Introduction
Malignant gliomas are the leading cause of central
nervous system tumor-related deaths and affect more than 50% of
glioma patients, and patients who are affected by glioblastoma
multiforme (GBM) have a poor prognosis, with a mean life expectancy
of less than 2 years (1,2). As the overall survival of GBM
patients has not improved markedly in the last 20 years, the
current gold standard for GBM treatment is only palliative,
including surgery, adjuvant radiotherapy, and temozolomide (TMZ)
chemotherapy. One of the major hurdles in the development of novel
treatment regimens for GBM is the challenge of translating
scientific knowledge from bench to bedside, mainly owing to the
fact that most research models only poorly replicate the tumor
behavior and, consequently, numerous drugs that perform well in
glioma models ultimately fail in clinical trials.
To date, malignant glioma cultures, particularly
publicly available cell lines, such as those obtained from the
American Type Culture Collection (ATCC), represent a useful
experimental tool for studies on glioma formation and progression,
discovery of new antitumor strategies, and for the identification
of molecular markers in response to conventional chemotherapies and
targeted agents showing clinical utility. In a search for articles
associated with original glioma cell cultures, it was found that
cell lines bearing the prefix 'U', such as U251, U87, and U138,
were established at Uppsala University, Sweden (3,4), and
cell lines bearing the prefix 'LN', such as LN-18, LN-229, and
LN-464, were established at the Neurosurgical Service of the
University Hospital (CHUV) in Lausanne, Switzerland (5,6). In
1999, Ishii et al (7)
demonstrated the diagnoses of the original tumors, clinical
features of the patients, the mutation status of the associated
genes (TP53, PTEN, CDKN2A), and the
subcutaneous tumorigenicity of 34 randomly chosen human glioma cell
lines, and this was a well comprehensive study of glioma cell
lines. Not all of these cell lines are equally used in the present
study, and the 19 cell lines of the 34 formed subcutaneous tumor
masses in nude mice were used relatively more widespread (7). The tumorigenicity of these GBM cell
lines is firmly associated with their popularity in cancer research
studies for its advantages in vivo in nude mice, thus,
tumorigenic cell lines as U87 and U251 are more widely used than
nontumorigenic cell lines such as HS683 and A172 (7,8).
Although the aforementioned cell lines have been
established and widely used for more than 40 years, they have
several drawbacks that need to be considered. For example, in
vitro subculture imposes a selection pressure on cell lines,
which could result in genetic drift over time. In addition,
long-term cultures represent a risk of cross-contamination with
other cell lines. The most commonly used cell line, U87, from the
ATCC, which has been passaged worldwide for more than 50 years, was
found to be different from the original U87 line established in the
Uppsala laboratory in 1966 or any other laboratory-established
glioma cell line (7,8). Furthermore, U373 and U251 were first
established as two different cell lines, but recent short-tandem
repeat (STR) identification confirmed that they have the same
origin (https://web.expasy.org/cellosaurus/CVCL_0021;
https://web.expasy.org/cellosaurus/CVCL_2219). Thus,
numerous journals require cell line STR authentication and primary
cell culture to ensure the reliability of the studies. Therefore,
new cell lines are urgently required to replace those old,
long-term-use cell lines.
During the past few years, several adherent primary
cultures from freshly resected tissues of patients with different
grades of glioma have been routinely conducted; one permanent and
tumorigenic cell line without any additional genetic modification,
named GWH04, was established from a 72-year-old female patient with
GBM. This cell line is suitable for the common culture medium and
has been sub-cultured more than 70 times, and cryopreserved at
different early-passage stages. In the present study, the
establishment of this primary GBM cell line was described and its
biological characteristics including proliferation rate, karyotype
analysis, half maximal inhibitory concentration (IC50)
of different chemotherapy drugs, and glioma-associated gene test,
as well as the pathological and histological characteristics of the
original tumor from the patient, and the intracranial xenografts in
nude mice of this cell line, were presented. This GWH04 cell line
(first named Fu) was deposited at the Bio-research Innovation
Center of Suzhou (BRICS; http://www.brics.ac.cn/cell/template/cell/cell_detail.html?id=3825),
and also preserved in the China Center for Type Culture Collection,
Wuhan, China (CCTCC, NO.C202163). Altogether, it will contribute to
the diversity of GBM cell lines and will be a useful tool for
studies of human GBM both in vitro and in vivo for
all researchers.
Materials and methods
Clinical history and characteristics
In the present study, 10 cases of primary cell
cultures from different GBM tumor tissues were comprehensively
investigated. Among which, GWH04 cells could well circumvent
replicative senescence and acquire the ability to sustain unlimited
proliferation in this culture medium. The detailed information of
the patient from whom GWH04 cells were derived is presented
below.
The 72-year-old woman was admitted to the
Neurosurgery Department of Tongji Hospital in November 2016 with a
10-day history of headache and left limb weakness, and 4 days of
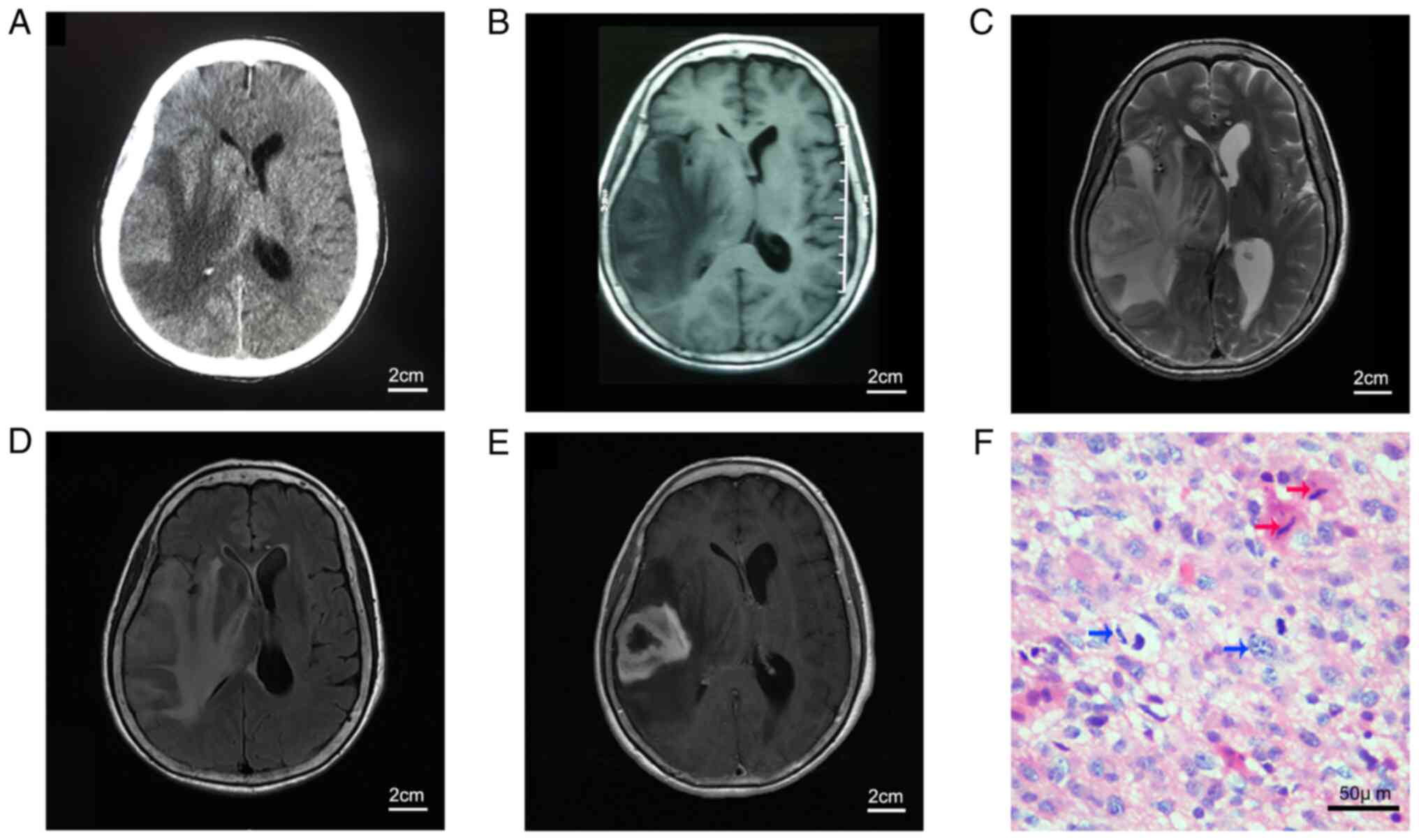
rapid aggravation since the initial onset. Computerized tomography
(CT) and magnetic resonance imaging (MRI) of the brain revealed a
lesion occupying the right temporal lobe with large areas of
surrounding edema (Fig. 1A-D).
Gadolinium-enhanced MRI of the head showed a solid, enhancing
lesion on the T1-weighted sequence, with the appearance of an
irregular ring (Fig. 1E). The
tumor was grossly microscopically resected during surgery, and a
histological diagnosis of GBM was made according to the 2007 World
Health Organization classification of tumors of the central nervous
system. However, the tumor recurred rapidly, and the patient died
within 3 months after surgery. Detailed information was given to
the relatives of the patient and informed consent was provided
prior to surgery. All samples were obtained from the Department of
Neurosurgery, Tongji Hospital, Huazhong University of Science and
Technology (Wuhan, China) after written informed consent was
obtained and according to the protocol approved (approval no.
