Introduction
Gastric cancer (GC) is characterized by insidious
onset, easy metastasis, and generally poor prognosis; the morbidity
and mortality of GC have been ranked fifth and third in the world,
respectively (1,2). Although intensive interventions
including radical resection and drug therapy are available for GC
patients, the overall 5-year survival rates of GC patients is still
only 25–30% (3). GC has been
identified as one of the most severe malignant tumors that endanger
human health. The fundamental reason for the poor prognosis
includes tumor metastasis, recurrence and drug resistance. It is
well known that the occurrence and development of GC is a
multifactorial, multistage, and multistep process involving the
activation of tumor-promoting factors and the inactivation of
tumor-suppressing factors. GC cells gradually adjust their
biological characteristics in an unfavorable environment through
complex molecular regulation mechanisms to promote tumor
proliferation and metastasis. Therefore, targeted drugs are the key
to effectively cure or improve long-term survival. However, the
effect of existing targeted agents, such as bevacizumab (4), cetuximab (5), and trastuzumab (6), are not completely satisfactory for
GC treatment. Therefore, finding pivotal molecular targets and
developing effective targeted drugs is an effective way to improve
current GC treatments.
The a disintegrin and metalloprotease (ADAM) protein
family, as important multi-domain transmembrane metalloproteinases,
plays important roles in activating Notch, epidermal growth factor
receptor (EGFR) and other signaling pathways which are related to
tumor progression by catalyzing the cleavage of cell surface
proteins (7). ADAM12, an
important member of the ADAM protein family, has been shown to
actively participate in various biological processes such as signal
transduction, cytoskeleton depolymerization, and micro-
environmental regulation by binding and activating targeted
proteins (8). The cellular effect
mediated by ADAM12 may be a key event in multiple biological and
pathological processes. Cumulated evidence indicates that ADAM12
can promote epithelial-mesenchymal transition (EMT) (9), metastasis (10), drug resistance (11) and cancer stemness maintenance
(12) in a variety of malignant
tumors. However, the biological role of ADAM12 in GC has not yet
been elucidated.
In the present study, bioinformatics methods and
experimental analyses were utilized to comprehensively investigate
the expression level, prognostic value, regulatory mechanism and
biological functions of ADAM12 in GC. Collectively, our results
identified ADAM12 as a tumor promoter, thus supporting its use as a
novel prognostic biomarker and therapeutic target in GC.
Materials and methods
Clinical tissue specimens
The tissues from 63 GC patients (mean age, 59.3±11.6
years; range, 35–82 years; 19 women and 44 men; stage I-II, 44
patients; stage III-IV, 19 patients) used for experiments were
obtained from the Affiliated Provincial Hospital of Anhui Medical
University between January 2016 to December 2017. This study was
approved by the Academic Committee of the Affiliated Provincial
Hospital of Anhui Medical University (certification no. 2019KY32)
and all patients provided written informed consent.
Immunohistochemistry (IHC) staining
analysis
Collected tissue specimens for IHC staining were
performed as previously described (13). The final IHC scores were
determined according to immunostaining intensity and positive cell
percentage. In detail, scores from 0 to 7 represented low
expression, and scores from 8 to 12 were regarded as high
expression. The staining procedure and results were independently
evaluated by two pathologists.
Public database analyses
Gene expression Profiling Interactive Analysis
(GEPIA) (14) and the GSE19826
dataset (15) obtained from the
GEO database were used to determine the expression level of ADAM12
in GC. Moreover, the prognostic value of ADAM12 in GC was also
confirmed by GEPIA database (Group cutoff for separating patients
into ADAM12 high and low expression groups was set as median).
To better understand the reasons for ADAM12
overexpression, we firstly used the MethSurv database (16) to evaluate the DNA methylation
status of ADAM12 in GC. Subsequently, starBase3.0 (17,18) was used to predict the targeted
miRNAs of ADAM12 using PITA (19), miRanda (20), and TargetScan (21) analyses. Thereafter, we analyzed
the correlation between ADAM12 expression and targeted miRNAs to
screen for miRNAs that were most suited to competing endogenous RNA
(ceRNA) conditions. In addition, we predicted the targeted long
non-coding RNAs (lncRNAs) of the microRNAs (miRNAs), which were
obtained from the previous analysis using miRNet2.0 (22) and starBase 3.0 (17,18). Sankey graph was constructed to
exhibit the ceRNA network (lncRNA-miRNA-mRNA) of ADAM12 using
d3Network package of R software version 4.0.2 (R Core Team,
www.r-project.org/).
The ADAM12 co-expression profiles were obtained from
the LinkedOmics database and the results were visualized as a
volcano plot and heatmaps (23).
These co-expressed genes of ADAM12 were then used to perform Gene
Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathways enrichment analysis. The GeneMANIA (24) and STRING (25) databases were mined to construct
the protein-protein interaction network of ADAM12 to identify the
potential interaction-partner of ADAM12.
The TIMER database is a user-friendly database that
can be used to comprehensively analyze tumor immune infiltration
(26). In our study, the 'gene
modules' were used to explore the relationship between ADAM12 and
immune cell infiltration levels in GC. In addition, we used the
'somatic copy number alteration module' of the TIMER tool to
determine the correlation between the genetic copy number variation
(CNV) of ADAM12 and the relative abundance of tumor-infiltrating
cells. The 'survival module' was used to detect the association
between clinical outcomes and immune cell infiltration in GC. The
'correlation module' was used to explore correlations between
ADAM12 expression and the expression of immune-infiltration cell
markers with tumor purity adjustment.
Cell culture and transfection
The human gastric mucosal cell line GES1 (control
cell line) and gastric cancer cell lines AGS, MKN28, MKN45, HGC27
and MKN1 were obtained from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China) and cultured in corresponding medium
with fetal bovine serum (FBS) [Biological Industries (BI), Israel].
Short tandem repeat analysis was performed to authenticate the cell
lines before the experiments. The stable GC cells with ADAM12
knockdown or overexpression and the corresponding control cells
were constructed by Tsingke Co., Ltd. and named as sh-NC,
sh-ADAM12, LV-Control or LV-ADAM12, respectively. In brief, the
lentiviral constructs for ADAM12 knockdown (shRNA-ADAM12) and
scramble control (sh-NC) were constructed into pLKO.1 neo (Addgene
#13425). All the plasmids were co-transfected with psPAX2 and
pMD2.G into 293T cells using EndoFectin™ transfection reagent
(GeneCopoeia), per the manufacturer's recommendations. The
supernatant was collected after culturing for 48 h. Subsequently,
the lentivirus supernatant was concentrated and the cells were
transfected with polybrene (8 µg/ml, GeneChem, Inc.). After
a 48-h infection, the transfected cells were cultured in medium
containing G418 (600 ug/ml) to select stable cells. The
transfection efficiency of the cells was verified by western
blotting. In addition, the miRNA mimics and negative control
(miR-NC) were purchased from GenePharma, Inc., and the transfection
procedures were performed using Lipofectamine 3000
(Invitrogen/Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's instructions. All the corresponding sequences are
listed in Table SI.
Western blot analysis
After lysing cells using RIPA buffer, the protein
concentration was determined using the BCA Kit (Beyotime Institute
of Biotechnology). Then, 20 µg protein from the samples was
separated by 10% SDS-PAGE gel followed by transferring to
0.45-µm PVDF membranes (Millipore, USA). Subsequently, the
PVDF membranes were incubated in blocking buffer (5% non-fat milk)
for 1 h at room temperature and then incubated with the primary
antibody overnight at 4°C, and the corresponding secondary antibody
was hybridized for 2 h at room temperature. The blotted proteins
was visualized with the help of the ECL detection system (Pierce
Biotech, USA), scanned with a Chemi-Doc System (Bio-Rad
Laboratories, Inc.) and analyzed using ImageJ software (https://imagej.net). Information regarding the primary
antibodies is listed in Table
SII.
Reverse transcription-quantitative PCR
(RT-qPCR)
The RNAeasy™ animal RNA extraction kit (cat. no.
