Introduction
Lung cancer is the leading cause of cancer-related
mortality in both genders and was responsible for 1.8 million
deaths worldwide in 2020. The incidence of lung cancer is rising
for females in North America and Northern Europe (1). Lung cancer is categorized into two
types, namely non-small cell lung cancer (NSCLC), the predominant
type that accounts for 85%, and SCLC (2). NSCLC has a five-year survival rate of
~20% (1), whereas SCLC has <5%
due to aggressive behavior and early metastasis (3). NSCLC may be sub-classified into three
groups based on histology: Squamous cell carcinoma, adenocarcinoma
and large cell carcinoma (4). The
treatment of lung cancer consists of surgery if detected early.
Chemo- and radiotherapy and targeted therapy are the cornerstones
for the treatment of SCLC and advanced NSCLC. However, a major
drawback of current treatments is the development of resistance
against chemo- and radiotherapy, as well as targeted therapies
(5,6), a resistance that may be due to
distinct subsets of cells within the tumor not being eliminated by
conventional treatment. These subpopulations are the
slow-proliferating cancer stem cells (CSCs). CSCs are characterized
by self-renewal, differentiation potential, tumorigenic potential
and high DNA repair capabilities (7). Challenges with CSCs are their ability
to evade therapy and cells of this type display tumorigenic
potential in vivo (8). To
date, CSCs have been identified in various solid tumor types
including, but not limited to, brain, breast, colorectal, prostate
and lung cancer (9–13). CSCs are frequently identified based
on their expression of surface markers and several markers have
been proposed for lung cancer, including CD44, CD90, CD133, CD166
and epithelial cell adhesion molecule (EPCAM) [reviewed in
(14)].
Auger electrons are emitted from radionuclides that
decay by electron capture or internal conversion (15) and have demonstrated promising
therapeutic potential in breast cancer, multiple myeloma and brain
cancer (16–19). The low energy (<1 keV) of Auger
electrons causes lethal damage to cancer cells when the emitter is
close to the DNA (15). The
electrons have a high linear energy transfer (~4–25 keV/µm) and
travel within nanometers. Therefore, they are sufficiently potent
to induce DNA double-strand breaks and eventually cell death, while
minimizing irradiation of non-targeted surrounding cells (15,20).
Labeling a thymidine analog with the Auger electron-emitting
radionuclide [125I], which is exclusively incorporated
into the DNA of proliferating cells during the S-phase, ensures a
close distance to the DNA (20).
Morgenroth et al (19) found that an Auger electron-emitting
thymidine analog induced highly efficient death of
CD133+ glioblastoma stem cells (17) and multiple myeloma stem cells. In
addition, Thisgaard et al (21) treated two immature glioblastoma
spheroid cultures with 5-[125I]iodo-2′-deoxyuridine
([125I]I-UdR) and observed a dose-dependent reduction in
cell survival. Furthermore, this inhibitory effect was also
observed in vivo in orthotopically xenografted
glioblastoma-bearing rats. Together, these results suggest that
Auger electron emitters may overcome the resistance of CSCs. In the
present study, the cellular responses to the thymidine analog
[125I]I-UdR in patient-derived primary lung cancer cells
grown as tumorspheres for CSC enrichment were evaluated.
Materials and methods
Establishment of primary cell
cultures
The primary cell cultures were established from
resected NSCLC lung tumors collected at Odense University Hospital
(Odense, Denmark) between February 2015 and July 2018. The
inclusion criterion was surgery for primary lung cancer and
exclusion criterion was prior radio- or chemotherapy. The Regional
Ethics Committee of Southern Denmark approved the protocol (no.
S-20140170). Tissue was washed twice with PBS containing 2%
penicillin/streptomycin (Thermo Fisher Scientific, Inc.) and minced
with scalpels. To prevent adherence, the tumorspheres were cultured
in flasks coated with poly(2-hydroxyethyl methacrylate (cat. no.
P3932; MilliporeSigma). The cells were grown as tumorspheres in a
serum-free medium at 37°C in a humidified incubator with 5%
CO2. The serum-free medium consisted of DMEM/F-12
nutrient mix, Glutamax™ supplemented with 1%
penicillin/streptomycin, 1% B27 (cat. no. 12587010), 20 ng/ml
epidermal growth factor (cat. no. PHG0311; all from Thermo Fisher
Scientific, Inc.) and 20 ng/ml basic fibroblast growth factor (cat.
no. 100-18B; Peprotech, Inc.). Non-adherent conditions in
serum-free medium were used to enrich CSCs and ensure homology to
the parental tumor (22,23). Cells were expanded by mechanical
dissociation or TrypLE™ Express (cat. no. 12605028; Thermo Fisher
Scientific, Inc.) and used until 25 passages.
Tumorsphere formation assay
Cells (1 cell/well in 50 µl medium) were seeded in
non-adherent 96-well plates (Deltalab). Wells containing only one
cell were validated by microscopy and included in the experiment.
Twice a week, 50 µl serum-free medium was added. After 21 days of
incubation, the number of wells with one tumorsphere (>50 µm
diameter) was determined microscopically (Leica DMIL LED; Leica
Microsystems).
Doubling time (DT)
Cells were seeded in non-adherent 24-well plates
(Deltalab) at a density of 5×104 cells/well in 2 ml and
counted every second day. The DT was calculated as
DT=t(log2)/(logNt-logN0), where t is the
culture time and N0 and Nt are the initial
and final cell numbers after seeding, respectively.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was purified from adherent cells or
tumorspheres using the RNeasy® Plus mini kit (cat. no.
74134; Qiagen GmbH) according to the manufacturer's protocol. The
adherent cells were trypsinized tumorspheres that were allowed to
grow in tissue culture-flasks in the presence of 10% fetal bovine
serum to support their growth as differentiated cells. The RNA
yield and quality were measured using the Qubit 4 Fluorometer
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocols. RNA samples were stored at −80°C. RNA was reverse
transcribed using the RevertAid Minus First-strand cDNA synthesis
kit (cat. no. EP0451; Thermo Fisher Scientific, Inc.) using
oligo(dT) primers (cat. no. SO131; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol, i.e., RNA was reverse
transcribed at 42°C for 60 min and the reaction was terminated by
heating to 70°C for 10 min. qPCR was performed with TaqMan Fast
Advanced Master Mix (Thermo Fisher Scientific, Inc.) and Taqman
assays for CD44 (Hs01075864_m1), prominin 1 (PROM1; Hs01009250_m1),
Thy-1 cell surface antigen (also known as CD90; Hs00174816_m1),
SOX2 (Hs01053049_s1), POU class 5 homeobox 1 (POU5F1; Hs0099632_g1)
and Nanog homeobox (NANOG; Hs04260366_g1) (Thermo Fisher
Scientific, Inc.). A total of 20 ng cDNA was used per reaction and
the qPCR cycling was performed on a QuantStudio 3 (Applied
Biosystems; Thermo Fisher Scientific, Inc.) as follows: 50°C for 2
min and 95°C for 2 min, and then 40 cycles of 95°C for 1 sec and
60°C for 20 sec. All reactions were performed in triplicate and
normalized to hypoxanthine phosphoribosyltransferase 1 (HPRT1;
Hs02800695_m1). Initially, five candidate reference genes (HPRT1,
RPLP0, B2M, GAPDH and ACTB) were tested and HPRT1 was the most
stable in the experiments when using the web-based analysis tool
RefFinder (24). Relative
quantification was performed using the ΔΔCq method
(25).
Proliferation
Cells were seeded in non-adherent 24-well plates
(1×105) and incubated with 10 µM
5-ethynyl-2′-deoxyuridine (EdU) (cat. no. BCK-EDU488;
MilliporeSigma) for 24 h. Subsequently, the cells were fixed with
3.7% formaldehyde (cat. no. 47608; MilliporeSigma) for 15 min at
room temperature and permeabilized in 0.5% Triton X-100 (cat. no.
T8787; MilliporeSigma) in PBS for 20 min at room temperature. The
cells were stained using the Click-iT EdU 488 Proliferation Kit
(cat. no. BCK-EDU488; MilliporeSigma) following the manufacturer's
protocol and counterstained with 10 µg/ml DAPI (cat. no. D8417;
MilliporeSigma) for 10 min. For each group, 300 cells were analyzed
by fluorescent microscopy using a Leica DM 2000 LED microscope
(Leica Microsystems). Quantification of EdU-positive and -negative
cells was performed manually using the ‘cell counter plugin’ in
ImageJ version 1.50i (National Institutes of Health).
Cellular uptake and DNA incorporation
of [125I]I-UdR
For each experimental condition, 100,000 cells were
incubated with 18.5 kBq/ml [125I]I-UdR [prepared as in
(26)] in 1 ml serum-free medium
for 1, 4 and 7 h in non-adherent 24-well plates. At each
time-point, the experiment was ended by washing cells twice with
400 µl cold PBS and twice with 400 µl 5% trichloroacetic acid
(TCA). The DNA was solubilized in 500 µl 1M NaOH. The radioactivity
in the TCA fractions and collected DNA was determined in a 2470
Wizard Automatic Gamma Counter (Perkin Elmer). Cellular uptake was
calculated as the sum of the radioactivity in the TCA fractions and
collected DNA relative to the added radioactivity (% of injected
dose/well). DNA incorporation was calculated as the percentage of
radioactivity in the DNA relative to the cellular uptake.
