Introduction
In previous years, a gradual increase has been
detected in the annual incidence of breast cancer worldwide.
Notably, breast cancer can be divided into several subtypes
according to the presence of receptors on the cell surface
(1). In 2018, there were
>2,000,000 new cases of breast cancer and >620,000 deaths
associated with breast cancer worldwide (2). Previous studies have reported that
20-30% of patients with breast cancer are diagnosed with distant
metastases at the time of primary diagnosis, and 25% of primary
non-metastatic cases eventually result in metastases. Despite
development being made in diagnostic and therapeutic methods, the
prognosis of patients with breast cancer remains unsatisfactory,
particularly concerning metastatic breast cancer (3). Moreover, the issues of drug
resistance and heterogeneity in breast cancer are often attributed
to treatment failure and tumor recurrence (4). In previous studies, several markers
have been used for the diagnostic and prognostic prediction of
breast cancer. For example, phosphorylated-STAT3 expression has
been reported to be associated with the survival and mammographic
density of patients with breast cancer (5). In addition, ceruloplasmin has been
shown to be correlated with immune infiltration and may serve as a
prognostic biomarker in breast cancer (6). However, a single biomarker does not
show better predictive performance compared with previously
existing predictive models, thus it is urgent to explore new
multi-gene models to improve diagnostic and prognostic predictions
in patients with breast cancer. Furthermore, identification of
novel and effective therapeutic targets and predictive models using
biomarkers may contribute to the comprehensive elucidation of the
potential mechanisms involved in the development and progression of
breast cancer, which would further aid in improving the overall
survival (OS) rate in patients with breast cancer.
It has previously been reported that various cancer
treatment methods can induce cell-specific programmed cell death,
which is known to be closely associated with tumor development and
progression (7). Ferroptosis, a
novel form of programmed cell death first identified in 2012
(8), has gained increasing
attention as a potential therapeutic pathway for cancer treatment.
Ferroptosis is characterized by iron-dependent lipid peroxide
accumulation, which differs from traditional apoptosis, autophagy
or necrosis in terms of morphology, biochemistry and genetics
(8). Numerous genes have
previously been identified as markers, inducers or inhibitors of
ferroptosis (9-11), collectively called
ferroptosis-related genes (FRGs), such as GPX4, CISD1 and NRF2.
Previous studies have identified a pivotal role of ferroptosis in
tumor progression and therapeutics (12-14).
Although the sensitivity of different types of tumor cells towards
ferroptosis are diverse, a combination of erastin (an inducer of
ferroptosis) and chemotherapeutics could improve curative effects
in various types of cancer, such as ovarian cancer (15), lung cancer (16) and breast cancer (17). Moreover, ferroptosis has been
reported to be associated with the prognosis of various types of
cancer, and ferroptosis-related prognostic models have been
constructed in glioma (18),
melanoma (19) and renal cell
carcinoma (20), further
indicating the potential value of FRGs as prognostic markers and
therapeutic targets in human cancer. Several reports have shown
that ferroptosis is strongly associated with breast cancer
(21,22), indicating that ferroptosis may be
considered an important biomarker in breast cancer. However, single
genes alone cannot comprehensively predict the diagnosis and
survival of patients with breast cancer. Hence, additional efforts
are required to establish ferroptosis-related predictive models for
prediction and treatment of breast cancer to further improve the
prognosis of patients with breast cancer.
The tumor immune microenvironment is typically
comprised of immune cells and immune-related molecules that are
present around the tumor (23,24),
highlighting the significant role of the immune system in
tumor-stroma interactions and the response towards immunotherapy.
In recent years, the association between immune cells and immune
molecules with iron metabolism has gained considerable attention
(25). Various types of immune
cells, including Th1 cells, natural killer (NK) T cells and
macrophages, have previously been shown to be associated with the
maintenance of iron homeostasis (26). Notably, ferroptosis in tumor cells
can increase the expression of tumor antigens that bind immune
cells, which further promotes the antitumor efficacy of
immunotherapy (27). However, the
role of ferroptosis in immunotherapy of breast cancer has not been
fully elucidated.
In the present study, a ferroptosis-related
prognostic model was constructed based on mRNA expression profiles
and clinical data of patients with breast cancer obtained from the
Gene Expression Omnibus GSE20685 cohort (28). The model was further validated
using data from the Molecular Taxonomy of Breast Cancer
International Consortium (METABRIC) cohort (29). Moreover, gene signature
characteristics in the tumor microenvironment were evaluated using
single-sample gene set enrichment analysis (ssGSEA) and immune
infiltration analysis. The results of the present study may provide
novel therapeutic targets for the management of breast cancer.
Additionally, the present study may assist in improving clinical
outcomes of patients with breast cancer when subjected to
personalized treatment.
Materials and methods
Data collection
The expression profiling of FRGs and the
corresponding clinical information of 327 patients with breast
cancer, including their age, TNM stage and survival information,
were obtained from the GSE20685 database (https://www.ncbi.nlm.nih.gov/geo/) and used as the
training cohort. Moreover, the gene expression and clinical
information of 1,904 patients with breast cancer from the METABRIC
database were downloaded from cBioPortal (https://www.cbioportal.org/) as previously reported
(30), which was used as the
validation cohort. FRGs were identified from the GeneCards database
(https://www.genecards.org/), FerrDb
database (http://www.zhounan.org/ferrdb/) and other related
literature (31,32). Consequently, a total of 314 FRGs
were included in the analysis.
Identification of breast cancer
subclasses
A filtering procedure was first conducted according
to a previous study (31). The low
median absolute deviation (MAD) value, a robust statistic to
measure the statistical deviation, was calculated for each
candidate gene. The genes with a MAD value <0.5 were excluded.
Subsequently, R software was used for further analysis (33), and the R packages were downloaded
from the R website (https://cran.r-project.org/mirrors.html). The
non-negative matrix factorization (NMF) clustering method was
applied using the 'NMF' R package (34) with the default parameters 'nrun=10'
and 'seed=1'. The best cluster number was selected as the
coexistence correlation coefficient k=2 (35). T-distributed stochastic neighbor
embedding (t-SNE) analysis was then performed to validate the
distribution of different groups using the 'Rtsne' R package
(https://github.com/jkrijthe/Rtsne).
The related parameters were: dims=2, perplexity=10, verbose=F,
max_iter=500, check_duplicates=F. Moreover, principal components
analysis (PCA) was performed to assess the differences in
expression between the subtypes.
Establishment and validation of the
prognostic predictive signature
Univariate Cox regression analysis was first
performed to screen the genes related to OS in patients with breast
cancer using the 'survival' package in R software (https://github.com/therneau/survival).
Genes that were P<0.05 were considered statistically significant
and were incorporated into the subsequent least absolute shrinkage
and selection operator (LASSO) Cox regression using the 'Glmnet'
package in R software (36). The
related parameters were: lasso family='cox', maxit=1000, nfold=10,
α=1. Subsequently, based on the multivariate Cox regression
analysis for these genes, a prognostic signature was constructed.
The prognostic risk score was calculated based on the regression
coefficients (β) in the multivariate Cox regression model and the
expression levels of the genes. The risk score calculation was as
follows: Risk score=(−0.854 × expression of YWHAE) + (−0.852 ×
expression of CD44) + (−0.683 × expression of HILPDA) + (−0.284 ×
expression of IFNG) + (−0.272 × expression of MYB) + (0.511 ×
expression of DBN1) + (0.554 × expression of HES1) + (0.652 ×
expression of HSPB1) + (1.107 × expression of SLC11A2). Receiver
operating characteristic (ROC) analysis was performed to determine
the optimal cut-off value, in order to divide the patients with
breast cancer into the high- and low-risk groups. Kaplan-Meier
(K-M) survival analysis followed by log-rank test were performed to
evaluate the prognosis between two groups in both the training
cohort and validation cohort. Moreover, the time-dependent ROC
curves were used to validate the sensitivity and accuracy of the
prognostic signature on OS in two cohorts.
Establishment of the nomogram model
Univariate Cox regression analysis was performed to
evaluate the prognostic values of the clinical information (i.e.,
age, grade and TNM stage) and risk score. Subsequently,
multivariate Cox regression analysis was applied to determine the
independent prognostic factors to predict the survival of patients
with breast cancer. By combining the TNM stage and risk score, a
nomogram was constructed using the survival rate and the 'RMS' R
package (https://github.com/harrelfe/rms). The related
parameters were: cmethod='KM', method='boot', u=time, B=1000.
According to the nomogram, the total point of each patient was
calculated. The calibration curve was employed to evaluate the
consistency between the actual and predicted survival rates. ROC
curve analysis was applied to validate the sensitivity and
specificity of the nomogram compared to that of a single
independent predictor for predicting OS. Decision curve analysis
(DCA) was performed using the 'RMDA' package (http://mdbrown.github.io/rmda/) with the
parameters 'family=binomial(link='logit')' to evaluate the clinical
predictive effect obtained by the nomogram compared to a single
independent prognostic predictor.
Estimation of immune infiltration
The estimation of stromal and immune cells in
malignant tumor tissues using expression data (ESTIMATE) algorithm
was applied to the GSE20685 and METABRIC cohorts to calculate the
immune score, stromal score and tumor purity, which reflected the
enrichment of immune and stromal cell gene signatures (37). The CIBERSORT analysis was performed
in two cohorts to assess the infiltration levels of 22 human immune
cell subpopulations using the 'CIBERSORT' R package with the
parameters 'perm=100' and 'QN=TRUE' (38). Subsequently, the fractions of 16
immune cells and the scores of 13 immune-related functions in two
cohorts were respectively calculated using the ssGSEA with the
'GSVA' R package (39). The
related parameters were: method='ssgsea', kcdf='Gaussian', abs.
ranking=TRUE. To predict whether each subgroup could benefit from
immunotherapy, the similarity of the gene expression profiles was
calculated between the subgroups and the previously published data
from patients with melanoma treated with immunotherapy based on the
SubMap analysis (40).
Prediction of potential drugs based on
drug-gene correlation analyses
The drug z-scores and the corresponding gene
expression of the NCI-60 cancerous cell lines were downloaded from
the CellMiner database (https://discover.nci.nih.gov/cellminer/loadDownload.do)
(41). A higher z-score is
indicative of higher sensitivity to the corresponding drug.
Subsequently, the Pearson correlation between gene expression and
drug sensitivity was analyzed. To perform this, the 'impute'
(https://bioconductor.org/packages/impute/) and 'limma'
R packages (42) were used for
data processing, and the 'ggplot2' (https://ggplot2.tidyverse.org) and 'ggpubr'
(https://rpkgs.datanovia.com/ggpubr/)
R packages were used for visualization. The drug information was
obtained from the DrugBank database (https://www.drugbank.ca/).
