Introduction
Lung cancer is one of the most common types of
malignant tumor (1). More than one
million individuals die of lung cancer every year worldwide,
accounting for 26-28% of the total number of cancer-associated
deaths, and the morbidity and mortality rates have risen to first
place among all tumors (2,3). Approximately 85% of lung cancer cases
are non-small cell lung cancer (NSCLC). The lack of effective
diagnosis at the early stages of lung cancer means that most
patients are already in the advanced stage at the time of
diagnosis. In addition, the poor sensitivity of advanced lung
cancer to chemotherapy drugs inevitably leads to poor prognosis,
with a five-year survival rate of only ~15.6% worldwide (4).
Gefitinib, considered the first-line drug for the
standard treatment of epidermal growth factor receptor
(EGFR)-mutated advanced NSCLC, inhibits the EGFR tyrosine kinase
activity (5). Mutations in EGFR
are observed in ~40 and 20% of patients with NSCLC in Asian and
non-Asian populations, respectively (6,7).
EGFR is mainly located on the plasma membrane of cells and belongs
to the receptor tyrosine kinase family. Once bound by EGF ligands,
EGFR kinase is activated, which induces the dimerizaiton of the
intracellular domains and then activates cell signal transduction
(8). EGFR is abnormally activated
and overexpressed in numerous different tumor types, including
NSCLC, thyroid cancer and colorectal cancer; therefore, EGFR has
become an important effector target for drug therapy for cancers
(7). The abnormal expression of
any protein downstream of EGFR-mediated signaling transduction
pathways promotes the erroneous activation of proliferation signals
in the cell nucleus, thus inducing unusual hyperplasia of cells.
Mutations in genes such as EGFR grant tumor cells the ability to
proliferate indefinitely; however, tumor growth must also have an
environment suitable for its growth. Tumor cells interact with the
body's own cells, including immune cells, vascular endothelial
cells and fibroblasts. These interactions constitute the tumor
microenvironment (TME), which is involved in tumor angiogenesis and
other processes. More importantly, the TME may help tumor cells
evade the body's normal immune cells. During tumorigenesis, not
only do the body's immune cells select mutant cells, but tumor
cells may also regulate normal immune cells. It has been reported
that the EGFR-mutated cancer cells are able to form an
immunosuppressive TME that reduces immune response of T cells
(9).
The B7 family is the most important member of T cell
costimulatory/inhibitory molecules and involved in B7/CD28
signaling. B7-1 (CD80) and B7-2 (CD86) were reported to stimulate
T-cell activation by binding to CD28 (10). Currently known members of the B7
family include B7-H1 [programmed death ligand 1 (PD-L1)], B7-H2
(ICOSL/B7h/B7RP-1), B7-H3, B7-DC (PD-L2), B7-H4 (B7x/B7S1), B7-H5
and six other members. These molecules all belong to the
immunoglobulin superfamily and have 20-39% identity (homology) in
their protein sequences (11).
They are expressed at different stages of different immune cells
and have different roles. B7 molecules participate in various
processes, such as the activation of T cells. Studies have
indicated that in mouse models, transfecting tumor cells with B7-1
or B7-2 may enhance T cell immunity, thereby inhibiting tumor
growth (12). Studies suggested
that the loss of B7-H5 may contribute to immune evasion in various
cancers, such as pancreatic cancer and NSCLC (13,14).
It has been reported that B7-H5 is constitutively expressed in
macrophages and may be induced on dendritic cells, and it was
additionally demonstrated that the B7-H5/CD28H [also known as
transmembrane and immunoglobulin domain containing 2] interaction
stimulated human T-cell growth and cytokine production (15). Previous studies by our group have
demonstrated that overexpression of B7H5/CD28H is associated with
unfavorable survival in human gastric cancer and pancreatic ductal
adenocarcinoma (16,17). Therefore, in the present study,
B7H5 was selected to further verify its effect in NSCLC cells.
In the present study, it was investigated whether
gefitinib is able to enhance the anti-tumor immune response against
EGFR-mutated NSCLC by upregulating B7H5 expression and activating T
cells via CD28H. It was also investigated whether in EGFR-mutant
NSCLC cells, over-activated EGFR downstream signaling pathways may
cause the decrease in the expression of B7-H5 on the tumor cell
surface and inhibit the activation of B7-H5/CD28H costimulatory
signals, thereby evading the attack of effector T cells.
Materials and methods
Cell culture
NSCLC cell lines (NCI-H1299, NCI-H358, HCC827 and
PC-9) were purchased from the American Type Culture Collection. The
identity of the NCI-H358 cells was confirmed using short tandem
repeat profiling. Among the cell lines used, NCI-H358 and NCI-H1299
harbor wild-type EGFR, while HCC827 and PC-9 cells harbor EGFR
mutations, both with 19 exon deletion and E746-A750 deletion
(18,19). PC-9 cells were cultured in
Dulbecco's modified Eagle's medium (GIBCO; Thermo Fisher
Scientific, Inc.), while NCI-H1299, HCC827 and NCI-H358 cells were
cultured in RPMI-1640 medium (GIBCO; Thermo Fisher Scientific,
Inc.). Both media contained 10% fetal bovine serum (FBS; GIBCO;
Thermo Fisher Scientific, Inc.) and all cells were cultured in an
incubator with 5% CO2 at 37°C.
Isolation of peripheral blood mononuclear
cells (PBMCs)
A blood sample taken using BD anticoagulant tubes
(BD Biosciences) was diluted with an equal volume of PBS, which was
used to dilute freshly collected peripheral blood of a healthy
subject (female; age, 35 years; Tongde Hospital of Zhejiang
Province, Hangzhou, China; November 2021). The blood was taken from
the vein in the antecubital region of the hand and arm. The diluted
blood was slowly added into a centrifuge tube containing the
lymphocyte separation solution (FICOLL) at a ratio of 1:0.5
(diluted blood/FICOLL solution). The samples were centrifuged at
400 × g at 20°C for 30 min. The middle white film was then
transferred into a new 50-ml centrifuge tube and mixed with 3-4
volumes of sterile PBS. The mixture was then centrifuged at 300 × g
for 10 min at 20°C and the supernatant was discarded. The cell
pellet was resuspended in RPMI-1640 medium with 10% FBS and then
cultured at 37°C.
Small interfering RNA (siRNA)
transfection
EGFR siRNA, Negative siRNA (negative control), CD28H
siRNA, EGFR pcDNA3.1-Flag-C and EGFR mutant plasmid (EGFR; mutation
site:L747-E749, A750P) were purchased from Guangzhou Ribobio Co.,
Ltd. The NCI-H358, NCI-H1299, HCC827, PC-9 cells and PBMCs were
seeded in a 6-well plate at 1×105 cells/well and
supplemented with 2 ml of the corresponding medium. When the cells
had reached 60-70% confluence, they were transfected with 50 nM
siRNA using the Lipofectamine® 2000 transfection reagent
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Cells were collected 48-72 h later for the subsequent
experiments. The siRNA sequences were as follows: EGFR siRNA 001
forward, 5′-GUA AUU AUG UGG UGA CAG ATT-3′ and reverse, 5′-UCU GUC
ACC ACA UAA UUA CTT-3′; EGFR siRNA 002 forward, 5′-CCU UAG CAG UCU
UAU CUA ATT-3′ and reverse, 5′-UUA GAU AAG ACU GCU AAG GTT-3′; EGFR
siRNA 003 forward, 5′-GGA ACU GGA UAU UCU GAA ATT-3′ and reverse,
5′-UUU CAG AAU AUC CAG UUC CTT-3′; B7H5 siRNA 001 forward, 5′-GUG
CCU GCA UCG UAG GAA UTT-3′ and reverse, 5′-AUU CCU ACG AUG CAG GCA
CTT-3′; B7H5 siRNA 002 forward, 5′-GGC ACG AUG UGA CCU UCU ATT-3′
and reverse, 5′-UAG AAG GUC ACA UCG UGC CTT-3′; B7H5 siRNA 002
forward, 5′-CAC GCC GUA UUC CCU GUA UTT-3′ and reverse, 5′-AUA CAG
GGA AUA CGG CGU GTT-3′; CD28H siRNA 001 forward, 5′-CCG UAG AGA UUC
CUG AGU UTT-3′ and reverse, 5′-AAC UCA GGA AUC UCU ACG GTT-3′;
CD28H siRNA 002 forward, 5′-CUC CGU GUU AAG UGG ACA ATT-3′ and
reverse, 5′-UUG UCC ACU UAA CAC GGA GTT-3′; CD28H siRNA 002
forward, 5′-UCU ACA GCA ACG UCC UAU ATT-3′ and reverse, 5′-UAU AGG
ACG UUG CUG UAG ATT-3′; negative control siRNA forward, 5′-UUC UCC
GAA CGU GUC ACG U-3′ and reverse, 5′-ACG UGA CAC GUU CGG AGA
A-3′.
