RPL27 contributes to colorectal cancer proliferation and stemness via PLK1 signaling
- Authors:
- Published online on: June 29, 2023 https://doi.org/10.3892/ijo.2023.5541
- Article Number: 93
-
Copyright: © Park et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
In 2020, colorectal cancer (CRC) had the second highest mortality rate worldwide and was the third most common type of carcinoma, accounting for 10.0% of mortality and 9.4% incidence of all carcinomas (1). The causative factors of CRC include excessive red and processed meat consumption, excessive drinking, smoking, inflammatory bowel disease, obesity, diabetes and family history of CRC (2-8). In addition, genetic factors are involved in the development of colorectal neoplasia, including activation of oncogenes and inactivation of tumor suppressors (9,10). Approximately 25% of patients with CRC are diagnosed at an advanced stage and patients in the early stages have a high chance of developing metastases (up to 50%) (11,12). Evidence regarding the biological involvement of metastasis has shown that CRC stem cells (CSCs), which are highly tumorigenic and self-renewing, serve a pivotal role in developing metastatic CRC (13,14).
Currently, CRC treatment primarily involves surgery, radiation therapy and chemotherapy (14). The management of early-stage CRC relies on laparoscopic colectomy. Depending on the tumor location and CRC stage (stage III or IV), neoadjuvant or adjuvant chemotherapy is prescribed with or without concurrent radiation therapy. If regional or distant metastases are discovered upon diagnosis, a combination of surgery and other therapeutic options is used. Conventional chemotherapy strategies include administration of 5-fluorouracil, irinotecan and oxaliplatin, which block DNA synthesis or replication, to decrease the CRC tumor burden (15). More recently, bevacizumab and cetuximab have been used for targeted CRC treatment. VEGF inhibitor bevacizumab exhibits anticancer effects by inhibiting angiogenesis. Cetuximab, an EGFR inhibitor, selectively binds the cell membrane region of EGFR and prevents its activation, thereby inhibiting intracellular signal transduction (16). Although reports on the aforementioned targeted agents have shown a significant increase in survival and greater efficiency in eliminating metastatic CRC, these treatments are associated with an increased risk of gastrointestinal, hematological and cardiac toxicities as well as severe cutaneous toxicity (17). Thus, there is a need to identify molecular targets that can support the development of alternative treatment options and improve the therapeutic index.
Ribosomes are key cellular organelles with numerous functions, including the translation of mRNA into proteins and regulation of cellular metabolism. Protein constituents, known as ribosomal proteins (RPs), have different functions across species. RPs serve an important role in ribosome synthesis and function and act as scaffolds to enhance catalytic ability of ribosomal (r)RNA to synthesize proteins. Some RPs also have important additional ribosomal functions, such as DNA repair, transcriptional regulation and apoptosis (18). In addition to these housekeeping functions, previous studies have demonstrated that ribosome biogenesis plays a key role in the cell cycle and that the upregulation of ribosome biogenesis can increase the risk of cancer onset (19,20); this is termed extra-ribosomal function. One mechanism underlying this process is that enhanced rRNA transcription increases mouse double minute 2 protein (MDM2)-mediated p53 degradation, thereby increasing cell proliferation (21). Accumulating evidence has demonstrated that RPs are functionally associated with cancer progression and stemness (22-26). Our previous studies reported that RPL9 and RPL17 are involved in CRC progression through a functional connection with inhibitor of DNA-binding protein 1 (Id-1)/NF-κB and NIMA related kinase 2 (NEK2)/β-catenin signaling axes, respectively (22,23). Other studies have also reported that RPL23 upregulation induces invasiveness in lung cancer cell lines (24), and RPL39 affects metaplastic breast cancer through the ADAR1/iNOS/STAT3 pathway (25) and that RPs promote plasticity and stemness induction in glioma cells (26).
Although ribosomal protein L27 (RPL27) is upregulated in CRC clinical samples (27), to the best of our knowledge, the oncogenic role of RPL27 and the potential underlying mechanisms have not yet been defined. The present study aimed to investigate the effect of RPL27 on CRC progression and determine whether RPL27 gains extra-ribosomal function during CRC development.
Materials and methods
Analysis of RPL27 expression in clinical CRC samples
RNA sequencing gene expression (Level 3) data were downloaded from The Cancer Genome Atlas-Colon Cancer (TCGA-COAD) database at the Genomic Data Commons data portal (https://portal.gdc.cancer.gov). Patients lacking RPL27 mRNA expression data were excluded. The dataset included 327 samples (286 colorectal tumors and 41 normal mucosa). The correlation between RPL27 and PLK1 mRNA expression was assessed by Pearson's correlation coefficient analysis.
Survival analysis
Overall survival (OS) data (death event and survival time) of 228 patients with CRC were downloaded from the UCSD Xena Browser (xena.ucsc.edu). OS was calculated from date of diagnosis and mortality. For survival analysis, the median expression of RPL27 gene was used to divide the patients into high- and low-expression groups. Survival curves were determined by univariate Kaplan-Meier estimators and analyzed by log-rank test.
Cell culture
Human CRC cell lines HCT116 and HT29 were purchased from the Korean Cell Line Bank (KCLB; Seoul, South Korea). KCLB performed cell line authentication using STR profiling. Both cell lines were cultured in RPMI-1640 medium (Welgene, Inc.) supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum (FBS) (Welgene, Inc.), in a 37°C humidified incubator with a mixture of 95% air and 5% CO2.
Small interfering RNA (siRNA) transfection
Before siRNA transfection, cells were plated at a density of 30%. After 24 h, cells were transfected with 15 nM negative control (NC) or RPL27 siRNA for 5 h using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and Opti-MEM (Thermo Fisher Scientific, Inc.) at room temperature. The sequences of NC siRNA (Bioneer) were as follows: 5′-ACG UGA CAC GUU CGG AGA A(UU)-3′ (sense) and 5′-UUC UCC GAA CGU GUC ACG U-3′ (antisense). The bracket indicates overhang. Three variants of RPL27-specific siRNA (RPL27-1 siRNA, cat. no. s12205; RPL27-2 siRNA, cat. no. 9273 and RPL27-3 siRNA, cat. no. 9361) were purchased from Ambion (Thermo Fisher Scientific, Inc.). The sequences of RPL27 siRNA were as follows: RPL27-1 sense, 5′-ACA AAA CUG UCG UCA AUA A-3′ and antisense, 5′-UUA UUG ACG ACA GUU UUG U-3′; RPL27-2 sense, 5′-GGA UGU CUU CAG AGA UCC U-3′ and antisense, 5′-AGG AUC UCU GAA GAC AUC C-3′ and RPL27-3 sense, 5′-GGU CAA GUU UGA AGA GAG A-3′ and antisense, 5′-UCU CUC UUC AAA CUU GAC C-3′.
MTT assay
Cell proliferation was assessed using MTT reagent according to the manufacturer's instructions (Duchefa Biochemie). HCT116 and HT29 cells (1×103) were transfected with siRNAs, as aforementioned. After 96 h, cells were treated with MTT reagent for 1 h. DMSO was added to dissolve the purple formazan. The optical density was evaluated at a wavelength of 540 nm using an Asys IVM 340 microplate reader (Biochrom, Ltd.).
Reverse transcription-quantitative (RT-q)PCR
The mRNA expression was quantified using RT-qPCR. Total RNA in HCT116 or HT29 cells was purified and reverse-transcribed to cDNA using the RNeasy Plus Mini (Qiagen GmbH) and PrimeScript II 1st strand cDNA Synthesis kits (Takara Bio, Inc.), respectively, according to the manufacturer's protocol. Amplification of each target gene was performed using TB Green (Takara Bio) and corresponding pair of primers (RPL27 forward, 5′-TGG CTG GAA TTG ACC GCT AC-3′ and reverse, 5′-CCT TGT GGG CAT TAG GTG ATT G-3′; GAPDH forward, 5′-ACA TCA AGA AGG TGG TGA AG-3′ and reverse, 5′-GGT GTC GCT GTT GAA GTC-3′; polo-like kinase 1 (PLK1) forward, 5′-CTC AAC ACG CCT CAT CCT C-3′ and reverse, 5′-GTG CTC GCT CAT GTA ATT GC-3′; CD133 forward, 5′-AGT CGG AAA CTG GCA GAT AGC-3′ and reverse, 5′-GGT AGT GTT GTA CTG GGC CAA T-3′; NANOG forward, 5′-CGA TCT CCT GAC CTT GT-3′ and reverse, 5′-CAC GCC TGT AAA TCC CA-3′; CD44 forward, 5′-CTG CCG CTT TGC AGG TGT A-3′ and reverse, 5′-CAT TGT GGG CAA GGT GCT ATT-3′; OCT4 forward, 5′-CTT GAA TCC CGA ATG GAA AGG G-3′ and reverse, 5′-GTG TAT ATC CCA GGG TGA TCC TC-3′ and c-MYC forward, 5′-AAT GAA AAG GCC CCC AAG GTA GTT ATC C-3′ and reverse, 5′-GTC GTT TCC GCA ACA AGT CCT CTT C-3′) synthesized by Genotech. Thermocycling conditions were as follows: 95°C for 60 sec, followed by 45 cycles at 95°C for 10 sec, one cycle at 60°C for 10 sec and one cycle at 72°C for 10 sec. Relative mRNA expression was assessed using a LightCycler 96 and quantified using the 2−ΔΔCq method (28) in LightCycler 96 software version 1.1 (Roche Diagnostics). In addition, mRNA levels were normalized to GAPDH as a control.