Cell culture
The human breast cancer cell lines MDA-MB-231
(HTB-26), MDA-MB-468 (HTB-132), MCF-7 (HTB-22) and T47D (HTB-133)
were obtained from the American Type Culture Collection. The cells
were cultured in DMEM (Macgene Biotechnology) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Macgene Biotechnology) in a standard
humidified incubator supplied with 5% CO2 at 37°C.
MTT assay
Erastin (10 or 20 µM; cat. no. HY-15763) and
ferrostatin-1 (Fer-1; 10 µM; cat. no. HY-100579) were
purchased from MedChemExpress. A total of 3×103 cells
were plated into 96-well plates and incubated overnight. After
treating the breast cancer cells (MDA-MB-231, MDA-MB-468, MCF-7 and
T47D) with the reagents erastin, ferrostatin-1, paclitaxel
(MedChemExpress), doxorubicin (MedChemExpress) or tamoxifen
(MilliporeSigma) at the indicated concentrations for 48 h at 37°C,
20 µl 5 mg/ml MTT was added to each well, and the cells were
further incubated for 4 h at 37°C, after which 200 µl DMSO
was added. The absorbance was measured at 490 nm using a microplate
reader (Bio-Rad Laboratories, Inc.).
Migration assay
A Transwell system (24 wells; pore size, 8
µm; a polycarbonate membrane) was used for the in
vitro migration assays. The breast cancer cells (MDA-MB-231,
MDA-MB-468, MCF-7 and T47D) were pretreated with 10 µM
erastin with or without 10 µM ferrostatin-1 for 24 h at
37°C. DMSO was used as vehicle. Subsequently, 1×105
cells were suspended in 200 µl serum-free medium and added
to the upper chamber, and 700 µl medium supplemented with
20% FBS was added to the lower chamber. Cells were incubated for
24-48 h, after which the cells that had attached to the lower
surface were fixed with methanol for 15 min and stained with 0.2%
crystal violet for 20 min at room temperature. Finally, the
migrated cells were imaged using a light microscope (Olympus
Corporation) and quantified.
Reactive oxygen species (ROS)
analysis
A reactive oxygen species assay kit (cat. no.
S0033S; Beyotime Institute of Biotechnology) was used to detect
cellular ROS levels. MDA-MB-231 and MDA-MB-468 cells were plated in
a 6-well plate. When cell density reached 80%, the cells were
treated with 10 or 20 µM erastin or 10 µM
ferrostatin-1 for 24 h at 37°C. The treated cells were collected
and washed three times with PBS. Serum-free DMEM supplemented with
2 µM DCFH-DA (1:5,000; Beyotime Institute of Biotechnology)
was added to the cells and incubated at 37°C for 30 min. After
incubation, the cells were washed and resuspended in 300 µl
PBS, and ROS accumulation in 10,000 cells was detected using a flow
cytometer (BD Accuri™ C6 Plus Flow Cytometer) and BD Accuri C6 Plus
software (BD Biosciences) with an excitation wavelength of 488 nm
and an emission wavelength of 525 nm. Analysis was performed using
FlowJo version 10.6.2 (FlowJo LLC).
Statistical analysis
Statistical analyses were performed using R, SPSS
version 19.0 (IBM Corp), and GraphPad Prism version 8 (GraphPad
Software, Inc.). Data from at least three independent experiments
are presented as the mean ± standard deviation. χ2 test
was used to evaluate the differences in clinicopathological
features between the two clusters of patients with breast cancer.
K-M survival analysis and the log-rank test were performed to
evaluate differences in the OS between the two groups. Univariate,
LASSO and multivariate Cox regression analyses were performed to
identify the independent prognostic factors. ROC curve analysis was
performed to assess the sensitivity and specificity of the
prognostic model. Unpaired Student's t-test was used to analyze the
differences between unpaired breast cancer tissues and normal
tissues in the databases. Comparisons in datasets containing
multiple groups were analyzed using a one-way ANOVA followed by a
post-hoc Dunnett's test. Pearson correlation analysis was used to
evaluate the correlation between gene expression and drug
sensitivity. P<0.05 was considered to indicate a statistically
significant difference.
Results
Classification of patients with breast
cancer based on FRGs
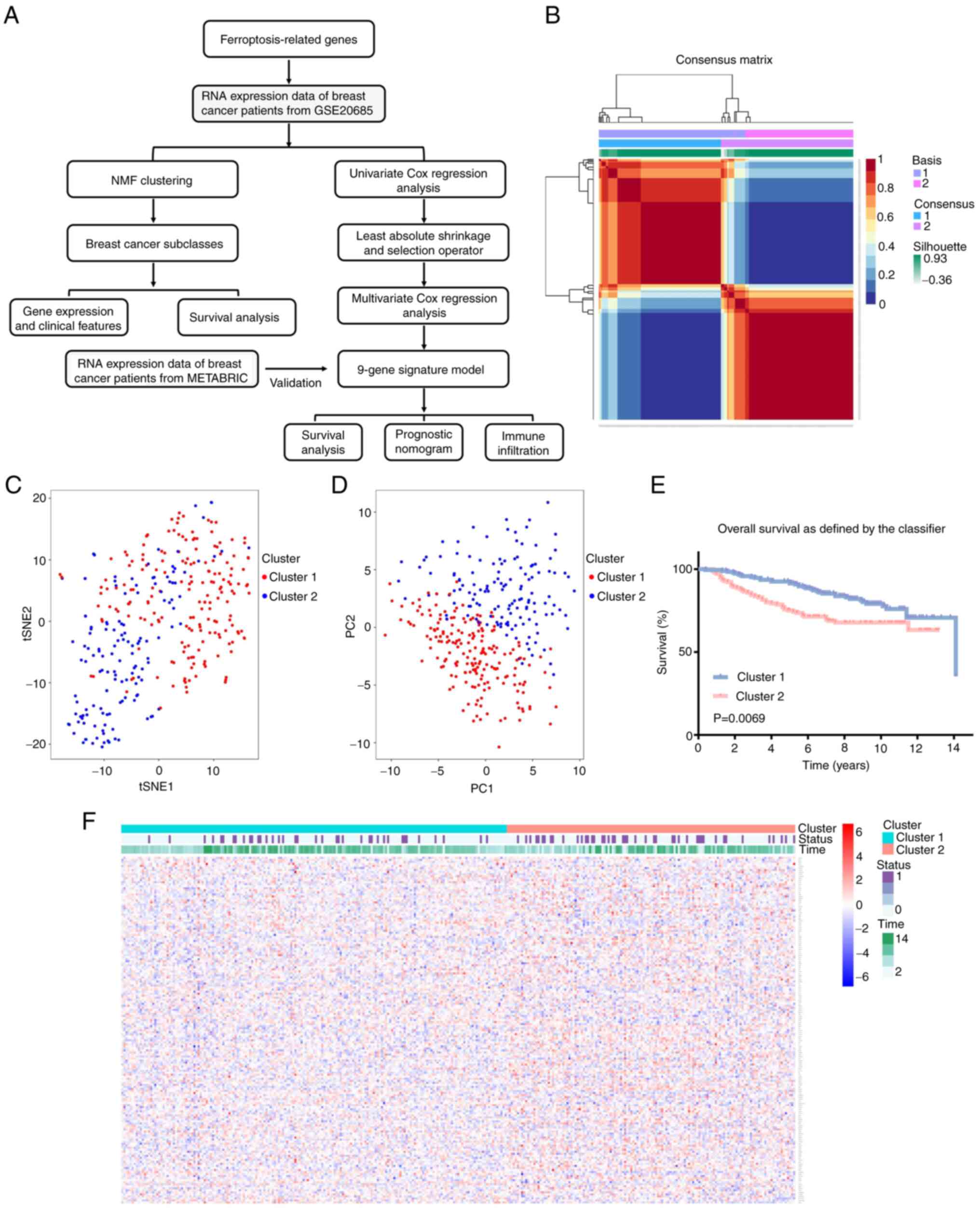
A flow chart of the study procedure is shown in
Fig. 1A. Following filtration
using MAD, a total of 171 genes were subjected to NMF analysis to
identify different groups of patients with breast cancer. To
identify the optimal k-value, cophenetic correlation coefficients
were calculated. For k=2, the consensus matrix heatmap
maintained a clear and sharp boundary, indicating that the samples
exhibited stable and robust clusters (Figs. 1B and S1). Consequently, the patients with
breast cancer were divided into two clusters, designated as cluster
1 and cluster 2. In particular, 187 and 140 samples were included
in cluster 1 (C1) and cluster 2 (C2), respectively. The results of
t-SNE analysis evidently confirmed that these two clusters were
largely in concordance with two-dimensional coordinate systems
(Fig. 1C). PCA analysis also
indicated that patients with breast cancer belonging to different
clusters were distributed in two directions (Fig. 1D). Furthermore, survival analysis
demonstrated that patients with breast cancer in cluster 2
exhibited a shorter survival time compared with patients in cluster
1 (Fig. 1E). The study further
analyzed the association between expression of FRGs and the
clinical status of each patient in the two clusters (Fig. 1F; Table SI). The results indicated
significant differences in the ferroptosis-related molecular
features between the two patient clusters. Clinical characteristics
of these two clusters are presented in Table SII. A χ2 test revealed
that there were more patients with an advanced TNM stage in cluster
2, suggesting that the expression of FRGs was closely associated
with tumor progression of patients with breast cancer.
