There were an estimated 19.3 million new cases and
10 million cancer-related deaths in 2020, causing a great burden
worldwide. The prevalence and mortality of cancer are also rapidly
increasing (1). Therefore, there
is an urgent need to develop effective cancer therapies. Although
immunotherapy, particularly cytotoxic T lymphocyte-associated
protein 4 inhibitors and programmed death 1 (PD-1)/programmed
death-ligand 1 (PD-L1) inhibitors have been proven to be effective
in cancer therapy, the presence of immune-mediated side effects
(e.g., myocarditis, colitis, pruritus, hepatitis) limits their use
in clinical practice (2,3). Furthermore, chemoresistance is
becoming a key obstacle for effective cancer therapy. Therefore,
more potential cancer targets should be identified to improve
future cancer therapy in addition to investigating better
combinatorial strategies for cancer therapy.
Histone methylation is a major type of
post-translational modification that has an important role in
epigenetic modification and contributes to numerous biological
processes, particularly carcinogenesis (4). Methyl groups may be added to the
side chains of arginine, lysine or histidine residues of histones
during histone methylation, among which methylation on lysine
residues is the most common (5).
In addition, the methylated lysine residues of histones may exhibit
mono-, di- or tri-methylated patterns (me1/me2/me3) (6–8).
The lysine (K)-specific demethylase (KDM) family of
proteins are histone demethylases that have the ability to remove
methyl groups from lysine residues, which are in turn involved in
numerous biological processes and diseases, such as development,
differentiation, neurological diseases and cancer (9). Histone lysine methylation and
demethylation are post-translational modifications that are highly
specific to the site and degree of methylation (6-8,10-14). The
presence of histone lysine demethylases has been debated for
numerous years, until lysine-specific demethylase 1 (LSD1/KDM1A)
was discovered. LSD1/KDM1A, which belongs to the flavin adenine
dinucleotide (FAD)-dependent lysine-specific histone demethylases,
was characterized as the first histone lysine demethylase with the
ability to mediate histone 3 lysine 4 (H3K4) demethylation
(15–17). Furthermore, the KDM2 to KDM8
families belong to the Jumonji (JmjC) domain-containing histone
demethylases. Similar to FAD-dependent lysine-specific histone
demethylases, JmjC domain-containing histone demethylases also
contribute to various biological processes by catalyzing
demethylation on histone lysine (18–24).
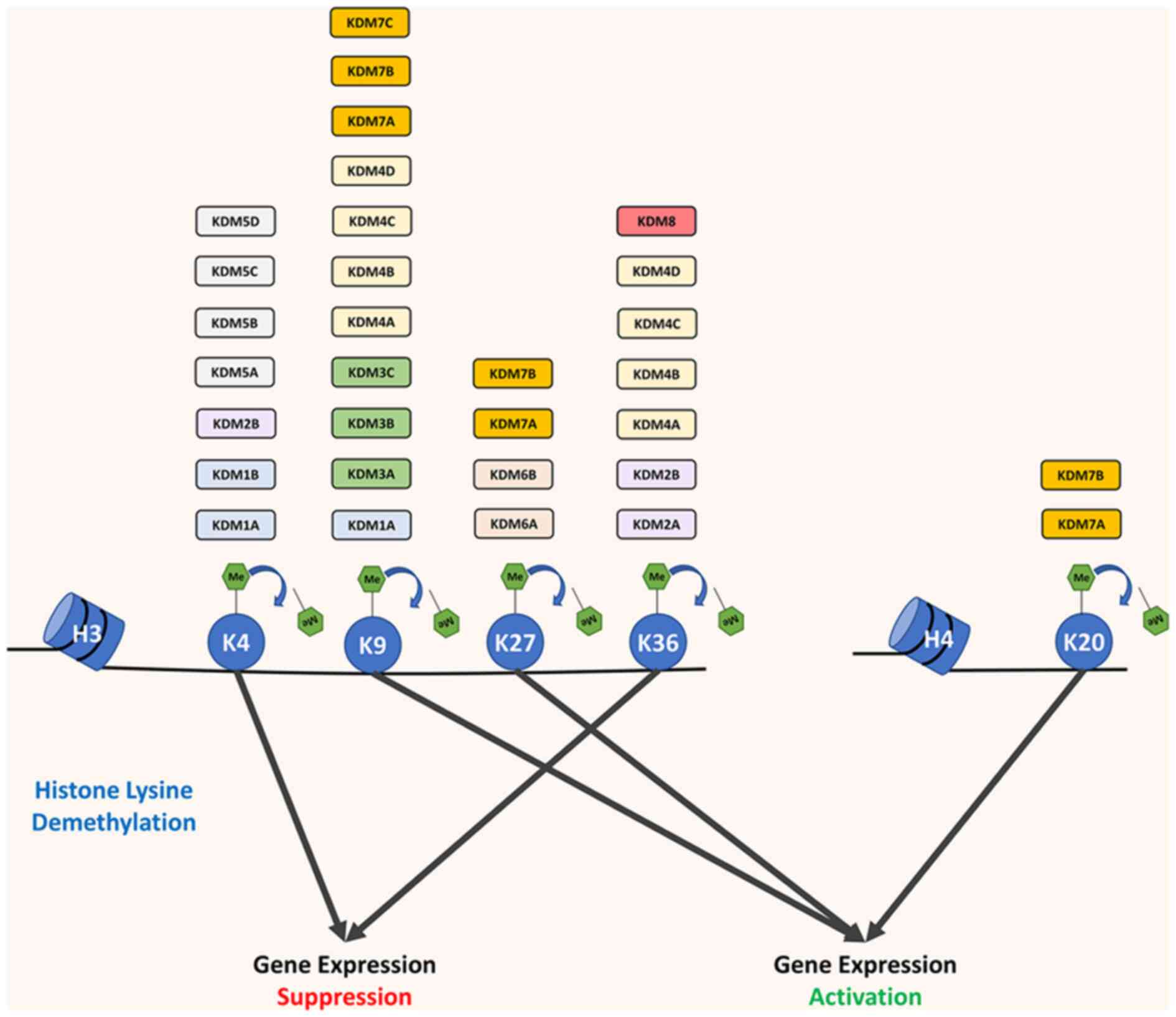
Different histone demethylases would target
different sites of histone lysine and demethylation on different
sites of histone lysine would have different effects on downstream
gene expression (Fig. 1). Since
methylated H3K4 and methylated H3K36 are activating factors for
gene expression, demethylation on H3K4 or H3K36 would repress
downstream gene expression (25–27). However, demethylation on H3K9,
H3K27 or H4K20 would contribute to downstream gene activation
(Fig. 1) (27).
Overall, histone lysine methylation is closely
associated with histone lysine demethylases and participates in
gene expression regulation. As a result, histone demethylation
performed by histone demethylases has an important role in numerous
biological processes, particularly cancer development. In the
present review, the role of histone lysine demethylases in cancer
is discussed and their potential as a target for cancer therapy is
further illustrated.
The KDM1 family consists of KDM1A (also named LSD1)
and KDM1B (also named LSD2). Both KDM1A and KDM1B have a
FAD-dependent amine oxidase domain and SWIRM domain. Furthermore,
KDM1A also contains a Tower domain, which is responsible for
protein interaction (28). The
FAD-dependent amine oxidase domain is responsible for removing a
methyl group from monomethylated (me1) or dimethylated (me2) lysine
residues, while the SWIRM domain is responsible for assisting
demethylation. Both KDM1A and KDM1B are able to catalyze the
demethylation of H3K4 with mono-methylation or di-methylation
(H3K4me1/me2) (29). However,
apart from demethylating H3K4me1/me2, KDM1A is also able to
catalyze H3K9me1/me2 demethylation (30,31).
In general, KDM1A was indicated to be overexpressed
and associated with poor prognosis in a variety of cancers,
indicating the oncogenic role of KDM1A (32–35). Therefore, numerous studies have
been performed to elucidate how KDM1A contributes to cancer
development and progression. First, KDM1A was reported to regulate
the cell cycle, which in turn modulated tumor growth. In an early
study, KDM1A was indicated to remove dimethylation at the K370 of
p53 to inhibit its interaction with p53 binding protein 1, thus
inhibiting apoptosis and promoting tumor cell growth (36). In addition, KDM1A-dependent
demethylation of myosin phosphatase target subunit 1 (MYPT1)
destabilized MYPT1 and reduced its expression level. Thus,
downregulation of MYPT1 led to retinoblastoma protein 1
phosphorylation, finally enhancing the G1/S transition of cancer
cells (37). In addition, KDM1A
has the ability to reduce hypoxia-inducible factor 1α (HIF-1α)
degradation and maintain HIF-1α protein levels, thus promoting
tumor growth (38). Furthermore,
it was recently reported that the immune landscape is regulated by
KDM1A by modulating the expression of immune checkpoint regulators
and related chemokines, such as PD-L1, C-C motif chemokine ligand
5, C-X-C motif chemokine ligand 9 (CXCL9) and CXCL10 (39). Apart from these three most studied
mechanisms, a variety of downstream genes regulated by KDM1A (E2F1,
STAT3 and AGO2) were identified to participate in cancer
development (35).
Unlike KDM1A, only a small number of studies have
identified the role of KDM1B in cancer. KDM1B is overexpressed in
cancers, such as breast cancer (40), colorectal cancer (41) and lung cancer (42), functions in tumor growth and
correlates with poor prognosis by catalyzing H3K4 demethylation
(43). According to the limited
literature, the ability of KDM1B to inhibit apoptosis in a
demethylation-dependent manner is the key mechanism for tumor
growth and progression (40,41,43,44).
KDM2A and KDM2B belong to the KDM2 family, the early
discovered JmjC domain-containing proteins. Both KDM2A and KDM2B
have a JmjC domain and one plant homeodomain (PHD) (29). However, KDM2A has H3K36me2
demethylation activity, while KDM2B demethylates H3K4me3 and
H3K36me2 (18,45). KDM2A and KDM2B were indicated to
be overexpressed in cancer tissues and contribute to tumor growth
and progression in various malignancies, including colorectal
cancer, gastric cancer, ovarian cancer and cervical cancer
(46–55).
KDM2A mediates H3K36me2 demethylation at the histone
deacetylase 3 (HDAC3) promoter, thereby suppressing HDAC3
expression and promoting carcinogenesis and invasiveness of lung
cancer (56). Similarly, KDM2A
was observed to repress dual-specificity phosphatase 3 (DUSP3)
expression through KDM2A-dependent H3K36me2 demethylation at the
DUSP3 promoter, which enhanced the ERK1/2 signaling pathway and
facilitated lung tumorigenesis (25). In breast cancer, KDM2A promoted
cancer stemness and angiogenesis through the upregulation of
signaling molecules, such as Jagged1 and Notch receptor 1 (NOTCH1),
in a demethylation-dependent manner, hence leading to poor
prognosis (57). Likewise, recent
research has also indicated that higher KDM2A expression in
cancer-associated fibroblasts is associated with advanced tumor
stage and poor survival in patients with breast cancer (58).
KDM2B was able to epigenetically suppress the
expression of Mps1 binding protein, an important component of the
Hippo pathway, contributing to the progression of pancreatic cancer
and leading to poor prognosis (59). In addition, among lung and
pancreatic cancer cell lines, KDM2B participates in TGF-β induced
epithelial-mesenchymal transition, contributing to cancer invasion
and metastasis (60). In
malignant hematopoiesis, knocking down of KDM2B markedly reduced
cell proliferation in vitro. Furthermore, knocking down
KDM2B delayed or even abrogated leukemogenesis in humanized
xenograft models (61). Several
studies have indicated the oncogenic role of KDM2B. However, one
study identified the tumor-suppressive effect of KDM2B, as
silencing of KDM2B triggered invasion of breast cancer cell lines
(62).
The KDM3 family is composed of three components:
KDM3A (also named JMJD1A), KDM3B (also named JMJD1B) and KDM3C
(also named JMJD1C). All of the three demethylases contain a JmjC
domain at the C terminal, a zinc-finger domain and an LXXLL motif.
The JmjC domain is responsible for histone demethylation, while the
zinc-finger domain and LXXLL motif are separately responsible for
DNA binding and nuclear receptor interaction (63). Among these demethylases, KDM3A and
KDM3B were observed to specifically demethylate H3K9me1/me2 in
vitro and in vivo, whereas KDM3C mainly demethylated
H3K9me2 (64–67). Most studies performed to date
indicate the oncogenic role of the KDM3 family in various cancer
types.