Viability assay
A total of 1,000 single cells were seeded in
quadruplicate in non-adherent 96-well plates in 50 µl serum-free
medium with 0.1-6.0 kBq/ml [125I]I-UdR in 50 µl medium.
As a control, the non-radioactive, but chemically identical
[127I]I-UdR (24 pg/ml; cat. no. I7125; MilliporeSigma)
corresponding to the mass concentration of 6 kBq/ml was also
tested. On day seven, the cell viability was evaluated by adding 13
µl CellTiter-Blue (cat. no. G8080; Promega Corporation) to each
well. Fluorescence was measured at 520 nm excitation/580–640 nm
emission in a GloMax Explorer (Promega Corporation).
Clonogenic assay
A total of 100 cells were seeded in non-adherent
24-well plates in 1 ml medium and incubated with 2.5 or 5 kBq/ml
[125I]I-UdR. The number of tumorspheres was evaluated
after 10 [lung cancer case no. 10 (LUC10) and LUC13] or 17 days
(LUC6) using a Leica DM IL LED microscope (Leica Microsystems).
Cell cycle analysis
A total of 100,000 cells were incubated with 2.5
kBq/ml [125I]I-UdR for seven days. The LUC6 tumorspheres
were mechanically dissociated by pipetting, and LUC10 and LUC13
were trypsinized, washed once with PBS and fixed in 70% ethanol at
−20°C overnight. After fixation, cells were washed once in PBS and
resuspended in 100 µl cell cycle reagent mix [20 µg/ml propidium
iodide (cat. no. P4170) and 10 mg/ml RNase A (cat. no.
10109142001); both from MilliporeSigma] and incubated in the dark
at room temperature for 30 min. Next, cells were washed in 100 µl
PBS and loaded into an A8 cassette (Chemometec). The cell cycle
distribution was measured by image cytometry in the Nucleocounter
NC-3000 (Chemometec).
Apoptosis and DNA damage
A total of 100,000 cells were incubated with 2.5
kBq/ml [125I]I-UdR for seven days, dissociated and
counted using the MUSE Count and Viability reagent (cat. no.
4000-0335; Luminex). Cells were resuspended in 50 µl 1% bovine
serum albumin (cat. no. A8022)/PBS for the apoptosis analysis and
mixed with 50 µl MUSE Annexin V and Dead reagent (cat. no.
4700-1485; Luminex). The samples were incubated at room temperature
in the dark for 20 min prior to analysis of 10,000 cells on the
Guava MUSE Cell Analyzer (Luminex).
DNA damage was analyzed using the MUSE H2AX
Activation Kit (cat. no. MCH200101; Luminex). In brief, dissociated
cells were resuspended in 50 µl 1X Assay buffer and 50 µl fixation
reagent for 5 min on ice. Subsequently, the cells were
permeabilized in 50 µl ice-cold permeabilization reagent for 5 min
on ice. Cells were incubated in 50 µl 1X Assay buffer containing 1
µl anti-H2A.X (cat. no. CS208162; Luminex) and 1 µl
anti-phosphorylated-histone H2A.X (phospho-H2AX; cat. no. CS208174;
Luminex) at room temperature in the dark for 30 min. Cells were
washed with 100 µl 1X Assay buffer and resuspended in 200 µl 1X
Assay buffer, and 1,000 cells were analyzed on the Guava MUSE Cell
Analyzer (Luminex).
Statistical analysis
Experiments were performed as three independent
replicates and descriptive statistics for quantitative measurements
comprised the mean ± standard error of the mean. One-way ANOVA was
used to compare means of sphere-formation, means of
EdU/DAPI-positive cells and means of cellular uptake and
incorporation (correction for multiple comparisons: Tukey).
Cellular uptake and incorporation were also evaluated by a
post-test for a linear trend. Differences between means in
viability and the effect of [125I]I-UdR on tumorsphere
growth were evaluated by one-way ANOVA (correction for multiple
comparisons: Dunnett/Bonferroni). Two-way ANOVA was applied to
compare the means from the cell cycle analysis (cell cycle phase
and treatment as independent factors; correction for multiple
comparisons: Ŝidák) and RT-qPCR analysis (gene and sample as
independent factors; correction for multiple comparisons: Tukey).
Differences between the mean values for cell death and DNA damage
were investigated by an unpaired t-test. P<0.05 was considered
as a threshold of statistical significance. Statistical tests were
performed using GraphPad Prism version 9.0 (GraphPad Software,
Inc.).
Results
Patient characteristics
Tissue was collected from 15 patients whose details
are provided in Table I. The median
patient age was 70 years (range, 57–85 years) and 60% were males.
No vital tumorspheres were formed in four of them, which were
therefore discarded. The remaining samples initially gave rise to
viable tumorspheres, but some samples were not susceptible to
long-term culture. In the end, only LUC6, LUC10 and LUC13 exhibited
stable unlimited exponential growth even in later passages.
 | Table I.Characteristics of patients and
tumorsphere formation ability of their samples. |
Table I.
Characteristics of patients and
tumorsphere formation ability of their samples.
| Patient code | Sex | Age (years) | Tumorsphere
formation | Histology |
|---|
| LUC1 | Female | 57 | +/- | Planocellular
carcinoma |
| LUC2 | Male | 70 | +/- | Mucinous
adenocarcinoma |
| LUC3 | Female | 77 | +/- | Adenocarcinoma |
| LUC4 | Female | 77 | - | Planocellular
carcinoma |
| LUC5 | Male | 70 | - | Adenocarcinoma |
| LUC6 | Male | 62 | + | Adenocarcinoma |
| LUC7 | Male | 68 | +/- | Papillary
adenocarcinoma |
| LUC8 | Male | 70 | +/- | Planocellular
carcinoma |
| LUC9 | Female | 85 | - | Planocellular
carcinoma |
| LUC10 | Female | 66 | + | Adenosquamous
carcinoma |
| LUC11 | Male | 76 | - | Adenocarcinoma |
| LUC12 | Male | 67 | +/- | Adenocarcinoma |
| LUC13 | Male | 67 | + | Pleomorphic
carcinoma |
| LUC14 | Female | 75 | +/- | Adenocarcinoma |
| LUC15 | Male | 74 | +/- | Planocellular
carcinoma |
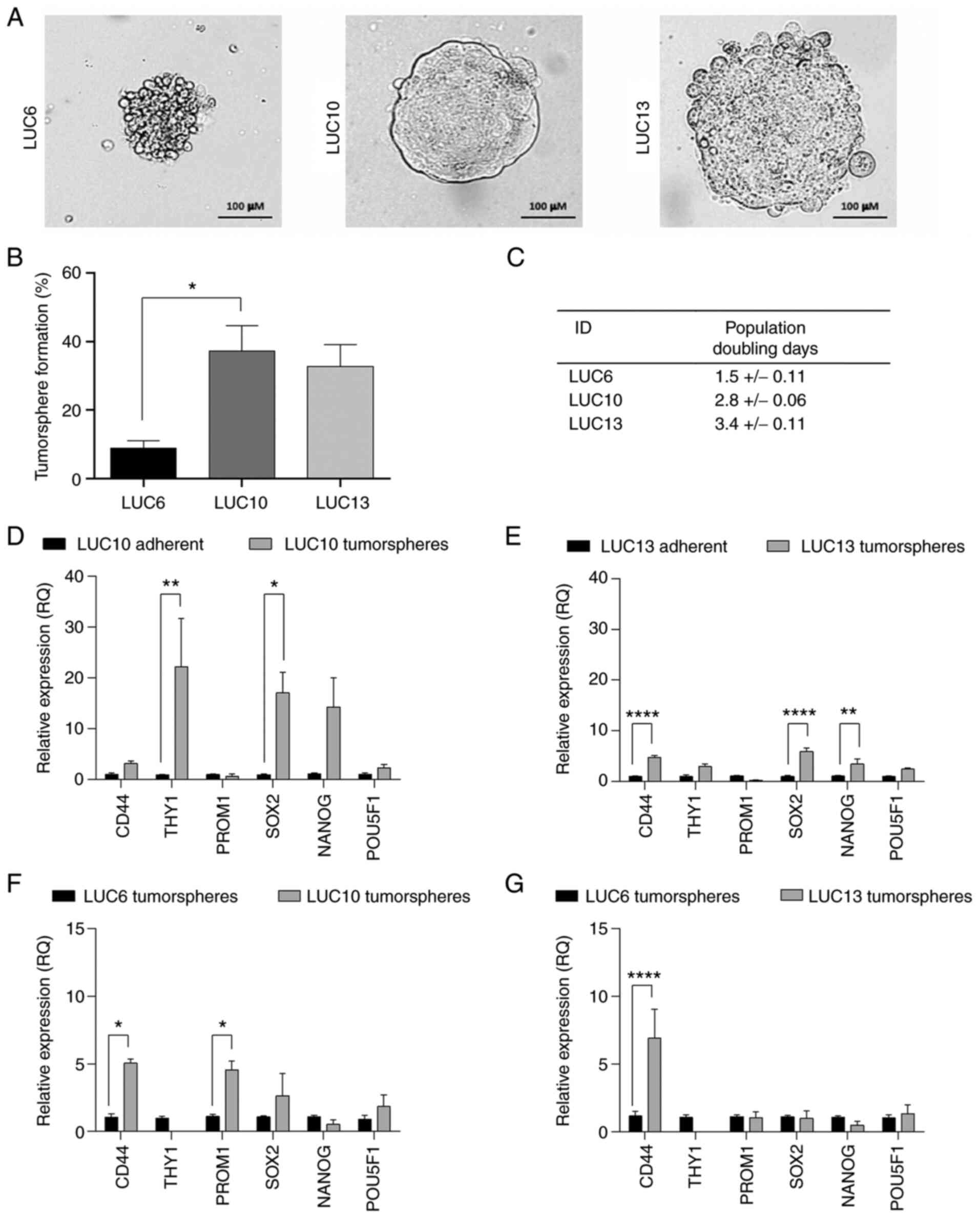
Tumorsphere morphology, tumorsphere
formation and population DT
LUC6, LUC10 and LUC13 all demonstrated the ability
to form vital tumorspheres (Fig.
