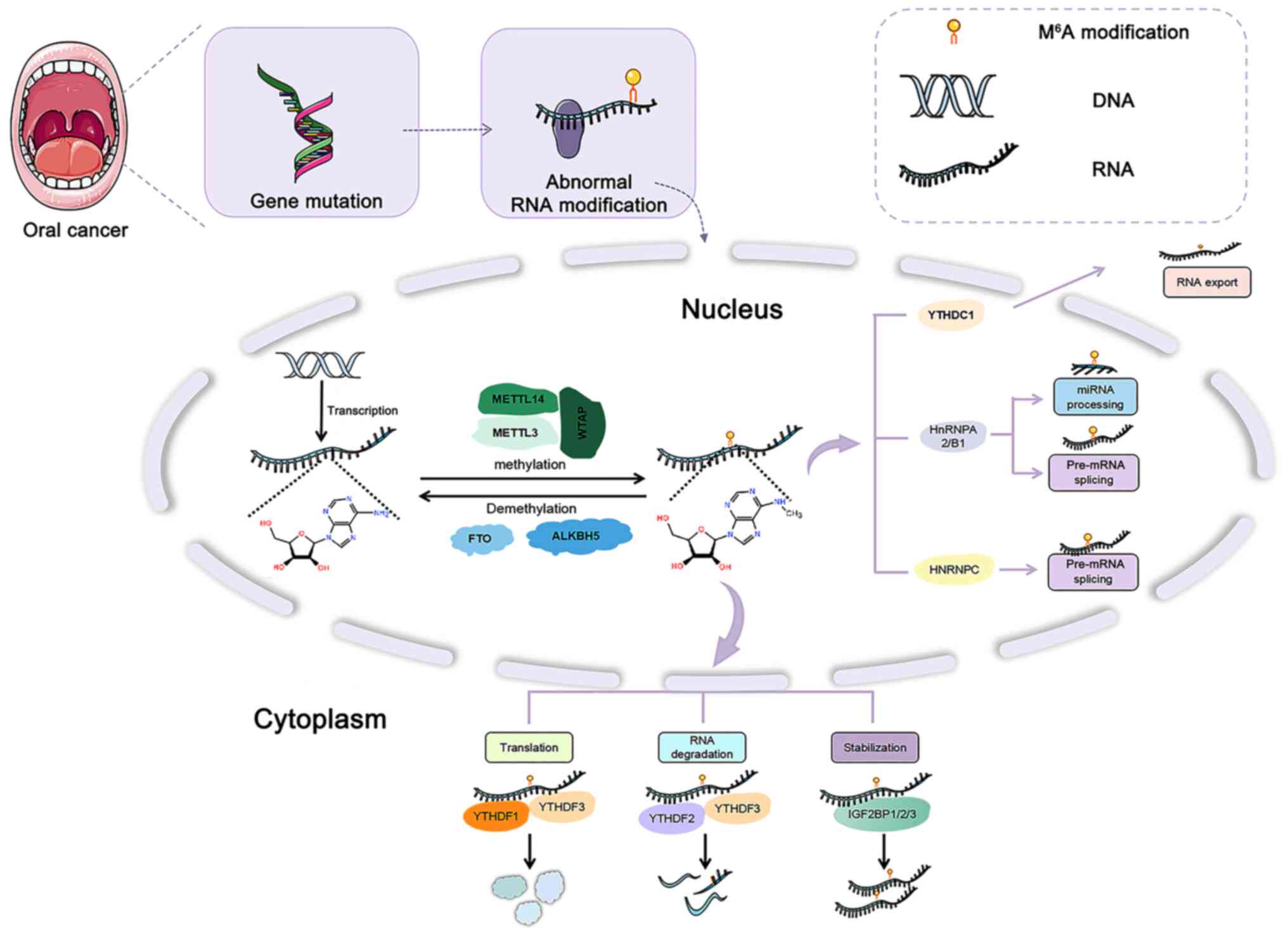
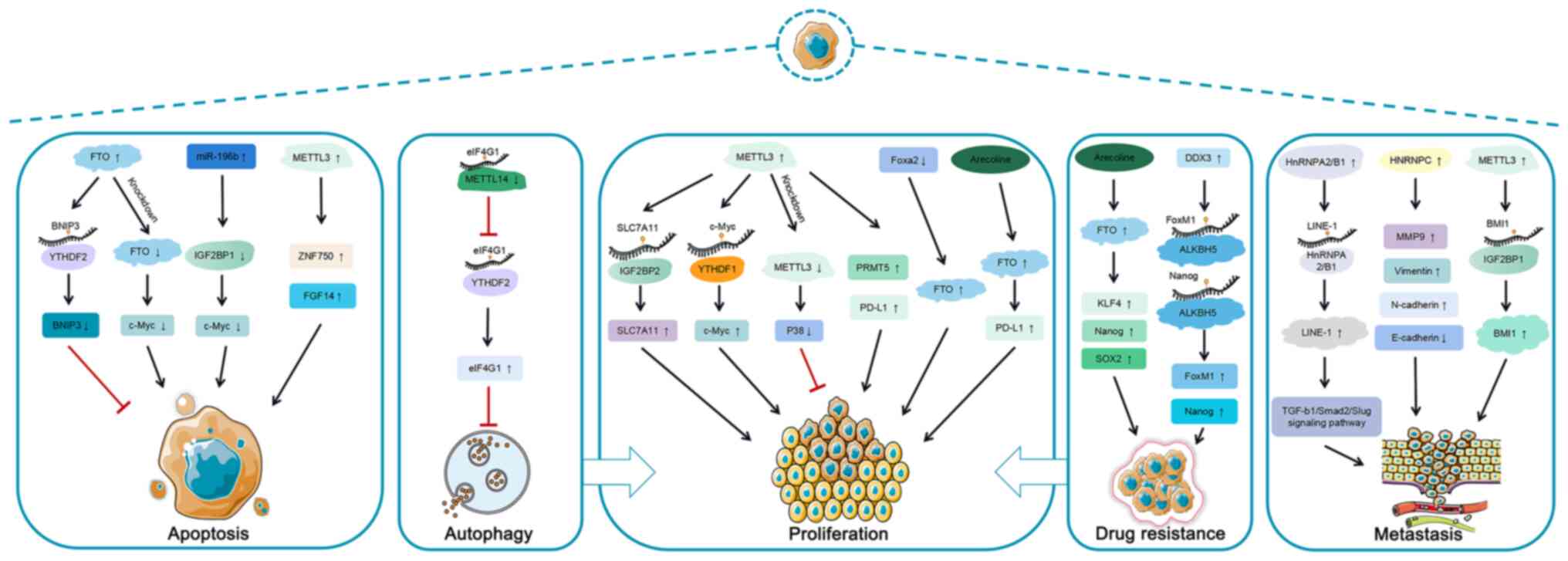
|
1
|
Chai AWY, Lim KP and Cheong SC:
Translational genomics and recent advances in oral squamous cell
carcinoma. Semin Cancer Biol. 61:71–83. 2020. View Article : Google Scholar
|
|
2
|
Huo XX, Wang SJ, Song H, Li MD, Yu H, Wang
M, Gong HX, Qiu XT, Zhu YF and Zhang JY: Roles of major RNA
adenosine modifications in head and neck squamous cell carcinoma.
Front Pharmacol. 12:7797792021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
D'souza S and Addepalli V: Preventive
measures in oral cancer: An overview. Biomed Pharmacother.
107:72–80. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pickering CR, Zhang J, Yoo SY, Bengtsson
L, Moorthy S, Neskey DM, Zhao M, Ortega Alves MV, Chang K, Drummond
J, et al: Integrative genomic characterization of oral squamous
cell carcinoma identifies frequent somatic drivers. Cancer Discov.
