Introduction
Colon cancer is the third most prevalent type of
cancer worldwide. Systemic treatment for patients with colon cancer
has expanded from chemotherapy to targeted therapy and
immunotherapy. Therefore, it is critical to explore new biomarkers
for the systematic treatment of colon cancer including targeted
therapy and immunotherapy.
DExD/H-Box Helicase 58 (DDX58) encodes retinoic
acid-inducible gene-I (RIG-I), which is an innate immune receptor
helicase (1-3). RIG-I is widely expressed in a variety
of tissues and cells and is involved in the production of
interferons to initiate innate antiviral immunity (4-6).
However, it has been indicated that RIG-I also participates in
cancer cellular activities. For example, the activation of RIG-I
signaling triggers apoptotic programs in tumor cells and activates
antitumor immunity (7,8). In hepatocellular carcinoma, a low
RIG-I expression is associated with a poor survival; that is, RIG-I
functions as a tumor suppressor by enhancing signal transducer and
activator of transcription (STAT)1 activation by competitively
binding the SH2 domain of STAT1 against its negative regulator SHP1
(9,10). In breast cancer, RIG-I activation
decreases tumor growth and metastasis (11,12).
Additionally, DDX58/RIG-I expression can also affect immune
response-based cancers, and CD4+ and CD8+
T-cells and T-cell-mediated effects enhance the antitumor efficacy
of RIG-I agonists (13). The
inhibition of the MAPK pathway mediates inflammatory reprogramming
and renders tumors sensitive to the targeted activation of RIG-I,
while a high MAPK activation has been shown to be associated with
the reduced presence of CD8+ T-cells (14). Overall, these findings suggest that
RIG-I plays more diverse roles in a variety of cellular activities,
and its function goes far beyond that of a pattern recognition
receptor. Therefore, on the basis that RIG-I exhibits
tumor-suppressive activity, the successful therapeutic delivery of
RIG-I agonists could induce tumor cell apoptosis and modulate the
tumor microenvironment.
To the best of our knowledge, there are no data
available to date on the clinical significance of RIG-I in colon
cancer. Therefore, the present study aimed to investigate the value
of RIG-I in colon cancer to address the urgent need to improve the
management of this tumor type. The second aim was to identify the
factors associated with RIG-I expression to characterize the
RIG-I-associated microenvironment and the immunotherapeutic options
of colon cancer.
In the present study, in vivo and in
vitro experiments were performed to explore the value and
signature of DDX58/RIG-I in colon cancer. It was found that a low
DDX58/RIG-I expression was associated with an increase in the
proliferation, migration and invasion of colon cancer cells. In
addition, the activation of DDX58/RIG-I suppressed the
proliferation of tumor cells by inhibiting
STAT3/cystathionine-γ-lyase (CSE) signaling in colon cancer. These
findings suggest that the activation of DDX58/RIG-I may be an
effective treatment strategy for colon cancer.
Materials and methods
Cell lines and animals
HCT8 and HCT116 human colon cancer cell lines and
FHC human normal colorectal mucosal cells were generously donated
by the Army Medical University (Chongqing, China). SW620 and LOVO
human colon cancer cell lines were generously donated by Tongji
University (Shanghai, China). SW480 human colon cancer cells were
generously donated by the School of Basic Medicine, Henan
University (Kaifeng, China). The cell lines (HCT116, CCL-247; HCT8,
CCL-244; LOVO, CCL-229; SW480, CCL-228; SW620, CCL-227; FHC,
CRL-1831) were purchased on the ATCC platform. Cell lines were
tested using short tandem repeat (STR) analysis as follows: DNA was
extracted and the STRs were amplified by multiplex PCR and
separated on a genetic analyzer. The signals were then analyzed
using GeneMapper software v4.1 (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The colon cancer cells, HCT8, SW480 and LOVO,
were used in RPMI-1640 medium (Thermo Fisher Scientific, Inc.), the
SW620 cells in Dulbecco's modified Eagle's medium (Thermo Fisher
Scientific, Inc.), the HCT116 cells in McCoy's 5A medium
(Biological Industries) and the FHC cells in DMEM/F-12 (Beijing
Solarbio Science & Technology Co., Ltd.). All cells were
cultured in a humidified atmosphere of 5% CO2 at
37°C.
Balb/c nude mice (n=12; specific pathogen-free;
male; age, 4-5 weeks) weighing 18-22 g were supplied by Beijing
Viton Lihua Experimental Animal Technology [certificate no. SCXK
(Jing) 2016-0006, no. 110011210107417128; Beijing, China]. The mice
were allowed free access to food and water and were housed in an
environment with a constant temperature and humidity (temperature,
24±1°C; humidity, 50-70%), ventilation rate (10-20 times/h), noise
(<40 db) and working illumination (250 lx) under a 12-h
light/dark cycle. The drinking water, food, and experimental
supplies were sterilized and disinfected. The present study was
approved by the Ethics Committee of the Medical School of Henan
University (HUSOM2020-301).
Tissue microarray analysis
A human colon cancer microarray (HColA180Su13)
containing 90 tumor samples and 90 para-carcinoma samples was
constructed by Shanghai Outdo Biotech Co., Ltd. The
clinicopathological characteristics of the tumor samples are
presented in Table I. Each tissue
was stained with anti-DDX58 antibody (diluted in PBS; cat. no.
20566-1-AP, supplier: ProteinTech Group, Inc.) as the primary
antibody (4°C, 16 h) and the secondary antibody (C2 biotin-labeled
sheep anti-mouse/rabbit IgG polymer, cat. no. KIT-9707, Fuzhou
Maixin Biotech. Co., Ltd.) was incubated at room temperature for 45
min with UltraSensitive SP reagent box (Maixin Inc.) Automatic
immunohistochemical staining was accomplished using Autostainer
Link 48 (Dako; Agilent Technologies, Inc.) The integral intensity
value was measured with Aperio ImageScope software v12.4.3 (Aperio,
Leica GmbH). All experiments were performed in accordance with
national ethical guidelines and with the approval from all patients
and the committee of Shanghai Outdo Biotech Co., Ltd.
 | Table IClinicopathological characteristics
of the tumor samples in the tissue microarray. |
Table I
Clinicopathological characteristics
of the tumor samples in the tissue microarray.
| Characteristic | No. of
patients |
|---|
| Age (years) | |
| ≤60 | 32 |
| >60 | 58 |
| Sex | |
| Male | 47 |
| Female | 43 |
| Tumor grade | |
| I, II and III | 76 |
| I-III, II-III, III
and III-IV | 14 |
| T stage | |
| T1/T2 | 11 |
| T3/T4 | 79 |
| N stage | |
| N0 | 61 |
| N1/N2 | 29 |
| M stage | |
| M0 | 87 |
| M1 | 3 |
| TNM stage | |
| I/II | 59 |
| III/IV | 31 |
| Tumor size
(cm) | |
| ≤5 | 12 |
| >5 | 78 |
The immunohistochemistry test score criteria were as
follows: i) Determination of the staining intensity: The whole
field of tissue points was observed under low magnification (x40
and x200) and the tissues were classified into weak positive,
medium positive and strong positive. Weak positive was light yellow
(+ or 1), medium positive was brownish yellow (++ or 2) and strong
positive was tan (+++ or 3). ii) Determination of the staining
positive rate: Firstly, tissue points were observed in the whole
field under a low-power lens of Aperio XT (Leica GmbH), and then
three fields with different staining intensity were selected for
interpretation under a high-power lens of AperioXT (Leica GmbH). If
staining was located in cytoplasm, 100 cells were randomly recorded
in each field, and the percentage of positive cells in 100 cells
was then recorded x1%. In the same manner, the percentage of
positive cells in the other two fields was x2% and x3%, and the
average of the positive staining rate of this tissue point was
obtained.
The data processing of human tissue chip was as
follows: The total score was obtained by the product of staining
score intensity and staining positive score, and the cut-off value
was then determined according to the total score to determine the
high, medium and low expression groups for survival. In the tissue
microarray, a DDX58 staining score <1 was considered as a low
protein expression, and a DDX58 staining score >1 and <1.5
was considered as a medium protein expression, and a DDX58 staining
score ≥1.5 was considered as a high protein expression.
Construction of DDX58-overexpressing
lentivirus
Full-length DDX58 cDNA (NM_014314) was cloned into
the Ubi-MCS-SV40-EGFP-IRES-puromycin vector (#63954-4; Shanghai
GeneChem Co., Ltd.) to construct DDX58-overexpressing lentivirus.
The DDX58-overexpressing lentivirus was used to infect the HCT8 and
HCT116 cells to obtain stable cells overexpressing DDX58.
Transfection was performed using HitransG P transfection reagent
(Shanghai GeneChem Co., Ltd.), with a lentiviral titer of 4E+8
TU/ml and vector titer of 1E+9 TU/ml. The time between transfection
and subsequent experiments was 2 weeks.
siRNA and plasmid transfection
For knockdown, HCT116/DDX58, HCT8/DDX58, HCT116,
HCT8 and SW620 cells in six-well plates were transfected with
scramble siRNA (Sc siRNA, 20 µM) or specific siRNA against human
DDX58 (20 µM Invitrogen; Thermo Fisher Scientific, Inc.) or
specific siRNA against human CSE (20 µM, Invitrogen; Thermo Fisher
Scientific, Inc.) or specific siRNA against human STAT3 (20 µM,
Invitrogen; Thermo Fisher Scientific, Inc.) using Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.). The
medium was replaced at 6 h post-transfection, and the silencing
efficiency was determined using western blot analysis at 48 h
post-transfection. The siRNA sequences were as follows: DDX58 siRNA
sense, 5'-CCG GCA CAG AAG UGU AUA UTT-3' and antisense, 5'-AUA UAC
ACU UCU GUG CCG GTT-3'; CSE-specific siRNA sense, 5'-GGU UUA GCA
GCC ACU GUA AdT dT-3' and antisense, 5'-UUA CAG UGG CUG CUA AAC CdT
dT-3'; STAT3-specific siRNA sense, 5'-CCC GUC AAC AAA UUA AGA AdT
dT-3' and antisense, 5'-UUC UUA AUU UGU UGA CGG GdT dT-3'; Sc siRNA
sense, 5'-UUC UCC GAA CGU GUC ACG UTT-3' and antisense, 5'-ACG UGA
CAC GUU CGG AGA ATT-3.
