Introduction
Lung cancer is a primary malignant tumor originating
from the bronchial mucosal epithelium, with 11.4% of morbidity and
18% of mortality in the world according to global cancer statistics
in 2020 (1), of which ~76% are
non-small cell lung cancer (NSCLC) and 13% are small cell lung
cancer according to clinicopathological characteristics (2). NSCLC mainly includes lung
adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) based
on histologic subtypes. LUAD is derived from pulmonary peripheral
tissue lesions, accounts for 40% of patients with lung cancer
(3) and is asymptomatic in the
early stage. LUSC is derived from proximal airway epithelial cells,
which are sensitive to chemoradiotherapy (4). LUAD and LUSC are both genetically
altered and associated with poor prognosis (5). The pathogenesis of lung cancer
remains poorly understood, and genetic factors and immune and
signaling pathways are involved in its occurrence and development
(6,7). Targeting these genes or signaling
pathways and regulating the immune system, numerous inhibitors or
targeted drugs have been developed to improve the treatment of lung
cancer in clinics (8,9). However, due to the tumor immune
microenvironment and its heterogeneity (10), susceptibility to drug resistance,
local invasion and distant metastasis (11), the five-year survival rate of lung
cancer is <15% (12,13). Therefore, it is crucial to
elucidate the pathogenesis of lung cancer, which will be helpful
for drug development to improve the survival of patients with lung
cancer.
The SERPINA3 gene, with 1,272 base sequences in
Homo sapiens, encodes a protein named
alpha-1-antichymotrypsin with a molecular weight of ~47 kDa.
SERPINA3 is a serpin peptidase inhibitor and a member of the acute
phase protein family that is synthesized in the liver and secreted
into the blood (14) and plays
essential roles in pathological processes (15), including the inflammatory response
(16,17), immunotherapy response (18), cardiovascular disease (19), and in neurodegenerative diseases
such as Alzheimer's syndrome (20). Notably, studies have reported that
SERPINA3 expression levels or glycosylation levels are abnormal in
tumors and could be a biomarker for the diagnosis of tumors such as
liver cancer (16,21). Moreover, it was found that the
expression level of SERPINA3 was also associated with the survival
of patients with cancer and may be a therapeutic target for tumor
treatment (16). For example,
Lara-Velazquez et al (22)
reported that knockdown of SERPINA3 inhibited tumor growth in
glioma in vitro and in vivo, suggesting that SERPINA3
could be a target for the treatment of glioma. Zhu et al
(23) found that SERPINA3 acted as
a tumor suppressor in liver cancer and inhibited the development
and metastasis of liver cancer, suggesting that SERPINA3 could be
used as a target for treatment intervention in liver cancer. Zhang
et al (24) reported that
overexpression of SERPINA3 promotes tumor invasion and migration in
triple-negative breast cancer cells.
As previously reported by the authors, it was found
that glycosylated SERPINA3 could be used as a candidate diagnostic
biomarker in early NSCLC and that it could also significantly
improve the specificity of carcinoembryonic antigen (25,26).
It was also discussed that SERPINA3 may be a biomarker for the
prognostic survival of patients with lung cancer (16). It was also found that SERPINA3
could be used to distinguish malignant from benign lung lesions
with an AUC value of 0.806 (Fig.
S1). To use SERPINA3 as a promising biomarker, it is necessary
to improve understanding of its effects on pathogenesis and the
molecular basis of lung cancer (27,28).
However, the detailed biological functions of SERPINA3 involvement
in the occurrence of lung cancer remain unknown.
In the present study, it was aimed to investigate
the biological roles of SERPINA3 in the pathogenesis of lung
cancer. Firstly, the expression of SERPINA3 in lung cancer was
measured with tissue samples and cell samples and then the effects
of SERPINA3 on the pathogenesis of lung cancer were explored
through the construction of stable SERPINA3-overexpressing lung
cancer cells at the cellular and animal levels. Moreover,
data-independent acquisition mass spectrometry (DIA-MS) for
quantitative proteomics detection was designed to explore the
regulatory mechanism of SERPINA3 involved in the development of
lung cancer. Furthermore, the differentially expressed proteins
(DEPs) regulated by overexpressed SERPINA3 in lung cancer cells
were analyzed and screened with bioinformatics analysis. Finally,
the DEPs were validated by western blotting (WB) in cell
experiments and in vivo models to investigate the possible
regulatory mechanism.
Materials and methods
Clinical samples collection
The patient was enrolled based on CT imaging data
and tumor biomarker screening results at the Suzhou Municipal
Hospital of Nanjing Medical University. The present study was
approved (approval no. KL901372) by the Ethics Committee of Suzhou
Municipal Hospital (Suzhou, China). Written informed consents were
obtained from all participants. All patients were diagnosed upon
histopathological analyses. The patients with lung cancer were
graded using the eighth edition of the TNM classification of the
International Association for the Study of Lung Cancer (IASLC).
Preoperative serum samples of 243 cases were selected and collected
to detect SERPINA3 expressions in serological level, of which 64
patients with benign diseases and 179 patients with lung cancer.
The fresh tissue samples were collected from patients with lung
cancer between June 2021 and June 2022, which were used to validate
the protein expression level of SERPINA3. The information on serum
and tissue samples is also listed in Table SI. The collected tissues were
frozen with liquid nitrogen, of which the tumor sizes were <3
cm, and the paracancerous tissues were 2 cm away from the tumor
tissues. The tissue samples were lysed with
radioimmunoprecipitation assay (RIPA) lysis buffer (cat. no.
P00138; Beyotime Institute of Biotechnology) containing 50 mM Tris
(pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate,
0.1% SDS and 1 mM phenylmethylsulfonyl fluoride, and ceramic beads
were added for automatic crushing. The supernatant, namely, tissue
samples' protein extract, was harvested after centrifugation at
4°C, 15,294 × g for 10 min with centrifugal machine (Eppendorf
5804R).
Cells and cell culture
The human lung cancer cell lines A549 (cat. no.
CL-0016), H157 (cat. no. CL-0388) and NCI-H1437 (cat. no. CL-0631)
were kindly provided by the Procell Life Science & Technology
Co., Ltd., and the NCI-H1975 cells (cat. no. BNCC340345) were
purchased from the BeNa Culture Collection. These cell lines were
detected by STR authentication. The cells were sustained in DMEM or
RPMI-1640 medium containing 10% fetal bovine serum (GIBCO,
Invitrogen; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin sulfate (Invitrogen; Thermo Fisher
Scientific, Inc.). All cells were cultivated at 37°C in a 5%
CO2 atmosphere.
Construction of the stable cell lines of
overexpressed SERPINA3 in lung cancer cells
To investigate the biological effects of SERPINA3 on
lung cancer, the stable cell lines of overexpressed SERPINA3 in
lung cancer cells were constructed, and the recombinant
overexpression vector and stable cells were prepared as previously
described (29,30). Briefly, the human SERPINA3 plasmid
(gene ID: 12; NCBI accession: NM_001085) was ligated into the
pCDH-CM V-MCS-EF1-copGFP-T2A-Puro control vector with the
restriction endonuclease XbaI site (TCT AGA) and NotI
site (GCG GCC GC). The lentivirus package was produced by
recombinant overexpression plasmid incubated with psPAX2 vector and
pMD2.G vector. The lentivirus vector carrying the empty-vector was
used as a negative control. The DNA was incubated with
lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for
20 min at room temperature and transfected into the 293T cells
(BeNa Culture Collection). Subsequently, the lentivirus was
harvested. The harvested lentivirus particles were used to infect
the three kinds of lung cancer cell lines for 48 h, and the stably
overexpressed SERPINA3 lung cancer cells were selected by culturing
in a medium containing 2 µg/ml puromycin.
Reverse transcription-quantitative PCR
(RT-qPCR)
The RT-qPCR experiments were used to detect whether
SERPINA3 was overexpressed in stable-transfected lung cancer cell
lines. Total RNA was extracted from the lung cancer cells after
overexpression of SERPINA3 with the TRIzol reagent (cat no.
15596026; Invitrogen; Thermo Fisher Scientific, Inc.) Then, cDNA
was synthesized from total RNA with 500 ng oligo (dT), 10 mM dNTPs
and the M-MLV reverse transcriptase (cat no. AE101-03; TransGen
Biotech Co., Ltd.) in 5X first chain synthetic buffer.
Thermocycling conditions for reverse transcription were as follows:
50 min at 37°C for cDNA synthesis and 15 min at 70°C for stopping
the reaction. All qPCR reactions contained 100 ng cDNA and
thermocycling conditions were as follows: 30 sec at 95°C for
initial denaturation, followed by forty cycles of 10 sec at 95°C
and 30 sec at 60°C and 30 sec at 72°C. Reactions were performed
with the 2X SYBR green mix (cat no. KK4601; Kapabiosystems, Inc.)
using a fluorescence quantitative PCR instrument (ABI 7500). The
PCR primers were synthesized from TsingKe Biological Technology and
the sequences were as follows: i) human SERPINA3 forward, 5′-TGG
TGC CCA TGA TGA GTT TG-3′ and reverse, 5′-AAC GCA CAA TGG TCC TTG
TC-3′; ii) human β-actin forward, 5′-CCT GGC ACC CAG CAC AAT-3′ and
reverse, 5′-GGG CCG GAC TCG TCA TAC-3′. The β-actin mRNA was used
as internal controls. The 2−ΔΔCq method (31) was applied to calculate the relative
expression of SERPINA3 gene.
Cell viability and cell proliferation
assays
The viability of the lung cancer cell lines after
overexpression of SERPINA3 was assessed using the Cell Counting
Kit-8 (CCK-8) assay according to the manufacturer's instructions.
Briefly, the A549 cells, H157 cells, and H1437 cells that stable
transfected overexpressed-SERPINA3 vector or empty-vector were
counted by hemocytometry and seeded at 3×103 cells per
well in 96-well culture plates and cultured at 37°C for 24 and 48
h. A total of 10 µl of CCK-8 reagent (cat no. MA0218; Dalian
Meilun Biology Technology Co., Ltd.) were added to each well and
incubated at 37°C for another 4 h. The absorbance was measured at
450 nm using a SMR60047 Smart Microplate Reader (USCNK Life Science
Co. Ltd.). The experimental samples were repeated three times.
The proliferation of the lung cancer cell lines
after overexpression of SERPINA3 was tested by the BeyoClick™ EdU
Cell Proliferation Kit with Alexa Fluor 488 (cat no. C0071S;
Beyotime Institute of Biotechnology, Inc.). Briefly, the cells were
seeded at 6×105 cells per well in six-well culture
plates and cultured at 37°C overnight. Then, the cells were
transfected with 3 µg overexpressed-SERPINA3 plasmid or
empty-vector with Highgene transfection reagents (cat no. RM09014;
ABclonal Technology Co., Ltd.) for 24 h. After transfection, the
cells were harvested and the DNA proliferation was determined by
incorporating of 5-ethynyl-2'-deoxyuridine (EdU) according to the
manufacturer's instructions. The nuclei were stained with 1 ml 1X
Hoechst 33342 per well at room temperature for 10 min away from
light, and the cells were visualized by a SOPTOP ICX41 fluorescence
microscope (Ningbo Sunny Instruments Co., Ltd.).
