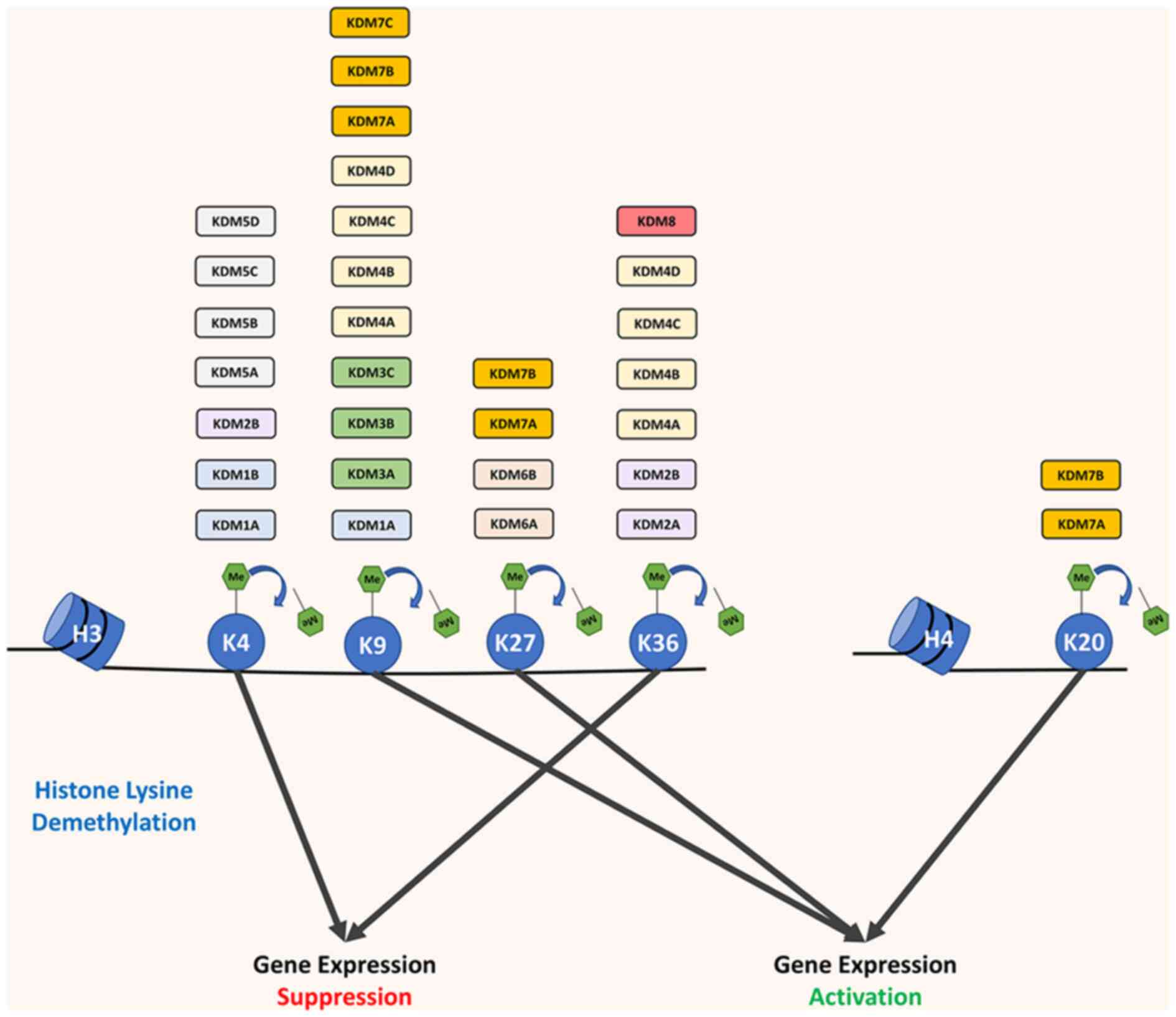
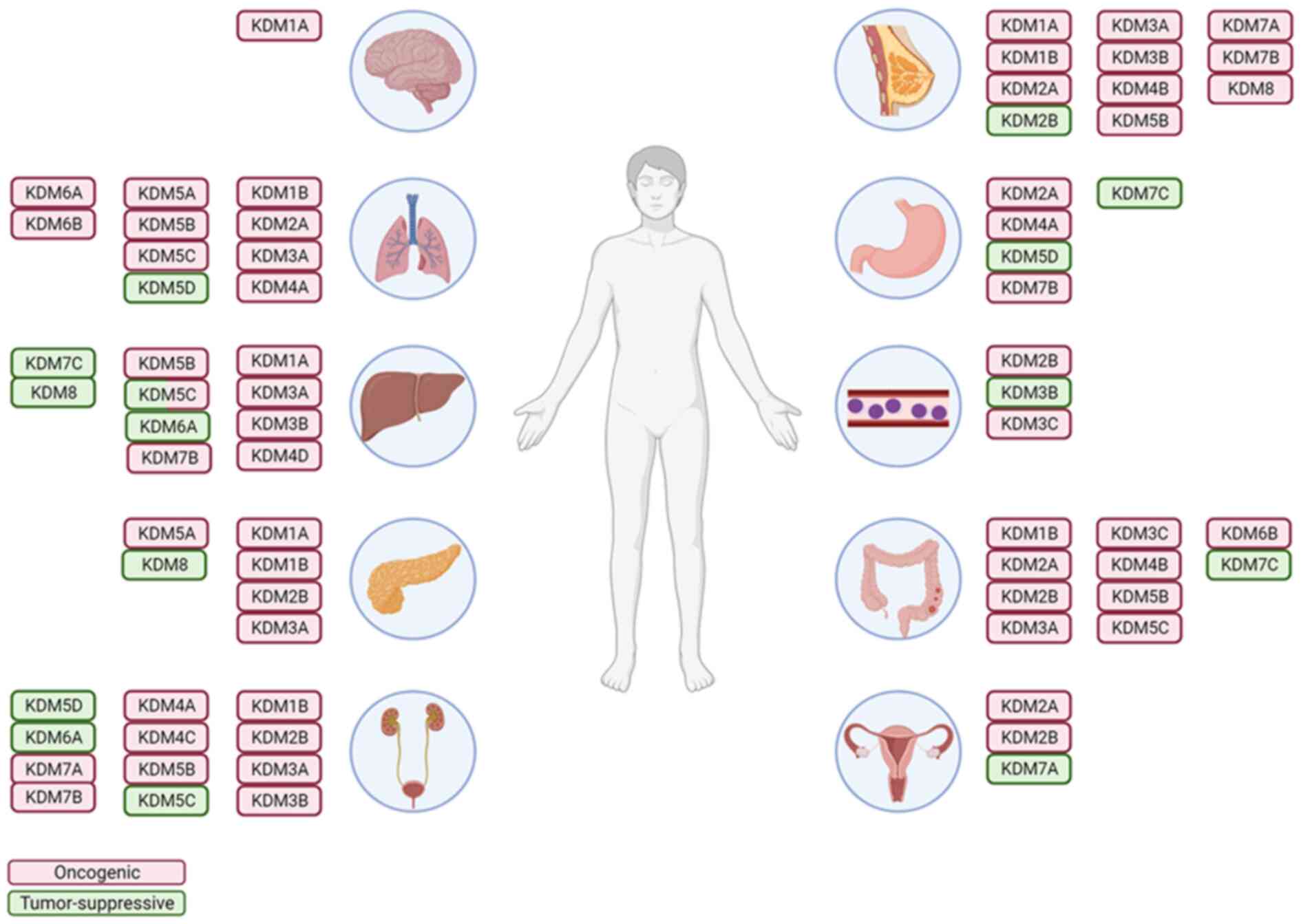
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kruger S, Ilmer M, Kobold S, Cadilha BL,
Endres S, Ormanns S, Schuebbe G, Renz BW, D'Haese JG, Schloesser H,
et al: Advances in cancer immunotherapy 2019-latest trends. J Exp
Clin Cancer Res. 38:2682019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thompson JA, Schneider BJ, Brahmer J,
Achufusi A, Armand P, Berkenstock MK, Bhatia S, Budde LE, Chokshi
S, Davies M, et al: Management of immunotherapy-related toxicities,
version 1.2022, NCCN clinical practice guidelines in oncology. J
Natl Compr Canc Netw. 20:387–405. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Michalak EM, Burr ML, Bannister AJ and
Dawson MA: The roles of DNA, RNA and histone methylation in ageing
and cancer. Nat Rev Mol Cell Biol. 20:573–589. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tachibana M, Sugimoto K, Nozaki M, Ueda J,
Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H and Shinkai Y:
G9a histone methyltransferase plays a dominant role in euchromatic
histone H3 lysine 9 methylation and is essential for early
embryogenesis. Genes Dev. 16:1779–1791. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peters AH, O'Carroll D, Scherthan H,
Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner
M, Kohlmaier A, et al: Loss of the Suv39h histone
methyltransferases impairs mammalian heterochromatin and genome
stability. Cell. 107:323–337. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee
JW, Verdine GL, Allis CD and Roeder RG: Regulation of MLL1 H3K4
methyltransferase activity by its core components. Nat Struct Mol
Biol. 13:713–719. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi Y: Histone lysine demethylases:
Emerging roles in development, physiology and disease. Nat Rev
Genet. 8:829–833. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Soares LM, He PC, Chun Y, Suh H, Kim T and
Buratowski S: Determinants of histone H3K4 methylation patterns.