In colorectal cancer, upregulation of KDM3A was
indicated to be associated with tumorigenesis, advanced stage and
poor prognosis (68,69). To achieve this effect, KDM3A
specifically demethylates H3K9me2, promoting Wnt/β-catenin pathway
activation in vitro (70).
In addition, H3K9me2 demethylation of the Hippo pathway was
facilitated by KDM3A and contributed to colorectal cancer
tumorigenesis (71). Among breast
cancers, KDM3A is essential for the tumorigenic growth of cancer
stem cells and promotes invasion by demethylating p53-K372me1 and
inhibiting p53 transcription (72). In another study focusing on breast
cancer, KDM3A was indicated to increase estrogen receptor (ER)
activity via demethylation of H3K9me2/1 and activation of ER target
genes, therefore facilitating tumor growth (73). Apart from colorectal cancer and
breast cancer, the oncogenic role of KDM3A was observed in prostate
cancer (74–76), lung cancer (77,78), pancreatic cancer (79), liver cancer (80,81) and Ewing sarcoma (82,83) through a variety of in vivo
and in vitro experiments.
To date, research on the relationship between KDM3B
and cancer is limited. In HepG2 cells, the expression of cyclin D1
decreased significantly, the cell cycle was mostly halted in the
G2/M phase and cell proliferation was reduced when KDM3B was
knocked down (84). In addition,
loss of KDM3B was associated with slower growth of
castration-resistant prostate cancer cells, although it did not
alter the androgen receptor signaling pathway (85). Recently, KDM3B was observed to
activate the Wnt/β-catenin signaling pathway, further enhancing the
invasion and metastasis of breast cancer (86). In hematopoietic malignancies,
KDM3B is recruited to the LIM domain-only protein 2 (lmo2) promoter
and transcriptionally activates lmo2, a hematopoietic oncogene that
promotes leukemogenesis (87). By
contrast, a recent study indicated that KDM3B is highly expressed
in patients with acute myeloid leukemia (AML) with favorable
prognoses, although KDM3B is highly expressed in hematopoietic
malignancies compared to solid tumors. Further examination
suggested that KDM3B has an important role in maintaining the
fusion protein promyelocytic leukemia/retinoic acid receptor-α
levels and the chromatin state during cell differentiation in a
demethylation-dependent manner, thus inhibiting acute promyelocytic
leukemia progression (88).
Of note, the oncogenic effects of KDM3C on solid
tumors and hematopoietic malignancies were identified, although
only a small number of studies on KDM3C exist. In AML, KDM3C was
able to be recruited by the fusion gene runt related transcription
factor 1 (RUNX1)/RUNX1 partner transcriptional co-repressor 1 and
demethylated H3K9me2, thus maintaining expression of the fusion
gene and its targeting genes, such as p21, fms related receptor
tyrosine kinase 1 and serine/threonine/tyrosine kinase 1, and
increasing AML cell proliferation (65). Similarly, the use of small
molecular modulators of KDM3C, which significantly decreased KDM3C
expression, was able to effectively inhibit AML cell growth
(89). Among esophageal and
colorectal cancers, KDM3C epigenetically sustained the expression
of yes-associated protein 1 (YAP1) and activating transcription
factor 2 separately and promoted tumor growth and metastasis
(90,91).
KDM4A (also named JMJD2A), KDM4B (also named
JMJD2B), KDM4C (also named JMJD2C), KDM4D (also named JMJD2D),
KDM4E (also named JMJD2E) and KDM4F belong to the KDM4 family. Of
these, KDM4A, KDM4B and KDM4C have catalytic JmjN and JmjC domains,
and non-catalytic PHD and Tudor domains, whereas KDM4D only has
catalytic domains (92). The KDM4
family has the ability to catalyze the demethylation of H3K9me2/me3
and H3K36me2/me3 (13,92). In a previous study, the KDM4
family, except for KDM4E and KDM4F with unclear functions in
cancer, is mainly overexpressed and acts as oncogenes in different
cancer cell lines and tissues (93–96).
KDM4A has a critical role in tumor growth and
invasion. KDM4A-mediated H3K9 demethylation has been reported to
contribute to androgen receptor activation and affect
transcriptional activation through demethylating H3K9me2/me3
(13). In another study,
researchers corroborated that KDM4A is responsible for the
epigenetic upregulation of YAP1 through recruitment by ETS variant
transcription factor 1, ultimately promoting tumor growth in
prostate cancer (97). In lung
cancers, KDM4A upregulated distal-less homeobox 5, thereby
activating the expression of the Myc gene and the downstream
Wnt/β-catenin signaling pathway to promote the growth, metastasis
and the occurrence of lung cancer (98). In gastric cancer, KDM4A was also
observed to promote tumor growth by suppressing apoptosis (99).
KDM4B is both functionally and structurally
homogeneous to KDM4A. However, the mechanism by which KDM4B
contributes to tumor growth, invasion and metastasis is not similar
to that of KDM4A. KDM4B was able to be upregulated by HIF-α,
further promoting G2/M phase transition by upregulating cyclin A1
(CCNA1) and downregulating WEE1 G2 checkpoint kinase. KDM4B was
also able to promote G1 phase transition by epigenetically
downregulating CCND1 through demethylating H3K9me2/me3, ultimately
promoting the proliferation of breast cancer (100). This process of KDM4B regulation
was also effective in promoting colorectal cancer growth (101). On the other hand, recent studies
have emphasized the significance of KDM4B in inducing glucose
uptake in tumor growth and progression (102,103). After the knockdown of KDM4B,
H3K9me3 levels at the promoter of glucose transporter 1 (GLUT1)
increased; thus, the expression of GLUT1 decreased, leading to a
reduction in glucose uptake in colon cancer cells (104).
KDM4C was able to remove the methyl group from
H3K9me2/me3. When accompanied by KDM1A, KDM4C contributed to
altering the expression of genes related to the androgen receptor
and promoting prostate carcinogenesis (105). A recent study indicated that
KDM4C served as an oncogene in glioblastoma with a dual function of
inactivating p53 by demethylating p53K372me1 and activating c-Myc
by directly binding to its promoter (106). In addition, similar to KDM4B,
KDM4C was also able to remove methyl groups of H3K9 on HIF-α and
promote tumor growth (107–109).
The KDM5 family includes four members, KDM5A (also
named JARID1A), KDM5B (also named JARID1B), KDM5C (also named
JARID1C), and KDM5D (also named JARID1D), having highly similar
structures. All members contain five domains: JmjC, JmjN, a zinc
finger an ARID (DNA-binding domain), as well as a PHD
(histone-binding domain) lining between JmjC and JmjN (115,116). Thus, all four members were able
to demethylate H3K4me2/me3 and participate in the epigenetic
regulation of biological processes related to cancer (117,118). However, Both KDM5A and KDM5B
have 3 PHD domains, while KDM5C and KDM5D have only 2 PHD domains.
Since the PHD domain is important for the binding of H3K4 with the
JmjC domain, KDM5C and KDM5D may exhibit poor catalytic function
and different effects in cancer compared to KDM5A and KDM5B
(29).
Compared to normal tissues, KDM5A is overexpressed
in cancer tissues and contributes to tumor growth and poor
prognosis. KDM5A has the ability to repress p27, a cyclin-dependent
kinase (CDK) inhibitor in cancer, trigger G1/S phase transition and
promote tumor malignancy (119–122). Furthermore, by demethylating
H3K4me2/me3 at the promoter, KDM5A was able to suppress the
expression of insulin-like growth factor 2 mRNA binding protein 2
and NOTCH2, facilitating tumor proliferation, invasion and
metastasis (123,124).
KDM5B mainly has oncogenic effects in cancers. In
breast cancer, KDM5B was indicated to be overexpressed and
associated with poor prognosis (125). Furthermore, it was indicated
that KDM5B suppressed BRCA1, caveolin 1 and homeobox A5 expression
by reducing H3K4me3 levels and facilitated G1 progression and tumor
growth in breast cell lines (126). Through activating the c-Met
signaling pathway or inhibiting p53 accumulation, KDM5B promoted
lung cancer cell aggressiveness (127,128). In addition, other studies also
indicated that knockdown of KDM5B led to cell cycle arrest at the
G1/S phase; the ability of KDM5B to influence tumor proliferation
by adjusting the cell cycle was identified in liver cancer, bladder
cancer and acute lymphoblastic leukemia (129–131). Furthermore, the oncogenic effect
of KDM5B in prostate cancer and colorectal cancer by demethylating
H3K4 was identified (132,133).
Unlike that of KDM5A and KDM5B, the role of KDM5C in
tumors has remained elusive. In clear-cell renal cell carcinoma
xenograft models, tumor cells highly expressing KDM5C were able to
significantly suppress tumor growth (134). Furthermore, patients with renal
cancer and KDM5C-inactivating mutations had shorter overall
survival, suggesting the tumor-suppressive role of KDM5C (135,136). Of note, the tumor-suppressive
effect of KDM5C was also observed in intrahepatic
cholangiocarcinoma (137).
However, KDM5C exhibits a tumor-promoting effect in other cancer
types. In lung cancer, KDM5C facilitates tumor proliferation and
metastasis by promoting H3K4me2 demethylation modification of the
promoter of miR-133a and downregulation of miR-133a (138). Furthermore, KDM5C highly
expressed in liver cancer was indicated to be associated with
distant metastasis and poor prognosis by demethylating at H3K4
(139). In colon cancer, KDM5C
was also observed to promote cell proliferation by demethylating
H3K4me2/me3 (140). In addition,
KDM5C upregulated ER expression and inhibited type I IFN expression
in a breast cancer cell line, and, as a result, promoting breast
carcinogenesis and cancer cell growth in a demethylase-independent
manner (141).
KDM5D was mainly observed to have a
tumor-suppressive effect. Upon specific knockdown of KDM5D, tumor
cell apoptosis was reduced and tumor proliferation was promoted in
a prostate cancer cell line (142). In addition, KDM5D repressed the
invasion-associated genes matrix metallopeptidase 1 (MMP1), MMP2,
MMP3 and MMP7 by demethylating H3K4me3, thus suppressing prostate
cancer invasion and metastasis (143). Apart from prostate cancer, the
mechanism of KDM5D to inhibit cancer cell growth and contributing
to a better prognosis through its demethylating activity was also
observed in gastric cancer and lung cancer (144–146).
KDM6A (also named UTX), KDM6B (also named JMJD3) and
KDM6C (also named UTY) belong to the KDM6 family. KDM6A is located
at the X chromosome, KDM6C is located at the Y chromosome and KDM6B
is located at chromosome 17 (147,148). All three contain the JmjC domain
and have the ability to catalyze the demethylation of H3K27me2/me3
(147,149), although KDM6C has relatively
poor catalytic activity compared to KDM6A and KDM6B (148).
Current evidence suggests both tumor-promoting and
tumor-suppressive effects of KDM6A and KDM6B in cancers. KDM6A
mutations frequently occur in various cancers. In hepatocellular
carcinoma, overexpression of KDM6A significantly suppressed
tumorigenesis (150). In
addition, by inhibiting enhancer of zeste 2 polycomb repressive
complex 2 subunit (EZH2)-mediated transcriptional repression
through catalyzing demethylation of H3K27me3, KDM6A acts as a tumor
suppressor in bladder cancer (151). However, a recent study
identified the oncogenic role of KDM6A and KDM6B by epigenetically
targeting stemness-controlling genes through demethylating
H3K27me3, which makes KDM6A and KDM6B important in maintaining
cancer cell stemness. At the same time, upregulation of KDM6B was
indicated to be strongly associated with a higher recurrence rate
and shorter survival in colorectal cancer (152). Furthermore, significantly
increasing the levels of H3K27me3 using GSK-J4, a KDM6 family
inhibitor, suppressed tumor growth in lung cancer mouse models,
indicating an oncogenic effect of KDM6A and KDM6B (153).
The KDM7 family is composed of KDM7A (also named
JHDM1D), KDM7B (also named PHF8) and KDM7C (also named PHF2). All
demethylases of the KDM7 family share the same composition,
containing a JmjC domain at the C-terminus and a PHD domain at the
N-terminus. The PHD domain binds to H3K4me3, while the JmjC domain
is responsible for binding to H3K9me2 (154). However, the structure of each
demethylase is slightly different, which may be the reason for the
different functions. Among the KDM7 family, KDM7C only catalyzes
H3K9me2 demethylation. However, KDM7A and KDM7B are able to
catalyze demethylation of H3K9me1/me2, H3K27 me1/me2 and H4K20me1
(155).
