Exosomes are a special class of nanoscale vesicles
that are secreted by most eukaryotic cells and exist extensively in
the extracellular space (1).
Exosomes may be found in a variety of body fluids, such as blood,
urine and saliva, and may carry a variety of molecules, such as
RNAs, DNAs, proteins and lipids (2). These are transported to the
appropriate cell and affect their biological function. Therefore,
exosomes are known as carriers of intercellular information
transfer. This is important for tumor progression (3).
Circular RNAs (circRNAs) are a class of noncoding
RNAs that widely exist in eukaryotic cells (4). They have specific biological
functions by regulating microRNA (miRNA) and proteins (5) and are closely related to the
occurrence, development and prognosis of various malignant tumors.
Their functions include acting as miRNA sponges (6), interactions with RNA-binding proteins
(7) and translating proteins
(8). Clinical studies have
indicated that certain circRNAs are significantly different in
terms of their expression between normal individuals and patients
with cancer, which means that circRNAs are expected to be new
diagnostic markers for cancer (9,10).
Studies have indicated that certain circRNAs are
enriched and highly expressed in exosomes, participate in
extracellular transport and are finally released into target cells,
regulating the biological behavior of target cells (11). These circRNAs that are loaded and
transported by exosomes are called exosomal circRNAs
(exo-circRNAs). Based on the characteristics of exosomes and
circRNAs, exo-circRNAs may have a more unique role in cancer
progression. In the present review, the origins and functions of
exosomes and circRNAs were briefly summarized and recent advances
in the molecular mechanisms of exo-circRNAs in cancer growth,
metastasis and drug resistance were highlighted. Exo-circRNAs are
expected to be potential neoteric cancer prediction markers and
therapeutic targets.
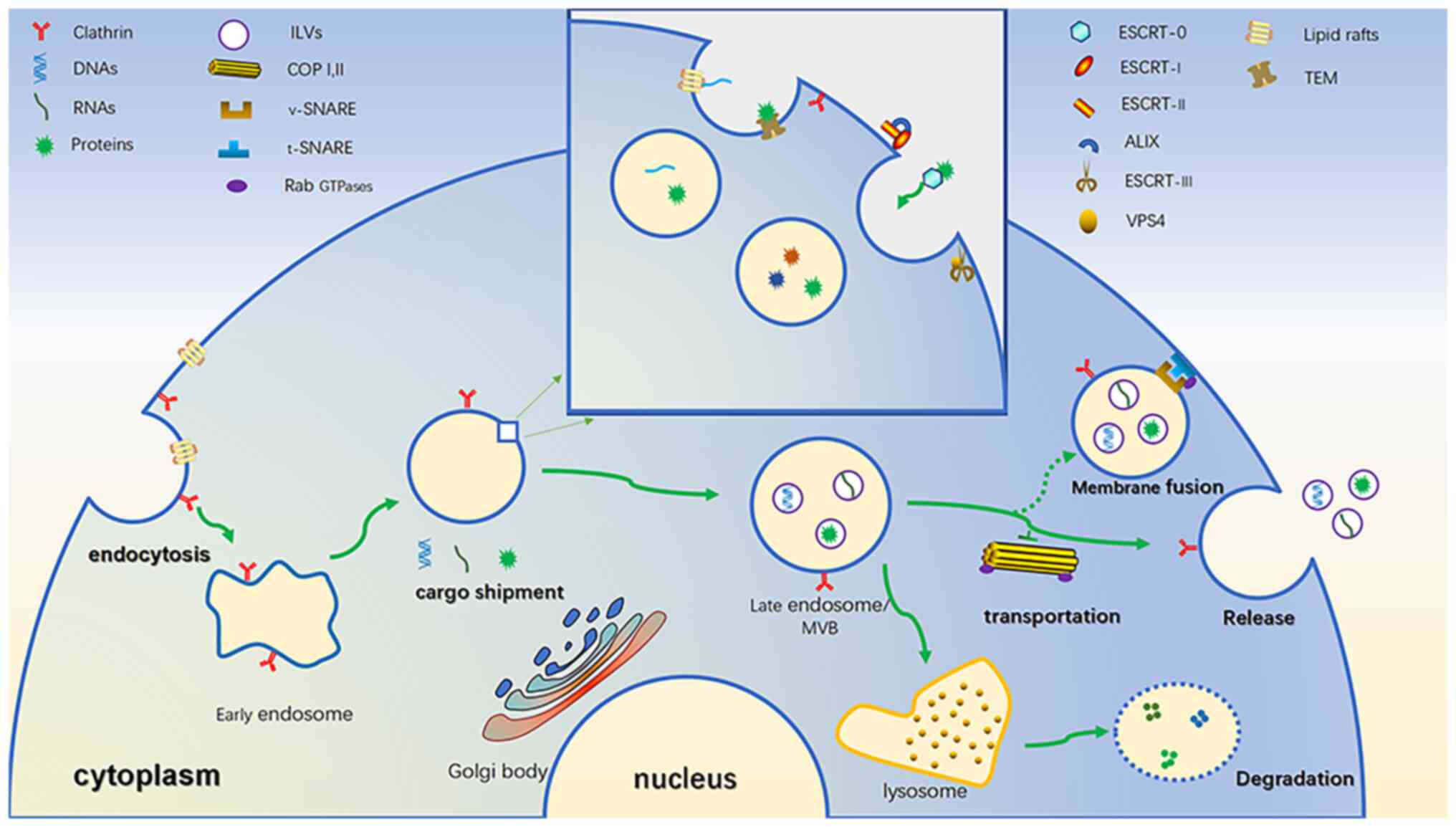
Exosomes (30–100 nm in diameter) are extracellular
vesicles that were first identified in 1987 (12). Other extracellular vesicles also
include microbubbles (50–2,000 nm in diameter), which are produced
by the plasma membrane to germinate and some of them are always
rich in certain proteins (13),
reverse transcription virus-like particles (90–100 nm in diameter),
directly by the plasma membrane of germination, containing an
endogenous retrovirus sequence (14) and apoptotic bodies (500–4,000 nm in
diameter, programmed cell death of package cell residue vesicles
(15,16). Unlike these, exosomes originate
from small vesicles formed by the endocytosis of cell membranes
(17). When the surface of the
cell membrane is rich in clathrin, it is easy to induce the inward
budding of the cell membrane to form endocytic vesicles (18). In addition, specific lipid rafts on
the cell membrane may also induce endocytosis, which is the source
of the early endosome (Fig. 1)
(19). Early endosomes may mediate
the inward transfer of numerous substances from the cytoplasm to
the endosome, which is similar to cargo shipment (20). With the assistance of the Golgi
body, the endoplasmic membrane spits inward and certain substances
in the cytoplasm are injected into it to form intraluminal vesicles
(ILVs) (21). Early endosomes are
thus transformed into late endosomes, also known as multivesicular
bodies, due to their abundance of ILVs (22). ILVs are the prototype of exosomes
(23). The most discussed
molecular mechanism is the endosomal sorting complex required for
transport (ESCRT) (24). ESCRT is
composed of four complexes (named ESCRT-0, I, II and III) and
related assistant proteins. Of these proteins, ESCRT-0 recruits
cargo in a ubiquitin-dependent manner, mainly as ubiquitin proteins
(25). ESCRT-I and ESCRT-II may
induce the inner body membrane to germinate inward and the helper
protein apoptosis-linked gene-2 interacting protein X is also
involved (24,26). ESCRT-III drives vesicles to be
released from the membrane and the auxiliary protein VPS4 is able
to assist ATP-dependent reactions to induce the dissociation of
activated ESCRT complexes and promote their recovery (27). In addition, there have been reports
of non-ESCRT-dependent shipments. For instance, miRNA may be loaded
in lipid rafts rich in ceramide on the endosomal membrane (19). In addition, studies including that
by Odorizzi et al (28)
found that in the early endosomal membrane of yeast cells, there
are specific regions called tetraspanin-enriched microdomains,
which consist of four transmembrane domains that form a stereotypic
tertiary structure and may interact with specific proteins in the
cytoplasm to promote ILV formation (Fig. 1) (29). Multivesicular bodies (MVBs)
experience two outcomes. One is that lysosomes conjugate and
degrade their lumen, which is the reason why exosomes were
initially known as garbage collectors (30). It may be that activating
conjugation of ISG15 by ubiquitin-like protein prevents the release
of exosomes and facilitates the fusion of MVBs and lysosomes
(31). Second, MVBs fuse with the
cell membrane, releasing ILVs into the extracellular space and
fluid circulation. These are called exosomes. This requires the
assistance of a variety of molecules, including coat protein
complex (COP) I and II, soluble NSF attachment protein receptors
(SNARE) and Rab GTPases (32).
Among them, COP I and II mainly mediate vesicle transport (33,34).
The Rab GTPase family is responsible for transporting MVBs to the
cell membrane, inducing vesicle budding, vesicle and organelle
mobility through cytoskeletal interactions and docking of vesicles
to their target compartment, leading to membrane fusion (23,35).
For instance, Rab35 mediates the docking of MVBs in nerve cells
with cell membranes (36). SNARE
is a membrane protein family and the SNARE in the transport vesicle
(known as v-SNARE) is paired with its corresponding SNARE binding
partner (known as t-SNARE), mediating the fusion of MVBs and the
cell membrane (Fig. 1) (32,37).
