Introduction
The local, dynamic tumor microenvironment (TME) is
composed of tumor, stromal and immune cells, and the products
secreted by these cells. The environment, which promotes tumor
survival, is characterized by low oxygen levels, a low pH and
immunosuppression, and is formed by the cooperative actions of
various cells and their metabolites (1). Tumor-associated macrophages (TAMs), a
type of immune cell in the human body, accounts for ~50% of all
cells in the complex TME (2). It
was previously considered that TAMs can inhibit tumor growth and
metastasis by directly killing tumor cells or presenting antigens.
However, accumulating research has been indicated that TAMs are not
antitumor cells, but rather cells that promote tumor cell growth,
invasion and metastasis, as well as angiogenesis in various types
of cancer, including breast cancer, by secreting a variety of
cytokines (3-5). In addition, the degree of TAM
infiltration is highly associated with tumor grade, stage and
patient prognosis (6-8).
Macrophages have an extremely high plasticity, and
can differentiate into different subsets and functional phenotypes
following stimulation by various signals in the TME; among the
differentiated phenotypes are the pro-inflammatory (M1) and
anti-inflammatory (M2) phenotypes. Toll-like receptor (TLR) ligands
[such as lipopolysaccharide (LPS) and interferon-γ (IFN-γ)] can
polarize macrophages toward the M1 phenotype and promote an
inflammatory response, which has a killing effect on tumor cells
(9). When macrophages are
stimulated by T-helper 2 (Th2) cell cytokines [such as interleukin
(IL)-4 and IL-13], they differentiate into the M2 phenotype and
thus inhibit the inflammatory response and promote tumor cell
progression (10). However, the
majority of TAMs in the hypoxic TME are M2-type macrophages (M2d
subtype) (11), which facilitate
tumor progression. Therefore, the reprogramming or polarization of
TAMs from an immunosuppressive phenotype (M2) to a classical
phenotype (M1) is considered a promising cancer treatment strategy
(12-14).
MicroRNAs (miRNAs or miRs) are small non-coding RNAs
of ~22 nucleotides that negatively regulate gene expression by
binding to the 3' untranslated regions (UTRs) of mRNAs to increase
mRNA degradation or block translation (15). Previous studies have demonstrating
that miRNAs are involved in regulating numerous cellular processes,
including metabolic homeostasis, cell proliferation and apoptosis
(16-18). In addition, miRNAs play crucial
roles in inflammation and immunity by balancing macrophage
phenotypes via targeting related molecules or affecting signaling
pathways (19-21). A widely reported tumor suppressor
miRNA, miR-382, plays a critical role in the occurrence and
development of several types of cancer (22-24).
However, the function of miR-382 in the TME and the underlying
mechanisms have not yet been reported, at least to the best of our
knowledge.
The present study examined the changes in miR-382
expression in TAMs associated with breast cancer and investigated
its roles in the regulation of TAM polarization and the underlying
mechanisms. It was found that miR-382 expression was downregulated
in TAMs associated with breast cancer. The overexpression of
miR-382 affected the mitochondrial function of TAMs and inhibited
their M2 polarization. It was also observed that TAMs
overexpressing miR-382 inhibited the invasion and migration of 4T1
breast cancer cells by reducing epithelial-mesenchymal
transformation (EMT). Furthermore, it was confirmed that peroxisome
proliferator-activated receptor γ coactivator-1α (PGC-1α) was the
downstream target of miR-382 and the aforementioned in vitro
results were verified in in vivo experiments. On the whole,
the present study discovered a novel (to the best of our knowledge)
mechanism that regulates macrophage plasticity, namely, that
miR-382 alters the metabolic state of macrophages by targeting
PGC-1α. Additionally, the findings presented herein reveal the role
of miR-382 in the breast cancer TME and contribute to the further
understanding of the polarization and transformation of TAMs.
Materials and methods
Clinical samples and isolation of primary
macrophages
Clinical samples and paracancerous tissues were
collected from 27 patients with breast cancer who underwent
modified radical mastectomy for breast cancer in The Second
Affiliated Hospital of Chongqing Medical University from May, 2018
to May, 2020 and for whom complete clinicopathological data were
available; these patients were diagnosed by post-operative
pathology. The 27 collected clinical breast cancer samples were
classified as luminal A (9 cases), luminal B (8 cases),
HER-2-positve (5 cases), or triple-negative (5 cases) according to
the clinicopathological data. The present study was approved by The
Ethics Committee of the Second Affiliated Hospital of Chongqing
Medical University (Chongqing, China; approval no. 99/2022), and
informed consent was obtained from all the patients. The collected
breast cancer tissues and matched paracancerous tissues (at least 2
cm at a distance from the tumor) were separated by Ficoll-Hypaque
density gradient centrifugation. The tissue suspensions were mixed
with saline (1:1) and then added to the surface of lymphocyte
separation medium (Beijing Solarbio Science & Technology Co.,
Ltd.) along the wall of the test tube. Centrifugation was performed
at 400 × g (1,500 rpm, 15 cm radius horizontal rotor) for 20 min at
room temperature. The annular white cell layers at the liquid
interface were extracted, and flow cytometry (Caliber Flow
Cytometer, BD Biosciences). was used to identify the proportion of
cells positive for the macrophage marker CD14 (FITC, 29943, Cell
Signaling Technology, Inc.). The results revealed that >70% of
the extracted macrophages were CD14-positive, which met the
detection standard (Fig. S1).
Primary mouse macrophages (PMs) were extracted from
the peritoneal cavity of 10 female BALB/c mice (aged 6 to 8 weeks,
weighing 18 to 22 g) and then cultured in RPMI-1640 medium
(supplemented with 10% FBS and 1% penicillin-streptomycin) in an
incubator with saturated humidity (37°C and 5% CO2). The
purity of the PMs was assessed using flow cytometry as described
below.
Animals
A total of 30 female BALB/c mice (aged 6 to 8 weeks,
weighing 18 to 22 g) were purchased from the Experimental Animal
Center, Chongqing Medical University, Chongqing, China. All
experiments involving animals were conducted in accordance with the
guidelines for the use of experimental animals at Chongqing Medical
University and were approved by The Ethics Committee of the Second
Affiliated Hospital of Chongqing Medical University (approval no.
99/2022).
Cell lines and cell culture
The 4T1 (SCSP-5056) mouse breast cancer cell line,
THP-1 (SCSP-567) human peripheral blood mononuclear cell line and
the RAW264.7 (SCSP-5036) mouse macrophage cell line were purchased
from The Cell Bank of Type Culture Collection of the Chinese
Academy of Sciences. The 4T1, THP-1 and RAW264.7 cells were
cultured in RPMI-1640 medium (supplemented with 10% FBS and 1%
penicillin-streptomycin) in an incubator with saturated humidity
(37°C and 5% CO2).
Induction of macrophage differentiation
in vitro
i) TAMs: PMs (5×105) were co-cultured
with 4T1 cells (5×105) in Transwell chambers (0.4
µm, Corning, Inc.) for 48 h. ii) M0-type macrophages:
RAW264.7 cells (5×105) were allowed to adhere to the
plates (6-well plates, Corning, Inc.), or THP-1 cells
(5×105) were stimulated with phorbol 12-myristate
13-acetate (PMA; 50 mg/ml). iii) M1-type macrophages: M0
macrophages were stimulated with 20 ng/ml IFN-γ (Beyotime
Biotechnology Co., Ltd.) and 500 ng/ml LPS (Beyotime Institute of
Biotechnology Co., Ltd.) for 48 h. iv) M2-type macrophages: M0
macrophages were stimulated with 20 ng/ml IL-4 (Beyotime Institute
of Biotechnology Co., Ltd.) for 48 h.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using
TRIzol® reagent (Mei5 Biotechnology Co., Ltd.) and then
reverse transcribed (PrimeScript RT reagent kit, Takara
Biotechnology, Co., Ltd.) into cDNA (reaction conditions: 42°C for
2 min followed by hold at 4°C; 37°C for 15 min, 85°C for 5 sec, and
holding at 4°C). For the detection of miR-382, miRNA was reverse
transcribed (miRNA First-Strand cDNA Synthesis, Tailing Reaction,
Sangon Biotech Co., Ltd.) into cDNA with the polyA tailing protocol
(reaction conditions: 37°C for 60 min, 85°C for 5 min, then holding
at 4°C). Subsequently, 1 µl cDNA was mixed with a 9
µl PCR assay mixture containing 0.4 µM of each primer
and 5 µl TB Green II (Takara Biotechnology, Co., Ltd.). PCR
was conducted using the Real-Time PCR Detection System (Bio-Rad
Laboratories, Inc.). β-actin was used as the internal reference for
tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, IL-10 and
transforming growth factor-β (TGF-β) expression, and U6 (sequence
not available) was used as the internal reference for miR-382
expression. The specific primer sequences used are presented in
Table SI. The reaction conditions
were 96°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and
55°C/60°C for 30 sec. After the reaction, the experimental results
were analyzed using the 2−ΔΔCq method (25).
Western blot analysis
All cells were collected in the logarithmic growth
phase, total protein was extracted using RIPA buffer (Beyotime
Institute of Biotechnology Co., Ltd.), and the protein
concentration was determined using a BCA kit (Beyotime Institute of
Biotechnology Co., Ltd. The proteins were then boiled (100°C, 10
min) and stored at -20°C. A gel was prepared according to the
molecular weight requirements (SDS-PAGE in 5% stacking gel and 10%
separating gel), and 30 µg protein were loaded into each
well of the gel. The proteins were then separated by
electrophoresis (constant voltage: 100 V; 100 min) and transferred
onto 0.2 µm PVDF membranes (constant current, 250 mA; 60-120
min). The membranes were incubated with blocking buffer (Beyotime
Biotechnology Co., Ltd.) at room temperature for 30 min, washed
with TBST. After blocking, the membranes were incubated with the
corresponding primary antibodies against GAPDH (1:1,000, ab181602,
Abcam), TNF-α (1:1,000, ab205587, Abcam), IL-1β (1:1,000, ab254360,
Abcam), TGF-β (1:1,000, ab215715, Abcam), IL-10 (1:1,000, ab9969,
Abcam), vimentin (1:1,000, ab92547, Abcam), E-cadherin (1:1,000,
ab40772, Abcam), nuclear respiratory factor-1 (NRF-1; 1:1,000,
12482-1-AP, Proteintech Group, Inc.), mitochondrial transcription
factor A (TFAM; 1:1,000, 22586-1-AP, Proteintech Group, Inc.) for
10 h at 4°C. The membranes were then washed three times with TBST
(10 min/wash) and then incubated with the corresponding secondary
anti-bodies (HRP-labeled, 1:5,000, A0208, Beyotime Institute of
Biotechnology Co., Ltd.) for 1 h on a shaking table at room
temperature. The membranes were then washed three times with TBST
(10 min/wash), incubated in the dark with ECL supersensitive
chemiluminescent solutions (Beyotime Institute of Biotechnology
Co., Ltd.) mixed at a ratio of 1:1, and imaged using an imaging
instrument (ChemiDoc Touch, Bio-Rad Laboratories, Inc.). The
densitometries of the bands were detected and analyzed using ImageJ
software (v1.8.0, National Institutes of Health).
