Introduction
Breast cancer is leading in terms of cancer
incidence and mortality in women worldwide (1). The mainstay of treatment is surgical
resection combined with chemo- and/or radiotherapy, plus adjunct
hormone therapy depending on the expression of hormonal receptors
(2,3). In spite of these established
strategies, a proportion of the patients progress to recurrence,
metastasis or resistance to cancer therapies, suggesting that
additional therapeutic strategies may be required to tailor for
individuals (4-6).
RAD51 recombinase has been shown to have a crucial
role in the repair of DNA damage, which may occur endogenously from
DNA replication stress in fast-growing cancer cells or exogenously
due to environmental challenges, including platinum-based
chemotherapy and radiotherapy that primarily cause DNA
double-strand breaks (7,8). Among pancreatic cell lines, RAD51
expression is lower in irradiation-sensitive CAPAN-1 cells,
compared to irradiation-resistant Panc-1 cells (9). In addition, RAD51 overexpression has
been reported in cell lines derived from different cancer types and
is associated with cellular resistance to irradiation and
chemotherapeutic drugs (10,11). As the function of RAD51 in DNA
damage repair is well-documented, the majority of research on RAD51
in breast cancer has focused on its expression level in the cell
nucleus, e.g. the expression of nuclear RAD51 in breast tumors has
been shown to be positively associated with tumor size and grade
(12), as well as lymph node
metastasis (13,14). However, discrepancies exist in
certain studies, with others finding the expression of nuclear
RAD51 in breast tumors inversely associated with grade and local
recurrence (15). These findings
point towards the possibility of a more elaborate function of RAD51
other than its canonical role in DNA damage repair within the cell
nucleus. For instance, increased cytoplasmic RAD51 expression has
been found to be a strong risk factor for developing brain
metastasis in patients with breast cancer compared to nuclear RAD51
expression (16), and breast
tumors expressing high cytoplasmic but low nuclear RAD51 has been
associated with adverse breast cancer progression (17).
In the present study, the impact of subcellular
RAD51 expression on breast cancer was investigated by analyzing the
association of differential RAD51 expression in various subcellular
localizations with clinicopathologic and prognostic outcomes in
patients with breast cancer. In addition, the roles of differential
RAD51 expression in breast cancer cell malignant behaviors were
explored in vitro.
Patients and methods
Patient specimens
All of the samples used in the present study were
de-identified samples from the hospital's biobank added with the
informed consent of the patients, but the requirement of informed
consent to include them in the present study was waived by the
ethics committee. Primary breast tumor specimens had been collected
from female patients diagnosed with invasive breast ductal
carcinoma and treated with surgical resection at Kaohsiung Medical
University Hospital (Kaohsiung, Taiwan) between January 2010 and
January 2017. The inclusion criteria were that the patients had no
history of cancer and were not simultaneously diagnosed with any
other type of cancer, and the exclusion criteria were a diagnosis
with benign breast conditions, ductal carcinoma in situ,
microinvasive carcinoma and rare histological tumor types. The
follow-up of the patients after treatment was up to 70 months
(median, 41 months). Clinical data used in this study were obtained
from the hospital's cancer registry with protocols approved by the
Institutional Review Board of Kaohsiung Medical University Hospital
[approval nos. KMUH-IRB-20130031 and KMUHIRB-E(I)-20180136] and
conducted in accordance with the Declaration of Helsinki.
Immunohistochemistry (IHC)
Paraffin-embedded and formalin-fixed tumor sections
prepared from the samples taken from surgically treated patients
with breast cancer were immunostained with anti-RAD51 antibodies by
the Bond-Max automated IHC stainer (Leica Microsystems GmbH)
according to the manufacturer's instructions and a previous study
by our group (18). The mouse
monoclonal antibody against human RAD51 (clone 14B4) for IHC
staining was obtained from GeneTex. The images of IHC staining were
captured with an Eclipse E600 microscope (Nikon Corp.), followed by
quantitation of the IHC staining with categorical scores according
to the percentage of positively stained cells (score 1, ≤25%; score
2, 26-50%; score 3, 51-75%; score 4, ≥76%), where samples with
scores 1 and 2 were further categorized as low RAD51 expression,
and those with scores 3 and 4 were categorized as high RAD51
expression for dichotomized comparisons (18,19). The IHC scoring was evaluated
independently by two trained investigators and confirmed by a
pathologist.
Cell culture
The human breast cancer cell lines MDA-MB-231 and
MCF-7 were purchased from the Bioresource Collection and Research
Center (Hsinchu, Taiwan) and maintained in DMEM (Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum
(Biological Industries) and 1% penicillin-streptomycin-amphotericin
B solution (Thermo Fisher Scientific, Inc.). The genotypes of these
human cell lines were authenticated by short tandem repeat
profiling (TopGen Biotechnology).
Gene overexpression and knockdown
Overexpression of RAD51 was conducted by
transfecting the breast cancer cell lines with prepackaged
lentiviral particles carrying pReceiver-Lv105 vector that expresses
full-length human RAD51 (accession no. NM_002875.4) or empty
pReceiver-Lv105 vector (GeneCopoeia) as a control. Knockdown of
RAD51 was conducted by transfecting the breast cancer cell lines
with prepackaged lentiviral particles carrying pLKO.1-puro vector
that expresses short hairpin RNA (shRNA) targeting human RAD51
(5′-CGC CCT TTA CAG AAC AGA CTA-3′) or targeting firefly luciferase
(5′-GCG GTT GCC AAG AGG TTC CAT-3′; National RNAi Core Facility,
Academia Sinica) as a control. The prepackaged lentiviral particles
were added to the corresponding cells in their cell culture medium
containing 8 µg/ml polybrene (Sigma-Aldrich; Merck KGaA).
After 48 h of transfection, 2 µg/ml puromycin
(Sigma-Aldrich; Merck KGaA) was added for selection. Surviving
cells were continuously maintained in the cell culture medium
containing 2 µg/ml puromycin until further experiments.
XTT cell viability assay
MDA-MB-231 or MCF-7 cells were seeded in 96-well
plates (5×103 cells/well) overnight and cultured for 72
h prior to the addition of XTT solution (Sigma-Aldrich; Merck KGaA)
according to the manufacturer's instructions. Cell viability was
assessed by measuring the optical density (OD) at the main
wavelength at 475 nm subtracted by the reference wavelength at 660
nm. To determine the half-maximal inhibitory concentration
(IC50), the cells were treated with a series of
different concentrations of cisplatin (min. to max., 0-160
µM) for 72 h prior to the addition of the XTT solution
followed by readout of the OD as described above. The
IC50 was calculated using GraphPad Prism 8
(Dotmatics).
Cell cycle distribution
MDA-MB-231 or MCF-7 cells were seeded in 6-well
plates (2×105 cells/well) under normal cell culture
conditions. At 80% confluence, the cells were collected and fixed
with 75% ethanol (Sigma-Aldrich; Merck KGaA) at -20°C overnight,
followed by staining the DNA contents with propidium iodide/RNase
Staining Buffer (BD Biosciences) for 30 min at 37°C. These cells
were then measured with a Cytomics FC 500 flow cytometer (Beckman
Coulter), and the distribution of the cell cycle phases was
analyzed by WinMDI 2.8 (http://www.cyto.purdue.edu/flowcyt/software.htm).
Transwell migration assay
MDA-MB-231 or MCF-7 cells were plated in Transwell
inserts with 8 µm pores (2×104 cells/insert in
500 µl serum-free cell culture medium per well) of the
24-well plate setting (Corning, Inc.), with the bottom wells
containing complete cell culture medium. After 24 h under normal
cell culture conditions, the cells remaining on the upper side of
the insert were removed with cotton swabs, while those that had
migrated to the underside of the insert were fixed with 4%
formaldehyde (Sigma-Aldrich; Merck KGaA) for 30 min at room
temperature, followed by staining with 0.05% crystal violet
(Sigma-Aldrich: Merck KGaA) for 15 min at room temperature. The
images of crystal violet staining were captured with an Eclipse Ti
Series microscope (Nikon Corp.) and analyzed by ImageJ version
1.53e [National Institutes of Health (NIH)].
Invadopodial invasion assay
MDA-MB-231 cells were plated onto 8-well Nunc
Lab-Tek glass chamber slides (5×103 cells/well; Thermo
Fisher Scientific, Inc.) that were pre-coated with Cy3-conjugated
gelatin (Merck Millipore) according to the procedures described in
the QCM Gelatin Invadopodia Assay (20). After 24 h under normal cell
culture conditions, the cells were fixed with 4% formaldehyde
(Sigma-Aldrich; Merck KGaA) for 30 min at room temperature,
followed by concurrent staining with 2 µg/ml FITC-conjugated
phalloidin and 1 µg/ml DAPI for 20 min at room temperature
to label filamentous actin (F-actin) and the cell nucleus,
respectively. The images of each well were captured by a Zeiss
Axioscope fluorescence microscope (Carl Zeiss GmbH) and gelatin
degradation was quantified as the degradation area of gelatin
normalized to the number of cells using ImageJ (NIH).
Western blot analysis
Protein was extracted using RIPA buffer and the
concentration of the protein was measured with a BCA protein assay
kit (Pierce™ BCA Protein Assay Kit; ThermoFisher). Equal amounts of
total protein extracted from MDA-MB-231 or MCF-7 cells (30
µg/sample) were separated by one-dimensional 10% SDS-PAGE
and subsequently transferred to PVDF membranes (PALL Life Sciences)
using the Mini-PROTEAN and Trans-Blot systems (Bio-Rad
Laboratories, Inc.). The membranes were blocked with 5% non-fat
milk (Anchor) for 1 h at room temperature, followed by sequential
incubation with primary antibodies overnight at 4°C and
species-matched horseradish peroxidase-conjugated secondary
antibodies (Thermo Fisher Scientific, Inc.) for 1 h at room
temperature. The signal of immunoreactive proteins was developed in
the presence of chemiluminescent reagents (Thermo Fisher
Scientific, Inc.) and acquired by the ChemiDoc XRS+ imaging system
(Bio-Rad Laboratories, Inc.). The primary antibodies used for
western blot were as follows: Mouse monoclonal antibodies against
human RAD51 (cat. no. 100469; 1:5,000 dilution; GeneTex), F-actin
(cat. no. 205; 1:500 dilution; Abcam) or Lamin A/C (cat. no. 4777;
1:2,000 dilution; Cell Signaling); and rabbit polyclonal antibodies
against human RAD51, GAPDH (cat. no. 100118; 1:60,000 dilution;
GeneTex) or α-tubulin (cat. no. 112141; 1:5,000 dilution;
GeneTex).