3:770–781. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mascolo M, Siano M, Ilardi G, Russo D,
Merolla F, Rosa G and Staibano S: Epigenetic disregulation in oral
cancer. Int J Mol Sci. 13:2331–2353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goldenberg D, Lee J, Koch WM, Kim MM,
Trink B, Sidransky D and Moon CS: Habitual risk factors for head
and neck cancer. Otolaryngol Head Neck Surg. 131:986–993. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guha N, Warnakulasuriya S, Vlaanderen J
and Straif K: Betel quid chewing and the risk of oral and
oropharyngeal cancers: A meta-analysis with implications for cancer
control. Int J Cancer. 135:1433–1443. 2014. View Article : Google Scholar
|
|
9
|
Herrero R, Castellsagué X, Pawlita M,
Lissowska J, Kee F, Balaram P, Rajkumar T, Sridhar H, Rose B,
Pintos J, et al: Human papillomavirus and oral cancer: The
International Agency for research on cancer multicenter study. J
Natl Cancer Inst. 95:1772–1783. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang W: mRNA methylation by NSUN2 in cell
proliferation. Wiley Interdiscip Rev RNA. 7:838–842. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Delaunay S and Frye M: RNA modifications
regulating cell fate in cancer. Nat Cell Biol. 21:552–559. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bonasio R, Tu S and Reinberg D: Molecular
signals of epigenetic states. Science. 330:612–616. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ling C and Rönn T: Epigenetics in human
obesity and type 2 diabetes. Cell Metab. 29:1028–1044. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barbieri I and Kouzarides T: Role of RNA
modifications in cancer. Nat Rev Cancer. 20:303–322. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Machnicka MA, Milanowska K, Osman Oglou O,
Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S,
Dunin-Horkawicz S, Rother KM, et al: MODOMICS: A data-base of RNA
modification pathways-2013 update. Nucleic Acids Res. 41(Database
Issue): D262–D267. 2013. View Article : Google Scholar
|
|
16
|
Wang T, Kong S, Tao M and Ju S: The
potential role of RNA N6-methyladenosine in cancer progression. Mol
Cancer. 19:882020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu ZX, Li LM, Sun HL and Liu SM: Link
between m6A modification and cancers. Front Bioeng Biotechnol.
6:892018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang X, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification in the
biological functions and diseases. Signal Transduct Target Ther.
6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Chen W, Gao Y, Song J, Gu Y, Zhang
J, Cheng X and Ai Y: FTO regulates arecoline-exposed oral cancer
immune response through PD-L1. Cancer Sci. 113:2962–2973. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu L, Li Q, Wang Y, Wang L, Guo Y, Yang R,
Zhao N, Ge N, Wang Y and Guo C: m6A methyltransferase
METTL3 promotes oral squamous cell carcinoma progression through
enhancement of IGF2BP2-mediated SLC7A11 mRNA stability. Am J Cancer
Res. 11:5282–5298. 2021.
|
|
21
|
Shi H, Wei J and He C: Where, when, and
how: Context-dependent functions of RNA methylation writers,
readers, and eras. Mol Cell. 74:640–650. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Malbec L, Zhang T, Chen YS, Zhang Y, Sun
BF, Shi BY, Zhao YL, Yang Y and Yang YG: Dynamic methylome of
internal mRNA N7-methylguanosine and its regulatory role
in translation. Cell Res. 29:927–941. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Saikia M, Fu Y, Pavon-Eternod M, He C and
Pan T: Genome-wide analysis of N1-methyl-adenosine modification in
human tRNAs. RNA. 16:1317–1327. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dominissini D, Nachtergaele S,
Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni
A, Salmon-Divon M, Clark WC, et al: The dynamic
N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature.
530:441–446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trixl L and Lusser A: The dynamic RNA
modification 5-methylcytosine and its emerging role as an
epitranscriptomic mark. Wiley Interdiscip Rev RNA. 10. pp.
e15102019, View Article : Google Scholar
|
|
26
|
Zhong H, Tang HF and Kai Y:
N6-methyladenine RNA modification (m6A): An emerging
regulator of metabolic diseases. Curr Drug Targets. 21:1056–1067.