Mol Cell. 68:773–785.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fodor BD, Kubicek S, Yonezawa M,
O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K,
Schotta G and Jenuwein T: Jmjd2b antagonizes H3K9 trimethylation at
pericentric heterochromatin in mammalian cells. Genes Dev.
20:1557–1562. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Z and Zhang Y: Maternal
H3K27me3-dependent autosomal and X chromosome imprinting. Nat Rev
Genet. 21:555–571. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cloos PA, Christensen J, Agger K, Maiolica
A, Rappsilber J, Antal T, Hansen KH and Helin K: The putative
oncogene GASC1 demethylates tri- and dimethylated lysine 9 on
histone H3. Nature. 442:307–311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klose RJ, Yamane K, Bae Y, Zhang D,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: The
transcriptional repressor JHDM3A demethylates trimethyl histone H3
lysine 9 and lysine 36. Nature. 442:312–316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vallianatos CN and Iwase S: Disrupted
intricacy of histone H3K4 methylation in neurodevelopmental
disorders. Epigenomics. 7:503–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clark EA, Wu F, Chen Y, Kang P, Kaiser UB,
Fang R and Shi YG: GR and LSD1/KDM1A-targeted gene activation
requires selective H3K4me2 demethylation at enhancers. Cell Rep.
27:3522–3532.e3. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tsukada Y, Fang J, Erdjument-Bromage H,
Warren ME, Borchers CH, Tempst P and Zhang Y: Histone demethylation
by a family of JmjC domain-containing proteins. Nature.
439:811–816. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Trojer P, Zhang J, Yonezawa M, Schmidt A,
Zheng H, Jenuwein T and Reinberg D: Dynamic histone H1 isotype 4
methylation and demethylation by histone lysine methyltransferase
G9a/KMT1C and the Jumonji domain-containing JMJD2/KDM4 proteins. J
Biol Chem. 284:8395–8405. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suzuki T, Ozasa H, Itoh Y, Zhan P, Sawada
H, Mino K, Walport L, Ohkubo R, Kawamura A, Yonezawa M, et al:
Identification of the KDM2/7 histone lysine demethylase subfamily
inhibitor and its antiproliferative activity. J Med Chem.
56:7222–7231. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Berry WL and Janknecht R: KDM4/JMJD2
histone demethylases: Epigenetic regulators in cancer cells. Cancer
Res. 73:2936–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Turberfield AH, Kondo T, Nakayama M,
Koseki Y, King HW, Koseki H and Klose RJ: KDM2 proteins constrain
transcription from CpG island gene promoters independently of their
histone demethylase activity. Nucleic Acids Res. 47:9005–9023.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian Z, Yao L, Shen Y, Guo X and Duan X:
Histone H3K9 demethylase JMJD1A is a co-activator of erythropoietin
expression under hypoxia. Int J Biochem Cell Biol. 109:33–39. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sui Y, Gu R and Janknecht R: Crucial
functions of the JMJD1/KDM3 epigenetic regulators in cancer. Mol
Cancer Res. 19:3–13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wagner KW, Alam H, Dhar SS, Giri U, Li N,
Wei Y, Giri D, Cascone T, Kim JH, Ye Y, et al: KDM2A promotes lung
tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin
Invest. 123:5231–5246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hughes AL, Kelley JR and Klose RJ:
Understanding the interplay between CpG island-associated gene
promoters and H3K4 methylation. Biochim Biophys Acta Gene Regul
Mech. 1863:1945672020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barski A, Cuddapah S, Cui K, Roh TY,
Schones DE, Wang Z, Wei G, Chepelev I and Zhao K: High-resolution
profiling of histone methylations in the human genome. Cell.
129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Y, Yang Y, Wang F, Wan K, Yamane K,
Zhang Y and Lei M: Crystal structure of human histone
lysine-specific demethylase 1 (LSD1). Proc Natl Acad Sci USA.
103:13956–13961. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arifuzzaman S, Khatun MR and Khatun R:
Emerging of lysine demethylases (KDMs): From pathophysiological
insights to novel therapeutic opportunities. Biomed Pharmacother.
129:1103922020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carnesecchi J, Forcet C, Zhang L,
Tribollet V, Barenton B, Boudra R, Cerutti C, Billas IM, Sérandour
AA, Carroll JS, et al: ERRα induces H3K9 demethylation by LSD1 to
promote cell invasion. Proc Natl Acad Sci USA. 114:3909–3914. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He Y, Zhao Y, Wang L, Bohrer LR, Pan Y,
Wang L and Huang H: LSD1 promotes S-phase entry and tumorigenesis
via chromatin co-occupation with E2F1 and selective H3K9
demethylation. Oncogene. 37:534–543. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y and Cao K: KDM1A promotes
immunosuppression in hepatocellular carcinoma by regulating PD-L1
through demethylating MEF2D. J Immunol Res. 2021:99650992021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hou X, Li Q, Yang L, Yang Z, He J, Li Q
and Li D: KDM1A and KDM3A promote tumor growth by upregulating cell
cycle-associated genes in pancreatic cancer. Exp Biol Med
(Maywood). 246:1869–1883. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim HS, Son BK, Kwon MJ, Kim DH and Min
KW: High KDM1A expression associated with decreased CD8+ T cells
reduces the breast cancer survival rate in patients with breast
cancer. J Clin Med. 10:11122021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Majello B, Gorini F, Saccà CD and Amente
S: Expanding the role of the histone lysine-specific demethylase
LSD1 in cancer. Cancers (Basel). 11:3242019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang J, Sengupta R, Espejo AB, Lee MG,
Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT,
Jenuwein T and Berger SL: p53 is regulated by the lysine
demethylase LSD1. Nature. 449:105–108. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cho HS, Suzuki T, Dohmae N, Hayami S,
Unoki M, Yoshimatsu M, Toyokawa G, Takawa M, Chen T, Kurash JK, et
al: Demethylation of RB regulator MYPT1 by histone demethylase LSD1
promotes cell cycle progression in cancer cells. Cancer Res.
71:655–660. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Saccà CD, Gorini F, Ambrosio S, Amente S,
Faicchia D, Matarese G, Lania L and Majello B: Inhibition of
lysine-specific demethylase LSD1 induces senescence in glioblastoma
cells through a HIF-1α-dependent pathway. Biochim Biophys Acta Gene
Regul Mech. 1862:535–546. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qin Y, Vasilatos SN, Chen L, Wu H, Cao Z,
Fu Y, Huang M, Vlad AM, Lu B, Oesterreich S, et al: Inhibition of
histone lysine-specific demethylase 1 elicits breast tumor immunity
and enhances antitumor efficacy of immune checkpoint blockade.