Subcellular fractionation and
immunoprecipitation
The cytoplasmic and nuclear protein fractions of
MDA-MB-231 cells were extracted using the Subcellular Protein
Fractionation Kit (Thermo Fisher Scientific, Inc.). Equal amounts
of each subcellular fraction (500 µg/sample) were added to
100 ml Protein A Mag Sepharose (Sigma-Aldrich; Merck KGaA)
conjugated with 5 µg of the RAD51 antibodies or F-actin
antibodies (as specified above) overnight at 4°C on a rotary tube
mixer (Thermo Fisher Scientific, Inc.). The immunoprecipitated
protein complex was then separated from the sepharose by boiling
for 10 min in Laemmli sample buffer (Bio-Rad Laboratories, Inc.)
and subjected to one-dimensional 10% SDS-PAGE for western blot
analysis according to the above-mentioned western blot protocol or
in-gel digestion of the proteins.
In-gel digestion and liquid
chromatography-tandem mass spectrometry (LC-MS/MS)
In-gel digestion and protein identification by
LC-MS/MS were conducted following previously reported procedures
(21). The protein band
identified from one-dimensional 10% SDS-PAGE by Coomassie Brilliant
Blue R-250 staining (Thermo Fisher Scientific, Inc.) was sliced and
de-stained with 25 mM ammonium bicarbonate (Sigma-Aldrich; Merck
KGaA) in 50% acetonitrile (Sigma-Aldrich; Merck KGaA) for 1 h at
room temperature. The sliced gel was then dehydrated in 100%
acetonitrile (Sigma-Aldrich; Merck KGaA) for 5 min and vacuum-dried
for 30 min at room temperature. In-gel digestion was conducted in
the presence of 0.5 µg trypsin (Sigma-Aldrich; Merck KGaA)
dissolved in 25 mM ammonium bicarbonate (Sigma-Aldrich; Merck KGaA)
for 16 h at 37°C. The digested peptide fragments were extracted
with 50 µl of a mixture containing 50% acetonitrile
(Sigma-Aldrich; Merck KGaA) and 0.1% trifluoroacetic acid
(Sigma-Aldrich; Merck KGaA). The extracted peptides were then
dissolved in a mixture of 0.1% formic acid (Sigma-Aldrich; Merck
KGaA) and 50% acetonitrile (Sigma-Aldrich; Merck KGaA), and
analyzed by LC-MS/MS at the Center for Research Resources and
Development, Kaohsiung Medical University (Kaohsiung, Taiwan).
Protein matching was conducted by a database search for the results
of LC-MS/MS spectra with the aid of Mascot (version 2.2; Matrix
Science) (https://www.matrixscience.com) (22).
Immunofluorescence
MDA-MB-231 cells grown on 8-well Nunc Lab-Tek II
chamber slides (Thermo Fisher Scientific, Inc.) were fixed with 4%
paraformaldehyde (Sigma-Aldrich; Merck KGaA) for 30 min at room
temperature, followed by incubation of the cells with a
permeabilization/blocking buffer containing 0.5% Triton-X 100
(Sigma-Aldrich; Merck KGaA) and 1% bovine serum albumin
(Sigma-Aldrich; Merck KGaA) in PBS for 1 h at room temperature. The
cells were then sequentially incubated with rabbit polyclonal
antibodies against human RAD51 (cat. no. 100469; 1:200 dilution;
GeneTex) overnight at 4°C and Alexa Fluor 555-conjugated donkey
polyclonal antibodies against rabbit IgG (cat. no. A32794; 1:500
dilution; Thermo Fisher Scientific, Inc.) for 1 h at room
temperature, followed by co-staining with 2 µg/ml
FITC-conjugated phalloidin (for F-actin) (cat. no. A12379; Thermo
Fisher Scientific, Inc.) and 1 µg/ml DAPI (AAT Bioquest)
(for the cell nuclei) for 20 min at room temperature. After the
cells were mounted onto coverslips with Mounting Media (Thermo
Fisher Scientific, Inc.), images of the cells were captured by
Zeiss Axioscope fluorescence microscope (Carl Zeiss GmbH) and
analyzed by ImageJ (NIH).
Statistical analysis
The statistics in this study were analyzed by SPSS
(version 25; IBM Corp.) or GraphPad Prism 8 (Dotmatics).
Kaplan-Meier survival analysis was conducted to determine
disease-free and overall survival of the patients, and differences
between survival curves were assessed with the log-rank test. The
association between RAD51 expression and clinicopathological
parameters was assessed by chi-square test. Hazard ratio (HR) and
95% confidence interval (CI) were derived from Cox regression
models. For the in vitro studies, the data were presented as
the mean ± standard deviation from three independent experiments
unless otherwise indicated, and the difference between experimental
and control groups was assessed by Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Association of cytoplasmic and nuclear
RAD51 expression with clinicopathologic parameters and patient
survival in breast cancer
To investigate the association of cytoplasmic (cyto)
and nuclear (nu) RAD51 expression with cancer progression, primary
tumors obtained from patients with surgically treated breast cancer
(age range, 28-86 years; mean ± SD, 51.54±11.42 years) were
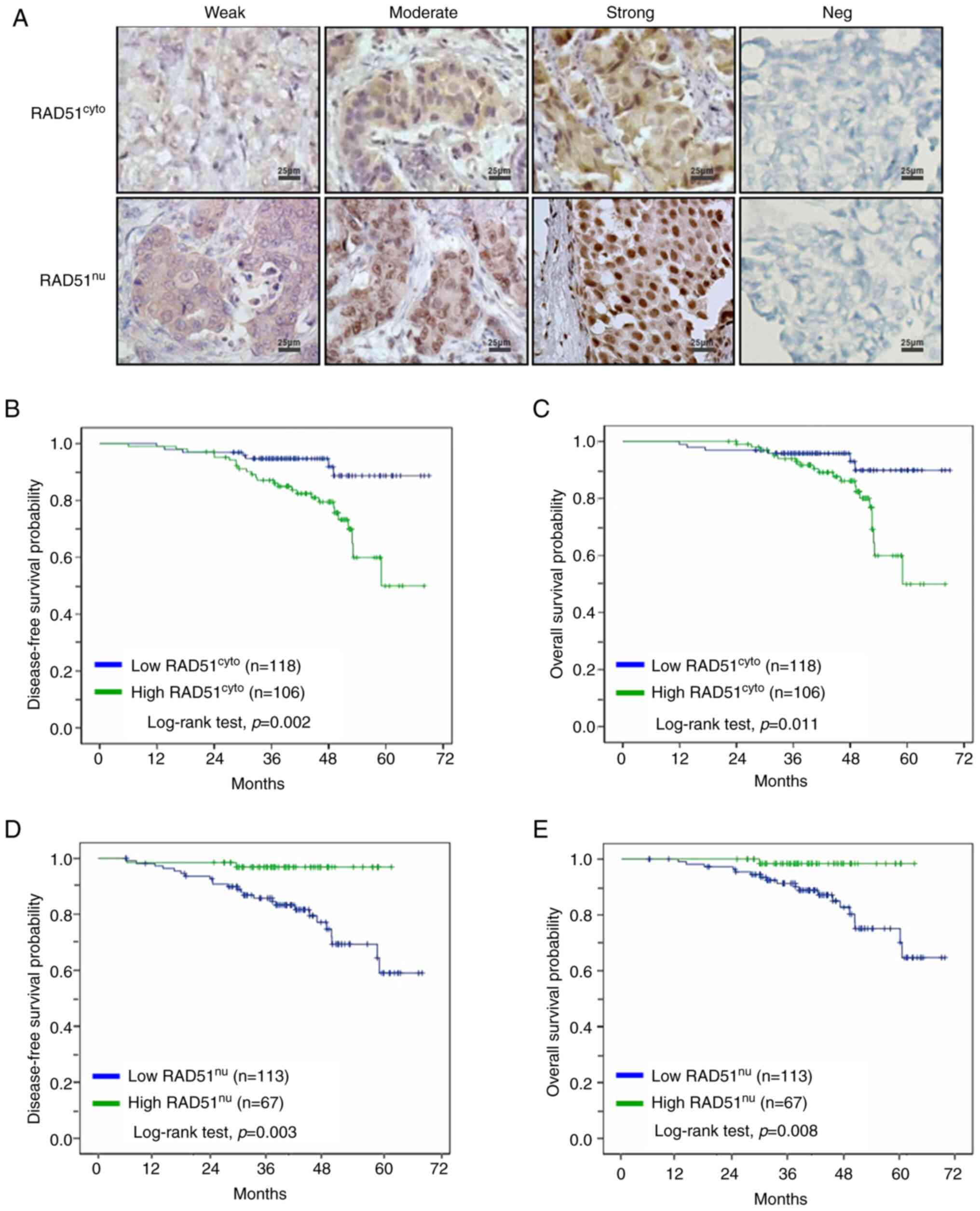
assessed by IHC (Fig. 1A), and
categorical scores were assigned according to the expression levels
of RAD51. In general, a heterogeneous expression pattern of RAD51
was observed across the tumor tissues (Fig. 1A). The initial screening was
conducted via classification of RAD51 expression into four groups
as reported previously (17),
including cytolow/nuhigh,
cytohigh/nuhigh,
cytolow/nulow and
cytohigh/nulow of RAD51 (n=148). Using this
analysis, consensus results of the association between subcellular
RAD51 expression and breast cancer progression were observed, such
as tumor size (P=0.022; Table
SI) and disease-free survival of patients with breast cancer
(P=0.007; Fig. S1A). In addition
to these, the current results revealed that cancer stage and lymph
node (LN) metastasis (P=0.001 for both; Table SI) and overall survival of the
patients (P=0.025; Fig. S1B)
were associated with subcellular RAD51 expression. Overall,
significant differences in patient survival, along with other
clinicopathological parameters, including tumor size, stage and LN
metastasis, were observed among the four patient groups according
to their status of subcellular RAD51 expression.
