Introduction
Head and neck squamous cell carcinoma (HNSCC) is one
of the most common malignancies in the world. Globally, >700,000
cases of HNSCC are newly diagnosed, and >300,000 patients with
HNSCC succumb to this disease each year (1). Despite multimodality treatments,
recurrence and metastasis of HNSCC develop in numerous patients.
Therefore, revealing the underlying molecular mechanism of HNSCC
progression and identifying new key targets for the diagnosis and
treatment of HNSCC are especially urgent.
Homeobox C10 (HOXC10) is a member of the HOX genes,
which include four clusters: HOXA, HOXB, HOXC, and HOXD (2). Accumulated evidence has indicated
that the HOX gene family is markedly expressed in various cancers
and plays an important role in cancer progression. For example,
pharmacologic inhibition of HOXA9 suppressed TWIST1-induced
aggressive cellular phenotypes of prostate cancer in vitro
and metastasis in vivo (3). Higher HOXB8 expression in ovarian
serous carcinoma effusions was associated with significantly
shorter overall survival in post-chemotherapy patients (4). Invasive and metastatic potential of
cancer cells were enhanced by HOXD3 through the TGF-β-dependent and
-independent pathways (5). The
HOXC10 gene is located on chromosome 12 and plays an important role
in embryonic morphogenesis and cellular identity (6). Pathiraja et al showed that
aromatase inhibitor treatment of breast cancer resulted in
downregulation of HOXC10 expression via methylation of HOXC10
promoter regions and confers inhibitors resistance (7). Moreover, another study reported that
HOXC10 overexpression promoted angiogenesis in glioma via
upregulation of VEGFA expression and interaction with PRMT5
(8). HOXC10 may become a useful
marker in the diagnosis or treatment evaluation of cancer. However,
to date, only a limited number of studies have focused on the role
of HOXC10 in the pathogenesis of HNSCC. The biologic mechanisms of
HOXC10 in HNSCC remain unclear, and further study is required.
In the present study, the expression and biological
functions of HOXC10 were determined using HNSCC samples and animal
models. In particular, the underlying mechanisms of HOXC10-induced
proliferation, invasion, and metastasis of HNSCC were also
elucidated. Furthermore, it was found that the
N6-methyladenosine (m6A) writer,
methyltransferase-like 3 (METTL3), and the m6A readers,
insulin like growth factor 2 mRNA binding protein (IGF2BP)1 and
IGF2BP3 participated in increasing the stability of the HOXC10
transcript in HNSCC.
Materials and methods
Antibodies and reagents
Antibodies used for western blot analysis were as
follows: Anti-HOXC10 (1:1,000; cat. no. A303-178A-M; Thermo Fisher
Scientific, Inc.), anti-β-actin (1:1,000; cat. no. ab8224; Abcam),
anti-E-cadherin (1:1,000; cat. no. 3195), anti-N-cadherin (1:1,000;
cat. no. 13116), anti-vimentin (1:1,000; cat. no. 5741) and
anti-Snail (1:1,000; cat. no. 3879; all from Cell Signaling
Technology, Inc.), anti-ADAM17 (1:1,000; cat. no. ab39162),
phosphorylated (p)-ERK1/2 (1:1,000; cat. no. ab278538), anti-ERK1/2
(1:1,000; cat. no. ab184699), anti-p-epidermal growth factor
receptor (EGFR) (1:1,000; cat. no. ab40815) and anti-EGFR (1:1,000;
cat. no. ab52894; all from Abcam), anti-RPS15A (1:1,000; cat. no.
A304-990A; Thermo Fisher Scientific, Inc.), anti-Axin2 (1:1,000;
cat. no. ab109307), anti-MMP7 (1:1,000; cat. no. ab207299),
anti-c-Myc (1:1,000; cat. no. ab32072), anti-β-catenin (1:1,000;
cat. no. ab32572), anti-histone H3 (1:1,000; cat. no. ab1791),
anti-METTL3 (1:1,000; cat. no. ab195352), anti-IGF2BP1 (1:1,000;
cat. no. ab184305) and anti-IMP3 (1:1,000; cat. no. ab177477; all
from Abcam). The following antibodies were used for
immunohistochemistry: Anti-HOXC10 (1:500; cat. no. ab153904;
Abcam), anti-E-cadherin (1:400; cat. no. 3195; Cell Signaling
Technology, Inc.), anti-vimentin (1:400; cat. no. 5741; Cell
Signaling Technology, Inc.), and anti-Snail (1:1,000; cat. no.
ab85936; Abcam). Anti-HOXC10 (1:100; cat. no. A303-178A) and
anti-RPS15A (1:100; cat. no. A304-990A; both from Thermo Fisher
Scientific, Inc.) were used for immunoprecipitation (IP).
Actinomycin D was obtained from MedChemExpress.
Patients and specimens
HNSCC tissues and para-carcinoma tissues were
obtained from 77 patients diagnosed with HNSCC from the Eye and ENT
Hospital, Fudan University (Shanghai, China), from November 2017 to
March 2022. There were 75 males and 2 female patients, aged 43-84
years, with a mean age (± standard deviation) of 64.4±9.3 years.
The inclusion criteria were as follows: Patients who were diagnosed
with primary HNSCC, had available tumor tissue samples suitable for
tissue microarray (TMA) construction, and had complete clinical and
pathological data. Patients who had received prior chemotherapy,
immunosuppressive therapy, or had severe comorbidities or medical
conditions were excluded from the study. The tissues were stored at
-80°C until their use. Neoplastic and matched normal tissues were
used for TMA construction. Pannoramic MIDI (3DHISTECH, Ltd.) was
then used to scan the microarray. The present study was authorized
by the Ethics Committee of the Eye and ENT Hospital of Fudan
University (approval. no. 2018036). These patients had received no
treatment before surgery. All patients provided written informed
consent.
Cell culture
The human HNSCC cell lines, Tu686 and FaDu (The Cell
Bank of Type Culture Collection of The Chinese Academy of
Sciences), were used in the present study. HuLa-PC (CRL-3342;
ATCC), a human normal laryngeal cell line, was also used in the
present study. Tu686 was cultured using RPMI-1640 medium (HyClone;
Cytiva) supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin.
FaDu and HuLa-PC was cultured in Dulbecco's modified Eagle's medium
(DMEM; HyClone; Cytiva) supplemented with 10% FBS and 1%
penicillin-streptomycin. All cells were placed in an incubator with
5% CO2 at 37°C. AMC-HN8 cells were a kind gift from
Professor Sang Yoon Kim of Samsung Medical Center (Seoul, Korea).
Tu212 and M4E cell lines were obtained from Central South
University (Changsa, China). AMC-HN8, Tu212 and M4E were cultured
using RPMI-1640 medium (HyClone; Cytiva) supplemented with 10% and
1% penicillin-streptomycin, and were placed in an incubator with 5%
CO2 at 37°C. For the mRNA stability assay, transfected
HNSCC cells were treated with Actinomycin D (5 µg/ml; cat.
no. S8964; Selleck Chemicals) at 37°C for 0, 3 and 6 h prior to RNA
isolation.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cell lines and tissues
using Trizol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and then was reverse-transcribed into cDNA using
a RevertAid First Strand cDNA Synthesis kit (cat. no. K1622; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. The resulting cDNA was amplified with a gene-specific
primer and a qPCR SYBR Green Master Mix kit (Shanghai Yeasen
Biotechnology Co., Ltd.). The PCR reaction involved an initial
denaturation step at 95°C for 5 min followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing at 55-60°C for 20 sec,
and extension at 72°C for 20 sec. The primer sequences are listed
in Table I. The relative
expression levels of the target genes were normalized to β-actin
and reported as 2−ΔΔCq (9). In addition, HNSCC cells were treated
with 5-AZA-CdR (cat. no. S1200; Selleck Chemicals) at 1 µM
for 48 h, and the HOXC10 mRNA level was examined using RT-qPCR.
 | Table ISequences of primers. |
Table I
Sequences of primers.
| Gene symbol | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| HOXC10 |
AGCCTCGCCCTCAACACCTATC |
GCAGCAGACATTCTCCTCCTTGAC |
| MMP7 |
GAGGATGAACGCTGGACGGATG |
AGGATCAGAGGAATGTCCCATACCC |
| Myc |
AGCAGCGACTCTGAGGAGGAAC |
TCCAGCAGAAGGTGATCCAGACTC |
| Axin2 |
CACCACCACCATTCGCAGTACC |
ACATGCTTCGTCGTCTGCTTGG |
| METTL3 |
TGCCTTTGCCAGTTCGTTAGTCTC |
ACTGACCTTCTTGCTCTGTTGTTCC |
| IGF2BP1 |
CACCCGAAACACCTGACTCCAAAG |
GCCATAGATTCTTCCCTGAGCCTTG |
| IGF2BP3 |
TCACTTCTATGCTTGCCAGGTTGC |
CCTTCTGTTGTTGGTGCTGCTTTAC |
| RPS15A |
AACCTCACAGGCAGGCTAAACAAG |
TGGCGGGATGGAAGCAGATTATTC |
| ADAM17 |
AGCAGATTCGCATTCTCAAGTCTCC |
GCAACATCTTCACATCCCAAGCATC |
| β-actin |
GCACTCTTCCAGCCTTCCTTCC |
GCGGATGTCCACGTCACACTTC |
Western blot analysis
Total proteins were extracted using RIPA lysis
buffer containing 1% phenylmethanesulfonyl fluoride (PMSF; Beyotime
Institute of Biotechnology). The protein concentration was detected
using a BCA protein assay (Beyotime Institute of Biotechnology). A
total of ~20 µg protein/lane was separated by 6-12% SDS-PAGE
and then transferred onto a PVDF membrane (MilliporeSigma).
Subsequently, the membranes were blocked using 5% nonfat milk for 1
h at room temperature, and then incubated with the primary
antibodies at 4°C overnight. The following day, secondary
antibodies: HRP-labeled goat anti-rabbit IgG (H+L) (cat. no. A0208;
1:1,000) and HRP-labeled goat anti-mouse IgG (H+L) (cat. no. A0216;
1:1,000), provided by Beyotime Institute of Biotechnology, were
used to incubate the membranes at room temperature for 1 h. Target
protein bands were finally visualized using an ECL system
(MilliporeSigma). β-actin served as the control.
Cell migration, invasion, and
proliferation assays
Cellular migration and invasion abilities were
determined in 24-well Transwell chambers (Corning, Inc.). For
migration assays, 5×104 cells with 200 µl of
serum-free media were added to the upper chamber, whereas 600
µl of complete media (RPMI-1640 medium supplemented with 10%
FBS) was added to the bottom chamber. After 24 h at 37°C, the cells
in the lower chambers were fixed and stained with crystal violet
(Beyotime Institute of Biotechnology) for 30 min at room
temperature. For the invasion assays, a polycarbonate membrane was
pre-coated with Matrigel (Corning, Inc.) at 37°C for 3 h. For
quantification, a light microscope (Leica Microsystems GmbH) was
used to analyze the number of cells.
5-Ethynyl-2′-deoxyuridine (EdU) assay was performed
to evaluate the ability of cellular proliferation based on the
BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 555 (cat.
no. C0075S; Beyotime Institute of Biotechnology). The transfected
cells were seeded into 24-well plates (5×103
cells/well). The cells were cultured in EdU (10 µM) for 2 h
at 37°C and then fixed with 4% paraformaldehyde for 10 min at room
temperature. Subsequently, the samples were stained with the kit at
room temperature for 1 h, according to the manufacturer's
instructions. Images were detected using a fluorescence microscope
(Leica Microsystems GmbH).
Co-immunoprecipitation (Co-IP) and
ubiquitination assays
Proteins were extracted using 450 µl IP lysis
buffer (Thermo Fisher Scientific, Inc.) containing 1% PMSF
(Beyotime Institute of Biotechnology). Following cell lysis the
supernatant was collected by centrifugation at 12,000-16,000 × g
for 10 min at 4°C) and then incubated with the primary antibodies
on a shaker at 4°C overnight. Subsequently, the samples were
incubated overnight with protein A/G magnetic beads (cat. no.
