Introduction
Lung cancer accounts for 11.6% of global
malignancies and it is the most common malignant tumor (1). Lung cancer mainly consists of two
subtypes; small cell lung cancer and non-small cell lung cancer
(NSCLC), accounting for 15 and 85% of lung cancer cases,
respectively (2). NSCLC is
divided into lung adenocarcinoma (LUAD), lung squamous cell
carcinoma (LUSC) and large cell carcinoma, with LUAD being the most
common type and its incidence increasing every year (3). The treatment options and survival
outcomes for patients with lung cancer are primarily based on
pathological staging, and patients with NSCLC generally have a poor
prognosis (4,5). As imaging technology advances and
increases awareness of health, there is a growing trend of
early-stage lung cancer diagnoses in an increasing number of
patients (6). There is increasing
evidence that early-stage LUAD can exhibit lymph node or
microscopic metastasis that is difficult to distinguish
pathologically (7). This can
significantly affect the prognosis of patients. It is crucial to
thoroughly investigate the underlying mechanisms of early
metastasis in order to discover effective biomarkers and enhance
personalized cancer treatments.
Solute carrier organic anion transporter family
member 4A1 (SLCO4A1) is a membrane or transmembrane protein
belonging to the SLCO family (8,9).
Specifically responsible for the transmembrane transport of
substances, it is an organic anion antiporter with apparent broad
substrate specificity. SLCO4A1 primarily facilitates the
transmembrane transport of substances that are independent of
Na+, including lipid-soluble drugs, thyroid and adrenal
hormones, and a selected few toxins (10). Studies have identified that
SLCO4A1 affects the chemosensitivity of cancer cells by influencing
the uptake of anticancer drugs. For example, the chemotherapeutic
drug cisplatin activates the membrane protein SLCO4A1, promoting
the proliferation and metastasis of primary small cell lung cancer
cells (11). Chen et al
(12) demonstrated the prognostic
value and tumor immune infiltration of SLCO4A1 in colon
adenocarcinoma. Moreover, overexpression of SLCO4A1 has been
observed in pancreatic cancer, thus holding immense potential as a
notable biomarker for targeted therapeutic approaches in the
management of this disease (13).
However, the expression pattern, biological functions and
prognostic value of SLCO4A1 in NSCLC remain unclear.
In the present study, bulk sequencing was carried
out on early-stage NSCLC tissues with lymph node metastasis to
identify biomarkers that influence NSCLC cell proliferation,
metastasis and prognosis. The expression levels of SLCO4A1 in NSCLC
tissues and cells were experimentally validated, demonstrating the
role of SLCO4A1 in promoting cell proliferation, migration and
invasion. Additionally, the downstream mechanisms involved were
explored. Subsequently, the tumor immune infiltration of SLCO4A1 in
NSCLC and its efficacy in immunotherapy have been thoroughly
investigated.
Materials and methods
Data source
To explore the molecular mechanisms involved in
lymph node metastasis in early stage NSCLC, RNA sequencing was
carried out using 16 cases of T1-stage NSCLC tissues, including
eight cases with and eight cases without lymph node metastasis. The
RNA sequencing data have been previously published (6) and the corresponding clinical
information is included in Table
SI. In addition, the transcriptomic data for 101 normal samples
and 909 cases of LUAD and LUSC along with their corresponding
clinical data were obtained from The Cancer Genome Atlas (TCGA)
database (https://portal.gdc.cancer.gov/). The research protocol
of the present study was approved (approval no. 2019-KY-255) by the
Institutional Ethics Committee of The First Affiliated Hospital of
Zhengzhou University (Zhengzhou, China). Written informed consent
was obtained from all participants.
Normalization and differential analysis
of RNA sequencing data
To eliminate batch effects among the 16 samples, the
raw sequencing data were normalized using the limma package
(https://bioconductor.org/packages/release/bioc/html/limma.html)
in the R software (version 4.3.1). To identify differentially
expressed genes (DEGs) among the T1 stage samples with and without
lymph node metastasis, the normalized data were subjected to
differential analysis using the DESeq2 package (https://bioconductor.org/packages/release/bioc/html/DESeq2.html).
A total of 40 DEGs were selected based on adjusted P-values and
log2-fold change (Table SII).
Subsequently, the ggplot2 package was used to generate a heatmap
and a volcano plot to visualize the results.
Constructing the risk score model
Using TCGA transcriptomic data and clinical
information, univariate Cox regression analysis was conducted to
identify genes among the 40 DEGs that significantly impact patient
survival (SLCO4A1, EPHA7, DCDC2 and IGF2). The glmnet package
(https://cran.r-project.org/web/packages/glmnet/index.html)
was used for Least Absolute Shrinkage and Selection Operator
(LASSO) regression analysis to reduce the impact of overfitting.
Subsequently, multivariable Cox regression analysis was conducted
to obtain the regression coefficients of independent prognostic
factors (Table SIII), thereby
establishing the risk score model. Based on the median value of the
risk score, TCGA-NSCLC samples were divided into high and low risk
groups. Finally, based on clinical information including sex, age,
Tumor-Node-Metastasis (TNM) stage and risk group, the nomogram was
constructed the to assess the 1-, 3- and 5-year survival rates of
patients with NSCLC. Calibration curves were used to evaluate the
applicability of the nomogram in clinical practice.
Enrichment analysis
Subsequently, to further explore the functionality
of the DEGs, 40 DEGs were imported into the FunRich software
(version 3.1.3) (14) for
enrichment analysis. Specific functionalities included 'biological
pathway', 'cellular component', 'molecular function' and
'biological process'. Pie charts were used to visualize the
results.
Machine learning
In order to identify key genes influencing early
stage NSCLC with lymph node metastasis from the pool of 40 DEGs,
three machine learning algorithms were employed for gene screening.
Support Vector Machine-Recursive Feature Elimination (SVM-RFE) is
an algorithm based on SVM and the maximum margin principle for
sequential feature selection. In the initial iteration, the SVM
model was trained and optimized using all features of the dataset.
Next, scores were calculated for each feature and sorted in
descending order. The feature set with the smallest score was
recorded and the feature with the smallest score was removed. This
process was iterated again until only one feature remained. Random
Forest (RF) is a suitable method that has no restrictions on
variable conditions and possesses high accuracy, sensitivity and
specificity. Furthermore, RF can be used for predicting continuous
variables and provides stable predictive outcomes (15). LASSO is a regression method that
performs variable selection and complexity adjustment
simultaneously while fitting a generalized linear model to improve
prediction accuracy (16). Venn
diagrams display the intersection of the genes identified by three
algorithms and the risk score model, which includes IGF2 and
SLCO4A1.
Prognosis and expression level of hub
genes
Kaplan-Meier plotter (http://kmplot.com/analysis/) is an authoritative
website for analyzing prognosis in various types of cancers. Lung
cancer prognosis using IGF2 and SLCO4A1 was investigated using the
aforementioned website. The overall survival (OS) in the high and
low IGF2 expression groups did not reveal statistical significance
(P=0.45), but SLCO4A1 demonstrated significant statistical
significance (P=7.1×10−6). Therefore, SLCO4A1 has
become the hub gene for subsequent research. Firstly,
immunohistochemical (IHC) analysis validated the protein expression
levels of SLCO4A1 in NSCLC tissues and normal tissues. Secondly,
reverse transcription-quantitative PCR (RT-qPCR) and western
blotting confirmed the expression levels of SLCO4A1 in NSCLC cell
lines.
Cell culture and transfection
The NSCLC cell lines NCI-H157, NCI-H1975, H1299 and
A549, along with the normal lung epithelial cell line BEAS-2B, were
obtained from Procell Life Science & Technology Co., Ltd. The
identification method of these cell lines was short tandem repeat
(STR) profiling. These cells were cultured using the following
conditions: i) RPMI-1640 medium supplemented with 10% fetal bovine
serum (both from Invitrogen; Thermo Fisher Scientific, Inc.); ii)
incubation in a cell culture incubator at 5% CO2/37°C.
The small interfering RNA (siRNA) that targeted SLCO4A1 was
obtained from Suzhou GenePharma Co., Ltd. (Infection concentration:
5.5 μl per 100,000 cells.). The sequences of all siRNAs are
included in Table SIV.
Lipofectamine™ 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
was used for transient transfection of these cell lines, following
the manufacturer's instructions. After 48 h of transfection, qPCR
and western blotting were performed to assess knockdown efficiency.
Based on the results, suitable siRNA sequences were selected for
further investigation.
RT-qPCR
RNA was extracted from cells using the Invitrogen
Ambion RNA extraction kit (Thermo Fisher Scientific, Inc.).
Subsequently, cDNA was synthesized according to the instructions of
the Bio-Rad Reverse Transcription Kit (Bole Life Sciences
Technology Co., Ltd., USA). cDNA, primers and SYBR Green dye (Wuhan
Servicebio Technology Co., Ltd.) were combined, and fluorescent
curves were obtained using qPCR (Thermo Fisher Scientific, Inc.).
The qPCR reaction process includes an initial denaturation for 10
min, followed by 40 cycles of denaturation for 10 sec, annealing
for 10 sec, and extension for 10 s. The method used to calculate
the expression level was the 2−ΔΔCq (17). The primer set for SLCO4A1 was
custom-designed and synthesized by Wuhan Saivell Biotechnology Co.,
Ltd. Primer sequences were as follows: SLCO4A1 forward, 5′-CTG CTC
GCC CGT CTA CAT TG-3′ and reverse, 5′-CCG AGG GTA ACC AAG GAT
GG-3′; and GAPDH forward, 5′-CTG GGC TAC ACT GAG CAC C-3′ and
reverse, 5′-AAG TGG TCG TTG AGG GCA ATG-3′.
(IHC)
The Department of Thoracic Surgery of The First
Affiliated Hospital of Zhengzhou University provided six pairs of
NSCLC and corresponding normal tissues for the present study.