To further compare the roles of differential RAD51
expression in the cytoplasm and in the cell nuclei of breast
tumors, independent analyses were performed for patients who were
dichotomously grouped into low vs. high expression of cytoplasmic
RAD51 (n=224) and nuclear RAD51 (n=180). It was found that the
patient group with high cytoplasmic RAD51 expression was associated
with increased cancer stage (P=0.003), grade (P=0.035), tumor size
(P=0.036) and LN metastasis (P=0.021; Table I). However, the association was
the inverse in the patient group with high nuclear RAD51
expression, as it was observed that the patient group with high
nuclear RAD51 expression was associated with decreased cancer stage
(P<0.001), tumor size (P=0.005) and LN metastasis (P=0.001;
Table II). In the survival
analysis, the patient group with high cytoplasmic RAD51 expression
was associated with poorer disease-free survival (P=0.002; Fig. 1B) and overall survival (P=0.011;
Fig. 1C) during a postoperative
follow-up period up to 72 months, whereas the patient group with
high nuclear RAD51 expression conversely showed positive
association with disease-free survival (P=0.003; Fig. 1D) and overall survival (P=0.008;
Fig. 1E) compared to their
corresponding low RAD51 expression groups. These results suggest
the utility of subcellular RAD51 in breast tumor tissues as
prognostic markers, with the notion that high cytoplasmic RAD51
expression was adversely associated with breast cancer progression,
such as increased cancer stage, tumor size and LN metastasis, along
with reduced patient survival; however, high nuclear RAD51
expression had the inverse effect.
 | Table IAssociation of cytoplasmic RAD51
expression with clinicopathologic characteristics in patients with
breast cancer (n=224). |
Table I
Association of cytoplasmic RAD51
expression with clinicopathologic characteristics in patients with
breast cancer (n=224).
| Variable | Total | Cytoplasmic RAD51
expression
| P-value |
|---|
| Low (n=118) | High (n=106) |
|---|
| Stage | | | | 0.003 |
| I | 79 (35.3) | 52 (44.1) | 27 (25.5) | |
| II | 83 (37.0) | 43 (36.4) | 40 (37.7) | |
| III | 62 (27.7) | 23 (19.5) | 39 (36.8) | |
| Grade | | | | 0.035 |
| 1 | 19 (8.4) | 15 (12.7) | 4 (3.8) | |
| 2 | 147 (65.6) | 77 (65.3) | 70 (66.0) | |
| 3 | 58 (25.9) | 26 (22.0) | 32 (30.2) | |
| Age, years | | | | 0.139 |
| ≤50 | 111 (49.6) | 64 (54.2) | 47 (44.3) | |
| >50 | 113 (50.4) | 54 (45.8) | 59 (55.7) | |
| BMI,
kg/m2 | | | | 0.671 |
| ≤24 | 128 (57.1) | 69 (58.5) | 59 (55.7) | |
| >24 | 96 (42.9) | 49 (41.5) | 47 (44.3) | |
| Tumor size, cm | | | | 0.036 |
| <2 | 114 (50.9) | 69 (58.5) | 45 (42.4) | |
| 2-5 | 90 (40.2) | 42 (35.6) | 48 (45.3) | |
| >5 | 20 (8.9) | 7 (5.9) | 13 (12.3) | |
| LN metastasis | | | | 0.021 |
| No | 132 (58.9) | 78 (66.1) | 54 (50.9) | |
| Yes | 92 (41.1) | 40 (33.9) | 52 (49.1) | |
| ER status | | | | 0.116 |
| Negative | 79 (35.3) | 36 (30.5) | 43 (40.6) | |
| Positive | 145 (64.7) | 82 (69.5) | 63 (59.4) | |
| PR status | | | | 0.076 |
| Negative | 96 (42.9) | 44 (37.3) | 52 (49.1) | |
| Positive | 128 (57.1) | 74 (62.7) | 54 (50.9) | |
| HER2 status | | | | 0.386 |
| Negative | 146 (65.2) | 80 (67.8) | 66 (62.3) | |
| Positive | 78 (34.8) | 38 (32.2) | 40 (37.7) | |
 | Table IIAssociation of nuclear RAD51
expression with clinicopathologic characteristics in patients with
breast cancer (n=180). |
Table II
Association of nuclear RAD51
expression with clinicopathologic characteristics in patients with
breast cancer (n=180).
| Variable | Total | Nuclear RAD51
expression
| P-value |
|---|
| Low (n=113) | High (n=67) |
|---|
| Stage | | | | <0.001 |
| I | 59 (32.8) | 25 (22.1) | 34 (50.8) | |
| II | 71 (39.4) | 50 (44.3) | 21 (31.3) | |
| III | 50 (27.8) | 38 (33.6) | 12 (17.9) | |
| Grade | | | | 0.218 |
| 1 | 14 (7.8) | 8 (7.1) | 6 (9.0) | |
| 2 | 119 (66.1) | 80 (70.8) | 39 (58.2) | |
| 3 | 47 (26.1) | 25 (22.1) | 22 (32.8) | |
| Age, years | | | | 0.203 |
| ≤50 | 91 (50.6) | 53 (46.9) | 38 (56.7) | |
| >50 | 89 (49.4) | 60 (53.1) | 29 (43.3) | |
| BMI,
kg/m2 | | | | 0.261 |
| ≤24 | 95 (52.8) | 56 (49.6) | 39 (58.2) | |
| >24 | 85 (47.2) | 57 (50.4) | 28 (41.8) | |
| Tumor size, cm | | | | 0.005 |
| <2 | 91 (50.6) | 47 (41.6) | 44 (65.7) | |
| 2-5 | 71 (39.4) | 51 (45.1) | 20 (29.8) | |
| >5 | 18 (10.0) | 15 (13.3) | 3 (4.5) | |
| LN metastasis | | | | 0.001 |
| No | 106 (58.9) | 56 (49.6) | 50 (74.6) | |
| Yes | 74 (41.1) | 57 (50.4) | 17 (25.4) | |
| ER status | | | | 0.727 |
| Negative | 62 (34.4) | 40 (35.4) | 22 (32.8) | |
| Positive | 118 (65.6) | 73 (64.6) | 45 (67.2) | |
| PR status | | | | 0.854 |
| Negative | 79 (43.9) | 49 (43.4) | 30 (44.8) | |
| Positive | 101 (56.1) | 64 (56.6) | 37 (55.2) | |
| HER2 status | | | | 0.503 |
| Negative | 121 (67.2) | 78 (69.0) | 43 (64.2) | |
| Positive | 59 (32.8) | 35 (31.0) | 24 (35.8) | |
The univariate and multivariate Cox regression
models were subsequently applied to analyze the utility of
parameters of clinicopathologic characteristics and differential
RAD51 expression as potential risk factors for patient survival. In
the univariate analysis, the parameters showing a strong
association with disease-free survival of the patients included
cancer stage (II vs. I, P=0.035; III vs. I, P=0.001) and
cytoplasmic RAD51 expression (high vs. low, P=0.004) (Table III). In the multivariate
analysis, the cancer stage (III vs. I, P=0.012) remained
significantly associated with disease-free survival of the patients
(Table III). In the univariate
analysis for overall survival, the parameters of cancer stage (II
vs. I, P=0.026; III vs. I, P=0.006) and cytoplasmic RAD51
expression (high vs. low, P=0.015) also showed a strong association
with patient survival, and the association of cancer stage (II vs.
I, P=0.048; III vs. I, P=0.017) with overall survival remained
significant in the multivariate analysis (Table IV). In these Cox regression
models, the association between nuclear RAD51 expression and
patient survival was not significant (Tables III and IV), suggesting that cytoplasmic RAD51
expression, rather than nuclear RAD51 expression, represented a
more influential risk regarding survival of patients with breast
cancer. In addition to these, it was observed that the histologic
grade, which is frequently associated with patient outcomes, showed
a trend of increasing hazard ratios but did not reach a
statistically significant level in the univariate and multivariate
analyses (Tables III and
IV); this discrepancy may be
resolved by further investigation with a larger magnitude of sample
size.
 | Table IIIUnivariate and multivariate Cox
regression analyses of disease-free survival in patients with
breast cancer. |
Table III
Univariate and multivariate Cox
regression analyses of disease-free survival in patients with
breast cancer.
| Variable | Univariate
| Multivariate
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Stage | | | | |
| II vs. I | 3.91
(1.10-13.88) | 0.035 | 3.29
(0.91-11.86) | 0.069 |
| III vs. I | 7.67
(2.26-26.07) | 0.001 | 5.12
(1.44-18.27) | 0.012 |
| Grade | | | | |
| 2 vs. 1 | 2.62
(0.63-10.88) | 0.186 | | |
| 3 vs. 1 | 2.39
(0.52-10.99) | 0.265 | | |
| Age (>50 vs. ≤50
years) | 1.79
(0.88-3.61) | 0.106 | | |
| BMI (>24 vs. ≤24
kg/m2) | 1.97
(0.99-3.94) | 0.055 | | |
| ER positive | 0.87
(0.73-1.03) | 0.111 | | |
| PR positive | 0.87
(0.73-1.04) | 0.117 | | |
| HER2 positive | 0.86
(0.67-1.11) | 0.254 | | |
| Cytoplasmic RAD51
(high vs. low) | 3.40
(1.47-7.86) | 0.004 | 2.26
(0.95-5.40) | 0.067 |
| Nuclear RAD51 (high
vs. low) | 0.28
(0.07-1.23) | 0.093 | | |
 | Table IVUnivariate and multivariate Cox
regression analyses of overall survival in patients with breast
cancer. |
Table IV
Univariate and multivariate Cox
regression analyses of overall survival in patients with breast
cancer.