LSKMAGAG02; MilliporeSigma). The beads were then washed 3 times
with lysis buffer (cat. no. 87788; Thermo Fisher Scientific, Inc.)
and then boiled at 100°C in loading buffer for 30 min for
subsequent western blot analysis assays.
For ubiquitination (Ubi) assays, transfected HNSCC
cells were treated with MG132 (5 µmol; Selleck Chemicals) at
37°C for 4 h before being harvested. The cell lysates were
immunoprecipitated with the RPS15A antibody (cat. no. A304-990A;
Thermo Fisher Scientific, Inc.) and analyzed by western blotting
with ubiquitin antibody (cat. no. sc8017; Santa Cruz Biotechnology,
Inc.) at 4°C overnight.
Transfection
For transient cell transfection, siRNAs against
human ADAM17 (5′-ACU UCA CAC UGU ACU CGC UTT-3′), RPS15A (5′-GAU
GAU GAA GCA UGG UUA CAU-3′), METTL3 (5′-CUG CAA GUA UGU UCA CUA
UGA-3′), IGF2BP1 (5′-GGC TCA GTA TGG TAC AGT A-3′), IGF2BP3 (5′-GCT
GAG AAG TCG ATT ACT A-3′) and the control siRNAs (5′-TTC TCC GAA
CGT GTC ACG T-3′) were obtained from Genomeditech (Shanghai) Co.,
Ltd. The RPS15A overexpression plasmid (Fig. S1A), ADAM17 overexpression plasmid
(Fig. S1B), and empty plasmids
were purchased from Genomeditech (Shanghai) Co., Ltd. The siRNAs
(50 nM) or plasmids (2 µg/ml) were transfected using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) at
37°C (for siRNAs, 48 h and plasmids, 6 h). For stable cell
transfection (at 37°C for 18-20 h), the HOXC10 overexpression and
knockdown lentivirus (5′-ACC TAG TGT CAA GGA GGA GAA-3′) were
obtained from Genomeditech (Shanghai) Co., Ltd. The 3rd generation
system was used and the interim cell line used was 293T (ATCC). The
quantity of lentiviral plasmid that was used was 20 µg, for
transfection, and the ratio used for the lentivirus, packaging and
envelope plasmids was: 3:1:1. The medium was collected by
centrifugation at 500 × g for 5 min at 4°C at a multiplicity of
infection of 10. The duration of transfection into the cells of
interest was 16 h and the time interval between transduction and
subsequent experimentation was ~2 weeks. To create stable cell
lines, the selection method used was puromycin (5 µg/ml),
and the concentration of puromycin used for maintenance was 2
µg/ml.
Immunofluorescence
The cells were fixed with 4% paraformaldehyde
(Beyotime Institute of Biotechnology) at room temperature for 15
min. and then permeabilized with 0.2% Triton X-100 (Beyotime
Institute of Biotechnology). Subsequently, the cells were blocked
using 1% BSA at room temperature for 1 h and then incubated with
the primary antibodies at 4°C overnight. The following day, the
cells were incubated with fluorescent secondary antibodies at room
temperature for 1 h. The stained cells were analyzed using a Leica
microscope (Leica Microsystems GmbH). The following primary
antibodies were used for immunofluorescence: Anti-HOXC10 (1:100;
cat. no. ab153904; Abcam), anti-E-cadherin (1:1,000; cat. no. 3195;
Cell Signaling Technology, Inc.), anti-vimentin (1:100; cat. no.
5741; Cell Signaling Technology, Inc.), anti-Snail (1:100; cat. no.
A5243; ABclonal), anti-RPS15A (1:100; cat. no. DF9117; Affinity
Biosciences), and anti-β-catenin (1:200; cat. no. ab32572; Abcam).
The fluorescent secondary antibodies used were as follows: Alexa
fluor 488-labeled goat anti-mouse IgG (H+L) (1:500; cat. no. A0428)
and Alexa fluor 555-labeled donkey anti-rabbit IgG (H+L) (1:500;
cat. no. A0453; both from Beyotime Institute of Biotechnology).
Immunohistochemistry
Tissues were fixed with 4% paraformaldehyde at room
temperature for 24 h. and then embedded in paraffin. The
paraffin-embedded tissues were sectioned into 4-µm thick
slices. Tissue slices were deparaffinized, rehydrated, blocked with
10% goat serum (cat. no. C0265; Beyotime Institute of
Biotechnology) at 37°C for 60 min, and then incubated overnight
with primary antibodies at 4°C. Subsequently, the slices were
incubated with secondary antibodies [HRP-labeled goat anti-mouse
IgG (H+L) (1:50; cat. no. A0216) and HRP-labeled goat anti-rabbit
IgG (H+L) (1:50; cat. no. A0208; both from Beyotime Institute of
Biotechnology] and stained with DAB. The slides were analyzed using
a light microscope (Leica Microsystems GmbH). The histochemistry
score (H-Score) value was calculated using Image-Pro Plus software
6.0 and used as an indicator of the level of protein.
Dual-luciferase reporter assay
The lentivirus-transfected cells were seeded in
24-well plates. ADAM17 promoter plasmids [Genomeditech (Shanghai)
Co., Ltd.] were transfected into these cells. After 48 h of
transfection using Lipofectamine 2000 (cat. no. 11668019; Thermo
Fisher Scientific, Inc., the relative luciferase activity of
reporter vectors was analyzed using a Dual-Luciferase Reporter Gene
Assay kit (cat. no. RG027; Beyotime Institute of Biotechnology)
following the manufacturer's protocols. The luciferase activity
against that of Renilla was measured.
RNA antisense purification (RAP)
assay
In this experiment, a RAP assay was performed using
a RAP kit (cat. no. Bes5103-1-S; Guangzhou Bersinbio Co., Ltd.),
following the manufacturer's protocols and as previously described
(10,11). The biotin-labeled specific probe
was used to pull down the target RNA. RNAs and proteins that
interacted with the target RNA were also obtained. The RNA products
were then analyzed by RT-qPCR, and the protein products were
analyzed by western blotting.
Methylated RNA immune precipitation
(MeRIP)
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The RNAs were cleaved
into RNA fragments. The fragmented RNAs were immunoprecipitated
with an anti-m6A antibody (included in a MeRIP kit; cat.
no. Bes5203-2; Bersinbio). Protein A/G magnetic beads (20 µl
per IP; included in the aforementioned kit) were added to the
mixture at 4°C, overnight. The beads were washed three times with
an elution buffer. The RNAs were extracted using
phenol-chloroform-isoamylol (included in the aforementioned kit;
25:24:1). The expression of the RNA was then analyzed by
RT-qPCR.
Animal experiments
A total of 20 male BALB/c nude mice (4-6 weeks old;
weight, 18-20 g) were obtained from the Shanghai SLAC Laboratory
Animal Co., Ltd. The present study was performed according to the
guidelines of the National Institutes of Health for the Care and
Use of Laboratory Animals (12).
Mice were housed in animal rooms with a 10-h light/14-h dark cycle
and at a constant temperature (22-27°C). Animals had free access to
standard rodent chow and water. For the subcutaneous xenograft
tumor model, transfected cells (5×106) were injected on
the sides of the flanks of mice (n=5 per group). Subsequently, 30
days later, the mice were euthanized by cervical dislocation
following anesthesia induced by intraperitoneal injection of 0.3%
sodium pentobarbital (30 mg/kg). Euthanasia was confirmed by
verifying respiratory and cardiac arrest, along with pupil
dilation, for a minimum of 10 min. Tumor size and tumor weight were
measured (max tumor diameter, 9.6 mm; max area, 82.56
mm2; max volume, 355.01 mm3). For the
pulmonary metastasis model, transfected cells (2×106)
were injected into the mouse tail veins (n=5 per group).
Subsequently, 60 days later, the mice were sacrificed, and lungs
were obtained. Finally, lung and tumor tissues were available for
H&E staining at room temperature for 5 min or
immunohistochemistry staining. All animal procedures were approved
(approval no. 202212201) by the Eye and ENT Hospital, Fudan
University (Shanghai, China).
RNA sequencing
Total RNA was extracted using the TRIzol reagent
(cat. no. 15596018CN; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RNA purity and
quantification were evaluated using a NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). RNA integrity
was assessed using the Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc.). The libraries were then constructed using
NEBNext®Ultra™ RNA Library Prep Kit for
Illumina® (New England BioLabs, Inc.) following
manufacturer's recommendations. The transcriptome sequencing and
analysis were conducted by Orizymes Biotechnologies (Shanghai) Co.,
Ltd. The libraries were sequenced on an Illumina HiSeq X Ten
platform and 150-bp paired-end reads were generated. Raw data (raw
reads) of fastq format (https://maq.sourceforge.net/fastq.shtml.) were firstly
processed using Trimmomatic (http://www.usadellab.org/cms/index.php?page=trimmomatic).
Clean data were obtained for downstream analyses by removing reads
containing adapter, reads containing ploy-N and low-quality reads
from raw data. The clean reads were mapped to the human genome
(hg38p13) using HISAT2 (https://ccb.jhu.edu/software/hisat2). The FPKM value
of each gene was calculated using Cufflinks (13), and the read counts of each gene
were obtained by HTSeq-count (14). Differential expression analysis
was performed using the DESeq (2012) R package (15). A P-value <0.05 and fold change
>2 or fold change <0.5 were set as the threshold for
significantly differential expression. The heatmap and volcano plot
were created using OmicStudio (https://www.omicstudio.cn).
ChIP-sequencing
DNA and protein cross-linking was achieved by adding
1% formaldehyde solution (cat. no. F8775; Sigma-Aldrich, Merck
KGaA), followed by quenching the reaction with 125 mM glycine
(included in a CHIP kit; cat. no. Bes5001; Bersinbio). Chromatin
was extracted from the cross-linked cells using lysis buffer
(included in a CHIP kit; cat. no. Bes5001; Bersinbio). The
chromatin DNA was sonicated to obtain fragments ranging from 100 to
500 base pairs using MinElute PCR Purification Kit (cat. no. 28004;
QIAGEN China Co., Ltd.). Immunoprecipitation of the chromatin DNA
and antibody-associated beads was performed by incubating them
overnight at 4°C. The immunoprecipitated protein-DNA complexes were
eluted from the Dynabeads (included in a CHIP kit; cat. no.
Bes5001; Bersinbio) using an optimal method (https://v2.fangcloud.com/share/bcc477b7f13badd6dc7c0163a4).
Sequencing libraries of the immunoprecipitated chromatin DNA were
generated using the NEBNext® Ultra™ DNA Library Prep Kit
for Illumina (New England BioLabs, Inc.). DNA purity and
quantification were evaluated using a NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). DNA integrity
was assessed using the Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc.). The libraries underwent end repair, adaptor
ligation, and removal of base U of the adaptor. After purification
and size selection, the libraries were quantified using Qubit and
the insert size was assessed using a high-sensitivity DNA chip. The
libraries (12 pM) were sequenced on an Illumina NovaSeq 6000
platform using NovaSeq 6000 SP reagent kit (100 cycles; cat. no.
2002746; Illumina Inc.), generating 150 bp paired-end reads. The
raw sequencing data in fastq format were processed using fastp
software [FASTP (version 0.20.1) (https://github.com/OpenGene/fastp] to obtain clean
reads by removing adapters, poly-N sequences, and low-quality
reads. The clean reads were then mapped to the genome using Bowtie2
(16) and peaks were called using
MACS2 (version 2.2.7.1) (17).
The called peaks were visualized using IGV software and annotated
using the ChIPseekerv (18)
package in R. De novo and known motifs were identified using
MEME-ChIP (version 4.9.1) (19),
and GO enrichment analysis was performed using clusterProfiler
(20).
Bioinformatic analyses
The pathways associated with HOXC10 in HNSCC were
analyzed by Gene set enrichment analysis (GSEA) analysis with the R
package ClusterProfiler (20). A
total of 520 patients with HNSCC from TCGA database (https://portal.gdc.cancer.gov/) were divided into
high and low expression groups based on HOXC10 expression, and the
the median expression level of HOXC10 was set as the cut-off.
Normalized enrichment score (NES), P-value, and false discovery
rate (FDR) for all variables and signatures were obtained by
running GSEA.
