation and the mechanism via which METTL5 could promote
specific selective translation was explored. It was hypothesized
that the METTL5-mediated upregulation of 18S rRNA
m6A1832 modification affected ribosome
structure, altering the affinity of ribosomes to mRNAs with
specific structural features or modifications. In addition, it was
verified that METTL5 and multifunctional methyltransferase subunit
TRMT112-like protein (TRMT112) may function together in pancreatic
cancer. Collectively, it may be proposed that the METTL5-mediated
increase in c-Myc translation facilitates tumorigenesis and could
represent a novel therapeutic strategy.
Materials and methods
Cell culture
Human pancreatic cancer cell lines, including PANC-1
(cat. no. CRL-1469), ASPC-1 (cat. no. CRL-1682) and BXPC-3 (cat.
no. CRL-1687), and the human pancreatic ductal epithelial cell line
HPDE6-C7 (cat. no. CVCL_0P38) were obtained from the American Type
Culture Collection. The cells were cultured in DMEM (cat. no.
11995040; Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS
(cat. no. 10099141; Gibco; Thermo Fisher Scientific, Inc.), 100
ng/ml streptomycin reagent (HyClone; Cytiva) and 100 U/ml
penicillin (HyClone; Cytiva). Cells were maintained in a humidified
atmosphere of 5% CO2 at 37°C.
Generation of stable cell lines
To generate stable cell lines transfected with
METTL5-overexpression vector (OE), METTL5mut-OE,
TRMT112-OE, short hairpin RNA (shRNA/sh) METTL5, shTRMT112,
METTL5-OE + TRMT112OE, METTL5-OE + shTRMT112 and TRMT112-OE +
shMETTL5, the coding DNA sequences (CDSs) of METTL5 and TRMT112
were subcloned into the pCDH-EF1α-MCS-T2A-Puro lentivirus vector
(cat. no. CD527A-1; System Biosciences, LLC) and the shRNA
sequences of METTL5 and TRMT112 were cloned into the
pLVX-shRNA2-BSD lentivirus vector (in which the ZsGreen gene
sequence is replaced with a BSD resistance gene sequence) (cat. no.
632179; Takara Bio USA, Inc.). For the METTL5mut CDS,
four amino acids (NPPF) from 126 to 129 were removed compared with
the wild-type CDS. 293T cells were transfected with
second-generation lentivirus packing system, consisting of the
aforementioned purified 0.5 µg/µl lentiviral plasmids, 0.5 µg/µl
pMD2.0, 0.5 µg/µl pspAX2 (the pMD2.0: pspAX2: lentivirus ratio was
1:3:4) using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.) for 72 h at 37°C. The supernatant containing
infectious lentivirus was then collected and transduced to PANC-1
cells for 48 h, at a multiplicity of infection of 10. Positive
overexpression and knockdown cells were selected by treatment with
2 µg/ml puromycin or 5 µg/ml blasticidin S for 10 days. shMETTL5-3
and shTRMT112-2 were chosen for subsequent experiments due to their
higher efficiency. The shRNA sequences for target gene knockdown
are shown in Table SI.
Bioinformatics analysis
The genes of interest, which were differentially
expressed in PDAC cells, were investigated in comparison with
adjacent normal non-neoplastic tissues using TCGA (http://gepia.cancer-pku.cn/; http://tcga-data.nci.nih.gov/tcga/). Overall survival,
METTL5 and TRMT112 expression analysis and correlation analysis
were examined using GEPIA (http://gepia.cancer-pku.cn/detail.php). The
m6A modification sites of c-Myc were predicted using the
website (https://www.cuilab.cn/sramp).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from PANC-1 cells using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RT was performed using
the HiScript III 1st Strand cDNA Synthesis kit (Vazyme Biotech Co.,
Ltd.) according to the manufacturer's protocol. qPCR was performed
using ChamQ SYBR Color qPCR Master mix (Vazyme Biotech Co., Ltd.)
and run on the ABI Prism 7900 system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The thermocycling protocol included an
initial denaturation step at 95°C for 3 min, followed by 40 cycles
of denaturation at 95°C for 15 sec, annealing at 60°C for 15 sec
and extension at 72°C for 30 sec. The 2−ΔΔCq values were
calculated to analyze the relative changes in gene expression
according to the previous reported method (27). β-actin was used as an internal
control. Primer sequences are shown in Table SII.
Western blotting
Proteins were extracted using cell lysis buffer RIPA
(Beyotime Institute of Biotechnology) and a protease and
phosphatase inhibitor cocktail (Beyotime Institute of
Biotechnology). A total of 20 µg protein/per lane was separated via
4–20% SDS-PAGE gels. The separated proteins were subsequently
transferred onto a membrane and blocked with 5% skim milk (Difco;
BD Biosciences) at room temperature for 2 h. The membranes were
incubated with appropriate primary antibodies at 4°C overnight.
Following primary incubation, membranes were incubated with
secondary antibodies for 2 h at room temperature (Table SIII). Protein bands were visualized
using a BeyoECL Moon Supersensitivity Detection kit (Beyotime
Institute of Biotechnology) according to the manufacturer's
protocol.
Ultraperformance liquid chromatography
tandem mass spectrometry (UPLC-MS/MS) analysis of
mononucleotides
A total of 1 µg purified 18S RNA was mixed with 0.1
unit Nuclease P1 (FUJIFILM Wako Pure Chemical Corporation) and 1.0
unit calf intestinal phosphatase (New England BioLabs, Inc.), in a
30-µl reaction volume adjusted with water, and incubated at 37°C
for 8 h. The digested RNA solutions were filtered using
ultra-filtration tubes (MW cutoff, 3 KDa; Pall Life Sciences), then
2 µl solution were injected into the UPLC-MS/MS system for
detection of m6A and A. The UPLC-MS/MS analysis was
performed with an Agilent 1290 UHPLC system coupled with a G6460
triple quadrupole mass spectrometer (Agilent Technologies, Inc.).
IF2 C18 column (100×2.1 mm I.D., 2.0 µm particle size; Shiseido
Co., Ltd.) was used for mononucleotide separation. A linear
gradient elution procedure was used for UPLC separation, elution
program settings were as follows: 0-1.5 min, 5.0% B; 1.6-10 min,
the proportion of B increased from 5 to 25%; 10.1-15.0 min, 100% B;
15.1-20 min, 5.0% B. Solvent A was an aqueous solution of 0.1%
formic acid, and solvent B was 100% methanol. The electrospray
ionization ion source was used for nucleotide ionization and
positive ion mode was selected for charged ion detection. A
multiple reaction monitoring mode was adopted: m/z 282.2-150.1 for
m6A (collision energy, 20 eV) and m/z 268.2-136.1 for A
(10 eV). The nebulization nitrogen was set at 40 psi with source
temperature set at 300°C and the flow-rate of desolvation nitrogen
was 9 l/min. Capillary voltage was set at 3,500 V. High purity
nitrogen (99.999%) was used as collision gas.