Oncogene. 38:390–405. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Katz TA, Vasilatos SN, Harrington E,
Oesterreich S, Davidson NE and Huang Y: Inhibition of histone
demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases
sensitivity to DNMT inhibitor-induced apoptosis in breast cancer
cells. Breast Cancer Res Treat. 146:99–108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cai S, Wang J, Zeng W, Cheng X, Liu L and
Li W: Lysine-specific histone demethylase 1B (LSD2/KDM1B) represses
p53 expression to promote proliferation and inhibit apoptosis in
colorectal cancer through LSD2-mediated H3K4me2 demethylation.
Aging (Albany NY). 12:14990–15001. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cao Y, Guo C, Yin Y, Li X and Zhou L:
Lysine-specific demethylase 2 contributes to the proliferation of
small cell lung cancer by regulating the expression of TFPI-2. Mol
Med Rep. 18:733–740. 2018.PubMed/NCBI
|
|
43
|
Kumar A, Kumari N, Sharma U, Ram S, Singh
SK, Kakkar N, Kaushal K and Prasad R: Reduction in H3K4me patterns
due to aberrant expression of methyltransferases and demethylases
in renal cell carcinoma: Prognostic and therapeutic implications.
Sci Rep. 9:81892019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Sun L, Luo Y and He S: Knockdown
of KDM1B inhibits cell proliferation and induces apoptosis of
pancreatic cancer cells. Pathol Res Pract. 215:1054–1060. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi
X, Qin B, Zeng L, Esteban MA, Pan G and Pei D: The histone
demethylases Jhdm1a/1b enhance somatic cell reprogramming in a
vitamin-C-dependent manner. Cell Stem Cell. 9:575–587. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kong Y, Zou S, Yang F, Xu X, Bu W, Jia J
and Liu Z: RUNX3-mediated up-regulation of miR-29b suppresses the
proliferation and migration of gastric cancer cells by targeting
KDM2A. Cancer Lett. 381:138–148. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang F, Liang S, Liu X, Han L, Wang J and
Du Q: LINC00460 modulates KDM2A to promote cell proliferation and
migration by targeting miR-342-3p in gastric cancer. Onco Targets
Ther. 11:6383–6394. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ou R, Zhu L, Zhao L, Li W, Tao F, Lu Y, He
Q, Li J, Ren Y and Xu Y: HPV16 E7-induced upregulation of KDM2A
promotes cervical cancer progression by regulating miR-132-radixin
pathway. J Cell Physiol. 234:2659–2671. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu DH, Yang J, Gao LK, Min J, Tang JM, Hu
M, Li Y, Li ST, Chen J and Hong L: Lysine demethylase 2A promotes
the progression of ovarian cancer by regulating the PI3K pathway
and reversing epithelial-mesenchymal transition. Oncol Rep.
41:917–927. 2019.PubMed/NCBI
|
|
50
|
Zhao Y, Chen X, Jiang J, Wan X, Wang Y and
Xu P: Epigallocatechin gallate reverses gastric cancer by
regulating the long noncoding RNA LINC00511/miR-29b/KDM2A axis.
Biochim Biophys Acta Mol Basis Dis. 1866:1658562020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xi C, Ye NY and Wang YB: LncRNA LINC01278
accelerates colorectal cancer progression via miR-134-5p/KDM2A
axis. Eur Rev Med Pharmacol Sci. 24:10526–10534. 2020.PubMed/NCBI
|
|
52
|
Kottakis F, Polytarchou C, Foltopoulou P,
Sanidas I, Kampranis SC and Tsichlis PN: FGF-2 regulates cell
proliferation, migration, and angiogenesis through an
NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell. 43:285–298. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tzatsos A, Paskaleva P, Ferrari F,
Deshpande V, Stoykova S, Contino G, Wong KK, Lan F, Trojer P, Park
PJ and Bardeesy N: KDM2B promotes pancreatic cancer via
polycomb-dependent and -independent transcriptional programs. J
Clin Invest. 123:727–739. 2013.PubMed/NCBI
|
|
54
|
Kuang Y, Lu F, Guo J, Xu H, Wang Q, Xu C,
Zeng L and Yi S: Histone demethylase KDM2B upregulates histone
methyltransferase EZH2 expression and contributes to the
progression of ovarian cancer in vitro and in vivo. Onco Targets
Ther. 10:3131–3144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sanches JGP, Song B, Zhang Q, Cui X,
Yabasin IB, Ntim M, Li X, He J, Zhang Y, Mao J, et al: The role of
KDM2B and EZH2 in regulating the stemness in colorectal cancer
through the PI3K/AKT pathway. Front Oncol. 11:6372982021.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dhar SS, Alam H, Li N, Wagner KW, Chung J,
Ahn YW and Lee MG: Transcriptional repression of histone
deacetylase 3 by the histone demethylase KDM2A is coupled to
tumorigenicity of lung cancer cells. J Biol Chem. 289:7483–7496.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen JY, Li CF, Chu PY, Lai YS, Chen CH,
Jiang SS, Hou MF and Hung WC: Lysine demethylase 2A promotes
stemness and angiogenesis of breast cancer by upregulating Jagged1.
Oncotarget. 7:27689–27710. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen JY, Li CF, Lai YS and Hung WC: Lysine
demethylase 2A expression in cancer-associated fibroblasts promotes
breast tumour growth. Br J Cancer. 124:484–493. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Quan M, Chen Z, Jiao F, Xiao X, Xia Q,
Chen J, Chao Q, Li Y, Gao Y, Yang H, et al: Lysine demethylase 2
(KDM2B) regulates hippo pathway via MOB1 to promote pancreatic
ductal adenocarcinoma (PDAC) progression. J Exp Clin Cancer Res.
39:132020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wanna-Udom S, Terashima M, Suphakhong K,
Ishimura A, Takino T and Suzuki T: KDM2B is involved in the
epigenetic regulation of TGF-β-induced epithelial-mesenchymal
transition in lung and pancreatic cancer cell lines. J Biol Chem.
296:1002132021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
van den Boom V, Maat H, Geugien M,
Rodríguez López A, Sotoca AM, Jaques J, Brouwers-Vos AZ, Fusetti F,
Groen RW, Yuan H, et al: Non-canonical PRC1.1 targets active genes
independent of H3K27me3 and is essential for leukemogenesis. Cell
Rep. 14:332–346. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Galbiati A, Penzo M, Bacalini MG,
Onofrillo C, Guerrieri AN, Garagnani P, Franceschi C, Treré D and
Montanaro L: Epigenetic up-regulation of ribosome biogenesis and
more aggressive phenotype triggered by the lack of the histone
demethylase JHDM1B in mammary epithelial cells. Oncotarget.
8:37091–37103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yoo J, Jeon YH, Cho HY, Lee SW, Kim GW,
Lee DH and Kwon SH: Advances in histone demethylase KDM3A as a
cancer therapeutic target. Cancers (Basel). 12:10982020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yamane K, Toumazou C, Tsukada Y,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: JHDM2A, a
JmjC-containing H3K9 demethylase, facilitates transcription
activation by androgen receptor. Cell. 125:483–495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen M, Zhu N, Liu X, Laurent B, Tang Z,
Eng R, Shi Y, Armstrong SA and Roeder RG: JMJD1C is required for
the survival of acute myeloid leukemia by functioning as a
coactivator for key transcription factors. Genes Dev. 29:2123–2139.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jiang Y, Li C, Wu Q, An P, Huang L, Wang
J, Chen C, Chen X, Zhang F, Ma L, et al: Iron-dependent histone 3
lysine 9 demethylation controls B cell proliferation and humoral
immune responses. Nat Commun. 10:29352019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Brauchle M, Yao Z, Arora R, Thigale S,
Clay I, Inverardi B, Fletcher J, Taslimi P, Acker MG, Gerrits B, et
al: Protein complex interactor analysis and differential activity
of KDM3 subfamily members towards H3K9 methylation. PLoS One.
8:e605492013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu J, Liang T and Zhangsun W: KDM3A is
associated with tumor metastasis and modulates colorectal cancer
cell migration and invasion. Int J Biol Macromol. 126:318–325.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang Z, Yang X, Liu C, Li X, Zhang B, Wang
B, Zhang Y, Song C, Zhang T, Liu M, et al: Acetylation of PHF5A
modulates stress responses and colorectal carcinogenesis through
alternative splicing-mediated upregulation of KDM3A. Mol Cell.
