Introduction
Hepatocellular carcinoma is the most common type of
primary liver cancer. Liver cancer is the second most common cause
of cancer-related death and the fifth most common type of cancer in
China (1). Patients in the early
and intermediate stages of the disease are treated with curative
intent through surgery and local ablation; however, the disease
develops rapidly and is associated with a high level of recurrence.
The advanced disease stages of liver cancer are treated by systemic
therapy (2,3). Multi-kinase inhibitors have been
approved as first-line therapy drugs (4). Sorafenib, regorafenib and lenvatinib,
which are multi-kinase inhibitors, have all been approved as
first-line treatments for primary hepatocellular carcinoma
(5); however, these drugs are
limited by poor overall survival, low objective response rate,
hepatotoxicity and high cost (6,7).
Developing new small-molecule drugs is thus of great clinical
significance in treating hepatocellular carcinoma.
Eupatorium lindleyanum DC. (EL), also called
'Yemazhui', is a traditional Chinese medicine that has been used to
treat cough and chronic bronchitis in China for several thousands
of years (8). Scientists have
isolated >100 compounds from EL, including sesquiterpenes,
diterpenoids, triterpenoids and volatile oils, and sesquiterpenes
are one of the most abundant ingredients. The present study
investigated the effect of the sesquiterpene lactone constituent
eupalinolide A (EA) on hepatocellular carcinoma. The constituents
of EL have several pharmacological effects, including
anti-bronchitis, antihypertensive, antiviral and anti-lung cancer
activities (9-11). Scientists have hypothesized that EL
may alter the cell metabolism to treat rat hyperlipidemia (12). Furthermore, it has been suggested
that EL extracts can exert antipyretic effects by inhibiting
myeloperoxidase activity in the liver (13). However, the effects of EL on
hepatocellular carcinoma remain largely unknown.
The present study investigated the effect and
mechanism of action of EA on hepatocellular carcinoma. The
pharmacological activity of EA on tumor growth was detected in
vivo in a xenograft model of human hepatocellular carcinoma
cells, whereas cell proliferation was evaluated in vitro
using hepatocarcinoma cell lines (MHCC97-L and HCCLM3). The effect
of EA on the cell cycle was then investigated, as well as on the
type of cell death, including apoptosis, necrosis and autophagy.
Cell migration was evaluated using the wound-healing and Transwell
assays. Inhibitors of cell death, reactive oxygen species (ROS) and
MAPK signaling pathways were used to identify the related
mechanisms. EA exerted an inhibitory effect on hepatocellular
carcinoma, highlighting its potential as a novel small-molecule
anticancer compound. The present study may improve the information
available regarding the effect of EA on hepatocellular carcinoma,
allowing for further investigation.
Materials and methods
Cell culture and treatment
Human hepatocellular carcinoma cells MHCC97-L and
HCCLM3 (Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd.) were
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.), and
incubated at 37°C in an atmosphere containing 5% CO2.
Media were supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.). EA was purchased from Chengdu Must
Bio-Technology Co., Ltd. and was dissolved in DMSO before being
used to treat the hepatocellular carcinoma cell lines MHCC97-L and
HCCLM3 at different concentrations (7, 14 and 28 µM) at 37°C
for 24, 48 or 72 h, according to the subsequent experiment. The
following inhibitors were used to treat the MHCC97-L and HCCLM3
cell lines for 30 min prior to EA treatment at 37°C: Z-VAD-FMK (20
µM; MedChemExpress LLC), necrostatin-1 (30 µM;
MedChemExpress LLC), 3-methyladenine (3-MA; 2 mM; MedChemExpress
LLC), N-acetylcysteine (NAC; 3 mM; MilliporeSigma) and PD98059 (20
µM; MedChemExpress LLC). DMSO was used to treat the control
group. All experiments were performed independently and in
triplicate.
Microscopic observation
Hepatocellular carcinoma cells (4×103
cells/well) were seeded into six-well cell culture plates and
incubated for 48 h after EA (7, 14 and 28 µM) and DMSO
treatment. The cells were observed under a light inverted
microscope (Leica Microsystems GmbH). Cell numbers were analyzed
using Image-Pro Plus 7.0 software (Media Cybernetics, Inc.). The
proliferation rate was calculated as follows: 100× (mean number of
cells in the experimental group/mean number of cells in control
group).
Cell Counting Kit 8 (CCK8) assay
The CCK8 (Dojindo Laboratories, Inc.) was used to
determine the viability of MHCC97-L and HCCLM3 cells. Cells were
seeded into 96-well culture plates at a density of 5,000
cells/well. EA was then added to the drug administration group at a
final concentration of 7, 14 and 28 µM, and the cells were
incubated for 24, 48 and 72 h at 37°C. CCK8 solution (10 µl)
was then added to each well and the cells were further incubated at
37° for 4 h. The OD value of the cells was measured at 450 nm using
a microplate reader.
5-bromo-2-deoxyuridine (BrdU)
staining
Cell proliferation was measured by BrdU staining. A
total of 1×104 cells were seeded in 24-well plates,
followed by the addition of DMEM containing DMSO or EA (28
µM), and were incubated at 37°C for 2 days. Subsequently,
the cells were incubated with 10 µg/ml BrdU (MilliporeSigma)
for 2 h at 37°C and fixed with 4% paraformaldehyde for 15 min at
room temperature. The cells were then treated with 2 M HCl at 37°C
for 30 min and blocked with 10% goat serum (OriGene Technologies,
Inc.) containing 0.3% Triton X-100 for 1 h at room temperature. The
cells were then incubated with BrdU primary antibody (1:1,000; cat.
no. ab6326; Abcam) at 4°C overnight and with an Alexa Fluor™
594-conjugated goat anti-rat IgG secondary antibody (1:1,000; cat.
no. A-11007; Thermo Fisher Scientific, Inc.) for 2 h at room
temperature. The cells were finally stained with DAPI for 15 min at
room temperature and observed under a fluorescence microscope.
BrdU-positive cells in three random fields were counted. The cells
were counted using Photoshop CC 2018 software (Adobe Systems,
Inc.).
Soft agar assay
The effect of EA on the colony-forming ability of
hepatocellular carcinoma cells was determined using the soft agar
assay. Briefly, 1.5 ml 2X RPMI-1640 (Gibco; Thermo Fisher
Scientific, Inc.) containing 0.6% agarose (MilliporeSigma) was
added to 6-well plates as base agar at 37°C for 1 h. A total of
103 cells and 28 µM EA were then added to the top
of the base agar and incubated at 37°C for 21 days. The
hepatocellular carcinoma cells were finally stained with MTT for 30
min at 37°C and were images were captured using a digital
camera.
Wound-healing assay
Hepatocellular carcinoma cells were seeded in a
6-well plate and incubated until they reached 90% confluence. The
cells were then scratched using a 100-µl sterile pipette tip
to form a linear wound, followed by the addition of serum-free DMEM
containing 7, 14 or 28 µM EA or DMSO. Cell migration across
the wounded area was observed at 6, 24 and 48 h and the extent of
wound closure was measured at the indicated time points (14). The cells were observed under a
light inverted microscope (Leica Microsystems GmbH). Cell migration
was analyzed using Image-Pro Plus 7.0 software. The wound closure
rate was calculated as follows: Wound closure (%)=100× [scratch
width at 0 h-scratch width at 6, 24 or 48 h)/(scratch width at 0
h)].
Cell migration
The migratory potential of hepatocellular carcinoma
cells was measured using Transwell chambers (pore size, 8
µm; Corning, Inc.). Briefly, the hepatocellular carcinoma
cells (2×105) were suspended in DMEM containing 1% FBS
and were seeded into the upper chamber. DMEM (500 µl)
containing 20% FBS was then added to the lower chamber to act as a
chemoattractant. The cells were incubated for 48 h with DMEM at
37°C containing 7, 14 or 28 µM EA, or DMSO. Cells on the
lower surface of the membrane were stained with 0.5% crystal violet
for 15 min at room temperature. The cells were observed under an
light inverted microscope (Leica Microsystems GmbH). Images of the
stained cells were randomly captured and the cells were counted
using the Image-Pro Plus 7.0 software. The migration rate was
calculated as follows: 100× (mean number of migrating cells in the
experimental group/mean number of migrating cells in control
group).
Western blot analysis
Western blotting was performed as previously
described (15). The cells were
treated with EA (7, 14 and 28 µM) for 24 h. The treated
cells were harvested and lysed on ice using RIPA lysis buffer
(Beyotime Institute of Biotechnology) containing protease and
phosphatase inhibitors (Roche Diagnostics). The protein
concentration of the cell lysate was measured using the BCA protein
assay kit (Beyotime Institute of Biotechnology). The cell lysate
was then denatured at 100°C for 5 min. Subsequently, 50 µg
protein extracts were subjected to SDS-PAGE on 10 and 12% gels and
were transferred onto a polyvinylidene difluoride membrane. The
membrane was blocked in 5% bovine serum albumin (MilliporeSigma) at
room temperature for 1 h, and was then incubated overnight at 4°C
with the following primary anti-bodies: E-cadherin (1:1,000; cat.
no. 14472), Vimentin (1:1,000; cat. no. 5741T), cleaved-PARP
(1:1,000; cat. no. 5625T), cleaved-caspase-3 (1:1,000; cat. no.
9661), autophagy-related 5 (Atg5; 1:1,000; cat. no. 12994T), p62
(1:1,000; cat. no. 8025T), MAPK family sampler kit (1:1,000; cat.
no. 9926T), p-ERK (1:1,000; cat. no. 4370T), p-JNK (1:1,000; cat.
no. 4668T), p-p38 (1:1,000; cat. no. 4511T), LC3B (1:1,000; cat.
no. 12741), Cyclin D1 (1:1,000; cat. no. 55506), fibronectin
(1:1,000; cat. no. 26836), CDK4 (1:1,000; cat. no. 12790), PARP
(1:1,000; cat. no. 9542), caspase-3 (1:1,000; cat. no. 9662) (all
Cell Signaling Technology, Inc.), CDK2 (1:500; cat. no. sc-6248),
Cyclin E1 (1:500; cat. no. sc-247) (both Santa Cruz Biotechnology,
Inc.), RIP1 (1:1,000; cat. no. CY6582), p-MLKL (1:1,000; cat. no.
CY7146), MLKL (1:1,000; cat. no. CY5493) (all Shanghai Abways
Biotechnology Co., Ltd.), N-cadherin (1:1,000; cat. no. AP71171),
ZEB1 (1:1,000; cat. no. AM1878A) (both from Abcepta Biotech Ltd.