Site-specific m6A
modification
A CRISPR-RfxCas13d-based fusion system was
established by fusion of RNA demethylase ALKBH5 (ALKBH5), an
m6A demethylase, and ALKBH5mut (H204A) to the
dCas13d protein (replaced the EGFP coding sequence of
pHAGE-IRES-puro-NLS-dRfxCas13d-EGFP-NLS-3×Flag plasmid to ALKBH5
gene coding sequence) by using a GSG flexible connection linker
(dCas13d-ALKBH5/ALKBH5mut). The
pHAGE-IRES-puro-NLS-dRfxCas13d-EGFP-NLS-3×Flag was a gift from
Ling-Ling Chen (Addgene plasmid no. 132411). In total, four
independent guide RNAs (gRNAs) were designed to lead dCas13d-ALKBH5
near the specific m6A modification sites. The gRNA1 and
gRNA2 were designed to lead dCas13d-ALKBH5 near the m6A
modification sites in the 5′untranslated region (UTR) and CDS
region, and gRNA3 and gRNA4 were designed to lead dCas13d-ALKBH5
near the m6A modification near the stop codon. Stably
transfected cell lines were established to directly remove the
m6A modifications in the 5′UTR and CDS region (closer to
the 5′UTR) of c-Myc mRNA [dCas13d-ALKBH5 + gRNAs (5′CDS)] and the
m6A near the stop codon of c-Myc was removed mRNA
[dCas13d-ALKBH5 + gRNAs (SC)] in PANC-1 cells, respectively. The
dCas13d-ALKBH5 + gRNAs (scramble) was used as the control. The
stable cell lines were established by nucleofection with 4 µg
dCas13d-ALKBH5 plasmids and 2 µg dRfxCas13d gRNA cloning backbone
(a gift from Patrick Hsu; Addgene plasmid no. 109053) using a
Nucleofector 2b Device (Nucleofection system; Lonza Group, Ltd.).
After the program was finished, cells were transferred to 6-well
plates with 1.5 ml culture medium. After 48 h, cells were harvested
for protein extraction and western blot assay. The gRNAs sequences
were designed using website: https://cas13design.nygenome.org/. The gRNA sequences
for targeting c-Myc m6A sites are listed in Table SIV.
Cell proliferation assay
The cell proliferative rate was analyzed using Cell
Counting Kit-8 (CCK-8) assay. PANC-1 cells were seeded in 96-well
plate at a density of 2×103 cells/well, and 10 µl of
CCK-8 (Cell Counting Kit-8, Beijing Solarbio Science &
Technology Co., Ltd.) reagent was added to each well. Following
incubation at 37°C for 2 h, the optical density was measured at 450
nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
PANC-1 cells were seeded in 6-well plates at a cell
density of 300 cells/well and maintained in humidified air
containing 5% CO2 at 37°C for 10-14 days. The cell
seeding density for shMETTL5 and shCtrl groups was 600 cells/well.
Cells were washed with PBS, fixed with 4% paraformaldehyde for 15
min and stained with 0.5% crystal violet for 15 min at room
temperature. After staining, images were captured and the number of
colonies (containing cells >50 cells) was counted manually.
Wound healing assay
A total of 2.5×105 PANC-1 cells were
seeded and cultured in a twelve-well plate to create a confluent
monolayer. When the confluency reached 70–80%, a horizontal scratch
was made using a sterile 200-µl microliter pipette tip. The medium
was removed and changed to fresh DMEM supplemented with 2% FBS.
Images of the selected areas were obtained at 0 h and subsequently
the culture was incubated at 37°C with 5% CO2. After 48
h, the selected wound area was imaged using a light microscope
(Leica Microsystems, Inc.), and the remaining wound size was
measured using ImageJ software (version 1.53e). Wound healing was
quantified by calculating the percent of wound closure. The % wound
closure=1-(wound surface area at the indicated time-point/initial
wound surface area). The assay was performed in triplicate.
Transwell invasion assay
Cell invasion assays were performed in Transwell
chambers pre-coated with Matrigel (8-µm Transwell inserts; BD
Biosciences). Cells were seeded in the upper chamber at a cell
density of 1×104 cells in serum-free DMEM, and the lower
chambers were filled with DMEM supplemented with 10% FBS as an
attractant. The cells were incubated at 37°C for 24 h. Then, the
cells on the lower surface were fixed in 4% paraformaldehyde for 15
min and stained with 0.1% crystal violet for 15 min at room
temperature. Stained cells were visualized under a light microscope
(Leica Microsystems, Inc.). For each insert, at least three
selected fields were selected randomly and counted.
In vivo tumorigenicity assays
A total of 36 Female Balb/c nude mice (6–8 weeks of
age; 20–25 g) were purchased from the Charles River Laboratories,
Inc. and housed under 12-h light/dark conditions with a standard
temperature of 18–23°C with humidity of 40–60%, and access to food
and water ad libitum. All animal experiments in the present
study were approved (approval no. HS202105004) by the Ethics
Committee of Beijing University of Technology (Beijing, China).
METTL5-OE and the corresponding control cells (2×106
cells) in 200 µl PBS were injected subcutaneously into the right
side of nude mice. At ~4 weeks after injection, mice were
sacrificed after anesthesia with 1% pentobarbital sodium (45 mg/kg,
intraperitoneally), then euthanized by cervical dislocation. The
tumor weight was measured and the volume was calculated at the end
point. The maximum tumor volume allowed by the Ethical Committee of
the Beijing University of Technology was 1.5 cm diameter or 300
mm3 per tumor.