Xylene and absolute ethanol were used to perform deparaffinization
and rehydration of tissue sections (5-μm) embedded in
paraffin, respectively. The antibodies were recovered using 1 mM
Tris Base EDTA Buffer (pH 9.0) at 121°C. The slides were then
immersed in 3% H2O2 for 30 min to suppress
the endogenous peroxidase and block protein activity. Subsequently,
the rabbit anti-SLCO4A1 primary antibody (1:200) was used and
slides were incubated overnight at 4°C. The next day, slides were
washed with 10% TBST before incubation with the secondary antibody
(1:400; cat. no. GB23204; Wuhan Servicebio Technology Co., Ltd.;
HRP-conjugated rabbit anti-goat IgG) for 1 h at room temperature
and stained with 3,3′-diaminobenzidine (brown) and hematoxylin
(blue). SLCO4A1 expression was quantified using the histochemical
score (H-score) to detect the differences in SLCO4A1 expression
among different tissues. The H-score was used as the quantitative
value to evaluate the relative expression level of the SLCO4A1
protein. Primary antibody information (cat. no. and supplier) is
included in Table SV. The
results were observed using a light microscope. In addition, the
Human Protein Atlas (HPA; https://www.proteinatlas.org/) database was used to
understand the expression levels of different proteins.
Western blotting
Protein was isolated using RIPA buffer supplemented
with a phosphatase inhibitor (Beijing Solarbio Science &
Technology Co., Ltd.), and the protein concentration was measured
using the BCA kit (Beijing Solarbio Science & Technology Co.,
Ltd.). Next, the cell lysates were obtained and separated on 15%
pre-cast gels (15 micrograms per lane), and then transferred onto
PVDF membranes (Bio-Rad Laboratories, Inc.). The PVDF membrane was
incubated at room temperature for 1 h in blocking buffer containing
5% BSA (Beijing Solarbio Science & Technology Co., Ltd.), and
then transferred to an incubator shaker at 4°C overnight with the
primary antibodies. Primary antibody information (cat. no.,
dilution and supplier) is included in Table SV. The membranes were washed
three times with 10% TBST for 5 min and then incubated with the
secondary antibody (HRP conjugated rabbit anti-goat IgG; 1:10,000)
at room temperature for 1 h. Following secondary antibody
incubation, the membranes were washed again and then completely
covered with a 1:1 mixture of (A) Luminol/Enhancer Reagent and (B)
Stabilized Reagent (Beijing Solarbio Science & Technology Co.,
Ltd.) to be visualized using the Amersham Imager 600.
Cell Counting Kit-8 (CCK-8) assay and
plate colony formation assay
Using the website Home For Researchers (https://www.home-for-researchers.com/static/index.html#/),
the involvement of SLCO4A1 in pathways related to cellular
proliferation was revealed. To confirm this, in vitro
functional assays including the CCK-8 and plate colony formation
assays were carried out.
The CCK-8 assay was conducted according to the
instructions of the CCK-8 assay kit (Beijing Solarbio Science &
Technology Co., Ltd.). After transfecting cells with siRNA for 48
h, cell suspensions were evenly distributed into 96-well plates,
with 100 μl per well and a seeding density of 4,000 cells.
Following cell adhesion to the culture plate, 10 μl CCK-8
reagent was added to every well, and the plate was subsequently
incubated in the dark at 37°C for 2 h. Subsequently, the optical
density at 450 nm was recorded using a microplate reader at 0, 24,
48, 72 and 96 h.
In a 6-well plate, the minimum number of cells
required for colony formation in the plate colony formation assay
was 500. Cells were evenly distributed into a 6-well plate after a
48-h long transfection, with every well containing 4,000 cells and
2 ml culture medium supplemented with 10% fetal bovine serum. After
culturing for 12 days, cells were fixed at room temperature with 4%
paraformaldehyde for 30 min and then stained with 0.1% crystal
violet for another 30 min. Afterwards, the colony numbers were
manually counted.
Transwell and wound healing assays
A Transwell culture chamber was placed into a
24-well plate, which separated it into the upper and lower
chambers. The upper chamber contained 5×104 cells and
200 μl RPMI-1640, while the lower chamber contained 500
μl culture medium with 10% fetal bovine serum. The migration
assay required a 24-h incubation period 37°C. By contrast, the
invasion assay required the addition of 10 μl basement
membrane gel in the upper chamber and a 48-h incubation period
37°C. Next, cells were fixed with 4% paraformaldehyde for 30 min
and stained with 0.1% crystal violet for 30 min. After wiping the
upper chamber membrane with a cotton swab, images of cells were
captured under a light microscope. ImageJ (version 1.8.0; National
Institutes of Health) was used for cell counting.
The control and experimental group cells were seeded
evenly in 6-well plates. When the cells reached ~90% confluency, a
sterile pipette tip was used to carefully create a straight scratch
on the cell monolayer. Subsequently, serum-free medium was added
for starvation culture at 37°C. It is important to ensure
consistent depth of the scratch and avoid damaging the underlying
culture plate. The initial position of the scratch and its position
at 48 h were observed and recorded using a light microscope.
Similarly, ImageJ was used to analyze the results of the wound
healing assay.
Tumor immunological analysis
A tumor immunological analysis on SLCOA41 was also
performed. Data were divided into the high and low expression
groups based on the median value of the SLCOA41 expression level.
This analysis involved evaluating immune cell infiltration,
assessing its relevance to immune checkpoint inhibitors, evaluating
the tumor microenvironment (TME) and measuring the tumor mutation
burden (TMB). Finally, the immune therapeutic efficacy of the
SLCOA41 high and low expression groups was evaluated in patients
with LUAD and LUSC. The transcriptomic expression data for NSCLC
were obtained from TCGA database. Specific immunological cell
markers were downloaded from the Molecular Signatures Database
(https://www.gsea-msigdb.org/gsea/msigdb/) for
subsequent analysis. Moreover, The ESTIMATE algorithm (https://bioinformatics.mdanderson.org/estimate/) was
used to evaluate tumor purity, and the CIBERSORTx algorithm
(https://cibersortx.stanford.edu/) for
immune cell analysis.
Statistical analysis
Data processing and visualization were carried out
using R (version 4.3.1) and SPSS (version 24; IBM Corp.). To
perform statistical analysis, normality testing was first conducted
using the Kolmogorov-Smirnov test and homogeneity of variances
testing using Levene's test. If the sample was divided into 2
groups, either paired t-test or independent t-test was used. For 3
groups, ANOVA was applied followed by LSD test for post hoc
comparisons if P<0.05. For 5 groups, ANOVA was used followed by
Bonferroni test for post hoc comparisons if P<0.05. In case of
non-normal data, Kruskal-Wallis test was utilized for rank sum
analysis. Additionally, the qPCR and CCK-8 assay results were
analyzed and visualized using GraphPad Prism (version 9.0;
Dotmatics). ImageJ (version 1.8.0; National Institutes of Health)
was used to analyze the Transwell and wound healing assay data. In
univariate and multivariate cox regression analysis, the
differences in categorical variables among groups were compared
using chi-square tests. Log-rank tests were used to compare
survival time differences among different groups in univariate
analysis. Significant variables in the univariate analysis were
further analyzed using Cox proportional hazards regression model
for multivariate analysis, and the hazard ratio (HR) with 95%
confidence interval (CI) were calculated as the effect size.
Spearman's was used for correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
Normalization and differential analysis
of sequencing data
To mitigate batch effects among samples, it was
imperative to normalize the raw sequencing data. Raw data
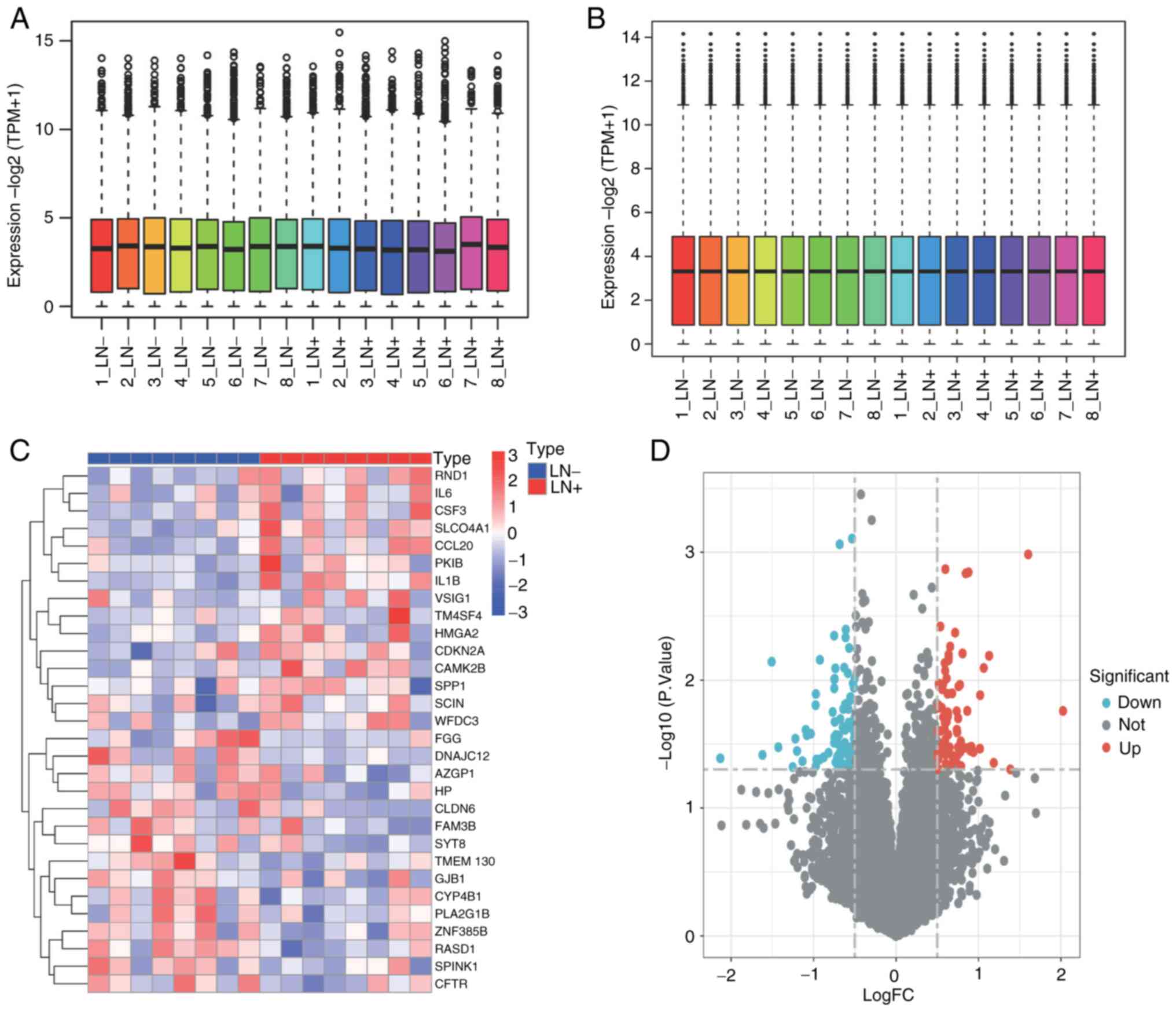
visualization before normalization is revealed in Fig. 1A; while raw data visualization
after normalization is demonstrated in Fig. 1B, significantly enhancing the
standardization and reliability of the raw data. Subsequently,
differential analysis on the data between T1 stage LUAD with and
without lymph node metastasis was performed, and 40 DEGs were
obtained. The heatmap (Fig. 1C)
and volcano plot (Fig. 1D)
display the distribution of DEGs.