Co.) and β-actin (1:1,000; cat. no. TA-09; OriGene Technologies,
Inc.). The membrane was subsequently incubated with horseradish
peroxidase-conjugated secondary antibodies [goat anti-mouse IgG
(cat. no. A0216) and goat anti-rabbit IgG (cat. no. A0208);
1:10,000; Beyotime Institute of Biotechnology] at room temperature
for 1 h. The protein bands were finally visualized using the
Clarity Western ECL Blotting Substrate (cat. no. 1705061; Bio-Rad
Laboratories, Inc.) and images were captured using a ChemiDoc MP
Imaging System (Bio-Rad Laboratories, Inc.). The integrated optical
density of the protein bands was measured using the Bio-Rad Image
Lab Software 5.2.1 (Bio-Rad Laboratories, Inc.), with β-actin
employed as the loading control.
Cell cycle analysis
Hepatocellular carcinoma cells were incubated for 48
h after EA (14 and 28 µM) at 37°C and DMSO treatment. The
treated cells at 70-80% confluence were fixed with 75% ethanol at
4°C and then incubated in 500 µl PBS containing PI and RNase
A (9:1) at 37°C for 60 min (Cell Cycle Kit; Nanjing KeyGen BioTech
Co., Ltd.). The cell samples were analyzed using DxFLEX flow
cytometry (Beckman Coulter, Inc.). The cell percentage in each
cycle was finally evaluated using Modfit-LT V 3.0 software (Verity
Software House).
Analysis of cell apoptosis
Hepatocellular carcinoma cells were cultured for 48
h after EA (7, 14 and 28 µM) and DMSO treatment. The cells
were then collected, washed with cold PBS and suspended in 200
µl binding buffer (Annexin V-FITC/PI apoptosis assay kit;
Neobioscience Technology Co., Ltd.) at a density of
5×105/ml. Subsequently, cells in binding buffer (195
µl) were incubated with 5 µl Annexin V-FITC at room
temperature in the dark for 10 min. The cells were then washed with
PBS, suspended in binding buffer, and mixed with 10 µl PI.
The mixture was gently mixed and analyzed by DxFLEX flow cytometry.
The apoptotic rate of the hepatocellular carcinoma cells was
finally determined using CytExpert 2.0 software (Beckman Coulter,
Inc.).
Transmission electron microscopy
Hepatocellular carcinoma cells were incubated for 48
h after EA (28 µM) and DMSO treatment. The cells at a
density of 106/ml were then collected into a 1.5 ml
Eppendorf tube containing trypsin and centrifuged at 100 × g for 5
min at room temperature. The supernatant was collected, and the
cells were fixed with 1 ml 2.5% glutaraldehyde fixing solution for
1 h at 4°C and treated with 1% OsO4 in PBS for 1 h at
4°C The cells were then dehydrated in ethanol, embedded in epoxy
resin and sliced into 90-nm sections. The sections were stained
with uranyl acetate for 5 min and lead citrate for 5 min at 37°.
Ultrathin sections of the cells were examined under a JEM1200EX
transmission electron microscope (Jeol Ltd.).
ROS detection analysis
Hepatocellular carcinoma cells were incubated for 48
h after EA (14 and 28 µM) and DMSO treatment. The cells
(4×103 cells/well) were seeded into six-well plates,
then collected, suspended in serum-free DMEM containing 10
µM DCFH-DA (Reactive Oxygen Species Assay Kit; Beyotime
Institute of Biotechnology) and incubated at 37°C for 20 min. The
cells were subsequently washed three times with serum-free cell
medium and analyzed by DxFLEX flow cytometry. The ROS content in
the hepatocellular carcinoma cells was finally analyzed using
CytExpert 2.0 software.
RNA sequencing and data analysis
HCCLM3 cells were harvested after 48 h of treatment
with DMSO and EA (14 and 28 µM) at 5% CO2 and
37°C for total RNA isolation using TRNzol Universal Total RNA
extraction reagent (cat. no. DP424, Tiangen Biotech Co., Ltd.). RNA
samples were submitted to Shanghai Personalbio Technology Co., Ltd.
for transcriptome sequencing and analysis. The concentration,
quality and integrity of RNA were determined using a NanoDrop
spectrophotometer (Thermo Fisher Scientific, Inc.). Integrity was
assessed by an Agilent 2100 Bioanalyzer using the RNA 6000 Nano kit
(cat. no. 5067-1511) (both Agilent Technologies Inc.).
Subsequently, 3 µg RNA was used as the input material for
RNA sample preparation. The experiment was performed according to
manufacturer's protocol of NEBNext Ultra II RNA Library Prep Kit
for Illumina (cat. no. E7775; New England Biolabs, Inc.).
Sequencing libraries were generated according to the following
steps. Firstly, mRNA was purified from total RNA using poly-T
oligo-attached magnetic beads. Fragmentation was carried out using
divalent cations under elevated temperature in an Illumina
proprietary fragmentation buffer (Illumina, Inc.). First strand
cDNA was synthesized using random oligonucleotides and Super Script
II. Second strand cDNA synthesis was subsequently performed using
DNA Polymerase I and RNase H. Remaining overhangs were converted
into blunt ends via exonuclease/polymerase activities and the
enzymes were removed. After adenylation of the 3' ends of the DNA
fragments, Illumina PE adapter oligonucleotides (Illumina, Inc.)
were ligated to prepare for hybridization. To select cDNA fragments
of the preferred 400-500 bp in length, the library fragments were
purified using the AMPure XP system (Beckman Coulter, Inc.).
DNA fragments with ligated adaptor molecules on both
ends were selectively enriched using Illumina PCR Primer Cocktail
(Illumina, Inc.) in a 15-cycle PCR reaction. Products were purified
(AMPure XP system) and quantified using the Agilent High
Sensitivity DNA assay on the 2100 Bioanalyzer system (both Agilent
Technologies, Inc.). The sequencing library was then sequenced on
the NovaSeq 6000 platform (Illumina, Inc.) by Shanghai Personal
Technology Co. Ltd. To select cDNA fragments of the preferred
400-500 bp in length, the library fragments were purified using the
AMPure XP system. The library was uniformly diluted to 2 nM and
PE150 mode sequencing was performed on the Illumina sequencer.
The original off-machine data were filtered for the
transcriptome analysis. The high-quality sequences obtained were
aligned with the human reference genome (Homo_sapiens.
GRCh38.dna.primary_assembly.fa). The expression levels of the genes
were calculated based on the comparison results, followed by
expression difference analysis between the EA (24 µM) and
control group, enrichment analysis and cluster analysis between EA
and Control groups. Gene read counts were estimated with HTSeq
(0.9.1) statistics (16). FPKM was
used to standardize the expression. The differential expression of
genes was analyzed using DESeq 1.30.0 (Bioconductor) (16,17).
The screening conditions were as follows: |log2FoldChange|>1 and
significant P<0.05. The R language heatmap (1.0.8) software
package was used to perform a bi-directional clustering analysis of
all different genes in the samples (16). The heatmap was developed according
to the expression levels of the same gene in different samples and
the expression patterns of different genes in the same sample. The
Euclidean method was used to calculate the distance and the
complete linkage method was performed for clustering.
ClusterProfiler (3.4.4) software was used to carry out the Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis of the differentially expressed genes, focusing on the
pathways with P<0.05 (18-20).
Animal studies
A total of 36 female nude mice (BALB/c; weight 18-20
g; age, 4 weeks) were purchased from Cavensbiogle and housed in a
specific pathogen-free room that was maintained at a constant
temperature (20-26°C) and humidity (40-70%). The mice were
maintained under a 12-h light/dark cycle with free access to food
and water. The animals were divided into three groups and the
number of mice per group was six; the number of mice in the
MHCC97-L group was 18 and the number of mice in the HCCLM3 group
was also 18. MHCC97-L and HCCLM3 cells (1×106 cells in
200 µl PBS) were subcutaneously injected into the nude mice.
The mice were then intraperitoneally injected with EA (30 or 60
mg/kg) and saline solution (Control) once per day for 3 weeks. The
body weight of the mice was monitored, and the tumor size was
measured to calculate the tumor volume, as follows: Volume=4π/3 ×
R3, where R=L + W/2; R refers to the radius of tumor, L
refers to the longest diameter of the tumor mass and W to the
longest transverse diameter perpendicular to the longest diameter.
Volume was calculated from day 12 to 26 (once every 2 days). The
mice were euthanized using CO2 (30% of the volume of the
euthanasia chamber was replaced with CO2 every minute)
on day 26 (the maximum tumor volume of the MHCC97-L group was
810.41 mm3 and the maximum tumor volume of the HCCLM3
group was 960.66 mm3), and the tumors were subsequently
excised and weighted.
Statistical analysis
Data were analyzed using GraphPad Prism 5.01
software (GraphPad Software, Inc.) and were presented as the mean ±
SD. All experiments were repeated at least three times. Two groups
were compared using an unpaired two-tailed Student's t-test.
One-way ANOVA followed by Tukey's test was used to assess and
compare the mean differences among multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
EA inhibits the proliferation of human
hepatocellular carcinoma cells in vitro
Hepatocellular carcinoma is characterized by rapid
development and high mortality (21). Effective inhibition of tumor growth
is therefore of great importance. The concentration of EA used in
the present study was chosen based on relevant studies of EL
extract and the IC50 of EA in MHCC97-L and HCCLM3 cells
(Fig. S2B) (22,23).
When the concentration of EA was ~10 µM, it could serve a
significant inhibitory effect on hepatocellular carcinoma cell
lines; therefore, the concentrations of EA used in the present
study were selected. In the present study, MHCC97-L and HCCLM3
cells were treated with different concentrations of EA (7, 14 and
28 µM) for 48 h to investigate the anti-proliferative
effects of EA on human hepatocellular carcinoma cells. DMSO was
used as the control. The viability of hepatocellular carcinoma
cells was examined through light morphological analysis, CCK8
assay, BrdU staining and soft agar assay. Microscopic examinations
revealed that EA altered their morphology in a dose-dependent
manner (Fig. 1A). Decreased cell
number, cell shrinkage, dispersed cell growth and pleomorphic cell
morphology were observed following EA treatment. Images of the
cells were captured and the number of cells was statistically
analyzed to obtain the proliferation rate. The proliferation rate
was measured by comparing the proliferation of the DMSO group with
that of the EA group and the proliferation rate was reduced
following EA treatment (Fig. 1B).
Notably, there was a significant decrease in the viability of the
two cell lines treated with EA after 24, 48 and 72 h compared with
that in the DMSO group (Fig. 1C).
DNA synthesis in cells treated with EA was also reduced (Fig. 1D and E). The soft agar assay
further revealed smaller and fewer colonies following EA treatment
(28 µM EA for 48 h) compared with the colonies in the DMSO
group (Fig. 1F and G). These
results suggested that EA markedly inhibited the viability and
proliferation of human hepatocellular carcinoma cells.