R0026, Beyotime Institute of Biotechnology) was used to extract
total intracellular RNA. mRNA and miRNA were reverse transcribed
into cDNA using a mRNA reverse transcription kit (Thermo Fisher
Scientific, Inc.) and miRNA reverse transcription kit (Vazyme
Biotech Co., Ltd.). SYBR-Green PCR Master Mix (Thermo Fisher
Scientific, Inc.) was used for RT-qPCR. Levels of miRNA were
normalized using small nuclear RNA U6, whereas ADAM12 levels were
normalized by GAPDH. The 2-ΔΔCq method was used to
calculate the difference in gene expression (27). The primers were synthesized by
Tsingke Co., Ltd. The specific sequences of the primers are shown
in Table SIII.
Molecular docking analysis
The structure of ADAM12 and FSTL3 were obtained from
AlphaFold prediction (https://alphafold.ebi.ac.uk/). Docking study of the
binding modes between ADAM12 and FSTL3 was conducted by using HDOCK
server (28–33). According to the docking score
provided by the HDOCK server, the best predicted binding mode was
selected to analyze the detailed interaction network between these
two proteins. The best predicted binding mode was visualized,
analyzed and mapped by using the PyMOL program version 2.4.0
(https://www.schrodinger.com/pymol).
Wound healing assay
The indicated HGC27 and AGS cells were seeded at a
density of 5×105 cells/well into 6-well plates and
cultured to achieve over 80% confluence. Subsequently, we created
scratches by using a 200-µl pipette tip to scrape
longitudinally in the center of the bottom of the well. After
washing twice with PBS, serum-free medium was added to the wells.
Images were captured with an optical microscope (magnification,
x40; Olympus, Japan) at 0 and 48 h after the scratch appeared.
Migration distance was calculated as: Migration distance=scratch
width observed at 0 h-scratch width observed at 48 h).
Transwell assay
The Transwell chamber was purchased from BD
Biosciences, USA. Approximately 5×104 indicated HGC27
and AGS cells were seeded in the upper chamber and 250 µl
serum-free medium was added, while the lower chamber was filled
with 750 µl complete medium. After 24 h of incubation, the
upper chamber was taken out and fixed with methanol for 10 min, and
then stained with 0.1% crystal violet at room temperature. The
number of transmembrane cells were counted under a microscope
(magnification, ×100; Olympus, Japan).
Statistical methods
Statistical analyses were performed using SPSS 21.0
software (IBM Corp.). The Student's t-test was used to analyze
differences between two variables, and differences among multiple
groups were analyzed using one-way ANOVA followed by Tukey's post
hoc test. Survival curves were constructed using the Kaplan-Meier
methods, and the P-value was obtained from the log-rank test.
Univariate and multivariate Cox regression analyses were performed
to evaluate risk factors for overall survival. A P-value of
<0.05 was considered to indicate a statistically significant
difference.
Results
ADAM12 expression and its prognostic
value in GC
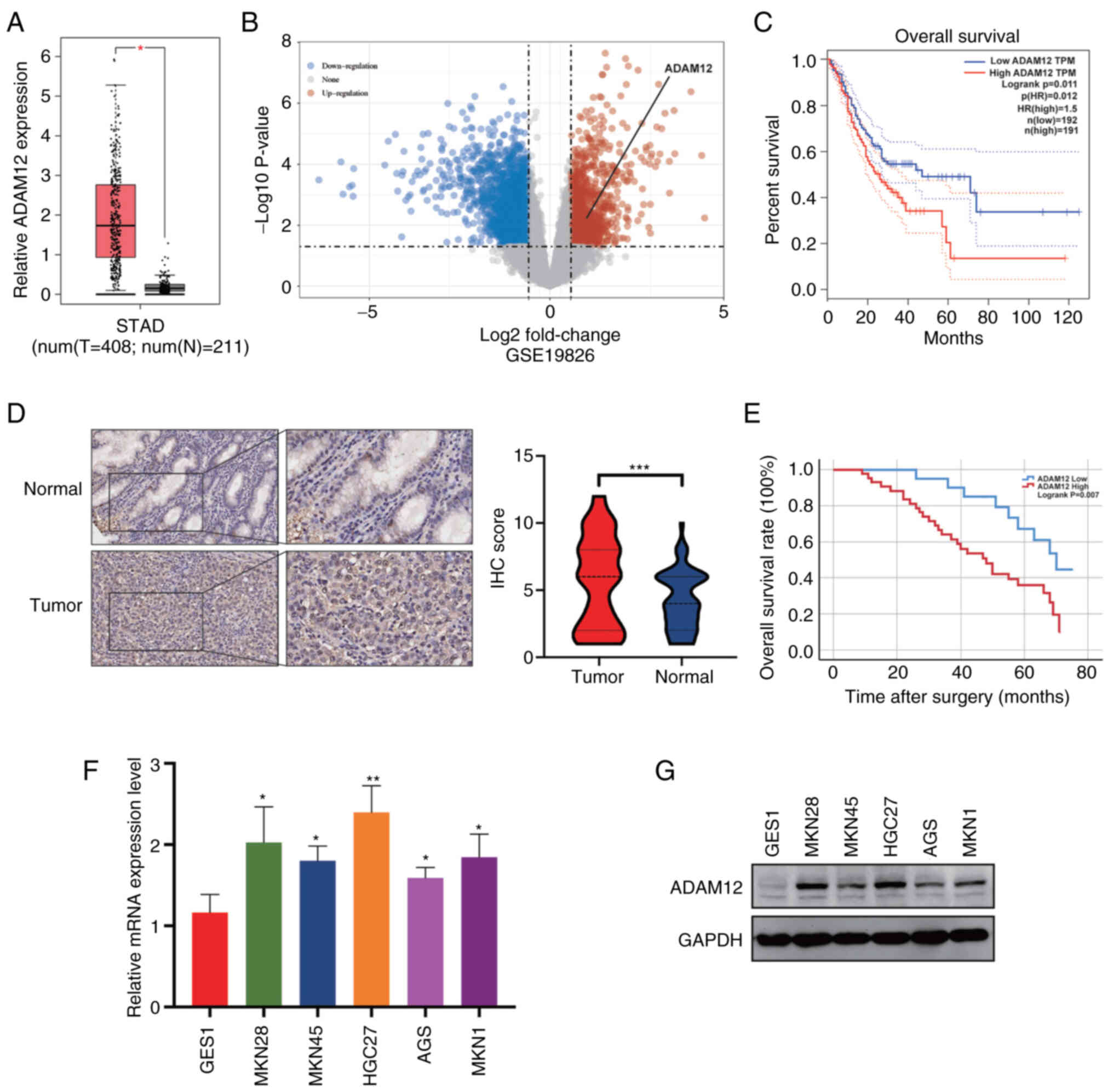
To identify the role of ADAM12 in GC, we performed
analysis to determine the expression level of ADAM12 in GC using
the GEPIA and GSE19826 dataset. As shown in Fig. 1A and B, the expression of ADAM12
was remarkedly increased in tumor tissues than that in normal
tissues. Moreover, the elevated expression of ADAM12 was strongly
associated with a poor prognosis in GC patients (Fig. 1C). To further determine the
expression level and prognostic value of ADAM12 in GC, IHC staining
was performed on a tissue microarray containing 63 pairs of GC
tissue and peri-tumor tissue (Fig.
S1). In addition, higher protein levels of ADAM12 were observed
in tumor tissues in comparison to peri-tumor tissues (Fig. 1D).