1A), one of the hallmarks of CSCs (27). LUC6 tumorspheres were loosely packed
with an irregular surface and clearly defined cells. The LUC10
tumorspheres were encapsulated with a membrane-like structure and
densely packed cells, whereas spheroid cells were more defined and
the tumorsphere-surface appeared more irregular in LUC13. CSCs are
also defined by their ability to self-renew, which is a functional
difference from non-CSCs (7).
Self-renewal may be assessed by the sphere formation assay, which
only considers tumorspheres formed from one single cell. The
results indicated that LUC10 (P<0.05) and LUC13 contained more
cells with self-renewing potential than LUC6 (Fig. 1B). The population DT was estimated
for the tumorspheres (Fig. 1C): The
DT of LUC6 was 1.5±0.11 days. LUC10 and LUC13 had population DTs of
2.8±0.06 and 3.4±0.11 days, respectively. The relative expression
levels of the stemness-related surface markers CD44, THY1 (CD90),
PROM1 (CD133), SOX2, NANOG and POU5F1 (Oct4) in LUC10 and LUC13
tumorspheres compared to adherent cells were also evaluated
(Fig. 1D and E). LUC10 cells grown
as tumorspheres exhibited significantly increased expression THY1
(P<0.01) and SOX2 (P<0.05) compared to adherent LUC10 cells
(Fig. 1D). Likewise, LUC13
tumorspheres displayed significantly increased expression of CD44
(P<0.0001), SOX2 (P<0.0001) and NANOG (P<0.01) compared to
adherent cells (Fig. 1E).
Adherently grown LUC10 and LUC13 expressed higher levels of PROM1;
however, the difference was not significant. Unfortunately, it was
not possible to grow adherent LUC6 cells, so instead, the
expression of LUC6 tumorspheres was compared to LUC10 and LUC13
tumorspheres (Fig. 1F and G). LUC6
expressed significantly lower levels of CD44 and PROM1 than LUC10
(P<0.05) and significantly lower levels of CD44 (P<0.0001)
than LUC13. These results support the notion that LUC10 and LUC13
contained more CSCs than LUC6.
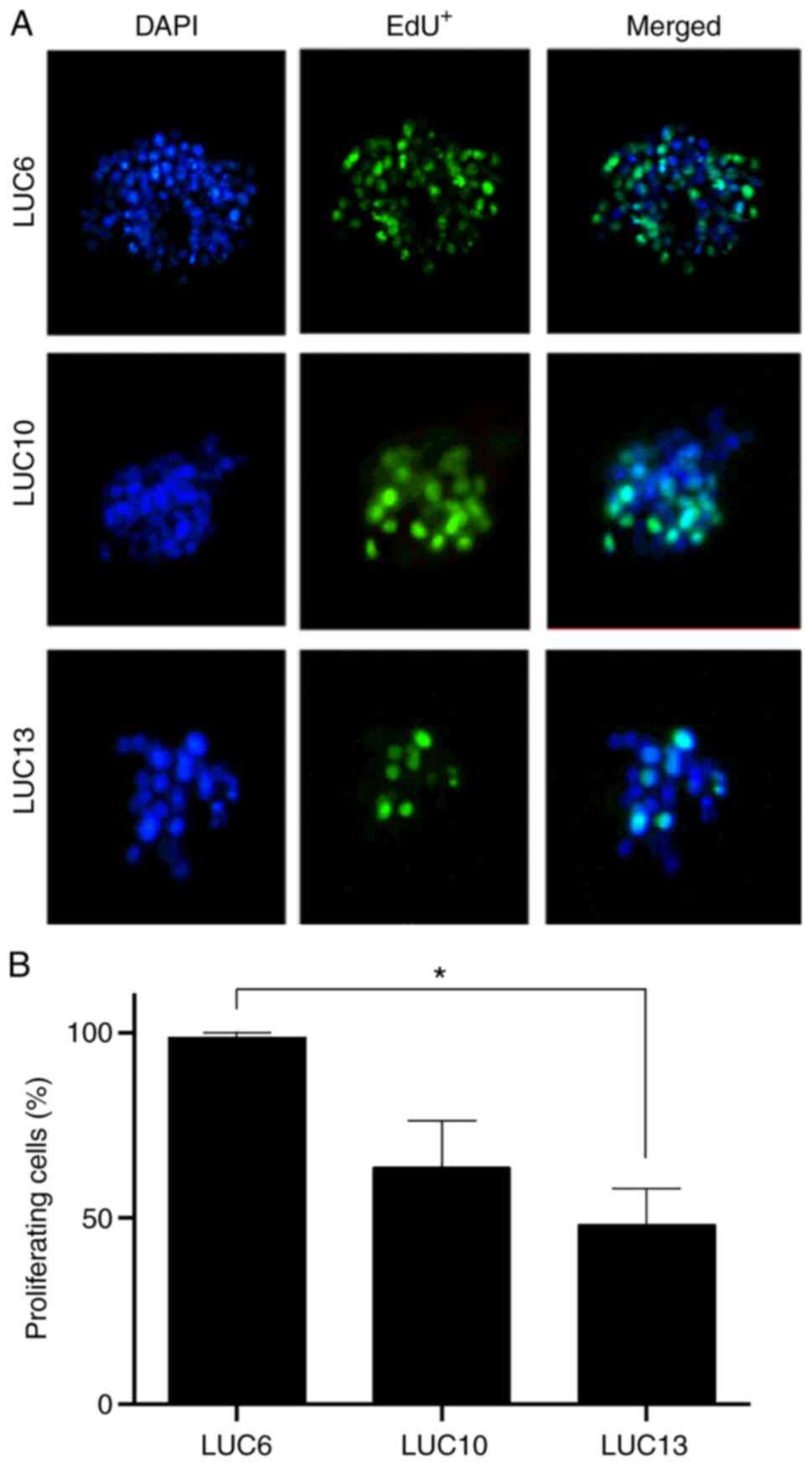
Proliferation
The thymidine analog EdU was used to determine the
proportion of proliferating cells in the tumorspheres (Fig. 2A). Most of the LUC6 cells in the
tumorspheres were EdU-positive (98.6±1.3%). Approximately half
(48.3±9.7%) of the LUC13 cells were proliferative, whereas
EdU-positive LUC10 cells accounted for 63.6±12.4% in the
tumorspheres (Fig. 2B).
Incorporation of EdU was present in all three samples within 24 h,
and correlated with the DT.