For sensitivity tests of osimertinib, the cells were
seeded at 1×104 cells per well in 96-well culture plates
overnight. After transfection of plasmids for 6 h, the cells were
treated with 0, 0.5, 1, 5 µM osimertinib (Shanghai
TopScience Co., Ltd.) for 24 h, then the cell viability was
determined with CCK-8 kits (cat no. CK18; Dojindo Laboratories,
Inc.) at 450 nm using a microplate reader (Thermo Scientific
Multiskan GO).
Cell growth assay
The growth of the lung cancer cell lines after
overexpression of SERPINA3 was detected by colony formation assay.
Briefly, the transfected cells were seeded in six-well culture
plates at 1×103 cells per well and cultured at 37°C. The
culture medium was changed once weekly and cultured for 14 days at
37°C. When the number of cells forming a colony was >50, the
cells were fixed and stained with 0.1% crystal violet (cat no.
C8470; Solarbio Beijing Solarbio Science & Technology Co.,
Ltd.) for 10 min at room temperature. Images of the colonies were
captured and quantified by ImageJ software v1.52 (National
Institutes of Health). The experimental samples were repeated three
times.
Cell apoptosis assay
The apoptosis rate of the lung cancer cell lines
after overexpression of SERPINA3 was evaluated using an Annexin
V-FITC/PI apoptosis detection kit (cat no. 401003; BestBio, Inc.)
according to the manufacturer's instructions. The transfected cells
were cultured for 24 h, and then harvested by low-speed
centrifugation and washed with ice-cold PBS. Next, 300 µl of
binding buffer was added to the suspended cells, and cells were
stained with 5 µl propidium iodide and 5 µl Annexin
V-FITC at room temperature for 15 min in the dark. The apoptotic
cells were tested by a BD FACSCalibur flow cytometer (BD
Biosciences). Finally, the results were interpreted using FlowJo
software v10.0 (FlowJo LLC).
Cell migration assay
The migration capacity of the lung cancer cell lines
after overexpression of SERPINA3 was detected by wound-healing
assays. Briefly, the transfected cells were seeded into six-well
plates at 1×106 cells per well and cultured with
serum-free medium at 37°C. After the cell density was near 90%, the
monolayer was scratched in a straight line using a new 10-µl
pipette tip across the center of the well and washed with sterile
PBS. The cells were further cultured with fresh medium for an
additional 48 h and were visualized at 0, 24 and 48 h under a
microscope. The cellular migration area was analyzed by ImageJ
software and the migration rate was calculated using the following
formula:
or
The experimental samples were repeated three
times.
Cell invasion assay
The invasion of the lung cancer cell lines after
overexpression of SERPINA3 was performed with Transwell assays
using 12-µm Transwell chambers. Briefly, the 24-well plates
were precoated with Matrigel (cat no. 356234; BD Biosciences) at
37°C for 2 h. The transfected A549, H157, or H1437 cells at
2.5×104 cells per well in 200 µl of serum-free
media were seeded in the upper chamber, and 500 µl medium
containing 10% FBS was into the lower chamber. The chambers were
inserted into the plates, and the cells were incubated at 37°C for
24 h. The cells were fixed with methanol for 20 min at room
temperature and stained with 0.1% crystal violet for 15 min at room
temperature. Then, the non-invasive cells in the upper chambers
were removed with cotton swabs, and images of the invasive cells
were captured under an inverted SOPTOP ICX41 fluorescence
microscope (Ningbo Sunny Instruments Co., Ltd.) using at least
three random fields of view. The experiment was repeated three
times. Image-Pro Plus software v6.0 (Media Cybernetics, Inc.) was
used to calculate the cellular invasion ability.
In vivo tumorigenic assay
The biological functions of SERPINA3 involved in the
occurrence of lung cancer were also evaluated at the animal level.
Five-week-old female BALB/c nude mice (n=12; weight, 16-17 g) were
purchased from the Experimental Animal Center of China Three Gorges
University (permission number: SCXK 2017-0012), and bred under
pathogen-free conditions (temperature, 18-22°C; humidity, 50-60%;
12/12-h lightdark cycle). The animals were fed an autoclaved rodent
diet ad libitum. The mice bedding, feed and water were
replaced every 2 days. All procedures followed the institutional
and national guidelines for the care and use of laboratory
animals.
The subcutaneous xenograft model of lung cancer was
constructed as previously described by Li et al (32). Briefly, stably transfected
empty-vector pCDH and stably transfected overexpressed-SERPINA3
A549 cells were suspended in 100 µl PBS and inoculated
subcutaneously 2×106 cells into the right flanks of nude
mice (6 mice in each group), respectively. The body weight of nude
mice was monitored every 2 days. After 9 days, the tumors were
measured every 2 days with a caliper, and tumor volume (V) was
calculated by measuring the length (L) and width (W) and applying
the formula V=(L × W2) × 0.5. On day 29, two mice were
found dead in each group. The cause of death may be due to the
marked body weight loss. When the mice began to succumb and were
immobile and rigid, and were not in a favorable mental state, the
body weight was very low, and certain tumors reached the 1-1.5 cm
in diameter, the mice were euthanized according to humane care of
animals, anesthetized by intraperitoneal injection of sodium
pentobarbital with 150 mg/kg (33). The lung and tumor tissues were
excised for further analysis.
Data-independent acquisition mass
spectrometry (DIA-MS) for quantitative proteomics detection
The mechanism of SERPINA3 involved in the
development of lung cancer was explored, and DIA-MS detection
identified the DEPs. Two replicates were performed for mass
spectrometry data. Firstly, the H157 cells transfected with
empty-vector and overexpressed-SERPINA3 were collected respectively
and quickly frozen with liquid nitrogen, then the cell samples were
further treated and analyzed by SpecAlly Life Technology Co., LTD
(Wuhan, China). The proteins from cell lysis were denatured,
reduced, and alkylated with a reaction solution containing 1%
sodium deoxycholate, 100 mM Tris-HCl with pH 8.5, 10 mM
trichloromethyl phosphate, and 40 mM chloroacetamide by one step at
60°C for 1 h, and digested with trypsin overnight at 37°C. The
digestion reaction was stopped by trifluoroacetic acid, and the
digested peptides were desalted, evaporated, and stored at -20°C
until used.
Secondly, DIA-MS detection and data acquisition was
performed using an Orbitrap Exploris 480 Mass Spectrometer coupled
with an EASY-NLC 1200 liquid phase LC-MS system, and the parameters
were referenced as previously described (34). Peptide samples were aspirated by an
autosampler and bound to a C18 analytical column for separation.
Mobile phase A consisted of 0.1% formic acid, mobile phase B
consisted of 0.1% formic acid, and 80% acetonitrile was used to
establish the analytical gradient. The flow rate was maintained at
300 nl/min. The mass spectrometry data was collected with DIA
model, and each cycle included one MS1 scan (Max IT was 30 ms, scan
range from 350 to 1,250 m/z) and 40 MS2 scans with variable windows
(Max IT=50 ms), and the collision energy was set to 30.
Finally, the DIA raw data files of MS detection were
analyzed by DIA-NN software v1.8. The human proteome reference
database in Uniprot (2021-03-12, containing 20381 protein
sequences; uniprot.org) was used for search
analysis. The spectrum libraries were obtained via the deep
learning algorithm in DIA-NN and MBR function, and the spectrum
libraries were used to extract the original DIA data to obtain
protein quantitative information. The final results were screened
by 1% false discovery rates for parent ion and protein levels. The
quality procedures including mass bias, enzyme digestion
efficiency, and missing data, have been carried out in the analysis
of MS detection data. In the MS data of the present study, the
distribution of missing enzyme sites was small, the enzyme
digestion efficiency was high, and the proportion of missing data
was <0.9%. The quantitative information of the proteome was used
for screening and subsequent analysis.
WB
The expression levels of SERPINA3, GAPDH,
speckle-type POZ protein (SPOP) and NF-κB p65 in lung cancer cells
or tumor tissue samples were detected by WB. Briefly, 15 µg
proteins from cell lysates or tissue lysates were measured using a
bicinchoninic acid (BCA) protein assay kit (Thermo Fisher
Scientific, Inc.) and were subjected to 12% SDS-PAGE. After
electrophoresis, the gels were transferred onto PVDF membranes, and
the membranes were blocked with 5% BSA (Biofroxx; neoFroxx) in 0.1%
TBST (0.1% Tween-20 in 1X TBS) at room temperature (RT) for 1 h.
The membranes were probed with polyclonal antibodies against
SERPINA3 (1:2,000; cat no. PAB015Hu01; Wuhan USCN Business Co.,
Ltd.), GAPDH (1:10,000; cat no. 60004-1-Ig; Proteintech Group,
Inc.), SPOP (1:10,000; cat no. A19578; ABclonal Technology Co.,
Ltd.) or NF-κB p65 (1:5,000; cat no. 10745-1-AP; Proteintech Group,
Inc.) as primary antibodies, overnight at 4°C. The membranes were
incubated with HRP-conjugated goat-anti-rabbit IgG as the secondary
antibody (1:5,000; cat no. 30000-0-AP; Proteintech Group, Inc.) for
1 h at RT and detected by Supersignal West Pico Chemiluminescent
HRP Substrate (Bio-Rad Laboratories, Inc.). Images of the data were
captured by a Vilber FUSION FX7 chemiluminescence imager. The
loading control sample in each gel was used as an internal standard
for quantification. Densitometric analysis of each band was
measured using ImageJ software, and the data statistics were
analyzed by GraphPad Prism v8.0 software (Dotmatics) with an
unpaired t-test.
Bioinformatics analysis
The SERPINA3 expression levels were detected in the
HUMAN PROTEIN ATLAS (HPA; https://www.proteinatlas.org/ENSG00000196136-SERPINA3).
The mRNA expression levels of SERPINA3 and correlation analysis
were detected using the Gene Expression Profiling Interactive
Analysis (GEPIA) database (http://gepia.cancer-pku.cn/). The Gene Expression
Omnibus (GEO) database was used for analysis of SERPINA3 expression
in the osimertinib-resistant NSCLC cell line NCI-H1975 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201608)
(35). The association between the
protein expression with tumor stage and immune subtypes in LUAD and
LUSC were performed using the TISIDB database (http://cis.hku.hk/TISIDB/). The correlation between
protein expression and the survival time of lung cancer patients
were tested by Kaplan-Meier Plotter database (http://kmplot.com/analysis/). The TargetScanHuman
database (https://www.targetscan.org/vert_72/) was used to
identify the target of SERPINA3 mRNA (TargetScanHuman 7.2 predicted
targeting of Human SERPINA3).
For MS data, the enrichment analysis of identified
245 proteins was analyzed by ShinyGO v0.76 (http://bioinfor-matics.sdstate.edu/go/), and the
histograms and dot bubble plots of enriched Gene Ontology (GO)
terms and pathways were acquired by W-bioinformatics (http://www.bioinformatics.com.cn/). The
identified 245 proteins were annotated by GO terms using WEGO
(https://biodb.swu.edu.cn/cgi-bin/wego/index.pl). The
heat map was analyzed with R v4.2.1 programming tools with the
gplots package. The protein-protein interaction networks were
constructed using STRING (https://string-db.org/), and the networks were
performed by Cytoscape v3.9.1 software (https://cytoscape.org/) (36).
