Introduction
Most bacteria and archaea have an adaptive defense
system termed the Clustered Regularly Interspaced Short Palindromic
Repeats (CRISPR) and CRISPR-associated sequence (Cas) genome
editing systems, which protect them from attack by phages, viruses,
plasmids and other foreign genetic materials (1,2).
With the discovery of the CRISPR/Cas system, recent advances in
genome editing technology hold great promise for improving the
environment, agriculture and human health (3-5).
The hallmark feature of CRISPR/Cas systems is the memory
acquisition of previous infections from mobile genetic elements
(MGEs) in the form of short sequences (spacers) incorporated into
the CRISPR array (6,7). The CRISPR array of adjacent regions
also consists of genes encoding Cas proteins. Cas proteins have a
notable role in CRISPR/Cas-driven immunity, including adaptation,
CRISPR RNA (crRNA) expression and interference (8,9).
More recently, the type VI CRISPR/Cas system, an
exclusively single-stranded RNA (ssRNA)-targeting platform, has
been developed (10,11). All the Cas13 subtype effectors are
functional crRNA-guided RNases with two definite and independent
catalytic centers. The pre-crRNA is processed by one catalytic
center and contains two R-X4-H motifs, termed higher
eukaryote and prokaryote nucleotide-binding (HEPN) domains,
responsible for the cleavage of ssRNA (12). The novel performance of Cas13
effectors has been used as a tool for targeting and manipulating
different RNAs, although practical applications of this technique
are still in their infancy (13-15).
Until recently, approaches for targeted cancer
therapy using small-molecule drugs and monoclonal antibodies have
become significant. However, certain complications of this
approach, such as off-target effects and the limited availability
of cancer-driven proteins, limit their broader application
(16,17). Furthermore, molecular analysis of
cancer using diagnostic methods is very expensive and is performed
using sophisticated equipment (18,19). To overcome these limitations of
cancer management, researchers are seeking a precise, efficient and
versatile platform for targeted cancer treatment (20,21).
CRISPR-mediated genetic manipulation has expanded
rapidly in recent years. RNA manipulation using the type VI
CRISPR/Cas13 system is a promising tool for cancer research,
diagnosis and therapy (22,23) (Fig.
1). Additionally, Cas13-based RNA detection and targeting
allows for the early monitoring of tumor markers from different
bodily fluids without the need for sophisticated instrumentation.
Furthermore, Cas13-based RNA editing offers significant prospects
for understanding drug resistance biology in addition to the
discovery of novel therapeutic targets (24). Cas13 can also be mutated to dead
