Introduction
Colorectal cancer (CRC) is the third most common
cause of cancer-related death worldwide (1). Notably, metastasis is common in CRC,
and is a major cause of morbidity and mortality. The liver is the
most frequent site of metastasis, often occurring as a single
metastatic site; however, ~20% of patients with CRC exhibit
peritoneal metastasis, often at multiple metastatic sites (2-4).
Compared with patients with oligometastasis which is characterized
by the localization of the disease to a few sites with the option
to use local ablative therapy (5),
patients with CRC and peritoneal metastasis have a poorer overall
survival (6-8). Therefore, preventing the development
of peritoneal metastasis could potentially improve CRC survival and
the chance of disease-free outcomes.
Peritoneal metastasis is a multi-step process
involving cellular proliferation, immune system evasion,
detachment, adhesion, invasion, translocation and colonization of
the peritoneal cavity (9).
Epithelial-mesenchymal transition (EMT) is a process in which cells
with an epithelial phenotype acquire a mesenchymal-like phenotype.
EMT is involved in cell proliferation and invasion, and in the
development of stem cell properties and therapeutic resistance,
thus aiding the spread of cancer cells from their original site to
distant organs (10-14).
A disintegrin and metalloproteases (ADAMs) are a
family of proteins that contain disintegrin and metalloproteinase
domains, and have adhesive and proteolytic functions (15). ADAMs are involved in the control of
a number of biological processes, such as cell migration, adhesion,
differentiation and proliferation (16,17).
In addition, dysregulated expression of ADAMs has been reported to
be a positive regulator of cancer progression in various types of
human cancer (18-22). ADAMs act as ectodomain sheddases,
and control the cleavage and release of biologically active
adhesion molecules, cytokines, chemokines, growth factors and
receptors from the membrane surface (23,24).
ADAM12 is a member of the ADAMs family that has been
implicated in various diseases, including rheumatoid arthritis,
asthma, Alzheimer's disease and cancer (25-27).
ADAM12 is overexpressed in various types of human cancer, including
breast, lung, stomach and liver cancer, and is associated with
tumor progression and poor prognoses (18-22).
ADAM12 overexpression has been shown to enhance proliferation,
inhibit apoptosis, and to be significantly associated with
metastases and poor survival in patients with CRC (28-31).
Furthermore, knockdown of ADAM12 has been demonstrated to have
potent antitumor activity in an in vivo mouse xenograft
model (28).
The aim of the present study was to evaluate the
molecular mechanism of ADAM12-induced EMT and the clinical
application of ADAM12 as a therapeutic target in treating
peritoneal metastases from CRC.
Materials and methods
Cell culture and transfection
Human CRC cell lines were provided by the American
Type Culture Collection. DLD1 (cat. no. CCL-221™) and SW480 (cat.
no. CCL-228™) cell lines were used in the present study. The cell
lines were maintained in high-glucose DMEM (cat. no. 11995065;
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(cat. no. 16000044; Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (cat. no. 15140122; Gibco; Thermo Fisher
Scientific, Inc.) in a 5% CO2 incubator (humidified
atmosphere) at 37°C. Overexpression of ADAM12 was performed by
inserting the full-length ADAM12 cDNA (GenBank accession no.
NM_021641.3) into the pcDNA6-myc vector (Invitrogen; Thermo Fisher
Scientific, Inc), which was used for transient or long-term ADAM12
overexpression. Empty-pcDNA6-myc vector was used as a negative
control of ADAM12 overexpression. Knockdown of ADAM12 was carried
out using ADAM12-pGFP-C-shLenti short hairpin RNA constructs (cat.
no. TL306850; Origene Technologies, Inc.) and ADAM12-small
interfering (si)RNA duplex (5′-GAC UAC AAC GGG AAA GCA A-3′;
Bioneer). Scrambled siRNA (AllStars Negative Control siRNA; cat.
no. 1027281) and scrambled shRNA control (cat. no. TR30021) were
purchased from Qiagen GmbH and Origene Technologies, Inc.,
respectively. To induce transient expression of genes, DLD1 and
SW480 cells were plated at a density of 1.5×106
cells/6well. Transfection of cells with the pcDNA6-myc vectors (500
ng/well) and siRNAs (100 pmol/well) was performed using 5 µl
Lipofectamine® 2000 and 5 µl Lipofectamine
RNAiMAX (cat. nos. 52887 and 56532; Invitrogen, Thermo Fisher
Scientific, Inc.), respectively. Briefly, the pcDNA6-myc vectors
and siRNAs were mixed with 5 µl Lipofectamine 2000 or 5
µl Lipofectamine RNAiMAX in 400 µl serum-free media,
respectively, and then the mixed solution was added to the plated
cells and incubated in a 5% CO2 at 37°C. After 6 h, the
medium was changed to fresh medium and transfected cells were
incubated for 24 or 48 h in a 5% CO2 incubator at 37°C
until in in vitro experiments were performed. To sustain
long-term gene expression, stable gene-transfected cells were
established. SW480 cells were plated at a density of
3×106 cells/well and the pcDNA6-myc vector (1,000
ng/well) and pGFP-C-shLenti shRNA (200 pmol/well) were transfected
into the cells using 5 µl Lipofectamine 2000 in a 5%
CO2 incubator at 37°C. The selection medium was changed
every 2-3 days until stable cell clones were formed. The next day,
the transfected cells were replaced with selection media
(containing 20 µg/ml blasticidin or 5 µg/ml
puromycin) and cultured in a 5% CO2 incubator at 37°C
for the selection of stable cell clones. After 7-10 days, the
stable cell clones formed were subcultured. The cells stably
expressing the genes were kept n selection media (with 10
µg/ml blasticidin or 2 µg/ml puromycin).
Cell proliferation assay
WST-1 reagent (cat. no. 11644807001; Roche
Diagnostics GmbH) was used to detect cell proliferation.
Transfected cells were plated in 96-well culture plates at a
density of 3×103 cells/well. After 1, 2, and 3 days,
cells were incubated with 10% WST-1 solution for 2 h at 37°C. The
absorbance was measured at 450 nm using an Infinite M200
spectrophotometer (Tecan Group, Ltd.). Three replicates of each
experiment were carried out.
Cell invasion assay
For the cell invasion assay, Transwell chambers
(pore size, 8 µm; cat. no. 3422; Corning, Inc.) were used.
The Transwell chambers were coated with 1% gelatin overnight at
37°C in serum-free medium and were then dried at room temperature
on a clean bench. Transfected cells (2×105) were
suspended in 100 µl serum-free medium containing 0.2% (w/v)
bovine serum albumin (BSA; Sigma-Aldrich; Merck KGaA) and seeded
into the upper chambers, and 400 µl 0.2% (w/v) BSA solution
supplemented with 20 µg/ml human plasma fibronectin (cat.
no. 341635; Calbiochem; Merck KGaA) was added to the lower
chambers. After cells were incubated for 24 h in a 5%
CO2 incubator at 37°C, the cells that invaded from the
upper chamber to the bottom surface of the lower chamber were fixed
with 4% paraformaldehyde for 10 min at room temperature. The cells
were then stained with Hemacolor® Rapid staining
solution (cat. no. 111955; Merck KGaA) for 10 sec at room
temperature. The stained cells on the bottom surface of the lower
chambers were counted using a light microscope; five randomly
selected fields were counted.
Cell migration assay
The migration of transfected cells was assessed
based on gap closure using Ibidi Culture Inserts (cat. no. 81176;
Ibidi GmbH). The transfected cells (5×104) were cultured
in Ibidi Culture Inserts for 1 day in serum-free medium containing
0.2% BSA at 37°C until 90-95% confluence was reached. The culture
inserts were gently removed to create a cell-free gap and images of
gap closure were captured under an inverted optic microscope after
24 and 48 h. Distance of gap closure was measured using Multi-Gauge
gel analysis software (version 3.0; FUJIFILM Wako Pure Chemical
Corporation). The initial wound width of the insert gap was
normalized to 10 mm, and gap closure was calculated in proportion
to the initial wound width of 10 mm.
In vivo tumor model experiments
As non-obese diabetic/severe combined
immunodeficiency (NOD/SCID) mice deficient in B, T and natural
killer cells are considered an ideal model for xenografts of human
cells and tissues, 6-week-old NOD/SCID male mice were purchased
from Charles River Laboratories, Inc. and were separated into four
groups (n=6). The mice were housed at 25°C and 50% humidity under a
12 h light/dark cycle, with ad libitum access to food and
water. For the in vivo tumorigenesis study, stable SW480
cells transfected with empty-pcDNA6-myc, ADAM12-pcDNA6-myc,
scrambled pGFP-C-shLenti or ADAM12-pGFP-C-shLenti were implanted
into the NOD/SCID mice. Stable cells (2×106/mice) were
diluted in 100 µ PBS and were intraperitoneally injected
into the mice as described in previous studies (32,33).
Each mouse was monitored, and changes in body weight were measured
twice per week. After 28 days, the mice were place in a closed
chamber and 5% isoflurane was delivered at a rate of 5 l/min with
O2. After the mice showed no movement and breathing,
cervical dislocation was performed to ensure death. A midline
laparotomy was performed for complete exploration. Peritoneal
tumors were isolated after laparotomy, images were captured and the
tumor nodules were weighed. The size and number of tumor nodules
were recorded after the resection. In addition, the peritoneal
carcinomatosis index (PCI) was calculated for each mouse to assess
the extent of peritoneal carcinomatosis. The abdominal cavity was
divided into 13 standard regions, as described by Sugarbaker
(34), adapted for rodents by
Klaver et al (35).
According to the size and number of tumor nodules, a score ranging
from 0 to 3 was assigned, as follows: 0, no macroscopic tumors; 1,
tumor nodules sized ≤2 mm; 2, tumor nodules sized 2-5 mm or more
than five tumor nodules; 3, tumor nodules sized >5 mm or more
than 10 tumor nodules. With a maximum score of 39, the PCI was
assigned by adding the scores from each of the 13 regions.
Protein isolation and western
blotting
The transfected cells or xenograft tumor tissue
samples were rinsed with phosphate-buffered saline (PBS) and lysed
in Pierce™ RIPA buffer (cat. no. 89900; Thermo Fisher Scientific,
Inc.) with Halt™ phosphatase and protease inhibitor cocktail (cat.
no. 1862495 and 1862209; Thermo Fisher Scientific, Inc.). The
quantification of proteins in the cell lysate was performed using a
BCA protein assay (cat. no. 23228; Thermo Fisher Scientific, Inc.).
Equal amounts (20 µg/lane) of protein lysate were separated
by electrophoresis on 8-12% polyacrylamide gels and transferred
onto Immobilon®-P transfer membranes (cat. no.