Statistical analysis
The receiver operator characteristic (ROC) curve was
constructed by SPSS v26.0 software (IBM Corp.) to analyze the AUC
value, sensitivity, and specificity for evaluating the diagnostic
performance of SERPINA3. To find the optimum efficiency, Youden
index was calculated using the formula: Youden index=sensitivity +
specificity-1 (25). The
sensitivity and specificity were confirmed when Youden index is
maximum.
The correlation between SERPINA3 mRNA expression and
LUAD or LUSC was performed by Spearman's correlation analysis. The
data of cytological experiments were statistically analyzed by
GraphPad Prism v8.0 software with an unpaired t-test, and P<0.05
was considered to indicate a statistically significant difference.
Data were expressed as the mean ± SD.
Results
The expression level of SERPINA3 is
decreased in lung cancer
Through bioinformatic analysis, the expression level
of SERPINA3 was measured in several normal tissues. It was found
that SERPINA3 mRNA levels were mainly expressed in the liver,
pancreas, prostate, urinary bladder, ovary and lung based on the
HPA database and GTEx database (Fig.
1A). The protein levels were mainly in the cerebellum, cervix,
prostate, liver, pancreas and lung (Fig. 1B). Using the GEPIA database based
on The Cancer Genome Atlas dataset and GTEx data, it was determined
that SERPINA3 mRNA expression in lung cancer tissues significantly
decreased in LUAD (n=347) and LUSC (n=338) compared with normal
tissues, as demonstrated in Fig.
1C, which was significantly correlated with tumor stage in LUSC
(Fig. 1D; P<0.001).
Furthermore, in experimental validation, the protein expression
level of SERPINA3 was significantly reduced in three lung cancer
tissues compared with paracancerous tissues by WB detection
(P=0.0324) (Fig. 1E and F). In
cancer cell lines, it was found that SERPINA3 mRNA was expressed in
skin, liver, lung, pancreatic, ovarian, bladder and prostate cancer
(Fig. 1G). In lung cancer cell
lines, SERPINA3 mRNA was transcribed at lower levels in A549, H157,
H1437 and H1975 cells (Fig. 1H).
It was also validated that the SERPINA3 protein expression levels
were reduced in three lung cancer cell lines (A549, H157 and H1975)
(Fig. 1I).
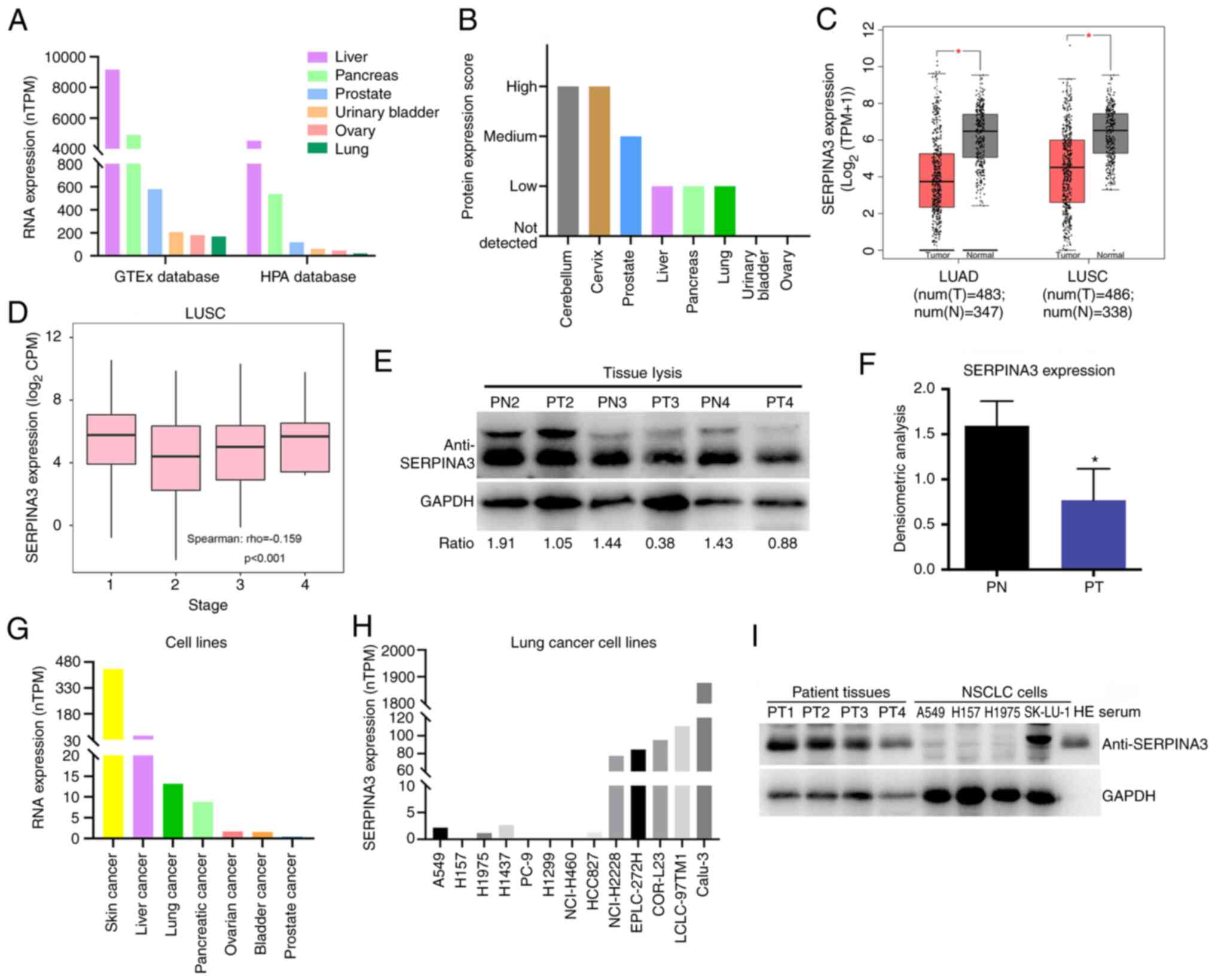 | Figure 1Analysis of SERPINA3 expression
levels in lung cancer. (A) Detection of the SERPINA3 mRNA
expression level in normal tissues with HPA database. (B) Analysis
of the SERPINA3 protein expression level in normal tissues with HPA
database. (C) The expression changes of SERPINA3 in lung cancer
tissues with the Gene Expression Profiling Interactive Analysis
database. Red represents tumor (T) tissues, and gray represents
normal (N) tissues. Parameter settings: |log2FC| cutoff:
1; P-value cutoff: 0.01; Log scale: yes, and the
log2(TPM + 1) was used for log-scale; Jitter size: 0.4;
Matched normal data: match The Cancer Genome Atlas normal and GTEx
data. (D) The correlation analysis between SERPINA3 protein and
tumor stage in LUSC. (E) WB analysis of SERPINA3 expression in lung
cancer tissues. (F) The densitometric analysis was measured using
ImageJ software. (G) The SERPINA3 mRNA expression level was
detected in cancer cell lines with the HPA database. (H) The
SERPINA3 mRNA expression level was detected in lung cancer cell
lines with HPA database. (I) WB analysis of SERPINA3 expression in
lung cancer cell lines. *P<0.05. SERPINA3, serpin
family A member 3; HPA, Human Protein Atlas; TPM, transcripts per
million; LUAD, lung adenocarcinoma; LUSC, lung squamous cell
carcinoma; CPM counts per million; WB, western blotting; PN,
paracancerous tissues; PT, patient tumors; WB, western
blotting. |
The TargetScanHuman database was also used to
analyze the reasons for regulating the SERPINA3 expression, which
was found that microRNA-137 (miR-137) is the only miRNA that could
bind to the 3′-untranslated region (UTR) of SERPINA3 mRNA as
revealed in Fig. S2
(TargetScanHuman 7.2 predicted targeting of Human SERPINA3), which
suggests that miR-137 could regulate SERPINA3 mRNA expression.
Overexpression of SERPINA3 suppresses
lung cancer at the cellular level
To investigate the role of SERPINA3 in the
pathogenesis of lung cancer, three lung cancer cell lines, A549,
H157 and H1437 cells that were stably transfected with SERPINA3
overexpression plasmids were constructed by lentiviral
transfection. The RT-qPCR results indicated that SERPINA3 was
significantly overexpressed in A549, H157 and H1437 cells (Fig. S3). Furthermore, the cytological
tests were performed after stable transfection of overexpressed
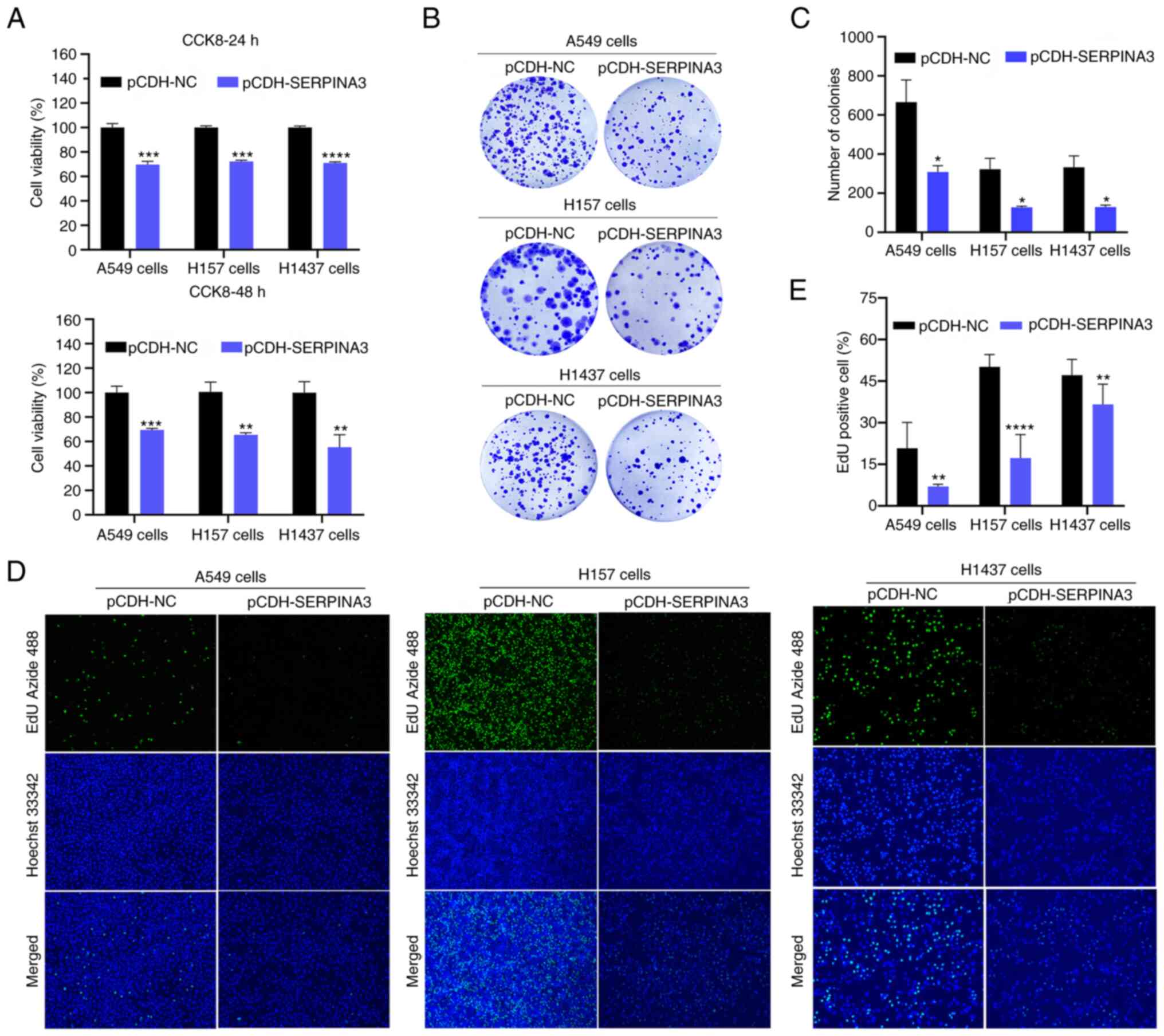
SERPINA3 in lung cancer cells. CCK-8 assays indicated that the cell
viability of the three lung cancer cell lines was significantly
reduced by ~30% when SERPINA3 was overexpressed compared with the
control group at 24 and 48 h (Fig.