Clonogenic assay
A total of 1×103 HCT116 or HT29 cells transfected with NC or RPL27 siRNA were seeded onto 6-well plates and incubated in RPMI-1640 medium at 37°C for 7-10 days until viable cells propagated to colonies for quantification. The colonies were fixed with 100% methanol for 15 min at room temperature and stained with 0.5% crystal violet for 30 min at room temperature. The number of colonies including >50 cells was counted manually under a light microscope (1× magnification).
Cell cycle analysis and apoptosis assay
CRC cells were plated in 100-mm dishes and harvested following 72 h transfection with control or target siRNA. To monitor cell cycle progression and apoptosis induction, the collected cells were stained with propidium iodide solution containing RNase A and FITC Annexin V Apoptosis Detection kit I (both BD Biosciences). The proportion of cells in each cell cycle phase and proportion of apoptotic cells were analyzed using a FACS Canto II flow cytometer and quantified using FACSDiva software 7.0 (both BD Bioscience). The total percentage of apoptotic cells were estimated by counting early + late apoptotic cells.
In vivo xenograft assay
To assess the therapeutic effect of RPL27 targeting in vivo, xenograft assay using 5 male BALB/c nude mice (age, 4 weeks; weight, 17.5-19.5 g; Orientbio) was performed. The housing conditions were as follows: 24°C, 50-60% humidity, 12:12 light/dark cycle and free access to food and water. HCT116 cells were transfected with 15 nM NC or RPL27-1 siRNA, as aforementioned. After 24 h, 1×106 cells were resuspended in RPMI-1640 medium (Welgene) containing Matrigel (Corning, Inc.) and injected subcutaneously into the left or right flank of mice. Tumor size was measured for 21 days at 2-3 day intervals using a Vernier Caliper and the tumor volume was calculated as width2 × length × 1/2 (n=5/treatment group). The mice were sacrificed at day 21 by CO2 euthanasia with flow rate of 30% volume/min. Death was confirmed by loss of heartbeat, breathing and pupil response. The tumor weight was then evaluated. All animal experiments were approved (approval no. KM-2021-03R1) by Keimyung University Institutional Animal Care and Use Committee. Humane experimental endpoints were the occurrence of tumor >10% of animal body weight, tumor size >2,000 mm3 and weight loss >20% of animal body weight.
RNA sequencing and functional network analysis
To compare the pattern of global gene expression between NC and RPL27-1 siRNA-treated CRC cells, RNA sequencing was performed as previously described (22). All sequencing data were deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE78195). Differentially expressed genes were selected based on at least 2-fold in CRC cells with RPL27 depletion were using Bootstrap ANOVA method (29). P≤0.001 were considered statistically significant. The significantly overlapping functional pathways of differentially expressed genes were analyzed using ingenuity pathway analysis (IPA, www.ingenuity.com).
Western blotting
HCT116 and HT29 cells were suspended in RIPA buffer containing 0.01% protease and phosphatase inhibitor cocktail (both Thermo Scientific Fisher, Inc.) 48 h after siRNA transfection. The amount of protein was quantified by Pierce BCA protein assay kit. Equal amounts (50 µg) of total protein were separated by 10-15% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Roche Diagnostics). The membranes were blocked at room temperature for 30 min with 5% milk/TBS + 0.05% Tween-20 and incubated overnight at 4°C with primary antibodies against RPL27 (cat. no. #PA5-88938; Invitrogen; Thermo Fisher Scientific, Inc.), PLK1 (cat. no. sc-17783; Santa Cruz Biotechnology, Inc.), phosphorylated (p-)CDC25C (cat. no. #9529; Cell Signaling Technology, Inc.), CDC25C (cat. no. sc-327; Santa Cruz Biotechnology, Inc.), CDK1 (cat. no. sc-54; Santa Cruz Biotechnology, Inc.), cyclin B1 (cat. no. sc-245; Santa Cruz Biotechnology, Inc.), β-actin (cat. no. sc-47778; Santa Cruz Biotechnology, Inc.), Flag (cat. no. F1804; Sigma Aldrich; Merck KGaA) and CD133 (cat. no. #64326; Cell Signaling Technology, Inc.). Horseradish peroxidase-conjugated anti-mouse (cat. no. 115-035-062; Jackson ImmunoResearch Laboratories, Inc.) and goat anti-rabbit IgG (cat. no. sc-2301; Santa Cruz Biotechnology, Inc.) were used as the secondary antibodies and incubated for 1 h at room temperature. Immuno-reactive bands were visualized using FUSIOM SOLO S (Vilber Lourmat) and quantified with ImageJ software (version 1.8.0, National Institutes of Health).
Migration and invasion assays
Migration and invasion assays were performed using Transwell migration assay (cat. no. #3422) and 24-well Matrigel invasion chambers (cat. no. #354480; both Corning, Inc.), respectively. siRNA-transfected cells were plated into the upper chamber with serum-free RPMI-1640 medium (Welgene, Inc.; 5×104 cells/well) and incubated for 24 h at 37°C. The lower chamber were filled with 10% FBS-containing RPMI-1640 medium (Welgene, Inc.). The cells were fixed and stained with 0.5% crystal violet for 30 min at room temperature. Images were captured at 100× magnification under a light microscope.
Overexpression of RPL27
To investigate the biological effects of RPL27 overexpression, HCT116 cells were cultured in a 24-well plate and transiently transfected with 0.8 µg/µl RPL27-expressing or pCMV6-entry vector (empty vector; both OriGene Technologies, Inc.) for 5 h using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and Opti-MEM (Thermo Fisher Scientific, Inc.) at 37°C. After 24 h, RPL27 expression was compared by RT-qPCR and western blotting, as aforementioned.
Sphere-forming assay
CRC cells were suspended in serum-free DMEM/F-12 (containing 2% B27, 4 ng/ml insulin, 10 ng/ml basic FGF and 20 ng/ml EGF; all Invitrogen; Thermo Fisher Scientific, Inc.) and 5×103 cells/well were plated on 24-well ultralow attachment plates (Corning, Inc.). Following 24 h culture at 37°C, 15 nM NC or RPL27-1 siRNA complexed with Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) was added to CRC cells. Sphere size and number were evaluated by iSolution Lite software (version 26.1, IMT i-Solution, Inc.) under a light microscope 8 days after siRNA transfection.
Isolation of CD133+ CSC population by flow cytometry
CD133 MicroBead kit (cat. no. 130-100-857) and AutoMACS Pro (both Miltenyi Biotec, Inc.) were used to isolate CD133+ HT29 CSC populations from HT29 parental cell culture according to the manufacturer's instructions. Magnetically labeled CD133+ and unlabeled CD133− cells were separately incubated with isotype control (mouse IgG2b APC conjugate; cat. no. 130-122-932; Miltenyi Biotec, Inc.) or CD133/2 antibody (anti-human APC conjugate; cat. no. 130-113-746; Miltenyi Biotec, Inc.). Separation efficiency was evaluated using a FACSCanto II flow cytometer and quantified using FACSDiva software 7.0 (both BD Biosciences).
Statistical analysis
All statistical analyses were repeated at least three times, except analysis of TCGA data and gene expression data. Unpaired Student's t test and one-way ANOVA with Dunnett's post hoc test were used for statistical comparison of two and multiple groups, respectively. Data are presented as the mean ± SEM. P<0.05 was considered to indicate a statistically significant difference.