Identification of prognostic FRGs in
breast cancer
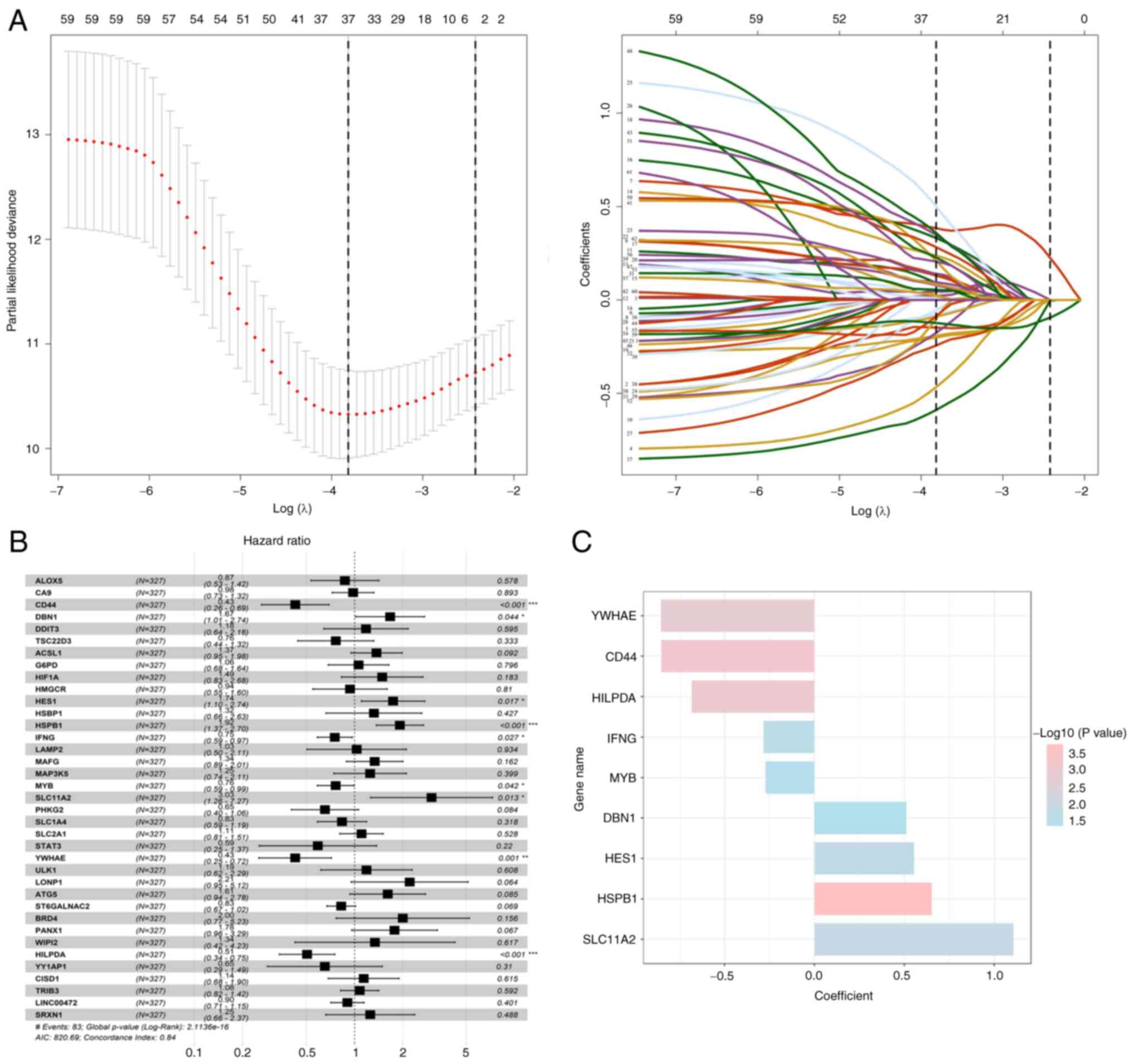
The study further evaluated the prognostic role of
FRGs in breast cancer. In the training cohort, 62 genes were
identified to be associated with OS, using univariate Cox
regression analysis (P<0.05; Table
SIII). Furthermore, 37 genes were considered to be associated
with the prognosis of breast cancer, based on the results of LASSO
regression analysis (Fig. 2A).
Multivariate Cox regression analysis was further performed, and
nine genes were finally selected for the construction of a
prognostic model on the basis of expression levels and regression
coefficients (Fig. 2B and C;
Table I). The risk scores for each
patient were calculated, and the patients were classified into
low-risk and high-risk groups based on the optimal cut-off value.
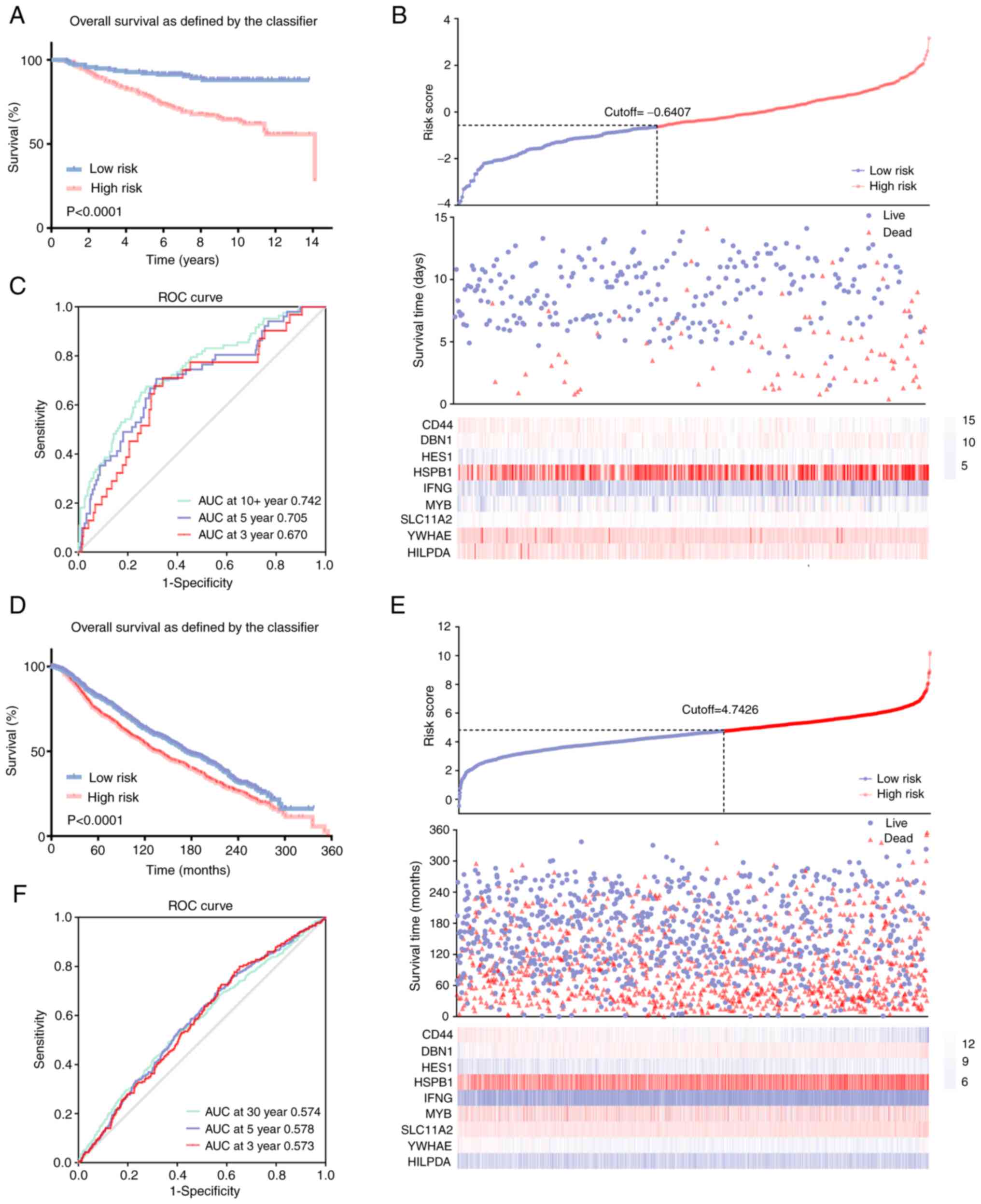
The results revealed that high-risk patients had a significantly
shorter OS and higher mortality rates compared with the low-risk
patients with breast cancer (Fig. 3A
and B). Notably, the area under the curve (AUC) values were
recorded to be 0.670, 0.705 and 0.742 in the time-dependent ROC at
3, 5 and 10 years, respectively (Fig.
3C), which indicated significant specificity and sensitivity of
the prognostic signature in the prediction of OS. The METABRIC
database was further used as an external validation cohort to
validate the predictive efficiency of the developed prognostic
model. To clarify the classification of the patients with breast
cancer into high-risk and low-risk groups, risk scores for the
patients from METABRIC were calculated, using the aforementioned
formula. In concordance with previous results, patients with breast
cancer in the high-risk group exhibited a significantly lower OS
and higher mortality rate compared with the patients in the
low-risk group (Fig. 3D and E).
Moreover, AUC values of 0.573, 0.578 and 0.574 were recorded for
the 3-, 5- and 10-year OS, respectively (Fig. 3F).
 | Table IP-values and regression coefficients
of the nine ferroptosis-related genes. |
Table I
P-values and regression coefficients
of the nine ferroptosis-related genes.