2A). Clonogenic assays revealed that the growth of the three
lung cancer cell lines was decreased after the overexpression of
SERPINA3 (Fig. 2B and C). The
fluorescence intensity was weakened when EdU was incorporated into
the SERPINA3-overexpressing group, indicating that the DNA
proliferation capacity was significantly decreased in these three
lung cancer cell lines after overexpression of SERPINA3 (P<0.01;
Fig. 2D and E).
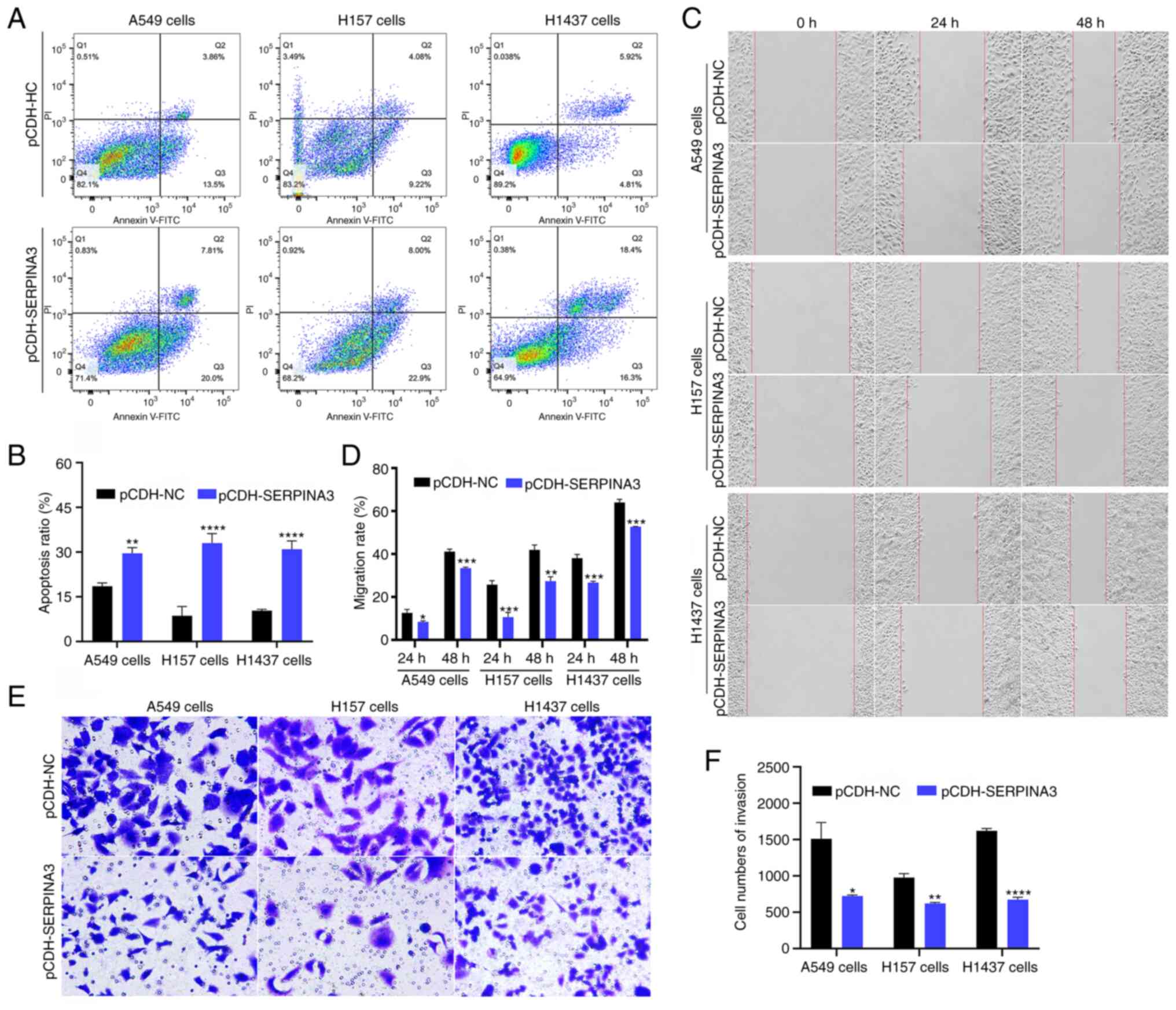
Moreover, cell apoptosis, migration and invasion
were also analyzed in the present study. The proportion of cell
apoptosis in the three lung cancer cell lines was significantly
higher in SERPINA3-overexpressing cells after flow cytometric
analysis (Fig. 3A and B;
P<0.01). The cell migration rate was assessed with wound-healing
assays, and the results showed that the stably transfected
SERPINA3-overexpressing cells migrated toward the scratch more
slowly in the three lung cancer cell lines at 24 and 48 h (Fig. 3C and D). The cell invasion ability
was suppressed after overexpression of SERPINA3 in three stable
lung cancer cell lines (Fig. 3E and
F; P<0.05). Thus, SERPINA3 may have antineoplastic roles in
lung cancer in vitro.
Overexpression of SERPINA3 enhances the
sensitivity of lung cancer cells to osimertinib
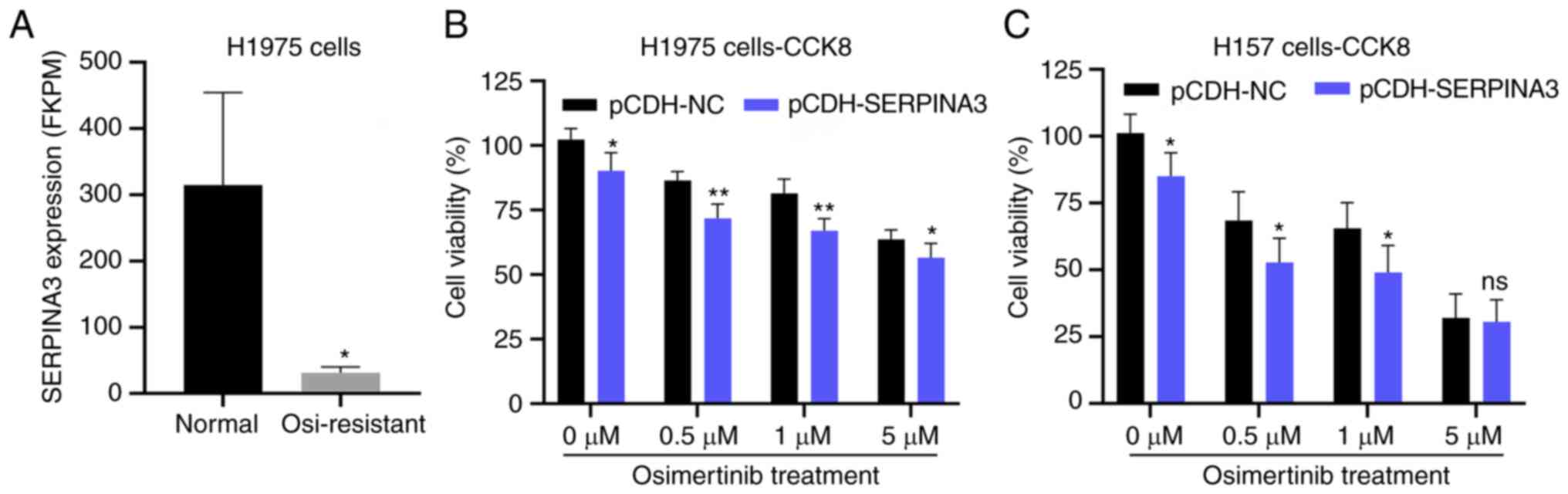
In addition, the Gene Expression Omnibus (GEO)
database (GSE201608) was mined and it was found that SERPINA3
expression was decreased ~10-fold in the osimertinib-resistant
NSCLC cell line NCI-H1975 in the aformentioned database (GSE201608)
(Fig. 4A); therefore, the
sensitivity of lung cancer cells to osimertinib after
overexpressing SERPINA3 was also detected. The cell viability
assays indicated that the H1975 and H157 cells, after transfection
of the SERPINA3 overexpression plasmid, were more sensitive to
osimertinib, and the H1975 cells were dose-dependent when
responding to osimertinib (P<0.05; Fig. 4B and C). However, the H1975 cells
showed resistance to osimertinib, maybe the treatment time of
osimertinib was not enough in this experiment.