In addition, for SW620 cells, DDX58 overexpression
plasmid (GV367-vector and GV367-hDDX58, Shanghai GeneChem Co.,
Ltd.) was used for transfection, for the HCT116, HCT8, SW620 cells,
STAT3 overexpression plasmid transfection was performed as follows:
pCMV-FLAG vector and pCMV-FLAG-hSTAT3 were provided as a gift from
Military Medical Sciences School. The plasmids (600 ng/µl) were
transiently transfected using Lipofectamine 2000®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. After 6 h, the cells were exposed
to fresh medium.
Cell viability and proliferation
assays
The cells (HCT8, HCT116 and SW620) were seeded into
96-well plates at a density of 1x104 cells per well for
48 h. Cell viability was evaluated by determining the number of
cells using MTS (MilliporeSigma) assay, and after 4 h, the
absorbance was measured at 570 nm by enzyme-labeled instrument
(PerkinElmer; Thermo Fisher Scientific, Inc.). For cell
proliferation assay, the HCT8, HCT116 and SW620 cells
(1x105/ml, 100 µl/well) were seeded in 96-well plates,
and cell proliferation was assessed according to the number of
EdU+ cells using an EdU assay kit (Guangzhou RiboBio
Co., Ltd.) that included the processing of EdU markers, cell
fixation, Apollo staining and DNA staining with the Hoechst 33342
reaction. Each experiment was performed in triplicate and repeated
three times. A microcope was used (U-LH100HG; Olympus Corporation)
to obtain images. ImageView (version 3.7) software was used for
processing analysis.
Scratch wound assay
The cell scratch wound assay is an in vitro
method used to examine cell migration (15). For this assay, the cells were grown
to 90% confluency in a six-well plate. A 10-µl pipette tip was used
to scratch a straight wound in cells in 35-mm dishes. The detached
cells were removed by washing with phosphate-buffered saline (PBS),
and serum-free medium was used to continue cell culture. After
using a digital camera (EOS450-D; China Canon Co. Ltd.) and
scraping at 0, 24 or 48 h, migration images were obtained and the
gap closing rates were compared. Adobe Photoshop CC software was
used for analysis.
Transwell assay
Transwell assays were performed to investigate cell
migration and invasion in 24-well Transwell chambers (pore size, 8
µm; Corning, Corning, Inc.). The Transwell chambers were covered
with or without Matrigel matrix (BD Biosciences) for invasion or
migration assays, respectively. The cells (1x105
cells/ml) were suspended in serum-free medium and inoculated in the
upper chamber, and medium containing 10% FBS was added to the lower
chamber. Following incubation for 24 h at 37°C, the cells were
fixed at room temperature with 4% cell fixative (Beijing Solarbio
Science & Technology Co., Ltd.) for 30 min and stained with
0.1% crystal violet (Beijing Solarbio Science & Technology Co.,
Ltd) for 15 min, then washed with water, dried and photographed as
previously described (16-18).
Western blot analysis
The cells or tissues were washed with ice-cold PBS
and lysed for 20 min on ice using RIPA lysis buffer (ProteinTech
Group, Inc.) with both protease and phosphatase inhibitors. Protein
concentrations were determined using a BCA protein assay kit
(Beijing Solarbio Science & Technology Co., Ltd.), and western
blot analysis was then performed using a standard protocol. A total
of 30 µg of protein was separated by 8% SDS-PAGE and transferred to
a polyvinylidene difluoride membrane (MilliporeSigma) at 70 mA for
2 h. The membrane was then blocked in 5% fat-free milk, and probed
with specific primary antibodies against DDX58, STAT3, p-STATS and
CSE at 4°C overnight. Following incubation with the secondary
antibody, the proteins were visualized using an K ECL Enhanced
Chemiluminescence kit (Kemix, Ltd.) and detected using a FluorChem
Q Multifluor System (ProteinSimple). Densitometric analysis was
performed using Image J 1.51j8 software (National Institutes of
Health). The antibodies used were as follows: DDX58 rabbit
polyclonal antibody (1:1,000, cat. no. 20566-1-AP, ProteinTech
Group, Inc.), STAT3 mouse monoclonal antibody (1:1,000, cat. no.
9139S, Cell Signaling Technology, Inc.), p-STAT3 rabbit polyclonal
antibody (1:1,000, cat. no. 9131S, Cell Signaling Technology,
Inc.), CSE rabbit polyclonal antibody (1:3,000, cat. no. F052208,
Abways Technology) and GAPDH mouse monoclonal antibody (1:100,000,
cat. no. 60004-1-lg, ProteinTech Group, Inc.), HRP-goat anti-rabbit
IgG (1:3,000, cat. no. SA00001-2, ProteinTech Group, Inc.),
HRP-goat anti-mouse IgG (1:3,000, cat. no. SA00001-1, ProteinTech
Group, Inc.)
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the cells using TRIzol
reagent (Ambion; Thermo Fisher Scientific, Inc.) and reverse
transcribed with HiScript II Q RT SuperMix for qPCR (Vazyme Biotech
Co., Ltd.) in accordance with a standard protocol (50°C, 15 min;
85°C, 5 sec). qPCR was performed on a SLAN-96S Real-Time PCR System
(Shanghai Hongshi Medical Technology Co., Ltd.) using hamQ
Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.). The PCR
conditions were as follows: 30 sec at 95°C pre-denaturation; 5 sec
at 95°C for denaturation, 30 sec at 60°C for annealing and
extension, a total of 40 cycles. The 2-ΔΔCq method was
used to evaluate the mRNA expression levels of genes. Please refer
to the literature for specific method (19). The following primers were used in
the RT-qPCR assay: DDX58 forward, 5'-AGA GCA CTT GTG GAC GCT TT-3'
and DDX58 reverse, 5'-AGC AAC TGA GGT GGC AAT CA-3'; and GAPDH
forward, 5'-GCA CCG TCA AGG CTG AGA AC-3' and GAPDH reverse, 5'-TGG
TGA AGA CGC CAG TGG A-3'.
Co-immunoprecipitation (Co-IP) assay
A total of 3x107 HCT116 cells were
treated with NP-40 lysis buffer (Beijing Solarbio Science &
Technology Co., Ltd.) and lysates were then incubated with DDX58
antibody-conjugate and protein A+G Agarose (Beyotime Institute of
Biotechnology) overnight at 4°C. Beads containing affinity-bound
proteins were washed three times with PBS buffer, followed by
elution with 1 ml NP-40 lysis buffer (Beijing Solarbio Science
& Technology Co., Ltd.) three times. All tubes were centrifuged
at 4°C at 1,500 x g for 5 min, the supernatant was discarded, 1 ml
cold PBS was added for washing; the tubes were centrifuged again at
1,500 x g for 1 min, and this was repeated three times; the beads
were then cleaned with NP-40 lysis solution three times. The method
was the same as that used for PBS washing. Finally, the supernatant
liquid was discarded, followed by the addition of 40 µl 2X sample
(Beijing Solarbio Science & Technology Co., Ltd.) to the IP
group, 25 µl 2X sample (Beijing Solarbio Science & Technology
Co., Ltd.) to the IgG group, and 5X sample (Beijing Solarbio
Science & Technology Co., Ltd.) to the Input group, and then
placed in a dry bath incubator (Hangzhou Aosheng Instruments Co.,
Ltd.) for 8 min. Following denaturation, proteins were separated on
sodium dodecyl sulfate-polyacrylamide gels and transferred onto
polyvinylidene difluoride membranes (MilliporeSigma) The membranes
were probed with anti-DDX58 (Protein Tech Group, Inc.) or
anti-STAT3 (Cell Signaling Technology, Inc.) antibodies. In this
part, western blot analysis was used to detect the expression level
of the target band. Primary antibody was incubated at 4°C, 11 rpm
for 14 h. Secondary antibody was incubated at 37°C, 60 rpm for 2 h.
The antibodies used were as follows: DDX58 rabbit polyclonal
antibody (1:1,000, cat. no. 20566-1-AP, ProteinTech Group, Inc.),
STAT3 mouse monoclonal antibody (1:1,000, cat. no. 9139S, Cell
Signaling Technology, Inc.). HRP-Goat Anti-Rabbit IgG (1:3,000,
cat. no. SA00001-2, ProteinTech Group, Inc.). HRP-Goat Anti-Mouse
IgG (1:3,000, cat. no. SA00001-1, ProteinTech Group, Inc.).
Animal experiments
Animal experiments were approved by the Ethics
Committee of Medical School of Henan University (HUSOM2020-301).
BALB/c nude mice were injected subcutaneously into their right
flanks with 3x106 HCT116 cells infected with empty
vector lentivirus or DDX58-overexpressing lentivirus. After 3
weeks, the experimental animals were first anesthetized (10%
chloral hydrate, 400 mg/kg, administered via intraperitoneal
injection). The mice were then sacrificed by cervical dislocation,
resulting in immediate death. The death of the experimental animals
was confirmed by the cessation of respiration, heartbeat, and pupil
and nerve reflex. The tumors were then harvested, weighed and lysed
using RIPA buffer for use in western blot analysis to detect the
levels of DDX58, STAT3, pSTAT3 and CSE, as described above.
