Human leukocyte antigen (HLA) is the expression
product of human major histocompatibility complex (MHC), which is
located on the short arm of chromosome 6p21.31 (1). HLA genes have a complex polygenic
architecture and are highly polymorphic (1). To date, based on the IPD-IMGT/HLA
database (https://www.ebi.ac.uk/ipd/imgt/hla/), >35,000
alleles of HLA have been identified, of which >15,000 functional
genes have been demonstrated to express proteins. The arrangement
of HLA genes on chromosomes is divided into three regions, namely
the class-I, -II and -III gene regions (2). HLA-I molecules are widely distributed
on the surface of human nucleated cells and participate in the
processing and presentation of endogenous antigen peptides
(3). The presentation of
endogenous antigens by HLA-I molecules involves three important
steps: i) The production of antigen polypeptides by proteasome
cleavage; ii) transportation of polypeptides to the endoplasmic
reticulum by transporter associated with antigen processing (TAP);
and iii) polypeptides bind to HLA-I molecule binding channels and
are transported to the cell surface for display (3). By contrast, HLA-II molecules are
predominantly expressed on the surface of antigen-presenting cells
and activated T cells (4). These
HLA-II molecules are mainly involved in the processing and
presentation of exogenous antigens (4). In the endoplasmic reticulum, HLA-II
molecules bind to a peptide chain named the invariant chain to form
a complex to prevent the binding of endogenous antigenic peptides
to HLA-II molecules. In this process, after the constant chain is
degraded by proteases, the exogenous antigen polypeptide binds to
the empty HLA-II molecule to form a complex that is expressed on
the cell surface (4). The immune
response of cytotoxic T cells has been demonstrated to be
restricted by HLA-I molecules (5).
By contrast, HLA-II molecules are involved in immune regulation
through the action of restricted helper T cells (5). HLA-III genes are located between the
class-II and class-I genes, and mainly include genes associated
with the complement system, and tumor necrosis factor (TNF) and
heat shock protein (HSP) genes (6,7). The
abnormal expression of HLA molecules has been demonstrated to
affect antigen processing and presentation, leading to immune
escape and lack of recognition of tumor cells by the body (8,9). In
recent years, an increasing number of studies have focused on HLAs
as potential targets for tumor immunotherapy (8-10).
The effective binding of HLA to antigen epitopes is the key to
immunotherapy. Tumor polypeptide vaccines targeting peptides
recognized jointly by HLA-I and HLA-II molecules can produce more
potent and long-lasting antitumor effects (10). In the present review, the roles and
mechanisms of HLAs in tumor immunotherapy are summarized.
HLA-I genes can be divided into three classical
groups (HLA-A, HLA-B and HLA-C) or the non-classical groups (for
example, HLA-E, HLA-F and HLA-G) (11). HLA-I molecules are composed of a
heavy chain (the α-chain) and a light chain (the β2m chain)
(12). The α-chain is encoded by
HLA-A, B and C alleles, and is divided into the extracellular,
transmembrane and intracellular regions (12). The extracellular region of the
α-chain is formed by the folding of the three functional regions
(namely, α1, α2 and α3), where CD8 binding sites are located on α3
(12). By contrast, the β2m chain,
encoded by the β2 microglobulin gene, is attached to the α3 region
by non-covalent bonds (13).
HLA-II molecules are heterodimers, located near to
the centromeres of chromosomes, which are composed of an αand a
β-chain (14). HLA-D alleles of
HLA-II are divided into HLA-DR, HLA-DQ and HLA-P isotypes, and
there are also DMA, DMB, latent membrane protein 2 (LMP2), LMP7,
TAP1 and TAP2 gene loci (15). At
least 36 genes have been mapped in the class III gene region, among
which the genes associated with the immune system include C4B, C4A,
C2, BF, TNF and HSP70, which encode the C4, C2, factor B, TNF-α,
TNF-β and HSP70 proteins (16-18).
In recent years, greater attention has been paid to
HLA-G and HLA-E, which belong to the group of non-classical HLA-I
molecules and are located at chromosome 6p21.3 (19,20).
During embryonic development, HLA-G is secreted by amniotic cells,
erythroid precursor cells and cytotrophoblasts (21); its main role is to inhibit the
function of maternal natural killer (NK) cells and to prevent NK
cells from attacking allogeneic fetuses with antigenicity (22). Compared with those in normal
pregnancy, the expression levels of HLA-G mRNA and protein in the
placenta of patients with pre-eclampsia and recurrent miscarriages
have been demonstrated to be decreased (23). HLA-G is expressed in immune
privilege sites, including the cornea, thymus, pancreatic islets,
erythroblasts and epithelioid progenitor cells, in healthy
individuals (24). By contrast,
HLA-E has been demonstrated to be mainly expressed on the surface
of endothelial cells, T and B lymphocytes, monocytes and
macrophages, and also at a high level in the amniotic membrane and
trophoblast cells at the maternal-fetal interface (25,26).
HLA-E molecules are the ligands of C-type lectin family receptors
(27). The inhibitory receptors of
C-type lectin family receptors are the CD94/NKG2A receptors, and
the activating receptors are the CD94/NKG2C receptors (28). Under physiological conditions, the
affinity between HLA-E and CD94/NKG2A is significantly higher than
that between HLA-E and CD94/NKG2C (28). Upregulation of HLA-E expression can
protect target cells from being killed by NK cells (29). HLA-G and HLA-E have been found to
be abnormally expressed in autoimmune diseases, viral infections
and inflammation (30-32).
HLA molecules have two main biological functions: A
target function and a recognition function (33). The antigenic specificity of the
HLA-I antigen lies in the specific amino acid sequence of the
antigenic determinant of the peptide chain (34,35).
The recognition function of HLA is associated with the unique
synergy of the immune response (36). Antibodies are produced in B cells,
although, in the majority of cases, macrophages and T lymphocytes
are required to participate in antibody production (8). After the antigen has been processed
by macrophages, the antigen information is transmitted to helper T
cells, which then transmit the information to B cells to ensure
that they can further secrete specific antibodies (37). During this process, helper T cells
not only recognize antigens on sensitized macrophages, but also
recognize whether the macrophages are consistent with their own
class II antigens (38). Helper T
cells are activated only when the haplotypes of macrophages and
helper T cells match consistently, and this mechanism ensures that
the immune response is regulated under strict genetic control
(39).
The majority of febrile non-hemolytic blood
transfusion reactions are caused by HLA, especially in patients who
have multiple blood transfusions (40,41).
HLA matching is the most important standard prior to organ
transplantation, and this is significant in terms of improving the
survival rate for patients with transplanted organs and ensuring
the success of organ transplantations for patients with high
sensitivity to anti-HLA antibodies (42-45).
The purpose of bone marrow transplantation (BMT) is to transplant
hematopoietic stem cells (46,47).
Stem cells have the ability to replicate and differentiate into
hematopoietic and immune active cells (48). BMT is associated with HLA and it is
therefore a requirement that the HLA-A, B, DR and DQ isotypes of
the donor and recipient are as compatible as possible (49).
The HLA complex is highly polymorphic, and the
probability of matching identical phenotypes among unrelated
individuals is extremely low. Therefore, this characteristic may be
used for individual identification in forensic medicine (50). The HLA complex is regarded as a
specific genetic marker that accompanies an individual for life
(50). To date, >60 diseases
have been found to be associated with HLA (51). The precise etiology or pathogenesis
underlying this involvement of HLA with disease remains unknown,
although it is known to be associated with immune abnormalities,
familial tendencies and environmental induction (51). The most notable example is that
>91% of white patients with ankylosing spondylitis carry the
HLA-B27 antigen (52). In
addition, 40% of patients with Hodgkin's are HLA-A1+
(53), 76% of patients with skin
pigmentation conditions are HLA-A3+ (54) and 40% of patients with Behcet's
disease are HLA-B5+ (55).
It can therefore be noted that the physical and
chemical properties and the biological functions of HLA, not only
have important theoretical significance, but they are also of great
biomedical value.
The degree of the association between HLA
polymorphism and tumor risk is affected by ethnicity, and even
results obtained from the same population can be quite different.
The results obtained from Spanish patients with liver disease
demonstrated that the frequency of the HLA-DR11 gene in hepatitis C
virus (HCV) carriers was significantly higher than that in patients
with end-stage liver disease and liver cancer (56). The frequency of the HLA-B18 allele
was significantly increased in patients with hepatocellular
carcinoma (HCC), although the HLA-B18 allele was found to be absent
in HCV carriers (56). Compared
with that in HCV carriers, the frequency of the HLA-A4 allele was
also found to be significantly higher in patients with HCC
(56). In a different study, the
frequency of the HLA-B15 antigen in Yugoslav patients with
HBsAg+ hepatoma was found to be significantly higher
than that in other chronic liver disease control groups and
hepatitis B virus carriers (57).
CW7, B8 and DR3 antigens were found to be significantly elevated in
Italian patients with HCC (58).
The frequencies of DRB1*07, DRB1*04 and DQB1*02 of Egyptian
patients with HCC were found to be significantly higher than those
of healthy controls, whereas the frequencies of DQB1*06 and DRB1*15
in the patient group were significantly lower than those in the
control group (59). These
findings suggested that DRB1*07, DRB1*04 and DQB1*02 are risk
factors for HCC, whereas DQB1*06 and DRB1*15 act as protective
factors (59). In a study by De Re
et al (60), the DRB1*1101
and DQB1*0301 haplotypes were found to be associated with the
natural clearance of HCV. In the same study, the DQB1*0301 allele
was also demonstrated to have a protective role in the development
of HCV infection associated with HCC, whereas DRB1*1101 exerted the
opposite effect (60).
The frequency of HLA-A1 and HLA-A2 in patients with
ovarian cancer was found to be higher than that in healthy
controls, although the frequency of HLA-A3 was lower than that in
healthy controls (61). HLA-A2:B8
haplotypes, such as A2, B5, DRB1*03, DRB1*04 and CW3, and HLA-II
haplotypes, namely DRB1*0301, DQA1*0501, DQB1*0201, DRB1*1001,
DQA1*0101 and DQB1*0501, were significantly higher compared with
those in the control group (62).