IPVH00010; MilliporeSigma). The blot membranes were incubated with
5% BSA solution at room temperature for 1 h and immunoblotted with
specific antibodies (1:1,000 dilution) overnight at 4°C. Antibodies
against ADAM12 (cat. no. ab28747), matrix metalloproteinase (MMP)2
(cat. no. ab37150) and MMP9 (cat. no. ab58803) were purchased from
Abcam. Antibodies against E-cadherin (cat. no. #14472), Snail (cat.
no. #3879), vimentin (cat. no. #5741), claudin-1 (cat. no. #4933),
integrin α5 (cat. no. #4705), integrin β1 (cat. no. #9699),
integrin β3 (cat. no. #13166), phosphorylated (p)-AKT (S473) (cat.
no. #4060), p-phosphoinositide-dependent protein kinase 1 (PDK1)
(S241) (cat. no. #3438), p-glycogen synthase kinase-3β (GSK-3β)
(S9) (cat. no. #9323), total AKT (cat. no. #4691), total PDK1 (cat.
no. #3062), total GSK-3β (cat. no. #9832) and Myc-tag (cat. no.
#2278) were obtained from Cell Signaling Technology, Inc.
Antibodies against β-tubulin (cat. no. sc-9104) and GAPDH (cat. no.
sc-25778) were purchased from Santa Cruz Biotechnology, Inc. The
blot membranes were washed four times with Tris-buffered
saline-0.1% Tween-20 (TBS-T) and were then incubated with a
horseradish peroxidase-conjugated secondary antibody (anti-rabbit,
cat. no. #7074, anti-mouse, cat. no. #7076; Cell Signaling,
Technology, Inc.) at 1:2,000 dilution for 1 h at room temperature.
Amersham ECL Prime Western Blotting Detection Reagent (cat no.
RPN2232SK; Cytiva) was used for blot development. Visualization of
specific bands was obtained using the LAS-400 luminescent image
analyzer (FUJIFILM Wako Pure Chemical Corporation).
Semi-quantification of specific bands was performed using
Multi-Gauge gel analysis software (version 3.0; FUJIFILM Wako Pure
Chemical Corporation).
Cell adhesion assay
For the cell adhesion assay, 96-well plates were
coated with the extracellular matrix (ECM) components collagen I
(cat. no. 354243; 40 µg/ml; Corning, Inc.) and fibronectin
(cat. no. 341635; 2 µg/ml; Calbiochem; Merck KGaA) for 1 h
at 37°C in a CO2 incubator. The ECM-coated wells were
then treated with 0.5% BSA for 30 min at 37°C. Suspended cells in
culture media were counted and an equal volume of 3×104
cells was seeded into the coated wells. Subsequently, the plate was
incubated for 20 min at 37°C in a CO2 incubator and
non-adherent cells were removed by washing with PBS. Cells attached
to the wells were incubated with 10% WST-1 solution at 37°C for 1 h
and the absorbance of the wells was measured at 450 nm using an
Infinite M200 spectrophotometer. Three replicates of each
experiment were carried out.
Patient and tissue samples
Tissue samples of primary tumor, non-metastatic and
metastatic lymph nodes were obtained from 113 patients (67 men and
46 women) with pathologically confirmed CRC who had surgery at
Chonnam National University Hwasun Hospital (Jeonnam, South Korea)
between April and December 2010. The average age of the patients
was 70.5 years (range, 38-94 years). Patients who had chemotherapy
or radiation therapy prior to surgery were excluded. Through
examination of the original pathology slides, tissue blocks were
chosen. The pathologic TNM stage of enrolled patients was assessed
using the standard TNM staging of the American Joint Committee on
Cancer criteria (36). Tissues
were fixed overnight at 4°C with 10% formalin buffer and dehydrated
with 70-100% alcohol at room temperature. Dehydrated tissues were
incubated in 100% xylene at 60°C for 30 min and embedded in
paraffin. Tissue blocks that demonstrated the intersection of the
normal colon epithelium and the tumor site were chosen. The present
study was approved by the Institutional Review Board (IRB) of
Chonnam National University Hwasun Hospital (IRB no.
CNUHH-2017-164). Written informed consent was obtained directly
from the patients or their caregivers.
Immunohistochemistry (IHC)
The paraffin-embedded tissues were sectioned and the
tissue sections (4 µm) was subjected to IHC. Tissue sections
were deparaffinized with 100% xylene after fixing for 5 min at 37°C
on glass slides. The sections were then rehydrated with alcohol
series (100-70%) and antigen retrieval was performed in a steam
cooker for 10 min using citrate buffer (pH 6.0; Dako; Agilent
Technologies, Inc.). After endogenous peroxidase was blocked with
Dako REAL peroxidase-blocking solution (cat. no. S2023; Dako;
Agilent Technologies, Inc.) for 30 min at room temperature, the
tissues were treated with Protein Block Serum-Free solution (Dako;
Agilent Technologies, Inc.) for 30 min at room temperature to block
non-specific antigens. Subsequently, the tissue slides were
submerged with primary ADAM12 (1:50; cat. no. ab28747; Abcam), and
E-cadherin (1:100; cat. no. #14472; Cell Signaling Technology,
Inc.) antibodies overnight at 4°C. The next day, color development
of the tissue slides was performed using the Dako REAL Envision
HRP/DAB detection system (cat. no. K5007; Dako; Agilent
Technologies, Inc.). Briefly, the tissue slides were washed four
times with TBS-T and were incubated with the Envision
HRP-conjugated secondary antibody (anti-rabbit/mouse) for 1 h at
room temperature. After washing, tissues were developed with enzyme
DAB solution for 10 min at room temperature and counterstained for
10 sec with hematoxylin (Millipore Sigma). The immunostaining of
ADAM12 and E-cadherin was evaluated using a semi-quantitative
scoring system that multiplied the degree of immunostaining
intensity by the distribution of positively stained cells. The
score of immunostaining intensity was classified as 0, negative; 1,
weak; 2, moderate; and 3, strong. The percentage distribution of
positively stained cells was scored as 1, 0-25%; 2, 26-50%; 3,
51-75%; and 4, >75%. The mean final score of 113 patient samples
served as the baseline for classifying positive and negative
expression groups. Patient samples with a final score of 1-7 were
regarded as negative and 8-12 were regarded as positive for ADAM12
and E-cadherin expression. Two independent observers assessed the
staining without information of the clinical outcome data. Sections
that were classified into different expression groups by two
evaluators were re-evaluated and carefully discussed for the
appropriate classification.
Statistical analysis
Statistical analyses were performed using SPSS
version 20.0 software (IBM Corporation) and GraphPad Prism version
9.5.1 (GraphPad Software; Dotmatics). Statistical analysis of
patient categorical data was performed using the χ2
test, Fisher's exact test or Wilcoxon signed rank test. Survival
curves were drawn using the Kaplan-Meier method and the statistical
significance of differences was examined by the log-rank test. The
in vitro experiments were performed in triplicate and data
are presented as the mean ± standard deviation. The data from mice
experiments are expressed as median and interquartile range.
Differences between the multiple samples were analyzed using a
one-way ANOVA and Tukey post hoc test, or Kruskal-Wallis and Dunn's
post hoc test, as appropriate. P<0.05 was considered to indicate
a statistically significant difference.
Results
ADAM12 promotes CRC cell
proliferation
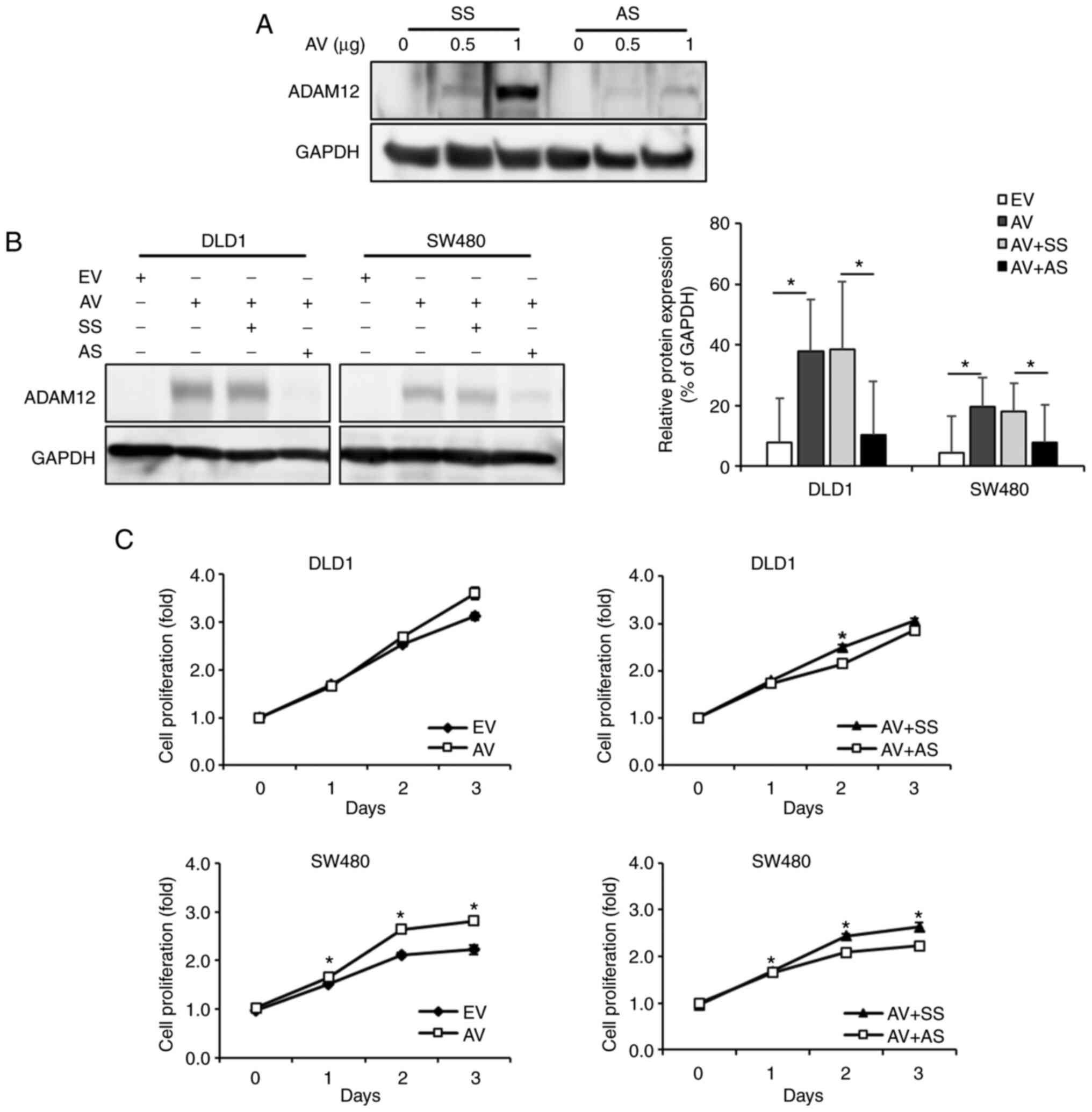
The protein expression levels of ADAM12 were
detected in DLD1 and SW480 cells by western blotting. Endogenous
ADAM12 expression was not detected in the human CRC cell lines DLD1
and SW480. Therefore, the present study induced overexpression and
knockout of ADAM12 to confirm its function in DLD1 and SW480 cells.
