Circular RNA (circRNA) is a novel and important
class of endogenous noncoding RNAs (ncRNAs) in addition to long
ncRNA and microRNA (miRNA) and was first discovered several decades
ago (1,2). Unlike linear RNAs, circRNAs are
covalently closed-loop molecules formed by back-splicing without 5′
to 3′ polarity or the poly(A) tail, which makes circRNAs difficult
to degrade by RNase and more stable in plasma and tissues than most
other ncRNAs (3,4). With the development of
high-throughput sequencing technologies and bioinformatic methods,
abundant circRNAs have been identified and demonstrated to have
high stability, sequence conservation and specific localization in
subcellular compartments (5).
Multiple studies have indicated that circRNAs have different
expression patterns and biological functions in different types of
cancer (6-8). Research on the mechanisms of circRNAs
in tumor development, as well as the diagnosis, treatment and
prognosis of cancer, has become a hotspot in recent years.
Lung cancer is one of the most common cancer types
worldwide. Although treatments are improving, the prognosis of lung
cancer remains poor and the five-year survival rate is still
<20% (9). Histologically, lung
cancer is usually divided into two categories: Non-small cell lung
cancer (NSCLC) and SCLC (10).
NSCLC, which includes lung adenocarcinoma (LUAD) and lung squamous
cell carcinoma (LUSC), accounts for >85% of lung cancers
(11). Clinically, the survival
rate of patients with early lung cancer is significantly higher
than that of patients with late-stage lung cancer (12). However, due to the occult symptoms
of early lung cancer, a large number of patients are not diagnosed
until reaching the intermediate and late stages. After diagnosis,
the treatments and prognosis vary for the different types of lung
cancer (13). Therefore, it is
necessary to improve the understanding of the molecular mechanisms
of different types of lung cancer to develop efficient and accurate
diagnostic and prognostic biomarkers and to identify different
therapeutic targets.
In recent studies, a variety of circRNAs have been
discovered in tumor tissues and blood samples and they have
important roles in the development and progression of lung cancer
(14). In the present review, the
expression of different circRNAs and their function in different
types of lung cancer were systematically summarized to understand
circRNAs as potential biomarkers for the diagnosis and
prognostication of different types of lung cancer. The emerging
roles of circRNAs and their biological functions in resistance to
clinical chemotherapy, targeted therapy, radiotherapy and
immunotherapy are also discussed in detail.
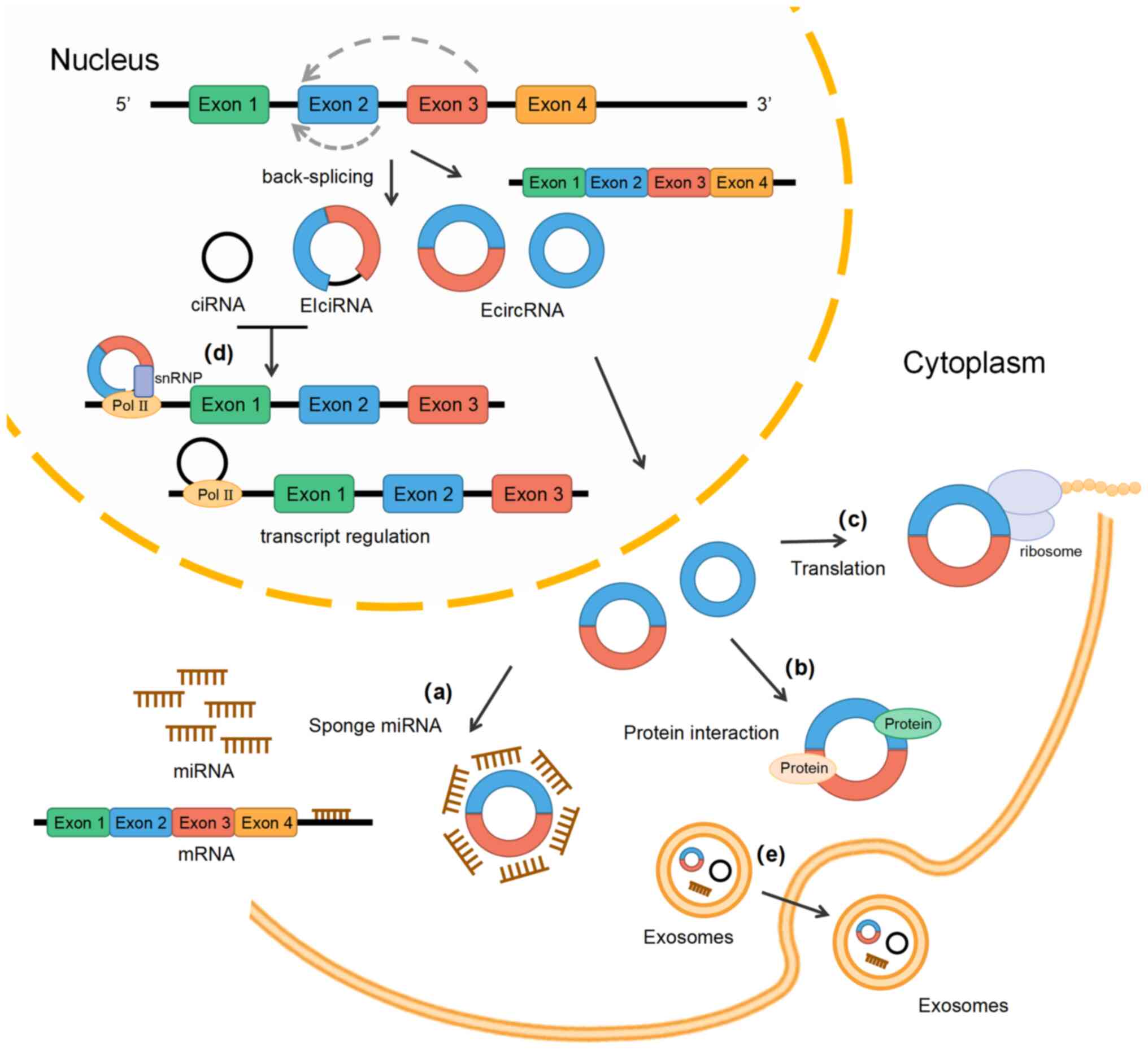
circRNAs are a class of noncoding RNA molecules that
form a loop structure via covalent bonds after back-splicing and
have no 5′ caps or 3′ poly(A) tails (15). Depending on their source, circRNAs
may be divided into exonic circRNAs (EcircRNAs), intronic circRNAs
(ciRNAs), and exon-intron circRNAs (EIciRNAs) (16) (Fig.
1). EcircRNAs consist of one or multiple exons and mainly
reside in the cytoplasm after biogenesis (17). EIciRNAs and ciRNAs mainly reside in
the nucleus. CiRNAs are composed of only introns, and EIciRNAs are
composed of exons and the introns located between the exons
(18). Among them, EcirRNAs are
the most common and account for ~80% of the total circRNAs
(19). EcircRNAs are transferred
into the cytoplasm after being synthesized in the nucleus and do
not only serve as miRNA sponges or interact with proteins to
perform various functions but may also be contained in exosomes
together with a large number of other nucleic acids, proteins and
lipids (20). A large number of
studies have indicated that circRNAs are stable and enriched in
exosomes (21-25), and the composition of circRNAs in
exosomes may be regulated by changes in the levels of related
miRNAs in donor cells, after which the molecular information is
transferred to recipient cells (26). Therefore, the predominant studies
are related to cytoplasmic EcircRNAs.
circRNAs are generated through different mechanisms,
including intron-pairing-driven circularization, RNA binding
protein (RBP)-driven circularization, exon-skipping lariat-driven
circularization and intron lariat-driven circularization (27). Intron pairing means that two
flanking introns located on both sides of the pre-mRNA exon/exons
have a connectable structure and are sufficiently close to each
other to form a secondary conformation via base pairing, which
enables the splicing site to conduct back-splicing and produce
EcirRNA or EIciRNA (19,28). RBPs are trans-acting factors that
are able to bind to specific motifs of pre-mRNAs and close the
flanking introns (29). RBP
protein dimerization promotes the formation of an RNA loop.
Muscleblind (MBL), heterogeneous nuclear ribonucleoprotein L and
quaking are common RBPs that have crucial roles in circRNA
biogenesis (29-31). Exon skipping means that one or more
exons of pre-mRNA are missing, which enables lariat
structure-driven circularization of circRNA (32). During circularization, the
spliceosome removes the introns in the lariat. If all intron
sequences are removed, EcircRNAs are formed, and if the introns are
not completely removed, EIciRNAs are generated. The intron lariat
model mainly drives the formation of ciRNAs, which mainly depends
on a 7-nt GU-rich sequence near the 5′ end splice site and an 11-nt
C-rich sequence near the branching point (4,33).
These two sequences combine with each other during the
back-splicing process and the branch point of the 3′ downstream
region is trimmed to form a stable ciRNA (34).
circRNAs are widely present in diverse cells and
have stable structures, conserved sequences and cell-tissue
expression specificity that determine their functions (35). The main functions of circRNAs
include miRNA sponging, interaction with proteins, gene
transcription regulation and translation templates (Fig. 1). Most circRNAs, particularly
EcircRNAs, have been proposed to exert their function as miRNA
sponges. These circRNAs contain miRNA response elements and may
competitively bind miRNAs, similar to 'sponge' effects, which may
prevent miRNA from interacting with mRNA in the 3′ untranslated
region and indirectly regulate downstream mRNA targets of miRNA
(36).
Certain circRNAs with introns that are found in the
nucleus, mainly ciRNAs and ElcircRNAs, may directly regulate host
gene transcription. ElcircRNAs or ciRNAs are able to bind to RNA
polymerase II (Pol II) in the nucleus and regulate gene
transcription (18). The ciRNA
ci-ankyrin repeat domain 52 (ANKRD52), which is derived from the
second intron region of the ANKRD52 gene and accumulates in the
nucleus, binds to elongation Pol II machinery and promotes the
transcriptional activity of Pol II, thus serving a cis regulatory
role for the parent gene (34). In
addition, EIciRNAs, e.g., circ-eukaryotic translation initiation
factor 3 subunit J (EIF3J) and circ-poly(A)-binding
protein-interacting protein 2 (PAIP2), are able to interact with U1
small nuclear ribonucleoprotein to form a complex, which binds to
the promoter of the parent gene along Pol II, promoting the
transcription of the parent genes EIF3J and PAIP2 and having a
positive regulatory role (19,37).
circRNAs may also change splicing patterns or mRNA stability by
binding to RBPs to form RNA-protein complexes (38).
Although circRNAs are noncoding RNAs, certain
studies have indicated that certain circRNAs have the potential to
encode functional polypeptides (39). One way to realize the translation
of circRNAs is to promote the direct binding of initiation factors
or ribosomes with translatable circRNAs through internal ribosome
entry site elements, such as circ-ZNF609 and circMBL (40,41).