A study by Kübler et al (62) also demonstrated that the HLA-A2:B8
and HLA-II haplotypes may be significantly associated with the
pathogenicity of ovarian cancer. The HLA-DRB1*04, HLA-DRB1*07,
HLA-DRB1*11 and HLA-DRB1*15 alleles are associated with the
occurrence of cervical squamous cell carcinoma and
HPV16+ cervical squamous cell carcinoma in women
(63). In a different study, the
alleles, HLA-DRB1*1402 and HLA-A*02 were demonstrated to decrease
the incidence of cervical cancer, whereas DRB1*1501 and DQA1*0102
increased the incidence of cervical cancer (64). A study by Guerini et al
(65) demonstrated that
HLA-DRB1*14 was significantly associated with the occurrence of
glioma. By contrast, the HLA-CW7, DRB1*1104, DRB1*1302, DQA1*0302
and DQB1*0604 haplotypes were significantly associated with the
occurrence of Kaposi's sarcoma (66). Reinders et al (67) demonstrated that HLA-B*35 could
inhibit the metastasis of head and neck squamous cell carcinoma,
whereas HLA-B*40 was associated with the occurrence of oral tumors,
and the HLA-B*40 and DRB1*13 haplotypes were susceptibility factors
for oral tumorigenesis (67). The
HLA polymorphisms associated with tumors are summarized in Table I.
The expression of HLA-G was demonstrated to be
highly correlated with tumor size, lymph node metastasis and the
Tumor-Node-Metastasis (TNM) stage of patients with breast cancer
(68). The survival rate of
patients with positive HLA-G expression was found to be lower than
that of patients with negative HLA-G expression (68). The level of serum HLA-G (sHLA-G) in
patients with breast cancer was demonstrated to be significantly
higher than that in healthy individuals, and this was found to be
significantly correlated with an increase in the
CD4+CD25highFoxp3+ to regulatory T
cells ratio (69). The mRNA and
protein levels of HLA-G in the tumor tissues of patients with
advanced ovarian cancer were demonstrated to be significantly
higher than those of early disease patients and healthy volunteers
(70). The expression of HLA-G was
also demonstrated to be associated with poor prognosis in patients
with ovarian cancer (70). In a
study by Lin et al (71),
the expression of HLA-G in non-small cell lung cancer (NSCLC) was
demonstrated to be significantly correlated with TNM stage, lymph
node metastasis and the immune response. In addition, it was
accompanied by an increase in the level of interleukin-10 (IL-10)
and the gene deletion of classical HLA-I (71). In a different study, HLA-G
expression in renal cancer tissue was demonstrated to be
significantly higher than that in adjacent normal tissue, and the
frequency of upregulated expression of HLA-G was significantly
higher than that of other HLA antigens (72). HLA-G expression in patients with
esophageal cancer, HCC and colorectal cancer has also been
demonstrated to be significantly higher than that in healthy
volunteers, and this was correlated with TNM stage, tumor
histological grade, invasion degree, lymph node status and the
immune response (73-75). The postoperative survival time of
patients with HCC and with HLA-G+ expression was
significantly shorter than that of patients with HLA-G−
expression. Furthermore, compared with the patients without HLA-G
expression, the recurrence rate of HCC was also higher in the
patients with HLA-G expression (74). The level of sHLA-G in patients with
colorectal cancer and NSCLC was also found to be significantly
higher than that in healthy volunteers, suggesting that sHLA-G may
be used as a diagnostic marker for malignant tumors (76,77).
The expression levels of the HLA-E and HLA-G
proteins and the levels of human papillomavirus (HPV) in cervical
cancer tissues were demonstrated to be higher than that in cervical
intraepithelial neoplasia (CIN) and chronic cervicitis tissues
(78). The expression levels of
HLA-E and HLA-G were positively correlated with differentiation,
CIN grade, TNM stage and HPV infection (78). The levels of sHLA-E and sHLA-G in
patients with gastric cancer were significantly higher than those
of the healthy volunteers (79).
In addition, the same study revealed that the level of HLA-E in
patients with stage III gastric cancer was significantly higher
than that in patients with stages I and II (79). HLA-E expression in HCC was found to
be higher than that in adjacent liver tissues, and this expression
was demonstrated to be positively correlated with tumor recurrence
(80). A study by Zhou et
al (81) revealed that the
distribution frequency of HLA-E*0103 in patients with breast cancer
was significantly higher than that in healthy controls, and the
risk of breast cancer was significantly increased in patients that
carried the HLA-E*0103 allele. The same study also demonstrated
that the sHLA-E level of patients with HLA-E*0103 was significantly
higher than that of healthy controls (81).
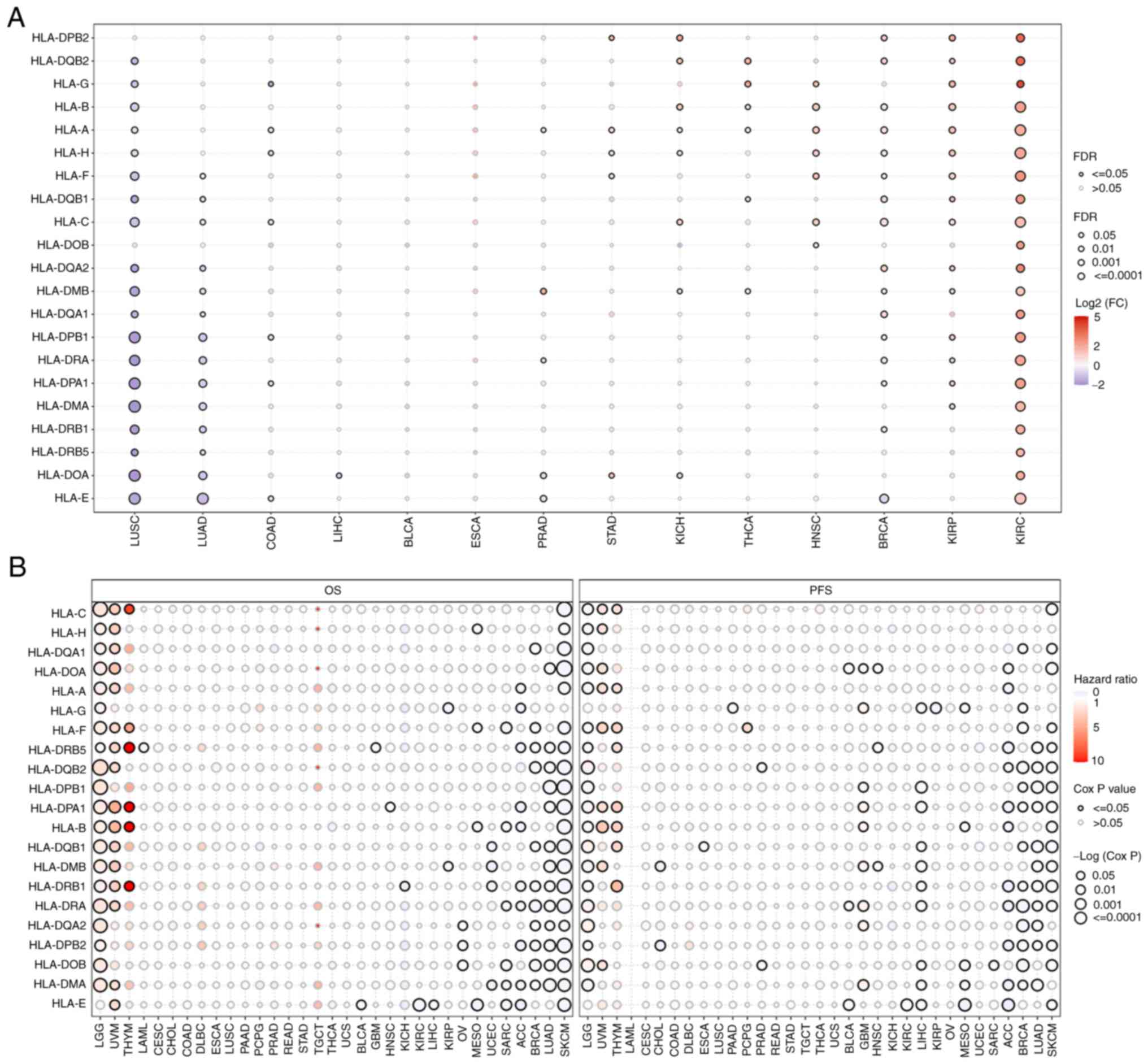
To avoid the bias of single studies due to ethnic
differences, the expression levels (Fig. 1A) and prognostic roles (Fig. 1B) of HLA subtypes in solid tumors
were analyzed. Gene Set Cancer Analysis (GSCA) (http://bioinfo.life.hust.edu.cn/GSCA)
was used to analyze and visualize the RNA-sequencing expression
profiles [fragments per kilobase of transcript per million (FPKM)]
(false discovery rate <0.05, log2 fold change >2)
and corresponding clinical information of 33 types of cancer in The
Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga).
The results obtained revealed that the majority of HLAs were
expressed at lower levels in human lung squamous cell carcinoma and
lung adenocarcinoma (LUAD), whereas the expression levels were
higher in kidney renal papillary cell carcinoma and kidney renal
clear cell carcinoma (Fig. 1A).
Most HLAs were found to be useful as favorable prognostic markers
for the overall survival and progression-free survival rates of
LUAD and skin cutaneous melanoma, although HLAs were generally
found to be unfavorable prognostic markers for lower-grade glioma,
uveal melanoma and thymoma (Fig.