An early study demonstrated that KDM7A acts as a
tumor suppressor by inhibiting the in vivo growth of B16 and
HeLa cells upon overexpression of KDM7A, even though this
suppressive effect was not prominent in vitro (156).
However, recent studies have discovered the oncogenic role of
KDM7A, since KDM7A was indicated to be upregulated in prostate
cancer tissue (157) and to
promote the migration and invasion of breast cancer cells in
vitro and in vivo (158). Therefore, the definitive role of
KDM7A remains to be determined.
By contrast, KDM7B was indicated to have an
oncogenic effect. KDM7B was determined to be associated with a
higher Gleason score and poor prognosis by comparing prostate
cancer tissue samples from 97 patients (159). In addition, KDM7B was indicated
to act as an oncogene by activating genes related to tumor
progression [PRKCA, ICAM-1, Snail (SNAI1), VIM and FIP200] in a
demethylase-dependent or demethylase-independent manner and promote
tumor progression in gastric cancer and hepatocellular carcinoma
(160–162). However, its
demethylase-catalyzing ability is also responsible for other
effects. By catalyzing demethylation of H3K4me3 and H3K9me2/1,
KDM7B activates the expression of SNAI1, which contributed to
breast cancer epithelial-to-mesenchymal transition, tumorigenesis
and metastasis (163). In
addition to SNAI1, forkhead box protein A2 was also epigenetically
upregulated by KDM7B through demethylating H3K9me1/me2, H3K27me2
and H4K20me1, further illustrating the oncogenic effect of KDM7B
(164).
By contrast, KDM7C acts as a tumor suppressor. KDM7C
expression was indicated to be downregulated in hepatocellular,
colon and stomach cancer tissues as compared with that in normal
tissues. Upregulation of KDM7C was associated with a favorable
prognosis in hepatocellular carcinoma and decreased tumor cell
migration (165). Another study
demonstrated that KDM7C demethylates H3K9me2 at p53 promoters,
resulting in activation of p53 transcription and suppression of
tumor growth (166).
KDM8 (also named JMJD5) has a JmjC domain and
β-barrel fold structure and has H3K36me2 demethylating activity
(167). However, the effect of
KDM8 in tumorigenesis has remained to be determined. In an early
study, KDM8 was indicated to be overexpressed in breast cancer
tissues, catalyzing H3K36me2 demethylation and leading to cyclin A1
overexpression. This results in the initiation of G2/M phase
transition and the promotion of tumor cell proliferation (168). In addition, downregulation of
KDM8 was indicated to inhibit tumor proliferation and metastasis in
oral cancer by upregulating the expression of p53 and E-cadherin
and downregulating the expression of N-cadherin and vimentin
(169). However, in a
large-scale, multi-cohort study of gene expression profiles in
several cancer types, KDM8 was indicated to be downregulated in
pancreatic cancer and liver cancer, and was reduced as the tumor
grade increased. Furthermore, the expression of KDM8 was negatively
correlated with the hypoxia score and the expression of cell cycle
genes (such as CCNA2, CCNB1, CDK1 and CDK2), indicating the
tumor-suppressive role of KDM8 (170). Therefore, further studies on
KDM8 are warranted.
Current cancer therapies include surgery,
chemotherapy, radiotherapy, targeted therapy and immunotherapy. For
each treatment modality, significant progress has been achieved in
the management of cancer. However, resistance to cancer therapy is
a major problem in cancer treatment. Targeting histone demethylases
not only has a critical role in tumor growth, invasion and
metastasis, but also in chemoresistance, radioresistance and
resistance to targeted therapy and immunotherapy (Table I). To date, most studies on the
effect of histone demethylases in cancer therapy resistance were
focused on the KDM1, KDM5 and KDM6 families.
Upregulation of KDM1A and KDM1B is associated with
chemoresistance and poor survival. In liver cancer, both KDM1A
knockdown and combination of KDM1A inhibitors with regorafenib
improved resistance to regorafenib (171). In breast cancer, KDM1A
overexpression was responsible for doxorubicin resistance (172) and regulation of the tumor
microenvironment, and contributed to the resistance against PD-1
inhibitors in vivo (39).
Similar to the role of KDM1A in chemoresistance, the downregulation
of KDM1B improved cisplatin resistance in ovarian cancer (173). In enzalutamide-resistant
prostate cancer, inhibition of KDM1B by tranylcypromine improved
enzalutamide resistance by decreasing androgen receptor-depending
anterior gradient 2 transcription epigenetically (174). Similarly, inhibition of KDM4B
epigenetically suppressed c-Myc transcription and enhanced the
efficacy of enzalutamide treatment in vitro and in
vivo (175).
To date, accumulating evidence has identified the
role of the KDM5 family in chemotherapy resistance of cancers
(117). An early study
demonstrated that breast cancer cells with KDM5A amplification
exhibited resistance to EGFR inhibitors (176). Furthermore, KDM5A also
contributed to temozolomide resistance in glioblastoma through
enhancing drug efflux, and knocking down KDM5A or using HDAC
inhibitors to suppress histone demethylases was able to resolve
temozolomide resistance (177).
Furthermore, KDM5B also contributes to chemoresistance.
Demethylation of H3K4, as a consequence of upregulation of KDM5B,
was observed in cisplatin-resistant gastric cancer cells (178). Knockdown of KDM5B resolved
multidrug resistance of melanoma in vivo by blocking the
mitochondrial respiratory chain (179) and enhancing the transition from
CD34- to CD34+ melanoma-propagating cell subpopulations that are
more sensitive to BRAF inhibitors through the demethylase-dependent
pathway (180). In addition to
chemoresistance, the KDM5 family also suppressed the sensitivity to
endocrine therapy in breast cancer (181). Furthermore, inhibiting the
expression of KDM5 family members in breast cancer cells increased
DNA damage accumulation through ionizing radiation. This phenomenon
suggested that breast cancer cell radiosensitivity may be improved
by knocking down KDM5 demethylase expression (182). Certain studies have demonstrated
that KDM5C aggravates drug resistance in colon cancer cells by
catalyzing H3K4me3 demethylation (183). In prostate cancer cells,
knocking down KDM5D led to reduced sensitivity to docetaxel. At the
same time, overexpression of KDM5D in prostate cancer cells
improved docetaxel sensitivity (184), demonstrating the effect of KDM5D
to improve chemoresistance, consistent with its tumor-suppressive
effect.
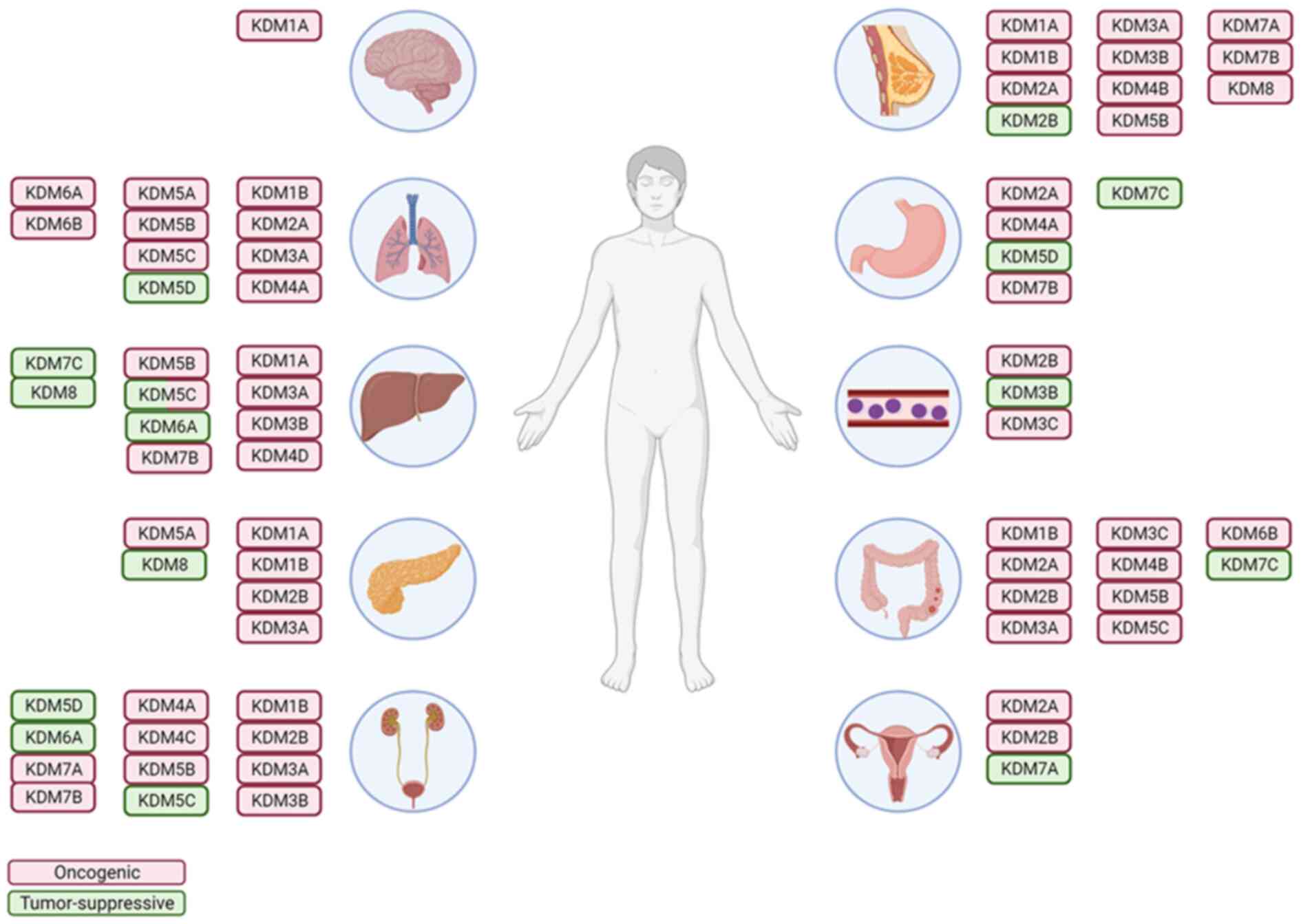
As the effects of KDMs on tumor growth, invasion and
metastasis are being discovered (Fig.
2 and Table I), several
inhibitors of KDMs have been identified or developed as novel
cancer treatment strategies. Numerous potent KDM1A inhibitors have
been developed and have demonstrated an excellent capacity to
inhibit cancer cell growth and metastasis (Table II) (191–193).
Inhibitors of the JmjC family, including ML324
(inhibitor of KDM4) and GSK-J4 (inhibitor of KDM6), achieved
excellent anti-tumor activity either alone or in combination
therapy in both cell lines and animal models (194,195). However, to date, no clinical
trial has been conducted to investigate the role of JmjC KDM family
inhibitors in cancer therapy (196,197). The major obstacle to the
therapeutic use of JmjC demethylase family inhibitors is the lack
of selective and potent inhibitors, which is possibly due to the
high similarity among catalytic domains (196–198). Under these circumstances, an
increasing number of effective and selective inhibitors of the JmjC
demethylases family, such as CBN209350 and purpurogallin analogs,
are being developed for cancer therapy (199,200).
TCP, a monoamine oxidase inhibitor used for
depression, irreversibly inhibits KDM1A. A recently completed phase
I clinical trial (NCT02273102) demonstrated that combined TCP and
all-trans retinoic acid (ATRA) therapy exhibited satisfactory
effects and acceptable safety by inhibiting KDM1A and sensitizing
AML cells to ATRA (201). In
addition, two further clinical trials (NCT02261779 and NCT02717884)
investigated the feasibility of using TCP in relapsed or refractory
AML, and a trial to assess the effect of TCP to sensitize ATRA in
patients with non-M3 AML is still recruiting.