Statistical analysis
The data are presented as the mean ± SD from at
least three independent experiments. Statistical analyses were
performed using GraphPad Prism 5.0 software (GraphPad Software,
Inc.). Significance levels between two groups were evaluated using
a two-tailed paired Student's t-test. The differences between
multiple groups were compared using a one-way or two-way ANOVA
followed by Tukey's post-hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
METTL5 is upregulated in human
pancreatic cancer
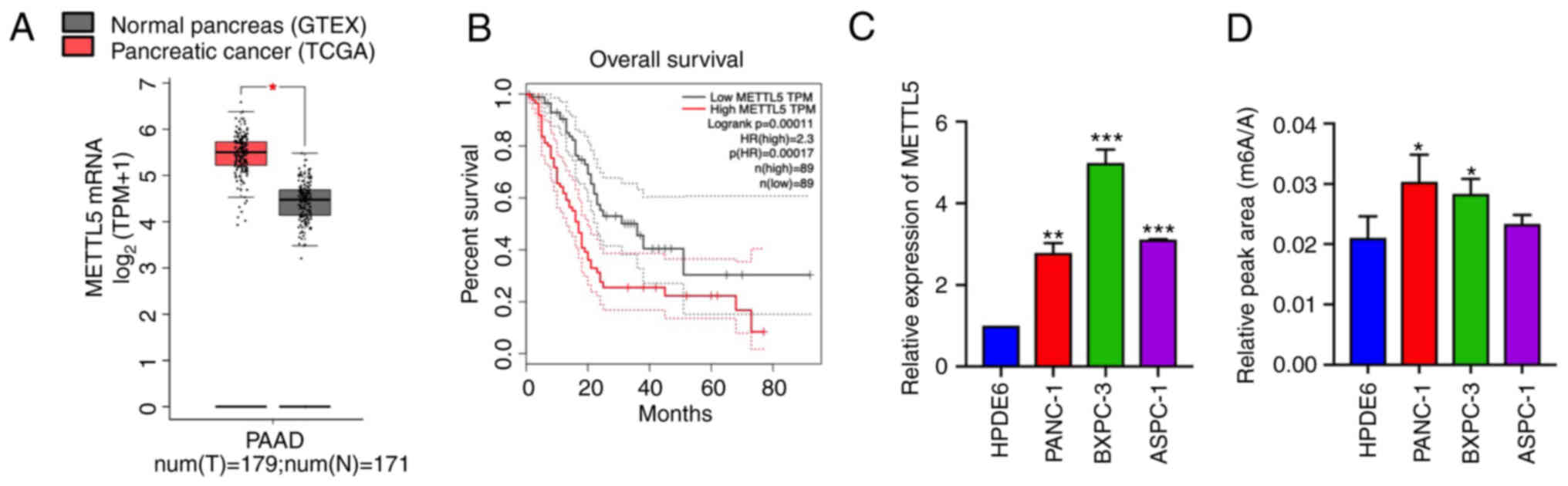
First, the expression of METTL5 in PDAC was
investigated. The Cancer Genome Atlas database showed that METTL5
expression was significantly elevated in pancreatic adenocarcinoma
(PAAD) tissues (n=179) compared with adjacent non-tumor tissues
(n=171; Fig. 1A). The increased
mRNA expression level of METTL5 was confirmed in three PDAC cell
lines (PANC-1, BxPC3 and ASPC1) and one pancreatic ductal
epithelial cell line (HPDE6-C7; Fig.
1C). Meanwhile, the elevated 18S rRNA m6A
methylation was detected in PDAC cells via UPLC-MS/MS, indicating
that increased rRNA methylation may affect tumorigenesis (Figs. 1D and S1A). Furthermore, a high level of METTL5
was associated with a poor overall survival rate (n=89; Fig. 1B). These results indicated that
METTL5 was upregulated in pancreatic cancer and may play an
important role in PDAC development.
METTL5 overexpression promotes
pancreatic cancer growth and invasion
To directly address the biological role of METTL5 in
PDAC, stable METTL5 overexpression and knockdown transfected cells
were established (Fig. 2A and B),
and a positive association between METTL5 and 18S rRNA
m6A level was confirmed in these cells (Figs. 2C and S1B). Cells overexpressing catalytically
inactive METTL5 (METTL5mut) showed no alteration in 18S
rRNA m6A compared with control cells (Fig. S1B). METTL5 overexpression
significantly stimulated proliferation, colony-forming capacity,
migration and invasion in PANC-1, ASPC-1 and BXPC-3 cells, whereas
knockdown of METTL5 exerted the opposite effects (Figs. 2D-G and S2A-D). METTL5 overexpression enhanced
tumor growth in xenotransplantation nude mice, specifically the
volume and weight were significantly increased in the METTL5
overexpression group (Fig. 2H).
Notably, METTL5mut cells had little effect on PDAC
progression, indicating that the oncogenic function of METTL5 in
PDAC was dependent on catalytic activity (Fig. 2I and J). Collectively, these results
demonstrated that METTL5 promoted cell proliferation, migration,
invasion and tumorigenesis in PDAC.
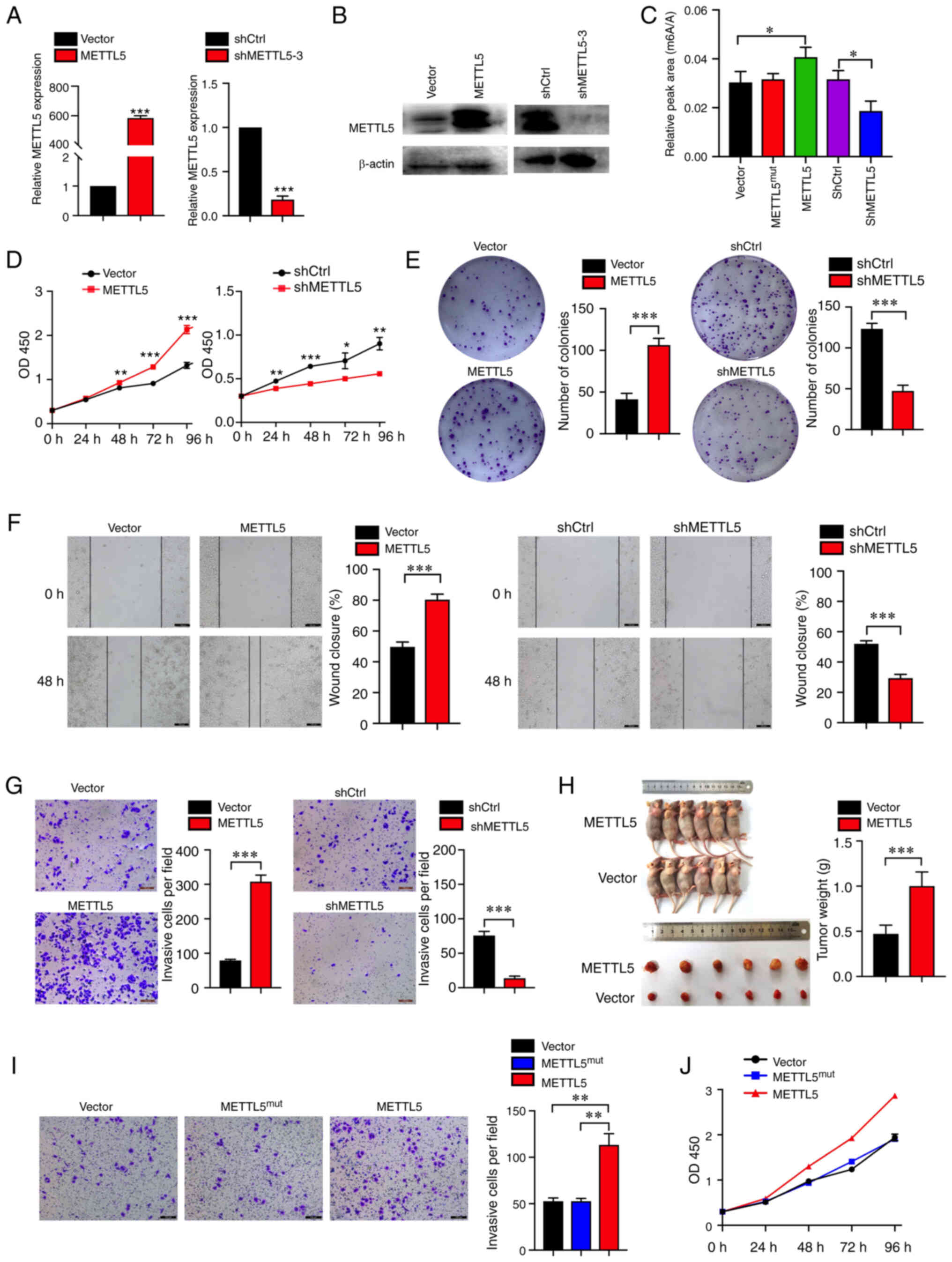 | Figure 2.Elevated METTL5 promotes pancreatic
cancer growth and invasion. (A) mRNA and (B) protein expression
level of METTL5 in PANC-1 cells expressing vector, exogenous
METTL5, shCtrl or shMETTL5. (C) The 18S rRNA m6A level
in cells was measured via liquid chromatography tandem mass
spectrometry and relative peak area (m6A/A) was
calculated. (D) Proliferation of PANC-1 cells transfected with
vector, exogenous METTL5, shCtrl or shMETTL5. (E) Representative
images of the colony formation assay in indicated cells. (F) Wound
healing and (G) Transwell assays were used to evaluate the
migratory and invasive potentials of METTL5 overexpression,
knockdown and corresponding control cells (Scale bar, 100 µm). (H)
The volume and weight analysis of subcutaneous tumors from the