For m6A site prediction, site-specific
RNA adenosine methyltransferase peaks (SRAMP; http://www.cuilab.cn/sramp/) and RMBase_v2.0
(http://rna.sysu.edu.cn/rmbase/) were
used.
Statistical analysis
GraphPad Prism 9 software (GraphPad Software, Inc.;
Dotmatics) and Stata 13.0 (StataCorp LP) were used for the
statistical analysis. All data represent the mean ± standard
deviation (SD) of triplicate repeats. Student's unpaired t-test was
used to compare two groups. The correlation between the two
proteins was assessed using Pearson's correlation coefficient.
Comparisons of three groups were performed using one-way analysis
of variance (ANOVA) with Tukey's multiple comparison test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
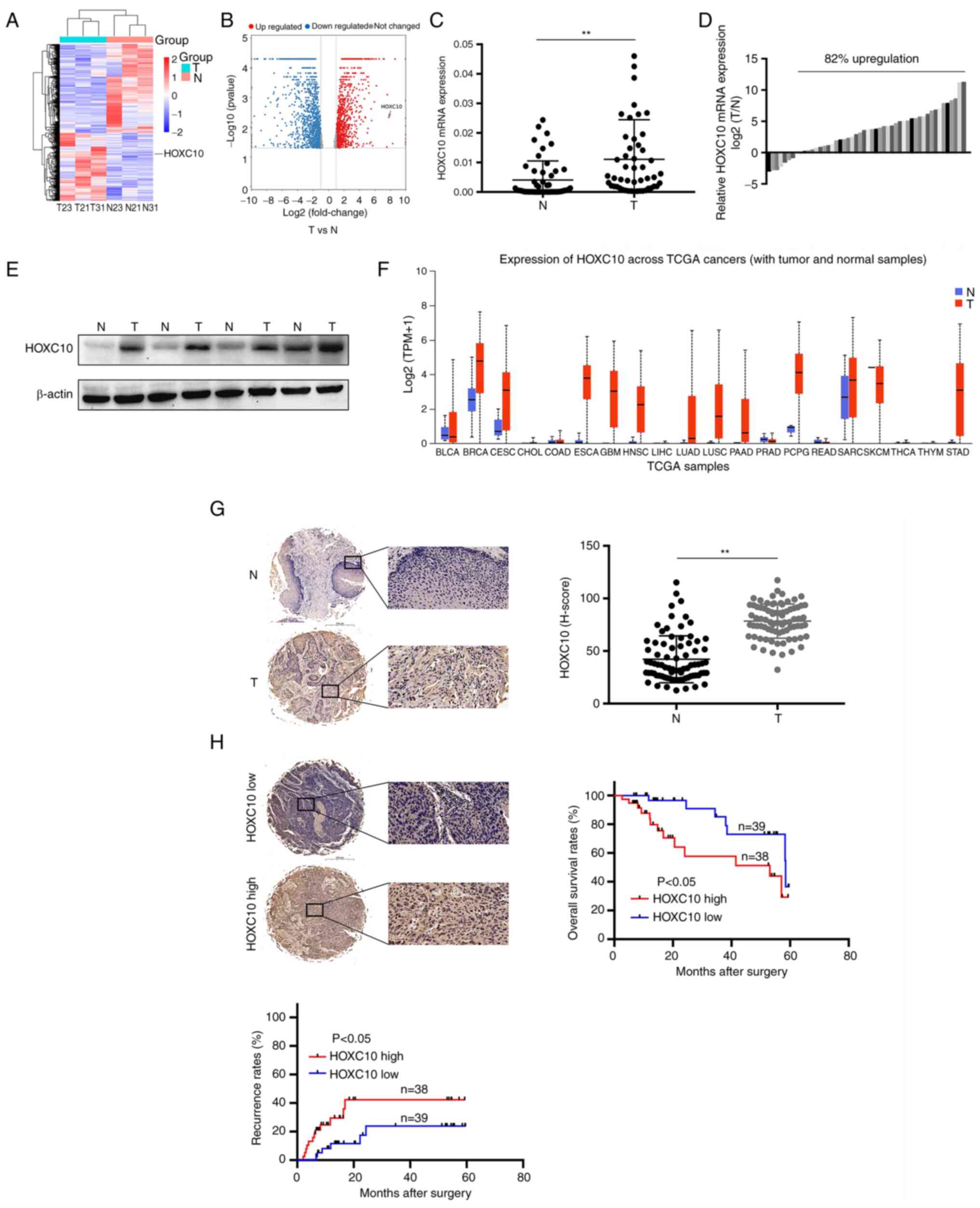
HOXC10 is overexpressed in HNSCC tissues
and is associated with a poor prognosis
The sum of three pairs of cancer and adjacent
tissues were first selected for RNA-seq and it was found that the
expression of HOXC10 in HNSCC was increased (Fig. 1A and B), which was consistent with
the results obtained from RT-qPCR and western blot analysis
(Fig. 1C-E). HOXC10 expression
was also upregulated in other cancer types, such as esophageal
cancer (ESCA) and cervical squamous cell carcinoma and endocervical
adenocarcinoma (CESC; Fig. 1F).
Immunohistochemical staining of tissues from 77 patients with HNSCC
revealed that HOXC10 expression was significantly higher in cancer
tissues than in adjacent tissues (Fig. 1G). A high level of HOXC10
expression was markedly associated with advanced TNM stage
(Table II), shorter survival
times, and a higher rate of recurrence (Fig. 1H). These results demonstrated that
HOXC10 is highly expressed in HNSCC tissues and is associated with
a poor prognosis in patients with HNSCC.
 | Table IIAssociation between HOXC10 and
clinicopathological characteristics in 77 patients with head and
neck squamous cell carcinoma. |
Table II
Association between HOXC10 and
clinicopathological characteristics in 77 patients with head and
neck squamous cell carcinoma.
| Variables | No. of patients
| P-value |
|---|
| 38
HOXC10high | 39
HOXC10low |
|---|
| Age (years) | | | |
| <65 | 17 | 21 | |
| ≥65 | 21 | 18 | 0.424 |
| Smoking | | | |
| Yes | 33 | 32 | |
| No | 5 | 7 | 0.562 |
| Drinking | | | |
| Yes | 19 | 24 | |
| No | 19 | 15 | 0.308 |
| Hypertension | | | |
| Yes | 13 | 12 | 0.747 |
| No | 25 | 27 | |
| Tumor size
(cm) | | | |
| <4 | 20 | 29 | 0.048 |
| ≥4 | 18 | 10 | |
| Lymphatic
metastasis | | | |
| Metastasis | 19 | 15 | |
| Nonmetastasis | 19 | 24 | 0.308 |
|
Differentiation | | | |
| Moderate to
poor | 29 | 29 | |
| High | 9 | 10 | 0.842 |
| Clinical
stages | | | |
| I - III | 14 | 23 | 0.016 |
| IV | 24 | 16 | |
In Table II,
clinical stage IV included 40 patients with HNSCC, of which 28 had
laryngeal cancer and 12 had hypopharyngeal cancer. It is crucial to
note that among the 40 stage IV patients included in the present
study, 38 were categorized as stage IVa, 2 were classified as stage
IVb, and there were no cases of stage IVc. The patients with
metastases are primarily characterized by neck lymph node
involvement, and notably, there were no instances of distant
metastases in the present study. Surgical management in cases of
neck lymph node metastasis involved radical neck lymph node
dissection, which has demonstrated the capability to achieve
substantial removal of metastatic regions in the neck. Furthermore,
the primary tumors of the stage IV patients were relatively large.
The post-operative pathology reports indicated that 2 patients had
positive margins. Thus, in the present study, surgery was found to
effectively remove a significant portion of the tumors in most
stage IV patients with HNSCC, but it may not completely remove
tumors in all patients with stage IV disease.
High levels of HOXC10 promote HNSCC
progression in vivo and in vitro
HOXC10 expression was revealed in five HNSCC cell
lines and a normal nasopharyngeal epithelial cell line. As shown in
Fig. 2A, HOXC10 expression in
cancer cells was higher than in normal cells. Furthermore, HOXC10
was transfected with shRNAs into FaDu cells and HOXC10 vectors into
Tu686 cells; transfection efficiencies were detected by RT-qPCR and
western blotting (Fig. 2B and C).
The migration and invasion assays revealed that HOXC10 knockdown
inhibited the motility of FaDu cells. The EdU assay showed that
knockdown of HOXC10 suppressed the cell growth rate of FaDu cells;
by contrast, HOXC10 upregulation significantly promoted the
migration, invasion, and growth rates of Tu686 cells (Fig. 2D-F). To analyze the function of
HOXC10 in vivo, a subcutaneous xenograft model was
established and a pulmonary metastasis model, as shown in Fig. 2G-J. FaDu-NC cells exhibited a
higher proliferation ability than FaDu-shRNA2 cells; beyond that,
the incidence of lung metastasis for FaDu-NC cells was higher than
that of FaDu-shRNA2 cells (Fig.
2K). These results revealed that HOXC10 upregulation promotes
HNSCC progression, both in vivo and in vitro.
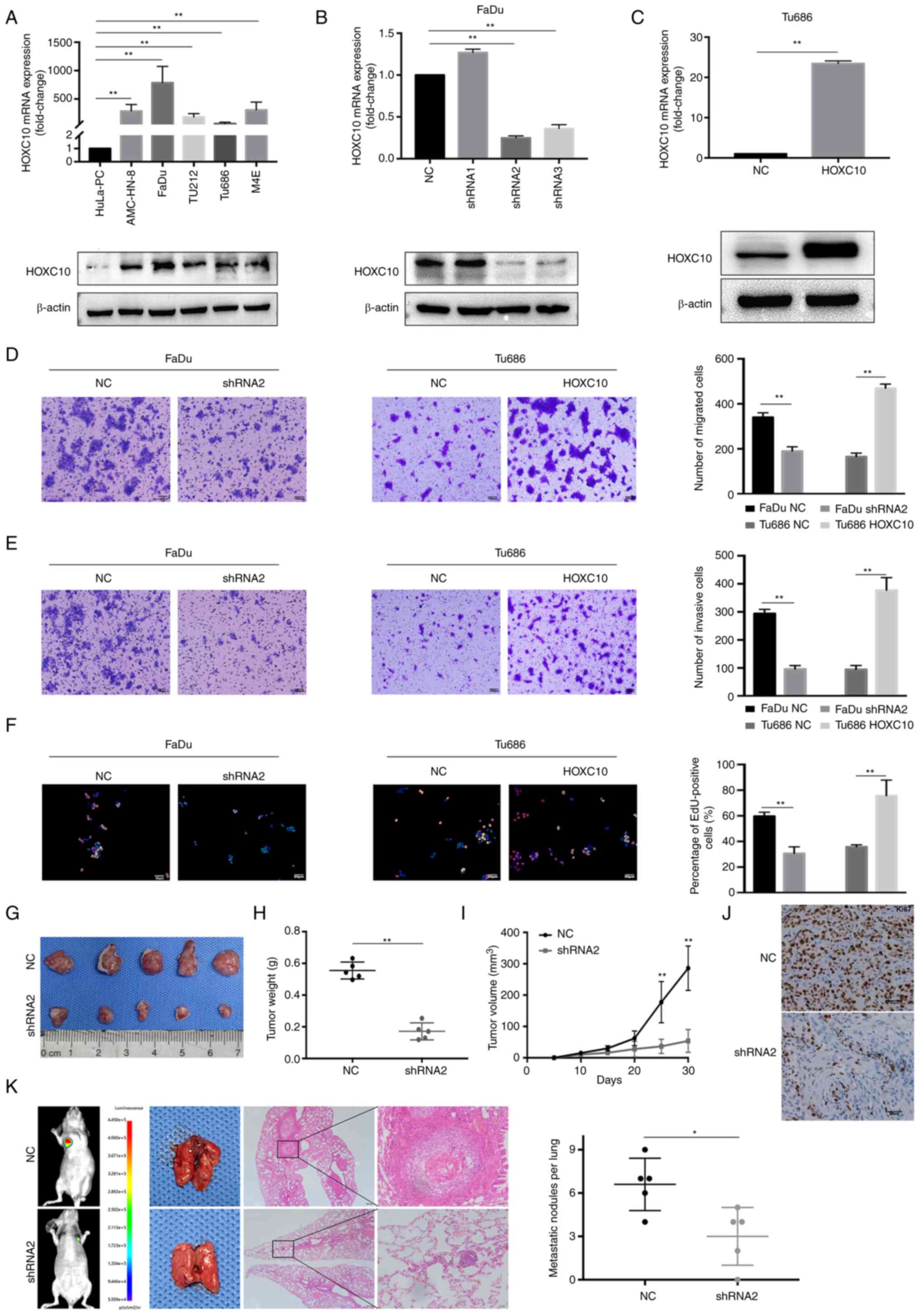 | Figure 2HOXC10 is involved in HNSCC cell
migration, invasion, and proliferation in vitro and in vivo.