Next, circulating exosomes search for specific receptor cells and
are internalized by receptor cells by phagocytosis, pinocytosis or
fusion with the cell membrane, possibly with the help of specific
mediators such as integrins, lipids, lectins and adhesion. Hoshino
et al (38) found that
exosome integrins α6β4 and α6β1 were associated with lung
metastasis.
It is worth noting that exosomes not only have an
important role in regulating normal physiological functions, such
as tissue regeneration (39),
immune surveillance, blood circulation (40) and stem cell plasticity. A large
number of studies have indicated that exosomes in tumors also have
an important role in regulating cell proliferation, metastasis and
drug resistance, and as biomarkers (41,42).
Exosomes derived from cancer cells are deemed important drivers of
pre-metastatic niche formation at distant organs, such as the
secreted ADAM17 transported via exosomes, which was indicated to
promote the migratory ability of colorectal cancer (CRC) cells by
cleaving the E-cadherin junction (42). Epithelial-mesenchymal transition
(EMT) is a process in which epithelial cells acquire mesenchymal
features. In cancer, EMT is associated with tumor initiation,
invasion, metastasis and resistance to therapy (43). Exosomes secreted by
cancer-associated fibroblasts in CRC enhance EMT of CRC cells,
thereby promoting CRC metastasis and drug resistance (44). Exosomes may affect recipient cells
through the engulfed cargos and exosomes have already emerged as
important mediators for intercellular communication (45). Osimertinib is a third-generation
epidermal growth factor receptor-tyrosine kinase inhibitor
(EGFR-TKI) approved for the treatment of patients with EGFR-mutant
non-small cell lung cancer (NSCLC). Current research indicated that
exosomal miR-210-3p may have a crucial role in resistance to
osimertinib in the tumor microenvironment of EGFR-mutant NSCLC
(46). In breast cancer cells,
extracellular long noncoding RNA small nucleolar RNA host gene 14
was able to be incorporated into exosomes and transmitted to
sensitive cells, thus disseminating trastuzumab resistance
(47). Therefore, exosomes may
have an important role in drug resistance by influencing the tumor
microenvironment and non-coding RNA. Cancer-secreted exosomes have
an effect on the exosome donor cells and support cancer growth and
metastasis (48). In multiple
myeloma (MM), exosomes derived from mesenchymal stromal cells
promote the progression of MM via LINCI00461 (49). Furthermore, breast cancer exosomes
have been indicated to promote cell proliferation and cancer
progression (50). Given that
exosomes have an important role in all aspects of tumors, exosomes
may be used as biomarkers to predict cancer progression in advance
and may have applications in tumor diagnosis, anti-tumor
chemotherapy and drug resistance prediction (51–53).
Exosomal miRNAs have been indicated to be highly sensitive and
specific in distinguishing healthy individuals from patients with
CRC (54). Exosome enrichment may
be found in the serum of patients with cisplatin-resistant gastric
cancer (GC) and the results suggest that exosomes derived from
drug-resistant cells may serve as a potential predictor of the
response to antitumor chemotherapy (53).
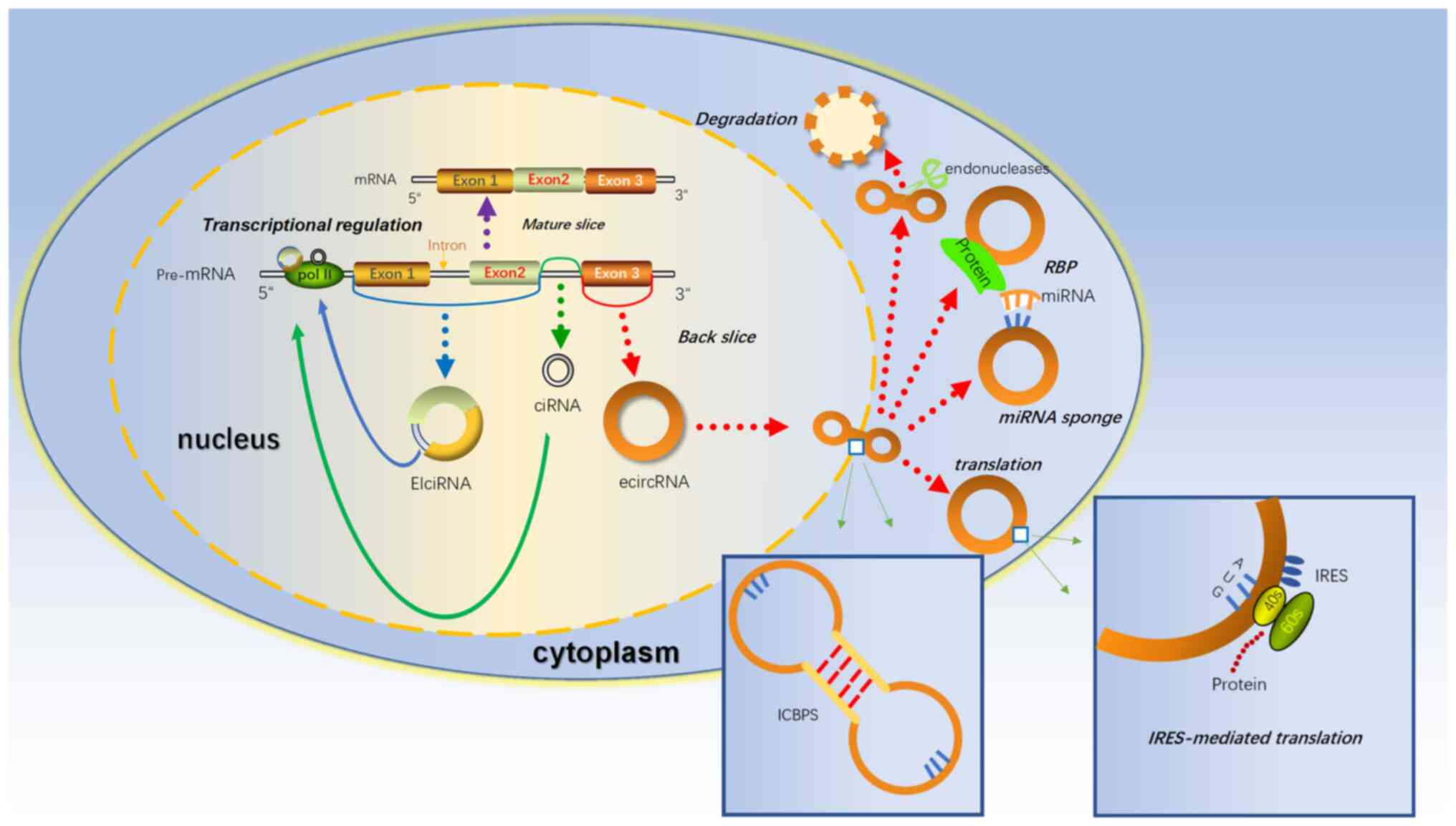
Covalently closed circRNAs, as the name suggests,
are a class of RNAs with a circular structure. In 1976, Sanger
et al (55) first described
viroids containing ‘single-stranded and covalently closed
circRNAs’. Under normal circumstances, the splicing sites of normal
precursor mRNAs are joined in a linear order, which generates
mature linear mRNAs with a complete 5′ cap and a 3′ tail after
modification (56). However, there
is still a special splicing mechanism called backsplicing, which
may join a 5′ splice site to an upstream splice acceptor (3′splice
site), resulting in the production of a circular RNA whose ends are
covalently linked by a 3′-5′ phosphodiester bond (57). Due to differences in splicing sites
and different cyclization mechanisms, certain intron sequences not
originally expressed to be contained in mature mRNAs may be
expressed in circRNAs (58).
Therefore, circRNAs may be classified into the following
categories: Exonic circRNAs (59),
intronic circRNAs (60)
exonic-intronic circRNAs (EIciRNAs) (Fig. 2) and circRNAs generated from tRNAs
(5,61). Due to the lack of a 5′ cap and a 3′
tail, they are strongly resistant to exonuclease and have a higher
stability compared with linear RNAs (62). Since they were first discovered in
viruses as early as the 1970s, circRNAs were originally thought to
be rare error-splicing sequences produced during RNA splicing.
However, with the recent advances in high-throughput sequencing and
bioinformatics, a wide variety of circRNAs have been detected and
identified (63). CircRNAs are
widely expressed in humans and other mammals and have high
repeatability. For instance, approximately 5–10% of circRNAs in the
human brain may also be expressed in the pig brain, which reflects
the high conservation of circRNAs (64). Due to the high abundance, relative
stability and high conservation of circRNAs, the possible functions
of circRNAs are gaining interest in the scientific community.
According to the difference in expression sites in cells, circRNAs
may be further divided into nuclear circRNAs and cytoplasmic
circRNAs. Due to the isolation of the nuclear membrane, these two
circRNAs may have completely different functions (65). An increasing number of studies have
indicated that circRNAs expressed in the cytoplasm mostly sponge
miRNAs and exert regulatory functions through competing endogenous
RNA (ceRNA) mechanisms (circRNAs may sponge miRNAs to influence the
stability of target RNAs or their translation, thus regulating gene
expression at the transcriptional level) (66). There are miRNA binding sites in
circRNAs, which may competitively bind miRNAs and reduce the
regulation of downstream target genes by miRNAs. For instance,
CIRS-7, the most adequately characterized circRNA with >70
conserved binding sites for miR-7 and highly stable expression in
numerous tissues, particularly in neuronal tissues, where it has
the potential to negatively regulate miR-7 expression and where
knockout of CIRS-7 would significantly enhance the miRNA-mediated
recruitment of the pluripotency gene AGO2 (67). In addition, recent studies have
indicated that cytoplasmic circRNAs may interact with proteins,
which are called RNA binding proteins (RBPs) (68). Similar to sponge miRNAs, circRNAs
also have regions that bind to specific proteins and circRNAs may
block active regions of proteins and inactivate them functionally.
