Introduction
Glioma is one of the most common types of brain
tumor, the main treatment methods for which include surgical
resection, radiotherapy and chemotherapy (1). However, because of tumor recurrence,
and resistance to radiotherapy and chemotherapy, most patients have
a poor prognosis, with a median overall survival time of 14.6-20.5
months worldwide (2). Glioma
recurrence and drug resistance are closely related to glioma stem
cells (GSCs) (3). Therefore, the
present study aimed to assess the characteristics and functions of
GSC, which may further reveal the causes of glioma occurrence,
development and recurrence, and provide novel ideas for
treatment.
N6-methyladenosine (m6A) is the most abundant
internal modification on RNA. It is dynamically and reversibly
regulated by methyltransferases and demethylases. m6A affects the
processing and metabolism of RNA, and regulates gene expression by
binding to m6A-reading proteins (4,5).
Abnormal m6A methylation on RNA is closely related to the
occurrence and development of glioma (6,7).
Cui et al (8) and
Visvanathan et al (9)
revealed that METTL3, METTL14 and FTO participate in regulating GSC
self-renewal and tumorigenesis. Recent research has shown that
knockdown of METTL3 reduces the self-renewal and proliferation of
GSCs (10,11), indicating that m6A methylation
serves a major role in regulating the self-renewal of GSCs. YTHDF1
is an m6A-binding protein that mediates m6A modification. In
addition, YTHDF1 maintains mRNA stability (12-14) and enhances mRNA translation
(15,16). Notably, YTHDF1 is significantly
upregulated in glioma tissues (17). Knockdown of YTHDF1 has been shown
to significantly inhibit glioblastoma (GBM) cell spheroidization,
invasion, and the expression of GSC markers CD133, NANOG and OCT4
(18). However, the molecular
mechanism underlying the regulatory effects of YTHDF1 on GSC
self-renewal and invasion remains unclear.
Long non-coding RNA (lncRNA) is a type of RNA that
contains >200 nucleotides, which has a major regulatory role in
the process of tumorigenesis through various molecular mechanisms
(19). m6A is distributed on mRNA
and widely distributed on non-coding RNAs, such as lncRNA (20). Notably, m6A affects the occurrence
and development of tumors by regulating the expression levels of
lncRNAs (21); for example, m6A
modification of lncDBET or DIAPH1-AS1 promotes the malignant
progression of bladder cancer (22) or nasopharyngeal carcinoma growth
and metastasis (23). LINC00900
is a newly discovered lncRNA. Wang et al (24) detected high expression levels of
LINC00900 in glioma, and revealed that LINC00900 is an m6A-related
prognostic lncRNA for primary GBM. YTHDF1 reads m6A motifs and
regulates the stability of the lncRNA THOR (25). Through preliminary research, it
was revealed that multiple m6A methylation sites were detected on
LINC00900 (SRAMP; http://www.cuilab.cn/sramp) and LINC00900 directly
binds to YTHDF1 (m6A2Target; http://rm2target.canceromics.org/#/home) (Fig. S1), suggesting that YTHDF1
upregulates LINC00900 expression by maintaining its stability, and
thus increases the viability and promotes self-renewal of GSCs.
The present study first investigated whether YTHDF1
regulates LINC00900 expression in an m6A-dependent manner, thereby
promoting GSC viability, invasion and self-renewal. In addition,
the mechanism by which LINC00900 upregulates STAT3 by sponging
microRNA (miR)-1205 was verified, further promoting GSC activity,
invasion, self-renewal and tumor growth. The present study aimed to
assess whether YTHDF1/LINC00900 could be considered a potential
therapeutic target for GBM.
Materials and methods
Clinical sample collection
Glioma tissues were collected from 20 patients
admitted to Wuxi No. 2 People's Hospital (Wuxi, China) for surgical
resection between January 2020 and January 2022. The 20 patients
included 12 men and 8 women, aged between 26 and 70 years old, with
an average age of 50.04±12.34 (mean ± SD) years. The patients did
not receive radiotherapy, chemotherapy, targeted therapy or any
other treatment that may affect the tumor, and did not have any
other tumors or systemic diseases. A total of 16 patients had GBM
[World Health Organization (WHO) grade IV] (26) and 4 patients had astrocytoma (WHO
grade III). Additionally, 20 tumor-adjacent normal brain tissues
(NBTs) were collected from the patients and were used as the
control. The present study was approved by the Ethics Committee of
Wuxi No. 2 People's Hospital (approval no. 2022-Y-116) and all
patients provided written informed consent.
Serum-free induction of GSCs
The following human GBM cell lines were used: A172
(cat. no. CL-0012; Wuhan Punosai Life Technology Co., Ltd.), SHG44
(cat. no. BH-0109468; Shanghai Bohu Biotechnology Co., Ltd.), U87
MG (cat. no. FH0162; GBM of unknown origin) and U251 (cat. no.
FH0159) (both from FuHeng Biology). A172, SHG44 and U87-MG cells
were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; cat. no. F0193; Shanghai Noning Biotechnology Co.,
Ltd.) and 100 U/ml penicillin/streptomycin (cat. no. G4003; Wuhan
Servicebio Technology, Co., Ltd.) at 37°C with 5% CO2.
U251 cells were cultured in DMEM/F12 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS and 100 U/ml
penicillin/streptomycin at 37°C with 5% CO2.
To induce differentiation, A172, SHG44, U87 MG and
U251 cells were resuspended in neurobasal medium (cat. no.
21103049; Gibco; Thermo Fisher Scientific, Inc.) containing 20
ng/ml bFGF (cat. no. C046), 20 ng/ml EGF (cat. no. C029), 1 mg/ml
heparin (cat. no. CK98) (all from Novoprotein Scientific, Inc.) and
100 U/ml penicillin/streptomycin. Cells were adjusted to
2×105/ml for culture and neurobasal medium was replaced
every 2-3 days. When the diameter of the glioma tumor spheres
reached 150-200 μm (after ~7 days), the glioma tumor spheres
were collected and digested in an Accutase solution (cat. no.
A6964; MilliporeSigma). A single cell suspension was prepared and
subcultured.
Cell transfection/infection
The miR-1205 mimics and mimics negative control (NC)
were synthesized by Shanghai GenePharma Co., Ltd., and 50 nM mimics
were mixed with Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) in OptiMEM (Gibco; Thermo Fisher
Scientific, Inc.) to form complexes at room temperature, which were
then transfected into the GSC-U87 and GSC-U251 cells at 37°C for a
2-3 day incubation, and RNA and protein extraction were performed
after 48 h.
LINC00900/YTHDF1 overexpression/short hairpin RNA
(shRNA) and control/shRNA-NC (sh-NC) lentiviruses
(Plvx-shRNA2-mcherry-T2A-Puro and pGMLVCMV-M
CS-3xflag-EF1-Zsgreen1-T2A-puro) were purchased from OLIGOBIO. To
obtain cells with overexpression or knock down of LINC00900 and
YTHDF1, GSC-U87 and GSC-U251 cells were infected with lentiviruses
(MOI=10) at 37°C for 48 h, and cultured in medium supplemented with
5 μg/ml puromycin after infection. The cells were collected
for further experiments at 2 weeks post-infection. The sequences of
the short hairpin (sh)RNAs and miRNA mimics are listed in Table I.
 | Table ISequences of the shRNAs and miRNA
mimics. |
Table I
Sequences of the shRNAs and miRNA
mimics.
| shRNA/mimics | Target sequence,
5′-3′ |
|---|
| sh-LINC00900-1 |
CCTGGCTAGTCAATCTTTATT |
| sh-LINC00900-2 |
GAGGGTCCAAGGTTGTTATTT |
| sh-LINC00900-3 |
GAGGTTACTGTGATGATTAAA |
| sh-NC2 |
CCTAAGGTTAAGTCGCCCTCGC |
| sh-YTHDF1-1 |
GTTCGTTACATCAGAAGGATA |
| sh-YTHDF1-2 |
CGCCGTCCATTGGATTTCCTT |
| sh-YTHDF1-3 |
AACCTCCATCTTCGACGACTT |
| sh-NC1 |
TTCTCCGAACGTGTCACGT |
| miR-1205
mimics |
UCUGCAGGGUUUGCUUUGAG |
| Mimics NC |
UCACAACCUCCUAGAAAGAGUAGA |
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from tissues or cells using
an RNA extraction kit (cat. no. R1200; Shanghai Yuduo Biotechnology
Co., Ltd.) in accordance with the manufacturer's instructions, and
the RNA concentration was measured using an ultraviolet
spectrophotometer. The extracted RNA was then reverse transcribed
into cDNA using a RT kit according to manufacturer's protocol (cat.
no. PC1703; Aidlab Biotechnologies Co., Ltd.), followed by qPCR
analysis using a SYBR Green PCR kit (cat. no. PC3302; Aidlab
Biotechnologies Co., Ltd.). The qPCR thermocycling conditions were
as follows: Denaturation at 94°C for 30 sec; followed by 30-35
cycles of denaturation at 94°C for 30 sec, annealing at 55-60°C for
30 sec, and extension at 72°C for 24 sec; and a final extension
step at 72°C for 5-10 min. The 2−ΔΔCq method (27) was used to analyze the relative
expression levels of the target gene, and GAPDH was used as
endogenous control. Primer sequences are listed in Table II.
 | Table IIPrimer sequences |
Table II
Primer sequences
| Gene | Primer sequence,
5′-3′ |
|---|
| LINC00900
(human) | F:
TGGTGTATGGATTGGATTTTGGTAG
R: CAGTGTTCTTGGTCGAGTTGCTCTT |
| STAT3 (human) | F:
TCAGTGAAAGCAGCAAAGAAGGAGG
R: AGGATAGAGATAGACCAGTGGAGAC |
| YTHDF1 (human) | F:
AAGTGGAAGGGGAAGTTTGATGT
R: ATGGAGGTTGTGTGCTTGTAGGA |
| GAPDH (human) | F:
GGAGCGAGATCCCTCCAAAAT
R: GGCTGTTGTCATACTTCTCATGG |
| miR-1205 | F:
ACACTCCAGCTGGGTCTGCAGGGTTTG
R: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTCAAAGC |
| U6 | F:
CTCGCTTCGGCAGCACA
R: AACGCTTCACGAATTTGCGT |
Western blotting
Total proteins were extracted from tissues or cells
using RIPA buffer (cat. no. P0013B; Beyotime Institute of
Biotechnology) in accordance with the manufacturer's instructions.