SB9200. SB9200 was obtained from
MedChemExpress (inarigivir soproxil, cat. no. HY-109035). The
HCT116 and SW620 cells were treated with SB9200 at a concentration
of 80 µM. Cell viability was detected using MTS assay. Cell
proliferation was detected using EdU assay. Wound healing was used
to detect cell migration. Cell migration and invasion was examined
using Transwell assay (all as described above).
Statistical analysis
Each experiment was repeated at least three times
and graphs were created using GraphPad Prism software 6.0 (GraphPad
Software Inc.). Statistical analyses were performed using SPSS 17.0
software (SPSS, Inc.). Data are expressed as the mean ± standard
deviation. Differences between multiple groups were analyzed using
one-way analysis of variance followed by Tukey's multiple
comparisons test. P<0.05 was considered to indicate a
statistically significant difference. The matched sample data in
Fig. 1B were compared using a
paired t-test and the data in Fig.
2D were compared used an unpaired t-test. The Kaplan-Meier
method was used for survival analysis, and the log-rank was used to
determine the differences in survival curves between groups.
Results
Low expression of DDX58 is associated
with a poor prognosis of patients with colon cancer
The activation of DDX58/RIG-I signaling in tumors is
a crucial component for checkpoint inhibitor-mediated immunotherapy
of cancer (20). In the present
study, the association between DDX58 expression and the
clinicopathological parameters and prognosis of patients with colon
cancer in a tissue microarray was analyzed. The results of
immunohistochemistry revealed that the level of DDX58 in cancer
tissues was slightly lower than that in adjacent tissues, with no
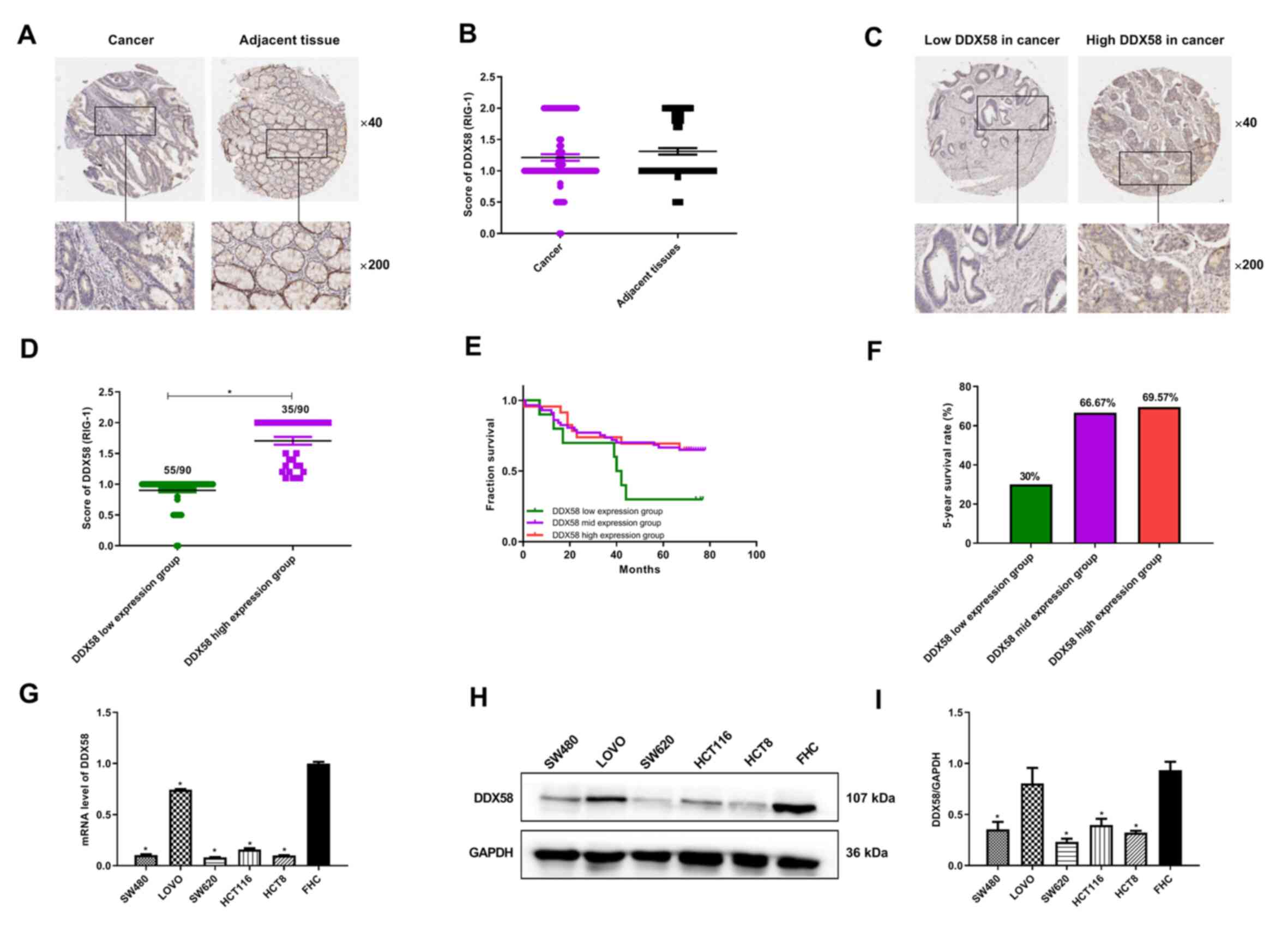
statistically significant difference (P=0.1866; Fig. 1A and B). However, it was observed
that DDX58 exhibited a low expression in 55 cases and a high
expression in 35 cases among the 90 human colon cancer tissues
(Fig. 1C and D). It was found that
patients with colon cancer with a low expression of DDX58 had a
lower 5-year survival rate than patients with a high expression of
DDX58 (Fig. 1E and F). However, no
notable associations were found between DDX58 expression and
patient age or sex, grade, TNM stage and tumor size (data no
shown). In addition, RT-qPCR and western blot analysis were
performed to detect the mRNA and protein levels of DDX58 in human
colon cancer cell lines and lower DDX58 levels were observed in the
majority of the colon cancer cell lines compared with FHC normal
colon epithelial cells (Fig.
1G-I). These data suggest that a low expression of DDX58
results in a poor prognosis in colon cancer and may represent a
potential prognostic biomarker for patients with colon cancer.
Overexpression of DDX58 inhibits the
proliferation, migration and invasion of colon cancer cells
In view of the low expression of DDX58 in the
majority of the colon cancer cells (while the expression level of
DDX58 in LOVO cells was high), and as SW620 and SW480 were derived
from the same patient, the classic colon cancer cell lines, HCT8,
HCT116 and SW620, were selected for use in subsequent experiments.
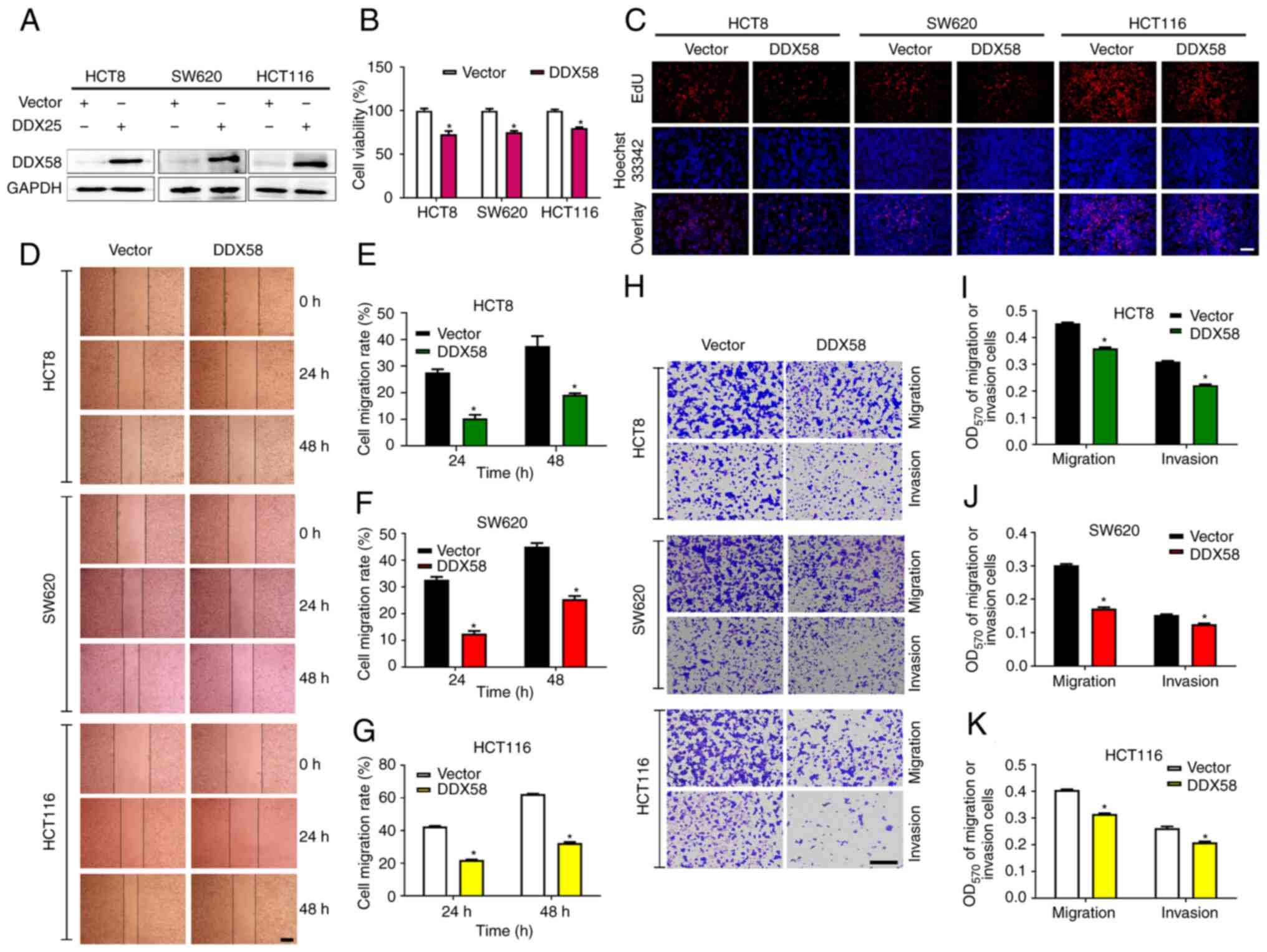
To evaluate the function of DDX58 in colon cancer cells, a plasmid
expressing DDX58 was first used to overexpress DDX58 expression in
colon cancer cells and the efficiency of the overexpression system
was examined using western blot analysis (Fig. 2A). The results of the MTS and EdU
assays revealed that the overexpression of DDX58 distinctly
decreased the viability and inhibited the proliferation of the
HCT8, HCT116 and SW620 cells (Fig. 2B
and C). Scratch and Transwell assays also demonstrated that the
overexpression of DDX58 significantly inhibited the migration and
invasion of HCT8, HCT116 and SW620 cells (Fig. 2D-K). These data thus suggested that
the activation of DDX58/RIG-I signaling inhibited the proliferation
and metastasis of colon cancer cells.