2020. View Article : Google Scholar
|
|
27
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yao ZT, Yang YM, Sun MM, He Y, Liao L,
Chen KS and Li B: New insights into the interplay between long
non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun
(Lond). 42:117–140. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alarcón CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du Y, Hou G, Zhang H, Dou J, He J, Guo Y,
Li L, Chen R, Wang Y, Deng R, et al: SUMOylation of the m6A-RNA
methyltransferase METTL3 modulates its function. Nucleic Acids Res.
46:5195–5208. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
van Tran N, Ernst FGM, Hawley BR, Zorbas
C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR,
Graille M and Lafontaine DLJ: The human 18S rRNA m6A
methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids
Res. 47:7719–7733. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar :
|
|
33
|
Wang X, Huang J, Zou T and Yin P: Human
m6A writers: Two subunits, 2 roles. RNA Biol.
14:300–304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang P, Doxtader KA and Nam Y: Structural
basis for cooperative function of Mettl3 and Mettl14
methyltransferases. Mol Cell. 63:306–317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D,
Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Structural basis of
N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature.
534:575–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou KI and Pan T: Structures of the m(6)A
methyltransferase complex: Two subunits with distinct but
coordinated roles. Mol Cell. 63:183–185. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu S, Li Q, Li G, Zhang Q, Zhuo L, Han X,
Zhang M, Chen X, Pan T, Yan L, et al: The mechanism of
m6A methyltransferase METTL3-mediated autophagy in
reversing gefitinib resistance in NSCLC cells by β-elemene. Cell
Death Dis. 11:9692020. View Article : Google Scholar
|
|
39
|
Wang F, Zhu Y, Cai H, Liang J, Wang W,
Liao Y, Zhang Y, Wang C and Hou J: N6-methyladenosine
methyltransferase METTL14-mediated autophagy in malignant
development of oral squamous cell carcinoma. Front Oncol.
11:7384062021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yue B, Song C, Yang L, Cui R, Cheng X,
Zhang Z and Zhao G: METTL3-mediated N6-methyladenosine modification
is critical for epithelial-mesenchymal transition and metastasis of
gastric cancer. Mol Cancer. 18:1422019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B,
Li C, Sun L, Qin J, Xu T, et al: METTL14-mediated
N6-methyladenosine modification of SOX4 mRNA inhibits tumor
metastasis in colorectal cancer. Mol Cancer. 19:1062020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu
HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al: METTL3 promote tumor
proliferation of bladder cancer by accelerating pri-miR221/222
maturation in m6A-dependent manner. Mol Cancer. 18:1102019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W,
Lu S, Xu D, Wu Y, Chen Q, et al: LNC942 promoting METTL14-mediated
m6A methylation in breast cancer cell proliferation and
progression. Oncogene. 39:5358–5372. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR
and Qian SB: Dynamic m(6)A mRNA methylation directs translational
control of heat shock response. Nature. 526:591–594. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N6-methyladenosine-modified RNA. Cell Res. 27:315–328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y,
Qi M, Lu Z, Shi H, Wang J, et al: Ythdc2 is an
N6-methyladenosine binding protein that regulates
mammalian spermatogenesis. Cell Res. 27:1115–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wu B, Su S, Patil DP, Liu H, Gan J,
Jaffrey SR and Ma J: Molecular basis for the specific and
multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun. 9:4202018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
He L, Li H, Wu A, Peng Y, Shu G and Yin G:
Functions of N6-methyladenosine and its role in cancer. Mol Cancer.
18:1762019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang
H, Hu Y, Qiu J, Pu L, Tang J and Wang X: HIF-1α-induced expression
of m6A reader YTHDF1 drives hypoxia-induced autophagy and
malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14
translation. Signal Transduct Target Ther. 6:762021. View Article : Google Scholar
|
|
52
|
Chen H, Yu Y, Yang M, Huang H, Ma S, Hu J,
Xi Z, Guo H, Yao G, Yang L, et al: YTHDF1 promotes breast cancer
progression by facilitating FOXM1 translation in an m6A-dependent
manner. Cell Biosci. 12:192022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y,
Cheng C, Li L, Pi J, Si Y, et al: The m6A reader YTHDF1 promotes
ovarian cancer progression via augmenting EIF3C translation.