(A) mRNA and protein levels of five HNSCC cell lines and a normal
nasopharyngeal epithelial cell line. (B and C) The efficiency of
transfection in the FaDu and Tu686 cell lines was verified by
reverse transcription-quantitative PCR and western blot analysis.
(D-F) Migration, invasion, and proliferation ability in transfected
HNSCC cells were assessed by Transwell migration and EdU assays.
Scale bar, 100 µm for D and E; scale bar, 50 µm for
F. (G) Tumors derived from nude mice were subcutaneously
transplanted with FaDu-NC and FaDu-shRNA2 cells. (H) Weight
measurement of tumors. (I) Growth curves of subcutaneous tumors.
(J) Ki67 expression was measured by immunohistochemistry. Scale
bar, 50 µm. (K) Nude mice tails were intravenously injected
with FaDu-NC and FaDu-shRNA2 cells to establish the pulmonary
metastasis models; representative images show pulmonary metastases.
Scale bar, 200 µm. **P<0.01. HOXC10, homeobox
C10; HNSCC, head and neck squamous cell carcinoma; EdU,
5-ethynyl-2′-deoxyuridine; NC, negative controls; sh, small
hairpin. |
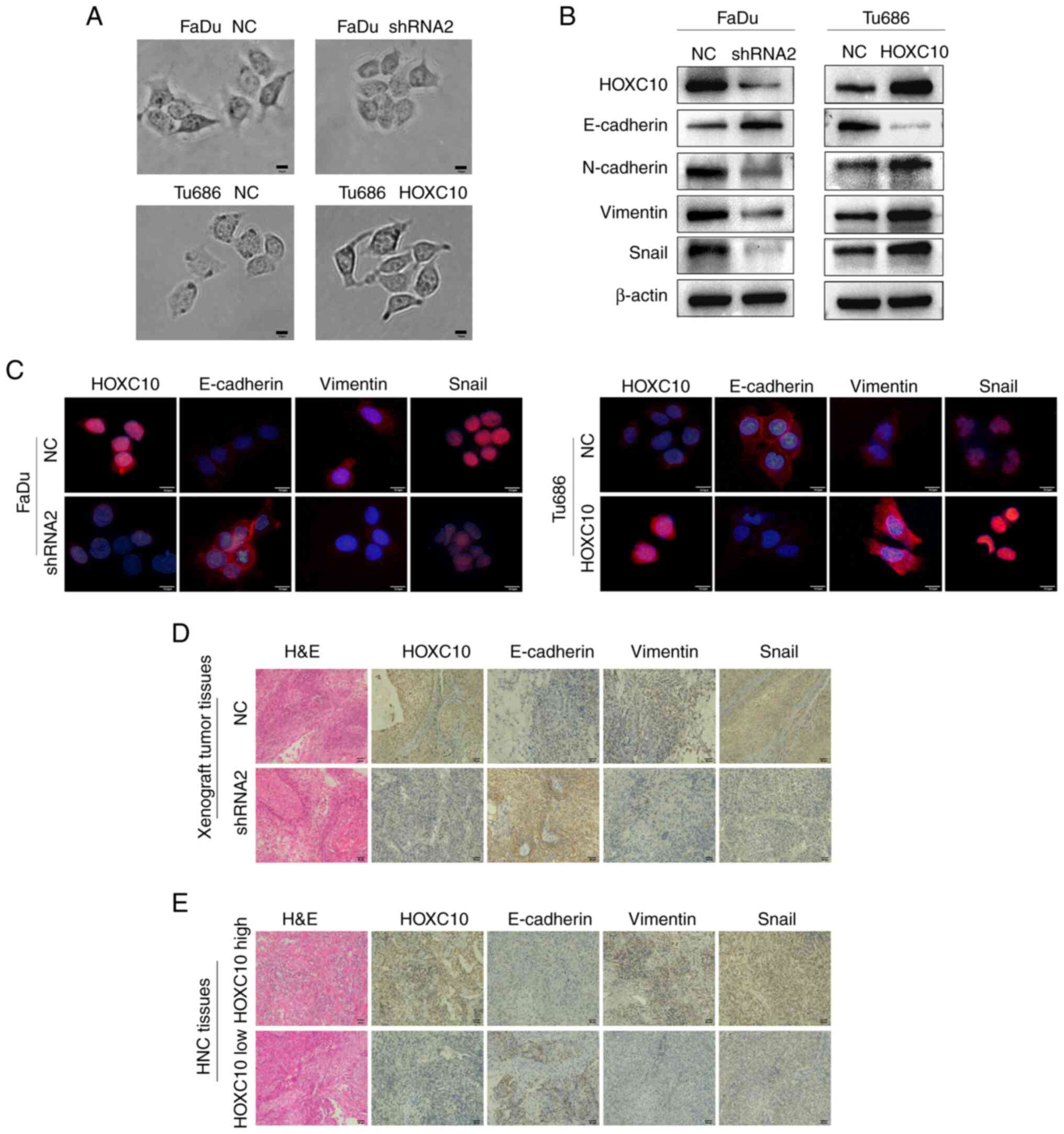
EMT is widely recognized as an important event
associated with cancer progression (21). The cellular morphology of the
transfected HNSCC cells was detected and it was observed that
FaDu-shRNA2 and Tu686-NC cells exhibited a cobblestone-like
appearance such as epithelial cells, while FaDu-NC and Tu686-HOXC10
cells exhibited a spindle-like morphology such as fibroblasts
(Fig. 3A). Next, it was found
that knockdown of HOXC10 enhanced E-cadherin expression but reduced
the expression of vimentin, N-cadherin and Snail in FaDu cells.
Conversely, HOXC10 overexpression brought about the opposite
changes in the expression of these EMT markers in Tu686 cells
(Fig. 3B), which was further
supported by immunofluorescence and immunohistochemical analyses
(Fig. 3C-E). Taken together,
these results indicated that HOXC10 could facilitate the
progression of HNSCC by inducing EMT.
ADAM17 is a direct target of HOXC10 in
HNSCC cells
To further investigate the target genes regulated by
HOXC10, chromatin immunoprecipitation sequencing (ChIP-seq) was
performed in Tu686-HOXC10 cells. To identify and annotate these
target genes, MACS2 was introduced for peak calling filtered with
the P-value and peak enrichment, and ChIPseeker was applied to
annotate the peak regions in gene promoter regions following the
promoter regions ranging from TSS 3,000 bp upstream to 3,000 bp
downstream (Fig. 4A). As shown in
Fig. 4B, peak regions in the
whole genome were distributed as follows: 9.26% enrichment in the
promoter regions, majority located introns (50.07%) and intergenic
regions (33.7%), minority located in other regions including 0.71%
downstream, 3.59% exons, 2.08% 3′UTR, and 0.58% 5′UTR. GO
enrichment revealed that HOXC10 played an important role in the
'epidermal growth factor receptor signaling pathway' (Fig. 4C). The metalloprotease, ADAM17 has
been reported to activate ligands of EGFR and contribute to tumor
progression (22). In the present
study, it was found that a putative HOXC10-binding peak at the
promoter region of the ADAM17 gene (Fig. 4D). Based on this, the potential
motif bound by HOXC10 was also identified with MEME motif software
(Fig. 4E). A dual-luciferase
reporter assay revealed that HOXC10 enhanced the transcriptional
activity of ADAM17 (Fig. 4F).
Knockdown of HOXC10 reduced the expression of ADAM17 in FaDu cells;
conversely, HOXC10 overexpression yielded the opposite effect in
Tu686 cells (Fig. 4G and H). A
previous study reported that ADAM17-EGFR signaling axis-dependent
ERK activation mediated the proliferation of collecting duct kidney
epithelial cells (23). In the
present study, it was revealed that ADAM17 treatment significantly
promoted the phosphorylation of EGFR and ERK1/2 in Tu686 cells
(Fig. 4I). HOXC10 knockdown in
FaDu cells suppressed the EGFR and ERK1/2 phosphorylation levels,
which were restored after ADAM17 overexpression; by contrast,
HOXC10 overexpression in Tu686 cells enhanced the phosphorylation
of EGFR and ERK1/2, which was inhibited after ADAM17 knockdown
(Fig. 4J). Correspondingly,
HOXC10 knockdown restrained the growth rate of FaDu cells, which
was regained after ADAM17 overexpression; HOXC10 overexpression
enhanced the growth rate of Tu686 cells, which was suppressed after
ADAM17 knockdown (Fig. 4K). Gene
Set Enrichment Analysis (GSEA), based on the TCGA dataset, also
indicated that HOXC10 expression was positively associated with
'DNA replication' (Fig. 4L).
Collectively, these data demonstrated that HOXC10 promotes the
proliferation of HNSCC via targeting the ADAM17/EGFR pathway.
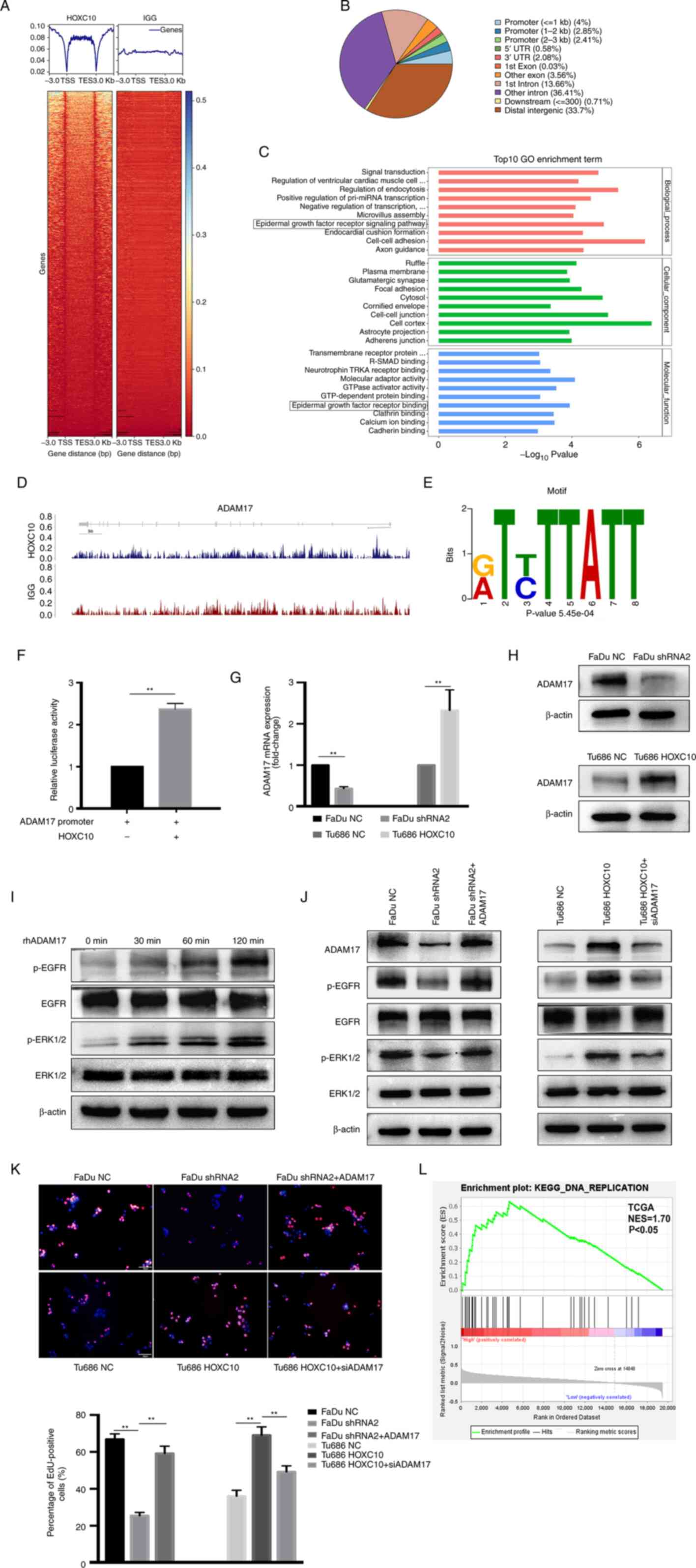 | Figure 4HOXC10 enhances the proliferation of
HNSCC cells by targeting ADAM17. (A) Heat map of chromatin
immunoprecipitation sequencing read densities around the
HOXC10-bound regions 3 kb upstream of the TSS and 3 kb downstream
of the TES. (B) Pie chart showing the distribution of HOXC10 peaks
across the genome. (C) GO enrichment analysis of the top 10
biological processes, cellular components, and molecular function
categories. (D) Distribution of HOXC10 binding peaks at ADAM17
promoters. (E) The ADAM17 promoter region contains a motif bound by
HOXC10. (F) Transcriptional activity of ADAM17 was assessed by dual
luciferase reporter assay. (G and H) Reverse transcription
quantitative-PCR and western blot analyses were used to detect the
expression of ADAM17 in the indicated cells. (I) Western blotting
showed phosphorylation levels of EGFR and ERK1/2 in Tu686 cells
treated with rhADAM17. (J) Phosphorylation levels of EGFR and
ERK1/2 in the indicated cells were analyzed by western blotting.