| Gene | P-value | Coefficient |
|---|
| YWHAE | 0.0013b | −0.854 |
| CD44 | 0.0006c | −0.852 |
| HILPDA | 0.0008c | −0.683 |
| IFNG | 0.0268a | −0.284 |
| MYB | 0.0422a | −0.272 |
| DBN1 | 0.0436a | 0.511 |
| HES1 | 0.0169a | 0.554 |
| HSPB1 | 0.0002c | 0.652 |
| SLC11A2 | 0.0133a | 1.107 |
Construction and validation of a
predictive nomogram for patients with breast cancer
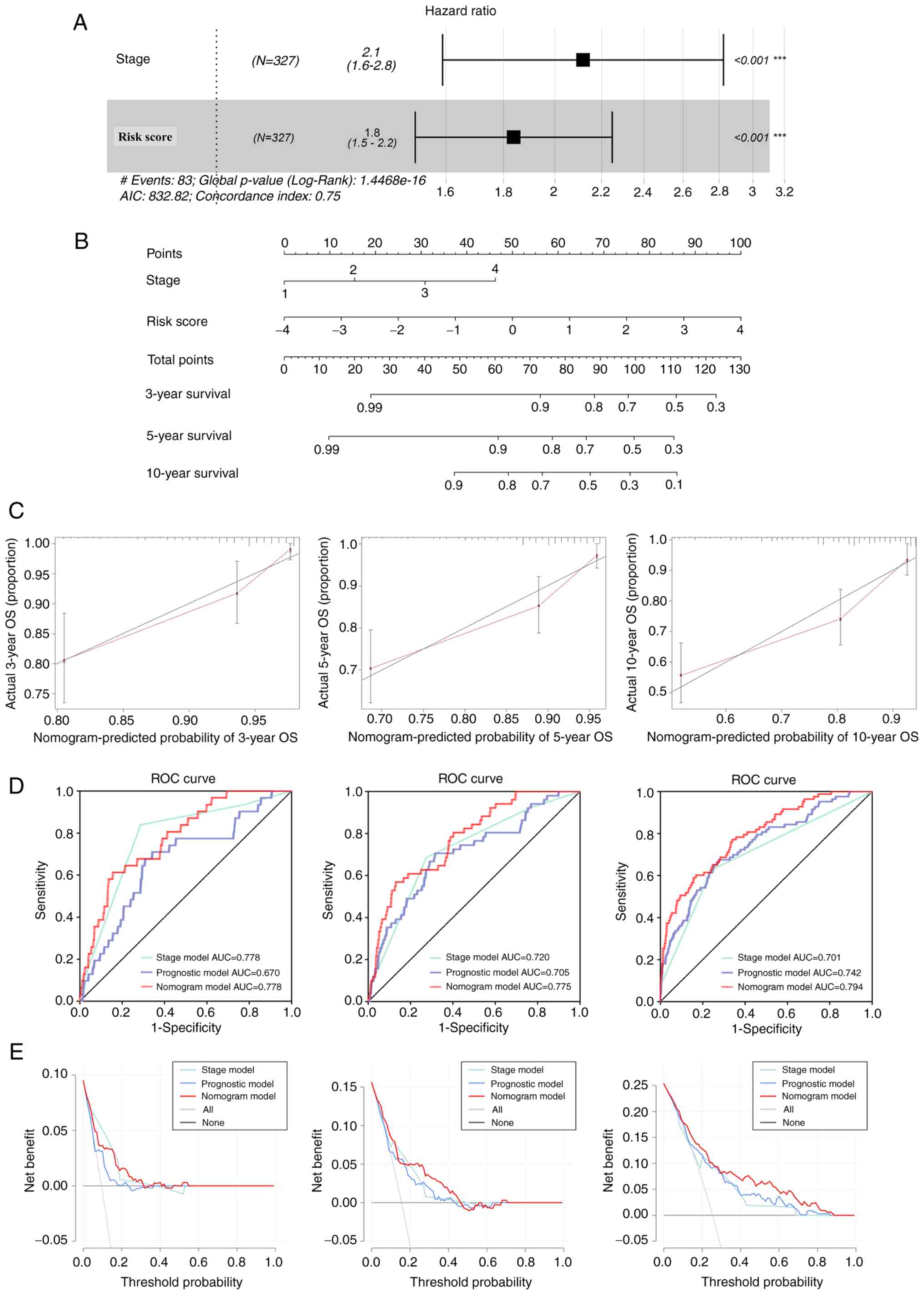
Univariate and multivariate Cox regression analyses
were performed to evaluate whether the prognostic value of the nine
gene model in the prediction of OS was independent of other
traditional clinicopathological parameters. The results of the
analysis indicated that the TNM stage and risk score of the
prognostic signature acted as independent predictors of OS
(Fig. 4A). C-index was recorded to
be 0.75. To quantify prediction results for individual survival
probability at 3, 5 and 10 years, a survival nomogram prediction
model was constructed (Fig. 4B).
The calibration curves showed an optimal agreement consistency
between observed and predicted OS rates at 3, 5 and 10 years
(Fig. 4C). Moreover, AUCs were
recorded to be 0.778, 0.775 and 0.794 with the nomogram for 3-, 5-
and 10-year OS, respectively. The model was considered to be
superior to a single independent predictive factor (Fig. 4D), which demonstrated the superior
predictive value of the nomogram. To further evaluate the
importance of the nomogram in clinical decision-making, DCA was
performed. DCA is a novel reliable evaluation tool that is used to
quantify the clinical value of a nomogram (43). The results of DCA indicated that
the nomogram provided optimal clinical decision-making benefits at
3, 5 and 10 years compared with a single independent predictive
factor (Fig. 4E). Additionally,
these results indicated that the nine-FRG prognostic model served
as a reliable prognostic indicator for patients with breast
cancer.
The present study also evaluated the mRNA expression
patterns of these FRGs in breast cancer according to METABRIC
database. The expression of HILPDA, IFNG, MYB, DBN1, HES1, HSPB1
and SLC11A2 was significantly higher in breast cancer tissues
compared with those in normal tissues. By contrast, the expression
levels of YWHAE and CD44 were significantly reduced in breast
cancer tissues as compared with in normal tissues (Fig. S2). These results indicated that
several prognostic FRGs may serve an oncogenic role in breast
cancer; however, the specific mechanisms regulated by these genes
require further study.
Comparison of immune infiltration between
high-risk and low-risk patients with breast cancer
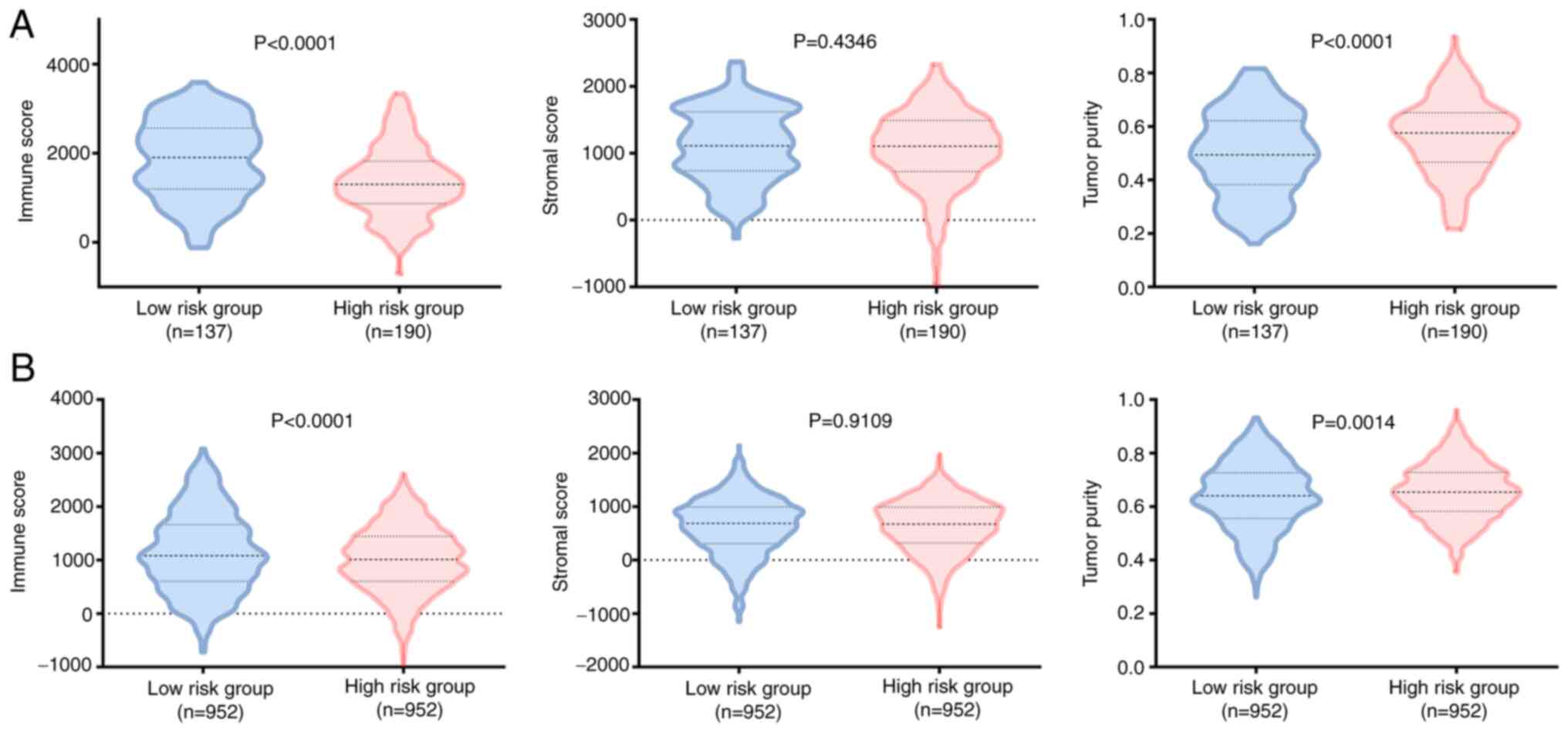
To evaluate the tumor heterogeneity between
high-risk and low-risk groups, stromal score, immune score and
tumor purity were calculated for both training and testing sets,
using the ESTIMATE algorithm. The results indicated that the
high-risk group exhibited higher tumor purity and lower immune
score in the training and testing sets, whereas no significant
differences were recorded between the stromal scores of the two
groups (Fig. 5). In view of the
significant differences recorded in the immune score for the two
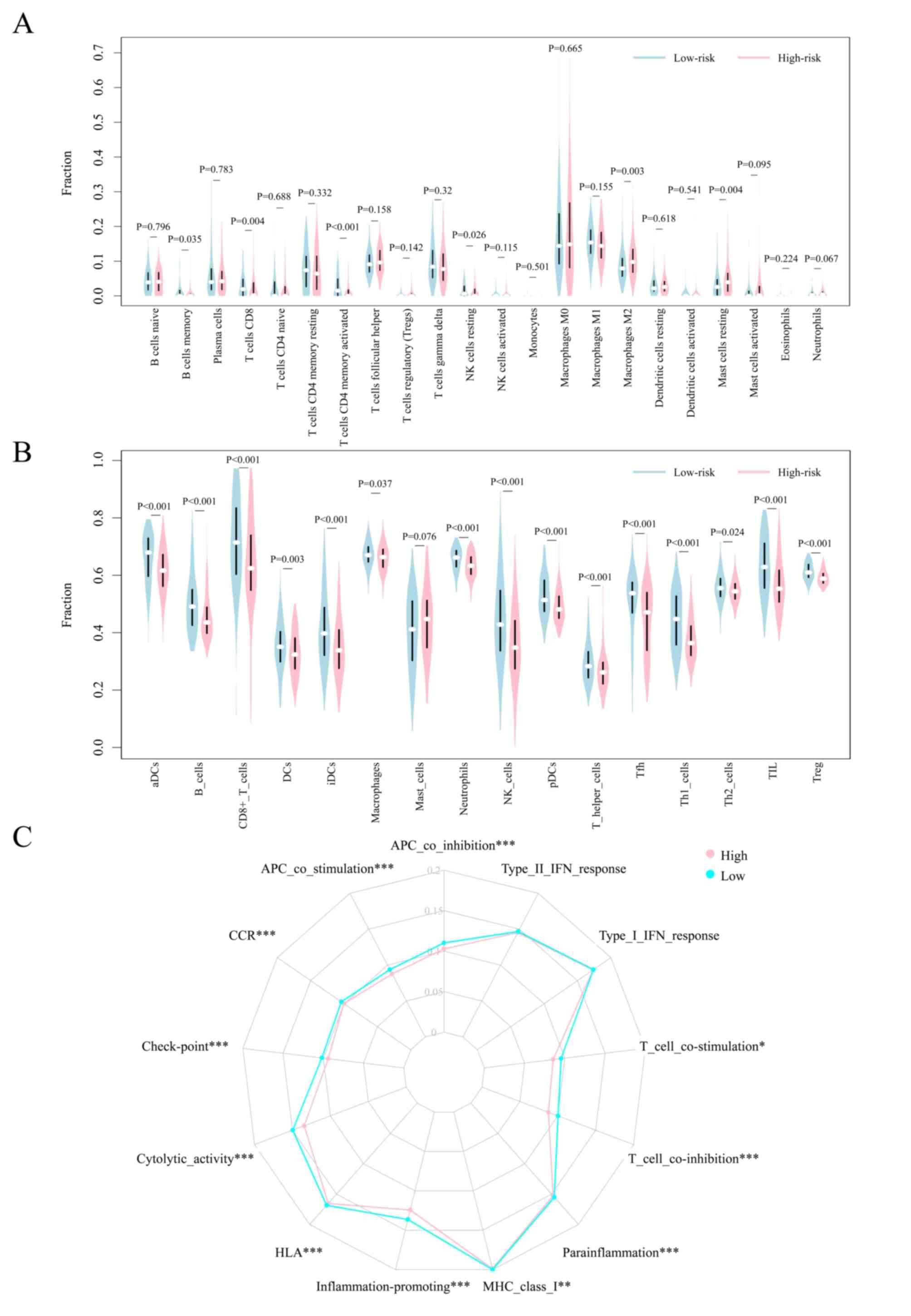
groups, the present study further evaluated immune infiltration
patterns for the 22 immune cell types in the patients with breast
cancer using the CIBERSORT algorithm. This may further aid in the
characterization of the immunological landscape.