Results
RPL27 overexpression correlates with poor survival of CRC patients
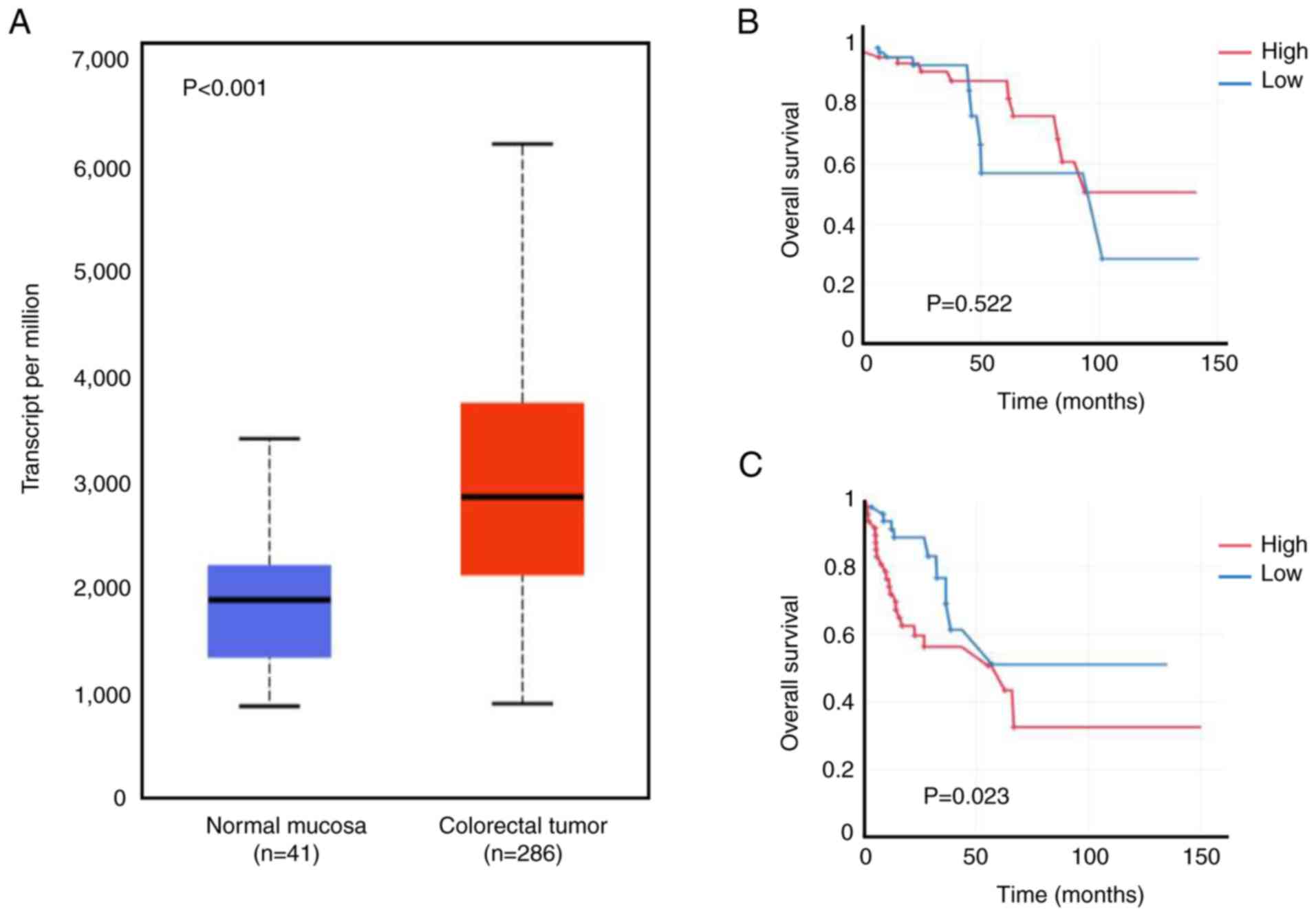
RPL27 is upregulated in CRC tissue (27); the present study verified its overexpression by accessing TCGA-Colon adenocarcinoma big data (286 colorectal tumors vs. 41 normal mucosa). Expression of RPL27 in CRC tumors was significantly higher than in the normal mucosa (Fig. 1A). It was analyzed whether increased expression affected survival of patients with CRC using TCGA data. RPL27 overexpression resulted in poorer overall survival in the stage III and IV than in stage I and II groups (Fig. 1B and C), implying that the biological and clinical impact of RPL27 expression increased as CRC progressed.
RPL27 silencing inhibits CRC cell proliferation
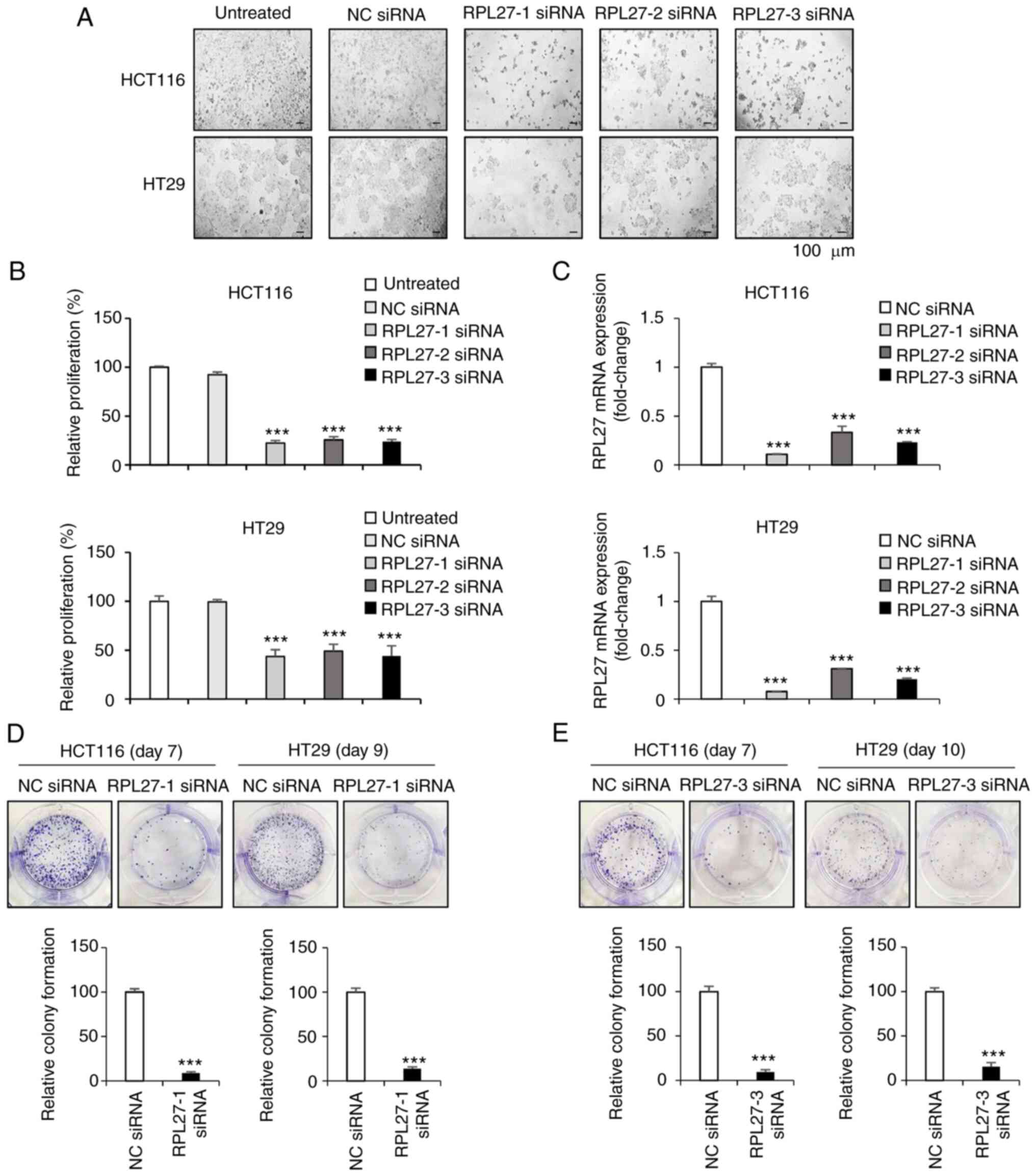
To investigate the effect of RPL27 silencing on cell proliferation, HCT116 and HT29 cells were transfected with NC or RPL27 siRNA. To select the siRNA with the highest treatment efficacy and target gene silencing ability, three sequences of RPL27-specific siRNAs (RPL27-1, -2 and -3) were tested. After 96 h, all three RPL27 siRNAs caused significant proliferation suppression in both HCT116 and HT29 cells, as determined by microscopic observation and MTT assay (Fig. 2A and B). Proliferation of cells treated with RPL27 siRNAs was inhibited by >50% compared with the control group. RT-qPCR was used to measure mRNA levels of RPL27 after transfection. All three siRNAs significantly inhibited the expression of RPL27 and this was greatest following RPL27-1 siRNA transfection (Fig. 2C). Similarly, treatment with RPL27-1 siRNA efficiently decreased colony formation by ~90 and 86% in HCT116 and HT29 cells, respectively, after 7 and 9 days of target siRNA treatment (Fig. 2D). The ability of RPL27-3 siRNA to inhibit colony formation in HCT116 and HT29 cells was similar to that of RPL27-1 siRNA (Fig. 2E). RPL27-1 siRNA was selected for all subsequent experiments. These results suggested that RPL27 was functionally involved in CRC tumor cell proliferation.