TJ-IBR20210119) by the Research Ethics Committee of Tongji
Hospital, Tongji Medical College, Huazhong University of Science
and Technology. All procedures involving human samples were in
accordance with the 1964 Helsinki Declaration and its later
amendments or comparable ethical standards.
Tumor tissue was acquired during the surgery for
paraffin tissue embedding, liquid nitrogen cryopreservation, and
primary cell culture.
Immunohistochemistry and pathological
examination of glioma tissues
Immediately after resection, the tumor samples were
fixed in 4% paraformaldehyde, dehydrated with an ethanol gradient,
permeabilized with xylene, and embedded in paraffin (25-28°C).
Paraffin-embedded tissue sections (4-µm thick) were
generated and stained with hematoxylin and eosin (H&E) at
25-28°C (Fig. 1F). Continuous
tissue sections were generated and immunohistochemically (IHC)
stained for glial fibrillary acidic protein (GFAP; cat. no.
ZM-0118), NeuN (cat. no. ZM-0352), Olig2 (cat. no. ZA-0561),
O6-methylguanine-DNA methyltransferase (MGMT; cat. no. ZM-0461),
Nestin (cat. no. ZA-0628), EMA (cat. no. ZM-0071), Syn (cat. no.
ZA-0263), c-Myc (cat. no. ZA-0658), EGFR (cat. no. ZM-0093), p53
(cat. no. ZM-0408) and Ki67 (cat. no. ZM-0167; all from OriGene
Technologies, Inc.). Antigen retrieval was performed in a microwave
with citrate buffer (pH 6.0), and the inactivation of endogenous
peroxidase was performed in 3% H2O2. Bovine
serum album (3%; cat. no. 01010010610; GENVIEW) was added to evenly
cover the tissue and block non-specific antigens for 30 min at
25-28°C. Then, the slides were incubated with primary working
solution antibodies at 25-28°C for 2 h. Then, the sections were
incubated with biotin-labeled secondary antibody (working solution;
cat. no. DS-0004; OriGene Technologies, Inc.) at 25-28°C for 50 min
and the final signals were developed using 3,3′-diaminobenzidine
substrate (R&D Systems, Inc.). The sections were analyzed by
optical microscopy after counterstaining with hematoxylin. All
procedures and subsequent histological diagnoses were performed in
the Pathology Department of Tongji Hospital.
Primary cell culture and
authentication
The tumor specimen (<1 cm3) was
obtained shortly after surgery and rinsed twice with
phosphate-buffered saline (PBS) to wash out residual red blood
cells. The specimen was prepared as a single cell suspension by
mechanical dissociation (1-2 mm) with sterile scissors, treated
with 0.125% trypsin (5), and
agitated with a horizontal shaker (180 rpm) until the tissue
granules became smaller. All suspensions were passed through a
sterile filter with a 200-µm pore diameter and centrifuged
(300 × g, 5 min, 4°C). The cell pellets were resuspended in
complete medium [10% fetal bovine serum (FBS) + Dulbecco's modified
Eagle's medium (DMEM)] and seeded on a 25 cm2 culture
flask as an adherent culture in a humidified incubator with 5%
CO2 at 37°C.
During the first five passages, only half of the
medium was changed each time. When a monolayer of primary tumor
cells was formed and reached 90% confluence, they were passaged
after 0.25% trypsin digestion, and cultured at 1:3-1:5 dilution in
a new flask. After subsequent passages, as with the U251 and U87
cell lines, all stably passaged cells were viably frozen in medium
plus 10% dimethyl sulfoxide and stored at −80°C (5).
For authentication, DNA from cultured cells and
primary tumor tissue was isolated using an AxyPrep Multisource
Genomic DNA Miniprep kit (Axygen; Corning, Inc.), and 20 STR loci
were examined and compared with the corresponding STR profile of
the tumor tissue. After bacterial and mycoplasma contamination
tests were confirmed negative, the GWH04 cell line (first named as
Fu) was deposited at BRICS, and also preserved in the CCTCC.
Immunofluorescence assay of GWH04
cells
Single cell suspensions of GWH04 were seeded into
24-well plates with round glass coverslips. After 24 h, the cells
were fixed with 4% paraformaldehyde at room temperature (25-28°C)
for 15 min, permeabilized with 0.25% Triton X-100 for 5 min, and
blocked with goat serum (5%, cat. no. WGAR1009-5; Wuhan Servicebio
Technology Co., Ltd.) for 1 h. Primary antibodies against GFAP
(1:1,000; cat. no. 3670; Cell Signaling Technologies, Inc.) and
Nestin (1:5,000; cat. no. 2280493; MilliporeSigma) were added and
incubated overnight at 4°C. Cells were rinsed with PBS, and
fluorescence-labeled secondary antibodies (1:100; cat. nos. GB21303
and GB25303; Wuhan Servicebio Technology Co., Ltd.) were added and
incubated at room temperature for 1 h. The cells were then stained
with DAPI (at 25-28°C) for 5 min, rinsed with PBS, sealed with a
mounting medium, and analyzed using a fluorescence microscope
(Olympus Corporation). To compare GFAP levels, cells cultured from
pilocytic astrocytoma tissue (grade I), grade II and grade III
gliomas were also stained for GFAP under similar staining
conditions as those used for GWH04 cells.
Cell lines of U87 and GL15
U87 cell line was purchased from the ATCC with
unknown origin (HTB-14), GL15 cell line was donated by Dr Håkan
Hedman, Umeå University Hospital, Sweden. Both of these two cell
lines were cultured in DMEM/high glucose supplemented with 10% FBS
(cat. no. 04-001; BioLegend, Inc.), 100 U/ml penicillin, and 0.1
mg/ml streptomycin in a humidified incubator with 5% CO2
at 37°C. The STR identification of the U87 used in the present
study was conducted in BRICS institution, and was in consistent
with the information presented in the website (https://web.expasy.org/cellosaurus/CVCL_0022).
Cell proliferation and soft agar colony
formation assay
Cells (U87, GL15 and the 10, 20, 50th passages of
GWH04) were seeded onto 96-well plates at 2×103 cells
per well in 100 µl suspensions in triplicate per day and
maintained in complete culture medium for 5 days. The cell
proliferation rate was measured using the Cell Counting Kit-8 assay
(CCK-8; cat. no. KJ800; Dojindo Laboratories, Kumamoto, Japan)
according to the manufacturer's instructions. At the time points of
attachment (day 0), 24, 48, 72, 96, and 120 h, 10 µl CCK-8
assay reagent was added to each well with 90 µl culture
medium and incubated for 1 h at 37°C. The absorbance was measured
with a microplate reader using a wavelength of 450 nm.
For soft agar assays, 0.5% Basal Medium Eagle (BME)
agar, containing 10% FBS, 2 mM L-glutamine, and 25 µg/ml
gentamicin, was added to six-well plates (3 ml/well), with
triplicates per cell line. The plates were then placed in an
incubator for 1 h to coagulate the semi-solid medium. Subsequently,
GWH04 cells were resuspended at a concentration of
9×103/ml in 0.33% BME agar containing 10% FBS, and
seeded at 1 ml/well. The cultures were maintained at 37°C in a 5%
CO2 incubator for 14 days, and the cell colonies (>25
µm) of bright field were observed via microscopy. The number
of colonies and their relative diameters were manually recorded and
analyzed.
IC50 tests
In the present study, TMZ (cat. no. HY-17364),
lomustine (CCNU; cat. no. HY-13669), irinotecan (SN-38; cat. no.
HY-13704), etoposide (VP-16; cat. no. HY-13629), 5-fluorouracil
(5-Fu; cat. no. HY-90006), oxaliplatin (cat. no. HY-17371),
gefitinib (cat. no. HY-50895), BIBR1532 (cat. no. HY-17353),
olaparib (cat. no. HY-10162) were selected for drug sensitivity
tests (all purchased from MedChemExpress). A total of 8-11
concentration gradients were set of each drug according to the
results of pre-experiments (TMZ: 1, 10, 20, 40, 80, 160, 320, 640,
1,280 and 2,560 µM; CCNU: 1, 2, 4, 8, 16, 32, 64, 128 and
256 µM; SN-38: 0.001, 0.01, 0.1, 1, 2, 5, 10, 20, 40, 80 and
160 nM; VP-16: 0.1, 1, 2, 5, 10, 20, 40 and 80 µM; 5-Fu:
0.1, 1, 2, 5, 10, 20, 40, 80, 160 and 320 µM; oxaliplatin:
0.01, 0.1, 1, 5, 10, 20, 40, 80, 160 and 320 µM; gefitinib:
0.1, 0.5, 1, 2, 5, 10, 20, 40, 80 and 100 µM; BIBR1532: 0.1,
1, 5, 10, 20, 40, 80, 160 and 320 µM; olaparib: 0.1, 1, 2,
5, 10, 20, 40, 80, 160 and 320 µM). GWH04, U87 and GL15
cells were seeded onto 96-well plates at 3×103 cells per
well in 100 µl suspensions in triplicate for each drug
concentration and maintained in complete culture medium for 24 h.
Then, the culture medium was changed with culture mediums
containing different drug concentrations and maintained for 5 days.