Silencing of DDX58 promotes the
proliferation, migration and invasion of colon cancer cells
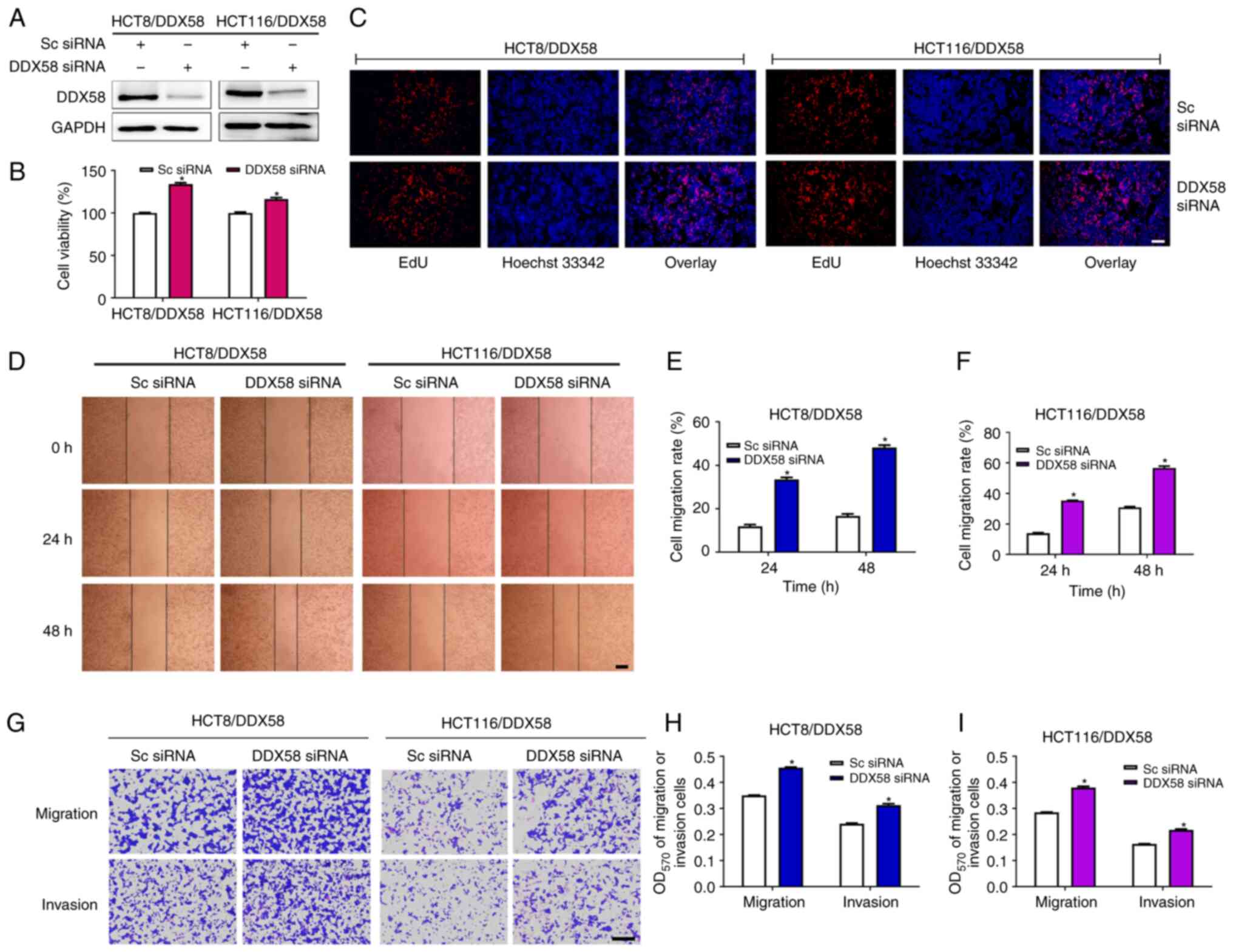
To further demonstrate the function of DDX58 in
colon cancer cells, specific DDX58 siRNA were used to silence the
DDX58 levels in colon cancer cells with the stable overexpression
of DDX58 (HCT8/DDX58 and HCT116/DDX58 cells). The results of
western blot analysis revealed the efficiency of the silencing of
DDX58 (Fig. 3A). Since SW620 cells
were constructed as an overexpressing transient cell model, the
HCT8/DDX58 and HCT116/DDX58 cells were selected for use to examine
the effects of DDX58 silencing on cell viability, proliferation,
migration and invasion. The results of MTS, EdU scratch wound and
Transwell assays indicated that the downregulation of DDX58
promoted the proliferation, migration and invasion of colon cancer
cells with the stable overexpression of DDX58 (Fig. 3B-I), which further demonstrated the
function of DDX58 in colon cancer cells.
Overexpression of DDX58 inhibits the
expression of CSE in colon cancer cells
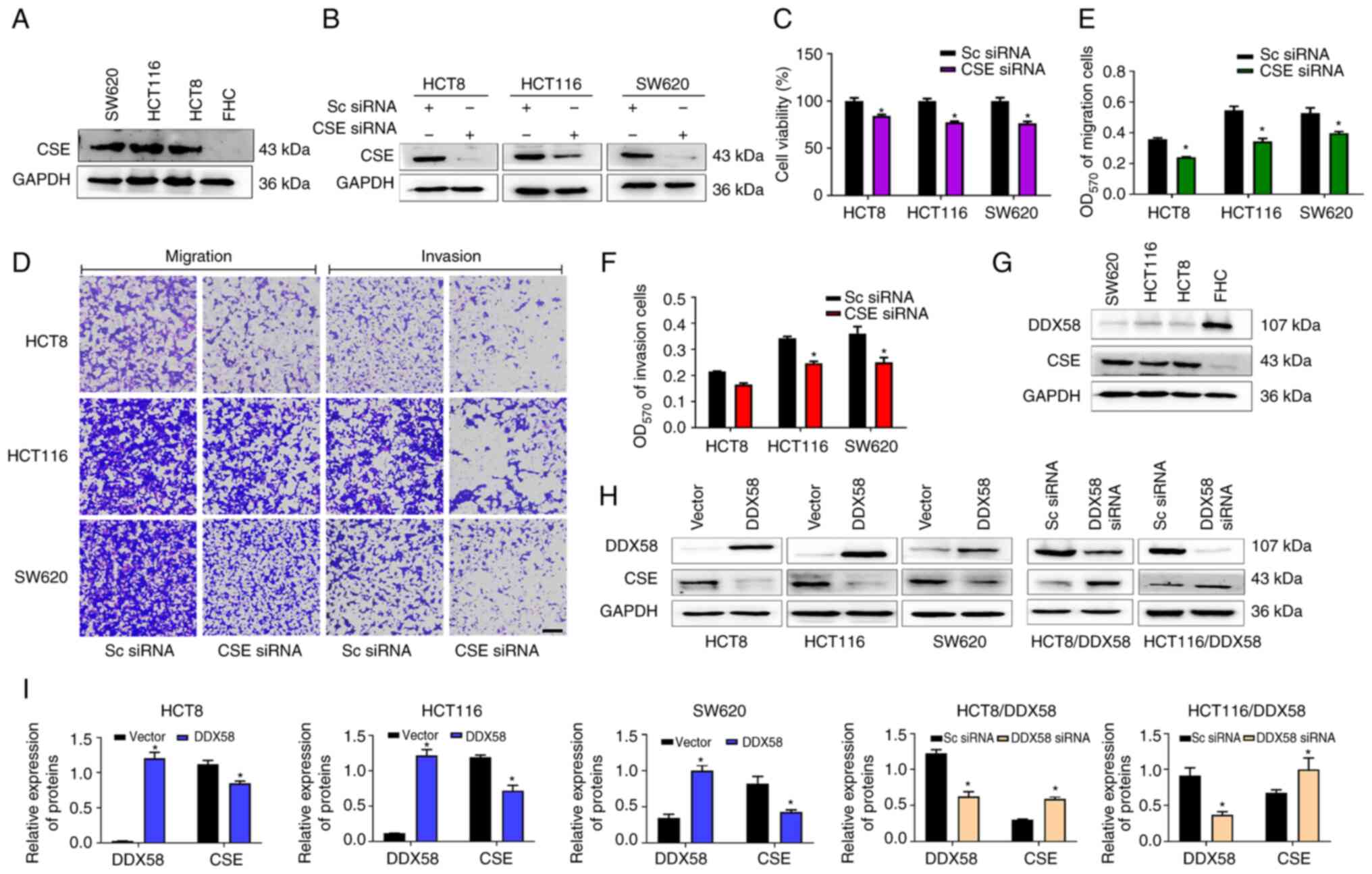
CSE, as a major endogenous H2S synthase,
plays critical roles in colon cancer processes, including
proliferation, migration and invasion (21-23).