Flow cytometric analyses
The PMs cells were collected from 6-well plates,
washed twice with PBS (Beyotime Institute of Biotechnology Co.,
Ltd.), and resuspended. After counting, the cells were fixed in 4%
paraformaldehyde for 10 min and then washed twice by resuspension
in PBS. Subsequently, 0.1% Triton was added for 5 min, and the
cells were washed twice with PBS. Following centrifugation (25°C,
200 × g, 5 min), cluster of differentiation CD86 (FITC, MA1-10300,
Thermo Fisher Scientific Co., Ltd.) and CD206 antibodies (APC,
17-2069-41, Thermo Fisher Scientific Co., Ltd.) were added to the
cells, which were incubated on ice in the dark for 30 min.
Following two washes with PBS, the supernatant was discarded, and
400 µl PBS were added. Following centrifugation (25°C, 200 ×
g, 5 min), the cells that had been incubated with the antibodies
were resuspended, and the cell polarization index was detected
using flow cytometry (Caliber Flow Cytometer, BD Biosciences). Data
were analyzed using FlowJo software (v10.8, Tree Star, Inc.).
Dual luciferase reporter assays
The 3'UTR of PGC-1α was cloned into the pSicheck2
vector (Promega Corporation). To produce the PGC-1α mutant
reporter, the PGC-1α 3'UTR sequence was mutated to eliminate
complementarity with miR-382. The luciferase reporter vector was
co-transfected with miR-382 mimic or miR-NC (50 nM, Thermo Fisher
Scientific, Inc.) into 293T cells (CRL-3216, ATCC) in 24-well
plates using Lipofectamine 2000® (Invitrogen, Thermo
Fisher Scientific, Inc.). At 24 h following transfection, Firefly
luciferase activity was normalized to Renilla luciferase
activity and determined using the two-enzyme GloMax-Multi detection
system (Promega Corporation).
Cell invasion assay
After coating the membrane with hydrated Matrigel
(BD Biosciences Co., Ltd.), the 4T1 cells were seeded into the
upper chamber (1×104 cells/well) with 100 µl
serum-free medium containing BSA (0.2%), and 500 µl of a
suspension containing PMs, TAMs, or TAMs overexpressing miR-382
(1×105 cells/ml, supplemented with 10% FBS) was added to
the lower chamber. After 24 h, the medium in the upper chamber was
discarded; the cells were fixed with 4% paraformaldehyde (30 min),
washed twice with PBS, stained with 0.1% crystal violet (Beyotime
Institute of Biotechnology Co., Ltd.) for 30 min at an ambient
temperature, and washed three times with PBS. The cells that did
not migrate from the upper chamber were removed using cotton swabs.
After air drying the chambers for 10 min, images of five randomly
selected fields were captured under a microscope (CX33, Olympus
Corporation), and the number of cells that had crossed the membrane
was counted.
Cell migration (wound healing) assay
A pen was used to draw six lines on the back of a
six-well plate. The 4T1 cells were evenly seeded in six-well plates
(5×105 cells/well), and macrophages from each group were
seeded in a Transwell chamber (5×105 cells/well) and
cultured separately until the cells attached to the well. After the
4T1 cells reached 90% confluency in the field of view, a
200-µl pipette tip was used to scratch an even horizontal
line in the cell monolayer. The cells were washed twice with PBS,
the floating cells were removed, serum-free medium was added, and
images were captured using an electron microscope (CX33, Olympus
Corporation), this time point was considered the 0 h time point.
For each group, Transwell chambers were moved to the corresponding
six-well plate for co-culture, and the medium was replaced with
serum-free medium. After 24 h, the upper chamber was removed, and
images were captured under a microscope (CX33, Olympus
Corporation).
ROS assay
Macrophages (1×105 cells/ml) in each
group were collected, washed and incubated with 10 µM
dihydroethidium-ROS probe (200 µl, FY17032, FEIYUBIO Co.,
Ltd.) for 30 min at 37°C in the dark. The cells were then washed
with PBS and resuspended in PBS. The mean fluorescence intensities
were measured within 30 min using a flow cytometer (Caliber Flow
Cytometer, BD Biosciences) and analyzed using FlowJo software
(v10.8, Tree Star, Inc.).
ATP measurements
Macrophages from each group were collected and
resuspended in PBS (1×106 cells/ml). ATP lysis buffer
(Beyotime Institute of Biotechnology Co., Ltd.) was added to the
cells, then mixed and incubated for 30 min on ice. The cells were
centrifuged at 12,000 × g for 5 min at 4°C and then the ATP level
in the supernatant was measured according to the specific steps of
the ATP assay kit (S0026, Beyotime Institute of Biotechnology Co.,
Ltd.). The luminescence signals were measured using a BCA protein
assay kit (P0010S, Beyotime Institute of Biotechnology Co., Ltd.)
and a BMG reader (Thermo Fisher Scientific, Inc.). Finally, the ATP
concentration was normalized to the protein concentration against
ATP standard solution (Beyotime Institute of Biotechnology Co.,
Ltd.).
Mitochondrial DNA (mtDNA) detection
Macrophages from each group were collected and
resuspended in PBS (5×105 cells). Total DNA was
extracted according to the steps of the GenElute™ Mammalian Genomic
DNA Miniprep Kit (G1N70, Sigma-Aldrich, Shanghai, Trading Co.,
Ltd.). The content of mtDNA was analyzed by qPCR using specific
indicators cytochrome b (Cytb) and beta-2 microglobulin [B2m;
cytochrome c oxidase subunit 4 (Cox4) was used as the
reference gene]. The specific primer sequences used are presented
in Table SI. Reaction conditions
were 96°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and
55°C/60°C for 30 sec. After the reaction, the experimental results
were analyzed using the 2−ΔΔCq method.
Construction of transfected cell
lines
The lentiviral vector (miR-382) used in the present
study mediates the green fluorescent protein GFP, the lentiviral
vector (PGC-1α) contains the red fluorescent protein RFP, and empty
control lentiviral vector were designed and synthesized by Shanghai
Genechem Co., Ltd. The miR-382-overexpression vector was sent for
sequencing and designated as GV369
(Ubi-MCS-SV40-EGFP-IRES-Puromycin). The PGC-1α-overexpression
vector was sent for sequencing and designated GV492
(Ubi-MCS-3FLAG-CBh-gcRFP-IRES-Puromycin). In brief, the lentiviral
vectors (miR-382 or PGC-1α) were transfected in the 2nd generation
transfection system into PMs cells (MOI:50) cultured in RMPI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS
(Hyclone; Cytiva) at 37°C with 5% CO2. After 72 h
culturing at 37°C, the transfection efficiency of the PMs was
determined by examining the fluorescence intensity under a
microscope (CX43, Olympus Corporation). Puromycin treatment at 2
µg/ml for 1 weeks at 37°C was used to select the stably
transfected cell lines before the efficiency was confirmed using
western blot analysis or RT-qPCR after the cells reached a
confluency of 80%. miR-382 inhibitor (100 nM, Thermo Fisher
Scientific, Inc.) was used to decrease the level of miR-382 in PMs
cells. In the rescue experiments, miR-382-overexpression vector and
PGC-1α-overexpression vector were co-transfected into PMs (MOI:50).
In total, at 24 h after transfection, the cells were harvested for
use in subsequent experiments.
Prediction of putative targets
To predict the potential targets of miR-382, the
following online software was applied: Starbase (https://starbase.sysu.edu.cn/), miRanda (http://www.microrna.org/). The predicted binding sites
are shown in Fig. 5C.
Animal welfare and euthanasia
It was ensured that the mice were provided with food
and water and all mice were kept in comfortable and clean cages
(constant temperature of 25°C with 30-40% humidity, 12 h light/dark
cycle, free access to food and water) in appropriate facilities
(SPF) with adequate space (4-5 mice/cage). All mice were in
narcotism by injecting 3% pentobarbital sodium into cavum abdominis
(40 mg/kg) before invasive operation. The following humane
endpoints were used: i) The animal was near death or immobile, or
did not respond to gentle stimuli; ii) dyspnea: Typical symptoms of
salivation and/or cyanosis; iii) diarrhea or urinary incontinence;
iv) 20% of their pre-trial weight loss; v) inability to eat or
drink; vi) the experimental animals should be euthanized if they
are obviously anxious or agitated or the tumor weight exceeds 10%
of the animal's own body weight; the maximum diameter of the
subcutaneous tumor should not exceed 20 mm; vii) paralysis,
persistent epilepsy or rigid behavior, etc.; viii) the area of
animal skin damage accounts for >30% of the whole body area, or
if infection and suppuration is observed; ix) other circumstances
in which a veterinary surgeon determines that a humane endpoint is
required.
For euthanasia, following deep anesthesia by
injecting 3% pentobarbital sodium into the cavum abdominis (40
mg/kg), the mice were placed in a euthanasia chamber filled with
CO2 at a rate of 50% of the replacement volume of the
chamber per minute. It was then confirmed that the mouse was
immobile, not breathing and with dilated pupils. The mice were
observed for a further 2 min to confirm their death.
Animal experiments
The 4T1 cells and PMs from each group were grown to
the logarithmic growth phase, harvested and then counted using a
hemocytometer (101010, Qiujin, Shanghai, Co., Ltd.). The 4T1 cells
and PMs in each group were mixed at a ratio of 1:4 and
subcutaneously injected into the fourth mammary fat pad of each
female BALB/c mouse (5×105 cells/mouse). The animal
experiment lasted for 30 days. A total of 30 female mice were used
in this experiment, the mice were randomly divided into three
groups with 10 mice in each group, and the number of accidental
deaths was 0 during the midway time point. All mice were euthanized
after the experiment as required. The behavior and health of mice
were observed every 1-2 days, and body weight and tumor growth were
assessed every 5 days. After 30 days, lung tissues and tumor
samples were harvested from the mice for use in hematoxylin and
eosin (H&E) staining, and immunohistochemical analysis.
H&E staining and
immunohistochemistry
Tumors derived from 4T1 cells and lung tissues were
removed and fixed in 4% paraformaldehyde (BL539A, Biosharp) for 24
h. Following tissue dewaxing, hematoxylin staining (hematoxylin was
added to the sections, incubated for 4 min at room temperature,
differentiated with 0.5% hydrochloric acid alcohol for 20 min,
re-blued with 0.5% ammonia for 30 sec, stained with eosin dye by
drop for 1 min, at a constant temperature; Beyotime Institute of
Biotechnology Co., Ltd.), dehydration and clearing (washed with 80%
ethanol for 10 sec, 95% ethanol for 10 sec, absolute ethanol for 5
min, absolute ethanol for 10 min, and xylene for twice, 10 min for
each time. Neutral gum was then added in a drop-wise manner to seal
the tablets, at a constant temperature; Beyotime Biotechnology Co.,
Ltd.), neutral gum was added to seal the sections. After air
drying, images were obtained under a microscope (CX33, Olympus
Corporation).