In addition, N6-methyladenosine (m6A) may also drive the
translation of circRNAs into polypeptides and a study strongly
suggested that m6A-containing RRACH sequences (R=G or A; H=A, C or
U) may be involved in the translation initiation of circRNAs
(42). A few specific circRNAs,
including circ-ZNF609 (40),
circNlgn (43), circMBL (44), circ-FBXW7 (45), circ-E-Cad (46), circRNA Rho GTPase activating
protein 35 (circARHGAP35) (47),
circMAPK1 (48), circPINTexon2
(49) and circRNA SNF2 histone
linker PHD RING helicase (50),
have been reported to act as protein templates. circPINTexon2, the
circular form of the long intergenic nonprotein-coding RNA
p53-induced transcript, may encode an 87-amino-acid peptide that is
able to suppress glioblastoma cell proliferation in vitro
and in vivo (49). However,
most of the functional proteins/peptides encoded by circRNAs remain
to be elucidated.
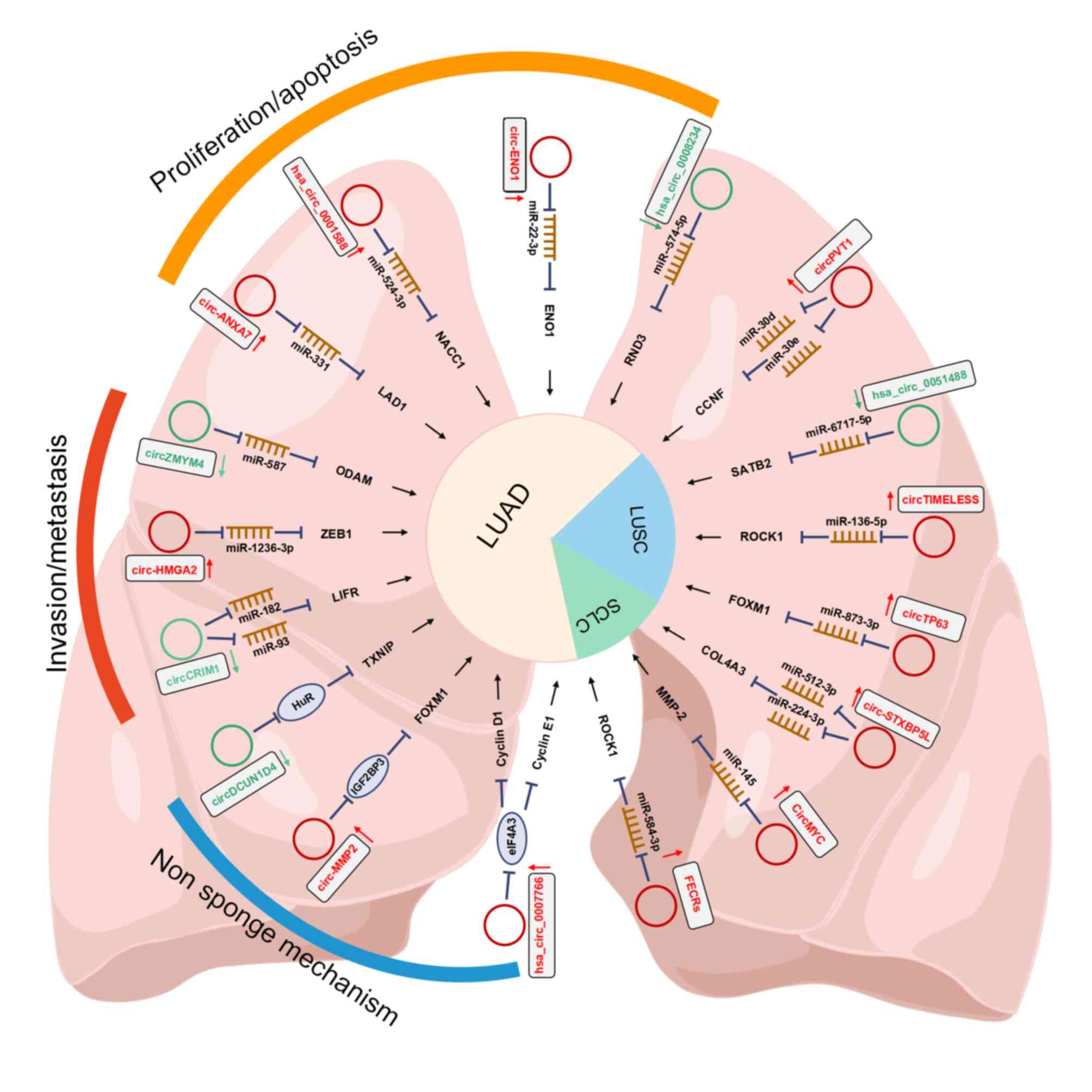
Different types of lung cancer have different
molecular mechanisms, therapeutic targets and prognoses.
Contemporary emerging evidence has indicated that circRNAs are
abnormally expressed and have pivotal roles in different lung
cancers (51). However, the
expression patterns and clinical significance of different circRNAs
in different types of lung cancer remain to be fully elucidated.
circRNAs confer a great diversity of important biological functions
by acting as miRNA sponges or interacting with proteins.
Considering the possible discriminative roles of circRNAs in
different types of lung cancer, their differential expression and
possible mechanisms in LUAD, LUSC and SCLC are summarized below and
are outlined in Fig. 2.
NSCLC is the leading cause of cancer-related death
and the major pathological type of lung cancer in China (52). Recent studies have focused on
investigating the roles of circRNAs in lung cancer and have
demonstrated the prospects of circRNAs as significant contributors
to tumor progression.
Numerous circRNAs were reported to be abnormally
expressed in NSCLC. Fluorescence in situ hybridization
assays indicated that circARHGAP10 was highly expressed in human
NSCLC samples compared to adjacent normal tissues and poor survival
was associated with high circARHGAP10 (circRNA Rho GTPase
activating protein 10) expression in NSCLC (53). Knockdown of circARHGAP10 suppressed
the proliferation and metastasis of NSCLC cells through a miRNA
sponge mechanism via the miR-159-5p/glucose transporter 1 axis.
Subsequently, circPTPRA (circRNA protein tyrosine phosphatase
receptor-type A) overexpression was found to cause an invasive
phenotype in NSCLC cell lines (54). A higher level of circPTPRA is
correlated with lower E-cadherin together with higher N-cadherin
and vimentin protein levels, indicating that circPTPRA may
accelerate invasion by promoting epithelial-mesenchymal transition
(EMT). Furthermore, circPTPRA was found to sponge miR-96-5p to
suppress EMT in NSCLC cells. By circRNA microarray,
hsa_circRNA_012515 expression was found to be significantly
upregulated in NSCLC tissues and cells (55), particularly in gefitinib-resistant
NSCLC cells.
These findings of recent studies demonstrated that
circRNA may have a promising role in NSCLC and highlighted their
novel biological functions and possible uses as diagnostic and
therapeutic targets. NSCLC primarily comprises LUAD and LUSC, which
exhibit nonidentical circRNA expression signatures (8). Thus, in the chapters below,
characteristic circRNAs in LUAD and LUSC were discussed.
Increased cell proliferation and blockage of
apoptosis are the main hallmarks of cancer; therefore, circRNAs
usually have roles in regulating the proliferation and apoptosis of
cancer cells (61,62). Several circRNAs were demonstrated
to be upregulated in LUAD and to regulate LUAD progression and
apoptosis. For instance, circ-ENO1 may promote proliferation and
EMT in LUAD via the miR-22-3p/ENO1 axis and inhibit apoptosis in
LUAD cells (56). Furthermore, the
silencing of circ-ENO1 prohibits the expression of ENO1 and
downregulates glycolysis in LUAD cells (56). Ladinin-1 (LAD1), a collagenous
anchoring filament protein, has been reported to be a marker of
aggressive malignancy in multiple cancers (63,64).
circRNA Annexin A7 (circ-ANXA7) is highly expressed in LUAD. It may
act as a sponge for miR-331 and indirectly upregulate LAD1 to
facilitate proliferation and suppress apoptosis in LUAD cells
(65). Furthermore, hsa_
circ_0001588 functions as a miR-524-3p sponge to increase nucleus
accumbens associated 1 expression and promote the malignant
progression of LUAD (66).
circ_0007766 mainly affects the malignant proliferation of LUAD and
significantly activates the cell cycle in the G0/G1 phase by
upregulating the expression of the cell cycle-related proteins
cyclin D1/cyclin E1/cyclin-dependent kinase 4 (67). No significant difference in the
percentage of apoptosis was found between LUAD with high and low
circ_0007766 expression (67).
circ_104640 and circFOXP1 were also demonstrated to promote LUAD
progression and decrease apoptosis. Jiang et al (68) found that circ_104640 affected the
expression of C-C motif chemokine ligand 20 by combining with
miR-145-5p and it had a tumor-promoting role. circ_104640 also
inhibited apoptosis in early-stage LUAD. Wnt family member 1
(WNT1), a member of the WNT family, is implicated in oncogenesis
and developmental processes and may inhibit cell apoptosis
(69,70). Downregulation of circFOXP1
inhibited cell proliferation and increased cell apoptosis in LUAD
cells via the circFOXP1/miR-185-5p/WNT1 axis (71).
Of note, several circRNAs were reported to be
downregulated in LUAD. Rho family GTPase 3 (RND3), a member of the
Rho GTPase family (72), is
associated with unfavorable prognosis of patients and participates
in functions such as cell proliferation and differentiation
(73). Jiang et al
(74) demonstrated that
circ_0008234 was downregulated in LUAD cells and exerted a
protective role in LUAD, since it may decelerate proliferation and
accelerate apoptosis of LUAD cells by sponging miR-574-5p and
subsequently inhibiting RND3. circRNA cMras was also decreased in
LUAD and suppressed cell proliferation by modulating the
miR-567/protein tyrosine phosphatase receptor type G (PTPRG) axis.
Transwell migration and invasion assays further suggested that
overexpression of circRNA cMras suppressed the migration and
invasion ability of LUAD cells by sponging miR-567 to upregulate
PTPRG (75). Dong et al
(76) found that circFBXW7 affects
patient prognosis and attenuates malignant progression in LUAD via
the miR-942-5p/BarH-like homeobox 2 axis. Further downregulated
circRNAs in LUAD are listed in detail in Table I.
The development of cancer invasion and metastases is
a leading cause of cancer-related death; therefore, it is essential
to prevent such malignant behaviors (77). Several studies have indicated that
certain circRNAs in LUAD promote metastasis and malignant
progression. For instance, circZMYM4 (circRNA zinc finger MYM type
4) inhibited growth and metastasis in LUAD by sponging miR-587 to
upregulate odontogenic ameloblast-associated (ODAM) expression
(78). Knockdown of ODAM reversed
the suppressive effect caused by circZMYM4 on cell proliferation,
migration and invasion abilities. Wang et al (60) found that circCRIM1 is associated
with favorable survival in LUAD and inhibits LUAD cell migration
and invasion. circCRIM1 functions as a ceRNA of miR-93 to promote
leukemia inhibitory factor receptor expression, which suppresses
PI3K/AKT signaling and functions as a suppressor of LUAD metastasis
(79,80). In addition, circ-HMGA2
(hsa_circ_0027446), circRNA high mobility group AT-hook 2 was
upregulated in LUAD cells, promoting metastasis and EMT via the
miR-1236-3p/zinc finger E-box binding homeobox 1 axis (81).