A BCA kit (cat. no. BL521A; Biosharp Life Sciences) was used to
measure the protein concentration. The proteins (25 μg) were
then separated by SDS-PAGE (Mini-PROTEAN® 3 cell;
Bio-Rad Laboratories, Inc.) on 8% gels and were transferred to a
PVDF membrane (cat. no. IPVH00010; MilliporeSigma). Subsequently,
5% dry skim milk was used to block the membranes at room
temperature for 3 h. The following primary antibodies were then
applied at 4°C overnight: Anti-YTHDF1 (1:1,000; cat. no. ab220162;
Abcam), anti-STAT3 (1:1,000; cat. no. ab68153; Abcam), anti-NANOG
(1:1,000; cat. no. ab109250; Abcam), anti-OCT4 (1:1,000; cat. no.
ab137427; Abcam) and anti-GAPDH (1:5,000; cat. no. 60004-1-Ig;
Proteintech Group, Inc.), followed by the secondary antibodies
(1:5,000; cat. nos. ZB2301 and ZB2305; OriGene Technologies, Inc.)
at room temperature for 2 h. Target protein bands were developed by
enhanced chemiluminescence (cat. no. ECL-0011; Beijing Dingguo
Changsheng Biotechnology Co., Ltd.) and detected using a
chemiluminescence imager (ChemiScope 5300 Pro; Shanghai Huanxi
Medical Equipment Co., Ltd.).
Methylated RNA immunoprecipitation
(MeRIP)-PCR
Total RNA was extracted from tissues, and the Magna
m6A RNA methylation immunoprecipitation kit (cat. no. C11051-1;
Guangzhou Ruibo Biotechnology Co., Ltd.) was used for MeRIP in
accordance with the manufacturer's instructions. The enriched RNA
was then analyzed by RT-qPCR as aforementioned.
Identification of GSCs by
immunofluorescence
The induced GSCs were collected, washed with PBS and
fixed with 4% paraformaldehyde for 30 min at 4°C. Then, the
anti-CD133 primary antibody (1:500; cat. no. 66666-1-Ig;
Proteintech Group, Inc.) was applied at 4°C overnight.
Subsequently, the corresponding Cy3-conjugated Affinipure Goat
Anti-Mouse IgG (H+L) secondary antibody (1:80; cat. no. SA00009-1;
Proteintech Group, Inc.) was applied at 37°C for 1 h and a Hoechst
33342 staining solution (10 μg/ml; cat. no. 875756-97-1;
MilliporeSigma) was applied for 10 min at room temperature. CD133
immunofluorescence was observed under a fluorescence microscope
(DP70; Olympus Corporation).
Cell viability assay
Suspensions of cells transfected with miRNA mimics
or infected with LINC00926/YTHDF1 overexpression/knockdown
lentiviruses were seeded in a 96-well plate at 2,000 cells/well.
After 24 h, 10 μl Cell Counting Kit (CCK)-8 solution (cat.
no. C0038; Beyotime Institute of Biotechnology) was added to each
well for 2 h at 37°C. A microplate reader (MK3; Thermo Fisher
Scientific, Inc.) was used to measure absorbance at 450 nm.
Transwell assay
GSCs (2×105) were collected and seeded in
the upper chamber of a 24-well Transwell system (pore size, 8
μm) precoated with Matrigel (cat. no. 354234; Shanghai
Yanhui Biotechnology Co., Ltd.) at 37°C for 1 h, and culture medium
containing 10% FBS was added to the lower chamber. Cells were
incubated for 24 h at 37°C, and then the medium in the upper
chamber was discarded. Cells were fixed with 4% 40 g/l
paraformaldehyde for 15 min at 4°C, washed with PBS and then
stained with 0.2% crystal violet at room temperature for 5 min.
Cells on the bottom membrane of the chamber were gently wiped with
a cotton swab. After drying, the chamber was observed under an
optical microscope (DFC300 FX; Leica Microsystems, Inc.) and cells
were counted.
Tumor sphere formation assay
A total of 2,000 GSCs/well were seeded in the well
of a 24-well plate in medium supplemented with 20 mg/ml B27 (cat.
no. 17504044; Gibco; Thermo Fisher Scientific, Inc.), 20 ng/ml EGF
and 20 ng/ml bFGF. EGF and bFGF were added to the medium twice a
week. After 2-3 weeks of culture, the number of spheres formed in
each well was observed under an optical microscope. Sphere
formation efficiency (SFE) was used to evaluate stem cell
self-renewal, as follows: SFE=number of cell spheres with a >75
μm diameter per well/total number of original seeded cells
in per well.
RNA pull-down assay
GSC-U87 and GSC-U251 cells were lysed using IP lysis
solution (cat. no. ZN2923; Beijing Baiao Leibo Technology Co.,
Ltd.) on ice for 15 min and the supernatant was collected by
centrifugation (4°C, 16,000 × g, 10 min). The biotin-labeled
LINC00900 probe and negative control probe were synthesized by
Guangzhou RiboBio Co., Ltd. Subsequently, 0.4 μg (50 pmol)
biotin-labeled RNA was incubated with 0.5 ml cell lysate at 4°C for
1 h. Then, 50 μl protein G agarose beads were added and
incubated at 4°C for 1 h. Agarose beads were centrifuged at 13,400
× g, for 30 sec at 4°C for precipitation and the supernatant was
collected into a clean tube. After washing, western blotting was
performed to detect the target protein as aforementioned.
RNA immunoprecipitation (RIP)
RIP was conducted using a RIP kit [cat. no.
GS-ET-006; Genseq; Hylegen Biotechnology (Shanghai) Co., Ltd.] in
accordance with the manufacturer's instructions. Cells were lysed
using 250 μl complete lysis buffer for 5 min, and 50
μl lysate was then incubated with IgG (1:100, cat. no.
36111ES10) or an anti-YTHDF1 antibody (1:500; cat. no. 31040ES50)
(both from Shanghai Yeasen Biotechnology Co., Ltd.). RNA was
extracted and purified, and RT-qPCR was used to measure target gene
expression, as aforementioned.
RNA stability analysis
Cells infected with LV-sh-NC and LV-sh-YTHDF1
lentiviruses [Hanheng Biotechnology (Shanghai) Co., Ltd.] were
seeded in 6-well plates. After 24 h of culture, the cells reached
70% confluence. The cells were then treated with 5 μg/ml
actinomycin D (cat. no. 1036; BioVision; Abcam) for 0, 2 or 4 h at
37°C, and the expression levels of LINC00900 were measured by
RT-qPCR as aforementioned.
Luciferase reporter assay
Prediction analysis (DIANA-LncBase v.2, http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=site/page&view=software;
and TargetScan, https://www.targetscan.org/vert_71/) revealed a
miR-1205-binding site in the LINC00900 transcript and STAT3 3′UTR.
LINC00900 and STAT3 3′-UTR wildtype (WT) and mutant type (MUT)
luciferase reporter genes (pmirGLO) were constructed by Beijing
Olinger Biotechnology Co., Ltd. and co-transfected with miR-1205
mimics or mimics NC into 293T cells (cat. no. JN15568; Shanghai
Jining Biotechnology Co., Ltd) by Lipofectamine 2,000. After 48 h,
the dual luciferase reporter gene detection kit (cat. no. E1910;
Promega Corporation) was used to detect the firefly luciferase
activity and Renilla luciferase activity. The ratio of
firefly luciferase to Renilla luciferase was calculated, and
the control group was set as 1 to calculate the relative luciferase
activity of different treatment groups.
Xenograft tumor model
Male nude specific pathogen-free mice (n=20; age,
3-4 weeks) were purchased from Beijing Huafukang Biotechnology Co.,
Ltd. The mice were maintained at 20-26°C and 50-60% relative
humidity. They were supplied with ad libitum access to
sufficient food/water and were kept under a 12-light/dark cycle.
The nude mice were divided into the following four groups
(n=5/group): LV-sh-NC1 + LV-sh-NC2, LV-sh-YTHDF1, LV-sh-LINC00900
and LV-sh-YTHDF1 + LV-sh-LINC00900. All mice were subcutaneously
injected with 5×106 GSC-U87 cells infected with
LINC00900/YTHDF1 knockdown lentiviruses. After 1 week, the tumor
growth curve and diameter were measured to calculate tumor volume
(1/2 × length × width2). All experiments were conducted
in accordance with protocols approved by the Medical Ethics
Committee of Wuxi No. 2 People's Hospital (Wuxi, China, approval
no. 2023-Y-200).
If the animals reached any of the following humane
endpoints they were sacrificed: The maximum tumor diameter exceeded
15 mm; animal weight loss reached ≤20%; the animal exhibited
cachexia or wasting syndrome; the size of the solid tumor exceeded
10% of body weight. The experimental duration of the present study
was 4 weeks, with the first week being used for adaptive feeding.