| Variable | Univariate
| Multivariate
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Stage | | | | |
| II vs. I | 5.55
(1.23-25.11) | 0.026 | 4.64
(1.01-21.24) | 0.048 |
| III vs. I | 8.10
(1.84-35.66) | 0.006 | 6.48
(1.41-29.88) | 0.017 |
| Grade | | | | |
| 2 vs. 1 | 3.48
(0.47-26.10) | 0.224 | | |
| 3 vs. 1 | 4.80
(0.58-39.86) | 0.146 | | |
| Age (>50 vs. ≤50
years) | 1.79
(0.83-3.89) | 0.140 | | |
| BMI (>24 vs. ≤24
kg/m2) | 1.78
(0.83-3.81) | 0.138 | | |
| ER positive | 0.87
(0.72-1.05) | 0.144 | | |
| PR positive | 0.85
(0.70-1.03) | 0.090 | | |
| HER2 positive | 0.97
(0.74-1.26) | 0.798 | | |
| Cytoplasmic RAD51
(high vs. low) | 3.10
(1.24-7.71) | 0.015 | 1.88
(0.71-4.94) | 0.202 |
| Nuclear RAD51 (high
vs. low) | 0.21
(0.03-1.64) | 0.137 | | |
Resistance to chemotherapy has been one of the main
obstacles in breast cancer management, yet clinical markers to
effectively predict responses to chemotherapy are still under
exploration (23,24). Therefore, in the present study,
the effects of subcellular RAD51 expression on patient survival
with regard to chemotherapy was further investigated in these
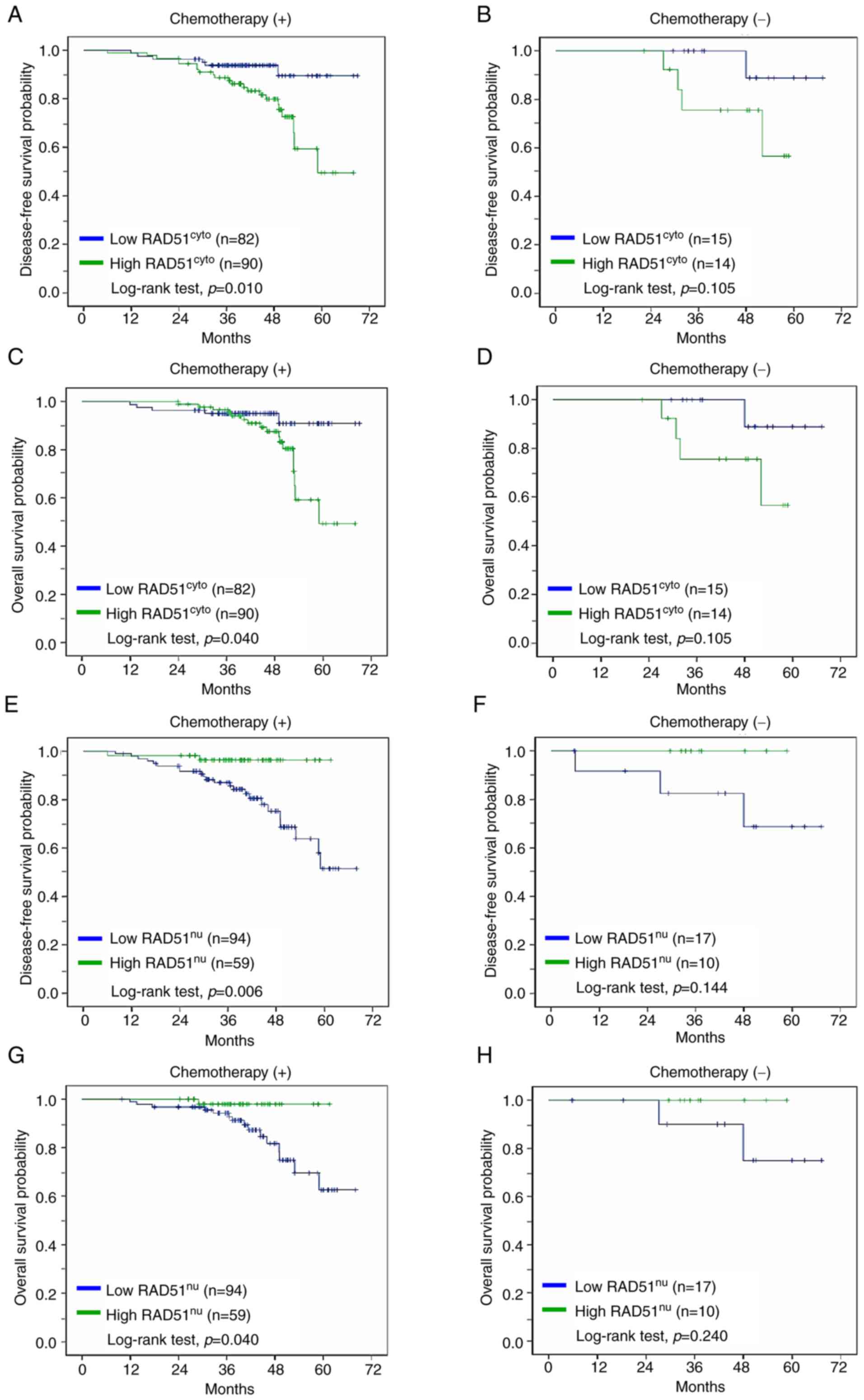
cohorts. As presented in Fig. 2A and
C, it was found that high cytoplasmic RAD51 expression was
associated with poorer disease-free survival (P=0.010) and overall
survival (P=0.040) in the patient group receiving adjuvant
chemotherapy, while the association of cytoplasmic RAD51 expression
with patient survival was not significant in the patient group
without chemotherapy (Fig. 2B and
D). An opposite trend was observed in the analysis for nuclear
RAD51 expression, where high nuclear RAD51 expression was in favor
of disease-free survival (P=0.006) and overall survival (P=0.040)
in the patient group receiving chemotherapy (Fig. 2E and G), while the significance of
association was not observed in the patient group without
chemotherapy (Fig. 2F and H).
These results indirectly suggest the potential of a different
prognostic role of subcellular RAD51 expression in consideration of
patients with breast cancer receiving adjuvant chemotherapy.
Effect of differential RAD51 expression
on in vitro breast cancer cell growth and chemoresistance
The roles of differential RAD51 expression on breast
cancer cell behaviors were investigated via the following in
vitro assays. Two widely studied human breast cancer cell
lines, MDA-MB-231 (basal type) and MCF-7 (luminal type A) (25), were transfected with lentiviral
particles that contain full-length RAD51 for gene overexpression or
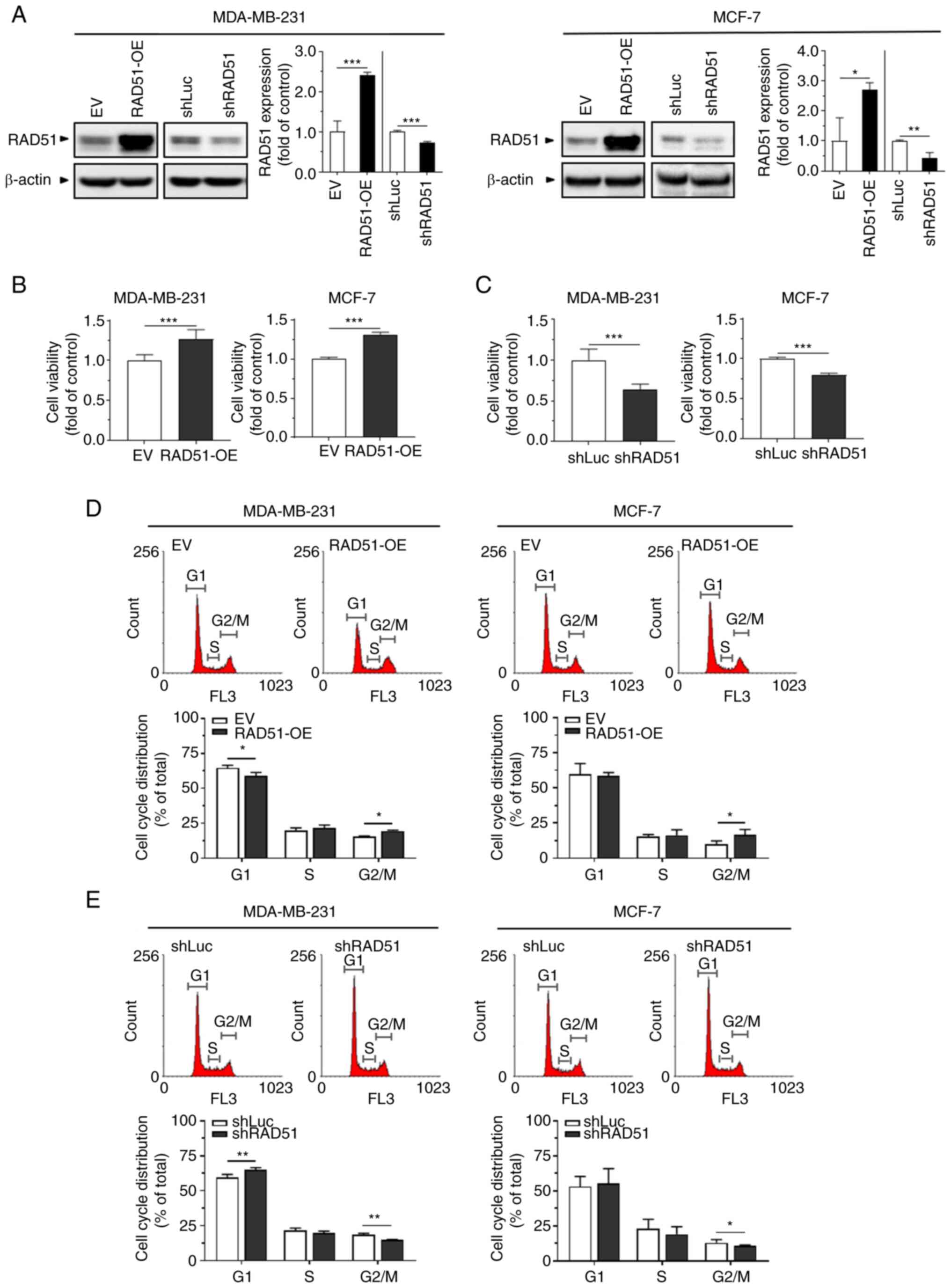
shRNA targeting RAD51 for gene knockdown (Fig. 3A). The XTT assay to determine
cancer cell growth revealed that overexpression of RAD51 enhanced
the proliferative ability of these breast cancer cells (Fig. 3B), whereas knockdown of RAD51
reduced their proliferative ability compared to their corresponding
controls (Fig. 3C). The cell
cycle distribution was further evaluated by flow cytometry, which
showed that both MDA-MB-231 and MCF-7 cells with overexpression of
RAD51 had an increased proportion of the G2/M phases (Fig. 3D). On the contrary, knockdown of
RAD51 led to a reduced proportion of the G2/M phases in these
breast cancer cells (Fig. 3E).