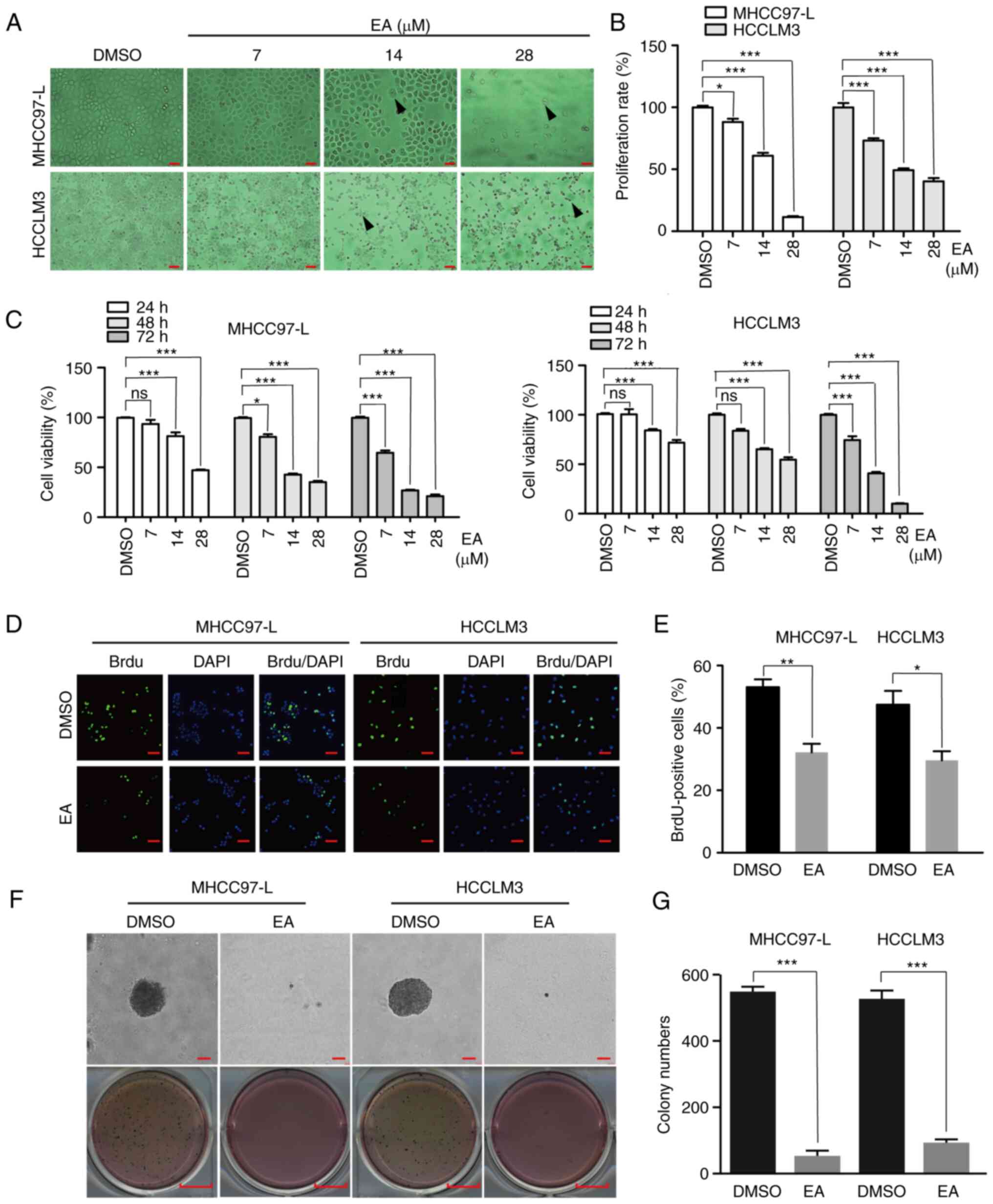 | Figure 1Effect of EA on the proliferation of
human hepatocellular carcinoma cells in vitro. (A)
Morphology of MHCC97-L and HCCLM3 cells after EA treatment for 48
h. Arrows indicate morphologically altered cells. Scale bar, 100
µm. (B) Proliferation rate of MHCC97-L and HCCLM3 cells
after EA treatment for 48 h. Scale bar, 100 mm. Representative of
biologically independent samples, n=5. (C) Viability of MHCC97-L
and HCCLM3 cells after EA treatment for 1, 2 or 3 days. At each
time point, DMSO was regarded as the control group. n=6. (D) Image
of MHCC97-L and HCCLM3 cells positive for BrdU staining after EA
treatment for 48 h. Scale bar, 100 µm. Representative of
biologically independent samples, n=3. (E) Quantification of
BrdU-positive MHCC97-L and HCCLM3 cells in (D). (F) Effects of EA
on colony formation in MHCC97-L and HCCLM3 cells. (G) Colony
numbers in (F) were quantified. Scale bar, 100 µm. n=3. All
data are shown as the mean ± SD. *P<0.05,
**P<0.01, ***P<0.001. EA, eupalinolide
A; BrdU, 5-bromo-2-deoxyuridine. |
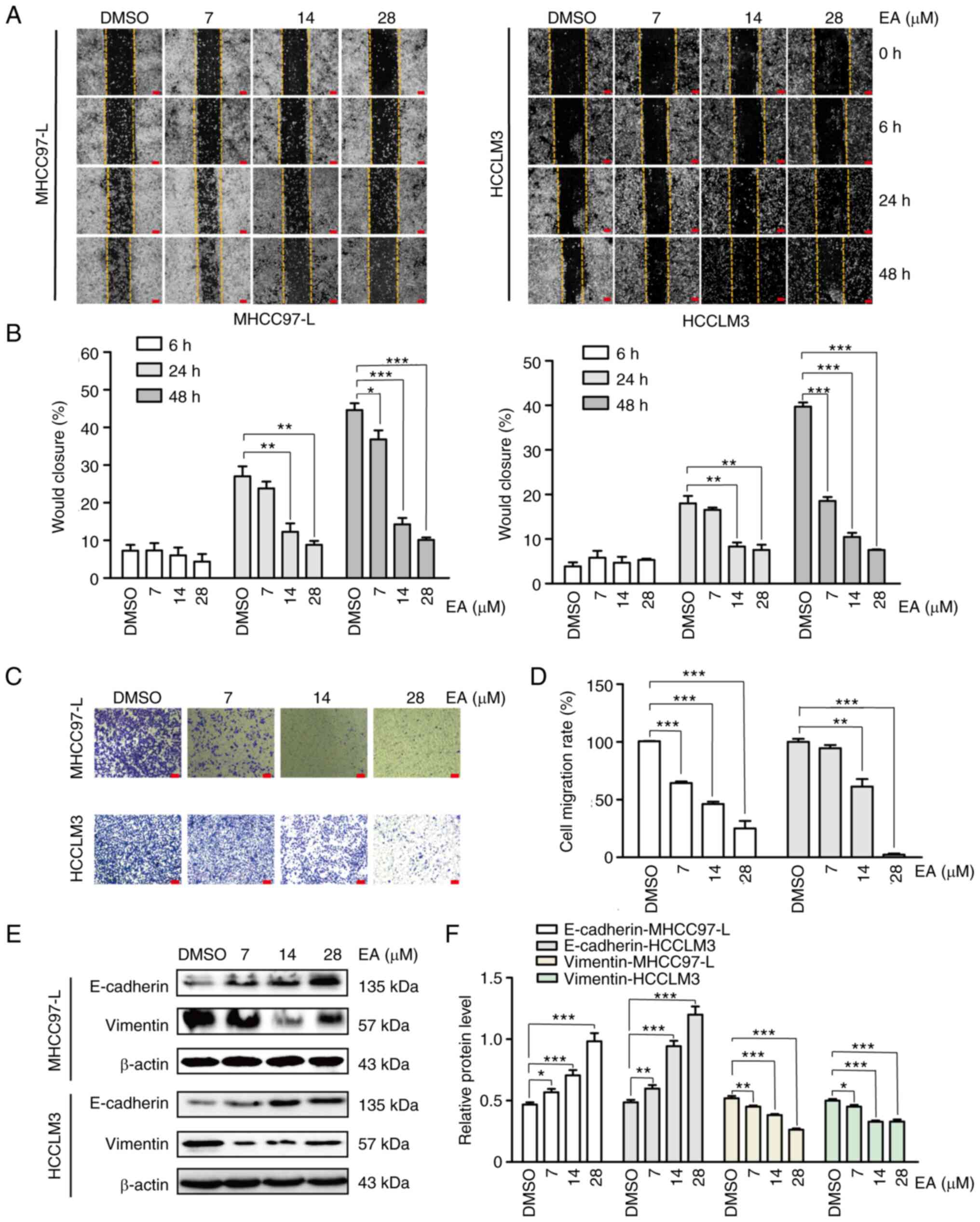
EA suppresses the migration of human
hepatocellular carcinoma cells
Surgical resection remains the main treatment method
for hepatocellular carcinoma and it significantly reduces the
mortality of patients with hepatocellular carcinoma; however, the
postoperative recurrence rate remains high at >70% (24). Therefore, the present study
assessed whether EA affected cell motility using the wound-healing
assay. Notably, there was a significant decrease in cell migration
following EA treatment for 24 and 48 h compared with that in the
DMSO group (Fig. 2A and B). The
results of Transwell assays were similar to the results of the
wound-healing assay (Fig. 2C and
D). E-cadherin, Vimentin, N-cadherin, fibronectin and ZEB1 are
major canonical markers of the epithelial-mesenchymal transition
(EMT), which serve crucial roles in the tumor migration process
(25). In the present study,
E-cadherin, N-cadherin, fibronectin and ZEB1 were significantly
upregulated, whereas Vimentin was significantly downregulated
following EA treatment in a dose-dependent manner (Figs. 2E and F, and S1A and B). These results suggested that
EA treatment reversed the EMT of human hepatocellular carcinoma
cells, thus strongly inhibiting their migration.