In the present study, to further confirm the role of CSE in colon
cancer, CSE siRNA was transfected into human colon cancer cells.
The results of western blot analysis, and MTS and Transwell assays
revealed that CSE was highly expressed in human colon cancer cells
(Fig. 4A) and the silencing of CSE
expression (Fig. 4B) inhibited the
proliferation, migration and invasion of HCT8, HCT116 and SW620
colon cancer cells (Fig. 4C-F). To
determine whether DDX58 regulates the progression of colon cancer
through CSE, the association between DDX58 and CSE expression in
colon cancer cells was first examined and it was found that DDX58
and CSE expression were negatively associated in colon cancer cells
(Fig. 4G). Subsequently, DDX58
overexpression plasmid was transfected into HCT8, HCT116 and SW620
cells, and it was observed that the CSE protein levels were
significantly decreased following the overexpression of DDX58
(Fig. 4H and I). Conversely, the
silencing of DDX58 led to a significant increase in the CSE protein
levels in the HCT-8/DDX58 and HCT116/DDX58 cells with the stable
overexpression of DDX58 (Fig. 4H and
I). The data demonstrated that DDX58 regulated CSE protein
expression in colon cancer cells and indicated that the
overexpression of DDX58 suppressed CSE signaling, and consequently
inhibited the proliferation, migration and invasion of colon cancer
cells.
STAT3 is involved in the regulation of
CSE expression in colon cancer cells
In a previous study by the authors, it was
demonstrated that STAT3 binds to the promoter of CSE to promote the
expression of CSE (24).
Therefore, in the present study, to explore the mechanisms
underlying altered CSE expression, it was first demonstrated that
STAT3 regulates the expression of CSE in colon cancer cells. The
HCT8, HCT116 and SW620 cells were transfected with STAT3 siRNA and
STAT3 overexpression plasmid. The results of western blot analyses
revealed that the silencing of STAT3 suppressed the expression of
CSE, while STAT3 overexpression reversed the STAT3 siRNA-induced
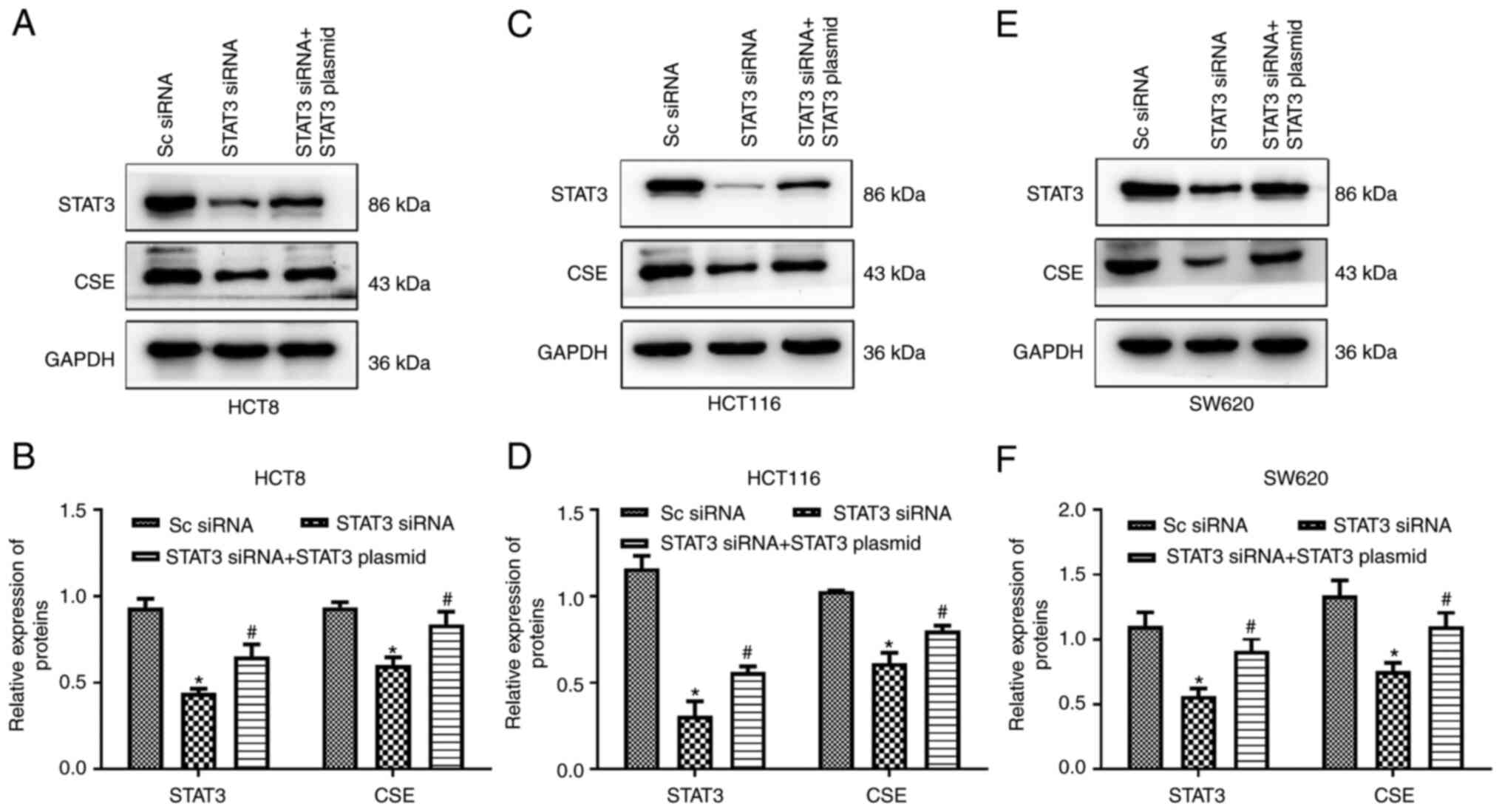
downregulation of CSE protein levels (Fig. 5), suggesting that STAT3 is involved
in the regulation of CSE expression in colon cancer cells. The
transfection efficiency of the STAT3 overexpression plasmid in
colon cancer cells was also examined (Fig. S1).
DDX58 regulates STAT3 by interacting with
STAT3 in colon cancer cells
To explore the role of STAT3 in the regulation of
CSE expression by DDX58 in colon cancer cells, the effects of DDX58
on STAT3 expression were first investigated. DDX58 overexpression
plasmid was transfected into the HCT8, HCT116 and SW620 cells, and
DDX58 siRNA was applied to the HCT8/DDX58 and HCT116/DDX58 cells.
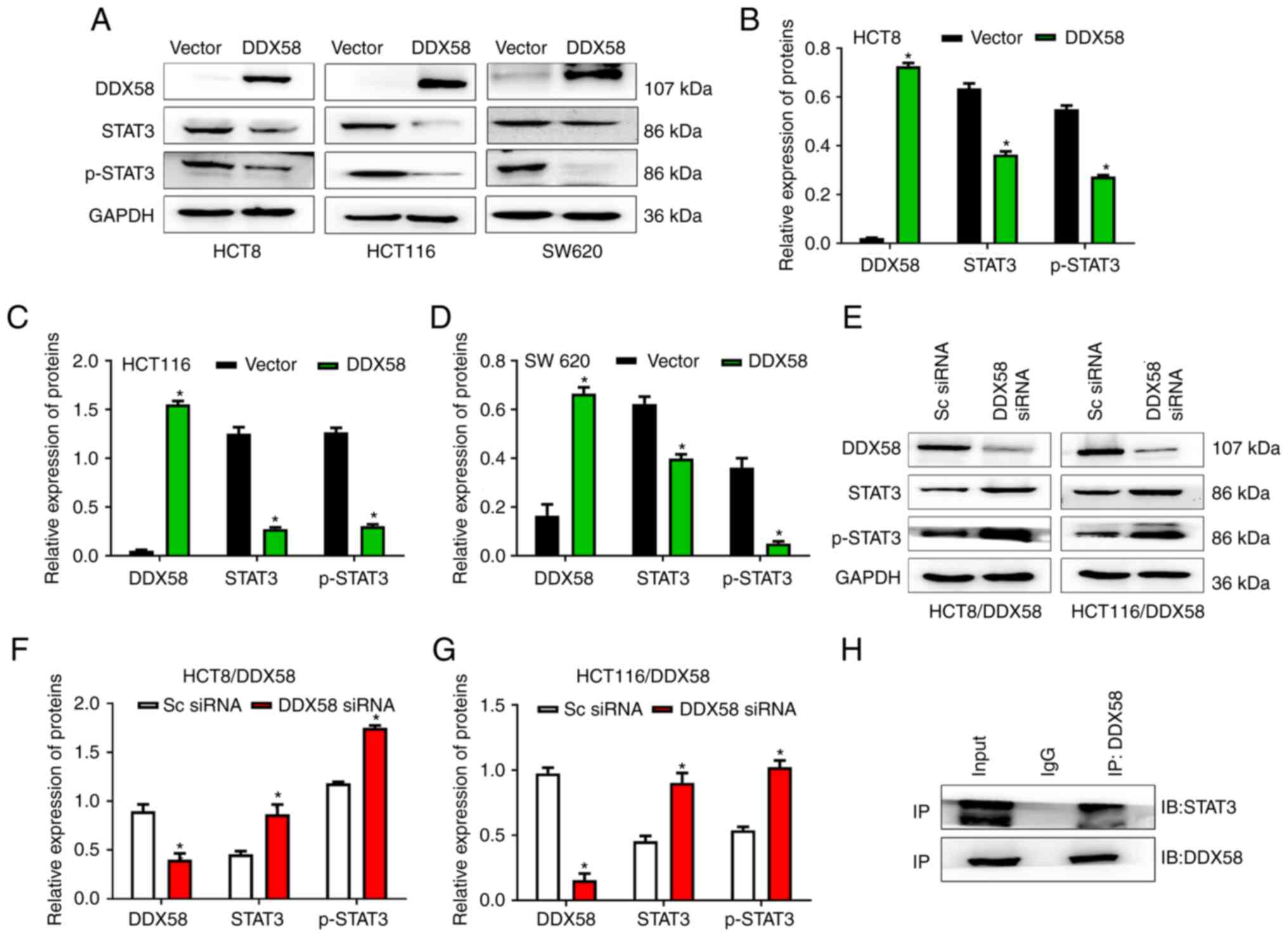
Western blot analyses revealed that the overexpression of DDX58
distinctly decreased the level of STAT3, and inhibited the
phosphorylation of STAT3 in the HCT8, HCT116 and SW620 cells
(Fig. 6A-D); however, the
silencing of DDX58 resulted in a significant increase in STAT3 and
p-STAT3 levels in the HCT-8/DDX58 and HCT116/DDX58 cells (Fig. 6E-G). These data indicated that
DDX58 regulates the expression and phosphorylation of STAT3.