(K) The proliferation ability in transfected HNSCC cells was
assessed by EdU assay. Scale bar, 50 µm. (L) GSEA, based on
the TCGA dataset, showed that HOXC10 expression was positively
associated with DNA replication. **P<0.01. HOXC10,
homeobox C10; HNSCC, head and neck squamous cell carcinoma; ADAM17,
a disintegrin and metalloproteinase 17; TSS, transcription start
site; TES, transcription end site; GO, Gene Ontology; EGFR,
epidermal growth factor receptor; EdU, 5-ethynyl-2′-deoxyuridine;
GSEA, Gene Set Enrichment Analysis; TCGA, The Cancer Genome Atlas
Program; KEGG, Kyoto Encyclopedia of Genes and Genomes. |
HOXC10 facilitates Wnt/β-catenin
signaling in HNSCC by interacting with RPS15A
To further explore the potential mechanism through
which HOXC10 promotes the invasion and metastasis of HNSCC, Co-IP
and mass spectrometric (MS) analyses were performed to identify the
underlying HOXC10-binding proteins in HNSCC cells. As shown in
Fig. 5A-C, 11 proteins were
identified among the candidate HOXC10-interacting partners. RPS15A
was selected for further investigation, as prior studies have
indicated that it is involved in the progression of gastric cancer
and colorectal cancer (24-25), but its role in HNSCC has not been
well studied. The interaction between HOXC10 and RPS15A was
verified by co-IP and colocalization assays (Fig. 5D and E). Moreover, changes in the
expression of HOXC10 did not influence RPS15A mRNA levels, whereas
HOXC10 knockdown significantly downregulated RPS15A protein levels
in FaDu cells, and overexpression of HOXC10 upregulated RPS15A
protein levels in Tu686 cells (Fig.
5F and G). This indicated that HOXC10 influenced RPS15A
expression at the protein level but not at the mRNA level.
Ubiquitin-mediated proteolysis regulates protein stability without
influencing mRNA levels and plays an important role in tumor
progression (26). In the present
study, it was hypothesized that the interaction between HOXC10 and
RPS15A may regulate RPS15A proteolysis and thereby influence the
protein level of RPS15A. As shown in Fig. 5H, ubiquitination assays showed
that knockdown of HOXC10 obviously increased RPS15A ubiquitination
after MG132 treatment; overexpression of HOXC10 produced the
opposite effect. A previous study reported that RPS15A promoted
angiogenesis in HCC by enhancing Wnt/β-catenin signaling (27). In the present study, GSEA based on
the TCGA dataset demonstrated that HOXC10 expression was positively
associated with genes upregulated by activation of the
Wnt/β-catenin pathway (Fig. 5I).
HOXC10 knockdown suppressed the expression levels of nuclear
β-catenin and its downstream target genes, such as MMP7, Myc, and
Axin2, which were partially restored by RPS15A overexpression; by
contrast, HOXC10 overexpression or RPS15A knockdown yielded the
opposite effect (Fig. 5J-M),
which was then further confirmed by a TOP/FOP-Flash luciferase
reporter assay (Fig. 5N).
Additionally, it was observed that RPS15A overexpression restored
cell migration and invasion impaired by HOXC10 knockdown in FaDu
cells. RPS15A knockdown abrogated cell migration and invasion
induced by HOXC10 overexpression in Tu686 cells (Fig. 5O and P). Taken together, these
results indicated that HOXC10 interacts with RPS15A to facilitate
the Wnt/β-catenin pathway activation and contribute to the invasion
of HNSCC.
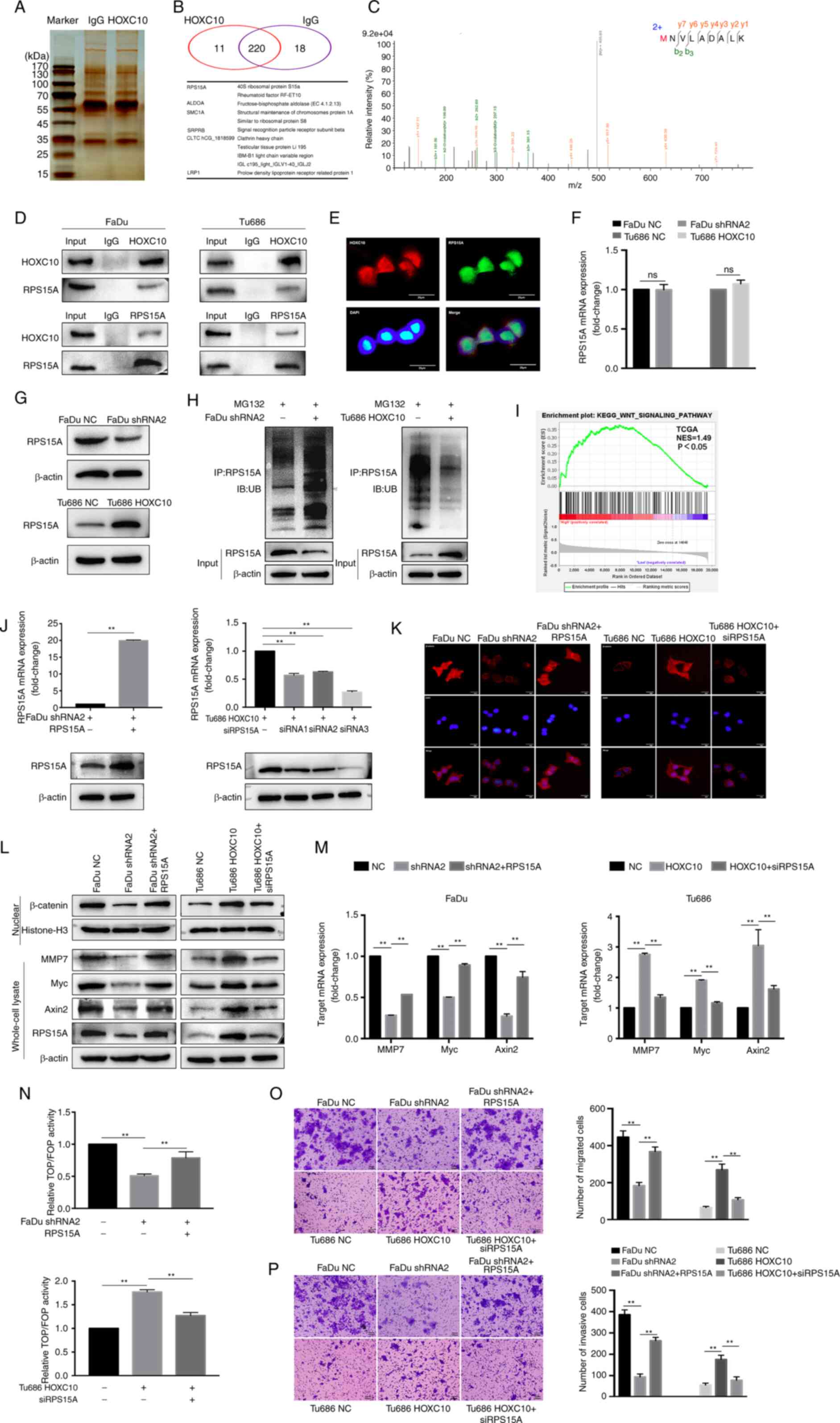 | Figure 5HOXC10 facilitates Wnt/β-catenin
signaling in HNSCC by interacting with RPS15A. (A-C) The binding
partners of HOXC10 were analyzed by the combination of Co-IP and
mass spectrometric analyses, and 11 proteins are presented. (D)
Co-IP and western blot analysis were used to verify the interaction
between HOXC10 and RPS15A. (E) Immunofluorescence was used to
identify the colocalization of HOXC10 and RPS15A. Scale bar, 25
µm. (F and G) RT-qPCR and western blot analysis of RPS15A
expression in HNSCC cells transfected with shRNA2 or HOXC10. (H)
Ubiquitination assay for the effects of HOXC10 on RPS15A
ubiquitination. (I) GSEA based on the TCGA dataset suggested that
HOXC10 expression was positively associated with the activation of
the Wnt/β-catenin pathway. (J) The efficiency of transfection in
the indicated cells was confirmed by RT-qPCR and western blotting.
(K) Immunofluorescence detection of β-catenin in the nucleus after
transfection with shRNA2, HOXC10, RPS15A, or siRPS15A in indicated
cells. Scale bar, 25 µm. (L) Western blot analysis was
performed to detect β-catenin, Myc, MMP7, Axin2, and RPS15A protein
in HNSCC cells transfected with shRNA2, HOXC10, RPS15A, or
siRPS15A. (M) Myc, MMP7, and Axin2 mRNA levels were analyzed by
RT-qPCR in transfected HNSCC cells. (N) The activity of the
Wnt/β-catenin pathway was analyzed by TOP/FOP-Flash luciferase
reporter assay. (O and P) Migration and invasion ability in
transfected HNSCC cells were assessed by Transwell assay. Scale
bar, 100 µm **P<0.01. HOXC10, homeobox C10;
HNSCC, head and neck squamous cell carcinoma; RPS15A, ribosomal
protein S15A; Co-IP, co-immunoprecipation; RT-qPCR, reverse
transcription quantitative-PCR; shRNA, short haipin RNA; siRPS15A,
small interfering RPS15A; GSEA, Gene Set Enrichment Analysis; TCGA,
The Cancer Genome Atlas Program. |
m6A modification is associated
with HOXC10 upregulation in HNSCC cells
Advances in tumor epigenetic regulation have shed
light on the role of m6A RNA methylation modification in
tumor development and progression (28); whether HOXC10 is modulated by
m6A RNA methylation in HNSCC cells remains unclear. We
found that the DNA methyltransferase inhibitor (5-Aza-CdR) failed
to upregulate HOXC10 expression (Fig.
6A). m6A modification has been reported to
preferentially locate in the consensus motif 'RRm6ACH'
(R=G or A; H=A, C or U) (29),
according to the results from online bioinformatics databases SRAMP
and RMBase v2.0. The sum of three potential m6A sequence
motifs were found in HOXC10 mRNA (Fig. 6B). MeRIP analysis showed that
m6A was highly enriched within these predicted
m6A sites (Fig. 6C).