74:1250–1263.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li J, Yu B, Deng P, Cheng Y, Yu Y, Kevork
K, Ramadoss S, Ding X, Li X and Wang CY: KDM3 epigenetically
controls tumorigenic potentials of human colorectal cancer stem
cells through Wnt/β-catenin signalling. Nat Commun. 8:151462017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang HY, Long QY, Tang SB, Xiao Q, Gao C,
Zhao QY, Li QL, Ye M, Zhang L, Li LY and Wu M: Histone demethylase
KDM3A is required for enhancer activation of hippo target genes in
colorectal cancer. Nucleic Acids Res. 47:2349–2364. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ramadoss S, Guo G and Wang CY: Lysine
demethylase KDM3A regulates breast cancer cell invasion and
apoptosis by targeting histone and the non-histone protein p53.
Oncogene. 36:47–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wade MA, Jones D, Wilson L, Stockley J,
Coffey K, Robson CN and Gaughan L: The histone demethylase enzyme
KDM3A is a key estrogen receptor regulator in breast cancer.
Nucleic Acids Res. 43:196–207. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Lee HY, Yang EG and Park H: Hypoxia
enhances the expression of prostate-specific antigen by modifying
the quantity and catalytic activity of Jumonji C domain-containing
histone demethylases. Carcinogenesis. 34:2706–2715. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fan L, Peng G, Sahgal N, Fazli L, Gleave
M, Zhang Y, Hussain A and Qi J: Regulation of c-Myc expression by
the histone demethylase JMJD1A is essential for prostate cancer
cell growth and survival. Oncogene. 35:2441–2452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wilson S, Fan L, Sahgal N, Qi J and Filipp
FV: The histone demethylase KDM3A regulates the transcriptional
program of the androgen receptor in prostate cancer cells.
Oncotarget. 8:30328–30343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Z, Xia J, Fang M and Xu Y: Epigenetic
regulation of lung cancer cell proliferation and migration by the
chromatin remodeling protein BRG1. Oncogenesis. 8:662019.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang F and Quan Q: The long non-coding RNA
SNHG4/microRNA-let-7e/KDM3A/p21 pathway is involved in the
development of non-small cell lung cancer. Mol Ther Oncolytics.
20:634–645. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Dandawate P, Ghosh C, Palaniyandi K, Paul
S, Rawal S, Pradhan R, Sayed AAA, Choudhury S, Standing D,
Subramaniam D, et al: The histone demethylase KDM3A, increased in
human pancreatic tumors, regulates expression of DCLK1 and promotes
tumorigenesis in mice. Gastroenterology. 157:1646–1659.e11. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nakatsuka T, Tateishi K, Kudo Y, Yamamoto
K, Nakagawa H, Fujiwara H, Takahashi R, Miyabayashi K, Asaoka Y,
Tanaka Y, et al: Impact of histone demethylase KDM3A-dependent AP-1
transactivity on hepatotumorigenesis induced by PI3K activation.
Oncogene. 36:6262–6271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang Y, Pan Q and Shao Z:
Tumor-suppressive role of microRNA-202-3p in hepatocellular
carcinoma through the KDM3A/HOXA1/MEIS3 pathway. Front Cell Dev
Biol. 8:5560042021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Parrish JK, Sechler M, Winn RA and
Jedlicka P: The histone demethylase KDM3A is a
microRNA-22-regulated tumor promoter in Ewing sarcoma. Oncogene.
34:257–262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Sechler M, Parrish JK, Birks DK and
Jedlicka P: The histone demethylase KDM3A, and its downstream
target MCAM, promote Ewing sarcoma cell migration and metastasis.
Oncogene. 36:4150–4160. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
An MJ, Kim DH, Kim CH, Kim M, Rhee S, Seo
SB and Kim JW: Histone demethylase KDM3B regulates the
transcriptional network of cell-cycle genes in hepatocarcinoma
HepG2 cells. Biochem Biophys Res Commun. 508:576–582. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Saraç H, Morova T, Pires E, McCullagh J,
Kaplan A, Cingöz A, Bagci-Onder T, Önder T, Kawamura A and Lack NA:
Systematic characterization of chromatin modifying enzymes
identifies KDM3B as a critical regulator in castration resistant
prostate cancer. Oncogene. 39:2187–2201. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hu A, Hong F, Li D, Xie Q, Chen K, Zhu L
and He H: KDM3B-ETF1 fusion gene downregulates LMO2 via the
WNT/β-catenin signaling pathway, promoting metastasis of invasive
ductal carcinoma. Cancer Gene Ther. 29:215–224. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kim JY, Kim KB, Eom GH, Choe N, Kee HJ,
Son HJ, Oh ST, Kim DW, Pak JH, Baek HJ, et al: KDM3B is the H3K9
demethylase involved in transcriptional activation of lmo2 in
leukemia. Mol Cell Biol. 32:2917–2933. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang X, Fan H, Xu C, Jiang G, Wang H and
Zhang J: KDM3B suppresses APL progression by restricting chromatin
accessibility and facilitating the ATRA-mediated degradation of
PML/RARα. Cancer Cell Int. 19:2562019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Xu X, Wang L, Hu L, Dirks WG, Zhao Y, Wei
Z, Chen D, Li Z, Wang Z, Han Y, et al: Small molecular modulators
of JMJD1C preferentially inhibit growth of leukemia cells. Int J
Cancer. 146:400–412. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Cai Y, Fu X and Deng Y: Histone
demethylase JMJD1C regulates esophageal cancer proliferation Via
YAP1 signaling. Am J Cancer Res. 7:115–124. 2017.PubMed/NCBI
|
|
91
|
Chen C, Aihemaiti M, Zhang X, Qu H, Sun
QL, He QS and Yu WB: Downregulation of histone demethylase JMJD1C
inhibits colorectal cancer metastasis through targeting ATF2. Am J
Cancer Res. 8:852–865. 2018.PubMed/NCBI
|
|
92
|
Lee DH, Kim GW, Jeon YH, Yoo J, Lee SW and
Kwon SH: Advances in histone demethylase KDM4 as cancer therapeutic
targets. FASEB J. 34:3461–3484. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Berry WL, Shin S, Lightfoot SA and
Janknecht R: Oncogenic features of the JMJD2A histone demethylase
in breast cancer. Int J Oncol. 41:1701–1706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kim TD, Fuchs JR, Schwartz E, Abdelhamid
D, Etter J, Berry WL, Li C, Ihnat MA, Li PK and Janknecht R:
Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon
cancer cells and identification of curcuminoids as JMJD2
inhibitors. Am J Transl Res. 6:236–247. 2014.PubMed/NCBI
|
|
95
|
Ye Q, Holowatyj A, Wu J, Liu H, Zhang L,
Suzuki T and Yang ZQ: Genetic alterations of KDM4 subfamily and
therapeutic effect of novel demethylase inhibitor in breast cancer.
Am J Cancer Res. 5:1519–1530. 2015.PubMed/NCBI
|
|
96
|
Li X and Dong S: Histone demethylase
JMJD2B and JMJD2C induce fibroblast growth factor 2: Mediated
tumorigenesis of osteosarcoma. Med Oncol. 32:532015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kim TD, Jin F, Shin S, Oh S, Lightfoot SA,
Grande JP, Johnson AJ, van Deursen JM, Wren JD and Janknecht R:
Histone demethylase JMJD2A drives prostate tumorigenesis through
transcription factor ETV1. J Clin Invest. 126:706–720. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sun S, Yang F, Zhu Y and Zhang S: KDM4A
promotes the growth of non-small cell lung cancer by mediating the
expression of Myc via DLX5 through the Wnt/β-catenin signaling
pathway. Life Sci. 262:1185082020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hu CE, Liu YC, Zhang HD and Huang GJ:
JMJD2A predicts prognosis and regulates cell growth in human
gastric cancer. Biochem Biophys Res Commun. 449:1–7. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yang J, Jubb AM, Pike L, Buffa FM, Turley
H, Baban D, Leek R, Gatter KC, Ragoussis J and Harris AL: The
histone demethylase JMJD2B is regulated by estrogen receptor alpha
and hypoxia, and is a key mediator of estrogen induced growth.