The protein expression levels of ADAM12 were increased with the
concentration of the transfected ADAM12-pcDNA6-myc construct and
this expression was knocked down by ADAM12-specific siRNA (Fig. 1A). The ADAM12-pcDNA6-myc construct
resulted in overexpression of ADAM12, whereas ADAM12-siRNA resulted
in the knockdown of ADAM12 expression in DLD1 and SW480 cells
(Fig. 1B). To assess the effects
of ADAM12 expression on cell proliferation, the WST-1 assay was
performed on days 1, 2 and 3 post-transfection with the
ADAM12-pcDNA6-myc construct or ADAM12-siRNA. Overexpression of
ADAM12 via ADAM12-pcDNA6-myc-transfection significantly increased
the proliferative ability of SW480 cells compared with cells
transfected with the empty-pcDNA6-myc (day 1, P=0.003; day 2,
P<0.001; day 3, P<0.001). Although it was not statistically
significant, overexpression of ADAM12 via ADAM12-pcDNA6-my
-transfection also increased the proliferative ability of DLD1
cells (day 1, P=0.578; day 2. P=0.080; day 3, P=0.129). By
contrast, ADAM12-siRNA-transfected SW480 cells exhibited
significantly lower proliferation compared with the scrambled
siRNA-transfected cells (day 1, P=0.031; day 2, P<0.001; day 3,
P<0.001), and ADAM12-siRNA-transfected DLD1 cells exhibited
significantly lower proliferation only on day 2 (day 1, P=0.358;
day 2, P=0.001; day 3, P=0.118), (Fig.
1C).
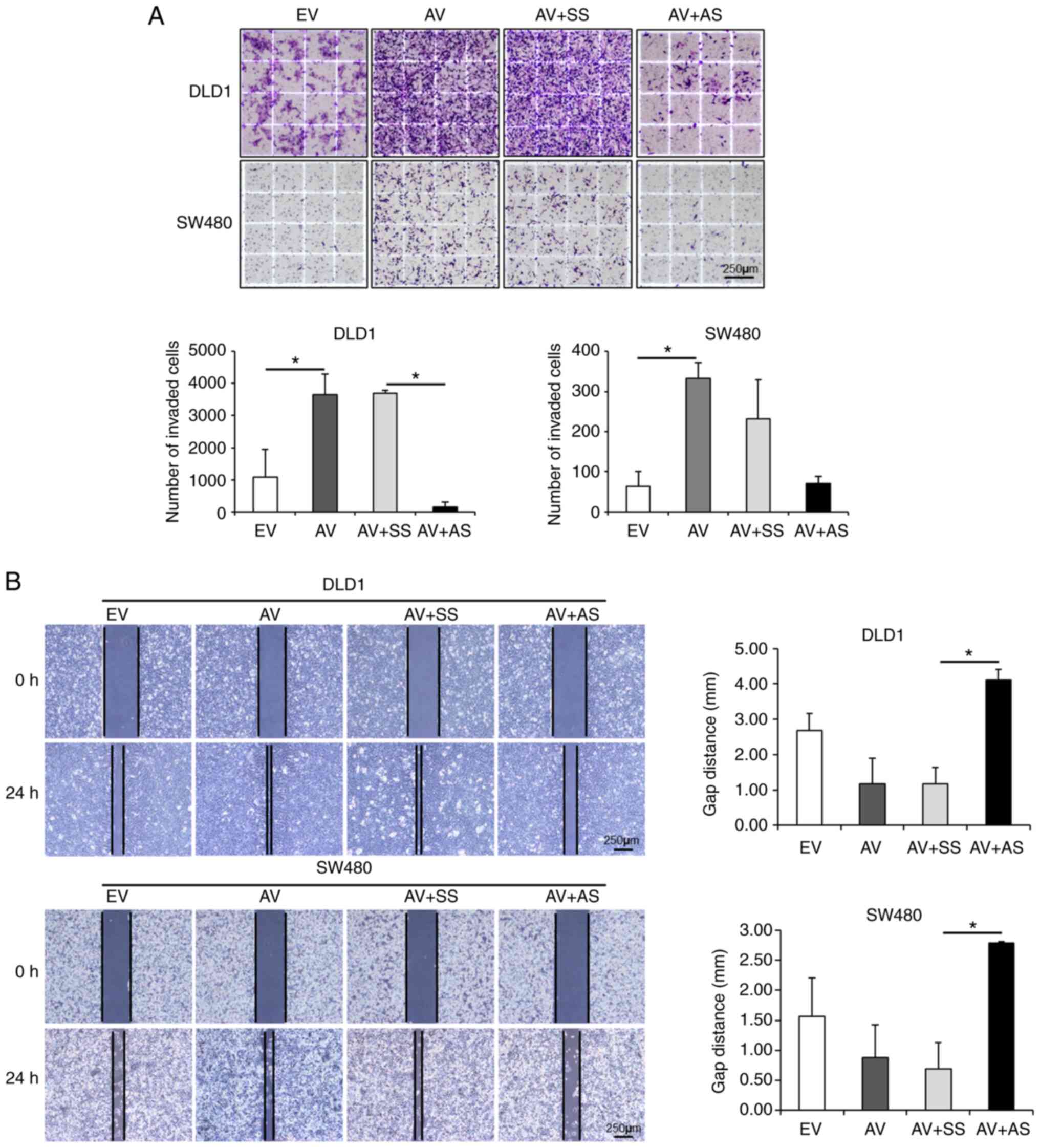
ADAM12 enhances the invasion and
migration of human CRC cells
The present study then assessed the effects of
ADAM12 on cell invasion. The number of invasive cells was
significantly increased in DLD1 and SW480 cells overexpressing
ADAM12 via ADAM12-pcDNA6-myc transfection compared with that in
empty-pcDNA6-myc-transfected cells (P<0.001 and P=0.011,
respectively). By contrast, the number of invasive
ADAM12-siRNA-transfected DLD1 was significantly decreased compared
with that in the scrambled siRNA-transfected cells (P<0.001),
whereas the number of invasive ADAM12-siRNA-transfected SW480 cells
was not significant (P=0.054; Fig.
2A). According to the cell migration assay, the artificial
wound gap became narrower in the ADAM12-pcDNA6-myc-transfected
cells compared with that in the empty-pcDNA6-myc-transfected cells
at 24 h in DLD1 cells and 48 h in SW480 cells, but this was not
statistically significant (P=0.232 and P=0.679, respectively). By
contrast, the artificial wound gap was significantly wider in the
ADAM12-siRNA-transfected cells compared with that in the scrambled
siRNA-transfected cells at 24 h in DLD1 cells and 48 h in SW480
cells (P=0.015 and P=0.048, respectively; Fig. 2B).
ADAM12 participates in the EMT process of
human CRC cells
To examine the relationship between EMT and ADAM12
expression in human CRC cells, cell adhesion assays were performed.
Cell adhesion capacity was assessed post-transfection with
ADAM12-pcDNA6-myc constructs or ADAM12-siRNA using the ECM factors
fibronectin and collagen I. Adhesion capacity to fibronectin and
collagen I was significantly elevated in
ADAM12-pcDNA6-myc-transfected DLD1 and SW480 cells compared with
that in empty-pcDNA6-myc-transfected cells (DLD1: both P<0.001;
SW480: P=0.038 and P=0.041, respectively; Fig. 3A). By contrast, the adhesion to
fibronectin and collagen I was significantly diminished in the
ADAM12-siRNA-transfected DLD1 and SW480 cells compared with that in
the scrambled siRNA-transfected cells (DLD1: P<0.001 and
P=0.002; SW480: P=0.008 and P<0.001).