We then analyzed the relationship between ADAM12
expression and clinicopathological factors. There was no
significant difference in sex, age, BMI, alcohol consumption,
venous invasion, operation time, TNM stage and postoperative
complications (P>0.05) between the ADAM12 high expression group
and ADAM12 low expression group (Table I). The Kaplan-Meier survival curve
demonstrated that the overall survival (OS) of patients with
elevated levels of ADAM12 expression was significantly shorter than
that of patients with low levels of ADAM12 expression (Fig. 1E). In regards to the disease-free
survival (DFS), the GC patients with high ADAM12 expression showed
a clear trend of adverse prognosis, although the difference was
insufficient to reach statistical significance (Fig. S2). Moreover, our results
demonstrated that ADAM12 expression level is an independent risk
factor for OS based on the results of the multivariate analysis
(Table II). The results from
RT-qPCR and western blotting showed that ADAM12 was significantly
overexpressed in five GC cell lines (MKN28, MKN45, HGC27, AGS and
MKN1) compared to the GES1 cell line. (Fig. 1F and G).
 | Table IAssociations between ADAM12 protein
expression (immunohistochemical staining) in gastric cancer and
various clinicopathological variables. |
Table I
Associations between ADAM12 protein
expression (immunohistochemical staining) in gastric cancer and
various clinicopathological variables.
| Variables | Total | ADAM12 expression
|
|---|
| Low (n=20) | High (n=43) | χ2 | P-value |
|---|
| Sex | | | | | |
| Female | 19 | 7 | 12 | 0.326 | 0.568 |
| Male | 44 | 13 | 31 | | |
| Age (years) | | | | | |
| ≤60 | 25 | 8 | 17 | 0.001 | 0.972 |
| >60 | 38 | 12 | 26 | | |
| BMI
(kg/m2) | | | | | |
| Normal | 34 | 9 | 25 | 0.949 | 0.330 |
| Abnormal | 29 | 11 | 18 | | |
| Alcohol
consumption | | | | | |
| No | 37 | 14 | 23 | 1.535 | 0.215 |
| Yes | 26 | 6 | 20 | | |
| Venous
invasion | | | | | |
| No | 33 | 10 | 23 | 0.067 | 0.796 |
| Yes | 30 | 10 | 20 | | |
| Operation time | | | | | |
| ≤3 h | 33 | 13 | 20 | 1.871 | 0.171 |
| >3 | 30 | 7 | 23 | | |
| TNM stage | | | | | |
| I–II | 44 | 16 | 28 | 1.436 | 0.231 |
| III–IV | 19 | 4 | 15 | | |
| Postoperative
complications | | | | | |
| No | 33 | 14 | 19 | 3.647 | 0.056 |
| Yes | 30 | 6 | 24 | | |
 | Table IIUnivariate and multivariate analysis
of the correlation between clinicopathological parameters and
overall survival of the patients with gastric cancer. |
Table II
Univariate and multivariate analysis
of the correlation between clinicopathological parameters and
overall survival of the patients with gastric cancer.
| Variables | Univariate analysis
| Multivariate
analysis
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Sex | 1.440 | 0.735-2.825 | 0.288 | | | |
| Age (years) | 1.826 | 0.836-3.989 | 0.131 | | | |
| BMI
(kg/m2) | 1.112 | 0.582-2.123 | 0.748 | | | |
| Consumption of
alcohol | 0.864 | 0.447-1.672 | 0.665 | | | |
| Operation time | 0.985 | 0.516-1.879 | 0.962 | | | |
| Venous
invasion | 3.509 | 1.705-7.221 | 0.001 | 3.877 | 1.853-8.112 | <0.001 |
| TNM stage | 2.639 | 1.375-5.063 | 0.004 | 2.441 | 1.252-4.757 | 0.009 |
| Postoperative
complications | 2.287 | 1.187-4.406 | 0.013 | 2.374 | 1.187-4.749 | 0.015 |
| ADAM12
expression | 2.720 | 1.270-5.827 | 0.010 | 2.315 | 1.076-4.984 | 0.032 |
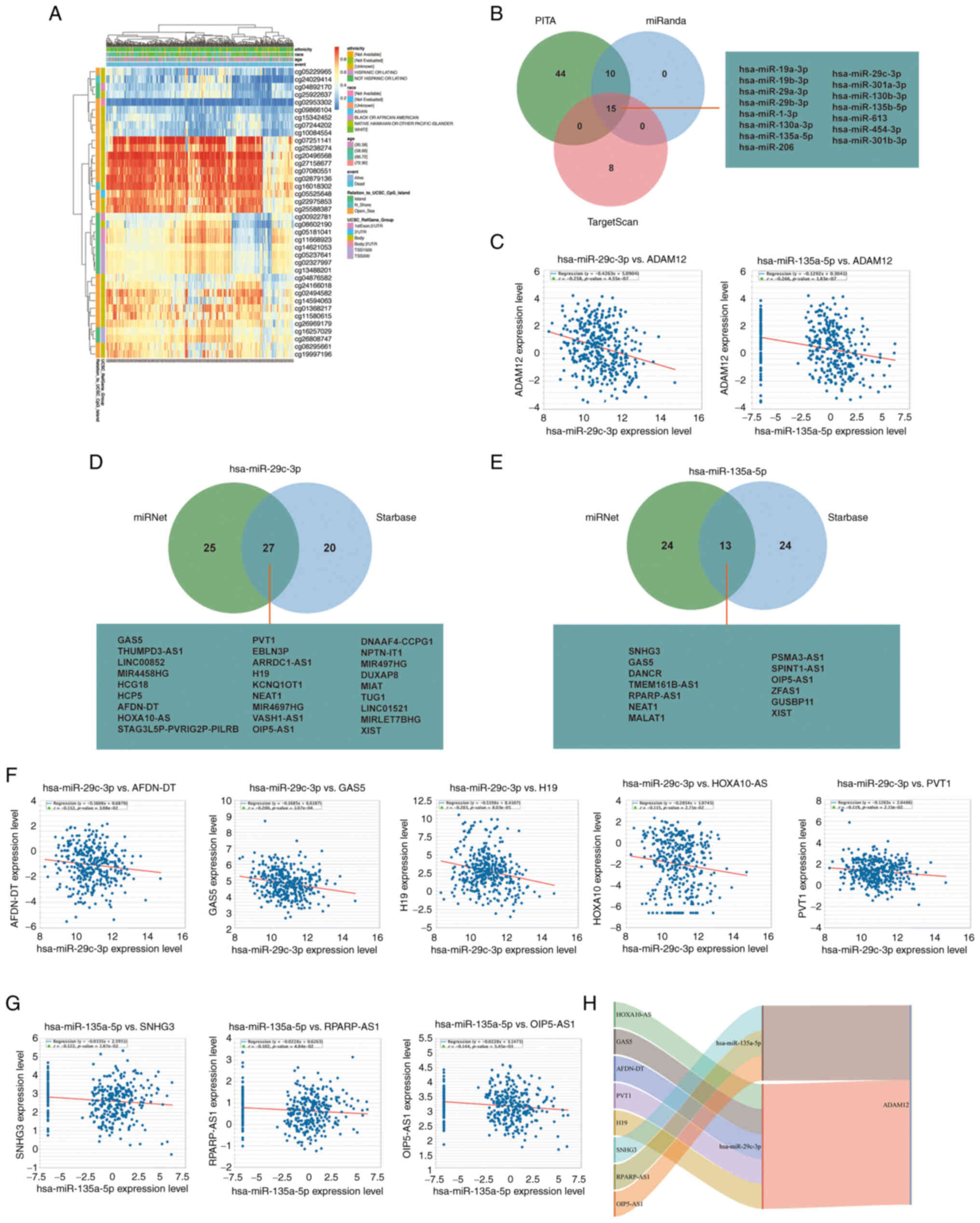
ADAM12 DNA methylation status and ceRNA
regulatory network in GC
Aberrant DNA methylation level and pattern resulted
in abnormal activation of proto-oncogenes (34). Therefore, we attempted to
investigate the DNA methylation status of ADAM12 in GC. As shown in
Fig. 2A, we determined that
cg05229965, cg24029414, cg04892170, cg25922637, cg02953302,
cg09866104, cg15342452, cg07244202, and cg10084554 sites exist with
lower levels of DNA methylation based on the data obtained from
TCGA-STAD. More importantly, cg04892170 and cg02953302 are located
in the region of the 1st exon and 5'UTR. Hypomethylation in these
regions may reveal the cause of the abnormally elevated ADAM12
expression.
Emerging evidence highlights that aberrant ceRNA
networks are important tumor promoters in human cancers. Therefore,
we attempted to construct the ceRNA network of ADAM12 in GC. Based
on Starbase3.0 database analysis, a series of target miRNAs of
ADAM12 were predicted. As shown in Fig. 2B, a total of 15 target miRNAs were
jointly predicted by PITA, miRanda, and TargetScan databases. Among
them, the expression of hsa-miR-29c-3p (R=-0.258, P<0.0001) and
hsa-miR-135a-5p (R=−0.266, P<0.0001) were negatively correlated
with the expression level of ADAM12 (Fig. 2C). To clarify the relationship of
hsa-miR-29c-3p and hsa-miR-135a-5p with ADAM12, the gain of
hsa-miR-29c-3p and hsa-miR-135a-5p were performed in HGC27 cells.