indicated groups. (I) Transwell assays of PANC-1 cells transfected
with vector, METTL5mut and METTL5. (J) Proliferation of
PANC-1 cells transfected with vector, METTL5mut and
METTL5. Data are shown as the mean ± SD (n=3). *P<0.05,
**P<0.01 and ***P<0.001. METTL5, methyltransferase
N6-adenosine; sh, short hairpin RNA; m6A, N6
methyladenosine; ctrl, control; mut, mutant. |
METTL5 overexpression enhances the
translation of c-Myc
METTL5-mediated 18S rRNA m6A modification
has been suggested to play an important role in promoting
translation (11). It was
hypothesized in the present study that the oncogenic function of
METTL5 in PDAC may be closely associated with its positive effect
on the translation of key target genes. Thus, the changes in
protein levels of several crucial oncogenes after METTL5
overexpression or knockdown were investigated to explore the
potential candidates. The results indicated that the expression of
c-Myc was positively regulated by METTL5 in PANC-1 cells (Fig. 3A). Notably, no significant
differences in the mRNA expression level of c-Myc were found
between METTL5 overexpression, knockdown and corresponding controls
(Fig. 3B). This indicated that the
regulation of METTL5 on c-Myc was mainly at the translation stage.
Elevated c-Myc has been identified to be a key driver of PDAC
(22). Therefore, it was
hypothesized that the oncogenic effect of METTL5 in PDAC may be
associated with the regulation of c-Myc translation. Thus, the
functional association between METTL5 and c-Myc was further
examined in PDAC, and it was demonstrated that the oncogenic
effects of METTL5 overexpression could be abolished by c-Myc
knockdown (Fig. S2E and F). These
data supported the hypothesis that METTL5 promoted pancreatic
cancer progression by enhancing the translation efficiency of c-Myc
mRNA. Additionally, this supported the notion that the
m6A modification plays an important role in mRNA
translation.
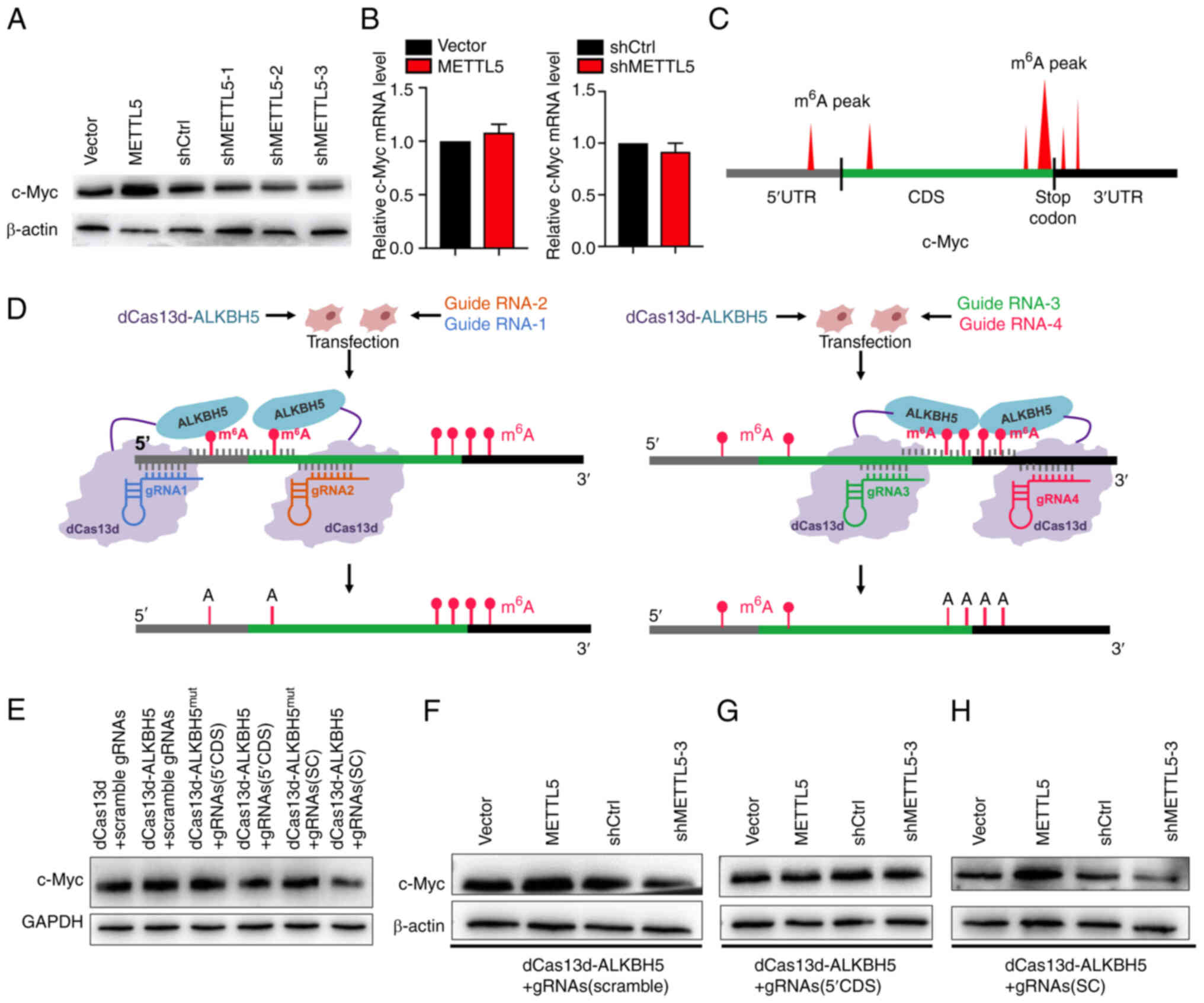 | Figure 3.METTL5-induced translation of c-Myc
is associated with m6A on c-Myc mRNA. (A) Protein
expression level of c-Myc in PANC-1 cells transfected with vector,
exogenous METTL5, shCtrl or shMETTL5. (B) The relative mRNA
expression levels of c-Myc in PANC-1 cells transfected with vector,
exogenous METTL5, shCtrl or shMETTL5. (C) Predicted m6A
modification sites on c-Myc mRNA. (D) Schematic representation of
the site-specific m6A modification removal. Stably
transfected cell lines were established to remove the
m6A modifications in the 5′UTR and CDS region (closer to
the 5′UTR) of c-Myc mRNA [dCas13d-ALKBH5 + gRNAs (5′CDS)] and the
m6A near the stop codon of c-Myc mRNA was removed
[dCas13d-ALKBH5 + gRNAs (SC)] in PANC-1 cells, respectively. The
gRNA1 and gRNA2 were designed to lead dCas13d-ALKBH5 near the
m6A modification sites in the 5′UTR and CDS region
(left), and gRNA3 and gRNA4 were designed to lead dCas13d-ALKBH5
near the m6A modification near the stop codon (right).
(E) c-Myc protein expression in stably transfected dCas13d-ALKBH5 +
gRNAs (5′CDS), dCas13d-ALKBH5 + gRNAs (SC),
dCas13d-ALKBH5mut + gRNAs (5′CDS),
dCas13d-ALKBH5mut + gRNAs (SC), dCas13d-ALKBH5 + gRNAs
(scramble) and dCas13d + gRNAs (scramble) cells. c-Myc protein
expression in (F) dCas13d-ALKBH5 + gRNAs (scramble) cells, (G)
dCas13d-ALKBH5 + gRNAs (5′CDS) cells and (H) dCas13d-ALKBH5 + gRNAs
(SC) cells transfected with vector, exogenous METTL5, shCtrl or
