Introduction
Rhabdomyosarcoma (RMS) is the commonest soft tissue
sarcoma in children and young adults (1). The two major subtypes are alveolar
RMS (ARMS) and embryonal RMS (ERMS) that have emerged based on
specific light microscopic features. Whereas ARMS cells are
distributed around an open central space thus resembling the
alveoli of a lung, ERMS cells resemble immature skeletal myoblasts
(2). Indeed, ARMS is molecularly
different from ERMS and is characterized by genetic translocations
of PAX3 or PAX7 and FOXO1. These fusion-positive ARMS are very
aggressive. By contrast, fusion-negative ARMS clinically and
molecularly resemble the less aggressive ERMS subtype (3). Although these subtypes are
distinguishable by genetics and prognosis they share an impaired
muscle differentiation phenotype and are probably derived from a
muscle or a mesenchymal progenitor cell (2,4).
The development of skeletal muscle from immature
precursors is partially driven by WNT/β-Catenin (canonical) and
β-Catenin independent (non-canonical) WNT signaling pathways.
Indeed, a number of WNT ligands and receptors take part not only in
embryonic myogenesis and skeletal muscle formation, but also in
muscle differentiation and expression of myogenic regulatory
factors [for a review see (5)].
WNT signaling is regulated by 19 secreted WNT
proteins that bind to more than 15 receptors or co-receptors. When
synthesized, WNT ligands are modified at the endoplasmic reticulum
and then transferred to the plasma membrane, where they are
secreted. How these secreted WNT ligands reach neighboring cells is
still a matter of debate. One model proposes a transport of the WNT
ligands by extracellular vesicles to the membrane receptor of other
cells, whereas the other model favors the direct contact between
cell-membrane-tethered WNT ligands and the receptor cells (6,7).
Of the WNT signaling pathways the canonical
WNT/β-Catenin pathway is the best characterized. In the inactive
stage β-Catenin is phosphorylated and ubiquitinated by a
destruction complex, which targets it for degradation by the
proteasome. Activation of the WNT/β-catenin pathway is induced by
binding of extracellular WNT ligands to Frizzled (FZD) and their
LRP5/6 co-receptors. This causes inhibition of the β-Catenin
destruction complex. Consequently, β-Catenin is stabilized,
accumulates in the cytoplasm and translocates to the nucleus, where
it triggers the expression of WNT target genes such as AXIN2
and cMYC (6,8).
The non-canonical β-Catenin-independent
WNT/Ca2+ or WNT/planar cell polarity (PCP) pathways are
activated mainly by WNT5A. Binding of WNT5A to FZD 2, 3, 4, 5 or 6
and their co-receptors ROR1 and ROR2 results in WNT/Ca2+
signaling (9). For induction of
PCP signaling, WNT5A must interact with ROR2 (10). Depending on the receptor context,
WNT5A also can either activate or inhibit canonical WNT/β-Catenin
signaling. Indeed, inhibition of β-Catenin accumulation by WNT5A is
observed in several cell lines (11).
Although WNT signaling is involved in muscle
development, its role in RMS remains to be elucidated. With respect
to canonical WNT/β-Catenin signaling, only a few studies have been
published. This may be due to the fact, that RMS does not show
mutations in relevant components of the pathway and only rarely
shows nuclear β-Catenin (12).
However, it has been reported that stimulation of RMS cell lines
with recombinant WNT3A leads to nuclear translocation of β-Catenin
and blocks cellular proliferation (13,14).
In addition, downregulation of the main inhibitor of WNT/β-Catenin
signaling Dickkopf1 leads to expression of active β-Catenin and
inhibition of proliferation and invasion of RMS tumor cells
(15). These results imply that
activation of WNT/β-Catenin signaling has antitumoral effects in
RMS. By contrast, data from our group show that activation of
WNT/β-Catenin or a β-Catenin knockdown barely affects
proliferation, apoptosis or myodifferentiation of RMS tumor cells.
In addition, RMS incidence, multiplicity or latency time are not
altered by a hypomorphic Wnt3a allele or a conditional
knockout of β-Catenin in genetically engineered mice (16,17).
Furthermore, genetic data show decreased RMS multiplicity on a
Wif1-deficient background (16). The latter experiment suggests that
WNTs normally bound by WIF1 inhibit RMS formation or growth, either
by blocking RMS initiation or by preventing the progression of
already initiated tumors. WIF1 has high affinity to WNT3A, WNT4,
WNT5A, WNT7A, WNT9A and WNT11 (18), of which WNT5A, WNT7A and WNT11
activate non-canonical WNT signaling pathways (19). Together with the fact that
canonical WNT signaling only serves a subordinate role in RMS
aggressiveness recent data suggest that non-canonical WNTs may have
antitumoral functions (16).
The present study investigated the basic functions
of WNT5A, the major ligand of non-canonical WNT signaling, in RMS.
Until now, alterations of WNT5A expression levels were reported as
a secondary phenomenon in RMS. Thus, in ERMS cells from p53/c-fos
double mutant mice WNT5A is downregulated in comparison with normal
myoblasts (13), whereas another
study shows higher WNT5A expression in ERMS compared with ARMS cell
lines (20), which is somewhat
contradictory. Moreover, WNT5A sequence variants have been
found in ARMS, but their effect on WNT5A function is unclear
(21).
To elucidate the role of WNT5A in RMS in more
detail, the present study investigated WNT5A expression in
human PAX3/FOXO1 positive ARMS and in ERMS. It also stably
overexpressed or knocked-down WNT5A in ERMS and fusion-positive
ARMS cell lines and investigated the growth behavior, migration and
differentiation of the cells. The present study also analyzed the
effect of WNT5A on β-Catenin stability.
Materials and methods
Biopsy specimens
For reverse transcription-quantitative (RT-q) PCR
analyses 20 RNA (10 ARMS and 10 ERMS) samples from the Cooperative
Weichteilsarkom Studiengruppe (CWS) tissue bank (Stuttgart,
Germany) were analyzed (for patient characteristics see Table SI). Muscle tissue from five
(anonymous) separate patients served as controls. For analysis of
WNT5A protein expression a tumor microarray (TMA) with 125 RMS
biopsies from the Pediatric Tumor Register, Kiel, Germany was used.
The histopathological features of all cases were centrally reviewed
by the late Professor I. Leuschner (Member of the CWS and Director
of the Pediatric Tumor Registry, Kiel, Germany). All patients were
treated according to CWS protocols (CWS-96 or CWS-2002P). TMA
studies and the use of normal muscle were authorized by the
approval 158/2009/b02; University of Tübingen, Tübingen, Germany;
April 2, 2009 within the framework of the CWS and that for the
RT-qPCR studies additionally by the approval 2017-802R-MA
(University of Heidelberg, University Medical Centre Mannheim).
Written informed consent, according to the Declaration of Helsinki,
was obtained from all patients or their legal guardians, depending
on the age of the patients.
In addition, two publicly available RMS datasets
with 103 (37 ARMS and 66 ERMS; dataset 1) (22) and 70 (34 ARMS and 36 ERMS; dataset
2) patients with RMS (3) were
analyzed. Note that samples with unclear diagnosis were omitted and
that only PAX3-FOXO1 translocation positive ARMS and
translocation negative ERMS were considered for analysis. As
described previously, a Custom CDF Version 20 with ENTREZ-based
gene definitions was used to annotate the arrays (23). The raw fluorescence intensity
values were normalized applying quantile normalization and RMS
background correction. All probes belonging to one gene (probeset)
were summarized to one intensity value. It was not necessary to
exclude or average the probesets for a pathway or gene ontology
analysis. Gene analysis was performed by SAS JMP10 Genomics,
version 6, from SAS (SAS Institute).
Cell lines and antibodies
The translocation positive ARMS cell lines CRL2061
(identical to Cellosaurus cell line SJCRH30) and RH30 and the ERMS
cell lines RD and TE671 were obtained from the American Type
Culture Collection (CRL2061 and RD) or the Leibniz Institute
DSMZ-German Collection of Microorganisms and Cell Cultures (RH30
and TE671). The cell lines CRL2061 and RH30 were verified for the
PAX3-FOXO1 fusion gene and all cell lines routinely for
mycoplasma contamination. ARMS cell lines were cultured in RPMI1640
and ERMS cell lines in DMEM, both supplemented with 10% (v/v) FCS
(MilliporeSigma). Culturing conditions were 37°C and 5%
CO2. For the experiments, cells were used during the
exponential growth phase. In addition, the cell lines RH30, RD and
TE671 were authenticated by STR profiling (Eurofins Genomics
Germany GmbH).
For protein detection, anti-human antibodies diluted
in TBS were applied. Source and dilutions of the antibodies are
listed in Table SII.
HRP-conjugated anti-mouse or anti-rabbit IgGs (Cell Signaling
Technology, Inc.) were used as secondary antibodies for western
blotting as detailed below.
Plasmids and cloning
The pcDNA3-WNT5a plasmid for transient expression
was from the Addgene Vector Database [Addgene plasmid cat. no.
35911 (24)]. For stable
overexpression, WNT5A from the pcDNA3-WNT5a plasmid was cloned in
the retroviral pBABE vector [Addgene plasmid cat. no. 1764
(25)]. WNT5A short hairpin
(sh)RNA (mature WNT5A targeting sequence 5′-GGA CGT TAA GAG ATA TTC
AAA-3′) cloned into the retroviral pGIPZ vector was purchased from
Horizon Discovery. Restriction enzymes (BamHI, EcoRI)
were purchased from New England BioLabs, Inc. Purification of
cloning products was performed with the QIAquick Gel Extraction kit
(Qiagen GmbH). Gene products were ligated with T4 ligase from New
England BioLabs, Inc. All plasmids were isolated with the EndoFree
Plasmid Maxi kit (Qiagen GmbH) according to the manufacturer's
instructions.