CircBIRC6 is enriched in the RBP-AGO2 complex and directly combines
with miR-34a and miR-145 to modulate and maintain the pluripotency
of human embryonic stem cells (hESCs) and suppress hESC
differentiation (69). By
contrast, nuclear circRNAs may exhibit more complex functions. To
date, the understanding of nuclear circRNAs, whose known functions
include the regulation of mRNA transcription and posttranscription,
is limited. Li et al (70)
found that EIciRNAs are incompletely spliced and have a retained
intron that allows them to interact with U1 small nuclear
ribonucleoprotein and promote the transcription of their host gene.
In addition, although circRNAs are called noncoding RNAs, a recent
study found that certain circRNAs encode proteins, and this type of
circRNA has a common feature, i.e. that it has at least one open
reading frame (ORF) and may be associated with polysomes.
Traditionally, eukaryotic mRNAs are always translated through
canonical cap-dependent translation (71). Due to the lack of a 5′ cap and 3′
end, circRNAs translation may only be started in a cap-independent
manner. One of the alternative mechanisms, internal ribosome entry
site (IRES)-mediated translation, has recently been indicated to
have a key regulatory role during mammalian development (72). For instance, circ-ZNF609 contains
an ORF, which has been proven by experiments to have a relatively
weak translation function (73).
Another important cap-independent translation mechanism is through
methylated adenosine residues in the form of N6-methyladenosines
(m6A) in the 5′ untranslated region (74). It has been reported that certain
short RNA elements containing m6A have IRES-like activity to
initiate circRNA translation. Yang et al (75) indicated that short RNA elements for
the most abundant m6A were enriched in circRNA sequences (Fig. 2). Recently, a noteworthy study
indicated that circRNA may not be ‘circular’: Sun et al
(76) found a large number of
internal complementary base pair sequences (ICBPS) in numerous
circRNAs, particularly in ‘extremely long circRNAs’ (>5,000 nt);
thus, they made an assumption that circRNA may not be simply
circular. They may contain a double-stranded structure. This
double-stranded structure has two states, ‘open’ and ‘closed’, and
the process may be reversible and regulated by the microenvironment
or other internal factors, such as the length of ICBPS, the binding
free energy, the distance between pairing fragments and the
secondary structure of RNA, or relevant RNA modifications, such as
m6A. The formation of a double-stranded structure compresses
circRNAs in space, which may help to facilitate circRNAs being
exported into the cytoplasm from the nucleus. Furthermore, the
double-stranded structure of circRNAs may make them more
susceptible to degradation by related enzymes, which may explain
how cells eliminate circRNAs (Fig.
2). Finally, since circRNAs are involved in numerous aspects of
gene expression regulation, the association of circRNAs with cancer
is considered to have great research prospects. An increasing
number of studies have indicated that circRNAs may act as tumor
suppressors or promoters in various human malignancies,
highlighting their potential as diagnostic biomarkers and
therapeutic targets for future treatment (77).
CircRNAs have long been considered noncoding RNAs
produced during cell transcription that may exist in the nucleus or
cytoplasm, and they mainly participate in gene regulation as
regulators. Their functions are mainly in the parental cells. In
their breakthrough study, Li et al (11) first revealed the enrichment of
circRNAs in exosomes using high-throughput sequencing and called
them exo-circRNAs. In addition, several reports have identified the
abundant and stable expression of circRNAs in extracellular
vesicles. At the same time, they found that exo-circRNAs were
enriched in CRC, LC, breast cancer and other cancer types, and
serum exo-circRNAs may distinguish cancer patients from healthy
individuals (11). This provided a
new way to study the mechanism of action of circRNAs. Exo-circRNAs
may have several possible mechanisms and characteristics. First,
exosome enrichment of specific circRNAs may be affected by their
parental transcription levels (78). Furthermore, exo-circRNAs have
tissue specificity, which may be related to the imbalance of gene
expression regulation in related tissues during the disease
process. Dou et al (79)
generated wild-type and mutant CRC cell lines and found
differentially expressed circRNAs by RNA-sequencing, some of which
exist in exosomes. Quantitative PCR analysis indicated that
circRNAs from exosomes expressed in wild-type and mutant cell lines
exhibit obvious differences, and circRNAs were more abundant in
exosomes than in cells, suggesting that exo-circRNAs may
selectively have certain organization for expression and tissue
specificity. More importantly, exo-circRNAs mainly regulate the
function of receptor cells by targeting relevant miRNAs in receptor
cells and by regulating downstream signaling pathways through ceRNA
mechanisms. For instance, exosomal circ-ZNF652 promotes the
proliferation, migration and invasion of hepatocellular carcinoma
(HCC) cells through the miR-29a-3p/GUCD1 axis (80). This provides an important way to
study the mechanism of the occurrence and progression of cancer. In
addition, Dou et al (79)
demonstrated that the circRNA content in exosomes is more abundant
than that in cells, and the expression of circRNA changes with KRAS
mutation. The following chapter will focus on the role of
exo-circRNAs in cancer progression.
The early detection and treatment of cancer is one
of the difficult problems in the medical field. More markers and
therapeutic targets are required for early cancer screening.
Exosomes have been recently identified as important mediators of
cancer metastasis, while circRNAs have been indicated to be
important regulators of tumor progression. When important vectors
are associated with important modulators, a novel approach to the
mechanism of tumor progression emerges. An increasing number of
studies have indicated that exo-circRNAs may have an important role
in the in situ growth, metastasis and drug resistance of
malignant tumors. It was also demonstrated that exo-circRNAs not
only have an important role in tumor diagnosis and prognosis as
markers, but also profoundly affect the in-situ growth,
metastasis, angiogenesis, drug resistance and treatment of
malignant tumors.
Unlimited growth is the most important
characteristic of malignant tumor cells. This involves a variety of
gene disorders in which exo-circRNAs may be involved. HCC is one of
the most common gastrointestinal malignancies and the main reason
for its occurrence is the imbalance of hepatocyte proliferation.
Its occurrence may be related to cirrhosis of the liver, viral
infection, fungal infection or long-term stimulation with chemical
factors (81). Under the
stimulation of these adverse factors, the expression of certain
exo-circRNAs may change, suggesting that exo-circRNAs may be
involved in the malignant transformation process of hepatocytes.
Dai et al (82) reported
that arsenite may promote the induction of malignant transformation
of human liver epithelial (L-02) cells. High expression of
circ-100284 was found in the transformed L-02 cell culture medium
and transformed cells transferred circ-100284 to normal L-02 cells
through exosomes, accelerating the cell cycle and promoting the
growth of normal L-02 cells. Additional mechanistic studies
indicated that exo-circRNAs may further increase the accumulation
of the cell cycle-related proteins EZH2 and cyclin-D1, accelerating
the G1/S transformation of the cell cycle and thus promoting l-02
cell proliferation (82). ATP
produced by glycolysis is a principal energy source for HCC. As
reported by Lai et al (83), circFBLIM1, which is highly
expressed in HCC serum exosomes, may reverse the inhibition of
miR-338 targeting LRP6 on HCC glycolysis, thereby promoting HCC
growth and anti-apoptotic ability. Furthermore, Zhang et al
(84) found that circ-DB was
upregulated in patients with HCC with high body fat and further
mechanistic studies indicated that exo-circ-DB promoted HCC growth
and reduced DNA damage by inhibiting miR-34A and activating the
USP7/cyclin A2 signaling pathway. By contrast, certain exo-circRNAs
are involved in the negative regulation of HCC. Chen et al
(85) reported that circ-0051443
was transmitted from normal cells to HCC cells through exosomes and
inhibited malignant biological behavior by promoting apoptosis and
blocking the cell cycle. Circ-0051443 may competitively bind with
miR-331-3P, releasing the more downstream apoptosis-related protein
BAK1 and accelerating cell apoptosis, thus curbing the unlimited
growth of hepatocellular carcinoma. In addition, Zhang et al
(86) also found downregulated
exosomal circGDI2 in oral squamous cell carcinoma, which may target
the miR-424-5P/SCAI axis to regulate the malignant growth of oral
squamous cell carcinoma cells. In CRC, Feng et al (87) found that circIFT80 was
significantly upregulated in serum exosomes of patients with CRC,
CRC tissues and CRC cell lines, and relevant functional tests
verified that knockdown of exosomal circIFT80 inhibited the growth
and cloning ability of CRC cells and increased cell apoptosis. They
also investigated the relevant mechanisms and found that the
circIFT80/miR-1236-3p/HOXB7 axis regulated the progression of CRC.