Patients with breast cancer belonging to the
high-risk group exhibited higher ratios of M2 macrophages and
resting mast cells, and lower ratios of memory B cells, CD8 T
cells, T cells, activated CD4 memory T cells and resting NK cells
in the GSE20685 cohort (Figs. 6A,
S3A and B). Moreover, regulatory
T cells (Tregs) were enriched in the high-risk group, whereas CD8 T
cells, resting CD4 memory T cells, activated CD4 memory T cells,
γδT cells, resting NK cells, activated NK cells and activated mast
cells were found to be enriched in the low-risk group in the
METABRIC cohort (Fig. S3C and D).
Although no significant differences were observed for M2
macrophages in the METABRIC cohort, the infiltration ratio was
found to be relatively higher in the high-risk group compared with
in the low-risk group. These results suggested that the poor
prognosis of high-risk patients may be partly attributed to the
immunosuppressive microenvironment.
To further analyze the immune status of the two
groups, the ssGSEA algorithm was used to analyze the enrichment of
additional types of immune cells, and related functions or
pathways. The results of the analysis indicated that the high-risk
group was associated with universally lower immune cell
infiltration in the GSE20685 cohort (Figs. 6B and S3E). This was consistent with the lower
immune scores recorded for the high-risk group. Moreover, the
functions of antigen presentation, cytokine-cytokine receptor
interaction, cytolytic activity, immune activation and immune
surveillance were found to be at lower levels in the high-risk
group (Figs. 6C and S3E). Comparative analysis in the
METABRIC cohort confirmed the differences recorded for
antigen-presenting cells, checkpoint molecules, macrophages,
neutrophils and Treg cells between the two risk groups (Fig. S4A-C). The results of the study
revealed that these FRGs may partly regulate tumor progression via
modulation of the patterns of immune cell infiltration. However,
further studies are needed to elucidate the underlying
mechanisms.
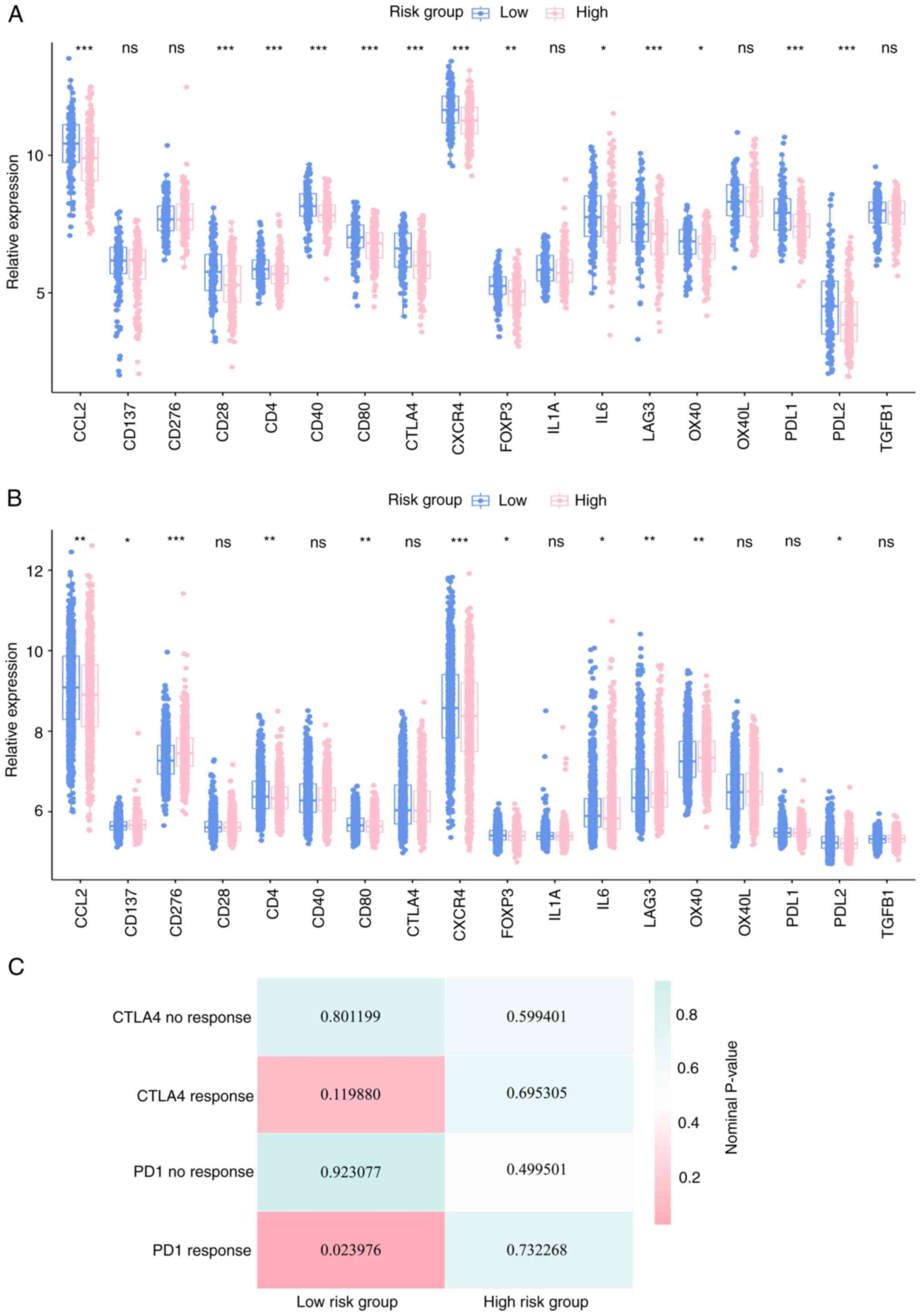
Previous studies have reported the importance of
checkpoint inhibitor-based immunotherapies (44,45).
The results of the present study also revealed a substantial
difference in the expression of several immune checkpoints between
the high-risk and low-risk groups in the two cohorts (Figs. 7A, B, S5A and B). Considering abnormal immune
infiltration patterns and differential expression of several immune
checkpoint genes in the high-risk and low-risk groups, the study
further assessed the probability of responding to immunotherapy.
SubMap analysis was used to compare expression profiles of the two
breast cancer sub-classes with a published dataset (40), which consisted of 47 patients with
melanoma that were subjected to treatment involving programmed cell
death protein-1 (PD-1) immune checkpoint inhibitor, or cytotoxic
T-lymphocyte-associated protein-4 (CTLA-4) immune checkpoint
inhibitor. The results of the SubMap analysis (46) indicated that the expression profile
of the low-risk group was significantly correlated with the PD-1
response group (P=0.023976), which indicated that patients in the
low-risk group would exhibit promising responses toward anti-PD-1
therapy (Fig. 7C).