RT-qPCR experiment confirmed the successful transfection efficiency
of hsa-miR-29c-3p and hsa-miR-135a-5p mimics (Fig. S3A). The western blotting
experiments demonstrated that hsa-miR-29c-3p and hsa-miR-135a-5p
can indeed inhibit the expression of ADAM12 in HGC27 cells
(Fig. S3B).
Subsequently, we used the miRNet and starBase online
databases to further predict the lncRNAs that could bind to the two
target miRNAs (hsa-miR-29c-3p and hsa-miR-135a-5p) (Fig. 2D and E). The ceRNA network
hypothesis indicated that there was a negative correlation between
lncRNAs and miRNAs. As shown in Fig.
2F and G, the expression levels of the 5 lncRNAs (AFDN-DT,
GAS5, H19, HOXA10-AS and PVT1) were negatively correlated with
hsa-miR-29c-3p, while 3 lncRNAs (SNHG3, RPARP-AS1 and OIP5-AS1)
were negatively correlated with the expression level of
hsa-miR-135a-5p. Therefore, we could construct 8 pairs of ceRNA
networks based on the results of the correlation analysis (Fig. 2H).
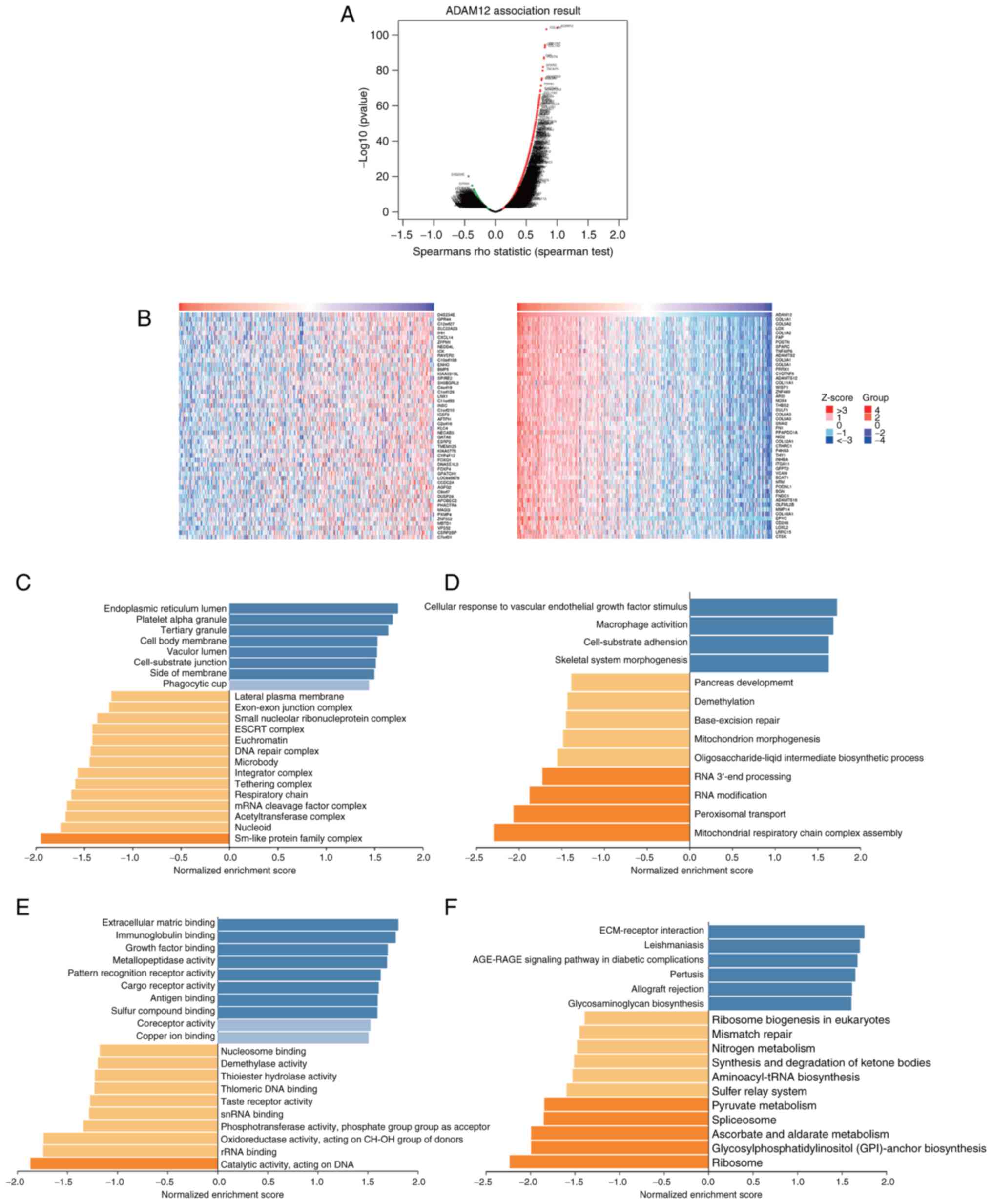
Enrichment analysis of the ADAM12 gene
co-expression profiles in GC
To better understand the biological function of
ADAM12 in GC, LinkedOmics was applied to perform a co-expression
gene profile analysis. As shown in Fig. 3A, a total of 9,307 genes were
found to be significantly positively correlated with ADAM12 (red
dots), while 10,919 genes showed significantly negative
correlations (green dots). The top 50 genes that were significantly
positively or negatively correlated with ADAM12 are shown on a heat
map (Fig. 3B).
Next, enrichment analysis was performed based on GO
functions and KEGG pathways using GSEA methods. The main biological
process was identified as 'endoplasmic reticulum lumen', 'platelet
alpha granule', while the most enriched molecular function was
'cellular response to vascular endothelial growth factor stimulus'
and 'macrophage activation' (Fig. 3C
and D). The top two cellular component terms were
'extracellular matrix binding' and 'immunoglobulin binding'
(Fig. 3E). KEGG pathway analysis
revealed that the most enriched pathways were 'ECM-receptor
interaction' and 'Leishmaniasis' (Fig. 3F).