At the beginning of the second week, the experiment was initiated
(subcutaneous injection of GSCs to establish a xenograft tumor
animal model) and animal status was observed daily. Tumor volume
was measured from the first week after tumor cell injection, every
3 days for 2 weeks. Notably, none of the mice succumbed during the
experimental process. At the end of the experiment, the mice were
euthanized using CO2 (100% CO2, 20 l/min;
total chamber volume, 40 l; displacement rate, 50% l/min), and to
verify death, breathing was assessed. In the present study, all
nude mice showed a maximum weight loss of <15% before
euthanasia.
Immunohistochemistry
After tumor formation, animals were euthanized, and
tumor tissues were harvested. The tumor tissues were fixed in 4%
paraformaldehyde at 4°C for 24 h and 4-μm paraffin-embedded
sections were prepared. The paraffin-embedded sections were
dehydrated in a descending series of ethanol, followed by antigen
repair with sodium citrate at 95-100°C, and blocking with
H2O2 at room temperature for 5-10 min and
goat serum (cat. no. 16210064; Thermo Fisher Scientific, Inc.) at
room temperature for 10 min. The sections were then stained with an
anti Ki-67 antibody (1:100; cat. no. TA500265; OriGene
Technologies, Inc.) at 4°C overnight and with a biotin-labeled
secondary antibody (cat. no. SAP-9100; OriGene Technologies, Inc.)
at 37°C for 30 min. After washing, Ki-67 staining was observed
under an optical microscope, and image analysis software (ImageJ
v1.8.0; National Institutes of Health) was used to
semiquantitatively evaluate the intensity and degree of Ki-67
staining.
TUNEL assay
The aforementioned paraffin-embedded tumor tissue
sections were subjected to apoptosis detection using the TUNEL cell
apoptosis assay kit (cat. no. C1091; Beyotime Institute of
Biotechnology) according to the manufacturer's instructions. Under
an optical microscope, five fields of view were observed. And the
brown yellow staining represented apoptotic cells. Apoptosis rate
was calculated as number of positive cells/total number of
cells.
Statistical analysis
GraphPad Prism 5 software (Dotmatics) was used for
statistical analysis. Each experiment was performed at least in
triplicate, and the data are presented as the mean ± SD.
Comparisons between two groups were performed using a two-tailed
unpaired Student's t-test, and multiple groups were analyzed by
one-way ANOVA followed by Tukey's post hoc test or mixed ANOVA
followed by Bonferroni's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
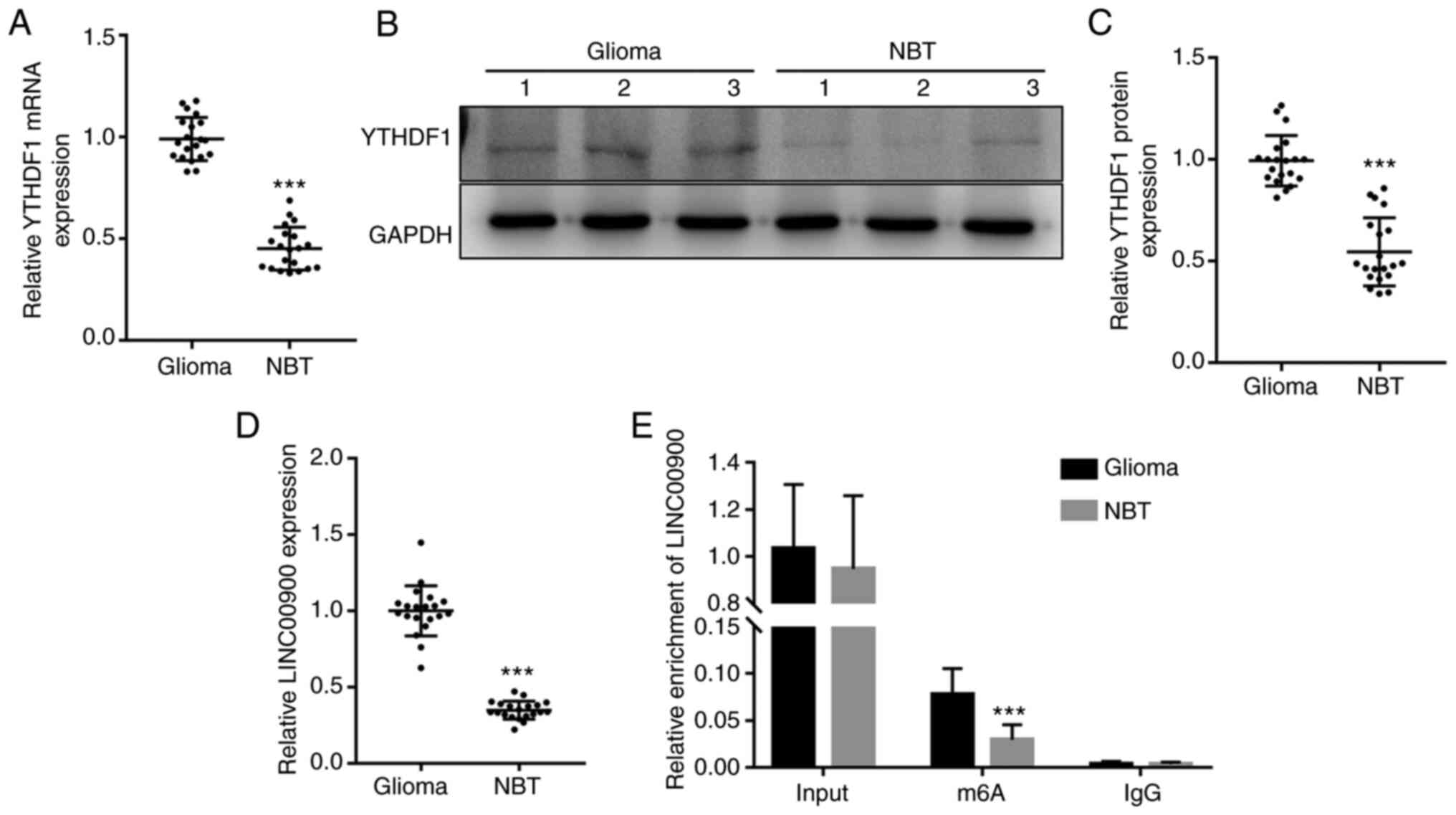
Abnormal expression of YTHDF1 and
LINC00900 in glioma tissue
RT-qPCR and western blotting were performed to
measure the expression levels of YTHDF1 in glioma tissue and NBT.
RT-qPCR showed that the mRNA expression levels of YTHDF1 were
significantly upregulated in glioma tissue (Fig. 1A). Consistent with the trend in
gene expression, the protein expression levels of YTHDF1 in glioma
tissues were also significantly higher than those in NBTs (Fig. 1B and C). Subsequently, RT-qPCR was
conducted to measure the expression levels of LINC00900 in glioma
tissue and NBT. As shown in Fig.
1D, the expression levels of LINC00900 in glioma tissues were
significantly higher than those in NBTs. A total of 16 patients had
GBM (WHO grade IV) (26) and 4
patients had astrocytoma (WHO grade III). As shown in Fig. S2, LINC00900 expression in the
glioma tissues of the two groups was significantly higher than that
in the adjacent tissues, and no significant difference was found in
the expression of LINC00900 between the two groups. Subsequently,
MeRIP-PCR analysis revealed that the LINC00900 transcript in glioma
tissues was enriched with m6A (Fig.
1E), suggesting that the high content of m6A in LINC00900
transcripts may be the cause of its abnormal expression in glioma
tissue.
Overexpression of YTHDF1 promotes
viability, invasion and self-renewal of GSCs
Four glioma cell lines (A172, U87, U251 and SHG44)
were selected to induce GSCs. GSCs were generated by serum-free
induction and, as shown in Fig.
S3A, stem cells were successfully induced and named GSC-U87 and
GSC-U251. GSC-A172 and GSC-SHG44 cells were also induced and are
shown in Fig. S4A. The GSCs were
identified by immunofluorescence (Figs. S3B and S4B), and western blotting
(Fig. S3C) of the stem cell
markers CD133, NANOG and OCT4. RT-qPCR and western blotting showed
that compared with that in A172, U87, U251 and SHG44 cells, YTHDF1
expression levels in GSC-A172, GSC-U87, GSC-U251 and GSC-SHG44
cells were significantly increased (Fig. 2A and B). U251 and U87 cells are
two commonly used glioma cell lines for in vitro
experiments, which were isolated from GBM and are often used as an
in vitro model of GBM. Therefore, these cell lines were
selected to study the effects of YTHDF1 on the viability, invasion
and self-renewal of GSCs. Subsequently, GSC-U87 and GSC-U251 cells
were infected with YTHDF1 overexpression or knockdown lentiviruses.
As shown in Fig. 2C-E, YTHDF1
overexpression significantly upregulated YTHDF1 mRNA and protein
expression levels, whereas YTHDF1 knockdown significantly
downregulated YTHDF1 mRNA and protein expression levels in GSC-U87
and GSC-U251 cells. Notably, shRNA-2 had the best knockdown
efficiency, and was therefore used in subsequent experiments. The
results of a CCK-8 assay showed that YTHDF1 overexpression
significantly increased GSC-U87 and GSC-U251 cell viability,
whereas knockdown of YTHDF1 significantly decreased GSC-U87 and
GSC-U251 cell viability (Fig.
2F). The Transwell assay showed that YTHDF1 overexpression
significantly increased the invasive ability of GSC-U87 and
GSC-U251 cells, whereas knockdown of YTHDF1 significantly decreased
the invasive ability of GSC-U87 and GSC-U251 cells (Fig. 2G and H). The tumor sphere
formation assay showed that YTHDF1 overexpression significantly
increased the tumor sphere formation ability of GSC-U87 and
GSC-U251 cells, whereas knockdown of YTHDF1 significantly decreased
the tumor sphere formation ability of GSC-U87 and GSC-U251 cells
(Fig. 2I and J). These results
suggested that YTHDF1 overexpression promoted the viability,
invasion and self-renewal of GSCs.