Anticancer effects of overexpressed
SERPINA3 in a mouse model of lung cancer
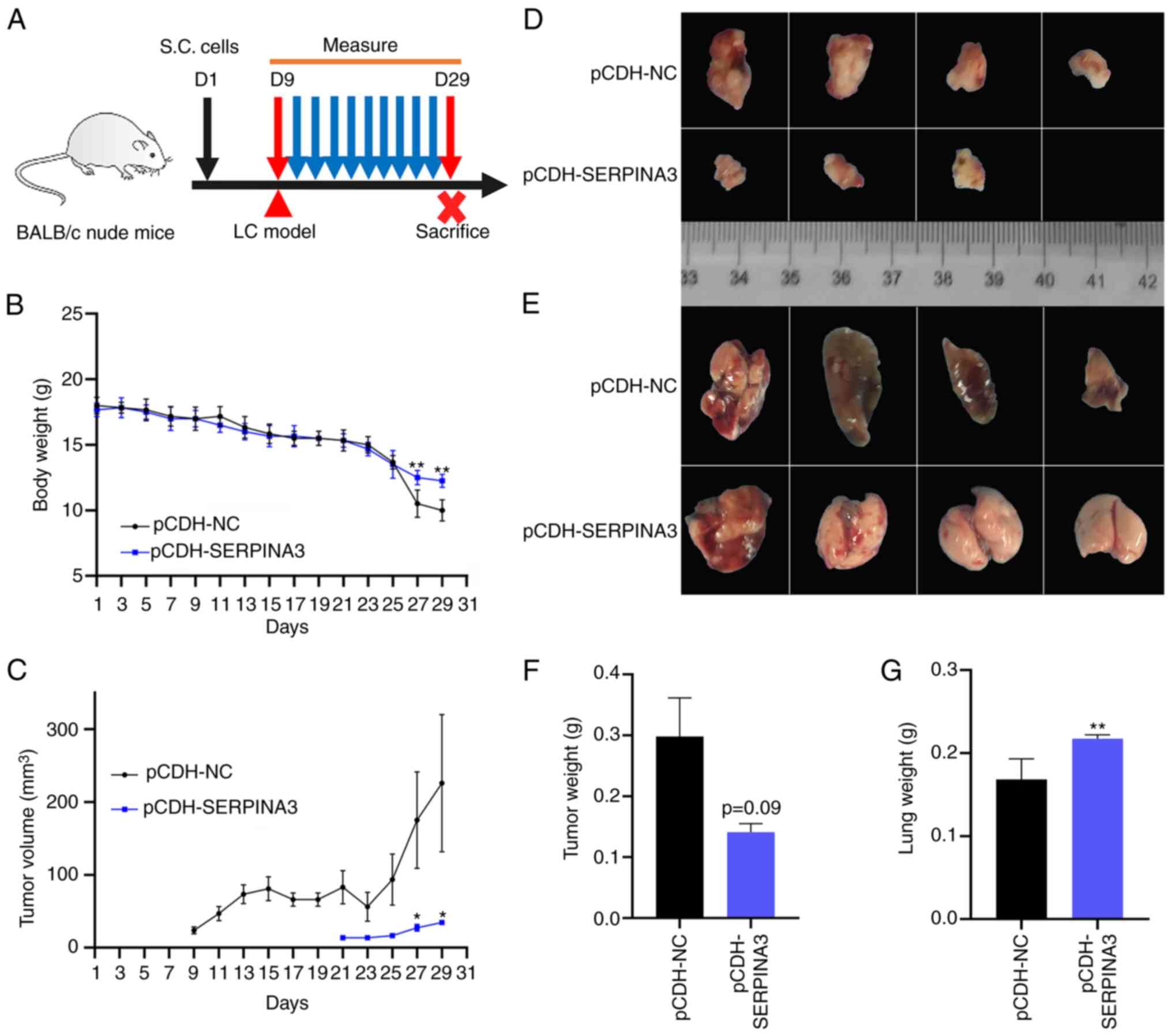
To monitor tumor growth and evaluate the antitumor
effect in vivo, a xenograft model of lung cancer with BALB/c
nude mice (n=6 per group) was also established. A schematic diagram
is shown in Fig. 5A. After a few
days of feeding, when the BALB/c nude mice weighed 17-18 g, they
were subcutaneously injected with stably transfected
SERPINA3-overexpressing or empty-vector A549 cells (D1) for
subsequent observation and monitoring, and the mice were sacrificed
on day 29 after injection. As shown in Fig. 5B, the body weight of tumor-bearing
mice decreased significantly and was very light on day 29, and the
average weight in the empty-vector group was 10 g (loss of 44.4%);
whereas the average weight in the SERPINA3-overexpressing group was
11.8 g (loss of 33.3%). As demonstrated in Fig. 5C, the tumor volume in the
empty-vector group gradually increased and the maximum diameter of
tumor reached 11 mm, whereas the tumor-bearing mice in the
SERPINA3-overexpressing group developed tumors on the 21st day
after injection of tumor cells, which appeared 12 days later than
in the empty-vector group. The tumors formed by the
SERPINA3-overexpressing cells were smaller in size than those
formed by the control cells (Fig.
5D). However, there was no significant difference in tumor
weight (P=0.09; Fig. 5F). In the
empty-vector group, the morphology of lung tissues was blackened
and changed, and the weight of the lung tissues was significantly
decreased compared with the SERPINA3-overexpressing group
(P=0.0083; Fig. 4E and G).
Overexpressed SERPINA3 upregulates SPOP
expression and inhibits NF-κB in lung cancer cells
To further explore the regulatory mechanism of
SERPINA3 involved in the development of lung cancer, the DEPs
regulated by SERPINA3 were measured with DIA-MS detection. The data
for quantitative proteomics detection are available via
ProteomeXchange with identifier PXD036119, and detailed information
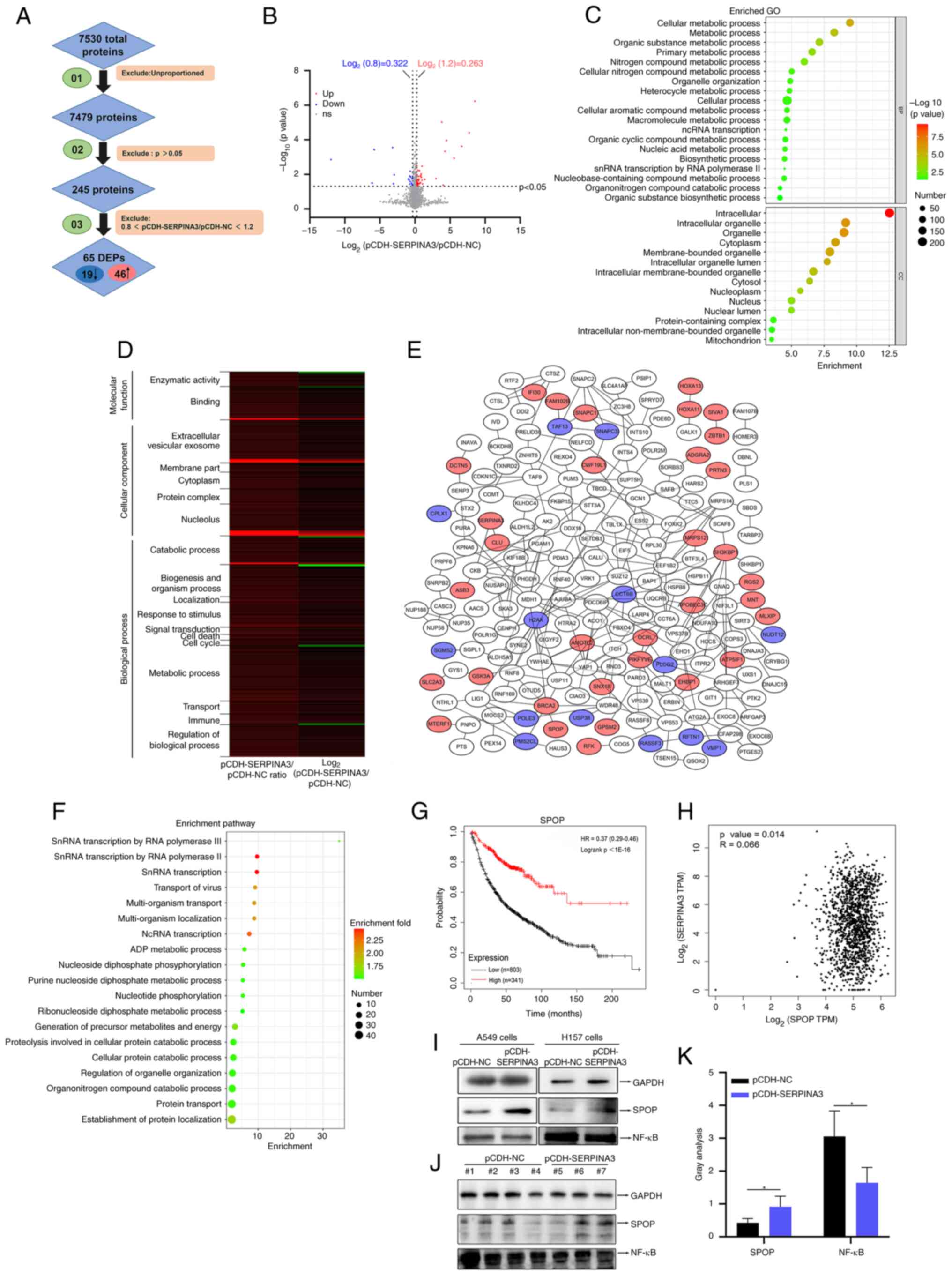
on the proteins identified by MS is shown in Table SII. A total of 7,530 proteins were
identified, and the proteins were filtered twice based on the
P-value and fold change, as shown in Fig. 6A. Finally, 245 proteins were
obtained, and 65 proteins were significantly differentially
expressed, of which 46 proteins were upregulated, and 19 proteins
were downregulated based on a ratio of pCDH-SERPINA3 to pCDH-NC
>1.2 or <0.8 (Table I). The
volcano diagram also shows 7479 proteins, of which downregulated
proteins are illustrated in blue and upregulated proteins are shown
in red (Fig. 6B). GO annotation
analysis was performed with 245 identified proteins, as
demonstrated in dot-bubble plots (Fig.
6C), and the data are provided in Table SIII. Of these, 18 functions were
screened from GO annotation and analyzed by a heatmap. The
identified proteins were related to the nucleolus, catabolic
process, response stimulation, signal transduction, cell death,
cell cycle, immune, metabolism, and other functions that were
significantly changed in lung cancer cells after SERPINA3
overexpression (Fig. 6D and
Table SIV). The protein-protein
interaction analysis was used by STRING, and the networks were
generated using Cytoscape software (Fig. 6E), which identified that SERPINA3
interacted with clusterin and creatine kinase B proteins, and the
original data are listed in Table
SV. The protein pathway enrichment analysis is also shown in
dot-bubble plots with 245 identified proteins (Fig. 6F), which are mainly involved in
transcription, multi-organism transport, protein localization and
metabolic processes, and the data are provided in Table SIII.
 | Table IList of the significant
differentially expressed proteins by data-independent acquisition
mass spectrometry detection. |
Table I
List of the significant
differentially expressed proteins by data-independent acquisition
mass spectrometry detection.
| Accession no.