Toxicity test in NSCLC/PBMCs
co-culture
Calcein acetoxymethyl ester (1 mg/ml; cat. no:
PJ693; Dojindo Laboratories, Inc.) was diluted in RPMI-1640 medium.
The toxicity assay was performed in a 96-well culture plate. A
gradient of diluted effector cells (E) and labeled target cells (T)
was added at 100 µl/well at E/T ratios of 50/1, 25/1,
12.5/1, 6.25/1, 3.13/1 and 1.56/1, with three replicate wells for
each ratio. At the same time, three replicate wells for the
spontaneous release group and three replicate wells for the
complete release group were set up. The E/T cells were incubated at
37°C in a 5% CO2 incubator for 4 h and then centrifuged
at 300 × g for 5 min at room temperature. The supernatant was
transferred into a new well. A multi-functional enzyme label
instrument was used to measure the fluorescence (excitation filter,
485 nm; emission filter, 530 nm). The percent-specific lysis was
calculated as follows: Percent-specific lysis=[(test
lysis-spontaneous lysis)/(maximum lysis-spontaneous lysis)]
×100.
Cell viability
NSCLC cells were seeded at 3,000 cells/well in
96-well plates. After the cells were completely attached to the
wells, the cells were treated with corresponding medium plus 1% FBS
for 24 h. Cells were then transfected with B7H5 siRNA or negative
controls for 6 h. The supernatant was discarded and cells were
incubated in fresh medium for 24 h. The cells were then incubated
with 100 µl Cell Counting Kit-8 (CCK-8; Dojindo
Laboratories, Inc.) solution per well for 2-3 h. Finally, the
absorbance at 460 nm was measured using an MRX II microplate reader
(Dynex).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from NSCLC cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
then reverse-transcribed to cDNA using a Reverse Transcription Kit
(cat. no. RR047A; Takara Bio, Inc.). The cDNA was then used as the
template for the qPCR step of the RT-qPCR protocol using a SYBR
Premix Ex Taq kit (Takara Bio, Inc.) on an Applied Biosystems
Real-time PCR System (Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 95°C for 30 sec, followed
by 40 cycles of denaturation at 95°C for 5 sec and annealing at
60°C for 30 sec. β-actin was used as a reference control. The
results were analyzed using the 2−ΔΔCq method (20). The primers used were as follows:
EGFR forward, 5′-CAG ATC GCA AAG GGC ATG AA-3′ and reverse, 5′-TTG
CCT CCT TCT GCA TGG TA-3′; B7H5 forward, 5′-AGG GGG CAT CTC TGT ACA
CT-3′ and reverse, 5′-GGC TGG ACT ATG CTC TCC AC-3′; β-actin
forward, 5′-GAC TTA GTT GCG TTA CAC CCT T-3′ and reverse, 5′-ACC
TTC ACC GTT CCA GTT TT-3′.
Western blot analysis
NSCLC cells were harvested and lysed with
radioimmunoprecipitation assay lysis buffer containing protease
inhibitors (cat. no. P0013C; Beyotime Institute of Biotechnology).
The concentration of the proteins was quantified using a
bicinchoninic acid protein assay (Sigma-Aldrich; Merck KGaA).
Subsequently, 40 µg of protein was separated using 10%
SDS-PAGE and then electrotransferred onto polyvinylidene fluoride
membranes (EMD Millipore). After blocking with 5% nonfat dry milk
(cat. no. 6342932; BD Biosciences) in Tris-buffered saline-Tween-20
buffer for 2 h at 37°C, the membranes were incubated with the
indicated primary antibodies, anti-EGFR and anti-B7H5 (cat. nos.
4267S and 54979T, respectively; 1:1,000 dilution; Cell Signaling
Technology, Inc.) or anti-CD28H [1:1,000 dilution; a kind gift from
Professor Yuwen Zhu (Department of Surgery, University of Colorado
Anschutz Medical Campus, Aurora, USA) (13,15)]
at 4°C overnight. The samples were then washed and incubated with
the corresponding secondary antibodies, anti-rabbit IgG HRP-linked
antibody (cat. no. 7074S; 1:2,000 dilution; Cell Signaling
Technology, Inc.) and anti-mouse IgG HRP-linked antibody (cat. no.
7076S; 1:2,000 dilution; Cell Signaling Technology, Inc.). GAPDH
was used as an internal control (cat. no. 2118S; 1:2,000 dilution;
Cell Signaling Technology, Inc.). The immunoreactive proteins were
visualized using an ECL kit (Bioworld Technology, Inc.). The
density of the bands was quantified using Image Lab 5.0 (Bio-Rad
Laboratories).
Immunofluorescence analysis
NSCLC cells subjected to different treatments were
washed with ice-cold PBS and then fixed using 4% paraformaldehyde
for 15 min at room temperature, incubated with 5% bovine serum
albumin at room temperature for 30 min, and then with primary
antibodies, anti-EGFR and anti-B7H5 (cat. nos. 4267S and 54979T;
respectively; 1:200 dilution; Cell Signaling Technology, Inc.) at
4°C overnight. The next day, the cells were washed with PBS,
incubated with corresponding secondary antibodies goat anti-rabbit
IgG (H+L) AF555 (cat. no. A32732; 1:2,000 dilution; Thermo Fisher
Scientific, Inc.) at room temperature for 2 h, incubated with DAPI
(Sigma-Aldrich; Merck KGaA) nuclear stain at room temperature for 2
min and washed using PBS twice. Finally, the cells were viewed and
imaged under an inverted fluorescence microscope (Olympus
Corporation).
Flow cytometric analysis
In brief, PBMCs in each group were collected into a
15-ml centrifuge tube at 300 × g and 4°C for 10 min. The cell
density was adjusted to 2×106/ml after discarding the
supernatant. The cells were washed with PBS containing 2% bovine
serum albumin (BSA; Biofroxx) 3 times, and then resuspended in 0.1
ml of ice-cold washing buffer and incubated with 1 µg
biotin-CD28H [a kind gift from Professor Yuwen Zhu (Department of
Surgery, University of Colorado Anschutz Medical Campus, Aurora,
USA) (13,15)] for 45 min at 4°C. The cells were
washed three times with 3 ml ice-cold washing buffer and then
incubated with 1 µg PE-Biotin (cat. no. 409004; BioLegend,
Inc.) for 30 min at 4°C. At the end of the incubation, the cells
were washed three times with 3 ml ice-cold washing buffer and
resuspended in 0.2 ml PBS for examination on a FACSCalibur flow
cytometer (BD Biosciences).
Tissue samples
A total of 55 pairs of NSCLC tumor tissues,
including 29 EGFR wild-type 26 EGFR Mut and their matched
paracancerous tissues, were obtained from the First Affiliated
Hospital of Huzhou University (Huzhou, China; January 2022) and the
detailed information of the patients is provided in Table SI (25 females and 30 males 30; age
>65 years, n=29; age <65 years, n=26; age range, 45-85 years;
median age, 65.43 years). The present study was approved by the
Medical Ethics Committee of the First Affiliated Hospital of Huzhou
University (Huzhou, China). All participants provided written
informed consent prior to using the tissues for scientific
research.
Immunohistochemical analysis
The tissue chip (project no. HLugA180Su02; Shanghai
Outdo Biotech Co., Ltd.) was baked in an oven at 63°C for 1 h,
followed by dewaxing in an automatic dyeing machine (ST5015; Leica
Microsystems). Antigens were retrieved by incubation with 0.1 M
citric acid at high pressure (50 kPa) at 110°C for 5 min. The chip
was blocked with 5% BSA at room temperature for 20 min and rinsed
with PBS three times. The primary antibody B7H5 (cat. no. 54979T;
1:300 dilution; Cell Signaling Technology, Inc.) was added dropwise
and the array was incubated at room temperature overnight at 4°C.
Next, the slide was rinsed with PBS three times, the secondary
antibody, anti-rabbit IgG HRP-linked antibody (cat. no. 7074S;
1:2,000 dilution; Cell Signaling Technology, Inc.), was added
dropwise, and the slide was incubated at room temperature for 30
min. Finally, 3,3′-diaminobenzidine solution was added dropwise and
the color development intensity was observed. Thereafter, the slide
was rinsed with tap water for 5 min and Hastelloy Hematoxylin
(Sigma-Aldrich; Merck KGaA) was added dropwise onto the slide,
followed by incubation for 1 min. The sample was then submerged in
0.25% hydrochloric acid alcohol for no less than 2 sec, rinsed with
tap water for >2 min, dried at room temperature and mounted for
a light microscopic (Olympus Corporation) observation.