Effectiveness of the risk score
model
The aim of the present study was to explore the
significant role of DEGs in NSCLC and develop novel prognostic
biomarkers based on their expression levels. Univariate Cox
regression analysis was carried out, and four genes were identified
within DEGs (SLCO4A1, EPHA7, DCDC2 and IGF2) that exhibited
significant associations with OS; this was further validated
through LASSO regression analysis (Fig. 2A and B). Then, multivariable Cox
regression analysis was carried out to determine the regression
coefficients of independent prognostic factors (Table SIII), and the risk score model
was constructed. Based on the median value of the risk score
calculated for all samples, every sample was classified into high
and low risk groups. Notably, a significant upregulation of
SLCO4A1, EPHA7, DCDC2 and IGF2 was observed in the high-risk group
(Fig. 2C). Furthermore, there was
a notable disparity in OS between patients with NSCLC and high-risk
scores and those patients with low risk scores, with the former
experiencing a significantly poorer prognosis (Fig. 2D). The analysis of receiver
operating characteristic curves revealed area under the curve
values of 0.589, 0.612 and 0.625 for patients at 1-, 3- and 5-year
intervals, respectively (Fig.
2E). The risk scores of every patient in TCGA-NSCLC dataset are
displayed in Fig. 2F. Moreover,
in order to validate the independence of the risk score model in
predicting OS time in patients with NSCLC, univariate Cox
regression analysis was performed on potential prognostic factors,
including sex, age, TNM stage and high/low risk group. TCGA-NSCLC
dataset was integrated into a nomogram model and the results
demonstrated that risk level, nodal (N) and tumor (T) stages were
identified as independent risk factors of OS (Fig. 2G). When comparing the nomogram
model with the ideal model, the calibration plot exhibited
favorable consistency between the predicted and observed 1-, 3- and
5-year OS rates (Fig. 2H).
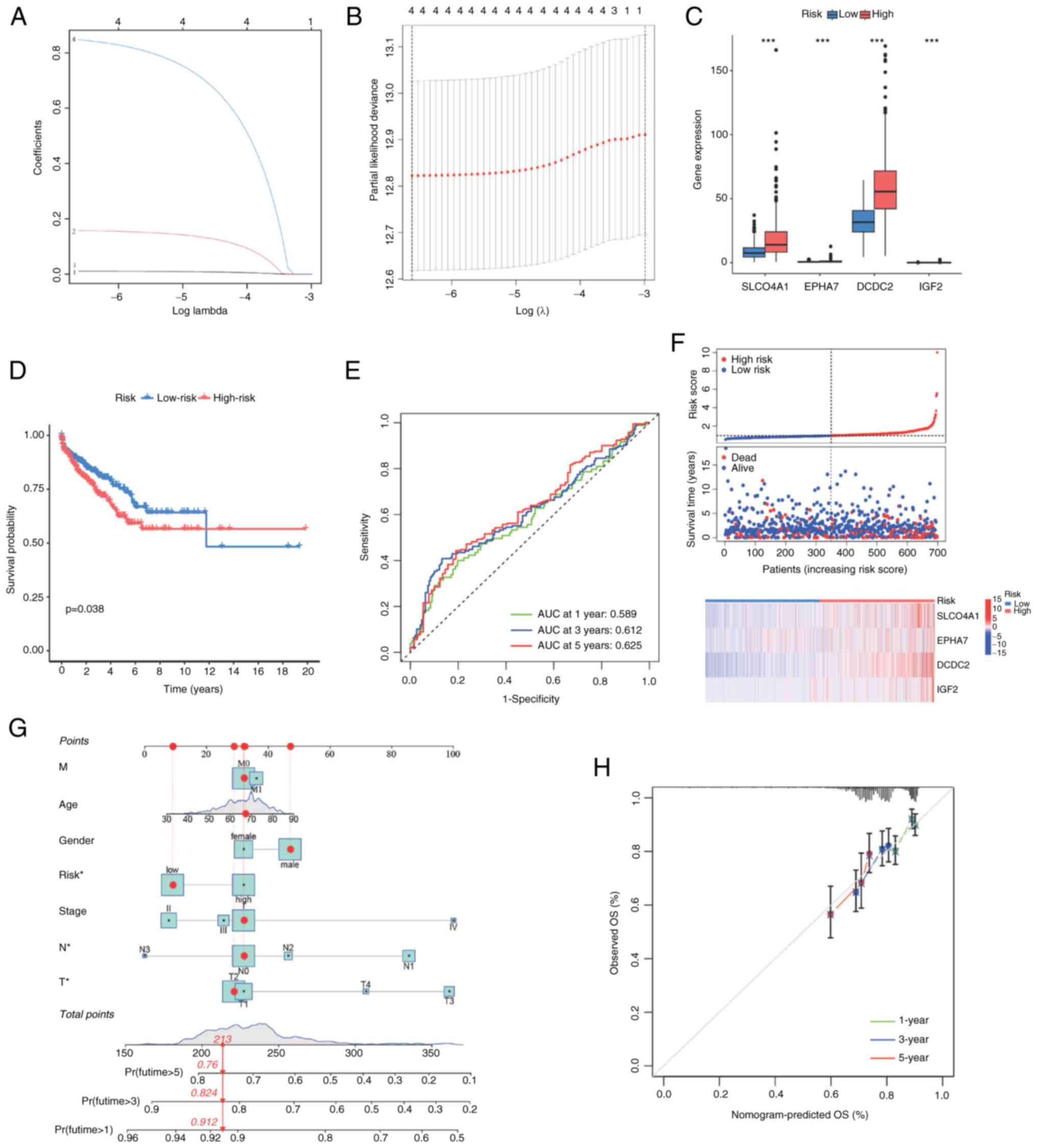 | Figure 2Constructing the risk score model.
(A) Coefficient profiles for the LASSO model. (B) Cross-validation
for parameter selection in the LASSO regression model. (C)
Expression levels of SLCO4A1, EPHA7, DCDC2 and IGF2 in the high
risk and low risk groups. (D) Survival analysis of the high risk
and low risk groups. (E) Receiver operating characteristic curve
prediction of survival at 1, 3 and 5 years. (F) Risk score and
survival time, and heatmap of SLCO4A1, EPHA7, DCDC2 and IGF2. (G)
Nomogram for predicting 1-, 3- and 5-year OS of patients with
NSCLC. (H) Calibration curve for the OS nomogram model in patients
with NSCLC. ***P<0.001. LASSO, Least Absolute
Shrinkage and Selection Operator; SLCO4A1, solute carrier organic
anion transporter family member 4A1; OS, overall survival; NSCLC,
non-small cell lung cancer. |
Enrichment analysis
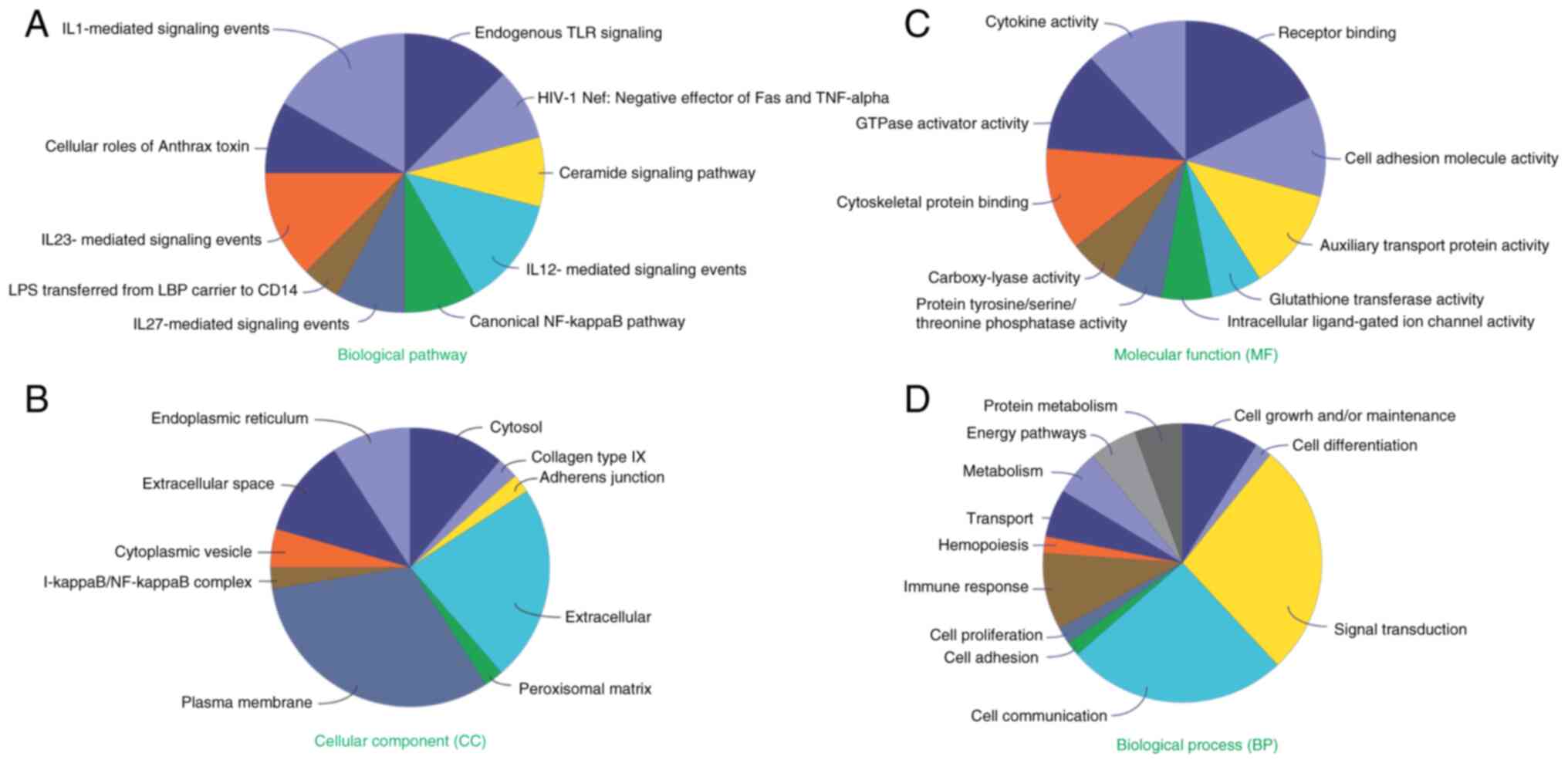
To gain deeper insights into the functional roles of
the 40 DEGs, enrichment analysis was performed in four major
domains: 'Biological pathway', 'cellular component', 'molecular
function' and 'biological process'. In terms of 'biological
pathway', these genes were primarily enriched in IL1-, IL-23- and
IL-12-mediated signaling events, and the canonical NF-κB pathway
(Fig. 3A). In 'cellular
component', these genes were mostly localized in the plasma
membrane, and extracellular region and cytosol (Fig. 3B). Regarding 'molecular function',
these genes were significantly enriched in 'cell adhesion molecule
activity', 'receptor binding', 'GTPase activator activity',
'cytokine activity' and 'cytoskeletal protein binding'. This
suggested their potential involvement in cellular migration and
deformation processes (Fig. 3C).