Association between ferroptosis,
proliferation, migration and drug resistance in breast cancer
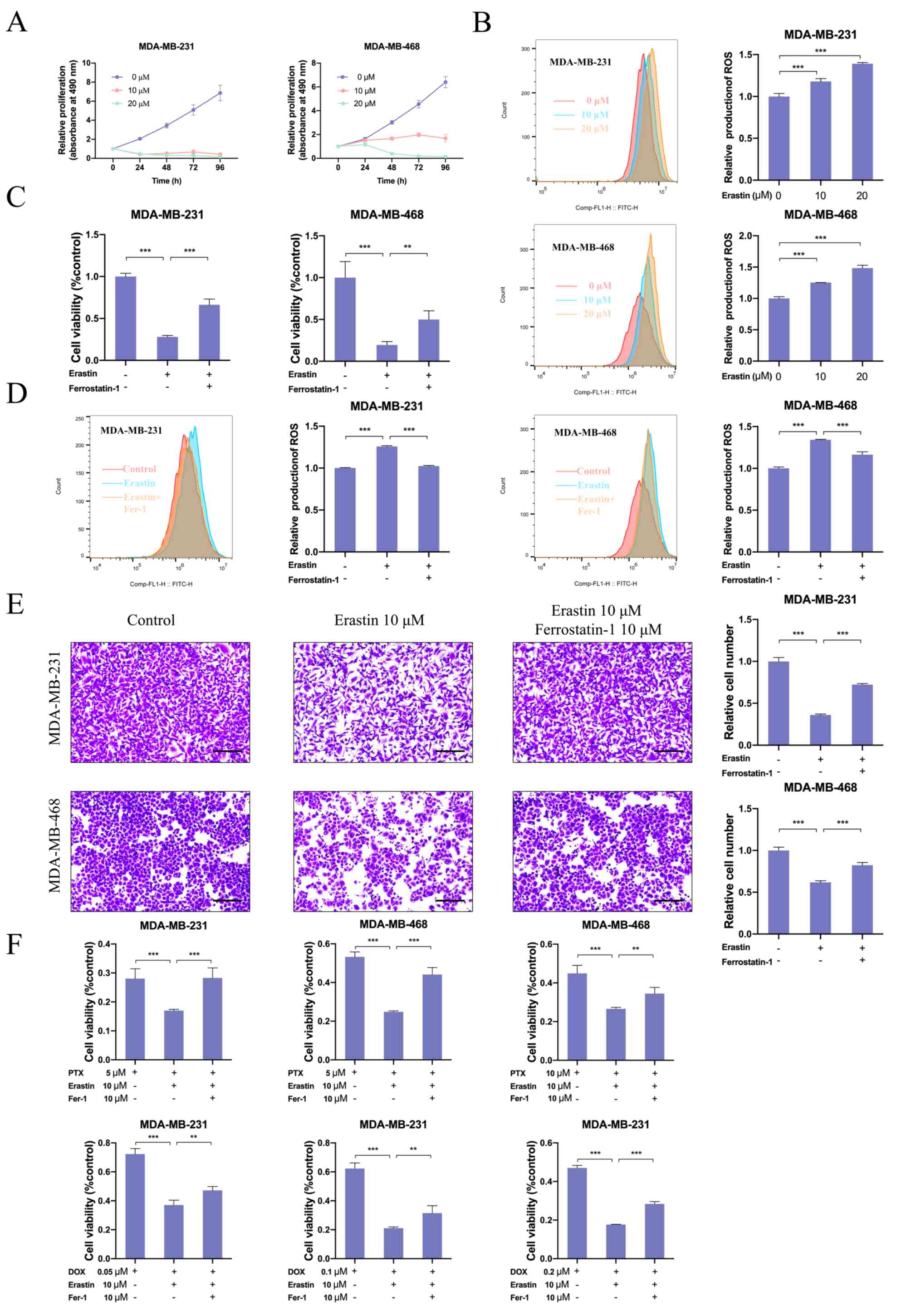
Erastin, an inducer of ferroptosis, is known to bind
and inhibit voltage-dependent anion channels (VDAC2/VDAC3) to
trigger iron-dependent cell death (8). The present study further evaluated
the effects of erastin on the development and progression of breast
cancer. The results indicated that erastin treatment could inhibit
breast cancer cell proliferation and promote the accumulation of
ROS in a dose-dependent manner (Figs.
8A, B, S6A and B). Moreover,
ferrostatin-1, a ferroptosis inhibitor, reversed the inhibition in
cell viability and decreased ROS production induced by erastin
(Figs. 8C, D, S6C and D). Furthermore, the migratory
ability of breast cancer cells treated with vehicle or erastin was
evaluated. When compared with vehicle-treated breast cancer cells,
the migratory ability of erastin-treated breast cancer cells was
inhibited. In comparison, ferrostatin-1 treatment could partially
rescue the migratory ability of breast cancer cells (Figs. 8E and S6E). The present study also evaluated
the effect of erastin on drug resistance of breast cancer cells.
Notably, erastin treatment significantly increased the cytotoxicity
of chemotherapeutics (paclitaxel and doxorubicin) and hormonal
agents (tamoxifen). In comparison, ferrostatin-1 attenuated the
activities of these drugs (Figs.
8F and S6F). These results
indicated that erastin inhibited proliferation, migration and
drug-resistance of breast cancer cells, which was mediated via
induction of ferroptosis.
Novel treatment against FRGs in breast
cancer
To screen potential molecular-targeted drugs for
breast cancer, the present study further analyzed the correlation
between the expression of FRGs and drug sensitivity on the basis of
NCI-60 drug z-scores and gene expression profiling data of cancer
cell lines obtained from the CellMiner database. The statistically
significant correlations between the drug z-scores and gene
expression (P<0.05) are listed in Table SIV. In general, a higher z-score
is indicative of the higher sensitivity of the cells towards the
corresponding drug. The representative Pearson's correlation dot
plots are presented in Fig. S7.
The drugs that were associated with at least four FRGs were
selected for potential therapeutic regimens. Subsequently, seven
FRG-related drugs were identified, which may be repurposed for
breast cancer. The detailed information regarding correlation
coefficients and drug applications obtained from DrugBank are
listed in Table SV. In
particular, nelarabine, which is known to be associated with MYB,
HES1, SLC11A2, HILPDA and YWHAE, is used in acute T-cell
lymphoblastic leukemia. This may be repurposed to treat breast
cancer.
Discussion
Ferroptosis is a novel oxidative, iron-dependent
form of cell death, which has attracted considerable attention in
recent years. Previous studies have shown that regulation of
ferroptosis can modulate cell proliferation, migration and drug
resistance in various types of cancer (47,48).
Additionally, it has been shown to serve a crucial role in tumor
progression and cancer therapeutics. However, the role of
ferroptosis in breast cancer has not been fully elucidated. The
present study systematically investigated the expression of FRGs,
and analyzed their association with OS and immune infiltration in
breast cancer.
Several prognostic models based on FRG signatures
have been constructed to explore prognosis-related biomarkers and
predict the prognosis of various types of cancer. A previous study
developed a 13-gene prognostic model (49), wherein AUCs were recorded to be
0.819, 0.815 and 0.891 for 1-, 3- and 5-year survival rates of
patients with acute myeloid leukemia. In another study, five FRGs
were used to construct a prognostic model with an AUC of 0.816, and
the nomogram exhibited good performance in the prediction of 3-year
survival rate of patients with thyroid cancer (50). However, the effect of FRGs in
prognostic prediction for patients with breast cancer has not been
fully evaluated. The present study established a
ferroptosis-related prognostic model of breast cancer based on the
GSE20685 database and the model was further validated using data
from the METABRIC database. Moreover, the results for univariate,
LASSO and multivariate regression analyses assisted in the
identification of a novel prognostic model that consisted of nine
FRGs. The nine-FRG prognostic model classified patients with breast
cancer into high-risk and low-risk groups. Survival analysis
demonstrated that prognosis of patients included in the high-risk
group was poorer compared with that in the low-risk group. ROC
analysis further confirmed the accuracy and sensitivity of the gene
signature. It has previously been reported that the
clinicopathological parameters age and TNM stage are commonly used
for clinical decision-making and prognostic prediction in breast
cancer (51). These clinical
features were included in the present study to perform univariate
and multivariate regression analysis, and the results indicated
that TNM stage and risk score acted as independent prognostic
indicators for OS. Subsequently, a novel prognostic nomogram was
constructed based on these two parameters, to estimate 3-, 5- and
10-year survival rates in patients with breast cancer. The AUCs of
the nomogram were recorded to be higher than those of FRGs or the
TNM stage alone, which was suggestive of the stability and
reliability of the nomogram in the prediction of the survival rate
in patients with breast cancer. AUCs were recorded to be 0.778,
0.775 and 0.794 with the nomogram for 3-, 5- and 10-year OS,
respectively, which had a higher predictive effect for breast
cancer than a single factor. However, given the incompleteness of
the clinical information obtained, the predictive nomogram was not
further validated in the validation dataset. A previous study also
used similar methods and did not validate the nomogram in the
validation dataset, demonstrating the feasibility and validity of
the present study (14). These
results indicated that the nine-FRGs prognostic model served as a
reliable prognostic indicator for patients with breast cancer.
The prognostic model consisted of nine FRGs that
could be roughly classified as protective factors (YWHAE, CD44,
HILPDA, IFNG and MYB) and risk factors (DBN1, HES1, HSPB1 and
SLC11A2) based on their regression coefficients for breast cancer
prognosis. YWHAE is a member of the 14-3-3 protein family, which
serves as a marker of ferroptosis. A previous study reported that
YWHAE was overexpressed in breast cancer tissues, and its
expression was associated with poor survival, which could further
promote cancer progression and chemoresistance in breast cancer
cells (52). CD44, a key marker of
cancer stemness, has previously been reported to be negatively
associated with ferroptosis. In a previous study, CD44
overexpression promoted the stability of SLC7A11, by enhancing the
interaction between SLC7A11 and OTUB1, which in turn resulted in
the suppression of ferroptosis in lung carcinoma cells (53). HILPDA is known to be an important
driver of ferroptosis for clear-cell carcinoma, which can enrich
polyunsaturated lipids and promote ferroptosis sensitivity
downstream of HIF-2α (54). IFNG,
released from immunotherapy-activated CD8+ T cells or
radiotherapy-activated ATM, can downregulate the expression of
SLC3A2 and SLC7A11, which can further assist in the promotion of
lipid peroxidation and ferroptosis that improve tumor control
(27,55). MYB is a well-known transcription
factor, which has a critical role in cellular metabolism (56,57),
including fatty acid metabolism, glucose-induced oxidative stress
and cysteine metabolism. It has been reported that MYB can
transcriptionally upregulate CDO1, which can result in a decrease
in GPX4 expression, leading to increased ferroptosis (58). Although the expression of partial
protective factors was reported to be upregulated in breast cancer
tissues in some studies, their expression levels may be
inconsistent with other databases or cohorts, and the detailed role
in breast cancer progression requires further exploration. Based on
a series of integrated analysis, the aforementioned five genes were
considered as potential protective factors in the present study,
which may draw a different conclusion from the analysis based on
single factors. In terms of risk factors, DBN1 is an actin-binding
protein, which acts as a ferroptosis regulator in pancreatic cancer
(32). High DBN1 expression has
been reported to be significantly associated with a poorer
prognosis and drug resistance in various types of cancer (59), including lung adenocarcinoma and
breast cancer. Elevated HES1 expression has also been previously
reported in multiple types of cancer, including ovarian cancer
(60), non-small cell lung cancer
(61) and breast cancer (62). Moreover, it was observed that high
expression of HES1 was associated with a poorer prognosis in
patients with breast cancer, and HES1 overexpression resulted in
enhanced proliferation, invasion and stemness in breast cancer
(62,63). HSPB1 is a member of the small heat
shock family of proteins, which can be induced by erastin treatment
in an HSF1-dependent manner in various cancer cells. In particular,
HSPB1 can reduce cellular iron uptake and lipid ROS production,
whereas HSPB1 knockdown has been shown to result in enhancement of
erastin-induced ferroptosis (64).