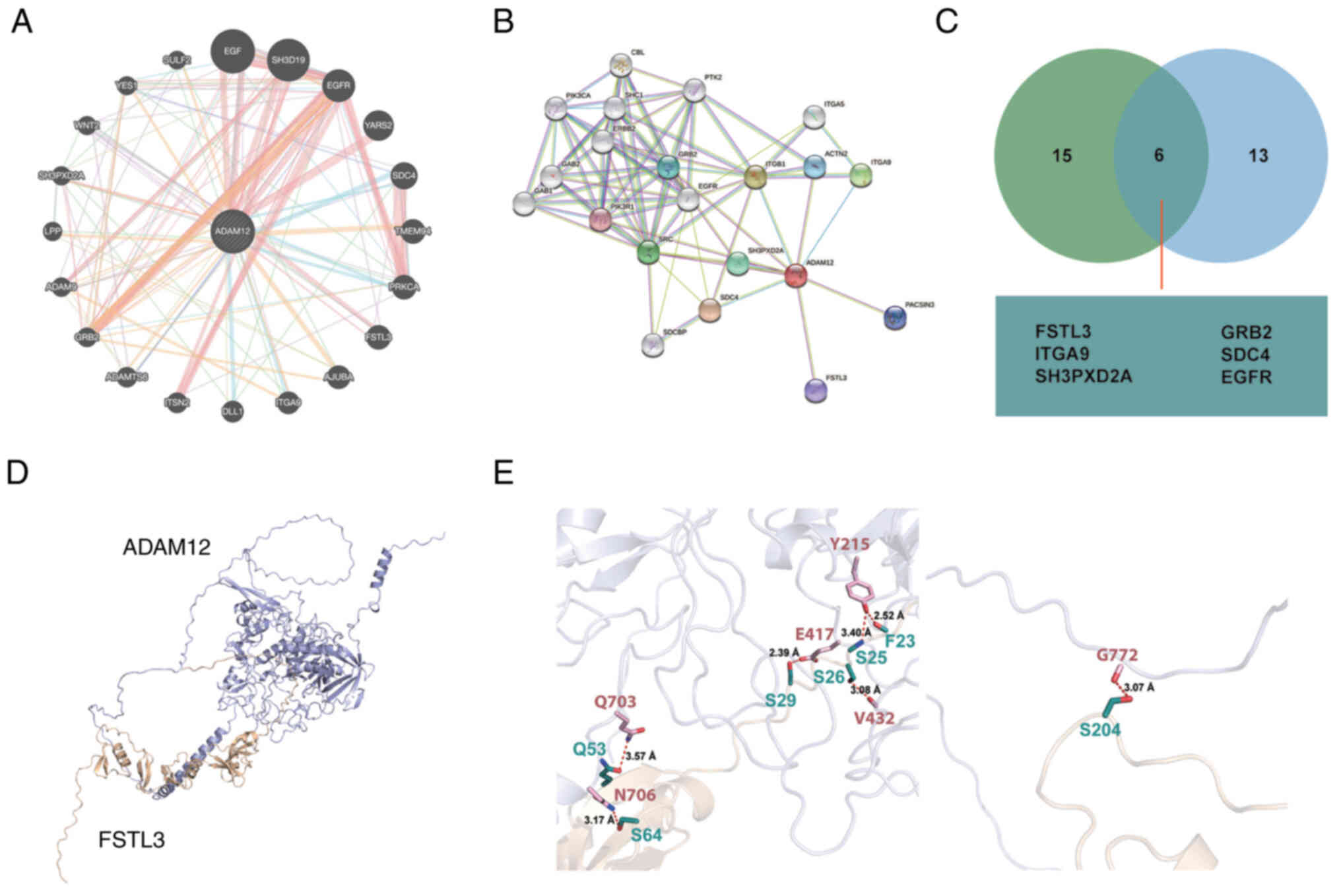
The protein-protein interaction network
of ADAM12
The interactions between proteins is the foundation
of many biological processes (35). To identify the potential binding
partner of ADAM12, the GeneMANIA and STRING data- bases were used
to construct the protein-protein interaction network of ADAM12
(Fig. 4A and B). Proteins,
including FSTL3, GRB2, ITGA9, SDC4, SH3PXD2A and EGFR, were
identified as potential common binding partners of ADAM12 through
the combined results obtained from the GeneMANIA and STRING
databases (Fig. 4C).
Previous studies suggest the interaction of ADAM12
with certain proteins, such as GRB2 (36), SH3PXD2A (37,38) and EGFR (39,40). However, the relationship between
ADAM12 and follistatin like 3 (FSTL3) has not been reported until
now. Due to the similarity in cell location and function of ADAM12
and FSTL3, we performed docking experiment to investigate the
binding mode between ADAM12 and FSTL3. As shown in Fig. 4D, the F23 and S29 of FSTL3 were
able to form hydro-bonding interactions with Y215 and E417 of
ADAM12, respectively. Moreover, the distances between the acceptor
and donor heavy atom of the two hydrogen bonds were both less than
3 Å, which indicates that these two hydrogen bonds are quite stable
(Fig. 4E, left panel). Moreover,
the Y215 and V432 of ADAM12 were also able to form hydrogen-bonding
interactions with S25 and S26 of FSTL3, respectively. Each of the
two residues, Q53 and S64 of FSTL3, were able to form
hydrogen-bonding interaction with the transmembrane helical region
of ADAM12. A hydrogen-bonding interaction was also found between
the S204 of FSTL3 and G772 of ADAM12 (Fig. 4E, right panel).
Key role of ADAM12 in GC cell migration,
invasion and EMT-like phenotype
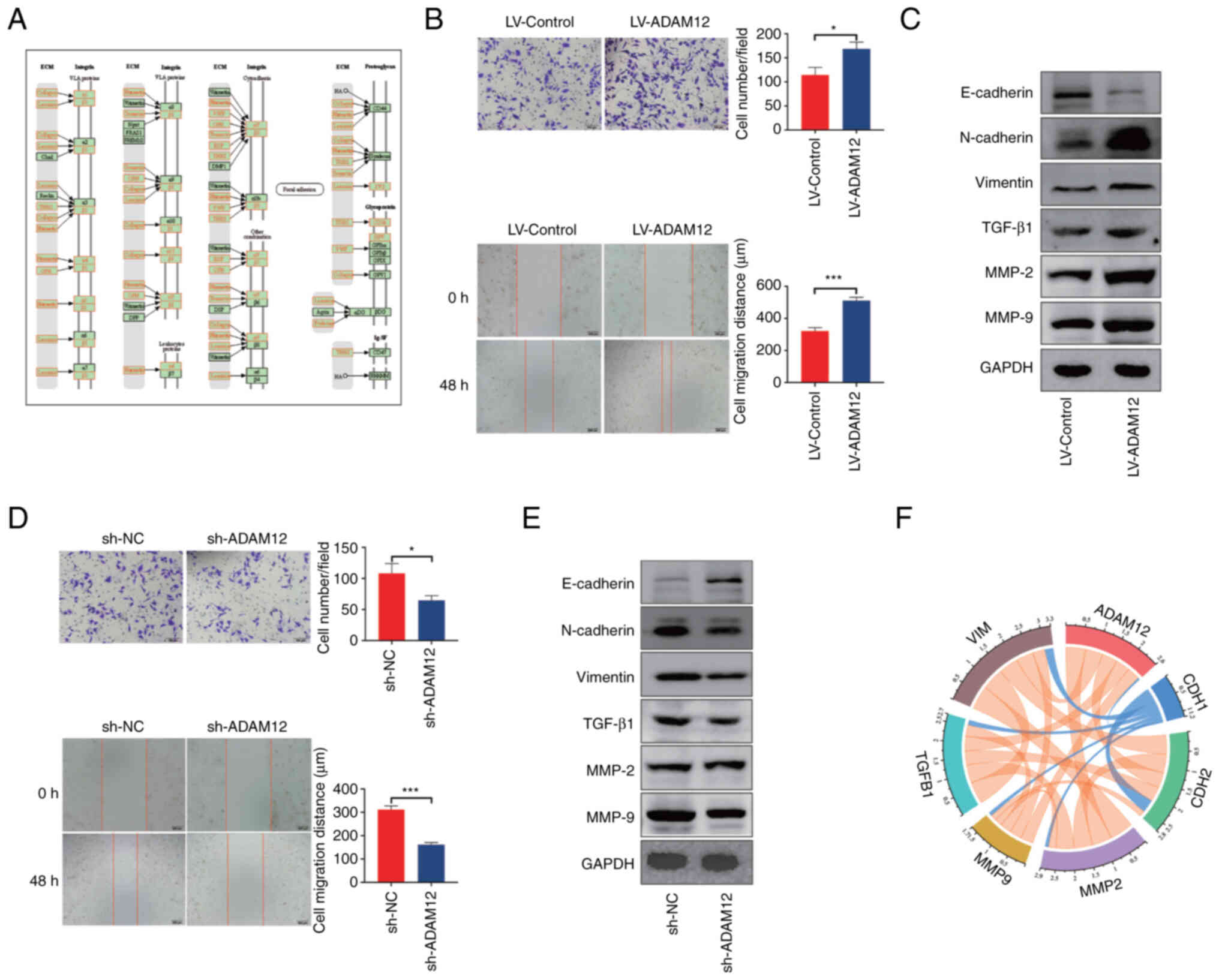
According to the results of our enrichment analysis,
ADAM12 may participate in the 'ECM-receptor interaction' signaling
pathway. As shown in Fig. 5A,
most of the molecules in this signaling pathway have a significant
relationship with the EMT phenotype. The analysis of the
interaction network indicated that ADAM12 may interact with FSTL3
to potentially promote the metastasis and EMT phenotype of tumors
(41,42). Therefore, we constructed stable
ADAM12 overexpression AGS cells and knockdown HGC27 cells (Fig. S4). The in vitro assay
demonstrated that ADAM12 overexpression significantly promoted the
migration and invasion abilities of AGS cells (Fig. 5B), while ADAM12 knockdown
significantly reduced the migration and invasion ability of HGC27
cells (Fig. 5D). In addition,
ADAM12 overexpression significantly downregulated the expression of
E-cadherin and upregulated the expression of TGF-β1, matrix
metalloproteinase (MMP)-2, MMP-9, N-cadherin and vimentin in AGS
cells (Fig. 5C). In contrast,
knockdown of ADAM12 markedly downregulated TGF-β1, MMP-2, MMP-9,
N-cadherin, and vimentin expression but promoted the expression of
E-cadherin protein in HGC27 cells (Fig. 5E).