Paraffin-embedded sections of 4T1 tumor tissues were
fixed with 4% paraformaldehyde (BL539A, Biosharp) for 24 h at a
constant temperature. The tumor tissues were then embedded in
paraffin and cut into 4-µm-thick slices and then transferred
to glass slides. After dewaxing and hydrating, the slides were
placed in citrate buffer (pH 6.0) in microwave (high fire) heating
mode for 5 min for antigen extraction. Endogenous peroxidase was
removed by 3% H2O2 after cooling at 25°C (10
min), Sections were processed at 4°C with CD206 (24595, 1:500, Cell
Signaling Technology, Inc.). After processing with secondary
antibodies (HRP-labeled, 1:5,000, A0208, Beyotime Institute of
Biotechnology Co., Ltd.) at 4°C overnight, sections were rinsed
three times using 0.1% Tween-20 TBST for 5 min, stained with 3,3
diaminobenzidine (DAB) for 10 min, and counterstained with
hematoxylin for 1 min in the room temperature. The samples were
placed on glass slides, and cell morphology was observed under
Aperio scanoscope (Leica Biosystems).
Statistical analysis
Data are presented as the mean ± SD. The differences
between paired samples were examined using a paired t-test.
Significant differences between independent samples were determined
using an unpaired Student's t-test. Multiple comparisons between
groups were analyzed using one-way ANOVA followed by Tukey's or
Dunnett's post hoc tests. All statistical analyses were performed
using SPSS for Windows version 24 (IBM Corp.). A value of P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-382 expression is downregulated in
TAMs and M2-polarized macrophages
To investigate the changes in miR-382 expression in
TAMs, TAMs and macrophages were isolated from breast cancer and
paracancerous tissues from 27 patients using density gradient
separation and miR-382 levels were examined using RT-qPCR. It was
found that the relative expression of miR-382 was significantly
downregulated in the TAMs from the majority of the breast cancer
tissues compared to those from control tissues (24/27, Fig. 1A). In the 27 patients, the relative
expression level of miR-382 in the TAMs from breast cancer tissues
was 1.26±0.77, whereas that in the TAMs from paracancerous tissues
was 3.42±2.66. Compared with that in macrophages from adjacent
tissues, the relative expression level of miR-382 in TAMs was
markedly decreased. These results suggest that miR-382 levels are
downregulated in TAMs within the breast cancer TME. To elucidate
whether the decreased expression of miR-382 in TAMs is related to
the stimulation of breast cancer cells, a Transwell co-culture
system was used to simulate the TME. The expression levels of
miR-382 in RAW264.7 cells (co-cultured with 4T1 cells for 48 h) and
THP-1 cells (co-cultured with MDA-MB-231 cells for 48 h) were
detected using RT-qPCR. It was observed that miR-382 expression in
RAW264.7 and THP-1 cells was downregulated to varying degrees
following 48 h of co-culture with breast cancer cells (Fig. 1B and C). It has been previously
demonstrated that TAMs are similar to M2-like macrophages in the
TME (11). The present study thus
induced RAW264.7 macrophages to differentiate into M1-type
macrophages by stimulation with LPS/IFN-γ and into M2-type
macrophages by stimulation with IL-4, and then detected the changes
in miR-382 expression during M1/M2 polarization. It was found that
miR-382 expression was downregulated in M2 macrophages, whereas it
was upregulated in M1-polarized macrophages (Fig. 1D). Since miR-382 is relatively
conserved in vertebrates (this conclusion was reached by searching
the sequence of miR-382 in different species on miRbase; https://www.mirbase.org/; it was found that the
complementary binding sequences presented are conserved in mice and
humans (AAGUUGUU)]. The present study investigated whether the same
trend exists in human macrophage lines. THP-1 cells were
differentiated into adherent macrophages by stimulation with PMA
and then induced to differentiate into M1 and M2 macrophages as
described above. Similar to the previous observation, miR-382
expression was downregulated in M2-polarized THP-1 cells (Fig. 1E). These experimental results
demonstrate that miR-382 expression is downregulated to varying
degrees in both TAMs and M2-polarized macrophages, and this
downregulation may be related to the stimulation of tumor
cells.
miR-382 affects the polarization of TAMs
and the secretion of inflammatory factors
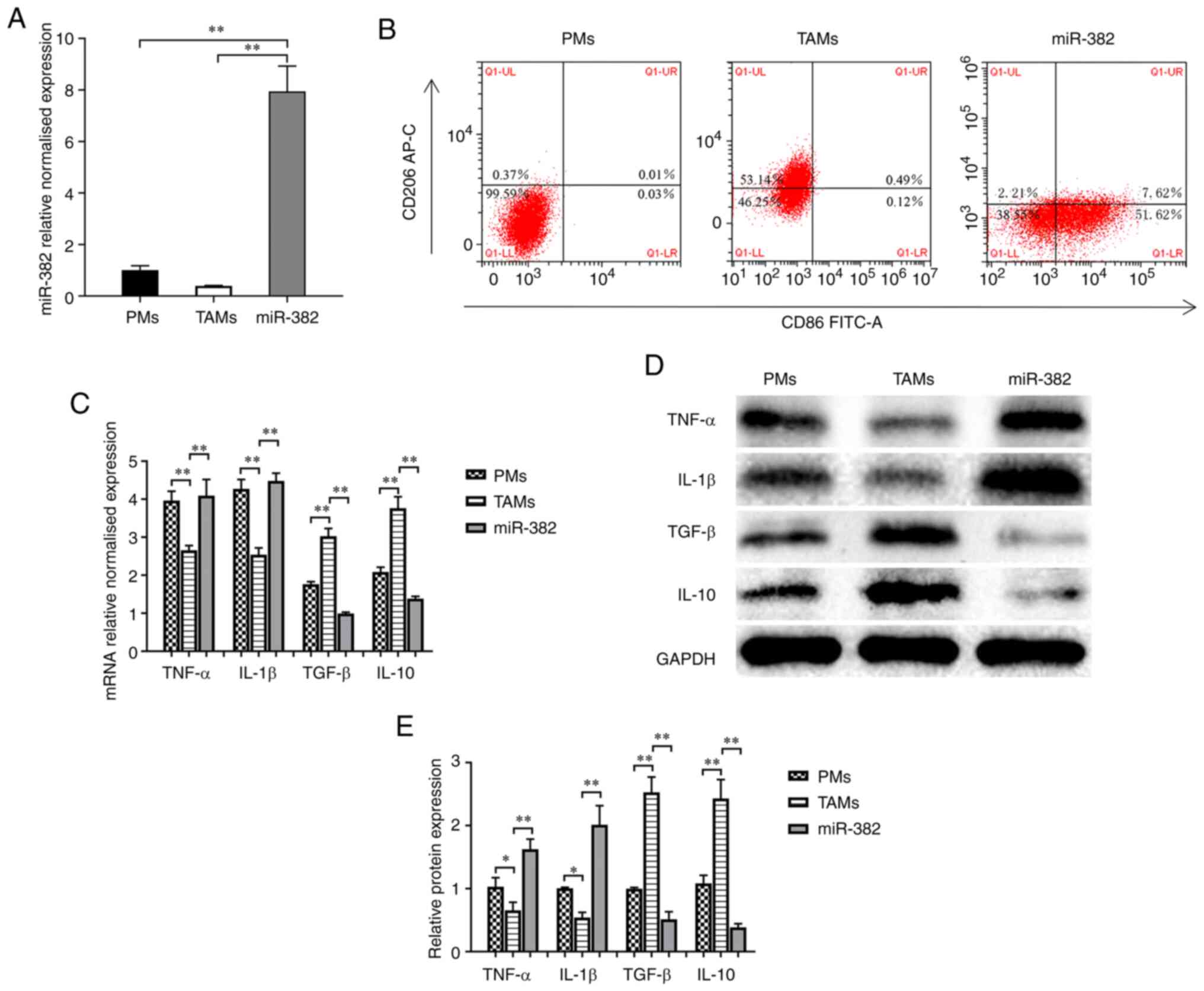
PMs were extracted and co-cultured with 4T1 cells
for 48 h to generate TAMs. The TAMs were transfected with
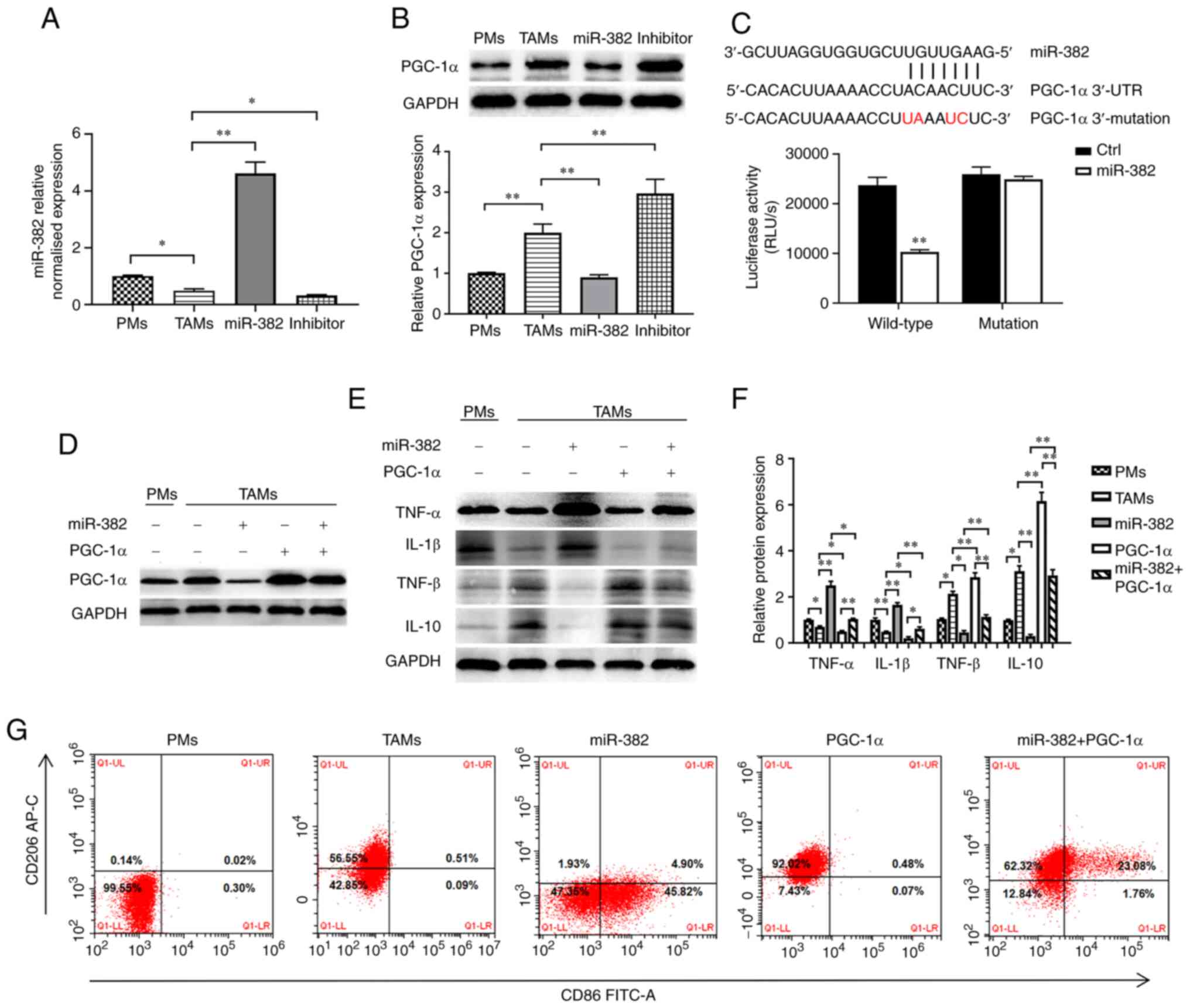
lentivirus overexpressing miR-382 (Fig. 2A) to determine whether miR-382
affects TAM plasticity. The protein levels of an M1-type macrophage
marker (CD86) and M2-type macrophage marker (CD206) were detected
using flow cytometry. The results revealed that compared with PMs
with an M0 phenotype, TAMs expressed decreased levels of CD86 (M1
marker) and increased levels of CD206 (M2 marker) (Fig. 2B). Compared with the TAM group, the
miR-382 group exhibited an increased expression of CD86 and a
decreased expression of CD206, and the TAM phenotype transition
from the M2 to the M1 phenotype following miR-382 overexpression
(Fig. 2B). It has been documented
that TAMs can produce a variety of tumor-related cytokines to
promote the progression of cancer (3-5). In
the present study, to determine whether miR-382 affects the
secretion of related inflammatory factors, the mRNA and protein
levels of M1-type secretory factors (TNF-α and IL-1β) and M2-type
secretory factors (TGF-β and IL-10) were detected using RT-qPCR and
western blot analysis. The results revealed that compared with PMs,
TAMs exhibited increased mRNA and protein levels of M2-type
secretory factors, and decreased levels of M1-type secretory
factors (Fig. 2C-E). Compared with
the control TAMs, TAMs with a high miR-382 expression exhibited an
increased mRNA and protein expression of M1-type secretory factors,
and a decreased expression of M2-type secretory factors (Fig. 2C-E). Thus, these results suggest
that a high miR-382 expression in TAMs affects the polarization
state of TAMs and the secretion of cytokines.