In addition, it is worth noting that circRNA may
also function via other distinct non-sponge mechanisms in LUAD
cells. Liang et al (82)
reported the suppression of LUAD cell metastasis in vitro
and in vivo. circDCUN1D4 (circRNA defective in cullin
neddylation 1 domain containing 4) interacts with
thioredoxin-interacting protein (TXNIP) mRNA directly through base
complementation to suppress metastasis and glycolysis in LUAD cells
in a TXNIP-dependent manner. Mechanistically, circDCUN1D4 enhances
the stability of TXNIP mRNA by facilitating the interaction between
the thioredoxin interacting protein protein and TXNIP as a
scaffold. In addition, circXPO1 (circRNA exportin 1) was
upregulated in LUAD tissues and was associated with unfavorable
overall survival (83). High
expression of circXPO1 may promote LUAD progression by binding with
insulin-like growth factor (IGF) 2 mRNA binding protein (IGF2BP)1
and enhancing catenin β1 mRNA stability. In addition, circ-MMP2
(circ-0039411) promotes malignant behaviors of LUAD cells by
recruiting IGF2BP3 (insulin-like growth factor II mRNA-binding
protein 3) to enhance the stability of forkhead box M1 (FOXM1)
mRNA. In addition, FOXM1 may also upregulate circ-0039411 and
induce nuclear translocation of β-catenin to form a positive
feedback loop (84). These studies
suggested that sponge and nonsponge circRNAs both function in
regulating the LUAD process.
Compared with LUAD, the dysregulated circRNAs and
their mechanistic underpinnings in LUSC remain largely elusive
(52). A small number of circRNAs
have been reported to be dysregulated in LUSC (Table II).
Furthermore, circRNA tumor protein p63 (circTP63)
was found to be upregulated in LUSC and silencing of circTP63
suppressed cell growth in LUSC (86). circTP63 may function as a miRNA
sponge for miR-873-3p and regulate FOXM1 expression. The expression
of hsa_circ_0000408 (circTIME-LESS) in LUSC was increased and
positively correlated with the TNM stage (87). A xenograft tumor growth assay
suggested that it acted as a tumor promoter in LUSC and influenced
LUSC proliferation and invasion via the miR-136-5p/Rho associated
coiled-coil containing protein kinase 1 (ROCK1) axis (87). In addition, hsa_circ_0051488
expression in cancer tissues was inversely correlated with tumor
diameter, lymphatic metastasis and TNM stage (88). Further experiments indicated that
circ0051488 may promote LUSC progression through the
miR6715/special AT-rich sequence-binding protein 2 (SATB2) axis,
suggesting that circ0051488/miR6715/SATB2 may be a potential
pathway for LUSC therapeutic intervention (88).
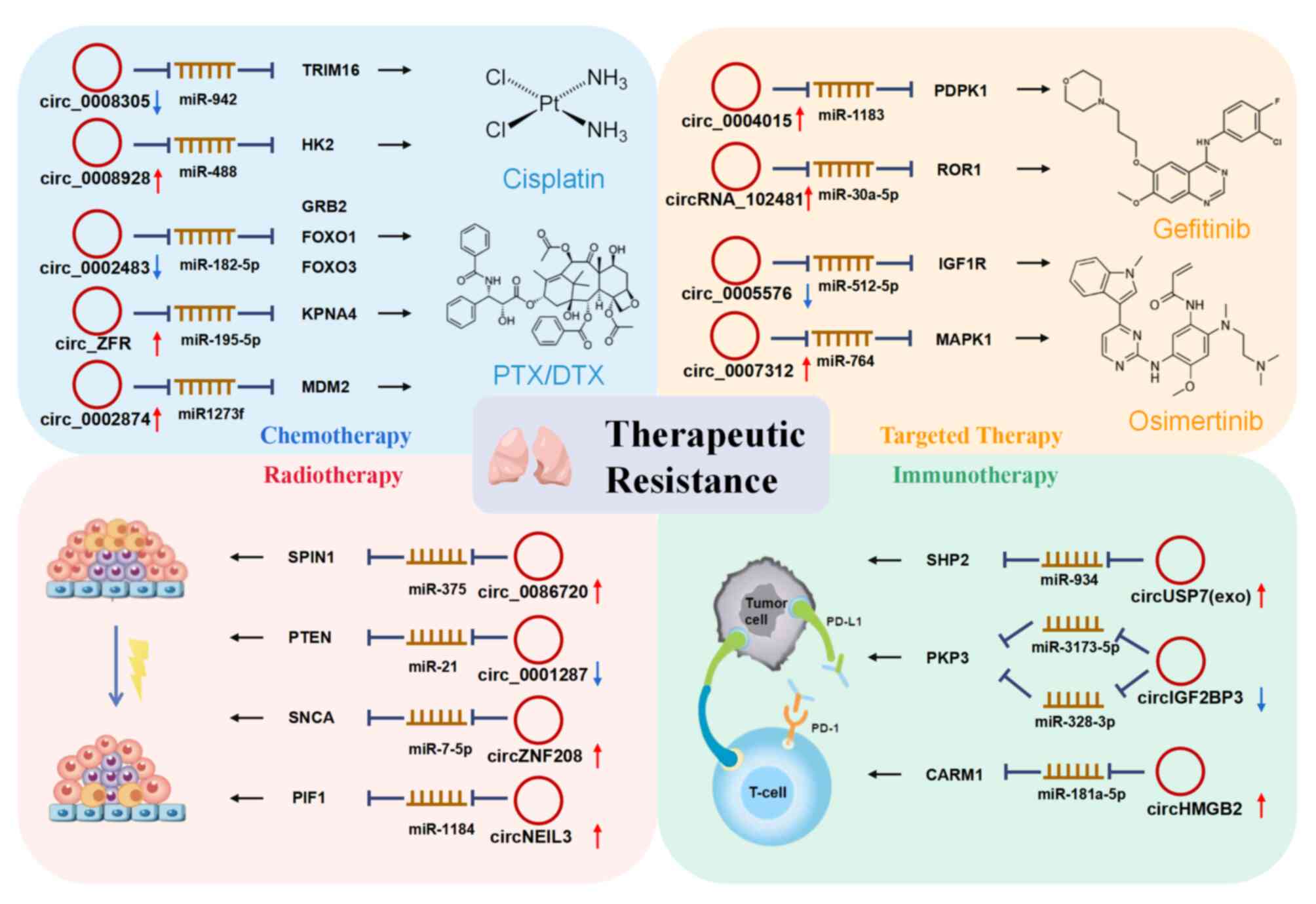
Recently, chemotherapeutic drugs, targeted therapy
and radiation have been commonly employed in lung cancer (93). Furthermore, immunotherapy has
recently been utilized more frequently. However, resistance is
increasingly obvious throughout the prolonged period of use and
limits the efficacy of therapy (94). The development of resistance
involves several mechanisms, and circRNA may be a promising target
to improve the therapeutic efficacy and act as diagnostic and
prognostic markers to improve treatment efficacy. The circRNAs that
are highly relevant to the efficacy of therapeutic treatment and
their mechanisms of action are presented in Table IV and Fig. 3.
The most commonly used chemotherapeutic agents
include platinum drugs [cisplatin (CDDP)], taxol derivatives
[paclitaxel (PTX), docetaxel (DTX)] and pemetrexed (95). circRNAs have a role in tumor
sensitivity and resistance to chemotherapy and act as potential
diagnostic biomarkers to predict the efficiency of chemotherapy
(96,97).
CDDP was reported to be the most efficient
chemotherapeutic drug for the treatment of NSCLC and SCLC (98). It mainly interacts with DNA and
activates signal transduction pathways, including ATR, p53, p73 and
MAPK, resulting in the apoptosis of cancer cells (99). Hsa_circ_0008305 (circPTK2)
expression was decreased in CDDP-resistant NSCLC tissues and cells,
and its overexpression significantly inhibited CDDP resistance in
NSCLC through the miR-942/tripartite motif containing 16 axis
(100). However, circ_0008928 was
highly expressed in CDDP-resistant NSCLC cells (101). Knockdown of circ_0008928 improved
CDDP sensitivity and accelerated cell proliferation, migration and
invasion via the miR-488/hexokinase 2 signaling pathway. In
addition, the expression of circ_0008928 was significantly
upregulated in serum exosomes of patients with CDDP-resistant
NSCLC, indicating its potential as a biomarker to predict the
resistance of NSCLC (101).
PTX resistance was also reported to be related to
circRNAs. Hsa_circ_0002483 was downregulated in PTX-resistant NSCLC
cells (102). Its overexpression
significantly enhanced the sensitivity of NSCLC cells to PTX.
circ_0002483 was demonstrated to sponge miR-182-5p and inhibit
growth factor receptor bound protein 2, forkhead box protein O1
(FOXO1) and FOXO3 expression, which may lead to increased
sensitivity to PTX (102). By
contrast, circRNA zinc-finger (circ_ZFR) and hsa_circ_0002874 were
demonstrated to promote PTX resistance in NSCLC (103). circ_ZFR was discovered to be
highly expressed in PTX-resistant NSCLC tissues and cell lines.
Mechanistically, circ_ZFR markedly upregulates the expression of
karyopherin subunit α4 by sponging miR-195-5p, leading to PTX
resistance, cell cycle progression, proliferation, migration and
invasion (103). In addition,
high expression of hsa_circ_0002874 regulated the miR1273f/mouse
double minute 2 protein/P53 signaling pathway to promote PTX
resistance in vitro and in vivo (104). Knockdown of hsa_circ_0002874
promoted PTX sensitivity and induced apoptosis in PTX-resistant
NSCLC cells (104).
DTX is also a useful taxane chemotherapeutic
medicine for lung cancer, for which resistance is a common problem
(105,106). Hsa_circ_0003998 was found to be
upregulated in LUAD tissue and DTX-resistant cell lines. Knockdown
of hsa_circ_0003998 significantly reversed DTX resistance in LUAD
cells by directly binding with miR-326 (107). In addition, in contrast to
DTX-sensitive tissues and cells, the 5-year survival rate of NSCLC
DTX-resistant patients was significantly lower and hsa_circ_0074027
(HC0074027) was significantly upregulated in DTX-resistant tissues
and cells (108).
Mechanistically, HC0074027 enhanced NSCLC DTX resistance in
vitro and in vivo via the miR-379-5p/insulin-like growth
factor 1 (IGF1) axis (108).
circRNAs may also modulate resistance to other
chemotherapeutics and combined chemotherapy, such as pemetrexed.