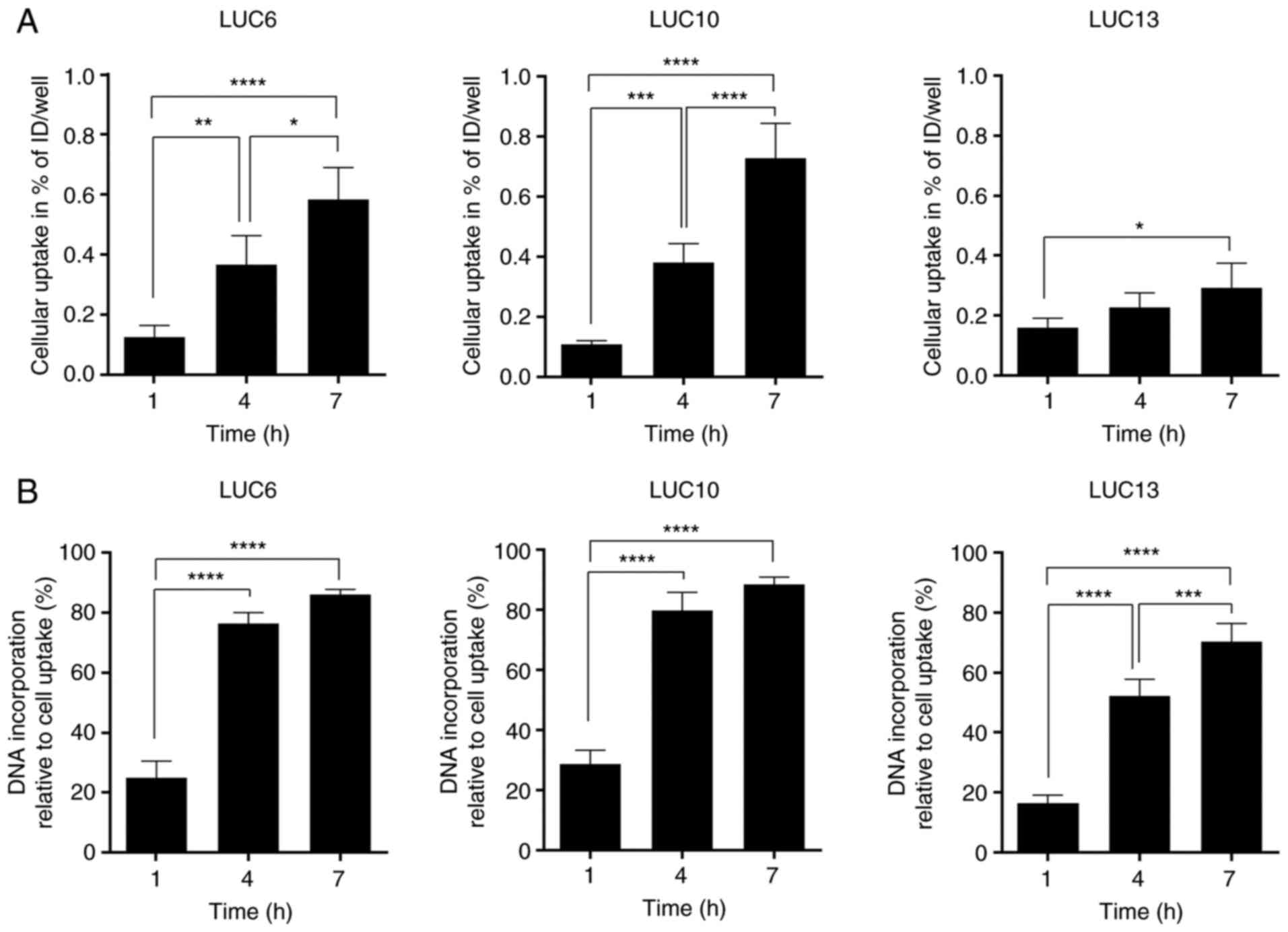
[125I]I-UdR uptake and
incorporation
Cellular uptake and DNA incorporation of
[125I]I-UdR were measured after 1, 4 and 7 h (Fig. 3). These time-points were chosen to
minimize cytotoxicity resulting from long exposure times (28). In addition, it was previously
reported that DNA incorporation was rapid within the first 4–5 h
and plateaued by 10 h (29).
Overall, the cellular uptake of [125I]I-UdR in LUC6 and
LUC10 increased significantly over time (post-test for linear
trend, P<0.0001). A significantly increased cellular uptake was
also seen for LUC13 between 1 and 7 h (P<0.05), although
[125I]I-UdR uptake was not doubled (post-test for linear
trend, P<0.05) (Fig. 3A).
[125I]I-UdR incorporation in LUC6 and
LUC10 increased significantly (P<0.0001) from ~28 to 79 and 88%,
respectively (post-test for linear trend between the time-points,
P<0.0001) (Fig. 3B). The DNA
incorporation in LUC13 increased significantly from 1 h, where 16%
[125I]I-UdR was incorporated, to 52 and 70% after 4 and
7 h, respectively (P<0.0001). Furthermore, the difference in the
incorporation of [125I]I-UdR from 4 to 7 h was also
significant (P<0.001; post-test for linear trend, P<0.0001).
Overall, DNA incorporation of [125I]I-UdR increased over
time; however, the level varied among the different
tumorspheres.
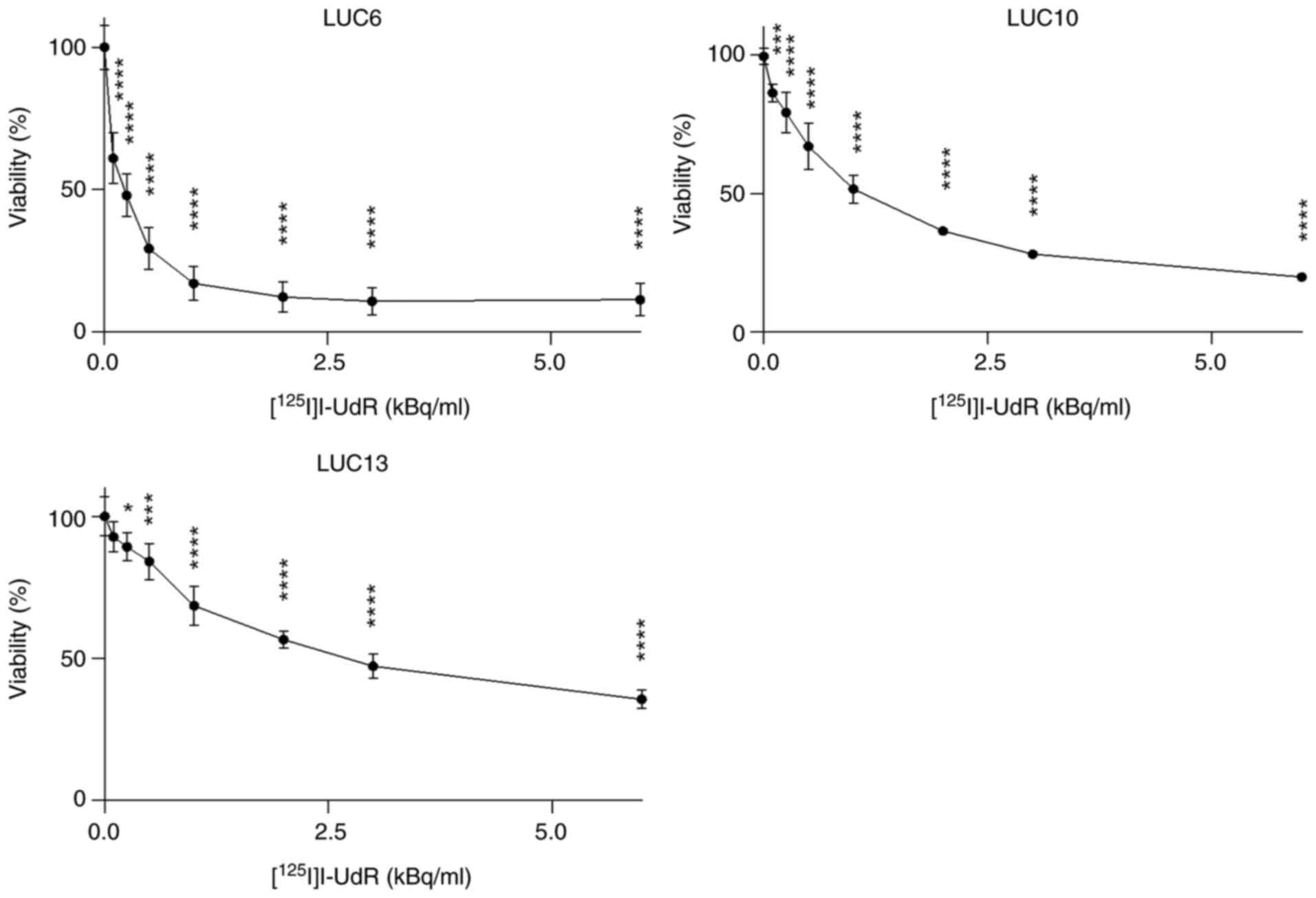
Viability
Next, the effect of [125I]I-UdR on the
viability of the tumorspheres was assessed by incubating with 0.1–6
kBq/ml [125I]I-UdR for seven days, followed by
CellTiter-Blue viability measurements (Fig. 4). The viability of LUC6
significantly decreased at all activities tested (P<0.0001)
compared to 0 kBq/ml [125I]I-UdR. The viability was ~50%
when LUC6 was treated with 0.25 kBq/ml [125I]I-UdR. The
viability of LUC6 did not decrease further when the activity
exceeded 2 kBq/ml. The viability of LUC10 also significantly
decreased at all tested activities of [125I]I-UdR
(P<0.0001; and 0.1 kBq/ml, P<0.001). LUC13 was more resistant
to [125I]I-UdR, as 3 kBq/ml was necessary to reduce the
viability to ~50%. Furthermore, the viability of LUC13 was still
>30% when treated with 6 kBq/ml. The non-radioactive and
chemically identical [127I]I-UdR did not decrease the
viability (results not shown). Overall, there was an activity
concentration-dependent decrease in viability; however, LUC6 was
more sensitive to [125I]I-UdR than LUC10 and LUC13.