(Uniprot database) | Gene | Description | Molecular weight
(kDa) |
pCDH-SERPINA3/pCDH-NC | P-value |
|---|
| Q0VAQ4 | SMAGP | Small cell adhesion
glycoprotein | 10.679 | 375.241 | 0.000 |
| P01011 | SERPINA3 |
Alpha-1-antichymotrypsin | 47.651 | 210.880 | 0.000 |
| P22079 | LPO |
Lactoperoxidase | 80.288 | 100.793 | 0.000 |
| P81274 | GPSM2 | G-protein-signaling
modulator 2 | 76.662 | 47.806 | 0.001 |
| O43309 | ZSCAN12 | Zinc finger and
SCAN domain-containing protein 12 | 70.222 | 22.863 | 0.000 |
| P51587 | BRCA2 | Breast cancer type
2 susceptibility protein | 384.202 | 19.397 | 0.000 |
| O43791 | SPOP | Speckle-type
POZ | 42.132 | 16.291 | 0.042 |
| P11169 | SLC2A3 | Solute carrier
family 2, facilitated glucose transporter member 3 | 53.924 | 14.177 | 0.000 |
| Q96PE1 | ADGRA2 | Adhesion G
protein-coupled receptor A2 | 142.647 | 7.983 | 0.018 |
| P41220 | RGS2 | Regulator of
G-protein signaling 2 | 24.382 | 2.630 | 0.020 |
| Q99551 | MTERF1 | Transcription
termination factor 1, mitochondrial | 45.778 | 2.098 | 0.036 |
| P31270 | HOXA11 | Homeobox protein
Hox-A11 | 34.486 | 2.020 | 0.003 |
| P24158 | PRTN3 | Myeloblastin | 27.807 | 2.004 | 0.022 |
| P13284 | IFI30 |
Gamma-interferon-inducible lysosomal thiol
reductase | 27.964 | 1.870 | 0.015 |
| Q9NRW3 | APOBEC3C | DNA
dC->dU-editing enzyme APOBEC-3C | 22.826 | 1.844 | 0.009 |
| Q8NHZ8 | CDC26 | Anaphase-promoting
complex subunit CDC26 | 9.777 | 1.791 | 0.012 |
| Q9Y2I7 | PIKFYVE |
1-phosphatidylinositol 3-phosphate
5-kinase | 237.136 | 1.729 | 0.013 |
| Q14CN4 | KRT72 | Keratin, type II
cytoskeletal 72 | 55.877 | 1.719 | 0.030 |
| Q969G6 | RFK | Riboflavin
kinase | 17.623 | 1.640 | 0.032 |
| Q86X95 | CIR1 | Corepressor
interacting with RBPJ 1 | 52.313 | 1.551 | 0.006 |
| Q5T8I3 | FAM102B | Protein
FAM102B | 39.308 | 1.466 | 0.019 |
| Q9Y2K1 | ZBTB1 | Zinc finger and BTB
domain-containing protein 1 | 82.016 | 1.451 | 0.036 |
| Q8N4S0 | CCDC82 | Coiled-coil
domain-containing protein 82 | 64.002 | 1.433 | 0.013 |
| P31271 | HOXA13 | Homeobox protein
Hox-A13 | 39.727 | 1.432 | 0.023 |
| Q9BTE1 | DCTN5 | Dynactin subunit
5 | 20.127 | 1.357 | 0.036 |
| Q9UII2 | ATP5IF1 | ATPase inhibitor,
mitochondrial | 12.249 | 1.347 | 0.033 |
| Q16533 | SNAPC1 | snRNA-activating
protein complex subunit 1 | 42.994 | 1.330 | 0.006 |
| O15304 | SIVA1 | Apoptosis
regulatory protein Siva | 18.695 | 1.321 | 0.036 |
| Q9HBE1 | PATZ1 | POZ-, AT hook-, and
zinc finger containing protein 1 | 74.060 | 1.318 | 0.038 |
| Q8N954 | GPATCH11 | G patch
domain-containing protein 11 | 33.277 | 1.316 | 0.047 |
| Q99583 | MNT | Max-binding protein
MNT | 62.300 | 1.290 | 0.041 |
| Q9Y575 | ASB3 | Ankyrin repeat and
S | 57.745 | 1.285 | 0.020 |
| P49840 | GSK3A | Glycogen synthase
kinase-3 alpha | 50.981 | 1.274 | 0.033 |
| Q96RF0 | SNX18 | Sorting
nexin-18 | 68.894 | 1.272 | 0.046 |
| O15235 | MRPS12 | 28S ribosomal
protein S12, mitochondrial | 15.173 | 1.267 | 0.006 |
| Q9Y2J4 | AMOTL2 | Angiomotin-like
protein 2 | 85.764 | 1.266 | 0.029 |
| Q9HAP2 | MLXIP | MLX-interacting
protein | 101.185 | 1.258 | 0.042 |
| Q01968 | OCRL | Inositol
polyphosphate 5-phosphatase OCRL | 104.205 | 1.251 | 0.021 |
| Q12979 | ABR | Active breakpoint
cluster region-related protein | 97.598 | 1.250 | 0.037 |
| Q96B97 | SH3KBP1 | SH3
domain-containing kinase-binding protein 1 | 73.126 | 1.240 | 0.031 |
| Q8NDI1 | EHBP1 | EH domain-binding
protein 1 | 140.017 | 1.238 | 0.011 |
| Q99956 | DUSP9 | Dual specificity
protein phosphatase 9 | 41.868 | 1.225 | 0.040 |
| P10909 | CLU | Clusterin | 52.495 | 1.217 | 0.024 |
| Q69YN2 | CWF19L1 | CWF19-like protein
1 | 60.619 | 1.209 | 0.039 |
| Q9Y4F5 | CEP170B | Centrosomal protein
of 170 kDa protein B | 171.688 | 1.206 | 0.036 |
| Q9H6A9 | PCNX3 | Pecanex-like
protein 3 | 222.039 | 1.204 | 0.003 |
| P17661 | DES | Desmin | 53.536 | 0.793 | 0.041 |
| Q5TCX8 | MAP3K21 | Mitogen-activated
protein kinase kinase kinase 21 | 113.957 | 0.790 | 0.016 |
| P16104 | H2AX | Histone H2AX | 15.145 | 0.786 | 0.043 |
| Q9NRF9 | POLE3 | DNA polymerase
epsilon subunit 3 | 16.860 | 0.743 | 0.018 |
| Q15543 | TAF13 | Transcription
initiation factor TFIID subunit 13 | 14.287 | 0.736 | 0.037 |
| Q2TB18 | ASTE1 | Protein asteroid
homolog 1 | 77.093 | 0.721 | 0.033 |
| P16885 | PLCG2 |
1-phosphatidylinositol 4,5-bisphosphate
phosphodiesterase gamma-2 | 147.870 | 0.650 | 0.014 |
| Q0P6H9 | TMEM62 | Transmembrane
protein 62 | 73.133 | 0.629 | 0.026 |
| Q96GC9 | VMP1 | Vacuole membrane
protein 1 | 46.238 | 0.620 | 0.007 |
| Q9BQG2 | NUDT12 | NAD-capped RNA
hydrolase NUDT12 | 52.076 | 0.601 | 0.013 |
| Q14699 | RFTN1 | Raftlin | 63.146 | 0.598 | 0.037 |
| O14810 | CPLX1 | Complexin-1 | 15.030 | 0.560 | 0.021 |
| Q8NB14 | USP38 | Ubiquitin
carboxyl-terminal hydrolase 38 | 116.546 | 0.528 | 0.019 |
| Q86WH2 | RASSF3 | Ras association
domain-containing protein 3 | 27.562 | 0.128 | 0.011 |
| Q68D20 | PMS2CL | Protein PMS2CL | 20.909 | 0.119 | 0.035 |
| Q92966 | SNAPC3 | snRNA-activating
protein complex subunit 3 | 46.753 | 0.110 | 0.000 |
| Q8NHU3 | SGMS2 |
Phosphatidylcholine:ceramide
cholinephosphotransferase 2 | 42.280 | 0.018 | 0.000 |
| Q7Z401 | DENND4A | C-myc
promoter-binding protein | 209.244 | 0.015 | 0.032 |
| Q92526 | CCT6B | T-complex protein 1
subunit zeta-2 | 57.821 | 0.000 | 0.001 |
According to the significance fold-change and
molecular weight of DEPs, whether their function is involved in
lung cancer, and whether they exist in commercial antibodies for
detection, SPOP was selected for further analysis. GEPIA database
analysis found that SPOP expression had a decreasing trend in LUSC
(Fig. S4A). Additionally, the
same was reported in LUAD by Dong et al (37). TISIDB (http://cis.hku.hk/TISIDB/) database analysis
demonstrated that SPOP had no significant correlation with tumor
stage in LUAD but had a significant correlation with tumor stage in
LUSC (Fig. S4B and C,
respectively), and SPOP expression was significantly associated
with immune subtype distribution (Fig. S4D and E). Kaplan-Meier Plotter
database analysis (http://kmplot.com/analysis/) indicated that the
survival time was shorter in lung cancer patients with lower SPOP
expression than in those with higher SPOP expression (Fig. 6G). GEPIA database analysis found
that SPOP expression was positively correlated with SERPINA3
expression in lung cancer (Fig.
6H). In the MS data of the present study, the protein
expression level of SPOP was significantly increased after SERPINA3
overexpression in H157 cells, as shown in Table I. Further validation with WB
revealed that SPOP expression was upregulated and NF-κb p65
expression was downregulated after overexpression of SERPINA3 in
A549 and H157 cells (Fig. 6I). In
mouse tumor tissues, SPOP expression was significantly increased
(P=0.0390), and NF-κB p65 expression (P=0.0396) was significantly
decreased (Fig. 6J and K).
Discussion
Elucidating the complex pathogenesis of lung cancer
is of great significance for drug development and improvement of
the survival rate, which is a promising study area. Aiming at gene
alterations, including epidermal growth factor receptor mutation,
Kirsten rat sarcoma viral homolog mutation (7,38),
epigenetic regulation (39,40)
and tumor microenvironment regulation (41), as well as uncontrolled signaling
pathways such as the MEK/ERK signaling pathway (42) and PI3K/AKT signaling pathway
(43), numerous drugs have been
developed and used for the treatment of lung cancer (44,45).
In addition, current research has found that certain novel genes
are involved in the occurrence and development of lung cancer; for
example, MG53 suppressed tumor progression by modulating G3BP2
activity in NSCLC (32), which is
a potential therapeutic target for lung cancer. Therefore,
elucidating the pathogenesis of lung cancer will be helpful for
lung cancer treatment.
In the present study, the biological effects of
SERPINA3 in the occurrence of lung cancer were clarified. In
vitro, a series of cytological experiments indicated that the
proliferation, growth, migration and invasion of lung cancer cells
were inhibited, and cell apoptosis was promoted after
overexpression of SERPINA3. In vivo, animal experiments
showed that tumor growth was slower in tumor-bearing mice after the
injection of SERPINA3 overexpressing A549 cells. In addition, it
was found that lung cancer cells were more sensitive to osimertinib
after SERPINA3 overexpression. The present study demonstrated that
overexpressed SERPINA3 could act against lung cancer, indicating
that SERPINA3 may have an antineoplastic role in lung cancer, and
SERPINA3 is also a noteworthy target for the treatment of lung
cancer. To date, no available agents targeting SERPINA3 have been
reported for tumor treatment.
SERPINA3 is expressed differently in different tumor
tissues, and the possible reasons have been reported in a previous
publication by the authors; abnormal expression in lung cancer may
be due to genetic alterations or immunological regulation (16). The gene changes of SERPINA3 were
different in tissue subtypes of lung cancer; for example, the point
mutation was 3.48% in LUAD and 2.61% in LUSC by the COSMIC database
in the authors' previous analysis (16), which may lead to abnormal
expression of SERPINA3 in LUAD and LUSC. In addition, immune cells
are closely related to lung cancer occurrence (46). The infiltrated immune cells, such
as CD8+ T cells, NK cells, neutrophils and macrophages
in the tumor, were significantly correlated with SERPINA3
expression in lung cancer (16).