RPL27 silencing suppresses cell cycle progression and induces apoptosis in CRC cells
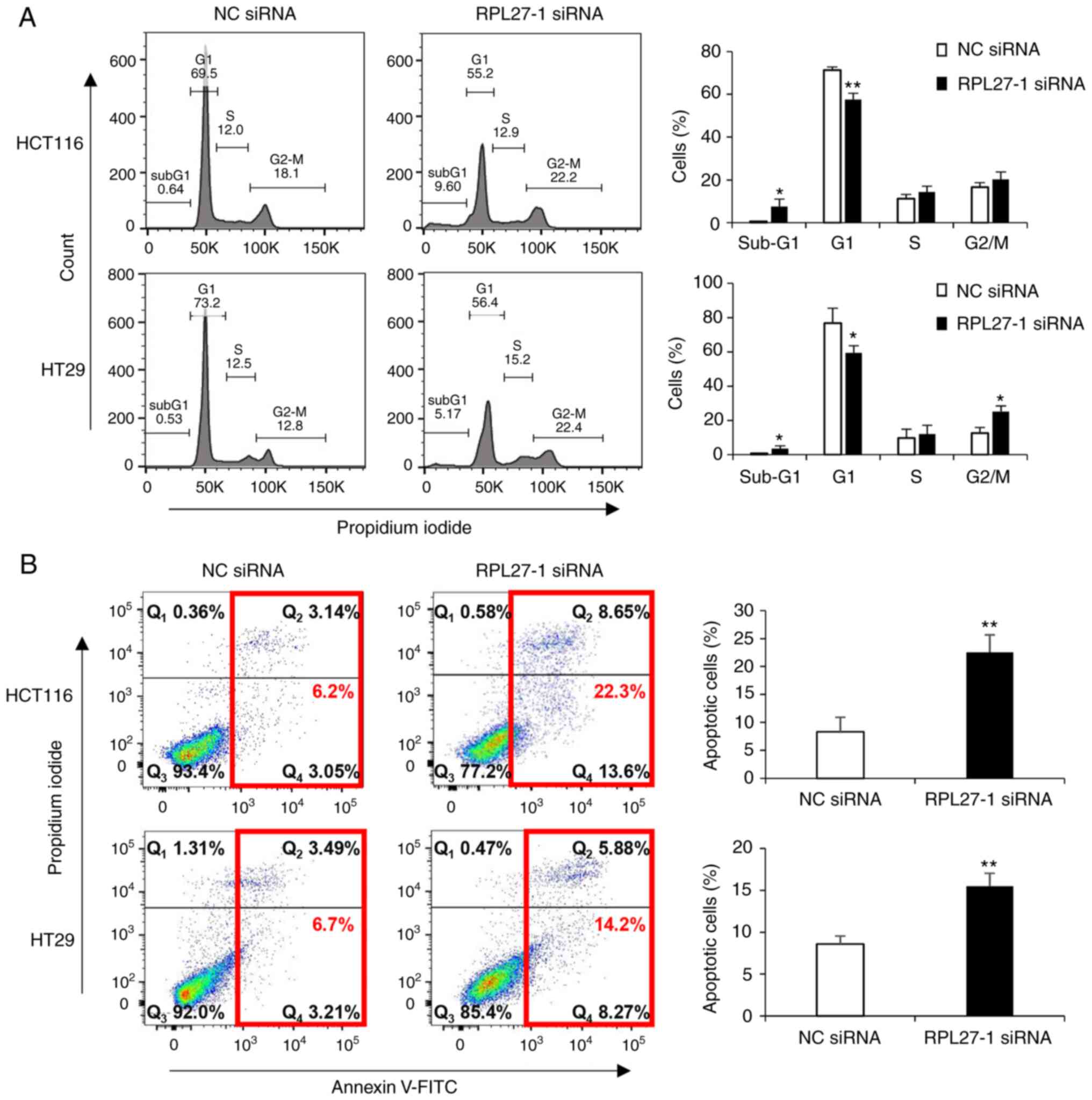
It was investigated whether the inhibition of CRC cell proliferation via RPL27 silencing was elicited by decreased cell cycle progression and/or the induction of apoptotic cell death. Flow cytometry analysis confirmed that RPL27-silenced cells displayed an increased proportion of sub-G1 phase in both HCT116 and HT29 cells (Fig. 3A). As the sub-G1 population contains apoptotic cells (30), apoptosis assay was performed by staining CRC cells with Annexin V. This showed that targeting RPL27 increased the number of apoptotic cells at 72 h after transfection (Fig. 3B). These results indicated that RPL27 silencing-mediated inhibition of CRC cell proliferation occurred due to decreased cell cycle progression and apoptosis induction.
RPL27 silencing suppresses CRC growth in vivo
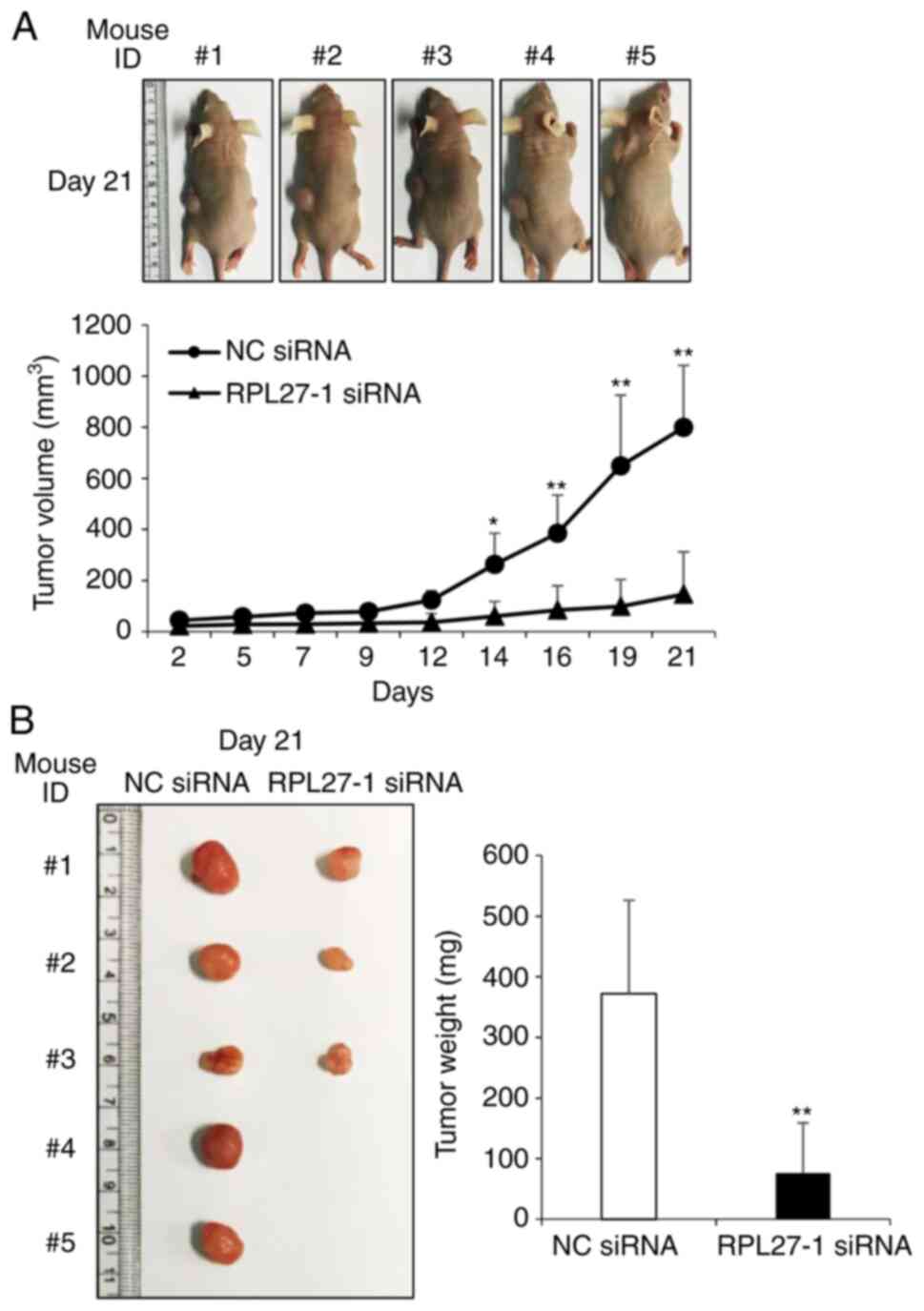
To investigate whether RPL27 targeting affects in vivo tumor growth, a xenograft mouse model was constructed. HCT116 cells were transfected with NC or RPL27-1 siRNA for 24 h and then injected into BALB/c nude mice. The size of the tumor was measured at 2-3 day intervals. After 14 days, tumors in the NC group were significantly larger than those in the RPL27 siRNA-group (Fig. 4A). After 21 days, a significant difference in weight was observed between the control and RPL27-silencing groups (Fig. 4A and B). These results showed that targeting RPL27 significantly decreased the growth of human CRC xenografts in vivo.