Effect of [125I]I-UdR on
clonogenic survival
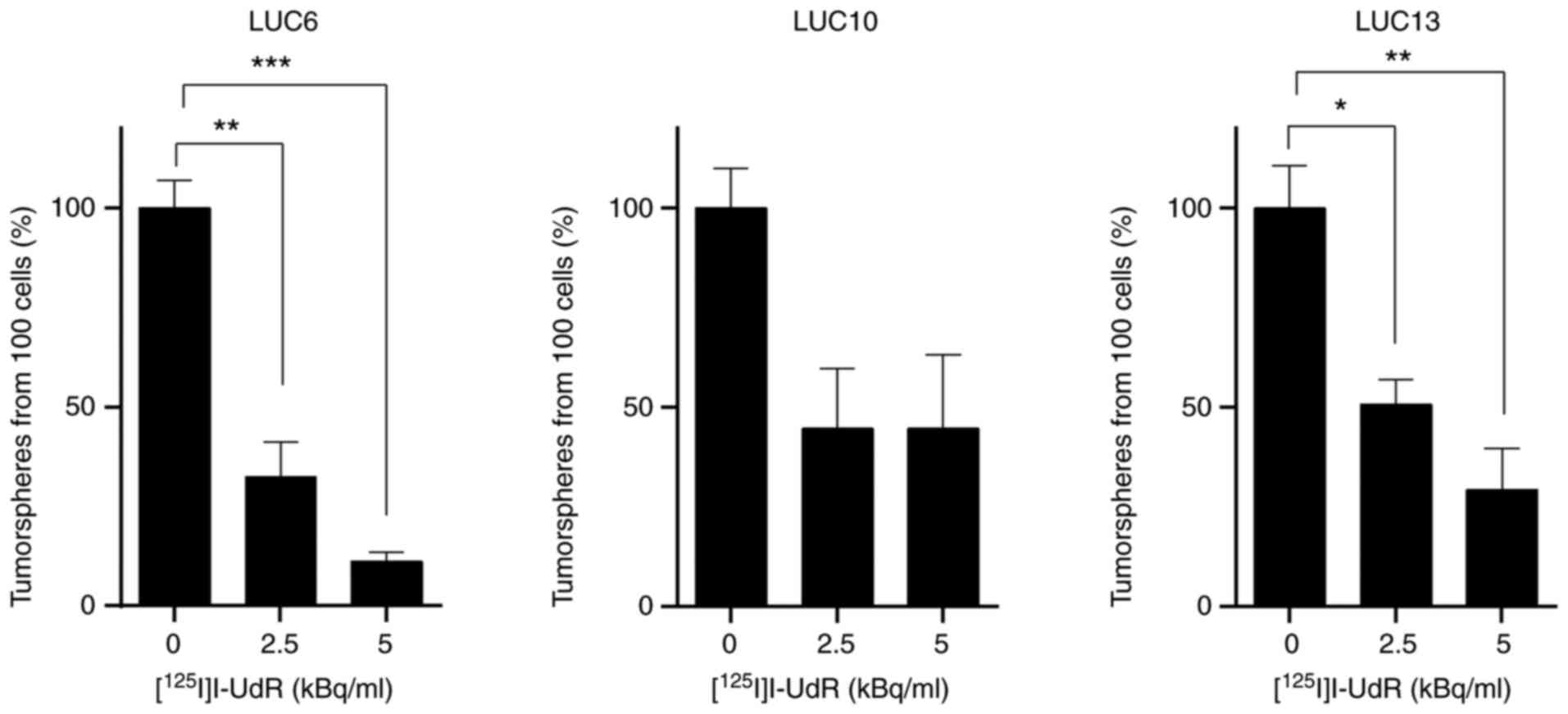
The effect of [125I]I-UdR on tumorsphere
formation was then investigated by incubating 100 cells with 0, 2.5
and 5 kBq/ml [125I]I-UdR (Fig. 5). Tumorspheres were counted
following 10 (LUC10 and LUC13) or 17 days (LUC6) after the addition
of [125I]I-UdR. The number of tumorspheres generated
from [125I]I-UdR-treated cells was normalized to that of
tumorspheres generated from untreated control cells. Treatment of
LUC6 with 2.5 kBq/ml led to a significant decrease in the number of
tumorspheres to 32.5±8.8% (P<0.01). Treatment with 5 kBq/ml
further reduced the formation of LUC6 tumorspheres significantly to
11.3±2.1% (P<0.001). The number of LUC10 tumorspheres decreased
to 44.6±14.9 and 44.6±18.6% when treated with 2.5 and 5 kBq/ml
[125I]I-UdR, respectively, yet not significantly.
Treatment with 2.5 kBq/ml decreased the number of LUC13
tumorspheres significantly to 50.8±6.2% (P<0.05) and following
treatment with 5 kBq/ml, only 29.3±10.3% tumorspheres were formed
(P<0.01). Overall, [125I]I-UdR decreased the ability
of LUC6 and LUC13 to form tumorspheres in a concentration-dependent
manner. By contrast, the ability of LUC10 to form tumorspheres did
not further decrease when treated with 5 kBq/ml compared to 2.5
kBq/ml [125I]I-UdR.
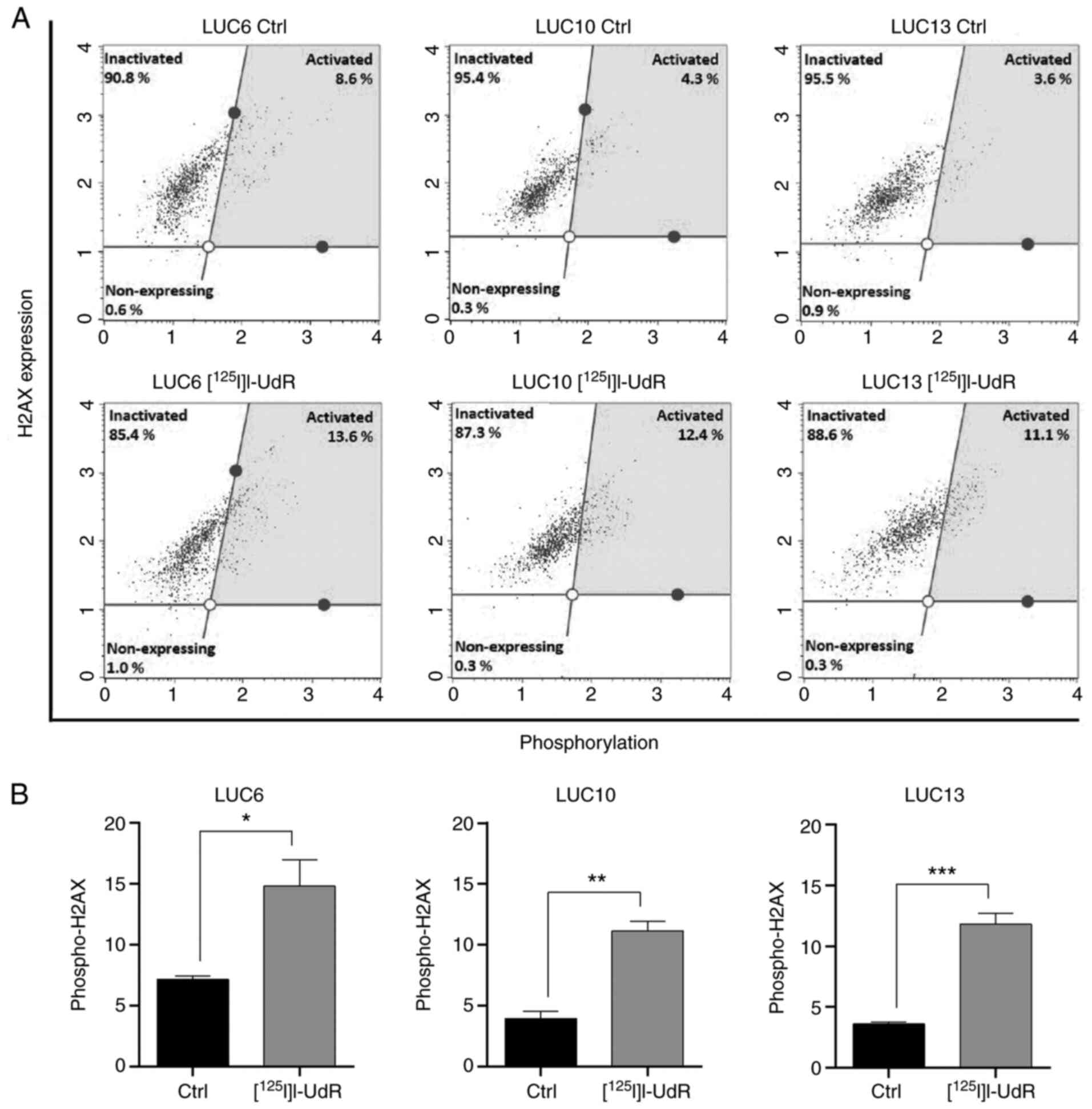
Radiation-induced DNA damage
The DNA-damaging effect of 2.5 kBq/ml
[125I]I-UdR was investigated by analyzing the
phosphorylation of H2AX as a marker of DNA double-strand breaks
(Fig. 6A) (30). The level of phospho-H2AX in LUC6
increased from 7.2±0.3 to 14.8±2.2% upon [125I]I-UdR
treatment (P<0.05). Exposure to [125I]I-UdR
significantly increased the percentage of phospho-H2AX in LUC10
from 3.9±0.6 to 11.1±0.8% (P<0.01). Likewise, the increase of
phospho-H2AX from 3.6±0.2 to 11.8±0.8% in LUC13 was significant
(P<0.001). [125I]I-UdR induced phospho-H2AX
activation at varying percentages; the most significant increase in
phospho-H2AX was observed in LUC10 and LUC13 (Fig. 6B).