In addition, the correlation analysis conducted
using the TIMER database demonstrated that ADAM12 expression was
associated with CDH2 (R=0.492, P=1.58e-24), MMP-2 (R= 0.628,
P=5.58e-43), MMP-9 (R= 0.407, P=1.59e-16), TGF-β1 (R=0.450,
P=2.83e-20), and vimentin expression (R=0.554, P=7.21e-32) but
negatively correlated with CDH1 expression (R=-0.061, P=2.23e-01),
which is consistent with the result of the in vitro
experiments (Fig. 5F). Based on
the above results, it was suggested that ADAM12 may regulate the
ability of metastasis and EMT in GC cells in vitro.
Correlation between ADAM12 and tumor
immune infiltration
Crosstalk between tumor cells and tumor-infiltrating
immune cells is extremely important for cancer development and
affects treatment outcomes (43).
Our results indicated that ADAM12 is involved in interactions
between cells and the extracellular matrix, as well as between
tumor cells. Therefore, we aimed to explore the role of ADAM12 in
the GC tumor immune environment. Based on the results of the TIMER
database analysis, we observed a positive correlation between
ADAM12 expression and infiltrating levels of CD8+ T
cells (R=0.116, P=2.54e-02), macrophages (R=0.29, P=1.28e-08),
neutrophils (R=0.359, P=9.44e-13), and dendritic cells (R=0.351,
P=3.57e-12) in GC (Fig. 6A). We
also analyzed the effect of ADAM12 copy number alternation (CNA) on
the infiltration level of six immune cells. As shown in Fig. 6B, the deep deletion, arm-level
deletion and arm-level gain of ADAM12 significantly affected the
level of infiltration level of B cells, CD8+ T cells,
CD4+ T cells, neutrophils, macrophages, and dendritic
cells in GC. In addition, macrophages, neutrophils, dendritic
cells, and ADAM12 were identified as factors involved with the
cumulative overall survival rate of GC patients (Fig. 6C).
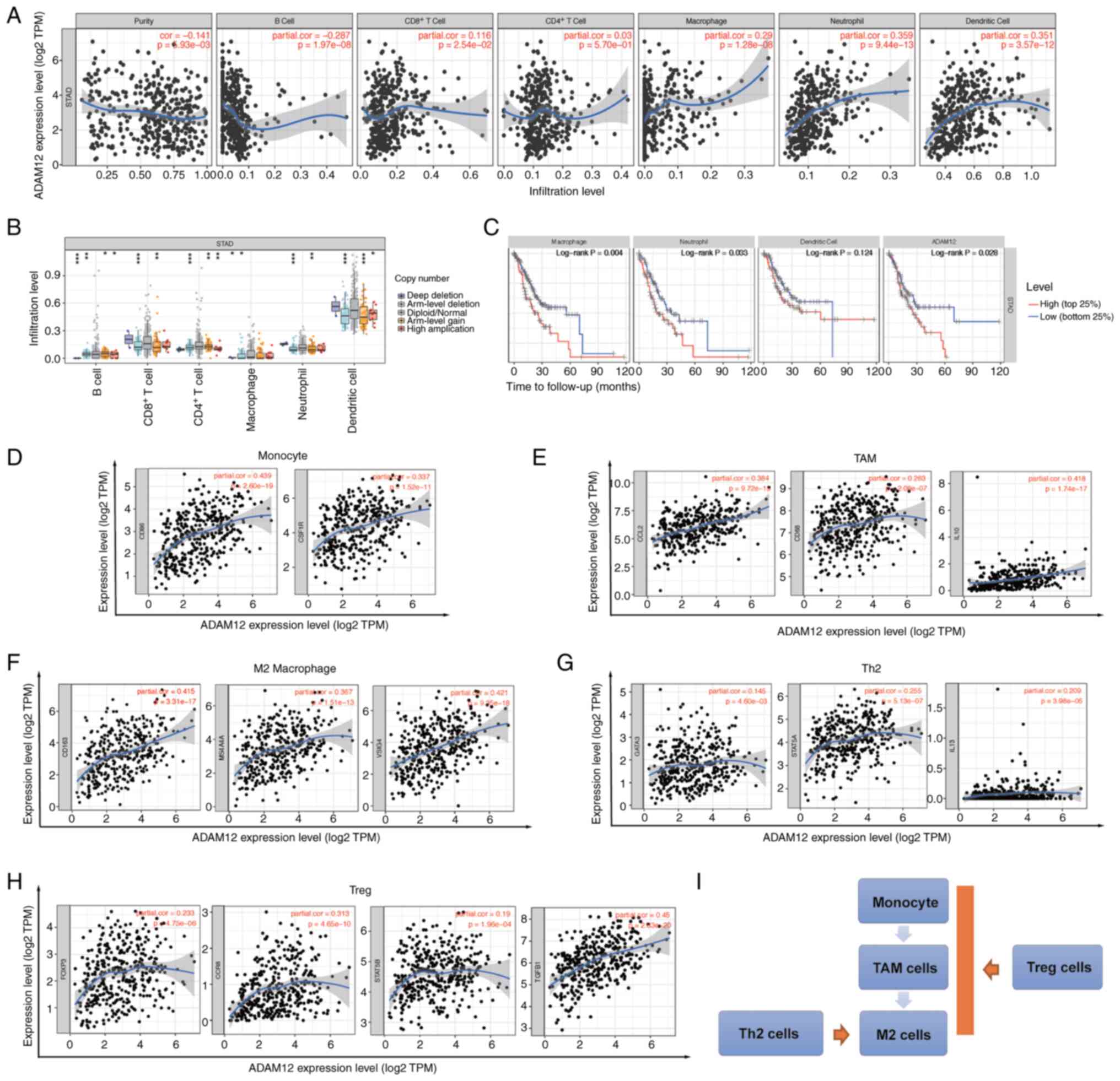 | Figure 6Correlation between ADAM12 expression
and immune cell infiltration in GC. (A) Correlation of ADAM12
expression with tumor purity and infiltrating levels of B cells,
CD8+ T cells, CD4+ T cells, macrophages,
neutrophils and dendritic cells in GC. (B) Effect of ADAM12 copy
number variation (CNV) on the immune infiltration level of B cells,
CD8+ T cells, CD4+ T cells, macrophages,
neutrophils and dendritic cells in GC. (C) The survival plot of
immune cell infiltration and ADAM12 expression on GC patients. (D)
Correlation between ADAM12 expression and markers of monocyte
cells. (E) Correlation between ADAM12 expression and markers of
tumor-associated macrophage (TAM) cells. (F) Correlation between
ADAM12 expression and markers of M2 cells. (G) Correlation between
ADAM12 expression and markers of Th2 macrophage cells. (H)
Correlation between ADAM12 expression and markers of Treg cells.
(I) The schematic diagram for the role of ADAM12 in immune
infiltration cells. ADAM12, a disintegrin and metalloprotease 12;
GC, gastric cancer. |
T cell and macrophage infiltration is strongly
associated with the clinical outcome of patients with malignant
tumors. Therefore, we decided to explore the correlation between
ADAM12 expression and the differentiation status of macrophages and
T cell subpopulation. As shown in Fig. 6D-F, ADAM12 was significantly
positively correlated with the cell markers CD86 (R=0.439,
P=2.60e-19) and CSF1R (R=0.337, P=1.52e-11) in monocytes, and the
cell markers CCL2 (R=0.384, P=9.72e-15), CD68 (R=0.263,
P=2.08e-07), IL10 (R=0.418, P=1.74e-17) in tumor-associated
macrophages (TAMs), as well as the M2 macrophage cell markers CD163
(R=0.415, P=3.31e-17), MS4A4A (R=0.367, P=1.51e-13), and VSIG4
(R=0.421, P=9.35e-18). It is well known that macrophages are
polarized into an M2 subpopulation upon Th2 and Treg cell
stimulation. Intriguingly, ADAM12 expression was significantly
positively associated with the expression of Th2 cell markers,
GATA3 (R=0.145, P=4.60e-03), STAT5A (R=0.255, P=5.13e-07), and IL13
(R=0.209, P=3.98e-05), and the Treg cell markers FOXP3 (R=0.233,
P=4.75e-06), CCR8 (R=0.313, P=4.65e-10), STAT5B (R=0.19,
P=1.96e-04), and TGF- β1 (R=0.45, P=2.63e-20) (Fig. 6G and H). Therefore, we speculated
that ADAM12 may further promote the polarization of M2 macrophages
by regulating the maturation of monocytes, TAMs, Th2 and Treg cells
in the tumor microenvironment (Fig.