Tissue microarray analysis
The tissue microarray included 87 pairs of tumor
tissues and their matched paracancerous tissues, which was analyzed
by Shanghai Outdo Biotech Co., Ltd. and the experimental procedures
were approved by the Shanghai Outdo Biotech Company ethics
committee (YB-M-05-02), which authorized the collection of tissue
samples from patients (39 females and 48 males; age >65 years,
n=36; age <65 years, n=51; age range, 44-84 years; median age,
63.07 years); the detailed information is listed in Table SII. An immunohistochemistry
protocol was used to analyze the expression of B7H5 and
subsequently examine its association with survival.
Statistical analysis
Values are expressed as the mean ± standard
deviation and data were statistically analyzed using GraphPad Prism
5 (GraphPad Software, Inc.). Unpaired Student's t-tests or one-way
ANOVA followed by Tukey's post-hoc test were used to analyze
differences between two groups or among multiple groups,
respectively. Kaplan-Meier survival analysis was used to compare
survival, while the log-rank test was performed to determine the
significance of differences between groups. All experiments were
performed as three independent replicates. P<0.05 was considered
to indicate a statistically significant difference.
Results
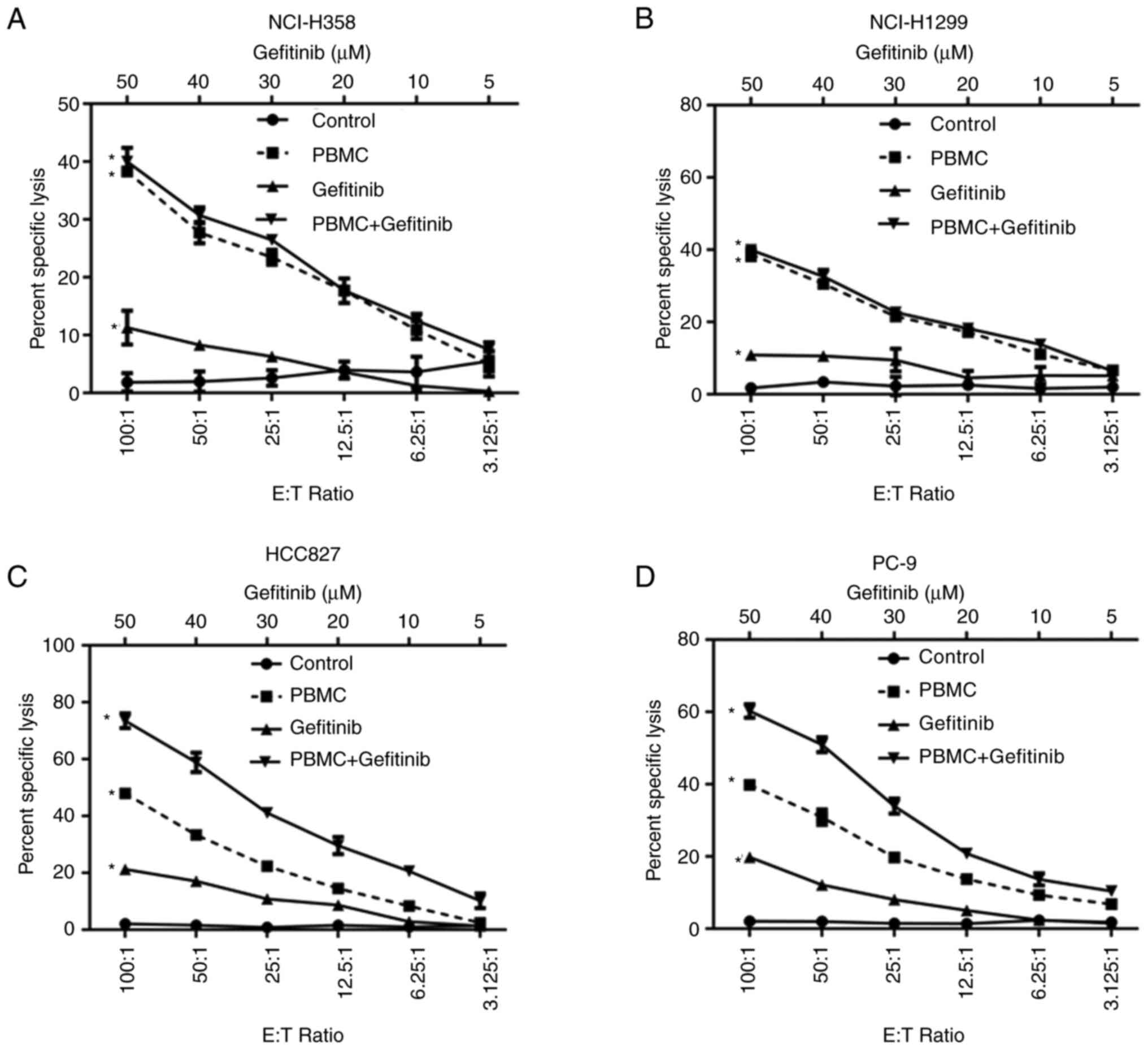
Gefitinib enhances the cytolytic capacity
of PBMCs toward EGFR-mutated NSCLC cells
To assess the anti-tumor cytotoxicity of gefitinib
and PBMCs toward NSCLC cells, an in vitro model was
established by co-culturing PBMCs with four different NSCLC cell
lines (NSC-H358, NCI-H1299, HCC827 and PC-9), respectively. The
NSCLC cells were stained with Calcein and then mixed with PBMCs at
different E/T ratios (100:1, 50:1, 25:1, 12.5:1 and 6.25:1). The
cytolytic capacity of PBMCs increased with an increasing E/T ratio
in all four cell lines. A 100:1 E/T ratio resulted in 40.01, 39.98,
47.93 and 39.87% lysis of NCI-H358, NCI-H1299, HCC827 and PC9
cells, respectively (Fig. 1A-D).
When NSCLC cells were treated with 50 mM gefitinib alone, 11.31 and
10.87% of HNC-H358 and NCI-H1299 cells expressing wild-type EGFR
were lysed, respectively, and 21.27 and 19.8% of HCC827 and PC-9
cells expressing mutant EGFR were lysed, respectively (Fig. 1A-D). Furthermore, the combination
of gefitinib and PBMCs significantly increased cytolysis from 47.93
and 39.87% to 73.51 and 60.28% in HCC827 and PC-9 cells,
respectively, compared with PBMCs alone, but had no significant
effects in HNC-H358 and NCI-H1299 cells [combination index: PC9,
0.77; HCC827, 0.588; NCI-H1299, 1.167; and NCI-H358, 1.316;
Fig. 1A-D]. The results indicated
that gefitinib confers better cytotoxicity in NSCLC cells that
carry EGFR mutations, particularly in the presence of PBMCs.