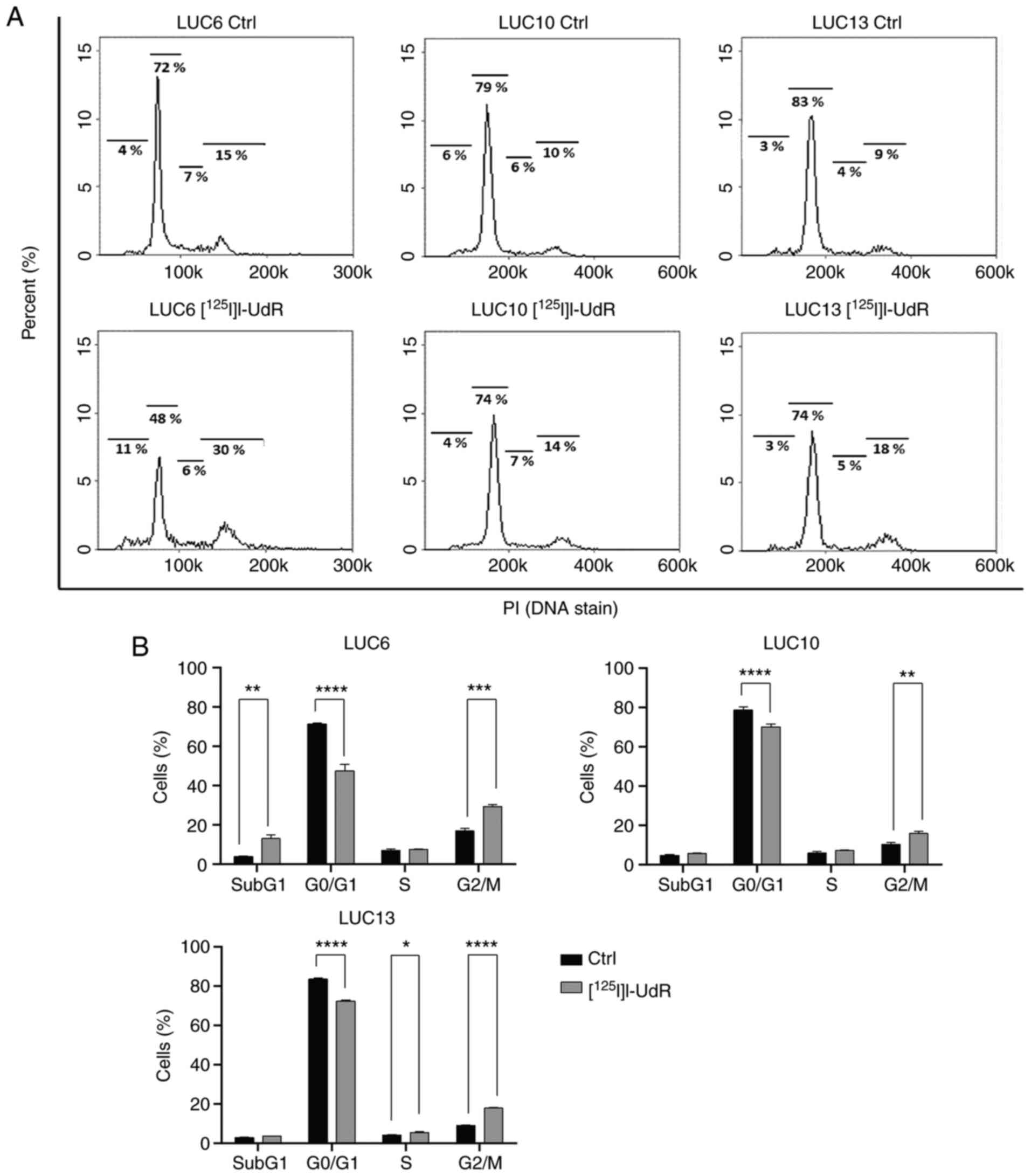
Cell cycle
Auger electrons are sufficiently potent to induce
DNA double-strand breaks and thereby cell-cycle arrest; thus, the
cell cycle distribution of control cells and cells treated with 2.5
kBq/ml [125I]I-UdR for seven days was analyzed (Fig. 7A). [125I]I-UdR
significantly increased the percentage of LUC6 in subG1 phase from
3.9±0.3 to 12.2±1.2% (P<0.01) and that in G2/M phase from
16.9±1.4 to 30.9±1.6% (P<0.001). Furthermore, the percentage of
LUC6 in G0/G1 phase decreased significantly from 71.3±0.6 to
47.5±1.6% (P<0.0001) upon [125I]I-UdR treatment. The
proportion of LUC10 in G0/G1 significantly decreased from 78.6±1.8
to 70.1±1.4% in response to [125I]I-UdR (P<0.0001).
Furthermore, [125I]I-UdR significantly increased LUC10
in G2/M-phase from 10.3±1.1 to 15.8±1.0% (P<0.01), but unlike
for LUC6, there were only minor increases in the proportion of
LUC10 in subG1 phase. The proportion of LUC13 in G0/G1 phase
significantly decreased from 83.6±0.4 to 72.3±0.6% when treated
with [125I]I-UdR (P<0.0001) and G2/M significantly
increased from 9.0±0.2 to 17.8±0.4% (P<0.001) as well as cells
in S (P<0.05; Fig. 7B). Overall,
[125I]I-UdR increased the subG1 and G2/M phase
populations and decreased the percentage of cells in G0/G1
phase.
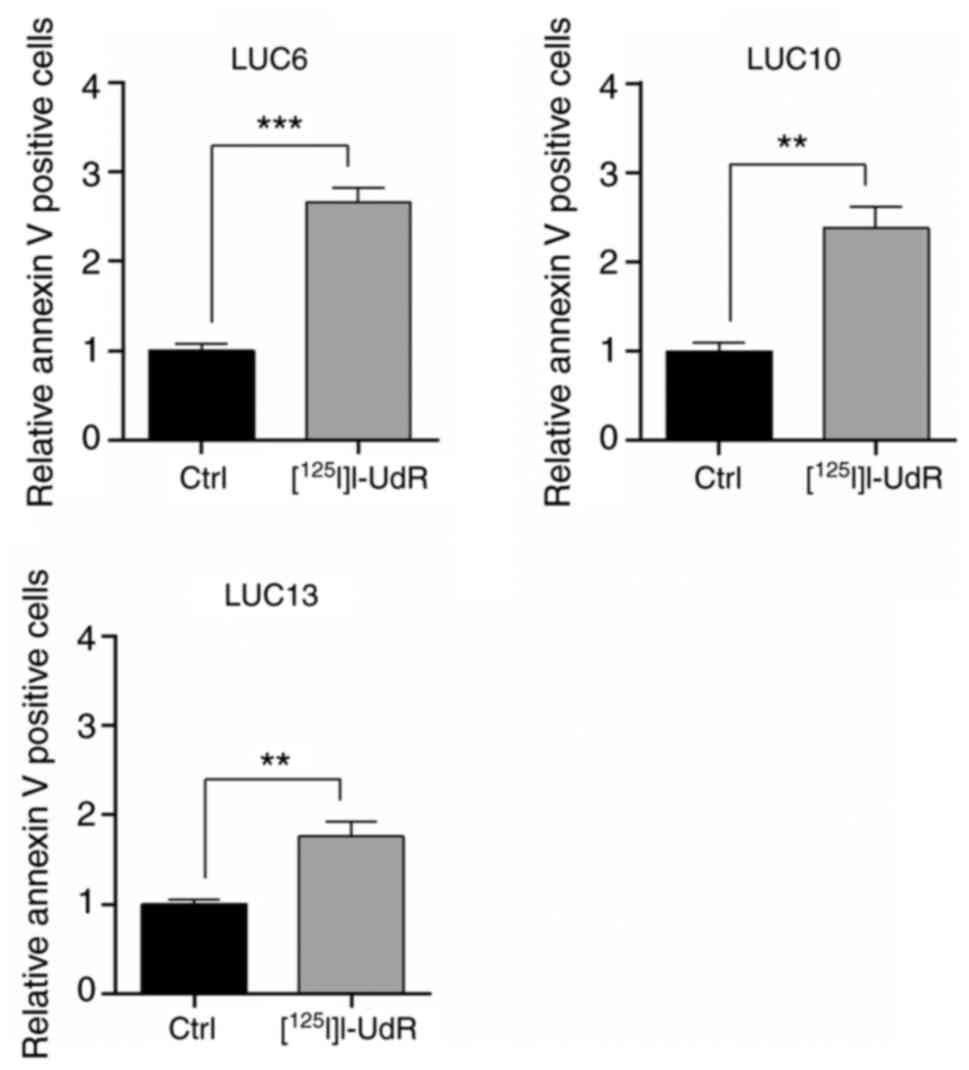
Radiation-induced apoptosis
Radiation-induced DNA damage may lead to apoptosis
if it remains unrepaired (31). In
the present study, Annexin V-positive cells (apoptotic) were
evaluated following treatment with 2.5 kBq/ml
[125I]I-UdR for 7 days (Fig.
8). [125I]I-UdR increased Annexin V-positive LUC6,
LUC10, and LUC13 significantly by 2.65±0.16-(P<0.001),
2.38±0.23-(P<0.01) and 1.85±0.16-(P<0.01) fold, respectively.
Apoptosis was highest in LUC6, corresponding to the results of the
cell cycle analysis, where a significant increase in subG1 was
observed in LUC6.
Discussion
The high recurrence rate of lung cancer is a major
clinical challenge and is associated with therapy-resistant CSCs
(7,8). The vast majority of anti-cancer
treatments are tested preclinically in vitro in adherent
monolayer cells, which may lead to insufficient efficacy in cancer
patients (32). In the present
study, patient-derived NSCLC tumorspheres were established and
characterized and the effects of the Auger electron-emitting
compound [125I]I-UdR, which previously proved effective
against CSCs from glioblastoma and multiple myeloma (17,19,21,26),
were evaluated. The tumorspheres were morphologically distinct and
differed concerning their proliferation rate and doubling time.