In addition, Luo et al (88) found a significant increase in
circulating exosomal circMYC in patients with nasopharyngeal cancer
(NPC) and demonstrated via cell experiments that circMYC
overexpression promoted NPC cell proliferation and enhanced cell
resistance to radiotherapy. They also demonstrated that
overexpression of circ-0000199 in SCC9 cells significantly promoted
cell growth and reduced cell apoptosis (89). It may be concluded that the high
expression of exo-circRNAs in malignant tumors may be caused by the
paracrine action of tumor cells on surrounding normal cells to make
normal cells malignant, or the transmission from tumor cells with a
high degree of malignancy to tumor cells with a low degree of
malignancy may increase the degree of malignancy. The
downregulation of exo-circRNAs in malignant tumors may have the
opposite effect. However, the specific transmission mechanism
remains elusive and requires further discussion.
Tumor metastasis has always been considered a poor
prognostic factor for malignant tumors. It is important to study
the mechanism of tumor metastasis to reduce the cancer stage and
improve prognosis. Numerous studies have suggested that serum
exo-circRNAs are important vectors for regulating tumor metastasis.
Tumor metastasis involves molecular mechanisms. Li et al
(90) indicated that exosomal
circ-PDE8A in pancreatic ductal adenocarcinoma (PDAC) has an
important role in tumor progression; circ-PDE8A is increased in
plasma from patients with PDAC and knockout of circ-PDE8A may
significantly reduce the invasion and migration of PDAC cells.
Further research confirmed that circ-PDE8A acts through a ceRNA
mechanism to adjust MACC1. Invasive growth of PDAC cells was
stimulated by the MACC/MET/ERK or AKT pathways (90). Li et al (91) reported that circ-IARS expression
was upregulated in plasma exosomes of patients with metastatic
pancreatic cancer. Furthermore, circ-IARS was found to enter human
umbilical vein endothelial cells (HUVECs) through exosomes and
promote tumor invasion and metastasis. Overexpression of circ-IARS
significantly downregulated the levels of miR-122 and Zo-1,
upregulated the levels of RhoA and RhoA-GTP, increased the
expression and adhesion of F-actin, enhanced endothelial monolayer
permeability and promoted tumor invasion and metastasis (91). EMT is a crucial cause of distant
metastasis of tumor cells. Chen et al (92) found that circPRMT5 was upregulated
in serum and urine exosomes from patients with urothelial carcinoma
of the bladder (UCB) and was significantly associated with tumor
metastasis. Their cohort study indicated that there may be a
circPRMT5/miR-30c/SNAIL1/E-cadherin pathway, which is of great
significance for promoting the metastasis of UCB (92). In CRC, hypoxic cancer cells tend to
have a higher metastatic potential than oxygen-rich cancer cells.
Yang et al (93) found that
circ-133 was enriched in the plasma exosomes of patients with CRC
and further studies proved that circ-133 from hypoxic cells were
delivered into normoxic cells and promoted cancer metastasis by
acting on the miR-133a/GEF-H1/RhoA axis. Similarly, Zhao et
al (94) reported that
exosomal circ-ABCC1 derived from CD133+ cells isolated from CRC
cells induced the metastasis of CRC cells, exacerbating the
malignant potential of CRC cells. In GC, Lu et al (95) determined that circ-RanGAP1 was
significantly upregulated in plasma exosomes of patients with GC at
the preoperative stage, and plasma exosomes derived from these
patients enhanced the migration and invasion of GC cells. In terms
of the mechanism, circ-RanGAP1 may mediate the miR-877-3P/VEGFA
axis to promote GC metastasis. Hui et al (96) also found that circNHSL1 was highly
expressed in exosomes derived from GC cells, and its knockdown
hindered the migration and invasion in vitro and inhibited
tumor growth in vivo via the miR-149-5p/YWHAZ axis in GC. In
LC, He et al (97) found
exo-circ-0056616 by researching circRNAs significantly associated
with chemokine receptor CXCR4 and confirmed its influence on the
migration and invasion ability of lung adenocarcinoma cells through
cell testing. Clinical sample analysis also confirmed that
exo-circ-0056616 is correlated with lymph node metastasis of lung
adenocarcinoma (97). Likewise,
Zhang et al (98) indicated
that circSATB2 was highly expressed in NSCLC cells, which may be
transferred by exosomes and promote the migration and invasion of
NSCLC cells by regulating fascin actin-bundling protein 1
expression via miR-326. In HCC, Wang et al (99) found high expression of circPTGR1 in
serum exosomes of patients with HCC with high metastasis potential.
They then cocultured serum exosomes from patients with HCC with
high metastasis potential with HCC cell lines, and silencing
circPTGR1 significantly inhibited cell migration and invasion.
Mechanistic studies revealed that circPTGR1 was able to target
miR449a (99). Of note, Huang
et al (100) also found
that circ-100338 was highly expressed in serum exosomes of patients
with HCC and Transwell invasion assays suggested that
overexpression or knockdown of exo-circ-100338 significantly
enhanced or reduced the invasion capacity of HCC cells.
Furthermore, circ-100338 may also affect the vascular permeability
of HUVECs and promote tumor metastasis (100). Peritoneal metastasis of ovarian
cancer is considered to be an important reason for the loss of
surgical opportunity in advanced ovarian cancer. Guan et al
(101) found that circPUM1 may
promote peritoneal metastasis of ovarian cancer in the form of
cancer-derived exosomes. Furthermore, Zong et al (102) also demonstrated that the highly
expressed circWHSC1 in ovarian cancer may act on peritoneal
mesothelial cells in exosome forms and promote peritoneal
dissemination. In conclusion, the specific mechanism by which
exo-circRNAs regulate tumor metastasis remains to be fully
clarified. It may be assumed that exosomal circRNAs may promote
tumor cell metastasis by mediating EMT and regulating the vascular
endothelial cell gap. In addition, it may be hypothesized that
exosomes carrying circRNAs may be released into the blood by remote
secretion and transported to metastatic foci or target cells for
long-distance transport to exert their role as exo-circRNAs in
distant metastasis of tumors. The specific mechanism still requires
further study.
Of note, certain powerful exo-circRNAs may modulate
certain key molecules to perform multiple functions. For instance,
vascular endothelial growth factor (VEGF) may promote vascular
permeability, proliferation and angiogenesis. Xie et al
(103) found high expression of
circSHKBP1 in serum exosomes of GC; however, the level of
circSHKBP1 in exosomes was significantly reduced after gastrectomy.
CircSHKBP1 was able to sponge miR-582-3p to increase HUR
expression, enhancing VEGF mRNA stability, in this way promoting
the proliferation, migration, invasion and angiogenesis of GC
cells. Furthermore, circSHKBP1 was able to directly bind to heat
shock protein (HSP)90 to inhibit the ubiquitin-dependent
degradation of HSP90, resulting in accelerated GC malignant
behavior (103). In addition,
Shang et al (104)
identified a novel CRC-derived exosomal circRNA, circPACRGL. It was
able to facilitate the expression of transforming growth factor-β1
(TGF-β1) through miR-142-3p/miR-506-3p. TGF-β belongs to a group of
TGF-β superfamilies that mainly regulate cell growth and
differentiation, and in the above study, TGF-β1 was indicated to
promote the proliferation, migration and invasion of CRC cells.
Furthermore, Fang et al (105) noticed the significance of circRNA
Rho GTPase activating protein 10 (circARHGAP10) and demonstrated
its high expression in serum-derived exosomes of patients with
NSCLC. Inhibition of circARHGAP10-mediated glycolysis repressed
proliferation, migration and invasion of NSCLC cells in
vitro, as well as curbed tumor growth in vivo through
the miR-638/FAM83F axis (Table I).
Beyond these examples, a large number of powerful exosomal circRNAs
remain to be discovered.
Drug therapy is important for the comprehensive
treatment of malignant tumors and drug resistance is a major
challenge in the treatment of advanced cancer. It has been
indicated that the expression of certain exo-circRNAs in the serum
of patients with drug resistance is severely disordered, indicating
that exo-circRNAs may be involved in the progression of drug
resistance in tumor cells. Platinum drugs are considered the most
classic chemotherapy drugs to kill tumor cells, and their phase I
clinical efficacy has been universally recognized. For instance,
cisplatin is mainly used to treat testicular cancer, ovarian cancer
and bladder cancer in clinical practice (106). However, with the progression of
advanced tumors, the sensitivity of numerous tumor cells to
platinum drugs is decreasing progressively and the mechanism is
still under study. Wang et al (107) first found that the expression of
pyruvate kinase isoenzyme 2 (PKM2) in oxaliplatin-resistant CRC
cells was significantly higher than that in oxaliplatin-sensitive
cells. PKM2 is a key enzyme in the glycolytic pathway and has
important significance for the rapid growth and chemical resistance
of tumor cells. In addition, they found high expression of ciRS-122
in serum exosomes of patients with CRC resistant to oxaliplatin and
discovered the ciRS-122/miR-122/PKM2 pathway. Through in
vitro and in vivo experiments, it was indicated that
oxaliplatin-resistant cell exosomes delivered ciRS-122 to sensitive
cells, thus promoting glycolysis and drug resistance through
sponging miR-122 and upregulating PKM2 (107). Hon et al (108) found high expression of
circ-0000338 in exosomes of HCT116 cells in CRC and knockout of
circ-0000338 improved the FOLFOX resistance of CRC cells. In
addition, Luo and Gui (109)
observed a significant increase of circFOXP1 in circulating
exosomes in patients with cisplatin (DDP)-resistant ovarian cancer
and mechanistic studies indicated that circFOXP1 regulated the
expression of CCAAT enhancer binding protein γ and formin-like 3 by
sponging miR-22 and miR-155-3P and that miR-22 and miR-150-3p
mimics may attenuate circFOXP1-mediated DDP resistance. Zhao et
al (110) also found that
circRNA-CDR1AS was downregulated in serum exosomes of patients with
DDP-resistant ovarian cancer. Subsequently, inhibition of CDR1AS
facilitated the expression of miR-1270, and miR-1270 exerted its
role via modulating the suppressor of cancer cell invasion (SCAI)
expression. In brief, CDR1AS sensitizes ovarian cancer to DDP by
regulating the miR-1270/SCAI signaling pathway. Similarly, Yao
et al (111) also found
DDP resistance in GC. In the serum and cells of patients with
DDP-resistant GC, exo-circ-PVT1 was upregulated, while miR-30a-5p
was downregulated. Mechanistic studies indicated that there was an
exosomal circ-PVT1/miR-30a-5P/YAP1 axis that regulated autophagy,
invasion and apoptosis in GC cells to promote DDP resistance.