In terms of 'biological process', these genes were mainly enriched
in 'cell communication', 'signal transduction' and 'immune
response'. They were also revealed to play a role in cell
proliferation, cell adhesion and cell growth and maintenance
(Fig. 3D), implying their
association with cell proliferation and metastasis.
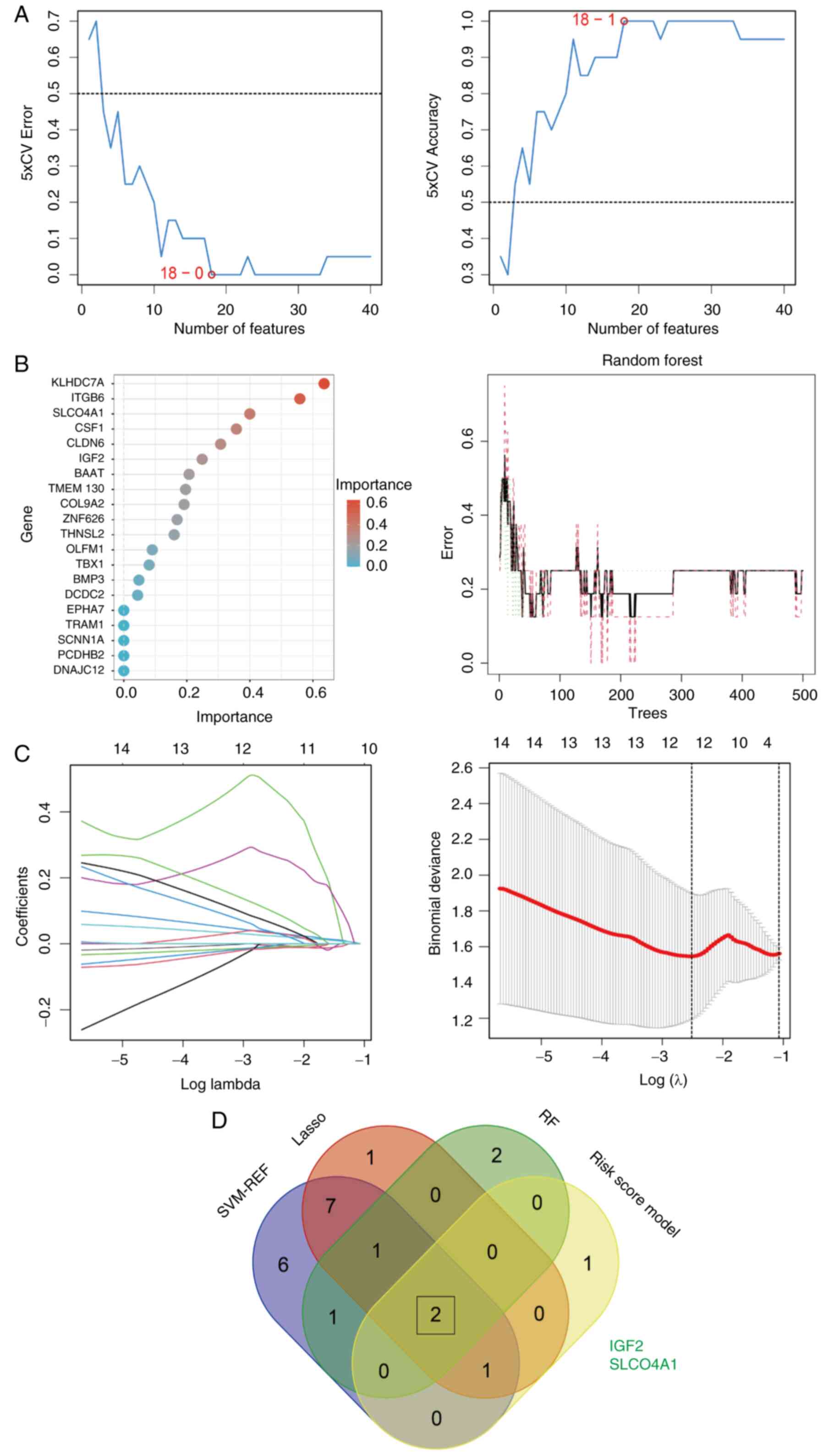
Obtaining hub genes
To identify the hub genes that influence early NSCLC
with lymph node metastasis among the 40 DEGs, three machine
learning algorithms were used for gene screening. The SVM-RFE
algorithm identified 18 key genes, including IL1B, CAMK2B,
CLDN6, C2CD4B, ARHGAP4, EPHA7, CMBL, RGS16, GSTO2, PPM1N, RASD1,
IGF2, CSF1, DNAJC12, SLCO4A1, TNF, CCL20 and OLFM1
(Fig. 4A). The RF algorithm
identified six key genes, including KLHDC7A, ITGB6, SLCO4A1,
CSF1, CLDN6 and IGF2 (Fig.
4B). The LASSO algorithm identified 12 key genes, including
RASD1, CMBL, IGF2, TBX1, EPHA7, ARHGAP4, CSF1, PPM1N, GSTO2,
C2CD4B, SLCO4A1 and CAMK2B (Fig. 4C). By incorporating the
aforementioned four genes that were used to construct the risk
score model and by applying a Venn diagram (Fig. 4D), two hub genes were obtained,
IGF2 and SLCO4A1.
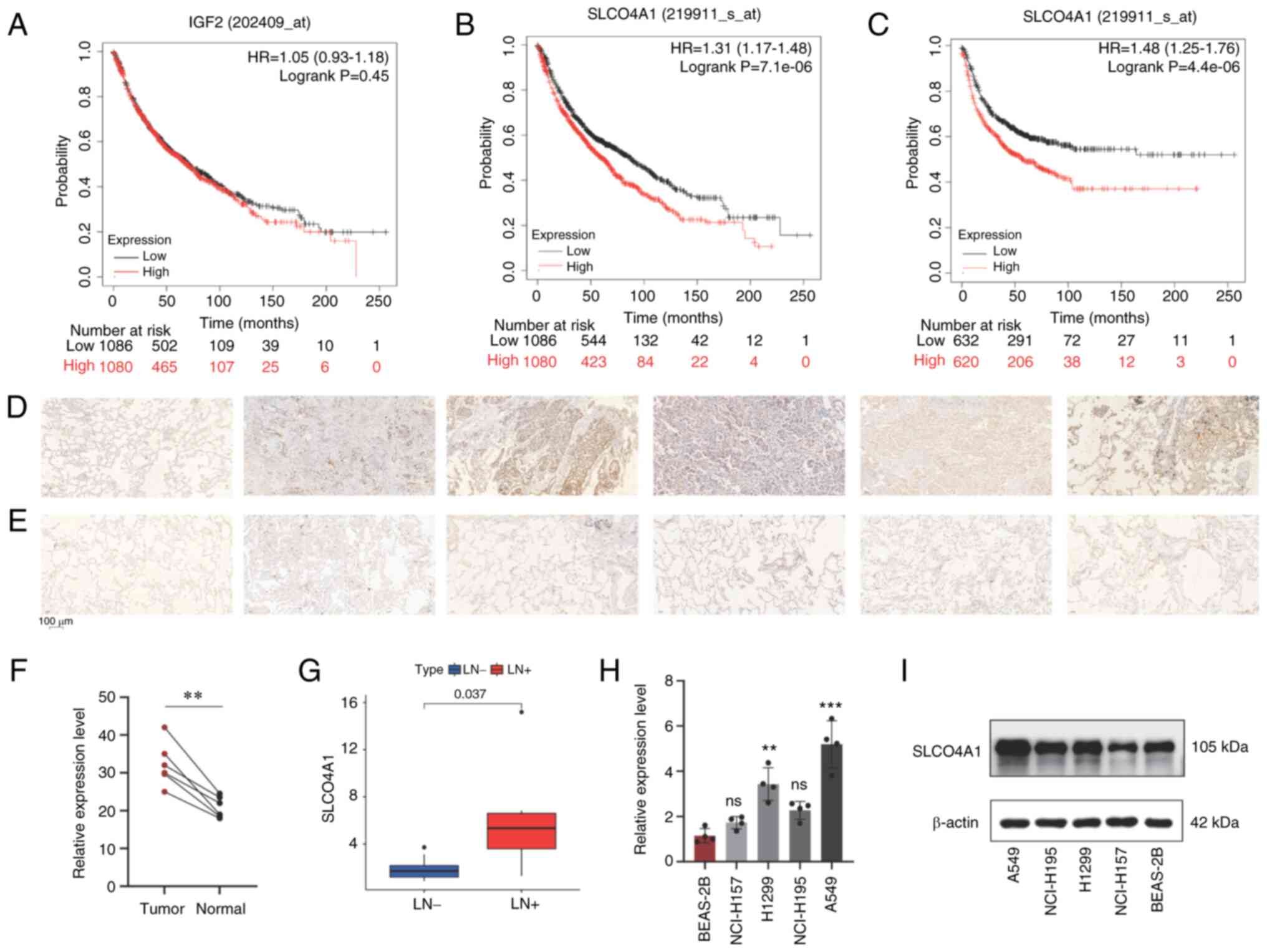
Prognosis of SLCO4A1 in patients with
NSCLC and expression level
Using Kaplan-Meier plotter, the impact of IGF2 and
SLCO4A1 on the survival of patients with NSCLC was investigated.