Taken together, the results suggest that elevation of RAD51
expression promoted breast cancer cell growth, which potentially
resulted from the progression of the cell cycle into the G2/M
phases.
To investigate whether the differential RAD51
expression is involved in the resistance of breast cancer cells to
chemotherapy, the effect of RAD51 overexpression or knockdown on
cell growth of the breast cancer cells that were treated with
cisplatin, a first-line therapeutics administered in breast and
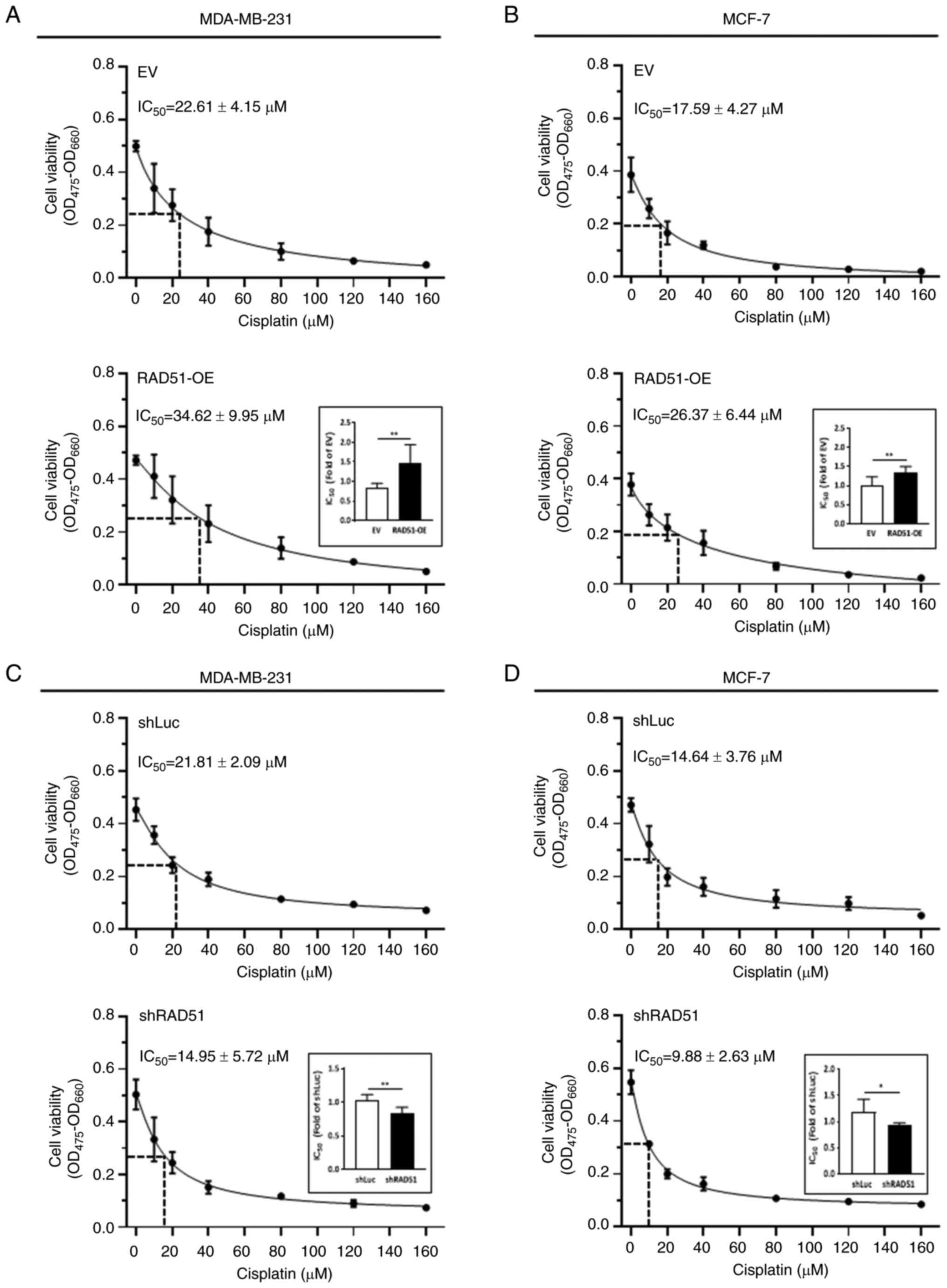
various tumors, was assessed (26). An increased shift of the
IC50 value of cisplatin was observed in both
RAD51-overexpressing MDA-MB-231 cells (Fig. 4A) and MCF-7 cells (Fig. 4B), whereas knockdown of RAD51
resulted in a reduced IC50 of cisplatin in these breast
cancer cells (Fig. 4C and D). The
shift of the IC50 for cisplatin treatment further
suggests that the differential RAD51 expression may be involved in
the regulation of cancer cell growth during chemotherapy, with the
notion that resistance to cisplatin treatment occurred in
RAD51-overexpressing breast cancer cells.
While the effect of cisplatin treatment on RAD51
subcellular localization has not been previously reported, to the
best of our knowledge, it was reported that curcumin induces DNA
damage and leads to RAD51 migrating to the nucleus (27). Previous studies also showed that
in HeLa and HCT116 cells, RAD51 redistributed from the cytoplasm to
the nucleus after exposure to ionizing radiation (28,29). In the current study, it was shown
that RAD51 overexpression led to increased cytoplasmic and nuclear
RAD51 expression and more surviving cells (chemoresistance) among
MDA-MB-231 and MCF-7 cells after cisplatin treatment (Figs. S2 and 4). On the other hand, RAD51 knockdown
led to decreased surviving cells (chemosensitivity) in MDA-MB-231
cells after cisplatin treatment. All of these data together suggest
that RAD51 expression levels are positively associated
chemoresistance and radioresistance in breast cancer cells.
Effect of differential RAD51 expression
on in vitro breast cancer cell migration and invasion ability
The effect of RAD51 expression on the in
vitro metastatic ability of breast cancer cells was
investigated by Transwell migration and invadopodial invasion
assays. As presented in Fig. 5A,
MDA-MB-231 and MCF-7 cells with overexpression of RAD51 exhibited
an increased ability of cell migration. On the contrary, knockdown
of RAD51 decreased the cell migration ability of these breast
cancer cells (Fig. 5B). In
addition to cell migration, increased ability of invadopodial cell
invasion was observed in MDA-MB-231 cells with overexpression of
RAD51, whereas knockdown of RAD51 repressed the cell invasion
ability (Fig. 5C). To further
explore the biological activity associated with subcellular RAD51
expression, the cytoplasmic and nuclear proteins from MDA-MB-231
cells with overexpression of RAD51, or those from control cells,
were fractionated prior to immunoprecipitation with anti-RAD51
antibodies. The fractionation result showed that endogenous RAD51
is expressed in both the cytoplasm and nuclei of MDA-MB-231 cells
and overexpression increased RAD51 expression in both cytoplasm and
nuclei (Fig. S2A). After
immunoprecipitation with anti-RAD51 antibodies, a clearly stained
band was shown above and near the molecular weight of 40 kDa in the
cytoplasmic fraction of RAD51-overexpressing MDA-MB-231 cells,
while there was no clear band in the nuclear fraction of
RAD51-overexpressing MDA-MB-231 cells or control cells (Fig. S2B), suggesting the presence of a
substantial protein interaction between cytoplasmic RAD51 and the
co-immunoprecipitated protein. Identification of the protein
sequences for this particular band by LC-MS/MS revealed that it
matched the sequences of β-actin (Fig. S2C), the major cytoplasmic isoform
of the actin family expressed in non-muscle cells (30). As β-actin is the essential subunit
of globular actin that forms F-actin in control of various types of
cell growth and motility (30-32), the potential of the protein
interaction between RAD51 and F-actin was further examined using
breast cancer cell models in the present study. As indicated in
Fig. 5D, overexpression of RAD51
in MDA-MB-231 cells did not affect the endogenous amount of F-actin
expression. However, immunoprecipitation with anti-F-actin
antibodies revealed that RAD51 was more abundantly
co-immunoprecipitated in RAD51-overexpressing MDA-MB-231 cells
compared to control cells (Fig.
5E). The increased protein interaction between RAD51 and
F-actin was also evidenced by immunofluorescence, where
RAD51-overexpressing MDA-MB-231 cells showed higher abundance of
co-localization between RAD51 and F-actin compared to control cells
(Fig. 5F). These results together
suggest that alteration of RAD51 expression was associated with the
migration and invasion ability of breast cancer cells, and the
interaction between cytoplasmic RAD51 and actin filaments was
potentially an association factor on these biological activities,
which warrants further investigation.