1B). Based on the current published data, the reasons why HLAs
play different roles in different types of cancer cannot be
explained. However, our hypothesis is that tumor heterogeneity and
the immune microenvironment are the main factors affecting
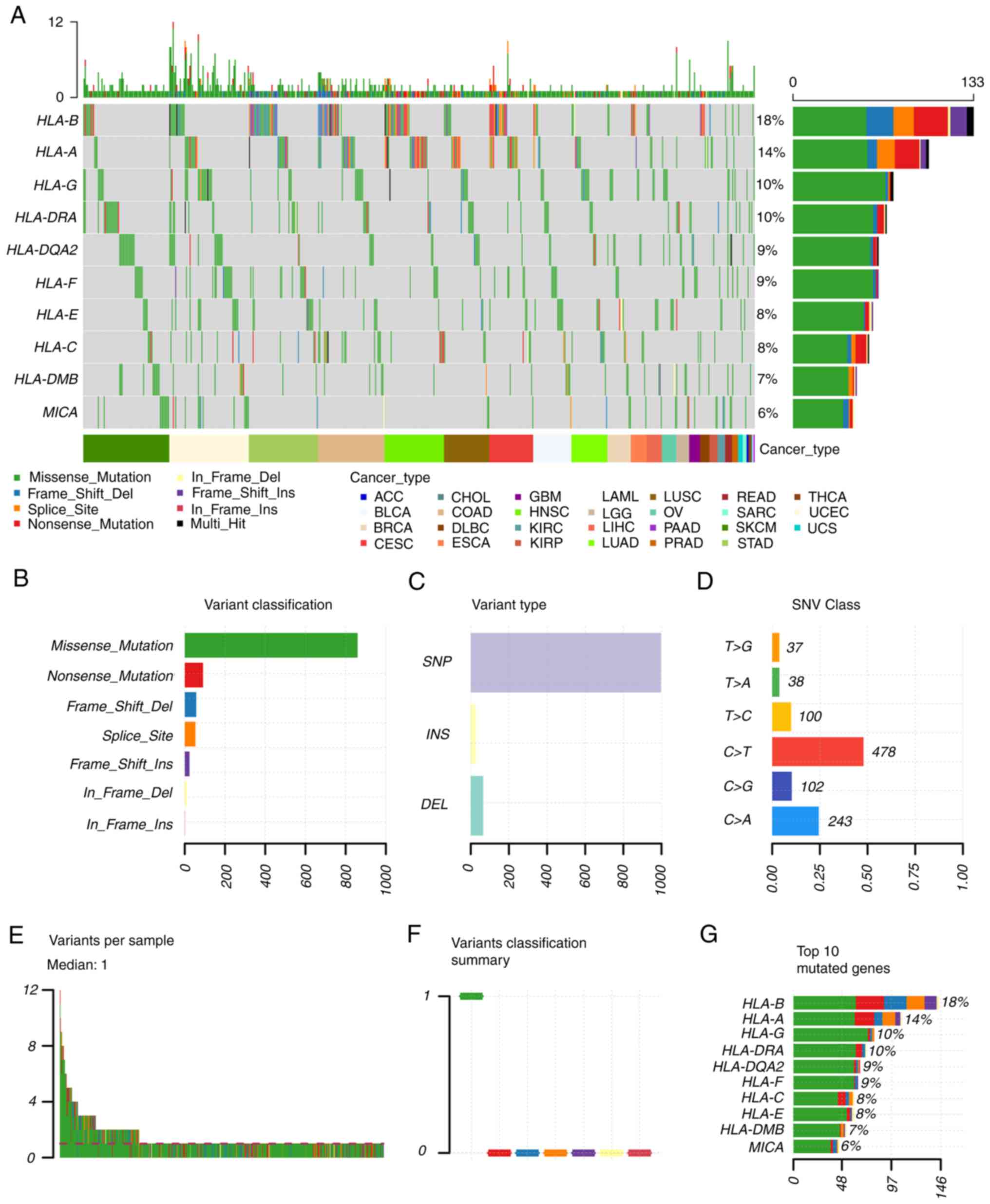
HLA-regulated tumor prognosis. In addition, the mutation status of
HLA in solid tumors was also analyzed (Fig. 2). The single nucleotide variation
(SNV) summary and oncoplot waterfall plot were generated by
GSCALite software (http://bioinfo.life.hust.edu.cn/web/GSCALite/) based
on 33 types of cancer in TCGA database. In cancer tissues, the
percentage variations of HLA-B, HLA-A, HLA-G and HLA-DRA were ≥10%.
The most common SNV class of HLAs in cancerous tissues was found to
be C>T.
Considered together, the available evidence has
demonstrated that HLA polymorphisms can affect the survival rate
of, and influence the curative effects on, patients with tumors,
and these may therefore be used as an indicator of individualized
treatment plans for these patients. Nonsynonymous mutations in the
HLA gene affect antigen presentation and the expression of HLA in
cancer cells, which therefore induces immune escape.
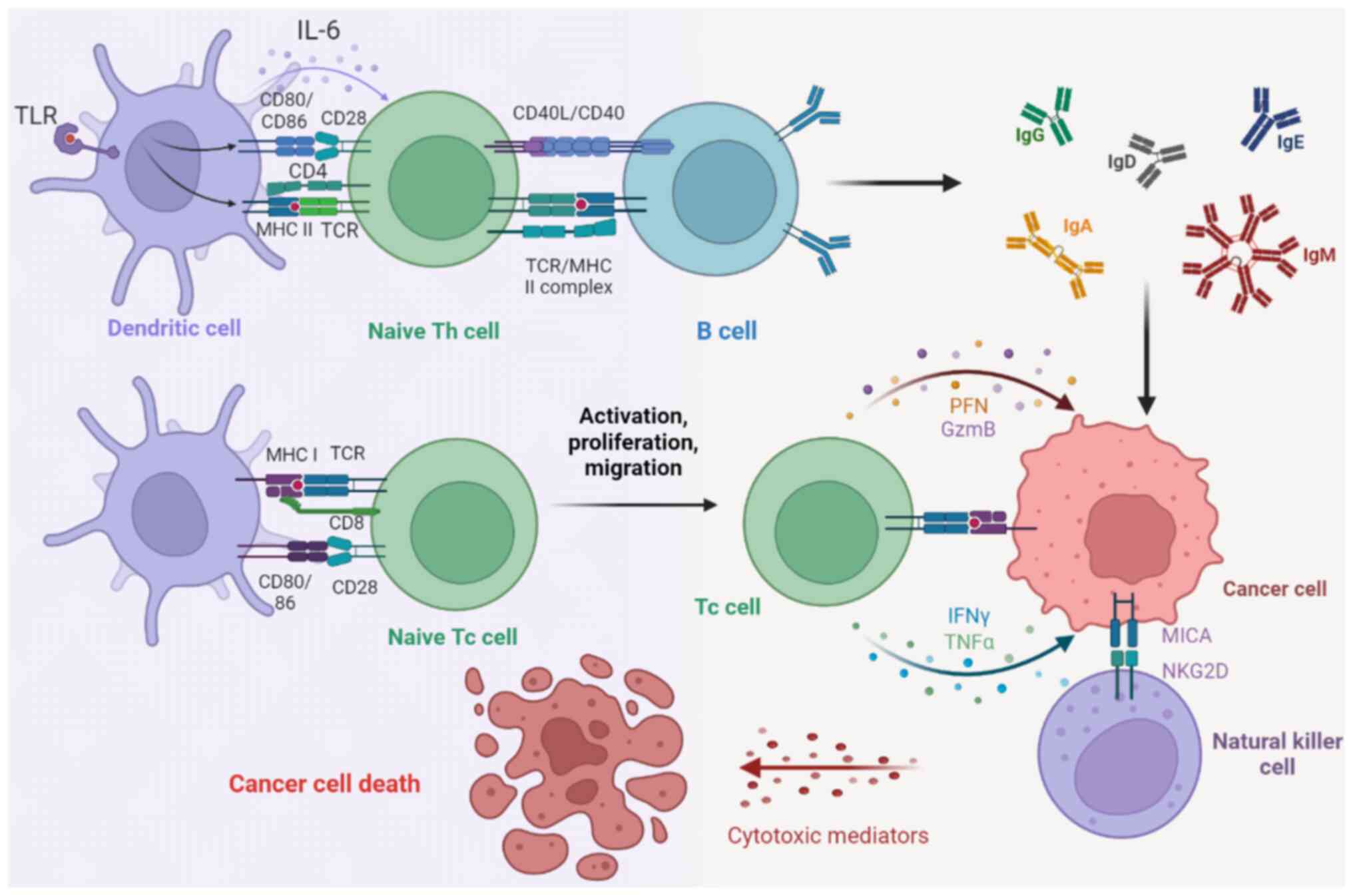
HLA-I molecules present endogenous antigenic
peptides to the cell surface, enabling the immune system to
recognize tumor-specific neoantigens and to eliminate them before
the tumor develops further (82).
The expression of the HLA-I gene in tumor cells is one of the
determinants of the recognition and destruction of tumor cells by
CD8+ cytotoxic T lymphocytes (CTLs) (83) (Fig.
3). However, mutation of the HLA-I gene often occurs in tumor
cells, resulting in a partial or total deletion of HLA-I on the
tumor surface, which decreases the secretion of perforin, Granzyme
B, IFN-γ and TNF-α from CTLs and promotes immune escape of tumor
cells (84) (Fig. 3). HLA-I gene expression on the
surface of pancreatic ductal gland tumor cells has been
demonstrated to improve the effect of antigen presentation, leading
to enhanced antitumor effects of T cells and consequently slower
tumor growth (85). In a study by
Yang et al (86), the
activation of the Wnt/β-catenin signaling pathway was demonstrated
to lead to a decrease in HLA-I gene expression, which in turn
decreased the CTL-mediated immune response to cancer cells. Tumor
cells have collectively selected the evolutionarily conserved and
lineage-specific functions of polycomb repressive complex 2 (PRC2)
to silence MHC class I molecular antigen processing and
presentation pathways, thereby escaping immune surveillance
(87). Notably, after
pharmacological inhibition of Drosophila zeste gene enhancer
human homolog 1 (EZH1) and EZH2, the PRC2-mediated silencing of MHC
class I molecule-dominated antigen processing pathway-associated
genes becomes reversible, thus re-establishing T cell-mediated
antitumor immunity (87). The
early embryonic transcription factor, double homeobox 4, which is
silenced in somatic tissues, is expressed in a variety of solid
tumors, where it inhibits HLA-I molecules to promote tumor immune
escape (88). Radiotherapy has
also been demonstrated to improve the efficacy of CD8+ T
cells in killing tumor cells via enhancement of the expression of
HLA-I molecules (89).
NK cells contribute significantly towards the
natural immune system and provide the first line of defense against
pathogens and tumors (90). These
cells not only serve an important role in the process of immune
surveillance, but also fulfill the functions of being antivirus and
anti-infection molecules, and killing tumor cells (91). Due to their property of being
unrestricted by MHC molecules, NK cells can recognize and kill
MHC-I-deficient tumor cells (92).