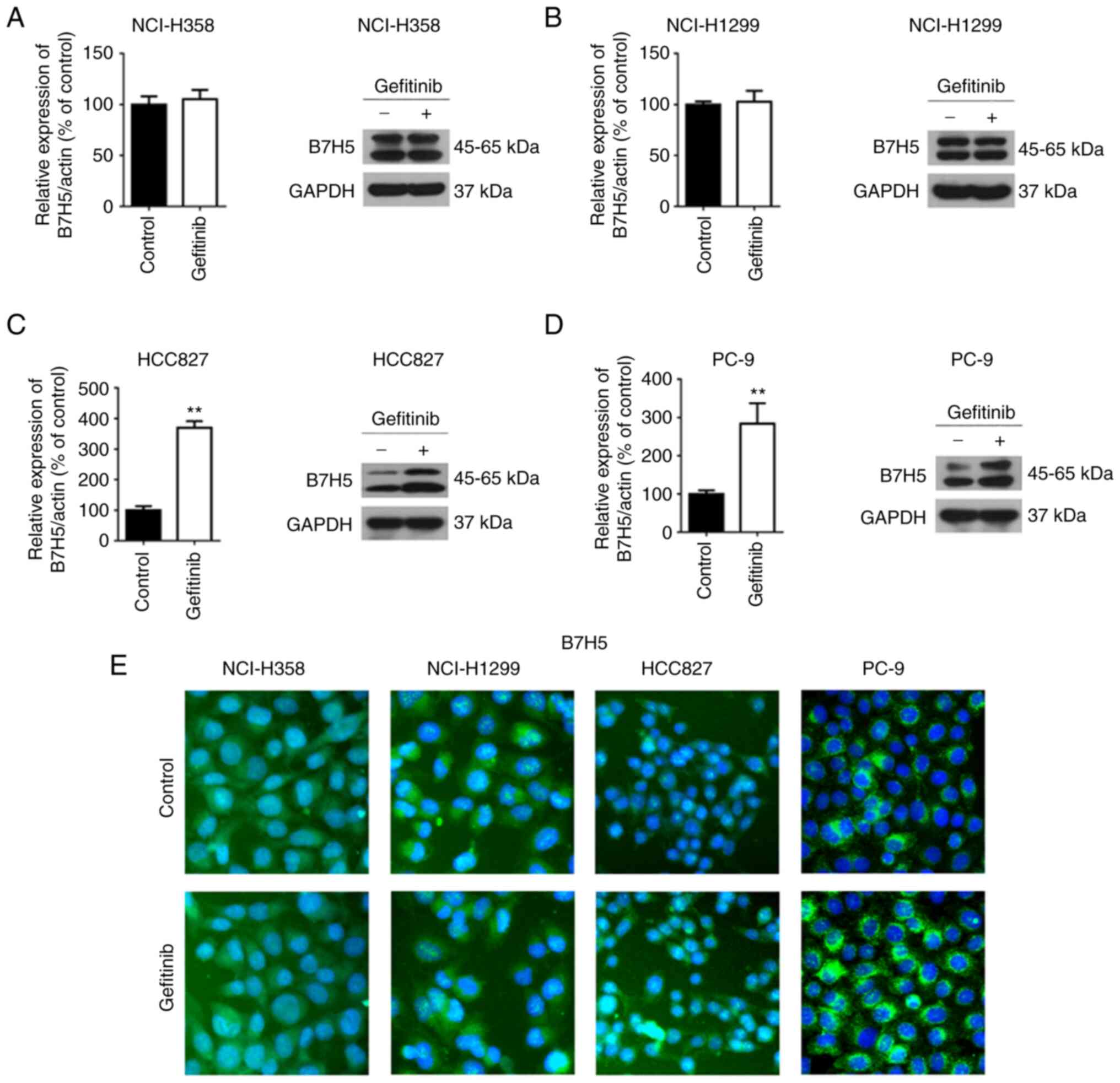
Gefitinib increases the expression of
B7H5 in EGFR-mutated NSCLC cells
To gain insight into the mechanisms by which
gefitinib produces enhanced toxicity against EGFR-mutated NSCLC
cells, the expression of B7H5 was examined using RT-qPCR, western
blot analysis and immunofluorescence. The results indicated that
the mRNA and protein levels of B7H5 were significantly upregulated
in EGFR-mutated NSCLC cells (HCC827 and PC-9), but unchanged in
wild-type EGFR NSCLC cell lines (NCI-H358 and NCI-H1299) after
gefitinib treatment (Fig. 2A-D).
Increased expression of B7H5 on the cell plasma membrane was
detected in both HCC827 and PC-9 cells, but not in NCI-H358 and
NCI-H1299 (Fig. 2E). Next, the
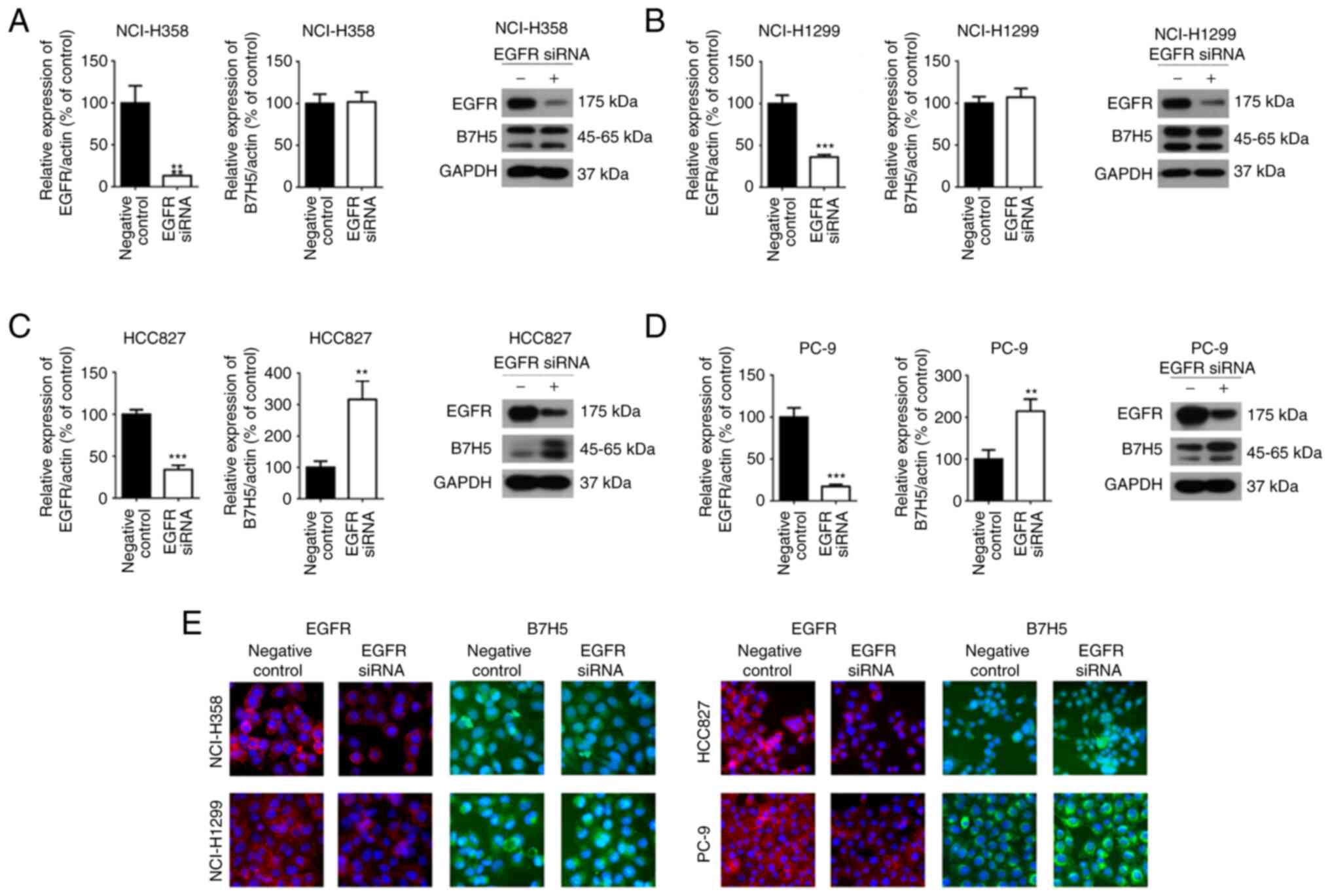
effects of EGFR siRNA on the expression of B7H5 in NSCLC cells were
examined. It was observed that the expression of B7H5 in
EGFR-mutant NSCLC cell lines (HCC827 and PC-9) was upregulated
after EGFR siRNA interference, but it was not affected in wild-type
EGFR NSCLC cell lines (NCI-H358 and NCI-H1299) (Fig. 3A-E). Regarding the application of
siRNA, three siRNAs were mixed together to perform the experiments.