However, this was not correlated with their ability to form
tumorspheres. Surface markers associated with CSCs were upregulated
in the tumorspheres compared to those in adherent cells.
Incorporation of [125I]I-UdR led to DNA double-strand
breaks, G2/M arrest and apoptosis, while samples from different
patients exhibited various degrees of sensitivity to Auger electron
irradiation.
Tumorspheres are non-adherent three-dimensional cell
cultures grown in serum-free medium. The technique is based on
self-renewal and anoikis resistance (22,32,33).
It enriches for cells with stemness features, including
self-renewal, unlimited growth abilities, tumorigenic potential
in vivo, ability to differentiate, high invasion capacity
and resistance to high doses of chemotherapy (22,33).
Tumorspheres are also more representative of the parental tumor
than cells grown in two-dimensional systems (22,23).
Furthermore, they are independent of surface markers, which thus
eliminates the use for unique markers whose identification in lung
CSCs remains challenging (34,35).
However, establishing primary stable long-term
tumorspheres may be challenging and the success rate in the present
study was only 20%. This is lower than the 35–40% previously
obtained for lung cancer (22,33),
which may be due to experimental design variables, such as the
tumor stage and genetics (36,37).
Certain cultures were only able to expand short-term, suggesting
that the tumorspheres depended on factors not provided by the
medium (37). Certain tumorspheres
were not sufficiently stable for propagation and may have
represented aggregates rather than true tumorspheres (38).
Overall, it was observed that LUC10 and LUC13
contained more CSCs than LUC6. This was supported by a tumorsphere
formation efficiency assay (i.e., one single cell/well) to
determine the frequency of CSCs in the population, where a higher
frequency in LUC10 and LUC13 was observed. CSCs are considered
slow-proliferating (36,39) and in the present study, it was also
observed that almost all of the LUC6 cells incorporated the
thymidine analog EdU, whereas the incorporation was lower in LUC10
and LUC13. These results were also supported by the analysis of
DTs, where it was determined that LUC6 had the shortest DT, again
supporting that LUC10 and LUC13 contain a higher frequency of CSCs
than LUC6. Furthermore, the RT-qPCR analysis indicated that LUC6
tumorspheres expressed lower levels of CSC-related genes than LUC10
and LUC13 tumorspheres.
When the expression of selected CSC-genes in LUC10
and LUC13 tumorspheres was compared with that in their adherent
counterparts, it was observed that the surface markers CD44 and
THY1 (CD90) and the stemness transcription factors NANOG and SOX2
were upregulated. Unexpectedly, PROM1 (CD133) and POU5F1 (Oct4)
were not significantly upregulated in the tumorspheres. Eramo et
al (33) previously identified
a CD133+ subpopulation in lung cancer that was able to
form tumorspheres and was tumorigenic in mice. By contrast,
Herreros-Pomares et al (22)
were not able to detect any PROM1 transcripts in eight
patient-derived tumorspheres and they did not observe that POU5F1
(Oct4) was significantly upregulated in tumorspheres. Furthermore,
Park et al (40) obtained
differences among NSCLC subtypes, e.g., there was no detectable
protein expression of CD133 and Oct4 in squamous cell
carcinoma.
All three tumorsphere cultures incorporated
[125I]I-UdR; the lowest uptake was, as expected,
observed in the slower proliferating LUC13 spheres. The uptake and
incorporation of thymidine analogs in cancer cells depend on human
nucleoside transporters (hNTs) and thymidine kinase (TK). hNTs are
upregulated in proliferating cells (41) and TK is also upregulated in CSCs
(42). De novo synthesis of
thymidine is catalyzed by thymidylate synthase and inhibition of
the enzyme increased uptake and incorporation of iodinated
thymidine analogs, also in CSCs (17,19).
[125I]I-UdR decreased the viability of
all three tumorsphere samples, with LUC6 being the most sensitive
and LUC10 and LUC13 the most resistant, results which were
supported by the clonogenic assay. The sphere formation assay is
based on the ability of CSCs to divide infinitely, whereas the
viability assay is based on the metabolic capacity of the cells.
However, LUC13 incorporated less [125I]I-UdR within the
7 h incorporation assay and exhibited a longer doubling time, which
may be reflected in the decreased response to Auger emission
compared to LUC10 and LUC6. LUC10, on the other hand, incorporated
the highest amount of [125I]I-UdR and was more resistant
than at least LUC6. However, based on the present results, it is
not possible to conclude whether LUC13 is more resistant than LUC10
or whether the result was due to decreased incorporation.
The tumorspheres exhibited increased phospho-H2AX,
G2/M phase arrest, as well as induction of apoptosis, which was
expected, since the emission of Auger electrons leads to DNA
double-strand breaks and cell cycle arrest through checkpoint
activation, which is essential in the radiation response, as it
provides cells with sufficient time to repair damaged DNA or
undergo apoptosis (15,20). Although it was observed that
[125I]I-UdR decreased viability in all three samples,
with LUC6 being the most sensitive, there was no considerable
difference in apoptosis induction among the three tumorsphere
specimens. Besides apoptosis, DNA damage may also lead to mitotic
catastrophe due to entering mitosis with damaged DNA and is
prevailing in cells lacking functional apoptotic pathways and is
frequently observed in epithelial cells (43).
It has previously been indicated that CSCs are
resistant to external radiation and that external radiation
increases the proportion of CSCs. This may be due to the
eradication of the radiation-sensitive non-stem cancer cells or, as
was indicated by previous studies, the induction of stem cell-like
properties in non-CSCs (44–47).
However, based on the clonogenic assay of the present study,
[125I]I-UdR did not increase the CSC frequency but also
targeted the CSCs, as also reported previously (17,19,21).
Limitations to the present study include that only
three tumorsphere samples were analyzed. Coincidentally, the
tumorspheres included in the present study were of three different
histological types, which challenges a comparison between histology
and response. Future work should include additional tumorspheres
and more ‘common’ histologies, such as adenocarcinoma and
planocellular carcinoma. This would allow for comparison of the
different histologies concerning CSC content and sensitivity to
Auger electrons. The experiments of the present study were
performed in vitro. The next steps should include the
characterization of the tumorspheres in vivo regarding CSC
content, expression of CSC markers, Auger electron therapy and
analyses of the response. In the present study, the response after
seven days was analyzed; this time-point was selected due to the
relatively long half-life of [125I] (60 days). However,
it may be worthwhile to analyze the response after a shorter
duration to understand the early cellular response. It may also be
interesting to elucidate the cell death response, e.g., the
involvement of caspases or mitotic catastrophe, the latter of which
was reported to be involved in the radiation response (43). Furthermore, sorting the cells into
CSCs and non-CSCs prior to Auger electron therapy would allow us to
analyze the response in distinct cell populations and not a mixed
population as was performed in the present study. However, as
described previously, identifying suitable CSC markers remains
challenging (34,35).
In conclusion, patient-derived long-term lung
tumorspheres enriched in CSCs were established and characterized.
The frequency of cells with sphere-forming potential varied, as
LUC10 and LUC13 contained a higher number than LUC6. The
tumorspheres with the highest frequency of CSCs exhibited slower
proliferation. The slower proliferating LUC10 and LUC13 were more
resistant to the Auger-emitting thymidine analog
[125I]I-UdR than LUC6.
Acknowledgements
The authors thank Consultant Karen Ege Olsen,
Department of Pathology, Odense University Hospital (Odense,
Denmark) and Professor Peter Bjørn Licht, Department of Thoracic,
Cardiac and Vascular Surgery, Odense University Hospital (Odense,
Denmark) for recruiting the patients.
Funding
This work was supported by grants from The Independent Research
Fund Denmark, Technology and Production (grant no. 7017-00303),
Director Emil C. Hertz and wife Inger Hertz' Foundation, Frode
Nygaards Foundation, Simon Fougner Hartmanns family Foundation,
Einar Willumsens Memorial Foundation, Odense University Hospital
Research Council, Eva and Henry Fraenkels Memorial Foundation, Aase
and Ejnar Danielsens Foundation, Brdr. Hartmann Foundation, Karen
S. Jensens Foundation and the Hede Nielsen family Foundation.
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
BBO and PFHC contributed to the conception and
design of the study. KLM performed the majority of the experiments.
KLM and BBO checked and confirmed the authenticity of the raw data.
KLM and BBO wrote the manuscript. BBO and PFHC revised the
manuscript. NL, OG, KLM, and BBO analyzed the experimental data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Surgical human lung cancer samples were obtained
with written informed consent. The Regional Ethics Committee of
Southern Denmark approved the protocol (no. S-20140170) according
to guidelines that complied with good clinical practice and the
Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heng WS, Gosens R and Kruyt FAE: Lung
cancer stem cells: Origin, features, maintenance mechanisms and
therapeutic targeting. Biochem Pharmacol. 160:121–133. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen J, Xu J, Wan T, Deng H and Li D:
High-sensitive detection of small-cell lung cancer cells based on
terminal deoxynucleotidyl transferase-mediated extension
polymerization aptamer probe. ACS Biomater Sci Eng. 7:1169–1180.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Leonetti A, Sharma S, Minari R, Perego P,
Giovannetti E and Tiseo M: Resistance mechanisms to osimertinib in
EGFR-mutated non-small cell lung cancer. Br J Cancer. 121:725–737.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
MacDonagh L, Gray SG, Breen E, Cuffe S,
Finn SP, O'Byrne KJ and Barr MP: Lung cancer stem cells: The root
of resistance. Cancer Lett. 372:147–156. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prabavathy D, Swarnalatha Y and Ramadoss
N: Lung cancer stem cells-origin, characteristics and therapy. Stem
Cell Investig. 5:62018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang J, Sun Z, Liu Y, Kong L, Zhou S, Tang
J and Xing HR: Comparison of tumor biology of two distinct cell
sub-populations in lung cancer stem cells. Oncotarget.