miR-382 can reverse the ability of TAMs
to promote 4T1 cell invasion, migration and EMT
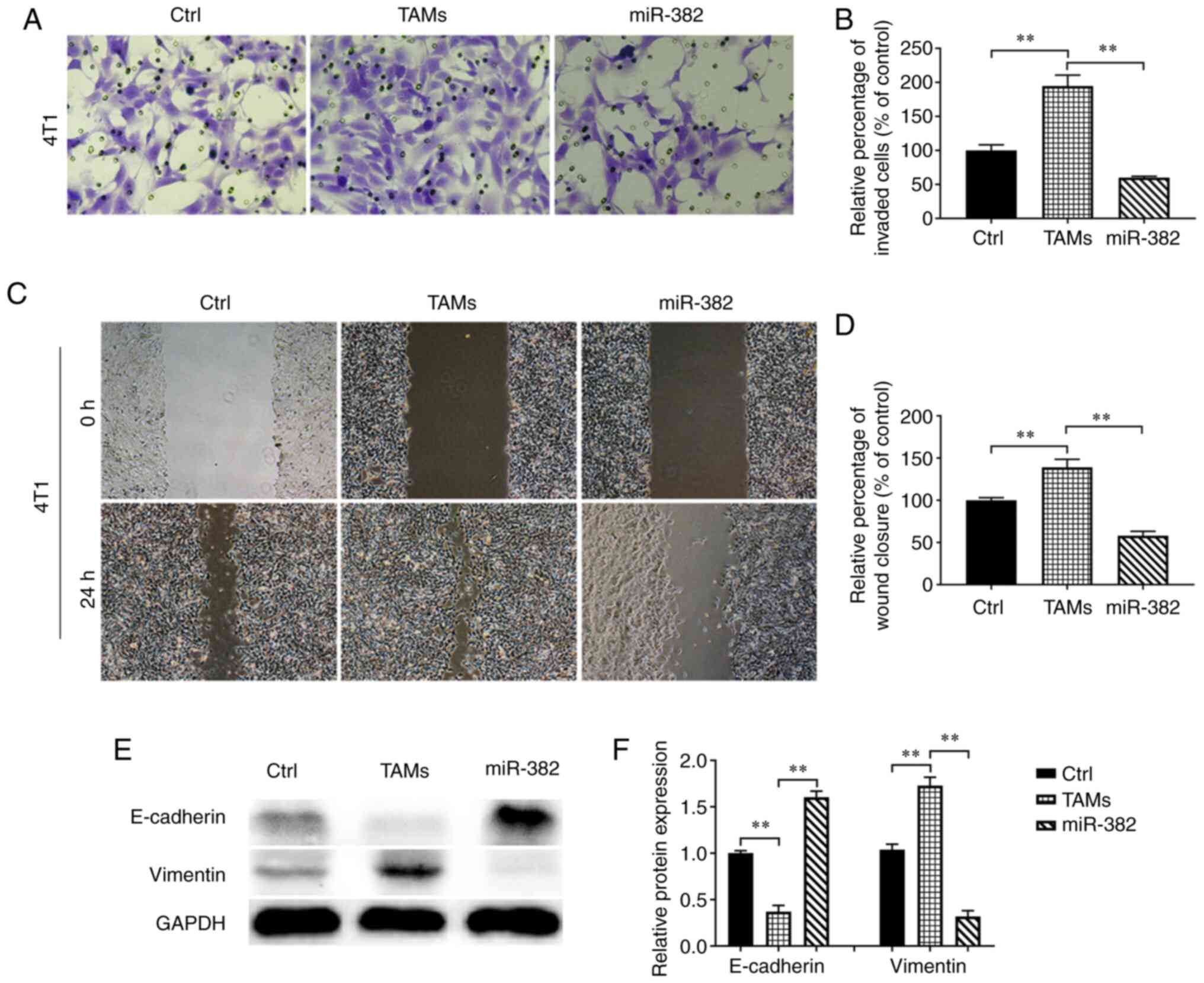
Proliferation, invasion and metastasis are the main
malignant behaviors of tumor cells. However, TAMs in the TME mostly
promote tumor progression (11).
In the present study, after confirming that miR-382 affects
cytokine secretion by TAMs, the effects of miR-382-overexpressing
TAMs on the invasion and migration of 4T1 breast cancer cells were
evaluated. The results revealed that compared with the control 4T1
cells, the 4T1 cells co-cultured with TAMs exhibited increased
invasion and migration abilities; however, these abilities were
reduced by co-culture with TAMs with overexpressing miR-382
(Fig. 3A-D). EMT is considered the
most critical step in promoting the progression of cancer to
metastatic disease (26), and TAMs
can induce EMT in tumor cells to promote cancer metastasis
(27). Researchers have found that
IL-10 and TGF-β produced by M2-type macrophages are involved in
various processes related to tumor progression, including effective
immunosuppression and the EMT of tumor cells (27-30).
In previous studies (31-33), it was found that miR-382
significantly reduced the amount of IL-10 and TGF-β secreted by
TAMs; it was thus hypothesized that miR-382 may interfere with EMT
in breast cancer cells. The expression of EMT markers, including an
epithelial marker (E-cadherin) and a stromal marker (vimentin), in
4T1 cells within each group was detected using western blot
analysis. It was found that compared with the control 4T1 cells,
the 4T1 cells co-cultured with TAMs expressed decreased levels of
E-cadherin and increased levels of vimentin. However, the 4T1 cells
co-cultured with miR-382-overexpressing TAMs expressed increased
levels of E-cadherin and decreased levels of vimentin (Fig. 3E and F). These results suggest that
TAMs promote the invasion, migration and EMT of 4T1 cells, while
miR-382 can reverse these promoting effects of TAMs.
An increased miR-382 expression reduces
the mitochondrial function of TAMs
Studies have noted that changes in energy metabolism
in macrophages can lead to a switch in polarization (34,35).
In the present study, to explore the mechanisms through which
miR-382 affects macrophage polarization, the effects of miR-382 on
energy metabolism in TAMs were investigated by detecting their
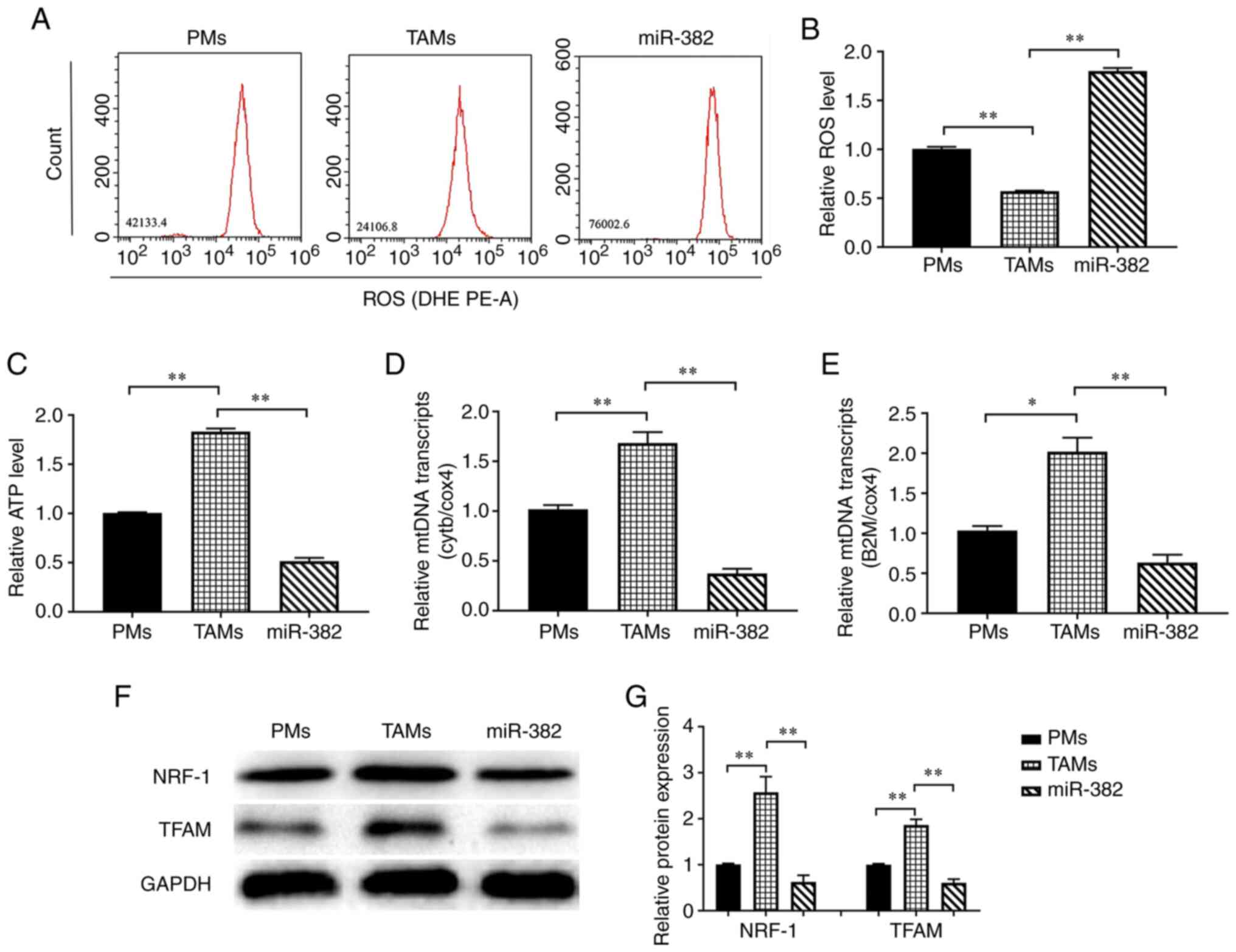
content of ATP and ROS. The results of flow cytometric analysis
revealed that the ROS levels were significantly lower in TAMs than
in PMs, whereas they were significantly higher in TAMs with high a
miR-382 expression (Fig. 4A and
B). The ATP measurements revealed that TAMs had a higher ATP
content than PMs, whereas the ATP levels were lower in TAMs with
overexpressing miR-382 than in control TAMs (Fig. 4C). Subsequently, the changes in the
expression of mtDNA genes and the representative transcripts, Cytb
and B2m, were detected. The results revealed that the transcript
levels of Cytb and B2m were higher in TAMs than in PMs, whereas
they were lower in TAMs overexpressing miR-382 than in control TAMs
(Fig. 4D and E). TFAM is a key
regulator of mitochondrial transcription initiation and mtDNA
replication (36,37), and NRF-1 stimulates the
transcription of nuclear coding genes that regulate mitochondrial
genome transcription and biogenesis (38). Therefore, the present study
examined the effects of miR-382 on the expression of the
mitochondrial function-related proteins, NRF-1 and TFAM. It was
found that the NRF-1 and TFAM levels were higher in TAMs than in
PMs, and were lower in miR-382-overexpressing TAMs (Fig. 4F and G). These results suggest that
TAMs have greater mitochondrial function than PMs, and the high
expression of miR-382 in TAMs may lead to reduced mitochondrial
biosynthesis or impaired mitochondrial function, resulting in a
reduced ATP production.