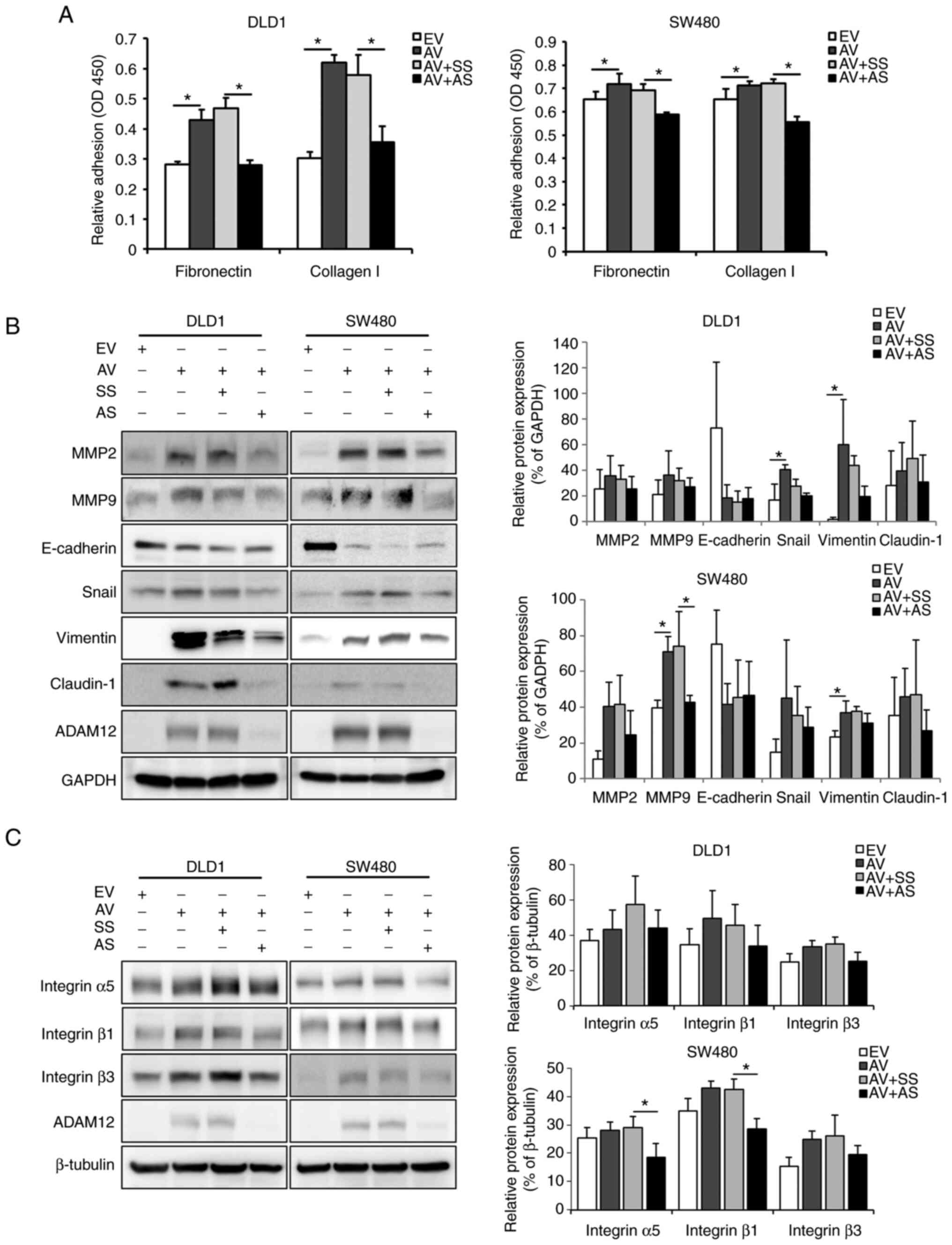 | Figure 3Effects of ADAM12 on the EMT process
in human colorectal cancer cells. (A) Cell adhesion was measured
after AV construct or AS transfection using two cell adhesion
substrates, fibronectin and collagen I. The adherent cells
underwent the WST-1 assay and adhesion was quantified by measuring
the absorbance at 450 nm using a plate reader. (B) Representative
western blot images and semi-quantification graphs of
EMT-associated proteins including MMP2, MMP9, E-cadherin, Snail,
vimentin and claudin-1 in AV-transfected or AS-transfected DLD1 or
SW480 cells. (C) Representative western blot images and
semi-quantification graphs of basement membrane proteins, including
integrin α5, integrin β1 and integrin β3 in AV-transfected or
AS-transfected DLD1 or SW480 cells. Data are presented as the mean
± SD (n=3). *P<0.05. ADAM12, a disintegrin and
metalloprotease 12; EMT, epithelial-mesenchymal transition; EV,
empty pcDNA6-myc vector; AV, ADAM12-pcDNA6-myc construct; SS,
scrambled siRNA; AS, ADAM12 siRNA; siRNA, small interfering RNA;
MMP, matrix metalloproteinase. |
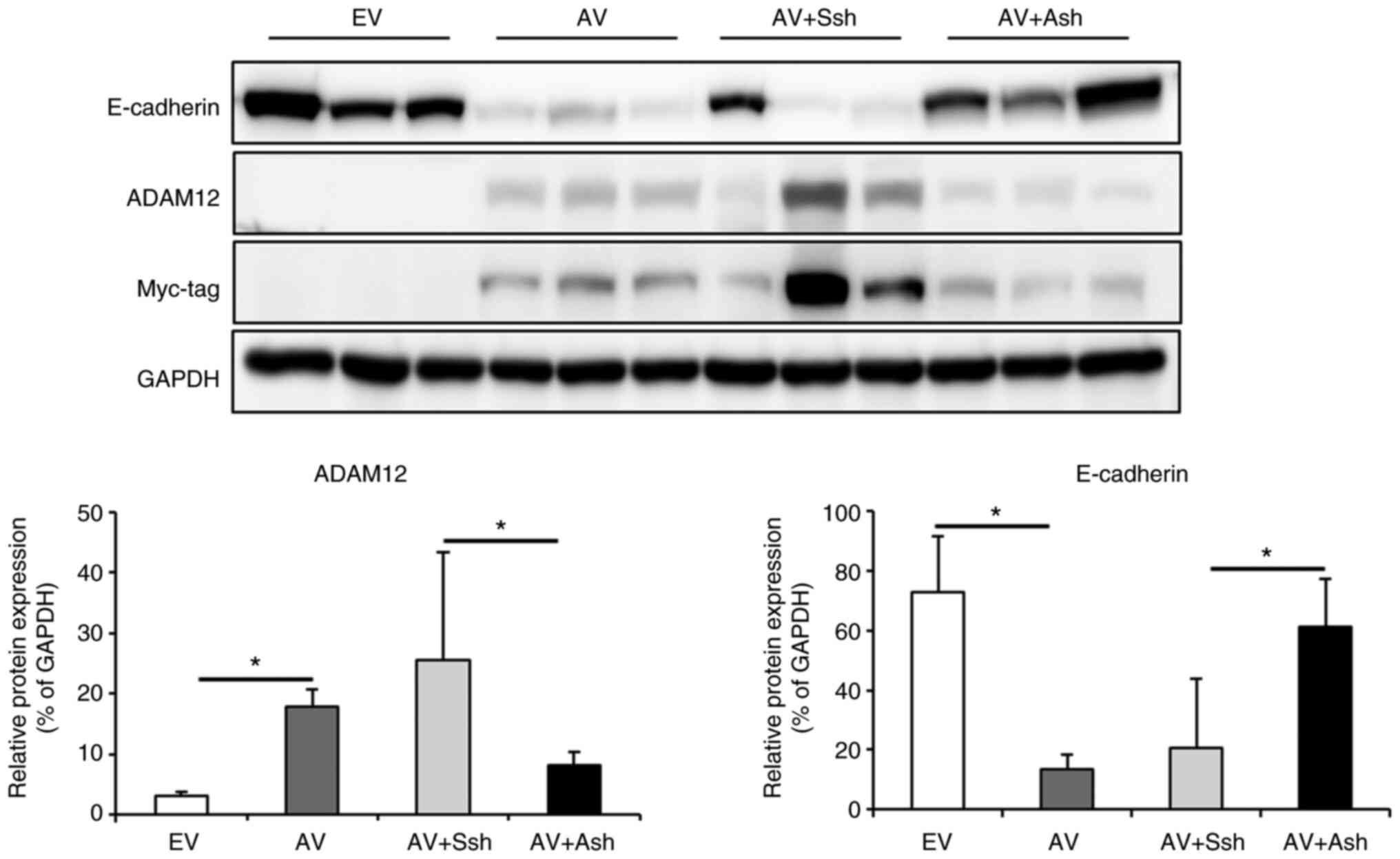
To confirm the association between ADAM12-induced
phenotypic changes and EMT in CRC cells, the expression levels of
the following EMT-related proteins were assessed: E-cadherin, MMP2,
MMP9, Snail, claudin-1 and vimentin. The protein expression levels
of vimentin, MMP2, MMP9, claudin-1 and Snail were increased, and
E-cadherin was decreased in the ADAM12-pcDNA6-myc-transfected cells
compared with those in the empty-pcDNA6-myc-transfected cells. The
protein expression levels of Snail (P=0.021) and vimentin (P=0.014)
were significantly increased in ADAM12-pcDNA6-myc-transfected DLD1
cells and, the protein expression levels of MMP9 (P=0.033) and
vimentin (P=0.036) were significantly increased in
ADAM12-pcDNA6-myc-transfected SW480 cells (Fig. 3B). By contrast, the protein
expression levels of vimentin, MMP2, MMP9, claudin-1 and Snail were
decreased, and E-cadherin was enhanced in the
ADAM12-siRNA-transfected cells compared with those in the scrambled
siRNA-transfected cells. Statistical significance was only shown
for MMP9 protein expression in ADAM12-siRNA-transfected SW480 cells
(P=0.035; Fig. 3B).
The expression levels of basement membrane (BM)
proteins (integrin α5, integrin β1 and integrin β3) were also
assessed. The expression levels of integrin α5, integrin β1 and
integrin β3 were higher in the ADAM12-pcDNA6-myc-transfected DLD1
and SW480 cells compared with those in the
empty-pcDNA6-myc-transfected cells, but this was not significant
(Fig. 3C). Conversely, the
expression levels of integrin α5, integrin β1 and integrin β3 were
lower in the ADAM12 siRNA-transfected DLD1 and SW480 cells compared
with those in the scrambled siRNA-transfected cells, Statistical
significance was only shown for integrin α5 (P=0.045) and integrin
β1 (P=0.007) protein expression in ADAM12-siRNA-transfected SW480
cells (Fig. 3C).
ADAM12 activates PI3K/PDK1/AKT and
PI3K/AKT/GSK-3β signaling pathways
The phosphorylation levels of PDK1, GSK-3β and AKT
were measured by western blotting to evaluate whether ADAM12
triggered intracellular signaling pathways in the human CRC cells.
The expression levels of p-PDK1, p-GSK-3β and p-AKT were higher in
the ADAM12-pcDNA6-myc-transfected DLD1 and SW480 cells compared
with those in the empty-pcDNA6-myc-transfected cells. Statistical
significance was found for p-AKT (P=0.048) and p-GSK-3β (P=0.022)
in ADAM12-pcDNA6-myc-transfected DLD1 cells, but no significant
differences were determined in SW480 cells (Fig. 4). By contrast, the expression
levels of p-PDK1, p-GSK-3β and p-AKT were lower in the ADAM12
siRNA-transfected DLD1 and SW480 cells than in the scrambled
siRNA-transfected cells. Statistical significance was only shown
for p-GSK-3β protein expression in ADAM12-siRNA-transfected DLD1
(P=0.039) and SW480 (P=0.015) cells (Fig. 4).
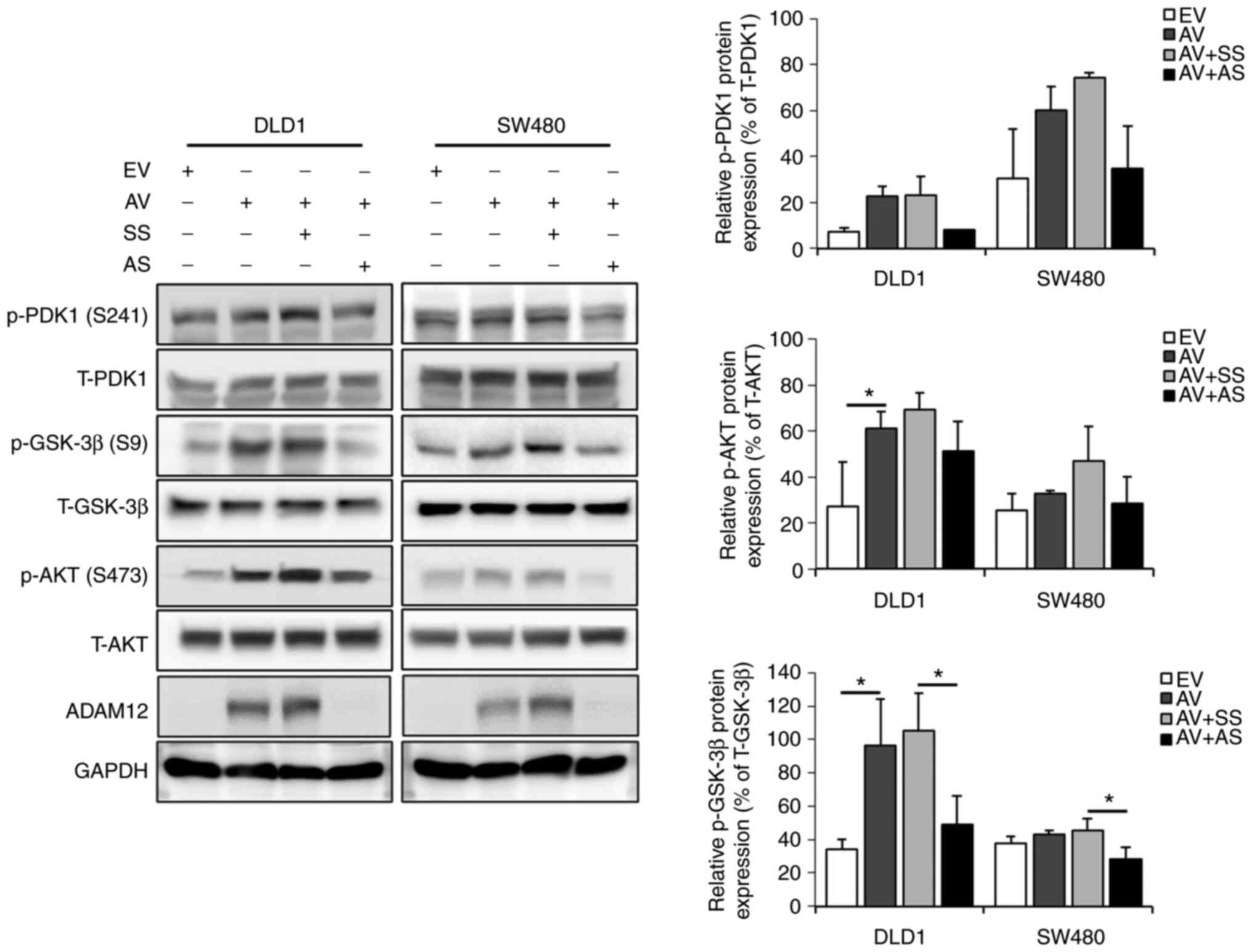 | Figure 4Effects of ADAM12 on oncogenic
signaling pathways in the human colorectal cancer cells.