Iadademstat (also called ORY-1001) is a selective
covalent KDM1A inhibitor, which is at the forefront of clinical
trials among all KDM1A inhibitors. In addition to the anti-cancer
effect of iadademstat in cancer cell lines (202,203), it also has good bioavailability
and significantly inhibits tumor growth in vivo (204). The first-in-human phase I study
of iadademstat (EudraCT 2013-002447-29) demonstrated a favorable
effect on relapsed or refractory AML with good safety, and one case
achieved complete remission (205). Subsequently, a phase II trial to
identify the effect of combined iadademstat and azacitidine was
launched (EudraCT 2018-000482-36). In addition, a phase I trial of
iadademstat in relapsed small cell lung cancer (SCLC) (NCT02913443)
was completed with 18 participants, although the results have not
been published.
GSK2879552 is a potent and selective small-molecule
KDM1A inhibitor that exhibits anti-cancer activity in numerous
cancer cell lines (206,207). All three clinical trials
investigating the safety and clinical viability of GSK2879552 in
AML (NCT02177812), SCLC (NCT02034123) and myelodysplastic syndrome
(NCT02929498) have been terminated due to frequent adverse events
and inadequate efficacy of cancer treatment. In the trial
NCT02034123, 83% of participants developed adverse events, while
100% of participants in NCT02177812 and NCT02929498 developed
adverse events. The most common adverse events were hematological
toxicity (such as thrombocytopenia or neutropenia) and fatigue.
However, the disease control rate in NCT02034123 was only 14%
(208).
CC-90011 is a potent, selective and reversible KDM1A
inhibitor developed by adding a fluorine substitution at the
3-position of benzonitrile (209). In a phase I study of non-Hodgkin
lymphoma, CC-90011 was indicated to be well-tolerated and this
clinical trial (NCT02875223) is still recruiting (210). In addition, a phase I functional
imaging study to assess the effect of CC-90011 on metastatic
castration-resistant prostate cancer (NCT04628988) is now
recruiting. Furthermore, a phase Ib, multi-center clinical trial
sponsored by Celgene was launched to demonstrate the safety and
efficacy of combining CC-90011 with cisplatin and etoposide in SCLC
(NCT03850067). Finally, a phase 2 clinical trial assessing the
safety and efficacy of CC-90011 in combination with nivolumab in
SCLC and squamous cell carcinoma (NCT04350463) by evaluating the
treatment response has recently started recruiting.
Previous reviews have provided insight into histone
demethylases in cancer, metabolic disease, regeneration,
inflammation and neurological diseases (29). At the same time, previous reviews
have also concluded on the role of KDMs in cancer and the
mechanisms by which KDMs participate in cancer development and
progression (92,211). It is evident that most histone
demethylases act as oncogenes in cancer development. However, the
effects of KDM3B, KDM5C, KDM6A, KDM6B and KDM8 are still under
debate, while KDM5D and KDM7C were proven to be tumor suppressive.
Furthermore, histone demethylases have been indicated to contribute
to chemoresistance and resistance to radiotherapy, targeted therapy
and immunotherapy. However, only a small number of studies have
illustrated how histone demethylases contribute to cancer therapy
resistance. Therefore, it is important to perform further studies
to answer this question. To date, several phase I clinical trials
have been launched to identify the safety and efficacy of histone
demethylase inhibitors in cancer therapy, whether combined with the
current standard of treatment or not. Certain inhibitors
demonstrated an ideal effect and most clinical trials for these
drugs are still recruiting, although all clinical trials of
GSK2879552 have already been terminated.
One important reason for the unclear effects of
certain KDMs on cancer is that catalytic domains other than JmjC of
these KDMs may also be involved in the biological processes;
however, how these domains interact with the cancer development
process remains largely elusive. Hence, further studies on the
effect and interaction of the catalytic domains in biological
processes should be performed to thoroughly illustrate the
regulatory mechanism between KDMs and cancers.
It is evident that histone demethylases have the
potential to be cancer therapeutics in the future; however,
additional studies should be performed to facilitate their wide use
in the clinic. On the basis of the success of KDM inhibitors
resolving therapy resistance in vitro and in vivo,
clinical trials examining the effect of KDM inhibitors on cancer
therapy resistance are expected. Following the termination of JmjC
KDM inhibitors, the development of more selective and potent
inhibitors is essential for further clinical application. On the
other hand, medication resolving the side effects of JmjC KDM
inhibitors is desired to ensure the application of these inhibitors
in the future. Recently, JIB-04, a histone lysine demethylase
inhibitor, has been successfully delivered to prostate cancer cells
and tumor spheroids by nanoparticles (212). Therefore, with the great success
of nanoparticle drug delivery systems, it is foreseeable that
delivering KDM inhibitors directly to tumors may reduce side
effects and enable their wide use in solid tumors.
Not applicable.
This review was funded by the National Natural Science
Foundation of China (grant no. 82172921), the Guangzhou Science and
Technology Program (Project Leader: SX), the Science and Technology
Program of Guangzhou (grant no. 201904010020) and the National Key
Clinical Specialty Construction Project (grant nos. 2021-2024 and
2022YW030009).
Not applicable.
Conceptualization: WD and JZ; original draft
writing: WD; review and editing: SX, JW and YL; supervision: SX, WJ
and YL; funding acquisition: JW. All authors have read and agreed
to the published version of the manuscript. Data authentication is
not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kruger S, Ilmer M, Kobold S, Cadilha BL,
Endres S, Ormanns S, Schuebbe G, Renz BW, D'Haese JG, Schloesser H,
et al: Advances in cancer immunotherapy 2019-latest trends. J Exp
Clin Cancer Res. 38:2682019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thompson JA, Schneider BJ, Brahmer J,
Achufusi A, Armand P, Berkenstock MK, Bhatia S, Budde LE, Chokshi
S, Davies M, et al: Management of immunotherapy-related toxicities,
version 1.2022, NCCN clinical practice guidelines in oncology. J
Natl Compr Canc Netw. 20:387–405. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Michalak EM, Burr ML, Bannister AJ and
Dawson MA: The roles of DNA, RNA and histone methylation in ageing
and cancer. Nat Rev Mol Cell Biol. 20:573–589. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tachibana M, Sugimoto K, Nozaki M, Ueda J,
Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H and Shinkai Y:
G9a histone methyltransferase plays a dominant role in euchromatic
histone H3 lysine 9 methylation and is essential for early
embryogenesis. Genes Dev. 16:1779–1791. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peters AH, O'Carroll D, Scherthan H,
Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner
M, Kohlmaier A, et al: Loss of the Suv39h histone
methyltransferases impairs mammalian heterochromatin and genome
stability. Cell. 107:323–337. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee
JW, Verdine GL, Allis CD and Roeder RG: Regulation of MLL1 H3K4
methyltransferase activity by its core components. Nat Struct Mol
Biol. 13:713–719. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi Y: Histone lysine demethylases:
Emerging roles in development, physiology and disease. Nat Rev
Genet. 8:829–833. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Soares LM, He PC, Chun Y, Suh H, Kim T and
Buratowski S: Determinants of histone H3K4 methylation patterns.
Mol Cell. 68:773–785.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fodor BD, Kubicek S, Yonezawa M,
O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K,
Schotta G and Jenuwein T: Jmjd2b antagonizes H3K9 trimethylation at
pericentric heterochromatin in mammalian cells. Genes Dev.
20:1557–1562. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Z and Zhang Y: Maternal
H3K27me3-dependent autosomal and X chromosome imprinting. Nat Rev
Genet. 21:555–571. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cloos PA, Christensen J, Agger K, Maiolica
A, Rappsilber J, Antal T, Hansen KH and Helin K: The putative
oncogene GASC1 demethylates tri- and dimethylated lysine 9 on
histone H3. Nature. 442:307–311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klose RJ, Yamane K, Bae Y, Zhang D,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: The
transcriptional repressor JHDM3A demethylates trimethyl histone H3
lysine 9 and lysine 36. Nature. 442:312–316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vallianatos CN and Iwase S: Disrupted
intricacy of histone H3K4 methylation in neurodevelopmental
disorders. Epigenomics. 7:503–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clark EA, Wu F, Chen Y, Kang P, Kaiser UB,
Fang R and Shi YG: GR and LSD1/KDM1A-targeted gene activation
requires selective H3K4me2 demethylation at enhancers. Cell Rep.
27:3522–3532.e3. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tsukada Y, Fang J, Erdjument-Bromage H,
Warren ME, Borchers CH, Tempst P and Zhang Y: Histone demethylation
by a family of JmjC domain-containing proteins. Nature.
439:811–816. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Trojer P, Zhang J, Yonezawa M, Schmidt A,
Zheng H, Jenuwein T and Reinberg D: Dynamic histone H1 isotype 4
methylation and demethylation by histone lysine methyltransferase
G9a/KMT1C and the Jumonji domain-containing JMJD2/KDM4 proteins. J
Biol Chem. 284:8395–8405. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suzuki T, Ozasa H, Itoh Y, Zhan P, Sawada
H, Mino K, Walport L, Ohkubo R, Kawamura A, Yonezawa M, et al:
Identification of the KDM2/7 histone lysine demethylase subfamily
inhibitor and its antiproliferative activity. J Med Chem.
56:7222–7231. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Berry WL and Janknecht R: KDM4/JMJD2
histone demethylases: Epigenetic regulators in cancer cells. Cancer
Res. 73:2936–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Turberfield AH, Kondo T, Nakayama M,
Koseki Y, King HW, Koseki H and Klose RJ: KDM2 proteins constrain
transcription from CpG island gene promoters independently of their
histone demethylase activity. Nucleic Acids Res. 47:9005–9023.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian Z, Yao L, Shen Y, Guo X and Duan X:
Histone H3K9 demethylase JMJD1A is a co-activator of erythropoietin
expression under hypoxia. Int J Biochem Cell Biol. 109:33–39. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sui Y, Gu R and Janknecht R: Crucial
functions of the JMJD1/KDM3 epigenetic regulators in cancer. Mol
Cancer Res. 19:3–13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wagner KW, Alam H, Dhar SS, Giri U, Li N,
Wei Y, Giri D, Cascone T, Kim JH, Ye Y, et al: KDM2A promotes lung
tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin
Invest. 123:5231–5246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hughes AL, Kelley JR and Klose RJ:
Understanding the interplay between CpG island-associated gene
promoters and H3K4 methylation. Biochim Biophys Acta Gene Regul
Mech. 1863:1945672020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barski A, Cuddapah S, Cui K, Roh TY,
Schones DE, Wang Z, Wei G, Chepelev I and Zhao K: High-resolution
profiling of histone methylations in the human genome. Cell.
129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Y, Yang Y, Wang F, Wan K, Yamane K,
Zhang Y and Lei M: Crystal structure of human histone
lysine-specific demethylase 1 (LSD1). Proc Natl Acad Sci USA.
103:13956–13961. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arifuzzaman S, Khatun MR and Khatun R:
Emerging of lysine demethylases (KDMs): From pathophysiological
insights to novel therapeutic opportunities. Biomed Pharmacother.
129:1103922020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carnesecchi J, Forcet C, Zhang L,
Tribollet V, Barenton B, Boudra R, Cerutti C, Billas IM, Sérandour
AA, Carroll JS, et al: ERRα induces H3K9 demethylation by LSD1 to
promote cell invasion. Proc Natl Acad Sci USA. 114:3909–3914. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He Y, Zhao Y, Wang L, Bohrer LR, Pan Y,
Wang L and Huang H: LSD1 promotes S-phase entry and tumorigenesis
via chromatin co-occupation with E2F1 and selective H3K9
demethylation. Oncogene. 37:534–543. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y and Cao K: KDM1A promotes
immunosuppression in hepatocellular carcinoma by regulating PD-L1
through demethylating MEF2D. J Immunol Res. 2021:99650992021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hou X, Li Q, Yang L, Yang Z, He J, Li Q
and Li D: KDM1A and KDM3A promote tumor growth by upregulating cell
cycle-associated genes in pancreatic cancer. Exp Biol Med
(Maywood). 246:1869–1883. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim HS, Son BK, Kwon MJ, Kim DH and Min
KW: High KDM1A expression associated with decreased CD8+ T cells
reduces the breast cancer survival rate in patients with breast
cancer. J Clin Med. 10:11122021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Majello B, Gorini F, Saccà CD and Amente
S: Expanding the role of the histone lysine-specific demethylase
LSD1 in cancer. Cancers (Basel). 11:3242019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang J, Sengupta R, Espejo AB, Lee MG,
Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT,
Jenuwein T and Berger SL: p53 is regulated by the lysine
demethylase LSD1. Nature. 449:105–108. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cho HS, Suzuki T, Dohmae N, Hayami S,
Unoki M, Yoshimatsu M, Toyokawa G, Takawa M, Chen T, Kurash JK, et
al: Demethylation of RB regulator MYPT1 by histone demethylase LSD1
promotes cell cycle progression in cancer cells. Cancer Res.