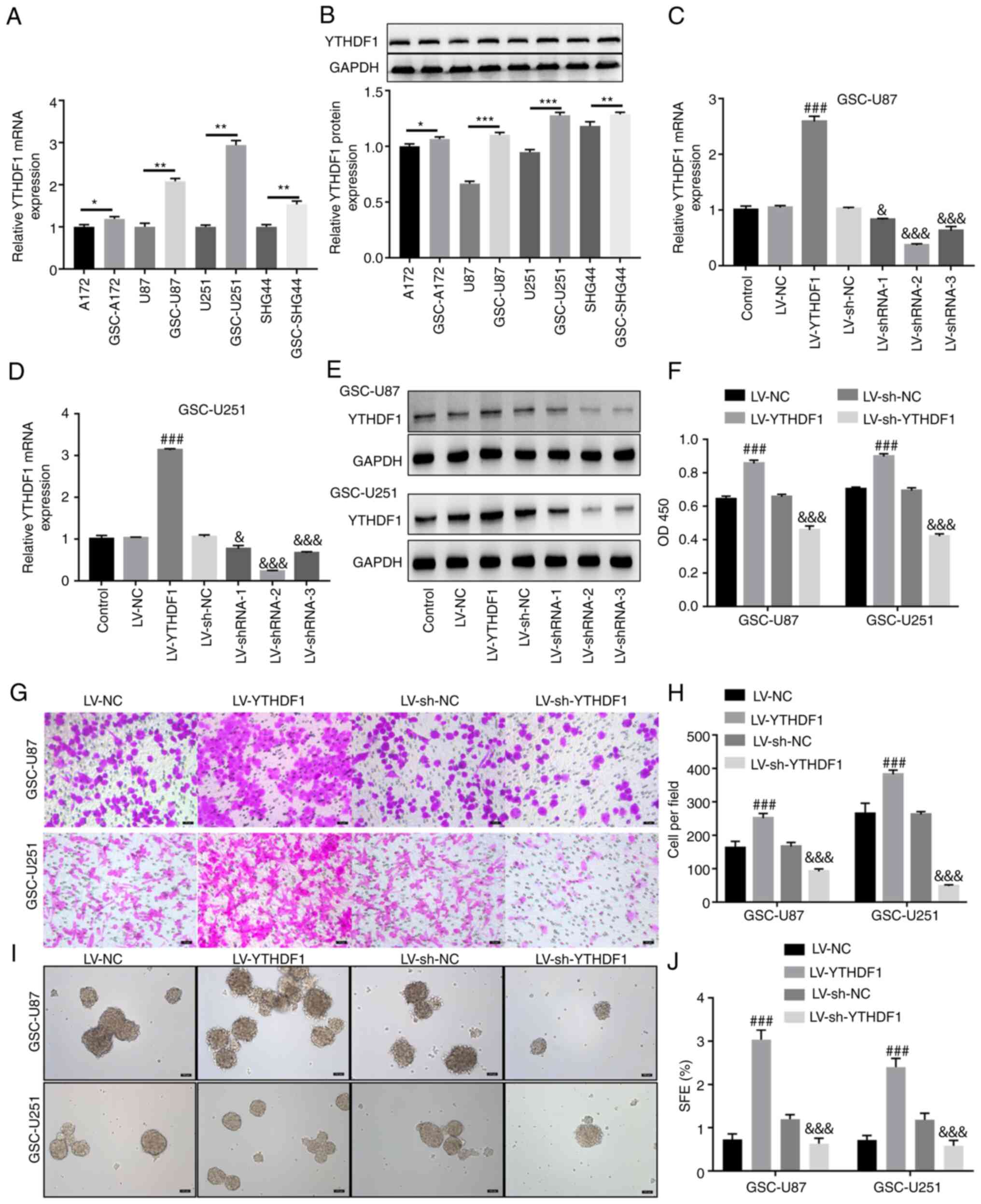 | Figure 2Overexpression of YTHDF1 promotes the
viability, invasion and self-renewal of GSCs. YTHDF1 overexpression
and LV-mediated knockdown were applied to GSC-U87 and GSC-U251
cells. (A) RT-qPCR and (B) western blotting was used to measure the
protein expression levels of YTHDF1 in U87, U251, GSC-U87 and
GSC-U251 cells. Overexpression and knockdown efficiencies assessed
by RT-qPCR in (C) GSC-U87 and (D) GSC-U251 cells. (E)
Overexpression and knockdown efficiencies assessed by western
blotting. (F) Cell Counting Kit-8 assay of GSC viability. (G)
Transwell assay of GSC invasion. Scale bars, 50 μm. (H)
Histogram of the number of invasive GSCs. (I) Tumor sphere
formation assay evaluated the self-renewal capability of GSCs.
Scale bars, 100 μm. (J) Histogram of the sphere formation
efficiency of GSCs. Data are presented as the mean ± SD.
*P<0.05 vs. A172, **P<0.01,
***P<0.001 vs. U87 or U251 or SHG44,
###P<0.001 vs. LV-NC; &P<0.05,
&&&P<0.001 vs. LV-sh-NC. GSCs, glioma
stem cells; LV, lentivirus; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; SFE, sphere formation efficiency;
sh, short hairpin. |
YTHDF1 upregulates LINC00900 expression
by enhancing its stability
To investigate whether YTHDF1 affects the expression
of LINC00900, YTHDF1 overexpression or knockdown was performed in
GSC cells. The results showed that the expression levels of
LINC00900 were increased in YTHDF1-overexpressing GSCs and were
decreased in YTHDF1-knockdown GSCs (Fig. 3A and B). RNA pull-down and RIP
assays were used to demonstrate the binding of the YTHDF1 protein
to LINC00900 in GSCs. As shown in Fig. 3C and D, the RNA pull-down assay
showed that the biotin-labeled LINC00900 (bio-LINC00900) pull-down
complex contained YTHDF1 protein. Moreover, in the RIP assay,
LINC00900 was detected in the YTHDF1 antibody pull-down complex. In
addition, there was a significant difference between anti-YTHDF1
and IgG groups, indicating that the enrichment of LINC00900 on the
YTHDF1 protein was significantly increased (Fig. 3E and F). These results identified
the binding relationship between LINC00900 and YTHDF1. Actinomycin
D can be absorbed by cells within a few minutes and preferentially
embedded into DNA sequences rich in GC, forming stable complexes
that inhibit the transcription process of all eukaryotic RNA
polymerase enzymes. After treating with actinomycin D for different
durations, the molecular level of target RNA (such as mRNA and
lncRNA) can be detected, the half-life of the target RNA can be
calculated, and its stability evaluated. Using actinomycin D, it
was revealed that LINC00900 expression was significantly reduced in
YTHDF1-knockdown GSCs (Fig. 3G and
H), indicating that YTHDF1 affected LINC00900 expression by
regulating its stability.