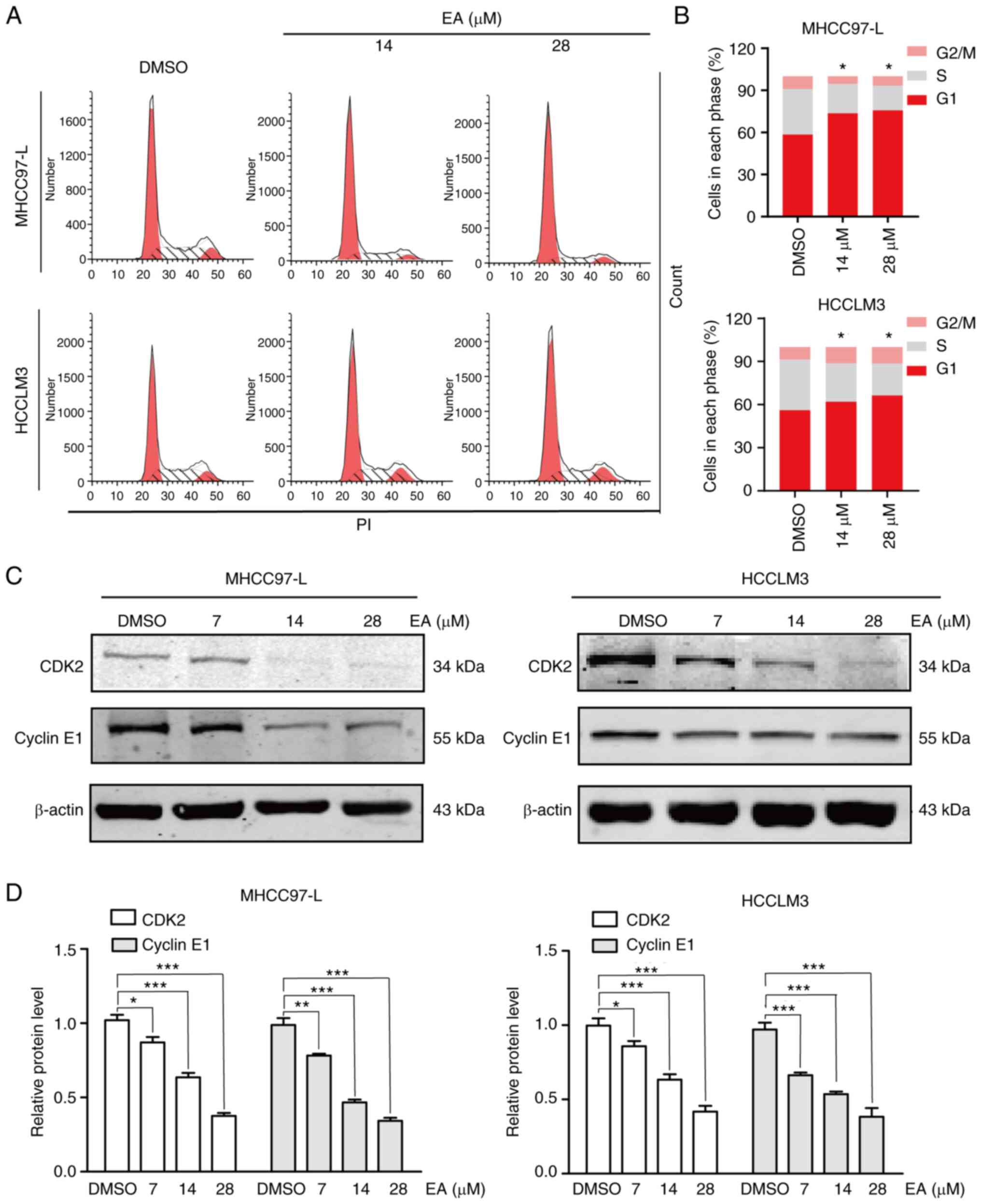
EA induces cell cycle arrest at the
G1 phase
The proliferation of tumor cells is determined by
cell cycle progression (26).
Antitumor drugs can be divided into cell cycle-specific and
non-specific drugs (27). In the
present study, the effect of EA on the cell cycle was examined
using flow cytometry. The percentage of MHCC97-L and HCCLM3 cells
in the G1 phase was significantly increased following
treatment with 14 and 28 µM EA for 48 h compared with the
control cells (Fig. 3A and B).
Notably, the expression levels of CDK2, CDK4, cyclin E1 and cyclin
D1, the key regulators for G1 phase transition (28), were downregulated following
treatment with 14 and 28 µM EA (Figs. 3C and D, and S1C and D). These results demonstrated
that EA induced G1-phase arrest by affecting the
expression of cell cycle regulatory proteins.
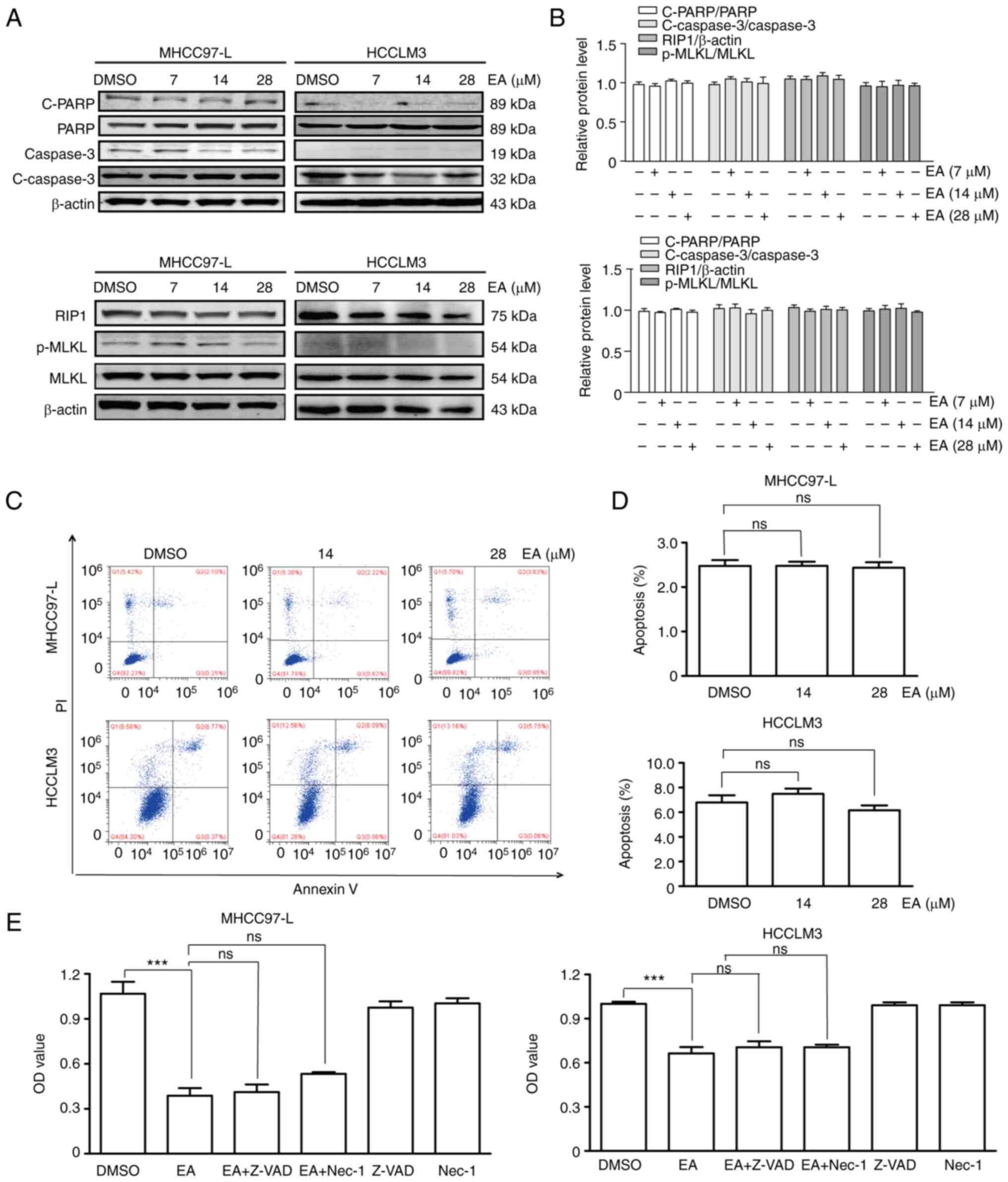
EA does not affect apoptosis and
necrosis
Apoptosis and necrosis are the major cell death
mechanisms (29), and numerous
antitumor drugs exert their effects by inducing apoptosis or
necrosis (30,31). The present study therefore
investigated whether EA treatment caused apoptosis and necrosis of
hepatocellular carcinoma cells by detecting the expression levels
of apoptosis- and necrosis-related proteins using western blot
analysis following treatment of the cells with 7, 14 and 28
µM EA for 48 h. Notably, EA treatment did not affect the
expression levels of cleaved-PARP and cleaved-caspase-3, which are
apoptosis-related proteins, nor those of RIP1 and p-MLKL, which are
necrosis-related proteins (Fig. 4A and
B). The Annexin V-FITC/PI apoptosis assay conducted using flow
cytometry further revealed that EA did not induce early or late
apoptosis (Fig. 4C and D).
Furthermore, treatment with the apoptosis inhibitor (Z-VAD-FMK) and
necrosis inhibitor (necrostatin-1) did not restore the EA-induced
decrease in cell viability (Fig.
4E). These results suggested that EA-induced inhibition of
human hepatocellular carcinoma cell viability was independent of
apoptotic and necroptotic factors.
EA induces autophagy and ROS activation
in human hepatocellular carcinoma cells
Autophagy is a classic type of programmed cell death
that participates in the functional modulation of cancer cells by
affecting Atg proteins (32). The
present study investigated whether EA served an anti-liver cancer
role by inducing autophagy. Notably, EA upregulated the expression
levels of LC3 II/I and Atg5, but downregulated p62/SQSTM1
expression, compared with those in the DMSO group (Fig. 5A-F). LC3 II/I, Atg5 and p62/SQSTM1
are autophagy biomarkers. Autophagosomes, which are two-layered
structures containing cytoplasmic components, were formed following
treatment with 28 µM EA (Fig.
5G). ROS regulate the autophagy pathway in cancer cells
(33-35). Notably, excessively high levels of
ROS can induce cell death through oxidative damage (36). In the present study, EA strongly
induced ROS production compared with in DMSO cells (Fig. 5H and I). Treatment with the
pharmacological inhibitor of autophagy 3-MA and anti-ROS reagent
NAC efficiently reversed the EA-induced inhibition of cell
viability and migration (Fig. 5J and
K). These results suggested that autophagy and ROS activation
potentially contributed to human hepatocellular carcinoma cell
death and defects in migration induced by EA.