To further explore the mechanisms underlying the
effects of DDX58 on STAT3, the interaction between DDX58 and STAT3
was investigated. DDX58 and STAT3 overexpression plasmids were
co-transfected into human HCT116 cells and Co-IP assay then was
performed. The results revealed that DDX58 interacted with STAT3 in
colon cancer cells (Fig. 6H),
which may affect the phosphorylation and dimerization of STAT3, and
may consequently prevent STAT3 from entering the nucleus, resulting
in suppression of the transcription and translation of the target
gene, CSE.
Overexpression of DDX58 inhibits tumor
growth and STAT3/CSE signaling in a nude mouse xenograft model of
colon cancer
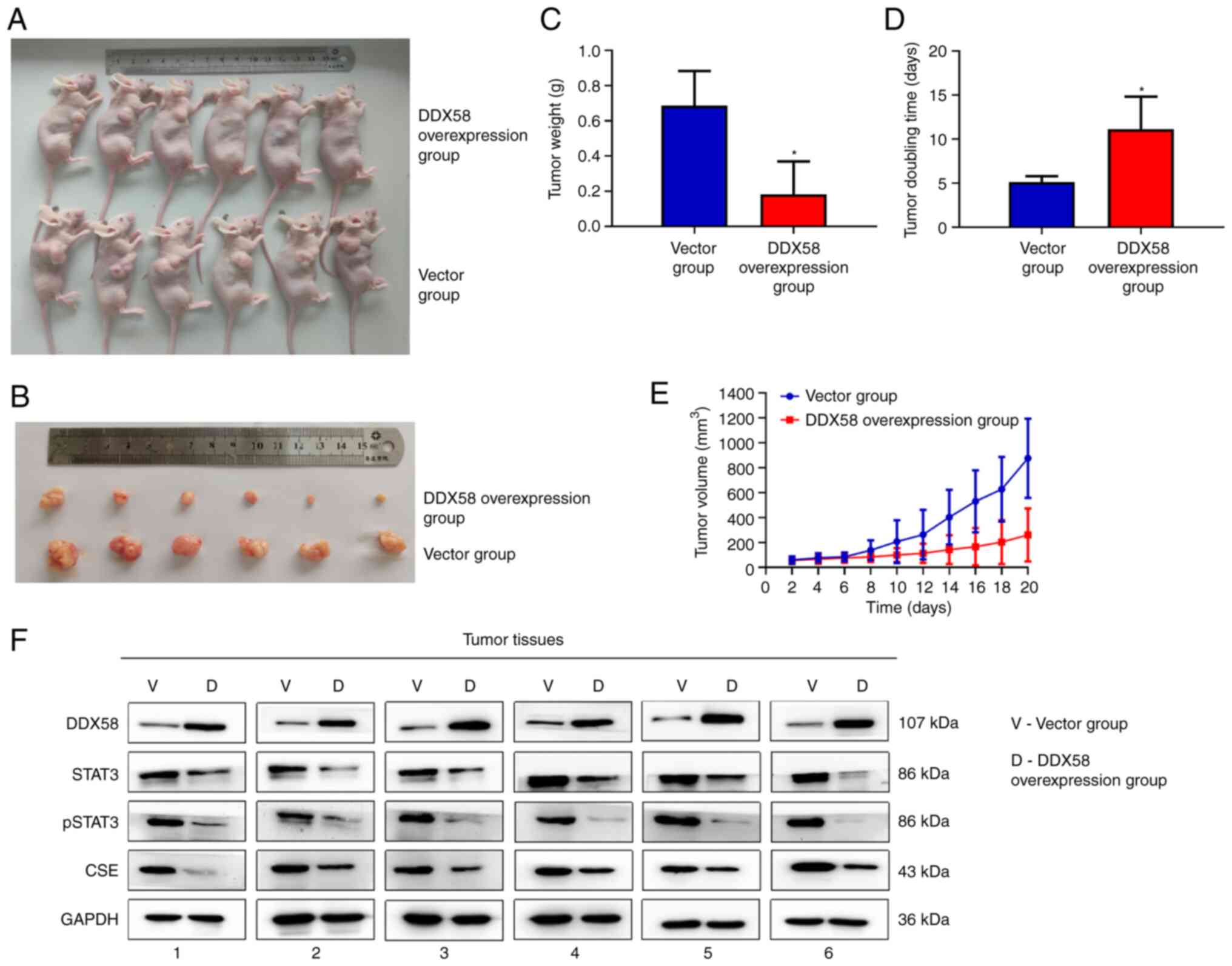
To investigate whether DDX58 influences the
progression of colon cancer in vivo, HCT116 cells with the
stable overexpression of DDX58 were injected subcutaneously into
the right flanks of nude mice to construct an axillary xenograft
tumor model to evaluate the effects of DDX58 on tumor growth. As
shown in Fig. 7A-E, the tumors
from the mice injected with HCT116 cells with the stable
overexpression of DDX58 were smaller in both size and weight, and
had a longer tumor doubling time compared with the vector group;
this indicated that the overexpression of DDX58 inhibited tumor
growth in the nude mouse xenograft model of colon cancer. Moreover,
it was found that the levels of STAT3, p-STAT3 and CSE were also
decreased in the tumors formed by HCT116/DDX58 cells compared with
the vector group (Fig. 7F). The
data suggest that the upregulation of DDX58 inhibits tumor growth
and STAT3/CSE signaling in a nude mouse xenograft model of colon
cancer.
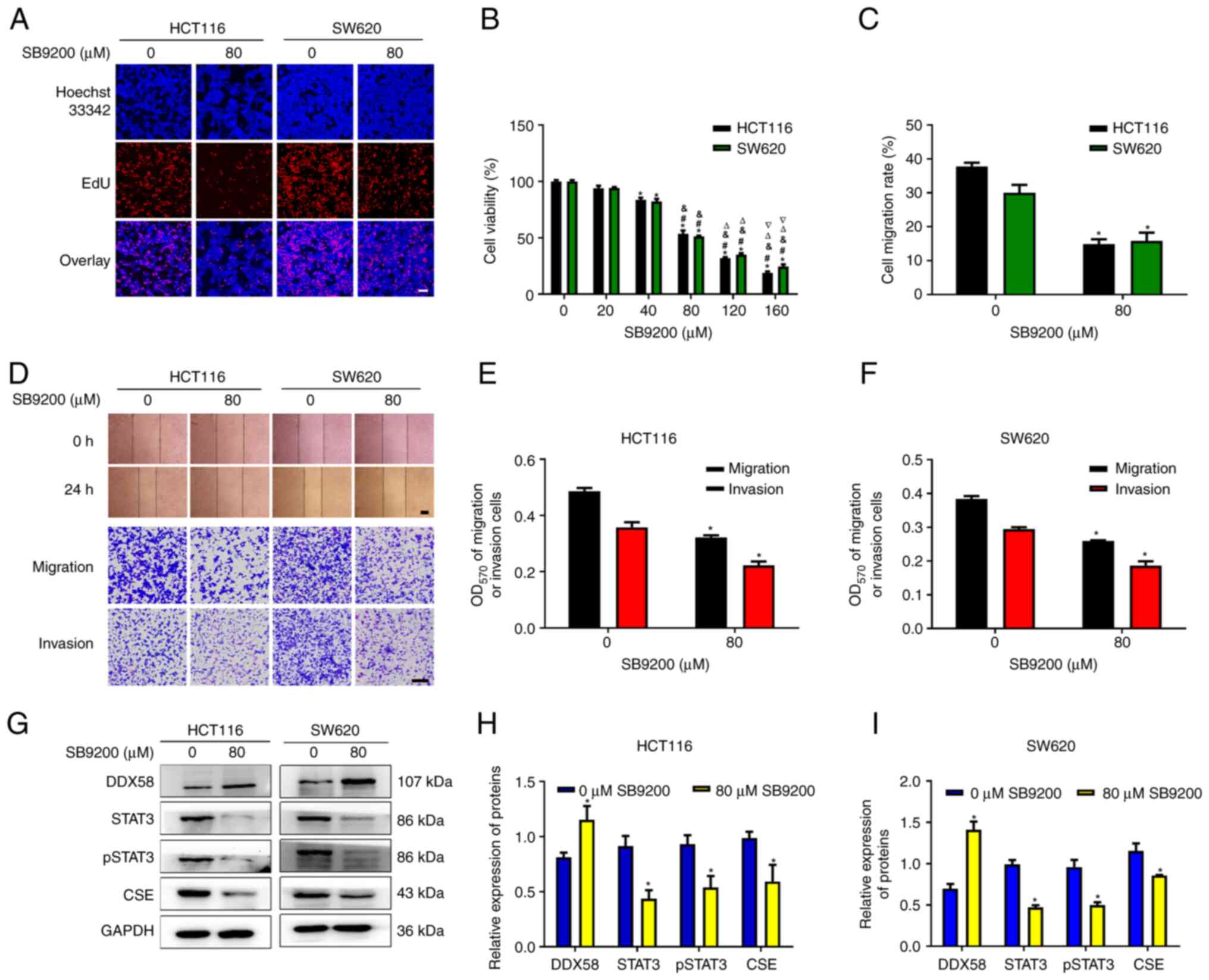
The DDX58 agonist, SB9200, induces the
inhibition of human colon cancer
SB9200 (inarigivir soproxil), a dinucleotide
antiviral compound, can activate intracellular innate immunity via
enabling DDX58/RIG-I. On the basis of the aforementioned results on
the function of DDX58 in colon cancer, the present study then
investigated the roles of SB9200 in colon cancer cells. Two classic
types of colon cancer cells, HCT116 and SW620 cells, were used for
subsequent analyses. The data demonstrated that SB9200
significantly reduced cell viability and the number of
EdU+ cells (Fig. 8A and
B), and inhibited the migration and invasion (Fig. 8C-F) of HCT116 and SW620 cells. We
also observed an increase in DDX58 protein expression, and a
decrease in STAT3, p-STAT3 and CSE expression in HCT116 and SW620
cells treated with SB9200 (Fig.