Cas13 (dCas13), an RNA-binding effector that can be easily
programmed for novel tasks. dCas13 is formed from native Cas13 by
the mutation of its HEPN domain residues, which are involved in RNA
cleavage (25). dCas13 has
significant applications when bound to other proteins for cell
imaging in its living state, RNA interaction mapping, mRNA
splicing, RNA base editing, RNA methylation/demethylation and
blocking other RNA-binding proteins (26,27) (Fig.
1).
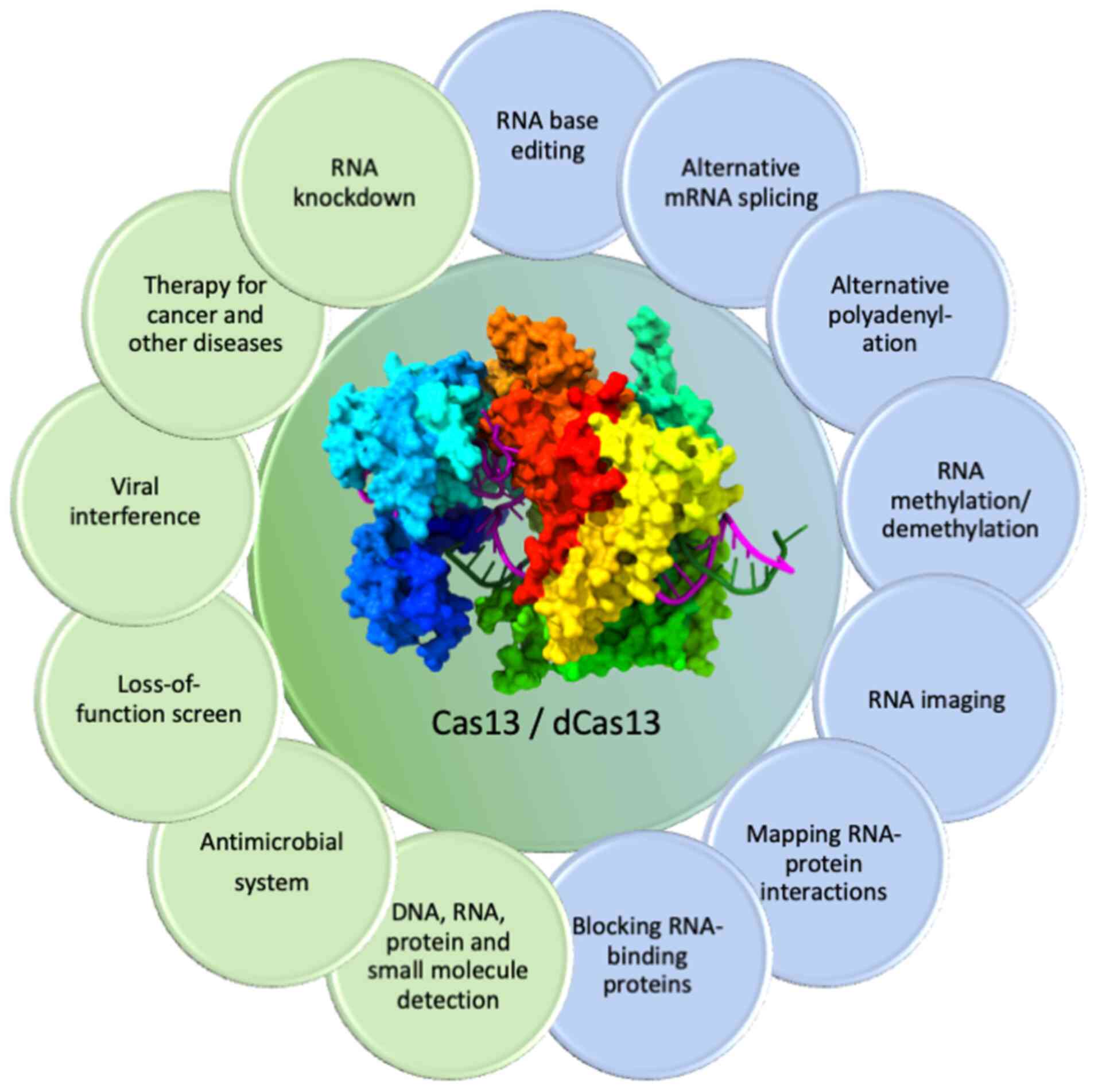 | Figure 1Some novel applications of Cas13 and
dCas13 in RNA targeting and editing. Activated Cas13 is employed
for different applications including disease therapeutics, RNA
knockdown, viral intervention, detection of biological agents,
loss-of-function screening and development of antimicrobials. The
RNA sequence can also be targeted by dCas13, which is catalytically
inactive and can be coupled to other proteins for the execution of
live cell RNA tracking and imaging, target mRNA
splicing/polyadenylation, single base editing,
methylation/demethylation of target RNA, RNA-binding protein
blocking and exploration of RNA-protein interactions. As an
example, for Cas representation, this figure shows the ternary
complex of LbuCas13a, downloaded from the PDB website https://www.rcsb.org/ (PDB ID: 5XWP) and was edited
using UCSF ChimeraX, version 1.8 (https://www.rbvi.ucsf.edu/chimerax). Cas13, Clustered
Regularly Interspaced Short Palindromic Repeats-associated sequence
13; dCas13, dead Cas13; PDB, protein data bank. |
The present review summarizes the recent updates on
the CRISPR/Cas13 system in view of its structural, mechanistic and
novel applications in cancer management. In addition, the present
review will be helpful in understanding CRISPR/ Cas13-related novel
techniques that can be used as innovative tools for cancer
diagnostics and targeted therapeutics. Furthermore, some
limitations and future prospects must be addressed before this
RNA-editing tool can be used more efficiently.
Classification of the CRISPR/Cas system
The CRISPR/Cas system in bacteria and archaea uses
three stages to defend against viruses or other MGEs: Protospacer
acquisition, expression and target recognition (4,28).
In the first stage, a protospacer is incorporated in the host
CRISPR locus as spacers between crRNA repeats (29). In the next stage, the expression
of Cas proteins occurs and the spacers are transcribed as pre-crRNA
repeats (1,29,30). In the third stage, the interaction
between the target site of some invading viruses or other MGEs
(protospacers) occurs with the help of crRNA (spacers), leading to
genome cleavage (29,31). Most CRISPR/ Cas systems use a
sequence-specific protospacer adjacent motif adjacent to the
crRNA-specific site within the target genome to distinguish self
from foreign genetic elements (32).