Zhang et al (47) have
reported that higher SERPINA3 expression was detected in urine of
patients with lung cancer at Stage IV. By contrast, it was found
that SERPINA3 expression decreased in sera of patients with lung
cancer at the early stage, subsequently recovered, and even
slightly increased in the late stage, which is probably due to the
complexity of regulation in the development of tumors, as
previously reported (25). In the
present study, the mRNA and protein expression levels of SERPINA3
in lung cancer tissues were decreased based on bioinformatics
analysis and experimental validation. In the current experiment,
the early-stage tissue samples were used for detection of the
expression level of SERPINA3 by WB, and the lung cancer tissues at
the late stage were not collected and detected. The different
expression level of SERPINA3 in tissues may be associated with
collected samples at various stages, tissue subtypes, glycosylation
modifications, or immunomodulation. However, the detailed mechanism
of abnormal SERPINA3 expression in lung cancer remains unclear and
needs further elucidation by experiments.
In addition, TargetScanHuman database analysis
indicated that miR-137 could regulate SERPINA3 mRNA expression. The
current study also found that miR-137 was a tumor suppressor in
human NSCLC (48,49). Chang et al (50) reported that upregulation of miR-137
expression could promote cell migration in lung cancer. Whether
miR-137 alters the cellular SERPINA3 expression levels in lung
cancer, resulting in SERPINA3 involvement in lung cancer
occurrence, has not been reported and needs further
exploration.
SPOP is a member of the E3 ubiquitin ligase complex
with a molecular weight of 42.132 kDa, which is an adaptor that
connects the cullin 3 RING ligase to its substrates for mediating
the ubiquitin-proteasome degradation pathway (51,52).
It was identified that SPOP played an essential role in
tumorigenesis and cancer progression (53,54);
for example, silencing SPOP accelerated the malignancy of breast
cancer (55), and SPOP suppressed
tumorigenesis during lung cancer progression (56,57).
SPOP inactivation, activation, amplification, and mislocalization
contribute to cancer pathogenesis, which is frequently mutated in
solid tumors; the mutation patterns and biological consequences are
different in cancers (58). The
regulation of SPOP expression at different levels includes DNA
methylation that affects transcription, miRNAs that affect
translation, and phosphorylation and self-ubiquitination that
affect post-transcriptional modifications (57). For example, the transcription
factor C/EBPα can bind the SPOP promoter; the combination of this
transcriptional regulatory element and DNA methylation regulates
the expression and function of SPOP in NSCLC (59). MiR-520b is upregulated in NSCLC,
and directly targets SPOP 3′-UTR and decreases SPOP expression to
promote proliferation and metastasis (60). In lung cancer, there are few
studies about the post-transcriptional modifications to regulate
the SPOP expression.
In the present study, the expression level of SPOP
increased 16-fold in lung cancer cells after SERPINA3
overexpression by DIA-MS detection, and it was further validated
that the expression of SPOP was upregulated with WB in lung cancer
cells and tumor tissues of mice. In addition, SPOP could promote
FAS-associated protein with death domain degradation and inhibit
NF-κB activity in NSCLC (61).
Previous studies have also indicated that inhibiting the NF-κB
signaling pathway is a promising strategy for treating lung cancer
(62,63). The present study demonstrated that
NF-κB p65 expression was downregulated in lung cancer. The
aforementioned results indicated that the overexpression of
SERPINA3 suppresses tumor progression by modulating SPOP/NF-κB in
lung cancer, suggesting that SERPINA3 is involved in the
pathogenesis of lung cancer.
However, there are limitations to the present study.
First, the reasons for SERPINA3 expression changes in lung cancer
are unknown. Second, the viability of lung cancer cells after
knockdown of SERPINA3 expression to explain the biological effects
of SERPINA3 in lung cancer was not assessed. In addition, whether
miR-137 alters SERPINA3 expression levels in lung cancer or whether
SERPINA3 directly interacts with SPOP to regulate its expression
remains unclear. These questions deserve further investigation.
In summary, the present results showed that SERPINA3
was reduced in lung cancer, and overexpressed SERPINA3 exerted
antitumoral properties at the cellular and animal levels, which
suppressed the tumor growth of lung cancer by activating SPOP
expression and inhibiting NF-κB in cell culture systems and in
vivo models. The present study revealed the antineoplastic role
of SERPINA3 in lung cancer and demonstrated that SERPINA3 is
involved in lung cancer occurrence, which will promote in-depth
elucidation of the pathogenesis of lung cancer and provide a new
direction for lung cancer treatment.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request. The mass spectrometry proteomics data have been deposited
to the ProteomeXchange Consortium via the PRIDE partner repository
with the dataset identifier PXD036119.
Authors' contributions
YJ and WW conceptualized the study. YJ, YZ and YC
developed methodology. YJ and WD performed formal analysis. YJ, AH,
JW, NL, XW and WD conducted investigation. AH and YG provided
resources. YJ and YG curated data. YJ and WD wrote the original
draft. WD and JP wrote, reviewed and edited the manuscript. YJ, YZ
and AH performed visualization. WD and JP supervised the study. YJ
and WD acquired funding. YJ, YZ, AH, YG and WD confirm the
authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The study was conducted according to the Declaration
of Helsinki. Human sample collection and animal experiments were
approved (approval no. KL901372) by the Research Ethics Board of
Nanjing Medical University and the Ethics Committee of Suzhou
Municipal Hospital. All the animal experiments were carried out
under the Guide for Care and Use of Laboratory Animals, and all
methods are reported in accordance with ARRIVE guidelines for
animal experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
SERPINA3
|
serpin family A member 3,
Alpha-1-antichymotrypsin
|
|
DIA-MS
|
data-independent acquisition mass
spectrometry
|
|
SPOP
|
speckle-type POZ protein
|
|
NSCLC
|
non-small cell lung cancer
|
|
LUAD
|
lung adenocarcinoma
|
|
LUSC
|
lung squamous cell carcinoma
|
|
DEPs
|
differentially expressed proteins
|
|
CCK-8
|
Cell Counting Kit-8
|
|
EdU
|
5-ethynyl-2′-deoxyuridine
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
Acknowledgments
The authors would like to thank Professor Hui Sun
(Wuhan University, Wuhan, China) for her helpful suggestions and
providing the SERPINA3 plasmid on the study.
Funding
The present study was supported by the National Natural Science
Foundation of China (NSFC) program (grant no. 32000908), the
Natural Science Foundation of Hubei Province program (grant no.
2020CFB417), the Hubei Key Laboratory of Edible Wild Plants
Conservation and Utilization (grant nos. EWPL202002, EWPL202106 and
EWPL202211), and the National innovation and entrepreneurship
training program for college students (grant no. 202210513012).
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Howlader N, Forjaz G, Mooradian MJ, Meza
R, Kong CY, Cronin KA, Mariotto AB, Lowy DR and Feuer EJ: The
effect of advances in lung-cancer treatment on population
mortality. N Engl J Med. 383:640–649. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kleczko EK, Kwak JW, Schenk EL and
Nemenoff RA: Targeting the complement pathway as a therapeutic
strategy in lung cancer. Front Immunol. 10:9542019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Su L, Chen M, Su H, Dai Y, Chen S and Li
J: Postoperative chemoradiotherapy is superior to postoperative
chemotherapy alone in squamous cell lung cancer patients with
limited N2 lymph node metastasis. BMC Cancer. 19:10232019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kinoshita T and Goto T: Molecular
Mechanisms of pulmonary fibrogenesis and its progression to lung
cancer: A review. Int J Mol Sci. 20:14612019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mascaux C, Tsao MS and Hirsch FR: Genomic
testing in lung cancer: Past, present, and future. J Natl Compr
Canc Netw. 16:323–334. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang M, Herbst RS and Boshoff C: Toward
personalized treatment approaches for non-small-cell lung cancer.
Nat Med. 27:1345–1356. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reck M, Carbone DP, Garassino M and
Barlesi F: Targeting KRAS in non-small-cell lung cancer: Recent
progress and new approaches. Ann Oncol. 32:1101–1110. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lim JU, Lee E, Lee SY, Cho HJ, Ahn DH,
Hwang Y, Choi JY, Yeo CD, Park CK and Kim SJ: Current literature
review on the tumor immune micro-environment, its heterogeneity and
future perspectives in treatment of advanced non-small cell lung
cancer. Transl Lung Cancer Res. 12:857–876. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Martinez-Ruiz C, Black JRM, Puttick C,
Hill MS, Demeulemeester J, Larose Cadieux E, Thol K, Jones TP,
Veeriah S, Naceur-Lombardelli C, et al: Genomic-transcriptomic
evolution in lung cancer and metastasis. Nature. 616:543–552. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garon EB, Hellmann MD, Rizvi NA, Carcereny
E, Leighl NB, Ahn MJ, Eder JP, Balmanoukian AS, Aggarwal C, Horn L,
et al: Five-Year overall survival for patients with advanced
non-small-cell lung cancer treated with pembrolizumab: Results from
the phase I KEYNOTE-001 study. J Clin Oncol. 37:2518–2527. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li T, Pan K, Ellinwood AK and Cress RD:
Survival trends of metastatic lung cancer in california by age at
diagnosis, gender, Race/Ethnicity, and histology, 1990-2014. Clin
Lung Cancer. 22:e602–e611. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fatima S, Gupta S, Khan AB, Rehman SU and
Jairajpuri MA: Identification and validation of two alternatively
spliced novel isoforms of human alpha-1-antichymotrypsin. Biochem
Biophys Res Commun. 628:25–31. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mateusz de Mezer, Rogalinski J, Przewozny
S, Chojnicki M, Niepolski L, Sobieska M and Przystanska A:
SERPINA3: Stimulator or Inhibitor of Pathological Changes.
Biomedicines. 11:1562023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin Y, Wang W, Wang Q, Zhang Y, Zahid KR,
Raza U and Gong Y: Alpha-1-antichymotrypsin as a novel biomarker
for diagnosis, prognosis, and therapy prediction in human diseases.
Cancer Cell Int. 22:1562022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim H, Leng K, Park J, Sorets AG, Kim S,
Shostak A, Embalabala RJ, Mlouk K, Katdare KA, Rose IVL, et al:
Reactive astrocytes transduce inflammation in a blood-brain barrier
model through a TNF-STAT3 signaling axis and secretion of alpha
1-antichymotrypsin. Nat Commun. 13:65812022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schneider MA, Rozy A, Wrenger S,
Christopoulos P, Muley T, Thomas M, Meister M, Welte T,
Chorostowska-Wynimko J and Janciauskiene S: Acute phase proteins as
early predictors for immunotherapy response in advanced NSCLC: An
explorative study. Front Oncol. 12:7720762022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brioschi M, Gianazza E, Agostoni P, Zoanni
B, Mallia A and Banfi C: Multiplexed mrm-based proteomics
identified multiple biomarkers of disease severity in human heart
failure. Int J Mol Sci. 22:2021. View Article : Google Scholar
|
|
20
|
Horta-López PH, Mendoza-Franco G,
Rodríguez-Cruz F, Torres-Cruz FM, Hernández-Echeagaray E,
Jarero-Basulto JJ, Rícny J, Florán Garduño B and Garcia-Sierra F:
Association of α-1-Antichymotrypsin expression with the development
of conformational changes of tau protein in Alzheimer's disease
brain. Neuroscience. 518:83–100. 2023. View Article : Google Scholar
|
|
21
|
Kim KH, Park GW, Jeong JE, Ji ES, An HJ,
Kim JY and Yoo JS: Parallel reaction monitoring with multiplex
immunoprecipitation of N-glycoproteins in human serum for detection
of hepatocellular carcinoma. Anal Bioanal Chem. 411:3009–3019.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lara-Velazquez M, Zarco N, Carrano A,
Phillipps J, Norton ES, Schiapparelli P, Al-Kharboosh R,
Rincon-Torroella J, Jeanneret S, Corona T, et al: Alpha
1-antichymotrypsin contributes to stem cell characteristics and
enhances tumorigenicity of glioblastoma. Neuro Oncol. 23:599–610.