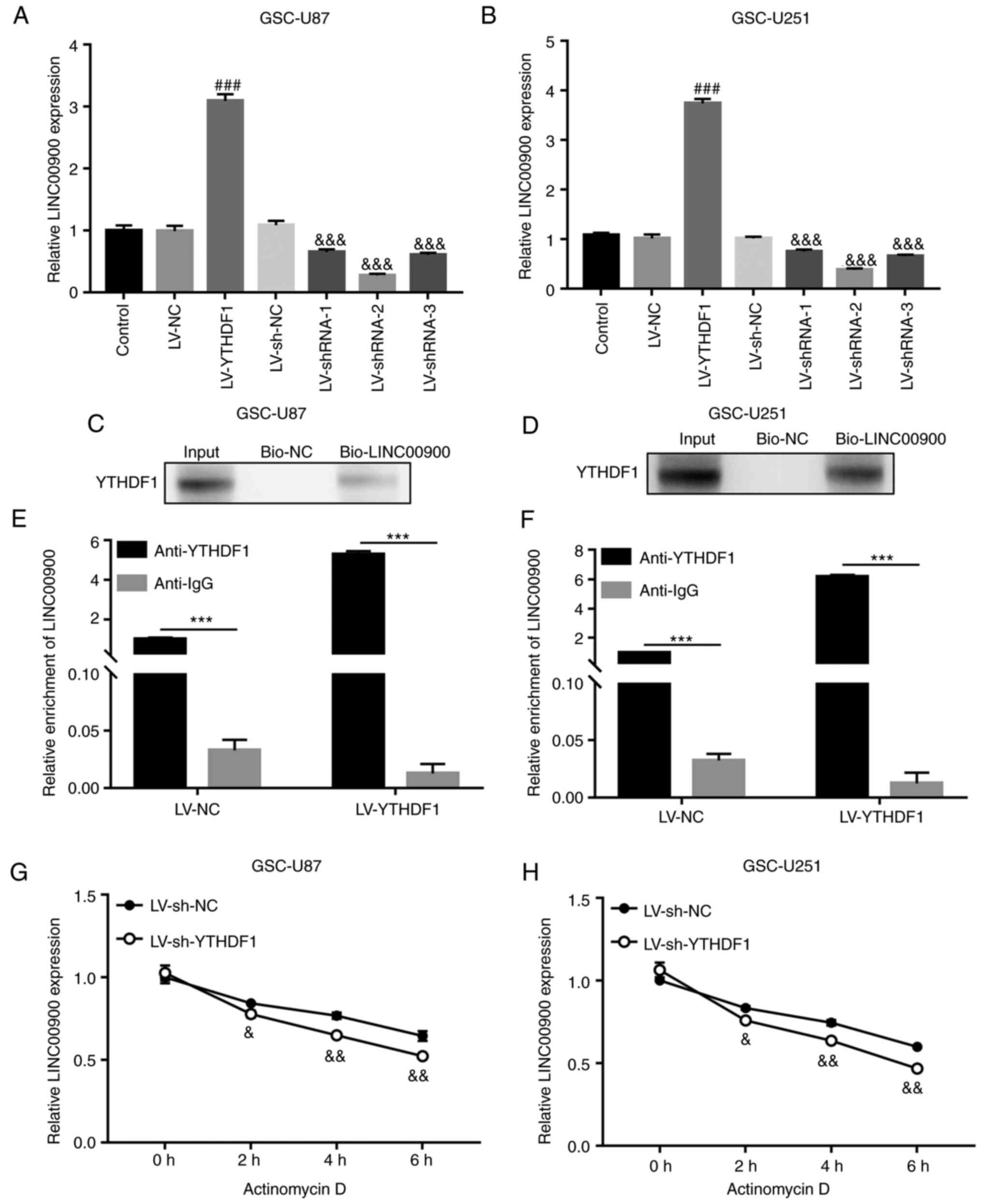 | Figure 3YTHDF1 upregulates LINC00900
expression by enhancing its stability. GSC-U87 and U251 cells were
infected with LVs for YTHDF1 overexpression or knockdown. LINC00900
expression was measured by RT-qPCR in (A) GSC-U87 And (B) GSC-U251
cells. RNA pull-down assays demonstrated direct binding of YTHDF1
protein to LINC00900 in (C) GSC-U87 and (D) GSC-U251 cells. RNA
immunoprecipitation assay indicated that YTHDF1 protein bound
directly to LINC00900 in (E) GSC-U87 and (F) GSC-U251 cells. Using
actinomycin D, the stability of LINC00900 was evaluated by RT-qPCR
in (G) GSC-U87 and (H) GSC-U251 cells. Data are presented as the
mean ± SD. ###P<0.001 vs. LV-NC,
***P<0.001, &P<0.05,
&&P<0.01,
&&&P<0.001 vs. LV-sh-NC. GSC, glioma stem
cell; LV, lentivirus; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; sh, short hairpin. |
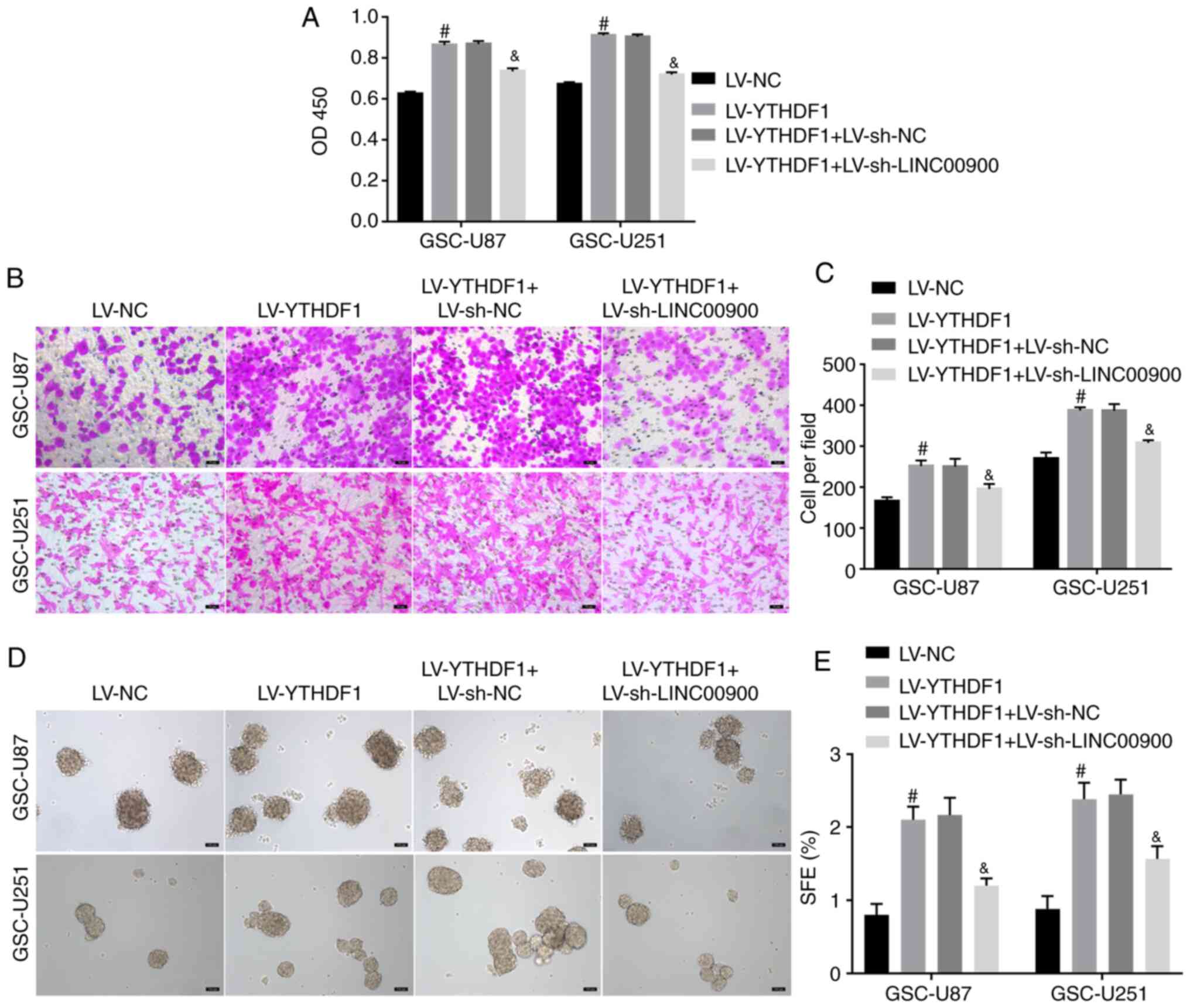
YTHDF1 promotes GSC viability, invasion
and self-renewal by upregulating LINC00900 expression
To further explore whether YTHDF1 regulates
LINC00900 expression and affects GSC functions, YTHDF1 was
overexpressed and LINC00900 was knocked down. Fig. S5A and B shows successful
knockdown of LINC00900 in GSC-U87 and GSC-U251 cells. The results
of a CCK-8 assay showed that YTHDF1 overexpression promoted
viability, whereas knockdown of LINC00900 reversed the enhancing
effect of YTHDF1 on viability (Fig.
4A). The Transwell assay showed that knockdown of LINC00900
reversed the effect of YTHDF1 on GSC invasion (Fig. 4B and C). The tumor sphere
formation assay showed that knockdown of LINC00900 reversed the
effect of YTHDF1 on GSC tumor sphere formation (Fig. 4D and E). These data indicated that
YTHDF1 promoted GSC viability, invasion and self-renewal by
upregulating LINC00900 expression.
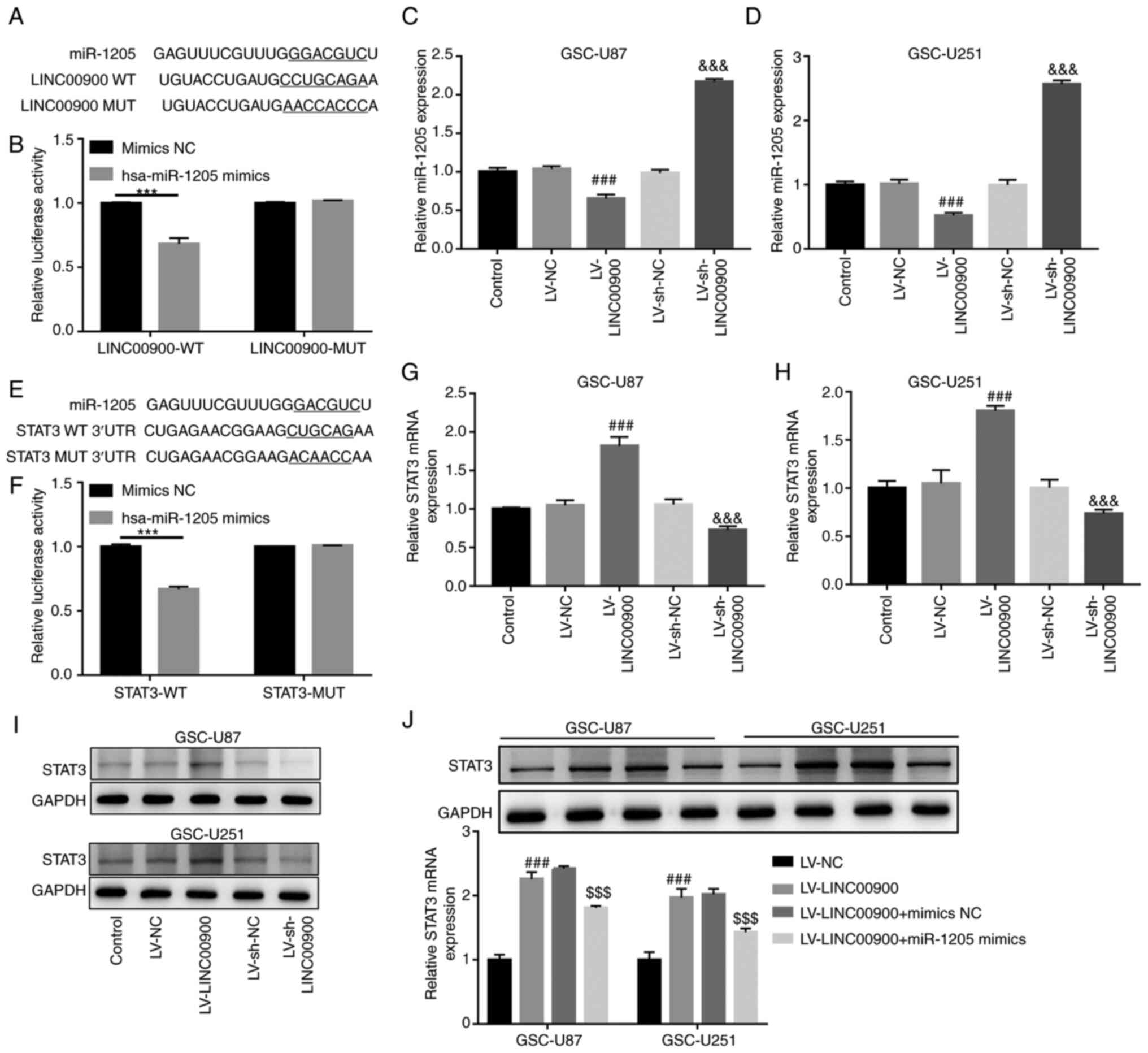
LINC00900 upregulates STAT3 expression by
sponging miR-1205
Prediction analysis using LncBase v.2 revealed a
miR-1205-binding site on the LINC00900 transcript (Fig. 5A). miR-1205 expression was
revealed to be significantly decreased in glioma tissue compared
with that in NBT (Fig. S6A). The
luciferase reporter assay confirmed the target binding relationship
between miR-1205 and LINC00900 (Fig.