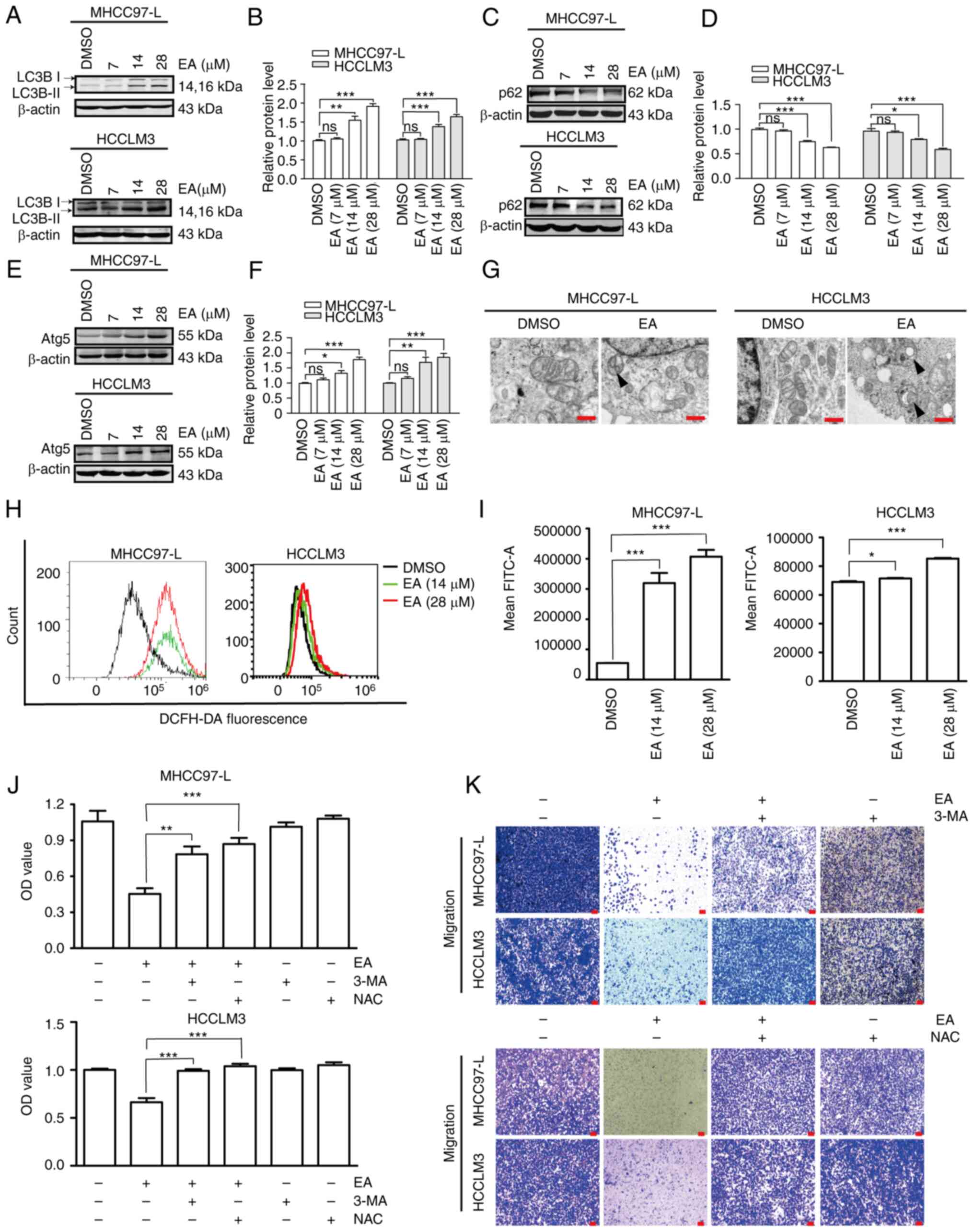 | Figure 5Effect of EA on the autophagy of
human hepatocellular carcinoma cells. (A-F) Western blot analysis
of the expression levels of autophagy-related proteins at 48 h in
MHCC97-L and HCCLM3 cells. β-actin was used as a control. (G)
Transmission electron microscopy of MHCC97-L and HCCLM3 cells
following EA treatment for 48 h. Arrow indicates autophagosomes.
Scale bar, 500 nm. (H and I) ROS production of MHCC97-L and HCCLM3
cells was measured by DFC fluorescence following EA treatment.
Representative of biologically independent samples, n=6. (J)
Viability of MHCC97-L and HCCLM3 cells treated with or without ROS
or autophagy inhibitors, as measured by Cell Counting Kit 8 assays.
Representative of biologically independent samples, n=6. (K)
Migratory ability of MHCC97-L and HCCLM3 cells treated with or
without ROS or autophagy inhibitors, as measured by Transwell
migration assays after EA treatment. All data are shown as the mean
± SD. *P<0.05, **P<0.01,
***P<0.001. EA, eupalinolide A; Atg5,
autophagy-related 5; 3-MA, 3-methyladenine; NAC, N-acetylcysteine;
ROS, reactive oxygen species. |
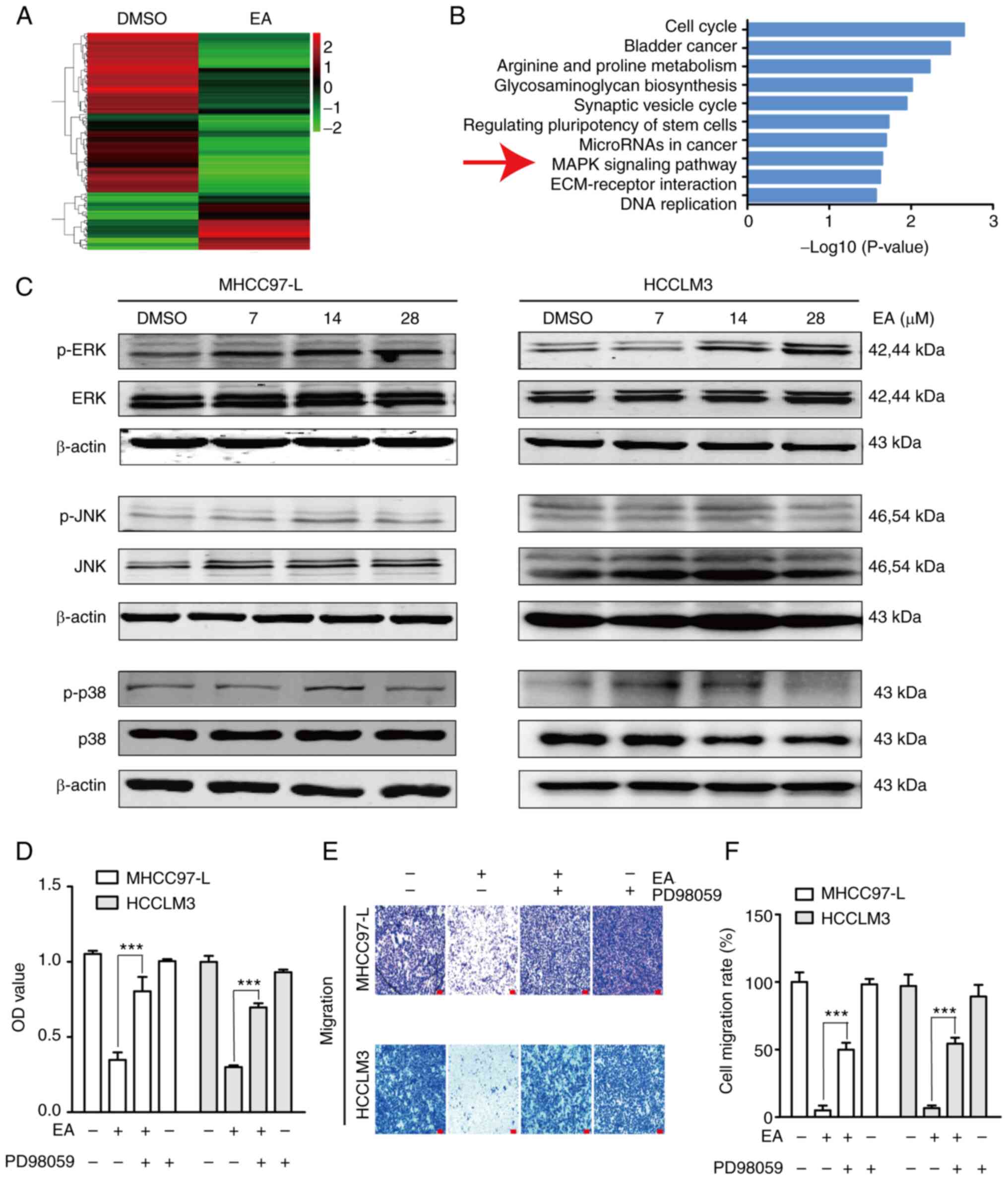
EA treatment activates the ERK/MAPK
signaling pathway in human hepatocellular carcinoma cells
Abnormal activation of the MAPK signaling pathway is
closely related to the occurrence, development and metastasis of
hepatocellular carcinoma (37). In
the present study, transcriptome analysis, which was conducted to
investigate the mechanism underlying EA-induced cell death and
migration, revealed that gene expression levels were significantly
altered after EA treatment compared with in the control group
(Fig. 6A). According to the KEGG
pathway enrichment analysis results of differentially expressed
genes, the top 10 pathways with the minimum P-value or the most
significant enrichment were selected (Fig. 6B). The present study focused on the
'MAPK signaling pathway'. Further validation of the transcriptome
sequencing data using western blotting revealed that EA
significantly upregulated p-ERK expression, but did not affect
p-p38 and p-JNK expression, in MHCC97-L and HCCLM3 cells compared
with in the DMSO group (Figs. 6C
and S2A). Furthermore, the
decreased cell viability and migration induced by EA were reversed
by ERK inactivation using PD98059 (Fig. 6E-G). These results suggested that
ERK signaling participated in EA-induced cell death and migration
inhibition.