Nucleic Acids Res. 48:3816–3831. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu L, Wu Y, Li Q, Liang J, He Q, Zhao L,
Chen J, Cheng M, Huang Z, Ren H, et al: METTL3 promotes
tumorigenesis and metastasis through BMI1 m6A
methylation in oral squamous cell carcinoma. Mol Ther.
28:2177–2190. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen
YX, Liu J, Luo XJ, Meng Q, Pu HY, et al: LncRNA LINRIS stabilizes
IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer.
Mol Cancer. 18:1742019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhu F, Yang T, Yao M, Shen T and Fang C:
HNRNPA2B1, as a m6A reader, promotes tumorigenesis and
metastasis of oral squamous cell carcinoma. Front Oncol.
11:7169212021. View Article : Google Scholar
|
|
57
|
Huang GZ, Wu QQ, Zheng ZN, Shao TR, Chen
YC, Zeng WS and Lv XZ: M6A-related bioinformatics analysis reveals
that HNRNPC facilitates progression of OSCC via EMT. Aging (Albany
NY). 12:11667–11684. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang F, Liao Y, Zhang M, Zhu Y, Wang W,
Cai H, Liang J, Song F, Hou C, Huang S, et al: N6-methyladenosine
demethyltransferase FTO-mediated autophagy in malignant development
of oral squamous cell carcinoma. Oncogene. 40:3885–3898. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li X, Xie X, Gu Y, Zhang J, Song J, Cheng
X, Gao Y and Ai Y: Fat mass and obesity-associated protein
regulates tumorigenesis of arecoline-promoted human oral carcinoma.
Cancer Med. 10:6402–6415. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang J, Qiao Y, Sun M, Sun H, Zie F, Chang
H, Wang Y, Song J, Lai S, Yang C, et al: FTO promotes colorectal
cancer progression and chemotherapy resistance via demethylating
G6PD/PARP1. Clin Transl Med. 12:e7722022.PubMed/NCBI
|
|
62
|
Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi
Y, He S and Shimamoto F: m6A demethylase ALKBH5 inhibits
pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation
and mediating Wnt signaling. Mol Cancer. 19:32020. View Article : Google Scholar
|
|
63
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang Y, Zhu GQ, Tian D, Zhou CW, Li N,
Feng Y and Zeng MS: Comprehensive analysis of tumor immune
microenvironment and prognosis of m6A-related lncRNAs in gastric
cancer. BMC Cancer. 22:3162022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Guo Y, Wang R, Li J, Song Y, Min J, Zhao
T, Hua L, Shi J, Zhang C, Ma P, et al: Comprehensive analysis of
m6A RNA methylation regulators and the immune microenvironment to
aid immunotherapy in pancreatic cancer. Front Immunol.
12:7694252021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu Z, Zhong J, Zeng J, Duan X, Lu J, Sun
X, Liu Q, Liang Y, Lin Z, Zhong W, et al: Characterization of the
m6A-associated tumor immune microenvironment in prostate cancer to
aid immunotherapy. Front Immunol. 12:7351702021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Visvanathan A, Patil V, Arora A, Hegde AS,
Arivazhagan A, Santosh V and Somasundaram K: Essential role of
METTL3-mediated m6A modification in glioma stem-like
cells maintenance and radioresistance. Oncogene. 37:522–533. 2018.