However, the immune function of NK cells is still regulated by MHC
class I-related chain A (MICA) (93). MICA is a non-classical HLA-I gene,
located between solute carrier superfamily 6 member 19 and HLA-B
genes, positioned 46 kb away from HLA-B with a total length of
11,772 bp, including 6 exons, and is highly polymorphic (94). It has been demonstrated that the
occurrence of multiple tumors is associated with the polymorphism
of MICA (95). MICA A9 is a
high-risk factor of gastric cancer (96), whereas MICA A6 has been
demonstrated to be a high-risk factor of oral squamous cell
carcinoma (97). The activated
receptor NKG2D fulfills an important role in the antitumor immunity
of NK cells (98,99). There is no currently recognized
signal transduction domain in the cytoplasmic region of NKG2D, and
the receptor mainly relies on YXXM, an immunoreceptor tyrosine
activation motif in the cytoplasmic region of DNAX-activating
protein 10 (DAP10), for its ability to activate signal transduction
(98,99). The NKG2D-DAP10 dimer binds to p85,
a subunit of phosphoinositide 3-kinase, and transmits an activation
signal (100). When MICA binds to
the NKG2D-DAP10 dimer, this leads to the activation of numerous
signal transduction pathways, such as those involving
lactaldehyde-derived lysine dimer, signal transducer and activator
of transcription 5, and AKT in effector cells, thereby activating
the antitumor immunity of NK cells (100). The sensitivity of
MICA+ leukemia cells to NK cells has been demonstrated
to be significantly higher than that of MICA− leukemia
cells (101). In situ
tumor cells of myeloma highly express MICA, although myeloma cells
with pleural effusion show little or no expression of MICA
(102). NK cells are only able to
recognize myeloma cells that kill tumor sites, and not myeloma
cells with pleural effusion (102). Another study published by Zhang
et al (103) revealed that
MICA on the surface of lung, colorectal and cervical cancer cells
can upregulate NKG2D activity and downregulate the expression of
the inhibitory receptors, NKG2A and KIR2DL1 (103). Although Vδ1+γδ T cells
also express NKG2D receptors, a low level of MICA expression is
insufficient to activate the Vδ1+γδ T cell immune
response (104). HeLa cells were
found to highly express the MICA membrane protein, while the
ovarian cancer cell line, SKOV3, expressed MICA plasma protein in
large quantities, which was unable to bind to NKG2D and activate
Vδ1+γδ T cells. Therefore, the killing effect of
Vδ1+γδ T cells on HeLa cells was found to be much
stronger than that of SKOV3 cells (105). For the present review, R
programming (version 4.1.1, https://cran.r-project.org/) was used to analyze and
visualize the RNA-sequencing expression profiles and corresponding
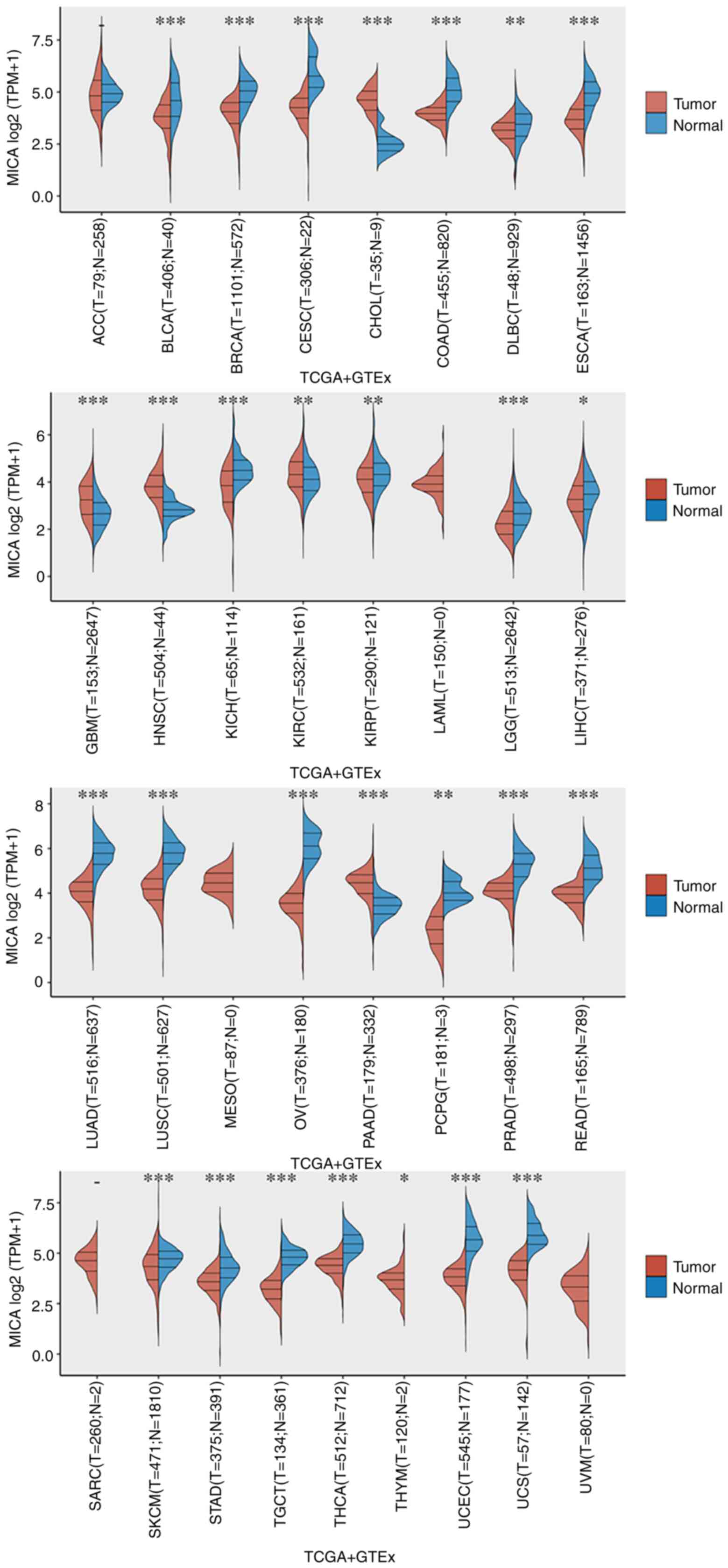
clinical information of the 33 types of cancer in TCGA database.
GTEx samples were collected from 54 non-diseased tissue sites
across nearly 1,000 individuals. Pan-cancer analysis revealed the
levels of MICA mRNA in solid tumors using the Wilcoxon rank-sum
test (Fig. 4). Compared with
tumoral tissues, higher levels of MICA mRNA were found in normal
tissues of most cancers, such as bladder urothelial carcinoma,
breast invasive carcinoma (BRCA), cervical squamous cell carcinoma
and endocervical adenocarcinoma, colon adenocarcinoma, esophageal
carcinoma, lung adenocarcinoma and lung squamous cell carcinoma
(Fig. 4). By contrast, higher MICA
expression was detected in tumoral tissues compared with in normal
tissues for cholangiocarcinoma (CHOL), glioblastoma multiforme,
kidney renal papillary cell carcinoma, head and neck squamous cell
carcinoma and pancreatic adenocarcinoma (Fig. 4). P<0.05 was considered to
indicate a statistically significant difference.
Tumor cells generally express MHC class I molecules,
and rarely express MHC class II molecules (106). Therefore, clinical research on
neoantigens and other types of tumor antigens has mainly focused on
MHC class I molecule-restricted antigens (106). However, given the in-depth
studies that have enabled an increase in the understanding of tumor
immunity, the regulatory effects of MHC class II molecular
restriction antigens on CD4+ T cells have been paid a
greater level of attention with respect to tumor immunity (107) (Fig.
3). The activation of toll-like receptor 2 leads to a decrease
in the expression of MHC class II molecules in microglia, an
inhibition of the proliferation and functional activation of
CD4+ T cells, and the promotion of immune escape of
microglia cells (108).
Interferon-γ stimulation was demonstrated to lead to the
upregulation of MHC class II molecules in chronic myeloid leukemia
(CML) stem cells (109). However,
the JAK1/2 inhibitor, ruxolitinib, inhibited the proliferation of
CML stem cells, resulting in a significant increase in the
expression level of MHC class II molecules (109). These results suggest that the
JAK-mediated signaling pathway is driven by cytokines provided by
CML cells and the immune microenvironment antagonizes MHC class II
molecule expression, thereby escaping immune surveillance. In a
different study, MHC class II molecules expressed in murine breast
tumor cells were found to promote the local activation of
CD4+ T cells, indirectly aiding the activation and
expansion of CD8+ T cells, delaying T-cell exhaustion
and inhibiting tumor cell proliferation (110). MHC class II molecules are
expected to stimulate more powerful and lasting immune responses,
either alone or in cooperation with MHC class I molecules. In
melanoma, colon cancer and breast cancer, tumor cells may be
recognized by CD4+ T cells instead of CD8+ T
cells, which suggests that, compared with MHC class I molecules
with a stricter restriction, tumor individualized neoantigens may
be more likely to bind to MHC class II molecules (111). Therefore, MHC class II molecules
have been demonstrated to be promising immunotherapeutic
targets.
Tumor cells express and secrete HLA-G, which
inhibits NK cell- and CTL-mediated cell lysis, thereby causing
tumor cells to escape from human immune surveillance and killing
(112). The mechanism of action
of HLA-G in tumors involves three steps of tumor immune escape:
Elimination, equilibrium and escape (113). In the elimination stage, HLA-G
expressed by tumor cells binds to the immunoglobulin-like
transcript 2 (ILT2) and ILT4 receptors on immune cells, inhibits
the cytolytic function of T and NK cells, and enables tumor cells
to escape recognition and elimination of immune cells (113). In addition, HLA-G can also
inhibit the secretion of cytokines, induce antigen-presenting cells
to secrete HLA-G and further inhibit immune function (113). In the equilibrium stage, most of
the tumor cells have been eliminated, and individual monoclonal
cells survive through mutation (113). At this stage, HLA-G expression
occurs at a low level, although it can still weaken the activity of
immune cells and promote the production of regulatory cells and
immunosuppressive factors, such as IL-10 (113). In the final escape stage, various
factors, such as hypoxia inducible factor 1 produced by hypoxia,
promote the expression of HLA-G, inhibiting the activity of immune
cells to the maximal possible extent, thereby causing rapid tumor
progression and local microenvironment hypoxia, which further
promotes the production of IL-10 and other cytokines, and
upregulates the expression of HLA-G to promote tumor formation
(113).
HLA-E fulfills an important role in regulating the
antitumor immune response, mainly by binding to the inhibitory
NKG2A receptors of NK cells or T cells (114). After NKG2A/CD94 binds to the
HLA-E/antigen polypeptide complex, it can cause the protein
tyrosine phosphatase, Src homology region 2 domain-containing
phosphatase-1, to be recruited to the vicinity of the
immunoreceptor tyrosine-based inhibitory motif in the intracellular
domain of NKG2A, thereby causing the signaling molecules upstream
and downstream of the pathway to undergo dephosphorylation, which
effectively inhibits NK cell activation (115). Tumor cells upregulate the
expression of HLA-E, interact with CD94/NKG2A and inhibit the
antitumor activity of IL-2 receptor-dependent CD8+ T
cells through inhibiting the proliferation, activity and secretion
of cytokines (116). The study
conducted by Chen et al (117) demonstrated that blocking NKG2A
could significantly enhance the killing ability of CD8+
T cells against tumor cells.
Further studies on the role and mechanism of HLA-G
and HLA-E in tumorigenesis and development will contribute towards
the application of HLA-G and HLA-E in the clinic, especially with
regards to clinical disease diagnosis, prognosis and
immunotherapy.