The culture medium was changed on the third day. Subsequently, cell
viability was assessed using CCK-8 according to the manufacturer's
instructions. Dose-response curves were plotted and the
IC50s were calculated using GraphPad Prism 8.
Detection of isocitrate dehydrogenase
(IDH mutational status), 1p/19q codeletions, MGMTp methylation,
telomerase reverse transcriptase (TERT) promoter mutation, and
BRAFV600E mutation
DNA from cultured GWH04 cells and liquid
nitrogen-frozen tumor tissues of the patient were extracted using
the QIAamp DNA mini kit (Qiagen GmbH) according to the
manufacturer's instructions. Next-generation sequencing was
conducted to analyze the mutational status of IDH1/2, 1p/19q
codeletions, TERT promoter mutation (C228T or C250T), and BRAF
mutation (V600E). As pyrosequencing is a highly accurate method to
analyze the changes at one or more CpG sites in the methylated
sequence, the detection of the methylation status of the MGMT
promoter region of GWH04 cells was conducted by Genetron (Beijing,
China) using PyroMark Q24 sequencer (Qiagen China Co., Ltd.), which
includes a complete software package for analyzing CpG site
methylation and built-in controls for confirming the completeness
of bisulfite processing.
Determination of glioma cell
tumorigenicity
A total of 5 male and 5 female BALB/c athymic nude
mice (age, 4-5 weeks; weight 19-21 g) were purchased from Hunan
Silaike Jingda Laboratory Animal Co. (Changsha, China) and housed
under specific pathogen-free conditions in a temperature- and
humidity-controlled environment (room temperature, 20-26°C;
humidity, 40-60%; 12-h light/dark cycle; free access to food and
water). All procedures in the animal experiments were approved
(approval no. TJ-A20161206) by the Ethical Committee of Tongji
Hospital, Tongji Medical College, Huazhong University of Science
and Technology (Wuhan, China) and were performed in accordance with
ARRIVE guidelines (https://arriveguidelines.org). The health and behavior
of the mice were monitored five days once. First, GWH04 cells were
harvested and injected subcutaneously into the right axilla of each
mouse (3×106 cells/mouse) to test the tumorigenicity. On
day 60, one mouse was found dead due to the tumor growth, and the
other tumor-burdened nude mice were euthanized through
intraperitoneal anesthesia of an overdose of pentobarbital sodium
(150 mg/kg). The death of the mice was verified by the presentation
of breath cessation and loss of heartbeat. Subsequently, the
intracranial tumorigenic capacity was assessed in 5 female mice as
this cell line was derived from a female patient. A short
longitudinal incision was made in the scalp to expose the calvarium
after anesthesia (pentobarbital sodium, intraperitoneal injection,
50 mg/kg). Then, a burr hole was made 0.5 mm posterior to the
bregma and 1.5 mm to the right of the sagittal suture. A Hamilton
syringe was introduced at a depth of 2 mm below the brain surface,
and 5×105 tumor cells (stably expressing luciferase)
were slowly injected into the brain. When the mice began to lose
weight and slow down actions, bioluminescence imaging indicated a
definite lesion in 3 mice. On day 60, the three mice were
euthanized as aforementioned and the brain tissues were surgically
harvested, measured, fixed in 4% paraformaldehyde overnight at
25-28°C, and embedded in paraffin. H&E and IHC staining (for
GFAP, Nestin, EMA, EGFR, CD31 and Ki67) was conducted, and the
results were compared with those for the respective stained patient
tumor tissue.
Whole-exome sequencing (WES) of GWH04
cells
DNA from GWH04 cells (15th passage) was extracted as
aforementioned for high-throughput next-generation sequencing.
Library construction and whole-exome capture of genomic DNA were
performed using the SureSelectXT Human All Exon V6 (Agilent
Technologies, Inc.) and MGIEasy Exome Universal Library Prep
Set-V1.0. The captured DNA was sequenced on an MGISEQ 2000 platform
(BGI-Shenzhen, China) with 150-bp paired-end sequencing. The human
genome data of Hg19 from the UCSC Genome Browser (http://genome.ucsc.edu/) was used as a reference, and
common mutated genes in GBM patients were analyzed in the cell
line.
Chromosome karyotype analysis
Harvesting for metaphase chromosome preparations was
performed by treating exponential growth phase cells with 0.25
µg/ml colchicine for 3 h. Then, enzymatic dispersal was
performed with trypsin/EDTA, and the preparation was centrifuged at
1,000 × g for 6 min (25-28°C). Preheated 0.075 mM KCl hypotonic
solution was added for 15 min at 37°C and fixed with 3:1
methanol-acetic acid (v:v; 25-28°C; 1 min). The pellets were fixed
twice at room temperature (25-28°C) for 10 min, and 500 µl
stationary liquid was added to resuspend the cells. Afterward, cell
droplets were spotted onto clean microscope glass slides that had
been soaked in iced water. The slides were immediately heated over
an alcohol lamp for 2 sec to allow the chromosomes to spread out.
Chromosome specimens were stained with Giemsa (cat. No. BA4219,
Baso Diagnostics, Inc.) for 10 min at 25-28°C according to the
manufacturer's instructions, and the chromosome numbers of M phase
cells were counted using an oil immersion lens (×100) under a
microscope (Nikon Corporation). Karyotypes were determined by
arranging all the photographed metaphases. The chromosomes were
classified according to the International System for Human
Cytogenetic Nomenclature (9).
Statistical analysis
All experiments were repeated at least twice with
consistent results. The differences of the growth rate on day 5 of
the cells, as well as the differences of the colony numbers and the
relatively diameters of the cells (GWH04, U87 and GL15) were
analyzed by one-way analysis of variance followed by a post hoc
test (Dunnett's; Fig. 3A, C and
D). All of these statistical analyses were performed using SPSS
Statistics version 19 (IBM Corp.) and the graphs were drawn using
GraphPad Prism 8 (GraphPad Software, Inc.). Alpha level was 0.05 in
the present study. P<0.05 was considered to indicate a
statistically significant difference. Dose-response curves of
different drugs and the analysis of the corresponding
IC50s were also performed using GraphPad Prism 8
(Fig. 3E).
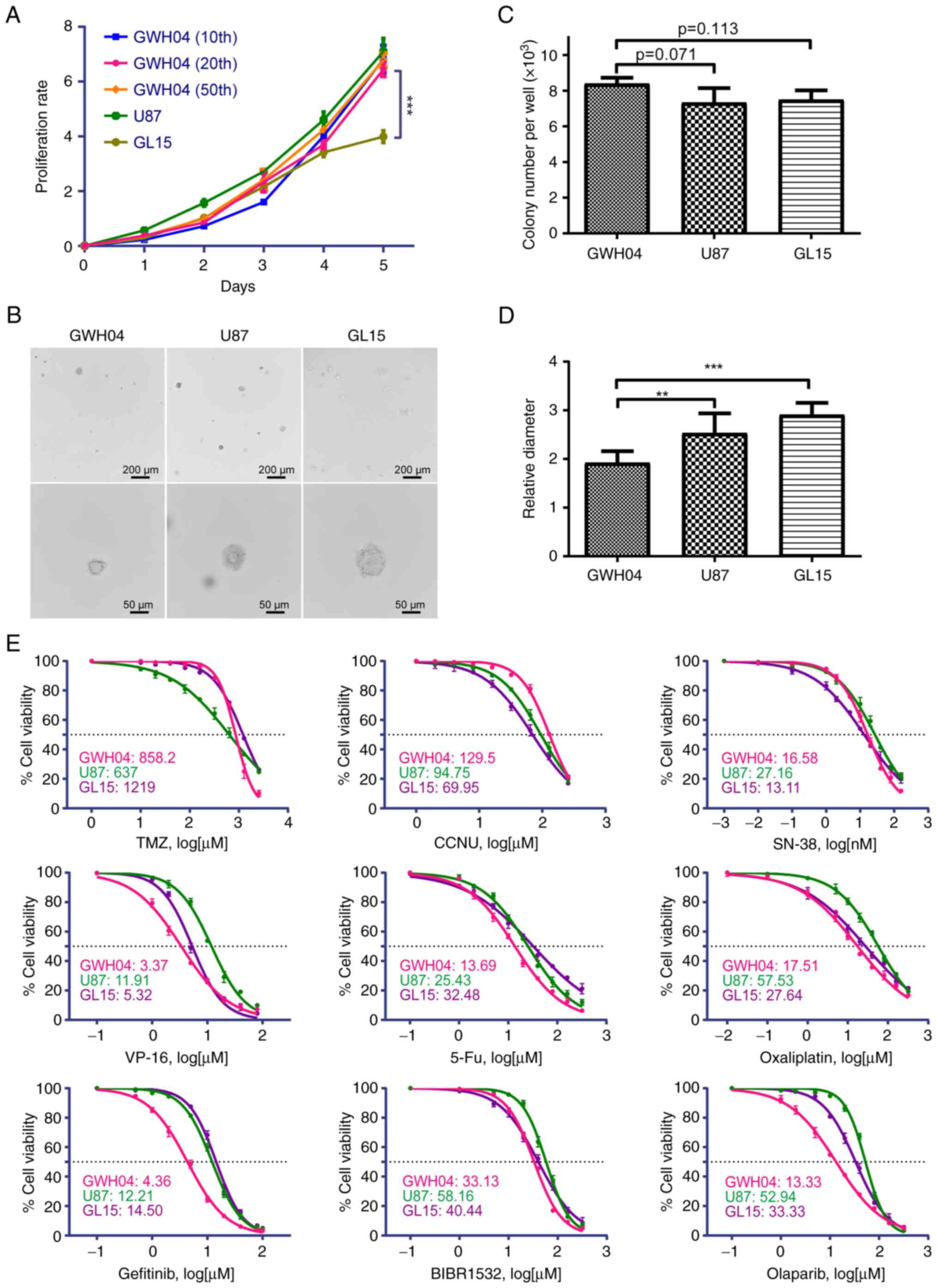 | Figure 3Proliferation rate,
anchorage-independent growth capacity and chemotherapy sensitivity
of several drugs of GWH04. (A) Analysis of proliferation rates with
Cell Counting Kit-8 in GWH04 (the 10, 20 and 50th passages), U87
and GL15 cells. (B) Anchorage-independent proliferation with colony
formation assays of GWH04, U87 and GL15 cells. Representative
images of soft agar colonies in each cell line are presented (Scale
bars, upper panel, 200 µm; lower panel, 50 µm). (C)
Quantitative analysis of colony numbers per well of each group (n=3
independent wells per cell line; data represent the mean ± SD of
three independent experiments). (D) Quantitative analysis of
diameters of 10 different clones showed that GWH04 cells formed
significantly smaller clones than that of U87 and GL15 cells (n=10
biologically independent colonies per cell line).