Stable WNT5A overexpression (WNT5AOE) or
knockdown (WNT5AKD)
The pBABE-WNT5A plasmid and the packaging plasmids
pUMVC and VSV-G [Addgene plasmids cat. no. 8449 and cat. no. 8454
(26)] were transfected into 293
cells (packaging cell line) using the calcium phosphate
transfection kit (MilliporeSigma) according to the manufacturer's
instructions. This procedure employs a sodium phosphate-containing
HEPES-buffered solution that is mixed with a calcium chloride
solution containing the DNA. This generates DNA-calcium phosphate
precipitates that are attached on the cell membrane and are
internalized into the cell. After sterile filtration, the
supernatant was used to transduce the RMS cell lines.
For transduction of the WNT5A shRNA expression
vector WNT5A-GIPZ, the vector was mixed with Trans-Lentiviral
packaging plasmid mix from Horizon Technologies according to the
manufacturer's protocol (Horizon Technologies) and transfected as
described above.
In all settings and if not stated otherwise, RMS
cells transduced with either the empty pBABE or the empty pGIPZ
vectors served as respective controls [CTR; please note that it was
decided to use an empty vector for the WNT5A shRNA validation
experiments, because an empty vector was also used for the WNT5A
overexpression experiments. In addition, empty vectors are
frequently used in knockdown experiments (27,28)]. pBABE and GIPZ plasmids both
contain a puromycin resistance cassette. Therefore, successfully
transduced RMS cells were selected with puromycin (MilliporeSigma)
at a concentration of 10 µg/ml. WNT5AOE or WNT5AKD was
verified by RT-qPCR and western blotting.
Transient WNT5A expression
Transient transfection of the pcDNA3-WNT5a plasmid
or the empty pcDNA3 vector was performed with the calcium phosphate
transfection kit (MilliporeSigma) according to the manufacturer's
instructions.
Cell viability and proliferation
assays
Cell viability were monitored by WST-1 and MTT
assay. For WST-1 or MTT assays, WNT5AKD, WNT5AOE and respective
control cells (CTR; transduced with the respective empty vectors)
were seeded in flat bottom 96-well plates at a density of
5×103 cells/well. After 48 h cell viability was verified
by adding 100 µl of WST-1 solution (Roche Diagnostics; WST-1
reagent was diluted 1:25 in cell culture medium) or by adding 20
µl MTT/well (5 mg/ml in sterile PBS; Carl Roth GmbH). After
an incubation for 3 h at 37°C, reduction of WST-1 by viable cells
was determined using a SynergyMx microtiter plate reader and the
Gen5 software (BioTek Instruments GmbH) at 450 nm and a reference
wavelength of 655 nm. For the MTT assay, the supernatant was
discarded and cells were lysed by adding 200 µl DMSO.
Reduction of MTT by viable cells was determined at 560 nm and a
reference wavelength of 620 nm.
Proliferation was measured by BrdU assay using the
Cell Proliferation ELISA BrdU kit from Roche Diagnostics. In this
assay, RMS cells were seeded in black 96-well plates with a clear
base at a density of 5×103 cells/well. Cells growing in
the exponential phase were treated with 10 µl/ml BrdU for 24
h. BrdU incorporation was measured according to the manufacturer's
instruction using a SynergyMx microtiter plate reader and Gen5 1.11
software (BioTek Instruments, Inc.). In both assays, and after a
settling time of 3 h, the cells were also incubated for 24 h with
200 ng/ml rWNT5A (R & D Systems) at 37°C in a 5% CO2
incubator.
Migration assays
The cell migration assay was performed in 24-well
plates (BD Falcon; Becton, Dickinson and Company) using 0.8
µm pore size inserts (BD Falcon; Becton, Dickinson and
Company). After 24 h of starvation, RMS cells were seeded into the
upper part of the insert at a density of 1×105 in a
total volume of 300 µl. The lower part of the wells was
filled with 500 µl medium supplemented with 10% FCS
(RPMI1604 medium for RH30 and DMEM medium for RD and TE671 cells).
The cells were allowed to migrate for 8 h (RH30) or 24 h (RD and
TE671) at 37°C in a 5% CO2 incubator. Thereafter the
media inside the insert was aspirated and cells that had not
migrated were scraped off the upper side of the inserts using a
cotton swab. The cells that had migrated to the lower side of the
inserts were washed once with PBS and stained with 5 µM
CellTracker Green CMFDA Dye (Thermo Fisher Scientific, Inc.) for 20
min at 37°C in a 5% CO2 incubator followed by a washing
step with PBS. Pictures of the migrated cells were taken by
fluorescence microscopy at 100-fold magnification (Olympus BX 60
with cellSens Dimension software 1.6; Olympus Corporation).
Migrated cells were quantified by counting the number of cells in
at least four individual fields per insert. Data shown represent
the mean of migrated cells of at least three independent
experiments + SD.
Cell migration was additionally verified by scratch
assays. For this purpose, RMS cells were seeded in 12-well plates
at a density of 2×105 cells/well. After a settling time
of 3 to 4 h, normal growth medium was replaced by starvation medium
supplemented with 1% FCS (without FCS the cells would have died)
and cells were cultured for another 24 h. After aspiration of the
medium, a cross-shaped scratch was made with a 200 µl
pipette tip. To remove any detached cells, each well was washed
twice with PBS and fresh starvation medium was added. Images were
captured with an inverse microscope (Leica Microsystems GmbH) after
0, 6, 12 and 24 h and analyzed with ImageJ (version 1.48t, National
Institutes of Health) and each scratch was measured at three
different fixed positions.
Sphere assay
Sphere assay was performed as previously described
(29). In brief, RMS cells were
seeded at a density of 1 cell/µl in ultra-low attachment
plates (MilliporeSigma) and cultured in neurobasal medium (Thermo
Fisher Scientific, Inc.) freshly supplemented with 2X B27 (Thermo
Fisher Scientific, Inc.), 100 U/ml penicillin-streptomycin (Gibco;
Thermo Fisher Scientific, Inc.), 10 ng/ml EGF (cat. no. 236-EG;
R&D Systems) and 20 ng/ml b-FGF (cat. no. 233-FB; R&D
Systems). Spheres were counted under the microscope and pictures
were taken using an inverse microscope (Leica Microsystems GmbH)
with phase contrast at ×10 magnification.
RT-qPCR
Total RNA from 90% confluent cells was isolated
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). cDNA was generated from 0.5 µg total RNA
using the PrimeScript RT Reagent kit (Takara Bio, Inc.). The
relative mRNA levels were detected by the Step one plus system
(Thermo Fisher Scientific, Inc.) using TB Green Premix Ex Taq II
(Takara Bio, Inc.). These steps were performed according to the
respective manufacturer's instructions. All primers were designed
with PRIMER3 and are listed in Table
SIII. The cycling conditions were 95°C for 10 min, followed by
40 cycles of 95°C for 15 sec and 60°C for 1 min. The quantification
was performed by the 2−ΔΔCq method (30). GAPDH was used as a reference
gene for relative quantification (please note that the reference
genes B2M, RPL13A and TBP for RMS were also
tested and it was found that all housekeeping genes yielded the
same results; data not shown). Respective primers are given in
Table SIII).
Western blotting
RIPA buffer (Thermo Fisher Scientific, Inc.)
complemented with proteinase inhibitor complete Tablets (EDTA-free;
Roche Diagnostics GmbH) was used for cell lysis and protein
isolation. For western blotting, 2×105 cells were seeded
in each well of a 6-well plate. After 24 h cells were harvested in
200 µl RIPA buffer. 20 µl of this lysate were mixed
with 5 µl loading buffer and loaded on the gel. Nuclear and
cytoplasmic proteins were separated by the nuclear and cytoplasmic
extraction kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Isolation of supernatant proteins was
performed from 1 ml medium with trichloroacetic acid 20% (Merck
KGaA) as previously described (31). Equivalent protein amounts
(determined by quantification of the respective loading controls on
the blots) were mixed with protein loading dye (Bio-Rad
Laboratories, Inc.), separated on 10% SDS-polyacrylamide gels and
transferred to a nitrocellulose membrane. Membranes were blocked at
room temperature with 5% milk powder (w/v) in TBST [TBS
supplemented with 0.05% Tween20 (v/v)] for 1 h, washed three times
for 10 min with TBS-T and incubated overnight at 4°C with the
primary antibody diluted in PBS supplemented with 0.02% (w/v)
sodium azide, 2% (w/v) BSA and 0.001% Tween20 (v/v). After washing
three times with TBS-T, membranes were incubated for 1 h at room
temperature with the respective secondary antibody diluted in 5%
milk powder in TBST. After washing three times with TBST, protein
bands were visualized with ECL Plus Substrate (Thermo Fisher
Scientific, Inc.) and the Fusion SL Imaging system (Peqlab
Biotechnologie GmbH). Data were quantified with ImageJ (version
1.48t, National Institutes of Health). Re-blot stripping solution
(MilliporeSigma) was used to strip the membrane. For details of the
antibodies used see Table
SII.
Statistical analysis
All analyses of transduced cell lines summarize the
results obtained from ≥3 independent experiments (each measured in
duplicates for RT-qPCR, BrdU, MTT and scratch assays) and are
represented as mean ± SD. If not indicated otherwise, the data were
analyzed by an unpaired Student's t-test (to determine if the
difference between two data sets is significant) or with one-way
ANOVA followed by Tukey's multiple comparison (to determine if the
differences between more than two data sets are significant) using
GraphPad Prism 6.0 (GraphPad Software, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
WNT5A is expressed in RMS
Initially, the present study assessed the WNT5A
expression profile in two publicly available ERMS/ARMS datasets
[dataset 1 and 2 published by Davicioni et al (22) and Williamson et al (3), respectively] and in 10 ARMS and 10
ERMS RMS samples (validation dataset) from the CWS study group
(32). Note that only
PAX3-FOXO1 positive ARMS were used for the analysis. The
results of the validation cohort were additionally compared with
WNT5A expression in normal skeletal muscle.
Analysis of WNT5A in dataset 1 and 2 revealed
a significantly higher expression level of WNT5A in ERMS
compared with ARMS (Fig. 1A). This
was similar in the validation dataset (Fig. 1B). In addition, the analysis of the
validation set shows that the WNT5A expression level of ARMS
resemble that of normal skeletal muscle (Fig. 1B). This suggests that WNT5A
mRNA expression is rather associated with the ERMS subtype.