As a crucial m6A methyltransferase, METTL3 has been
reported to play a significant role in catalyzing m6A
formation (30); RAP-western blot
assays indicated that HOXC10 mRNA interacts with METTL3 (Fig. 6D). In addition, it was revealed
that METTL3 expression was higher in HNSCC cells compared with
HuLa-PC cells, and METTL3 knockdown reduced m6A levels
of HOXC10 mRNA (Fig. 6E and F).
Notably, RNA stability assays demonstrated that METTL3 knockdown
significantly decreased the stability of HOXC10 mRNA under
actinomycin D treatment; knockdown of METTL3 resulted in a decrease
in HOXC10 expression (Fig. 6G and
H). These findings indicated that METTL3-mediated
m6A modification is associated with the upregulation of
HOXC10 in HNSCC, probably by increasing the stability of its
transcript.
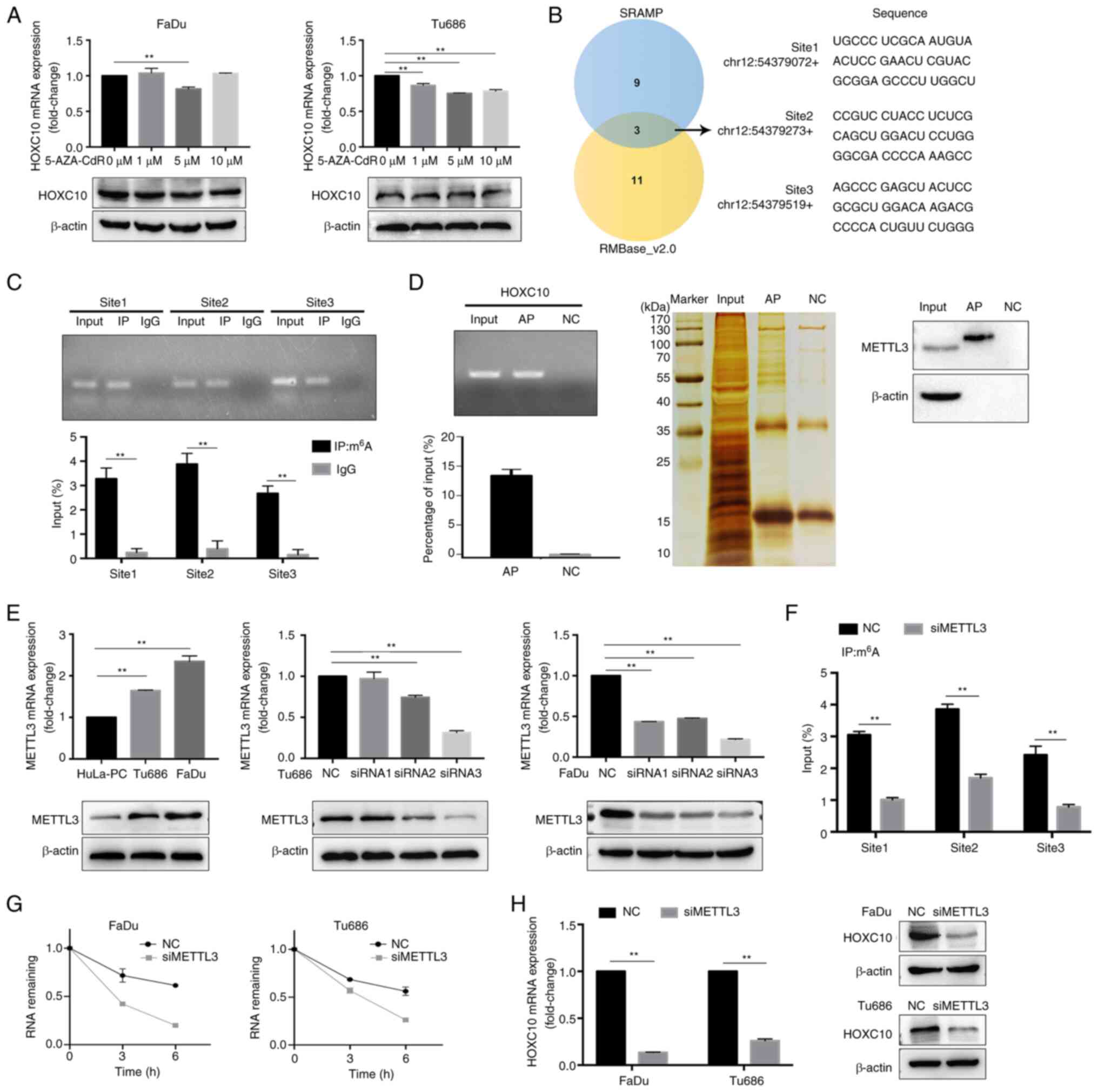 | Figure 6HOXC10 is modulated by m6A
RNA methylation. (A) HNSCC cells were treated with 5-AZA-CdR at
different concentrations for 48 h, and the HOXC10 mRNA level was
examined using RT-qPCR. (B) Predicted m6A sites in
HOXC10 mRNA from overlapping results of SRAMP and RMBase_ v2.0. (C)
m6A RIP-qPCR analysis showed that m6A was
highly enriched within the predicted m6A sites in FaDu
cells. (D) RAP-western blot assays indicated that HOXC10 mRNA
interacted with METTL3. (E) HOXC10 mRNA and protein levels were
analyzed by RT-qPCR and western blotting in the indicated HNSCC
cells. (F) MeRIP assay for m6A-modified HOXC10 mRNA in
FaDu cells transfected with si-NC or si-METTL3. (G) HNSCC cells
with METTL3 knockdown were treated with actinomycin D (5
µg/ml) for the indicated time points, and the HOXC10 mRNA
level was examined by RT-qPCR. (H) HOXC10 mRNA and protein levels
in HNSCC cells with METTL3 knockdown were detected by RT-qPCR and
western blotting. **P<0.01. HOXC10, homeobox C10;
m6A, N6-methyladenosine; HNSCC, head
and neck squamous cell carcinoma; RT-qPCR, reverse transcription
quantitative-PCR; RIP-qPCR, RNA immune precipitation-quantitative
PCR; RAP, RNA antisense purification; METTL3,
methyltransferase-like 3; MeRIP, methylated RNA immune
precipitation; si-, small interfering; NC, negative control. |
Previous research reported that IGF2BPs could
recognize m6A modications in mRNA transcripts and
enhance the stability and translation of these mRNAs (31); therefore, it was investigated
whether IGF2BPs participate in the recognition and stabilization of
m6A-modified HOXC10 mRNA, and thereby regulate HOXC10
expression. As shown in Fig. 7A,
the RAP-western blot assays showed that IGF2BP1 and IGF2BP3 bound
to HOXC10 mRNA. IGF2BP1 or IGF2BP3 expression was higher in HNSCC
cells compared to HuLA-PC cells (Fig.
7B-C). IGF2BP1 or IGF2BP3 knockdown decreased the stability of
HOXC10 mRNA at different degrees after actinomycin D treatment
(Figs. S2 and 7D). Moreover, knockdown of IGF2BP1 or
IGF2BP3 resulted in a significant decrease in HOXC10 expression at
both the mRNA and protein levels (Fig. 7E). These results indicated that
IGF2BP1 and IGF2BP3 recognize and stabilize m6A-tagged
HOXC10 mRNA. Finally, the results of Pearson's correlation
analysis, RT-qPCR and western blotting indicated that the
expression level of HOXC10 was positively correlated with the
levels of IGF2BP1, IGF2BP3, and METTL3 in HNSCC tissues, which was
further supported by immunohistochemistry analysis in the tissue
microarray (Fig. 7F-K).
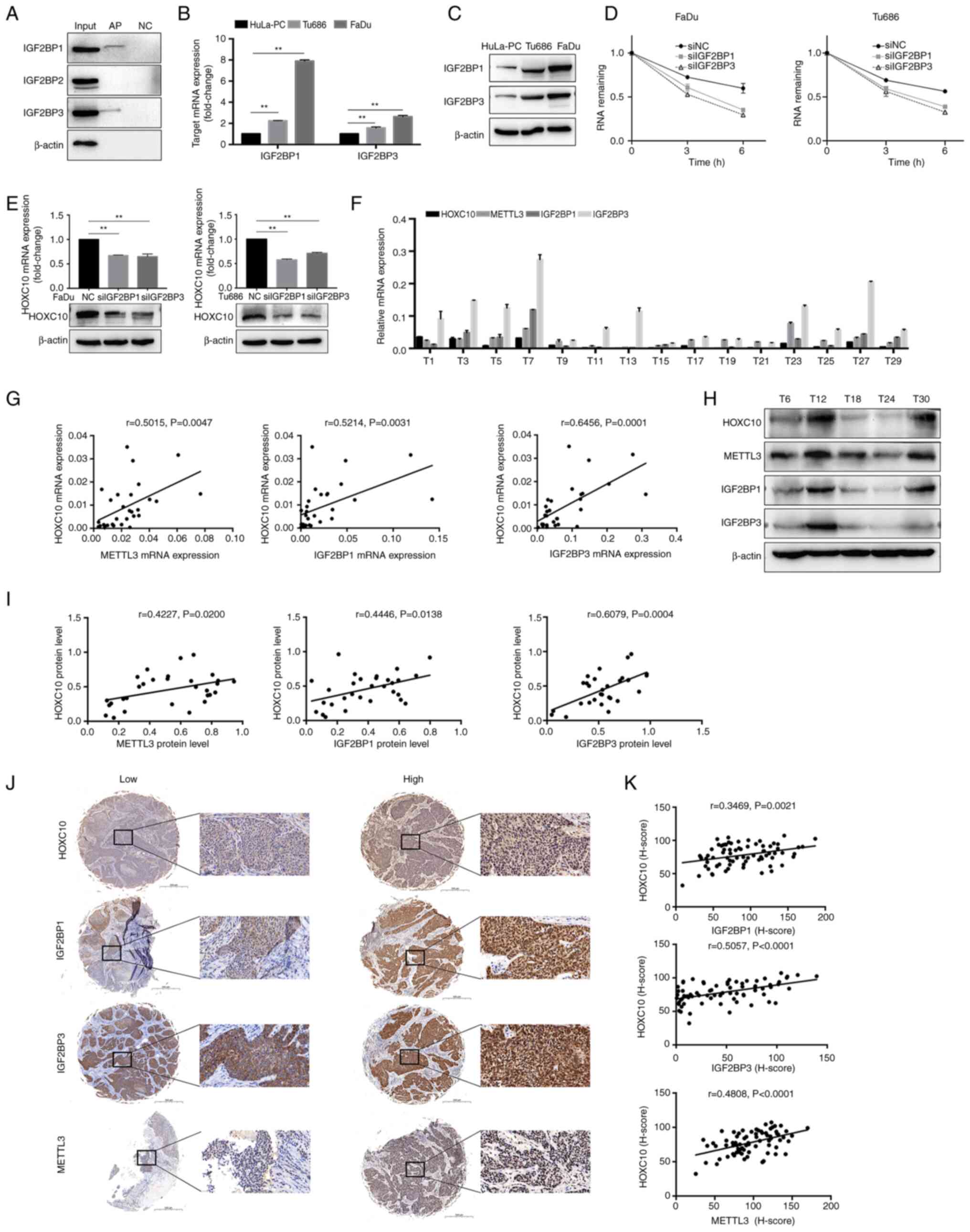 | Figure 7IGF2BP1 and IGF2BP3 participated in
the recognition and stabilization of m6A-modified HOXC10
mRNA. (A) The RAP-western blot assays indicated that HOXC10 mRNA
interacted with IGF2BP1 and IGF2BP3. (B and C) IGF2BP1 and IGF2BP3
mRNA and protein levels were analyzed by RT-qPCR and western
blotting in the indicated HNSCC cells. (D) HNSCC cells with IGF2BP1
or IGF2BP3 knockdown were treated with actinomycin D (5
µg/ml) for the indicated time points, and the HOXC10 mRNA
level was examined by RT-qPCR. (E) HOXC10 mRNA and protein levels
in HNSCC cells with IGF2BP1 or IGF2BP3 knockdown were detected by
RT-qPCR and western blotting. (F) The mRNA expression of HOXC10,
IGF2BP1, IGF2BP3, and METTL3 in 30 HNSCC tumor tissues was detected
by RT-qPCR. A representative image is shown. (G) Pearson's
correlation analysis between HOXC10 and IGF2BP1, IGF2BP3, or METTL3
was performed at the mRNA level. (H) The protein expression of
HOXC10, IGF2BP1, IGF2BP3, and METTL3 in 30 HNSCC tumor tissues was
detected by western blot analysis, and representative images are
shown. (I) Pearson's correlation analysis between HOXC10 and
IGF2BP1, IGF2BP3, or METTL3 was performed at the protein level. (J)
Representative immunostaining images of HOXC10, IGF2BP1, IGF2BP3,
and METTL3 in HNSCC tumor tissues. Scale bar, 500 µm. The
enlarged image scale bar, 50 µm. (K) Correlation analysis of
H-scores of HOXC10, IGF2BP1, IGF2BP3, and METTL3 using the
Pearson's correlation coefficient. **P<0.01. IGF2BP1,
insulin like growth factor 2 mRNA binding protein 1; IGF2BP3,
insulin like growth factor 2 mRNA binding protein 3;
m6A, N6-methyladenosine; HOXC10,
homeobox C10; RAP, RNA antisense purification; RT-qPCR, reverse
transcription quantitative-PCR; HNSCC, head and neck squamous cell
carcinoma; METTL3, methyltransferase-like 3; NC, negative
control. |
Discussion
The present study demonstrated that HOXC10 is
significantly elevated in HNSCC tissues, that a high level of
HOXC10 is associated with the malignant phenotype of HNSCC, and
that it is also an important prognostic factor for patients with
HNSCC. The experimental approach combining CHIP-seq with MS
analysis revealed the underlying mechanism by which HOXC10 promoted
ADAM17 expression by binding its promoter region, and ADAM17/EGFR
pathway activation facilitated the proliferation of HNSCC.