71:655–660. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Saccà CD, Gorini F, Ambrosio S, Amente S,
Faicchia D, Matarese G, Lania L and Majello B: Inhibition of
lysine-specific demethylase LSD1 induces senescence in glioblastoma
cells through a HIF-1α-dependent pathway. Biochim Biophys Acta Gene
Regul Mech. 1862:535–546. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qin Y, Vasilatos SN, Chen L, Wu H, Cao Z,
Fu Y, Huang M, Vlad AM, Lu B, Oesterreich S, et al: Inhibition of
histone lysine-specific demethylase 1 elicits breast tumor immunity
and enhances antitumor efficacy of immune checkpoint blockade.
Oncogene. 38:390–405. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Katz TA, Vasilatos SN, Harrington E,
Oesterreich S, Davidson NE and Huang Y: Inhibition of histone
demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases
sensitivity to DNMT inhibitor-induced apoptosis in breast cancer
cells. Breast Cancer Res Treat. 146:99–108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cai S, Wang J, Zeng W, Cheng X, Liu L and
Li W: Lysine-specific histone demethylase 1B (LSD2/KDM1B) represses
p53 expression to promote proliferation and inhibit apoptosis in
colorectal cancer through LSD2-mediated H3K4me2 demethylation.
Aging (Albany NY). 12:14990–15001. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cao Y, Guo C, Yin Y, Li X and Zhou L:
Lysine-specific demethylase 2 contributes to the proliferation of
small cell lung cancer by regulating the expression of TFPI-2. Mol
Med Rep. 18:733–740. 2018.PubMed/NCBI
|
|
43
|
Kumar A, Kumari N, Sharma U, Ram S, Singh
SK, Kakkar N, Kaushal K and Prasad R: Reduction in H3K4me patterns
due to aberrant expression of methyltransferases and demethylases
in renal cell carcinoma: Prognostic and therapeutic implications.
Sci Rep. 9:81892019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Sun L, Luo Y and He S: Knockdown
of KDM1B inhibits cell proliferation and induces apoptosis of
pancreatic cancer cells. Pathol Res Pract. 215:1054–1060. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi
X, Qin B, Zeng L, Esteban MA, Pan G and Pei D: The histone
demethylases Jhdm1a/1b enhance somatic cell reprogramming in a
vitamin-C-dependent manner. Cell Stem Cell. 9:575–587. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kong Y, Zou S, Yang F, Xu X, Bu W, Jia J
and Liu Z: RUNX3-mediated up-regulation of miR-29b suppresses the
proliferation and migration of gastric cancer cells by targeting
KDM2A. Cancer Lett. 381:138–148. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang F, Liang S, Liu X, Han L, Wang J and
Du Q: LINC00460 modulates KDM2A to promote cell proliferation and
migration by targeting miR-342-3p in gastric cancer. Onco Targets
Ther. 11:6383–6394. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ou R, Zhu L, Zhao L, Li W, Tao F, Lu Y, He
Q, Li J, Ren Y and Xu Y: HPV16 E7-induced upregulation of KDM2A
promotes cervical cancer progression by regulating miR-132-radixin
pathway. J Cell Physiol. 234:2659–2671. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu DH, Yang J, Gao LK, Min J, Tang JM, Hu
M, Li Y, Li ST, Chen J and Hong L: Lysine demethylase 2A promotes
the progression of ovarian cancer by regulating the PI3K pathway
and reversing epithelial-mesenchymal transition. Oncol Rep.
41:917–927. 2019.PubMed/NCBI
|
|
50
|
Zhao Y, Chen X, Jiang J, Wan X, Wang Y and
Xu P: Epigallocatechin gallate reverses gastric cancer by
regulating the long noncoding RNA LINC00511/miR-29b/KDM2A axis.
Biochim Biophys Acta Mol Basis Dis. 1866:1658562020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xi C, Ye NY and Wang YB: LncRNA LINC01278
accelerates colorectal cancer progression via miR-134-5p/KDM2A
axis. Eur Rev Med Pharmacol Sci. 24:10526–10534. 2020.PubMed/NCBI
|
|
52
|
Kottakis F, Polytarchou C, Foltopoulou P,
Sanidas I, Kampranis SC and Tsichlis PN: FGF-2 regulates cell
proliferation, migration, and angiogenesis through an
NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell. 43:285–298. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tzatsos A, Paskaleva P, Ferrari F,
Deshpande V, Stoykova S, Contino G, Wong KK, Lan F, Trojer P, Park
PJ and Bardeesy N: KDM2B promotes pancreatic cancer via
polycomb-dependent and -independent transcriptional programs. J
Clin Invest. 123:727–739. 2013.PubMed/NCBI
|
|
54
|
Kuang Y, Lu F, Guo J, Xu H, Wang Q, Xu C,
Zeng L and Yi S: Histone demethylase KDM2B upregulates histone
methyltransferase EZH2 expression and contributes to the
progression of ovarian cancer in vitro and in vivo. Onco Targets
Ther. 10:3131–3144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sanches JGP, Song B, Zhang Q, Cui X,
Yabasin IB, Ntim M, Li X, He J, Zhang Y, Mao J, et al: The role of
KDM2B and EZH2 in regulating the stemness in colorectal cancer
through the PI3K/AKT pathway. Front Oncol. 11:6372982021.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dhar SS, Alam H, Li N, Wagner KW, Chung J,
Ahn YW and Lee MG: Transcriptional repression of histone
deacetylase 3 by the histone demethylase KDM2A is coupled to
tumorigenicity of lung cancer cells. J Biol Chem. 289:7483–7496.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen JY, Li CF, Chu PY, Lai YS, Chen CH,
Jiang SS, Hou MF and Hung WC: Lysine demethylase 2A promotes
stemness and angiogenesis of breast cancer by upregulating Jagged1.
Oncotarget. 7:27689–27710. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen JY, Li CF, Lai YS and Hung WC: Lysine
demethylase 2A expression in cancer-associated fibroblasts promotes
breast tumour growth. Br J Cancer. 124:484–493. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Quan M, Chen Z, Jiao F, Xiao X, Xia Q,
Chen J, Chao Q, Li Y, Gao Y, Yang H, et al: Lysine demethylase 2
(KDM2B) regulates hippo pathway via MOB1 to promote pancreatic
ductal adenocarcinoma (PDAC) progression. J Exp Clin Cancer Res.
39:132020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wanna-Udom S, Terashima M, Suphakhong K,
Ishimura A, Takino T and Suzuki T: KDM2B is involved in the
epigenetic regulation of TGF-β-induced epithelial-mesenchymal
transition in lung and pancreatic cancer cell lines. J Biol Chem.
296:1002132021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
van den Boom V, Maat H, Geugien M,
Rodríguez López A, Sotoca AM, Jaques J, Brouwers-Vos AZ, Fusetti F,
Groen RW, Yuan H, et al: Non-canonical PRC1.1 targets active genes
independent of H3K27me3 and is essential for leukemogenesis. Cell
Rep. 14:332–346. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Galbiati A, Penzo M, Bacalini MG,
Onofrillo C, Guerrieri AN, Garagnani P, Franceschi C, Treré D and
Montanaro L: Epigenetic up-regulation of ribosome biogenesis and
more aggressive phenotype triggered by the lack of the histone
demethylase JHDM1B in mammary epithelial cells. Oncotarget.
8:37091–37103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yoo J, Jeon YH, Cho HY, Lee SW, Kim GW,
Lee DH and Kwon SH: Advances in histone demethylase KDM3A as a
cancer therapeutic target. Cancers (Basel). 12:10982020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yamane K, Toumazou C, Tsukada Y,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: JHDM2A, a
JmjC-containing H3K9 demethylase, facilitates transcription
activation by androgen receptor. Cell. 125:483–495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen M, Zhu N, Liu X, Laurent B, Tang Z,
Eng R, Shi Y, Armstrong SA and Roeder RG: JMJD1C is required for
the survival of acute myeloid leukemia by functioning as a
coactivator for key transcription factors. Genes Dev. 29:2123–2139.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jiang Y, Li C, Wu Q, An P, Huang L, Wang
J, Chen C, Chen X, Zhang F, Ma L, et al: Iron-dependent histone 3
lysine 9 demethylation controls B cell proliferation and humoral
immune responses. Nat Commun. 10:29352019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Brauchle M, Yao Z, Arora R, Thigale S,
Clay I, Inverardi B, Fletcher J, Taslimi P, Acker MG, Gerrits B, et
al: Protein complex interactor analysis and differential activity
of KDM3 subfamily members towards H3K9 methylation. PLoS One.
8:e605492013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu J, Liang T and Zhangsun W: KDM3A is
associated with tumor metastasis and modulates colorectal cancer
cell migration and invasion. Int J Biol Macromol. 126:318–325.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang Z, Yang X, Liu C, Li X, Zhang B, Wang
B, Zhang Y, Song C, Zhang T, Liu M, et al: Acetylation of PHF5A
modulates stress responses and colorectal carcinogenesis through
alternative splicing-mediated upregulation of KDM3A. Mol Cell.
74:1250–1263.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li J, Yu B, Deng P, Cheng Y, Yu Y, Kevork
K, Ramadoss S, Ding X, Li X and Wang CY: KDM3 epigenetically
controls tumorigenic potentials of human colorectal cancer stem
cells through Wnt/β-catenin signalling. Nat Commun. 8:151462017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang HY, Long QY, Tang SB, Xiao Q, Gao C,
Zhao QY, Li QL, Ye M, Zhang L, Li LY and Wu M: Histone demethylase
KDM3A is required for enhancer activation of hippo target genes in
colorectal cancer. Nucleic Acids Res. 47:2349–2364. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ramadoss S, Guo G and Wang CY: Lysine
demethylase KDM3A regulates breast cancer cell invasion and
apoptosis by targeting histone and the non-histone protein p53.
Oncogene. 36:47–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wade MA, Jones D, Wilson L, Stockley J,
Coffey K, Robson CN and Gaughan L: The histone demethylase enzyme
KDM3A is a key estrogen receptor regulator in breast cancer.
Nucleic Acids Res. 43:196–207. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Lee HY, Yang EG and Park H: Hypoxia
enhances the expression of prostate-specific antigen by modifying
the quantity and catalytic activity of Jumonji C domain-containing
histone demethylases. Carcinogenesis. 34:2706–2715. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fan L, Peng G, Sahgal N, Fazli L, Gleave
M, Zhang Y, Hussain A and Qi J: Regulation of c-Myc expression by
the histone demethylase JMJD1A is essential for prostate cancer
cell growth and survival. Oncogene. 35:2441–2452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wilson S, Fan L, Sahgal N, Qi J and Filipp
FV: The histone demethylase KDM3A regulates the transcriptional
program of the androgen receptor in prostate cancer cells.
Oncotarget. 8:30328–30343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Z, Xia J, Fang M and Xu Y: Epigenetic
regulation of lung cancer cell proliferation and migration by the
chromatin remodeling protein BRG1. Oncogenesis. 8:662019.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang F and Quan Q: The long non-coding RNA
SNHG4/microRNA-let-7e/KDM3A/p21 pathway is involved in the
development of non-small cell lung cancer. Mol Ther Oncolytics.
20:634–645. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Dandawate P, Ghosh C, Palaniyandi K, Paul
S, Rawal S, Pradhan R, Sayed AAA, Choudhury S, Standing D,
Subramaniam D, et al: The histone demethylase KDM3A, increased in
human pancreatic tumors, regulates expression of DCLK1 and promotes
tumorigenesis in mice. Gastroenterology. 157:1646–1659.e11. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nakatsuka T, Tateishi K, Kudo Y, Yamamoto
K, Nakagawa H, Fujiwara H, Takahashi R, Miyabayashi K, Asaoka Y,
Tanaka Y, et al: Impact of histone demethylase KDM3A-dependent AP-1
transactivity on hepatotumorigenesis induced by PI3K activation.
Oncogene. 36:6262–6271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang Y, Pan Q and Shao Z:
Tumor-suppressive role of microRNA-202-3p in hepatocellular
carcinoma through the KDM3A/HOXA1/MEIS3 pathway. Front Cell Dev
Biol. 8:5560042021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Parrish JK, Sechler M, Winn RA and
Jedlicka P: The histone demethylase KDM3A is a
microRNA-22-regulated tumor promoter in Ewing sarcoma. Oncogene.