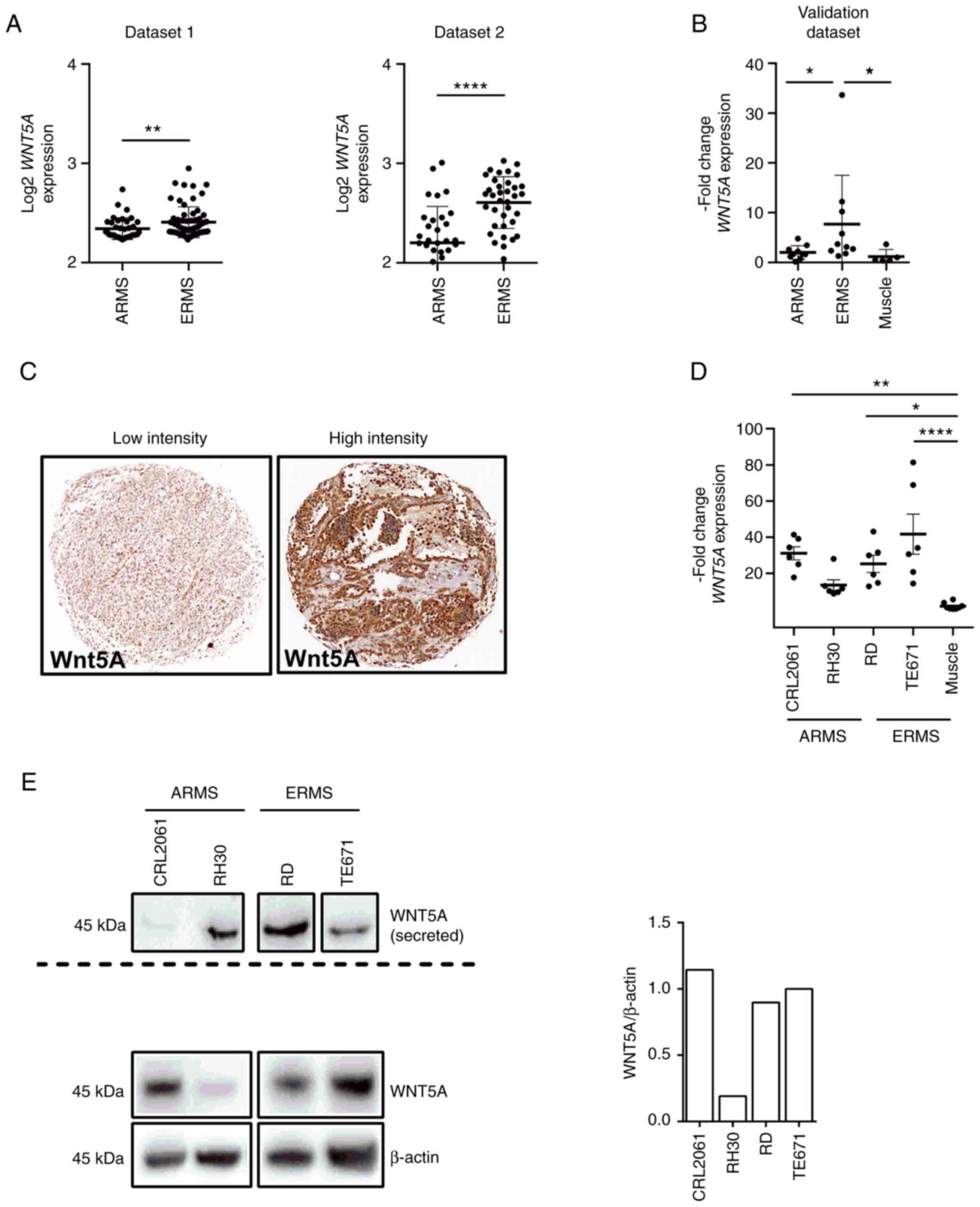 | Figure 1WNT5A expression in human RMS.
WNT5A expression level (A) of ARMS and ERMS from two
publicly available microarray datasets [see (22) and (3) for datasets 1 and 2, respectively] and
(B) of 10 ARMS, 10 ERMS and five normal skeletal muscle samples in
the validation data set from the CWS study group measured by
reverse transcription-quantitative PCR. (C) Immunohistochemical
analysis of human RMS showing low or high expression of WNT5A when
using the WNT5A monoclonal antibody (3D10) from Thermo Fisher
Scientific, Inc (magnification, 50×). (D) WNT5A mRNA expression
(three independent experiments each measured in duplicates) of the
human ARMS cell lines CRL2061 and RH30 and ERMS cell lines RD and
TE671 in comparison with normal skeletal muscle, which was set=1.
(E) Western blot analysis of WNT5A protein levels in the
supernatant (top left; please note the quantification of secreted
WNT5A using HSC70, which is occasionally secreted by tumor cells
(58), was not possible because
RMS cell lines apparently do not secrete HSC70) and on another gel
in the respective whole cell lysates (bottom left) of RMS tumor
cell lines. β-Actin detection served as loading control for whole
cell lysates (bottom) and the right panel shows the respective
quantification. Horizontal lines in (A-B) are mean ± SD; bars in
(D) are also ± SD. Statistical analyses in (A-B) were performed by
non-parametric Mann-Whitney tests and that in (D) by one-way ANOVA.
*P<0.05, **P<0.01,
****P<0.0001. RMS, rhabdomyosarcoma; ARMS, alveolar
RMS; ERMS, embryonal RMS. |
The present study also examined a TMA with 125 RMS
for WNT5A expression by immunohistochemical analysis. After quality
control, 41 ERMS and 7 fusion-positive ARMS samples were evaluable
(all tumors, which showed mitoses but were Ki67 negative were
excluded). The analyses revealed that all RMS samples were positive
for WNT5A although to a variable intensity. Unfortunately, tumor-
and stromal cells could not be clearly distinguished so that a
scoring (which would have been multiplication of the percentage of
WNT5A positive cells by staining intensity) was not possible.
Nevertheless, it was found that 4, 23 and 14 ERMS express WNT5A at
low, intermediate and high intensity, respectively and that 2 ARMS
showed intermediate and 5 ARMS showed high WNT5A intensity
(Fig. 1C shows a tumor with low
and one with high WNT5A intensity).
The present study also analyzed the expression of
some WNT5A-associated receptors (i.e., FZD2, FZD4,
FZD5, ROR1 and ROR2) in datasets 1 and 2. In
both sets, ERMS show higher expression of FZD2 and
FZD4 compared with ARMS. In addition, ROR1 and
probably also ROR2 seem to be elevated in ERMS (Table SIV).
Similarly, the PAX3/FOXO1 positive ARMS cell
lines CRL2061 and RH30 and the ERMS cell lines RD and TE671
expressed WNT5A mRNA (Fig.
1D). In addition, the ARMS cell line RH30 and the ERMS cell
lines RD und TE671 expressed and secreted WNT5A (Fig. 1E; top left shows WNT5A protein in
the supernatant of the cells; bottom left shows cellular WNT5A
protein expression and the respective loading control).
Furthermore, the mRNA levels in general correlate with the
respective WNT5A protein levels (compare Fig. 1D and E bottom right). Note that the
WNT5A antibody detects a protein of the correct size (i.e., 45 kDa)
in the supernatant of WNT5A producing fibroblasts, in media
supplemented with 200 ng/ml rWNT5A (R&D Systems, Inc.) and in
supernatants of RMS cells transiently transfected with WNT5A
(Fig. S1A and B). However, it
does not detect a protein in the supernatants of L-cells or WNT3A
producing L-cells (Fig. S1A).
Therefore, the anti-WNT5A antibody can be considered specific.
Finally, all cell lines produce β-Catenin and FZD5
protein and most of them FZD2 and ROR2, whereas ROR1 is rather
expressed by ERMS cell lines (Fig.
S1C). This indicated that ERMS cell lines express all WNT5A
receptors analyzed (i.e., FZD2, FZD5, ROR1 and ROR2), whereas ARMS
cell lines occasionally lack expression of these receptors.