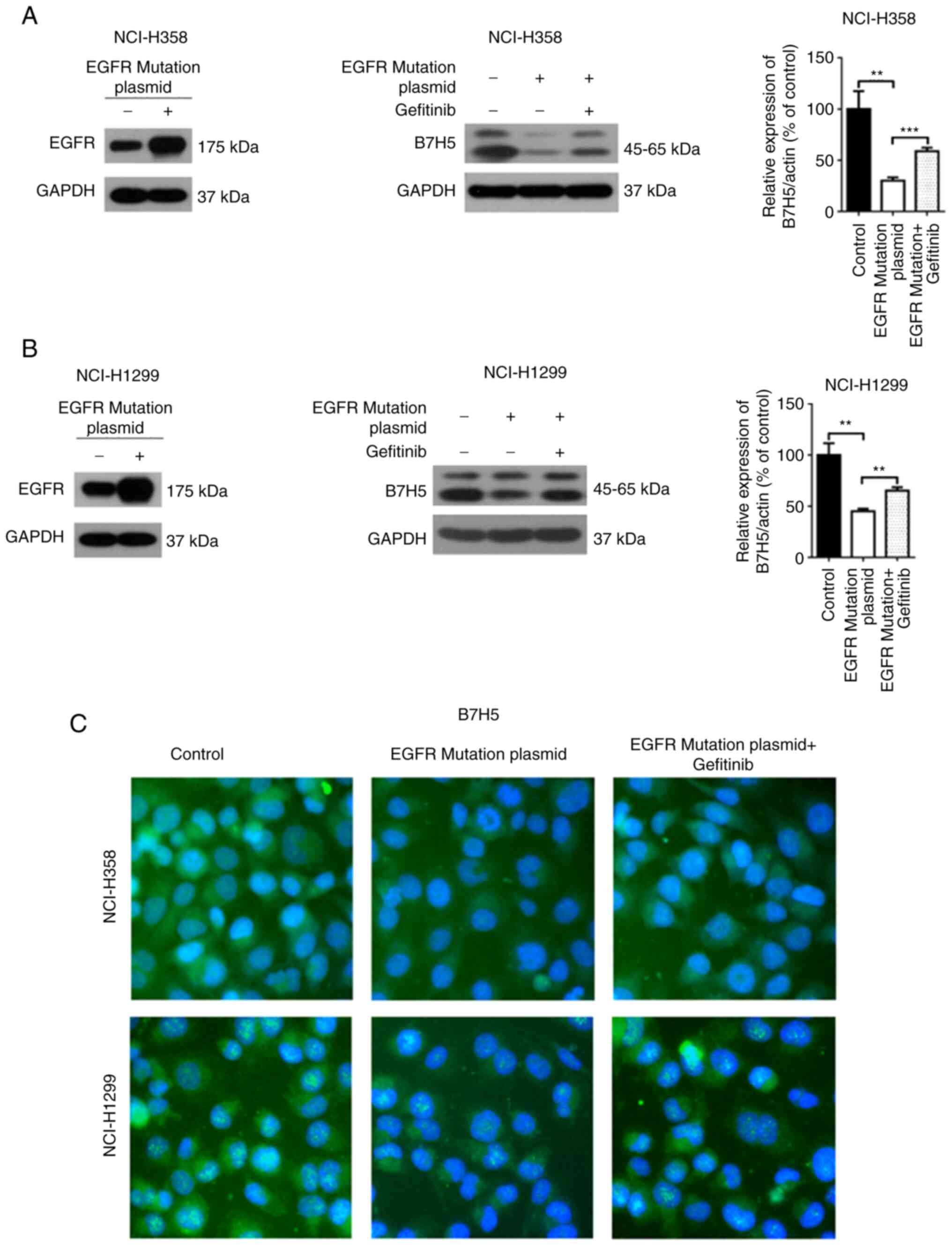
Furthermore, the expression of B7H5 was measured in EGFR wild-type
NSCLC cell lines transfected with EGFR mutant plasmid (L747-E749,
A750P) and it was indicated that the B7H5 mRNA levels were reduced
after expression of the EGFR mutants (Fig. 4A-C). Gefitinib treatment increased
the expression of B7H5 in these cells expressing EGFR mutants
(Fig. 4A-C).
B7H5 increases the cytolytic activity of
PBMCs in EGFR-mutated NSCLC cells after treatment with
gefitinib
It was then evaluated whether increased B7H5
expression is the cause of the enhanced cytotoxicity in
EGFR-mutated cells treated with gefitinib. First, cell viability
was measured in NSCLC cell lines with wild-type EGFR (NCI-H358 and
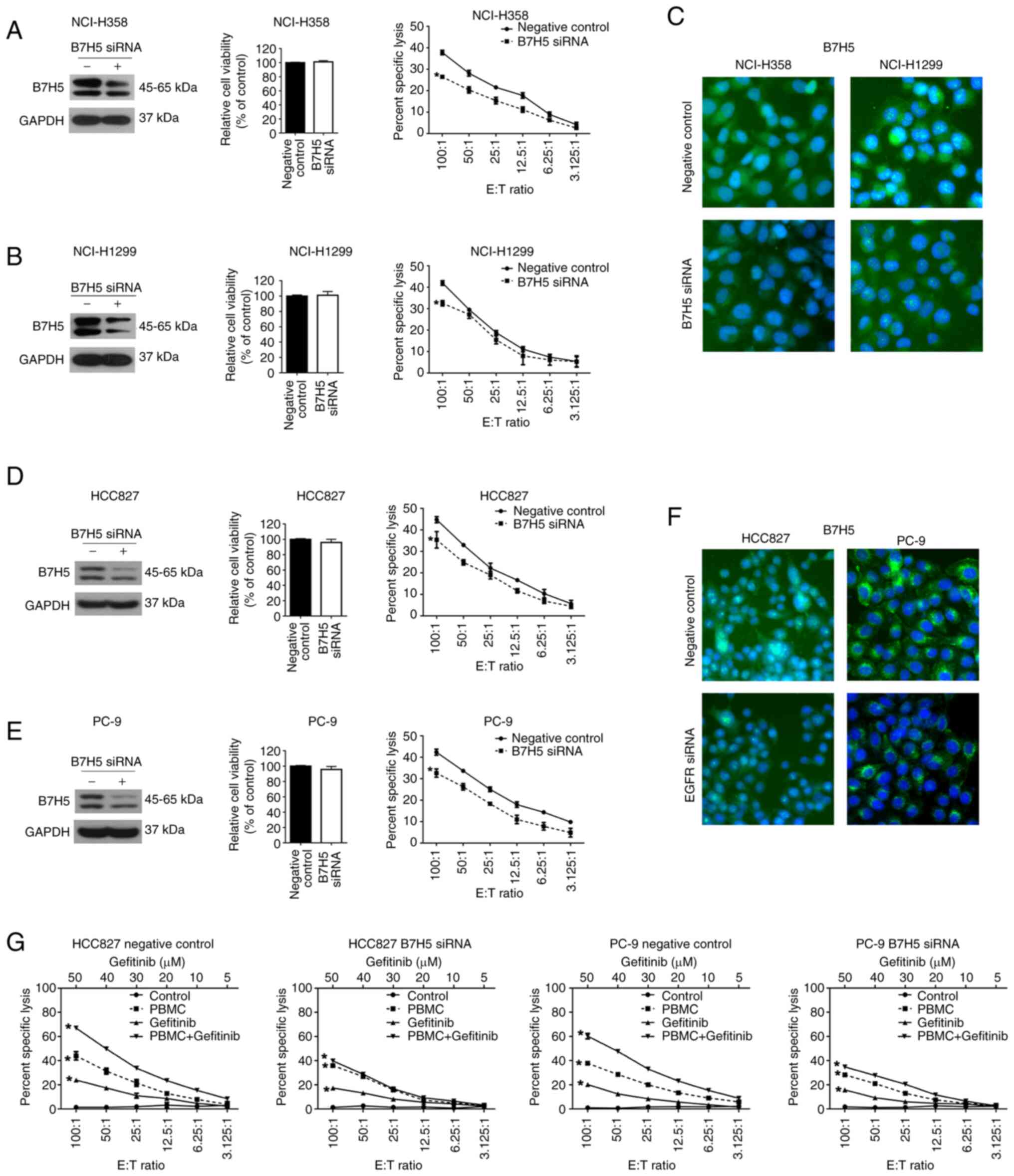
NCI-H1299) after transfection with B7H5 siRNA. As presented in
Fig. 5A-C, the efficiency of siRNA
interference of B7H5 was verified by western blot and
immunofluorescence analyses. B7H5 knockdown had no significant
effects on cell viability in these two cell lines. Of note, the
PBMC-induced cytolysis decreased in NCI-H358 and NCI-H1299 cells
after B7H5 siRNA treatment. Subsequently, B7H5 expression was
reduced by siRNA silencing in EGFR-mutated NSCLC cell lines (HCC827
and PC-9). The knockdown effects were confirmed by western blot and
immunofluorescence analysis. However, reduced expression of B7H5 in
HCC827 and PC-9 cells had no effects on cell viability (Fig. 5D-F). PBMC-mediated toxicity in
HCC827 and PC-9 cells was reduced when B7H5 expression was
decreased (Fig. 5D and E). The
results suggested that B7H5 expression is not related to the
viability of NSCLC cells, but its expression is associated with the
PBMC-mediated immune response. Given that gefitinib enhanced
PBMC-mediated cytotoxicity in EGFR-mutated NSCLC cell lines, it was
next investigated whether B7H5 is involved in this improved
immunoregulation. When B7H5 expression was reduced in EGFR-mutated
NSCLC cells co-cultured with PBMCs, the gefitinib-enhanced toxicity
was not different from that of the control (Fig. 5G). These findings indicated that
gefitinib-induced immune toxicity toward EGFR-mutated NSCLC cells
is mediated by upregulation of B7H5.