8G-I). These data indicated that SB9200 induced the inhibition
of human colon cancer cells, and further demonstrated the function
of DDX58 in colon cancer cells, and the possible use of SB9200 as a
therapeutic agent for colon cancer.
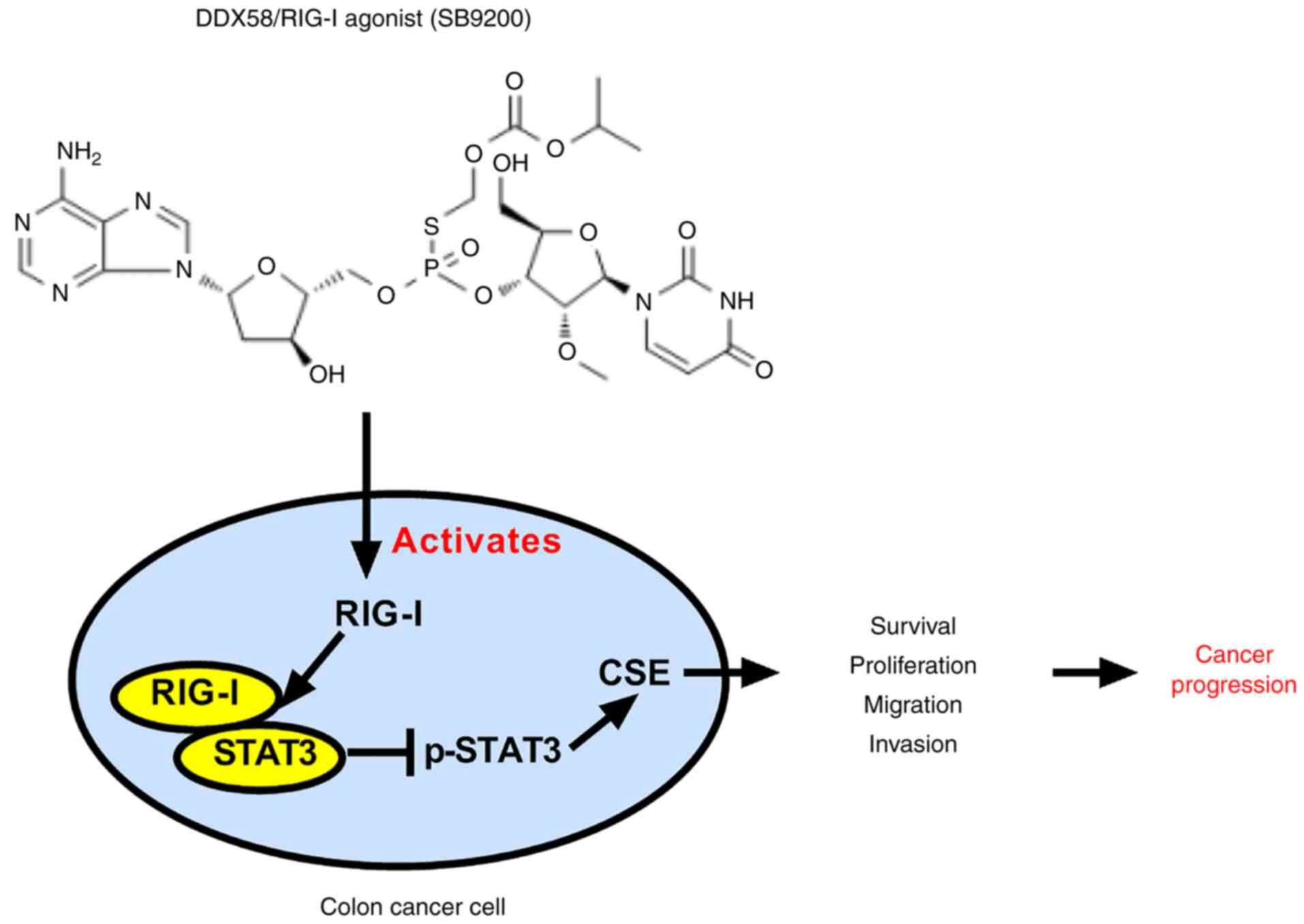
The inhibitory effects of DDX58 and its activator,
SB9200, on the growth of colon cancer cells by affecting the
DDX58/pSTAT3/CSE pathway are summarized in the schematic diagram in
Fig. 9.
Discussion
Colon cancer is one of the most common
gastrointestinal malignancies and the third most common type of
cancer in the United States (25).
However, even though the application of chemotherapeutic agents and
targeted therapies and the clinical advances in early detection and
surgery have been shown to be relatively effective in patients with
colon cancer, the 5-year survival rate of patients with metastatic
colon cancer remains low (26).
Moreover, some patients with colon cancer eventually develop
metastasis during treatment (27,28),
which most occurs due to the ineffectiveness of standard
treatments. Therefore, there is an urgent need for the development
of more effective treatment options. Immunotherapy has shown
promise in the treatment of colon cancer (20).
DDX58/RIG-I is a natural immune receptor helicase
that recognizes double-stranded virus RNA and engages the
production of interferons to initiate innate antiviral immunity.
Studies have demonstrated that the activation of RIG-I can induce
immunogenic cell death in tumor cells (12,29,30),
which indicates that it plays a critical role in colon cancer. In
the present study, it was found that a low expression of DDX58
resulted in a poor prognosis in colon cancer. It was further
confirmed that the overexpression of DDX58 in colon cancer cells
attenuated the proliferation, migration and invasion of colon
cancer cells in vitro and tumor growth in vivo, while
the opposite was observed following the silencing of DDX58. These
data demonstrated the tumor suppressor function of DDX58 in colon
cancer. In addition, we observed that the expression level of DDX58
was low in a series of colon cancer cells, while the expression
level in LOVO cells was higher than that of other colon cancer
cells, which may be related to the characteristics of the cells
themselves. For example, LOVO cells were found to proliferate
slowly, multiply for a long period of time, and tolerate fewer
passages than other cell lines (data not shown).
CSE is a major endogenous H2S synthase
that plays a crucial role in colon cancer (21-23).
In the present study, the role of CSE overexpression in promoting
colon cancer progression was further confirmed. To identify whether
the DDX58 regulation of colon cancer is related to CSE, the
association between DDX58 and CSE in colon cancer cells was
investigated and the regulatory effects of DDX58 on CSE protein
were demonstrated.
STAT3, as a transcriptional mediator of oncogenic
signals, is constantly activated in the majority of human cancers
(31). An increased pSTAT3
expression has been shown to be inversely associated with the
survival of patients with colon cancer (32). Therefore, STAT3 plays a crucial
role in the progression of colon cancer. Moreover, a previous study
by the authors demonstrated that STAT3 binds to the promoter of CSE
to promote the expression of CSE (24). Herein, it was also proven that
STAT3 was involved in the regulation of CSE expression in colon
cancer cells. Thus, to further explore the underlying mechanisms of
the regulation of CSE by DDX58, the effects of DDX58 on the
transcriptional activity of STAT3 were determined, and it was found
that DDX58 affected the phosphorylation of STAT3 and consequently
affected the transcriptional activity of STAT3, which also
demonstrated that STAT3 may mediate DDX58 to regulate CSE. In
addition, it was also demonstrated that DDX58 interacted with STAT3
in colon cancer cells, and consequently affected the
phosphorylation and dimerization of STAT3, resulting in the
suppression of the transcription and translation of its target
gene, CSE. Furthermore, the inhibitory effect of DDX58 upregulation
on STAT3/CSE signaling was also observed in a nude mouse xenograft
model of colon cancer.