WNT5A inhibits cellular proliferation and
migration and may decrease expression of MYOD, DES and MYOG in RMS
cells
To investigate the effect of WNT5A on RMS growth
behavior and differentiation, the present study stably
overexpressed or knocked down WNT5A in the ARMS cell line RH30 and
in the ERMS cell lines RD and TE671 (WNT5AOE or WNT5AKD,
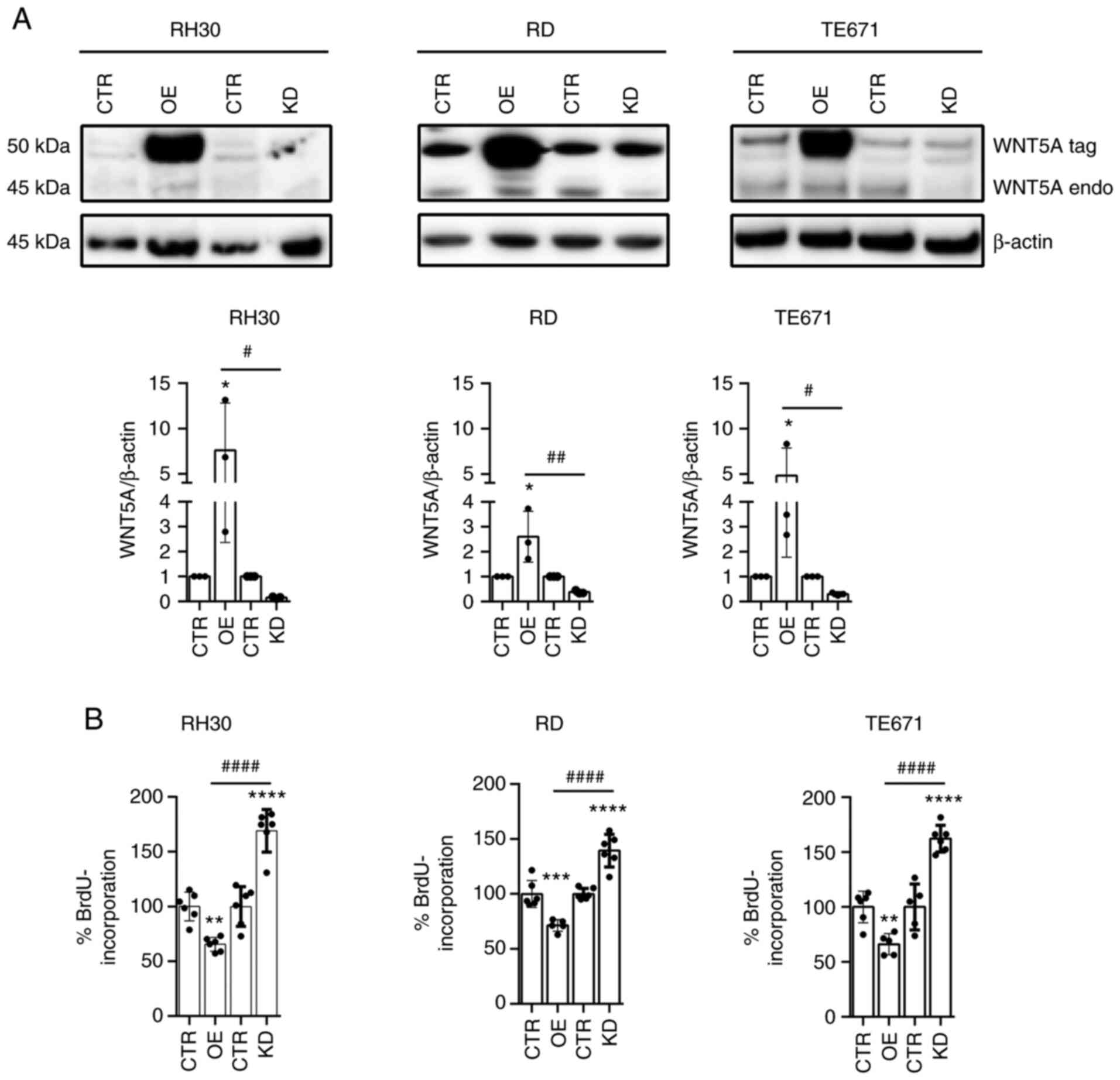
respectively; Fig. 2A). Note that
WNT5A is V5-tagged (24) and
therefore is larger than the endogenous WNT5A protein. Also note
that the used monoclonal anti-Wnt5A antibody gave an unspecific
band of around 55 kDa that overlaped with the tagged WNT5A protein.
The present study was not able to stably overexpress WNT5A in the
ARMS cell line CRL2061. Therefore, all subsequent experiments were
performed with RH30, RD and TE671 cells, which all express and
secrete WNT5A (Fig. 1D and E).
At least two different cell batches, i.e., cells
that have been transduced at different time points, were collected.
Each batch was analyzed for WNT5A expression and for cell
viability by MTT assay. As shown in Fig. S2A and B, all batches showed an
almost identical WNT5A mRNA level or proliferative behavior
and thus the first batches were chosen for further experiments.
Notably, when compared with cells transduced with
the respective vector control, WNT5AOE significantly inhibited
cellular proliferation of all RMS cell lines (Fig. 2B; for cell viability see Fig. S3A). By contrast, the WNT5AKD
enhances the respective parameters (Fig. 2B; for cell viability see Fig. S3A). Similarly, when respective
parental cells were incubated with rWNT5A, proliferative capacity
and viability of the cells were inhibited (Fig. S3B and C). However, this was only
seen in RH30 and RD cells, but not in TE671 cells. This suggests
that both membrane-tethered and extracellular WNT5A is responsible
for these effects.
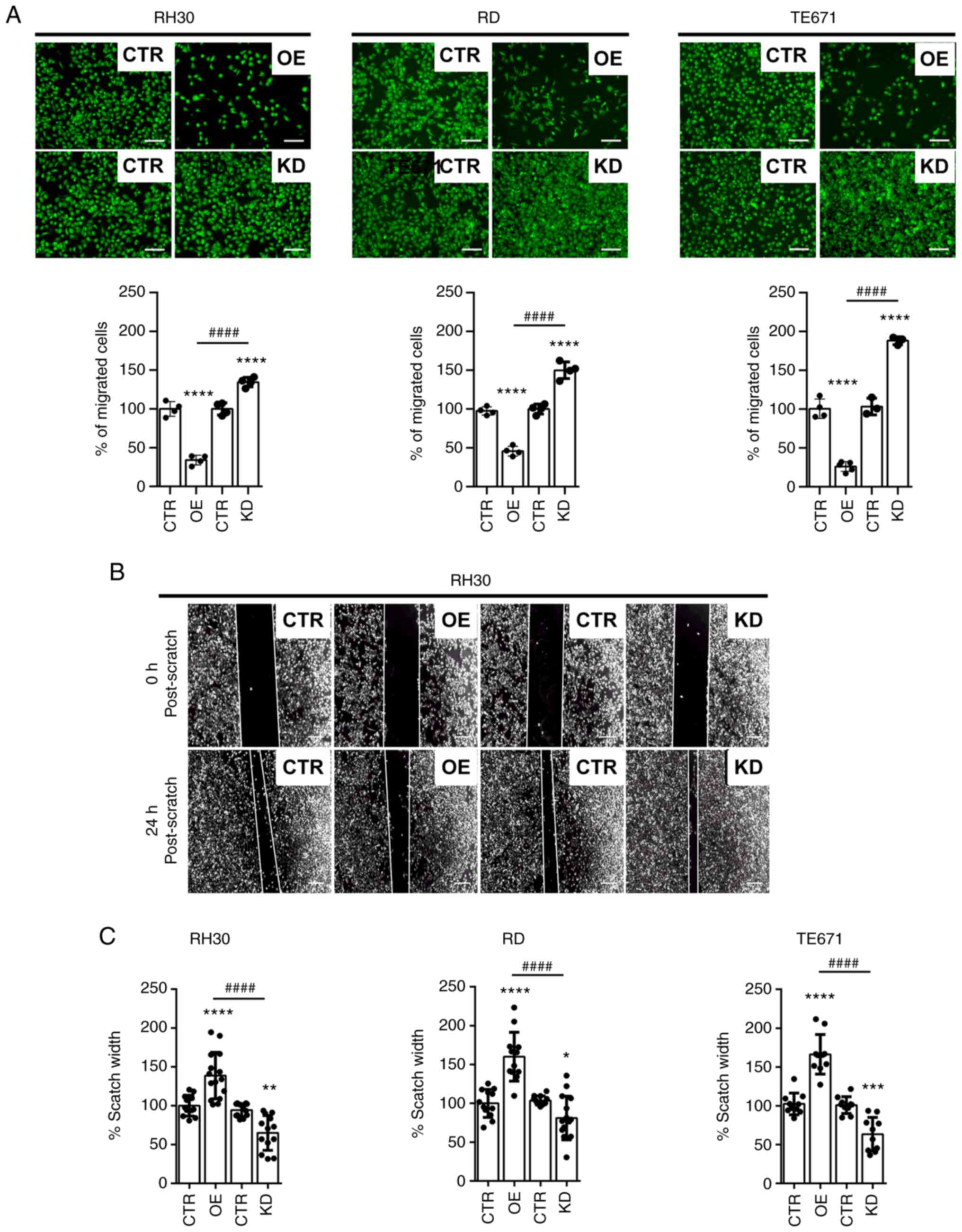
Next, the influence of WNT5A on migration of RMS
cells was investigated by Transwell assay. The data showed that the
migratory capacity of RH30, RD and TE671 cells with a WNT5AOE is
significantly lower compared with the vector controls, whereas that
of WNT5AKD is significantly higher (Fig. 3A). This was confirmed by a scratch
assay (Fig. 3B shows
representative pictures of RH30 cells and Fig. 3C shows quantification of the
scratch widths of all cell lines; note that the scratch width of
WNT5AOE cells is higher and that of WNT5AKD cells is smaller). This
demonstrated that WNT5A can inhibit the migratory capacity of RMS
cell lines.
As WNT signaling serves an important role in
myogenesis, the influence of WNT5A on the expression of myogenic
markers was also analyzed. These were myoblast determination
protein 1 (MYOD), myogenin (MYOG) and desmin (DES), which are
correlated with a higher growth rate of RMS in zebrafish (33). Indeed, MYOD has an unappreciated
and dominant oncogenic role to regulate human RMS growth (33). The results of the present study
showed that WNT5AOE significantly decreased MYOD in all cell lines.
WNT5AOE also decreased DES and MYOG protein, which was significant
for RD and RH30 cells, respectively (Fig. 4). By contrast, the WNT5AKD
significantly increased MYOG and DES protein levels in all analyzed
cell lines (Fig. 4). Together,
these data indicated that WNT5A may decrease the expression of
MYOD, MYOG and DES in RMS cells, which goes along with concomitant
inhibition of proliferation and migration. However, differences in
muscle marker expression between the two control cell lines (CTR)
was also detected, which were transduced with the backbones pBABE
and pGIPZ for WNT5AO and WNT5AKD cells, respectively (Fig. S4). Therefore, these data need to
be looked at with care, although other parameters such as
WNT5A expression and cellular proliferation were identical
to each other and in most cases also to the untransduced parental
cell lines (see Fig. S4 for more
details).