6I).
Discussion
Cumulated evidence has demonstrated important roles
for ADAM family proteins in tumor formation and progression
(44). ADAM12, an important
member of the ADAM family, promotes the proliferation and
metastasis of tumor cells by participating in biological processes
such as enzyme catalysis, cell-cell binding and cell signal
transduction (8). Although
previous studies have shown that ADAM12 is highly expressed in GC
(45), the clinical value and
biological function of ADAM12 in GC have not yet been fully
elucidated.
In the present study, it was demonstrated that the
expression level of ADAM12 was elevated in GC. Moreover, GC
patients with high ADAM12 expression demonstrated a poorer
prognosis. To determine the potential causes of ADAM12 elevation,
the DNA methylation status and ceRNA regulatory network of ADAM12
were investigated, respectively. Intriguingly, two hypomethylation
sites were identified in the ADAM12 promoter region, indicating a
potential cause of ADAM12 overexpression. In addition, we
constructed eight vital ceRNA networks of ADAM12 using
bioinformatics analysis. Although previous studies have shown that
ADAM12 may be regulated by a ceRNA network (9,46),
our newly discovered ceRNA networks need further experimental
verification.
To gain further insights into the biological
functions of ADAM12 in GC, GO and KEGG enrichment analyses of
ADAM12 co-expressed genes in GC were conducted. We found that they
were mainly involved in 'macrophage activation', 'extracellular
matrix binding', 'immunoglobulin binding' and 'ECM-receptor
interaction', indicating the potential role of ADAM12 in tumor
metastasis and tumor immune infiltration. Interestingly, the
protein-protein interaction network revealed that follistatin like
3 (FSTL3) may be a potential partner of ADAM12. Previous studies
have emphasized that FSTL3 can enhance tumor cell metastasis
(41,42) and the polarization of macrophages
and fibroblasts by forming an inhibitory immune microenvironment
(47). Therefore, it is clear
that ADAM12 and FSTL3 perform similar functions, which may imply a
mutual adjustment relationship between them. The subsequent
molecular docking model was used to predict potential binding sites
between them to further determine their interactions.
Next, we attempted to determine whether ADAM12 plays
a crucial role in GC metastasis and tumor immune invasion. A stable
overexpression cell line, LV-ADAM12-AGS, and a knockdown cell line,
sh-ADAM12-HGC27 were constructed. As expected, wound healing and
Transwell experiments confirmed that ADAM12 could enhance the
migration and metastatic abilities of GC cells. In addition, the
expression of epithelial-mesenchymal transition (EMT)-related
markers were assessed and this demonstrated that ADAM12 plays an
important role in regulating EMT. This is similar to the results of
a previous study conducted on pituitary tumors (48). Therefore, we speculated that
ADAM12 may promote the migration and metastasis of GC cells by
inducing EMT.
The role of ADAM12 in GC immune infiltration was
another focus of our investigation. In the present study, the
expression level of ADAM12 was significantly correlated with
multiple immune cell infiltration in GC. Our results based on data
mining indicated that ADAM12 expression was significantly
positively correlated with M2 macrophages. In addition, a positive
correlation was demonstrated between ADAM12 and monocytes, tumor
associated macrophages (TAMs), Th2 and Treg cells by analyzing the
correlation between immune cell-specific markers and ADAM12
expression levels. It is well known that TAMs derived from
circulating monocyte populations can be differentiated into M2
macrophages under the stimulation of Treg and Th2 cells (49,50). Therefore, we speculated that
ADAM12, as a ubiquitously expressed transmembrane protein in tumor
cells, may participate in the recruitment of various immune cells
(including monocytes, TAM, Th2 and Treg cells), which in turn
promotes the polarization and maturation of M2 macrophages. M2
macrophages are known to be a necessary factor in favoring tumor
metastasis and immunosuppression. These results suggest the
promoting role of ADAM12 in tumorigenesis and progression from
another perspective, indicating the complex function of ADAM12 in
tumors.
There are still some limitations to the present
study. Firstly, the small sample size resulted in an unclear
relationship between ADAM12 expression levels and disease-free
survival (DFS) in gastric cancer patients. The collection of GC
tissue samples and follow-up data in subsequent studies is required
to further analyze the correlation between ADAM12 and DFS.
Secondly, the present study was based on public data mining and
bioinformatic analysis. The available data was limited and the
results still require corresponding experimental verification.
Furthermore, sufficient data was lacking to explore the role of
ADAM12 in regulating immune cell recruitment, and more
sophisticated experimental designs are required to validate these
promising results.
In the present study, we demonstrated that ADAM12
expression is frequently upregulated and correlated with a poor
prognosis of GC patients. Promoter hypomethylation and aberrant
ceRNA network regulation may contribute to the dysregulation of
ADAM12 expression. The results of enrichment analysis and
protein-protein interaction network indicated that ADAM12 is
probably involved in a variety of pivotal biological processes
regulating GC metastasis and the immune microenvironment. Through
further experimental analysis, we successfully confirmed that
ADAM12 significantly enhances the invasion and metastatic abilities
of GC cells. More importantly, it was found that ADAM12 potentially
plays a crucial role in tumor immune infiltration, especially the
polarization of M2 cells. In conclusion, our results highlight that
ADAM12 is a vital tumor promoter and a potential therapeutic target
for GC.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ, WJ and XH designed the study and wrote the
manuscript. HZ, HZ, BT and ZZ performed the experiments. WJ, JH and
XH performed patient recruitment and sample collection. HZ, WJ, JH
and XH analyzed and verified the integrity of the data. All authors
contributed to the article and approved the submitted version. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The studies involving human participants were
reviewed and approved by the Medical Ethics Committee of the
Affiliated Provincial Hospital of Anhui Medical University
(certification no. 2019KY32). The patients/participants provided
their written informed consent to participate in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This work was supported by the Natural Science Foundation of
Anhui Province (1908085MH282).
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rawla P and Barsouk A: Epidemiology of
gastric cancer: Global trends, risk factors and prevention. Prz
Gastroenterol. 14:26–38. 2019.PubMed/NCBI
|
|
3
|
Thrift AP and El-Serag HB: Burden of
gastric cancer. Clin Gastroenterol Hepatol. 18:534–542. 2020.
View Article : Google Scholar
|
|
4
|
Shen L, Li J, Xu J, Pan H, Dai G, Qin S,
Wang L, Wang J, Yang Z, Shu Y, et al: Bevacizumab plus capecitabine
and cisplatin in Chinese patients with inoperable locally advanced
or metastatic gastric or gastroesophageal junction cancer:
Randomized, double-blind, phase III study (AVATAR study). Gastric
Cancer. 18:168–176. 2015. View Article : Google Scholar :
|
|
5
|
Lordick F, Kang YK, Chung HC, Salman P, Oh
SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V, et
al: Capecitabine and cisplatin with or without cetuximab for
patients with previously untreated advanced gastric cancer
(EXPAND): A randomised, open-label phase 3 trial. Lancet Oncol.
14:490–499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thuss-Patience PC, Shah MA, Ohtsu A, Van
Cutsem E, Ajani JA, Castro H, Mansoor W, Chung HC, Bodoky G,
Shitara K, et al: Trastuzumab emtansine versus taxane use for
previously treated HER2-positive locally advanced or metastatic
gastric or gastro-oesophageal junction adenocarcinoma (GATSBY): An
international randomised, open-label, adaptive, phase 2/3 study.