34:257–262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Sechler M, Parrish JK, Birks DK and
Jedlicka P: The histone demethylase KDM3A, and its downstream
target MCAM, promote Ewing sarcoma cell migration and metastasis.
Oncogene. 36:4150–4160. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
An MJ, Kim DH, Kim CH, Kim M, Rhee S, Seo
SB and Kim JW: Histone demethylase KDM3B regulates the
transcriptional network of cell-cycle genes in hepatocarcinoma
HepG2 cells. Biochem Biophys Res Commun. 508:576–582. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Saraç H, Morova T, Pires E, McCullagh J,
Kaplan A, Cingöz A, Bagci-Onder T, Önder T, Kawamura A and Lack NA:
Systematic characterization of chromatin modifying enzymes
identifies KDM3B as a critical regulator in castration resistant
prostate cancer. Oncogene. 39:2187–2201. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hu A, Hong F, Li D, Xie Q, Chen K, Zhu L
and He H: KDM3B-ETF1 fusion gene downregulates LMO2 via the
WNT/β-catenin signaling pathway, promoting metastasis of invasive
ductal carcinoma. Cancer Gene Ther. 29:215–224. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kim JY, Kim KB, Eom GH, Choe N, Kee HJ,
Son HJ, Oh ST, Kim DW, Pak JH, Baek HJ, et al: KDM3B is the H3K9
demethylase involved in transcriptional activation of lmo2 in
leukemia. Mol Cell Biol. 32:2917–2933. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang X, Fan H, Xu C, Jiang G, Wang H and
Zhang J: KDM3B suppresses APL progression by restricting chromatin
accessibility and facilitating the ATRA-mediated degradation of
PML/RARα. Cancer Cell Int. 19:2562019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Xu X, Wang L, Hu L, Dirks WG, Zhao Y, Wei
Z, Chen D, Li Z, Wang Z, Han Y, et al: Small molecular modulators
of JMJD1C preferentially inhibit growth of leukemia cells. Int J
Cancer. 146:400–412. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Cai Y, Fu X and Deng Y: Histone
demethylase JMJD1C regulates esophageal cancer proliferation Via
YAP1 signaling. Am J Cancer Res. 7:115–124. 2017.PubMed/NCBI
|
|
91
|
Chen C, Aihemaiti M, Zhang X, Qu H, Sun
QL, He QS and Yu WB: Downregulation of histone demethylase JMJD1C
inhibits colorectal cancer metastasis through targeting ATF2. Am J
Cancer Res. 8:852–865. 2018.PubMed/NCBI
|
|
92
|
Lee DH, Kim GW, Jeon YH, Yoo J, Lee SW and
Kwon SH: Advances in histone demethylase KDM4 as cancer therapeutic
targets. FASEB J. 34:3461–3484. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Berry WL, Shin S, Lightfoot SA and
Janknecht R: Oncogenic features of the JMJD2A histone demethylase
in breast cancer. Int J Oncol. 41:1701–1706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kim TD, Fuchs JR, Schwartz E, Abdelhamid
D, Etter J, Berry WL, Li C, Ihnat MA, Li PK and Janknecht R:
Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon
cancer cells and identification of curcuminoids as JMJD2
inhibitors. Am J Transl Res. 6:236–247. 2014.PubMed/NCBI
|
|
95
|
Ye Q, Holowatyj A, Wu J, Liu H, Zhang L,
Suzuki T and Yang ZQ: Genetic alterations of KDM4 subfamily and
therapeutic effect of novel demethylase inhibitor in breast cancer.
Am J Cancer Res. 5:1519–1530. 2015.PubMed/NCBI
|
|
96
|
Li X and Dong S: Histone demethylase
JMJD2B and JMJD2C induce fibroblast growth factor 2: Mediated
tumorigenesis of osteosarcoma. Med Oncol. 32:532015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kim TD, Jin F, Shin S, Oh S, Lightfoot SA,
Grande JP, Johnson AJ, van Deursen JM, Wren JD and Janknecht R:
Histone demethylase JMJD2A drives prostate tumorigenesis through
transcription factor ETV1. J Clin Invest. 126:706–720. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sun S, Yang F, Zhu Y and Zhang S: KDM4A
promotes the growth of non-small cell lung cancer by mediating the
expression of Myc via DLX5 through the Wnt/β-catenin signaling
pathway. Life Sci. 262:1185082020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hu CE, Liu YC, Zhang HD and Huang GJ:
JMJD2A predicts prognosis and regulates cell growth in human
gastric cancer. Biochem Biophys Res Commun. 449:1–7. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yang J, Jubb AM, Pike L, Buffa FM, Turley
H, Baban D, Leek R, Gatter KC, Ragoussis J and Harris AL: The
histone demethylase JMJD2B is regulated by estrogen receptor alpha
and hypoxia, and is a key mediator of estrogen induced growth.
Cancer Res. 70:6456–6466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Fu L, Chen L, Yang J, Ye T, Chen Y and
Fang J: HIF-1α-induced histone demethylase JMJD2B contributes to
the malignant phenotype of colorectal cancer cells via an
epigenetic mechanism. Carcinogenesis. 33:1664–1673. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li H, Lan J, Wang G, Guo K, Han C, Li X,
Hu J, Cao Z and Luo X: KDM4B facilitates colorectal cancer growth
and glucose metabolism by stimulating TRAF6-mediated AKT
activation. J Exp Clin Cancer Res. 39:122020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tan J, Wang HL, Yang J, Liu QQ, Li CM,
Wang YQ, Fu LN, Gao QY, Chen YX and Fang JY: JMJD2B-induced amino
acid alterations enhance the survival of colorectal cancer cells
under glucose-deprivation via autophagy. Theranostics.
10:5763–5777. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Fu LN, Wang YQ, Tan J, Xu J, Gao QY, Chen
YX and Fang JY: Role of JMJD2B in colon cancer cell survival under
glucose-deprived conditions and the underlying mechanisms.
Oncogene. 37:389–402. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wissmann M, Yin N, Müller JM, Greschik H,
Fodor BD, Jenuwein T, Vogler C, Schneider R, Günther T, Buettner R,
et al: Cooperative demethylation by JMJD2C and LSD1 promotes
androgen receptor-dependent gene expression. Nat Cell Biol.
9:347–353. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Lee DH, Kim GW, Yoo J, Lee SW, Jeon YH,
Kim SY, Kang HG, Kim DH, Chun KH, Choi J and Kwon SH: Histone
demethylase KDM4C controls tumorigenesis of glioblastoma by
epigenetically regulating p53 and c-Myc. Cell Death Dis. 12:892021.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Luo W, Chang R, Zhong J, Pandey A and
Semenza GL: Histone demethylase JMJD2C is a coactivator for
hypoxia-inducible factor 1 that is required for breast cancer
progression. Proc Natl Acad Sci USA. 109:E3367–E3376. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Yang D, Xu T, Fan L, Liu K and Li G:
microRNA-216b enhances cisplatin-induced apoptosis in osteosarcoma
MG63 and SaOS-2 cells by binding to JMJD2C and regulating the
HIF1α/HES1 signaling axis. J Exp Clin Cancer Res. 39:2012020.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wu X, Deng Y, Zu Y and Yin J: Histone
demethylase KDM4C activates HIF1α/VEGFA signaling through the
costimulatory factor STAT3 in NSCLC. Am J Cancer Res. 10:491–506.
2020.PubMed/NCBI
|
|
110
|
Hu F, Li H, Liu L, Xu F, Lai S, Luo X, Hu
J and Yang X: Histone demethylase KDM4D promotes gastrointestinal
stromal tumor progression through HIF1β/VEGFA signalling. Mol
Cancer. 17:1072018. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Peng K, Zhuo M, Li M, Chen Q, Mo P and Yu
C: Histone demethylase JMJD2D activates HIF1 signaling pathway via
multiple mechanisms to promote colorectal cancer glycolysis and
progression. Oncogene. 39:7076–7091. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhuo M, Chen W, Shang S, Guo P, Peng K, Li
M, Mo P, Zhang Y, Qiu X, Li W and Yu C: Inflammation-induced JMJD2D
promotes colitis recovery and colon tumorigenesis by activating
Hedgehog signaling. Oncogene. 39:3336–3353. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Peng K, Kou L, Yu L, Bai C, Li M, Mo P, Li
W and Yu C: Histone demethylase JMJD2D interacts with β-catenin to
induce transcription and activate colorectal cancer cell
proliferation and tumor growth in mice. Gastroenterology.
156:1112–1126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Li M, Deng Y, Zhuo M, Zhou H, Kong X, Xia
X, Su Z, Chen Q, Guo P, Mo P, et al: Demethylase-independent
function of JMJD2D as a novel antagonist of p53 to promote liver
cancer initiation and progression. Theranostics. 10:8863–8879.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Pilka ES, James T and Lisztwan JH:
Structural definitions of Jumonji family demethylase selectivity.
Drug Discov Today. 20:743–749. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Horton JR, Engstrom A, Zoeller EL, Liu X,
Shanks JR, Zhang X, Johns MA, Vertino PM, Fu H and Cheng X:
Characterization of a linked Jumonji domain of the KDM5/JARID1
family of histone H3 lysine 4 demethylases. J Biol Chem.
291:2631–2646. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Plch J, Hrabeta J and Eckschlager T: KDM5
demethylases and their role in cancer cell chemoresistance. Int J
Cancer. 144:221–231. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Yang GJ, Zhu MH, Lu XJ, Liu YJ, Lu JF,
Leung CH, Ma DL and Chen J: The emerging role of KDM5A in human
cancer. J Hematol Oncol. 14:302021. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Teng YC, Lee CF, Li YS, Chen YR, Hsiao PW,
Chan MY, Lin FM, Huang HD, Chen YT, Jeng YM, et al: Histone
demethylase RBP2 promotes lung tumorigenesis and cancer metastasis.
Cancer Res. 73:4711–4721. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Liang X, Zeng J, Wang L, Fang M, Wang Q,
Zhao M, Xu X, Liu Z, Li W, Liu S, et al: Histone demethylase
retinoblastoma binding protein 2 is overexpressed in hepatocellular
carcinoma and negatively regulated by hsa-miR-212. PLoS One.
8:e697842013. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Yang GJ, Wang W, Mok SWF, Wu C, Law BYK,
Miao XM, Wu KJ, Zhong HJ, Wong CY, Wong VKW, et al: Selective
inhibition of lysine-specific demethylase 5A (KDM5A) using a
rhodium(III) complex for triple-negative breast cancer therapy.
Angew Chem Int Ed Engl. 57:13091–13095. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yang GJ, Ko CN, Zhong HJ, Leung CH and Ma
DL: Structure-Based discovery of a selective KDM5A inhibitor that
exhibits anti-cancer activity via inducing cell cycle arrest and
senescence in breast cancer cell lines. Cancers (Basel). 11:922019.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Lin W, Watanabe H, Peng S, Francis JM,
Kaplan N, Pedamallu CS, Ramachandran A, Agoston A, Bass AJ and
Meyerson M: Dynamic epigenetic regulation by menin during
pancreatic islet tumor formation. Mol Cancer Res. 13:689–698. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Oser MG, Sabet AH, Gao W, Chakraborty AA,
Schinzel AC, Jennings RB, Fonseca R, Bonal DM, Booker MA, Flaifel
A, et al: The KDM5A/RBP2 histone demethylase represses NOTCH
signaling to sustain neuroendocrine differentiation and promote
small cell lung cancer tumorigenesis. Genes Dev. 33:1718–1738.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Yamamoto S, Wu Z, Russnes HG, Takagi S,
Peluffo G, Vaske C, Zhao X, Moen Vollan HK, Maruyama R, Ekram MB,
et al: JARID1B is a luminal lineage-driving oncogene in breast
cancer. Cancer Cell. 25:762–777. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Yamane K, Tateishi K, Klose RJ, Fang J,
Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P
and Zhang Y: PLU-1 is an H3K4 demethylase involved in
transcriptional repression and breast cancer cell proliferation.