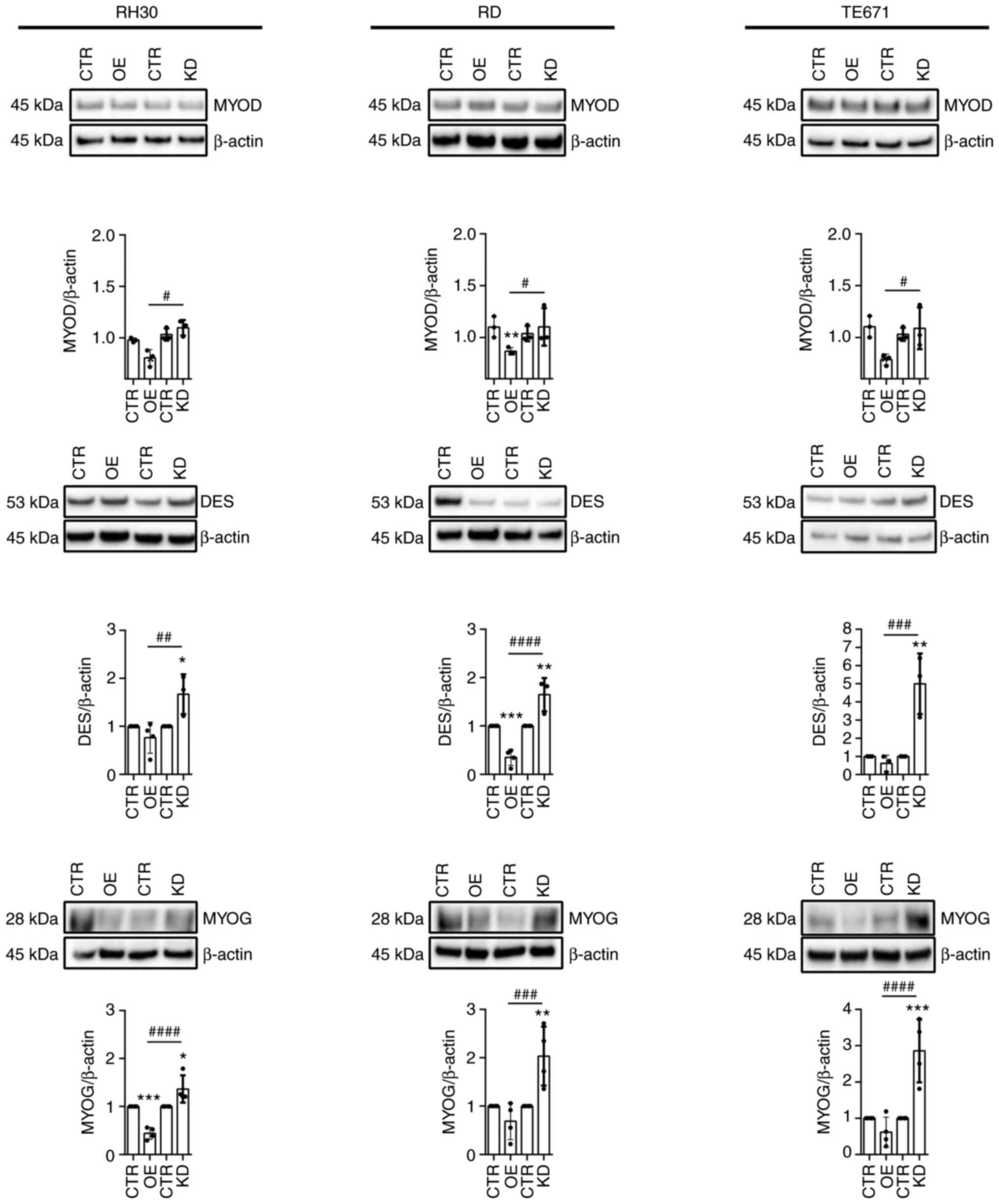 | Figure 4Expression of muscle markers in
WNT5AOE and WNT5AKD RMS cell lines. Western blot analyses (top)
along with the respective protein quantification (bottom) of the
muscle markers MYOD, DES and MYOG of the human ARMS cell line RH30
and ERMS cell lines RD and TE671 stably overexpressing WNT5A or
stably expressing a WNT5A shRNA to induce a WNT5A knockdown. Cell
lines transduced with the respective empty vectors (pBABE or pGIPZ)
served as controls. Detection of β-Actin served as loading control.
For protein quantification from ≥3 independent experiments, values
of CTR were set to 1. Bars are mean ± SD; statistical analysis was
performed by one-way ANOVA. *indicates significance vs.
CTR; #indicates significance between OE and KD.
*/#P<0.05,
**/##P<0.01,
***/###P<0.001,
####P<0.0001. RMS, rhabdomyosarcoma; ARMS, alveolar
RMS; ERMS, embryonal RMS; CTR, control; OE, overexpressing; KD,
knockdown. |
WNT5A inhibits sphere formation of RMS
cells
It has been demonstrated that ERMS cell lines RD and
TE671 and RH30 cells form rhabdospheres. Since rhabdospheres show a
much higher tumorigenicity than adherent cells (29,34),
the rhabdosphere colony formation assay is a powerful assay to
analyze the self-renewal potential of RMS and to identify signaling
pathways, which are essential for growth and maintenance of these
stem cell-like and tumor-propagating RMS cells. After 7 days in
culture, all cell lines had robustly formed rhabdospheres,
independently of whether they were genetically manipulated or not.
However, WNT5AOE spheres of all cell lines were smaller compared
with control or WNT5AKD spheres. This phenomenon was already
observed after 1 week in culture (Fig.
5A, shown by images in the upper panel). Notably, rhabdosphere
numbers of WNT5AOE cells were also significantly lower compared
with the control, whereas the numbers of WNT5AKD cells were
significantly higher (Fig. 5A,
lower panels). For RH30 cells, this characteristic was already
observed after 1 week in culture (Fig.
5A, lower left panel) and for RD rhabdospheres after 2 weeks in
culture (Fig. 5A, lower middle
panel). To some extend this was also observed with TE671 cells
after 8 weeks (Fig. 5A, lower
right panel). Together, these data indicate that WNT5A inhibit
self-renewal potential and maintenance of tumor-propagating RMS
cells.
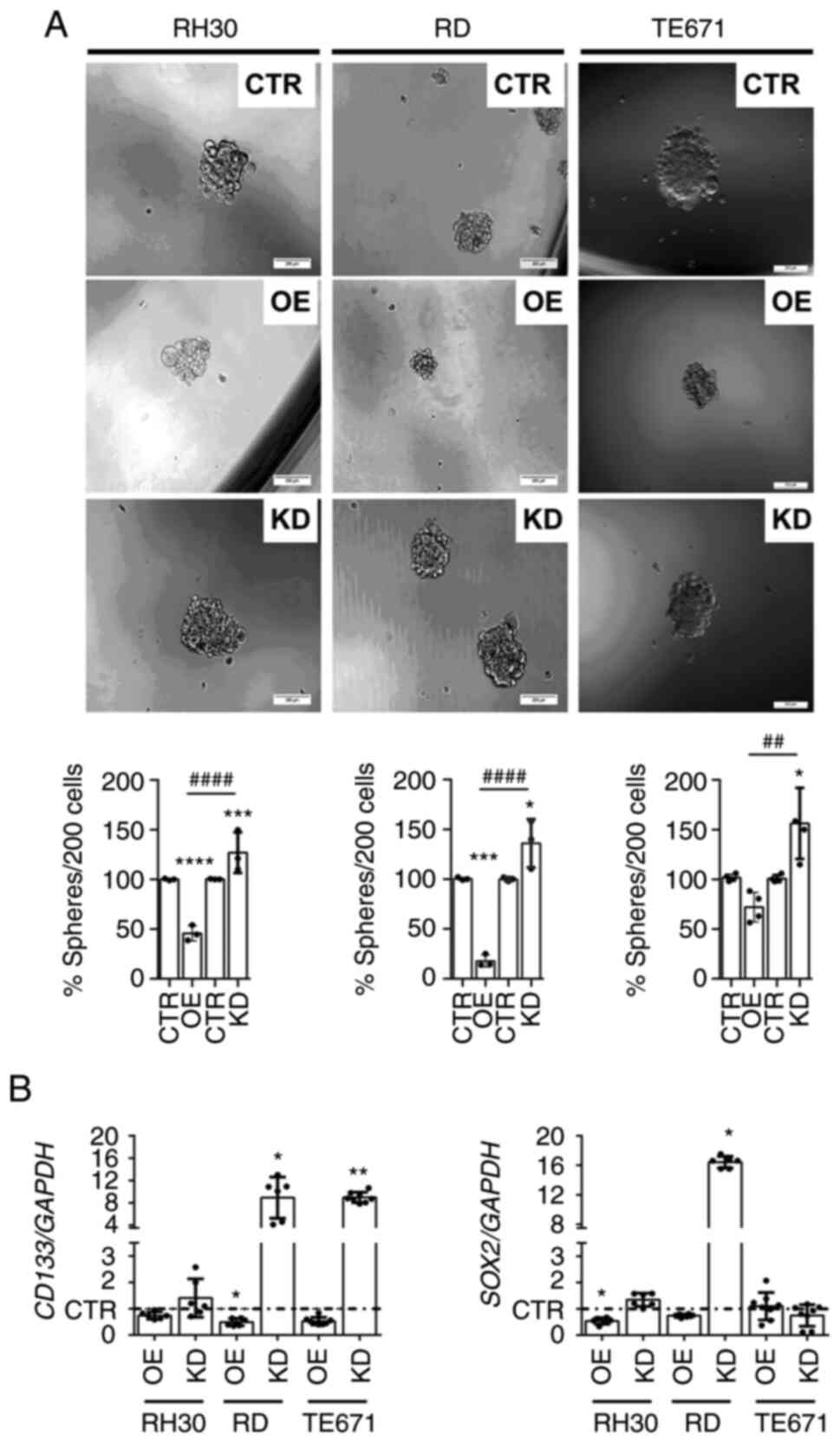 | Figure 5Sphere formation capacity and stem
cell marker expression of WNT5AOE and WNT5AKD RMS cell lines. (A)
Representative phase contrast pictures (top) and quantification
(bottom) of sphere cultures and (B) Reverse
transcription-quantitative PCR-based quantification of CD133
and SOX2 expression levels of 2D-cultured human ARMS cell
line RH30 and ERMS cell lines RD and TE671 stably overexpressing
WNT5A or stably expressing a WNT5A shRNA to induce a WNT5A
knockdown in relation to cell lines transduced with the respective
empty vectors (pBABE or pGIPZ) served as controls (CTR). (A)
Pictures were taken after 1-week sphere culture (top),
quantification was performed after 1-, 2- or 8-weeks sphere culture
of RH30, RD or TE671 cells, respectively (bottom) of 5 independent
experiments. Scale bars=200 µm. (B) Results represent data
from ≥3 independent experiments each measured in duplicates. For
sphere quantification, values of CTR were set to 100% in (A). CTR
values of the gene expression analyses were set to 1 and are
presented as a dashed line to ensure greater clarity (B). Bars are
mean ± SD; statistical analysis in (A) was performed by one-way
ANOVA and that in (B) by Student's t-test. *indicates
significance vs. CTR; #indicates significance between OE
and KD. *P<0.05,
**/##P<0.01, ***P<0.001,
****/####P<0.0001. OE, overexpressing; KD,
knockdown. RMS, rhabdomyosarcoma; ARMS, alveolar RMS; ERMS,
embryonal RMS; sh, short hairpin; CTR, control. |
The present study also analyzed the expression of
the stem cell markers CD133 and SOX2, which both
serve a role in RMS colony formation (29,35).