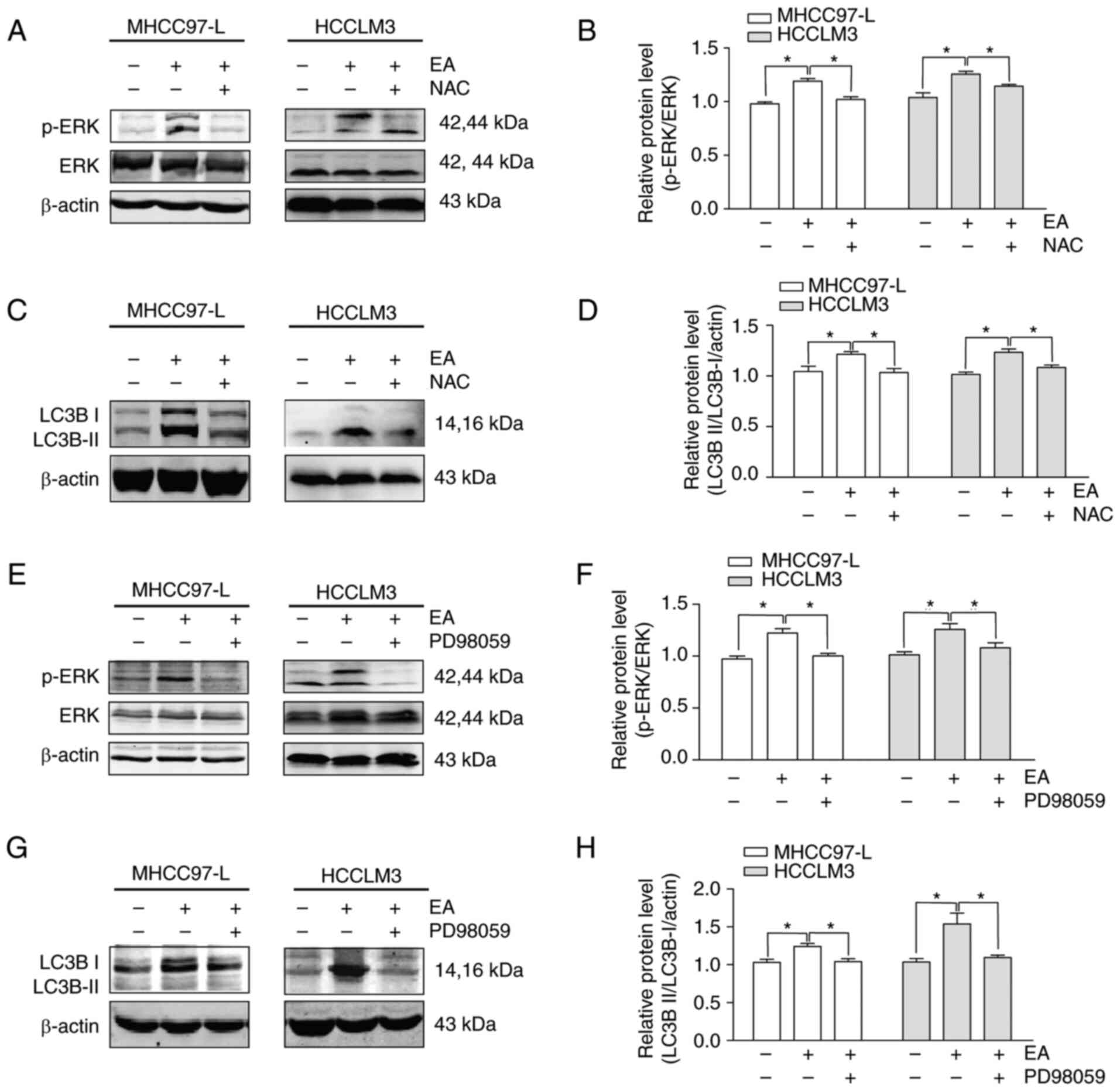
EA induces autophagy via the ROS/ERK
signaling pathway
A ROS scavenger (NAC) and ERK-specific inhibitor
(PD98059) were used to investigate the interplay between ROS and
ERK in EA-induced autophagy using western blotting. Pretreatment
with NAC significantly reduced the activation of p-ERK and LC3 II/I
induced by EA in both MHCC97-L and HCCLM3 cells, indicating that
ROS was an upstream signal of EA and induced ERK activation and
autophagy (Fig. 7A-D). Notably,
the ERK-specific inhibitor PD98059 also suppressed the EA-induced
increased expression of p-ERK and LC3 II/I in both MHCC97-L and
HCCLM3 cells (Fig. 7E-H). These
data suggested that EA induced autophagy by activating ROS
production and ERK signaling.
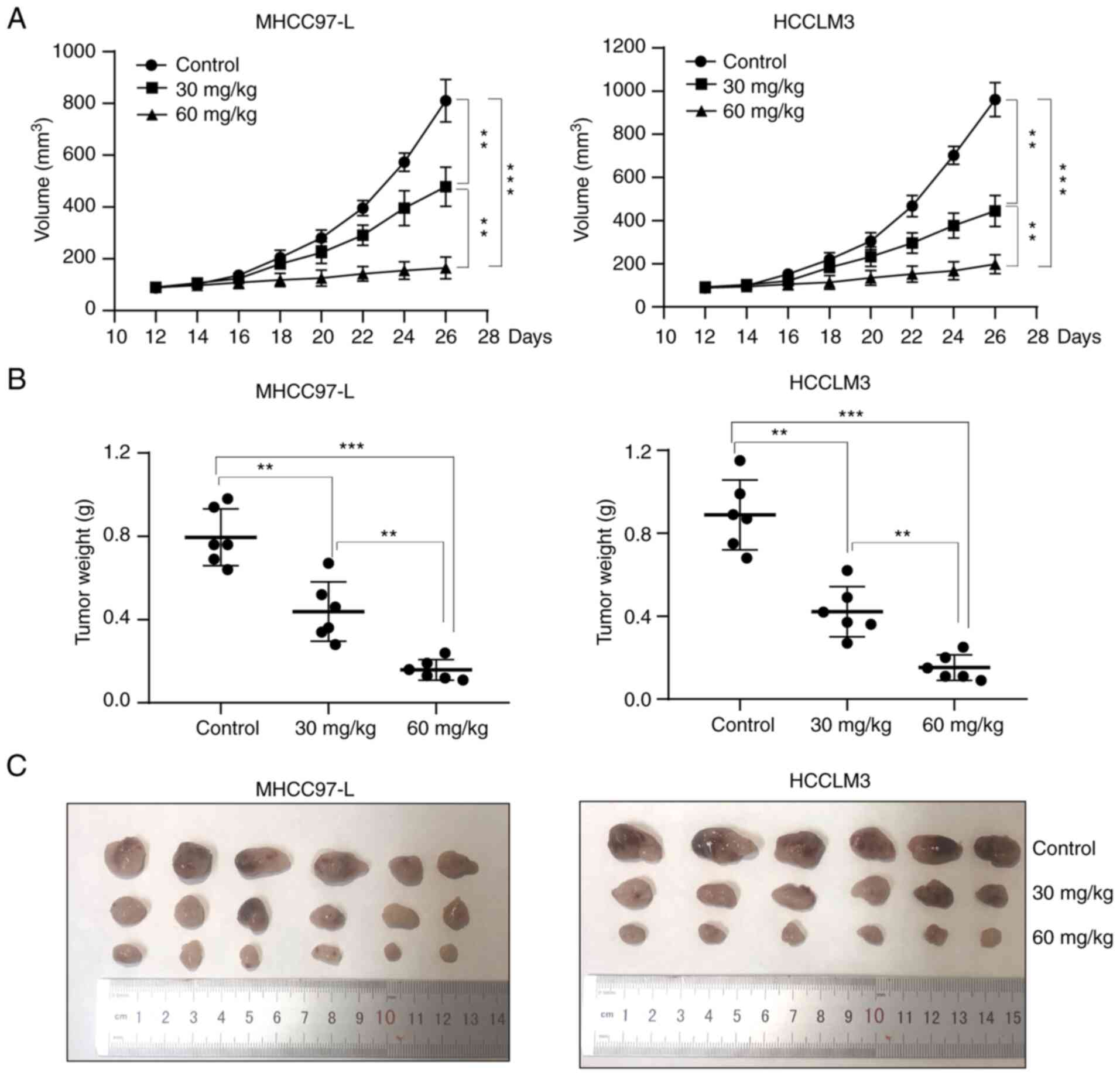
EA inhibits tumor growth in vivo
In vivo assays using nude mice were further
performed to determine whether EA inhibited the growth of
hepatocellular carcinoma. Nude mice were subcutaneously inoculated
with 1×106 hepatocellular carcinoma cells. The mice were
subsequently intraperitoneally injected with EA or DMSO (control)
daily for 21 days, followed by an assessment of tumor growth
following cessation of treatment. Mice treated with EA exhibited a
smaller tumor volume and weight than mice in the control group
(Fig. 8A-C), thus indicating that
EA markedly inhibited tumor growth in vivo.
Discussion
The present study investigated the effect of EA on
hepatocellular carcinoma. In vitro, MHCC97-L and HCCLM3 cell
lines were selected because MHCC97-L is a human hepatocellular
carcinoma cell line with low metastatic potential and HCCLM3 is a
human hepatocellular carcinoma cell line with high metastatic
potential. In addition, MHCC97-L and HCCLM3 cell lines have been
adopted in a number of studies (38,39).
The present study revealed that EA inhibited the
growth of hepatocellular carcinoma cells in vivo and in
vitro. The in vitro results demonstrated that EA caused
cell cycle arrest at the G1 phase and induced cell
autophagy via the ROS/ERK signaling pathway. By contrast, treatment
with a ROS scavenger, ERK inhibitor or autophagy inhibitor reversed
the decrease in cell viability and migration induced by EA. The ROS
scavenger induced a nearly complete recovery of the proliferative
and migratory ability of hepatocellular carcinoma cells. The
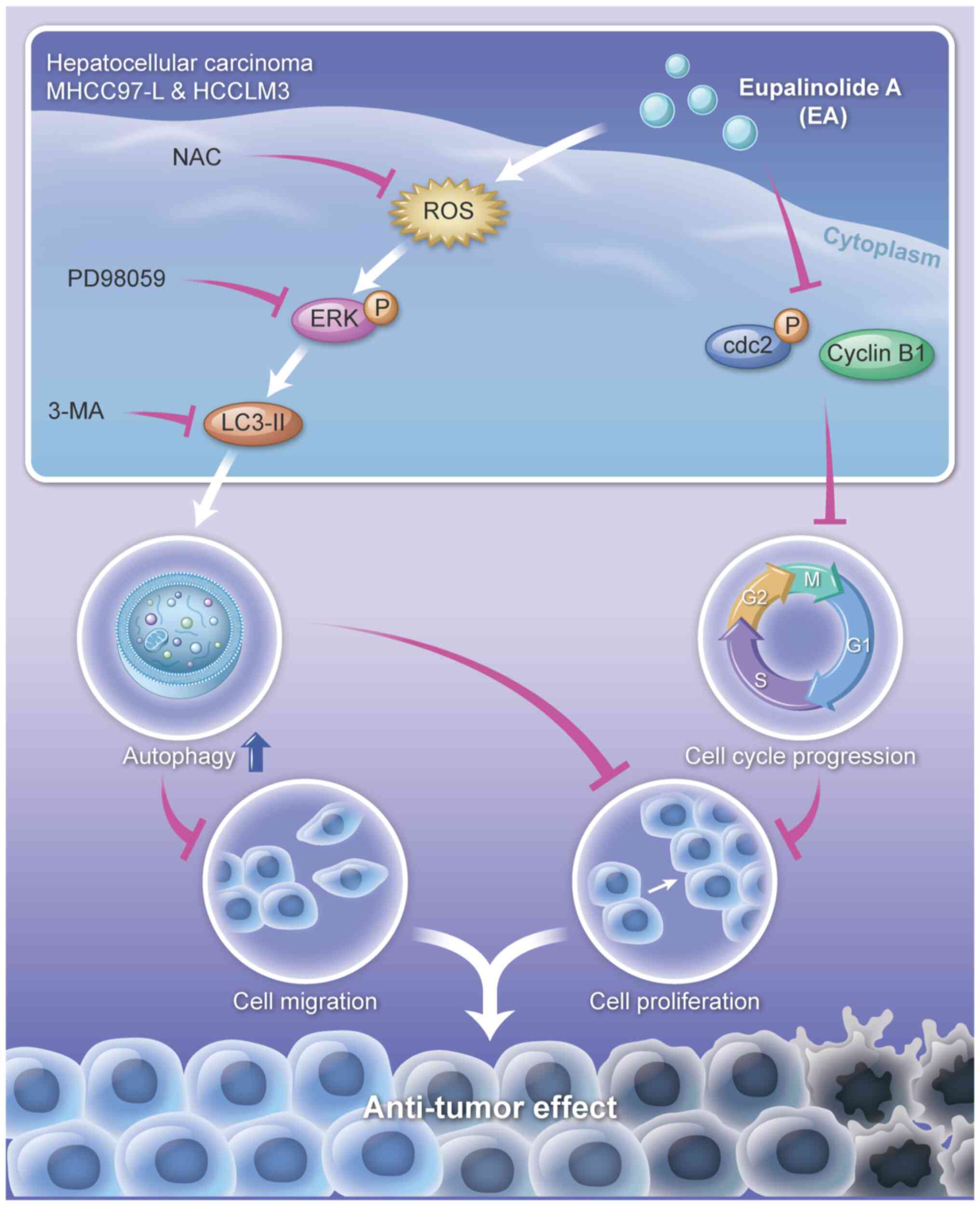
anti-hepatocellular carcinoma mechanism of EA is summarized in
Fig. 9.