In recent years, the number and diversity of CRISPR/
Cas systems have significantly increased. The most updated
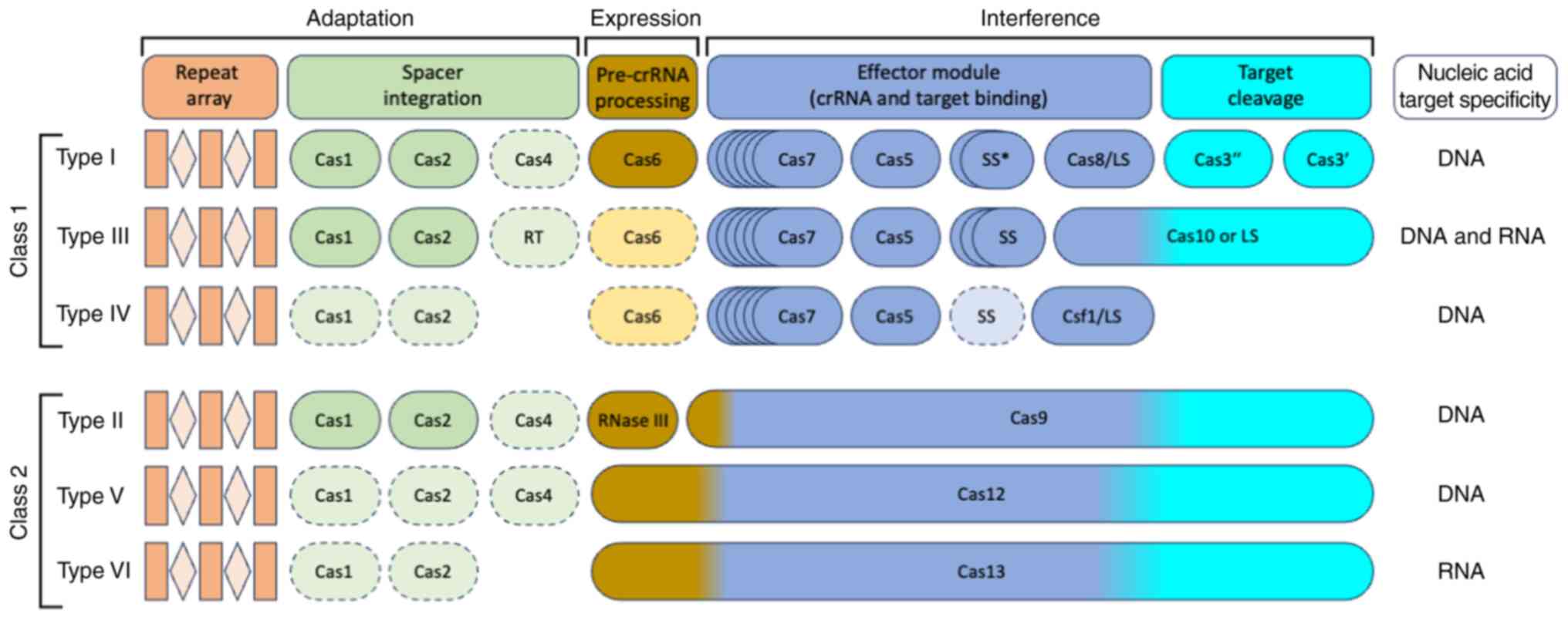
classification system for this genome editing tool consists of two
classes (class 1 and class 2), six types (type I to type VI) and 33
subtypes (33). The CRISPR/Cas
system classification is primarily based on the composition of the
Cas protein and the divergence of the sequence between the effector
modules (34,35). In the class 1 system, several
effector proteins are required for RNA-guided target cleavage.
However, only one RNA-guided endonuclease is required in class 2
systems to cleave foreign DNA/RNA sequences (34,36,37). The class 1 system of this genome
editing tool is divided into three types: Types I, III and IV,
while the class 2 system is classified as types II, V and VI
(34,38,39) (Fig.