8:96852–96864. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
10
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: Prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim CF, Jackson EL, Woolfenden AE,
Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT and Jacks T:
Identification of bronchioalveolar stem cells in normal lung and
lung cancer. Cell. 121:823–835. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Raniszewska A, Kwiecień I, Rutkowska E,
Rzepecki P and Domagala-Kulawik J: Lung cancer stem cells-origin,
diagnostic techniques and perspective for therapies. Cancers
(Basel). 13:29962021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kassis AI: Molecular and cellular
radiobiological effects of Auger emitting radionuclides. Radiat
Prot Dosimetry. 143:241–247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pirovano G, Jannetti SA, Carter LM,
Sadique A, Kossatz S, Guru N, Demétrio De Souza França P, Maeda M,
Zeglis BM, Lewis JS, et al: Targeted brain tumor radiotherapy using
an Auger emitter. Clin Cancer Res. 26:2871–2881. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morgenroth A, Vogg AT, Ermert K,
Zlatopolskiy B and Mottaghy FM: Hedgehog signaling sensitizes
glioma stem cells to endogenous nano-irradiation. Oncotarget.
5:5483–5493. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chan C, Fonge H, Lam K and Reilly RM:
Effectiveness and normal tissue toxicity of Auger electron (AE)
radioimmunotherapy (RIT) with
[111In]In-Bn-DTPA-nimotuzumab in mice with
triple-negative or trastuzumab-resistant human breast cancer
xenografts that overexpress EGFR. Nucl Med Biol. 80-81:37–44. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morgenroth A, Vogg AT, Zlatopolskiy BD,
Siluschek M, Oedekoven C and Mottaghy FM: Breaking the
invulnerability of cancer stem cells: Two-step strategy to kill the
stem-like cell subpopulation of multiple myeloma. Mol Cancer Ther.
13:144–153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Balagurumoorthy P, Xu X, Wang K, Adelstein
SJ and Kassis AI: Effect of distance between decaying (125)I and
DNA on Auger-electron induced double-strand break yield. Int J
Radiat Biol. 88:998–1008. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thisgaard H, Halle B, Aaberg-Jessen C,
Olsen BB, Therkelsen AS, Dam JH, Langkjær N, Munthe S, Någren K,
Høilund-Carlsen PF and Kristensen BW: Highly effective
Auger-electron therapy in an orthotopic glioblastoma xenograft
model using convection-enhanced delivery. Theranostics.
6:2278–2291. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herreros-Pomares A, de-Maya-Girones JD,
Calabuig-Fariñas S, Lucas R, Martínez A, Pardo-Sánchez JM, Alonso
S, Blasco A, Guijarro R, Martorell M, et al: Lung tumorspheres
reveal cancer stem cell-like properties and a score with prognostic
impact in resected non-small-cell lung cancer. Cell Death Dis.
10:6602019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee J, Kotliarova S, Kotliarov Y, Li A, Su
Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al:
Tumor stem cells derived from glioblastomas cultured in bFGF and
EGF more closely mirror the phenotype and genotype of primary
tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xie F, Xiao P, Chen D, Xu L and Zhang B:
miRDeepFinder: A miRNA analysis tool for deep sequencing of plant
small RNAs. Plant Mol Biol. Jan 31–2012.(Epub ahead of print).
View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Madsen KL, Therkelsen ASN, Langkjær N,
Olsen BB and Thisgaard H: Auger electron therapy of glioblastoma
using [125I]5-iodo-2′-deoxyuridine and concomitant
chemotherapy-evaluation of a potential treatment strategy. Nucl Med
Biol. 96-97:35–40. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shigdar S, Lin J, Li Y, Yang CJ, Wei M,
Zhus Y, Liu H and Duan W: Cancer stem cell targeting: The next
generation of cancer therapy and molecular imaging. Ther Deliv.
3:227–244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lawrence TS, Davis MA, Maybaum J, Stetson
PL and Ensminger WD: The effect of single versus double-strand
substitution on halogenated pyrimidine-induced radiosensitization
and DNA strand breakage in human tumor cells. Radiat Res.
123:192–198. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dupertuis YM, Vazquez M, Mach JP, De
Tribolet N, Pichard C, Slosman DO and Buchegger F:
Fluorodeoxyuridine improves imaging of human glioblastoma
xenografts with radiolabeled iododeoxyuridine. Cancer Res.
61:7971–7977. 2001.PubMed/NCBI
|
|
30
|
Rogakou EP, Pilch DR, Orr AH, Ivanova VS
and Bonner WM: DNA double-stranded breaks induce histone H2AX
phosphorylation on serine 139. J Biol Chem. 273:5858–5868. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rothkamm K, Krüger I, Thompson LH and
Löbrich M: Pathways of DNA double-strand break repair during the
mammalian cell cycle. Mol Cell Biol. 23:5706–5715. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lv D, Hu Z, Lu L, Lu H and Xu X:
Three-dimensional cell culture: A powerful tool in tumor research
and drug discovery. Oncol Lett. 14:6999–7010. 2017.PubMed/NCBI
|
|
33
|
Eramo A, Lotti F, Sette G, Pilozzi E,
Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C and De
Maria R: Identification and expansion of the tumorigenic lung
cancer stem cell population. Cell Death Differ. 15:504–514. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qiu X, Wang Z, Li Y, Miao Y, Ren Y and
Luan Y: Characterization of sphere-forming cells with stem-like
properties from the small cell lung cancer cell line H446. Cancer
Lett. 323:161–170. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bertolini G, Roz L, Perego P, Tortoreto M,
Fontanella E, Gatti L, Pratesi G, Fabbri A, Andriani F, Tinelli S,
et al: Highly tumorigenic lung cancer CD133+ cells display
stem-like features and are spared by cisplatin treatment. Proc Natl
Acad Sci USA. 106:16281–16286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pece S, Tosoni D, Confalonieri S, Mazzarol
G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG and Di Fiore
PP: Biological and molecular heterogeneity of breast cancers
correlates with their cancer stem cell content. Cell. 140:62–73.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim SY, Lee JY, Kim DH, Joo HS, Yun MR,
Jung D, Yun J, Heo SG, Ahn BC, Park CW, et al: Patient-derived
cells to guide targeted therapy for advanced lung adenocarcinoma.
Sci Rep. 9:199092019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yeon SE, No da Y, Lee SH, Nam SW, Oh IH,
Lee J and Kuh HJ: Application of concave microwells to pancreatic
tumor spheroids enabling anticancer drug evaluation in a clinically
relevant drug resistance model. PLoS One. 8:e733452013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roesch A, Fukunaga-Kalabis M, Schmidt EC,
Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T
and Herlyn M: A temporarily distinct subpopulation of slow-cycling
melanoma cells is required for continuous tumor growth. Cell.
141:583–594. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Park E, Park SY, Sun PL, Jin Y, Kim JE,
Jheon S, Kim K, Lee CT, Kim H and Chung JH: Prognostic significance
of stem cell-related marker expression and its correlation with
histologic subtypes in lung adenocarcinoma. Oncotarget.
7:42502–42512. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Plotnik DA, Emerick LE, Krohn KA, Unadkat
JD and Schwartz JL: Different modes of transport for 3H-thymidine,
3H-FLT, and 3H-FMAU in proliferating and nonproliferating human
tumor cells. J Nucl Med. 51:1464–1471. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tsunekuni K, Konno M, Haraguchi N, Koseki
J, Asai A, Matsuoka K, Kobunai T, Takechi T, Doki Y, Mori M and
Ishii H: CD44/CD133-positive colorectal cancer stem cells are
sensitive to trifluridine exposure. Sci Rep. 9:148612019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Eriksson D and Stigbrand T:
Radiation-induced cell death mechanisms. Tumour Biol. 31:363–372.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Al-Assar O, Muschel RJ, Mantoni TS,
McKenna WG and Brunner TB: Radiation response of cancer stem-like
cells from established human cell lines after sorting for surface
markers. Int J Radiat Oncol Biol Phys. 75:1216–1225. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ghisolfi L, Keates AC, Hu X, Lee DK and Li
CJ: Ionizing radiation induces stemness in cancer cells. PLoS One.
7:e436282012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lagadec C, Vlashi E, Della Donna L,
Dekmezian C and Pajonk F: Radiation-induced reprogramming of breast
cancer cells. Stem Cells. 30:833–844. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Y, Li W, Patel SS, Cong J, Zhang N,
Sabbatino F, Liu X, Qi Y, Huang P, Lee H, et al: Blocking the
formation of radiation-induced breast cancer stem cells.
Oncotarget. 5:3743–3755. 2014. View Article : Google Scholar : PubMed/NCBI
|