Several types of traditional Chinese medicine have
been explored for their use in the treatment of cancer (40,41).
Our research team has aimed to identify a new drug for treating
liver cancer. EL has been widely applied to treat respiratory
diseases, because of its antihypertensive, antiviral and anti-tumor
effects. In the present study, EA markedly reduced the
proliferation of MHCC97-L and HCCLM3 cells. EA, especially at 28
µM, led to a number of floating cells, a change in cell
morphology and death of most MHCC97-L and HCCLM3 cells. These
results demonstrated that EA had a powerful hepatocellular
carcinoma cell-killing ability in vitro.
Chemotactic migration is vital for the invasion of
cancer cells into the surrounding tissue, resulting in tumor
metastasis (42). In the present
study, the migratory ability of hepatocellular carcinoma cells was
inhibited by EA, especially at 28 µM. Notably, the invasive
and metastatic ability of carcinoma cells has been reported to be
promoted by EMT (43). E-cadherin
has a vital role in maintaining the integrity of intercellular
junctions in cell adhesion. Loss of E-cadherin expression can lead
to a loss of contact, increased cell motility and cancer
advancement (44). Vimentin has
been shown to be overexpressed in gastrointestinal tumors, and
usually aggravates tumor growth and invasion, leading to poor
prognosis (45). In the present
study, EA inhibited EMT by increasing the expression levels of
E-cadherin and decreasing those of Vimentin. In addition, EA can
affect the expression of other EMT-related proteins, including
N-cadherin, fibronectin and ZEB1. EA thus inhibited the migration
of hepatocellular carcinoma cells by reversing EMT progression.
Most human somatic cells experience a fixed life
cycle through a highly coordinated and regulated process; however,
faults occur during the cell cycle in cancer cells, resulting in
unchecked cell growth (46).
Numerous anticancer drugs, such as vincristine and colchicine,
induce cell cycle arrest (47,48).
In the present study, EA arrested the cell cycle at the
G1 phase, which is similar to the effect of vinblastine,
by regulating the cell cycle phase checkpoint proteins such as
Cyclin E and CDK2 (49). These
results indicated that EA exerted an anticancer effect by arresting
the cell cycle.
Apoptosis and necrosis are classic cell death
mechanisms and promising targets in anticancer therapy (40,50,51).
In the present study, EA had no significant effect on apoptosis and
necrosis. Moreover, apoptosis and necrosis inhibitors did not
reverse the EA-induced decrease in cell viability. These results
suggested that EA induced cell death independent of apoptosis and
necrosis.
Autophagy and ROS are involved in cell survival and
cell death pathways. A number of anticancer drugs activate
autophagy and ROS signaling (52,53).
Autophagy is a dynamic process of protein degradation that usually
occurs during nutrient deprivation. Autophagy serves dual roles in
cancer by acting as a tumor suppressor and a tumor promoter that
accelerates the growth of established tumors (54). Autophagy is thus a common target
for drugs used to treat cancer. The LC3 II/I ratio is used to
estimate the level of autophagy. p62/SQSTM1 binds directly to
Atg8/LC3 to facilitate the degradation of ubiquitinated proteins
aggregated by autophagy. The overexpression of Atg5 can also
enhance autophagy (55,56). In the present study, EA
significantly activated the autophagy signal biomarker LC3 II/I and
Atg5, and suppressed the expression of p62. Furthermore, EA induced
autophagy in hepatocellular carcinoma cells, which was verified by
electron microscopic observation. Autophagy is categorized into
macro-autophagy, micro-autophagy and chaperone-mediated autophagy
(57). EA-induced autophagy
belongs to macro-autophagy, which is characterized by the formation
of autophagosomes and autolysosomes. ROS is an important upstream
molecule that regulates cancer cell death and survival; excessive
levels of ROS can cause oxidative damage to cancer cells (58). In the present study, EA
significantly increased ROS generation in hepatocellular carcinoma
cells; however, treatment with the ROS scavenger NAC and
autophagy-specific inhibitor 3-MA reversed EA-induced inhibition of
cell proliferation and migration. These results indicated that ROS
and autophagy were involved in EA-induced cell death.
A number of signals are involved in regulating ROS
and autophagy in cancer. The MAPK signaling pathway, a down-stream
pathway of ROS, is known to have an essential role in inducing
autophagy (59,60). In the present study, there was an
increase in the phosphorylation of ERK. The specific inhibitor
PD98059 also partially reversed EA-induced inhibition of cell
viability and migration. Furthermore, the ROS scavenger NAC almost
abolished the effect of EA on p-ERK and LC3 II/I. The in
vivo study also indicated that EA inhibited the growth of
hepatocellular carcinoma xenografts.
EL mainly contains flavonoids, sesquiterpenoids,
diterpenoids, triterpenoids, volatile oils, organic acids, amino
acids, trace elements and other bioactive substances, and has been
used for the clinical treatment of tracheitis. In recent years,
numerous scientists have focused on studying the role of EL in
cancer. EL extracts, including eupalinolide O, eupalinolide J and
other sesquiterpene lactones, have been reported to serve vital
roles in breast and prostate cancer (10,23,61).
In the present study, EA inhibited liver cancer in vitro and
in vivo. These findings indicated that EL and other similar
sesquiterpene lactones may be potential anticancer drugs. However,
the mechanisms of other sesquiterpene lactones in cancer should be
investigated further. EA is a promising candidate for treating
hepatocellular carcinoma, as the present study indicated that it
may induce cell autophagy via the ROS/ERK signaling pathway in
vitro and in vivo.
Supplementary Data
Availability of data and materials
The RNA sequencing datasets used and/or analyzed
during the current study are available from https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA805012.
The non-sequencing data are available from the corresponding author
on reasonable request.
Authors' contributions
GL, CZ and YZ conceived and designed the study. FD,
ZC, TW, LP, WL, LJ, HL and JL performed the experiments. WD, HZ,
JM, MH, YW, XD, DL and PH acquired, analyzed and interpretated the
data. GZ and YP conducted statistical analysis. GL, CZ and YZ
confirmed the authenticity of all the raw data. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The animal experiments were reviewed and approved
(approval no. 2022-SCSL-KY-001) by the medical ethics committee of
the Affiliated Hospital of Chongqing Three Gorges Medical College
(Chongqing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors thank Sagene eBioart for their help in
generating Fig. 9.
Funding
This work was supported by the Science and Technology Research
Program of Chongqing Municipal Education Commission (grant nos.
KJZD-K201902701 and KJQN201802702), the Chongqing Natural Science
Foundation of Chongqing Science and Technology Bureau (grant no.
cstc2019jcyj-msxmX0607), the Natural Science Research Program of
Chongqing Three Gorges Medical College (grant nos. 2018xzz01 and
2019XZZ004), the Chongqing Key Disciplines of Traditional Chinese
Medicine (Basic Theory of Traditional Chinese Medicine)
Construction Project (grant no. 16, 2021), the Young and
middle-aged Top-notch Talent Support Program of Chongqing Three
Gorges Medical College, Bayu scholar program, and Chongqing
University Innovation Group Project (grant no. CXQT20030).
References
|
1
|
Zheng RS, Zhang SW, Zeng HM, Wang SM, Sun
KX, Chen R, Li L, Wei WQ and He J: Cancer incidence and mortality
in China, 2016. J Nat Cancer Cent. 2:1–9. 2022. View Article : Google Scholar
|
|
2
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu CY, Chen KF and Chen PJ: Treatment of
liver cancer. Cold Spring Harb Perspect Med. 5:a0215352015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen S, Cao Q, Wen W and Wang H: Targeted
therapy for hepatocellular carcinoma: Challenges and opportunities.
Cancer Lett. 460:1–9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Llovet JM, Montal R, Sia D and Finn RS:
Molecular therapies and precision medicine for hepatocellular
carcinoma. Nat Rev Clin Oncol. 15:599–616. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Gao ZH and Qu XJ: The adverse
effects of sorafenib in patients with advanced cancers. Basic Clin
Pharmacol Toxicol. 116:216–221. 2015. View Article : Google Scholar
|
|
7
|
Heo YA and Syed YY: Regorafenib: A review
in hepatocellular carcinoma. Drugs. 78:951–958. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang X, Ma S, Lai F, Wang Y and Lou C:
Traditional applications, phytochemistry, and pharmacological
activities of Eupatorium lindleyanum DC.: A comprehensive review.
Front Pharmacol. 8:5771242020. View Article : Google Scholar
|
|
9
|
Chi D: Study on the antihypertensive
effects of water decoction of Eupatorium lindleyanum on
spontaneously hypertensive rats and corresponding mechanism. China
Pharm. 27:3502–3504. 2016.
|
|
10
|
Lou C, Chen Y, Zhang J, Yang B and Zhao H:
Eupalinolide J suppresses the growth of triple-negative breast
cancer cells via targeting STAT3 signaling pathway. Front
Pharmacol. 10:10712019. View Article : Google Scholar :
|
|
11
|
Peng Y, Dou J, Huang F and Qian D:
Antiviral effects of Fufang Yemazhui capsule in vitro and in vivo.