**P<0.01 and ***P<0.001. (E) The
corresponding dose-response curves of GWH04, U87 and GL15 cells
exposed to increasing concentrations of different drugs, and the
IC50s of each drug were presented. IC50, half
maximal inhibitory concentration. |
Results
Primary culture and identification of
GWH04 cells
CT and MRI scans of the brain tumor and H&E
staining of the paraffin-embedded tumor tissue of the patient are
shown in Fig. 1. The
histopathological diagnosis was GBM, based on the H&E staining,
which showed numerous nuclear atypia, mitoses, and microvascular
proliferation and different IHC staining items (Fig. S1). During the culture, GWH04 cells
began to proliferate significantly faster in the 7th generation,
and cell morphology was intended to be stable, as shown in passage
10 (P10) and passage 50 (P50), compared with passage 1 (P1) and 2
(P2) (Fig. 2A). Immunofluorescence
staining was performed in the 10th generation, showing that GWH04
cells were positive for GFAP and Nestin expression (Fig. 2B). However, this GFAP expressed in
GWH04 cells is markedly blurry even with longer exposure times when
compared with GFAP expression in cultured cells digested from grade
I-III gliomas (Fig. S2), despite
these grade I-III glioma cells couldn't circumvent replicative
senescence in the later culture. As GFAP is a marker of
differentiated astrocytes and Nestin is a marker of neuroepithelial
stem cells, GWH04 is a GBM cell line derived from dedifferentiated
glial cells. A previous STR study of 482 different human tumor cell
lines used in China showed that up to 96 cell lines were
misidentified (10), and Nature
research journals, AACR publications, and certain other scientific
publishers currently require cell identification based on DNA
analysis of the samples and cell lines used. In the present study,
STR authentications of 20 locations (covering the 9 loci demanded
by the ATCC) from the GWH04 cells and the corresponding tumor
tissue were tested and a complete match was observed (Table I; Fig.
2C). Moreover, the STR profile differed from that of all cell
lines available in different cell banks (http://cellresource.cn/str/default.aspx).
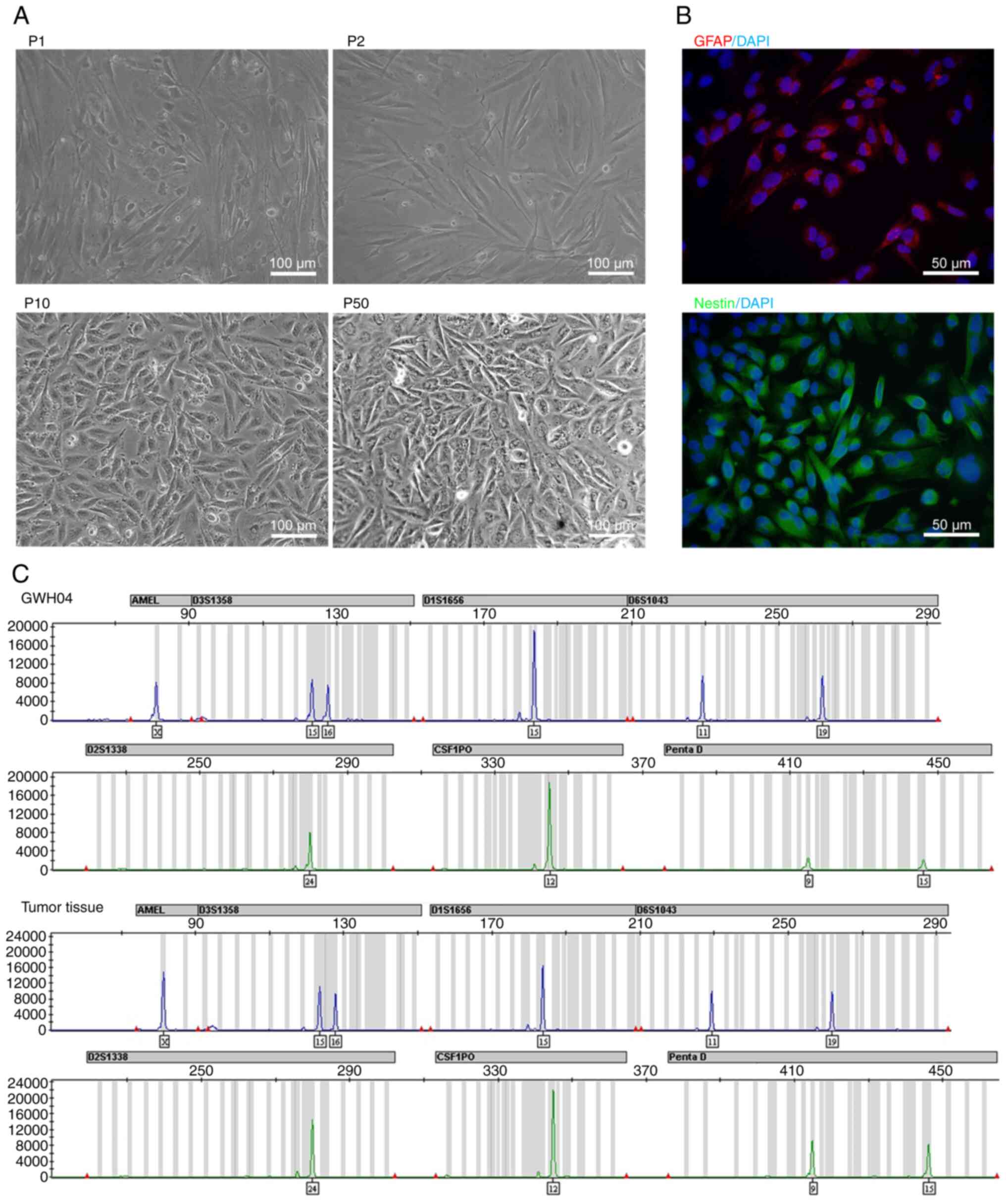 | Figure 2Bright field morphology,
immunofluorescent staining and STR identification of GWH04. (A)
Bright field morphology of GWH04 cells in the 1st, 2nd, 10th and
50th passage (P1, P2, P10, P50; scale bars, 100 µm). (B) The
immunofluorescence staining of GFAP and Nestin. DAPI is a nuclear
dye used as counterstain in immunofluorescence (magnification,
×400; scale bars, 50 µm). (C) Part of STR loci
identification map of GWH04 cells and the corresponding tumor
tissue. STR, short-tandem repeat. |
 | Table IInformation of 20 STR loci of GWH04
cells and the corresponding tumor tissue. |
Table I
Information of 20 STR loci of GWH04
cells and the corresponding tumor tissue.
| STR loci | GWH04 | Tumor tissue from
patient |
|---|
| Amelogenin | X, X | X, X |
| D3S1358 | 15, 16 | 15, 16 |
| D1S1656 | 15, 15 | 15, 15 |
| D6S1043 | 11, 19 | 11, 19 |
| D13S317 | 9, 9 | 9, 9 |
| PentaE | 12, 18 | 12, 18 |
| D16S539 | 10, 12 | 10, 12 |
| D2S1338 | 24, 24 | 24, 24 |
| CSF1PO | 12, 12 | 12, 12 |
| PentaD | 9, 15 | 9, 15 |
| TH01 | 7, 9 | 7, 9 |
| vWA | 14, 17 | 14, 17 |
| D21S11 | 30, 30 | 30, 30 |
| D7S820 | 11,12 | 11,12 |
| D5S818 | 12, 13 | 12, 13 |
| TPOX | 8, 9 | 8, 9 |
| D8S1179 | 12, 13 | 12, 13 |
| D12S391 | 22, 23 | 22, 23 |
| D19S433 | 14, 14 | 14, 14 |
| FGA | 22, 23 | 22, 23 |
Genetic diagnoses of GWH04 cells
According to the 2016 World Health Organization
Classification of Tumors of the Central Nervous System (11), genetic diagnosis is recommended to
identify the mutational status of IDH1/2, TERT
promoter, and BRAF, as well as 1p/19q co-deletions and MGMT
promoter region methylation, which can help to predict prognosis
and decide direct treatment options. In the present study, the
GWH04 cells were harvested (P7) and the DNA was extracted for
detection. The results were presented in Table II. The current consensus is that
MGMT expression is inhibited by the methylation of MGMT promoter,
and lower MGMT expression is associated with the TMZ-sensitivity.