Beyond the role of exo-circRNAs in platinum-based chemotherapy-drug
resistance, Han et al (112) found circ-HIPK3 to be obviously
increased in temozolomide (TMZ)-resistant glioma cells and their
exosomes. Exosomal circ-HIPK3 was able to aggravate TMZ resistance
by regulating the miR-421/ZIC5 axis in TMZ-resistant glioma.
Similarly, Ding et al (113) found that exosomal circNFIX
enhanced the resistance to TMZ in glioma at least in part by
sponging miR-132. In addition to chemotherapy drug resistance,
exo-circRNAs also have a role in immune drug resistance, endocrine
drug resistance and targeted drug resistance. Luo and Gui (114) found that circMYC expression in
circulating exosomes was significantly higher in
bortezomib-resistant patients with multiple myeloma than in
nondrug-resistant patients. Ma et al (115) revealed a novel serum
exosomes-based circ-0002130, which was able to target miR-498 to
regulate key molecules of the glycolytic pathway, including glucose
transporter 1, hexokinase-2 and lactate dehydrogenase A, thereby
facilitating osimertinib resistance, which is closely related to
glycolysis in NSCLC. Hu et al (116) found that exosomal circ-UBE2D2 is
able to bind miR-200A-3P to enhance tamoxifen resistance in breast
cancer. Zhang et al (117)
found that exo-derived circUHRF1 secreted by HCC cells inhibited
natural killer-cell function by upregulating the expression of
TIM-3 by sponging miR-449C-5p, driving the resistance to anti-PD1
immunotherapy (Table II). In
general, exo-circRNAs have great research prospects in the drug
resistance of malignant tumors. In recent years, an increasing
number of exosomal circRNAs have been found to be dysregulated in
drug-resistant tumor cells, which may be involved in the
development of drug resistance in tumors. Currently, the known
mechanism is mainly that exosomal circRNAs act as sponges of miRNAs
and regulate related genes and signaling pathways involved with
downstream drug targets through ceRNA mechanisms. Other relevant
mechanisms remain to be elucidated.
The low efficiency of cancer diagnosis and prognosis
is a problem faced by all clinicians in diagnosis and treatment.
Researchers urgently need to discover a more stable and accurate
indicator to diagnose cancer and judge prognosis compared to
traditional diagnosis and treatment indicators. Studies have
indicated that there are significantly differentially expressed
circRNA lineages in peripheral blood exosomes from certain tumor
patients, suggesting that specific circRNAs may be detected after
peripheral blood exosome extraction, which may be used as follow-up
molecular markers to diagnose or treat this disease (118). Tumor cells may spread circRNAs by
excreting them into exosomes. They may transmit biological
information to specific cells to establish efficient phenotypic
transmission to induce cancer. For instance, Yang et al
(119) isolated serum exosomes
from patients with NSCLC and found that exo-circRNA_102481 was able
to enhance ROR1 expression, thereby promoting EGFR-TKI resistance
in NSCLC. Exo-circRNA_102481 may be used as a therapeutic target
and biomarker for the diagnosis of EGFR-TKI resistance in NSCLC
(119). RNA-sequencing analysis
indicated that exo-circRNAs in exosomes were at least 2-fold
enriched compared to parental cells, for example, in patients with
CRC, colon cancer patients were able to be distinguished from
healthy controls by exo-circRNAs, which not only lays the
foundation for the development of exosomal cancer biomarkers, but
also suggests the potential biological functions of exo-circRNAs
(11). Furthermore, exosomes may
cross the blood-brain barrier and are readily available in almost
all types of human biological fluids. Li et al (120) demonstrated the presence of
abundant circRNAs in both high-grade astrocytoma (HGA) cells and
HGA cell-derived exosomes. This also means that exo-circRNA is a
promising biomarker for HGA.
Despite great advances in surgery, therapeutic
radiotherapy, selection and chemotherapy in recent years, the
prognosis of cancer patients remains unsatisfactory. In the study
of Liu et al (121) in
esophageal squamous cell carcinoma (ESCC), the expression of serum
exosomal hsa_circ_0026611 was significantly upregulated in ESCC
with lymph node metastasis, and it was a predictor of ESCC
prognosis. In studies on CRC, the upregulated exosomal circular
RNA-circCOG2 was indicated to promote CRC proliferation, migration
and invasion through the miR-1305/TGF-β2/SMAD3 pathway. circCOG2 is
associated with poor prognosis and may be used as a therapeutic
target for CRC (122).
Cancer-related cachexia, which may be associated
with the excessive use of fat and skeletal muscle, is one of the
causes of death in patients with advanced tumors. Regarding the
role of circRNAs in cancer cachexia, there is only one study in GC,
which reports that ciRS-133 (circRNA hsa_circ_0010522) not only
promotes the browning of white adipose tissue (WAT), but also
aggravates cachexia in mice with tumor implantation. Zhang et
al (123) found that
exocrines from GC cells were able to deliver CIRS-133 to
preadipocytes, promoting the browning of WAT by activating PRDM16
and inhibiting miR-133. Of note, Luo et al (124) found that patients with HCC with
higher levels of circulating exo-circAKT3 had higher rates of tumor
recurrence. There are numerous exo-circRNAs that simultaneously
affect multiple biological functions of malignant tumor cells by
mechanisms that remain to be elucidated.
Exosomes are special extracellular vesicles with
small inner diameters. They may act as carriers of proteins, RNA,
DNA, lipids and other substances, participate in intercellular
communication and carry the genetic information of donor cells to
recipient cells to regulate the biological functions of recipient
cells. CircRNAs are a type of noncoding RNA with a high abundance,
conservation and relative stability. With increasing research on
circRNAs, they are no longer regarded as the wrong splicing
sequence produced in the transcription process. Instead, circRNAs
may act as sponges of miRNAs, bind proteins and act as regulators.
In addition, certain circRNAs may specifically bind to
transcriptional initiation sequences and regulate RNA
transcription. There are even circRNAs with ORFs that may encode
proteins under certain circumstances. Research in general on the
mechanisms of circRNAs has been mainly performed in parental cells,
but this cannot fully explain the role of circRNAs in distant
metastasis of tumors. Of note, differentially expressed circRNAs
were found in the serum of tumor patients compared to normal
subjects. Further studies revealed that exo-circRNAs, as a new
‘combination’, may be involved in tumor metastasis, in situ
growth, drug resistance and even tumor recurrence.
Exo-circRNAs are a new form of circRNAs. CircRNAs
may be expressed not only in donor cells but also in receptor cells
by acting as ‘outcomers’. In summary, exo-circRNAs released into
the extracellular microenvironment may be ingested by surrounding
cells through paracrine signaling, regulating the in situ
growth of malignant tumor cells, exacerbating the malignancy of
tumor cells and even promoting the malignant transformation of
adjacent normal cells. As demonstrated by Wu et al (125), exosomes from cholangiocarcinoma
cells enhanced circ-0000284 expression and were able to be
transferred directly from cholangiocarcinoma cells to surrounding
normal cells via exosomes, and in this way stimulate the malignant
biological behavior of surrounding normal cells, including
proliferation and metastasis particularly. Furthermore,
exo-circRNAs may enter the humoral circulation and be absorbed by
distant specific target cells via remote secretion, thus indirectly
affecting the metastatic ability of tumors. In addition, drug
resistance has always been a manifestation of malignant tumor cells
and circRNA disorders may be involved. Indications for the
involvement of circulating exo-circRNAs in extensive drug
resistance have been identified. When drugs enter the humoral
circulation, they select the corresponding target cells to exert a
role, while exo-circRNAs may selectively bind to the receptor cells
targeted by drugs and enhance or weaken drug sensitivity by
regulating the downstream signaling pathways. In other aspects,
exo-circRNAs are also involved in tumor recurrence and immune
escape, and the mechanisms remain to be further studied.