RPL27 silencing downregulates PLK1 expression in CRC cells
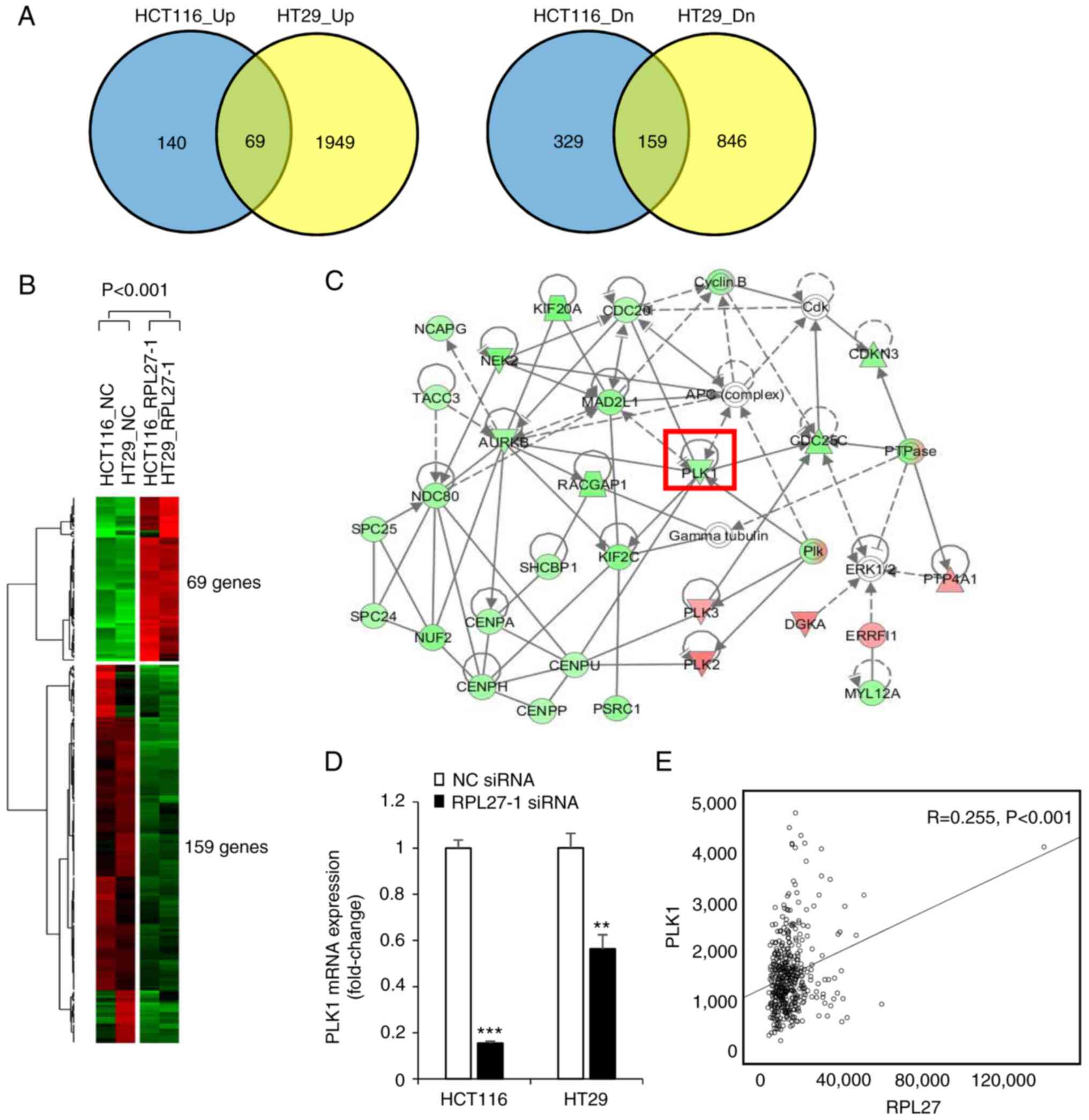
To determine molecular mechanisms by which RPL27 silencing induces phenotypical changes, RNA sequencing was performed to compare patterns of global gene expression in RPL27-silenced HCT116 and HT29 cells with those in control cells transfected with NC siRNA. When at least a 2-fold change was defined, global gene expression analysis revealed that RPL27 silencing resulted in up- and downregulation of 697 RNA transcripts in HCT116 and 3,023 RNA transcripts in HT29 cells (Fig. 5A). Overlapping these two gene sets generated 228 commonly dysregulated genes (69 up- and 159 downregulated genes), which were considered to be common RPL27-silencing signatures (Fig. 5A and B). Subsequent IPA showed that 228 mRNA transcripts were functionally enriched in the top five networks (Fig. S1). Expression of PLK1, which is known to play an important role in mitotic cell cycle progression and migration (31), was downregulated by RPL27 silencing (Fig. 5C). The decrease in PLK1 mRNA expression was validated by RT-qPCR in both HCT116 and HT29 cells (Fig. 5D). This association was confirmed in TCGA data: RPL27 mRNA expression was positively correlated with PLK1 expression (R=0.255) in CRC tissue (Fig. 5E). These findings suggested that the function of RPL27 was associated with PLK1 expression.