WNT5AOE inhibited expression of CD133 in all RMS cell lines,
which was significant for RD WNT5AOE cells. By contrast,
CD133 is overexpressed in WNT5AKD cells, which was
significant for RD WNT5AKD and TE671 WNT5AKD cells (Fig. 5B, left panel). In addition,
SOX2 expression is significantly lower in RH30 WNT5AOE cells
and is significantly higher in RD WNT5AKD cells (Fig. 5B). These data indicate that WNT5A
can modulate the expression of stem cell markers in RMS cells.
WNT5A destabilizes or downregulates
β-Catenin
As WNT5A can destabilize β-Catenin (11,36,37),
the present study next analyzed the quantity and distribution of
active non-phosphorylated β-Catenin protein in WNT5AOE and WNT5AKD
cells. As shown in Fig. 6A the
β-Catenin protein level was slightly decreased in WNT5AOE cells,
whereas it was unequivocally increased in WNT5AKD cells (Fig. 6A). When the distribution of
β-Catenin was investigated it appeared that both the cytoplasmic
level of active β-Catenin and the nuclear level (at least in RH30
and TE671 cells) were decreased in WNT5AOE cells, whereas they were
elevated in WNT5AKD cells compared with the control (Fig. 6B, upper and lower panel). This is
different at the mRNA level. As shown in Fig. 6B, WNT5AOE did not block
transcription of β-Catenin mRNA CTNNB1 in any of the cell
lines (Fig. 6B; indeed, in RD
WNT5AOE cells CTNNB1 is rather induced). By contrast,
WNT5AKD did not induce CTNNB1 in RH30 and RD cells. The
exception was TE671 cells, in which the WNT5AKD resulted in
upregulation of CTNNB1 mRNA. This, by contrast to all other
cell lines, accompanies significant induction of the β-Catenin
downstream target cMYC. By contrast, the expression of
AXIN2 was not much altered by WNT5AOE or WNT5AKD (Fig. 6B) and if so, then AXIN2
expression was upregulated.
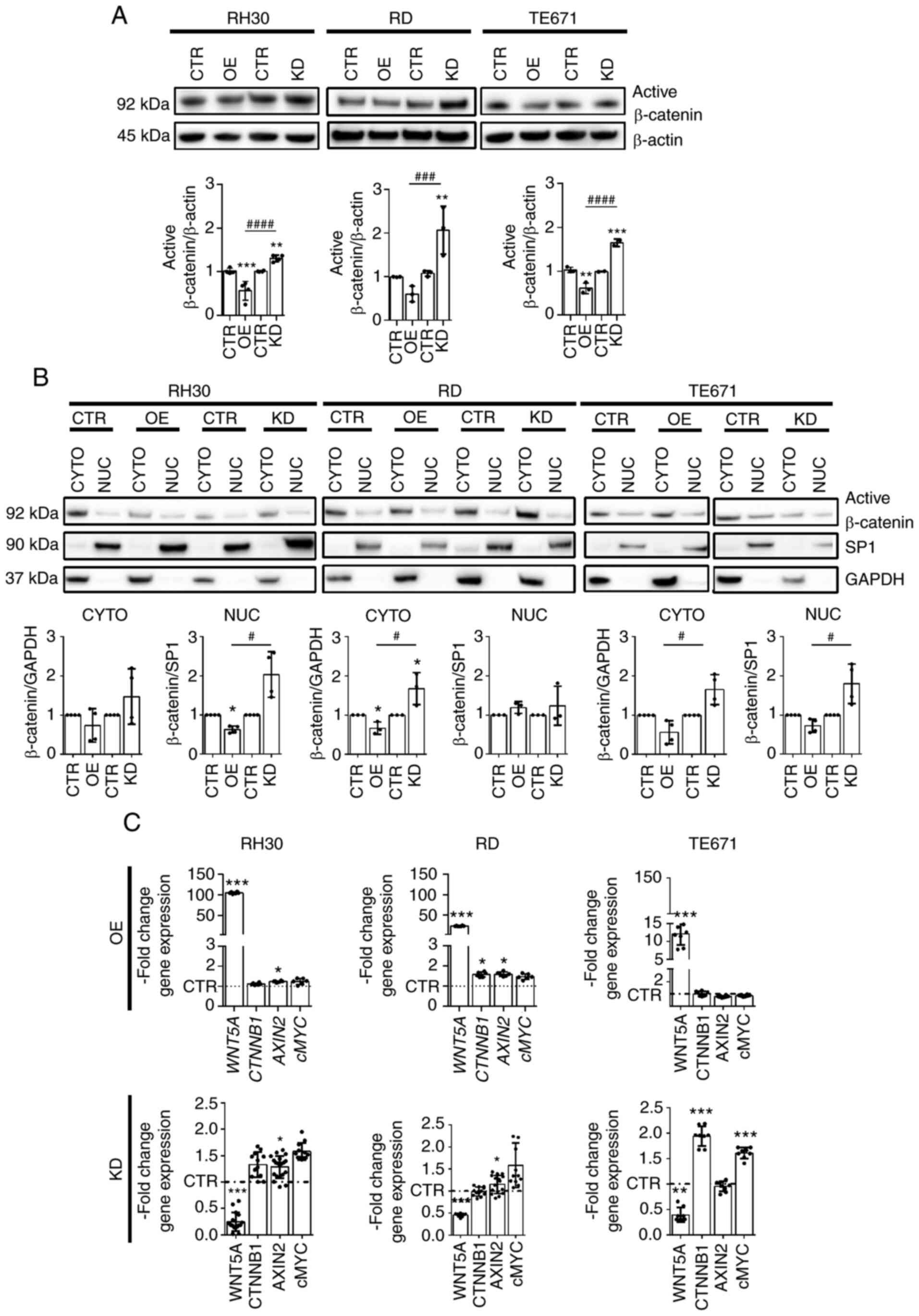 | Figure 6Stability of β-Catenin protein in
WNT5AOE and WNT5AKD RMS cell lines. (A and B) Representative
western blot analyses (top) along with the respective protein
quantification (from ≥3 independent experiments; bottom) of active
β-Catenin protein levels in (A) whole cell lysates and (B)
cytoplasmic and nuclear fractions and (C) Reverse
transcription-quantitative PCR analyses of WNT5A,
CTNNB1, AXIN2 and cMYC expression of the human
ARMS cell line RH30 and ERMS cell lines RD and TE671 stably
overexpressing WNT5A or stably expressing a WNT5A shRNA to induce a
WNT5A knockdown in relation to cell lines transduced with the
respective empty vectors (pBABE or pGIPZ), which served as
controls. Detection of (A) β-Actin for whole lysates, (B) SP1 for
nuclear and GAPDH for cytoplasmic fractions served as loading
controls. (C) Gene expression levels were analyzed in ≥3
independent expriments each measured in duplicates and normalized
to GAPDH expression levels. For protein and transcript
quantification, values of CTR were set to 1 and in (C) the CTR
values are presented as a dashed line to ensure greater clarity.
Bars are mean ± SD; statistical analysis in (A) and (B) was
performed by one-way ANOVA and in (C) by Student's t-test.
*indicates significance vs. CTR; #indicates
significance between OE and KD. */#P<0.05,
**P<0.01, ***/###P<0.001,
####P<0.0001. OE, overexpressing; KD, knockdown;
CYTO, cytoplasmic; NUC, nuclear; sh, short hairpin; CTR,
control. |
In summary, these data demonstrate that WNT5AOE
decreased the β-Catenin protein level in all RMS cell lines,
whereas the WNT5AKD increased it. In almost all settings, this
β-Catenin regulation is not correlated with alterations of the
respective CTNNB1 mRNA levels, indicating that WNT5A can
destabilize β-Catenin protein. The exception is TE671 WNT5AKD
cells, in which the increase in β-Catenin protein is associated
with increased CTNNB1 transcription. This shows that,
depending on the cellular context, WNT5A can also modulate
CTNNB1 transcription.
Discussion
WNT signaling pathways control a number of
developmental processes and contribute to a variety of human
cancers. Numerous studies on the WNT/β-Catenin pathway have been
published and also encompass those in childhood tumors (17,38-40).
However, knowledge about the role of WNT5A and non-canonical WNT
signaling and the interaction with canonical WNT/β-Catenin pathway
in these tumors is sparse, particularly in RMS.
WNT5A can have either tumor suppressive or tumor
promoting functions, which depends on the tumor entity. For
example, WNT5A is a reliable marker for aggressive growth behavior
and invasiveness of nasopharyngeal cancer (41), gastric cancer (42), oral squamous cell carcinoma
(43) and melanoma (44,45),
whereas it has tumor suppressive activity in thyroid carcinoma
(46), prostate cancer (47), colorectal (48) and breast cancer (49). Indeed, it has been hypothesized
that the function of WNT5A as an oncogene or a tumor suppressor
gene is dependent on the receptor context of the respective tumors
(45,46,50).
The present study revealed that cell
membrane-tethered and most likely also secreted WNT5A in RMS is
rather tumor suppressive, since it inhibited proliferation,
migration, the expression of stemness markers (e.g., CD133)
and concomitantly sphere formation of RMS cells. These conclusions
were based on in vitro experiments using RMS cell lines that
overexpress WNT5A (WNT5AOE) or harbor a WNT5A knockdown (WNT5AKD).