However, the CpG islands of the MGMT promoter were found to
be unmethylated in this cell line, which results in the
TMZ-resistance. Moreover, the TERT promoter region harbored
a C228T mutation, which is frequently mutated and considered as a
driver gene of GBM. Thus, the mutation status of Table II in this case indicates a poor
prognosis of the patient and usually calls for a more aggressive
treatment.
 | Table IIResults of WHO recommended genetic
testing for gliomas. |
Table II
Results of WHO recommended genetic
testing for gliomas.
| Tested items | Results |
|---|
| MGMT
promoter methylation | Unmethylated |
| 1p deletion | Intact |
| 19q deletion | Deleted |
| IDH1 R132
mutation | No mutation |
| IDH2 R172
mutation | No mutation |
| TERT
promoter C228T mutation | No mutation |
| TERT
promoter C250T mutation | Mutated |
| BRAF V600E
mutation | No mutation |
With the identification of these most commonly
mutated genes associated with the formation of gliomas at GWH04
establishment, this cell line could be a useful tool in GBM
research for the discovery of new antitumor compounds and the
assessment of drug sensitivity, resistance, and toxicity
biomarkers, as well as the identification of targeted agents
showing clinical utility.
Cell proliferation and soft agar colony
formation assays of GWH04 cells
The proliferation rate and tumorigenicity are basic
features of primary cell lines, which are closely related to their
popularity in cancer research. CCK-8 assays of GWH04 cells (the 10,
20 and 50th passage) were performed to detect the proliferation
rates of different passages, which was compared with that of U87
and GL15 cells. During the first 2 days, GWH04 cells grew slower
than U87 and GL15 cells, but this rate rapidly increased in the
following 3 days, indicating that the growth of GWH04 cells is more
density-dependent (Fig. 3A). The
growth rates at the fifth day were of no significance among
different passages of GWH04 cells and U87 cells (n=3; 10th GWH04
vs. U87, P=0.364; 20th GWH04 vs. U87, P=0.054; 50th GWH04 vs. U87,
P=0.365), but were significantly faster of different passages of
GWH04 cells than GL15 cells (n=3; 10th GWH04 vs. GL15, P<0.001;
20th GWH04 vs. GL15, P<0.001; 50th GWH04 vs. GL15, P<0.001).
Soft agar colony formation assays can preliminarily judge the
tumorigenicity of cells in vitro. Thus, to examine the clone
formation capacity of GWH04 cells, parallel experiments were
conducted with U87 and GL15 cell lines. The assays showed that the
colony formation number of GWH04 cells was slightly higher than
that of U87 and GL15, but was not significantly different (Fig. 3B and C; GWH04 vs. U87, P=0.071;
GWH04 vs. GL15, P=0.113). By contrast, the average diameter of
GWH04 cells was significantly smaller than that of U87 and GL15
cells (Fig. 3B and D; GWH04 vs.
U87, P=0.004; GWH04 vs. GL15, P<0.001), indicating that this
newly established cell line possesses a higher clone formation
capacity, but a lower proliferation rate of anchorage-independent
growth than U87 and GL15 cells.
IC50 tests of different
drugs
TMZ is the first line chemotherapy strategy for GBM
treatment in recent decades (12),
and CCNU has been demonstrated to provide a benefit for GBM
patients in terms of both progression-free survival and overall
survival, despite increased adverse events (13,14).
Numerous patients with advanced GBM may have treatment with
bevacizumab, irinotecan, etoposide (VP-16), 5-Fu, and cisplatin as
the second line or the rescue treatment strategy, even though not
all pre-clinical studies have presented satisfactory therapeutic
results (15-18). As the amplification and activation
of EGFR are the most commonly RTK aberrations in GBMs (19), and GWH04 cells possess TERT
promoter mutation and BRCA1 mutation (presented below), gefitinib
(EGFR inhibitor), BIBR1532 (telomerase inhibitor) and Olaparib
(PARP inhibitor, which shows standalone activity against
BRCA1-deficient breast cancer cell lines) were also selected.
In the present study, GWH04, U87 and GL15 cells were
simultaneously exposed to TMZ, CCNU, SN-38 (an active metabolite of
irinotecan), VP-16, 5-Fu, oxaliplatin, gefitinib, BIBR1532 and
Olaparib. The growth rate inhibition metrics with each condition
after 5 days of drug exposure and fitted dose-response curves of
different drugs, as well as the IC50s, are presented in
Fig. 3E. When compared with U87
and GL15, GWH04 appears to be more sensitively to VP-16, 5-Fu,
oxaliplatin and olaparib in vitro (Fig. 3E). The IC50 of GWH04
cells is 858.2 µM treated with TMZ in vitro, which
indicates definite TMZ-resistance when compared with T98G cells
(800 µM) (20).
Furthermore, the unmethylated promoter region of MGMT also
indicated resistance to TMZ treatment. Thus, GWH04 is a primary
TMZ-resistant cell line.
Subcutaneous and intracranial
tumorigenicity
Not every cell line can form xenograft tumors in
nude mice; for example, U251 and U87 cells are tumorigenic, but
HS683 and A172 are not (7,8). Indeed, in a study of 34 randomly
selected human glioma cell lines, only 19 (56%) were able to
generate tumors in immunocompromised mice (7). Combined with the observation that
only ~10% of GBMs will generate permanent/immortal cell lines in
vitro when cultured in standard medium (7), it is clear that tumorigenicity in
immunocompromised mice and immortalization in vitro are
distinct phenotypes.
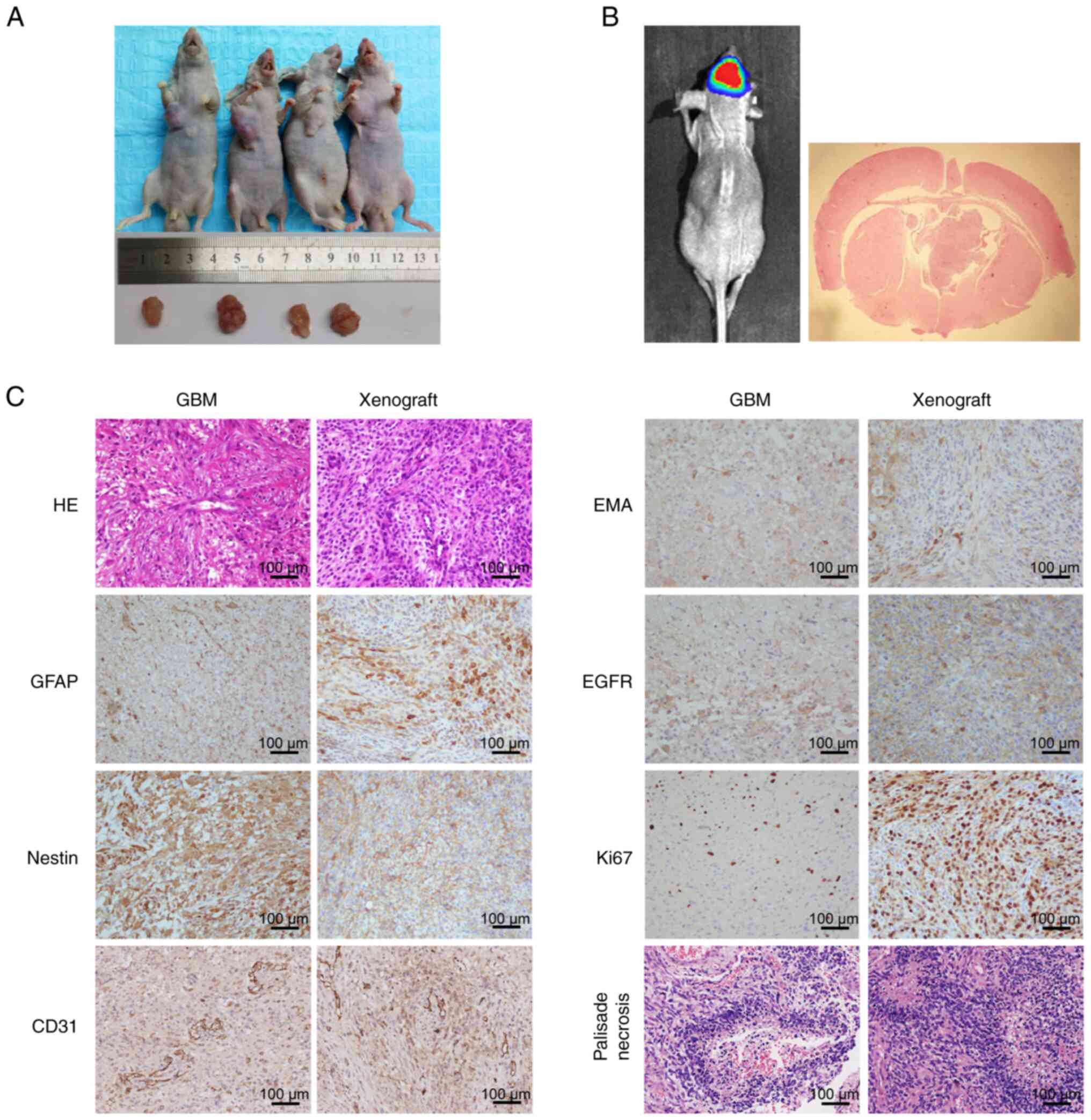
To determine whether GWH04 cells possess tumorigenic
capacity in vivo, subcutaneous and intracranial injection
were conducted. The subcutaneous tumor-burdened nude mice and the
harvested tumors on day 60 were presented in Fig. 4A, which confirmed the well
tumorigenic capacity of this cell line. For intracranial injected
mice, when obviously poor general conditions were detected, the
bioluminescence imaging indicated a definite lesion in 3 mice.