During the process of T cells initiating the immune
reaction, the recognition of weak binding peptides by T-cell
receptors may lead to an unstable interaction between the peptide
and the MHC molecule antigen-binding groove, thereby causing tumor
immune escape (118). The
analysis and screening of tumor antigen peptides that efficiently
bind to MHC molecules is of great significance for the rapid
development of safe and effective tumor vaccines (119). Mass spectrometric analysis of
tumor antigen peptides has been used to identify effective tumor
neoantigen peptides from tumor cells or tissues (120). Oncolytic viruses induce tumor
cells to produce new MHC class I molecular ligands to activate
CD8+ T-cell responses (121). A peptide MHC class I molecule
IgG-antibody fusion protein-redirected vaccine was demonstrated to
target lung cancer cells in vivo and endow them with viral
antigenicity, thereby activating antiviral CD8+ T cells
induced by the vaccine and inhibiting tumor growth (122). Tumor vaccines are designed to
target tumor neoantigens as a part of MHC molecule-binding
neoantigen immunotherapy (123).
Duperret et al (124) used
DNA vaccines to target tumor neoantigens for preclinical research
and found that the optimized tumor neoantigens were immunogenic and
mainly produced MHC I molecule-restricted CD8+ T-cell
responses. DNA neoantigen vaccines have also been demonstrated to
induce therapeutic antitumor immune responses in vivo, and
the neoantigen-specific T cells expanded in vivo were
demonstrated to kill tumor cells in vitro (125). At present, research on tumor
antigens, including neoantigens, has mainly focused on MHC class I
molecules, but with the gradual deepening of the understanding of
tumor immunity and the accumulation of clinical and experimental
data, researchers are also gradually shifting their attention
towards CD4+ T cells and MHC class II molecular antigen
peptides (126). The selective
pressure against MHC class II-restricted neoantigens is stronger
than that against MHC class I molecules, indicating that the immune
monitoring of CD4+ T cells is an important limiting
factor in tumorigenesis and development (127). MHC class II molecule-restricted
neoantigens can be recognized from their own tumor-infiltrating
lymphocytes from patients with metastatic CHOL, and CD4+
T-cell reinfusion treatment has been performed to effectively
alleviate the condition of patients (128).
Immune checkpoint inhibitors (ICIs) fight cancer by
reactivating the adaptive immune system of the patient (129). Selective autophagy of MHC class I
molecules mediated by the selective autophagy substrate
receptor/ATG8 interacting protein neighbor of BRCA1 is a novel
mechanism that has been discovered, which enables pancreatic ductal
gland tumor cells to evade immunity (130). Autophagy or lysosomal inhibition
can restore the expression of MHC class I molecules, enhance the
immune effect of antitumor T cells and improve the response to
immune checkpoint blockade therapy (131). Friedman et al (132) found that the decreased expression
of MHC class I molecules in patients with endometrial cancer is a
potential mechanism through which patients develop resistance to
ICIs. The induced expression of MHC class I molecules further
increases the therapeutic potential of ICIs in breast cancer
(133). In a different study, the
expression of chemokine (CXC motif) ligand 14 was demonstrated to
inhibit human HPV+ cervical cancer by restoring the
expression of MHC class I molecules on tumor cells and promoting
antigen-specific CD8+ T-cell responses (134). Taken together, these studies have
collectively demonstrated that the increased expression of MHC
class I molecules can enhance the efficacy of ICIs in treating
tumors.
In the present review, the relationship between HLA
polymorphism and tumorigenesis has been considered, and the key
role and mechanism of HLA in tumor immunity and treatment has been
focused on. Effective tumor immunotherapy is based on the synergy
of various components of the immune system. In order to establish
effective immunotherapy for patients with tumors, NK cells,
CD4+ T cells and CD8+ T cells have an
indispensable role. Although tumor vaccines and adoptive cell
therapy based on MHC-restricted antigens have achieved significant
efficacy in preclinical and clinical studies, they still cannot
completely cure tumors, and the efficacy needs to be strengthened
further. Previous studies have demonstrated that HLA serves an
important role in the occurrence and development of a variety of
tumors, including in gastric, lung and liver cancer. Due to the
high degree of polymorphism of HLA, the research results obtained
on the association between HLA and human tumors have not proven to
be entirely consistent, and future studies incorporating big data
analysis are required. In addition, recent studies demonstrated
that HLA affects the survival rate of patients undergoing ICI
therapy (135-137). These studies have suggested that
HLAs could be used to improve the efficacy of individualized
treatments of patients with cancer in the future.
All data generated or analyzed during this study are
included in the published article.
This review was conceptualized by PX; DHL, FFM and
MA performed the software analysis; MA, DHL and PX were responsible
for the original draft preparation; FFM and PX wrote the review and
subsequently edited it; PX supervised the project and was also the
project administrator; PX was responsible for the funding
acquisition. DHL, FFM, MA and PX confirm the authenticity of all
the raw data. All authors, read and approved the final version of
the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This study was supported by The National Natural Scientific
Foundation of China (grant no. 81972784) and the 'Double
First-Class' Disciplinary Construction Project of Jinzhou Medical
University.
|
1
|
Mungall AJ, Palmer SA, Sims SK, Edwards
CA, Ashurst JL, Wilming L, Jones MC, Horton R, Hunt SE, Scott CE,
et al: The DNA sequence and analysis of human chromosome 6. Nature.
425:805–811. 2003.
|
|
2
|
Robinson J, Barker DJ, Georgiou X, Cooper
MA, Flicek P and Marsh SGE: IPD-IMGT/HLA Database. Nucleic Acids
Res. 48(D1): D948–D955. 2020.
|
|
3
|
Urban RG, Chicz RM and Hedley ML: The
discovery and use of HLA-associated epitopes as drugs. Crit Rev
Immunol. 17:387–397. 1997.
|
|
4
|
Blees A, Januliene D, Hofmann T, Koller N,
Schmidt C, Trowitzsch S, Moeller A and Tampé R: Structure of the
human MHC-I peptide-loading complex. Nature. 551:525–528. 2017.
|
|
5
|
Reith W, LeibundGut-Landmann S and
Waldburger JM: Regulation of MHC class II gene expression by the
class II transactivator. Nat Rev Immunol. 5:793–806. 2005.
|
|
6
|
Boegel S, Löwer M, Bukur T, Sorn P, Castle
JC and Sahin U: HLA and proteasome expression body map. BMC Med
Genomics. 11:362018.
|
|
7
|
Liu B, Shao Y and Fu R: Current research
status of HLA in immune-related diseases. Immun Inflamm Dis.
9:340–350. 2021.
|
|
8
|
Blum JS, Wearsch PA and Cresswell P:
Pathways of antigen processing. Annu Rev Immunol. 31:443–473.
2013.
|
|
9
|
Hazini A, Fisher K and Seymour L:
Deregulation of HLA-I in cancer and its central importance for
immunotherapy. J Immunother Cancer. 9:e0028992021.
|
|
10
|
Marcu A, Bichmann L, Kuchenbecker L,
Kowalewski DJ, Freudenmann LK, Backert L, Mühlenbruch L, Szolek A,
Lübke M, Wagner P, et al: HLA Ligand Atlas: A benign reference of
HLA-presented peptides to improve T-cell-based cancer
immunotherapy. J Immunother Cancer. 9:e0020712021.
|
|
11
|
Djajadiningrat RS, Horenblas S, Heideman
DA, Sanders J, de Jong J and Jordanova ES: Classic and nonclassic
HLA class I expression in penile cancer and relation to HPV status
and clinical outcome. J Urol. 193:1245–1251. 2015.
|
|
12
|
Cruz FM, Chan A and Rock KL: Pathways of
MHC I cross-presentation of exogenous antigens. Semin Immunol.
66:1017292023.
|
|
13
|
Bradley P: Structure-based prediction of T
cell receptor:peptide-MHC interactions. Elife. 12:e828132023.
|
|
14
|
Sarri CA, Giannoulis T, Moutou KA and
Mamuris Z: HLA class II peptide-binding-region analysis reveals
funneling of polymorphism in action. Immunol Lett. 238:75–95.
2021.
|
|
15
|
Klein J and Sato A: The HLA system. First
of two parts. N Engl J Med. 343:702–709. 2000.
|
|
16
|
Mišunová M, Svitálková T, Pleštilová L,
Kryštufková O, Tegzová D, Svobodová R, Hušáková M, Tomčík M, Bečvář
R, Závada J, et al: Molecular markers of systemic autoimmune
disorders: The expression of MHC-located HSP70 genes is
significantly associated with autoimmunity development. Clin Exp
Rheumatol. 35:33–42. 2017.
|
|
17
|
Ota M, Katsuyama Y, Kimura A, Tsuchiya K,
Kondo M, Naruse T, Mizuki N, Itoh K, Sasazuki T and Inoko H: A
second susceptibility gene for developing rheumatoid arthritis in
the human MHC is localized within a 70-kb interval telomeric of the
TNF genes in the HLA class III region. Genomics. 71:263–270.
2001.
|
|
18
|
Cucca F, Zhu ZB, Khanna A, Cossu F, Congia
M, Badiali M, Lampis R, Frau F, De Virgiliis S, Cao A, et al:
Evaluation of IgA deficiency in Sardinians indicates a
susceptibility gene is encoded within the HLA class III region.
Clin Exp Immunol. 111:76–80. 1998.
|
|
19
|
Arnaiz-Villena A, Suarez-Trujillo F,
Juarez I, Rodríguez-Sainz C, Palacio-Gruber J, Vaquero-Yuste C,
Molina-Alejandre M, Fernández-Cruz E and Martin-Villa JM: Evolution
and molecular interactions of major histocompatibility complex
(MHC)-G, -E and -F genes. Cell Mol Life Sci. 79:4642022.
|
|
20
|
Gulati R, Kavadichanda GC, Mariaselvam CM,
Kumar G and Negi VS: Association of HLA-G, HLA-E and HLA-B*27 with
susceptibility and clinical phenotype of enthesitis related
arthritis (ERA). Hum Immunol. 82:615–620. 2021.
|
|
21
|
Xu X, Zhou Y and Wei H: Roles of HLA-G in
the maternal-fetal immune microenvironment. Front Immunol.
11:5920102020.
|
|
22
|
Rajagopalan S and Long EO: A Human
Histocompatibility Leukocyte Antigen (HLA)-G-Specific receptor
expressed on all natural killer cells. J Exp Med. 189:1093–1100.
1999.
|
|
23
|
Mosaferi E, Alizadeh Gharamaleki N,
Farzadi L, Majidi J, Babaloo Z, Kazemi T, Ramezani M, Tabatabaei M,
Ahmadi H, Aghebati Maleki L and Baradaran B: The Study of HLA-G
gene and protein expression in patients with recurrent miscarriage.