Of note, exo-circRNAs also have important clinical
value in the early screening of tumor patients. Pan et al
(126) upregulated exosomal
hsa-circ-0004771 in patients with CRC and the upregulation
amplitude increased significantly with the progression of the CRC
stage, which has been regarded as a new potential diagnostic
biomarker for CRC. Shao et al (127) also demonstrated that exosomal
hsa_circ_0065149 was downregulated in patients with GC and the
degree of its downregulation was significantly associated with the
stage, survival time and lymph node metastasis of patients, which
may be used as an important screening indicator for early GC. In
recent studies, it was indicated that exo-circRNAs have important
value in a new technique called liquid biopsy. This is an important
discovery that is expected to be a new diagnostic criterion for
malignant tumors. Fluid biopsies are easy to obtain blood samples
taken at any point in time to monitor the disease status of
patients who have a tumor by analyzing tumor cells or tumor cell
products in the blood sample (128). Previously, pathological diagnosis
of biopsy samples has always been considered the ‘gold standard’
for the diagnosis of malignant tumors, but it has certain
disadvantages. First, biopsy is an invasive procedure and compared
with noninvasive or minimally invasive procedures, patient
compliance is insufficient. Furthermore, for patients with tumors
in deep locations or small tumor lesions, biopsy is likely to give
false-negative diagnoses, resulting in missed diagnoses. In
addition, biopsies are usually taken from the primary tumor and
reflect its current characterization at the time of sampling.
However, due to the heterogeneity of the tumor, it is possible to
fail to detect certain features, even the most aggressive
subclones. By contrast, liquid biopsy is a minimally invasive
examination that may dynamically reflect the degree of malignancy
of the tumor, particularly for certain patients with small primary
tumors in the early stage, and its diagnostic value will be
magnified (129). However, the
most critical step in liquid biopsy technology, and the most
important challenge, is the selection of diagnostic markers. The
current introduction of exo-circRNAs indicates that, due to their
performance in early screening, they have potential as diagnostic
markers in cancer.
Despite these encouraging developments, numerous
deficiencies remain in the current research on the mechanisms of
exo-circRNAs, and there are still numerous difficulties and
challenges in the process of their formal incorporation into
clinical applications. In terms of the mechanism, exo-circRNAs
mainly exert their role through the ceRNA mechanism, which may be
because exosomes are mainly phagocytic cytoplasmic circRNAs, thus
acting according to the ceRNA mechanism of circRNAs in the
cytoplasm. In addition, it is traditionally thought that, as
circRNAs involved in transcriptional and posttranscriptional
regulation are mainly present in the nucleus, it is difficult for
nuclear circRNAs to reach the cytoplasm due to the isolation of the
nuclear membrane; thus, exo-circRNAs may hardly have the role of
regulating transcription and translation. The latest report of
circRNAs that may not be ‘circular’ also proposed a new hypothesis
for the study of exo-circRNAs, i.e., nuclear circRNAs may also be
transported to target cells by exosomes and regulate the biological
behavior of target cells at the gene level, which may better
explain the powerful function of exo-circRNAs. More research is
required to confirm this. Finally, exosomes carrying circRNAs
secrete circulating fluid, the circRNAs are released in the target
cell and then function in combination with miRNAs or proteins;
thus, it may be suspected that if a new form exists in donor cells,
the circRNAs will combine with both miRNAs and proteins in the form
of a composition commonly taken in by exosomes. After the same mode
of transportation, circRNAs are released in the target cell and
then, through specific circRNA degradation, dissociative circRNAs
and a combination of miRNAs and proteins, the function of specific
miRNAs and proteins in parental cells may be modified. All these
findings provide different ideas for future research on
exo-circRNAs.
In recent years, exo-circRNAs have been a hot field
in cancer research. The present review began with the origin and
function of exosomes and then introduced the origin and function of
circRNAs. Subsequently, the concept of exo-circRNAs and the
differentially expressed exo-circRNAs most known cancers were
introduced. The functions of exo-circRNAs in tumors were
classified, including tumor cell growth, metastasis and malignant
tumor drug resistance. The mechanisms by which exo-circRNAs
regulate tumor functions were summarized. Finally, the deficiencies
and limitations of current exo-circRNA research were proposed and
guidance for the future direction of exo-circRNAs was provided. It
may be expected that exo-circRNAs become a hot topic for future
research and emerge as potential new diagnostic markers and
therapeutic targets for cancer.
Not applicable.
The present study was supported in part by the National Natural
Science Foundation of China (grant nos. 82073133 and 82203486),
Scientific research project of Jiangsu Health Committee (grant no.
ZDA2020005), ‘Six Talents Peak’ High-level Talent Project of
Jiangsu Province (grant no. WSW-050) and Xuzhou Medical Leading
Talents Training Project (grant no. XWRCHT20210034)
Not applicable.
JS and TJ provided direction and guidance
throughout the preparation of this manuscript. JC, JW and LL
collected and analyzed studies and were major contributors in
writing and editing the manuscript. CZ, YZ, ZX and XS reviewed and
revised the manuscript. All authors read and approved the final
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Vestad B, Llorente A, Neurauter A, Phuyal
S, Kierulf B, Kierulf P, Skotland T, Sandvig K, Haug KBF and
Øvstebø R: Size and concentration analyses of extracellular
vesicles by nanoparticle tracking analysis: A variation study. J
Extracell Vesicles. 6:13440872017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mathieu M, Martin-Jaular L, Lavieu G and
Théry C: Specificities of secretion and uptake of exosomes and
other extracellular vesicles for cell-to-cell communication. Nat
Cell Biol. 21:9–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wortzel I, Dror S, Kenific CM and Lyden D:
Exosome-mediated metastasis: Communication from a distance. Dev
Cell. 49:347–360. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and
Chen M: Circular RNA: An emerging key player in RNA world. Brief
Bioinform. 18:547–557. 2017.PubMed/NCBI
|
|
6
|
Peng F, Gong W, Li S, Yin B, Zhao C, Liu
W, Chen X, Luo C, Huang Q, Chen T, et al: circRNA_010383 acts as a
sponge for miR-135a, and its downregulated expression contributes
to renal fibrosis in diabetic nephropathy. Diabetes. 70:603–615.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X,
Zhong J, Zhao Z, Zhao K, Liu D, et al: Circular RNA-encoded
oncogenic E-cadherin variant promotes glioblastoma tumorigenicity
through activation of EGFR-STAT3 signalling. Nat Cell Biol.
23:278–291. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881.e13. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su C, Han Y, Zhang H, Li Y, Yi L, Wang X,
Zhou S, Yu D, Song X, Xiao N, et al: CiRS-7 targeting miR-7
modulates the progression of non-small cell lung cancer in a manner
dependent on NF-κB signalling. J Cell Mol Med. 22:3097–3107. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Johnstone RM, Adam M, Hammond JR, Orr L
and Turbide C: Vesicle formation during reticulocyte maturation.
Association of plasma membrane activities with released vesicles
(exosomes). J Biol Chem. 262:9412–9420. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cocucci E, Racchetti G and Meldolesi J:
Shedding microvesicles: Artefacts no more. Trends Cell Biol.
19:43–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bronson DL, Fraley EE, Fogh J and Kalter
SS: Induction of retrovirus particles in human testicular tumor
(Tera-1) cell cultures: An electron microscopic study. J Natl
Cancer Inst. 63:337–339. 1979.PubMed/NCBI
|
|
15
|
Akers JC, Gonda D, Kim R, Carter BS and
Chen CC: Biogenesis of extracellular vesicles (EV): Exosomes,
microvesicles, retrovirus-like vesicles, and apoptotic bodies. J
Neurooncol. 113:1–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ihara T, Yamamoto T, Sugamata M, Okumura H
and Ueno Y: The process of ultrastructural changes from nuclei to
apoptotic body. Virchows Arch. 433:443–447. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Milane L, Singh A, Mattheolabakis G,
Suresh M and Amiji MM: Exosome mediated communication within the
tumor microenvironment. J Control Release. 219:278–294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pols MS and Klumperman J: Trafficking and
function of the tetraspanin CD63. Exp Cell Res. 315:1584–1592.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Janas T, Janas MM, Sapoń K and Janas T:
Mechanisms of RNA loading into exosomes. FEBS Lett. 589:1391–1398.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
D'Souza-Schorey C and Schorey JS:
Regulation and mechanisms of extracellular vesicle biogenesis and
secretion. Essays Biochem. 62:125–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hessvik NP and Llorente A: Current
knowledge on exosome biogenesis and release. Cell Mol Life Sci.
75:193–208. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sotelo JR and Porter KR: An electron
microscope study of the rat ovum. J Biophys Biochem Cytol.
5:327–342. 1959. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kowal J, Tkach M and Théry C: Biogenesis
and secretion of exosomes. Curr Opin Cell Biol. 29:116–125. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wollert T and Hurley JH: Molecular
mechanism of multivesicular body biogenesis by ESCRT complexes.
Nature. 464:864–869. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Frankel EB and Audhya A: ESCRT-dependent
cargo sorting at multivesicular endosomes. Semin Cell Dev Biol.
74:4–10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Teo H, Perisic O, González B and Williams
RL: ESCRT-II, an endosome-associated complex required for protein
sorting: Crystal structure and interactions with ESCRT-III and
membranes. Dev Cell. 7:559–569. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schöneberg J, Pavlin MR, Yan S, Righini M,
Lee IH, Carlson LA, Bahrami AH, Goldman DH, Ren X, Hummer G, et al:
ATP-dependent force generation and membrane scission by ESCRT-III
and Vps4. Science. 362:1423–1428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Odorizzi G, Babst M and Emr SD: Fab1p
PtdIns(3)P 5-kinase function essential for protein sorting in the
multivesicular body. Cell. 95:847–858. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hemler ME: Tetraspanin functions and
associated microdomains. Nat Rev Mol Cell Biol. 6:801–811. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jian X, He H, Zhu J, Zhang Q, Zheng Z,
Liang X, Chen L, Yang M, Peng K, Zhang Z, et al: Hsa_circ_001680
affects the proliferation and migration of CRC and mediates its
chemoresistance by regulating BMI1 through miR-340. Mol Cancer.