shMETTL5. Data are shown as the mean ± SD (n=3). METTL5,
methyltransferase N6-adenosine; sh, short hairpin RNA;
m6A, N6 methyladenosine; CDS, coding DNA sequence; gRNA,
guide RNA; ALKBH5, RNA demethylase ALKBH5; UTR, untranslated
region; mut, mutant; SC, stop codon. |
However, according to the present data, not all mRNA
translations were regulated by METTL5 (Fig. S3). Previous studies have also
mentioned that METTL5-mediated 18S rRNA m6A may
fine-tune the translation of a particular subset of mRNA (14,28),
instead of influencing the basal translation activity of ribosomes.
Therefore, the underlying mechanism via which METTL5 regulates the
translation of specific genes was further explored.
METTL5-mediated translation of c-Myc
is associated with m6A on c-Myc mRNA
m6A is the most abundant RNA modification
located on mRNA with a clear positioning bias (1,4). Thus,
it was speculated whether METTL5-mediated translation regulation of
specific mRNAs was related to m6A modifications of these
mRNAs, including the abundance and distribution characteristics. A
previous study reported that m6A peaks of most genes
were predominantly localized near stop codons (29). EGFR, tafazzin, MAP kinase-activated
protein kinase 2 and DNA (cytosine-5)-methyltransferase 3A have all
been found to have one or more m6A peaks near the stop
codon, whereas c-Myc has more broadly distributed m6A
across multiple exons (29).
Therefore, it was hypothesized in the present study that the
m6A modifications of c-Myc, in addition to the
modification near the stop codon, may be associated with its
specific regulation by METTL5. Accordingly, the predicted
m6A modification sites of c-Myc were investigated using
bioinformatics. In addition to the four m6A modification
sites near the terminal region of the CDS, c-Myc was also found to
have two distinct m6A modification sites near the
translation initiation site, one in the 5′UTR and another in the
CDS region near the 5′UTR (Fig.
3C).
To gain insight into the involvement of
m6A modifications on c-Myc mRNA in METTL5-mediated c-Myc
translation regulation, stably transfected cell lines were
established by applying a CRISPR-dRfxCas13d-based fusion system to
fuse dCas13d with an m6A demethylase ALKBH5
(dCas13d-ALKBH5) (30–32), the m6A modifications in
the 5′UTR and CDS region (close to the 5′UTR) of c-Myc mRNA (named
5′CDS) were directly removed and the m6A near the stop
codon of c-Myc mRNA (named SC) was removed in PANC-1 cells,
respectively, in the presence of a specific gRNA (Fig. 3D). Cells overexpressing dCas13d
fused with a catalytically inactive ALKBH5
(dCas13d-ALKBH5mut) were used as a negative control.
c-Myc expression was decreased in both dCas13d-ALKBH5 + gRNAs
(5′CDS) and dCas13d-ALKBH5 + gRNAs (SC) cells compared with control
cells, indicating that the removal of m6A on mRNA at
these two regions affected the expression level of c-Myc (Fig. 3E), and also demonstrated that this
system was successful in deleting the m6A at target
sites of c-Myc mRNA.
Subsequently, METTL5 was overexpressed or knocked
down in dCas13d-ALKBH5 + gRNAs (5′CDS), dCas13d-ALKBH5 + gRNAs (SC)
and dCas13d-ALKBH5 + gRNAs (scramble) cells, and the c-Myc protein
levels were compared to evaluate the effects of these
m6A alterations on the METTL5-mediated c-Myc translation
regulation. METTL5 overexpression and knockdown without
m6A intervention significantly mediated c-Myc
translation levels (Fig. 3F).