Adv Pharm Bull. 9:70–75. 2019.
|
|
24
|
Zhuang B, Shang J and Yao Y: HLA-G: An
important mediator of maternal-fetal immune-tolerance. Front
Immunol. 12:7443242021.
|
|
25
|
Camilli G, Cassotta A, Battella S,
Palmieri G, Santoni A, Paladini F, Fiorillo MT and Sorrentino R:
Regulation and trafficking of the HLA-E molecules during
monocyte-macrophage differentiation. J Leukoc Biol. 99:121–130.
2016.
|
|
26
|
Ruibal P, Franken KLMC, van Meijgaarden
KE, Walters LC, McMichael AJ, Gillespie GM, Joosten SA and
Ottenhoff THM: Discovery of HLA-E-Presented Epitopes: MHC-E/Peptide
Binding and T-Cell Recognition. Methods Mol Biol. 2574:15–30.
2022.
|
|
27
|
Lo Monaco E, Sibilio L, Melucci E,
Tremante E, Suchànek M, Horejsi V, Martayan A and Giacomini P:
HLA-E: Strong association with beta2-microglobulin and surface
expression in the absence of HLA class I signal sequence-derived
peptides. J Immunol. 181:5442–5450. 2008.
|
|
28
|
Prašnikar E, Perdih A and Borišek J: What
a difference an amino acid makes: An All-Atom simulation study of
nonameric peptides in inhibitory HLA-E/NKG2A/CD94 immune complexes.
Front Pharmacol. 13:9254272022.
|
|
29
|
Li D, Brackenridge S, Walters LC, Swanson
O, Harlos K, Rozbesky D, Cain DW, Wiehe K, Scearce RM, Barr M, et
al: Mouse and human antibodies bind HLA-E-leader peptide complexes
and enhance NK cell cytotoxicity. Commun Biol. 5:2712022.
|
|
30
|
Morandi F, Rizzo R, Fainardi E,
Rouas-Freiss N and Pistoia V: Recent advances in our understanding
of HLA-G Biology: Lessons from a wide spectrum of human diseases. J
Immunol Res. 2016:43264952016.
|
|
31
|
Bertol BC, Dias FC, da Silva DM, Zambelli
Ramalho LN and Donadi EA: Human Antigen Leucocyte (HLA)-G and HLA-E
are differentially expressed in pancreatic disorders. Hum Immunol.
80:948–954. 2019.
|
|
32
|
Seliger B: The non-classical antigens of
HLA-G and HLA-E as diagnostic and prognostic biomarkers and as
therapeutic targets in transplantation and tumors. Clin Transpl.
465–472. 2013.
|
|
33
|
Pagliuca S, Gurnari C, Rubio MT, Visconte
V and Lenz TL: Individual HLA heterogeneity and its implications
for cellular immune evasion in cancer and beyond. Front Immunol.
13:9448722022.
|
|
34
|
Rognan D, Krebs S, Kuonen O, Lamas JR,
López de Castro JA and Folkers G: Fine specificity of antigen
binding to two class I major histocompatibility proteins (B*2705
and B*2703) differing in a single amino acid residue. J Comput
Aided Mol Des. 11:463–478. 1997.
|
|
35
|
Schendel DJ, Reinhardt C, Nelson PJ, Maget
B, Pullen L, Bornkamm GW and Steinle A: Cytotoxic T lymphocytes
show HLA-C-restricted recognition of EBV-bearing cells and
allorecognition of HLA class I molecules presenting self-peptides.
J Immunol. 149:2406–2414. 1992.
|
|
36
|
Neefjes J, Jongsma ML, Paul P and Bakke O:
Towards a systems understanding of MHC class I and MHC class II
antigen presentation. Nat Rev Immunol. 11:823–836. 2011.
|
|
37
|
Rastogi I, Jeon D, Moseman JE, Muralidhar
A, Potluri HK and McNeel DG: Role of B cells as antigen presenting
cells. Front Immunol. 13:9549362022.
|
|
38
|
Erb P, Feldmann M and Hogg N: Role of
macrophages in the generation of T helper cells. IV. Nature of
genetically related factor derived from macrophages incubated with
soluble antigens. Eur J Immunol. 6:365–372. 1976.
|
|
39
|
Erb P and Feldmann M: The role of
macrophages in the generation of T-helper cells. II. The genetic
control of the macrophage-T-cell interaction for helper cell
induction with soluble antigens. J Exp Med. 142:460–472. 1975.
|
|
40
|
Takeuchi C, Ohto H, Miura S, Yasuda H, Ono
S and Ogata T: Delayed and acute hemolytic transfusion reactions
resulting from red cell antibodies and red cell-reactive HLA
antibodies. Transfusion. 45:1925–1929. 2005.
|
|
41
|
Chen Y, Huang XJ, Wang Y, Liu KY, Chen H,
Chen YH, Zhang XH, Wang FR, Han W, Wang JZ, et al: Febrile reaction
associated with the infusion of haploidentical peripheral blood
stem cells: Incidence, clinical features, and risk factors.
Transfusion. 55:2023–2031. 2015.
|
|
42
|
Gavroy B, Timmermans T, Van Caenegem O,
Mastrobuoni S, Jacquet L, Latinne D and Poncelet AJ: Significance
of HLA-matching and anti-HLA antibodies in heart transplant
patients receiving induction therapy? Transpl Immunol.
75:1017062022.
|
|
43
|
Rennie TJW, Battle RK, Abel AA, McConnell
S, McLaren R, Phelan PJ, Geddes C, Padmanabhan N, Clancy MJ, Little
AM and Turner DM: Comparison of kidney transplant outcomes in HLA
compatible and incompatible transplantation: A national cohort
study. Nephrology (Carlton). 27:962–972. 2022.
|
|
44
|
Chen R, Yi H, Zhen J, Fan M, Xiao L, Yu Q,
Yang Z, Ning L, Deng Z and Chen G: Donor with HLA-C2 is associated
with acute rejection following liver transplantation in Southern
Chinese. HLA. 100:133–141. 2022.
|
|
45
|
Kim HE, Yang YH, Paik HC, Jeong SJ, Kim
SY, Park MS and Lee JG: The Assessment and outcomes of
crossmatching in lung transplantation in Korean patients. J Korean
Med Sci. 37:e1772022.
|
|
46
|
Halleck F, Khadzhynov D, Liefeldt L,
Schrezenmeier E, Lehner L, Duerr M, Schmidt D, Bamoulid J, Lachmann
N, Waiser J, et al: Immunologic outcome in elderly kidney
transplant recipients: Is it time for HLA-DR matching? Nephrol Dial
Transplant. 31:2143–2149. 2016.
|
|
47
|
Arcuri LJ, Kerbauy MN, Kerbauy LN, Santos
FPS, Ribeiro AAF and Hamerschlak N: ATG in HLA-matched, peripheral
blood, hematopoietic cell transplantation in acute myeloid leukemia
and myelodysplastic syndrome: A secondary analysis of a CIBMTR
database. Transplant Cell Ther. 29:40.e1–40.e4. 2023.
|
|
48
|
Steens J and Klein D: HOX genes in stem
cells: Maintaining cellular identity and regulation of
differentiation. Front Cell Dev Biol. 10:10029092022.
|
|
49
|
Olerup O, Möller E and Persson U: HLA-DP
incompatibilities induce significant proliferation in primary mixed
lymphocyte cultures in HLA-A, -B, -DR and -DQ compatible
individuals: Implications for allogeneic bone marrow
transplantation. Tissue Antigens. 36:194–202. 1990.
|
|
50
|
Jin DQ and Zheng XF: HLA gene polymorphism
and forensic medicine. Fa Yi Xue Za Zhi. 19:51–53. 2003.In
Chinese.
|
|
51
|
Ritzmann SE: HLA patterns and disease
associations. JAMA. 236:2305–2309. 1976.
|
|
52
|
Hanson A and Brown MA: Genetics and the
causes of ankylosing spondylitis. Rheum Dis Clin North Am.
43:401–414. 2017.
|
|
53
|
Abdou AM, Gao X, Cozen W, Cerhan JR,
Rothman N, Martin MP, Davis S, Schenk M, Chanock SJ, Hartge P, et
al: Human leukocyte antigen (HLA) A1-B8-DR3 (8.1) haplotype, tumor
necrosis factor (TNF) G-308A, and risk of non-Hodgkin lymphoma.
Leukemia. 24:1055–1058. 2010.
|
|
54
|
Baxter KR and Opremcak EM: Panretinal
acute multifocal placoid pigment epitheliopathy: A novel posterior
uveitis syndrome with HLA-A3 and HLA-C7 association. J Ophthalmic
Inflamm Infect. 3:292013.
|
|
55
|
Mohammad-Ebrahim H, Kamali-Sarvestani E,
Mahmoudi M, Beigy M, Karami J, Ahmadzadeh N and Shahram F:
Association of killer cell immunoglobulin-like receptor (KIR) genes
and their HLA ligands with susceptibility to Behçet's disease.
Scand J Rheumatol. 47:155–163. 2018.
|
|
56
|
López-Vázquez A, Rodrigo L and
López-Larrea C: Association of killer immunoglobulin-like receptors
and their HLA Class I ligands with progression of chronic hepatitis
C virus infection. Tissue Antigens. 69(Suppl 1): S241–S242.
2007.
|
|
57
|
Golubovic G, Stajic M, Stolic I, Nikolic
JA, Neskovic AN and Pandey L: Histocompatibility antigens in
patients with hepatocellular carcinoma. Z Gastroenterol. 34:15–20.
1996.
|
|
58
|
Ricci G, Colombo C, Ghiazza B, Porta C,
Moroni M and Illeni MT: HLA-A, B, C, DR and DQ expression and
hepatocellular carcinoma: Study of 205 Italian subjects. Cancer
Lett. 98:121–125. 1995.
|
|
59
|
El-Chennawi FA, Auf FA, Metwally SS,
Mosaad YM, El-Wahab MA and Tawhid ZE: HLA-class II alleles in
Egyptian patients with hepatocellular carcinoma. Immunol Invest.
37:661–674. 2008.
|
|
60
|
De Re V, Caggiari L, Talamini R, Crovatto
M, De Vita S, Mazzaro C, Cannizzaro R, Dolcetti R and Boiocchi M:
Hepatitis C virus-related hepatocellular carcinoma and B-cell
lymphoma patients show a different profile of major
histocompatibility complex class II alleles. Hum Immunol.
65:1397–1404. 2004.
|
|
61
|
Gamzatova Z, Villabona L, van der Zanden
H, Haasnoot GW, Andersson E, Kiessling R, Seliger B, Kanter L,
Dalianis T, Bergfeldt K and Masucci GV: Analysis of HLA class I-II
haplotype frequency and segregation in a cohort of patients with
advanced stage ovarian cancer. Tissue Antigens. 70:205–213.