Although it is not currently known if cell membrane-tethered and
secreted WNT5A acts through similar mechanisms, the WNT5A-mediated
tumor suppressive effects are similar to those achieved in colon
cancer (51) and hepatocellular
carcinoma cells (52), in which
WNT5AOE effectively inhibits cell proliferation. Furthermore, also
in accordance with the present study, Cheng et al (51) observed reduced migration of colon
cancer cells after WNT5AOE and increased migration after WNT5AKD.
In addition, the authors showed an antagonizing effect of WNT5A on
the canonical WNT/β-Catenin pathway with decreased expression of
cytoplasmic/nuclear β-Catenin after WNT5AOE (51). Therefore, at first glance, the
WNT5A-induced inhibition of the WNT/β-Catenin pathway seemed to be
associated with a decrease in cell proliferation and migration in
both colon cancer and RMS cells. However, this is rather not the
case, since a recent study showed decreased proliferation with
concomitant activation of WNT/β-Catenin signaling in RMS cell lines
deficient for DKK1 (15) and since
we recently showed that canonical WNT signaling and β-Catenin do
not affect aggressiveness of RMS cell lines or in a murine RMS
model (17).
WNT5A may also inhibit the expression of muscle
differentiation markers, such as MYOD, DES and MYOG. Although not
all results were significant and muscle marker expression was
apparently influenced by transduction of the empty vectors, the
present study found that WNT5AOE in general downregulated these
factors, whereas WNT5AKD increased their expression at least on
protein level (Fig. 3B). These
data supported a tumor suppressive role of WNT5A, because at least
MYOD has an unexpected oncogenic function in RMS (33).
As already mentioned, WNT5A-mediated inhibition of
the canonical WNT signaling pathway depends on the expression of
specific receptors and is highly complex. Until now, we did not
analyze the non-canonical signaling pathways that are activated in
RMS cells by WNT5A. However, it has been shown that WNT5A-mediated
β-Catenin degradation can involve the induction of the E3
ubiquitin-protein ligase SIAH2 (53). Furthermore, an interaction of WNT5A
with FZD2 can activate WNT/Ca2+ signaling and can
concomitantly block binding of WNT3A by internalization of FZD2,
which results in inhibition of β-Catenin accumulation and thus
inhibition of canonical WNT Signaling (11). Moreover, binding of WNT5A to FZD4,
which normally induces β-Catenin-dependent signaling, can be
blocked in the presence of ROR2 (54) that is also involved in activation
of WNT5A/JNK signaling (55).
Notably, RH30, RD and TE671 cell lines express ROR2 and FZD2
(Fig. S1D). Because
WNT5A-mediated activation of these receptors may involve
destabilization of β-Catenin (11,36,37),
it will be useful to investigate whether these receptors and their
downstream signaling pathways are indeed involved in WNT5A-mediated
inhibition of canonical WNT/β-Catenin signaling in the RMS cell
lines RD and RH30. In TE671 cells, on the other hand, WNT5A also
appeared to regulate β-Catenin on transcriptional level, because
CTNNB1 mRNA (and its target cMYC) was significantly
upregulated in TE671 WNT5AKD cells.
In the present study, WNT5A also counteracted RMS
stemness because it is negatively associated with RMS sphere
formation and CD133 expression. This is also seen in colon
cancer cells, in which WNT5A is frequently silenced. In this
tumor entity, reactivation of WNT5A leads to inhibition of tumor
cell clonogenicity, which is due to antagonizing the WNT/β-Catenin
pathway (56,57). However, in RMS this mechanism is
rather unlikely, because WNT3A does not alter RMS sphere formation
(17).
Together, the present study revealed WNT5A
expression in RMS in vivo and a tumor suppressive function
of WNT5A in one ARMS and two ERMS cell lines in vitro, where
WNT5A also inhibited migration and stem cell properties. This
accompanied a decrease in MYOD, MYOG and DES expression and of
active β-Catenin. How these processes are regulated and whether
canonical and/or non-canonical WNT pathways are involved is
currently unclear and more mechanistic data are needed for an
improved understanding of the role of WNT5A in RMS. Thus, it
remains to be analyzed if secreted WNT5A affects tumor
proliferation via other mechanisms compared with cell
membrane-tethered WNT5A. It also will be useful to investigate
whether the WNT5A-induced changes in muscle marker and β-Catenin
expression are directly involved in the anti-tumoral activities of
WNT5A.
Supplementary Data
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
NR, HH and KK conceived the present study. NR, JB
and KK were responsible for the methodology and investigation NR,
JB and KK. HH and KK wrote the manuscript, which was reviewed and
edited by AU and AM. HH and KK were responsible for project
administration and funding acquisition. NR, KK and HH confirm the
authenticity of all the raw data. All authors reviewed and approved
the final manuscript.
Ethics approval and consent to
participate
TMA studies and the use of normal muscle were
authorized by the approval 158/2009/b02; University of Tübingen,
Tübingen, Germany; April 2, 2009 within the framework of the CWS
and that for the RT-qPCR studies additionally by the approval
2017-802R-MA (University of Heidelberg, University Medical Centre
Mannheim). Written informed consent, according to the Declaration
of Helsinki, was obtained from all patients or their legal
guardians, depending on the age of the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors thank Ms. Anke Frommhold and Ms. Ina
Heß (Institute of Human Genetics, University Medical Center
Goettingen) for excellent technical assistance.
Funding
The present study was funded by the grant no. 2017.110.1 from
the Wilhelm-Sander-Stiftung to HH and KSK and by the grant no. DFG
HA 2197/9-2 from the DFG to HH.
References
|
1
|
Dagher R and Helman L: Rhabdomyosarcoma:
An overview. Oncologist. 4:34–44. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Skapek SX, Ferrari A, Gupta AA, Lupo PJ,
Butler E, Shipley J, Barr FG and Hawkins DS: Rhabdomyosarcoma. Nat
Rev Dis Primers. 5:12019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Williamson D, Missiaglia E, de Reyniès A,
Pierron G, Thuille B, Palenzuela G, Thway K, Orbach D, Laé M,
Fréneaux P, et al: Fusion gene-negative alveolar rhabdomyosarcoma
is clinically and molecularly indistinguishable from embryonal
rhabdomyosarcoma. J Clin Oncol. 28:2151–2158. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hettmer S and Wagers AJ: Muscling in:
Uncovering the origins of rhabdomyosarcoma. Nat Med. 16:171–173.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Girardi F and Le Grand F: Wnt signaling in
skeletal muscle development and regeneration. Prog Mol Biol Transl
Sci. 153:157–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martinez-Font E, Pérez-Capó M, Vögler O,
Martin-Broto J, Alemany R and Obrador-Hevia A: WNT/β-catenin
pathway in soft tissue sarcomas: New therapeutic opportunities?
Cancers (Basel). 13:55212021. View Article : Google Scholar
|
|
7
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Clevers H and Nusse R: Wnt/beta-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De A: Wnt/Ca2+ signaling pathway: A brief
overview. Acta Biochim Biophys Sin (Shanghai). 43:745–756. 2011.
View Article : Google Scholar
|
|
10
|
Thiele S, Rachner TD, Rauner M and
Hofbauer LC: WNT5A and its receptors in the bone-cancer dialogue. J
Bone Miner Res. 31:1488–1496. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sato A, Yamamoto H, Sakane H, Koyama H and
Kikuchi A: Wnt5a regulates distinct signalling pathways by binding
to frizzled2. EMBO J. 29:41–54. 2010. View Article : Google Scholar :
|
|
12
|
Bouron-Dal Soglio D, Rougemont AL, Absi R,
Giroux LM, Sanchez R, Barrette S and Fournet JC: Beta-catenin
mutation does not seem to have an effect on the tumorigenesis of
pediatric rhabdomyosarcomas. Pediatr Dev Pathol. 12:371–373. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh S, Vinson C, Gurley CM, Nolen GT,
Beggs ML, Nagarajan R, Wagner EF, Parham DM and Peterson CA:
Impaired Wnt signaling in embryonal rhabdomyosarcoma cells from
p53/c-fos double mutant mice. Am J Pathol. 177:2055–2066. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Annavarapu SR, Cialfi S, Dominici C, Kokai
GK, Uccini S, Ceccarelli S, McDowell HP and Helliwell TR:
Characterization of Wnt/β-catenin signaling in rhabdomyosarcoma.
Lab Invest. 93:1090–1099. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Giralt I, Gallo-Oller G, Navarro N,
Zarzosa P, Pons G, Magdaleno A, Segura MF, Sábado C, Hladun R,
Arango D, et al: Dickkopf-1 inhibition reactivates Wnt/β-catenin
signaling in rhabdomyosarcoma, induces myogenic markers in vitro
and impairs tumor cell survival in vivo. Int J Mol Sci.
22:129212021. View Article : Google Scholar
|
|
16
|
Nitzki F, Cuvelier N, Dräger J, Schneider
A, Braun T and Hahn H: Hedgehog/patched-associated rhabdomyosarcoma
formation from delta1-expressing mesodermal cells. Oncogene.
35:2923–2931. 2016. View Article : Google Scholar
|
|
17
|
Ragab N, Viehweger F, Bauer J, Geyer N,
Yang M, Seils A, Belharazem D, Brembeck FH, Schildhaus HU, Marx A,
et al: Canonical WNT/β-catenin signaling plays a subordinate role
in rhabdomyosarcomas. Front Pediatr. 6:3782018. View Article : Google Scholar
|
|
18
|
Surmann-Schmitt C, Widmann N, Dietz U,
Saeger B, Eitzinger N, Nakamura Y, Rattel M, Latham R, Hartmann C,
von der Mark H, et al: Wif-1 is expressed at cartilage-mesenchyme
interfaces and impedes Wnt3a-mediated inhibition of chondrogenesis.
J Cell Sci. 122:3627–3637. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
von Maltzahn J, Chang NC, Bentzinger CF
and Rudnicki MA: Wnt signaling in myogenesis. Trends Cell Biol.