Mol Cell. 25:801–812. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Tortelli TC, Tamura RE, de Souza Junqueira
M, da Silva Mororó J, Bustos SO, Natalino RJM, Russell S, Désaubry
L, Strauss BE and Chammas R: Metformin-induced chemosensitization
to cisplatin depends on P53 status and is inhibited by Jarid1b
overexpression in non-small cell lung cancer cells. Aging (Albany
NY). 13:21914–21940. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Kuo KT, Huang WC, Bamodu OA, Lee WH, Wang
CH, Hsiao M, Wang LS and Yeh CT: Histone demethylase JARID1B/KDM5B
promotes aggressiveness of non-small cell lung cancer and serves as
a good prognostic predictor. Clin Epigenetics. 10:1072018.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wang D, Han S, Peng R, Jiao C, Wang X,
Yang X, Yang R and Li X: Depletion of histone demethylase KDM5B
inhibits cell proliferation of hepatocellular carcinoma by
regulation of cell cycle checkpoint proteins p15 and p27. J Exp
Clin Cancer Res. 35:372016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Hayami S, Yoshimatsu M, Veerakumarasivam
A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue
H, et al: Overexpression of the JmjC histone demethylase KDM5B in
human carcinogenesis: Involvement in the proliferation of cancer
cells through the E2F/RB pathway. Mol Cancer. 9:592010. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Huang YQ, Zou Y, Zheng RJ and Ma XD:
Down-regulation of JARID1B expression inhibits cell proliferation,
induces apoptosis and blocks cell cycle in human acute
lymphoblastic leukemia cells. Eur Rev Med Pharmacol Sci.
22:1366–1373. 2018.PubMed/NCBI
|
|
132
|
Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z,
Ma Y, Yu Y, Lin H, Chen AP and Chen CD: JARID1B is a histone H3
lysine 4 demethylase up-regulated in prostate cancer. Proc Natl
Acad Sci USA. 104:19226–19231. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Yan G, Li S, Yue M, Li C and Kang Z:
Lysine demethylase 5B suppresses CC chemokine ligand 14 to promote
progression of colorectal cancer through the Wnt/β-catenin pathway.
Life Sci. 264:1187262021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Niu X, Zhang T, Liao L, Zhou L, Lindner
DJ, Zhou M, Rini B, Yan Q and Yang H: The von Hippel-Lindau tumor
suppressor protein regulates gene expression and tumor growth
through histone demethylase JARID1C. Oncogene. 31:776–786. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Rondinelli B, Rosano D, Antonini E,
Frenquelli M, Montanini L, Huang D, Segalla S, Yoshihara K, Amin
SB, Lazarevic D, et al: Histone demethylase JARID1C inactivation
triggers genomic instability in sporadic renal cancer. J Clin
Invest. 125:4625–4637. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zheng Q, Li P, Zhou X, Qiang Y, Fan J, Lin
Y, Chen Y, Guo J, Wang F, Xue H, et al: Deficiency of the
X-inactivation escaping gene KDM5C in clear cell renal cell
carcinoma promotes tumorigenicity by reprogramming glycogen
metabolism and inhibiting ferroptosis. Theranostics. 11:8674–8691.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhang B, Zhou BH, Xiao M, Li H, Guo L,
Wang MX, Yu SH and Ye QH: KDM5C represses FASN-mediated lipid
metabolism to exert tumor suppressor activity in intrahepatic
cholangiocarcinoma. Front Oncol. 10:10252020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zhang Q, Xu L, Wang J, Zhu X, Ma Z, Yang
J, Li J, Jia X and Wei L: KDM5C expedites lung cancer growth and
metastasis through epigenetic regulation of MicroRNA-133a. Onco
Targets Ther. 14:1187–1204. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Ji X, Jin S, Qu X, Li K, Wang H, He H, Guo
F and Dong L: Lysine-specific demethylase 5C promotes
hepatocellular carcinoma cell invasion through inhibition BMP7
expression. BMC Cancer. 15:8012015. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Lin H, Ma N, Zhao L, Yang G and Cao B:
KDM5c promotes colon cancer cell proliferation through the
FBXW7-c-Jun regulatory axis. Front Oncol. 10:5354492020. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Shen HF, Zhang WJ, Huang Y, He YH, Hu GS,
Wang L, Peng BL, Yi J, Li TT, Rong R, et al: The dual function of
KDM5C in both gene transcriptional activation and repression
promotes breast cancer cell growth and tumorigenesis. Adv Sci
(Weinh). 8:20046352021. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Jangravi Z, Tabar MS, Mirzaei M,
Parsamatin P, Vakilian H, Alikhani M, Shabani M, Haynes PA,
Goodchild AK, Gourabi H, et al: Two splice variants of Y
chromosome-located lysine-specific demethylase 5D have distinct
function in prostate cancer cell line (DU-145). J Proteome Res.
14:3492–3502. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Li N, Dhar SS, Chen TY, Kan PY, Wei Y, Kim
JH, Chan CH, Lin HK, Hung MC and Lee MG: JARID1D is a suppressor
and prognostic marker of prostate cancer invasion and metastasis.
Cancer Res. 76:831–843. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Shen X, Hu K, Cheng G, Xu L, Chen Z, Du P
and Zhuang Z: KDM5D inhibit epithelial-mesenchymal transition of
gastric cancer through demethylation in the promoter of Cul4A in
male. J Cell Biochem. 120:12247–12258. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Cai LS, Chen QX, Fang SY, Lian MQ, Lian MJ
and Cai MZ: ETV4 promotes the progression of gastric cancer through
regulating KDM5D. Eur Rev Med Pharmacol Sci. 24:2442–2451.
2020.PubMed/NCBI
|
|
146
|
Willis-Owen SAG, Domingo-Sabugo C, Starren
E, Liang L, Freidin MB, Arseneault M, Zhang Y, Lu SK, Popat S, Lim
E, et al: Y disruption, autosomal hypomethylation and poor male
lung cancer survival. Sci Rep. 11:124532021. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Schulz WA, Lang A, Koch J and Greife A:
The histone demethylase UTX/KDM6A in cancer: Progress and puzzles.
Int J Cancer. 145:614–620. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Walport LJ, Hopkinson RJ, Vollmar M,
Madden SK, Gileadi C, Oppermann U, Schofield CJ and Johansson C:
Human UTY(KDM6C) is a male-specific N-methyl lysyl demethylase. J
Biol Chem. 289:18302–18313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Sengoku T and Yokoyama S: Structural basis
for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev.
25:2266–2277. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Li Y, Yang J, Zhang X, Liu H and Guo J:
KDM6A suppresses hepatocellular carcinoma cell proliferation by
negatively regulating the TGF-β/SMAD signaling pathway. Exp Ther
Med. 20:2774–2782. 2020.PubMed/NCBI
|
|
151
|
Ler LD, Ghosh S, Chai X, Thike AA, Heng
HL, Siew EY, Dey S, Koh LK, Lim JQ, Lim WK, et al: Loss of tumor
suppressor KDM6A amplifies PRC2-regulated transcriptional
repression in bladder cancer and can be targeted through inhibition
of EZH2. Sci Transl Med. 9:eaai83122017. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Zhang J, Ying Y, Li M, Wang M, Huang X,
Jia M, Zeng J, Ma C, Zhang Y, Li C, et al: Targeted inhibition of
KDM6 histone demethylases eradicates tumor-initiating cells via
enhancer reprogramming in colorectal cancer. Theranostics.
10:10016–10030. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Romero OA, Vilarrubi A, Alburquerque-Bejar
JJ, Gomez A, Andrades A, Trastulli D, Pros E, Setien F, Verdura S,
Farré L, et al: SMARCA4 deficient tumours are vulnerable to
KDM6A/UTX and KDM6B/JMJD3 blockade. Nat Commun. 12:43192021.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Chaturvedi SS, Ramanan R, Waheed SO,
Karabencheva-Christova TG and Christov CZ: Structure-function
relationships in KDM7 histone demethylases. Adv Protein Chem Struct
Biol. 117:113–125. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Park SY, Park JW and Chun YS: Jumonji
histone demethylases as emerging therapeutic targets. Pharmacol
Res. 105:146–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Osawa T, Muramatsu M, Wang F, Tsuchida R,
Kodama T, Minami T and Shibuya M: Increased expression of histone
demethylase JHDM1D under nutrient starvation suppresses tumor
growth via down-regulating angiogenesis. Proc Natl Acad Sci USA.
108:20725–20729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Lee KH, Hong S, Kang M, Jeong CW, Ku JH,
Kim HH and Kwak C: Histone demethylase KDM7A controls androgen
receptor activity and tumor growth in prostate cancer. Int J
Cancer. 143:2849–2861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Zhang Z, Chen B, Zhu Y, Zhang T, Zhang X,
Yuan Y and Xu Y: The Jumonji domain-containing histone demethylase
homolog 1D/lysine demethylase 7A (JHDM1D/KDM7A) is an epigenetic
activator of RHOJ transcription in breast cancer cells. Front Cell
Dev Biol. 9:6643752021. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Tong D, Liu Q, Liu G, Yuan W, Wang L, Guo
Y, Lan W, Zhang D, Dong S, Wang Y, et al: The HIF/PHF8/AR axis
promotes prostate cancer progression. Oncogenesis. 5:e2832016.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Li S, Sun A, Liang X, Ma L, Shen L, Li T,
Zheng L, Shang W, Zhao W and Jia J: Histone demethylase PHF8
promotes progression and metastasis of gastric cancer. Am J Cancer
Res. 7:448–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Zhou W, Gong L, Wu Q, Xing C, Wei B, Chen
T, Zhou Y, Yin S, Jiang B, Xie H, et al: PHF8 upregulation
contributes to autophagic degradation of E-cadherin,
epithelial-mesenchymal transition and metastasis in hepatocellular
carcinoma. J Exp Clin Cancer Res. 37:2152018. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Tseng LL, Cheng HH, Yeh TS, Huang SC, Syu
YY, Chuu CP, Yuh CH, Kung HJ and Wang WC: Targeting the histone
demethylase PHF8-mediated PKCα-Src-PTEN axis in HER2-negative
gastric cancer. Proc Natl Acad Sci USA. 117:24859–24866. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Shao P, Liu Q, Maina PK, Cui J, Bair TB,
Li T, Umesalma S, Zhang W and Qi HH: Histone demethylase PHF8
promotes epithelial to mesenchymal transition and breast
tumorigenesis. Nucleic Acids Res. 45:1687–1702. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Liu Q, Pang J, Wang LA, Huang Z, Xu J,
Yang X, Xie Q, Huang Y, Tang T, Tong D, et al: Histone demethylase
PHF8 drives neuroendocrine prostate cancer progression by
epigenetically upregulating FOXA2. J Pathol. 253:106–118. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Fu Y, Liu M, Li F, Qian L, Zhang P, Lv F,
Cheng W and Hou R: miR-221 promotes hepatocellular carcinoma cells
migration via targeting PHF2. Biomed Res Int. 2019:43714052019.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Lee KH, Park JW, Sung HS, Choi YJ, Kim WH,
Lee HS, Chung HJ, Shin HW, Cho CH, Kim TY, et al: PHF2 histone
demethylase acts as a tumor suppressor in association with p53 in
cancer. Oncogene. 34:2897–2909. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Del Rizzo PA, Krishnan S and Trievel RC:
Crystal structure and functional analysis of JMJD5 indicate an
alternate specificity and function. Mol Cell Biol. 32:4044–4052.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Hsia DA, Tepper CG, Pochampalli MR, Hsia
EY, Izumiya C, Huerta SB, Wright ME, Chen HW, Kung HJ and Izumiya
Y: KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1
coding region to regulate cancer cell proliferation. Proc Natl Acad
Sci USA. 107:9671–9676. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Yao Y, Zhou WY and He RX: Down-regulation
of JMJD5 suppresses metastasis and induces apoptosis in oral
squamous cell carcinoma by regulating p53/NF-κB pathway. Biomed
Pharmacother. 109:1994–2004. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Chang WH, Forde D and Lai AG: Dual
prognostic role of 2-oxoglutarate-dependent oxygenases in ten
cancer types: Implications for cell cycle regulation and cell
adhesion maintenance. Cancer Commun (Lond). 39:232019. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Wu LW, Zhou DM, Zhang ZY, Zhang JK, Zhu
HJ, Lin NM and Zhang C: Suppression of LSD1 enhances the cytotoxic
and apoptotic effects of regorafenib in hepatocellular carcinoma
cells. Biochem Biophys Res Commun. 512:852–858. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Verigos J, Karakaidos P, Kordias D,
Papoudou-Bai A, Evangelou Z, Harissis HV, Klinakis A and Magklara
A: The histone demethylase LSD1/ΚDM1A mediates chemoresistance in
breast cancer via regulation of a stem cell program. Cancers
(Basel). 11:15852019. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Lee YK, Lim J, Yoon SY, Joo JC, Park SJ
and Park YJ: Promotion of cell death in cisplatin-resistant ovarian
cancer cells through KDM1B-DCLRE1B modulation. Int J Mol Sci.