PGC-1α is the downstream target of
miR-382 and partially reverses the inhibitory effects of miR-382 on
the M2 polarization of TAMs
The present study identified PGC-1α as a potential
target gene of miR-382 through an online prediction tool
(https://starbase.sysu.edu.cn/ and
http://www.microrna.org). PGC-1α is a
transcriptional coactivator that promotes mitochondrial
biosynthesis and enhances respiratory capacity and oxidative
phosphorylation (OXPHOS) (39).
First, the present study used RT-qPCR and western blot analysis to
investigate the association between the expression levels of
miR-382 and PGC-1α in TAMs following transfection with
miR-382-overexpressing lentivirus or miR-382 inhibitor. It was
found that PGC-1α expression was higher in TAMs than in PMs and
that PGC-1α expression was negatively associated with miR-382
expression (Fig. 5A and B). In
addition, to confirm whether PGC-1α is regulated by the direct
binding of miR-382 to its 3'UTR, a dual luciferase assay we
performed. The recombinant plasmid was co-transfected with miR-382
mimics into 293T cells, and luciferase activity was detected. These
co-transfection experiments revealed that miR-382 significantly
reduced luciferase activity in cells expressing luc-PGC-1α-3'UTR,
whereas it had a minimal effect in those expressing the mutant
sequence (Fig. 5C). Although it
was demonstrated that PGC-1α is the target gene of miR-382, these
results do not prove that the changes induced by miR-382 in TAMs
are achieved by targeting PGC-1α. To further ascertain whether
miR-382 regulates TAM polarization by targeting PGC-1α, a
lentiviral vector (PGC-1α) was constructed, PGC-1α was
differentially expressed in miR-382-overexpressing TAMs (Fig. 5D), and the polarization of TAMs was
detected. The experimental results revealed that TAMs transfected
with miR-382-overexpression lentivirus exhibited a decreased
secretion of M1 cytokines and an increased secretion of M2
cytokines. Moreover, the results of flow cytometric analysis
revealed the decreased expression of CD86 and the increased
expression of CD206 (Fig. 5E-G).
However, in TAMs overexpressing both miR-382 and PGC-1α, the high
expression of M1-type cytokines was blocked, the release of M2-type
cytokines was increased, and the increased CD86 expression and
decreased CD206 expression induced by the overexpression of miR-382
were partially reversed (Fig.
5E-G). On the whole, these data suggest that PGC-1α is the
downstream target of miR-382 and that the ectopic expression of
PGC-1α in TAMs can reverse the inhibitory effects of miR-382 on the
M2 polarization of TAMs.
PGC-1α reverses the changes in TAM
mitochondrial function induced by miR-382 overexpression
The afore-mentioned experimental results suggested
that miR-382 may affect TAM polarization by altering mitochondrial
function. Therefore, the role of PGC-1α in this process was then
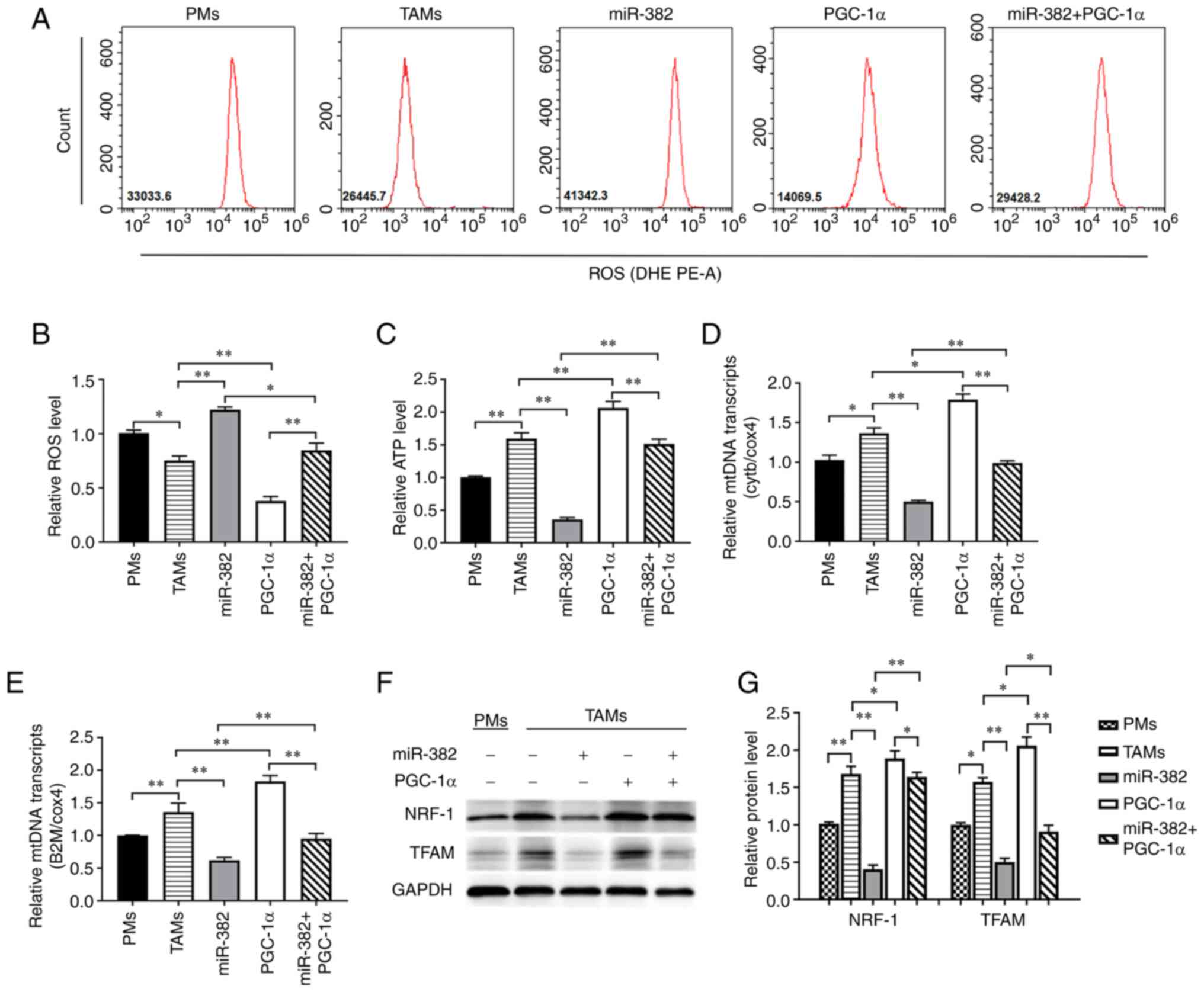
investigated. The results revealed that a high PGC-1α expression
decreased intracellular ROS levels and increased ATP levels.
However, Compared with the miR-382 overexpression group, TAMs with
overexpressing both miR-382 and PGC-1α, the ROS levels were
inhibited to a certain extent, while the ATP levels were relatively
restored (Fig. 6A-C). Through
RT-qPCR analysis, it was found that PGC-1α increased the mtDNA
transcript levels, represented by Cytb and B2m, and partially
reversed the downregulation induced by the overexpression of
miR-382 (Fig. 6D and E).
Similarly, PGC-1α increased the expression of the mitochondrial
function-related proteins, NRF-1 and TFAM, and restored the changes
induced by miR-382 overexpression to a certain extent (Fig. 6F and G). These results indicate
that PGC-1α can reverse the changes in TAM mitochondrial function
induced by miR-382 overexpression.
PGC-1α restores the ability of TAMs to
promote the biological properties of breast cancer cells
After demonstrating that PGC-1α is the target of
miR-382, the role of PGC-1α in the tumorigenic properties of TAMs
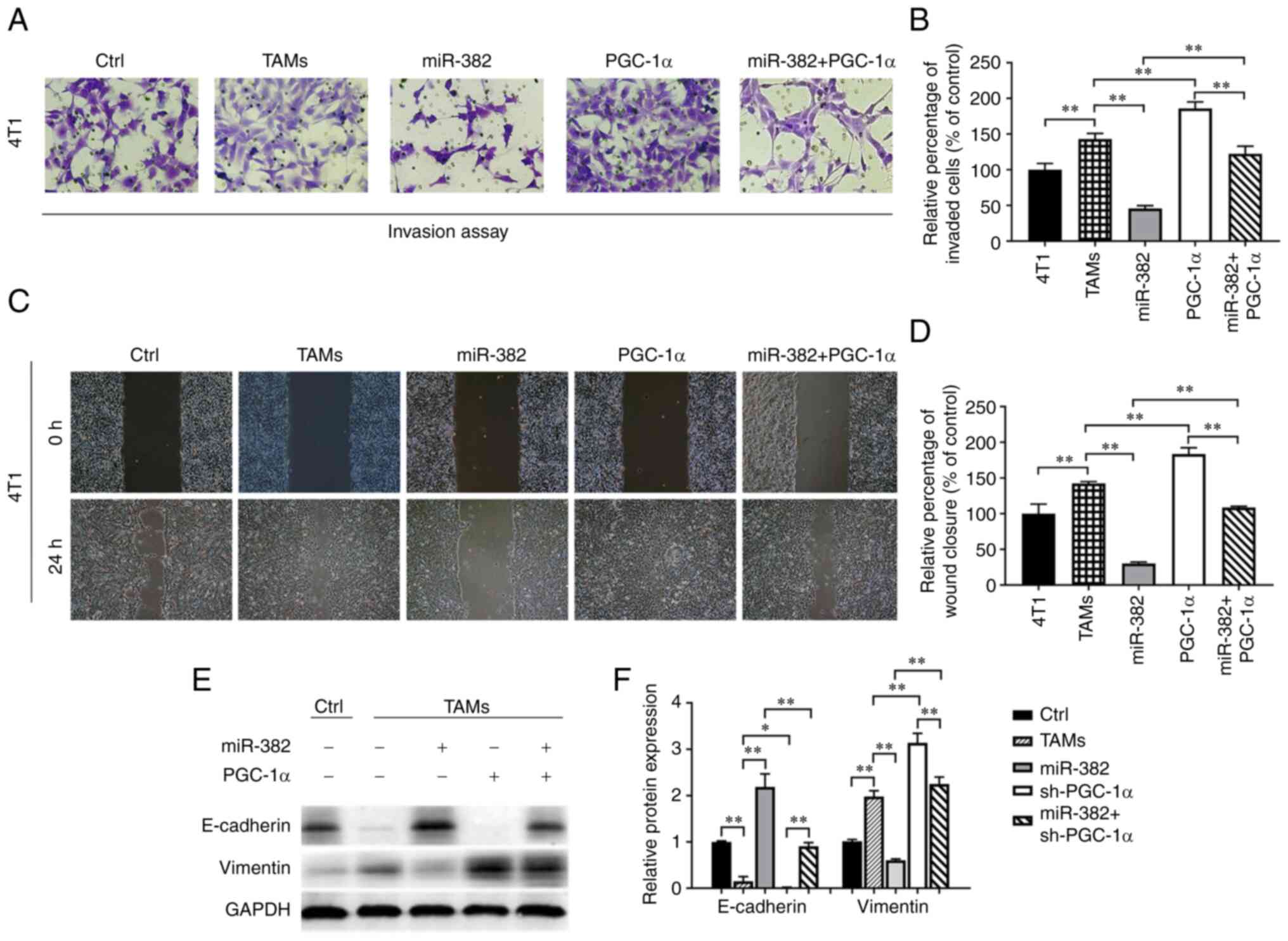
was investigated. The experimental results revealed that the
overexpression of PGC-1α enhanced the ability of TAMs to promote
the invasion and migration of breast cancer cells, while the
overexpression of both miR-382 and PGC-1α restored these abilities
of TAMs to their baseline levels (Fig.