The results revealed that the expression level of IGF2 did not have
an effect on the survival of patients with NSCLC (Fig. 5A). Compared with the SLCO4A1 low
expression group, patients with high expression of SLCO4A1 had a
shorter OS time (Fig. 5B).
Additionally, the high expression group exhibited poorer first
progression (Fig. 5C).
Multi-cohort studies, including results from TCGA and Gene
Expression Omnibus databases, demonstrated that patients with NSCLC
and high expression of SLCO4A1 have a shorter survival time
(Fig. S1A). IHC results
identified that SLCO4A1 is significantly upregulated in NSCLC
tissues (Fig. 5D) compared with
normal lung tissues (Fig. 5E),
and has statistical significance (Fig. 5F). HPA database is a globally
renowned authoritative protein expression database. Dara clearly
indicated that SLCO4A1 can be used for prognosis, and high
expression is unfavorable in lung cancer. The HPA database showed
that patients with NSCLC and high expression of SLCO4A1 protein
have a poorer prognosis (Fig.
S1B). Next, through the analysis of the RNA sequencing data, it
was identified that SLCO4A1 is significantly upregulated in NSCLC
tissues with lymph node metastasis (Fig. 5G). Furthermore, RT-qPCR results
revealed a significant upregulation of SLCO4A1 in NSCLC cell lines
compared with normal alveolar epithelial cells (Fig. 5H). Western blotting results also
demonstrated similar findings (Fig.
5I). It is noteworthy that the upregulation of SLCO4A1 was most
pronounced in the A549 and H1299 cell lines, which served as the
selected cell lines for further investigations. In addition, using
TCGA-NSCLC expression and clinical data, univariate and
multivariate Cox regression analyses were performed, and the
results indicated that the expression level of SLCO4A1 can be an
independent prognostic factor affecting the survival of patients
with NSCLC (Table I).
 | Table IBy utilizing The Cancer Genome Atlas
data, it was demonstrated that solute carrier organic anion
transporter family member 4A1 can serve as an independent
prognostic factor influencing the survival of patients with
non-small cell lung cancer. |
Table I
By utilizing The Cancer Genome Atlas
data, it was demonstrated that solute carrier organic anion
transporter family member 4A1 can serve as an independent
prognostic factor influencing the survival of patients with
non-small cell lung cancer.
| Clinicopathological
characteristics | Number | Univariate analysis
| Multivariate
analysis
|
|---|
| Survival time [ x
(95% CI), days] | X2 | P-value |
WaldX2 | Hazard ratio (95%
CI) | P-value |
|---|
| Sex | | | 3.481 | 0.062 | | | |
| Female | 279 | 2510
(1405.098-3614.902) | | | | | |
| Male | 409 | 2811
(2166.342-3455.658) | | | | | |
| Age | | | 0.419 | 0.518 | | | |
| ≤60 | 179 | 3636
(1348.902-5923.098) | | | | | |
| >60 | 509 | 2524
(2003.433-3044.567) | | | | | |
| SLCO4A1 | | | 11.620 | 0.001 | | | |
| Low | 346 | 4299
(1615.569-6982.431) | | | | | |
| High | 342 | 2167
(1586.389-2747.611) | | | | 1.000 | |
| T stage | | | 1.767 | 0.622 | 8.760 | 1.606
(1.173-2.197) | 0.003 |
| T1 | 218 | 4299
(1316.619-7281.381) | | | | | |
| T2 | 369 | 2524
(1946.891-3101.109) | | | | | |
| T3 | 79 | 3636
(158.210-7113.790) | | | | | |
| T4 | 22 | - | | | | | |
| N stage | | | 0.619 | 0.892 | | | |
| N0 | 469 | 3636
(2222.747-5049.253) | | | | | |
| N1 | 142 | 2589
(1596.966-3581.034) | | | | | |
| N2 | 62 | 2510
(1317.644-3702.356) | | | | | |
| N3 | 15 | - | | | | | |
| M stage | | | 12.263 | <0.001 | | | |
| M0 | 506 | 2811
(1813.422-3808.578) | | | | 1.000 | |
| M1 | 182 | 1344
(0-2877.617) | | | 3.329 | 0.705
(0.484-1.026) | 0.068 |
| Stage | | | 55.740 | <0.001 | | | |
| I | 383 | 4299
(1476.846-7121.154) | | | | 1.000 | |
| II | 189 | 2065
(1146.779-2983.221) | | | 4.739 | 1.477
(1.040-2.098) | 0.029 |
| III | 98 | 2510
(1149.926-3870.074) | | | 0.105 | 1.086
(0.659-1.789) | 0.745 |
| IV | 18 | 669
(555.617-782.383) | | | 22.499 | 4.674
(2.471-8.839) | <0.001 |
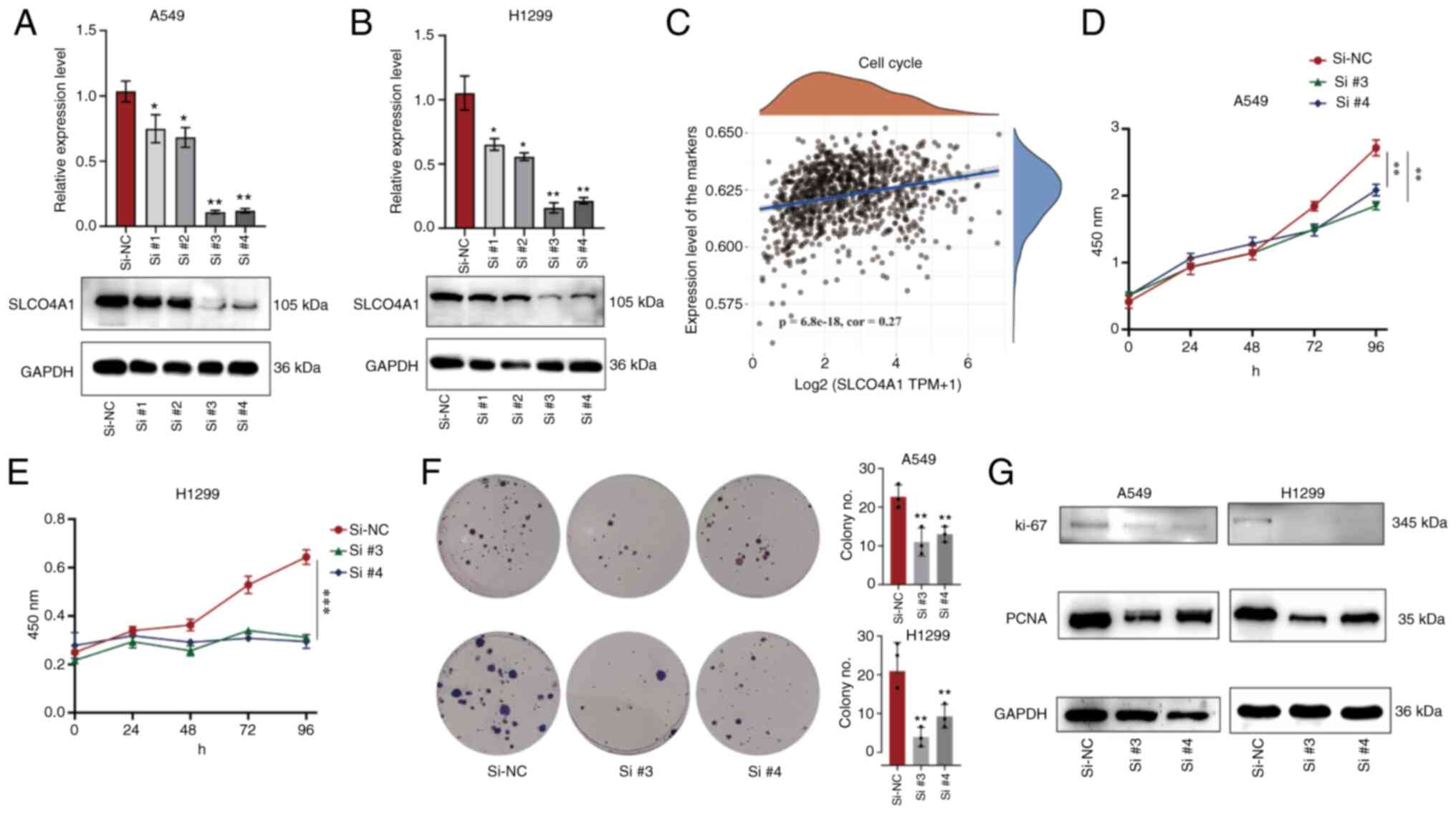
Knockdown SLCO4A1 inhibits the
proliferation of NSCLC cells
During the knockdown assay of SLCO4A1, Si#3 and Si#4
exhibited the most effective knockdown efficiency in A549 cells
(Fig. 6A) and H1299 cells
(Fig. 6B), rendering them the
focus of subsequent analysis. In addition, it was identified that
the expression level of SLCO4A1 in NSCLC was positively correlated
with the expression levels of cell cycle-related pathway molecules
(Fig. 6C), and this correlation
was statistically significant. The CCK-8 proliferation assay
demonstrated that knockdown of SLCO4A1 could inhibit the
proliferation ability of A549 cells (Fig. 6D) and that of H1299 cells
(Fig. 6E); the plate colony
formation assay revealed similar results (Fig. 6F). Meanwhile, knockdown of SLCO4A1
led to suppression of the protein expression levels of the cell
cycle and proliferation markers Ki-67 and PCNA compared with the
control group (Fig. 6G).