Representative western blot images and semi-quantification graphs
for the phosphorylation levels of PDK1, GSK-3β and AKT in
AV-transfected or AS-transfected DLD1 or SW480 cells. Data are
presented as the mean ± SD (n=3). *P<0.05. ADAM12, a
disintegrin and metalloprotease 12; PDK1,
phosphoinositide-dependent protein kinase 1; GSK-3β, glycogen
synthase kinase-3β; EV, empty pcDNA6-myc vector; AV,
ADAM12-pcDNA6-myc construct; SS, scrambled siRNA; AS, ADAM12 siRNA;
siRNA, small interfering RNA; p-, phosphorylated; T-, total. |
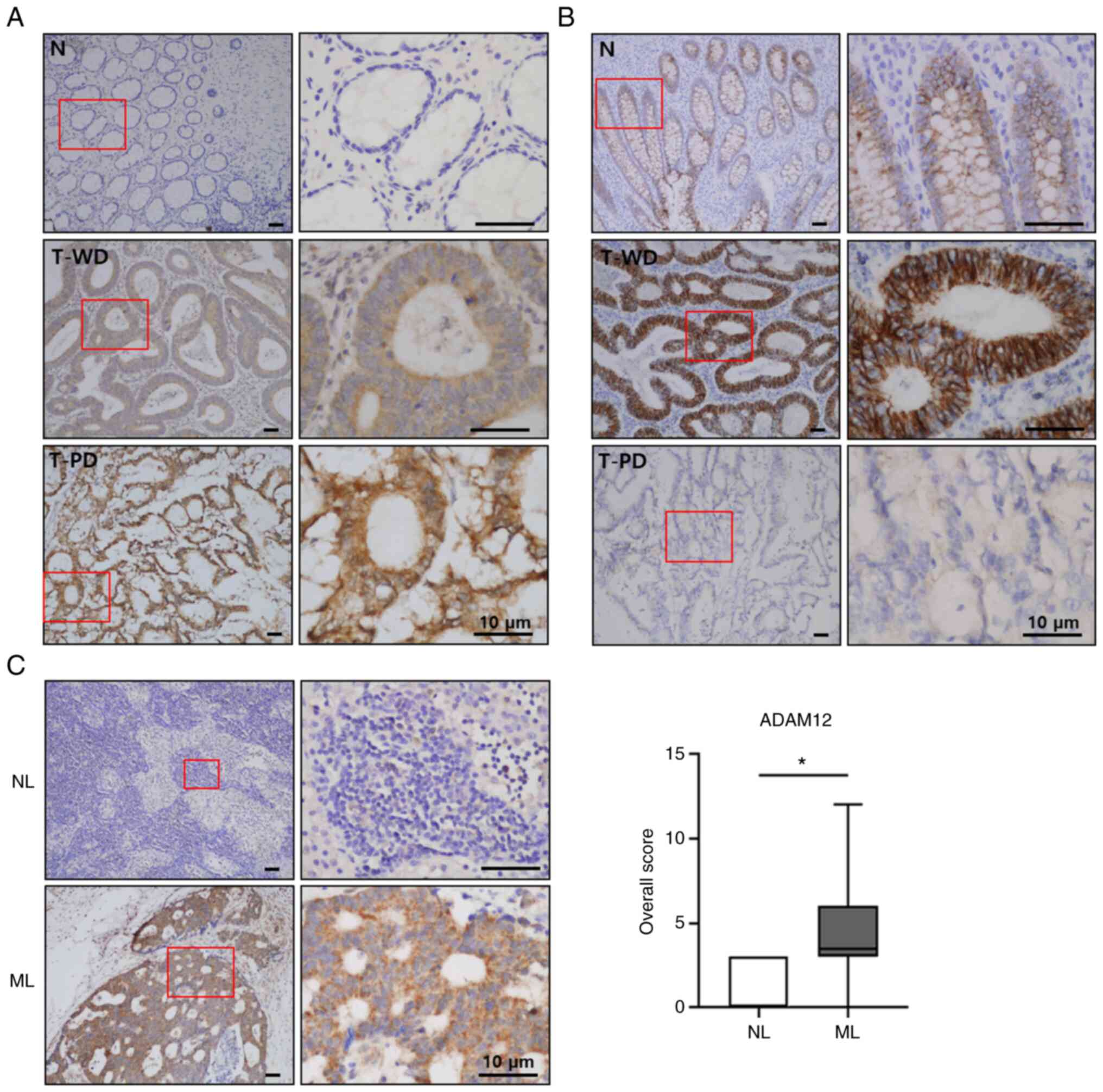
ADAM12 and E-cadherin expression is
associated with the metastasis of CRC
In order to validate the findings of the CRC cell
line studies, the protein expression levels of ADAM12 and
E-cadherin were detected in the human CRC tissues using IHC. From
the same individuals, CRC tissue, normal colonic mucosa, and
metastatic or non-metastatic lymph node tissues were obtained via
surgical specimens and endoscopic biopsies. ADAM12 was weakly or
not expressed in the normal colorectal mucosa, whereas ADAM12 was
predominantly expressed in the cytoplasm of CRC cells but was not
found in the tumor stroma (Fig.
5A). In the mucosa of noncancerous areas, the epithelial cells
showed high membranous E-cadherin expression at cell-cell
boundaries, which is the normal localization of this intercellular
adhesion molecule, which served as an internal positive control.
The stroma and nuclei of normal epithelium mucosa did not exhibit
E-cadherin expression. Generally, a normal colon epithelium
displays strong E-cadherin expression. The CRC tissues were
positive for E-cadherin expression and it was predominantly
associated with cell-cell boundaries as in the normal epithelium
(Fig. 5B). The expression of
ADAM12 was significantly increased in the metastatic lymph node
tissues compared with in the non-metastatic lymph node tissues
(P=0.003; Fig. 5C).
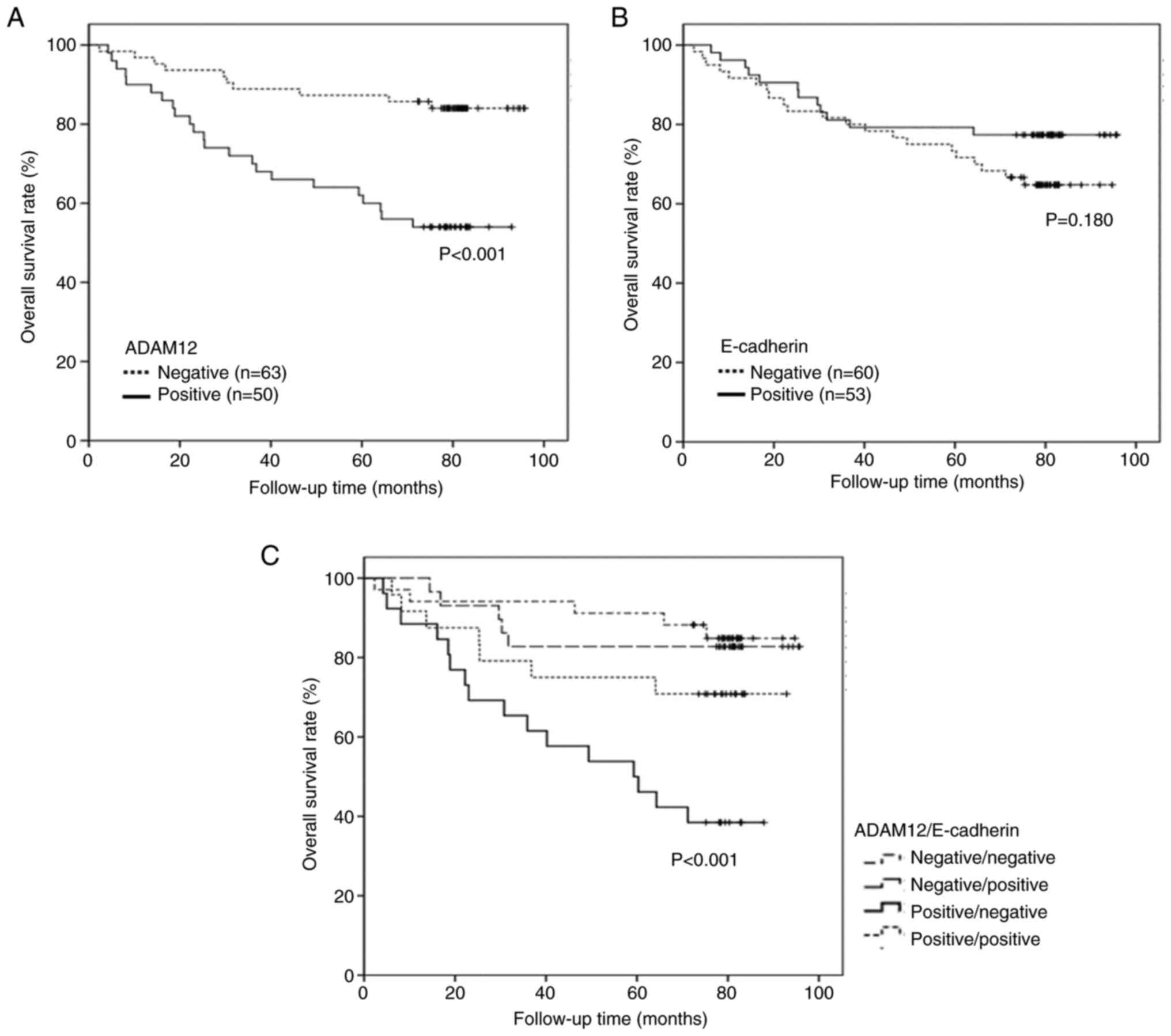
Association between ADAM12 or E-cadherin
expression and clinicopathological features
The association between ADAM12 or E-cadherin
expression and the clinicopathological features of patients are
summarized in Table I. Advanced
cancer stage, lymph node metastasis, distant metastasis and poor
survival (P=0.016, P=0.048, P=0.022 and P<0.001, respectively)
were significantly associated with positive ADAM12 expression
(Table I; Fig. 6A). Lack of E-cadherin expression
showed significant association with undifferentiated tumors,
perineural invasion, advanced cancer stage and lymph node
metastasis, but was not associated with poor survival (P=0.026,
P=0.002, P=0.007, P=0.018 and P=0.180, respectively; Table I; Fig.