SLC11A2 is a proton-dependent iron importer of Fe2+,
which has previously been shown to be physiologically important for
cellular uptake of iron. A previous study reported higher
expression of SLC11A2 in MCF-7 cells compared with in MCF-12A cells
(65). Notably, the present study
revealed that most of these prognostic FRGs were differentially
expressed between breast cancer tissues and normal tissues;
however, classical FRGs, such as ACSL4 and GPX4, were not included
in the FRGs model. It was hypothesized that on the one hand, ACSL4
and GPX4 may have little independent prognostic value, independent
of the FRGs model. On the other hand, genes in the FRGs model are
potentially upstream and downstream of the key ferroptosis genes,
partly replacing the functions of the key ferroptosis genes in the
model for breast cancer. Therefore, in general, as crucial
components associated with tumor progression or modulating
treatment sensitivity, these nine FRGs have considerable potential
as therapeutic targets or biomarkers in breast cancer.
Immunotherapy, involving immune checkpoint
inhibitors, cyclin-dependent kinase inhibitors and dendritic cell
vaccines, is widely applied in the treatment of various types of
cancer (66). However, it has been
reported that immunosuppressive mechanisms may be initiated during
the development and progression of several types of cancer, to
circumvent antitumor immune responses (67). These immunosuppressive mechanisms
include the increased infiltration of immunosuppressive cells and
molecules, and the enrichment of low-immunogenic cancer cells. The
results of the present study obtained from the two datasets
revealed that there was a considerable difference in the types of
immune cells and the infiltration ratio of immune cells between
high-risk and low-risk groups, indicating that the FRGs may
regulate tumor progression partly via modulation of the patterns of
immune cell infiltration. The low-risk group exhibited a higher
immune score, and it was enriched with multiple immune cells and
immune-related pathways. This further indicated that patients in
the low-risk group may exhibit a better immune status and immune
function. Notably, a higher proportion of M2 macrophages and Treg
cells indicated that a stronger immunosuppressive effect may be
responsible for the poor prognosis in the high-risk group.
Moreover, increased expression of several immune checkpoints
indicated that patients in the low-risk group may benefit more from
immune checkpoint inhibitors, as indicated by the SubMap analysis.
Previous studies have also highlighted the promising role of
ferroptosis in cancer immunotherapy. In particular, GPX4 may
facilitate activation of stimulator-of-interferon genes, which can
further promote the initiation of an innate immune response against
microbial infection and tumors (68). CD8+ T cells have been
reported to enhance sensitization towards ferroptosis, via
secretion of IFNγ in cancer cells (27). Therefore, targeting the tumor
ferroptosis pathway in combination with immunotherapy may serve as
a novel therapeutic strategy for cancer management. However,
further investigation is required to elucidate the immunomodulatory
role of ferroptosis in antitumor immunity.
Erastin is a classical inducer of ferroptosis, which
can directly bind to VDAC2/VDAC3 to alter the permeability of the
outer mitochondrial membrane, thus leading to a decreased rate of
NADH oxidation and increased ROS production, thereby inducing
ferroptosis (69,70). The present study revealed that
erastin treatment could inhibit the proliferation and migration of
breast cancer cells, which was mediated via induction of
ferroptosis. Notably, these effects could be attenuated by
ferrostatin-1 treatment. It has previously been reported that
resistance to chemotherapeutics can result in therapeutic failure
and a poor prognosis in patients with breast cancer (71). The present study demonstrated that
erastin treatment could increase the sensitivity of breast cancer
cells towards chemotherapeutics and hormonal agents, which
indicated that a ferroptosis inducer may be used as a potential
combinatorial treatment strategy for the treatment of breast
cancer. The underlying mechanisms responsible for drug sensitivity
should be investigated in the future.
The present study has some limitations. First, with
the continuous advances in ferroptosis research, an increasing
number of FRGs may be identified and analyzed in the future,
leading to different analysis results and prognostic models.
Second, limited by the clinical information available in the
databases used, stratification analysis by molecular subtypes,
therapies or clinical stage could not be performed. Third, the
predictive model needs to be validated in large-scale studies
before application in clinical practice. Fourth, non-tumor cell
lines were not used to evaluate the cytotoxicity of the ferroptosis
inducer. Finally, future in vivo and in vitro studies
need to be conducted to validate the study, and further explore the
specific function and mechanism of genes in the model.
In conclusion, the present study identified a novel
prognostic model based on nine FRGs. This model could independently
predict the prognosis of patients with breast cancer. In addition,
the present study provided novel insights into the roles of
ferroptosis in the progression and treatment of breast cancer, and
revealed an important avenue that may be utilized for the treatment
of breast cancer. Further investigations are required to elucidate
the functional roles and underlying mechanisms of these FRGs in
breast cancer.
Supplementary Data
Availability of data and materials
The datasets used were obtained from the Gene
Expression Omnibus (accession no. GSE20685, https://www.ncbi.nlm.nih.gov/geo/) and METABRIC
databases (downloaded from cBioPortal, https://www.cbioportal.org/). The data used for
prediction of potential drugs was obtained from the CellMiner and
DrugBank databases. The other datasets used and/or analyzed during
the current study are available from the corresponding author on
reasonable request.
Authors' contributions
YZ and QY conceived and designed the experiments.
YZ, YL, YW, FY and XK performed the experiments and analyzed the
data. YZ and QY wrote and revised the manuscript. YZ and YL confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This work was supported by the National Key Research and
Development Program (grant no. 2020YFA0712400), the Special
Foundation for Taishan Scholars (grant no. ts20190971) and the
National Natural Science Foundation of China (grant nos. 81874119,
81902695, 82072912, and 82002785).
References
|
1
|
Radenkovic S, Konjevic G, Isakovic A,
Stevanovic P, Gopcevic K and Jurisic V: HER2-positive breast cancer
patients: Correlation between mammographic and pathological
findings. Radiat Prot Dosimetry. 162:125–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tao Z, Shi A, Lu C, Song T, Zhang Z and
Zhao J: Breast cancer: Epidemiology and etiology. Cell Biochem
Biophys. 72:333–338. 2015. View Article : Google Scholar
|
|
4
|
Feng Y, Spezia M, Huang S, Yuan C, Zeng Z,
Zhang L, Ji X, Liu W, Huang B, Luo W, et al: Breast cancer
development and progression: Risk factors, cancer stem cells,
signaling pathways, genomics, and molecular pathogenesis. Genes
Dis. 5:77–106. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Radenkovic S, Konjevic G, Gavrilovic D,
Stojanovic-Rundic S, Plesinac-Karapandzic V, Stevanovic P and
Jurisic V: pSTAT3 expression associated with survival and
mammographic density of breast cancer patients. Pathol Res Pract.
215:366–372. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen F, Han B, Meng Y, Han Y, Liu B, Zhang
B, Chang Y, Cao P, Fan Y and Tan K: Ceruloplasmin correlates with
immune infiltration and serves as a prognostic biomarker in breast
cancer. Aging (Albany NY). 13:20438–20467. 2021. View Article : Google Scholar
|
|
7
|
Lee SY, Ju MK, Jeon HM, Jeong EK, Lee YJ,
Kim CH, Park HG, Han SI and Kang HS: Regulation of tumor
progression by programmed necrosis. Oxid Med Cell Longev.
2018:35374712018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seibt TM, Proneth B and Conrad M: Role of
GPX4 in ferroptosis and its pharmacological implication. Free Radic
Biol Med. 133:144–152. 2019. View Article : Google Scholar
|
|
10
|
Yuan H, Li X, Zhang X, Kang R and Tang D:
CISD1 inhibits ferroptosis by protection against mitochondrial
lipid peroxidation. Biochem Biophys Res Commun. 478:838–844. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dodson M, Castro-Portuguez R and Zhang DD:
NRF2 plays a critical role in mitigating lipid peroxidation and
ferroptosis. Redox Biol. 23:1011072019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ooko E, Saeed ME, Kadioglu O, Sarvi S,
Colak M, Elmasaoudi K, Janah R, Greten HJ and Efferth T:
Artemisinin derivatives induce iron-dependent cell death
(ferroptosis) in tumor cells. Phytomedicine. 22:1045–1054. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu ZH, Tang Y, Yu H and Li HD: The role of
ferroptosis in breast cancer patients: A comprehensive analysis.
Cell Death Discov. 7:932021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang D, Wei G, Ma J, Cheng S, Jia L, Song
X, Zhang M, Ju M, Wang L, Zhao L, et al: Identification of the
prognostic value of ferroptosis-related gene signature in breast
cancer patients. BMC Cancer. 21:6452021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sato M, Kusumi R, Hamashima S, Kobayashi
S, Sasaki S, Komiyama Y, Izumikawa T, Conrad M, Bannai S and Sato
H: The ferroptosis inducer erastin irreversibly inhibits system xc-
and synergizes with cisplatin to increase cisplatin's cytotoxicity
in cancer cells. Sci Rep. 8:9682018. View Article : Google Scholar
|
|
16
|
Guo J, Xu B, Han Q, Zhou H, Xia Y, Gong C,
Dai X, Li Z and Wu G: Ferroptosis: A novel anti-tumor action for
cisplatin. Cancer Res Treat. 50:445–460. 2018. View Article : Google Scholar :
|
|
17
|
Li Z, Chen L, Chen C, Zhou Y, Hu D, Yang
J, Chen Y, Zhuo W, Mao M, Zhang X, et al: Targeting ferroptosis in
breast cancer. Biomark Res. 8:582020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie Y, Xiao Y, Liu Y, Lu X, Wang Z, Sun S,
Liu L, Tang X, Xiao H and Liu H: Construction of a novel
radiosensitivity- and ferroptosis-associated gene signature for
prognosis prediction in gliomas. J Cancer. 13:2683–2693. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yue Z, Sun J and Shi L: Construction and
validation of a 6-Ferroptosis related gene signature for prognosis
and immune landscape prediction in melanoma. Front Genet.
13:8875422022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xing XL, Liu Y, Liu J, Zhou H, Zhang H,
Zuo Q, Bu P, Duan T, Zhou Y and Xiao Z: Comprehensive analysis of
ferroptosis- and immune-related signatures to improve the prognosis
and diagnosis of kidney renal clear cell carcinoma. Front Immunol.
13:8513122022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sui S, Xu S and Pang D: Emerging role of
ferroptosis in breast cancer: New dawn for overcoming tumor
progression. Pharmacol Ther. 232:1079922022. View Article : Google Scholar
|
|
22
|
Wu M, Zhang X, Zhang W, Chiou YS, Qian W,
Liu X, Zhang M, Yan H, Li S, Li T, et al: Cancer stem cell
regulated phenotypic plasticity protects metastasized cancer cells
from ferroptosis. Nat Commun. 13:13712022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Konjevic GM, Vuletić AM, Mirjacic
Martinovic KM, Larsen AK and Jurisic VB: The role of cytokines in
the regulation of NK cells in the tumor environment. Cytokine.