Mao and Xu (109) reported that
cerebellar degeneration-related protein 1 transcript (CDR1-AS) is
highly expressed in patients with pemetrexed- and CDDP-insensitive
LUAD, and silencing CDR1-AS significantly sensitized LUAD cells to
PTX and CDDP via the epidermal growth factor receptor (EGFR)/PI3K
signaling pathway. circRNA planning target volume 1 (circPVT1) was
highly expressed in CDDP- and pemetrexed-resistant LUAD tissues,
while knockdown of circPVT1 resensitized chemoresistant LUAD cells
(110). In addition, subsequent
experiments revealed that circPVT1 knockdown resulted in decreased
ATP binding cassette subfamily C member 1 expression through
miR-145-5p and inhibited resistance to drugs including doxorubicin,
etoposide and vincristine (110,111). These studies on drug resistance
will help elucidate the mechanisms of chemoresistance and may
provide a therapeutic target for lung cancer.
Recently, targeted therapies for the management of
lung cancer have provided promising results (112). Of note, several circRNAs were
reported to cause resistance to targeted drugs. EGFR is currently
the most favored target in lung cancer. Gefitinib was the first
EGFR inhibitor, approved in 2003. Hsa_circRNA_012515 was
particularly highly expressed in peripheral blood samples of
patients with gefitinib-resistant NSCLC compared to the sensitive
group, demonstrating that upregulation of hsa_circRNA_012515 may be
a mechanism leading to gefitinib resistance in patients with NSCLC
(55). Furthermore,
hsa_circRNA_012515 exhibited high diagnostic accuracy as a
biomarker for efficient targeted therapy [area under the receiver
operating characteristic (ROC) curve (AUC)=0.89] (55). Similarly, hsa_circ_0004015
expression regulated proliferation and invasion and contributed to
gefitinib resistance in NSCLC cells via the
miR-1183/3-phosphoinositide dependent protein kinase-1 signaling
pathway (96). Of note, exosomal
circRNA_102481 was upregulated in NSCLC with EGFR-tyrosine kinase
inhibitor (TKI) resistance and it was able to promote EGFR-TKI
(gefitinib and erlotinib)-resistant NSCLC cell proliferation and
inhibit cell apoptosis by regulating the miR-30a-5p/receptor
tyrosine kinase like orphan receptor 1 axis (113). A total of 58 patients treated
with gefitinib or erlotinib were collected to validate these
results and circRNA_102481 was demonstrated to be significantly
upregulated in patients with EGFR-TKI resistance (113).
Osimertinib is a third-generation EGFR inhibitor
against the acquired gefitinib-resistant mutation EGFR T790 M.
However, resistance to osimertinib has also been observed and
circRNAs are also involved. Hsa_circ_0005576 was markedly increased
in osimertinib-resistant LUAD cells. Knockdown of hsa_circ_0005576
recovered osimertinib sensitivity via the miR-512-5p/IGF1 receptor
pathway (114). In addition,
hsa_circ_0007312 (circ7312) was positively correlated with
osimertinib IC50 values and xenograft experiments
indicated that knockdown of circ7312 decreased resistance to
osimertinib in vivo. Mechanistically, circ7312 caused
osimertinib resistance in LUAD cells by sponging miR-764 to
upregulate MAPK1 and suppress pyroptosis and apoptosis (115).
Studies have indicated that circRNAs also regulate
cancer radiotherapy resistance. Colony-formation experiments
indicated that overexpression of circ_0001287 significantly
inhibited the survival of NSCLC cells after irradiation at
different ionizing radiation (IR) intensities (116), and knockdown of circ_0001287
promoted radioresistance, indicating that circ_0001287 was a vital
regulator of radiotherapy resistance. In addition, knockdown of
circ_0086720 was observed to enhance radiosensitivity and prevent
the growth of NSCLC (117).
circ_0086720 expression was upregulated in radioresistant NSCLC
tissues and inhibited the radiation sensitivity of NSCLC by
regulating the miR-375/spindling 1 axis (117). Astrocyte elevated gene-1 (AEG-1)
was indicated to significantly induce the apoptotic rate of NSCLC
cells after radiation (118).
circMTDH.4 is able to regulate the radioresistance of NSCLC cells
to IR by sponging miR-630 to regulate AEG-1 expression (118). Knockdown of circRNA Nei
endonuclease VIII-like 3 (circNEIL3) efficiently enhanced the
sensitivity to radiation treatment. After irradiation, knockdown of
circNEIL3 in LUAD cells led to increased efficacy of pyroptosis
through the DNA damage pathway via the
miR-1184/phytochrome-interacting factor 1 axis (119). This evidence indicates that
circNEIL3 may target pyroptosis in LUAD cells and may have the
potential to enhance the efficacy of radiotherapy in lung cancer.
Furthermore, knockdown of circZNF208 inhibited DNA synthesis and
decreased radioresistance in NSCLC cells with radioresistance to
IR. However, circZNF208 knockdown was not able to change the
radiosensitivity of NSCLC cells to carbon ions, which provides a
new perspective to further explore the mechanisms of circRNAs in
association with different types of irradiation (120).
Immunotherapy, particularly immune checkpoint
inhibitors, has emerged in recent years as an important treatment
option for numerous different cancers and has been successful for
certain types of cancer. However, it was reported that with the use
of anti-PD1, the majority of patients inevitably acquire resistance
after several cycles of treatment (121), and circRNAs were involved in the
resistance to these immunotherapies. In a humanized mouse model,
tumors with circRNA ubiquitin specific protease 7 (circUSP7)
overexpression exhibited elevated exosomal circUSP7 in the serum
and obvious resistance to anti-PD1 immunotherapy. The expression of
plasma exosomal circUSP7 in patients with NSCLC was found to be
negatively correlated with CD8+ T-cell infiltration in the tumor
and increased levels of circUSP7 predicted a poor clinical outcome.
circUSP7 was also found to promote CD8+ T-cell dysfunction via the
circUSP7/miR-934/Src homology-2-containing protein tyrosine
phosphatase 2 axis (122). These
results may be helpful for predicting the efficacy of immune
checkpoint therapy in patients with NSCLC.
circIGF2BP3 (hsa_circ_0079587) was also observed to
be increased in the tumor microenvironment and negatively
correlated with CD8+ T-cell infiltration (123). Elevated circIGF2BP3 led to
compromised antitumor immunity in an immunocompetent mouse and CD8+
T cells were indispensable in this effect. Further examination
indicated that circIGF2BP3 also restricted the efficacy of
anti-PD-1 treatment through miR-181a/coactivator associated
arginine methyltransferase 1 (CARM1) in patients with NSCLC. CARM1
was reported to promote the type 1 interferon response and the
sensitivity of the tumor to T-cell-dependent immune attack
(124). Overexpression of
circIGF2BP3 elicits high PD-L1 expression via the plakophilin-3/OTU
deubiquitinase 1 axis and damages anti-PD-1 treatment-mediated CD8+
T-cell recruitment and tumor regression (123). Furthermore, circHMGB2 was
demonstrated to increase the proliferation of LUAD and LUSC cells
and regulate anti-PD-1 resistance (125). These studies help elucidate the
mechanism of resistance to immunotherapy in lung cancer and may
also provide a potential therapeutic target for lung cancer.
The analysis of circRNAs enables the construction of
a reliable system and demonstrates their potential for providing
new diagnostic and prognostic biomarkers (126). Following the rapid development of
high-throughput transcriptome analysis techniques, liquid biopsy
assays have finally been implemented in clinical practice (127). circRNA molecules have the
characteristics of high abundance, stability, sequence
conservation, tissue specificity and a long half-life, and distinct
differences are exhibited in peripheral blood and tissue from
cancer patients (128).
Therefore, circRNAs have been increasingly recognized as promising
and specific tumor biomarkers. Due to the convenience and low
injury risk for patients, combined with the detectability of
circRNA in peripheral body fluids (126), the use of circRNA as a
significant biomarker for lung cancer is promising.
To help diagnose lung cancer, circRNAs should be
easily detected in the periphery. Compared to normal human
bronchial epithelial cells, circSATB2 was highly expressed in NSCLC
cells and knockdown of circSATB2 was able to markedly decrease the
cell proliferation, migration and invasive capacity of NSCLC cells,
signifying that circSATB2 has the potential to predict the presence
of NSCLC (129). Furthermore,
circSATB2 was also able to be specifically and sensitively detected
in serum exosomes from patients with lung cancer, supporting its
use as a blood biomarker for convenient clinical diagnosis of
NSCLC. In addition, patients with NSCLC exhibited higher serum
circRNA_001846 levels than healthy participants (130). circRNAs was also able to predict
the level of lymph node metastasis, tumor differentiation and TNM
stage. Furthermore, in the ROC analysis, the AUC was 0.678, which
revealed the specificity of circRNA_001846 in diagnosing patients
with NSCLC.
Tumor-educated platelets have emerged as rich
sources of cancer-related RNA profiles in liquid biopsies and are
also applicable for cancer detection (131). A total of 411 circRNAs
differentially expressed between patients with NSCLC and healthy
controls were identified, and circNRIP1 was the most significantly
differentially expressed circRNA in platelets between patients with
NSCLC and healthy controls. In addition, downregulation of
circNRIP1 is associated with an advanced tumor stage. This suggests
that circNRIP1 is a potential diagnostic biomarker for NSCLC
(132). All of these studies
indicated the potential of circRNAs as noninvasive biomarkers for
diagnosing lung cancer.
Prognostic evaluation is an important part of
formulating an optimal treatment, which is of great significance
for improving the medical effects and the quality of life of
patients. Studies have confirmed that a variety of circRNAs have
high specificity in respiratory diseases and are closely related to
the survival of patients with lung cancer, such as circ-ANXA7
(65), circRNA ArfGAP with SH3
domain ankyrin repeats and PH domain 1 (133), and hsa_circ_0001715 (134). These circRNAs may be used as
independent prognostic indicators for patients with lung
cancer.
In addition to circRNAs in the tumor tissue, several
circRNAs upregulated in the blood of patients with lung cancer may
also efficiently predict the progression of the disease. Luo et
al (137) observed that the
plasma level of oncogenic circRNA hsa_circ_0000190 (C190) was
highly expressed in patients with NSCLC and the AUC indicated that
the diagnostic accuracy of hsa_circ_0000190 was good, particularly
in the later stages (III-IV), and it was a potentially valuable
blood-based biomarker to assess the prognosis of NSCLC. At the same
time, exosomal circRNA FLI1 exonic circular RNA (FECR), exo-FECR
were detected at relatively high levels in the serum of patients
with SCLC compared with other cancers, and longer disease remission
periods were observed in patients with lower exo-FECR1 compared to
patients with higher exo-FECR1. This evidence suggests that
exo-FECR1 may serve as a useful clinical indicator to monitor
disease progression and predict survival outcomes in SCLC (92).