Cancer Res. 70:6456–6466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Fu L, Chen L, Yang J, Ye T, Chen Y and
Fang J: HIF-1α-induced histone demethylase JMJD2B contributes to
the malignant phenotype of colorectal cancer cells via an
epigenetic mechanism. Carcinogenesis. 33:1664–1673. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li H, Lan J, Wang G, Guo K, Han C, Li X,
Hu J, Cao Z and Luo X: KDM4B facilitates colorectal cancer growth
and glucose metabolism by stimulating TRAF6-mediated AKT
activation. J Exp Clin Cancer Res. 39:122020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tan J, Wang HL, Yang J, Liu QQ, Li CM,
Wang YQ, Fu LN, Gao QY, Chen YX and Fang JY: JMJD2B-induced amino
acid alterations enhance the survival of colorectal cancer cells
under glucose-deprivation via autophagy. Theranostics.
10:5763–5777. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Fu LN, Wang YQ, Tan J, Xu J, Gao QY, Chen
YX and Fang JY: Role of JMJD2B in colon cancer cell survival under
glucose-deprived conditions and the underlying mechanisms.
Oncogene. 37:389–402. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wissmann M, Yin N, Müller JM, Greschik H,
Fodor BD, Jenuwein T, Vogler C, Schneider R, Günther T, Buettner R,
et al: Cooperative demethylation by JMJD2C and LSD1 promotes
androgen receptor-dependent gene expression. Nat Cell Biol.
9:347–353. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Lee DH, Kim GW, Yoo J, Lee SW, Jeon YH,
Kim SY, Kang HG, Kim DH, Chun KH, Choi J and Kwon SH: Histone
demethylase KDM4C controls tumorigenesis of glioblastoma by
epigenetically regulating p53 and c-Myc. Cell Death Dis. 12:892021.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Luo W, Chang R, Zhong J, Pandey A and
Semenza GL: Histone demethylase JMJD2C is a coactivator for
hypoxia-inducible factor 1 that is required for breast cancer
progression. Proc Natl Acad Sci USA. 109:E3367–E3376. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Yang D, Xu T, Fan L, Liu K and Li G:
microRNA-216b enhances cisplatin-induced apoptosis in osteosarcoma
MG63 and SaOS-2 cells by binding to JMJD2C and regulating the
HIF1α/HES1 signaling axis. J Exp Clin Cancer Res. 39:2012020.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wu X, Deng Y, Zu Y and Yin J: Histone
demethylase KDM4C activates HIF1α/VEGFA signaling through the
costimulatory factor STAT3 in NSCLC. Am J Cancer Res. 10:491–506.
2020.PubMed/NCBI
|
|
110
|
Hu F, Li H, Liu L, Xu F, Lai S, Luo X, Hu
J and Yang X: Histone demethylase KDM4D promotes gastrointestinal
stromal tumor progression through HIF1β/VEGFA signalling. Mol
Cancer. 17:1072018. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Peng K, Zhuo M, Li M, Chen Q, Mo P and Yu
C: Histone demethylase JMJD2D activates HIF1 signaling pathway via
multiple mechanisms to promote colorectal cancer glycolysis and
progression. Oncogene. 39:7076–7091. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhuo M, Chen W, Shang S, Guo P, Peng K, Li
M, Mo P, Zhang Y, Qiu X, Li W and Yu C: Inflammation-induced JMJD2D
promotes colitis recovery and colon tumorigenesis by activating
Hedgehog signaling. Oncogene. 39:3336–3353. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Peng K, Kou L, Yu L, Bai C, Li M, Mo P, Li
W and Yu C: Histone demethylase JMJD2D interacts with β-catenin to
induce transcription and activate colorectal cancer cell
proliferation and tumor growth in mice. Gastroenterology.
156:1112–1126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Li M, Deng Y, Zhuo M, Zhou H, Kong X, Xia
X, Su Z, Chen Q, Guo P, Mo P, et al: Demethylase-independent
function of JMJD2D as a novel antagonist of p53 to promote liver
cancer initiation and progression. Theranostics. 10:8863–8879.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Pilka ES, James T and Lisztwan JH:
Structural definitions of Jumonji family demethylase selectivity.
Drug Discov Today. 20:743–749. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Horton JR, Engstrom A, Zoeller EL, Liu X,
Shanks JR, Zhang X, Johns MA, Vertino PM, Fu H and Cheng X:
Characterization of a linked Jumonji domain of the KDM5/JARID1
family of histone H3 lysine 4 demethylases. J Biol Chem.
291:2631–2646. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Plch J, Hrabeta J and Eckschlager T: KDM5
demethylases and their role in cancer cell chemoresistance. Int J
Cancer. 144:221–231. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Yang GJ, Zhu MH, Lu XJ, Liu YJ, Lu JF,
Leung CH, Ma DL and Chen J: The emerging role of KDM5A in human
cancer. J Hematol Oncol. 14:302021. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Teng YC, Lee CF, Li YS, Chen YR, Hsiao PW,
Chan MY, Lin FM, Huang HD, Chen YT, Jeng YM, et al: Histone
demethylase RBP2 promotes lung tumorigenesis and cancer metastasis.
Cancer Res. 73:4711–4721. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Liang X, Zeng J, Wang L, Fang M, Wang Q,
Zhao M, Xu X, Liu Z, Li W, Liu S, et al: Histone demethylase
retinoblastoma binding protein 2 is overexpressed in hepatocellular
carcinoma and negatively regulated by hsa-miR-212. PLoS One.
8:e697842013. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Yang GJ, Wang W, Mok SWF, Wu C, Law BYK,
Miao XM, Wu KJ, Zhong HJ, Wong CY, Wong VKW, et al: Selective
inhibition of lysine-specific demethylase 5A (KDM5A) using a
rhodium(III) complex for triple-negative breast cancer therapy.
Angew Chem Int Ed Engl. 57:13091–13095. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yang GJ, Ko CN, Zhong HJ, Leung CH and Ma
DL: Structure-Based discovery of a selective KDM5A inhibitor that
exhibits anti-cancer activity via inducing cell cycle arrest and
senescence in breast cancer cell lines. Cancers (Basel). 11:922019.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Lin W, Watanabe H, Peng S, Francis JM,
Kaplan N, Pedamallu CS, Ramachandran A, Agoston A, Bass AJ and
Meyerson M: Dynamic epigenetic regulation by menin during
pancreatic islet tumor formation. Mol Cancer Res. 13:689–698. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Oser MG, Sabet AH, Gao W, Chakraborty AA,
Schinzel AC, Jennings RB, Fonseca R, Bonal DM, Booker MA, Flaifel
A, et al: The KDM5A/RBP2 histone demethylase represses NOTCH
signaling to sustain neuroendocrine differentiation and promote
small cell lung cancer tumorigenesis. Genes Dev. 33:1718–1738.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Yamamoto S, Wu Z, Russnes HG, Takagi S,
Peluffo G, Vaske C, Zhao X, Moen Vollan HK, Maruyama R, Ekram MB,
et al: JARID1B is a luminal lineage-driving oncogene in breast
cancer. Cancer Cell. 25:762–777. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Yamane K, Tateishi K, Klose RJ, Fang J,
Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P
and Zhang Y: PLU-1 is an H3K4 demethylase involved in
transcriptional repression and breast cancer cell proliferation.