7A-D). Furthermore, changes in the expression levels of EMT
markers in 4T1 cells were detected using western blot analysis. The
results revealed that TAMs overexpressing PGC-1α induced a decrease
in E-cadherin expression and an increase in vimentin expression in
4T1 cells; additionally, the overexpression of PGC-1α partially
reversed the ability of TAMs also overexpressing miR-382 to inhibit
EMT (Fig. 7E and F). These results
suggest that PGC-1α can restore the ability of TAMs to promote the
biological properties of breast cancer cells.
miR-382 inhibits the metastasis of 4T1
breast cancer cells by reducing the polarization of M2 macrophages
in vivo
After identifying the role of miR-382 in regulating
the M1/M2 polarization of TAMs in vitro, its role in
vivo was investigated, particularly in inhibiting the invasion
and migration of breast cancer cells. Murine macrophages were
transfected with miR-382-overexpressing lentivirus and mixed with
4T1 cells at a 4:1 ratio. This cell mixture was subcutaneously
inoculated into BALB/c mice, and changes in breast tumor growth
were monitored. After 30 days, tumor tissues and lung tissues were
resected from the mice, and the formation of metastatic foci and
the number of CD206-positive cells were detected by H&E
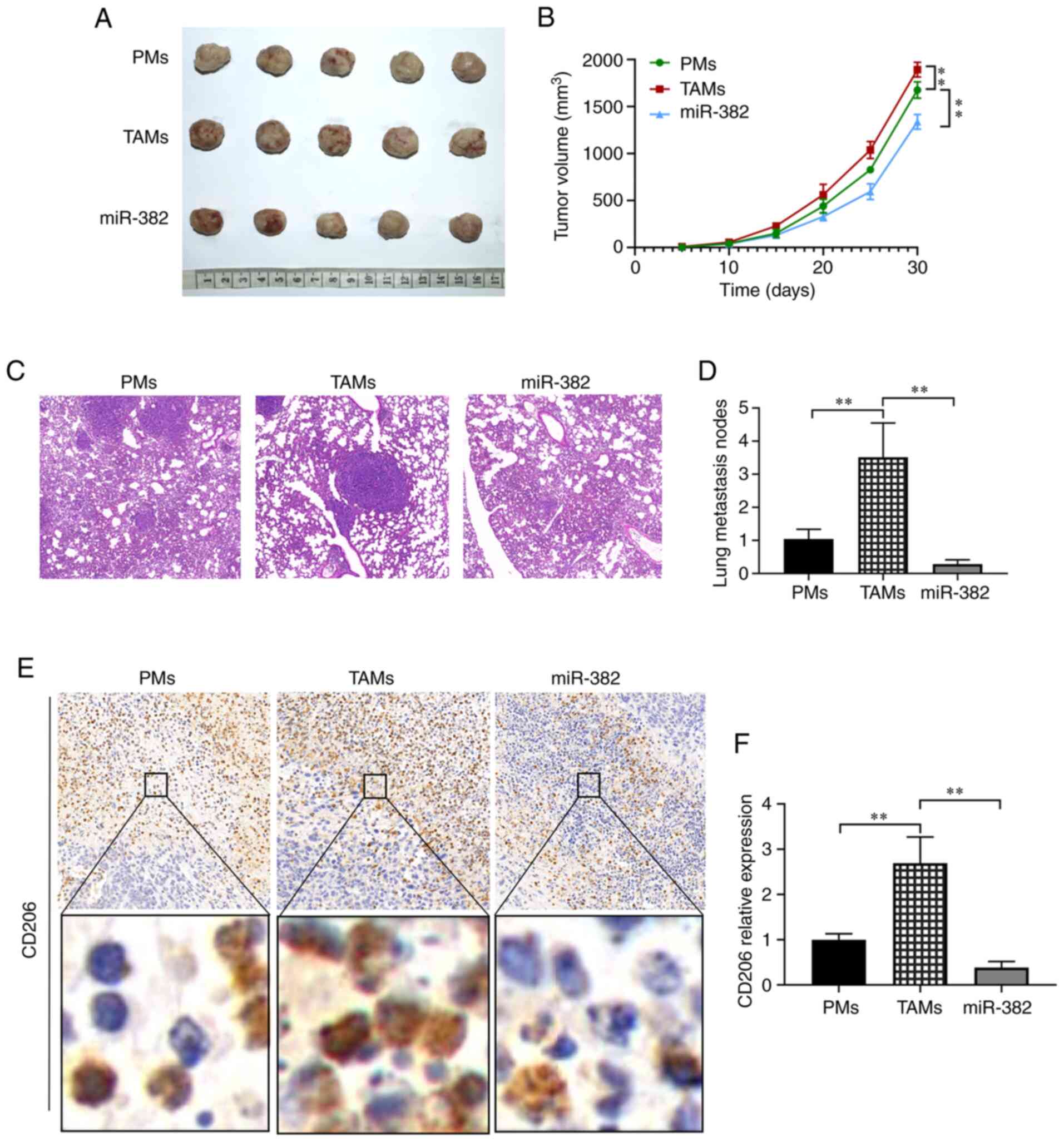
staining and immunohistochemistry. The experimental results
revealed that compared with the control group, the TAM group
exhibited larger tumors and a more rapid growth rate, while the
parameters of tumor size and growth rate were inhibited in the
miR-382 overexpression group (Fig. 8A
and B). To further explore whether miR-382 can inhibit the
metastasis of breast cancer in vivo, H&E staining of the
mouse lung tissues was conducted. The results revealed that the
number of lung metastases was 1.0±0.29 in the control group,
3.51±1.0 in the TAM group and 0.29±0.13 in the miR-382 group, which
represented a significant decrease (Fig. 8C and D). To determine whether
miR-382 inhibits the growth and metastasis of 4T1 breast tumors by
inhibiting M2-type macrophages, the expression levels of CD206 in
tumor tissues were examined by immunohistochemistry. The data
revealed that the relative expression level of CD206 was 1.0±0.13
in the control group, 2.69±0.58 in the TAM group and 0.38±0.13 in
the miR-382 group, representing a significant decrease (Fig. 8E and F). The results obtained in
vivo were consistent with the afore-mentioned results obtained
in vitro. On the whole, these data suggest that miR-382 can
also inhibit the M2 polarization of TAMs in vivo, thereby
preventing 4T1 tumor cell growth and metastasis.
Discussion
Breast cancer is a highly heterogeneous malignancy
that is common among females. According to the latest survey data
(2020) of the International Agency for Research on Cancer (IARC),
breast cancer accounted for 30% of female cancer cases worldwide,
ranking first among female malignancies, and the incidence of
breast cancer is gradually increasing (40). Malignant breast tumors, which are
highly aggressive, often metastasize to distant organs, such as the
lungs and brain, and metastasis is the main cause of mortality
among patients with breast cancer. A number of studies have
confirmed that the transformation of macrophages from the M1
phenotype to the M2 phenotype is a key event in tumor progression,
and blocking this process is considered a promising therapeutic
strategy for inhibiting tumor progression (12-14).
Recently, several strategies targeting M2-differentiated
macrophages have been tested in clinical settings, such as blocking
TAM activation (41), inhibiting
TAM differentiation (42) and
reprogramming TAMs to differentiate into M1-type macrophages
(43). However, the molecular
mechanisms underlying TAM transformation in the complex TME are not
yet fully understood.
miRNAs are principal regulators of gene expression
that function through the post-transcriptional regulation of target
genes in several immune cells, including macrophages (44,45).
A variety of miRNAs have been confirmed to be involved in
macrophage polarization. For instance, miR-19a-3p induces
macrophage polarization by downregulating Fra-1 proto-oncogene
expression (46). Let-7c promotes
the M2 polarization of TAMs by targeting CCAAT-enhancer-binding
protein δ (47). miR-146a
participates in the regulation of macrophage polarization by
inhibiting the Notch1 signaling pathway (48). Previous studies have confirmed that
miR-382 plays a tumor suppressive role in a variety of cancer
types, including breast cancer (22-24),
and related studies have indicated that miR-382 may play critical
roles in regulating tissue inflammation and T-cell differentiation
(49,50). Among the molecules and signaling
pathways reportedly targeted by miR-382, the TLR4/MyD88/NF-κB
(49) and Akt/mTOR axes (51) play key roles in the polarization of
TAMs. Nevertheless, whether miR-382 is involved in the regulation
of TAM plasticity has not yet been reported, at least to the best
of our knowledge. Previous studies by the authors found that
miR-382 expression was significantly reduced in TAMs, which affects
the polarization state of macrophages (31-33,52).
The present study further explored the mechanisms of action and
possible targets.
Metabolic reprogramming is a principal feature of
malignant tumor, immune and mesenchymal cells in the TME, and is
also the material basis of macrophage activation (53). TAMs exhibit metabolic
heterogeneity, and their unique metabolic reprogramming process
promotes the invasion and metastasis of tumor cells. The majority
of TAMs in the TME are of the M2 phenotype, and their metabolic
characteristics include enhanced mitochondrial OXPHOS and fatty
acid β-oxidation (54), as well as
the increased expression of anti-inflammatory cytokines, such as
IL-10, and the decreased expression of pro-inflammatory molecules,
such as iNOS and TNF-α. However, the M2 phenotype is powered by
energy, and the levels of Arg-1, adenosine monophosphate activated
protein kinase and
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 are increased
(34,55). Thus, metabolic alterations are not
only characteristic of TAM subsets, but also a prerequisite for
proper polarization and the regulation of inflammation. The present
study investigated whether mitochondrial biogenesis within TAMs is
involved in miR-382-mediated macrophage polarization. Of note, it
was found that the overexpression of miR-382 reduced the ATP
content, mtDNA transcript levels, and TFAM and NRF-1 (related to
mitochondrial biogenesis) protein levels in TAMs. These results
indicate that miR-382 overexpression reduces the mitochondrial
function of TAMs and may lead to changes in macrophage polarization
by altering the patterns of energy metabolism.