In addition, it was found that a dinucleotide
antiviral compound (SB9200), as a DDX58/RIG-I agonist, induced the
inhibition of human colon cancer by affecting the DDX58/pSTAT3/CSE
pathway (Fig. 9), which further
demonstrated the tumor suppressive function of DDX58 in colon
cancer cells, and the possible use of SB9200 as a therapeutic agent
for colon cancer.
In conclusion, the present study revealed a critical
tumor suppressor molecule, namely DDX58/RIG-I, in colon cancer. The
activation of DDX58/RIG-I inhibited the proliferation of tumor
cells by regulating STAT3/CSE signaling in colon cancer. This
suggests that the development of DDX58/RIG-I agonists may prove to
be a novel and effective therapeutic strategy for patients with
colon cancer.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TW and KL conceived and designed the experiments.
YD, HF and XH performed the experiments. YL, WZ, XZ, CY and WG
analyzed the data and produced the figures. YD and TW wrote the
manuscript. All authors have read and approved the manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved. YD, HF and XH confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
All experiments were performed in accordance with
national ethical guidelines, all patient consents were obtained and
with the approval from the committee of Shanghai Outdo Biotech Co.,
Ltd. (HCA18ol0Su13). The animal experiments were approved by the
Ethics Committee of the Medical School of Henan University
(HUSOM2020-301).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study supported by the grants from the National
Natural Science Foundation of China (no. 82072726), as well as the
Natural Science Foundation of Henan Province in China (no.
202300410079).
References
|
1
|
Kell AM and Gale M Jr: RIG-I in RNA virus
recognition. Virology. 479-480:110–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hur S: Double-stranded RNA sensors and
modulators in innate immunity. Annu Rev Immunol. 37:349–375. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Streicher F and Jouvenet N: Stimulation of
innate immunity by host and viral RNAs. Trends Immunol.
40:1134–1148. 2019. View Article : Google Scholar
|
|
4
|
Prasov L, Bohnsack BL, El Husny AS, Tsoi
LC, Guan B, Kahlenberg JM, Almeida E, Wang H, Cowen EW, De Jesus
AA, et al: DDX58(RIG-I)-related disease is associated with
tissuespecific interferon pathway activation. J Med Genet.
59:294–304. 2022. View Article : Google Scholar
|
|
5
|
Wu S, Lin J, Fu Y and Ou Q: RIG-I enhances
interferon-alpha response by promoting antiviral protein expression
in patients with chronic hepatitis B. Antivir Ther. 23:575–583.
2018. View
Article : Google Scholar
|
|
6
|
Loo YM and Gale M Jr: Immune signaling by
RIG-Ilike receptors. Immunity. 34:680–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Besch R, Poeck H, Hohenauer T, Senft D,
Häcker G, Berking C, Hornung V, Endres S, Ruzicka T, Rothenfusser S
and Hartmann G: Proapoptotic signaling induced by RIG-I and MDA-5
results in type I interferon-independent apoptosis in human
melanoma cells. J Clin Invest. 119:2399–2411. 2009.PubMed/NCBI
|
|
8
|
Duewell P, Steger A, Lohr H, Bourhis H,
Hoelz H, Kirchleitner SV, Stieg MR, Grassmann S, Kobold S, Siveke
JT, et al: RIG-I-like helicases induce immunogenic cell death of
pancreatic cancer cells and sensitize tumors toward killing by
CD8(+). T cells Cell Death Differ. 21:1825–1837. 2014. View Article : Google Scholar
|
|
9
|
Hou J, Zhou Y, Zheng Y, Fan J, Zhou W, Ng
IO, Sun H, Qin L, Qiu S, Lee JM, et al: Hepatic RIG-I predicts
survival and interferon-alpha therapeutic response in
hepatocellular carcinoma. Cancer Cell. 25:49–63. 2014. View Article : Google Scholar
|
|
10
|
Xu XX, Wan H, Nie L, Shao T, Xiang LX and
Shao JZ: RIG-I: A multifunctional protein beyond a pattern
recognition receptor. Protein Cell. 9:246–253. 2018. View Article : Google Scholar :
|
|
11
|
Elion DL, Jacobson ME, Hicks DJ, Rahman B,
Sanchez V, Gonzales-Ericsson PI, Fedorova O, Pyle AM, Wilson JT and
Cook RS: Therapeutically active RIG-I agonist induces immunogenic
tumor cell killing in breast cancers. Cancer Res. 78:6183–6195.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jacobson ME, Wang-Bishop L, Becker KW and
Wilson JT: Delivery of 5'-triphosphate RNA with endosomolytic
nanoparticles potently activates RIG-I to improve cancer
immunotherapy. Biomater Sci. 7:547–559. 2019. View Article : Google Scholar
|
|
13
|
Ellermeier J, Wei J, Duewell P, Hoves S,
Stieg MR, Adunka T, Noerenberg D, Anders HJ, Mayr D, Poeck H, et
al: Therapeutic efficacy of bifunctional siRNA combining TGF-β1
silencing with RIG-I activation in pancreatic cancer. Cancer Res.
73:1709–1720. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brägelmann J, Lorenz C, Borchmann S,
Nishii K, Wegner J, Meder L, Ostendorp J, Ast DF, Heimsoeth A,
Nakasuka T, et al: MAPK-pathway inhibition mediates inflammatory
reprogramming and sensitizes tumors to targeted activation of
innate immunity sensor RIG-I. Nat Commun. 12:55052021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang S, Yang C, Yu F, Ding W, Hu Y, Cheng
F, Zhang F, Guan B, Wang X, Lu L and Rao J: Endoplasmic reticulum
resident oxidase ERO1-Lalpha promotes hepatocellular carcinoma
metastasis and angiogenesis through the S1PR1/STAT3/VEGF-A pathway.
Cell Death Dis. 9:11052018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Shi H, Liu Y, Zhang W, Duan X, Li
M, Shi X and Wang T: Cystathionine-γ-lyase promotes the metastasis
of breast cancer via the VEGF signaling pathway. Int J Oncol.
55:473–487. 2019.PubMed/NCBI
|
|
17
|
Wang L, Shi H, Zhang XY, Zhang XL, Liu Y,
Kang W, Shi X and Wang T: I157172, a novel inhibitor of
cystathionine γ-lyase, inhibits growth and migration of breast
cancer cells via SIRT1-mediated deacetylation of STAT3. Oncol Rep.
41:427–436. 2019.
|
|
18
|
Nitti M, Piras S, Marinari UM, Moretta L,
Pronzato MA and Furfaro AL: HO-1 induction in cancer progression: A
matter of cell adaptation. Antioxidants (Basel). 6:292017.
View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C(T)). method Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Heidegger S, Wintges A, Stritzke F, Bek S,
Steiger K, Koenig PA, Göttert S, Engleitner T, Öllinger R, Nedelko
T, et al: RIG-I activation is critical for responsiveness to
checkpoint blockade. Sci Immunol. 4:eaau89432019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fan K, Li N, Qi J, Yin P, Zhao C, Wang L,
Li Z and Zha X: Wnt/β-catenin signaling induces the transcription
of cystathionine-gamma-lyase, a stimulator of tumor in colon
cancer. Cell Signal. 26:2801–2808. 2014. View Article : Google Scholar
|
|
22
|
Oláh G, Módis K, Törö G, Hellmich MR,
Szczesny B and Szabo C: Role of endogenous and exogenous nitric
oxide, carbon monoxide and hydrogen sulfide in HCT116 colon cancer
cell proliferation. Biochem Pharmacol. 149:186–204. 2018.
View Article : Google Scholar :
|
|
23
|
Cao X, Ding L, Xie ZZ, Yang Y, Whiteman M,
Moore PK and Bian JS: A review of hydrogen sulfide synthesis,
metabolism, and measurement: Is modulation of hydrogen sulfide a
novel therapeutic for cancer? Antioxid Redox Signal. 31:1–38. 2019.
View Article : Google Scholar :
|
|
24
|
You J, Shi X, Liang H, Ye J, Wang L, Han
H, Fang H, Kang W and Wang T: Cystathionine--lyase promotes process
of breast cancer in association with STAT3 signaling pathway.
Oncotarget. 8:65677–65686. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar
|
|
26
|
Lichtenstern CR, Ngu RK, Shalapour S and
Karin M: Immunotherapy, inflammation and colorectal cancer. Cells.
9:6182020. View Article : Google Scholar :
|
|
27
|
Vatandoust S, Price TJ and Karapetis CS:
Colorectal cancer: Metastases to a single organ. World J
Gastroenterol. 21:11767–11776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reddy TP, Khan U, Burns EA and Abdelrahim
M: Chemotherapy rechallenge in metastatic colon cancer: A case
report and literature review. World J Clin Oncol. 11:959–967. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Elion DL and Cook RS: Harnessing RIG-I and
intrinsic immunity in the tumor microenvironment for therapeutic
cancer treatment. Oncotarget. 9:29007–29017. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang X, Muthusamy V, Fedorova O, Kong Y,
Kim DJ, Bosenberg M, Pyle AM and Iwasaki A: Intratumoral delivery
of RIG-I agonist SLR14 induces robust antitumor responses. J Exp
Med. 216:2854–2868. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei N, Li J, Fang C, Chang J, Xirou V,
Syrigos NK, Marks BJ, Chu E and Schmitz JC: Targeting colon cancer
with the novel STAT3 inhibitor bruceantinol. Oncogene.
38:1676–1687. 2019. View Article : Google Scholar
|
|
32
|
Heichler C, Scheibe K, Schmied A, Geppert
CI, Schmid B, Wirtz S, Thoma OM, Kramer V, Waldner MJ, Büttner C,
et al: STAT3 activation through IL-6/IL-11 in cancer-associated
fibroblasts promotes colorectal tumour development and correlates
with poor prognosis. Gut. 69:1269–1282. 2020. View Article : Google Scholar
|