Then, the mice were euthanized, the brain tissue was surgically
harvested and analyzed by H&E and IHC staining (Fig. 4B and C). Both mouse intracranial
xenografts and primary tumor tissues of the patient positively
expressed GFAP, Nestin, EMA, EGFR, Ki67 and CD31 (Fig. 4C). Ki67 expression indicated that
the proliferation status was markedly more active in the mouse
intracranial xenograft than in the primary tumor of the patient
(Fig. 4C), probably owing to the
fact that in vitro culture enriched the cells with stronger
proliferative ability. The histological features of microvascular
hyperplasia and palisade necrosis were of highly consistence
between the GBM tissue of the patient and the tumorigenic tissue in
nude mice (Fig. 4C). These results
indicated the culture system faithfully recapitulated and
maintained the expression status of the primary tumor and also
indicate the substantial tumorigenicity of GWH04 cells. In
conclusion, GWH04 cell line will be a useful tool for both in
vitro and in vivo GBM research.
Mutated genes associated with
tumorigenicity in GWH04 cells
Apart from the several genetic factors recommended
for analysis to support diagnosis and treatment, numerous other
gene mutations are firmly related to the occurrence and development
of gliomas. Ishii et al (7)
studied tumor suppressor genes, namely TP53, p16,
p14ARF, and PTEN, in 34 randomly selected human
glioma cells and found that mutations and deletions occurred at the
following frequencies: TP53 (76.5%), p14ARF (64.7%),
p16 (64.7%) and PTEN (73.5%). Another study
demonstrated that ~86% of GBM samples harbor at least one genetic
event in the core RTK/PI3K pathway, such as in EGFR,
ERBB2, PDGFRA, or MET (21). To investigate the mutational status
of these important genes and other associated genes in GWH04 cells,
WES was performed. GWH04 cells were found to possess mutated
TP53, PTEN, PDGFRA, ERBB2,
BRCA1, NF1, and MLH1, and wild-type
CDKN2A, PIK3R1, PIK3CA, Rb1 and
MET. The sequencing and gene mutation data are shown in
Table III.
 | Table IIIList of certain mutated genes based
on whole-exome sequencing of GWH04 cells. |
Table III
List of certain mutated genes based
on whole-exome sequencing of GWH04 cells.
| Gene | Location | Mutation type | Transcript |
|---|
| TP53 | 17p13.1 |
missense_variant |
ENST00000269305:p.Tyr205Cys/c.614A>G |
| PTEN | 10q23.31 | stop_gain |
ENST00000371953:p.Tyr315*/c.945T>A |
| PDGFRA | 4q12 |
missense_variant |
ENST00000257290:p.Ser478Pro/c.1432T>C |
| ERBB2 | 17q12 |
missense_variant |
ENST00000269571:p.Ser423Gly/c.1267A>G |
| BRCA1 | 17q21.31 |
missense_variant |
ENST00000471181:p.Glu1038Gly/c.3113A>G |
| NF1 | 17q11.2 | stop_lost |
ENST00000471572:p.Ter640Argext*?/c.1918T>C |
| MLH1 | 3p22.2 |
missense_variant |
ENST00000231790:p.Val384Asp/c.1151T>A |
Chromosome karyotype of GWH04 cells
In addition to heterogeneity in the biological and
molecular features of GBM, chromosomal instability (CIN) is a
frequently occurring event in cancer that involves numerical
abnormalities of chromosomes and large-scale structural
alterations. Previous studies have demonstrated that GBM cell lines
have hyperdiploid karyotypes and exhibit glioma-specific
chromosomal abnormalities, such as gain of chromosome 7 and loss of
chromosome 10 (4,9). The range of possible chromosome
numbers of certain GBM cell lines based on previous studies was
compiled and presented in Table
IV.
 | Table IVAbnormal chromosome numbers in glioma
cell lines from previous studies. |
Table IV
Abnormal chromosome numbers in glioma
cell lines from previous studies.
| Cell line | Number of
chromosomes | Number of
karyotyped cells | (Refs.) |
|---|
| GL15 | 75-90 | 20 | (9) |
| GL22 | 56-73 | 10 | (9) |
| K308 | 54-108 | 100 | (22) |
| ANGM-CSS | 88-91 | - | (23) |
| SHG140 | 55 | 1 | (24) |
| LN18 | 70-80 | - | (5) |
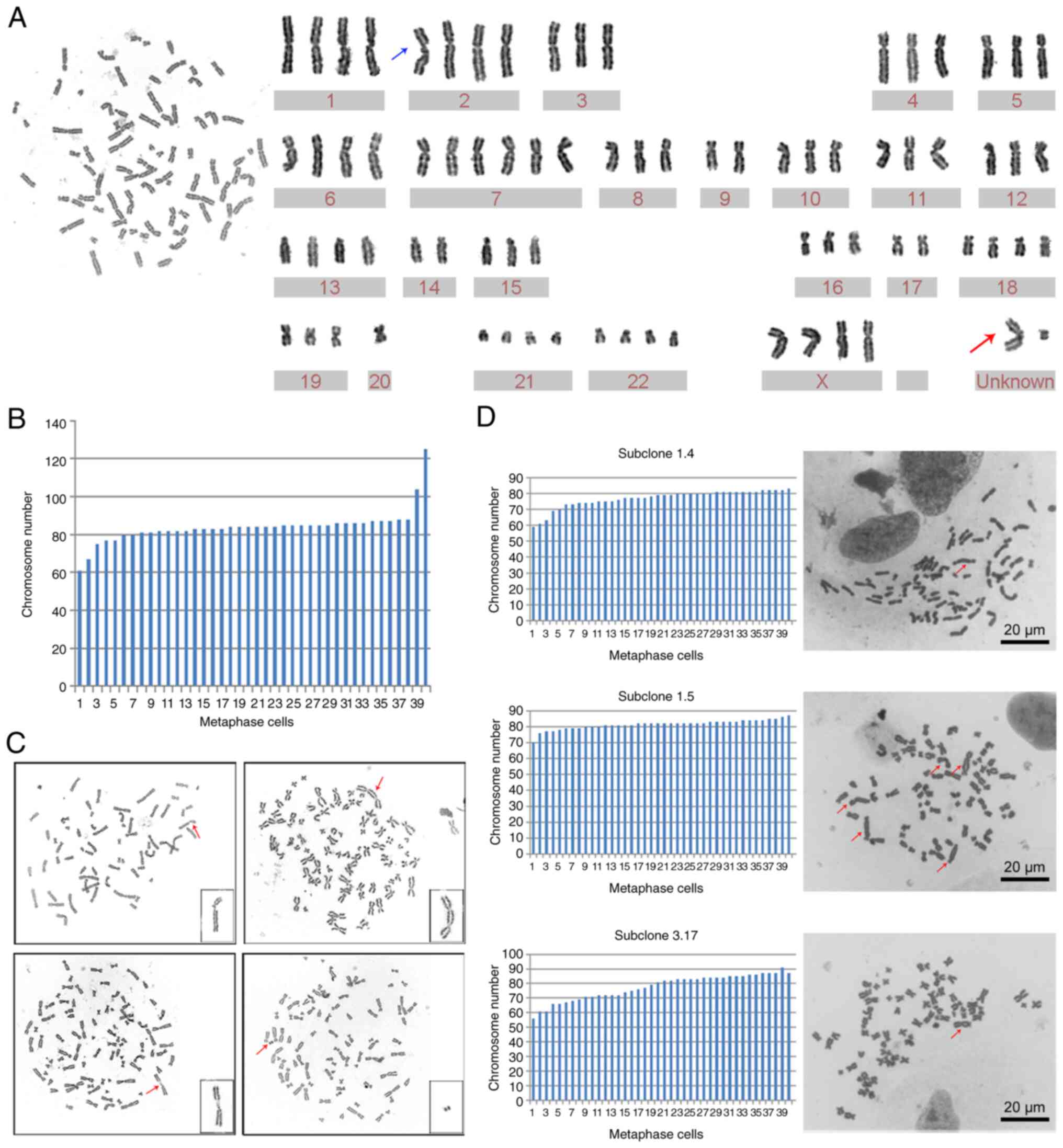
To determine whether GWH04 cells had CIN, the 10th
passage cells were harvested for G-banding karyotype analysis and
G-banding metaphase images were obtained. The chromosomal number
ranged from 61 to 125 while counting 40 karyotypes, and the first
and third quartiles were 82 and 86, respectively (Fig. 5A). Chromosome karyotype pairing
showed that extra copies of different chromosomes were rather
common, and aberrantly structured chromosomes presented with two
unknown chromosomes in one karyotype analysis (Fig. 5B). Considering that reported glioma
cell lines are all hyperdiploid (5,9,22,23),
it was concluded that this is a common phenomenon in glioma and
glioma cell lines. In addition to chromosome number abnormalities,
chromosomal structural abnormalities, such as broken chromosomes,
double centromere chromosomes, homogeneous staining regions (HSR),
and double microbodies, were presented in other karyotype analyses
of GWH04 cells (Fig. 5C). These
changes in the overall chromosomal configuration expand our
understanding of the CIN and heterogeneity of tumor cells.