2). In the type I system, the Cas3 signature gene is located at
the CRISPR/Cas locus; it encodes a large protein possessing
helicase activity to unwind RNA-DNA and DNA-DNA duplexes (40,41). The type II system is one of the
most well-known and widely used CRISPR/Cas systems, as only a
single multidomain protein targets and cleaves double-stranded DNA
(dsDNA) in this system (42,43). In the type III CRISPR/Cas system,
Cas10 acts as a signature gene that encodes a multidomain protein
for the target detection and cleavage of single-stranded DNA
(ssDNA) (44,45). The type IV CRISPR/Cas system has
no Cas nucleases and integrases as typically seen in other CRISPR
systems, but it does have CRISPR-associated splicing factor 1, and
deeper details about its function have yet to be explored (34,46). In the type V CRISPR/Cas system,
Cas12 acts as a signature gene, known as CRISPR from Prevotella and
Francisella 1 (Cpf1), C2c1 (class 2, candidate 1) or C2c3 (class 2,
candidate 3) proteins, that encodes the RuvC domain, which cleaves
both ssDNA and dsDNA (47,48).
The type VI CRISPR/Cas system contains Cas13 (C2c2), which encodes
the nucleotide binding domain (HEPN) of higher eukaryotes and
prokaryotes and can cleave ssRNA (49,50) (Fig.
2).
Type VI CRISPR/Cas system
Cas13 is a single effector protein found in the type
VI CRISPR/ Cas system with the ability to process crRNA, recognize
invaders and trigger the immune system (25,51) (Figs.
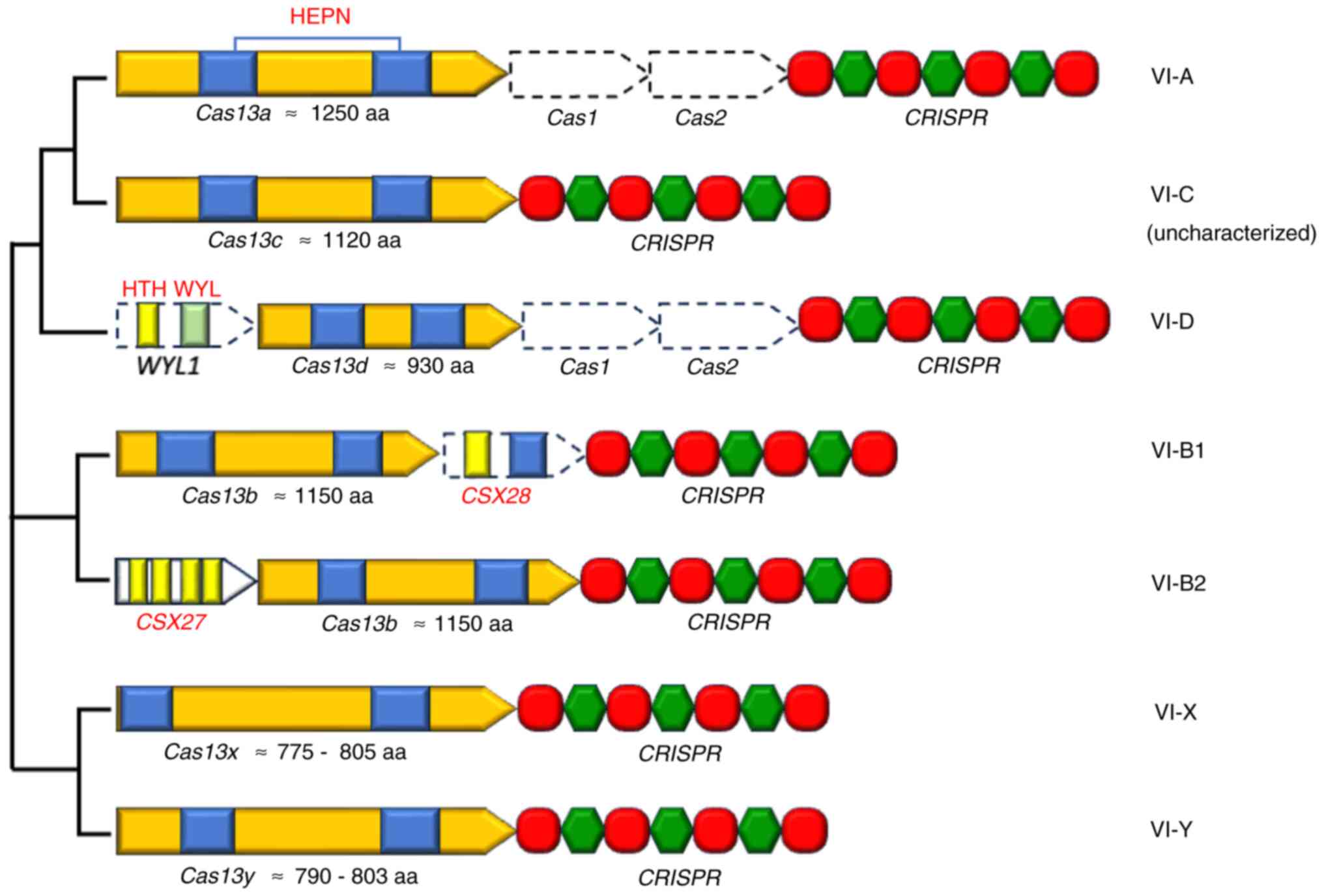
1 and 2). To date, six
distinct type VI subtypes have been identified: VI-A to VI-D,
Cas13X and Cas13Y (39,52) (Fig.