2007.
|
|
62
|
Kübler K, Arndt PF, Wardelmann E, Krebs D,
Kuhn W and van der Ven K: HLA-class II haplotype associations with
ovarian cancer. Int J Cancer. 119:2980–2985. 2006.
|
|
63
|
Yang YC, Chang TY, Lee YJ, Su TH, Dang CW,
Wu CC, Liu HF, Chu CC and Lin M: HLA-DRB1 alleles and cervical
squamous cell carcinoma: Experimental study and meta-analysis. Hum
Immunol. 67:331–340. 2006.
|
|
64
|
Schiff MA, Apple RJ, Lin P, Nelson JL,
Wheeler CM and Becker TM: HLA alleles and risk of cervical
intraepithelial neoplasia among Southwestern American Indian women.
Hum Immunol. 66:1050–1056. 2005.
|
|
65
|
Guerini FR, Agliardi C, Zanzottera M,
Delbue S, Pagani E, Tinelli C, Boldorini R, Car PG, Veggiani C and
Ferrante P: Human leukocyte antigen distribution analysis in North
Italian brain Glioma patients: An association with HLA-DRB1*14. J
Neurooncol. 77:213–217. 2006.
|
|
66
|
Masala MV, Carcassi C, Cottoni F, Mulargia
M, Contu L and Cerimele D: Classic Kaposi's sarcoma in Sardinia HLA
positive and negative associations. Int J Dermatol. 44:743–745.
2005.
|
|
67
|
Reinders J, Rozemuller EH, Otten HG, van
der Veken LT, Slootweg PJ and Tilanus MG: HLA and MICA associations
with head and neck squamous cell carcinoma. Oral Oncol. 43:232–240.
2007.
|
|
68
|
He X, Dong DD, Yie SM, Yang H, Cao M, Ye
SR, Li K, Liu J and Chen J: HLA-G expression in human breast
cancer: Implications for diagnosis and prognosis, and effect on
allocytotoxic lymphocyte response after hormone treatment in vitro.
Ann Surg Oncol. 17:1459–1469. 2010.
|
|
69
|
Zidi I, Kharrat N, Sebai R, Zidi N, Ben
Yahia H, Bouaziz A, Rifi H, Mezlini A and Rizzo R: Pregnancy and
breastfeeding: A new theory for sHLA-G in breast cancer patients?
Immunol Res. 64:636–639. 2016.
|
|
70
|
Jung YW, Kim YT, Kim SW, Kim S, Kim JH,
Cho NH and Kim JW: Correlation of human leukocyte antigen-G (HLA-G)
expression and disease progression in epithelial ovarian cancer.
Reprod Sci. 16:1103–1111. 2013.
|
|
71
|
Lin A, Zhu CC, Chen HX, Chen BF, Zhang X,
Zhang JG, Wang Q, Zhou WJ, Hu W, Yang HH, et al: Clinical relevance
and functional implications for human leukocyte antigen-g
expression in non-small-cell lung cancer. J Cell Mol Med.
14:2318–2329. 2010.
|
|
72
|
Tronik-Le Roux D, Renard J, Vérine J,
Renault V, Tubacher E, LeMaoult J, Rouas-Freiss N, Deleuze JF,
Desgrandschamps F and Carosella ED: Novel landscape of HLA-G
isoforms expressed in clear cell renal cell carcinoma patients. Mol
Oncol. 11:1561–1578. 2017.
|
|
73
|
Zheng J, Xu C, Chu D, Zhang X, Li J, Ji G,
Hong L, Feng Q, Li X, Wu G, et al: Human leukocyte antigen G is
associated with esophageal squamous cell carcinoma progression and
poor prognosis. Immunol Lett. 161:13–19. 2014.
|
|
74
|
Catamo E, Zupin L, Crovella S, Celsi F and
Segat L: Non-classical MHC-I human leukocyte antigen (HLA-G) in
hepatotropic viral infections and in hepatocellular carcinoma. Hum
Immunol. 75:1225–1231. 2014.
|
|
75
|
Kaprio T, Sariola H, Linder N, Lundin J,
Kere J, Haglund C and Wedenoja S: HLA-G expression correlates with
histological grade but not with prognosis in colorectal carcinoma.
HLA. 98:213–217. 2021.
|
|
76
|
Jiao F, Zhou J, Sun H, Song X and Song Y:
Plasma soluble human leukocyte antigen G predicts the long-term
prognosis in patients with colorectal cancer. Transl Cancer Res.
9:4011–4019. 2020.
|
|
77
|
Cao M, Yie SM, Liu J, Ye SR, Xia D and Gao
E: Plasma soluble HLA-G is a potential biomarker for diagnosis of
colorectal, gastric, esophageal and lung cancer. Tissue Antigens.
78:120–128. 2011.
|
|
78
|
Ferguson R, Ramanakumar AV, Richardson H,
Tellier PP, Coutlée F, Franco EL and Roger M: Human leukocyte
antigen (HLA)-E and HLA-G polymorphisms in human papillomavirus
infection susceptibility and persistence. Hum Immunol. 72:337–341.
2011.
|
|
79
|
Dutta N, Majumder D, Gupta A, Mazumder DN
and Banerjee S: Analysis of human lymphocyte antigen class I
expression in gastric cancer by reverse transcriptase-polymerase
chain reaction. Hum Immunol. 66:164–169. 2005.
|
|
80
|
Chen A, Shen Y, Xia M, Xu L, Pan N, Yin Y,
Miao F, Shen C, Xie W and Zhang J: Expression of the nonclassical
HLA class I and MICA/B molecules in human hepatocellular carcinoma.
Neoplasma. 58:371–376. 2011.
|
|
81
|
Zhou Y, Wu Z, Tang Y and Jia T: HLA-E gene
polymorphisms and plasma soluble HLA-E levels and their
relationship with genetic susceptibility to breast cancer. Xi Bao
Yu Fen Zi Mian Yi Xue Za Zhi. 31:524–527. 2015.In Chinese.
|
|
82
|
Mouchess ML and Anderson M: Central
tolerance induction. Curr Top Microbiol Immunol. 373:69–86.
2014.
|
|
83
|
Goulmy E, Termijtelen A, Bradley BA and
van Rood JJ: Y-antigen killing by T cells of women is restricted by
HLA. Nature. 266:544–545. 1977.
|
|
84
|
Dersh D, Hollý J and Yewdell JW: A few
good peptides: MHC class I-based cancer immunosurveillance and
immunoevasion. Nat Rev Immunol. 21:116–128. 2021.
|
|
85
|
Ryschich E, Nötzel T, Hinz U, Autschbach
F, Ferguson J, Simon I, Weitz J, Fröhlich B, Klar E, Büchler MW and
Schmidt J: Control of T-cell-mediated immune response by HLA class
I in human pancreatic carcinoma. Clin Cancer Res. 11(2 Pt 1):
498–504. 2005.
|
|
86
|
Yang W, Li Y, Gao R, Xiu Z and Sun T: MHC
class I dysfunction of glioma stem cells escapes from CTL-mediated
immune response via activation of Wnt/β-catenin signaling pathway.
Oncogene. 39:1098–1111. 2020.
|
|
87
|
Burr ML, Sparbier CE, Chan KL, Chan YC,
Kersbergen A, Lam EYN, Azidis-Yates E, Vassiliadis D, Bell CC,
Gilan O, et al: An evolutionarily conserved function of polycomb
silences the MHC class I antigen presentation pathway and enables
immune evasion in cancer. Cancer Cell. 36:385–401.e8. 2019.
|
|
88
|
Chew GL, Campbell AE, De Neef E, Sutliff
NA, Shadle SC, Tapscott SJ and Bradley RK: DUX4 suppresses MHC
class I to promote cancer immune evasion and resistance to
checkpoint blockade. Dev Cell. 50:658–671.e7. 2019.
|
|
89
|
Zebertavage LK, Alice A, Crittenden MR and
Gough MJ: Transcriptional upregulation of NLRC5 by radiation drives
STING- and interferon-independent MHC-I expression on cancer cells
and T cell cytotoxicity. Sci Rep. 10:73762020.
|
|
90
|
Björkström NK, Strunz B and Ljunggren HG:
Natural killer cells in antiviral immunity. Nat Rev Immunol.
22:112–123. 2022.
|
|
91
|
Höglund P and Brodin P: Current
perspectives of natural killer cell education by MHC class I
molecules. Nat Rev Immunol. 10:724–734. 2010.
|
|
92
|
Wolf NK, Kissiov DU and Raulet DH: Roles
of natural killer cells in immunity to cancer, and applications to
immunotherapy. Nat Rev Immunol. 23:90–105. 2023.
|
|
93
|
Ferrari de Andrade L, Tay RE, Pan D, Luoma
AM, Ito Y, Badrinath S, Tsoucas D, Franz B, May KF Jr, Harvey CJ,
et al: Antibody-mediated inhibition of MICA and MICB shedding
promotes NK cell-driven tumor immunity. Science. 359:1537–1542.
2018.
|
|
94
|
Jarduli LR, Sell AM, Reis PG, Sippert EÂ,
Ayo CM, Mazini PS, Alves HV, Teixeira JJ and Visentainer JE: Role
of HLA, KIR, MICA, and cytokines genes in leprosy. Biomed Res Int.
2013:9898372013.
|
|
95
|
Chen D and Gyllensten U: MICA
polymorphism: Biology and importance in cancer. Carcinogenesis.
35:2633–2642. 2014.
|
|
96
|
Lo SS, Lee YJ, Wu CW, Liu CJ, Huang JW and
Lui WY: The increase of MICA gene A9 allele associated with gastric
cancer and less schirrous change. Br J Cancer. 90:1809–1813.
2004.
|
|
97
|
Chung-Ji L, Yann-Jinn L, Hsin-Fu L,
Ching-Wen D, Che-Shoa C, Yi-Shing L and Kuo-Wei C: The increase in
the frequency of MICA gene A6 allele in oral squamous cell
carcinoma. J Oral Pathol Med. 31:323–328. 2002.
|
|
98
|
Moretta L, Bottino C, Cantoni C, Mingari
MC and Moretta A: Human natural killer cell function and receptors.
Curr Opin Pharmacol. 1:387–391. 2001.
|
|
99
|
Moretta A, Parolini S, Castriconi R,
Bottino C, Vitale M, Sivori S and Millo R: Function and specificity
of human natural killer cell receptors. Eur J Immunogenet.