Targeting RPL27 decreases metastatic potential and stemness in CRC
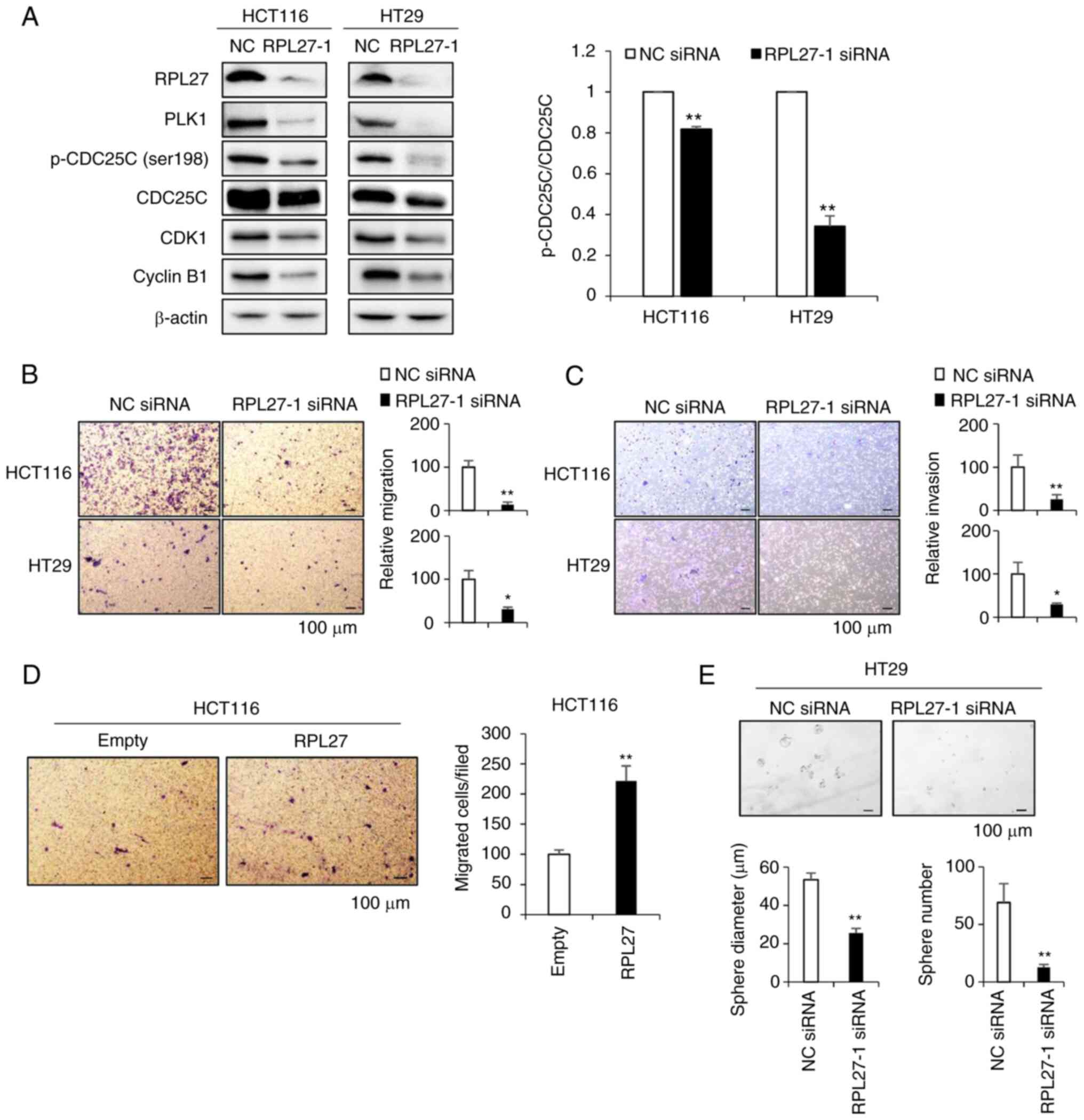
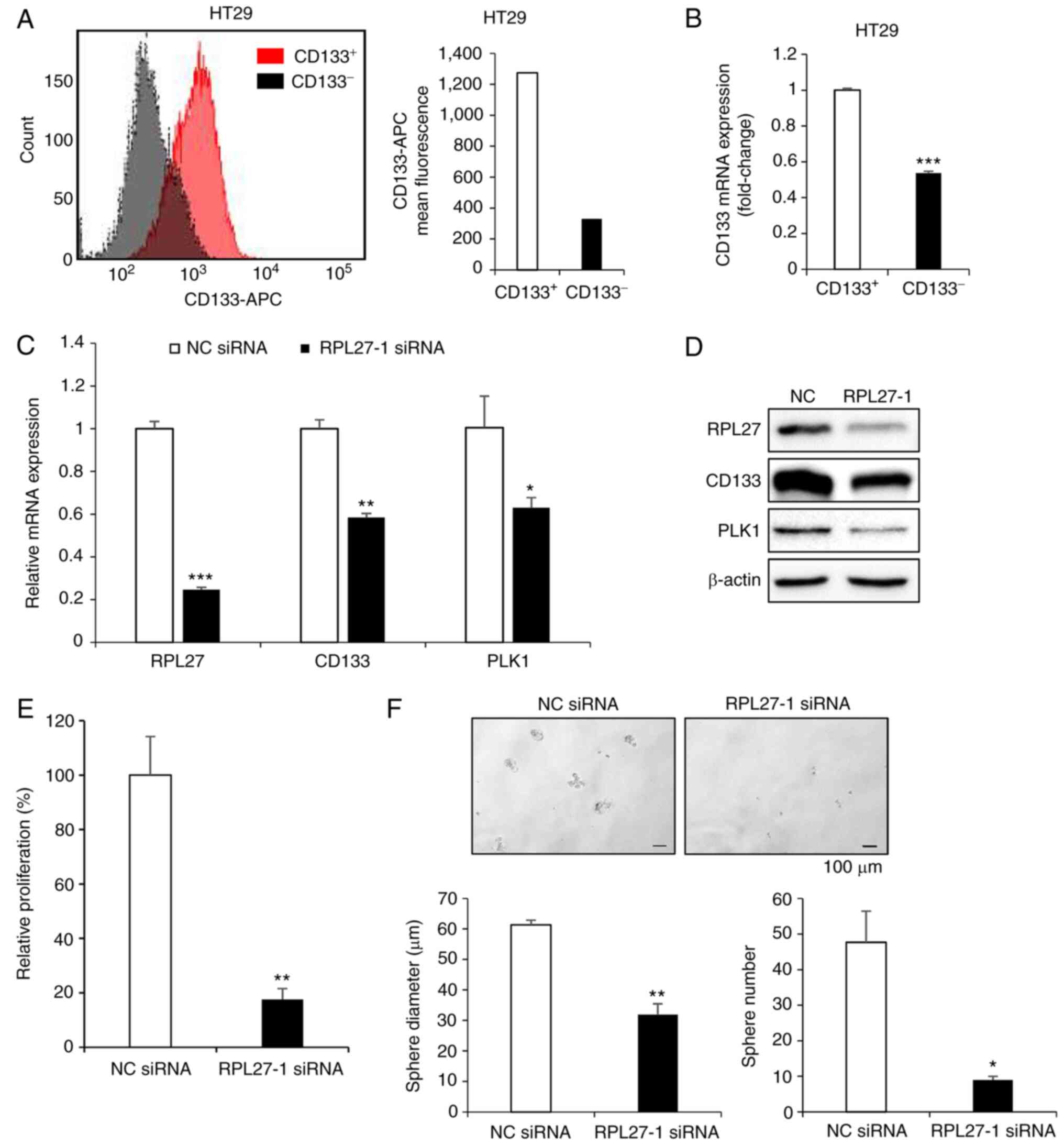
Consistent with transcriptional profiling of RPL27-silenced cells, the protein expression of PLK1 was also decreased (Fig. 6A). PLK1 phosphorylates CDC25C at serine 198 residue for subsequent cyclin B1/CDK1 activation and mitotic entry (32). Here, silencing of RPL27 decreased the levels of p-CDC25C (ser198) and CDC25C (Fig. 6A). When normalized to CDC25C expression, silencing of RPL27 decreased phosphorylation of CDC25C by ~20 and 65% in HCT116 and HT29 cell lines, respectively, and decreased levels CDK1 and cyclin B1 protein, which are downstream effectors of PLK1 signaling that induce transition of G2/M (33). PLK1 silencing leads to decreased migration and invasion of various types of cancer, including CRC (31,34-36). Therefore, it was investigated whether targeting RPL27 inhibited metastatic potential of CRC cells using migration and invasion assay. Silencing of RPL27 effectively decreased migration (Fig. 6B) and invasion (Fig. 6C) abilities of both HCT116 and HT29 cells. Next, it was determined whether RPL27 overexpression could reverse the effect of RPL27 silencing on CRC migration and if RPL27 silencing could inhibit CRC stemness. Overexpression assay was performed in HCT116 cells as the basal expression of RPL27 protein in HCT116 cells was lower than that in HT29 cells (Fig. S2A). As expected, HCT116 cells transfected with RPL27-expressing vector (Fig. S2B and C) exhibited improved migration ability compared with control vector-transfected cells (Fig. 6D). The levels of CSC marker CD133 and the efficiency of CSC sphere formation are associated (37). To verify the functional involvement of RPL27 in CRC stemness, HT29 cells were used as expression of CD133 in HT29 cells is significantly higher than in HCT116 cells (38). RPL27-1 siRNA suppressed sphere formation in HT29 parental cells compared with NC siRNA (Fig. 6E). PLK1 inhibitor decreased CD133+ CSC proliferation (39). Given the functional significance of PLK1 signaling in CSC (40) and suppressed PLK1 expression following RPL27 silencing, it was investigated whether RPL27 targeting affected CSC proliferation and stemness. CD133+ CSC population of HT29 cells were sorted from the CD133− population by FACS using APC-conjugated CD133 antibody (Fig. 7A); successful separation was confirmed by demonstrating that expression of CD133 mRNA in the CD133+ population was higher than that in the CD133− population (Fig. 7B). RPL27 silencing decreased levels of CD133 and PLK1 in CD133+ CSCs (Fig. 7C and D) and significantly decreased the proliferation rate and sphere-forming capacity of CD133+ cells (Fig. 7E and F). Taken together, these data indicated that RPL27 was functionally associated with CRC stemness via PLK1 signaling.