METTL5 overexpression and knockdown with the removal of
m6A near the translation initiation site failed to
maintain translation increase and inhibition, indicating that
METTL5-mediated regulation of c-Myc translation was greatly
weakened by the loss of m6A deposition at the 5′UTR and
CDS region (near the 5′UTR) of c-Myc mRNA (Fig. 3G). On the other hand,
METTL5-mediated c-Myc translation alteration was only slightly
affected by the removal of the four m6A modifications
near the stop codon (Fig. 3H). It
was suspected that the presence of m6A on 18S rRNA
caused subtle changes in the structure of rRNA that affect the
translation efficiency of specific mRNAs, while the distribution
characteristics of m6A on mRNAs determined the
significance of the influence. Collectively, the m6A
modifications of c-Myc mRNA played a critical role in specific
selective translation increase of c-Myc by METTL5 in PANC-1
cells.
METTL5 and TRMT112 may function
together in pancreatic cancer
TRMT112 acts as a cofactor for multiple
methyltransferases and is involved in the modification of tRNA,
rRNA and various proteins (33).
METTL5 has been shown to form a METTL5-TRMT112 m6A rRNA
methyltransferase complex to gain metabolic stability in cells
(10). In addition, as a
coactivator of METTL5, TRMT112 is essential for the enzymatic
activity of METTL5, and may synergistically catalyze 18S rRNA
m6A methylation (28).
In summary, TRMT112 plays an important role in stabilizing METTL5
and METTL5 activity. Compared with normal tissues, the expression
of TRMT112 in PAAD tissue was also significantly upregulated
(Fig. 4A), and associated with the
expression pattern of METTL5 in PDAC (Fig. 4B). Therefore, it was hypothesized
that the function of METTL5 in PDAC may also be associated with the
involvement of its coactivator TRMT112.
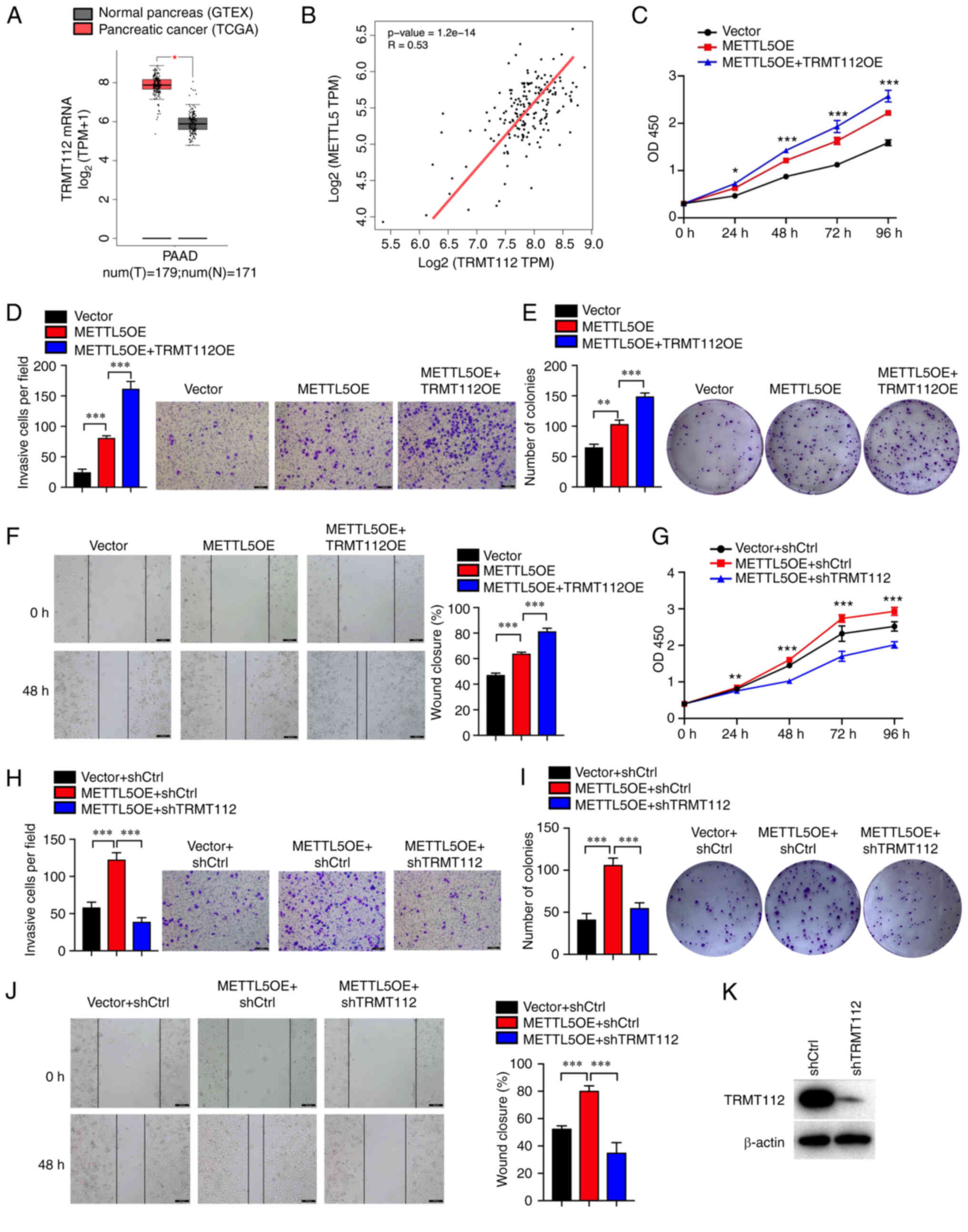 | Figure 4.METTL5 and TRMT112 may function
together in pancreatic cancer. (A) Expression of TRMT112 in
pancreatic cancer according to TCGA database. (B) Analysis of the
possible association between TRMT112 and METTL5 mRNA expression in
pancreatic cancer tissues according to TCGA database. The combined
overexpression of TRMT112 and METTL5 further promoted (C) cell
proliferation, (E) colony formation capacity, and (F) migratory and
(D) invasive abilities compared with METTL5 overexpression alone in
PANC-1 cells. TRMT112 knockdown abolished the effect METTL5
overexpression on (G) proliferation, (I) colony formation capacity,
(J) migration and (H) invasion of PANC-1 cells. (K) Protein level
of TRMT112 in PANC-1 cells expressing shCtrl and shTRMT112. Data
are shown as the mean ± SD (n=3). *P<0.05, **P<0.01 and
***P<0.001. METTL5, methyltransferase N6-adenosine; TRMT112,
multifunctional methyltransferase subunit TRMT112-like protein;
TCGA, The Cancer Genome Atlas; GTEX, Genotype-Tissue Expression;
PAAD, pancreatic adenocarcinoma; ctrl, control; OE, overexpression;
TPM, transcripts per million. |
To further investigate the underlying mechanisms of
METTL5 and TRMT112, their abilities to influence cell proliferation
and invasion were compared by using established stable cell lines
transfected with different combinations of vectors (METTL5-OE,
TRMT112-OE, shMETTL5, shTRMT112 and relative controls).