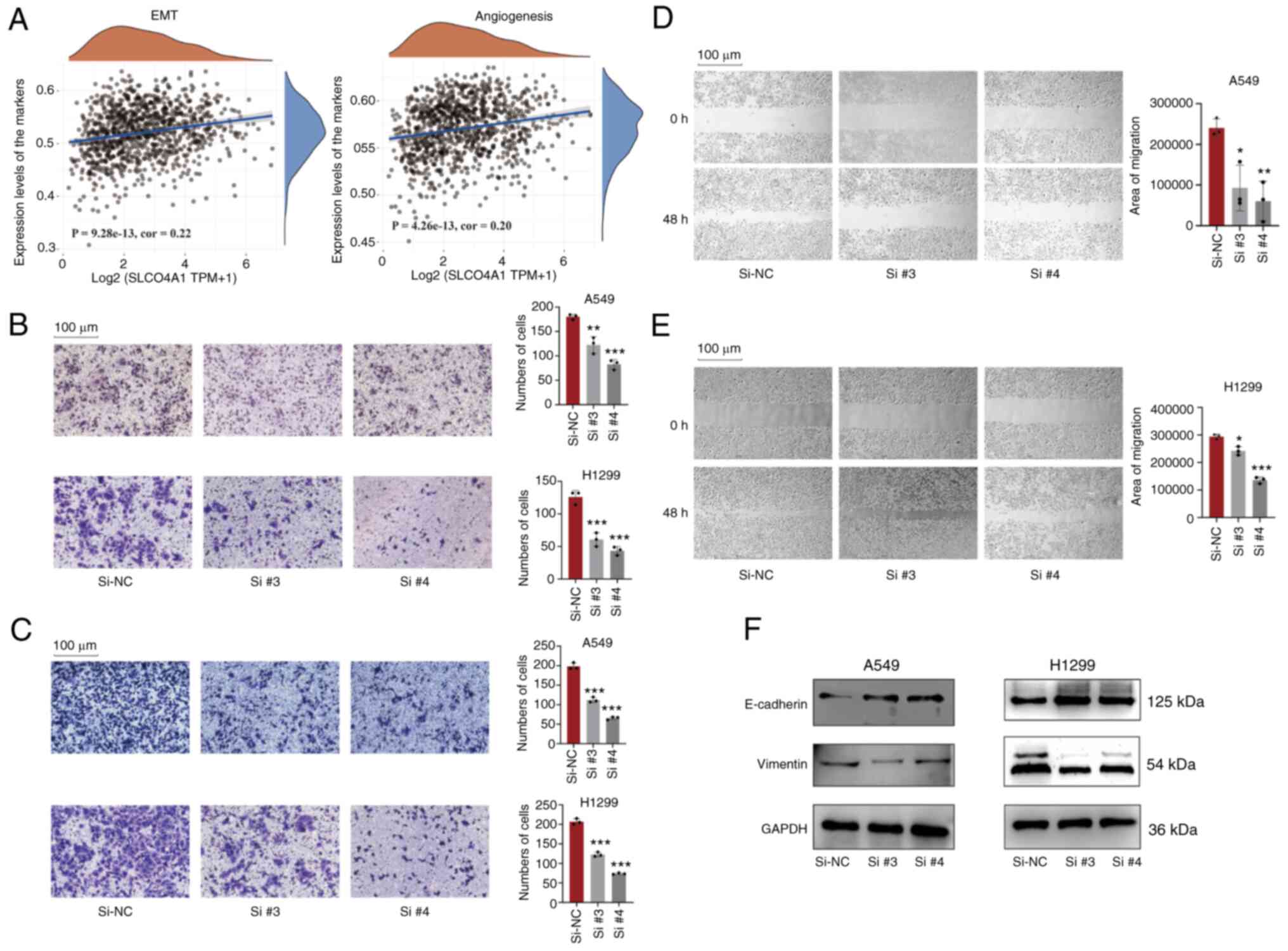
Knockdown of SLCO4A1 inhibits migration
and invasion of NSCLC cells
It was found that the expression level of SLCO4A1
was associated with metastasis phenotype markers, such as
epithelial-mesenchymal transition (EMT) and angiogenesis pathways
(Fig. 7A). The migration assay
(Fig. 7B) and the wound healing
assay (Fig. 7D and E) indicated
that the knockdown of SLCO4A1 significantly inhibited the migration
ability of A549 and H1299 cells. Similarly, the invasion assay
indicated that the knockdown of SLCO4A1 significantly inhibited the
invasion ability of A549 and H1299 cells (Fig. 7C). In addition, the knockdown of
SLCO4A1 affected the protein expression levels of EMT
pathway-related molecules, including E-cadherin and vimentin
(Fig. 7F). The expression level
of N-cadherin was not detected.
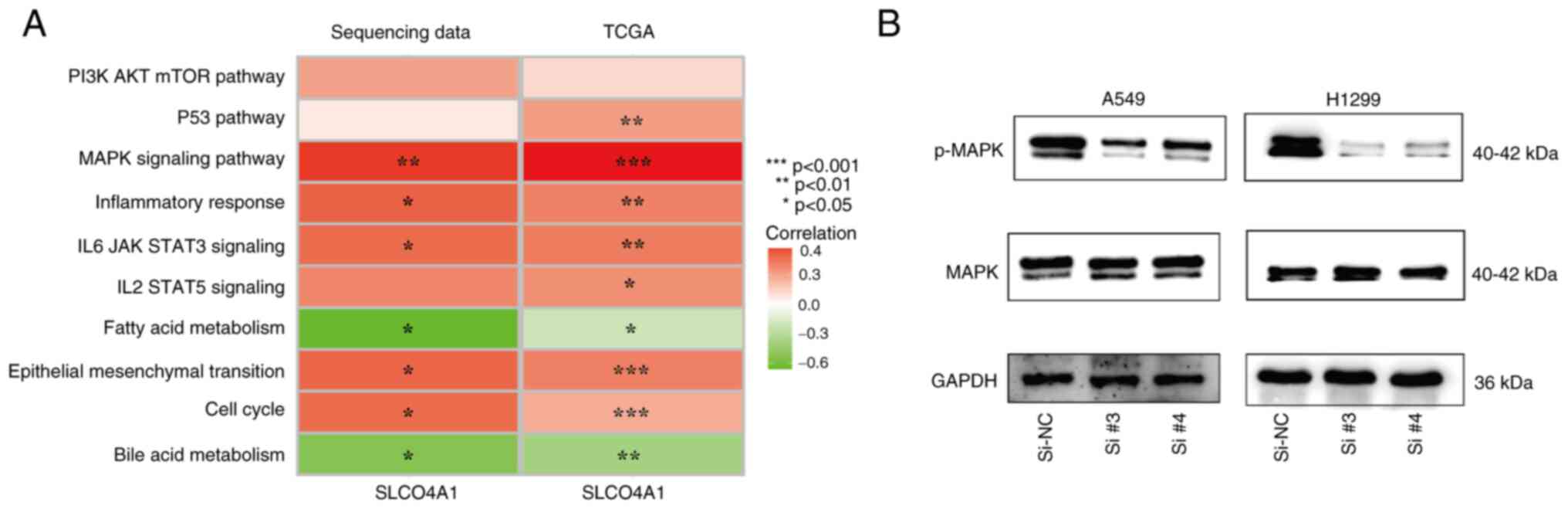
Exploration of the downstream mechanism
of SLCO4A1
Markers for different pathways were obtained from
the Molecular Signatures Database. To explore the downstream
mechanisms of SLCO4A1, single sample Gene Set Enrichment Analysis
(ssGSEA) was performed on the sequencing data. The results revealed
that the expression level of SLCO4A1 was positively associated with
the MAPK signaling pathway, inflammatory response, IL-6/JAK/STAT3
signaling and the EMT pathway, and these associations were
statistically significant. Conversely, SLCO4A1 demonstrated a
negative association with fatty acid metabolism and the bile acid
metabolism pathway. It is noteworthy that the association between
SLCO4A1 and the MAPK signaling pathway was the strongest and had
the most significant P-value (Fig.
8A). To enhance the credibility of the findings of the present
study, ssGSEA was also performed on TCGA-NSCLC transcriptomic data
and similar results were obtained. Western blotting identified that
the knockdown of SLCO4A1 significantly inhibited the expression of
the phosphorylated MAPK molecule (Fig. 8B), Therefore, there is compelling
evidence to support the notion that SLCO4A1 exerts an influence on
the proliferation and metastasis of NSCLC cells through the
modulation of molecules associated with the MAPK signaling
pathway.