19:202020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Villarroya-Beltri C, Baixauli F,
Mittelbrunn M, Fernández-Delgado I, Torralba D, Moreno-Gonzalo O,
Baldanta S, Enrich C, Guerra S and Sánchez-Madrid F: ISGylation
controls exosome secretion by promoting lysosomal degradation of
MVB proteins. Nat Commun. 7:135882016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cai H, Reinisch K and Ferro-Novick S:
Coats, tethers, Rabs, and SNAREs work together to mediate the
intracellular destination of a transport vesicle. Dev Cell.
12:671–682. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Barlowe C, Orci L, Yeung T, Hosobuchi M,
Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M and
Schekman R: COPII: A membrane coat formed by Sec proteins that
drive vesicle budding from the endoplasmic reticulum. Cell.
77:895–907. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Béthune J, Wieland F and Moelleken J:
COPI-mediated transport. J Membr Biol. 211:65–79. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stenmark H: Rab GTPases as coordinators of
vesicle traffic. Nat Rev Mol Cell Biol. 10:513–525. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hsu C, Morohashi Y, Yoshimura S,
Manrique-Hoyos N, Jung S, Lauterbach MA, Bakhti M, Grønborg M,
Möbius W, Rhee J, et al: Regulation of exosome secretion by Rab35
and its GTPase-activating proteins TBC1D10A-C. J Cell Biol.
189:223–232. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rothman JE: Mechanisms of intracellular
protein transport. Nature. 372:55–63. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Su N, Hao Y, Wang F, Hou W, Chen H and Luo
Y: Mesenchymal stromal exosome-functionalized scaffolds induce
innate and adaptive immunomodulatory responses toward tissue
repair. Sci Adv. 7:eabf72072021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamashita T, Takahashi Y, Nishikawa M and
Takakura Y: Effect of exosome isolation methods on physicochemical
properties of exosomes and clearance of exosomes from the blood
circulation. Eur J Pharm Biopharm. 98:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shin S, Park YH, Jung SH, Jang SH, Kim MY,
Lee JY and Chung YJ: Urinary exosome microRNA signatures as a
noninvasive prognostic biomarker for prostate cancer. NPJ Genom
Med. 6:452021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun J, Lu Z, Fu W, Lu K, Gu X, Xu F, Dai
J, Yang Y and Jiang J: Exosome-derived ADAM17 promotes liver
metastasis in colorectal cancer. Front Pharmacol. 12:7343512021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS,
Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ, et al: CAFs secreted
exosomes promote metastasis and chemotherapy resistance by
enhancing cell stemness and epithelial-mesenchymal transition in
colorectal cancer. Mol Cancer. 18:912019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li X, Li K, Li M, Lin X, Mei Y, Huang X
and Yang H: Chemoresistance transmission via exosome-transferred
MMP14 in pancreatic cancer. Front Oncol. 12:8446482022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hisakane K, Seike M, Sugano T, Yoshikawa
A, Matsuda K, Takano N, Takahashi S, Noro R and Gemma A:
Exosome-derived miR-210 involved in resistance to osimertinib and
epithelial-mesenchymal transition in EGFR mutant non-small cell
lung cancer cells. Thorac Cancer. 12:1690–1698. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dong H, Wang W, Chen R, Zhang Y, Zou K, Ye
M, He X, Zhang F and Han J: Exosome-mediated transfer of
lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast
cancer. Int J Oncol. 53:1013–1026. 2018.PubMed/NCBI
|
|
48
|
Zhang Z, Xing T, Chen Y and Xiao J:
Exosome-mediated miR-200b promotes colorectal cancer proliferation
upon TGF-β1 exposure. Biomed Pharmacother. 106:1135–1143. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Deng M, Yuan H, Liu S, Hu Z and Xiao H:
Exosome-transmitted LINC00461 promotes multiple myeloma cell
proliferation and suppresses apoptosis by modulating microRNA/BCL-2
expression. Cytotherapy. 21:96–106. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang P, Zhou H, Lu K, Lu Y, Wang Y and
Feng T: Exosome-mediated delivery of MALAT1 induces cell
proliferation in breast cancer. Onco Targets Ther. 11:291–299.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li X, Chen C, Wang Z, Liu J, Sun W, Shen
K, Lv Y, Zhu S, Zhan P, Lv T and Song Y: Elevated exosome-derived
miRNAs predict osimertinib resistance in non-small cell lung
cancer. Cancer Cell Int. 21:4282021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Song Z, Mao J, Barrero RA, Wang P, Zhang F
and Wang T: Development of a CD63 aptamer for efficient cancer
immunochemistry and immunoaffinity-based exosome isolation.
Molecules. 25:55852020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jing X, Xie M, Ding K, Xu T, Fang Y, Ma P
and Shu Y: Exosome-transmitted miR-769-5p confers cisplatin
resistance and progression in gastric cancer by targeting CASP9 and
promoting the ubiquitination degradation of p53. Clin Transl Med.
12:e7802022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Han L, Shi WJ, Xie YB and Zhang ZG:
Diagnostic value of four serum exosome microRNAs panel for the
detection of colorectal cancer. World J Gastrointest Oncol.
13:970–979. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Greene J, Baird AM, Casey O, Brady L,
Blackshields G, Lim M, O'Brien O, Gray SG, McDermott R and Finn SP:
Circular RNAs are differentially expressed in prostate cancer and
are potentially associated with resistance to enzalutamide. Sci
Rep. 9:107392019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wilusz JE: A 360° view of circular RNAs:
From biogenesis to functions. Wiley Interdiscip Rev RNA.
9:e14782018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lu Z, Filonov GS, Noto JJ, Schmidt CA,
Hatkevich TL, Wen Y, Jaffrey SR and Matera AG: Metazoan tRNA
introns generate stable circular RNAs in vivo. RNA. 21:1554–1565.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kun-Peng Z, Xiao-Long M, Lei Z, Chun-Lin
Z, Jian-Ping H and Tai-Cheng Z: Screening circular RNA related to
chemotherapeutic resistance in osteosarcoma by RNA sequencing.
Epigenomics. 10:1327–1346. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Bach DH, Lee SK and Sood AK: Circular RNAs
in Cancer. Mol Ther Nucleic Acids. 16:118–129. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Barrett SP and Salzman J: Circular RNAs:
Analysis, expression and potential functions. Development.
143:1838–1847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. 98:87–97. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yu CY, Li TC, Wu YY, Yeh CH, Chiang W,
Chuang CY and Kuo HC: The circular RNA circBIRC6 participates in
the molecular circuitry controlling human pluripotency. Nat Commun.
8:11492017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang Y and Wang Z: Efficient backsplicing
produces translatable circular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 Is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)A promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Sun H, Wu Z, Liu M, Yu L, Li J, Ding X and
Jin H: CircRNA may not be ‘circular’. bioRxiv.
2020.2009.2027.315275. 2020.
|
|
77
|
Patop IL and Kadener S: circRNAs in
cancer. Curr Opin Genet Dev. 48:121–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Bao C, Lyu D and Huang S: Circular RNA
expands its territory. Mol Cell Oncol. 3:e10844432015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG
and Zhang B: Circular RNAs are down-regulated in KRAS mutant colon
cancer cells and can be transferred to exosomes. Sci Rep.
6:379822016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li Y, Zang H, Zhang X and Huang G:
Exosomal Circ-ZNF652 promotes cell proliferation, migration,
invasion and glycolysis in hepatocellular carcinoma via
miR-29a-3p/GUCD1 axis. Cancer Manag Res. 12:7739–7751. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Dai X, Chen C, Yang Q, Xue J, Chen X, Sun
B, Luo F, Liu X, Xiao T, Xu H, et al: Exosomal circRNA_100284 from
arsenite-transformed cells, via microRNA-217 regulation of EZH2, is
involved in the malignant transformation of human hepatic cells by
accelerating the cell cycle and promoting cell proliferation. Cell
Death Dis. 9:4542018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lai Z, Wei T, Li Q, Wang X, Zhang Y and
Zhang S: Exosomal circFBLIM1 promotes hepatocellular carcinoma
progression and glycolysis by regulating the miR-338/LRP6 axis.
Cancer Biother Radiopharm. Sep 9–2020.(Epub ahead of print).
|
|
84
|
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu
K, Fan Q, Li J, Ning T, Tian F, et al: Exosome circRNA secreted
from adipocytes promotes the growth of hepatocellular carcinoma by
targeting deubiquitination-related USP7. Oncogene. 38:2844–2859.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen W, Quan Y, Fan S, Wang H, Liang J,
Huang L, Chen L, Liu Q, He P and Ye Y: Exosome-transmitted circular
RNA hsa_circ_0051443 suppresses hepatocellular carcinoma
progression. Cancer Lett. 475:119–128. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang Y, Tang K, Chen L, Du M and Qu Z:
Exosomal CircGDI2 suppresses oral squamous cell carcinoma
progression through the regulation of MiR-424-5p/SCAI axis. Cancer
Manag Res. 12:7501–7514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang
Y and Cui G: circIFT80 functions as a ceRNA of miR-1236-3p to
promote colorectal cancer progression. Mol Ther Nucleic Acids.