Furthermore, HOXC10 interacted with RPS15A and enhanced RPS15A
protein expression, which activated Wnt/β-catenin pathways and
contributed to invasion and metastasis of HNSCC. Additionally,
m6A writer METTL3 regulated the m6A
modification of the HOXC10 transcript, and m6A readers
IGF2BP1 and IGF2BP3 participated in the recognizing and stabilizing
of m6A-tagged HOXC10 mRNA.
HOXC10 is a member of the HOX genes, which are a
family of homeodomain transcription factors including HOXA, HOXB,
HOXC, and HOXD (2). Previous
studies have revealed that aberrant HOX gene expression plays a
crucial role in the progression of human cancer. For example, the
HOXA4/HOXB3 gene expression signature may be a biomarker of
recurrence of high-grade serous ovarian cancer after cytoreductive
surgery and adjuvant chemotherapy (32). The growth and metastasis of breast
cancer may be regulated by the HMGA2/TET1/HOXA9 signaling pathway
(33). HOXD9 promoted the
invasion and metastasis of gastric cancer cells by transcriptional
activation of RUFY3 (34). IL-1β
induced HOXC10 upregulation and then promoted HCC metastasis by
transactivating PDPK1 and VASP expression (35). In the present study, it was
demonstrated that HOXC10 is overexpressed in HNSCC, and that HOXC10
overexpression endowed HNSCC cells with enhancement of
proliferation, migration, and invasion abilities.
ADAM17, known as tumor necrosis factor-α
(TNFα)-converting enzyme (TACE), is responsible for protease-driven
shedding of membrane-tethered cytokines, cell surface receptors,
and growth factors; among these, EGFR family ligands are included
(36). The large number of
substrates processed by ADAM17 renders it a key coordinator of
numerous physiological and pathological processes, especially those
related to the occurrence and development of cancer. For example,
ADAM17 is overexpressed in diverse cancers, including colon
carcinoma, breast cancer, hepatocellular carcinoma, and gastric
cancer (37-40). ADAM17-mediated EGFR ligand
shedding facilitated cancer cell invasion promoted by macrophages
(22). Genetic,
antibody-mediated, or pharmacological methods targeting ADAM17
prevented formation of metastases in the lung, suppressed tumor
growth in various cancers, and have become a new strategy for
advanced-stage cancer therapy (41-43). In the present study, to further
investigate target genes regulated by HOXC10, ChIP-seq was
performed. The results revealed that a putative HOXC10 binding site
located at the promoter region of ADAM17, HOXC10, could enhance the
transcriptional activity of ADAM17. Activation of ADAM17 by
carcinogenic forms of Src has been reported to help promote
tumorigenesis by enhancing signaling via EGFR and ERK in an
autocrine and paracrine manner (44). Silencing ADAM17 may repress the
activity of the EGFR/ERK pathway to reduce the proliferation of
keloid fibroblasts (45).
Consistently, the experiments that were conducted in the present
study revealed that HOXC10 enhanced ADAM17 expression, increased
the ADAM17-activated EGFR-ERK pathway, and contributed to the
proliferation of HNSCC.
In order to explore the potential mechanism of
HOXC10 in the progression of HNSCC more thoroughly, MS analysis was
also applied, apart from ChIP-seq. Co-IP and MS confirmed a direct
interaction between HOXC10 and RPS15A proteins. Notably, HOXC10
influenced RPS15A expression at the protein level but not the mRNA
level. Previous studies have indicated that ubiquitin-mediated
proteolysis regulates protein stability without influencing mRNA
levels and plays an important role in tumor progression (46). In the present study,
overexpression of HOXC10 decreased RPS15A ubiquitination and
increased the protein level of RPS15A. HOXC10 itself does not
belong to deubiquitinating enzymes; the HOXC10/RPS15A complex may
influence the interaction between RPS15A and E3 ligase and thereby
affect ubiquitin-mediated proteolysis of RPS15A. In future
research, the mechanism through which HOXC10 regulates the
ubiquitylation level of RPS15A, will be further analyzed. A
previous study reported that RPS15A enhanced angiogenesis in HCC by
enhancing Wnt/β-catenin signaling (27). RPS15A knockdown downregulated
β-catenin expression and blocked the activation of Wnt signaling
(47). In the present study,
HOXC10 was demonstrated to influence RPS15A expression, HOXC10
knockdown supressed Wnt/β-catenin pathway activation, and RPS15A
overexpression restored cell migration and invasion impaired by
HOXC10 knockdown.
It is widely known that chemical modifications to
human RNAs are involved in numerous pathophysiological processes,
including cancer. Notably, N6-methyladenosine
(m6A) modification, one of the most abundant
post-translational mRNA internal modifications, participated in all
stages of the RNA life cycle, such as RNA production and stability
(48). m6A
modification is a dynamic and reversible biological process
regulated by 'writers' (METTL3, METTL14, and WTAP), 'erasers' (FTO
and ALKBH5), and 'readers' (YTH domain proteins and IGF2BPs)
(49). The deposition of
m6A is encoded by writers (methyltransferases), which
catalyze the formation of m6A. Erasers (demethylases)
selectively remove the m6A code. Readers (specific
RNA-binding proteins) decode m6A methylation and
modulate the m6A progression (49). 'Writers', 'erasers', and 'readers'
are frequently dysregulated in human cancers, which influence the
expression of oncogene transcripts and oncoproteins (49). However, the relationship between
m6A modification and the oncogenic role of HOXC10 in
HNSCC remains unclear. Recently, Wu et al (50) demonstrated that METTL3 alleviated
human mesenchymal stem cell senescence through m6A
modification-dependent stabilization of the MIS12 transcript.
Herein, the results of the present stydy showed that m6A
methylation was enriched within HOXC10 in HNSCC cells; moreover,
METTL3 regulated the m6A modification, and IGF2BP1 and
IGF2BP3 participated in recognizing and stabilizing the
m6A-modified HOXC10 transcript, thus affecting its mRNA
expression. These results indicated that the upregulation of HOXC10
in HNSCC may be attributed to m6A modification.
In the present study, the importance of HPV
infection status is acknowledged as a clinically relevant
parameter. However, a limitation due to the absence of routine
preoperative HPV testing must be noted in the clinical protocol of
the present study, which prevented the authors from including HPV
data in Table II. Future studies
with HPV infection status assessment could provide valuable
insights into its potential implications, and its incorporation in
the analyses of the present study will be considered when such data
becomes available.
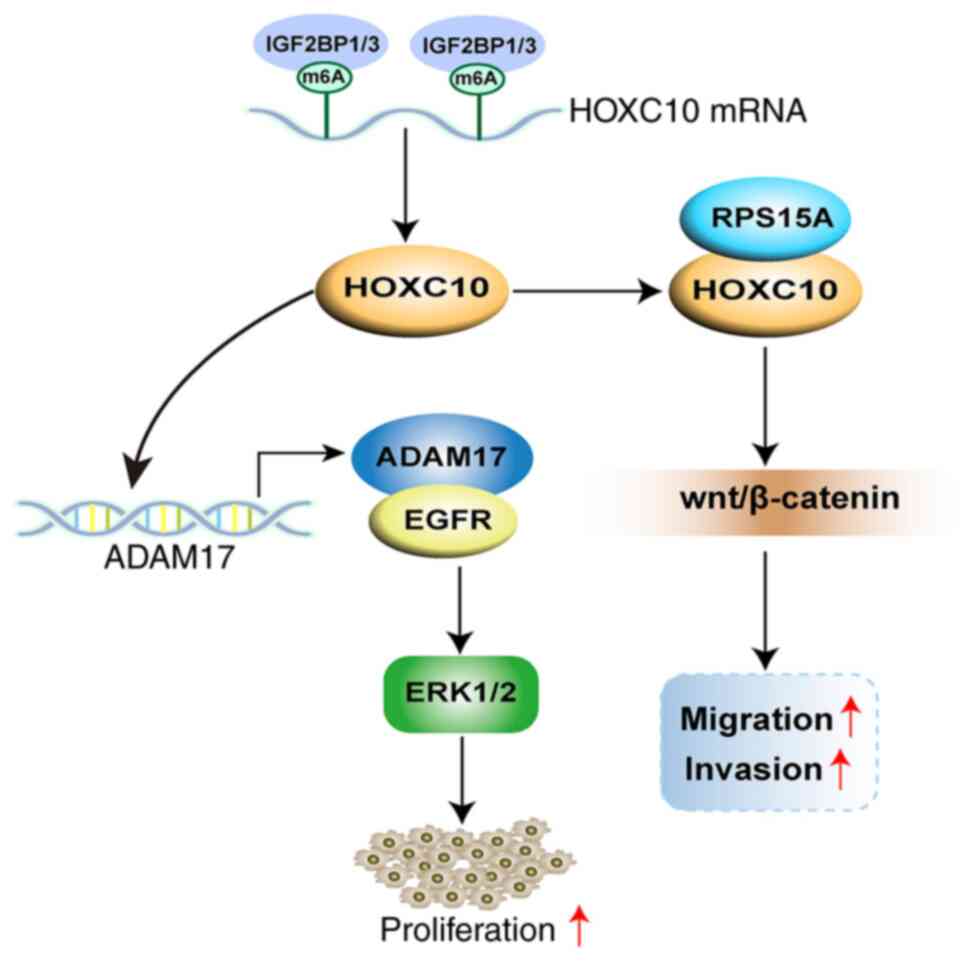
In conclusion, HOXC10 was demonstrated to be
overexpressed in HNSCC, and m6A modification-mediated
HOXC10 upregulation promoted proliferation and invasion of HNSCC
cells by co-activation of ADAM17/EGFR and Wnt/β-catenin signaling
(Fig. 8). Therefore, HOXC10 may
be a novel marker and a potential therapeutic strategy for
HNSCC.
Supplementary Data
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request. The original RNA-seq data has been deposited in the NCBI
Sequence Read Archive (SRA) with the associated accession number
PRJNA1033223 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1033223).
The original CHIP-seq data has been deposited in the NCBI SRA with
the associated accession number PRJNA1033205 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1033205).
Authors' contributions
LZ and CX conceived and designed the project. YZ, QH
and CW conducted the experiments. YZ, QH, CW collected the clinical
samples and data. YZ, QH and CW performed the statistical analysis.
LZ, CX and YX provided technical and material support. YX and YG
performed language editing. LZ, YZ, YG and XY reviewed and revised
the manuscript. YZ, YG, XY and YX assembled the figures. YG and XY
confirm the authenticity of all the raw data. All authors approved
the final version of the manuscript.