View Article : Google Scholar
|
|
68
|
Small W Jr, Bacon MA, Bajaj A, Chuang LT,
Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR,
Viswanathan AN and Gaffney DK: Cervical cancer: A global health
crisis. Cancer. 123:2404–2412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R,
Wang YY and Zhe H: FTO regulates the chemo-radiotherapy resistance
of cervical squamous cell carcinoma (CSCC) by targeting β-catenin
through mRNA demethylation. Mol Carcinog. 57:590–597. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sheng H, Li Z, Su S, Sun W, Zhang X, Li L,
Li J, Liu S, Lu B, Zhang S and Shan C: YTH domain family 2 promotes
lung cancer cell growth by facilitating 6-phosphogluconate
dehydrogenase mRNA translation. Carcinogenesis. 41:541–550. 2020.
View Article : Google Scholar
|
|
71
|
Ding RB, Chen P, Rajendran BK, Lyu X, Wang
H, Bao J, Zeng J, Hao W, Sun H, Wong AH, et al: Molecular landscape
and subtype-specific therapeutic response of nasopharyngeal
carcinoma revealed by integrative pharmacogenomics. Nat Commun.
12:30462021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang P, He Q, Lei Y, Li Y, Wen X, Hong M,
Zhang J, Ren X, Wang Y, Yang X, et al: m6A-mediated
ZNF750 repression facilitates nasopharyngeal carcinoma progression.
Cell Death Dis. 9:11692018. View Article : Google Scholar
|
|
73
|
Jin S, Li M, Chang H, Wang R, Zhang Z,
Zhang J, He Y and Ma H: The m6A demethylase ALKBH5 promotes tumor
progression by inhibiting RIG-I expression and interferon alpha
production through the IKKε/TBK1/IRF3 pathway in head and neck
squamous cell carcinoma. Mol Cancer. 21:972022. View Article : Google Scholar
|
|
74
|
Yu D, Pan M, Li Y, Lu T, Wang Z, Liu C and
Hu G: RNA N6-methyladenosine reader IGF2BP2 promotes lymphatic
metastasis and epithelial-mesenchymal transition of head and neck
squamous carcinoma cells via stabilizing slug mRNA in an
m6A-dependent manner. J Exp Clin Cancer Res. 41:62022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Hirayama M, Wei FY, Chujo T, Oki S, Yakita
M, Kobayashi D, Araki N, Takahashi N, Yoshida R, Nakayama H and
Tomizawa K: FTO demethylates cyclin D1 mRNA and controls cell-cycle
progression. Cell Rep. 31:1074642020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Vu LP, Pickering BF, Cheng Y, Zaccara S,
Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al:
The N6-methyladenosine (m6A)-forming enzyme
METTL3 controls myeloid differentiation of normal hematopoietic and
leukemia cells. Nat Med. 23:1369–1376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jiang F, Tang X, Tang C, Hua Z, Ke M, Wang
C, Zhao J, Gao S, Jurczyszyn A, Janz S, et al: HNRNPA2B1 promotes
multiple myeloma progression by increasing AKT3 expression via
m6A-dependent stabilization of ILF3 mRNA. J Hematol Oncol.
14:542021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Han H, Fan G, Song S, Jiang Y, Qian C,
Zhang W, Su Q, Xue X, Zhuang W and Li B: piRNA-30473 contributes to
tumorigenesis and poor prognosis by regulating m6A RNA methylation
in DLBCL. Blood. 137:1603–1614. 2021. View Article : Google Scholar
|
|
79
|
Tang D, Kang R, Berghe TV, Vandenabeele P
and Kroemer G: The molecular machinery of regulated cell death.