117:30–40. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jurisic V: Multiomic analysis of cytokines
in immuno-oncology. Expert Rev Proteomics. 17:663–674. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ni S, Yuan Y, Kuang Y and Li X: Iron
metabolism and immune regulation. Front Immunol. 13:8162822022.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ganz T and Nemeth E: Iron homeostasis in
host defence and inflammation. Nat Rev Immunol. 15:500–510. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang W, Green M, Choi JE, Gijon M, Kennedy
PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al:
CD8+ T cells regulate tumour ferroptosis during cancer
immunotherapy. Nature. 569:270–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kao KJ, Chang KM, Hsu HC and Huang AT:
Correlation of microarray-based breast cancer molecular subtypes
and clinical outcomes: Implications for treatment optimization. BMC
Cancer. 11:1432011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu Q, Tang X, Zhu W, Li Q, Zhang X and Li
H: The potential prognostic role of oligosaccharide-binding
fold-containing protein 2A (OBFC2A) in triple-negative breast
cancer. Front Oncol. 11:7514302021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhuo S, Chen Z, Yang Y, Zhang J, Tang J
and Yang K: Clinical and biological significances of a
ferroptosis-related gene signature in glioma. Front Oncol.
10:5908612020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tang R, Hua J, Xu J, Liang C, Meng Q, Liu
J, Zhang B, Yu X and Shi S: The role of ferroptosis regulators in
the prognosis, immune activity and gemcitabine resistance of
pancreatic cancer. Ann Transl Med. 8:13472020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
R Core Team: 2012, R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: ISBN: 3-900051-07-0URL http://www.R-project.org/.
|
|
34
|
Gaujoux R and Seoighe C: A flexible R
package for nonnegative matrix factorization. BMC Bioinformatics.
11:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brunet JP, Tamayo P, Golub TR and Mesirov
JP: Metagenes and molecular pattern discovery using matrix
factorization. Proc Natl Acad Sci USA. 101:4164–4169. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Friedman J, Hastie T and Tibshirani R:
Regularization paths for generalized linear models via coordinate
descent. J Stat Softw. 33:1–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Becht E, Giraldo NA, Lacroix L, Buttard B,
Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautes-Fridman
C, Fridman WH, et al: Estimating the population abundance of
tissue-infiltrating immune and stromal cell populations using gene
expression. Genome Biol. 17:2182016. View Article : Google Scholar
|
|
39
|
Rooney MS, Shukla SA, Wu CJ, Getz G and
Hacohen N: Molecular and genetic properties of tumors associated
with local immune cytolytic activity. Cell. 160:48–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Roh W, Chen PL, Reuben A, Spencer CN,
Prieto PA, Miller JP, Gopalakrishnan V, Wang F, Cooper ZA, Reddy
SM, et al: Integrated molecular analysis of tumor biopsies on
sequential CTLA-4 and PD-1 blockade reveals markers of response and
resistance. Sci Transl Med. 9:eaah35602017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Reinhold WC, Sunshine M, Liu H, Varma S,
Kohn KW, Morris J, Doroshow J and Pommier Y: CellMiner: A web-based
suite of genomic and pharmacologic tools to explore transcript and
drug patterns in the NCI-60 cell line set. Cancer Res.
72:3499–3511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vickers AJ, Cronin AM, Elkin EB and Gonen
M: Extensions to decision curve analysis, a novel method for
evaluating diagnostic tests, prediction models and molecular
markers. BMC Med Inform Decis Mak. 8:532008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Michielin O, Lalani AK, Robert C, Sharma P
and Peters S: Defining unique clinical hallmarks for immune
checkpoint inhibitor-based therapies. J Immunother Cancer.
10:e0030242022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hubert P, Roncarati P, Demoulin S, Pilard
C, Ancion M, Reynders C, Lerho T, Bruyere D, Lebeau A, Radermecker
C, et al: Extracellular HMGB1 blockade inhibits tumor growth
through profoundly remodeling immune microenvironment and enhances
checkpoint inhibitor-based immunotherapy. J Immunother Cancer.
9:e0019662021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang C, Huang X, Liu Z, Qin W and Wang C:
Metabolism-associated molecular classification of hepatocellular
carcinoma. Mol Oncol. 14:896–913. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu
N, Shi Y, Chen L, Xiao D, Yu F, et al: Long noncoding RNA LINC00336
inhibits ferroptosis in lung cancer by functioning as a competing
endogenous RNA. Cell Death Differ. 26:2329–2343. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gai C, Yu M, Li Z, Wang Y, Ding D, Zheng
J, Lv S, Zhang W and Li W: Acetaminophen sensitizing
erastin-induced ferroptosis via modulation of Nrf2/heme oxygenase-1
signaling pathway in non-small-cell lung cancer. J Cell Physiol.
235:3329–3339. 2020. View Article : Google Scholar
|
|
49
|
Cui Z, Fu Y, Yang Z, Gao Z, Feng H, Zhou
M, Zhang L and Chen C: Comprehensive analysis of a ferroptosis
pattern and associated prognostic signature in acute myeloid
leukemia. Front Pharmacol. 13:8663252022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang Y, Yang J, Chen S, Wang W and Teng L:
Identification and validation of a prognostic signature for thyroid
cancer based on ferroptosis-related genes. Genes (Basel).
13:9972022. View Article : Google Scholar
|
|
51
|
Fan R, Chen Y, Nechuta S, Cai H, Gu K, Shi
L, Bao P, Shyr Y, Shu XO and Ye F: Prediction models for breast
cancer prognosis among Asian women. Cancer. 127:1758–1769. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang YF, Lee YC, Wang YY, Wang CH, Hou MF
and Yuan SF: YWHAE promotes proliferation, metastasis, and
chemoresistance in breast cancer cells. Kaohsiung J Med Sci.
35:408–416. 2019.PubMed/NCBI
|
|
53
|
Liu T, Jiang L, Tavana O and Gu W: The
Deubiquitylase OTUB1 mediates ferroptosis via stabilization of
SLC7A11. Cancer Res. 79:1913–1924. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zou Y, Palte MJ, Deik AA, Li H, Eaton JK,
Wang W, Tseng YY, Deasy R, Kost-Alimova M, Dančík V, et al: A
GPX4-dependent cancer cell state underlies the clear-cell
morphology and confers sensitivity to ferroptosis. Nat Commun.
10:16172019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lang X, Green MD, Wang W, Yu J, Choi JE,
Jiang L, Liao P, Zhou J, Zhang Q, Dow A, et al: Radiotherapy and
immunotherapy promote tumoral lipid oxidation and ferroptosis via
synergistic repression of SLC7A11. Cancer Discov. 9:1673–1685.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lee YH, Kim HS, Kim JS, Yu MK, Cho SD,
Jeon JG and Yi HK: C-myb regulates autophagy for pulp vitality in
glucose oxidative stress. J Dent Res. 95:430–438. 2016. View Article : Google Scholar
|
|
57
|
Minekura H, Kang MJ, Inagaki Y, Suzuki H,
Sato H, Fujino T and Yamamoto TT: Genomic organization and
transcription units of the human acyl-CoA synthetase 3 gene. Gene.
278:185–192. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hao S, Yu J, He W, Huang Q, Zhao Y, Liang
B, Zhang S, Wen Z, Dong S, Rao J, et al: Cysteine dioxygenase 1
mediates erastin-induced ferroptosis in human gastric cancer cells.
Neoplasia. 19:1022–1032. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Alfarsi LH, El Ansari R, Masisi BK, Parks
R, Mohammed OJ, Ellis IO, Rakha EA and Green AR: Integrated
analysis of key differentially expressed genes identifies DBN1 as a
predictive marker of response to endocrine therapy in luminal
breast cancer. Cancers (Basel). 12:15492020. View Article : Google Scholar
|
|
60
|
Wang X, Fu Y, Chen X, Ye J, Lü B, Ye F, Lü
W and Xie X: The expressions of bHLH gene HES1 and HES5 in advanced
ovarian serous adenocarcinomas and their prognostic significance: A
retrospective clinical study. J Cancer Res Clin Oncol. 136:989–996.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Konishi J, Kawaguchi KS, Vo H, Haruki N,
Gonzalez A, Carbone DP and Dang TP: Gamma-secretase inhibitor
prevents Notch3 activation and reduces proliferation in human lung
cancers. Cancer Res. 67:8051–8057. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li X, Cao Y, Li M and Jin F: Upregulation
of HES1 promotes cell proliferation and invasion in breast cancer
as a prognosis marker and therapy target via the AKT pathway and
emt process. J Cancer. 9:757–766. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li X, Li Y, Du X, Wang X, Guan S, Cao Y,
Jin F and Li F: HES1 promotes breast cancer stem cells by elevating
slug in triple-negative breast cancer. Int J Biol Sci. 17:247–258.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X,
Wang H, Cao L and Tang D: HSPB1 as a novel regulator of ferroptotic
cancer cell death. Oncogene. 34:5617–5625. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jiang XP, Elliott RL and Head JF:
Manipulation of iron transporter genes results in the suppression
of human and mouse mammary adenocarcinomas. Anticancer Res.
30:759–765. 2010.PubMed/NCBI
|
|
66
|
Emens LA: Breast cancer immunotherapy:
Facts and hopes. Clin Cancer Res. 24:511–520. 2018. View Article : Google Scholar :
|
|
67
|
Dunn GP, Bruce AT, Ikeda H, Old LJ and
Schreiber RD: Cancer immunoediting: From immunosurveillance to
tumor escape. Nat Immunol. 3:991–998. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Jia M, Qin D, Zhao C, Chai L, Yu Z, Wang
W, Tong L, Lv L, Wang Y, Rehwinkel J, et al: Redox homeostasis
maintained by GPX4 facilitates STING activation. Nat Immunol.
21:727–735. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Maldonado EN and Lemasters JJ: Warburg
revisited: Regulation of mitochondrial metabolism by
voltage-dependent anion channels in cancer cells. J Pharmacol Exp
Ther. 342:637–641. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yagoda N, von Rechenberg M, Zaganjor E,
Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM,
Boniface JJ, et al: RAS-RAF-MEK-dependent oxidative cell death
involving voltage-dependent anion channels. Nature. 447:864–868.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ponde NF, Zardavas D and Piccart M:
Progress in adjuvant systemic therapy for breast cancer. Nat Rev
Clin Oncol. 16:27–44. 2019. View Article : Google Scholar
|