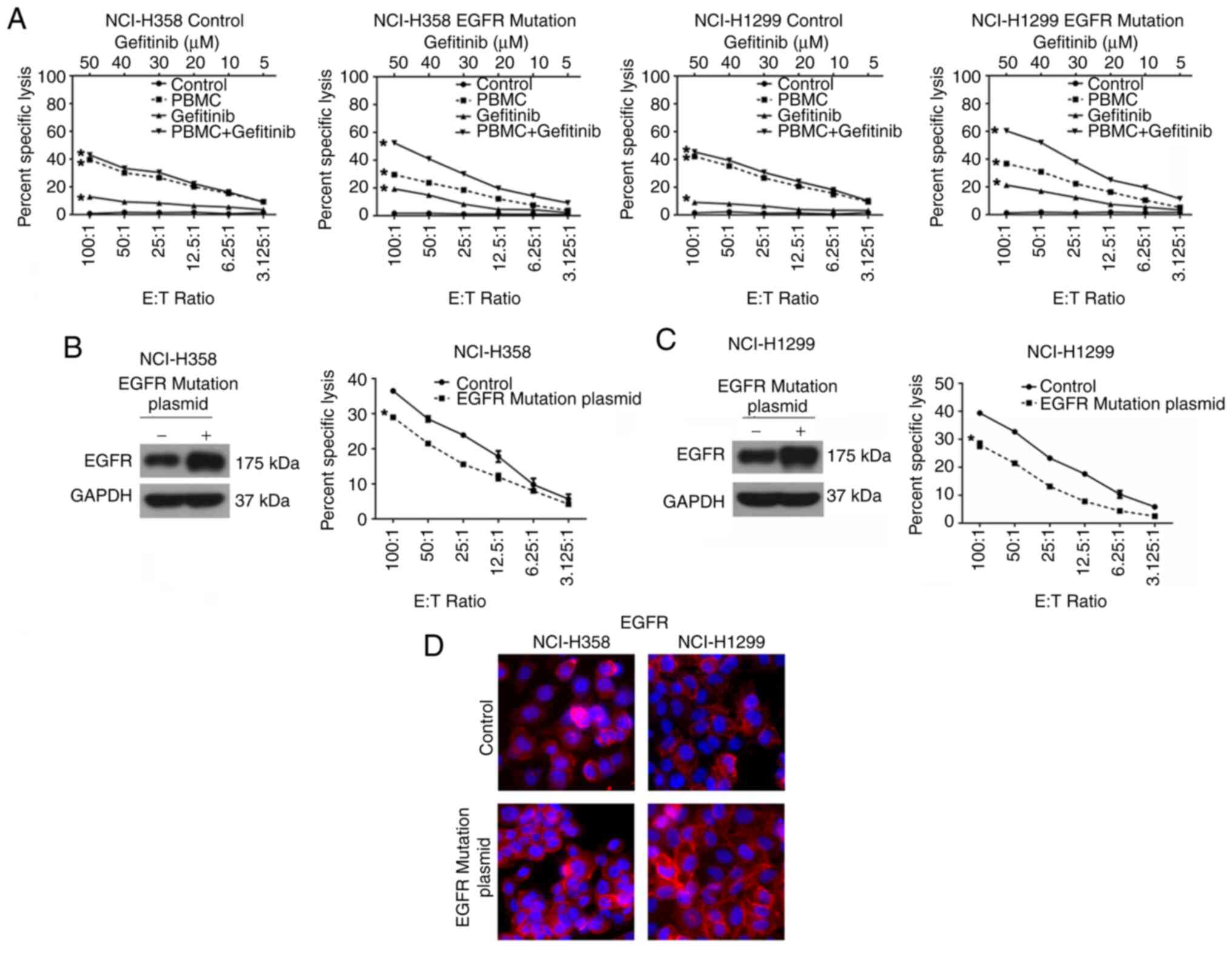
NCI-H358 and NCI-H1299 cells were then transfected
with plasmids expressing either wild-type or mutant EGFR, and these
cells were then cultured with PBMCs. Consistent with the results in
EGFR-mutated cell lines, gefitinib significantly enhanced the
PBMC-mediated cytolysis in cells transfected with EGFR mutants, but
not in cells expressing wild-type EGFR (Fig. 6A-D).
CD28H knockdown in PBMCs decreases
PBMC-mediated cytotoxicity in EGFR-mutated NSCLC cells
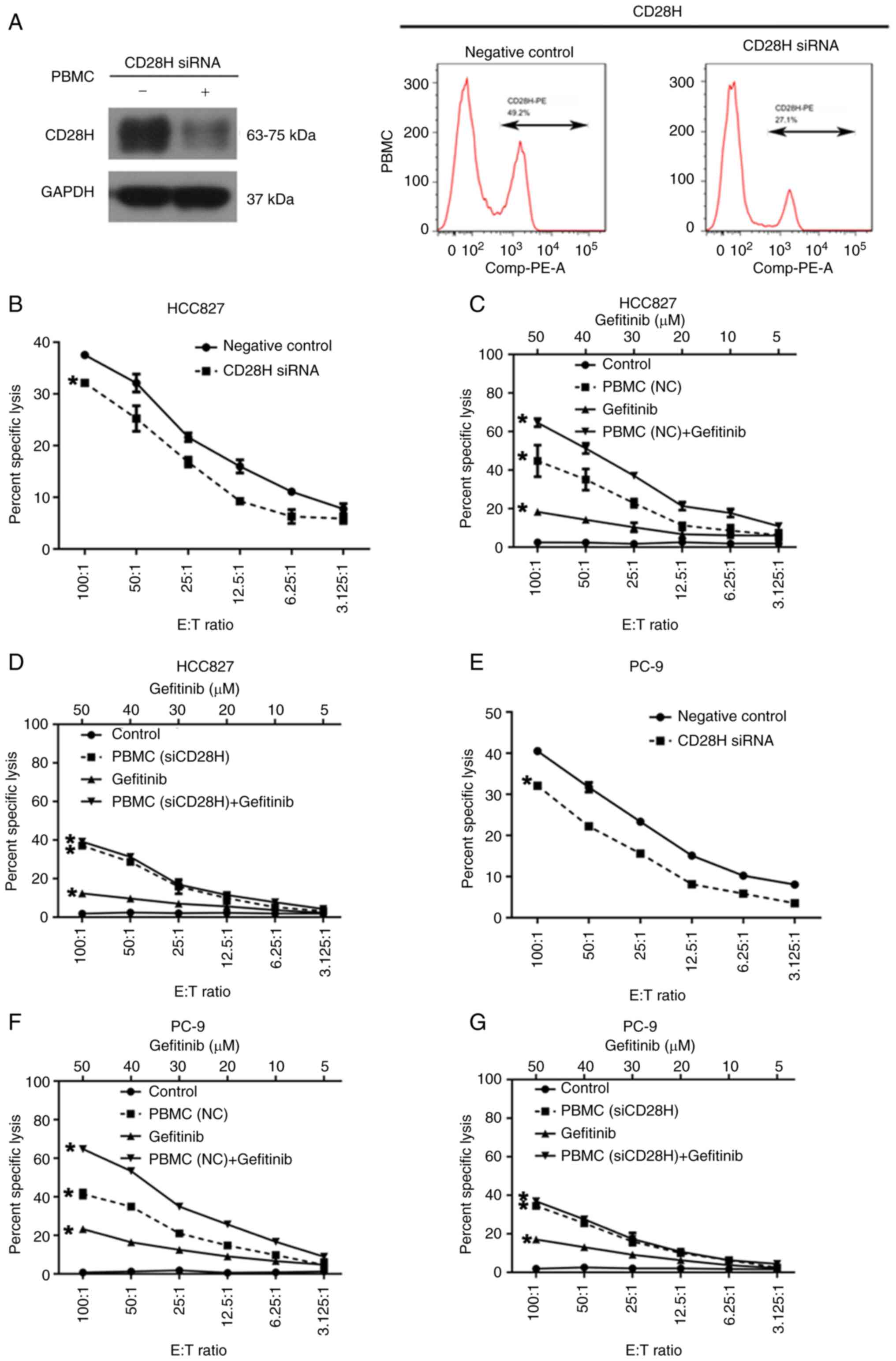
It has been previously reported that B7H5 is the
ligand for CD28H and the interaction between B7H5 and CD28H
stimulates the T cell response (16); therefore, the present study aimed
to determine whether the B7H5/CD28H signaling pathway is involved
in PBMCs-mediated cytotoxicity in EGFR-mutated NSCLC cells. Western
blot and flow cytometric analyses confirmed that the expression of
CD28H was reduced in PBMCs treated with CD28H siRNA (Fig. 7A). The cytotoxic activity of PBMCs
against EGFR-mutated NSCLC cell lines prior to and after CD28H
siRNA interference was compared in the in vitro NSCLC/PBMCs
co-culture model. PBMC-mediated cell lysis in HCC827 and PC-9 cells
decreased significantly when PBMCs were transfected with CD28H
siRNA (Fig. 7B and C).