20:24432019. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Tang D, He J, Dai Y, Geng X, Leng Q, Jiang
H, Sun R and Xu S: Targeting KDM1B-dependent miR-215-AR-AGR2-axis
promotes sensitivity to enzalutamide-resistant prostate cancer.
Cancer Gene Ther. 29:543–557. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Tang DE, Dai Y, He JX, Lin LW, Leng QX,
Geng XY, Fu DX, Jiang HW and Xu SH: Targeting the KDM4B-AR-c-Myc
axis promotes sensitivity to androgen receptor-targeted therapy in
advanced prostate cancer. J Pathol. 252:101–113. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Hou J, Wu J, Dombkowski A, Zhang K,
Holowatyj A, Boerner JL and Yang ZQ: Genomic amplification and a
role in drug-resistance for the KDM5A histone demethylase in breast
cancer. Am J Transl Res. 4:247–256. 2012.PubMed/NCBI
|
|
177
|
Banelli B, Carra E, Barbieri F, Würth R,
Parodi F, Pattarozzi A, Carosio R, Forlani A, Allemanni G, Marubbi
D, et al: The histone demethylase KDM5A is a key factor for the
resistance to temozolomide in glioblastoma. Cell Cycle.
14:3418–3429. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Xu W, Zhou B, Zhao X, Zhu L, Xu J, Jiang
Z, Chen D, Wei Q, Han M, Feng L, et al: KDM5B demethylates H3K4 to
recruit XRCC1 and promote chemoresistance. Int J Biol Sci.
14:1122–1132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Roesch A, Vultur A, Bogeski I, Wang H,
Zimmermann KM, Speicher D, Körbel C, Laschke MW, Gimotty PA,
Philipp SE, et al: Overcoming intrinsic multidrug resistance in
melanoma by blocking the mitochondrial respiratory chain of
slow-cycling JARID1B(high) cells. Cancer Cell. 23:811–825. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Liu X, Zhang SM, McGeary MK, Krykbaeva I,
Lai L, Jansen DJ, Kales SC, Simeonov A, Hall MD, Kelly DP, et al:
KDM5B promotes drug resistance by regulating melanoma-propagating
cell subpopulations. Mol Cancer Ther. 18:706–717. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Hinohara K, Wu HJ, Vigneau S, McDonald TO,
Igarashi KJ, Yamamoto KN, Madsen T, Fassl A, Egri SB, Papanastasiou
M, et al: KDM5 histone demethylase activity links cellular
transcriptomic heterogeneity to therapeutic resistance. Cancer
Cell. 34:939–953.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Pippa S, Mannironi C, Licursi V, Bombardi
L, Colotti G, Cundari E, Mollica A, Coluccia A, Naccarato V, La
Regina G, et al: Small molecule inhibitors of KDM5 histone
demethylases increase the radiosensitivity of breast cancer cells
overexpressing JARID1B. Molecules. 24:17392019. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Lin H, Yang G, Yu J, Wang J, Li Q, Guo S
and Cao B: KDM5c inhibits multidrug resistance of colon cancer cell
line by down-regulating ABCC1. Biomed Pharmacother. 107:1205–1209.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Komura K, Jeong SH, Hinohara K, Qu F, Wang
X, Hiraki M, Azuma H, Lee GS, Kantoff PW and Sweeney CJ: Resistance
to docetaxel in prostate cancer is associated with androgen
receptor activation and loss of KDM5D expression. Proc Natl Acad
Sci USA. 113:6259–6264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
He C, Sun J, Liu C, Jiang Y and Hao Y:
Elevated H3K27me3 levels sensitize osteosarcoma to cisplatin. Clin
Epigenetics. 11:82019. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Wang Q, Chen X, Jiang Y, Liu S, Liu H, Sun
X, Zhang H, Liu Z, Tao Y, Li C, et al: Elevating H3K27me3 level
sensitizes colorectal cancer to oxaliplatin. J Mol Cell Biol.
12:125–137. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Mathur R, Sehgal L, Havranek O, Köhrer S,
Khashab T, Jain N, Burger JA, Neelapu SS, Davis RE and Samaniego F:
Inhibition of demethylase KDM6B sensitizes diffuse large B-cell
lymphoma to chemotherapeutic drugs. Haematologica. 102:373–380.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Zhang C, Shen L, Zhu Y, Xu R, Deng Z, Liu
X, Ding Y, Wang C, Shi Y, Bei L, et al: KDM6A promotes imatinib
resistance through YY1-mediated transcriptional upregulation of
TRKA independently of its demethylase activity in chronic
myelogenous leukemia. Theranostics. 11:2691–2705. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Macedo-Silva C, Miranda-Goncalves V,
Lameirinhas A, Lencart J, Pereira A, Lobo J, Guimarães R, Martins
AT, Henrique R, Bravo I and Jerónimo C: JmjC-KDMs KDM3A and KDM6B
modulate radioresistance under hypoxic conditions in esophageal
squamous cell carcinoma. Cell Death Dis. 11:10682020. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Xu S, Fan L, Jeon HY, Zhang F, Cui X,
Mickle MB, Peng G, Hussain A, Fazli L, Gleave ME, et al:
p300-mediated acetylation of histone demethylase JMJD1A prevents
its degradation by ubiquitin ligase STUB1 and enhances its activity
in prostate cancer. Cancer Res. 80:3074–3087. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
McAllister TE, England KS, Hopkinson RJ,
Brennan PE, Kawamura A and Schofield CJ: Recent progress in histone
demethylase inhibitors. J Med Chem. 59:1308–1329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Fang Y, Liao G and Yu B: LSD1/KDM1A
inhibitors in clinical trials: Advances and prospects. J Hematol
Oncol. 12:1292019. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Fu DJ, Li J and Yu B: Annual review of
LSD1/KDM1A inhibitors in 2020. Eur J Med Chem. 214:1132542021.
View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Kleszcz R, Skalski M, Krajka-Kuźniak V and
Paluszczak J: The inhibitors of KDM4 and KDM6 histone lysine
demethylases enhance the anti-growth effects of erlotinib and
HS-173 in head and neck cancer cells. Eur J Pharm Sci.
166:1059612021. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Illiano M, Conte M, Salzillo A, Ragone A,
Spina A, Nebbioso A, Altucci L, Sapio L and Naviglio S: The KDM
inhibitor GSKJ4 triggers CREB downregulation via a protein kinase A
and proteasome-dependent mechanism in human acute myeloid leukemia
cells. Front Oncol. 10:7992020. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Yang GJ, Wu J, Miao L, Zhu MH, Zhou QJ, Lu
XJ, Lu JF, Leung CH, Ma DL and Chen J: Pharmacological inhibition
of KDM5A for cancer treatment. Eur J Med Chem. 226:1138552021.
View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Baby S, Gurukkala Valapil D and
Shankaraiah N: Unravelling KDM4 histone demethylase inhibitors for
cancer therapy. Drug Discov Today. 26:1841–1856. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Varghese B, Del Gaudio N, Cobellis G,
Altucci L and Nebbioso A: KDM4 involvement in breast cancer and
possible therapeutic approaches. Front Oncol. 11:7503152021.
View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Carter DM, Specker E, Małecki PH,
Przygodda J, Dudaniec K, Weiss MS, Heinemann U, Nazaré M and Gohlke
U: Enhanced properties of a benzimidazole benzylpyrazole lysine
demethylase inhibitor: Mechanism-of-action, binding site analysis,
and activity in cellular models of prostate cancer. J Med Chem.
64:14266–14282. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Souto JA, Sarno F, Nebbioso A, Papulino C,
Álvarez R, Lombino J, Perricone U, Padova A, Altucci L and de Lera
ÁR: A new family of Jumonji C domain-containing KDM inhibitors
inspired by natural product purpurogallin. Front Chem. 8:3122020.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Tayari MM, Santos HGD, Kwon D, Bradley TJ,
Thomassen A, Chen C, Dinh Y, Perez A, Zelent A, Morey L, et al:
Clinical responsiveness to all-trans retinoic acid is potentiated
by LSD1 inhibition and associated with a quiescent transcriptome in
myeloid malignancies. Clin Cancer Res. 27:1893–1903. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Cuyàs E, Gumuzio J, Verdura S, Brunet J,
Bosch-Barrera J, Martin-Castillo B, Alarcón T, Encinar JA, Martin
ÁG and Menendez JA: The LSD1 inhibitor iadademstat (ORY-1001)
targets SOX2-driven breast cancer stem cells: A potential
epigenetic therapy in luminal-B and HER2-positive breast cancer
subtypes. Aging (Albany NY). 12:4794–4814. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Wang T, Zhang F and Sun F: ORY-1001, a
KDM1A inhibitor, inhibits proliferation, and promotes apoptosis of
triple negative breast cancer cells by inactivating androgen
receptor. Drug Dev Res. 83:208–216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Maes T, Mascaró C, Tirapu I, Estiarte A,
Ciceri F, Lunardi S, Guibourt N, Perdones A, Lufino MMP,
Somervaille TCP, et al: ORY-1001, a potent and selective covalent
KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell.
33:495–511.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Salamero O, Montesinos P, Willekens C,
Pérez-Simón JA, Pigneux A, Récher C, Popat R, Carpio C, Molinero C,
Mascaró C, et al: First-in-human phase I study of iadademstat
(ORY-1001): A first-in-class lysine-specific histone demethylase 1A
inhibitor, in relapsed or refractory acute myeloid leukemia. J Clin
Oncol. 38:4260–4273. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Huang M, Chen C, Geng J, Han D, Wang T,
Xie T, Wang L, Wang Y, Wang C, Lei Z and Chu X: Targeting KDM1A
attenuates Wnt/β-catenin signaling pathway to eliminate
sorafenib-resistant stem-like cells in hepatocellular carcinoma.
Cancer Lett. 398:12–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Lillico R, Lawrence CK and Lakowski TM:
Selective DOT1L, LSD1, and HDAC class I inhibitors reduce HOXA9
expression in MLL-AF9 rearranged leukemia cells, but dysregulate
the expression of many histone-modifying enzymes. J Proteome Res.
17:2657–2667. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Bauer TM, Besse B, Martinez-Marti A, Trigo
JM, Moreno V, Garrido P, Ferron-Brady G, Wu Y, Park J, Collingwood
T, et al: Phase I, open-label, dose-escalation study of the safety,
pharmacokinetics, pharmacodynamics, and efficacy of GSK2879552 in
relapsed/refractory SCLC. J Thorac Oncol. 14:1828–1838. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Kanouni T, Severin C, Cho RW, Yuen NY, Xu
J, Shi L, Lai C, Del Rosario JR, Stansfield RK, Lawton LN, et al:
Discovery of CC-90011: A potent and selective reversible inhibitor
of lysine specific demethylase 1 (LSD1). J Med Chem.
63:14522–14529. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Hollebecque A, Salvagni S, Plummer R,
Isambert N, Niccoli P, Capdevila J, Curigliano G, Moreno V,
Martin-Romano P, Baudin E, et al: Phase I study of lysine-specific
demethylase 1 inhibitor, CC-90011, in patients with advanced solid
tumors and relapsed/refractory non-Hodgkin lymphoma. Clin Cancer
Res. 27:438–446. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Sterling J, Menezes SV, Abbassi RH and
Munoz L: Histone lysine demethylases and their functions in cancer.
Int J Cancer. Oct 31–2020.(Epub ahead of print). doi:
10.1002/ijc.33375. PubMed/NCBI
|
|
212
|
Oner E, Kotmakci M, Baird AM, Gray SG,
Debelec Butuner B, Bozkurt E, Kantarci AG and Finn SP: Development
of EphA2 siRNA-loaded lipid nanoparticles and combination with a
small-molecule histone demethylase inhibitor in prostate cancer
cells and tumor spheroids. J Nanobiotechnology. 19:712021.
View Article : Google Scholar : PubMed/NCBI
|