Discussion
The present study confirmed that RPL27 was upregulated during CRC development and that its overexpression negatively affected survival of patients with CRC patient. To verify whether RPL27 exhibited extra-ribosomal function in CRC progression, in vitro and in vivo experiments were performed, which demonstrated that RPL27 served an important role in CRC development. The present study confirmed inhibition of cell proliferation and colony formation and induction of apoptosis induced by RPL27 silencing. In vivo experiments also showed that inhibiting RPL27 expression suppressed the growth of CRC xenografts. The present study aimed to identify the molecular mechanism by which RPL27 depletion inhibited CRC proliferation. Of the 228 commonly dysregulated genes following RPL27 silencing, the present study investigated the association between PLK1 and extra-ribosomal function of RPL27 as PLK1 is involved in both mitosis and cancer cell stemness and preventing metastasis is crucial in managing caner severity (31,34-36,39,40).
PLK1, a serine/threonine protein kinase, is overexpressed in numerous types of cancer, including colorectal, breast, renal, hepatocellular and lung cancer (41-46). Its functions include mitosis entry and G2/M checkpoint control, centrosome and cell cycle regulation, spindle assembly and chromosome separation regulation and promotion of DNA replication and cytokinesis (47). Previous studies have shown that PLK1 depletion induces apoptosis in cancer cells (48,49) and PLK1 expression correlates with tumor size and invasion and lymphatic metastasis in CRC (31). Therefore, PLK1 has been used as a therapeutic target for cancer treatment. To the best of our knowledge, however, the functional association between RPL27 and PLK1 has not yet been revealed. In cell cycle regulation, PLK1 controls the activity of the CDK1/cyclin B1 complex, which serves a vital role in the transition to the G2/M phase of the cell cycle via CDC25C phosphorylation (32,50). Consistent with previous observations (33,51), the present study showed that the protein levels of PLK1, p-CDC25C (ser198), CDK1 and cyclin B1 were decreased after 48 h RPL27 silencing, suggesting cell cycle arrest in the G2/M phase.
Identifying target genes functionally associated with CSC biology may provide novel treatment options against CRC to prevent secondary tumor formation. In previous studies, PLK1 inhibition led to proliferation inhibition in colon cancer, normal pediatric neural and breast CSCs (39,52,53). CD133 functions as a marker for CSC isolation and identification in CRC (54). In the present study, RPL27 silencing blocked cell cycle progression by inactivating PLK1 signaling and decreased the sphere-forming capacity of both the parental CRC cell culture and isolated CD133+ CSC cells. The downregulation of PLK1 is a key molecular response that promotes RPL27 silencing-mediated proliferation inhibition and stemness reduction in CRC. These molecular alterations may partially explain how targeting RPL27 inhibits proliferation and stemness of CRC cells and provide a basis for development of RPL27 inhibitors. To the best of our knowledge, the present study is the first to report that the function of RPL27 is associated with CRC proliferation and stemness. However, the present study did not fully elucidate how silencing RPL27 affects PLK1 expression. This should be explored in future studies. In addition to PLK1, there may be other factors that affect oncogenic function of RPL27 or mediate its association with the PLK1 signaling pathway. Future studies should investigate the specific regulatory mechanisms by which RPL27 controls expression of PLK1 in both CRC cells and CSCs and whether RPL27 is involved via different mechanisms throughout CRC progression.
In summary, the present study defined a novel role of RPL27 in augmenting CRC progression and promoting stemness and identified RPL27 silencing-induced molecular alterations, which indicate its potential therapeutic efficacy. Disrupted PLK1 signaling was observed in parental cell culture and isolated CD133+ CSCs with RPL27 depletion. Taken together, the present findings demonstrated the potential of RPL27 as a therapeutic target for both primary CRC treatment and metastasis control.
Supplementary Data
Availability of data and materials
The RNA sequencing datasets generated and/or analyzed during the current study are available in the GEO repository, accession number no. GSE78195 (ncbi.nlm.nih. gov/geo/query/acc.cgi?acc=GSE78195).
Authors' contributions
SP, CHC and YL conceived the study, designed the experiments and wrote the manuscript. SP, DS, EJ, JYP, JL and SK performed the experiments and analyzed data. BJ, JIK and SI designed experimental methods SP constructed figures. CHC and YL supervised the study. CHC acquired funding. CHC and YL confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were approved (approval no. KM-2021-03R1; 31 March 2021) by Keimyung University Institutional Animal Care and Use Committee (Daegu, South Korea).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Bisa Research Grant of Keimyung University (grant no. 20170014).
References
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Chan DS, Lau R, Aune D, Vieira R, Greenwood DC, Kampman E and Norat T: Red and processed meat and colorectal cancer incidence: Meta-analysis of prospective studies. PLoS One. 6:e204562011. View Article : Google Scholar : PubMed/NCBI | |
|
Fedirko V, Tramacere I, Bagnardi V, Rota M, Scotti L, Islami F, Negri E, Straif K, Romieu I, La Vecchia C, et al: Alcohol drinking and colorectal cancer risk: An overall and dose-response meta-analysis of published studies. Ann Oncol. 22:1958–1972. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Jo A and Oh H: Incidence of colon cancer related to cigarette smoking and alcohol consumption in adults with metabolic syndrome: Prospective cohort study. J Korean Acad Nurs. 49:713–723. 2019.In Korean. View Article : Google Scholar | |
|
von Roon AC, Reese G, Teare J, Constantinides V, Darzi AW and Tekkis PP: The risk of cancer in patients with Crohn's disease. Dis Colon Rectum. 50:839–855. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Ma Y, Yang Y, Wang F, Zhang P, Shi C, Zou Y and Qin H: Obesity and risk of colorectal cancer: A systematic review of prospective studies. PLoS One. 8:e539162013. View Article : Google Scholar : PubMed/NCBI | |
|
Jiang Y, Ben Q, Shen H, Lu W, Zhang Y and Zhu J: Diabetes mellitus and incidence and mortality of colorectal cancer: A systematic review and meta-analysis of cohort studies. Eur J Epidemiol. 26:863–876. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Taylor DP, Burt RW, Williams MS, Haug PJ and Cannon-Albright LA: Population-based family history-specific risks for colorectal cancer: A constellation approach. Gastroenterology. 138:877–885. 2010. View Article : Google Scholar | |
|
Fearon ER and Vogelstein B: A genetic model for colorectal tumorigenesis. Cell. 61:759–767. 1990. View Article : Google Scholar : PubMed/NCBI | |
|
Bahnassy AA, Zekri AR, Salem SE, Abou-Bakr AA, Sakr MA, Abdel-Samiaa AG and Al-Bradei M: Differential expression of p53 family proteins in colorectal adenomas and carcinomas: Prognostic and predictive values. Histol Histopathol. 29:207–216. 2014. | |
|
Mody K, Baldeo C and Bekaii-Saab T: Antiangiogenic therapy in colorectal cancer. Cancer J. 24:165–170. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Chibaudel B, Tournigand C, Bonnetain F, Richa H, Benetkiewicz M, André T and de Gramont A: Therapeutic strategy in unresectable metastatic colorectal cancer: An updated review. Ther Adv Med Oncol. 7:153–169. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
O'Brien CA, Pollett A, Gallinger S and Dick JE: A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 445:106–110. 2007. View Article : Google Scholar | |
|
Kozovska Z, Gabrisova V and Kucerova L: Colon cancer: Cancer stem cells markers, drug resistance and treatment. Biomed Pharmacother. 68:911–916. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Braun MS and Seymour MT: Balancing the efficacy and toxicity of chemotherapy in colorectal cancer. Ther Adv Med Oncol. 3:43–52. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Soueidy C, Skaff S, Stephan F and Kattan J: Cetuximab severe cutaneous toxicity a gateway for bacteremia: Case report. Anticancer Drugs. 34:187–189. 2023. View Article : Google Scholar | |