PGC-1α is a transcriptional coactivator that
promotes mitochondrial biosynthesis and enhances respiratory
capacity (39). The level of
OXPHOS is closely related to the number and function of
mitochondria, which are tightly regulated by PGC-1α. High levels of
PGC-1α in melanoma increase the ability of mitochondria to fight
oxidative stress (56), and PGC-1α
promotes the distant metastasis of cancer cells by enhancing
mitochondrial function in invasive breast cancer (57). Through prediction and analysis, the
present study found that PGC-1α is a potential target of miR-382.
Furthermore, a dual luciferase assay confirmed that PGC-1α was
directly bound by miR-382. Therefore, it was hypothesized that the
changes in TAM polarization and metabolism induced by miR-382
overexpression may be achieved by targeting PGC-1α. In verifying
this hypothesis, it was found that the ectopic expression of PGC-1α
in TAMs overexpressing miR-382 partially reversed the changes in
TAM polarization and restored the expression of the M2 cytokines,
TGF-β and IL-10. In addition, the overexpression of PGC-1α
partially restored ATP production and mtDNA transcript levels, as
well as the expression of the mitochondrial biogenesis-related
proteins, TFAM and NRF-1. More importantly, the overexpression of
PGC-1α restored the ability of TAMs to promote the invasion and
migration of breast cancer cells. Taken together, these results
suggest that miR-382 suppresses the invasion and metastasis of 4T1
cells by reversing M2 polarization and inhibiting the mitochondrial
function of TAMs, and that these effects are achieved by targeting
PGC-1α. Notably, PGC-1α did not completely reverse the
miR-382-induced changes in TAMs, suggesting that miR-382 may also
function through other mechanisms or pathways. In addition, PGC-1α
is a co-activator of peroxisome proliferator-activated receptor γ
(PPARγ), an important mediator of TAM polarization (53). Studies have demonstrated that PPARγ
plays a crucial role in the the maturation of alternately activated
macrophages, and its activation enables human monocytes to
differentiate into replacement M2 macrophages (53,58).
These results suggest that the regulation of TAMs by PGC-1α may
also be achieved through the PPARγ signaling pathway. These
hypotheses warrant further investigation and verification in future
studies.
There is substantial evidence to indicate that the
degree of TAM infiltration into tumors is related to metastatic
potential. In fact, by reshaping the TME and relaxing the
extracellular matrix of tumor cells, TAMs allow tumor cells to
separate from the tumor mass and spread, leading to distant
metastasis (12-14). In addition, cytokines (such as
IL-10 and TGF-β) secreted by TAMs promote the detachment of tumor
cells from the primary site by affecting EMT, which is considered
one of the key steps in distant metastasis (27-30).
In the present study, it was found that miR-382 inhibited TGF-β and
IL-10 secretion by TAMs, and increased the expression of an
epithelial marker (E-cadherin) and decreased the expression of an
interstitial marker (vimentin) in breast cancer cells. Moreover,
the invasive and migratory abilities of 4T1 breast cancer cells
were inhibited to varying degrees. Of note, in the BALB/c mouse
model, it was found that TAMs overexpressing miR-382 inhibited the
growth of 4T1 breast cancer cells and the formation of lung
metastases. Furthermore, the results of immunohistochemical
analysis revealed that the number of CD206-positive cells among
TAMs overexpressing miR-382 was significantly lower than that among
the control TAMs. These results are consistent with those obtained
from the in vitro experiments. Previous research has
revealed that other target molecules of miR-382 (such as CXCL12)
may also affect tumor metastasis by affecting macrophage
recruitment to tumor sites (59).
However, the present study, no significant difference was observed
in the degree of TAM infiltration. In general, the in vivo
experiments confirmed that the inhibitory effects of miR-382 on 4T1
breast cancer cell metastasis were achieved by regulating
macrophage plasticity.
In conclusion, the present study revealed an
association between miR-382 expression in TAMs and breast cancer
metastasis, and a novel mechanism through which miR-382 may
regulate TAM polarization by altering the metabolic state through
targeting PGC-1α. Tumor cells induced a decrease in miR-382
expression in TAMs, which relieved the inhibition of downstream
PGC-1α; PGC-1α then further induced the M2 polarization of TAMs by
altering their metabolic state. Finally, the TGF-β and IL-10
secreted by M2-type TAMs promoted EMT and the distant metastasis of
breast cancer cells. These results may be valuable for the
development of effective prevention and treatment strategies for
metastatic breast cancer from the perspective of the metabolic
reprogramming of the TME.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JM and HZ designed the outline of the study. All
authors (HZ, MG, XJ, MD, YW, YL, ZL and JM) conducted the
experiments and performed the data analysis. HZ and MG were
involved in the collation of the experimental data. HZ and XJ wrote
the manuscript. MG, MD and YW supervised the study and manuscript
revision. YL, ZL and JM confirm the authenticity of all the raw
data. All authors have read and agreed to the published version of
the manuscript.
Ethics approval and consent to
participate
The research involving human tissues and animals was
approved by The Ethics Committee of the Second Affiliated Hospital
of Chongqing Medical University (approval no. 99/2022).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the 'Kuanren
meritocrat' Backbone Talents Project of The Second Affiliated
Hospital of Chongqing Medical University (no. KY2019G016), the
Project of Science and Technology Innovation for Social and
Livelihood of Chongqing Science and Technology Commission (no.
CSTC2018jscx-msyhu0020) and the Project of Science and Technology
Plan of Yuzhong District, Chongqing (no. 20160104).
Abbreviations:
|
TAMs
|
tumor-associated macrophages
|
|
TME
|
tumor microenvironment
|
|
TLR
|
Toll-like receptor
|
|
LPS
|
lipopolysaccharide
|
|
INF-γ
|
interferon-γ
|
|
IL
|
interleukin
|
|
Th2 cell
|
T-helper 2 cell
|
|
miRNA/miR
|
microRNA
|
|
UTRs
|
untranslated regions
|
|
EMT
|
epithelial-mesenchymal transition
|
|
PMs
|
primary mouse macrophages
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
TNF-α
|
tumor necrosis factor-α
|
|
TGF-β
|
transforming growth factor-β
|
|
ROS
|
reactive oxygen species
|
|
TFAM
|
mitochondrial transcription factor
A
|
|
NRF-1
|
nuclear respiratory factor-1
|
|
PGC-1α
|
peroxisome proliferator-activated
receptor γ coactivator-1α
|
|
OXPHOS
|
oxidative phosphorylation
|
|
CD
|
cluster of differentiation
|
|
PPARγ
|
peroxisome proliferator-activated
receptor γ
|
|
iNOS
|
inducible nitric oxide synthase
|
|
mtDNA
|
mitochondrial DNA
|
|
Cytb
|
cytochrome b
|
|
B2m
|
beta-2 microglobulin
|
|
Cox4
|
cytochrome c oxidase subunit
4
|
References
|
1
|
Martin JD, Fukumura D, Duda DG, Boucher Y
and Jain RK: Reengineering the tumor microenvironment to alleviate
hypoxia and overcome cancer heterogeneity. Cold Spring Harb
Perspect Med. 6:a0270942016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang Q, Liang X, Ren T, Huang Y, Zhang H,
Yu Y, Chen C, Wang W, Niu J, Lou J and Guo W: The role of
tumor-associated macrophages in osteosarcoma
progression-therapeutic implications. Cell Oncol (Dordr).
44:525–539. 2021. View Article : Google Scholar
|
|
3
|
Lao L, Fan S and Song E: Tumor associated
macrophages as therapeutic targets for breast cancer. Adv Exp Med
Biol. 1026:331–370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu Z, Zhang H, Chen B, Liu X, Zhang S,
Zong Z and Gao M: PD-L1-mediated immunosuppression in glioblastoma
is associated with the infiltration and M2-polarization of
tumor-associated macrophages. Front Immunol. 11:5885522020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cersosimo F, Lonardi S, Bernardini G,
Telfer B, Mandelli GE, Santucci A, Vermi W and Giurisato E:
Tumor-associated macrophages in osteosarcoma: From mechanisms to
therapy. Int J Mol Sci. 21:52072020. View Article : Google Scholar :
|
|
6
|
Macciò A, Gramignano G, Cherchi MC, Tanca
L, Melis L and Madeddu C: Role of M1-polarized tumor-associated
macrophages in the prognosis of advanced ovarian cancer patients.
Sci Rep. 10:60962020PubMed/NCBI
|
|
7
|
Wang X, Li X and Wang X: Identification of
immune microenvironment subtypes that predicted the prognosis of
patients with ovarian cancer = Identification of immune
microenvironment subtypes that predicted the prognosis of patients
with ovarian cancer. J Cell Mol Med. 25:4053–4061. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Honkanen TJ, Tikkanen A, Karihtala P,
Mäkinen M, Väyrynen JP and Koivunen JP: Prognostic and predictive
role of tumour-associated macrophages in HER2 positive breast
cancer. Sci Rep. 9:109612019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mills CD, Kincaid K, Alt JM, Heilman MJ
and Hill AM: M-1/M-2 macrophages and the Th1/Th2 paradigm. J
Immunol. 164:6166–6173. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gordon S and Martinez FO: Alternative
activation of macrophages: Mechanism and functions. Immunity.
32:593–604. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lewis CE and Pollard JW: Distinct role of
macrophages in different tumor microenvironments. Cancer Res.
66:605–612. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Komohara Y, Fujiwara Y, Ohnishi K and
Takeya M: Tumor-associated macrophages: Potential therapeutic
targets for anti-cancer therapy. Adv Drug Deliv Rev. 99:180–185.
2016. View Article : Google Scholar
|
|
13
|
Mantovani A, Marchesi F, Malesci A, Laghi
L and Allavena P: Tumour-associated macrophages as treatment
targets in oncology. Nat Rev Clin Oncol. 14:399–416. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shu Y and Cheng P: Targeting
tumor-associated macrophages for cancer immunotherapy. Biochim
Biophys Acta Rev Cancer. 1874:1884342020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Davis BN and Hata A: Regulation of
MicroRNA biogenesis: A miRiad of mechanisms. Cell Commun Signal.
7:182009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fathullahzadeh S, Mirzaei H, Honardoost
MA, Sahebkar A and Salehi M: Circulating microRNA-192 as a
diagnostic biomarker in human chronic lymphocytic leukemia. Cancer
Gene Ther. 23:327–332. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Keshavarzi M, Sorayayi S, Jafar Rezaei M,
Mohammadi M, Ghaderi A, Rostamzadeh A, Masoudifar A and Mirzaei H:
MicroRNAs-based imaging techniques in cancer diagnosis and therapy.