Furthermore, after CD28H expression was inhibited in PBMCs,
gefitinib treatment had no effect on the cytolysis of HCC827 and
PC-9 cells co-cultured with PBMCs (Fig. 7D-G), suggesting that CD28H is
required for gefitinib-enhanced PBMC cytotoxicity against
EGFR-mutated NSCLC cells.
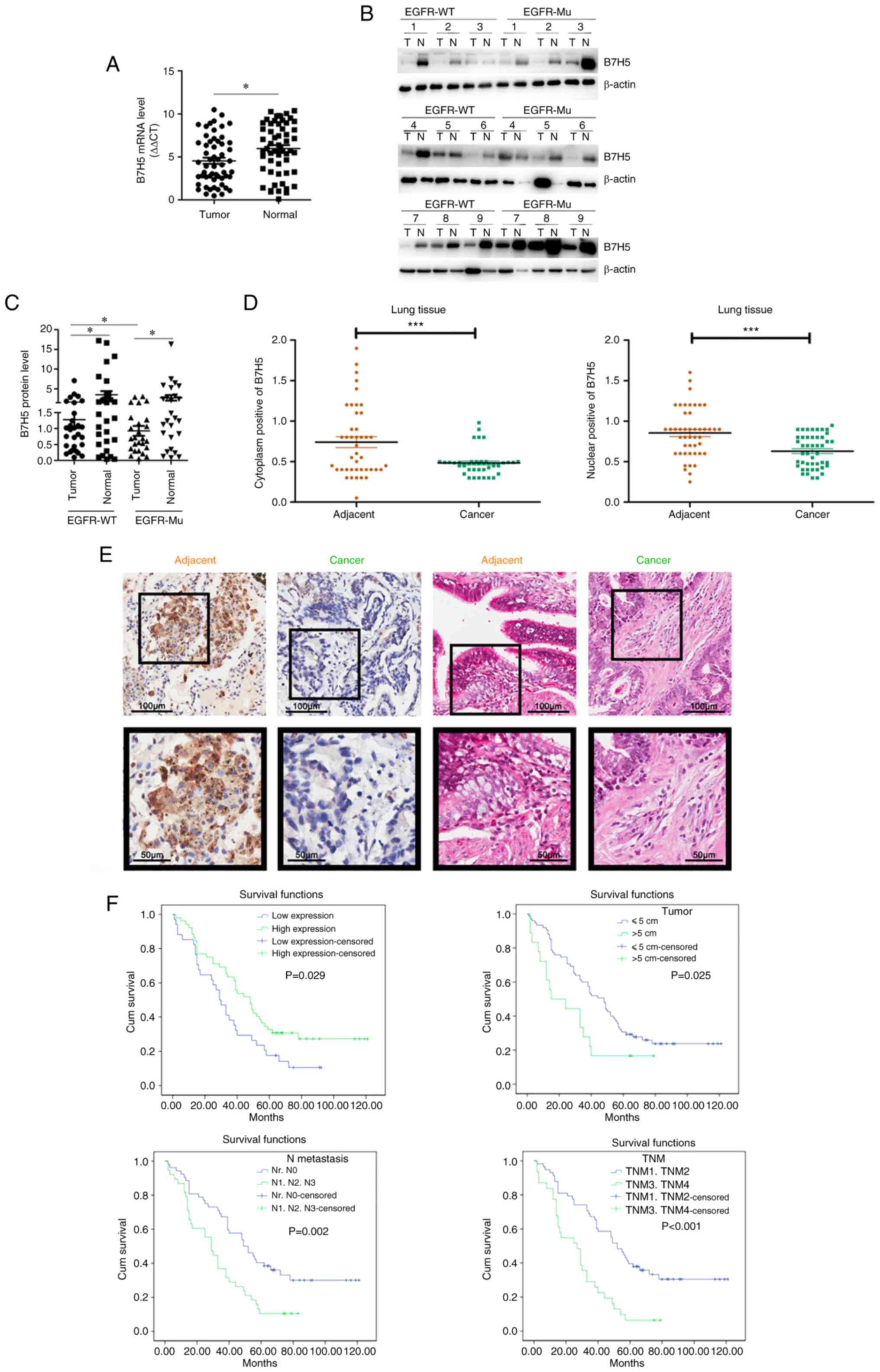
Expression of B7H5 is lower in NSCLC
tumors than that in normal tissue
The expression of B7H5 was determined in lung tumor
tissues and matched paracancerous tissues collected from 55
different patients, including 29 patients carrying wild-type EGFR
and 26 patients carrying mutations in EGFR. The results indicated
that B7H5 expression was lower in the tumor tissues compared with
that in the matched paracancerous tissues in both the wild-type and
mutated EGFR groups (Fig. 8A-C).
Further tissue microarray analysis indicated that the expression of
B7H5 was lower in lung tumor tissue compared with that in the
adjacent tissue (Fig. 8D).
Immunohistochemical analysis also indicated that the expression of
B7H5 in lung cancer tissues was lower than that in adjacent tissues
(Fig. 8E). Kaplan-Meier analysis
suggested that patients with certain characteristics, such as low
B7H5 and high B7H5 expression, tumor diameter >5 cm, TNM3/TNM4
or metastasis stages of N1, N2 or N3, had a shorter overall
survival time (Fig. 8F).
Discussion
EGFR is a glycoprotein belonging to the tyrosine
kinase receptor family that is inextricably linked to the
occurrence and development of various solid tumors, as well as
tumor invasion and metastasis. EGFR signaling pathways are
frequently abnormally activated or their members are highly
expressed in cancer, which indicates a correlation between tumor
progression and EGFR-associated gene expression. When ATP binds to
the ATP binding site on the tyrosine kinase domain, an EGFR-TK
inhibitor is able to block the phosphorylation and activation of
EGFR tyrosine kinase, which delays EGFR signal transduction,
restrains cell proliferation and accelerates apoptosis, thus
inhibiting the tumor. Recently, immune cells in the TME have been
identified as immunological effector cells that serve an important
role in tumor recognition and immune defense to attack tumor cells.
Thus, immunotherapy has an important role in the treatment of
NSCLC. It may activate the immune system by killing tumor cells
that have escaped previous immunological surveillance (21). The results of the present study
confirmed that gefitinib was able to increase the cytotoxicity of
PBMCs to kill EGFR-mutant NSCLC cells. Transfection with the
mutated EGFR plasmid reduced the cytotoxicity of PBMCs in wild-type
NSCLC cells; however, gefitinib treatment partially recovered the
cytotoxicity of PBMCs in wild-type NSCLC cells expressing mutant
EGFR. These results demonstrated that gefitinib was able to inhibit
the overactivation of EGFR in EGFR-mutated NSCLC cells and enhance
the toxicity of PBMCs against NSCLC cells.
Immune checkpoint therapy is a novel treatment that
targets regulatory pathways in T cells to enhance anti-tumor immune
responses. The B7/CD28 axis, the first discovered T-cell
ligand/receptor complex, has been studied extensively (22). The B7/CD28 family has an important
role in cancer pathogenesis. It has been reported that the
B7-H5/CD28H pathway has co-inhibitory and co-stimulatory functions
in immune signaling (15,17,23).
High B7-H5 expression induced a more potent immune reaction
following co-culture with T cells in pancreatic ductal
adenocarcinoma (PDAC) cell culture and high expression of B7-H5 was
able to improve the prognosis of patients with PDAC (16). The present findings indicated that
the expression of B7H5 in EGFR-mutant NSCLC cells was upregulated
after gefitinib treatment; however, B7H5 expression was not changed
in wild-type NSCLC cells treated with gefitinib. For wild-type
NSCLC cells, B7H5 was downregulated relative to the control group
only after transfection with the EGFR mutation plasmid and it was
upregulated after EGFR mutation combined with gefitinib treatment,
leading to an enhanced killing effect of PBMCs on wild-type NSCLC
cells. The results also indicated that reduced B7H5 expression by
siRNA in NSCLC cells decreased the cytotoxicity of PBMCs.
Furthermore, B7H5 knockdown suppressed the gefitinib-enhanced
cytotoxicity by PBMCs in the EGFR-mutant NSCLC cells. These
findings suggested that B7H5 expression is upregulated under
gefitinib treatment, causing an improved immune response in
PBMCs.