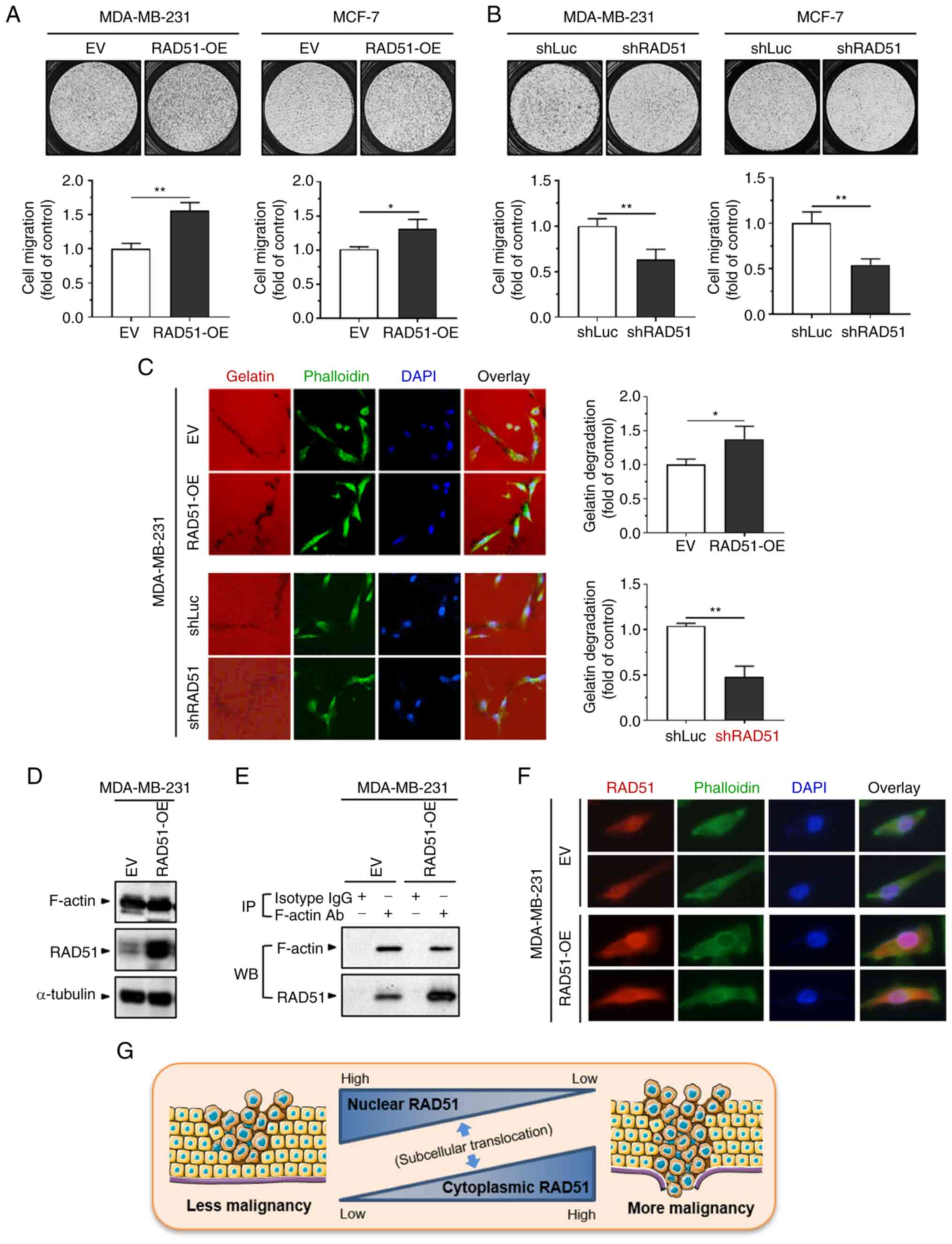 | Figure 5Effect of differential RAD51
expression on breast cancer cell migration and invasion, and the
protein interaction with F-actin. MDA-MB-231 and MCF-7 cells with
(A) overexpression of RAD51 or (B) knockdown of RAD51 and their
corresponding controls were cultured for 24 h in Transwell inserts,
followed by the procedures of the Transwell migration assay. The
cell images were generated by NIS-Elements Imaging Software 5.0
(Nikon Corp.). (C) MDA-MB-231 cells with overexpression of RAD51 or
knockdown of RAD51 and their corresponding controls were cultured
for 24 h on glass chamber slides pre-coated with Cy3-conjugated
gelatin (red), followed by the procedures of the invadopodial
invasion assay. FITC-conjugated phalloidin (green) and DAPI (blue)
were applied to detect F-actin and the cell nucleus, respectively
(magnification, ×200). Data were presented as the mean ± standard
deviation from three independent experiments. P-values were
determined by two-sided Student's t-test between experimental and
control groups. *P<0.05; **P<0.01s. The
cell images were created by AxioVision Software 4.8 (Carl Zeiss
GmbH). (D) The protein expression in MDA-MB-231 cells with
overexpression of RAD51 and their controls was analyzed by western
blot. (E) Immunoprecipitation with anti-F-actin antibodies or
isotype IgG was performed using the protein extracts of MDA-MB-231
cells with overexpression of RAD51 and their controls, followed by
western blot analysis of the protein expression. The blot images
were created by Image Lab Software 6.1 (Bio-Rad Laboratories,
Inc.). (F) Immunofluorescence with anti-RAD51 antibodies was
conducted on MDA-MB-231 cells with overexpression of RAD51 and
their controls. Alexa Fluor 555-conjuagted donkey polyclonal
antibodies against rabbit IgG was applied to detect RAD51 (red),
and FITC-conjugated phalloidin (green) and DAPI (blue) were applied
to detect F-actin and the cell nucleus, respectively. The cell
images were created by AxioVision Software 4.8 (Carl Zeiss GmbH)
(magnification, ×400). (G) Schematic diagram of the impact of
cytoplasmic vs. nuclear RAD51 on breast cancer malignancy. The
current study suggests a pro-oncogenic role of cytoplasmic RAD51 in
breast cancer progression. On the other hand, the canonical role of
nuclear RAD51 in DNA damage repair may prevent breast tumors from
further malignant transformation caused by genomic instability. The
differential impact of subcellular RAD51 on breast malignancy may
result from the translocation of RAD51 that occurs in a dynamic
manner regulated by a network of RAD51 interaction proteins, such
as BRCA1/2 and RAD51C. EV, empty vector; RAD51-OE, overexpression
of RAD51; shLuc, knockdown of firefly luciferase; shRAD51,
knockdown of RAD51; F-actin, filamentous actin; Ab, antibody; IP,
immunoprecipitation; WB, western blot. |
Discussion
RAD51 represents one of the potential DNA damage
repair-associated therapeutic candidates in breast cancer (8,33).
However, the bilocation nature of RAD51 distributed in both the
cytoplasm and the cell nucleus suggests that its role may extend
further beyond DNA damage repair (13,15-17,34). In the present study, the
differential expression of subcellular RAD51 in breast tumors
revealed that elevated expression of cytoplasmic RAD51 was
associated with adverse breast cancer progression and clinical
outcomes, including increased cancer stage, grade, tumor size, LN
metastasis, as well as poor disease-free and overall survival. In
addition, elevated expression of cytoplasmic RAD51 in breast tumors
was associated with poor disease-free and overall survival in the
patients who received adjuvant chemotherapy, but this association
was not significant in those without adjuvant chemotherapy. By
contrast, elevated expression of nuclear RAD51 in breast tumors
showed an opposite role for these clinical outcomes in comparison
to cytoplasmic RAD51. The present clinical data support the notion
of a distinct role between these two subcellular localizations of
RAD51 in breast cancer progression and prognosis (16,17). Stratification of patient groups
according to differential subcellular RAD51 location may therefore
allow a more personalized approach to breast cancer evaluation.
Manipulation of the RAD51 expression level in human
breast cancer cell lines has unveiled several roles of RAD51 in
cancer cell growth and metastatic ability (35,36). For instance, knockdown of RAD51 in
MDA-MB-231 cells was shown to reduce cell migration ability,
whereas overexpression of RAD51 in BT549 and Hs578T cells promoted
cell migration (14). In
addition, increased chemoresistance to the poly(ADP-ribose)
polymerase (PARP) inhibitor ABT-888 (Veliparib) (37) was observed in RAD51-overexpressing
Hs578T cells (38). Another study
using a brain metastasis-favored MDA-MB-231 subline, MDA-MB-231-BR
(39), with RAD51 overexpression
showed resistant phenotypes following chemotherapeutic treatment
with doxorubicin (40).
Furthermore, knockdown of RAD51 restored treatment sensitivity with
HER2-targeted trastuzumab (Herceptin) (41) in SKBR3 and JIMT-1 cells (42). The current in vitro data
using MDA-MB-231 and MCF-7 cells further unveiled that
overexpression of RAD51 promoted malignant behaviors in these
breast cancer cells, including enhanced cell growth with
progressive G2/M cell cycle accumulation, enhanced metastatic
ability in a Transwell migration assay and invadopodial invasion,
and resistance to chemotherapeutic treatment with cisplatin. On the
contrary, knockdown of RAD51 suppressed the malignant behaviors in
MDA-MB-231 and MCF-7 cells. These results provide further evidence
for the pro-oncogenic effect of RAD51 on breast cancer cells.
Whether the differential RAD51 expression also affects treatment
responses to first-line chemotherapeutic agents for breast cancer,
such as anthracyclines and taxanes, warrants further investigation.
In addition, caution should be taken regarding the potential
off-target effects resulting from the application of shRNA, which
may be further evaluated by additional experiments with the use of
shRNA sequences against other coding regions of RAD51 for
comparison. The complementation experiments by re-expressing RAD51
in breast cancer cells carrying knockdown of RAD51 may also be
applied to demonstrate the specificity of shRNA. Furthermore, while
the effect of differential RAD51 expression on breast cancer cell
growth was examined by XTT assay in the current study, application
of another approach to assess cell growth, such as measurement of
the doubling time by direct counting of the cell number, may
further confirm the results derived from this study.