Subclone analysis
The karyotype analysis of GWH04 cells at the 10th
passage demonstrated the prevalence of chromosome number
abnormalities and chromosomal structural abnormalities. To
understand in an improved way the dynamic CIN of GWH04 cells,
monoclonal culture derived from these cells and karyotype analyses
of three subclones (subclone 1.4, 1.5, and 3.17) were conducted and
compared. The chromosome numbers of the three subclones were not
exactly the same, with a narrow range of fluctuations and more
common dicentric chromosomes compared with results based on GWH04
cells. The chromosome numbers of 40 metaphase karyotypes from
subclones 1.4, 1.5, and 3.17 cells exhibited ranges of 59-83,
70-87, and 55-91, with 7.5% (3/40), 40% (16/40), and 20% (8/40)
positive for dicentric chromosomes, respectively (Fig. 5D). As genetic and genomic
aberrations are the primary cause of cancer, chromosome
mis-segregation leads to aneuploidy, and aberrant chromosomes
provide cancer cells with a mechanism to lose tumor suppressor loci
and gain extra copies of oncogenes, which will definitely increase
the mutation rates and malignancy of offspring cells.
Discussion
Cell culture and cell lines are the fundamental and
most powerful tools in cancer research. They are often used in
cancer biology studies and for the discovery of new anti-tumor
compounds, drug sensitivity and resistance, toxicity biomarkers,
and targeted agents showing clinical utility. However, the
accumulation of genetic aberrations in cancer cell lines often
occurs with increasing passage numbers, making it impossible for
numerous preclinical studies to translate into clinical
application. Therefore, establishing new primary cancer cell lines
derived from primary tumor tissues is of crucial importance for
different cancer research studies.
In recent years, primary culture conditions for GBM
have mainly been high-glucose DMEM or RPMI-1640 supplemented with
10% FBS for 2D adhesion culture and serum-free culture medium
(neurobasal medium + B27 + EGF + FGF) for 3D suspension sphere
culture, as the latter maintains more stem cell features (24-26).
Different culture conditions were selected according to different
study goals. Compared with sphere culture cell lines, adherent
cultures are easy to manipulate for common experimental purposes,
such as transfection, transduction and drug screening. Adherent
tumor cell lines are also amenable to growth in sphere cultures and
are commonly switched to sphere cultures for specific assays. In
the present study, culture from 10 GBM tissues of different
patients was conducted in the same culture medium (high-glucose
DMEM + 10% FBS), but only GWH04 cells from patient 4 could
circumvent replicative senescence and acquire the ability to
sustain unlimited proliferation in this culture medium. As the
genetic backgrounds of different GBMs are not determined when
establishing primary cultures from fresh patient-derived tissues,
high efficiencies can result from culturing samples with various
media conditions that differ in their combinations of growth
factors.
For the identification of GWH04 cells,
immunofluorescence staining of GFAP and Nestin was conducted. GFAP,
a glial fibrillary acidic protein, is a marker of astrocyte
activation. Although GFAP can be readily detected in most
astrocytic glioma and GBM cells, only the most morphologically
differentiated cells express it, whereas more primitive and
anaplastic cells do not (4).
Nestin, an intermediate filament protein that is specifically
expressed in neuroepithelial stem cells, is a marker of neural stem
cells. It is normally expressed in the neuroepithelium during the
early stage of embryonic development. For normal brain tissue and
normal astrocytic cells, Nestin expression is negative. In the
present study, immunofluorescence staining showed clear Nestin
expression with a distinct sense of cytoskeleton fibers, which
indicated firm derivation from tumor cells, whereas the GFAP
expression pattern was blurry even with longer exposure times when
compared with GFAP expression in cultured cells digested from grade
I-III gliomas (stained under the same conditions). However, this
did not affect protein expression in the mouse intracranial
xenografts of this cell line. Bigner et al (4) showed that only two of the 15 GBM cell
lines (U251 MG and U251 MG sp) yielded positive immunofluorescent
reactions for GFAP, and another study showed that, in the early
passages, ~30% of GL15 and 40% of GL22 cells exhibit GFAP
immunoreactivity in the fibrils of the cytoplasm (9). In conclusion, Nestin staining is more
reliable than GFAP staining when identifying GBM-derived tumor
cells.
TP53 is a typical tumor suppressor gene
associated with several biological processes, such as DNA damage
repair, apoptosis and proliferation (27). Indeed, among all genes in human
cancer cell genomes examined to date, TP53 is the most
frequently mutated gene and is mutated in almost one-third of all
human tumors (28). Ishii et
al (7) confirmed preliminary
observations that the incidence of TP53 mutations is markedly
higher in GBM cell lines (>75%) than in gliomas (25-35%).
Moreover, as TERT encodes a highly specialized reverse
transcriptase, which adds hexamer repeats to the 3′-end of
chromosomes (29), somatic
mutations in the TERT promoter region (C228T or C250T)
increase telomerase activity leading to preservation of telomeres,
thereby allowing tumors to avoid the induction of senescence
(30). As GWH04 cells possess
mutations in both TP53 and TERT promoter region (C228T), it
is reasonable to consider that the accommodation of GWH04 cells
both in vitro and in vivo may result from the
combination of different mutations. With the continuous
optimization of current technology and culture conditions, primary
culture is expected to improve in the future.
Genome instability is a common characteristic of
tumor cells. Notably, GWH04 cells were found to have chromosome
number abnormalities and structural abnormalities. Karyotype
analysis of several subclones illustrated the dynamic chromosome
instability of these cells. The increased tolerance to chromosomal
segregation errors could contribute to the association between
hyperploidy and genomic complexity, and may reflect the progressive
adaptation of self-renewing cells to the microenvironment in
vivo and their culture conditions in vitro (31). When considering chromosomal
structural abnormalities, double microbodies and HSR were presented
in certain G-banding metaphase images. It has been reported that
double microbodies are large extracellular DNA (ecDNA), and ecDNA
has been demonstrated to be rather common in GBM and to be
associated with oncogene amplification and targeted therapy
resistance (32-34). HSR was also reported to be
associated with amplified oncogenes, such as MYC, EGFR and ERBB2
(35). Therefore, exploring
relevant therapy from the perspective of aneuploidy and chromosomal
abnormalities, may be effective for treating GBM.
Given the common use of several cell lines to
illustrate the function and contribution of a single molecule
and/or mutation to cancer, it is of great importance to learn more
about the mutation background of each cell line. If a study is
based on several cell lines with similar mutational landscapes and
the cell lines are from early passages with a high similarity to
the primary tumor, it is expected that the transition time from
preclinical to clinical research will be shortened. As GWH04 cells
are presented with complete genetic information and cell biological
characteristics, readers could have a deeper insight of it. To
date, this cell line has demonstrated to be easy to manipulate for
common experimental purposes with strong stability in various
experimental studies, such as transfection, transduction and drug
screening in vitro, as well as to form xenografts in nude
mice in vivo. Any researcher interested in GBM study can
obtain this cell line from either institution of BRICS or CCTCC.
This will facilitate the exploration of molecular mechanisms and
the screening and evaluation of antitumor drugs in GBM studies.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The WES-seq dataset generated and/or analyzed during the
current study is available in the NCBI repository (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA806683).
Authors' contributions
FM and DG discussed the project and designed the
research. FM and BW confirm the authenticity of all the raw data.
FC and FM put forward the ideas of this article, conducted the
primary culture and wrote the manuscript. BW revised the
manuscript. FC and XW performed the karyotype analysis and animal
experiments. YL and PP conducted the data acquisition and analysis
from CCK-8 and soft agar colony formation assays. CH collected the
clinical samples and conducted the follow-up record collections. SX
and BW performed the pathological examination of tumor tissue and
xenografts. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Detailed information was given to the relatives of
the patient and informed consent was provided prior to surgery. All
samples were obtained from the Department of Neurosurgery, Tongji
Hospital, Huazhong University of Science and Technology (Wuhan,
China) after written informed consent was obtained and according to
the protocol approved (approval no. TJ-IBR20210119) by the Research
Ethics Committee of Tongji Hospital, Tongji Medical College,
Huazhong University of Science and Technology. All procedures
involving human samples were in accordance with the 1964 Helsinki
Declaration and its later amendments or comparable ethical
standards.
All procedures in the animal experiments were
approved (approval no. TJ-A20161206) by the Ethical Committee of
Tongji Hospital, Tongji Medical College, Huazhong University of
Science and Technology and were performed in accordance with ARRIVE
guidelines (36) (https://arriveguidelines.org).
Patient consent for publication
The relatives of the patient provided written
informed consent before the surgery for the study of this case and
for the publication of the data and accompanying images. The right
of the patient to anonymity and privacy was fully respected.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Professors Zhang
Songling and Yu Changjie at the BRICS for the identification of
this cell line.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81874086).