In recent years, numerous studies have focused on
circRNAs, revealing diverse molecular mechanisms and functional
roles in lung cancer. As mentioned above, circRNAs exhibit the
characteristics of structural specificity and biological stability,
which is helpful for diagnosis, drug resistance monitoring,
treatment evaluation and prognostication of lung cancer. Liquid
biopsy detection technology for circRNAs is of great potential
clinical value. At present, although numerous strategies have been
used to treat lung cancer, such as chemotherapy, radiotherapy,
surgery, immunotherapy and targeted therapy, in most patients with
NSCLC, treatment resistance is still an important reason for poor
treatment effects or relapse. There is a close relationship between
circRNAs and the treatment resistance of NSCLC. In the future,
circRNAs may be used to screen and predict treatment efficacy in
populations treated with chemotherapy, radiotherapy and
immunotherapy, as well as the sensitivity to and tolerance of these
treatments, to provide a theoretical basis for the clinical
optimization of treatment strategies for NSCLC.
circRNAs function by acting as miRNA molecule
sponges, protein decoys, gene transcriptional regulators or
translation templates and have important roles in gene expression
and signaling pathways involved in a variety of biological
processes and diseases. With the advancement of high-throughput
sequencing and related research, the functions of circRNAs continue
to be revealed. Of note, circRNAs are considered to be conserved,
stable and abundant, and frequently exhibit tissue specificity.
Based on these findings, circRNAs hold extraordinary promise as
diagnostic, prognostic and predictive biomarkers. Due to their
stability, circRNAs may be present in exosomes and blood plasma,
and their stable presence in exosomes and plasma provides a more
convenient way to diagnose cancer compared to tumor tissues.
However, numerous challenges and controversies
remain that require to be resolved to move this field forward. The
present review indicated that most studies mainly focused on LUAD,
while the current understanding of circRNAs in LUSC and SCLC
remains limited. SCLC has a rapid development process and a high
degree of malignancy, and it requires more thorough research on its
mechanisms in the future to improve patient prognosis. In addition,
most circRNAs exert their effects as miRNA sponges. Whether the
miRNA sponge function is the primary function of circRNAs remains
controversial. In addition, current research on circRNAs mainly
focuses on lung cancer tissue or cell lines, but there remains a
gap in the study of circRNAs in body fluids, which may be more
feasible and convenient in the clinic. Furthermore, the current
research is mostly aimed at single circRNAs. It is necessary to
construct lung cancer-related circRNA networks to obtain a full
view of their roles in lung cancer development before these results
become clinically applicable.
In conclusion, in addition to the diagnostic and
prognostic value of circRNAs in lung cancer, the construction of
circRNAs and the continuous development and maturity of circRNA
interference technology make it possible to use circRNAs to alter
biological regulation. There will be additional breakthroughs in
the field of circRNAs in the future, which will offer profound
clinical diagnostic and therapeutic applications.
Not applicable.
Conceptualization: SX and LC. Compilation of
literature: FW and CTY. Article writing and editing: FW and CTY.
Figures: FW and CTY. Supervision: SX and LC. All authors have read
and approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This work was supported by the National Natural Science
Foundation of China (grant nos. 81471569, 31870910 and 82071789)
and Shanghai Committee of Science and Technology (15QA1404700).
|
1
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Suzuki H and Tsukahara T: A view of
pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci.
15:9331–9342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
5
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ahmadov U, Bendikas MM, Ebbesen KK,
Sehested AM, Kjems J, Broholm H and Kristensen LS: Distinct
circular RNA expression profiles in pediatric ependymomas. Brain
Pathol. 31:387–392. 2021. View Article : Google Scholar :
|
|
7
|
Smid M, Wilting SM, Uhr K,
Rodríguez-González FG, de Weerd V, Prager-Van der Smissen WJC, van
der Vlugt-Daane M, van Galen A, Nik-Zainal S, Butler A, et al: The
circular RNome of primary breast cancer. Genome Res. 29:356–366.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu
Y, Liu X, Cheng W, Wei YQ, Peng Y and Li W: RNA-Seq profiling of
circular RNA in human lung adenocarcinoma and squamous cell
carcinoma. Mol Cancer. 18:1342019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 World Health Organization
classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oser MG, Niederst MJ, Sequist LV and
Engelman JA: Transformation from non-small-cell lung cancer to
small-cell lung cancer: Molecular drivers and cells of origin.
Lancet Oncol. 16:e165–e172. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ettinger DS, Wood DE, Aisner DL, Akerley
W, Bauman J, Chirieac LR, D'Amico TA, DeCamp MM, Dilling TJ,
Dobelbower M, et al: Non-small cell lung cancer, version 5.2017,
NCCN clinical practice guidelines in oncology. J Natl Compr Canc
Netw. 15:504–535. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schneider BJ, Ismaila N, Aerts J, Chiles
C, Daly ME, Detterbeck FC, Hearn JWD, Katz SI, Leighl NB, Levy B,
et al: Lung cancer surveillance after definitive curative-intent
therapy: ASCO guideline. J Clin Oncol. 38:753–766. 2020. View Article : Google Scholar
|
|
14
|
Kristensen LS, Jakobsen T, Hager H and
Kjems J: The emerging roles of circRNAs in cancer and oncology. Nat
Rev Clin Oncol. 19:188–206. 2022. View Article : Google Scholar
|
|
15
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Misir S, Wu N and Yang BB: Specific
expression and functions of circular RNAs. Cell Death Differ.
29:481–491. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen I, Chen CY and Chuang TJ: Biogenesis,
identification, and function of exonic circular RNAs. Wiley
Interdiscip Rev RNA. 6:563–579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang
W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs:
Biogenesis, effect and application in human diseases. Mol Cancer.
18:1162019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang XY, Huang ZL, Huang J, Xu B, Huang
XY, Xu YH, Zhou J and Tang ZY: Exosomal circRNA-100338 promotes
hepatocellular carcinoma metastasis via enhancing invasiveness and
angiogenesis. J Exp Clin Cancer Res. 39:202020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ,
Shi GM, Cai JB and Ke AW: Cancer cell-derived exosomal circUHRF1
induces natural killer cell exhaustion and may cause resistance to
anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer.
19:1102020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y,
Chen X, Chen Y, Xu C, Hu Y, et al: Exosome-derived circTRPS1
promotes malignant phenotype and CD8+ T cell exhaustion in bladder
cancer microenvironments. Mol Ther. 30:1054–1070. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fanale D, Taverna S, Russo A and Bazan V:
Circular RNA in exosomes. Adv Exp Med Biol. 1087:109–117. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li C, Ni YQ, Xu H, Xiang QY, Zhao Y, Zhan
JK, He JY, Li S and Liu YS: Roles and mechanisms of exosomal
non-coding RNAs in human health and diseases. Signal Transduct
Target Ther. 6:3832021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang
P, Cotter MB, Bowden M, Lis RT, Zhao SG, et al: Genome-wide CRISPR
screen identifies HNRNPL as a prostate cancer dependency regulating
RNA splicing. Proc Natl Acad Sci USA. 114:E5207–E5215. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Barrett SP, Wang PL and Salzman J:
Circular RNA biogenesis can proceed through an exon-containing
lariat precursor. Elife. 4:e075402015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Eger N, Schoppe L, Schuster S, Laufs U and
Boeckel JN: Circular RNA splicing. Adv Exp Med Biol. 1087:41–52.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maass PG, Glažar P, Memczak S, Dittmar G,
Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC and
Rajewsky N: A map of human circular RNAs in clinically relevant
tissues. J Mol Med (Berl). 95:1179–1189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu D and Xu AD: Mini review: Circular RNAs
as potential clinical biomarkers for disorders in the central
nervous system. Front Genet. 7:532016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pamudurti NR, Patop IL, Krishnamoorthy A,
Bartok O, Maya R, Lerner N, Ashwall-Fluss R, Konakondla JVV, Beatus
T and Kadener S: circMbl functions in cis and in trans to regulate
gene expression and physiology in a tissue-specific fashion. Cell
Rep. 39:1107402022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Du WW, Xu J, Yang W, Wu N, Li F, Zhou L,
Wang S, Li X, He AT, Du KY, et al: A neuroligin isoform translated
by circNlgn contributes to cardiac remodeling. Circ Res.
129:568–582. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar :
|
|
46
|
Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X,
Zhong J, Zhao Z, Zhao K, Liu D, et al: Circular RNA-encoded
oncogenic E-cadherin variant promotes glioblastoma tumorigenicity
through activation of EGFR-STAT3 signalling. Nat Cell Biol.
23:278–291. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Y, Chen B, Zhao J, Li Q, Chen S, Guo T,
Li Y, Lai H, Chen Z, Meng Z, et al: HNRNPL circularizes ARHGAP35 to
produce an oncogenic protein. Adv Sci (Weinh). 8:20017012021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S,
Xuan Z, Xie L, Qiu S, He Z, et al: A novel protein encoded by
circMAPK1 inhibits progression of gastric cancer by suppressing
activation of MAPK signaling. Mol Cancer. 20:662021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ghafouri-Fard S, Dinger ME, Maleki P,
Taheri M and Hajiesmaeili M: Emerging role of circular RNAs in the
pathobiology of lung cancer. Biomed Pharmacother. 141:1118052021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jin M, Shi C, Yang C, Liu J and Huang G:
Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in
NSCLC through regulation of the miR-150-5p/GLUT-1 axis. Mol Ther
Nucleic Acids. 18:219–231. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wei S, Zheng Y, Jiang Y, Li X, Geng J,
Shen Y, Li Q, Wang X, Zhao C, Chen Y, et al: The circRNA circPTPRA
suppresses epithelial-mesenchymal transitioning and metastasis of
NSCLC cells by sponging miR-96-5p. EBioMedicine. 44:182–193. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fu Y, Huang L, Tang H and Huang R:
hsa_circRNA_012515 Is Highly expressed in NSCLC patients and
affects its prognosis. Cancer Manag Res. 12:1877–1886. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhou J, Zhang S, Chen Z, He Z, Xu Y and Li
Z: CircRNA-ENO1 promoted glycolysis and tumor progression in lung
adenocarcinoma through upregulating its host gene ENO1. Cell Death
Dis. 10:8852019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan
Y, Kong X, Bu J, Liu M and Xu S: circRNA-002178 act as a ceRNA to
promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis.
11:322020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang Z, Pei H, Liang H, Zhang Q, Wei L,
Shi D, Chen Y and Zhang J: Construction and analysis of a
circRNA-Mediated ceRNA network in lung adenocarcinoma. Onco Targets
Ther. 14:3659–3669. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yao Y, Hua Q and Zhou Y: CircRNA
has_circ_0006427 suppresses the progression of lung adenocarcinoma
by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/β-catenin
signaling pathway. Biochem Biophys Res Commun. 508:37–45. 2019.
View Article : Google Scholar
|
|
60
|
Wang L, Liang Y, Mao Q, Xia W, Chen B,
Shen H, Xu L, Jiang F and Dong G: Circular RNA circCRIM1 inhibits
invasion and metastasis in lung adenocarcinoma through the microRNA
(miR)-182/miR-93-leukemia inhibitory factor receptor pathway.