Transfection of PANC-1-shTRMT112 cells was confirmed by analyzing
the relative protein level of TRMT112 (Fig. 4K). As expected, the oncogenic
function of METTL5 in PDAC was significantly enhanced when TRMT112
was overexpressed simultaneously (Fig.
4C-F), and exchange with each other led to similar trends
(Fig. S4). In addition, the
oncogenic effects of METTL5 could be abolished by TRMT112 knockdown
(Fig. 4G-J), and the oncogenic
effects of TRMT112 could also be abolished by METTL5 knockdown
(Fig. S5). These results suggested
that METTL5 and TRMT112 may function together in pancreatic
cancer.
Discussion
rRNA is highly abundant in the total RNA of cells,
and rRNA modifications play an important role in regulating
ribosome structure and functions (7). METTL5 is a methyltransferase that
specifically catalyzes 18S rRNA m6A1832,
which is located in a critical position in the decoding center, and
therefore suggests its potential importance in translation
regulation (10). However, the
underlying mechanism via which METTL5-mediated translation
regulation of specific genes, and its biological functions are
largely undefined. To the best of our knowledge, the present study
showed for the first time that METTL5 functioned as an oncogene,
significantly promoting cell proliferation, migration, invasion and
tumorigenesis in pancreatic cancer. The oncogenic function of
METTL5 may involve an increase in c-Myc translation, while the
oncogenic effect of METTL5 overexpression could be abolished by
c-Myc knockdown. Mechanistically, it was suggested that
m6A modifications at the 5′UTR and CDS region (near the
5′UTR) of c-Myc mRNA played a critical role in the specific
selective translation regulation of c-Myc by METTL5. In addition,
it was further validated that METTL5 and its cofactor TRMT112 may
function together in pancreatic cancer. Thus, the METTL5/c-Myc
pathway in PDAC was proposed, which may represent a potential
therapeutic strategy for treatment.
The 18S rRNA m6A1832
modification is located in a critical position in the decoding
center, which indicates its importance in mRNA translation
processes. A recent study indicated that METTL5 promotes
translation initiation, and the METTL5-mediated
m6A1832 modification may encourage the
decoding center to interact with mRNA undergoing active translation
(11). In the present study, it was
verified that METTL5 enhanced the translation of c-Myc, which
supported the speculation that METTL5-mediated m6A
modification on rRNA plays an important role in translation
regulation. However, the present study and previous study observed
that not all mRNA translations were regulated by METTL5 (28). The mechanism via which METTL5
regulates the translation of a particular subset of mRNA is not
clear. It was suggested in the current study that the
METTL5-mediated upregulation of 18S rRNA
m6A1832 modification affected ribosome
structure, altering the affinity of ribosomes to mRNAs with
specific structural features or modifications. Thus, site-specific
m6A modification was directly removed on c-Myc to test
whether METTL5-mediated translation regulation of c-Myc was related
to m6A modifications of c-Myc mRNAs, including the
abundance and distribution characteristics. The results showed that
METTL5-mediated c-Myc translation was associated with the
m6A modifications on c-Myc mRNA, in which m6A
modifications at the 5′UTR and CDS region (near the 5′UTR) of c-Myc
mRNA played a critical role in the specific translation regulation
by METTL5. Meanwhile, METTL5-mediated c-Myc translation alteration
was only slightly affected by the removal of the four
m6A modifications near the stop codon. It was indicated
that the METTL5-mediated upregulation of 18S rRNA
m6A1832 modification affected ribosome
structure, altering the affinity of ribosomes to the mRNAs with
specific structural features conferred by the m6A
modifications. In this case, the closer the m6A
modification site to the initial region of the translation on the
mRNA, the more significant the impact will be. This may be the
reason that METTL5-mediated c-Myc translation alteration was still
significant after the removal of the four m6A
modifications near the stop codon. In conclusion, it was suggested
that the presence of m6A on 18S rRNA causes subtle
changes in the structure of rRNA that affect the translation
efficiency of specific mRNAs, while the distribution
characteristics of m6A on mRNAs determined the
significance of the influence. Notably, the functions of METTL5 in
the translation of c-Myc may be cell-type specific. The abundance
of m6A modification of c-Myc mRNA varies in different
cells, which may lead to different regulatory effects of METTL5.
Further research is required to verify whether this underlying
mechanism is widely applicable to METTL5-mediated translation
regulation in other genes.
As a coactivator of METTL5, TRMT112 is essential for
the stabilization and enzymatic activity of METTL5 and may
synergistically catalyze 18S rRNA m6A methylation
(10). In the present study, it was
verified that the oncogenic function of METTL5 in PDAC was
associated with the involvement of its coactivator TRMT112.
Additionally, the significant effect of TRMT112 interference on the
carcinogenic function of METTL5 suggested that the role of METTL5
in cancer may depend on its catalytic activity. However, it could
not be excluded that METTL5 also promotes cancer progression
through other mechanisms, such as interactions with key proteins.
It will be interesting and important to further investigate the
potential molecular mechanisms of METTL5 in cancer in the
future.
To conclude, to the best of our knowledge, the
present study revealed for the first time a distinct oncogenic role
of METTL5 in pancreatic cancer. It was demonstrated that METTL5
promoted c-Myc translation, while the location and abundance of
c-Myc mRNA m6A modifications played a critical role in
the specific translation regulation by METTL5. It was further
validated that TRMT112, as a cofactor of METTL5, is required for
the oncogenic functions of METTL5 in pancreatic cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81702802) and the programs of the
Beijing Municipal Education Commission (grant no.
KM201910005005).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HH conceptualized the study. HH, HL, RP, AAK, HZ and
SW performed the experiments. YZ performed the animal experiments.
HL and YZ analyzed the data. HH and XL were involved in writing the
manuscript. XL and HH supervised the overall project. All authors
have read and agreed to the published version of the manuscript. HL
and XL confirm the authenticity of the raw data.