References
|
1
|
Wen PY and Reardon DA: Neuro-oncology in
2015: rogress in glioma diagnosis, classification and treatment.
Nat Rev Neurol. 12:69–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Noushmehr H, Weisenberger DJ, Diefes K,
Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP,
Bhat KP, et al: Identification of a CpG island methylator phenotype
that defines a distinct subgroup of glioma. Cancer Cell.
17:510–522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re32016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bigner DD, Bigner SH, Pontén J, Westermark
B, Mahaley MS, Ruoslahti E, Herschman H, Eng LF and Wikstrand CJ:
Heterogeneity of genotypic and phenotypic characteristics of
fifteen permanent cell lines derived from human gliomas. J
Neuropathol Exp Neurol. 40:201–229. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Diserens AC, de Tribolet N, Martin-Achard
A, Gaide AC, Schnegg JF and Carrel S: Characterization of an
established human malignant glioma cell line: LN-18. Acta
Neuropathol. 53:21–28. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Van Meir E, Sawamura Y, Diserens AC, Hamou
MF and de Tribolet N: Human glioblastoma cells release interleukin
6 in vivo and in vitro. Cancer Res. 50:6683–6688. 1990.PubMed/NCBI
|
|
7
|
Ishii N, Maier D, Merlo A, Tada M,
Sawamura Y, Diserens AC and Van Meir EG: Frequent co-alterations of
TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human
glioma cell lines. Brain Pathol. 9:469–479. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Van Meir EG, Kikuchi T, Tada M, Li H,
Diserens AC, Wojcik BE, Huang HJ, Friedmann T, de Tribolet N and
Cavenee WK: Analysis of the p53 gene and its expression in human
glioblastoma cells. Cancer Res. 54:649–652. 1994.PubMed/NCBI
|
|
9
|
Bocchini V, Casalone R, Collini P, Rebel G
and Lo Curto F: Changes in glial fibrillary acidic protein and
karyotype during culturing of two cell lines established from human
glioblastoma multiforme. Cell Tissue Res. 265:73–81. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bian X, Yang Z, Feng H, Sun H and Liu Y: A
combination of species identification and STR profiling identifies
cross-contaminated cells from 482 human tumor cell lines. Sci Rep.
7:97742017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al: Effects of radiotherapy with concomitant and
adjuvant temozolomide versus radiotherapy alone on survival in
glioblastoma in a randomised phase III study: 5-Year analysis of
the EORTC-NCIC trial. Lancet Oncol. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Herrlinger U, Tzaridis T, Mack F,
Steinbach JP, Schlegel U, Sabel M, Hau P, Kortmann RD, Krex D,
Grauer O, et al: Lomustine-temozolomide combination therapy versus
standard temozolomide therapy in patients with newly diagnosed
glioblas-0toma with methylated MGMT promoter (CeTeG/NOA-09): A
randomised, open-label, phase 3 trial. Lancet. 393:678–688. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Killock D: Lomustine-temozolomide
combination efficacious in newly diagnosed glioblastoma. Nat Rev
Clin Oncol. 16:2732019.
|
|
15
|
Wick W, Gorlia T, Bendszus M, Taphoorn M,
Sahm F, Harting I, Brandes AA, Taal W, Domont J, Idbaih A, et al:
Lomustine and bevacizumab in progressive glioblastoma. New Engl J
Med. 377:1954–1963. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Friedman HS, Prados MD, Wen PY, Mikkelsen
T, Schiff D, Abrey LE, Yung WK, Paleologos N, Nicholas MK, Jensen
R, et al: Bevacizumab alone and in combination with irinotecan in
recurrent glioblastoma. J Clin Oncol. 27:4733–4740. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jeremic B, Grujicic D, Jevremovic S,
Stanisavljevic B, Milojevic L, Djuric L and Mijatovic L:
Carboplatin and etoposide chemotherapy regimen for recurrent
malignant glioma: A phase II study. J Clin Oncol. 10:1074–1077.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Buckner JC, Ballman KV, Michalak JC,
Burton GV, Cascino TL, Schomberg PJ, Hawkins RB, Scheithauer BW,
Sandler HM, Marks RS, et al: Phase III trial of carmustine and
cisplatin compared with carmustine alone and standard radiation
therapy or accelerated radiation therapy in patients with
glioblastoma multiforme: North central cancer treatment group
93-72-52 and southwest oncology group 9503 trials. J Clin Oncol.
24:3871–3879. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Snuderl M, Fazlollahi L, Le LP, Nitta M,
Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD,
Betensky RA, et al: Mosaic amplification of multiple receptor
tyrosine kinase genes in glioblastoma. Cancer Cell. 20:810–817.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yi GZ, Huang G, Guo M, Zhang X, Wang H,
Deng S, Li Y, Xiang W, Chen Z, Pan J, et al: Acquired temozolomide
resistance in MGMT-deficient glioblastoma cells is associated with
regulation of DNA repair by DHC2. Brain. 142:2352–2366. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McLendon R, Friedman A, Bigner D, Van Meir
EG, Brat DJ, Mastrogianakis GM, Olson JJ, Mikkelsen T, Lehman N,
Aldape K, et al: Comprehensive genomic characterization defines
human glioblastoma genes and core pathways. Nature. 455:1061–1068.
2008. View Article : Google Scholar
|
|
22
|
Yao PS, Kang DZ, Wang XF, Lin RY and Ye
ZC: Cell-density-dependent manifestation of partial characteristics
for neuronal precursors in a newly established human gliosarcoma
cell line. In Vitro Cell Dev Biol Anim. 51:345–352. 2015.
View Article : Google Scholar
|
|
23
|
Notarangelo A, Trombetta D, D'Angelo V,
Parrella P, Palumbo O, Storlazzi CT, Impera L, Muscarella LA, La
Torre A, Affuso A, et al: Establishment and genetic
characterization of ANGM-CSS, a novel, immortal cell line derived
from a human glioblastoma multiforme. Int J Oncol. 44:717–724.
2014. View Article : Google Scholar
|
|
24
|
Li Y, Sun T, Chen Z, Shao Y, Huang Y and
Zhou Y: Characterization of a new human astrocytoma cell line
SHG140: Cell proliferation, cell phenotype, karyotype, STR markers
and tumorigenicity analysis. J Cancer. 12:371–378. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Wang H, Sun T, Chen J, Guo L, Shen
H, Du Z and Zhou Y: Biological characteristics of a new human
glioma cell line transformed into A2B5(+) stem cells. Mol Cancer.
14:752015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Man J, Yu X, Huang H, Zhou W, Xiang C,
Huang H, Miele L, Liu Z, Bebek G, Bao S and Yu JS: Hypoxic
induction of vasorin regulates notch1 turnover to maintain glioma
stem-like cells. Cell Stem Cell. 22:104–118.e6. 2018. View Article : Google Scholar :
|
|
27
|
Aubrey BJ, Strasser A and Kelly GL:
Tumor-suppressor functions of the TP53 pathway. Cold Spring Harb
Perspect Med. 6:a0260622016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bouaoun L, Sonkin D, Ardin M, Hollstein M,
Byrnes G, Zavadil J and Olivier M: TP53 variations in human
cancers: New lessons from the IARC TP53 database and genomics data.
Hum Mutat. 37:865–876. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cesare AJ and Reddel RR: Alternative
lengthening of telomeres: Models, mechanisms and implications. Nat
Rev Genet. 11:319–330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Labussière M, Di Stefano AL, Gleize V,
Boisselier B, Giry M, Mangesius S, Bruno A, Paterra R, Marie Y,
Rahimian A, et al: TERT promoter mutations in gliomas, genetic
associations and clinico-pathological correlations. Br J Cancer.
111:2024–2032. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dewhurst SM, McGranahan N, Burrell RA,
Rowan AJ, Grönroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P,
Ward RL, et al: Tolerance of whole-genome doubling propagates
chromosomal instability and accelerates cancer genome evolution.
Cancer Discov. 4:175–185. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim H, Nguyen NP, Turner K, Wu S, Gujar
AD, Luebeck J, Liu J, Deshpande V, Rajkumar U, Namburi S, et al:
Extrachromosomal DNA is associated with oncogene amplification and
poor outcome across multiple cancers. Nat Genet. 52:891–897. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Koche RP, Rodriguez-Fos E, Helmsauer K,
Burkert M, MacArthur IC, Maag J, Chamorro R, Munoz-Perez N,
Puiggròs M, Dorado Garcia H, et al: Extrachromosomal circular DNA
drives oncogenic genome remodeling in neuroblastoma. Nat Genet.
52:29–34. 2020. View Article : Google Scholar :
|
|
34
|
Nathanson DA, Gini B, Mottahedeh J,
Visnyei K, Koga T, Gomez G, Eskin A, Hwang K, Wang J, Masui K, et
al: Targeted therapy resistance mediated by dynamic regulation of
extrachromosomal mutant EGFR DNA. Science. 343:72–76. 2014.
View Article : Google Scholar :
|
|
35
|
Turner KM, Deshpande V, Beyter D, Koga T,
Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, et al:
Extrachromosomal oncogene amplification drives tumour evolution and
genetic heterogeneity. Nature. 543:122–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Percie du Sert N, Ahluwalia A, Alam S,
Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U,
Emerson M, et al: Reporting animal research: Explanation and
elaboration for the ARRIVE guidelines 2.0. PLoS Biol.
18:e30004112020. View Article : Google Scholar : PubMed/NCBI
|