Mol Cell. 25:801–812. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Tortelli TC, Tamura RE, de Souza Junqueira
M, da Silva Mororó J, Bustos SO, Natalino RJM, Russell S, Désaubry
L, Strauss BE and Chammas R: Metformin-induced chemosensitization
to cisplatin depends on P53 status and is inhibited by Jarid1b
overexpression in non-small cell lung cancer cells. Aging (Albany
NY). 13:21914–21940. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Kuo KT, Huang WC, Bamodu OA, Lee WH, Wang
CH, Hsiao M, Wang LS and Yeh CT: Histone demethylase JARID1B/KDM5B
promotes aggressiveness of non-small cell lung cancer and serves as
a good prognostic predictor. Clin Epigenetics. 10:1072018.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wang D, Han S, Peng R, Jiao C, Wang X,
Yang X, Yang R and Li X: Depletion of histone demethylase KDM5B
inhibits cell proliferation of hepatocellular carcinoma by
regulation of cell cycle checkpoint proteins p15 and p27. J Exp
Clin Cancer Res. 35:372016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Hayami S, Yoshimatsu M, Veerakumarasivam
A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue
H, et al: Overexpression of the JmjC histone demethylase KDM5B in
human carcinogenesis: Involvement in the proliferation of cancer
cells through the E2F/RB pathway. Mol Cancer. 9:592010. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Huang YQ, Zou Y, Zheng RJ and Ma XD:
Down-regulation of JARID1B expression inhibits cell proliferation,
induces apoptosis and blocks cell cycle in human acute
lymphoblastic leukemia cells. Eur Rev Med Pharmacol Sci.
22:1366–1373. 2018.PubMed/NCBI
|
|
132
|
Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z,
Ma Y, Yu Y, Lin H, Chen AP and Chen CD: JARID1B is a histone H3
lysine 4 demethylase up-regulated in prostate cancer. Proc Natl
Acad Sci USA. 104:19226–19231. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Yan G, Li S, Yue M, Li C and Kang Z:
Lysine demethylase 5B suppresses CC chemokine ligand 14 to promote
progression of colorectal cancer through the Wnt/β-catenin pathway.
Life Sci. 264:1187262021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Niu X, Zhang T, Liao L, Zhou L, Lindner
DJ, Zhou M, Rini B, Yan Q and Yang H: The von Hippel-Lindau tumor
suppressor protein regulates gene expression and tumor growth
through histone demethylase JARID1C. Oncogene. 31:776–786. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Rondinelli B, Rosano D, Antonini E,
Frenquelli M, Montanini L, Huang D, Segalla S, Yoshihara K, Amin
SB, Lazarevic D, et al: Histone demethylase JARID1C inactivation
triggers genomic instability in sporadic renal cancer. J Clin
Invest. 125:4625–4637. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zheng Q, Li P, Zhou X, Qiang Y, Fan J, Lin
Y, Chen Y, Guo J, Wang F, Xue H, et al: Deficiency of the
X-inactivation escaping gene KDM5C in clear cell renal cell
carcinoma promotes tumorigenicity by reprogramming glycogen
metabolism and inhibiting ferroptosis. Theranostics. 11:8674–8691.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhang B, Zhou BH, Xiao M, Li H, Guo L,
Wang MX, Yu SH and Ye QH: KDM5C represses FASN-mediated lipid
metabolism to exert tumor suppressor activity in intrahepatic
cholangiocarcinoma. Front Oncol. 10:10252020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zhang Q, Xu L, Wang J, Zhu X, Ma Z, Yang
J, Li J, Jia X and Wei L: KDM5C expedites lung cancer growth and
metastasis through epigenetic regulation of MicroRNA-133a. Onco
Targets Ther. 14:1187–1204. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Ji X, Jin S, Qu X, Li K, Wang H, He H, Guo
F and Dong L: Lysine-specific demethylase 5C promotes
hepatocellular carcinoma cell invasion through inhibition BMP7
expression. BMC Cancer. 15:8012015. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Lin H, Ma N, Zhao L, Yang G and Cao B:
KDM5c promotes colon cancer cell proliferation through the
FBXW7-c-Jun regulatory axis. Front Oncol. 10:5354492020. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Shen HF, Zhang WJ, Huang Y, He YH, Hu GS,
Wang L, Peng BL, Yi J, Li TT, Rong R, et al: The dual function of
KDM5C in both gene transcriptional activation and repression
promotes breast cancer cell growth and tumorigenesis. Adv Sci
(Weinh). 8:20046352021. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Jangravi Z, Tabar MS, Mirzaei M,
Parsamatin P, Vakilian H, Alikhani M, Shabani M, Haynes PA,
Goodchild AK, Gourabi H, et al: Two splice variants of Y
chromosome-located lysine-specific demethylase 5D have distinct
function in prostate cancer cell line (DU-145). J Proteome Res.
14:3492–3502. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Li N, Dhar SS, Chen TY, Kan PY, Wei Y, Kim
JH, Chan CH, Lin HK, Hung MC and Lee MG: JARID1D is a suppressor
and prognostic marker of prostate cancer invasion and metastasis.
Cancer Res. 76:831–843. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Shen X, Hu K, Cheng G, Xu L, Chen Z, Du P
and Zhuang Z: KDM5D inhibit epithelial-mesenchymal transition of
gastric cancer through demethylation in the promoter of Cul4A in
male. J Cell Biochem. 120:12247–12258. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Cai LS, Chen QX, Fang SY, Lian MQ, Lian MJ
and Cai MZ: ETV4 promotes the progression of gastric cancer through
regulating KDM5D. Eur Rev Med Pharmacol Sci. 24:2442–2451.
2020.PubMed/NCBI
|
|
146
|
Willis-Owen SAG, Domingo-Sabugo C, Starren
E, Liang L, Freidin MB, Arseneault M, Zhang Y, Lu SK, Popat S, Lim
E, et al: Y disruption, autosomal hypomethylation and poor male
lung cancer survival. Sci Rep. 11:124532021. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Schulz WA, Lang A, Koch J and Greife A:
The histone demethylase UTX/KDM6A in cancer: Progress and puzzles.
Int J Cancer. 145:614–620. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Walport LJ, Hopkinson RJ, Vollmar M,
Madden SK, Gileadi C, Oppermann U, Schofield CJ and Johansson C:
Human UTY(KDM6C) is a male-specific N-methyl lysyl demethylase. J
Biol Chem. 289:18302–18313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Sengoku T and Yokoyama S: Structural basis
for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev.
25:2266–2277. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Li Y, Yang J, Zhang X, Liu H and Guo J:
KDM6A suppresses hepatocellular carcinoma cell proliferation by
negatively regulating the TGF-β/SMAD signaling pathway. Exp Ther
Med. 20:2774–2782. 2020.PubMed/NCBI
|
|
151
|
Ler LD, Ghosh S, Chai X, Thike AA, Heng
HL, Siew EY, Dey S, Koh LK, Lim JQ, Lim WK, et al: Loss of tumor
suppressor KDM6A amplifies PRC2-regulated transcriptional
repression in bladder cancer and can be targeted through inhibition
of EZH2. Sci Transl Med. 9:eaai83122017. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Zhang J, Ying Y, Li M, Wang M, Huang X,
Jia M, Zeng J, Ma C, Zhang Y, Li C, et al: Targeted inhibition of
KDM6 histone demethylases eradicates tumor-initiating cells via
enhancer reprogramming in colorectal cancer. Theranostics.
10:10016–10030. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Romero OA, Vilarrubi A, Alburquerque-Bejar
JJ, Gomez A, Andrades A, Trastulli D, Pros E, Setien F, Verdura S,
Farré L, et al: SMARCA4 deficient tumours are vulnerable to
KDM6A/UTX and KDM6B/JMJD3 blockade. Nat Commun. 12:43192021.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Chaturvedi SS, Ramanan R, Waheed SO,
Karabencheva-Christova TG and Christov CZ: Structure-function
relationships in KDM7 histone demethylases. Adv Protein Chem Struct
Biol. 117:113–125. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Park SY, Park JW and Chun YS: Jumonji
histone demethylases as emerging therapeutic targets. Pharmacol
Res. 105:146–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Osawa T, Muramatsu M, Wang F, Tsuchida R,
Kodama T, Minami T and Shibuya M: Increased expression of histone
demethylase JHDM1D under nutrient starvation suppresses tumor
growth via down-regulating angiogenesis. Proc Natl Acad Sci USA.