Genomic instability is a hallmark of cancer,
resulting from dysregulated DNA damage repair (43-45). The involvement of RAD51 in breast
cancer progression may depend on its subcellular localization, with
high cytoplasmic RAD51 or low nuclear RAD51 in breast tumors
associated with poorer clinical outcomes, as shown in the present
study and previous reports (16,17). An explanation for these clinical
observations may relate to RAD51 translocation from the cell
nucleus to the cytoplasm, hence the enhancement of genomic
instability due to inappropriate DNA damage repair (Fig. 5G). For instance, the localization
of RAD51 and BRCA1, a RAD51-interacting protein (46), was found to increase in the
cytoplasm of breast cancer cells via activation of AKT signaling,
leading to a phenotype of reduced nuclear localization of these two
proteins and decreased DNA damage repair ability (47). The subcellular localization of
RAD51 has been shown to be associated with multiple
RAD51-interacting proteins, including BRCA1/2 (46,48,49) and RAD51 paralog RAD51C (50). Of note, RAD51 does not contain a
nuclear localization signal (NLS) but a nuclear export signal
(NES), and therefore, its nuclear translocation relies on the
NLS-containing interaction proteins, such as BRCA1/2 and RAD51C
(28,46,51). The presence of NES on RAD51 may
partly explain the abundance of RAD51 in the cytoplasm of cancer
cells expressing mutated BRCA, which commonly shows compromised
nuclear localization of both BRCA and RAD51, leading to genomic
instability (52). In addition,
the NES of RAD51 can be specifically masked when interacting with
intact BRCA2 to retain itself in the cell nucleus, suggesting that
the nuclear RAD51 concertedly participates in the BRCA tumor
suppressor network via protein interactions (51,53).
In contrast to nuclear RAD51, the biological
activity of cytoplasmic RAD51 has remained largely elusive. In the
present study, β-actin was identified as a novel RAD51-interacting
protein, and this protein interaction occurred in particular within
the subcellular localization of cytoplasmic RAD51. Evidence was
also provided to show that overexpression of RAD51 resulted in an
increase of invadopodia, an F-actin-based cytoskeletal remodeling
for cancer invasion (54), and
that overexpression of RAD51 co-localized with F-actin in the
cytoplasm of breast cancer cells. β-Actin has been shown to be the
monomeric component for F-actin formation and the process is
intricately regulated by a number of actin interaction proteins, as
alteration of F-actin formation may have a critical influence on
cancer progression and metastasis (32,55,56). The current findings not only add
cytoplasmic RAD51 and β-actin to the growing pool of
protein-protein interaction, but also raise the possibility for a
potential role of the protein interaction between cytoplasmic RAD51
and actin filaments in breast cancer. Future studies are required
to elucidate whether alteration of this protein interaction
impinges on breast cancer cell malignancy.
In the current study, one limitation is that the
role of overall RAD51 expression but not changes in the subcellular
location of RAD51 was examined in the in vitro experiments.
RAD51 itself has no NLS and its subcellular localization is
influenced by its interaction with other proteins, including
BRCA1/2 (46,48,49) and RAD51 paralog RAD51C (50). To avoid the inconclusive result
from manipulation of subcellular localization of RAD51 by
overexpression or downregulation of BRCA1/2 or RAD51C, adding NLS
to the RAD51 gene to manipulate its subcellular localization may be
an alternative approach. Another limitation is that the majority of
patients recruited in the present study received adjuvant
chemotherapy, leading to a significantly reduced observation number
for those without adjuvant chemotherapy. Therefore, further
increments of the observation number for patients without adjuvant
chemotherapy are required to confirm the prognostic role of
subcellular RAD51 in this population of patients with breast
cancer. Furthermore, simultaneous evaluation of RAD51 expression in
the cytoplasmic and nuclear localization may provide additional
information for the prognostic role of subcellular RAD51. For
instance, future studies may consider analyzing treatment outcomes
according to both subcellular RAD51 expression levels, such as
cytohigh/nulow vs.
cytolow/nuhigh of RAD51 expression in
patients with breast cancer. The third limitation of the present
study is that the effect of cisplatin treatment on the subcellular
location of RAD51 and the interaction between cytoplasmic RAD51 and
F-actin in transfected cells was not investigated and is worthy of
further investigation in a future study.
For future recommendations, with the increasing use
of PARP inhibitors in cancer treatment, there is an unmet clinical
need to implement homologous recombination deficiency (HRD) testing
in clinical practice. In the coming years, RAD51 recombinase
protein may offer additional dimensions by functioning as a
biomarker for HRD tests (57).
In conclusion, in the present study, the role of
subcellular RAD51 in breast cancer was evaluated by clinical
measurements, unveiling that elevated cytoplasmic RAD51 expression
was associated with adverse clinicopathologic features and outcomes
for the patients, whereas elevated nuclear RAD51 expression had the
inverse effect on these clinical parameters. The results of the
in vitro investigation were in accordance with the
cancer-promoting effect of RAD51, where the malignant behaviors of
breast cancer cells were enhanced by overexpression of RAD51 but
repressed by knockdown of RAD51. These findings together suggest a
pro-oncogenic role of cytoplasmic RAD51 in contrast to nuclear
RAD51, which may provide insight for the further development of
therapeutic strategies for breast cancer. For instance,
stratification of patients with breast cancer by differential
subcellular RAD51 expression may benefit them in terms of
chemotherapeutic selection and improvement of personalized
prognosis.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YYW, KHC, ACH, SL, PYC, YCW, MFH and SSFY conceived
and designed the study. YYW, KHC, ACH, PYC and SSFY performed the
experiments. YYW, ACH, SL, YCW, MFH, and SSFY analyzed and
interpreted the data. YYW, KHC and ACH wrote the manuscript. SL,
PYC, YCW, MFH and SSFY critically revised the manuscript. YYW and
SSFY confirm the authenticity of all the raw data. All authors have
reviewed and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Institutional Review
Board of Kaohsiung Medical University Hospital [Kaohsiung, Taiwan;
approval no. KMUH-IRB-20130031 and KMUHIRB-E(I)-20180136] and
conducted in accordance with the Declaration of Helsinki. All of
the samples used in the present study were de-identified samples
from the hospital's biobank added with the informed consent of the
patients, but the requirement of informed consent to include them
in the present study was waived by the ethics committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The analysis of clinical data was performed with the
assistance of Dr Yi-Chen Lee (Department of Anatomy, School of
Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan). Dr
Shyh-Horng Chiou (Quantitative Proteomics Center, Center for
Research Resources and Development, Kaohsiung Medical University,
Kaohsiung, Taiwan) performed the LC-MS/MS analysis. The schematic
presented in Fig. 5G was
generated using the illustration elements from Servier Medical Art
(https://smart.servier.com), which is in
compliance with the terms of the Creative Commons Attribution 3.0
Unported License (https://creativecommons.org/licenses/by/3.0/).
Funding
This work was supported by grants from the National Science and
Technology Council (grant nos. NSTC 112-2314-B-037-120 and NSTC
112-2314-B-037-112-MY3) and Center for Intelligent Drug Systems and
Smart Bio-devices (IDS2B) from the Featured Areas Research Center
Program within the framework of the Higher Education Sprout Project
by the Ministry of Education in Taiwan. This work was also
supported by grants from Kaohsiung Medical University Hospital
[grant nos. KMUH110-0R43, KMUH111-1R37, KMUH-DK(A)110001 and
KMUH-DK(A)112001] and Kaohsiung Medical University [grant nos.
KMU-DK(A)111005, KMU-DK(A)112006, NYCU-KMU-111-I002,
NYCU-KMU-112-I005, NSYSU-KMU-112-P04, KMU-TC112A03-5], Taiwan.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moo TA, Sanford R, Dang C and Morrow M:
Overview of breast cancer therapy. PET Clin. 13:339–354. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Trayes KP and Cokenakes SEH: Breast cancer
treatment. Am Fam Physician. 104:171–178. 2021.PubMed/NCBI
|
|
4
|
Martinez-Perez C, Turnbull AK, Ekatah GE,
Arthur LM, Sims AH, Thomas JS and Dixon JM: Current treatment
trends and the need for better predictive tools in the management
of ductal carcinoma in situ of the breast. Cancer Treat Rev.
55:163–172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fahad Ullah M: Breast cancer: Current
perspectives on the disease status. Adv Exp Med Biol. 1152:51–64.
2019. View Article : Google Scholar
|
|
6
|
Giridhar KV and Liu MC: Available and
emerging molecular markers in the clinical management of breast
cancer. Expert Rev Mol Diagn. 19:919–928. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nickoloff JA: Targeting replication stress
response pathways to enhance genotoxic chemo- and radiotherapy.
Molecules. 27:47362022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Z, Jia R, Wang L, Yang Q, Hu X, Fu Q,
Zhang X, Li W and Ren Y: The emerging roles of Rad51 in cancer and
its potential as a therapeutic target. Front Oncol. 12:9355932022.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee MG, Lee KS and Nam KS: The association
of changes in RAD51 and survivin expression levels with the proton
beam sensitivity of Capan-1 and Panc-1 human pancreatic cancer
cells. Int J Oncol. 54:744–752. 2019.
|
|
10
|
Connell PP, Jayathilaka K, Haraf DJ,
Weichselbaum RR, Vokes EE and Lingen MW: Pilot study examining
tumor expression of RAD51 and clinical outcomes in human head
cancers. Int J Oncol. 28:1113–1119. 2006.PubMed/NCBI
|
|
11
|
Luzhna L, Golubov A, Ilnytskyy S, Chekhun
VF and Kovalchuk O: Molecular mechanisms of radiation resistance in
doxorubicin-resistant breast adenocarcinoma cells. Int J Oncol.
42:1692–1708. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maacke H, Opitz S, Jost K, Hamdorf W,
Henning W, Krüger S, Feller AC, Lopens A, Diedrich K, Schwinger E
and Stürzbecher HW: Over-expression of wild-type Rad51 correlates
with histological grading of invasive ductal breast cancer. Int J
Cancer. 88:907–913. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu J, Wang N and Wang YJ: XRCC3 and RAD51
expression are associated with clinical factors in breast cancer.