6B). ADAM12 expression was not associated with E-cadherin
expression (P=0.835; Table II).
According to the combined analysis of ADAM12 and E-cadherin
expression, positive ADAM12 and negative E-cadherin expressions
were significantly associated with poorer survival than other
expression statuses of both proteins (P<0.001; Table III; Fig. 6C).
 | Table IAssociation between ADAM12 or
E-cadherin expression and the clinicopathological parameters of
patients with colorectal cancer. |
Table I
Association between ADAM12 or
E-cadherin expression and the clinicopathological parameters of
patients with colorectal cancer.
| Parameters | Total (n=113) | ADAM12
| E-cadherin
|
|---|
| Negative
(n=63) | Positive
(n=50) | P-value | Negative
(n=60) | Positive
(n=53) | P-value |
|---|
| Age, years | | | | 0.962 | | | 0.864 |
| <70.5 | 50 | 28 | 22 | | 27 | 23 | |
| ≥70.5 | 63 | 35 | 28 | | 33 | 30 | |
| Sex | | | | 0.160 | | | 0.871 |
| Male | 67 | 41 | 26 | | 36 | 31 | |
| Female | 46 | 22 | 24 | | 24 | 22 | |
| Tumor size, cm | | | | 0.973 | | | 0.731 |
| <4.5 | 68 | 38 | 30 | | 37 | 31 | |
| ≥4.5 | 45 | 25 | 20 | | 23 | 22 | |
| Histological
type | | | | 0.849 | | | 0.026 |
|
Differentiated | 101 | 56 | 45 | | 50 | 51 | |
|
Undifferentiated | 12 | 7 | 5 | | 10 | 2 | |
| Lymphovascular
invasion | | | | 0.730 | | | 0.167 |
| Negative | 92 | 52 | 40 | | 46 | 46 | |
| Positive | 21 | 11 | 10 | | 14 | 7 | |
| Perineural
invasion | | | | 0.093 | | | 0.002 |
| Negative | 75 | 46 | 29 | | 32 | 43 | |
| Positive | 38 | 17 | 21 | | 28 | 10 | |
| Stage | | | | 0.016 | | | 0.007 |
| I/II | 55 | 37 | 18 | | 22 | 33 | |
| III/IV | 58 | 26 | 32 | | 38 | 20 | |
| Depth of
invasion | | | | 0.315 | | | 0.834 |
| T1/T2 | 26 | 18 | 8 | | 12 | 14 | |
| T3/T4 | 87 | 45 | 42 | | 48 | 39 | |
| Lymph node
metastasis | | | | 0.048 | | | 0.018 |
| N0 | 57 | 37 | 20 | | 24 | 33 | |
| N1-3 | 56 | 26 | 30 | | 36 | 20 | |
| Distant
metastasis | | | | 0.022 | | | 0.167 |
| M0 | 92 | 56 | 36 | | 46 | 46 | |
| M1 | 21 | 7 | 14 | | 14 | 7 | |
 | Table IIAssociation between ADAM12 and
E-cadherin expression in human colorectal cancer. |
Table II
Association between ADAM12 and
E-cadherin expression in human colorectal cancer.
| ADAM12
expression | E-cadherin
expression
| P-value |
|---|
| Positive
(n=53) | Negative
(n=60) |
|---|
| Positive
(n=50) | 24 | 26 | 0.835 |
| Negative
(n=63) | 29 | 34 | |
 | Table IIIAssociation between survival and
ADAM12 or E-cadherin expression in human colorectal cancer. |
Table III
Association between survival and
ADAM12 or E-cadherin expression in human colorectal cancer.
| Parameters | Total (n=113) | Survival, months
| P-value |
|---|
| Mean ± SD | Range |
|---|
| ADAM12
expression | | | | <0.001 |
| Negative | 63 | 85.61±3.14 | 2.3-95.7 | |
| Positive | 50 | 64.31±4.82 | 4.2-92.9 | |
| E-cadherin
expression | | | | 0.180 |
| Negative | 60 | 73.63±4.12 | 2.3-94.7 | |
| Positive | 53 | 79.74±4.17 | 6.1-95.7 | |
| ADAM12/E-cadherin
expression | | | | <0.001 |
|
Negative/Negative | 34 | 86.57±3.85 | 2.3-94.7 | |
|
Negative/Positive | 29 | 83.43±5.02 | 14.4-95.7 | |
|
Positive/Negative | 26 | 54.09±6.19 | 4.2-87.9 | |
|
Positive/Positive | 24 | 73.29±6.57 | 6.1-92.9 | |
ADAM12 enhances metastasis of human CRC
cells in an in vivo mouse xenograft model
To investigate the role of ADAM12 in tumor
metastasis in vivo, human CRC SW480 cells transfected with
empty-pcDNA6-myc, ADAM12-pcDNA6-myc, scrambled pGFP-C-shLenti or
ADAM12-pGFP-C-shLenti were injected into the NOD/SCID mice. After
28 days, the mice injected with the ADAM12-pcDNA6-myc-transfected
cells exhibited a significantly higher number of peritoneal
nodules, PCI and tumor weight compared with those in the mice
injected with the empty-pcDNA6-myc-transfected cells (P=0.035,
P=0.007 and P=0.001, respectively; Fig. 7A-D). By contrast, the mice injected
with the ADAM12-pGFP-C-shLenti-transfected cells showed
significantly lower PCI and tumor weight compared with those in the
mice injected with the scrambled pGFP-C-shLenti-transfected cells
(P=0.034 and P=0.039, respectively; Fig. 7A-D).
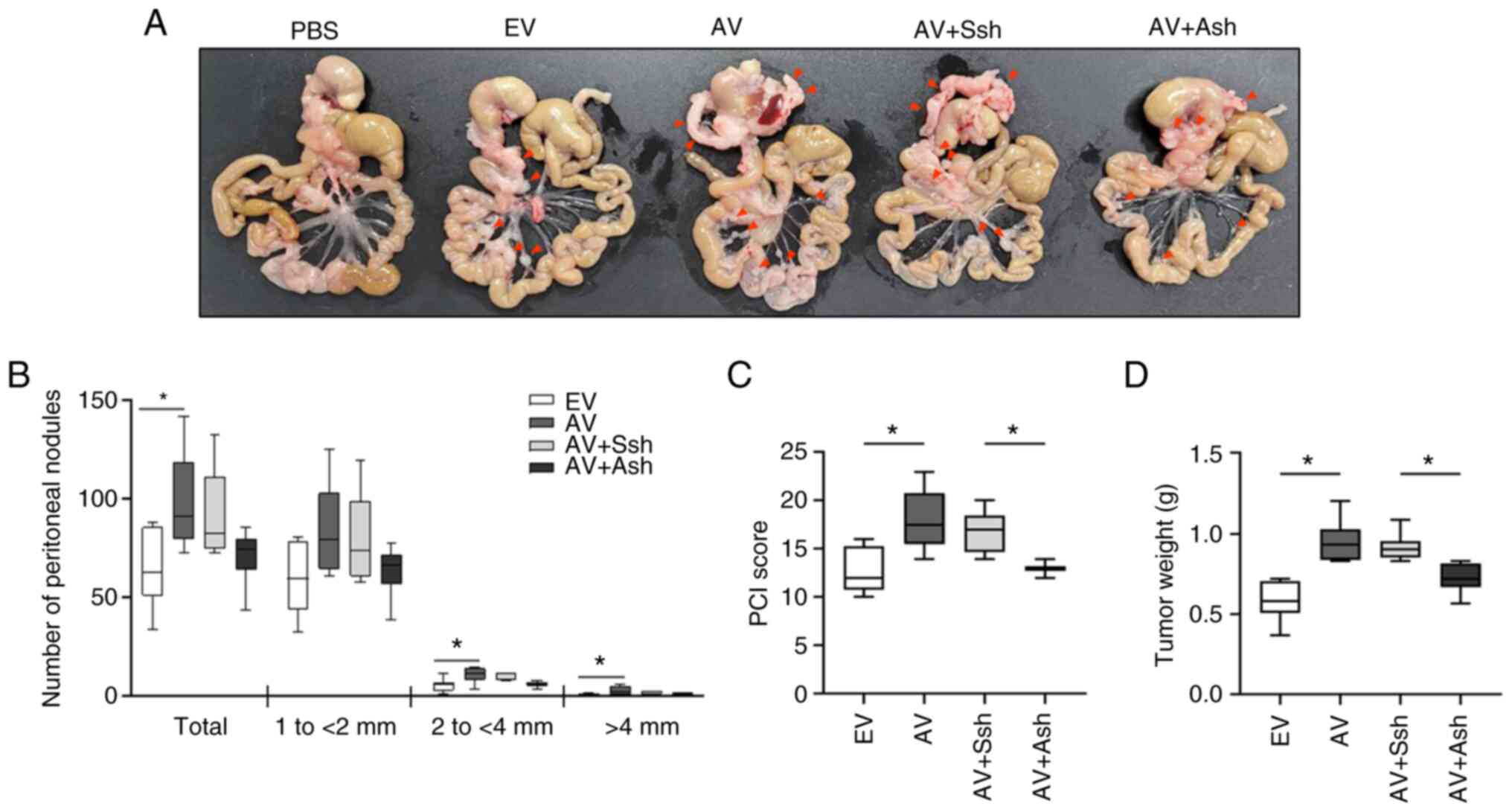 | Figure 7Effects of ADAM12 on the metastasis
of human CRC cells in the mouse model of peritoneal metastasis. The
same number of EV-, AV construct-, Ssh- or Ash-transfected SW480
cells were injected intraperitoneally into non-obese
diabetic/severe combined immunodeficiency mice (n=6/group). (A)
Representative images of peritoneal nodules in the mice injected
with EV-, AV-, Ssh- and Ash-transfected cells. After 28 days, the
(B) number of peritoneal nodules, (C) PCI and (D) tumor weight in
the mice injected with the AV-transfected cells were higher than in
the mice injected with the EV-transfected cells. By contrast, the
number of peritoneal nodules, PCI and tumor weight in the mice
injected with the Ash-transfected cells were lower than in the mice
injected with the Ssh-transfected cells. Data are presented as the
median ± interquartile range. *P<0.05. ADAM12, a
disintegrin and metalloprotease 12; EV, empty pcDNA6-myc vector;
AV, ADAM12-pcDNA6-myc construct; Ssh, scrambled pGFP-C-shLenti
vector; Ash, ADAM12-pGFP-C-shLenti construct; PCI, peritoneal
carcinomatosis index. |
The present study also investigated whether ADAM12
affected the EMT process in vivo via western blotting. Since
ADAM12 overexpression was induced using a Myc-tagged ADAM12 vector,
the Myc-tag antibody was used to confirm that the mouse tumor
tissue was stable SW480 cell-derived tumor tissue. The mice
injected with the ADAM12-pcDNA6-myc-transfected cells exhibited
significantly increased ADAM12 expression and decreased E-cadherin
expression compared with that in the mice injected with the
empty-pcDNA6-myc-transfected cells (P=0.043 and P=0.005,
respectively; Fig. 8). Conversely,
the mice injected with the ADAM12-pGFP-C-shLenti-transfected cells
exhibited significantly decreased ADAM12 expression and increased
E-cadherin expression compared with that in the mice injected with
the scrambled pGFP-C-shLenti-transfected cells (P=0.004 and
P=0.040, respectively; Fig. 8).