18:375–387. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Luo Y, Ma J, Liu F, Guo J and Gui R:
Diagnostic value of exosomal circMYC in radioresistant
nasopharyngeal carcinoma. Head Neck. 42:3702–3711. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Luo Y, Liu F, Guo J and Gui R:
Upregulation of circ_0000199 in circulating exosomes is associated
with survival outcome in OSCC. Sci Rep. 10:137392020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li Z, Yanfang W, Li J, Jiang P, Peng T,
Chen K, Zhao X, Zhang Y, Zhen P, Zhu J and Li X: Tumor-released
exosomal circular RNA PDE8A promotes invasive growth via the
miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett.
432:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen
K, Liu H, Bi H, Liu X and Li X: Circular RNA IARS (circ-IARS)
secreted by pancreatic cancer cells and located within exosomes
regulates endothelial monolayer permeability to promote tumor
metastasis. J Exp Clin Cancer Res. 37:1772018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging mir-30c to induce epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Yang H, Zhang H, Yang Y, Wang X, Deng T,
Liu R, Ning T, Bai M, Li H, Zhu K, et al: Hypoxia induced exosomal
circRNA promotes metastasis of colorectal cancer via targeting
GEF-H1/RhoA axis. Theranostics. 10:8211–8226. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhao H, Chen S and Fu Q: Exosomes from
CD133+ cells carrying circ-ABCC1 mediate cell stemness
and metastasis in colorectal cancer. J Cell Biochem. 121:3286–3297.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie
JW, Wang JB, Lin JX, Chen QY, Cao LL, et al: Circular RNA
circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to
facilitate gastric cancer invasion and metastasis. Cancer Lett.
471:38–48. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Hui C, Tian L and He X: Circular RNA
circNHSL1 contributes to gastric cancer progression through the
miR-149-5p/YWHAZ axis. Cancer Manag Res. 12:7117–7130. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
He F, Zhong X, Lin Z, Lin J, Qiu M, Li X
and Hu Z: Plasma exo-hsa_circRNA_0056616: A potential biomarker for
lymph node metastasis in lung adenocarcinoma. J Cancer.
11:4037–4046. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu
M, Dai X, Zhou H, Zhu J, Zhang H and Jiang Y: Circular RNA
circSATB2 promotes progression of non-small cell lung cancer cells.
Mol Cancer. 19:1012020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo
J, Zhang Y, Li H, Zhang Q, Yang Y and Chen G: Three isoforms of
exosomal circPTGR1 promote hepatocellular carcinoma metastasis via
the miR449a-MET pathway. EBioMedicine. 40:432–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Huang XY, Huang ZL, Huang J, Xu B, Huang
XY, Xu YH, Zhou J and Tang ZY: Exosomal circRNA-100338 promotes
hepatocellular carcinoma metastasis via enhancing invasiveness and
angiogenesis. J Exp Clin Cancer Res. 39:202020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Guan X, Zong ZH, Liu Y, Chen S, Wang LL
and Zhao Y: circPUM1 promotes tumorigenesis and progression of
ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther
Nucleic Acids. 18:882–892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zong ZH, Du YP, Guan X, Chen S and Zhao Y:
CircWHSC1 promotes ovarian cancer progression by regulating MUC1
and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer
Res. 38:4372019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F,
Ma P, Jiang H, Wu X, Shu Y and Xu T: Exosomal circSHKBP1 promotes
gastric cancer progression via regulating the miR-582-3p/HUR/VEGF
axis and suppressing HSP90 degradation. Mol Cancer. 19:1122020.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Shang A, Gu C, Wang W, Wang X, Sun J, Zeng
B, Chen C, Chang W, Ping Y, Ji P, et al: Exosomal circPACRGL
promotes progression of colorectal cancer via the
miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer. 19:1172020.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Fang K, Chen X, Qiu F, Xu J, Xiong H and
Zhang Z: Serum-derived exosomes-mediated circular RNA ARHGAP10
modulates the progression of non-small cell lung cancer through the
miR-638/FAM83F axis. Cancer Biother Radiopharm. 37:96–110.
2020.PubMed/NCBI
|
|
106
|
Farrell N: Platinum formulations as
anticancer drugs clinical and pre-clinical studies. Curr Top Med
Chem. 11:2623–2631. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wang X, Zhang H, Yang H, Bai M, Ning T,
Deng T, Liu R, Fan Q, Zhu K, Li J, et al: Exosome-delivered circRNA
promotes glycolysis to induce chemoresistance through the
miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 14:539–555.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal
R and Abu N: Extracellular vesicle-derived circular RNAs confers
chemoresistance in colorectal cancer. Sci Rep. 9:164972019.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Luo Y and Gui R: Circulating exosomal
circFoxp1 confers cisplatin resistance in epithelial ovarian cancer
cells. J Gynecol Oncol. 31:e752020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhao Z, Ji M, Wang Q, He N and Li Y:
Circular RNA Cdr1as upregulates SCAI to suppress cisplatin
resistance in ovarian cancer via miR-1270 suppression. Mol Ther
Nucleic Acids. 18:24–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yao W, Guo P, Mu Q and Wang Y:
Exosome-derived Circ-PVT1 contributes to cisplatin resistance by
regulating autophagy, invasion, and apoptosis via miR-30a-5p/YAP1
axis in gastric cancer cells. Cancer Biother Radiopharm.
36:347–359. 2021.PubMed/NCBI
|
|
112
|
Han C, Wang S, Wang H and Zhang J:
Exosomal circ-HIPK3 facilitates tumor progression and temozolomide
resistance by regulating miR-421/ZIC5 axis in glioma. Cancer
Biother Radiopharm. 36:537–548. 2021.PubMed/NCBI
|
|
113
|
Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z,
Zhang G, Gu J and Kang D: Exosome-mediated transfer of circRNA
CircNFIX enhances temozolomide resistance in glioma. Cancer Lett.
479:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Luo Y and Gui R: Circulating exosomal
circMYC is associated with recurrence and bortezomib resistance in
patients with multiple myeloma. Turk J Haematol. 37:248–262. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Ma J, Qi G and Li L: A novel serum
exosomes-based biomarker hsa_circ_0002130 facilitates
osimertinib-resistance in non-small cell lung cancer by sponging
miR-498. Onco Targets Ther. 13:5293–5307. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Hu K, Liu X, Li Y, Li Q, Xu Y, Zeng W,
Zhong G and Yu C: Exosomes mediated transfer of Circ_UBE2D2
enhances the resistance of breast cancer to tamoxifen by binding to
MiR-200a-3p. Med Sci Monit. 26:e9222532020. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ,
Shi GM, Cai JB and Ke AW: Cancer cell-derived exosomal circUHRF1
induces natural killer cell exhaustion and may cause resistance to
anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer.
19:1102020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Xia D and Gu X: Plasmatic exosome-derived
circRNAs panel act as fingerprint for glioblastoma. Aging (Albany
NY). 13:19575–19586. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Yang B, Teng F, Chang L, Wang J, Liu DL,
Cui YS and Li GH: Tumor-derived exosomal circRNA_102481 contributes
to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small
cell lung cancer. Aging (Albany NY). 13:13264–13286. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Li P, Xu Z, Liu T, Liu Q, Zhou H, Meng S,
Feng Z, Tang Y, Liu C, Feng J, et al: Circular RNA sequencing
reveals serum exosome circular RNA panel for high-grade astrocytoma
diagnosis. Clin Chem. 68:332–343. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Liu S, Lin Z, Rao W, Zheng J, Xie Q, Lin
Y, Lin X, Chen H, Chen Y and Hu Z: Upregulated expression of serum
exosomal hsa_circ_0026611 is associated with lymph node metastasis
and poor prognosis of esophageal squamous cell carcinoma. J Cancer.
12:918–926. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Gao L, Tang X, He Q, Sun G, Wang C and Qu
H: Exosome-transmitted circCOG2 promotes colorectal cancer
progression via miR-1305/TGF-β2/SMAD3 pathway. Cell Death Discov.
7:2812021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Luo Y, Liu F and Gui R: High expression of
circulating exosomal circAKT3 is associated with higher recurrence
in HCC patients undergoing surgical treatment. Surg Oncol.
33:276–281. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Wu G, Sun Y, Xiang Z, Wang K, Liu B, Xiao
G, Niu Y, Wu D and Chang C: Preclinical study using circular RNA 17
and micro RNA 181c-5p to suppress the enzalutamide-resistant
prostate cancer progression. Cell Death Dis. 10:372019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Pan B, Qin J, Liu X, He B, Wang X, Pan Y,
Sun H, Xu T, Xu M, Chen X, et al: Identification of serum exosomal
hsa-circ-0004771 as a novel diagnostic biomarker of colorectal
cancer. Front Genet. 10:10962019. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao
B, Ye G and Guo J: Hsa_circ_0065149 is an indicator for early
gastric cancer screening and prognosis prediction. Pathol Oncol
Res. 26:1475–1482. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Poulet G, Massias J and Taly V: Liquid
biopsy: General concepts. Acta Cytol. 63:449–455. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Mader S and Pantel K: Liquid biopsy:
Current status and future perspectives. Oncol Res Treat.
40:404–408. 2017. View Article : Google Scholar : PubMed/NCBI
|