5B). Subsequently, LINC00900 was successfully overexpressed in
GSC-U251 and GSC-U87 cells (Fig. S5A
and B). The results revealed that LINC00900 overexpression
significantly inhibited miR-1205 expression, whereas knockdown of
LINC00900 upregulated miR-1205 expression (Fig. 5C and D), implying that LINC00900
negatively regulated miR-1205 expression.
Prediction analysis using TargetScan also revealed a
miR-1205-binding site on the STAT3 3′-UTR (Fig. 5E). The mRNA and protein expression
levels of STAT3 were significantly increased in glioma tissue
compared with those in NBT (Fig. S6B
and C). The luciferase reporter assay confirmed the target
binding relationship between miR-1205 and the STAT3 3′-UTR
(Fig. 5F). In addition, it was
revealed that LINC00900 overexpression significantly upregulated
STAT3 expression, whereas knockdown of LINC00900 inhibited STAT3
expression (Fig. 5G-I), implying
that LINC00900 positively regulated STAT3 expression. Furthermore,
LINC00900 and miR-1205 were overexpressed in GSCs. Fig. S5C and D show successful
overexpression of miR-1205 in GSC-U87 and GSC-U251 cells. The
miR-1205 mimics reversed the upregulating effect of LINC00900 on
STAT3 expression (Fig. 5J), which
indicated that LINC00900 may upregulate STAT3 expression by
sponging miR-1205.
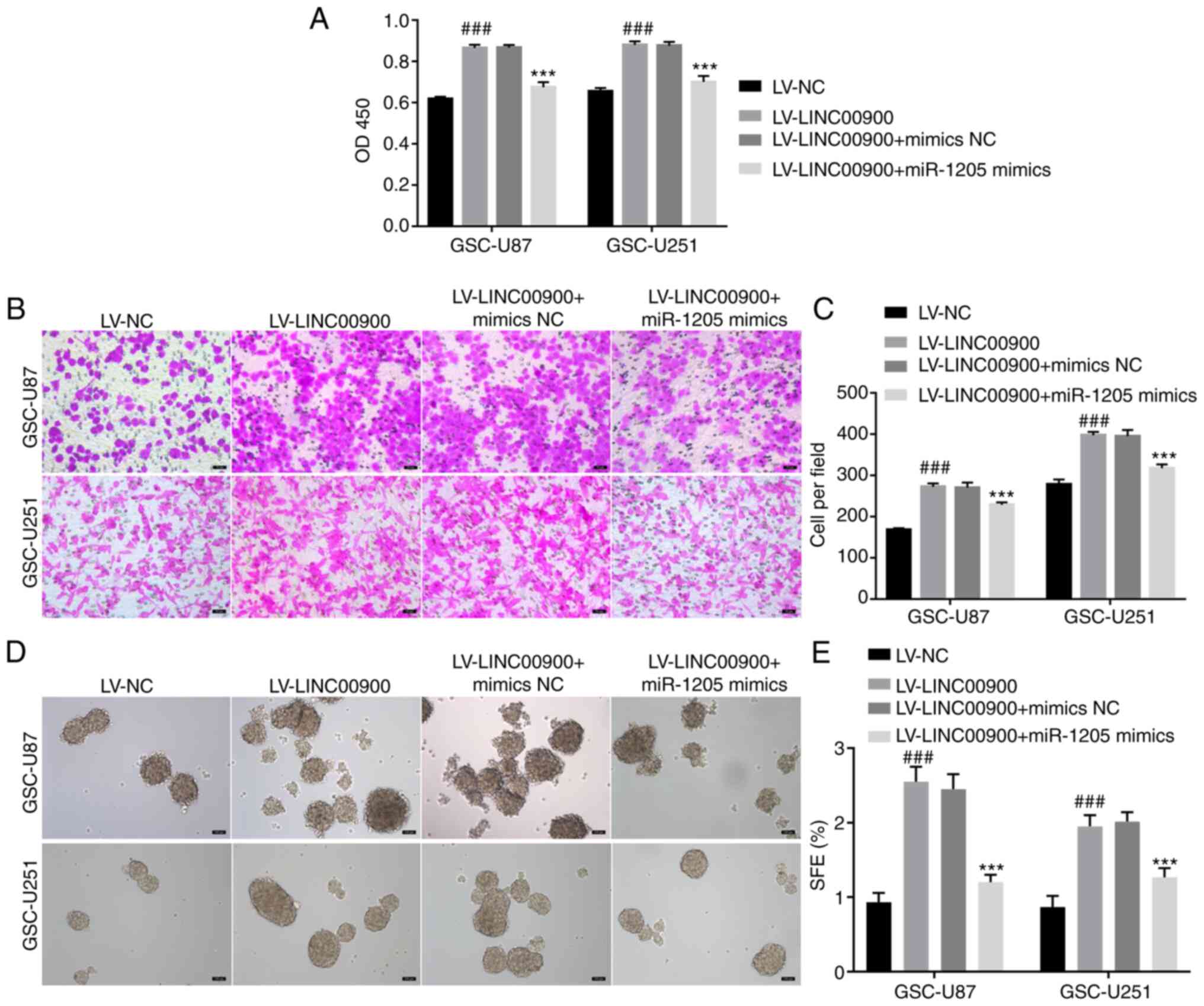
LINC00900 promotes GSC viability,
invasion, self-renewal and tumor growth by regulating the
miR-1205/STAT3 axis
To further explore whether LINC00900 regulates the
miR-1205/STAT3 axis and thus affects GSC functions, LINC00900 and
miR-1205 were overexpressed in GSCs. The transfection efficiency of
LINC00900 and miR-1205 is shown in Fig. S5. The results of a CCK-8 assay
showed that LINC00900 overexpression promoted viability, whereas
the miR-1205 mimics reversed the inducing effect of LINC00900 on
cell viability (Fig. 6A). The
Transwell assay showed that the miR-1205 mimics reversed the effect
of LINC00900 on GSC invasion (Fig. 6B
and C). The tumor sphere formation assay showed that the
miR-1205 mimics also reversed the effects of LINC00900 on GSC tumor
sphere formation (Fig. 6D and
E).
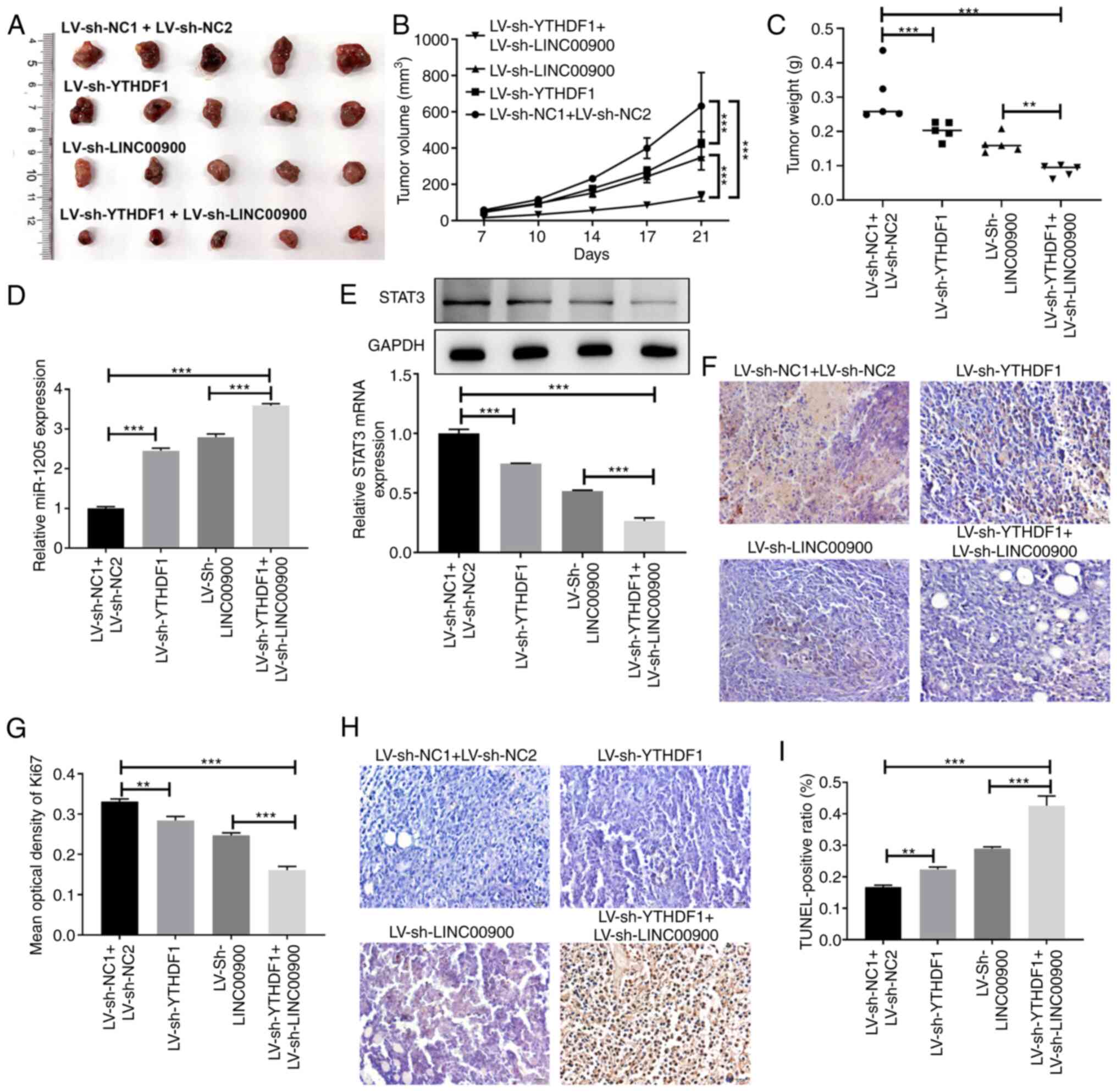
Analysis of xenografted tumors in nude mice showed
that inhibition of YTHDF1 or LINC00900 slowed tumor growth rate,
and the tumor volume and weight of the simultaneous YTHDF1 and
LINC00900 inhibition group were lower than those in the single
inhibition groups (Fig. 7A-C).