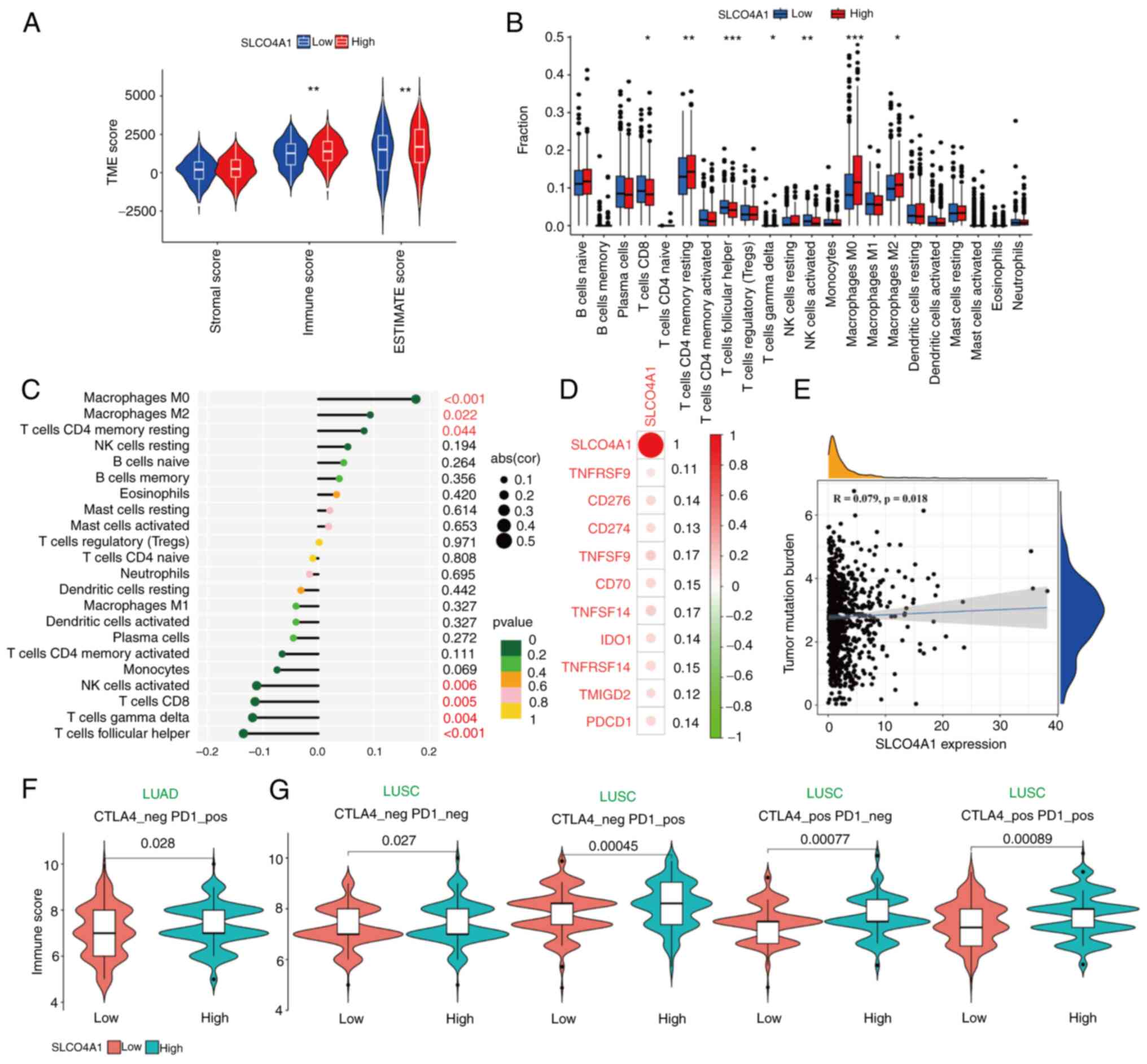
SLCO4A1 is involved in the regulation of the tumor
immune microenvironment in NSCLC and is predictive of immune
therapy response. Given the well-documented significance of the TME
in NSCLC tumorigenesis and drug resistance (18,19), an investigation was carried out to
determine the potential involvement of SLCO4A1 in the regulation of
the NSCLC TME. The ESTIMATE algorithm is a method for evaluating
tumor purity based on expression data. By calculating scores for
stromal and immune cells, a significant positive association was
observed between high expression of SLCO4A1 and immune cell
infiltration levels. This finding aligns with the results from
immune cell analysis using the CIBERSORTx algorithm, which also
demonstrated a positive association between high expression of
SLCO4A1 and varying levels of immune cell infiltration (Fig. 9A and B). Moreover, immune cell
correlation analysis revealed a positive association between
SLCO4A1 expression and macrophages M0 and M2, and resting
CD4+ memory T cells. By contrast, SLCO4A1 expression
exhibited a negative association with activated natural killer
cells, and CD8+, γδ and follicular helper T cells
(Fig. 9C). Immunotherapy
checkpoint inhibitor (ICB) correlation analysis demonstrated a
positive association between SLCO4A1 expression and ICBs including
TNFRSF9, CD276, CD274, TNFSF9, CD70, TNFSF14, IDO1, TNFRSF14,
TMIGD2 and PDCD1 (Fig.
9D). TMB is also an important indicator influencing the
efficacy of immunotherapy. Similarly, a positive correlation was
identified between SLCO4A1 expression and TMB (r=0.079; P=0.018;
Fig. 9E). Lastly, the
immunotherapy efficacy of SLCO4A1 expression in patients with NSCLC
was evaluated. In CTLA4+ and PD1+ patients
with LUAD, those with high SLCO4A1 expression exhibited higher
immune scores (Fig. 9F). In
patients with LUSC, regardless of CTLA4 and PD1 status (−/−,
+/+, +/− or −/+), patients with high SLCO4A1
expression demonstrated higher immune scores (Fig. 9G). These findings suggested that
patients with NSCLC and high SLCO4A1 expression may have improved
immunotherapy efficacy.
Discussion
Over an extended period of time, scientists have
persistently pursued investigations on tumor-specific biomarkers,
leading to remarkable breakthroughs. Through diverse methodologies,
numerous oncogenes and tumor suppressor genes have been unveiled,
thereby enabling their gradual integration into targeted treatments
for various types of cancer, including NSCLC. These advancements
have notably enhanced the prognosis and quality of life of patients
(20-22). At present, NSCLC persistently
retains the highest cancer-related mortality rate, with its
incidence and mortality rate showing a worrisome upward trend
(23,24). Lymph node metastasis significantly
affects the prognosis of NSCLC. It has been indicated that N0
patients have a median OS of 83.7 months, whereas N1 and N2
patients have only 48.0 and 37.9 months. Patients diagnosed with
occult N2 micro-metastases after surgery exhibit lower survival
rates (36%) and an elevated risk of disease recurrence (25).
At present, lymph node metastasis in NSCLC is
influenced by various factors, in which the size and extent of the
primary tumor (T stage) is one of the most influential factors.
Among the categories of T stage (occult, T1, T2, T3 and T4), T1 is
considered as early-stage lung cancer. However, it has been
suggested that T1 tumors are unlikely to have lymph node
metastasis, ignoring the occurrence of lymph node metastasis in T1
stage NSCLC patients. Furthermore, it is comparatively easy to
obtain specimens of T1 stage compared with other stages. Therefore,
in the present study, the NSCLC patients with T1 stage were
selected.
In the present study, the occurrence of lymph node
metastasis in T1 stage NSCLC patients served as the starting point
for the research. Sequencing was conducted on tissues from T1 stage
NSCLC cases and comprehensive analysis was performed, identifying
the hub gene SLCO4A1. Through experimental validation, SLCO4A1 was
established as an independent prognostic factor and the occurrence
of lymph node metastasis in early-stage NSCLC was confirmed. The
present study provided research value and evidence for the early
diagnosis and treatment of NSCLC. Inhibiting SLCO4A1expression at
an early stage will greatly improve the prognosis of patients with
NSCLC.
SLCO4A1 belongs to the organic anion transporter
family and exhibits apparent broad substrate specificity (10). Organic anion transporting peptides
are widely expressed across mammalian tissues and play a vital role
in facilitating the cellular uptake of diverse amphipathic organic
compounds. One plausible mechanism used to achieve this is through
their function as anion exchangers (8). Organic anion transporting peptides
are a well-established group of 12 transmembrane domain
glycoproteins; their expression is typically observed in various
organs. In terms of function, organic anion transporting peptides
play a crucial role in transporting a diverse array of amphipathic
organic compounds. These compounds include bile salts, steroids,
adrenal hormones and numerous organic drugs. Notably, this
transport mechanism is independent of sodium (10,26,27). It has been demonstrated that
abnormalities in SLCO4A1 have diverse impacts on various tumors.
Increased expression of SLCO4A1 in prostate and thyroid cancer has
been associated with an unfavorable prognosis (28,29). A previous study indicated that
SLCO4A1 may influence the accumulation of anticancer drugs in
specific cancer cells, thereby exerting an antitumor effect
(30). The presence of organic
anion transmembrane transporters facilitates the uptake of numerous
essential drugs and hormones, consequently affecting the
distribution of drugs and the concentration of drugs within cells
(31).
The presence of lymph node metastasis in most tumors
has been shown to have a marked effect on the survival time and
quality of life of patients. Takada et al (32) reported that early lymph node
metastasis in breast cancer is associated with the infiltration of
tumor-infiltrating lymphocytes, thereby promoting tumor
progression. In the current study, the sequencing data of early
lymph node metastasis NSCLC tissues were screened for SLCO4A1. The
expression level of SLCO4A1 was assessed in NSCLC tissues,
revealing that high expression of SLCO4A1 is associated with
reduced OS in patients with NSCLC. Experimental findings further
supported this by demonstrating that the knockdown of SLCO4A1
inhibits NSCLC cell proliferation, migration and invasion. ssGSEA
revealed that SLCO4A1 primarily affects downstream proliferation
and migration phenotypes through the MAPK signaling pathway.
Furthermore, the significant role of SLCO4A1 in tumor immunity and
immunotherapy was also uncovered.
In the field of cancer research, the role of SLCO4A1
in tumors has not been evaluated. The present study, to the best of
the authors' knowledge, is the first to report the influence of
SLCO4A1 on NSCLC progression and to identify SLCO4A1 as an
independent prognostic biomarker, providing novel molecules for
investigating NSCLC progression. Furthermore, conjoint analysis of
multiple databases was used for the first time to demonstrate that
SLCO4A1 can act as a prognostic biomarker in patients with NSCLC,
which renders the conclusion more authentic and reliable. Finally,
three machine learning algorithms, a new analytical method, were
used in the present study, which is another innovation point.