Cancer Sci. 110:2960–2972. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Andersen MH, Becker JC and Straten PT:
Regulators of apoptosis: Suitable targets for immune therapy of
cancer. Nat Rev Drug Discov. 4:399–409. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kroemer G and Pouyssegur J: Tumor cell
metabolism: Cancer's Achilles' heel. Cancer Cell. 13:472–482. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Teixeira JC, de Filippo C, Weihmann A,
Meneu JR, Racimo F, Dannemann M, Nickel B, Fischer A, Halbwax M,
Andre C, et al: Long-term balancing selection in LAD1 maintains a
missense trans-species polymorphism in humans, chimpanzees, and
bonobos. Mol Biol Evol. 32:1186–1196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Roth L, Srivastava S, Lindzen M, Sas-Chen
A, Sheffer M, Lauriola M, Enuka Y, Noronha A, Mancini M, Lavi S, et
al: SILAC identifies LAD1 as a filamin-binding regulator of actin
dynamics in response to EGF and a marker of aggressive breast
tumors. Sci Signal. 11:eaan09492018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang Y: circ-ANXA7 facilitates lung
adenocarcinoma progression via miR-331/LAD1 axis. Cancer Cell Int.
21:852021. View Article : Google Scholar
|
|
66
|
Sun Z: Circular RNA hsa_circ_0001588
promotes the malignant progression of lung adenocarcinoma by
modulating miR-524-3p/NACC1 signaling. Life Sci. 259:1181572020.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang S, Xia W, Dong G, Xu W, Li M and Xu
L: Cyclic RNA molecule circ_0007766 promotes the proliferation of
lung adenocarcinoma cells by up-regulating the expression of Cyclin
D1/CyclinE1/CDK4. Zhongguo Fei Ai Za Zhi. 22:271–279. 2019.In
Chinese. PubMed/NCBI
|
|
68
|
Jiang W, Zhang C, Kang Y, Li G, Feng Y and
Ma H: The roles and mechanisms of the circular RNA circ_104640 in
early-stage lung adenocarcinoma: A potential diagnostic and
therapeutic target. Ann Transl Med. 9:1382021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kahn M: Wnt signaling in stem cells and
cancer stem cells: A tale of two coactivators. Prog Mol Biol Transl
Sci. 153:209–244. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li O, Kang J, Zhang JJ, Wang J, Hu LW, Li
L, Sun YY, Bai Y, Wei QQ, Yan YP and Yi X: Circle RNA FOXP1
promotes cell proliferation in lung cancer by regulating
miR-185-5p/Wnt1 signaling pathway. Eur Rev Med Pharmacol Sci.
24:6767–6778. 2020.PubMed/NCBI
|
|
72
|
Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J
and Settleman J: Identification of a novel human Rho protein with
unusual properties: GTPase deficiency and in vivo farnesylation.
Mol Cell Biol. 16:2689–2699. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yang X, Wang T, Lin X, Yue X, Wang Q, Wang
G, Fu Q, Ai X, Chiang DY, Miyake CY, et al: Genetic deletion of
Rnd3/RhoE results in mouse heart calcium leakage through
upregulation of protein kinase A signaling. Circ Res. 116:e1–e10.
2015. View Article : Google Scholar :
|
|
74
|
Jiang W, He Y, Ma Z, Zhang Y, Zhang C,
Zheng N and Tang X: hsa_circ_0008234 inhibits the progression of
lung adenocarcinoma by sponging miR-574-5p. Cell Death Discov.
7:1232021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yu C, Tian F, Liu J, Su M, Wu M, Zhu X and
Qian W: Circular RNA cMras inhibits lung adenocarcinoma progression
via modulating miR-567/PTPRG regulatory pathway. Cell Prolif.
52:e126102019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Dong Y, Qiu T, Xuan Y, Liu A, Sun X, Huang
Z, Su W, Du W, Yun T, Wo Y, et al: circFBXW7 attenuates malignant
progression in lung adenocarcinoma by sponging miR-942-5p. Transl
Lung Cancer Res. 10:1457–1473. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Eccles SA and Welch DR: Metastasis: Recent
discoveries and novel treatment strategies. Lancet. 369:1742–1757.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yuan DF, Wang HR, Wang ZF, Liang GH, Xing
WQ and Qin JJ: CircRNA CircZMYM4 inhibits the growth and metastasis
of lung adenocarcinoma via the miR-587/ODAM pathway. Biochem
Biophys Res Commun. 580:100–106. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kosfeld A, Brand F, Weiss AC, Kreuzer M,
Goerk M, Martens H, Schubert S, Schäfer AK, Riehmer V, Hennies I,
et al: Mutations in the leukemia inhibitory factor receptor (LIFR)
gene and Lifr deficiency cause urinary tract malformations. Hum Mol
Genet. 26:1716–1731. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Luo Q, Wang C, Jin G, Gu D, Wang N, Song
J, Jin H, Hu F, Zhang Y, Ge T, et al: LIFR functions as a
metastasis suppressor in hepatocellular carcinoma by negatively
regulating phosphoinositide 3-kinase/AKT pathway. Carcinogenesis.
36:1201–1212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yu Z, Zhu X, Li Y, Liang M, Liu M, Liu Z,
Qin L, Wu X, Du K, Liu L, et al: Circ-HMGA2 (hsa_circ_0027446)
promotes the metastasis and epithelial-mesenchymal transition of
lung adenocarcinoma cells through the miR-1236-3p/ZEB1 axis. Cell
Death Dis. 12:3132021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Liang Y, Wang H, Chen B, Mao Q, Xia W,
Zhang T, Song X, Zhang Z, Xu L, Dong G and Jiang F: circDCUN1D4
suppresses tumor metastasis and glycolysis in lung adenocarcinoma
by stabilizing TXNIP expression. Mol Ther Nucleic Acids.
23:355–368. 2020. View Article : Google Scholar
|
|
83
|
Huang Q, Guo H, Wang S, Ma Y, Chen H, Li
H, Li J, Li X, Yang F, Qiu M, et al: A novel circular RNA,
circXPO1, promotes lung adenocarcinoma progression by interacting
with IGF2BP1. Cell Death Dis. 11:10312020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Lv X, Huang H, Feng H and Wei Z: Circ-MMP2
(circ-0039411) induced by FOXM1 promotes the proliferation and
migration of lung adenocarcinoma cells in vitro and in vivo. Cell
Death Dis. 11:4262020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Xu J, Shu Y, Xu T, Zhu W, Qiu T, Li J,
Zhang M, Xu J, Guo R, Lu K, et al: Microarray expression profiling
and bioinformatics analysis of circular RNA expression in lung
squamous cell carcinoma. Am J Transl Res. 10:771–783.
2018.PubMed/NCBI
|
|
86
|
Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang
C, Li B, Qin M, Yang C, He J, et al: circTP63 functions as a ceRNA
to promote lung squamous cell carcinoma progression by upregulating
FOXM1. Nat Commun. 10:32002019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang W, Shi J, Cheng C and Wang H:
CircTIMELESS regulates the proliferation and invasion of lung
squamous cell carcinoma cells via the miR-136-5p/ROCK1 axis. J Cell
Physiol. 235:5962–5971. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Xiao M, Cui S, Zhang L, Yu T, Zhang G, Li
L, Cai Y, Jin C, Yang J, Wu S, et al: Benzo[a]pyrene diol
epoxide-induced transformed cells identify the significance of
hsa_circ_0051488, a ERCC1-derived circular RNA in pulmonary
squamous cell carcinoma. Mol Carcinog. 60:684–701. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sabari JK, Lok BH, Laird JH, Poirier JT
and Rudin CM: Unravelling the biology of SCLC: Implications for
therapy. Nat Rev Clin Oncol. 14:549–561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhang C, Zhang B, Yuan B, Chen C, Zhou Y,
Zhang Y, Sheng Z, Sun N and Wu X: RNA-Seq profiling of circular
RNAs in human small cell lung cancer. Epigenomics. 12:685–700.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yang X, Tao L, Xu Y, Li S, Yang W, Wang L
and Zhu J: CircMYC promotes proliferation, migration, invasion and
inhibits apoptosis of small cell lung cancer by targeting
miR-145/matrix metallopeptidase 2 axis. Bioengineered.
13:10552–10563. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y,
Pan X, Zhang X, Zhou L, Yu D, et al: FLI1 exonic circular RNAs as a
novel oncogenic driver to promote tumor metastasis in small cell
lung cancer. Clin Cancer Res. 25:1302–1317. 2019. View Article : Google Scholar
|
|
93
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Joseph NA, Chiou SH, Lung Z, Yang CL, Lin
TY, Chang HW, Sun HS, Gupta SK, Yen L, Wang SD and Chow KC: The
role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR
resistance and epithelial-to-mesenchymal transition of lung
adenocarcinoma cells. J Hematol Oncol. 11:742018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Gerber DE and Schiller JH: Maintenance
chemotherapy for advanced non-small-cell lung cancer: New life for
an old idea. J Clin Oncol. 31:1009–1020. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhou Y, Zheng X, Xu B, Chen L, Wang Q,
Deng H and Jiang J: Circular RNA hsa_circ_0004015 regulates the
proliferation, invasion, and TKI drug resistance of non-small cell
lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys
Res Commun. 508:527–535. 2019. View Article : Google Scholar
|
|
97
|
Feng B, Zhou H, Wang T, Lin X, Lai Y, Chu
X and Wang R: Insights into circRNAs: Functional roles in lung
cancer management and the potential mechanisms. Front Cell Dev
Biol. 9:6369132021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Schiller JH, Harrington D, Belani CP,
Langer C, Sandler A, Krook J, Zhu J and Johnson DH; Eastern
Cooperative Oncology Group: Comparison of four chemotherapy
regimens for advanced non-small-cell lung cancer. N Engl J Med.
346:92–98. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Siddik ZH: Cisplatin: Mode of cytotoxic
action and molecular basis of resistance. Oncogene. 22:7265–7279.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wang Y, Wu Y and Xie S: CircPTK2 inhibits
cell cisplatin (CDDP) resistance by targeting miR-942/TRIM16 axis
in non-small cell lung cancer (NSCLC). Bioengineered. 13:3651–3664.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Shi Q, Ji T, Ma Z, Tan Q and Liang J:
Serum exosomes-based biomarker circ_0008928 regulates cisplatin
sensitivity, tumor progression, and glycolysis metabolism by
miR-488/HK2 axis in cisplatin-resistant nonsmall cell lung
carcinoma. Cancer Biother Radiopharm. Mar 3–2021.Epub ahead of
print.
|
|
102
|
Li X, Yang B, Ren H, Xiao T, Zhang L, Li
L, Li M, Wang X, Zhou H and Zhang W: Hsa_circ_0002483 inhibited the
progression and enhanced the Taxol sensitivity of non-small cell
lung cancer by targeting miR-182-5p. Cell Death Dis. 10:9532019.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li J, Fan R and Xiao H: Circ_ZFR
contributes to the paclitaxel resistance and progression of
non-small cell lung cancer by upregulating KPNA4 through sponging
miR-195-5p. Cancer Cell Int. 21:152021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Xu J, Ni L, Zhao F, Dai X, Tao J, Pan J,
Shi A, Shen Z, Su C and Zhang Y: Overexpression of hsa_circ_0002874
promotes resistance of non-small cell lung cancer to paclitaxel by
modulating miR-1273f/MDM2/p53 pathway. Aging (Albany NY).