Compared with tumors in the LV-sh NC1 + LV-sh NC2 group mice,
miR-1205 expression was increased in tumors in the LV-sh-YTHDF1 and
LV-sh-LINC00900 group mice, whereas STAT3 expression was reduced
(Fig. 7D and E). Additionally,
compared with tumors in the LV-sh NC1 + LV-sh NC2 group mice, Ki-67
expression in tumors in the LV-sh-YTHDF1 and LV-sh-LINC00900 group
mice was decreased (Fig. 7F and
G), whereas the proportion of TUNEL-positive cells was
increased (Fig. 7H and I). These
findings indicated that LINC00900 promoted GSC viability, invasion,
self-renewal and tumor growth by regulating the miR-1205/STAT3
axis.
Discussion
Functional studies have shown that m6A
regulator-mediated m6A modification serves a major role in glioma
progression (28). For example,
the m6A methylase METTL3 enhances the stability of MALAT1 via m6A
modification and activates NF-κB to promote malignant progression
of IDH-wildtype glioma (29).
Furthermore, the m6A demethylase ALKBH5 maintains the
tumorigenicity of GBM stem-like cells by sustaining FOXM1
expression and cell proliferation (30). The m6A reader YTHDF2 facilitates
UBXN1 mRNA decay by recognizing METTL3-mediated m6A modification to
activate NF-κB and promote malignant progression of glioma
(31). The present study
investigated a glioma-related m6A regulator, the m6A-reading
protein YTHDF1, which has been shown to increase glioma cell
viability in vitro and promote tumor formation in
vivo (32). The present study
observed increased expression of YTHDF1 in glioma tissues, which
was consistent with a previous report (18). Moreover, it was revealed that the
expression of YTHDF1 in GSCs was significantly higher than that in
glioma cells. Overexpression of YTHDF1 promoted GSC viability,
invasion and self-renewal, whereas knockdown of YTHDF1 expression
inhibited GSC viability, invasion and self-renewal. These results
suggested that YTHDF1 serves a role in the self-renewal and
invasiveness of GSCs. The present study also assessed the molecular
mechanism underlying the regulatory effects of YTHDF1 on the
self-renewal and invasion of GSCs.
m6A affects the occurrence and development of tumors
by regulating lncRNA expression (21). YTHDF proteins, including YTHDF1,
YTHDF2 and YTHDF3, exhibit the same m6A-binding site in mRNA and
co-mediate degradation of mRNA containing the m6A modification
(33,34). YTHDF1 is highly expressed in GBM
cells (35), and it positively
regulates GBM cell proliferation, drug resistance and cancer stem
cell-like characteristics (18).
The present study showed that the expression levels of YTHDF1 in
GSC-U87 and GSC-U251 cells was higher than those U87 and U251
cells, and overexpression of YTHDF1 promoted the viability,
invasion and self-renewal of GSCs.
LINC00900 is a newly discovered lncRNA that is
highly expressed in glioma, and is an m6A-related prognostic lncRNA
of primary GBM (24). The present
study revealed that LINC00900 was upregulated in glioma tissue,
which is in agreement with a previous report (24). A total of 20 glioma tissues were
collected, among which 16 patients had GBM (WHO grade IV) and 4
patients had astrocytoma (WHO grade III). To clarify differential
expression of LINC00900 in tumor types, the 20 cancer tissues and
adjacent tissues were divided into two groups by the tumor type,
GBM and astrocytoma. In the future, the clinical sample size should
be expanded for further analysis.
The present study also revealed that LINC00900 had a
high level of m6A modification in glioma tissues. RIP and RNA
pull-down assays showed that LINC00900 directly bound to YTHDF1.
Actinomycin D treatment confirmed that YTHDF1 upregulated LINC00900
expression by maintaining its stability. Rescue experiments showed
that LINC00900 knockdown significantly reversed the promoting
effect of YTHDF1 on the viability, invasion and self-renewal of
GSCs. Collectively, these results indicated that YTHDF1 upregulated
the expression of LINC00900 by maintaining its stability, and
promoted the viability and self-renewal of GSCs.
LncRNAs serve a carcinogenic role by acting as
endogenous molecular sponges to compete with miRNA and affect
target mRNA (36). For example,
lncRNA BCYRN1 inhibits glioma tumorigenesis by competitively
binding to miR-619-5p to regulate CUEDC2 expression (37). LncRNA PVT1 facilitates
tumorigenesis and progression of glioma via regulation of the
miR-128-3p/GREM1 axis (38).
Notably, miR-1205 is downregulated in glioma tissues and cells, and
multiple circular (circ)RNAs [circ-UBAP2 (39), circMAN2B2 (40), circ_0001982 (41) and circ_0034642 (42)] function as sponges of miR-1205 to
promote glioma progression. Through bioinformatics prediction, a
miR-1205-binding site was identified on the LINC00900 transcript.
The results of a luciferase reporter assay confirmed the target
binding relationship between miR-1205 and LINC00900. Further rescue
experiments showed that miR-1205 mimics significantly reversed the
promoting effect of LINC00900 on the viability, invasion and
self-renewal of GSCs. Thus, these results indicated that LINC00900
promotes the viability and self-renewal of GSCs by regulating
miR-1205.
STAT3 is a member of the STAT protein family, and
has an important role in transmitting signals from cytokines and
growth factors (43). Activated
STAT3 is associated with tumor occurrence and cancer progression by
promoting the transcription of genes that control tumor cell
viability, and inhibit apoptosis, vascularization and the cell
cycle (44,45). For example, STAT3 promotes tumor
progression in glioma by inducing FOXP1 transcription (46), and STAT3 inhibition promotes GBM
cell apoptosis through MYC (47).
In the present study, bioinformatics prediction revealed a
miR-1205-binding site on the STAT3 3′-UTR. The results of a
luciferase reporter assay confirmed the target binding relationship
between miR-1205 and the STAT3 3′-UTR. Further rescue experiments
showed that the miR-1205 mimics significantly reversed the
enhancing effect of LINC00900 on STAT3 expression in GSCs. Taken
together, these results indicated that LINC00900 upregulates STAT3
by sponging miR-1205.
In conclusion, the present study indicated that
YTHDF1 promotes the viability, invasion and self-renewal of GSCs by
regulating the LINC00900/miR-1205/STAT3 axis. Therefore, the
YTHDF1/LINC00900/miR-1205/STAT3 axis may be a new therapeutic
target for glioma.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YuZ and YiZ designed the research and drafted the
manuscript. YuZ, YiZ, YaZ and ZL participated in the experiments.
XZ performed the data analysis and revised the manuscript. YuZ and
XZ confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The ethics approval for studies on human tissue was
approved by the Ethics Committee of Wuxi No. 2 People's Hospital
(approval no. 2022-Y-116). All patients signed an informed consent
form. The animal study was approved by the Medical Ethics Committee
of Wuxi No. 2 People's Hospital (approval no. 2023-Y-200).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Xu S, Tang L, Li X, Fan F and Liu Z:
Immunotherapy for glioma: Current management and future
application. Cancer Lett. 476:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rong L, Li N and Zhang Z: Emerging
therapies for glioblastoma: Current state and future directions. J
Exp Clin Cancer Res. 41:1422022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma Q, Long W, Xing C, Chu J, Luo M, Wang
HY, Liu Q and Wang RF: Cancer stem cells and immunosuppressive
microenvironment in glioma. Front Immunol. 9:29242018. View Article : Google Scholar
|
|
4
|
Zhao LY, Song J, Liu Y, Song CX and Yi C:
Mapping the epigenetic modifications of DNA and RNA. Protein Cell.
11:792–808. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible
m6A RNA methylation. Nat Rev Genet. 15:293–306. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Du J, Ji H, Ma S, Jin J, Mi S, Hou K, Dong
J, Wang F, Zhang C, Li Y and Hu S: m6A regulator-mediated
methylation modification patterns and characteristics of immunity
and stemness in low-grade glioma. Brief Bioinform. 22:bbab0132021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tao N, Wen T, Li T, Luan L, Pan H and Wang
Y: Interaction between m6A methylation and noncoding RNA in glioma.