Chin Tradit Pat Med. 30:650–654. 2008.
|
|
12
|
Chen WY, Qin J, He HX, Li Q, Wang KJ and
Zhou YD: Effects of lindley eupatorium herb total flavonoid on
blood lipid metabolism in experimental hyperlipidemic rats. J Third
Mil Med Univ. 31:1589–1591. 2009.
|
|
13
|
Cu CJ, Ren HL and Wu TW: Study on the
chemical constituents and antipyretic action of Eupatorium
lindleyanum DC. Nat Prod Res Dev. 5:816–821. 2015.
|
|
14
|
Wang J, Wu M, Zheng D, Zhang H, Lv Y,
Zhang L, Tan HS, Zhou H, Lao YZ and Xu HX: Garcinol inhibits
esophageal cancer metastasis by suppressing the p300 and TGF-β1
signaling pathways. Acta Pharmacol Sin. 41:82–92. 2020. View Article : Google Scholar
|
|
15
|
Zhang YH, Pan LH, Pang Y, Yang JX, Lv MJ,
Liu F, Qu XF, Chen XX, Gong HJ, Liu D and Wei Y: GDF11/BMP11 as a
novel tumor marker for liver cancer. Exp Ther Med. 15:3495–3500.
2018.PubMed/NCBI
|
|
16
|
Yue SR, Tan YY, Zhang L, Zhang BJ, Jiang
FY, Ji G, Liu BC and Wang RR: Gynostemma pentaphyllum
polysaccharides ameliorate non-alcoholic steatohepatitis in mice
associated with gut microbiota and the TLR2/NLRP3 pathway. Front
Endocrinol (Lausanne). 13:8850392022. View Article : Google Scholar
|
|
17
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Li M, Li L, Qian G, Wang Y, Chen
Z, Liu J, Fang C, Huang F, Guo D, et al: β-arrestin 2 as an
activator of cGAS-STING signaling and target of viral immune
evasion. Nat Commun. 11:60002020. View Article : Google Scholar
|
|
20
|
Wang Y, Qian G, Zhu L, Zhao Z, Liu Y, Han
W, Zhang X, Zhang Y, Xiong T, Zeng H, et al: HIV-1 Vif suppresses
antiviral immunity by targeting STING. Cell Mol Immunol.
19:108–121. 2022. View Article : Google Scholar
|
|
21
|
Piñero F, Dirchwolf M and Pessôa MG:
Biomarkers in hepatocellular carcinoma: Diagnosis, prognosis and
treatment response assessment. Cells. 9:13702020. View Article : Google Scholar :
|
|
22
|
Yang B, Zhao Y, Lou C and Zhao H:
Eupalinolide O, a novel sesquiterpene lactone from Eupatorium
lindleyanum DC., induces cell cycle arrest and apoptosis in human
MDA-MB-468 breast cancer cells. Oncol Rep. 36:2807–2813. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang B, Shen JW, Zhou DH, Zhao YP, Wang
WQ, Zhu Y and Zhao HJ: Precise discovery of a STAT3 inhibitor from
Eupatorium lindleyanum and evaluation of its activity of
anti-triple-negative breast cancer. Nat Prod Res. 33:477–485. 2019.
View Article : Google Scholar
|
|
24
|
Bekki Y, Von Ahrens D, Takahashi H,
Schwartz M and Gunasekaran G: Recurrent intrahepatic
cholangiocarcinoma-review. Front Oncol. 11:7768632021. View Article : Google Scholar
|
|
25
|
Nijkamp MM, Span PN, Hoogsteen IJ, van der
Kogel AJ, Kaanders JH and Bussink J: Expression of E-cadherin and
vimentin correlates with metastasis formation in head and neck
squamous cell carcinoma patients. Radiother Oncol. 99:344–348.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alenzi FQ: Links between apoptosis,
proliferation and the cell cycle. Br J Biomed Sci. 61:99–102. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Johansson M and Persson JL: Cancer
therapy: Targeting cell cycle regulators. Anticancer Agents Med
Chem. 8:723–731. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Honda R, Lowe ED, Dubinina E, Skamnaki V,
Cook A, Brown NR and Johnson LN: The structure of cyclin E1/CDK2:
Implications for CDK2 activation and CDK2-independent roles. EMBO
J. 24:452–463. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol
Cancer. 14:482015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Denicourt C and Dowdy SF: Medicine.
Targeting apoptotic pathways in cancer cells. Science.
305:1411–1413. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Amaravadi RK and Thompson CB: The roles of
therapy-induced autophagy and necrosis in cancer treatment. Clin
Cancer Res. 13:7271–7279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Poillet-Perez L, Despouy G,
Delage-Mourroux R and Boyer-Guittaut M: Interplay between ROS and
autophagy in cancer cells, from tumor initiation to cancer therapy.
Redox Biol. 4:184–192. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park HS, Han JH, Park JW, Lee DH, Jang KW,
Lee M, Heo KS and Myung CS: Sodium propionate exerts anticancer
effect in mice bearing breast cancer cell xenograft by regulating
JAK2/STAT3/ROS/p38 MAPK signaling. Acta Pharmacol Sin.
42:1311–1323. 2021. View Article : Google Scholar :
|
|
35
|
He C, Lu S, Wang XZ, Wang CC, Wang L,
Liang SP, Luo TF, Wang ZC, Piao MH, Chi GF and Ge PF: FOXO3a
protects glioma cells against temozolomide-induced DNA double
strand breaks via promotion of BNIP3-mediated mitophagy. Acta
Pharmacol Sin. 42:1324–1337. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schieber M and Chandel NS: ROS function in
redox signaling and oxidative stress. Curr Biol. 24:E453–R462.
2014. View Article : Google Scholar
|
|
37
|
Delire B and Stärkel P: The Ras/MAPK
pathway and hepatocarcinoma: Pathogenesis and therapeutic
implications. Eur J Clin Invest. 45:609–623. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou SL, Zhou ZJ, Hu ZQ, Huang XW, Wang Z,
Chen EB, Fan J, Cao Y, Dai Z and Zhou J: Tumor-associated
neutrophils recruit macrophages and T-regulatory cells to promote
progression of hepatocellular carcinoma and resistance to
sorafenib. Gastroenterology. 150:1646–1658.e17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu F, Yang LY, Li YF, Ou DP, Chen DP and
Fan C: Novel role for epidermal growth factor-like domain 7 in
metastasis of human hepatocellular carcinoma. Hepatology.
50:1839–1850. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang YS, Shen Q and Li J: Traditional
Chinese medicine targeting apoptotic mechanisms for esophageal
cancer therapy. Acta Pharmacol Sin. 37:295–302. 2016. View Article : Google Scholar :
|
|
41
|
Liu Y, Yang S, Wang K, Lu J, Bao X, Wang
R, Qiu Y, Wang T and Yu H: Cellular senescence and cancer: Focusing
on traditional Chinese medicine and natural products. Cell Prolif.
53:e128942020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yamaguchi H, Wyckoff J and Condeelis J:
Cell migration in tumors. Curr Opin Cell Biol. 17:559–564. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chaffer CL, San Juan BP, Lim E and
Weinberg RA: EMT, cell plasticity and metastasis. Cancer Metastasis
Rev. 35:645–654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mendonsa AM, Na TY and Gumbiner BM:
E-cadherin in contact inhibition and cancer. Oncogene.
37:4769–4780. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target for cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Suski JM, Braun M, Strmiska V and Sicinski
P: Targeting cell-cycle machinery in cancer. Cancer Cell.
39:759–778. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Camplejohn RS: A critical review of the
use of vincristine (VCR) as a tumour cell synchronizing agent in
cancer therapy. Cell Tissue Kinet. 13:327–335. 1980.PubMed/NCBI
|
|
48
|
Kumar A, Sharma PR and Mondhe DM:
Potential anticancer role of colchicine-based derivatives: An
overview. Anticancer Drugs. 28:250–262. 2017. View Article : Google Scholar
|
|
49
|
Chen TK, Tang MS, Jiang CX and Zeng BX:
Effect of vincristine on proliferation and apoptosis of human
gastric cancer BGC cells and its mechanism. Chin Tradit Herb Drugs.
46:72015.
|
|
50
|
Pfeffer CM and Singh ATK: Apoptosis: A
target for anticancer therapy. Int J Mol Sci. 19:4482018.
View Article : Google Scholar :
|
|
51
|
Karsch-Bluman A, Feiglin A, Arbib E, Stern
T, Shoval H, Schwob O, Berger M and Benny O: Tissue necrosis and
its role in cancer progression. Oncogene. 38:1920–1935. 2019.
View Article : Google Scholar
|
|
52
|
Azad MB, Chen Y and Gibson SB: Regulation
of autophagy by reactive oxygen species (ROS): Implications for
cancer progression and treatment. Antioxid Redox Signal.
11:777–790. 2009. View Article : Google Scholar
|
|
53
|
Gibson SB: A matter of balance between
life and death: Targeting reactive oxygen species (ROS)-induced
autophagy for cancer therapy. Autophagy. 6:835–837. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang ZJ, Chee CE, Huang S and Sinicrope
FA: The role of autophagy in cancer: Therapeutic implications. Mol
Cancer Ther. 10:1533–1541. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pankiv S, Clausen TH, Lamark T, Brech A,
Bruun JA, Outzen H, Øvervatn A, Bjørkøy G and Johansen T:
p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of
ubiquitinated protein aggregates by autophagy. J Biol Chem.
282:24131–2445. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH,
Kam TI, Jung S and Jung YK: Overexpression of Atg5 in mice
activates autophagy and extends lifespan. Nat Commun. 4:23002013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Moloney JN and Cotter TG: ROS signalling
in the biology of cancer. Semin Cell Dev Biol. 80:50–64. 2018.
View Article : Google Scholar
|
|
59
|
Liu Y and Fan D: Ginsenoside Rg5 induces
G2/M phase arrest, apoptosis and autophagy via regulating
ROS-mediated MAPK pathways against human gastric cancer. Biochem
Pharmacol. 168:285–304. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fan J, Ren D, Wang J, Liu X, Zhang H, Wu M
and Yang G: Bruceine D induces lung cancer cell apoptosis and
autophagy via the ROS/MAPK signaling pathway in vitro and in vivo.
Cell Death Dis. 11:1262020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wu Z, Xu X, Dai L, Wang Y, Yang B, Zhao H
and Lou C: Eupalinolide J induces apoptosis, cell cycle arrest,
mitochondrial membrane potential disruption and DNA damage in human
prostate cancer cells. J Toxicol Sci. 45:15–23. 2020. View Article : Google Scholar : PubMed/NCBI
|