In the present study, T1 stage NSCLC patients served
as the starting point to investigate NSCLC progression mechanism,
breaking traditional understanding in the field. With the
advancement of imaging technology, increasing lung nodules are
being diagnosed, especially in T1 stage lesions, but traditional
views suggest that early-stage NSCLC are unlikely to have lymph
node metastasis, ignoring the occurrence of lymph node metastasis
in early-stage NSCLC patients. In the current study, focus was
addressed on T1 stage NSCLC patients, confirming that lymph node
metastasis can occur even in cases where the primary tumor is
extremely small. From the point of clinic application, evidence was
provided for the occurrence of lymph node metastasis in early-stage
patients. If patients undergo chest CT scans that detect
early-stage lesions (T1 stage) and exhibit increased SLCO4A1
levels, it will be easier to evaluate the metastasis and prognosis
of the patients and apply early intervention and treatment.
Supplementary Data
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The datasets generated and/or
analyzed during the current study are available in the Gene
Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68465;
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157010)
and The Cancer Genome Atlas repositories (https://portal.gdc.cancer.gov/). The RNA sequencing
data and clinical information for the 16 patient samples analyzed
in the present study can be found in (https://www.frontiersin.org/articles/10.3389/fcell.2021.739358/full#supplementary-material).
Authors' contributions
SHL, ZHL, ZYGu and LH designed and conducted the
experiments and wrote the manuscript. ZYGe and FL conducted the
western blot analysis. BW, YLS, YFX and BWL conducted the cell
proliferation assay and Transwell assay. YMX conducted qPCR and
western blot. YQ conceptualized and supervised all aspects of the
studies. All authors contributed to the article, read and approved
the final version of the manuscript. SHL and YQ confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved (approval no.
2019-KY-255) by the Ethics Committee of The First Affiliated
Hospital of Zhengzhou University (Zhengzhou, China). Written
informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present was supported by the Cultivation Project of Henan
Health Science and Technology Innovation Talents (grant no.
YXKC2022014, YQRC2023011) and the Henan Provincial Science and
Technology Development Project (grant nos. 222102310239,
LHGJ20210286 and LHGJ20230246).
References
|
1
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018.
|
|
2
|
Sher T, Dy GK and Adjei AA: Small cell
lung cancer. Mayo Clin Proc. 83:355–367. 2008.
|
|
3
|
Noguchi M, Morikawa A, Kawasaki M, Matsuno
Y, Yamada T, Hirohashi S, Kondo H and Shimosato Y: Small
adenocarcinoma of the lung. Histologic characteristics and
prognosis. Cancer. 75:2844–2852. 1995.
|
|
4
|
Xu W, Chen B, Ke D and Chen X: TRIM29
mediates lung squamous cell carcinoma cell metastasis by regulating
autophagic degradation of E-cadherin. Aging (Albany NY).
12:13488–13501. 2020.
|
|
5
|
Caiola E, Iezzi A, Tomanelli M, Bonaldi E,
Scagliotti A, Colombo M, Guffanti F, Micotti E, Garassino MC,
Minoli L, et al: LKB1 Deficiency Renders NSCLC Cells Sensitive to
ERK Inhibitors. J Thorac Oncol. 15:360–370. 2020.
|
|
6
|
Dong B, Wu C, Huang L and Qi Y:
Macrophage-Related SPP1 as a potential biomarker for early lymph
node metastasis in lung adenocarcinoma. Front Cell Dev Biol.
9:7393582021.
|
|
7
|
Martinez-Zayas G, Almeida FA, Yarmus L,
Steinfort D, Lazarus DR, Simoff MJ, Saettele T, Murgu S, Dammad T,
Duong DK, et al: Predicting lymph node metastasis in non-small cell
lung cancer: Prospective external and temporal validation of the
HAL and HOMER Models. Chest. 160:1108–1120. 2021.
|
|
8
|
Tamai I, Nezu J, Uchino H, Sai Y, Oku A,
Shimane M and Tsuji A: Molecular identification and
characterization of novel members of the human organic anion
transporter (OATP) family. Biochem Biophys Res Commun. 273:251–260.
2000.
|
|
9
|
Leuthold S, Hagenbuch B, Mohebbi N, Wagner
CA, Meier PJ and Stieger B: Mechanisms of pH-gradient driven
transport mediated by organic anion polypeptide transporters. Am J
Physiol Cell Physiol. 296:C570–C582. 2009.
|
|
10
|
Hagenbuch B and Meier PJ: Organic anion
transporting polypeptides of the OATP/SLC21 family: Phylogenetic
classification as OATP/SLCO superfamily, new nomenclature and
molecular/functional properties. Pflugers Arch. 447:653–665.
2004.
|
|
11
|
Brenner S, Klameth L, Riha J, Schölm M,
Hamilton G, Bajna E, Ausch C, Reiner A, Jäger W, Thalhammer T and
Buxhofer-Ausch V: Specific expression of OATPs in primary small
cell lung cancer (SCLC) cells as novel biomarkers for diagnosis and
therapy. Cancer Lett. 356(2 Pt B): 517–524. 2015.
|
|
12
|
Chen X, Yi G, Zhou Y, Hu W, Xi L, Han W
and Wang F: Prognostic biomarker SLCO4A1 is correlated with tumor
immune infiltration in colon adenocarcinoma. Mediators Inflamm.
2023:49264742023.
|
|
13
|
Hays A, Apte U and Hagenbuch B: Organic
anion transporting polypeptides expressed in pancreatic cancer may
serve as potential diagnostic markers and therapeutic targets for
early stage adenocarcinomas. Pharm Res. 30:2260–2269. 2013.
|
|
14
|
Benito-Martin A and Peinado H: FunRich
proteomics software analysis, let the fun begin! Proteomics.
15:2555–2556. 2015.
|
|
15
|
Ellis K, Kerr J, Godbole S, Lanckriet G,
Wing D and Marshall S: A random forest classifier for the
prediction of energy expenditure and type of physical activity from
wrist and hip accelerometers. Physiol Meas. 35:2191–2203. 2014.
|
|
16
|
Kang J, Choi YJ, Kim IK, Lee HS, Kim H,
Baik SH, Kim NK and Lee KY: LASSO-Based machine learning algorithm
for prediction of lymph node metastasis in T1 Colorectal Cancer.
Cancer Res Treat. 53:773–783. 2021.
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
18
|
Horvath L, Thienpont B, Zhao L, Wolf D and
Pircher A: Overcoming immunotherapy resistance in non-small cell
lung cancer (NSCLC)-novel approaches and future outlook. Mol
Cancer. 19:1412020.
|
|
19
|
Liu WJ, Du Y, Wen R, Yang M and Xu J: Drug
resistance to targeted therapeutic strategies in non-small cell
lung cancer. Pharmacol Ther. 206:1074382020.
|
|
20
|
Lee EY and Muller WJ: Oncogenes and tumor
suppressor genes. Cold Spring Harb Perspect Biol.
2:a0032362010.
|
|
21
|
Stella GM, Luisetti M, Pozzi E and
Comoglio PM: Oncogenes in non-small-cell lung cancer: Emerging
connections and novel therapeutic dynamics. Lancet Respir Med.
1:251–261. 2013.
|
|
22
|
Thomas A, Liu SV, Subramaniam DS and
Giaccone G: Refining the treatment of NSCLC according to
histological and molecular subtypes. Nat Rev Clin Oncol.
12:511–526. 2015.
|
|
23
|
Cai Z and Liu Q: Understanding the Global
Cancer Statistics 2018: Implications for cancer control. Sci China
Life Sci. 64:1017–1020. 2021.
|
|
24
|
Barta JA, Powell CA and Wisnivesky JP:
Global epidemiology of lung cancer. Ann Glob Health. 85:82019.
|
|
25
|
Bille A, Woo KM, Ahmad U, Rizk NP and
Jones DR: Incidence of occult pN2 disease following resection and
mediastinal lymph node dissection in clinical stage I lung cancer
patients. Eur J Cardiothorac Surg. 51:674–679. 2017.
|
|
26
|
Kobayashi D, Nozawa T, Imai K, Nezu J,
Tsuji A and Tamai I: Involvement of human organic anion
transporting polypeptide OATP-B (SLC21A9) in pH-dependent transport
across intestinal apical membrane. J Pharmacol Exp Ther.
306:703–708. 2003.
|
|
27
|
König J, Seithel A, Gradhand U and Fromm
MF: Pharmacogenomics of human OATP transporters. Naunyn
Schmiedebergs Arch Pharmacol. 372:432–443. 2006.
|
|
28
|
Wang XS, Wu SL, Peng Z and Zhu HH: SLCO4A1
is a Prognosis-Associated Biomarker Involved in Neutrophil-Mediated
Immunity in Thyroid Cancer. Int J Gen Med. 14:9615–9628. 2021.
|
|
29
|
Wright JL, Kwon EM, Ostrander EA,
Montgomery RB, Lin DW, Vessella R, Stanford JL and Mostaghel EA:
Expression of SLCO transport genes in castration-resistant prostate
cancer and impact of genetic variation in SLCO1B3 and SLCO2B1 on
prostate cancer outcomes. Cancer Epidemiol Biomarkers Prev.
20:619–627. 2011.
|
|
30
|
Buxhofer-Ausch V, Secky L, Wlcek K,
Svoboda M, Kounnis V, Briasoulis E, Tzakos AG, Jaeger W and
Thalhammer T: Tumor-specific expression of organic
anion-transporting polypeptides: Transporters as novel targets for
cancer therapy. J Drug Deliv. 2013:8635392013.
|
|
31
|
Stieger B and Hagenbuch B: Organic
anion-transporting polypeptides. Curr Top Membr. 73:205–532.
2014.
|
|
32
|
Takada K, Kashiwagi S, Asano Y, Goto W,
Kouhashi R, Yabumoto A, Morisaki T, Shibutani M, Takashima T,
Fujita H, et al: Prediction of lymph node metastasis by
tumor-infiltrating lymphocytes in T1 breast cancer. BMC Cancer.
20:5982020.
|