24:455–468. 1997.
|
|
100
|
Wu J, Song Y, Bakker AB, Bauer S, Spies T,
Lanier LL and Phillips JH: An activating immunoreceptor complex
formed by NKG2D and DAP10. Science. 285:730–732. 1999.
|
|
101
|
Danier AC, de Melo RP, Napimoga MH and
Laguna-Abreu MT: The role of natural killer cells in chronic
myeloid leukemia. Rev Bras Hematol Hemoter. 33:216–220. 2011.
|
|
102
|
Nwangwu CA, Weiher H and Schmidt-Wolf IGH:
Increase of CIK cell efficacy by upregulating cell surface MICA and
inhibition of NKG2D ligand shedding in multiple myeloma. Hematol
Oncol. 35:719–725. 2017.
|
|
103
|
Zhang C, Tian ZG, Zhang J, Feng JB, Zhang
JH and Xu XQ: The negative regulatory effect of IFN-gamma on
cognitive function of human natural killer cells. Zhonghua Zhong
Liu Za Zhi. 26:324–327. 2004.In Chinese.
|
|
104
|
Kuroda H, Saito H and Ikeguchi M:
Decreased number and reduced NKG2D expression of Vδ1 γδ T cells are
involved in the impaired function of Vδ1 γδ T cells in the tissue
of gastric cancer. Gastric Cancer. 15:433–439. 2012.
|
|
105
|
Qi J, Peng P, Dai MH, Li YH, Cui LX and He
W: Cytotoxicity of MICA-reactive V delta 1 gamma delta T cells
towards epithelial tumor cells. Zhongguo Yi Xue Ke Xue Yuan Xue
Bao. 26:1–7. 2004.In Chinese.
|
|
106
|
Lv D, Khawar MB, Liang Z, Gao Y and Sun H:
Neoantigens and NK Cells: 'Trick or Treat' the Cancers? Front
Immunol. 13:9318622022.
|
|
107
|
Bertolino P and Rabourdin-Combe C: The MHC
class II-associated invariant chain: A molecule with multiple roles
in MHC class II biosynthesis and antigen presentation to CD4+ T
cells. Crit Rev Immunol. 16:359–379. 1996.
|
|
108
|
Qian J, Luo F, Yang J, Liu J, Liu R, Wang
L, Wang C, Deng Y, Lu Z, Wang Y, et al: TLR2 promotes glioma immune
evasion by downregulating MHC class II molecules in microglia.
Cancer Immunol Res. 6:1220–1233. 2018.
|
|
109
|
Tarafdar A, Hopcroft LE, Gallipoli P,
Pellicano F, Cassels J, Hair A, Korfi K, Jørgensen HG, Vetrie D,
Holyoake TL and Michie AM: CML cells actively evade host immune
surveillance through cytokine-mediated downregulation of MHC-II
expression. Blood. 129:199–208. 2017.
|
|
110
|
McCaw TR, Li M, Starenki D, Cooper SJ, Liu
M, Meza-Perez S, Arend RC, Buchsbaum DJ, Forero A and Randall TD:
The expression of MHC class II molecules on murine breast tumors
delays T-cell exhaustion, expands the T-cell repertoire, and slows
tumor growth. Cancer Immunol Immunother. 68:175–188. 2019.
|
|
111
|
Halder T, Pawelec G, Kirkin AF, Zeuthen J,
Meyer HE, Kun L and Kalbacher H: Isolation of novel HLA-DR
restricted potential tumor-associated antigens from the melanoma
cell line FM3. Cancer Res. 57:3238–3244. 1997.
|
|
112
|
Liu L, Wang L, Zhao L, He C and Wang G:
The Role of HLA-G in tumor escape: Manipulating the phenotype and
function of immune cells. Front Oncol. 10:5974682020.
|
|
113
|
Curigliano G, Criscitiello C, Gelao L and
Goldhirsch A: Molecular pathways: Human leukocyte antigen G
(HLA-G). Clin Cancer Res. 19:5564–5571. 2013.
|
|
114
|
Salomé B, Sfakianos JP, Ranti D, Daza J,
Bieber C, Charap A, Hammer C, Banchereau R, Farkas AM, Ruan DF, et
al: NKG2A and HLA-E define an alternative immune checkpoint axis in
bladder cancer. Cancer Cell. 40:1027–1043.e9. 2022.
|
|
115
|
Maeda A, Kawamura T, Ueno T, Usui N,
Eguchi H and Miyagawa S: The suppression of inflammatory
macro-phage-mediated cytotoxicity and proinflammatory cytokine
production by transgenic expression of HLA-E. Transpl Immunol.
29:76–81. 2013.
|
|
116
|
Chua HL, Serov Y and Brahmi Z: Regulation
of FasL expression in natural killer cells. Hum Immunol.
65:317–327. 2004.
|
|
117
|
Chen X, Lin Y, Yue S, Yang Y, Wang X, Pan
Z, Yang X, Gao L, Zhou J, Li Z, et al: Differential expression of
inhibitory receptor NKG2A distinguishes disease-specific exhausted
CD8+ T cells. MedComm. 2020:3e1112022.
|
|
118
|
Schumacher T, Bunse L, Pusch S, Sahm F,
Wiestler B, Quandt J, Menn O, Osswald M, Oezen I, Ott M, et al: A
vaccine targeting mutant IDH1 induces antitumour immunity. Nature.
512:324–327. 2014.
|
|
119
|
Li Y, Jiang W and Mellins ED: TCR-like
antibodies targeting autoantigen-mhc complexes: A mini-review.
Front Immunol. 13:9684322022.
|
|
120
|
Schachner LF, Phung W, Han G, Darwish M,
Bell A, Mellors JS, Srzentic K, Huguet R, Blanchette C and Sandoval
W: High-Throughput, quantitative analysis of peptide-exchanged MHCI
Complexes by native mass spectrometry. Anal Chem. 94:14593–14602.
2022.
|
|
121
|
Pourchet A, Fuhrmann SR, Pilones KA,
Demaria S, Frey AB, Mulvey M and Mohr I: CD8(+) T-cell immune
evasion enables oncolytic virus immunotherapy. EBioMedicine.
5:59–67. 2016.
|
|
122
|
Fischer C, Munks MW, Hill AB, Kroczek RA,
Bissinger S, Brand V, Schmittnaegel M, Imhof-Jung S, Hoffmann E,
Herting F, et al: Vaccine-induced CD8 T cells are redirected with
peptide-MHC class I-IgG antibody fusion proteins to eliminate tumor
cells in vivo. MAbs. 12:18348182020.
|
|
123
|
Eshkiki ZS, Agah S, Tabaeian SP, Sedaghat
M, Dana F, Talebi A and Akbari A: Neoantigens and their clinical
applications in human gastrointestinal cancers. World J Surg Oncol.
20:3212022.
|
|
124
|
Duperret EK, Perales-Puchalt A, Stoltz R,
G H H, Mandloi N, Barlow J, Chaudhuri A, Sardesai NY and Weiner DB:
A Synthetic DNA, multi-neoantigen vaccine drives predominately MHC
Class I CD8+ T-cell responses, impacting tumor challenge. Cancer
Immunol Res. 7:174–182. 2019.
|
|
125
|
Tran E: Neoantigen-Specific T cells in
adoptive cell therapy. Cancer J. 28:278–284. 2022.
|
|
126
|
Alspach E, Lussier DM, Miceli AP,
Kizhvatov I, DuPage M, Luoma AM, Meng W, Lichti CF, Esaulova E,
Vomund AN, et al: MHC-II neoantigens shape tumour immunity and
response to immunotherapy. Nature. 574:696–701. 2019.
|
|
127
|
Marty Pyke R, Thompson WK, Salem RM,
Font-Burgada J, Zanetti M and Carter H: Evolutionary Pressure
against MHC Class II binding cancer mutations. Cell.
175:416–428.e13. 2018.
|
|
128
|
Robbins PF: Tumor-Infiltrating lymphocyte
therapy and neoantigens. Cancer J. 23:138–143. 2017.
|
|
129
|
Jiang S, Geng S, Luo X, Zhang C, Yu Y,
Cheng M, Zhang S, Shi N and Dong M: Biomarkers of related driver
genes predict anti-tumor efficacy of immune checkpoint inhibitors.
Front Immunol. 13:9957852022.
|
|
130
|
Huang X, Zhang X, Bai X and Liang T:
Eating self for not be eaten: Pancreatic cancer suppresses
self-immunogenicity by autophagy-mediated MHC-I degradation. Signal
Transduct Target Ther. 5:942020.
|
|
131
|
Münz C: Canonical and non-canonical
functions of the autophagy machinery in MHC restricted antigen
presentation. Front Immunol. 13:8688882022.
|
|
132
|
Friedman LA, Bullock TN, Sloan EA, Ring KL
and Mills AM: MHC class I loss in endometrial carcinoma: A
potential resistance mechanism to immune checkpoint inhibition. Mod
Pathol. 34:627–636. 2021.
|
|
133
|
Deng Y, Xia X, Zhao Y, Zhao Z, Martinez C,
Yin W, Yao J, Hang Q, Wu W, Zhang J, et al: Glucocorticoid receptor
regulates PD-L1 and MHC-I in pancreatic cancer cells to promote
immune evasion and immunotherapy resistance. Nat Commun.
12:70412021.
|
|
134
|
Westrich JA, Vermeer DW, Silva A, Bonney
S, Berger JN, Cicchini L, Greer RO, Song JI, Raben D, Slansky JE,
et al: CXCL14 suppresses human papillomavirus-associated head and
neck cancer through antigen-specific CD8+ T-cell responses by
upregulating MHC-I expression. Oncogene. 38:7166–7180. 2019.
|
|
135
|
Zheng X, Sun Y, Li Y, Ma J, Lv Y, Hu Y,
Zhou Y and Zhang J: A novel tongue squamous cell carcinoma cell
line escapes from immune recognition due to genetic alterations in
HLA Class I Complex. Cells. 12:352022.
|
|
136
|
Aparicio B, Repáraz D, Ruiz M, Llopiz D,
Silva L, Vercher E, Theunissen P, Tamayo I, Smerdou C, Igea A, et
al: Identification of HLA class I-restricted immunogenic
neoantigens in triple negative breast cancer. Front Immunol.
13:9858862022.
|
|
137
|
Gorris MAJ, van der Woude LL, Kroeze LI,
Bol K, Verrijp K, Amir AL, Meek J, Textor J, Figdor CG and de Vries
IJM: Paired primary and metastatic lesions of patients with
ipilimumab-treated melanoma: High variation in lymphocyte
infiltration and HLA-ABC expression whereas tumor mutational load
is similar and correlates with clinical outcome. J Immunother
Cancer. 10:e0043292022.
|