13:5986–6009. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Joshi M, Liu X and Belani CP: Taxanes,
past, present, and future impact on non-small cell lung cancer.
Anticancer Drugs. 25:571–583. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Chen J, Liu X, Xu Y, Zhang K, Huang J, Pan
B, Chen D, Cui S, Song H, Wang R, et al: TFAP2C-activated MALAT1
modulates the chemoresistance of docetaxel-resistant lung
adenocarcinoma cells. Mol Ther Nucleic Acids. 14:567–582. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yu W, Peng W, Sha H and Li J:
Hsa_circ_0003998 promotes chemoresistance via modulation of miR-326
in lung adenocarcinoma cells. Oncol Res. 27:623–628. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zheng S, Wang C, Yan H and Du Y: Blocking
hsa_circ_0074027 suppressed non-small cell lung cancer
chemoresistance via the miR-379-5p/IGF1 axis. Bioengineered.
12:8347–8357. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Mao Y and Xu R: Circular RNA CDR1-AS
contributes to pemetrexed and cisplatin chemoresistance through
EGFR/PI3K signaling pathway in lung adenocarcinoma. Biomed
Pharmacother. 123:1097712020. View Article : Google Scholar
|
|
110
|
Zheng F and Xu R: CircPVT1 contributes to
chemotherapy resistance of lung adenocarcinoma through
miR-145-5p/ABCC1 axis. Biomed Pharmacother. 124:1098282020.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Mirski SE, Gerlach JH and Cole SP:
Multidrug resistance in a human small cell lung cancer cell line
selected in adriamycin. Cancer Res. 47:2594–2598. 1987.PubMed/NCBI
|
|
112
|
Desai A and Adjei AA: FGFR signaling as a
target for lung cancer therapy. J Thorac Oncol. 11:9–20. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Yang B, Teng F, Chang L, Wang J, Liu DL,
Cui YS and Li GH: Tumor-derived exosomal circRNA_102481 contributes
to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small
cell lung cancer. Aging (Albany NY). 13:13264–13286. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Liu S, Jiang Z, Xiao P, Li X, Chen Y, Tang
H, Chai Y, Liu Y, Zhu Z, Xie Q, et al: Hsa_circ_0005576 promotes
osimertinib resistance through the miR-512-5p/IGF1R axis in lung
adenocarcinoma cells. Cancer Sci. 113:79–90. 2022. View Article : Google Scholar
|
|
115
|
Dai C, Ma Z, Si J, An G, Zhang W, Li S and
Ma Y: Hsa_ circ_0007312 promotes third-generation epidermal growth
factor receptor-tyrosine kinase inhibitor resistance through
pyroptosis and apoptosis via the MiR-764/MAPK1 axis in lung
adenocarcinoma cells. J Cancer. 13:2798–2809. 2022. View Article : Google Scholar :
|
|
116
|
Zhang CC, Li Y, Feng XZ and Li DB:
Circular RNA circ_0001287 inhibits the proliferation, metastasis,
and radiosensitivity of non-small cell lung cancer cells by
sponging microRNA miR-21 and up-regulating phosphatase and tensin
homolog expression. Bioengineered. 12:414–425. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Jin Y, Su Z, Sheng H, Li K, Yang B and Li
S: Circ_0086720 knockdown strengthens the radiosensitivity of
non-small cell lung cancer via mediating the miR-375/SPIN1 axis.
Neoplasma. 68:96–107. 2021. View Article : Google Scholar
|
|
118
|
Li YH, Xu CL, He CJ, Pu HH, Liu JL and
Wang Y: circ-MTDH.4/miR-630/AEG-1 axis participates in the
regulation of proliferation, migration, invasion, chemoresistance,
and radioresistance of NSCLC. Mol Carcinog. 59:141–153. 2020.
View Article : Google Scholar
|
|
119
|
Zhang T, Wu DM, Luo PW, Liu T, Han R, Deng
SH, He M, Zhao YY and Xu Y: CircNEIL3 mediates pyroptosis to
influence lung adenocarcinoma radiotherapy by upregulating PIF1
through miR-1184 inhibition. Cell Death Dis. 13:1672022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Liu B, Li H, Liu X, Li F, Chen W, Kuang Y,
Zhao X, Li L, Yu B, Jin X and Li Q: CircZNF208 enhances the
sensitivity to X-rays instead of carbon-ions through the
miR-7-5p/SNCA signal axis in non-small-cell lung cancer cells. Cell
Signal. 84:1100122021. View Article : Google Scholar
|
|
121
|
Horn L, Spigel DR, Vokes EE, Holgado E,
Ready N, Steins M, Poddubskaya E, Borghaei H, Felip E, Paz-Ares L,
et al: Nivolumab versus docetaxel in previously treated patients
with advanced non-small-cell lung cancer: Two-year outcomes from
two randomized, open-label, phase III trials (CheckMate 017 and
CheckMate 057). J Clin Oncol. 35:3924–3933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D,
Long X, Lin K, Lu F, Xu JJ and Wu YB: Cancer cell-derived exosomal
circUSP7 induces CD8+ T cell dysfunction and anti-PD1
resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol
Cancer. 20:1442021. View Article : Google Scholar
|
|
123
|
Liu Z, Wang T, She Y, Wu K, Gu S, Li L,
Dong C, Chen C and Zhou Y: N6-methyladenosine-modified
circIGF2BP3 inhibits CD8+ T-cell responses to facilitate
tumor immune evasion by promoting the deubiquitination of PD-L1 in
non-small cell lung cancer. Mol Cancer. 20:1052021. View Article : Google Scholar
|
|
124
|
Kumar S, Zeng Z, Bagati A, Tay RE, Sanz
LA, Hartono SR, Ito Y, Abderazzaq F, Hatchi E, Jiang P, et al:
CARM1 inhibition enables immunotherapy of resistant tumors by dual
action on tumor cells and T cells. Cancer Discov. 11:2050–2071.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zhang LX, Gao J, Long X, Zhang PF, Yang X,
Zhu SQ, Pei X, Qiu BQ, Chen SW, Lu F, et al: The circular RNA
circHMGB2 drives immunosuppression and anti-PD-1 resistance in lung
adenocarcinomas and squamous cell carcinomas via the
miR-181a-5p/CARM1 axis. Mol Cancer. 21:1102022. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs as
a new class of putative biomarkers in human blood. PLoS One.
10:e01412142015. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Ye D, Gong M, Deng Y, Fang S, Cao Y, Xiang
Y and Shen Z: Roles and clinical application of exosomal circRNAs
in the diagnosis and treatment of malignant tumors. J Transl Med.
20:1612022. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Chen HH, Zhang TN, Wu QJ, Huang XM and
Zhao YH: Circular RNAs in lung cancer: Recent advances and future
perspectives. Front Oncol. 11:6642902021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu
M, Dai X, Zhou H, Zhu J, Zhang H and Jiang Y: Circular RNA
circSATB2 promotes progression of non-small cell lung cancer cells.
Mol Cancer. 19:1012020. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Yang F, Ma C, Qiu J, Feng X and Yang K:
Identification of circRNA_001846 as putative non-small cell lung
cancer biomarker. Bioengineered. 12:8690–8697. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Best MG, Sol N, In 't Veld SGJG, Vancura
A, Muller M, Niemeijer AN, Fejes AV, Tjon Kon Fat LA, Huis In 't
Veld AE, Leurs C, et al: Swarm intelligence-enhanced detection of
non-small-cell lung cancer using tumor-educated platelets. Cancer
Cell. 32:238–252.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
D'Ambrosi S, Visser A, Antunes-Ferreira M,
Poutsma A, Giannoukakos S, Sol N, Sabrkhany S, Bahce I, Kuijpers
MJE, Oude Egbrink MGA, et al: The analysis of platelet-derived
circRNA repertoire as potential diagnostic biomarker for non-small
cell lung cancer. Cancers (Basel). 13:46442021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC,
Xin HY, Mao L, Luo CB, Yu SY, Huang XW, et al: Circular RNA
sequencing identifies CircASAP1 as a key regulator in
hepatocellular carcinoma metastasis. Hepatology. 72:906–922. 2020.
View Article : Google Scholar
|
|
134
|
Lu GJ, Cui J, Qian Q, Hou ZB, Xie HY, Hu
W, Hao KK, Xia N and Zhang Y: Overexpression of hsa_circ_0001715 is
a potential diagnostic and prognostic biomarker in lung
adenocarcinoma. Onco Targets Ther. 13:10775–10783. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhang ZY, Gao XH, Ma MY, Zhao CL, Zhang YL
and Guo SS: CircRNA_101237 promotes NSCLC progression via the
miRNA-490-3p/MAPK1 axis. Sci Rep. 10:90242020. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Hong Y, Si J, Xiao B, Xiong Y, Dai C, Yang
Y, Li S and Ma Y: circ_0000567/miR-421/TMEM100 axis promotes the
migration and invasion of lung adenocarcinoma and is associated
with prognosis. J Cancer. 13:1540–1552. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Luo YH, Yang YP, Chien CS, Yarmishyn AA,
Ishola AA, Chien Y, Chen YM, Huang TW, Lee KY, Huang WC, et al:
Plasma level of circular RNA hsa_circ_0000190 correlates with tumor
progression and poor treatment response in advanced lung cancers.
Cancers (Basel). 12:17402020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Feng D, Xu Y, Hu J, Zhang S, Li M and Xu
L: A novel circular RNA, hsa-circ-0000211, promotes lung
adenocarcinoma migration and invasion through sponging of
hsa-miR-622 and modulating HIF1-α expression. Biochem Biophys Res
Commun. 521:395–401. 2020. View Article : Google Scholar
|
|
139
|
Xu Y, Yu J, Huang Z, Fu B, Tao Y, Qi X,
Mou Y, Hu Y, Wang Y, Cao Y, et al: Circular RNA hsa_circ_0000326
acts as a miR-338-3p sponge to facilitate lung adenocarcinoma
progression. J Exp Clin Cancer Res. 39:572020. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Wang L, Li M and Lian R: Depleting
hsa_circ_0000567 suppresses acquired gefitinib resistance and
proliferation of lung adenocarcinoma cells through regulating the
miR-377-3p/ZFX axis: An in vitro and in vivo study. Histol
Histopathol. 37:637–654. 2022.PubMed/NCBI
|
|
141
|
Zuo Y, Shen W, Wang C, Niu N and Pu J:
Circular RNA Circ-ZNF609 promotes lung adenocarcinoma proliferation
by modulating miR-1224-3p/ETV1 signaling. Cancer Manag Res.
12:2471–2479. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Huang C, Yue W, Li L, Li S, Gao C, Si L,
Qi L, Cheng C, Lu M, Chen G, et al: Circular RNA hsa-circ-000881
suppresses the progression of lung adenocarcinoma in vitro via a
miR-665/PRICKLE2 axis. Ann Transl Med. 9:4982021. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Mao Y and He JX, Zhu M, Dong YQ and He JX:
Circ0001320 inhibits lung cancer cell growth and invasion by
regulating TNFAIP1 and TPM1 expression through sponging miR-558.