CD28 has been recognized as a major co-stimulatory
receptor that specializes in priming pan-naïve T cells, and
promoting both T cell division and cytokine production,
particularly IL-2, in secondary lymphoid organs (7). CD28 signaling prevents T-cell anergy,
the unresponsiveness status of T cells to antigen challenge
(10,17). The results of the present study
indicated that transfection of a CD28 siRNA into PBMCs blocked the
gefitinib-induced high toxicity of PBMCs toward EGFR-mutated NSCLC.
These results indicated that the B7H5/CD28H interaction is involved
in the boosted immune response of PBMCs after EGFR-mutated cells
were treated with gefitinib. Furthermore, our group is also
considering examining and further screening how the mutant EGFR
signal enters cells and specifically regulates B7H5 or what
signaling pathways are involved. This will comprise a large number
of signaling pathways and the workload is high, which will be the
focus of research by our group in the future. Furthermore, deep
research will be performed to verify whether mutant EGFR activates
any specific signaling that is not affected by the wild-type
protein.
In conclusion, the present study indicated that
inhibition of mutated EGFR by gefitinib was able to activate the
B7-H5/CD28H signaling pathway and increase the cytotoxicity of
PBMCs against NSCLC cells. Furthermore, the expression of B7-H5 is
associated with improved prognosis in NSCLC. The present findings
indicated that the B7-H5/CD28H pathway may be a potential
immunotherapeutic target in NSCLC.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW and WX conceived the study. HG, XZ, SX and TW
performed the experiments. DX, YC and DC analyzed the data. XW
wrote the manuscript. All authors have read and approved the final
version of the manuscript. XW and WX confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
This study was approved by the Medical Ethics
Committee of the First Affiliated Hospital of Huzhou University
(Huzhou, China; approval no. 2022KYLL043). All participants and/or
their legal guardian provided written informed consent prior to use
of their tissues for scientific research.
Patient consent for publication
All participants and/or their legal guardian
provided written informed consent prior to use of their tissues for
scientific research.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors thank Professor Yuwen Zhu (Department of
Surgery, University of Colorado Anschutz Medical Campus, Aurora,
USA) for providing antibodies/biotin-CD28H reagents.
Funding
This study was funded by Huzhou Science and Technology Fund
(grant nos. 2018GY04 and 2017GYB09), the National Natural Science
Foundation of China (grant nos. 81870377, 81970570 and 82172361),
the Scientific Technology Projects of Health and Medicine of
Zhejiang Province (grant no. 2020KY940), Zhejiang Province
Traditional Medical Science Fund Project of China (grant no.
2020ZB036) and Zhejiang Province Medical and Health Science (grant
no. 2019RC128).
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Dyba T, Randi G, Bettio M, Gavin A, Visser O and Bray F: Cancer
incidence and mortality patterns in Europe: Estimates for 40
countries and 25 major cancers in 2018. Eur J Cancer. 103:356–387.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu C, Li M, Meng H, Liu Y, Niu W, Zhou Y,
Zhao R, Duan Y, Zeng Z, Li X, et al: Analysis of status and
countermeasures of cancer incidence and mortality in China. Sci
China Life Sci. 62:640–647. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nasim F, Sabath BF and Eapen GA: Lung
cancer. Med Clin North Am. 103:463–473. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Relli V, Trerotola M, Guerra E and Alberti
S: Abandoning the notion of non-small cell lung cancer. Trends Mol
Med. 25:585–594. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Noronha V, Patil VM, Joshi A, Menon N,
Chougule A, Mahajan A, Janu A, Purandare N, Kumar R, More S, et al:
Gefitinib versus gefitinib plus pemetrexed and carboplatin
chemotherapy in EGFR-mutated lung cancer. J Clin Oncol. 38:124–136.
2020. View Article : Google Scholar
|
|
6
|
Gibson AJW, D'Silva A, Elegbede AA, Tudor
RA, Dean ML, Bebb DG and Hao D: Impact of Asian ethnicity on
outcome in metastatic EGFR-mutant non-small cell lung cancer. Asia
Pac J Clin Oncol. 15:343–352. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harrison PT, Vyse S and Huang PH: Rare
epidermal growth factor receptor (EGFR) mutations in non-small cell
lung cancer. Semin Cancer Biol. 61:167–179. 2020. View Article : Google Scholar :
|
|
8
|
Gelatti ACZ, Drilon A and Santini FC:
Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in
epidermal growth factor receptor (EGFR) mutation-positive non-small
cell lung cancer (NSCLC). Lung Cancer. 137:113–122. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dong ZY, Zhang JT, Liu SY, Su J, Zhang C,
Xie Z, Zhou Q, Tu HY, Xu CR, Yan LX, et al: EGFR mutation
correlates with uninflamed phenotype and weak immunogenicity,
causing impaired response to PD-1 blockade in non-small cell lung
cancer. Oncoimmunology. 6:e13561452017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhong C, Lang Q, Yu J, Wu S, Xu F and Tian
Y: Phenotypical and potential functional characteristics of
different immune cells expressing CD28H/B7-H5 and their
relationship with cancer prognosis. Clin Exp Immunol. 200:12–21.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y, Zheng Q and Jin L: The role of B7
family molecules in maternal-fetal immunity. Front Immunol.
11:4582020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kang JH, Jung MY and Leof EB: B7-1 drives
TGF-β stimulated pancreatic carcinoma cell migration and expression
of EMT target genes. PLoS One. 14:e02220832019. View Article : Google Scholar
|
|
13
|
Byers JT, Paniccia A, Kaplan J, Koenig M,
Kahn N, Wilson L, Chen L, Schulick RD, Edil BH and Zhu Y:
Expression of the novel costimulatory molecule B7-H5 in pancreatic
cancer. Ann Surg Oncol. 22(Suppl 3): S1574–S1579. 2015. View Article : Google Scholar
|
|
14
|
Dong Z, Zhang L, Xu W and Zhang G: EGFR
may participate in immune evasion through regulation of B7-H5
expression in non-small cell lung carcinoma. Mol Med Rep.
18:3769–3779. 2018.PubMed/NCBI
|
|
15
|
Zhu Y, Yao S, Iliopoulou BP, Han X,
Augustine MM, Xu H, Phennicie RT, Flies SJ, Broadwater M, Ruff W,
et al: B7-H5 costimulates human T cells via CD28H. Nat Commun.
4:20432013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Q, Wang J, Chen W, Zhang Q, Wei T,
Zhou Y, Xu X, Bai X and Liang T: B7-H5/CD28H is a co-stimulatory
pathway and correlates with improved prognosis in pancreatic ductal
adenocarcinoma. Cancer Sci. 110:530–539. 2019. View Article : Google Scholar :
|
|
17
|
Hu C, Xu Z, Chen S, Lv H, Wang Y, Wang X,
Mo S, Shi C, Wei S, Hu L, et al: Overexpression of B7H5/CD28H is
associated with worse survival in human gastric cancer. J Cell Mol
Med. 24:1360–1369. 2020. View Article : Google Scholar :
|
|
18
|
Das AK, Sato M, Story MD, Peyton M, Graves
R, Redpath S, Girard L, Gazdar AF, Shay JW, Minna JD and Nirodi CS:
Non-small-cell lung cancers with kinase domain mutations in the
epidermal growth factor receptor are sensitive to ionizing
radiation. Cancer Res. 66:9601–9608. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Steiner P, Joynes C, Bassi R, Wang S,
Tonra JR, Hadari YR and Hicklin DJ: Tumor growth inhibition with
cetuximab and chemotherapy in non-small cell lung cancer xenografts
expressing wild-type and mutated epidermal growth factor receptor.
Clin Cancer Res. 13:1540–1551. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trick AY, Chen FE, Schares JA, Freml BE,
Lor P, Yun Y and Wang TH: High resolution estimates of relative
gene abundance with quantitative ratiometric regression PCR
(qRR-PCR). Analyst. 146:6463–6469. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jia Y, Li X, Jiang T, Zhao S, Zhao C,
Zhang L, Liu X, Shi J, Qiao M, Luo J, et al: EGFR-targeted therapy
alters the tumor microenvironment in EGFR-driven lung tumors:
Implications for combination therapies. Int J Cancer.
145:1432–1444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ni L and Dong C: New B7 family checkpoints
in human cancers. Mol Cancer Ther. 16:1203–1211. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao R, Chinai JM, Buhl S, Scandiuzzi L,
Ray A, Jeon H, Ohaegbulam KC, Ghosh K, Zhao A, Scharff MD and Zang
X: HHLA2 is a member of the B7 family and inhibits human CD4 and
CD8 T-cell function. Proc Natl Acad Sci USA. 110:9879–9884. 2013.
View Article : Google Scholar : PubMed/NCBI
|