J Cell Biochem. 118:4121–4128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Khalife H, Skafi N, Fayyad-Kazan M and
Badran B: MicroRNAs in breast cancer: New maestros defining the
melody. Cancer Genet. 246-247:18–40. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu G and Abraham E: MicroRNAs in immune
response and macrophage polarization. Arterioscler Thromb Vasc
Biol. 33:170–177. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Essandoh K, Li Y, Huo J and Fan GC:
MiRNA-mediated macrophage polarization and its potential role in
the regulation of inflammatory response. Shock. 46:122–131. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Graff JW, Dickson AM, Clay G, McCaffrey AP
and Wilson ME: Identifying functional microRNAs in macrophages with
polarized phenotypes. J Biol Chem. 287:21816–21825. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang S, Ge W, Zou G, Yu L, Zhu Y, Li Q,
Zhang Y, Wang Z and Xu T: MiR-382 targets GOLM1 to inhibit
metastasis of hepatocellular carcinoma and its down-regulation
predicts a poor survival. Am J Cancer Res. 8:120–131.
2018.PubMed/NCBI
|
|
23
|
Xu M, Jin H, Xu CX, Sun B, Song ZG, Bi WZ
and Wang Y: miR-382 inhibits osteosarcoma metastasis and relapse by
targeting Y box-binding protein 1. Mol Ther. 23:89–98. 2015.
View Article : Google Scholar :
|
|
24
|
Yao H, Xia D, Li ZL, Ren L, Wang MM, Chen
WS, Hu ZC, Yi GP and Xu L: MiR-382 functions as tumor suppressor
and chemosensitizer in colorectal cancer. Biosci Rep.
39:BSR201804412019. View Article : Google Scholar :
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Taki M, Abiko K, Ukita M, Murakami R,
Yamanoi K, Yamaguchi K, Hamanishi J, Baba T, Matsumura N and Mandai
M: Tumor immune microenvironment during epithelial-mesenchymal
transition. Clin Cancer Res. 27:4669–4679. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye XZ, Xu SL, Xin YH, Yu SC, Ping YF, Chen
L, Xiao HL, Wang B, Yi L, Wang QL, et al: Tumor-associated
microglia/macrophages enhance the invasion of glioma stem-like
cells via TGF-β1 signaling pathway. J Immunol. 189:444–453. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chaudhury A, Hussey GS, Ray PS, Jin G, Fox
PL and Howe PH: TGF-beta-mediated phosphorylation of hnRNP E1
induces EMT via transcript-selective translational induction of
Dab2 and ILEI. Nat Cell Biol. 12:286–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cai J, Xia L, Li J, Ni S, Song H and Wu X:
Tumor-associated macrophages derived TGF-β-induced epithelial to
mesenchymal transition in colorectal cancer cells through
Smad2,3-4/Snail signaling pathway. Cancer Res Treat. 51:252–266.
2019. View Article : Google Scholar
|
|
30
|
Liu CY, Xu JY, Shi XY, Huang W, Ruan TY,
Xie P and Ding JL: M2-polarized tumor-associated macrophages
promoted epithelial-mesenchymal transition in pancreatic cancer
cells, partially through TLR4/IL-10 signaling pathway. Lab Invest.
93:844–854. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lei Y, Zhou H, Dai M, Jin X and Ming J:
Effects of Mir-382 on biological characteristics of triple negative
breast cancer cell line 4T1 through PGC-1α. J Third Mil Med Univ.
41:1415–1422. 2019.
|
|
32
|
Jin X, Ni TG, Wang N, Luo H, Lei Y and
Ming J: Mechanism of mitochondria-mediated PGC-1α in macrophage
polarization. J Third Mil Med Univ. 41:56–62. 2019.
|
|
33
|
Dai M, Jin X, Lei YY and Ming: Effect of
miR-382 overexpressing tumor-associated macrophages on biological
properties of triple-negative breast cancer 4T1 cells. J Third Mil
Med Univ. 40:1375–1382. 2018.
|
|
34
|
Hobson-Gutierrez SA and Carmona-Fontaine
C: The metabolic axis of macrophage and immune cell polarization.
Dis Model Mech. 11:dmm0344622018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Krawczyk CM, Holowka T, Sun J, Blagih J,
Amiel E, DeBerardinis RJ, Cross JR, Jung E, Thompson CB, Jones RG
and Pearce EJ: Toll-like receptor-induced changes in glycolytic
metabolism regulate dendritic cell activation. Blood.
115:4742–4749. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao M, Wang Y, Li L, Liu S, Wang C, Yuan
Y, Yang G, Chen Y, Cheng J, Lu Y and Liu J: Mitochondrial ROS
promote mitochondrial dysfunction and inflammation in ischemic
acute kidney injury by disrupting TFAM-mediated mtDNA maintenance.
Theranostics. 11:1845–1863. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Campbell CT, Kolesar JE and Kaufman BA:
Mitochondrial transcription factor A regulates mitochondrial
transcription initiation, DNA packaging, and genome copy number.
Biochim Biophys Acta. 1819:921–929. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Klinge CM: Estrogenic control of
mitochondrial function. Redox Biol. 31:1014352020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cruz-Bermúdez A, Laza-Briviesca R,
Vicente-Blanco RJ, García-Grande A, Coronado MJ, Laine-Menéndez S,
Palacios-Zambrano S, Moreno-Villa MR, Ruiz-Valdepeñas AM, Lendinez
C, et al: Cisplatin resistance involves a metabolic reprogramming
through ROS and PGC-1α in NSCLC which can be overcome by OXPHOS
inhibition. Free Radic Biol Med. 135:167–181. 2019. View Article : Google Scholar
|
|
40
|
Ferlay J, Colombet M, Soerjomataram I,
Parkin DM, Piñeros M, Znaor A and Bray F: Cancer statistics for the
year 2020: An overview. Int J Cancer. Apr 5–2021.Epub ahead of
print. View Article : Google Scholar
|
|
41
|
Butowski N, Colman H and De Groot JF:
Orally administered colony stimulating factor 1 receptor inhibitor
PLX3397 in recurrent glioblastoma: An Ivy Foundation Early Phase
Clinical Trials Consortium phase II study. Neuro Oncol. 18:557–564.
2016. View Article : Google Scholar :
|
|
42
|
Edelman MJ, Wang X, Hodgson L, Cheney RT,
Baggstrom MQ, Thomas SP, Gajra A, Bertino E, Reckamp KL, Molina J,
et al: Phase III randomized, placebo-controlled, double-blind trial
of celecoxib in addition to standard chemotherapy for advanced
non-small-cell lung cancer with cyclooxygenase-2 overexpression:
CALGB 30801 (Alliance). J Clin Oncol. 35:2184–2192. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Reid T, Oronsky B, Scicinski J, Scribner
CL, Knox SJ, Ning S, Peehl DM, Korn R, Stirn M, Carter CA, et al:
Safety and activity of RRx-001 in patients with advanced cancer: A
first-in-human, open-label, dose-escalation phase 1 study. Lancet
Oncol. 16:1133–1142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chatterjee B, Saha P, Bose S, Shukla D,
Chatterjee N, Kumar S, Tripathi PP and Srivastava AK: MicroRNAs: As
critical regulators of tumor-associated macrophages. Int J Mol Sci.
21:71172020. View Article : Google Scholar
|
|
45
|
Gholamin S, Mirzaei H, Razavi SM,
Hassanian SM, Saadatpour L, Masoudifar A, ShahidSales S and Avan A:
GD2-targeted immunotherapy and potential value of circulating
microRNAs in neuroblastoma. J Cell Physiol. 233:866–879. 2018.
View Article : Google Scholar
|
|
46
|
Yang J, Zhang Z, Chen C, Liu Y, Si Q,
Chuang TH, Li N, Gomez-Cabrero A, Reisfeld RA, Xiang R and Luo Y:
MicroRNA-19a-3p inhibits breast cancer progression and metastasis
by inducing macrophage polarization through downregulated
expression of Fra-1 proto-oncogene. Oncogene. 33:3014–3023. 2014.
View Article : Google Scholar
|
|
47
|
Banerjee S, Xie N, Cui H, Tan Z, Yang S,
Icyuz M, Abraham E and Liu G: MicroRNA let-7c regulates macrophage
polarization. J Immunol. 190:6542–6549. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang C, Liu XJ, Qun Zhou, Xie J, Ma TT,
Meng XM and Li J: MiR-146a modulates macrophage polarization by
inhibiting Notch1 pathway in RAW264.7 macrophages. Int
Immunopharmacol. 32:46–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lei J, Fu Y, Zhuang Y, Zhang K and Lu D:
miR-382-3p suppressed IL-1β induced inflammatory response of
chondrocytes via the TLR4/MyD88/NF-κB signaling pathway by directly
targeting CX43. J Cell Physiol. 234:23160–23168. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang J, Wang F, Argyris E, Chen K, Liang
Z, Tian H, Huang W, Squires K, Verlinghieri G and Zhang H: Cellular
microRNAs contribute to HIV-1 latency in resting primary CD4+ T
lymphocytes. Nat Med. 13:1241–1247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang X, Xue N, Zhao S, Shi Y, Ding X and
Fang Y: Upregulation of miR-382 contributes to renal fibrosis
secondary to aristolochic acid-induced kidney injury via PTEN
signaling pathway. Cell Death Dis. 11:6202020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou H, Wang Y, Lin Z and Ming J:
Mir-382-5p affects the polarization of breast cancer
tumor-associated macrophages through the Akt/mTOR signaling
pathway. J Third Mil Med Univ. 43:1358–1365. 2021.
|
|
53
|
Chawla A: Control of macrophage activation
and function by PPARs. Circ Res. 106:1559–1569. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Niu Z, Shi Q, Zhang W, Shu Y, Yang N, Chen
B, Wang Q, Zhao X, Chen J, Cheng N, et al: Caspase-1 cleaves PPARγ
for potentiating the pro-tumor action of TAMs. Nat Commun.
8:7662017. View Article : Google Scholar
|
|
55
|
Liu Y, Xu R, Gu H, Zhang E, Qu J, Cao W,
Huang X, Yan H, He J and Cai Z: Metabolic reprogramming in
macrophage responses. Biomark Res. 9:12021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Vazquez F, Lim JH, Chim H, Bhalla K,
Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM
and Puigserver P: PGC1α expression defines a subset of human
melanoma tumors with increased mitochondrial capacity and
resistance to oxidative stress. Cancer Cell. 23:287–301. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Bost F and Kaminski L: The metabolic
modulator PGC-1α in cancer. Am J Cancer Res. 9:198–211. 2019.
|
|
58
|
Abdalla HB, Napimoga MH, Lopes AH, de
Macedo Maganin AG, Cunha TM, Van Dyke TE and Clemente Napimoga JT:
Activation of PPAR-γ induces macrophage polarization and reduces
neutrophil migration mediated by heme oxygenase 1. Int
Immunopharmacol. 84:1065652020. View Article : Google Scholar
|
|
59
|
Zhang X, Zhang Y, Meng Q, Sun H, Wu S, Xu
J, Yun J, Yang X, Li B, Zhu H, et al: MicroRNA-382-5p is involved
in pulmonary inflammation induced by fine particulate matter
exposure. Environ Pollut. 262:1142782020. View Article : Google Scholar : PubMed/NCBI
|