Lancet Oncol. 18:640–653. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saha N, Robev D, Himanen JP and Nikolov
DB: ADAM proteases: Emerging role and targeting of the
non-catalytic domains. Cancer Lett. 467:50–57. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kveiborg M, Albrechtsen R, Couchman JR and
Wewer UM: Cellular roles of ADAM12 in health and disease. Int J
Biochem Cell Biol. 40:1685–1702. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang X and Xie X, Liu P, Yang L, Chen B,
Song C, Tang H and Xie X: Adam12 and lnc015192 act as ceRNAs in
breast cancer by regulating miR-34a. Oncogene. 37:6316–6326. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luo ML, Zhou Z, Sun L, Yu L, Sun L, Liu J,
Yang Z, Ran Y, Yao Y and Hu H: An ADAM12 and FAK positive feedback
loop amplifies the interaction signal of tumor cells with
extracellular matrix to promote esophageal cancer metastasis.
Cancer Lett. 422:118–128. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X, Wang Y, Gu J, Zhou D, He Z, Wang X
and Ferrone S: ADAM12-L confers acquired 5-fluorouracil resistance
in breast cancer cells. Sci Rep. 7:96872017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Duhachek-Muggy S, Qi Y, Wise R, Alyahya L,
Li H, Hodge J and Zolkiewska A: Metalloprotease-disintegrin ADAM12
actively promotes the stem cell-like phenotype in claudin-low
breast cancer. Mol Cancer. 16:322017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu H, Wang G, Zhu H and Xu A: ITGA5 is a
prognostic biomarker and correlated with immune infiltration in
gastrointestinal tumors. BMC Cancer. 21:2692021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan
DW, Tang HM and Peng ZH: Upregulated INHBA expression is associated
with poor survival in gastric cancer. Med Oncol. 29:77–83. 2012.
View Article : Google Scholar
|
|
16
|
Modhukur V, Iljasenko T, Metsalu T, Lokk
K, Laisk-Podar T and Vilo J: MethSurv: A web tool to perform
multivariable survival analysis using DNA methylation data.
Epigenomics. 10:277–288. 2018. View Article : Google Scholar
|
|
17
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2017. View Article : Google Scholar
|
|
18
|
Zhou KR, Liu S, Cai L and Bin L: The
Encyclopedia of RNA Interactomes (ENCORI): The Encyclopedia of RNA
Interactomes, 2021.
|
|
19
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang L, Zhou G, Soufan O and Xia J:
miRNet 2.0: Network-based visual analytics for miRNA functional
analysis and systems biology. Nucleic Acids Res. 48:W244–W251.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar :
|
|
24
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, Jensen LJ and Mering CV: STRING v11: Protein-protein association
networks with increased coverage, supporting functional discovery
in genome-wide experimental datasets. Nucleic Acids Res.
47:D607–D613. 2019. View Article : Google Scholar
|
|
26
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Remmert M, Biegert A, Hauser A and Söding
J: HHblits: Lightning-fast iterative protein sequence searching by
HMM-HMM alignment. Nature Methods. 9:173–175. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pearson WR and Lipman DJ: Improved tools
for biological sequence comparison. Proc Natl Acad Sci USA.
85:2444–2448. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sievers F, Wilm A, Dineen D, Gibson TJ,
Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al:
Fast, scalable generation of high-quality protein multiple sequence
alignments using Clustal Omega. Mol Syst Biol. 7:5392011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Larkin MA, Blackshields G, Brown NP,
Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm
A, Lopez R, et al: Clustal W and Clustal X version 2.0.
Bioinformatics. 23:2947–2948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Martí-Renom MA, Stuart AC, Fiser A,
Sánchez R, Melo F and Sali A: Comparative protein structure
modeling of genes and genomes. Annu Rev Biophys Biomol Struct.
29:291–325. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The protein
data bank. Nucleic Acids Res. 28:235–242. 2000. View Article : Google Scholar
|
|
34
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu X, Salokas K, Tamene F, Jiu Y,
Weldatsadik RG, Öhman T and Varjosalo M: An AP-MS- and
BioID-compatible MAC-tag enables comprehensive mapping of protein
interactions and subcellular localizations. Nat Commun. 9:11882018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Suzuki A, Kadota N, Hara T, Nakagami Y,
Izumi T, Takenawa T, Sabe H and Endo T: Meltrin alpha cytoplasmic
domain interacts with SH3 domains of Src and Grb2 and is
phosphorylated by v-Src. Oncogene. 19:5842–5580. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Malinin NL, Wright S, Seubert P, Schenk D
and Griswold-Prenner I: Amyloid-beta neurotoxicity is mediated by
FISH adapter protein and ADAM12 metalloprotease activity. Proc Natl
Acad Sci USA. 102:3058–3063. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Harold D, Jehu L, Turic D, Hollingworth P,
Moore P, Summerhayes P, Moskvina V, Foy C, Archer N, Hamilton BA,
et al: Interaction between the ADAM12 and SH3MD1 genes may confer
susceptibility to late-onset Alzheimer's disease. Am J Med Genet B
Neuropsychiatr Genet. 144B:448–452. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang R, Godet I, Yang Y, Salman S, Lu H,
Lyu Y, Zuo Q, Wang Y, Zhu Y, Chen C, et al: Hypoxia-inducible
factor-dependent ADAM12 expression mediates breast cancer invasion
and metastasis. Proc Natl Acad Sci USA. 118:e20204901182021.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Díaz B, Yuen A, Iizuka S, Higashiyama S
and Courtneidge SA: Notch increases the shedding of HB-EGF by
ADAM12 to potentiate invadopodia formation in hypoxia. J Cell Biol.
201:279–292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Y, Tian M, Liu W, Wang D, Zhou Z, Pei
Q, Huang Y, Tan F and Güngör C: Follistatin-Like 3 enhances
invasion and metastasis via β-catenin-mediated EMT and aerobic
glycolysis in colorectal cancer. Front Cell Dev Biol. 9:6601592021.
View Article : Google Scholar
|
|
42
|
Dai ZT, Xiang Y, Zhang XY, Zong QB, Wu QF,
Huang Y, Shen C, Li JP, Ponnambalam S and Liao XH: Regulation of
follistatin-like 3 expression by miR-486-5p modulates gastric
cancer cell proliferation, migration and tumor progression. Aging
(Albany NY). 13:20302–20318. 2021. View Article : Google Scholar
|
|
43
|
Wang J, Li D, Cang H and Guo B: Crosstalk
between cancer and immune cells: Role of tumor-associated
macrophages in the tumor microenvironment. Cancer Med. 8:4709–4721.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Duffy MJ, McKiernan E, O'Donovan N and
McGowan PM: Role of ADAMs in cancer formation and progression. Clin
Cancer Res. 15:1140–1144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Carl-McGrath S, Lendeckel U, Ebert M,
Roessner A and Röcken C: The disintegrin-metalloproteinases ADAM9,
ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol.
26:17–24. 2005.
|
|
46
|
Wang H, Wu J and Guo W: SP1-Mediated
upregulation of lncRNA LINC01614 Functions a ceRNA for miR-383 to
facilitate glioma progression through regulation of ADAM12. Onco
Targets Ther. 13:4305–4318. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang C, Cao F, Huang S and Zheng Y:
Follistatin-like 3 correlates with lymph node metastasis and serves
as a biomarker of extracellular matrix remodeling in colorectal
cancer. Front Immunol. 12:7175052021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang J, Zhang Z, Li R, Mao F, Sun W, Chen
J, Zhang H, Bartsch JW, Shu K and Lei T: ADAM12 induces EMT and
promotes cell migration, invasion and proliferation in pituitary
adenomas via EGFR/ERK signaling pathway. Biomed Pharmacother.
97:1066–1077. 2018. View Article : Google Scholar
|
|
49
|
Shapouri-Moghaddam A, Mohammadian S,
Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi
A, Afshari JT and Sahebkar A: Macrophage plasticity, polarization,
and function in health and disease. J Cell Physiol. 233:6425–6440.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Weirather J, Hofmann UD, Beyersdorf N,
Ramos GC, Vogel B, Frey A, Ertl G, Kerkau T and Frantz S:
Foxp3+ CD4+ T cells improve healing after
myocardial infarction by modulating monocyte/macrophage
differentiation. Circ Res. 115:55–67. 2014. View Article : Google Scholar : PubMed/NCBI
|