108:20725–20729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Lee KH, Hong S, Kang M, Jeong CW, Ku JH,
Kim HH and Kwak C: Histone demethylase KDM7A controls androgen
receptor activity and tumor growth in prostate cancer. Int J
Cancer. 143:2849–2861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Zhang Z, Chen B, Zhu Y, Zhang T, Zhang X,
Yuan Y and Xu Y: The Jumonji domain-containing histone demethylase
homolog 1D/lysine demethylase 7A (JHDM1D/KDM7A) is an epigenetic
activator of RHOJ transcription in breast cancer cells. Front Cell
Dev Biol. 9:6643752021. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Tong D, Liu Q, Liu G, Yuan W, Wang L, Guo
Y, Lan W, Zhang D, Dong S, Wang Y, et al: The HIF/PHF8/AR axis
promotes prostate cancer progression. Oncogenesis. 5:e2832016.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Li S, Sun A, Liang X, Ma L, Shen L, Li T,
Zheng L, Shang W, Zhao W and Jia J: Histone demethylase PHF8
promotes progression and metastasis of gastric cancer. Am J Cancer
Res. 7:448–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Zhou W, Gong L, Wu Q, Xing C, Wei B, Chen
T, Zhou Y, Yin S, Jiang B, Xie H, et al: PHF8 upregulation
contributes to autophagic degradation of E-cadherin,
epithelial-mesenchymal transition and metastasis in hepatocellular
carcinoma. J Exp Clin Cancer Res. 37:2152018. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Tseng LL, Cheng HH, Yeh TS, Huang SC, Syu
YY, Chuu CP, Yuh CH, Kung HJ and Wang WC: Targeting the histone
demethylase PHF8-mediated PKCα-Src-PTEN axis in HER2-negative
gastric cancer. Proc Natl Acad Sci USA. 117:24859–24866. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Shao P, Liu Q, Maina PK, Cui J, Bair TB,
Li T, Umesalma S, Zhang W and Qi HH: Histone demethylase PHF8
promotes epithelial to mesenchymal transition and breast
tumorigenesis. Nucleic Acids Res. 45:1687–1702. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Liu Q, Pang J, Wang LA, Huang Z, Xu J,
Yang X, Xie Q, Huang Y, Tang T, Tong D, et al: Histone demethylase
PHF8 drives neuroendocrine prostate cancer progression by
epigenetically upregulating FOXA2. J Pathol. 253:106–118. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Fu Y, Liu M, Li F, Qian L, Zhang P, Lv F,
Cheng W and Hou R: miR-221 promotes hepatocellular carcinoma cells
migration via targeting PHF2. Biomed Res Int. 2019:43714052019.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Lee KH, Park JW, Sung HS, Choi YJ, Kim WH,
Lee HS, Chung HJ, Shin HW, Cho CH, Kim TY, et al: PHF2 histone
demethylase acts as a tumor suppressor in association with p53 in
cancer. Oncogene. 34:2897–2909. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Del Rizzo PA, Krishnan S and Trievel RC:
Crystal structure and functional analysis of JMJD5 indicate an
alternate specificity and function. Mol Cell Biol. 32:4044–4052.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Hsia DA, Tepper CG, Pochampalli MR, Hsia
EY, Izumiya C, Huerta SB, Wright ME, Chen HW, Kung HJ and Izumiya
Y: KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1
coding region to regulate cancer cell proliferation. Proc Natl Acad
Sci USA. 107:9671–9676. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Yao Y, Zhou WY and He RX: Down-regulation
of JMJD5 suppresses metastasis and induces apoptosis in oral
squamous cell carcinoma by regulating p53/NF-κB pathway. Biomed
Pharmacother. 109:1994–2004. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Chang WH, Forde D and Lai AG: Dual
prognostic role of 2-oxoglutarate-dependent oxygenases in ten
cancer types: Implications for cell cycle regulation and cell
adhesion maintenance. Cancer Commun (Lond). 39:232019. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Wu LW, Zhou DM, Zhang ZY, Zhang JK, Zhu
HJ, Lin NM and Zhang C: Suppression of LSD1 enhances the cytotoxic
and apoptotic effects of regorafenib in hepatocellular carcinoma
cells. Biochem Biophys Res Commun. 512:852–858. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Verigos J, Karakaidos P, Kordias D,
Papoudou-Bai A, Evangelou Z, Harissis HV, Klinakis A and Magklara
A: The histone demethylase LSD1/ΚDM1A mediates chemoresistance in
breast cancer via regulation of a stem cell program. Cancers
(Basel). 11:15852019. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Lee YK, Lim J, Yoon SY, Joo JC, Park SJ
and Park YJ: Promotion of cell death in cisplatin-resistant ovarian
cancer cells through KDM1B-DCLRE1B modulation. Int J Mol Sci.
20:24432019. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Tang D, He J, Dai Y, Geng X, Leng Q, Jiang
H, Sun R and Xu S: Targeting KDM1B-dependent miR-215-AR-AGR2-axis
promotes sensitivity to enzalutamide-resistant prostate cancer.
Cancer Gene Ther. 29:543–557. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Tang DE, Dai Y, He JX, Lin LW, Leng QX,
Geng XY, Fu DX, Jiang HW and Xu SH: Targeting the KDM4B-AR-c-Myc
axis promotes sensitivity to androgen receptor-targeted therapy in
advanced prostate cancer. J Pathol. 252:101–113. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Hou J, Wu J, Dombkowski A, Zhang K,
Holowatyj A, Boerner JL and Yang ZQ: Genomic amplification and a
role in drug-resistance for the KDM5A histone demethylase in breast
cancer. Am J Transl Res. 4:247–256. 2012.PubMed/NCBI
|
|
177
|
Banelli B, Carra E, Barbieri F, Würth R,
Parodi F, Pattarozzi A, Carosio R, Forlani A, Allemanni G, Marubbi
D, et al: The histone demethylase KDM5A is a key factor for the
resistance to temozolomide in glioblastoma. Cell Cycle.
14:3418–3429. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Xu W, Zhou B, Zhao X, Zhu L, Xu J, Jiang
Z, Chen D, Wei Q, Han M, Feng L, et al: KDM5B demethylates H3K4 to
recruit XRCC1 and promote chemoresistance. Int J Biol Sci.
14:1122–1132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Roesch A, Vultur A, Bogeski I, Wang H,
Zimmermann KM, Speicher D, Körbel C, Laschke MW, Gimotty PA,
Philipp SE, et al: Overcoming intrinsic multidrug resistance in
melanoma by blocking the mitochondrial respiratory chain of
slow-cycling JARID1B(high) cells. Cancer Cell. 23:811–825. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Liu X, Zhang SM, McGeary MK, Krykbaeva I,
Lai L, Jansen DJ, Kales SC, Simeonov A, Hall MD, Kelly DP, et al:
KDM5B promotes drug resistance by regulating melanoma-propagating
cell subpopulations. Mol Cancer Ther. 18:706–717. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Hinohara K, Wu HJ, Vigneau S, McDonald TO,
Igarashi KJ, Yamamoto KN, Madsen T, Fassl A, Egri SB, Papanastasiou
M, et al: KDM5 histone demethylase activity links cellular
transcriptomic heterogeneity to therapeutic resistance. Cancer
Cell. 34:939–953.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Pippa S, Mannironi C, Licursi V, Bombardi
L, Colotti G, Cundari E, Mollica A, Coluccia A, Naccarato V, La
Regina G, et al: Small molecule inhibitors of KDM5 histone
demethylases increase the radiosensitivity of breast cancer cells
overexpressing JARID1B. Molecules. 24:17392019. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Lin H, Yang G, Yu J, Wang J, Li Q, Guo S
and Cao B: KDM5c inhibits multidrug resistance of colon cancer cell
line by down-regulating ABCC1. Biomed Pharmacother. 107:1205–1209.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Komura K, Jeong SH, Hinohara K, Qu F, Wang
X, Hiraki M, Azuma H, Lee GS, Kantoff PW and Sweeney CJ: Resistance
to docetaxel in prostate cancer is associated with androgen
receptor activation and loss of KDM5D expression. Proc Natl Acad
Sci USA. 113:6259–6264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
He C, Sun J, Liu C, Jiang Y and Hao Y:
Elevated H3K27me3 levels sensitize osteosarcoma to cisplatin. Clin
Epigenetics. 11:82019. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Wang Q, Chen X, Jiang Y, Liu S, Liu H, Sun
X, Zhang H, Liu Z, Tao Y, Li C, et al: Elevating H3K27me3 level
sensitizes colorectal cancer to oxaliplatin. J Mol Cell Biol.