2021. View Article : Google Scholar :
|
|
23
|
Zhu H, Liu Q, Tang J, Xie Y, Xu X, Huang
R, Zhang Y, Jin K and Sun B: Alpha1-ACT functions as a tumour
suppressor in hepatocellular carcinoma by inhibiting the
PI3K/AKT/mTOR signalling pathway via activation of PTEN. Cell
Physiol Biochem. 41:2289–2306. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Tian J, Qu C, Peng Y, Lei J, Li
K, Zong B, Sun L and Liu S: Overexpression of SERPINA3 promotes
tumor invasion and migration, epithelial-mesenchymal-transition in
triplenegative breast cancer cells. Breast Cancer. 28:859–873.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jin Y, Wang J, Ye X, Su Y, Yu G, Yang Q,
Liu W, Yu W, Cai J, Chen X, et al: Identification of GlcNAcylated
alpha-1-antichymotrypsin as an early biomarker in human
non-small-cell lung cancer by quantitative proteomic analysis with
two lectins. Br J Cancer. 114:532–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jin Y, Yang Y, Su Y, Ye X, Liu W, Yang Q,
Wang J, Fu X, Gong Y and Sun H: Identification a novel clinical
biomarker in early diagnosis of human non-small cell lung cancer.
Glycoconj J. 36:57–68. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seijo LM, Peled N, Ajona D, Boeri M, Field
JK, Sozzi G, Pio R, Zulueta JJ, Spira A, Massion PP, et al:
Biomarkers in lung cancer screening: Achievements, promises, and
challenges. J Thorac Oncol. 14:343–357. 2019. View Article : Google Scholar
|
|
28
|
Wadowska K, Bil-Lula I, Trembecki L and
Sliwinska-Mosson M: Genetic markers in lung cancer diagnosis: A
Review. Int J Mol Sci. 21:45692020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kang K, Niu B, Wu C, Hua J and Wu J: The
construction and application of lentiviral overexpression vector of
goat miR-204 in testis. Res Vet Sci. 130:52–58. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu H, Cheng Q, Xu DS, Wang W, Fang Z, Xue
DD, Zheng Y, Chang AH and Lei YJ: Overexpression of CXCR7
accelerates tumor growth and metastasis of lung cancer cells.
Respir Res. 21:2872020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
32
|
Li H, Lin PH, Gupta P, Li X, Zhao SL, Zhou
X, Li Z, Wei S, Xu L, Han R, et al: MG53 suppresses tumor
progression and stress granule formation by modulating G3BP2
activity in non-small cell lung cancer. Mol Cancer. 20:1182021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Laferriere CA and Pang DS: Review of
intraperitoneal injection of sodium pentobarbital as a method of
euthanasia in laboratory rodents. J Am Assoc Lab Anim Sci.
59:254–263. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang F, Ge W, Ruan G, Cai X and Guo T:
Data-Independent acquisition mass spectrometry-based proteomics and
software tools: A glimpse in 2020. Proteomics. 20:e19002762020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Murakami Y, Kusakabe D, Watari K, Kawahara
A, Azuma K, Akiba J, Taniguchi M, Kuwano M and Ono M: AXL/CDCP1/SRC
axis confers acquired resistance to osimertinib in lung cancer. Sci
Rep. 12:89832022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dong Y, Zhang D, Cai M, Luo Z, Zhu Y, Gong
L, Lei Y, Tan X, Zhu Q and Han S: SPOP regulates the DNA damage
response and lung adenocarcinoma cell response to radiation. Am J
Cancer Res. 9:1469–1483. 2019.PubMed/NCBI
|
|
38
|
Rubio K, Romero-Olmedo AJ, Sarvari P,
Swaminathan G, Ranvir VP, Rogel-Ayala DG, Cordero J, Gunther S,
Mehta A, Bassaly B, et al: Non-canonical integrin signaling
activates EGFR and RAS-MAPK-ERK signaling in small cell lung
cancer. Theranostics. 13:2384–2407. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fan T, Zhu M, Muhammad S, Xiao C, Li S,
Tian H, Liu Y, Xue L, Zheng B, Li C, et al: H3K4me3-related lncRNAs
signature and comprehensive analysis of H3K4me3 regulating tumor
immunity in lung adenocarcinoma. Respir Res. 24:1222023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu J, Feng J, Zhang Q, He Y, Xu C, Wang C
and Li W: Epigenetic regulation of stem cells in lung cancer
oncogenesis and therapy resistance. Front Genet. 14:11208152023.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Srivastava S, Mohanty A, Nam A, Singhal S
and Salgia R: Chemokines and NSCLC: Emerging role in prognosis,
heterogeneity, and therapeutics. Semin Cancer Biol. 86:233–246.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yuan J, Ju Q, Zhu J, Jiang Y, Yang X, Liu
X, Ma J, Sun C and Shi J: RASSF9 promotes NSCLC cell proliferation
by activating the MEK/ERK axis. Cell Death Discov. 7:1992021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jin Y, Chen Y, Tang H, Hu X, Hubert SM, Li
Q, Su D, Xu H, Fan Y, Yu X, et al: Activation of PI3K/AKT pathway
is a potential mechanism of treatment resistance in small cell lung
cancer. Clin Cancer Res. 28:526–539. 2022. View Article : Google Scholar
|
|
44
|
Lu S, Zhou J, Jian H, Wu L, Cheng Y, Fan
Y, Fang J, Chen G, Zhang Z, Lv D, et al: Befotertinib (D-0316)
versus icotinib as first-line therapy for patients with
EGFR-mutated locally advanced or metastatic non-small-cell lung
cancer: A multicentre, open-label, randomised phase 3 study. Lancet
Respir Med. S2213-2600:00183-22023.
|
|
45
|
Ghiringhelli F, Bibeau F, Greillier L,
Fumet JD, Ilie A, Monville F, Lauge C, Catteau A, Boquet I, Majdi
A, et al: Immunoscore immune checkpoint using spatial quantitative
analysis of CD8 and PD-L1 markers is predictive of the efficacy of
anti-PD1/PD-L1 immunotherapy in non-small cell lung cancer.
EBioMedicine. 92:1046332023. View Article : Google Scholar
|
|
46
|
Stankovic B, Bjorhovde HAK, Skarshaug R,
Aamodt H, Frafjord A, Muller E, Hammarstrom C, Beraki K, Baekkevold
ES, Woldbaek PR, et al: Immune cell composition in human non-small
cell lung cancer. Front Immunol. 9:31012018. View Article : Google Scholar
|
|
47
|
Zhang Y, Li Y, Qiu F and Qiu Z:
Comparative analysis of the human urinary proteome by 1D SDS-PAGE
and chip-HPLC-MS/MS identification of the AACT putative urinary
biomarker. J Chromatogr B Analyt Technol Biomed Life Sci.
878:3395–401. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nuzzo S, Catuogno S, Capuozzo M, Fiorelli
A, Swiderski P, Boccella S, de Nigris F and Esposito CL:
Axl-Targeted Delivery of the Oncosuppressor miR-137 in
Non-small-Cell Lung Cancer. Mol Ther Nucleic Acids. 17:256–263.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang B, Liu T, Wu T, Wang Z, Rao Z and
Gao J: microRNA-137 functions as a tumor suppressor in human
non-small cell lung cancer by targeting SLC22A18. Int J Biol
Macromol. 74:111–8. 2015. View Article : Google Scholar
|
|
50
|
Chang TH, Tsai MF, Gow CH, Wu SG, Liu YN,
Chang YL, Yu SL, Tsai HC, Lin SW, Chen YW, et al: Upregulation of
microRNA-137 expression by Slug promotes tumor invasion and
metastasis of non-small cell lung cancer cells through suppression
of TFAP2C. Cancer Lett. 402:190–202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li XM, Wu HL, Xia QD, Zhou P, Wang SG, Yu
X and Hu J: Novel insights into the SPOP E3 ubiquitin ligase: From
the regulation of molecular mechanisms to tumorigenesis. Biomed
Pharmacother. 149:1128822022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shi L, Yan Y, He Y, Yan B, Pan Y, Orme JJ,
Zhang J, Xu W, Pang J and Huang H: Mutated SPOP E3 ligase promotes
17betaHSD4 protein degradation to drive androgenesis and prostate
cancer progression. Cancer Res. 81:3593–3606. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang H, Jin X and Huang H: Deregulation
of SPOP in cancer. Cancer Res. 83:489–499. 2023. View Article : Google Scholar
|
|
54
|
Zhou P, Chang WY, Gong DA, Huang LY, Liu
R, Liu Y, Xia J, Wang K, Tang N and Huang AL: O-GlcNAcylation of
SPOP promotes carcinogenesis in hepatocellular carcinoma. Oncogene.
42:725–736. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wei C, Liu Y, Liu X, Cheng J and Fu J,
Xiao X, Moses RE, Li X and Fu J: The speckle-type POZ protein
(SPOP) inhibits breast cancer malignancy by destabilizing TWIST1.
Cell Death Discov. 8:3892022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Song Y, Xu Y, Pan C, Yan L, Wang ZW and
Zhu X: The emerging role of SPOP protein in tumorigenesis and
cancer therapy. Mol Cancer. 19:22020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang X and Zhu Q: SPOP in Cancer:
Phenomena, mechanisms and its role in therapeutic implications.
Genes (Basel). 13:20512022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cuneo MJ and Mittag T: The ubiquitin
ligase adaptor SPOP in cancer. FEBS J. 286:3946–3958. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yao S, Chen X, Chen J, Guan Y, Liu Y, Chen
J and Lv X: Speckle-type POZ protein functions as a tumor
suppressor in non-small cell lung cancer due to DNA methylation.
Cancer Cell Int. 18:2132018. View Article : Google Scholar
|
|
60
|
Liu X, Liu J, Zhang X, Tong Y and Gan X:
MiR-520b promotes the progression of non-small cell lung cancer
through activating Hedgehog pathway. J Cell Mol Med. 23:205–215.
2019. View Article : Google Scholar
|
|
61
|
Luo J, Chen B, Gao CX, Xie HK, Han CN and
Zhou CC: SPOP promotes FADD degradation and inhibits NF-κB activity
in non-small cell lung cancer. Biochem Biophys Res Commun.
504:289–294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang Y, Zhang J, Li YJ, Yu NN, Liu WT,
Liang JZ, Xu WW, Sun ZH, Li B and He QY: MEST promotes lung cancer
invasion and metastasis by interacting with VCP to activate NF-κB
signaling. J Exp Clin Cancer Res. 40:3012021. View Article : Google Scholar
|
|
63
|
Rasmi RR, Sakthivel KM and Guruvayoorappan
C: NF-κB inhibitors in treatment and prevention of lung cancer.
Biomed Pharmacother. 130:1105692020. View Article : Google Scholar
|