Cell Res. 29:347–364. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Koren E and Fuchs Y: Modes of regulated
cell death in cancer. Cancer Discov. 11:245–265. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhi Y, Zhang S, Zi M, Wang Y, Liu Y, Zhang
M, Shi L, Yan Q, Zeng Z, Ziong W, et al: Potential applications of
N6-methyladenosine modification in the prognosis and treatment of
cancers via modulating apoptosis, autophagy, and ferroptosis. Wiley
Interdiscip Rev RNA. 13. pp. e17192022, View Article : Google Scholar
|
|
82
|
Wong RS: Apoptosis in cancer: From
pathogenesis to treatment. J Exp Clin Cancer Res. 30:872011.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun
L, Wang Y, Li X, Xiong XF, Wei B, et al: RNA N6-methyladenosine
demethylase FTO promotes breast tumor progression through
inhibiting BNIP3. Mol Cancer. 18:462019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Rebucci M, Sermeus A, Leonard E, Delaive
E, Dieu M, Fransolet M, Arnould T and Michiels C: miRNA-196b
inhibits cell proliferation and induces apoptosis in HepG2 cells by
targeting IGF2BP1. Mol Cancer. 14:792015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mizushima N and Levine B: Autophagy in
human diseases. N Engl J Med. 383:1564–1576. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Mizushima N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell. 147:728–741. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Dikic I and Elazar Z: Mechanism and
medical implications of mammalian autophagy. Nat Rev Mol Cell Biol.
19:349–364. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Amaravadi RK, Kimmelman AC and Debnath J:
Targeting autophagy in cancer: Recent advances and future
directions. Cancer Discov. 9:1167–1181. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ferro F, Servais S, Besson P, Roger S,
Dumas JF and Brisson L: Autophagy and mitophagy in cancer metabolic
remodelling. Semin Cell Dev Biol. 98:129–138. 2020. View Article : Google Scholar
|
|
90
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chen Y and Gibson SB: Three dimensions of
autophagy in regulating tumor growth: Cell survival/death, cell
proliferation, and tumor dormancy. Biochim Biophys Acta Mol Basis
Dis. 1867:1662652021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Lei G, Zhuang L and Gan B: Targeting
ferroptosis as a vulnerability in cancer. Nat Rev Cancer.
22:381–396. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Li T, Wang Y, Xiang X and Chen C:
Development and validation of a ferroptosis-related lncRNAs
prognosis model in oral squamous cell carcinoma. Front Genet.
13:8479402022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Gong Y, Fan Z, Luo G, Yang C, Huang Q, Fan
K, Cheng H, Jin K, Ni Q, Yu X and Liu C: The role of necroptosis in
cancer biology and therapy. Mol Cancer. 18:1002019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Pasparakis M and Vandenabeele P:
Necroptosis and its role in inflammation. Nature. 517:311–320.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li J, Huang S, Zeng L, Li K, Yang L, Gao
S, Guan C, Zhang S, Lao X, Liao G and Liang Y: Necroptosis in head
and neck squamous cell carcinoma: Characterization of
clinicopathological relevance and in vitro cell model. Cell Death
Dis. 11:3912020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Shi J, Liu Z and Xu Q: Tumor necrosis
factor receptor-associated factor 6 contributes to malignant
behavior of human cancers through promoting AKT ubiquitination and
phosphorylation. Cancer Sci. 110:1909–1920. 2019.PubMed/NCBI
|
|
98
|
Lan H, Liu Y, Liu J, Wang X, Guan Z, Du J
and Jin K: Tumor-associated macrophages promote oxaliplatin
resistance via METTL3-mediated m6A of TRAF5 and
necroptosis in colorectal cancer. Mol Pharm. 18:1026–1037. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yu P, Zhang X, Liu N, Tang L, Peng C and
Chen X: Pyroptosis: Mechanisms and diseases. Signal Transduct
Target Ther. 6:1282021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yue E, Tuguzbaeva G, Chen X, Qin Y, Li A,
Sun X, Dong C, Liu Y, Yu Y, Zahra SM, et al: Anthocyanin is
involved in the activation of pyroptosis in oral squamous cell
carcinoma. Phytomedicine. 56:286–294. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wu L, Liu G, He YW, Chen R and Wu ZY:
Identification of a pyroptosis-associated long non-coding RNA
signature for predicting the immune status and prognosis in skin
cutaneous melanoma. Eur Rev Med Pharmacol Sci. 25:5597–5609.
2021.PubMed/NCBI
|
|
102
|
Deshpande A, Sicinski P and Hinds PW:
Cyclins and cdks in development and cancer: A perspective.