12:125–137. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Mathur R, Sehgal L, Havranek O, Köhrer S,
Khashab T, Jain N, Burger JA, Neelapu SS, Davis RE and Samaniego F:
Inhibition of demethylase KDM6B sensitizes diffuse large B-cell
lymphoma to chemotherapeutic drugs. Haematologica. 102:373–380.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Zhang C, Shen L, Zhu Y, Xu R, Deng Z, Liu
X, Ding Y, Wang C, Shi Y, Bei L, et al: KDM6A promotes imatinib
resistance through YY1-mediated transcriptional upregulation of
TRKA independently of its demethylase activity in chronic
myelogenous leukemia. Theranostics. 11:2691–2705. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Macedo-Silva C, Miranda-Goncalves V,
Lameirinhas A, Lencart J, Pereira A, Lobo J, Guimarães R, Martins
AT, Henrique R, Bravo I and Jerónimo C: JmjC-KDMs KDM3A and KDM6B
modulate radioresistance under hypoxic conditions in esophageal
squamous cell carcinoma. Cell Death Dis. 11:10682020. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Xu S, Fan L, Jeon HY, Zhang F, Cui X,
Mickle MB, Peng G, Hussain A, Fazli L, Gleave ME, et al:
p300-mediated acetylation of histone demethylase JMJD1A prevents
its degradation by ubiquitin ligase STUB1 and enhances its activity
in prostate cancer. Cancer Res. 80:3074–3087. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
McAllister TE, England KS, Hopkinson RJ,
Brennan PE, Kawamura A and Schofield CJ: Recent progress in histone
demethylase inhibitors. J Med Chem. 59:1308–1329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Fang Y, Liao G and Yu B: LSD1/KDM1A
inhibitors in clinical trials: Advances and prospects. J Hematol
Oncol. 12:1292019. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Fu DJ, Li J and Yu B: Annual review of
LSD1/KDM1A inhibitors in 2020. Eur J Med Chem. 214:1132542021.
View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Kleszcz R, Skalski M, Krajka-Kuźniak V and
Paluszczak J: The inhibitors of KDM4 and KDM6 histone lysine
demethylases enhance the anti-growth effects of erlotinib and
HS-173 in head and neck cancer cells. Eur J Pharm Sci.
166:1059612021. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Illiano M, Conte M, Salzillo A, Ragone A,
Spina A, Nebbioso A, Altucci L, Sapio L and Naviglio S: The KDM
inhibitor GSKJ4 triggers CREB downregulation via a protein kinase A
and proteasome-dependent mechanism in human acute myeloid leukemia
cells. Front Oncol. 10:7992020. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Yang GJ, Wu J, Miao L, Zhu MH, Zhou QJ, Lu
XJ, Lu JF, Leung CH, Ma DL and Chen J: Pharmacological inhibition
of KDM5A for cancer treatment. Eur J Med Chem. 226:1138552021.
View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Baby S, Gurukkala Valapil D and
Shankaraiah N: Unravelling KDM4 histone demethylase inhibitors for
cancer therapy. Drug Discov Today. 26:1841–1856. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Varghese B, Del Gaudio N, Cobellis G,
Altucci L and Nebbioso A: KDM4 involvement in breast cancer and
possible therapeutic approaches. Front Oncol. 11:7503152021.
View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Carter DM, Specker E, Małecki PH,
Przygodda J, Dudaniec K, Weiss MS, Heinemann U, Nazaré M and Gohlke
U: Enhanced properties of a benzimidazole benzylpyrazole lysine
demethylase inhibitor: Mechanism-of-action, binding site analysis,
and activity in cellular models of prostate cancer. J Med Chem.
64:14266–14282. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Souto JA, Sarno F, Nebbioso A, Papulino C,
Álvarez R, Lombino J, Perricone U, Padova A, Altucci L and de Lera
ÁR: A new family of Jumonji C domain-containing KDM inhibitors
inspired by natural product purpurogallin. Front Chem. 8:3122020.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Tayari MM, Santos HGD, Kwon D, Bradley TJ,
Thomassen A, Chen C, Dinh Y, Perez A, Zelent A, Morey L, et al:
Clinical responsiveness to all-trans retinoic acid is potentiated
by LSD1 inhibition and associated with a quiescent transcriptome in
myeloid malignancies. Clin Cancer Res. 27:1893–1903. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Cuyàs E, Gumuzio J, Verdura S, Brunet J,
Bosch-Barrera J, Martin-Castillo B, Alarcón T, Encinar JA, Martin
ÁG and Menendez JA: The LSD1 inhibitor iadademstat (ORY-1001)
targets SOX2-driven breast cancer stem cells: A potential
epigenetic therapy in luminal-B and HER2-positive breast cancer
subtypes. Aging (Albany NY). 12:4794–4814. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Wang T, Zhang F and Sun F: ORY-1001, a
KDM1A inhibitor, inhibits proliferation, and promotes apoptosis of
triple negative breast cancer cells by inactivating androgen
receptor. Drug Dev Res. 83:208–216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Maes T, Mascaró C, Tirapu I, Estiarte A,
Ciceri F, Lunardi S, Guibourt N, Perdones A, Lufino MMP,
Somervaille TCP, et al: ORY-1001, a potent and selective covalent
KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell.
33:495–511.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Salamero O, Montesinos P, Willekens C,
Pérez-Simón JA, Pigneux A, Récher C, Popat R, Carpio C, Molinero C,
Mascaró C, et al: First-in-human phase I study of iadademstat
(ORY-1001): A first-in-class lysine-specific histone demethylase 1A
inhibitor, in relapsed or refractory acute myeloid leukemia. J Clin
Oncol. 38:4260–4273. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Huang M, Chen C, Geng J, Han D, Wang T,
Xie T, Wang L, Wang Y, Wang C, Lei Z and Chu X: Targeting KDM1A
attenuates Wnt/β-catenin signaling pathway to eliminate
sorafenib-resistant stem-like cells in hepatocellular carcinoma.
Cancer Lett. 398:12–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Lillico R, Lawrence CK and Lakowski TM:
Selective DOT1L, LSD1, and HDAC class I inhibitors reduce HOXA9
expression in MLL-AF9 rearranged leukemia cells, but dysregulate
the expression of many histone-modifying enzymes. J Proteome Res.
17:2657–2667. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Bauer TM, Besse B, Martinez-Marti A, Trigo
JM, Moreno V, Garrido P, Ferron-Brady G, Wu Y, Park J, Collingwood
T, et al: Phase I, open-label, dose-escalation study of the safety,
pharmacokinetics, pharmacodynamics, and efficacy of GSK2879552 in
relapsed/refractory SCLC. J Thorac Oncol. 14:1828–1838. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Kanouni T, Severin C, Cho RW, Yuen NY, Xu
J, Shi L, Lai C, Del Rosario JR, Stansfield RK, Lawton LN, et al:
Discovery of CC-90011: A potent and selective reversible inhibitor
of lysine specific demethylase 1 (LSD1). J Med Chem.
63:14522–14529. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Hollebecque A, Salvagni S, Plummer R,
Isambert N, Niccoli P, Capdevila J, Curigliano G, Moreno V,
Martin-Romano P, Baudin E, et al: Phase I study of lysine-specific
demethylase 1 inhibitor, CC-90011, in patients with advanced solid
tumors and relapsed/refractory non-Hodgkin lymphoma. Clin Cancer
Res. 27:438–446. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Sterling J, Menezes SV, Abbassi RH and
Munoz L: Histone lysine demethylases and their functions in cancer.
Int J Cancer. Oct 31–2020.(Epub ahead of print). doi:
10.1002/ijc.33375. PubMed/NCBI
|
|
212
|
Oner E, Kotmakci M, Baird AM, Gray SG,
Debelec Butuner B, Bozkurt E, Kantarci AG and Finn SP: Development
of EphA2 siRNA-loaded lipid nanoparticles and combination with a
small-molecule histone demethylase inhibitor in prostate cancer
cells and tumor spheroids. J Nanobiotechnology. 19:712021.
View Article : Google Scholar : PubMed/NCBI
|