|
Hu CY, Chan W, Delclos GP and Du XL: Adjuvant chemotherapy and risk of gastrointestinal, hematologic, and cardiac toxicities in elderly patients with stage III colon cancer. Am J Clin Oncol. 35:228–236. 2012. View Article : Google Scholar | |
|
Cai Y, Zhang C, Hao L, Chen J, Xie P and Chen Z: Systematic identification of seven ribosomal protein genes in bighead carp and their expression in response to microcystin-LR. J Toxicol Sci. 41:293–302. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Montanaro L, Treré D and Derenzini M: Nucleolus, ribosomes, and cancer. Am J Pathol. 173:301–310. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Derenzini M, Montanaro L and Trerè D: Ribosome biogenesis and cancer. Acta Histochem. 119:190–197. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Donati G, Bertoni S, Brighenti E, Vici M, Treré D, Volarevic S, Montanaro L and Derenzini M: The balance between rRNA and ribosomal protein synthesis up- and downregulates the tumour suppressor p53 in mammalian cells. Oncogene. 30:3274–3288. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Baik IH, Jo GH, Seo D, Ko MJ, Cho CH, Lee MG and Lee YH: Knockdown of RPL9 expression inhibits colorectal carcinoma growth via the inactivation of Id-1/NF-κB signaling axis. Int J Oncol. 49:1953–1962. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Ko MJ, Seo YR, Seo D, Park SY, Seo JH, Jeon EH, Kim SW, Park KU, Koo DB, Kim S, et al: RPL17 promotes colorectal cancer proliferation and stemness through ERK and NEK2/β-catenin signaling pathways. J Cancer. 13:2570–2583. 2022. View Article : Google Scholar : | |
|
Liu F, Li Y, Yu Y, Fu S and Li P: Cloning of novel tumor metastasis-related genes from the highly metastatic human lung adenocarcinoma cell line Anip973. J Genet Genomics. 34:189–195. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Dave B, Gonzalez DD, Liu ZB, Li X, Wong H, Granados S, Ezzedine NE, Sieglaff DH, Ensor JE, Miller KD, et al: Role of RPL39 in metaplastic breast cancer. J Natl Cancer Inst. 109:djw2922016. View Article : Google Scholar | |
|
Hide T, Shibahara I, Inukai M, Shigeeda R and Kumabe T: Ribosomes and ribosomal proteins promote plasticity and stemness induction in glioma cells via reprogramming. Cells. 11:21422022. View Article : Google Scholar : PubMed/NCBI | |
|
Sim EU, Bong IP, Balraj P, Tan SK, Jamal R, Sagap I, Nadeson S, Rose IM and Lom PKM: A preliminary study of differentially expressed genes in Malaysian colorectal carcinoma cases. Trop Life Sci Res. 17:19–37. 2006. | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar | |
|
McHugh ML: Multiple comparison analysis testing in ANOVA. Biochem Med (Zagreb). 21:203–209. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Riccardi C and Nicoletti I: Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc. 1:1458–1461. 2006. View Article : Google Scholar | |
|
Han DP, Zhu QL, Cui JT, Wang PX, Qu S, Cao QF, Zong YP, Feng B, Zheng MH and Lu AG: Polo-like kinase 1 is overexpressed in colorectal cancer and participates in the migration and invasion of colorectal cancer cells. Med Sci Monit. 18:BR237–BR246. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Gutteridge RE, Ndiaye MA, Liu X and Ahmad N: Plk1 inhibitors in cancer therapy: From laboratory to clinics. Mol Cancer Ther. 15:1427–1435. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Bhosale PB, Vetrivel P, Ha SE, Kim HH, Heo JD, Won CK, Kim SM and Kim GS: Iridin Induces G2/M phase cell cycle arrest and extrinsic apoptotic cell death through PI3K/AKT signaling pathway in AGS gastric cancer cells. Molecules. 26:28022021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Z, Zhang G, Gao Z, Li S, Li Z, Bi J, Liu X, Li Z and Kong C: Comprehensive analysis of differentially expressed genes associated with PLK1 in bladder cancer. BMC Cancer. 17:8612017. View Article : Google Scholar : PubMed/NCBI | |
|
Song R, Hou G, Yang J, Yuan J, Wang C, Chai T and Liu Z: Effects of PLK1 on proliferation, invasion and metastasis of gastric cancer cells through epithelial-mesenchymal transition. Oncol Lett. 16:5739–5744. 2018.PubMed/NCBI | |
|
Wu ZY and Wei N: Knockdown of PLK1 inhibits invasion and promotes apoptosis in glioma cells through regulating autophagy. Eur Rev Med Pharmacol Sci. 22:2723–2733. 2018.PubMed/NCBI | |
|
Gao J, Yang T, Wang X, Zhang Y, Wang J, Zhang B, Tang D, Liu Y, Gao T, Lin Q, et al: Identification and characterization of a subpopulation of CD133+ cancer stem-like cells derived from SK-UT-1 cells. Cancer Cell Int. 21:1572021. View Article : Google Scholar | |
|
Asadzadeh Z, Mansoori B, Mohammadi A, Kazemi T, Mokhtarzadeh A, Shanehbandi D, Hemmat N, Derakhshani A, Brunetti O, Safaei S, et al: The combination effect of Prominin1 (CD133) suppression and Oxaliplatin treatment in colorectal cancer therapy. Biomed Pharmacother. 137:1113642021. View Article : Google Scholar : PubMed/NCBI | |
|
Francescangeli F, Patrizii M, Signore M, Federici G, Di Franco S, Pagliuca A, Baiocchi M, Biffoni M, Ricci Vitiani L, Todaro M, et al: Proliferation state and polo-like kinase1 dependence of tumorigenic colon cancer cells. Stem Cells. 30:1819–1830. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Grinshtein N, Datti A, Fujitani M, Uehling D, Prakesch M, Isaac M, Irwin MS, Wrana JL, Al-Awar R and Kaplan DR: Small molecule kinase inhibitor screen identifies polo-like kinase 1 as a target for neuroblastoma tumor-initiating cells. Cancer Res. 71:1385–1395. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Takahashi T, Sano B, Nagata T, Kato H, Sugiyama Y, Kunieda K, Kimura M, Okano Y and Saji S: Polo-like kinase 1 (PLK1) is overexpressed in primary colorectal cancers. Cancer Sci. 94:148–152. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Golsteyn RM, Lane HA, Mundt KE, Arnaud L and Nigg EA: The family of polo-like kinases. Prog Cell Cycle Res. 2:107–114. 1996.PubMed/NCBI | |
|
Maire V, Némati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, Gravier E, Marty-Prouvost B, De Koning L, Lang G, et al: Polo-like kinase 1: A potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 73:813–823. 2013. View Article : Google Scholar | |
|
Zhang G, Zhang Z and Liu Z: Polo-like kinase 1 is overexpressed in renal cancer and participates in the proliferation and invasion of renal cancer cells. Tumour Biol. 34:1887–1894. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
He ZL, Zheng H, Lin H, Miao XY and Zhong DW: Overexpression of polo-like kinase1 predicts a poor prognosis in hepatocellular carcinoma patients. World J Gastroenterol. 15:4177–4182. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Yoon HE, Kim SA, Choi HS, Ahn MY, Yoon JH and Ahn SG: Inhibition of Plk1 and Pin1 by 5′-nitro-indirubinoxime suppresses human lung cancer cells. Cancer Lett. 316:97–104. 2012. View Article : Google Scholar | |
|
van de Weerdt BC and Medema RH: Polo-like kinases: A team in control of the division. Cell Cycle. 5:853–864. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Liu X and Erikson RL: Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci USA. 100:5789–5794. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Spänkuch B, Kurunci-Csacsko E, Kaufmann M and Strebhardt K: Rational combinations of siRNAs targeting Plk1 with breast cancer drugs. Oncogene. 26:5793–5807. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Gavet O and Pines J: Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol. 189:247–259. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Hsiao YC, Hsieh YS, Kuo WH, Chiou HL, Yang SF, Chiang WL and Chu SC: The tumor-growth inhibitory activity of flavanone and 2′-OH flavanone in vitro and in vivo through induction of cell cycle arrest and suppression of cyclins and CDKs. J Biomed Sci. 14:107–119. 2007. View Article : Google Scholar | |
|
Kamijo T: Role of stemness-related molecules in neuroblastoma. Pediatr Res. 71:511–515. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Hu K, Law JH, Fotovati A and Dunn SE: Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells. Breast Cancer Res. 14:R222012. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang H, Li W, Nan F, Ren F, Wang H, Xu Y and Zhang F: MicroRNA expression profile of colon cancer stem-like cells in HT29 adenocarcinoma cell line. Biochem Biophys Res Commun. 404:273–278. 2011. View Article : Google Scholar |