Ethics approval and consent to
participate
All animal procedures and patient experiments were
approved (approval no. 202212201) by the Eye and ENT Hospital,
Fudan University (Shanghai, China). The present study was approved
(approval no. 2018036) by the Ethics Committee of the Fudan
University Eye and ENT Hospital. All patients provided written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (grant no. 81972529), the
Science and Technology Commission of Shanghai Municipality (grant
no. 19411961300), and the Shanghai Sailing Program (grant no.
23YF1404700).
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Taketani T, Taki T, Shibuya N, Kikuchi A,
Hanada R and Hayashi Y: Novel NUP98-HOXC11 fusion gene resulted
from a chromosomal break within exon 1 of HOXC11 in acute myeloid
leukemia with t(11;12)(p15;q13). Cancer Res. 62:4571–4574.
2002.PubMed/NCBI
|
|
3
|
Malek R, Gajula RP, Williams RD, Nghiem B,
Simons BW, Nugent K, Wang H, Taparra K, Lemtiri-Chlieh G, Yoon AR,
et al: TWIST1-WDR5-Hottip regulates Hoxa9 chromatin to facilitate
prostate cancer metastasis. Cancer Res. 77:3181–3193. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stavnes HT, Holth A, Don T, Kærn J,
Vaksman O, Reich R, Trope' CG and Davidson B: HOXB8 expression in
ovarian serous carcinoma effusions is associated with shorter
survival. Gynecol Oncol. 129:358–363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miyazaki YJ, Hamada J, Tada M, Furuuchi K,
Takahashi Y, Kondo S, Katoh H and Moriuchi T: HOXD3 enhances
motility and invasiveness through the TGF-beta-dependent and
-independent pathways in A549 cells. Oncogene. 21:798–808. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
de Stanchina E, Gabellini D, Norio P,
Giacca M, Peverali FA, Riva S, Falaschi A and Biamonti G: Selection
of homeotic proteins for binding to a human DNA replication origin.
J Mol Biol. 299:667–680. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pathiraja TN, Nayak SR, Xi Y, Jiang S,
Garee JP, Edwards DP, Lee AV, Chen J, Shea MJ, Santen RJ, et al:
Epigenetic reprogramming of HOXC10 in endocrine-resistant breast
cancer. Sci Transl Med. 6:229ra2412014. View Article : Google Scholar
|
|
8
|
Tan Z, Chen K, Wu W, Zhou Y, Zhu J, Wu G,
Cao L, Zhang X, Guan H, Yang Y, et al: Overexpression of HOXC10
promotes angiogenesis in human glioma via interaction with PRMT5
and upregulation of VEGFA expression. Theranostics. 8:5143–5158.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
10
|
Wang SH, Zhu XL, Wang F, Chen SX, Chen ZT,
Qiu Q, Liu WH, Wu MX, Deng BQ, Xie Y, et al: LncRNA H19 governs
mitophagy and restores mitochondrial respiration in the heart
through Pink1/Parkin signaling during obesity. Cell Death Dis.
12:5572021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang H, Zhu Q, Ji Y, Wang M, Zhang Q, Liu
W, Li R, Zhang J, Xu P, Song X and Lv C: hucMSCs treatment prevents
pulmonary fibrosis by reducing circANKRD42-YAP1-mediated mechanical
stiffness. Aging (Albany NY). 15:5514–5534. 2023.PubMed/NCBI
|
|
12
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the care and use of laboratory animals. 8th
edition. National Academies Press; Washington, DC: 2011
|
|
13
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Anders S, Pyl PT and Huber W: HTSeq-a
python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar
|
|
15
|
Love M, Anders S and Huber W: Differential
analysis of RNA-Seq data at the gene level using the DESeq2
package. Heidelberg: European Molecular Biology Laboratory (EMBL);
2013, https://bioconductor.org/help/course-materials/2013/EMBOBGI/DESeq2_parathyroid.pdf.
|
|
16
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Liu T, Meyer CA, Eeckhoute J,
Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W and
Liu XS: Model-based analysis of ChIP-Seq (MACS). Genome Biol.
9:R1372008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu G, Wang LG and He QY: ChIPseeker: An
R/Bioconductor package for ChIP peak annotation, comparison, and
visualization. Bioinformatics. 31:2382–2383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bailey TL, Boden M, Buske FA, Frith M,
Grant CE, Clementi L, Ren J, Li WW and Noble WS: MEME SUITE: Tools
for motif discovery and searching. Nucleic Acids Res. 37:W202–W208.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang Y, Hong W and Wei X: The molecular
mechanisms and therapeutic strategies of EMT in tumor progression
and metastasis. J Hematol Oncol. 15:1292022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gnosa SP, Puig Blasco L, Piotrowski KB,
Freiberg ML, Savickas S, Madsen DH, Auf dem Keller U, Kronqvist P
and Kveiborg M: ADAM17-mediated EGFR ligand shedding directs
macrophage-promoted cancer cell invasion. JCI Insight.
7:e1552962022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Beck Gooz M, Maldonado EN, Dang Y, Amria
MY, Higashiyama S, Abboud HE, Lemasters JJ and Bell PD: ADAM17
promotes proliferation of collecting duct kidney epithelial cells
through ERK activation and increased glycolysis in polycystic
kidney disease. Am J Physiol Renal Physiol. 307:F551–F559. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu T, Zhang J, Chen H, Bianba T, Pan Y,
Wang X, Jiang Y and Yang Z: PSMC2 promotes the progression of
gastric cancer via induction of RPS15A/mTOR pathway. Oncogenesis.
11:122022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng Z, Cui H, Wang Y and Yao W:
Downregulation of RPS15A by miR-29a-3p attenuates cell
proliferation in colorectal carcinoma. Biosci Biotechnol Biochem.
83:2057–2064. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wei CY, Zhu MX, Yang YW, Zhang PF, Yang X,
Peng R, Gao C, Lu JC, Wang L, Deng XY, et al: Downregulation of
RNF128 activates Wnt/β-catenin signaling to induce cellular EMT and
stemness via CD44 and CTTN ubiquitination in melanoma. J Hematol
Oncol. 12:212019. View Article : Google Scholar
|
|
27
|
Guo P, Wang Y, Dai C, Tao C, Wu F, Xie X,
Yu H, Zhu Q, Li J, Ye L, et al: Ribosomal protein S15a promotes
tumor angiogenesis via enhancing Wnt/β-catenin-induced FGF18
expression in hepatocellular carcinoma. Oncogene. 37:1220–1236.
2018. View Article : Google Scholar
|
|
28
|
Wu Y, Yang X, Chen Z, Tian L, Jiang G,
Chen F, Li J, An P, Lu L, Luo N, et al: m6A-induced
lncRNA RP11 triggers the dissemination of colorectal cancer cells
via upregulation of Zeb1. Mol Cancer. 18:872019. View Article : Google Scholar
|
|
29
|
Csepany T, Lin A, Baldick CJ Jr and Beemon
K: Sequence specificity of mRNA N6-adenosine methyltransferase. J
Biol Chem. 265:20117–20122. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol
Oncol. 13:1172020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Miller KR, Patel JN, Zhang Q, Norris EJ,
Symanowski J, Michener C, Sehouli J, Braicu I, Destephanis DD,
Sutker AP, et al: HOXA4/HOXB3 gene expression signature as a
biomarker of recurrence in patients with high-grade serous ovarian
cancer following primary cytoreductive surgery and first-line
adjuvant chemotherapy. Gynecol Oncol. 149:155–162. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun M, Song CX, Huang H, Frankenberger CA,
Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C and Rosner
MR: HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer
growth and metastasis. Proc Natl Acad Sci USA. 110:9920–9925. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu H, Dai W, Li J, Xiang L, Wu X, Tang W,
Chen Y, Yang Q, Liu M, Xiao Y, et al: HOXD9 promotes the growth,
invasion and metastasis of gastric cancer cells by transcriptional
activation of RUFY3. J Exp Clin Cancer Res. 38:4122019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dang Y, Chen J, Feng W, Qiao C, Han W, Nie
Y, Wu K, Fan D and Xia L: Interleukin 1β-mediated HOXC10
overexpression promotes hepatocellular carcinoma metastasis by
upregulating PDPK1 and VASP. Theranostics. 10:3833–3848. 2020.
View Article : Google Scholar :
|
|
36
|
Grötzinger J, Lorenzen I and Düsterhöft S:
Molecular insights into the multilayered regulation of ADAM17: The
role of the extracellular region. Biochim Biophys Acta Mol Cell
Res. 1864:2088–2095. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Blanchot-Jossic F, Jarry A, Masson D,
Bach-Ngohou K, Paineau J, Denis MG, Laboisse CL and Mosnier JF:
Up-regulated expression of ADAM17 in human colon carcinoma:
Co-expression with EGFR in neoplastic and endothelial cells. J
Pathol. 207:156–163. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao MQ, Kim BG, Kang S, Choi YP, Yoon JH
and Cho NH: Human breast cancer-associated fibroblasts enhance
cancer cell proliferation through increased TGF-α cleavage by
ADAM17. Cancer Lett. 336:240–246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW,
Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, et al: MicroRNA-122, a
tumor suppressor microRNA that regulates intrahepatic metastasis of
hepatocellular carcinoma. Hepatology. 49:1571–1582. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shou ZX, Jin X and Zhao ZS: Upregulated
expression of ADAM17 is a prognostic marker for patients with
gastric cancer. Ann Surg. 256:1014–1022. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bolik J, Krause F, Stevanovic M, Gandraß
M, Thomsen I, Schacht SS, Rieser E, Müller M, Schumacher N, Fritsch
J, et al: Inhibition of ADAM17 impairs endothelial cell necroptosis
and blocks metastasis. J Exp Med. 219:e202010392022. View Article : Google Scholar
|
|
42
|
Ye J, Yuen SM, Murphy G, Xie R and Kwok
HF: Anti-tumor effects of a 'human & mouse cross-reactive'
anti-ADAM17 antibody in a pancreatic cancer model in vivo. Eur J
Pharm Sci. 110:62–69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang L, Chen J, Quan J and Xiang D:
Rosmarinic acid inhibits proliferation and migration, promotes
apoptosis and enhances cisplatin sensitivity of melanoma cells
through inhibiting ADAM17/EGFR/AKT/GSK3β axis. Bioengineered.
12:3065–3076. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Maretzky T, Zhou W, Huang XY and Blobel
CP: A transforming Src mutant increases the bioavailability of EGFR
ligands via stimulation of the cell-surface metalloproteinase
ADAM17. Oncogene. 30:611–618. 2011. View Article : Google Scholar
|
|
45
|
Le X and Fan YF: ADAM17 regulates the
proliferation and extracellular matrix of keloid fibroblasts by
mediating the EGFR/ERK signaling pathway. J Plast Surg Hand Surg.
57:129–136. 2023. View Article : Google Scholar
|
|
46
|
Peng R, Zhang PF, Yang X, Wei CY, Huang
XY, Cai JB, Lu JC, Gao C, Sun HX, Gao Q, et al: Overexpression of
RNF38 facilitates TGF-β signaling by ubiquitinating and degrading
AHNAK in hepatocellular carcinoma. J Exp Clin Cancer Res.
38:1132019. View Article : Google Scholar
|
|
47
|
Liang J, Liu Y, Zhang L, Tan J, Li E and
Li F: Overexpression of microRNA-519d-3p suppressed the growth of
pancreatic cancer cells by inhibiting ribosomal protein
S15A-mediated Wnt/β-catenin signaling. Chem Biol Interact. 304:1–9.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lan Q, Liu PY, Haase J, Bell JL,
Hüttelmaier S and Liu T: The critical role of RNA m6A
methylation in cancer. Cancer Res. 79:1285–1292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wu Z, Shi Y, Lu M, Song M, Yu Z, Wang J,
Wang S, Ren J, Yang YG, Liu GH, et al: METTL3 counteracts premature
aging via m6A-dependent stabilization of MIS12 mRNA.
Nucleic Acids Res. 48:11083–11096. 2020. View Article : Google Scholar : PubMed/NCBI
|