Ethics approval and consent to
participate
All animal experiments in the present study were
approved (approval no. HS202105004) by the Ethics Committee of
Beijing University of Technology (Beijing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartosovic M, Molares HC, Gregorova P,
Hrossova D, Kudla G and Vanacova S: N6-methyladenosine demethylase
FTO targets pre-mRNAs and regulates alternative splicing and 3′-end
processing. Nucleic Acids Res. 45:11356–11370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)A promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu N, Dai Q, Zheng G, He C, Parisien M
and Pan T: N(6)-methyladenosine-dependent RNA structural switches
regulate RNA-protein interactions. Nature. 518:560–564. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Natchiar SK, Myasnikov AG, Kratzat H,
Hazemann I and Klaholz BP: Visualization of chemical modifications
in the human 80S ribosome structure. Nature. 551:472–477. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sergiev PV, Aleksashin NA, Chugunova AA,
Polikanov YS and Dontsova OA: Structural and evolutionary insights
into ribosomal RNA methylation. Nat Chem Biol. 14:226–235. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma H, Wang X, Cai J, Dai Q, Natchiar SK,
Lv R, Chen K, Lu Z, Chen H, Shi YG, et al: N(6-)Methyladenosine
methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat
Chem Biol. 15:88–94. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pinto R, Vagbo CB, Jakobsson ME, Kim Y,
Baltissen MP, O'Donohue MF, Guzmán UH, Małecki JM, Wu J, Kirpekar
F, et al: The human methyltransferase ZCCHC4 catalyses
N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids
Res. 48:830–846. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Tran N, Ernst FGM, Hawley BR, Zorbas
C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR,
Graille M and Lafontaine DLJ: The human 18S rRNA m6A
methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids
Res. 47:7719–7733. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rong B, Zhang Q, Wan J, Xing S, Dai R, Li
Y, Cai J, Xie J, Song Y, Chen J, et al: Ribosome 18S m(6)A
methyltransferase METTL5 promotes translation initiation and breast
cancer cell growth. Cell Rep. 33:1085442020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ignatova VV, Stolz P, Kaiser S, Gustafsson
TH, Lastres PR, Sanz-Moreno A, Cho YL, Amarie OV, Aguilar-Pimentel
A, Klein-Rodewald T, et al: The rRNA m(6)A methyltransferase METTL5
is involved in pluripotency and developmental programs. Genes Dev.
34:715–729. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Liang Y, Lin R, Xiong Q, Yu P, Ma
J, Cheng M, Han H, Wang X, Wang G, et al: Mettl5 mediated 18S rRNA
N6-methyladenosine (m6A) modification controls stem cell fate
determination and neural function. Genes & Diseases. 9:268–274.
2022. View Article : Google Scholar
|
|
14
|
Xing M, Liu Q, Mao C, Zeng H, Zhang X,
Zhao S, Chen L, Liu M, Shen B, Guo X, et al: The 18S rRNA m(6) A
methyltransferase METTL5 promotes mouse embryonic stem cell
differentiation. EMBO Rep. 21:e498632020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leismann J, Spagnuolo M, Pradhan M,
Wacheul L, Vu MA, Musheev M, Mier P, Andrade-Navarro MA, Graille M,
Niehrs C, et al: The 18S ribosomal RNA m(6) A methyltransferase
Mettl5 is required for normal walking behavior in Drosophila. EMBO
Rep. 21:e494432020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoshida GJ: Emerging roles of Myc in stem
cell biology and novel tumor therapies. J Exp Clin Cancer Res.
37:1732018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
AlSultan D, Kavanagh E, O'Grady S, Eustace
AJ, Castell A, Larsson LG, Crown J, Madden SF and Duffy MJ: The
novel low molecular weight MYC antagonist MYCMI-6 inhibits
proliferation and induces apoptosis in breast cancer cells. Invest
New Drugs. 39:587–594. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Destefanis F, Manara V and Bellosta P: Myc
as a regulator of ribosome biogenesis and cell competition: A link
to cancer. Int J Mol Sci. 21:40372020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu H, Li F and Zhu R: miR-338-5p inhibits
cell growth and migration via inhibition of the METTL3/m6A/c-Myc
pathway in lung cancer. Acta Biochim Biophys Sin (Shanghai).
53:304–316. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shan H, Cao Y, Xiao X, Liu M and Wu Y, Zhu
Q, Xu H, Lei H, Yao Z and Wu Y: YL064 activates
proteasomal-dependent degradation of c-Myc and synergistically
enhances the anti-tumor activity of ABT-199 in diffuse large B cell
lymphoma. Signal Transduct Target Ther. 5:1162020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang G, Xiong G, Feng M, Zhao F, Qiu J,
Liu Y, Cao Z, Wang H, Yang J, You L, et al: OLR1 promotes
pancreatic cancer metastasis via increased c-Myc expression and
transcription of HMGA2. Mol Cancer Res. 18:685–697. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong Y, Yang L, Xiong F, He Y, Tang Y,
Shi L, Fan S, Li Z, Zhang S, Gong Z, et al: Long non-coding RNA
AFAP1-AS1 accelerates lung cancer cells migration and invasion by
interacting with SNIP1 to upregulate c-Myc. Signal Transduct Target
Ther. 6:2402021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shams R, Aghdaei HA, Behmanesh A, Sadeghi
A, Zali M, Salari S and Padrón JM: MicroRNAs targeting MYC
expression: Trace of hope for pancreatic cancer therapy. A
systematic review. Cancer Manag Res. 12:2393–2404. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hessmann E, Schneider G, Ellenrieder V and
Siveke JT: MYC in pancreatic cancer: Novel mechanistic insights and
their translation into therapeutic strategies. Oncogene.
35:1609–1618. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang XS, He JR, Yu S and Yu J:
Methyltransferase-like 3 promotes the proliferation of acute
myeloid leukemia cells by regulating N6-methyladenosine Levels of
MYC. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 40:308–314. 2018.(In
Chinese). PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen H, Liu Q, Yu D, Natchiar K, Zhou C,
Hsu CH, Hsu PH, Zhang X, Klaholz B, Gregory RI, et al: METTL5, an
18S rRNA-specific m6A methyltransferase, modulates expression of
stress response genes. bioRxiv. doi: 10.1101/2020.04.27.064162.
|
|
29
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)A methyltransferase METTL3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wilson C, Chen PJ, Miao Z and Liu DR:
Programmable m(6)A modification of cellular RNAs with a
Cas13-directed methyltransferase. Nat Biotechnol. 38:1431–1440.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Konermann S, Lotfy P, Brideau NJ, Oki J,
Shokhirev MN and Hsu PD: Transcriptome engineering with
RNA-targeting type VI-D CRISPR effectors. Cell. 173:665–676, e614.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li S, Li X, Xue W, Zhang L, Yang LZ, Cao
SM, Lei YN, Liu CX, Guo SK, Shan L, et al: Screening for functional
circular RNAs using the CRISPR-Cas13 system. Nat Methods. 18:51–59.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liger D, Mora L, Lazar N, Figaro S, Henri
J, Scrima N, Buckingham RH, van Tilbeurgh H, Heurgué-Hamard V and
Graille M: Mechanism of activation of methyltransferases involved
in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res.
44:14822016. View Article : Google Scholar : PubMed/NCBI
|