These data suggested that ADAM12 may promote the EMT process and
has a potential implication for the peritoneal metastasis of
CRC.
Discussion
CRC metastases occur via direct extension,
vascular/lymphatic spread, portal venous spread and peritoneal
dissemination. Metastases significantly affect the mortality rate
and pose a significant challenge for cancer therapies (2-4,6-8).
ADAM12 expression and EMT are associated with cancer
metastasis and poor prognoses in various types of cancer, including
CRC (10-14,18-22,28-31).
ADAM12 has been reported to induce EMT, which can lead to tumor
formation and metastasis in various types of cancer, including
gastric cancer, pituitary adenoma and breast cancer, indicating its
oncogenic role in carcinogenesis (37-43).
However, the relationship between ADAM12 and EMT in CRC metastasis
remains unclear.
In the present study, ADAM12 overexpression was
associated with enhanced proliferation, migration, invasion and
adhesion of CRC cells. Cancer metastasis is a complex process,
including the detachment of metastatic cells from the primary
tumor, the spread of cells to distant organs or tissues via blood
and lymphatic vasculature, and the settlement and colonization of
cells at distant organs or tissues via migration, invasion and
adhesion processes (44). These
metastatic processes are interconnected and are affected by a
number of genetic and epigenetic alterations of proto-oncogenes and
tumor suppressor genes. The results of this study suggested that
ADAM12 may contribute to CRC metastasis via the induction of
oncogenic phenotypes.
The EMT process is aberrantly reactivated in cancer,
causing epithelial cells to lose their junctions and polarity, and
gain mesenchymal properties and invasive abilities (10-14).
Additionally, cancer metastasis is significantly influenced by the
tumor microenvironment, including the ECM, chemokines, growth
factors and MMPs (45). The ECM
serves as a crucial barrier against the potentially pathogenic
migration of cells, and is crucial for maintaining tissue function
and integrity (46). However,
altered expression of ECM macromolecules in the tumor
microenvironment can affect cancer cell proliferation and survival,
adhesion and migration (47). In
addition, the BM is a specialized sheet-like ECM structure located
under the endothelial and epithelial tissues, which is composed of
proteins including collagen, laminin and integrin (11,48,49).
The breakdown of normal ECM and BM is an essential part of
carcinogenesis and metastasis, acting as a key driver for cancer
progression (11,50). MMPs are known to cleave components
of the BM and participate in EMT-related processes (11,49).
In the present study, there was an increase in the expression
levels of mesenchymal markers, such as vimentin, MMP2, MMP9,
claudin-1 and Snail, and a decrease in the expression levels of the
epithelial marker E-cadherin in CRC cells overexpressing ADAM12.
These results are in concordance with the protein expression levels
revealed to be associated with EMT in previous studies (10,13,14).
In addition, the present study revealed that the expression levels
of BM proteins, such as integrin α5 and integrin β1, were increased
in the CRC cells overexpressing ADAM12. It has been proven in
preclinical trials that inhibiting the ligand of these integrins
using antagonists, such as arginylglycylaspartic acid, can result
in tumor inhibition (11). The
results of the present study indicated that ADAM12 may participate
in the EMT process, and could be a potential target for cancer
metastasis prevention and inhibition.
The present study investigated how ADAM12 stimulated
several intracellular signaling pathways that regulate EMT, in
order to clarify the mechanisms that may lead to an ADAM12
overexpression-induced increase in EMT. ADAM12 overexpression led
to increased phosphorylation levels of PDK1, GSK-3β and AKT.
Activation of the PI3K/AKT signaling pathway is closely linked to
cancer invasion, migration and EMT, indicating that it has a role
in the aggressiveness of malignancies and is a well-known driver of
tumorigenesis (51,52). PDK1 is one of the key components of
the PI3K/AKT signaling pathway. The activation of PIK sends a
signal to PDK1, which leads to activation of the AKT signaling
pathway (53,54). GSK-3β act as the central hub that
orchestrates signals from the PI3K/AKT signaling pathway to
initiate regulatory effects on cancer initiation, EMT and
resistance to therapies. PI3K/AKT/GSK-3β signaling is critical for
tumor metastasis via modulating the EMT induction in multiple types
of cancer (55,56). The results of the present study
indicated that ADAM12 may be associated with the EMT process of
human CRC cells by activating the PI3K/PDK1/AKT and PI3K/AKT/GSK-3β
signaling pathways.
Previous studies have reported that upregulation of
ADAM12 is associated with poor survival (29,30),
whereas reduced expression of E-cadherin has been revealed to be
associated with poor prognosis in patients with CRC (13). In the present study, upregulated
ADAM12 expression was observed in the CRC and metastatic lymph node
tissues compared with in normal mucosa and non-metastatic lymph
node tissues. Increased ADAM12 expression was significantly
associated with advanced cancer stage, lymph node metastasis,
distant metastasis and poor survival. Reduced E-cadherin expression
was significantly associated with undifferentiated tumors,
perineural invasion, advanced cancer stage and lymph node
metastasis, but was not associated with poor survival. In addition,
both increased ADAM12 and reduced E-cadherin expression were
significantly associated with poorer survival compared with other
expression levels of both proteins.
Finally, based on the results of in vitro and
human clinical data, the present study investigated the molecular
mechanism of EMT mediated by ADAM12 in the metastasis of human CRC
in an in vivo mouse tumor model. The number of peritoneal
nodules, tumor weight and PCI in the mice injected with the
ADAM12-pcDNA6-myc-transfected cells were significantly higher than
those in the mice injected with the empty-pcDNA6-myc-transfected
cells. By contrast, tumor weight and PCI in the mice injected with
the ADAM12-pGFP-C-shLenti-transfected cells were significantly
lower than those in the mice injected with the scrambled
pGFP-C-shLenti-transfected cells. Subsequently, the present study
investigated whether ADAM12 affected the EMT process in the in
vivo tumor model via western blotting. The mice injected with
ADAM12-pcDNA6-myc-transfected cells showed significantly decreased
E-cadherin expression compared with that in the mice injected with
the empty-pcDNA6-myc-transfected cells. The mice injected with the
ADAM12-pGFP-C-shLenti-transfected cells showed increased E-cadherin
expression compared with that in the mice injected with the
scrambled pGFP-C-shLenti-transfected cells.
In conclusion, ADAM12 overexpression contributed to
CRC metastasis by regulating EMT. Furthermore, ADAM12 knockdown
exhibited potent anti-metastatic activity in a mouse model of
peritoneal metastasis showing the possibility of ADAM12 as a
therapeutic target in the treatment and prevention of CRC
metastasis.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HHO and YEJ conceived and designed the present
study. HHO, YLP and SYP performed the experiments. HHO and YLP
collected and analyzed the data. HHO and YEJ wrote, reviewed and/or
revised the manuscript. HHO and YLP confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The patient sample study complied with the
guidelines and protocols approved by the Institutional Review Board
(IRB) of Chonnam National University Hwasun Hospital (IRB no.
CNUHH-2017-164). Written informed consent was obtained prior to
sample collection from all of the participants who agreed to the
use of their samples in scientific research, and procedures were
conducted according to The Declaration of Helsinki. All animal
experiments were performed in accordance with Chonnam National
University Medical School's guidelines for animal experiments, and
the protocols were approved by Chonnam National University Medical
School's Animal Care Committee (approval no. CNU
IACUC-H-2021-14).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hossain MS, Karuniawati H, Jairoun AA,
Urbi Z, Ooi DJ, John A, Lim YC, Kibria KMK, Mohiuddin AKM, Ming LC,
et al: Colorectal cancer: A review of carcinogenesis, global
epidemiology, current challenges, risk factors, preventive and
treatment strategies. Cancers (Basel). 14:17322022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sullivan BA, Noujaim M and Roper J: Cause,
epidemiology, and histology of polyps and pathways to colorectal
cancer. Gastrointest Endosc Clin N Am. 32:177–194. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mattiuzzi C, Sanchis-Gomar F and Lippi G:
Concise update on colorectal cancer epidemiology. Ann Transl Med.
7:6092019. View Article : Google Scholar
|
|
5
|
Van Cutsem E, Cervantes A, Adam R, Sobrero
A, Van Krieken JH, Aderka D, Aguilar EA, Bardelli A, Benson A,
Bodoky G, et al: ESMO consensus guidelines for the management of
patients with metastatic colorectal cancer. Ann Oncol.
27:1386–1422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bijelic L, Ramos I and Goeré D: The
landmark series: Surgical treatment of colorectal cancer peritoneal
metastases. Ann Surg Oncol. 28:4140–4150. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sugarbaker PH: Prevention and treatment of
peritoneal metastases: A comprehensive review. Indian J Surg Oncol.
10:3–23. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parikh MS, Johnson P, Romanes JP, Freitag
HE, Spring ME, Garcia-Henriquez N and Monson JRT: Cytoreductive
surgery and hyperthermic intraperitoneal chemotherapy for
colorectal peritoneal metastases: A systematic review. Dis Colon
Rectum. 65:16–26. 2022. View Article : Google Scholar
|
|
9
|
Kciuk M, Gielecińska A, Budzinska A,
Mojzych M and Kontek R: Metastasis and MAPK Pathways. Int J Mol
Sci. 23:38472022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
LeBleu VS and Thiery JP: The continuing
search for causality between epithelial-to-mesenchymal transition
and the metastatic fitness of carcinoma cells. Cancer Res.