PLoS One. 8:e721042013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wiegmans AP, Al-Ejeh F, Chee N, Yap PY,
Gorski JJ, Da Silva L, Bolderson E, Chenevix-Trench G, Anderson R,
Simpson PT, et al: Rad51 supports triple negative breast cancer
metastasis. Oncotarget. 5:3261–3272. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Soderlund K, Skoog L, Fornander T and
Askmalm MS: The BRCA1/BRCA2/Rad51 complex is a prognostic and
predictive factor in early breast cancer. Radiother Oncol.
84:242–251. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sosinska-Mielcarek K, Duchnowska R,
Winczura P, Badzio A, Majewska H, Lakomy J, Pęksa R, Pieczyńska B,
Radecka B, Dębska S, et al: Immunohistochemical prediction of brain
metastases in patients with advanced breast cancer: The role of
Rad51. Breast. 22:1178–1183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alshareeda AT, Negm OH, Aleskandarany MA,
Green AR, Nolan C, TigHhe PJ, Madhusudan S, Ellis IO and Rakha EA:
Clinical and biological significance of RAD51 expression in breast
cancer: A key DNA damage response protein. Breast Cancer Res Treat.
159:41–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiu WC, Fang PT, Lee YC, Wang YY, Su YH,
Hu SC, Chen YK, Tsui YT, Kao YH, Huang MY and Yuan SF: DNA Repair
Protein Rad51 induces tumor growth and metastasis in esophageal
squamous cell carcinoma via a p38/Akt-Dependent pathway. Ann Surg
Oncol. 27:2090–2101. 2020. View Article : Google Scholar
|
|
19
|
Yuan SS, Hou MF, Hsieh YC, Huang CY, Lee
YC, Chen YJ and Lo S: Role of MRE11 in cell proliferation, tumor
invasion, and DNA repair in breast cancer. J Natl Cancer Inst.
104:1485–1502. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Lu LL, Wen D, Liu DL, Dong LL, Gao
DM, Bian XY, Zhou J, Fan J and Wu WZ: MiR-612 regulates invadopodia
of hepatocellular carcinoma by HADHA-mediated lipid reprogramming.
J Hematol Oncol. 13:122020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee MY, Huang CH, Kuo CJ, Lin CL, Lai WT
and Chiou SH: Clinical proteomics identifies urinary CD14 as a
potential biomarker for diagnosis of stable coronary artery
disease. PLoS One. 10:e01171692015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cottrell JS: Protein identification using
MS/MS data. J Proteomics. 74:1842–1851. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nedeljkovic M and Damjanovic A: Mechanisms
of chemotherapy resistance in triple-negative breast cancer-how we
can rise to the challenge. Cells. 8:9572019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Prihantono and Faruk M: Breast cancer
resistance to chemotherapy: When should we suspect it and how can
we prevent it? Ann Med Surg. 70:1027932021. View Article : Google Scholar
|
|
25
|
Holliday DL and Speirs V: Choosing the
right cell line for breast cancer research. Breast Cancer Res.
13:2152011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guney Eskiler G, Sahin E, Deveci Ozkan A,
Cilingir Kaya OT and Kaleli S: Curcumin induces DNA damage by
mediating homologous recombination mechanism in triple negative
breast cancer. Nutr Cancer. 72:1057–1066. 2020. View Article : Google Scholar
|
|
28
|
Gildemeister OS, Sage JM and Knight KL:
Cellular redistribution of Rad51 in response to DNA damage: Novel
role for Rad51C. J Biol Chem. 284:31945–31952. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mladenov E, Anachkova B and Tsaneva I:
Sub-nuclear localization of Rad51 in response to DNA damage. Genes
Cells. 11:513–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dugina VB, Shagieva GS and Kopnin PB:
Biological role of actin isoforms in mammalian cells. Biochemistry.
84:583–592. 2019.PubMed/NCBI
|
|
31
|
Bunnell TM, Burbach BJ, Shimizu Y and
Ervasti JM: β-Actin specifically controls cell growth, migration,
and the G-actin pool. Mol Biol Cell. 22:4047–4058. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Suresh R and Diaz RJ: The remodelling of
actin composition as a hallmark of cancer. Transl Oncol.
14:1010512021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gourley C, Balmana J, Ledermann JA, Serra
V, Dent R, Loibl S, Pujade-Lauraine E and Boulton SJ: Moving from
poly (ADP-ribose) polymerase inhibition to targeting DNA repair and
DNA damage response in cancer therapy. J Clin Oncol. 37:2257–2269.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Honrado E, Osorio A, Palacios J, Milne RL,
Sánchez L, Díez O, Cazorla A, Syrjakoski K, Huntsman D, Heikkilä P,
et al: Immunohistochemical expression of DNA repair proteins in
familial breast cancer differentiate BRCA2-associated tumors. J
Clin Oncol. 23:7503–7511. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Klein HL: The consequences of Rad51
overexpression for normal and tumor cells. DNA Repair. 7:686–693.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gachechiladze M, Skarda J, Soltermann A
and Joerger M: RAD51 as a potential surrogate marker for DNA repair
capacity in solid malignancies. Int J Cancer. 141:1286–1294. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stodtmann S, Nuthalapati S, Eckert D,
Kasichayanula S, Joshi R, Bach BA, Mensing S, Menon R and Xiong H:
A population pharmacokinetic meta-analysis of veliparib, a PARP
inhibitor, across phase 1/2/3 trials in cancer patients. J Clin
Pharmacol. 61:1195–1205. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wiegmans AP, Yap PY, Ward A, Lim YC and
Khanna KK: Differences in expression of key DNA damage repair genes
after epigenetic-Induced BRCAness dictate synthetic lethality with
PARP1 inhibition. Mol Cancer Ther. 14:2321–2331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoneda T, Williams PJ, Hiraga T, Niewolna
M and Nishimura R: A bone-seeking clone exhibits different
biological properties from the MDA-MB-231 parental human breast
cancer cells and a brain-seeking clone in vivo and in vitro. J Bone
Miner Res. 16:1486–1495. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Woditschka S, Evans L, Duchnowska R, Reed
LT, Palmieri D, Qian Y, Badve S, Sledge G Jr, Gril B, Aladjem MI,
et al: DNA double-strand break repair genes and oxidative damage in
brain metastasis of breast cancer. J Natl Cancer Inst.
106:dju1452014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J and Xu B: Targeted therapeutic
options and future perspectives for HER2-positive breast cancer.
Signal Transduct Target Ther. 4:342019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nam S, Chang HR, Jung HR, Gim Y, Kim NY,
Grailhe R, Seo HR, Park HS, Balch C, Lee J, et al: A pathway-based
approach for identifying biomarkers of tumor progression to
trastuzumab-resistant breast cancer. Cancer Lett. 356:880–890.
2015. View Article : Google Scholar
|
|
43
|
Negrini S, Gorgoulis VG and Halazonetis
TD: Genomic instability-an evolving hallmark of cancer. Nat Rev Mol
Cell Biol. 11:220–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schild D and Wiese C: Overexpression of
RAD51 suppresses recombination defects: A possible mechanism to
reverse genomic instability. Nucleic Acids Res. 38:1061–1070. 2010.
View Article : Google Scholar :
|
|
45
|
Chen CC, Feng W, Lim PX, Kass EM and Jasin
M: Homology-directed repair and the role of BRCA1, BRCA2, and
related proteins in genome integrity and cancer. Annu Rev Cancer
Biol. 2:313–336. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Scully R, Xie A and Nagaraju G: Molecular
functions of BRCA1 in the DNA damage response. Cancer Biol Ther.
3:521–527. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Plo I, Laulier C, Gauthier L, Lebrun F,
Calvo F and Lopez BS: AKT1 inhibits homologous recombination by
inducing cytoplasmic retention of BRCA1 and RAD51. Cancer Res.
68:9404–9412. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yuan SS, Lee SY, Chen G, Song M, Tomlinson
GE and Lee EY: BRCA2 is required for ionizing radiation-induced
assembly of Rad51 complex in vivo. Cancer Res. 59:3547–3551.
1999.PubMed/NCBI
|
|
49
|
Davies AA, Masson JY, McIlwraith MJ,
Stasiak AZ, Stasiak A, Venkitaraman AR and West SC: Role of BRCA2
in control of the RAD51 recombination and DNA repair protein. Mol
Cell. 7:273–282. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Suwaki N, Klare K and Tarsounas M: RAD51
paralogs: Roles in DNA damage signalling, recombinational repair
and tumorigenesis. Semin Cell Dev Biol. 22:898–905. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jeyasekharan AD, Liu Y, Hattori H,
Pisupati V, Jonsdottir AB, Rajendra E, Lee M, Sundaramoorthy E,
Schlachter S, Kaminski CF, et al: A cancer-associated BRCA2
mutation reveals masked nuclear export signals controlling
localization. Nat Struct Mol Biol. 20:1191–1198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Paul A and Paul S: The breast cancer
susceptibility genes (BRCA) in breast and ovarian cancers. Front
Biosci. 19:605–618. 2014. View
Article : Google Scholar
|
|
53
|
Zhao W, Wiese C, Kwon Y, Hromas R and Sung
P: The BRCA tumor suppressor network in chromosome damage repair by
homologous recombination. Annu Rev Biochem. 88:221–245. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Paterson EK and Courtneidge SA:
Invadosomes are coming: New insights into function and disease
relevance. FEBS J. 285:8–27. 2018. View Article : Google Scholar :
|
|
55
|
Izdebska M, Zielinska W, Grzanka D and
Gagat M: The role of actin dynamics and actin-binding proteins
expression in epithelial-to-mesenchymal transition and its
association with cancer progression and evaluation of possible
therapeutic targets. Biomed Res Int. 2018:45783732018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nersesian S, Williams R, Newsted D, Shah
K, Young S, Evans PA, Allingham JS and Craig AW: Effects of
modulating actin dynamics on HER2 cancer cell motility and
metastasis. Sci Rep. 8:172432018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
van Wijk LM, Nilas AB, Vrieling H and
Vreeswijk MPG: RAD51 as a functional biomarker for homologous
recombination deficiency in cancer: A promising addition to the HRD
toolbox? Expert Rev Mol Diagn. 22:185–199. 2022. View Article : Google Scholar
|