Oncogene. 24:2909–2915. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Thakur C and Chen F: Connections between
metabolism and epigenetics in cancers. Semin Cancer Biol. 57:52–58.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Xu T, Zhang W, Chai L, Liu C, Zhang S and
Xu T: Methyltransferase-like 3-induced N6-methyladenosine
upregulation promotes oral squamous cell carcinoma by through p38.
Oral Dis. Sep 3–2021.Epub ahead of print. View Article : Google Scholar
|
|
105
|
Zhao W, Cui Y, Liu L, Ma X, Qi X, Wang Y,
Liu Z, Ma S, Liu J and Wu J: METTL3 facilitates oral squamous cell
carcinoma tumorigenesis by enhancing c-Myc stability via
YTHDF1-mediated m6A modification. Mol Ther Nucleic
Acids. 20:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Ai Y, Liu S, Luo H, Wu S, Wei H, Tang Z,
Li X, Lv X and Zou C: METTL3 intensifies the progress of oral
squamous cell carcinoma via modulating the m6A amount of PRMT5 and
PD-L1. J Immunol Res. 2021:61495582021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar
|
|
108
|
Trédan O, Galmarini CM, Patel K and
Tannock IF: Drug resistance and the solid tumor microenvironment. J
Natl Cancer Inst. 99:1441–1454. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Shriwas O, Priyadarshini M, Samal SK, Rath
R, Panda S, Das Majumdar SK, Muduly DK, Botlagunta M and Dash R:
DDX3 modulates cisplatin resistance in OSCC through ALKBH5-mediated
m6A-demethylation of FOXM1 and NANOG. Apoptosis.
25:233–246. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Bellmunt J, Powles T and Vogelzang NJ: A
review on the evolution of PD-1/PD-L1 immunotherapy for bladder
cancer: The future is now. Cancer Treat Rev. 54:58–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Li N, Kang Y, Wang L, Huff S, Tang R, Hui
H, Agrawal K, Gonzalez GM, Wang Y, Patel SP and Rana TM: ALKBH5
regulates anti-PD-1 therapy response by modulating lactate and
suppressive immune cell accumulation in tumor microenvironment.
Proc Natl Acad Sci USA. 117:20159–20170. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Chen J, Lu T, Zhong F, Lv Q, Fang M, Tu Z,
Ji Y, Li J and Gong X: A signature of N6-methyladenosine
regulator-related genes predicts prognoses and immune responses for
head and neck squamous cell carcinoma. Front Immunol.
13:8098722022. View Article : Google Scholar
|
|
114
|
Shu CW, Weng JR, Chang HW, Liu PF, Chen
JJ, Peng CC, Huang JW, Lin WY and Yen CY: Tribulus terrestris fruit
extract inhibits autophagic flux to diminish cell proliferation and
metastatic characteristics of oral cancer cells. Environ Toxicol.
36:1173–1180. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Kumar VB, Lin SH, Mahalakshmi B, Lo YS,
Lin CC, Chuang YC, Hsieh MJ and Chen MK: Sodium danshensu inhibits
oral cancer cell migration and invasion by modulating p38 signaling
pathway. Front Endocrinol (Lausanne). 11:5684362020. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Balaji S, Terrero D, Tiwari AK, Ashby CR
Jr and Raman D: Alternative approaches to overcome chemoresistance
to apoptosis in cancer. Adv Protein Chem Struct Biol. 126:91–122.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Luo R, Xie L, Lin Y, Shao J and Lin Z:
Oxymatrine suppresses oral squamous cell carcinoma progression by
suppressing CXC chemokine receptor 4 in an m6A
modification decrease dependent manner. Oncol Rep. 48:1772022.
View Article : Google Scholar
|
|
118
|
Gong J, Wang C, Zhang F and Lan W: Effects
of allocryptopine on the proliferation and epithelial-mesenchymal
transition of oral squamous cell carcinoma through m6A mediated
hedgehog signaling pathway. J Environ Pathol Toxicol Oncol.
41:15–24. 2022. View Article : Google Scholar : PubMed/NCBI
|