22:602–609. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kephart JJ, Tiller RG, Crose LE, Slemmons
KK, Chen PH, Hinson AR, Bentley RC, Chi JT and Linardic CM:
Secreted frizzled-related protein 3 (SFRP3) is required for
tumorigenesis of PAX3-FOXO1-positive alveolar rhabdomyosarcoma.
Clin Cancer Res. 21:4868–4880. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen X, Stewart E, Shelat AA, Qu C,
Bahrami A, Hatley M, Wu G, Bradley C, McEvoy J, Pappo A, et al:
Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer
cell. 24:710–724. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Davicioni E, Anderson MJ, Finckenstein FG,
Lynch JC, Qualman SJ, Shimada H, Schofield DE, Buckley JD, Meyer
WH, Sorensen PH and Triche TJ: Molecular classification of
rhabdomyosarcoma-genotypic and phenotypic determinants of
diagnosis: A report from the Children's oncology group. Am J
Pathol. 174:550–564. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dräger J, Simon-Keller K, Pukrop T, Klemm
F, Wilting J, Sticht C, Dittmann K, Schulz M, Leuschner I, Marx A
and Hahn H: LEF1 reduces tumor progression and induces
myodifferentiation in a subset of rhabdomyosarcoma. Oncotarget.
8:3259–3273. 2017. View Article : Google Scholar :
|
|
24
|
Najdi R, Proffitt K, Sprowl S, Kaur S, Yu
J, Covey TM, Virshup DM and Waterman ML: A uniform human Wnt
expression library reveals a shared secretory pathway and unique
signaling activities. Differentiation. 84:203–213. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Morgenstern JP and Land H: Advanced
mammalian gene transfer: High titre retroviral vectors with
multiple drug selection markers and a complementary helper-free
packaging cell line. Nucleic Acids Res. 18:3587–3596. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stewart SA, Dykxhoorn DM, Palliser D,
Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et
al: Lentivirus-delivered stable gene silencing by RNAi in primary
cells. RNA. 9:493–501. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Czarnek M, Sarad K, Karaś A, Kochan J and
Bereta J: Non-targeting control for MISSION shRNA library silences
SNRPD3 leading to cell death or permanent growth arrest. Mol Ther
Nucleic Acids. 26:711–731. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weng Y, Shi Y, Xia X, Zhou W, Wang H and
Wang C: A multi-shRNA vector enhances the silencing efficiency of
exogenous and endogenous genes in human cells. Oncol Lett.
13:1553–1562. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Walter D, Satheesha S, Albrecht P,
Bornhauser BC, D'Alessandro V, Oesch SM, Rehrauer H, Leuschner I,
Koscielniak E, Gengler C, et al: CD133 positive embryonal
rhabdomyosarcoma stem-like cell population is enriched in
rhabdospheres. PLoS One. 6:e195062011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Link AJ and LaBaer J: Trichloroacetic acid
(TCA) precipitation of proteins. Cold Spring Harb Protoc.
2011:993–994. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Simon-Keller K, Paschen A, Hombach AA,
Ströbel P, Coindre JM, Eichmüller SB, Vincent A, Gattenlöhner S,
Hoppe F, Leuschner I, et al: Survivin blockade sensitizes
rhabdomyosarcoma cells for lysis by fetal acetylcholine
receptor-redirected T cells. Am J Pathol. 182:2121–2131. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tenente IM, Hayes MN, Ignatius MS,
McCarthy K, Yohe M, Sindiri S, Gryder B, Oliveira ML, Ramakrishnan
A, Tang Q, et al: Myogenic regulatory transcription factors
regulate growth in rhabdomyosarcoma. Elife. 6:e192142017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Deel MD, Slemmons KK, Hinson AR, Genadry
KC, Burgess BA, Crose LES, Kuprasertkul N, Oristian KM, Bentley RC
and Linardic CM: The transcriptional coactivator TAZ is a potent
mediator of alveolar rhabdomyosarcoma tumorigenesis. Clin Cancer
Res. 24:2616–2630. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Slemmons KK, Crose LES, Riedel S,
Sushnitha M, Belyea B and Linardic CM: A novel notch-YAP circuit
drives stemness and tumorigenesis in embryonal rhabdomyosarcoma.
Mol Cancer Res. 15:1777–1791. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamamoto H, Yoo SK, Nishita M, Kikuchi A
and Minami Y: Wnt5a modulates glycogen synthase kinase 3 to induce
phosphorylation of receptor tyrosine kinase Ror2. Genes Cells.
12:1215–1223. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mikels A, Minami Y and Nusse R: Ror2
receptor requires tyrosine kinase activity to mediate Wnt5A
signaling. J Biol Chem. 284:30167–30176. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pedersen EA, Menon R, Bailey KM, Thomas
DG, Van Noord RA, Tran J, Wang H, Qu PP, Hoering A, Fearon ER, et
al: Activation of Wnt/β-catenin in ewing sarcoma cells antagonizes
EWS/ETS function and promotes phenotypic transition to more
metastatic cell states. Cancer Res. 76:5040–5053. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu T, Wang LN, Tang DR and Sun FY: SOST
silencing promotes proliferation and invasion and reduces apoptosis
of retinoblastoma cells by activating Wnt/β-catenin signaling
pathway. Gene Ther. 24:399–407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mavila N: Thundimadathil J. The emerging
roles of cancer stem cells and Wnt/beta-catenin signaling in
hepatoblastoma. Cancers (Basel). 11:14062019. View Article : Google Scholar
|
|
41
|
Qin L, Yin YT, Zheng FJ, Peng LX, Yang CF,
Bao YN, Liang YY, Li XJ, Xiang YQ, Sun R, et al: WNT5A promotes
stemness characteristics in nasopharyngeal carcinoma cells leading
to metastasis and tumorigenesis. Oncotarget. 6:10239–10252. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu JJ, Zhang YJ, Xu R, Du J, Hu Z, Yang
L, Chen Y, Zhu Y and Gu L: PI3K/Akt-dependent phosphorylation of
GSK3β and activation of RhoA regulate Wnt5a-induced gastric cancer
cell migration. Cell Signal. 25:447–456. 2013. View Article : Google Scholar
|
|
43
|
Prgomet Z, Axelsson L, Lindberg P and
Andersson T: Migration and invasion of oral squamous carcinoma
cells is promoted by WNT5A, a regulator of cancer progression. J
Oral Pathol Med. 44:776–784. 2015. View Article : Google Scholar
|
|
44
|
Sinnberg T, Levesque MP, Krochmann J,
Cheng PF, Ikenberg K, Meraz-Torres F, Niessner H, Garbe C and Busch
C: Wnt-signaling enhances neural crest migration of melanoma cells
and induces an invasive phenotype. Mol Cancer. 17:592018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Weeraratna AT, Jiang Y, Hostetter G,
Rosenblatt K, Duray P, Bittner M and Trent JM: Wnt5a signaling
directly affects cell motility and invasion of metastatic melanoma.
Cancer Cell. 1:279–288. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kremenevskaja N, von Wasielewski R, Rao
AS, Schöfl C, Andersson T and Brabant G: Wnt-5a has tumor
suppressor activity in thyroid carcinoma. Oncogene. 24:2144–2154.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Thiele S, Göbel A, Rachner TD, Fuessel S,
Froehner M, Muders MH, Baretton GB, Bernhardt R, Jakob F, Glüer CC,
et al: WNT5A has anti-prostate cancer effects in vitro and reduces
tumor growth in the skeleton in vivo. J Bone Miner Res. 30:471–480.
2015. View Article : Google Scholar
|
|
48
|
Castell A and Larsson LG: Targeting MYC
translation in colorectal cancer. Cancer Discov. 5:701–703. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Säfholm A, Tuomela J, Rosenkvist J, Dejmek
J, Härkönen P and Andersson T: The Wnt-5a-derived hexapeptide
Foxy-5 inhibits breast cancer metastasis in vivo by targeting cell
motility. Clin Cancer Res. 14:6556–6563. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Blanc E, Roux GL, Bénard J and Raguénez G:
Low expression of Wnt-5a gene is associated with high-risk
neuroblastoma. Oncogene. 24:1277–1283. 2005. View Article : Google Scholar
|
|
51
|
Cheng R, Sun B, Liu Z, Zhao X, Qi L, Li Y
and Gu Q: Wnt5a suppresses colon cancer by inhibiting cell
proliferation and epithelial-mesenchymal transition. J Cell
Physiol. 229:1908–1917. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang T, Liu X and Wang J: Up-regulation of
Wnt5a inhibits proliferation and migration of hepatocellular
carcinoma cells. J Cancer Res Ther. 15:904–908. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Topol L, Jiang X, Choi H, Garrett-Beal L,
Carolan PJ and Yang Y: Wnt-5a inhibits the canonical Wnt pathway by
promoting GSK-3-independent beta-catenin degradation. J Cell Biol.
162:899–908. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mikels AJ and Nusse R: Purified Wnt5a
protein activates or inhibits beta-catenin-TCF signaling depending
on receptor context. PLoS Biol. 4:e1152006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Oishi I, Suzuki H, Onishi N, Takada R,
Kani S, Ohkawara B, Koshida I, Suzuki K, Yamada G, Schwabe GC, et
al: The receptor tyrosine kinase Ror2 is involved in non-canonical
Wnt5a/JNK signalling pathway. Genes Cells. 8:645–654. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Osman J, Bellamkonda K, Liu Q, Andersson T
and Sjölander A: The WNT5A agonist foxy5 reduces the number of
colonic cancer stem cells in a xenograft mouse model of human
colonic cancer. Anticancer Res. 39:1719–1728. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ying J, Li H, Yu J, Ng KM, Poon FF, Wong
SC, Chan AT, Sung JJ and Tao Q: WNT5A exhibits tumor-suppressive
activity through antagonizing the Wnt/beta-catenin signaling, and
is frequently methylated in colorectal cancer. Clin Cancer Res.
14:55–61. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nirdé P, Derocq D, Maynadier M, Chambon M,
Basile I, Gary-Bobo M and Garcia M: Heat shock cognate 70 protein
secretion as a new growth arrest signal for cancer cells. Oncogene.
29:117–127. 2010. View Article : Google Scholar :
|