Cell Death Discov. 8:2832022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m(6)A RNA methylation regulates
the self-renewal and tumorigenesis of glioblastoma stem cells. Cell
Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Visvanathan A, Patil V, Arora A, Hegde AS,
Arivazhagan A, Santosh V and Somasundaram K: Essential role of
METTL3-mediated m(6)A modification in glioma stem-like cells
maintenance and radioresistance. Oncogene. 37:522–533. 2018.
View Article : Google Scholar
|
|
10
|
Shi J, Zhang P, Dong X, Yuan J, Li Y, Li
S, Cheng S, Ping Y, Dai X and Dong J: METTL3 knockdown promotes
temozolomide sensitivity of glioma stem cells via decreasing MGMT
and APNG mRNA stability. Cell Death Discov. 9:222023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
You J, Tao B, Peng L, Peng T, He H, Zeng
S, Han J, Chen L, Xia X, Yang X and Zhong C: Transcription factor
YY1 mediates self-renewal of glioblastoma stem cells through
regulation of the SENP1/METTL3/MYC axis. Cancer Gene Ther.
30:683–693. 2023. View Article : Google Scholar
|
|
12
|
Sun Y, Dong D, Xia Y, Hao L, Wang W and
Zhao C: YTHDF1 promotes breast cancer cell growth, DNA damage
repair and chemoresistance. Cell Death Dis. 13:2302022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He Y, Wang W, Xu X, Yang B, Yu X, Wu Y and
Wang J: Mettl3 inhibits the apoptosis and autophagy of chondrocytes
in inflammation through mediating Bcl2 stability via
Ythdf1-mediated mA modification. Bone. 154:1161822022. View Article : Google Scholar
|
|
14
|
Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen
X, Sun L, Zhan S, Chen L, Cheng C, et al: RNA m A methylation
regulates sorafenib resistance in liver cancer through
FOXO3-mediated autophagy. EMBO J. 39:e1031812020. View Article : Google Scholar
|
|
15
|
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y,
Cheng C, Li L, Pi J, Si Y, et al: The m6A reader YTHDF1 promotes
ovarian cancer progression via augmenting EIF3C translation.
Nucleic Acids Res. 48:3816–3831. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang S, Gao S, Zeng Y, Zhu L, Mo Y, Wong
CC, Bao Y, Su P, Zhai J, Wang L, et al: N6-Methyladenosine reader
YTHDF1 promotes ARHGEF2 translation and RhoA signaling in
colorectal cancer. Gastroenterology. 162:1183–1196. 2022.
View Article : Google Scholar
|
|
17
|
Deng X, Sun X, Hu Z, Wu Y, Zhou C, Sun J,
Gao X and Huang Y: Exploring the role of m6A methylation regulators
in glioblastoma multiforme and their impact on the tumor immune
microenvironment. FASEB J. 37:e231552023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yarmishyn AA, Yang YP, Lu KH, Chen YC,
Chien Y, Chou SJ, Tsai PH, Ma HI, Chien CS, Chen MT and Wang ML:
Musashi-1 promotes cancer stem cell properties of glioblastoma
cells via upregulation of YTHDF1. Cancer Cell Int. 20:5972020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang MC, Ni JJ, Cui WY, Wang BY and Zhuo
W: Emerging roles of lncRNA in cancer and therapeutic
opportunities. Am J Cancer Res. 9:1354–1366. 2019.PubMed/NCBI
|
|
20
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang H, Weng H and Chen J: M(6)a
modification in coding and non-coding RNAs: Roles and therapeutic
implications in cancer. Cancer Cell. 37:270–288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu P, Fan B, Othmane B, Hu J, Li H, Cui
Y, Ou Z, Chen J and Zu X: m6A-induced lncDBET promotes the
malignant progression of bladder cancer through FABP5-mediated
lipid metabolism. Theranostics. 12:6291–6307. 2022. View Article : Google Scholar :
|
|
23
|
Li ZX, Zheng ZQ, Yang PY, Lin L, Zhou GQ,
Lv JW, Zhang LL, Chen F, Li YQ, Wu CF, et al: WTAP-mediated
m6A modification of lncRNA DIAPH1-AS1 enhances its
stability to facilitate nasopharyngeal carcinoma growth and
metastasis. Cell Death Differ. 29:1137–1151. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang W, Li J, Lin F, Guo J and Zhao J:
Identification of N(6)-methyladenosine-related lncRNAs for patients
with primary glioblastoma. Neurosurg Rev. 44:463–470. 2021.
View Article : Google Scholar
|
|
25
|
Liu H, Xu Y, Yao B, Sui T, Lai L and Li Z:
A novel N6-methyladenosine (m6A)-dependent fate decision for the
lncRNA THOR. Cell Death Dis. 11:6132020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu W, Klockow JL, Zhang M, Lafortune F,
Chang E, Jin L, Wu Y and Daldrup-Link HE: Glioblastoma multiforme
(GBM): An overview of current therapies and mechanisms of
resistance. Pharmacol Res. 171:1057802021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Zhang G, Zheng P, Lv Y, Shi Z and Shi F:
m6A regulatory gene-mediated methylation modification in glioma
survival prediction. Front Genet. 13:8737642022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chang YZ, Chai RC, Pang B, Chang X, An SY,
Zhang KN, Jiang T and Wang YZ: METTL3 enhances the stability of
MALAT1 with the assistance of HuR via m6A modification and
activates NF-κB to promote the malignant progression of
IDH-wildtype glioma. Cancer Lett. 511:36–46. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m(6)A demethylase
ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by
sustaining FOXM1 expression and cell proliferation program. Cancer
Cell. 31:591–606.e596. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chai RC, Chang YZ, Chang X, Pang B, An SY,
Zhang KN, Chang YH, Jiang T and Wang YZ: YTHDF2 facilitates UBXN1
mRNA decay by recognizing METTL3-mediated m(6)A modification to
activate NF-κB and promote the malignant progression of glioma. J
Hematol Oncol. 14:1092021. View Article : Google Scholar
|
|
32
|
Xu C, Yuan B, He T, Ding B and Li S:
Prognostic values of YTHDF1 regulated negatively by mir-3436 in
glioma. J Cell Mol Med. 24:7538–7549. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zaccara S and Jaffrey S: A unified model
for the function of YTHDF proteins in regulating mA-modified mRNA.
Cell. 181:1582–1595.e1518. 2020. View Article : Google Scholar
|
|
34
|
Zou Z, Sepich-Poore C, Zhou X, Wei J and
He C: The mechanism underlying redundant functions of the YTHDF
proteins. Genome Biol. 24:172023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cong P, Wu T, Huang X, Liang H, Gao X,
Tian L, Li W, Chen A, Wan H, He M, et al: Identification of the
role and clinical prognostic value of target genes of m6A RNA
methylation regulators in glioma. Front Cell Dev Biol.
9:7090222021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu G, Li H, Ji W, Gong H, Jiang Y, Ji G
and Liu G: Construction of a ceRNA network in glioma and analysis
of its clinical significance. BMC Genomics. 22:7222021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mu M, Niu W, Zhang X, Hu S and Niu C:
LncRNA BCYRN1 inhibits glioma tumorigenesis by competitively
binding with miR-619-5p to regulate CUEDC2 expression and the
PTEN/AKT/p21 pathway. Oncogene. 39:6879–6892. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fu C, Li D, Zhang X, Liu N, Chi G and Jin
X: LncRNA PVT1 facilitates tumorigenesis and progression of glioma
via regulation of miR-128-3p/GREM1 axis and BMP signaling pathway.
Neurotherapeutics. 15:1139–1157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Li T and Wang B: Circ-UBAP2
functions as sponges of miR-1205 and miR-382 to promote glioma
progression by modulating STC1 expression. Cancer Med.
10:1815–1828. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiong J, Wang T, Tang H, Lv Z and Liang P:
Circular RNA circMAN2B2 facilitates glioma progression by
regulating the miR-1205/S100A8 axis. J Cell Physiol.
234:22996–23004. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma Z, Ma J, Lang B, Xu F, Zhang B and Wang
X: Circ_0001982 up-regulates the expression of E2F1 by adsorbing
miR-1205 to facilitate the progression of glioma. Mol Biotechnol.
65:466–476. 2023. View Article : Google Scholar
|
|
42
|
Yang M, Li G, Fan L, Zhang G, Xu J and
Zhang J: Circular RNA circ_0034642 elevates BATF3 expression and
promotes cell proliferation and invasion through miR-1205 in
glioma. Biochem Biophys Res Commun. 508:980–985. 2019. View Article : Google Scholar
|
|
43
|
Hirano T, Ishihara K and Hibi M: Roles of
STAT3 in mediating the cell growth, differentiation and survival
signals relayed through the IL-6 family of cytokine receptors.
Oncogene. 19:2548–2556. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zou S, Tong Q, Liu B, Huang W, Tian Y and
Fu X: Targeting STAT3 in cancer immunotherapy. Mol Cancer.
19:1452020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee H, Jeong AJ and Ye SK: Highlighted
STAT3 as a potential drug target for cancer therapy. BMB Rep.
52:415–423. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun X, Wang J, Huang M, Chen T, Chen J,
Zhang F, Zeng H, Xu Z and Ke Y: STAT3 promotes tumour progression
in glioma by inducing FOXP1 transcription. J Cell Mol Med.
22:5629–5638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang H, Tao Z, Feng M, Li X, Deng Z, Zhao
G, Yin H, Pan T, Chen G, Feng Z, et al: Dual PLK1 and STAT3
inhibition promotes glioblastoma cells apoptosis through MYC.
Biochem Biophys Res Commun. 533:368–375. 2020. View Article : Google Scholar : PubMed/NCBI
|