82:1467–1469. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Banerjee S, Lo WC, Majumder P, Roy D,
Ghorai M, Shaikh NK, Kant N, Shekhawat MS, Gadekar VS, Ghosh S, et
al: Multiple roles for basement membrane proteins in cancer
progression and EMT. Eur J Cell Biol. 101:1512202022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han L, Wang S, Wei C, Fang Y, Huang S, Yin
T, Xiong B and Yang C: Tumour microenvironment: A non-negligible
driver for epithelial-mesenchymal transition in colorectal cancer.
Expert Rev Mol Med. 23:e162021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ni Q, Li M and Yu S: Research progress of
epithelial-mesenchymal transition treatment and drug resistance in
colorectal cancer. Technol Cancer Res Treat.
21:153303382210812192022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang N, Ng AS, Cai S, Li Q, Yang L and
Kerr D: Novel therapeutic strategies: Targeting
epithelial-mesenchymal transition in colorectal cancer. Lancet
Oncol. 22:e358–e368. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wolfsberg TG, Straight PD, Gerena RL,
Huovila AP, Primakoff P, Myles DG and White JM: ADAM, a widely
distributed and developmentally regulated gene family encoding
membrane proteins with a disintegrin and metalloprotease domain.
Dev Biol. 169:378–383. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Edwards DR, Handsley MM and Pennington CJ:
The ADAM metalloproteinases. Mol Aspects Med. 29:258–289. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reiss K and Saftig P: The 'a disintegrin
and metalloprotease' (ADAM) family of sheddases: Physiological and
cellular functions. Semin Cell Dev Biol. 20:126–137. 2009.
View Article : Google Scholar
|
|
18
|
Ma B, Ma Q, Jin C, Wang X, Zhang G, Zhang
H, Seeger H and Mueck AO: ADAM12 expression predicts clinical
outcome in estrogen receptor-positive breast cancer. Int J Clin Exp
Pathol. 8:13279–13283. 2015.
|
|
19
|
Pan J, Huang Z, Zhang Y and Xu Y: ADAM12
as a clinical prognostic indicator associated with tumor immune
infiltration in lung adenocarcinoma. DNA Cell Biol. 41:410–423.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu H, Jiang W, Zhu H, Hu J, Tang B, Zhou
Z and He X: Elevation of ADAM12 facilitates tumor progression by
enhancing metastasis and immune infiltration in gastric cancer. Int
J Oncol. 60:512022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Du S, Sun L, Wang Y, Zhu W, Gao J, Pei W
and Zhang Y: ADAM12 is an independent predictor of poor prognosis
in liver cancer. Sci Rep. 12:66342022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mendaza S, Ulazia-Garmendia A,
Monreal-Santesteban I, Córdoba A, de Azúa YR, Aguiar B, Beloqui R,
Armendáriz P, Arriola M, Martín-Sánchez E and Guerrero-Setas D:
ADAM12 is a potential therapeutic target regulated by
hypomethylation in triple-negative breast cancer. Int J Mol Sci.
21:9032020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Reiss K, Leitzke S, Seidel J, Sperrhacke M
and Bhakdi S: Scramblases as regulators of proteolytic ADAM
function. Membranes (Basel). 12:1852022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zadka L, Kulus MJ and Piatek K: ADAM
protein family-its role in tumorigenesis, mechanisms of
chemoresistance and potential as diagnostic and prognostic factors.
Neoplasma. 65:823–839. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kveiborg M, Albrechtsen R, Couchman JR and
Wewer UM: Cellular roles of ADAM12 in health and disease. Int J
Biochem Cell Biol. 40:1685–1702. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jacobsen J and Wewer UM: Targeting ADAM12
in human disease: Head, body or tail? Curr Pharm Des. 15:2300–2310.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nyren-Erickson EK, Jones JM, Srivastava DK
and Mallik S: A disintegrin and metalloproteinase-12 (ADAM12):
Function, roles in disease progression, and clinical implications.
Biochim Biophys Acta. 1830:4445–4455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park YL, Park SY, Oh HH, Chung MW, Hong
JY, Kim KY, Myung DS, Cho SB, Lee WS, Kim HS and Joo YE: A
disintegrin and metalloprotease 12 promotes tumor progression by
inhibiting apoptosis in human colorectal cancer. Cancers (Basel).
13:19272021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang Z, Lai H, Liao J, Cai J, Li B, Meng
L, Wang W, Mo X and Qin H: Upregulation of ADAM12 is associated
with a poor survival and immune cell infiltration in colon
adenocarcinoma. Front Oncol. 11:7292302021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ten Hoorn S, Waasdorp C, van Oijen MGH,
Damhofer H, Trinh A, Zhao L, Smits LJH, Bootsma S, van Pelt GW,
Mesker WE, et al: Serum-based measurements of stromal activation
through ADAM12 associate with poor prognosis in colorectal cancer.
BMC Cancer. 22:3942022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mochizuki S, Ao T, Sugiura T, Yonemura K,
Shiraishi T, Kajiwara Y, Okamoto K, Shinto E, Okada Y and Ueno H:
Expression and function of a disintegrin and metalloproteinases in
cancer-associated fibroblasts of colorectal cancer. Digestion.
101:18–24. 2020. View Article : Google Scholar
|
|
32
|
Yao Y, Zhou Y, Su X, Dai L, Yu L, Deng H,
Gou L and Yang J: Establishment and characterization of
intraperitoneal xenograft models by co-injection of human tumor
cells and extracellular matrix gel. Oncol Lett. 10:3450–3456. 2015.
View Article : Google Scholar
|
|
33
|
Bastiaenen VP, Klaver CEL, van der Heijden
MCS, Nijman LE, Lecca MC, Tanis PJ, Lenos KJ and Vermeulen L: A
mouse model for peritoneal metastases of colorectal origin
recapitulates patient heterogeneity. Lab Invest. 100:1465–1474.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sugarbaker PH: Intraperitoneal
chemotherapy and cytoreductive surgery for the prevention and
treatment of peritoneal carcinomatosis and sarcomatosis. Semin Surg
Oncol. 14:254–261. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Klaver YL, Hendriks T, Lomme RM, Rutten
HJ, Bleichrodt RP and de Hingh IH: Intraoperative hyperthermic
intraperitoneal chemotherapy after cytoreductive surgery for
peritoneal carcinomatosis in an experimental model. Br J Surg.
97:1874–1880. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Amin MB, Edge SB, Greene FL, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC Cancer Staging Manual. Springer
International Publishing; 2018
|
|
37
|
Chen N, Zhu X, Zhu Y, Shi J, Zhang J, Tang
C and Chen J: The regulatory relationship and function of LncRNA
FAM225A-miR-206-ADAM12 in gastric cancer. Am J Transl Res.
13:8632–8652. 2021.PubMed/NCBI
|
|
38
|
Huang X and Xie X, Liu P, Yang L, Chen B,
Song C, Tang H and Xie X: Adam12 and lnc015192 act as ceRNAs in
breast cancer by regulating miR-34a. Oncogene. 37:6316–6326. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dekky B, Ruff M, Bonnier D, Legagneux V
and Théret N: Proteomic screening identifies the zonula occludens
protein ZO-1 as a new partner for ADAM12 in invadopodia-like
structures. Oncotarget. 9:21366–21382. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang J, Zhang Z, Li R, Mao F, Sun W, Chen
J, Zhang H, Bartsch JW, Shu K and Lei T: ADAM12 induces EMT and
promotes cell migration, invasion and proliferation in pituitary
adenomas via EGFR/ERK signaling pathway. Biomed Pharmacother.
97:1066–1077. 2018. View Article : Google Scholar
|
|
41
|
Eckert MA, Santiago-Medina M, Lwin TM, Kim
J, Courtneidge SA and Yang J: ADAM12 induction by Twist1 promotes
tumor invasion and metastasis via regulation of invadopodia and
focal adhesions. J Cell Sci. 130:2036–2048. 2017.PubMed/NCBI
|
|
42
|
Ruff M, Leyme A, Le Cann F, Bonnier D, Le
Seyec J, Chesnel F, Fattet L, Rimokh R, Baffet G and Théret N: The
disintegrin and metalloprotease ADAM12 is associated with
TGF-β-induced epithelial to mesenchymal transition. PLoS One.
10:e01391792015. View Article : Google Scholar
|
|
43
|
Li H, Duhachek-Muggy S, Dubnicka S and
Zolkiewska A: Metalloproteinase-disintegrin ADAM12 is associated
with a breast tumor-initiating cell phenotype. Breast Cancer Res
Treat. 139:691–703. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guan X: Cancer metastases: Challenges and
opportunities. Acta Pharm Sin B. 5:402–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Neophytou CM, Panagi M, Stylianopoulos T
and Papageorgis P: The role of tumor microenvironment in cancer
metastasis: Molecular mechanisms and therapeutic opportunities.
Cancers (Basel). 13:20532021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Henke E, Nandigama R and Ergün S:
Extracellular matrix in the tumor microenvironment and its impact
on cancer therapy. Front Mol Biosci. 6:1602019. View Article : Google Scholar
|
|
47
|
Popova NV and Jücker M: The functional
role of extracellular matrix proteins in cancer. Cancers (Basel).
14:2382022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Devergne O, Sun GH and Schüpbach T:
Stratum, a homolog of the human GEF Mss4, partnered with Rab8,
controls the basal restriction of basement membrane proteins in
epithelial cells. Cell Rep. 18:1831–1839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Horejs CM: Basement membrane fragments in
the context of the epithelial-to-mesenchymal transition. Eur J Cell
Biol. 95:427–440. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liao Z, Tan ZW, Zhu P and Tan NS:
Cancer-associated fibroblasts in tumor microenvironment-Accomplices
in tumor malignancy. Cell Immunol. 343:1037292019. View Article : Google Scholar
|
|
51
|
Deng S, Leong HC, Datta A, Gopal V, Kumar
AP and Yap CT: PI3K/AKT signaling tips the balance of cytoskeletal
forces for cancer progression. Cancers (Basel). 14:16522022.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Peng Y, Wang Y, Zhou C, Mei W and Zeng C:
PI3K/Akt/mTOR pathway and its role in cancer therapeutics: Are we
making headway? Front Oncol. 12:8191282022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Di Blasio L, Gagliardi PA, Puliafito A and
Primo L: Serine/Threonine kinase 3-phosphoinositide-dependent
protein kinase-1 (PDK1) as a key regulator of cell migration and
cancer dissemination. Cancers (Basel). 9:252017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gagliardi PA, Puliafito A and Primo L:
PDK1: At the crossroad of cancer signaling pathways. Semin Cancer
Biol. 48:27–35. 2018. View Article : Google Scholar
|
|
55
|
Lin J, Song T, Li C and Mao W: GSK-3β in
DNA repair, apoptosis, and resistance of chemotherapy, radiotherapy
of cancer. Biochim Biophys Acta Mol Cell Res. 1867:1186592020.
View Article : Google Scholar
|
|
56
|
Nagini S, Sophia J and Mishra R: Glycogen
synthase kinases: Moonlighting proteins with theranostic potential
in cancer. Semin Cancer Biol. 56:25–36. 2019. View Article : Google Scholar
|