Hum Cell. 34:468–477. 2021. View Article : Google Scholar
|
|
144
|
Shen HY, Shi LX, Wang L, Fang LP, Xu W, Xu
JQ, Fan BQ and Fan WF: Hsa_circ_0001361 facilitates the progress of
lung adenocarcinoma cells via targeting miR-525-5p/VMA21 axis. J
Transl Med. 19:3892021. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Lu T, Qiu T, Han B, Wang Y, Sun X, Qin Y,
Liu A, Ge N and Jiao W: Circular RNA circCSNK1G3 induces HOXA10
signaling and promotes the growth and metastasis of lung
adenocarcinoma cells through hsa-miR-143-3p sponging. Cell Oncol
(Dordr). 44:297–310. 2021. View Article : Google Scholar
|
|
146
|
Zhou H, Huang X, Yang X, Jiang F, Shao F,
Shi W, Huang K, Pan J, Zhang Y, Chen J and Wang Y: CircRAPGEF5
promotes the proliferation and metastasis of lung adenocarcinoma
through the miR-1236-3p/ZEB1 axis and serves as a potential
biomarker. Int J Biol Sci. 18:2116–2131. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Cao L, Zhou X, Ding X and Gao D: Knockdown
of circ-PVT1 inhibits the progression of lung adenocarcinoma and
enhances the sensitivity to cisplatin via the miR-429/FOXK1
signaling axis. Mol Med Rep. 24:6842021. View Article : Google Scholar :
|
|
148
|
Yao Y, Hua Q, Zhou Y and Shen H: CircRNA
has_circ_0001946 promotes cell growth in lung adenocarcinoma by
regulating miR-135a-5p/SIRT1 axis and activating Wnt/β-catenin
signaling pathway. Biomed Pharmacother. 111:1367–1375. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zhu L, Guo T, Chen W, Lin Z, Ye M and Pan
X: CircMMD_007 promotes oncogenic effects in the progression of
lung adenocarcinoma through microRNA-197-3p/protein tyrosine
phosphatase non-receptor type 9 axis. Bioengineered. 13:4991–5004.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Wan J, Ding G, Zhou M, Ling X and Rao Z:
Circular RNA hsa_circ_0002483 promotes growth and invasion of lung
adenocarcinoma by sponging miR-125a-3p. Cancer Cell Int.
21:5332021. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Li H and Liu L: Zinc moderates circular
RNA CircFOXP1 expression in order to regulate ferroptosis during
lung adenocarcinoma. Chem Biol Interact. 352:1097602022. View Article : Google Scholar
|
|
152
|
Wang HL, Wang HR, Liang Y, Hu AN, Enguita
FJ, Zhou XG and Dong J: Hsa_circ_0006571 promotes spinal metastasis
through sponging microRNA-138 to regulate sirtuin 1 expression in
lung adenocarcinoma. Transl Lung Cancer Res. 9:2411–2427. 2020.
View Article : Google Scholar
|
|
153
|
Yang Y, Fan X, Nie Y, Liu D, Zhu D, Wu K,
Zhang Y, Li W, Tian X, Wang H and Fan Y: CircTUBGCP3 facilitates
the tumorigenesis of lung adenocarcinoma by sponging miR-885-3p.
Cancer Cell Int. 21:6512021. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Ma D, Liu H, Qin Y, Li D, Cui Y, Li L, He
J, Chen Y and Zhou X: Circ_0007142/miR-186/FOXK1 axis promoted lung
adenocarcinoma progression. Am J Transl Res. 12:4728–4738.
2020.PubMed/NCBI
|
|
155
|
Zhang B, Chen M, Jiang N, Shi K and Qian
R: A regulatory circuit of circ-MTO1/miR-17/QKI-5 inhibits the
proliferation of lung adenocarcinoma. Cancer Biol Ther.
20:1127–1135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Chen M, Huang X, Li L, Huang M, Cai R and
Liao X: A regulatory axis of circ_0008193/miR-1180-3p/TRIM62
suppresses proliferation, migration, invasion, and Warburg effect
in lung adenocarcinoma cells under hypoxia. Med Sci Monit.
26:e9229002020.PubMed/NCBI
|
|
157
|
Wang M, Ma M, Yang Y, Li C, Wang Y, Sun X,
Wang M, Sun Y and Jiao W: Overexpression of hsa_circ_0008274
inhibited the progression of lung adenocarcinoma by regulating
HMGA2 via sponging miR-578. Thorac Cancer. 12:2258–2264. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Chen J, Xu S, Chen S, Zong Z, Han X, Zhao
Y and Shang H: CircPUM1 promotes the malignant behavior of lung
adenocarcinoma by regulating miR-326. Biochem Biophys Res Commun.
508:844–849. 2019. View Article : Google Scholar
|
|
159
|
Wang X, Zhu X, Zhang H, Wei S, Chen Y,
Chen Y, Wang F, Fan X, Han S and Wu G: Increased circular RNA
hsa_circ_0012673 acts as a sponge of miR-22 to promote lung
adenocarcinoma proliferation. Biochem Biophys Res Commun.
496:1069–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan
X, Han S and Wu G: hsa_circ_0013958: A circular RNA and potential
novel biomarker for lung adenocarcinoma. FEBS J. 284:2170–2182.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Zan SJ, Zhao Y, Fang T and Chen K:
Expression of circular RNA hsa_circ_0014130 in lung adenocarcinoma
cell lines and its effect on proliferation and invasion of lung
adenocarcinoma cell line. Zhonghua Bing Li Xue Za Zhi. 48:934–939.
2019.In Chinese. PubMed/NCBI
|
|
162
|
Yao Y, Zhou Y and Hua Q: circRNA
hsa_circ_0018414 inhibits the progression of LUAD by sponging
miR-6807-3p and upregulating DKK1. Mol Ther Nucleic Acids.
23:783–796. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Ying X, Zhu J and Zhang Y: Circular RNA
circ-TSPAN4 promotes lung adenocarcinoma metastasis by upregulating
ZEB1 via sponging miR-665. Mol Genet Genomic Med. 7:e9912019.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Li X, Su S, Ye D, Yu Z, Lu W and Liu L:
Hsa_circ_0020850 promotes the malignant behaviors of lung
adenocarcinoma by regulating miR-326/BECN1 axis. World J Surg
Oncol. 20:132022. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Xin T, Li S, Zhang Y, Kamali X, Liu H and
Jia T: circRNA Hsa_circ_0020850 silence represses the development
of lung adenocarcinoma via regulating miR-195-5p/IRS2 axis. Cancer
Manag Res. 12:10679–10692. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Wu S, Li H, Lu C, Zhang F, Wang H, Lu X
and Zhang G: Aberrant expression of hsa_circ_0025036 in lung
adenocarcinoma and its potential roles in regulating cell
proliferation and apoptosis. Biol Chem. 399:1457–1467. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Liu M, Wang P, Sui X, Ding F, Liu L, Gao Z
and Cheng Z: Circular RNA circABCC4 regulates lung adenocarcinoma
progression via miR-3186-3p/TNRC6B axis. J Cell Biochem.
121:4226–4238. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Yang Y, Zhang Y, Ding X, Ren Y, Wei B, Lin
Z, Nie Y and Fan Y: Construction and analysis of the ceRNA network
hsa_circ_0031968/miR-3611/GCG in lung adenocarcinoma. Ann Transl
Med. 9:17572021. View Article : Google Scholar
|
|
169
|
Wang Y, Ren F, Sun D, Liu J, Liu B, He Y,
Pang S, Shi B, Zhou F, Yao L, et al: CircKEAP1 suppresses the
progression of lung adenocarcinoma via the miR-141-3p/KEAP1/NRF2
axis. Front Oncol. 11:6725862021. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Zhou Q and Sun Y: Circular RNA cMras
suppresses the progression of lung adenocarcinoma through
ABHD5/ATGL axis using NF-κB signaling pathway. Cancer Biother
Radiopharm. Aug 19–2020.Epub ahead of print.
|
|
171
|
Sui MH, Zhang WW, Geng DM and Sun DJ:
CircPRKCI regulates proliferation, migration and cycle of lung
adenocarcinoma cells by targeting miR-219a-5p-regulated CAMK1D. Eur
Rev Med Pharmacol Sci. 25:1899–1909. 2021.PubMed/NCBI
|
|
172
|
Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z,
Xu W, Zhang E, Wang J, Fang T, et al: The circular RNA circPRKCI
promotes tumor growth in lung adenocarcinoma. Cancer Res.
78:2839–2851. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Xu L, Ma Y, Zhang H, Lu QJ, Yang L, Jiang
GN and Liao WL: HMGA2 regulates circular RNA ASPH to promote tumor
growth in lung adenocarcinoma. Cell Death Dis. 11:5932020.
View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Wang Y, Wo Y, Lu T, Sun X, Liu A, Dong Y,
Du W, Su W, Huang Z and Jiao W: Circ-AASDH functions as the
progression of early stage lung adenocarcinoma by targeting
miR-140-3p to activate E2F7 expression. Transl Lung Cancer Res.
10:57–70. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Du J, Zhang G, Qiu H, Yu H and Yuan W: The
novel circular RNA circ-CAMK2A enhances lung adenocarcinoma
metastasis by regulating the miR-615-5p/fibronectin 1 pathway. Cell
Mol Biol Lett. 24:722019. View Article : Google Scholar :
|
|
176
|
Gao N and Ye B: Circ-SOX4 drives the
tumorigenesis and development of lung adenocarcinoma via sponging
miR-1270 and modulating PLAGL2 to activate WNT signaling pathway.
Cancer Cell Int. 20:22020. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Shi J, Lv X, Zeng L, Li W, Zhong Y, Yuan
J, Deng S, Liu B, Yuan B, Chen Y, et al: CircPVT1 promotes
proliferation of lung squamous cell carcinoma by binding to
miR-30d/e. J Exp Clin Cancer Res. 40:1932021. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Wang L, Xu C, Wang C, Gong W, Zhang K,
Chen Q, Zhou S and Qi T: Circ-PAX2 promotes proliferation and
metastasis by absorbing miR-186 in lung cancer cells. Int J Clin
Exp Pathol. 11:3793–3801. 2018.PubMed/NCBI
|
|
179
|
Yu M, Tian Y, Wu M, Gao J, Wang Y, Liu F,
Sheng S, Huo S and Bai J: A comparison of mRNA and circRNA
expression between squamous cell carcinoma and adenocarcinoma of
the lungs. Genet Mol Biol. 43:e202000542020. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang
J, Huang X, Liang S, Chen R, Chen S and Guo L: Circular RNA cESRP1
sensitises small cell lung cancer cells to chemotherapy by sponging
miR-93-5p to inhibit TGF-β signalling. Cell Death Differ.
27:1709–1727. 2020. View Article : Google Scholar
|