3). The functional characterization of the VI-A, VI-B and VI-D
subtypes is now understood in more detail in relation to their
respective effectors compared with the other subtypes (53). However, there are other subtypes
of this system, such as Cas13e and Cas13i (54).
A single feature common to Cas13 proteins from
different subtypes is the shared inclusion of two HEPN domains
(39,55). This effector protein has a bilobed
shape, wherein RNA nuclease activity is attributed to one lobe and
RNA target recognition to the other. Cas13 can detect a target RNA
in a population of non-target RNAs with femtomolar sensitivity
(56). Binding of the target RNA
to Cas13 induces conformational changes in the nuclease lobe, which
contains two HEPN domains. This conformational change results in a
stable composite RNase pocket that mediates the cleavage of the
target RNA and bystander RNA (57-59), resulting in the inhibition of
invading DNA viruses (60,61).
The different subtypes of Cas effectors in the type VI CRISPR/Cas
system are elaborated below.
CRISPR/Cas13a
Cas13a, an effector of the VI-A CRISPR/ Cas system,
was previously termed class 2, candidate 2 (C2c2) and was first
discovered in 2015 using a computational pipeline by Shmakov et
al (62). In contrast to the
other CRISPR/Cas loci, the Cas1 and Cas2 genes are absent in the
type VI-A loci, but it does contain a CRISPR array along with a
Cas13 gene (Fig. 3). This CRISPR
array is notably unstructured and heterogeneous, with 35-39 bp
direct repeats (35,62). Cas13a is currently a
well-understood type VI effector protein with >1,000 amino
acids, and its primary structure lacks noticeable similarity with
other known Cas effectors (15,62,63). Based on cleavage preference and
non-cognate crRNAs, the currently recognized Cas13a orthologs are
distributed as either A-cleaving or U-cleaving (64,65). pre-crRNA maturation is not
essential for RNA interference; however, it can enhance activity by
releasing crRNA from the CRISPR array (64).
The domain organization of several Cas13a and crRNAs
in complex with activator RNA (aRNA) have been reported. Some
well-known examples include Lachnospiraceae bacterium Cas13a
(LbaCas13a), Leptotrichia shahii Cas13a (66) and Leptotrichia buccalis
Cas13a (LbuCas13a) (51,65) (Fig.
4). Cas13a is predominantly α-helical, possessing a recognition
(REC) lobe and nuclease (NUC) lobe (Fig. 4A). However, the 3D structure
notably differs at the levels of architectural and domain
organization (51,65,66). The REC lobe consists of a strongly
basic cleft, which accommodates the repeat region of crRNA between
the N-terminal domain (NTD) and Helical-1 domain (Fig. 4C and D). The NUC lobe comprises
the HEPN-1, Helical-2, HEPN-2 and Helical-3 domains (Fig. 4A and B). The Helical-2 domain
divides HEPN-1 further into two distinct subdomains, HEPN-1I and
HEPN-1II (51,65). Cas13a undergoes a significant
conformational rearrangement upon crRNA binding and forms a compact
structure. This complex facilitates aRNA binding by closing the
crRNA-binding channel (51)
(Fig. 4E and F). In both
U-cleaving and A-cleaving Cas13a effectors, recognition of the
repeat region of the crRNA occurs in a sequence- and
structure-specific manner. It is well known that Cas13a is a single
effector protein performing the crRNA processing and interferences
steps without the help from other accessory proteins. For pre-crRNA
processing, critical catalytic residues have been identified for
all three Cas13a orthologs, but the A-cleaving LbaCas13a presents a
more detailed acid-base mechanism (51,65,66).
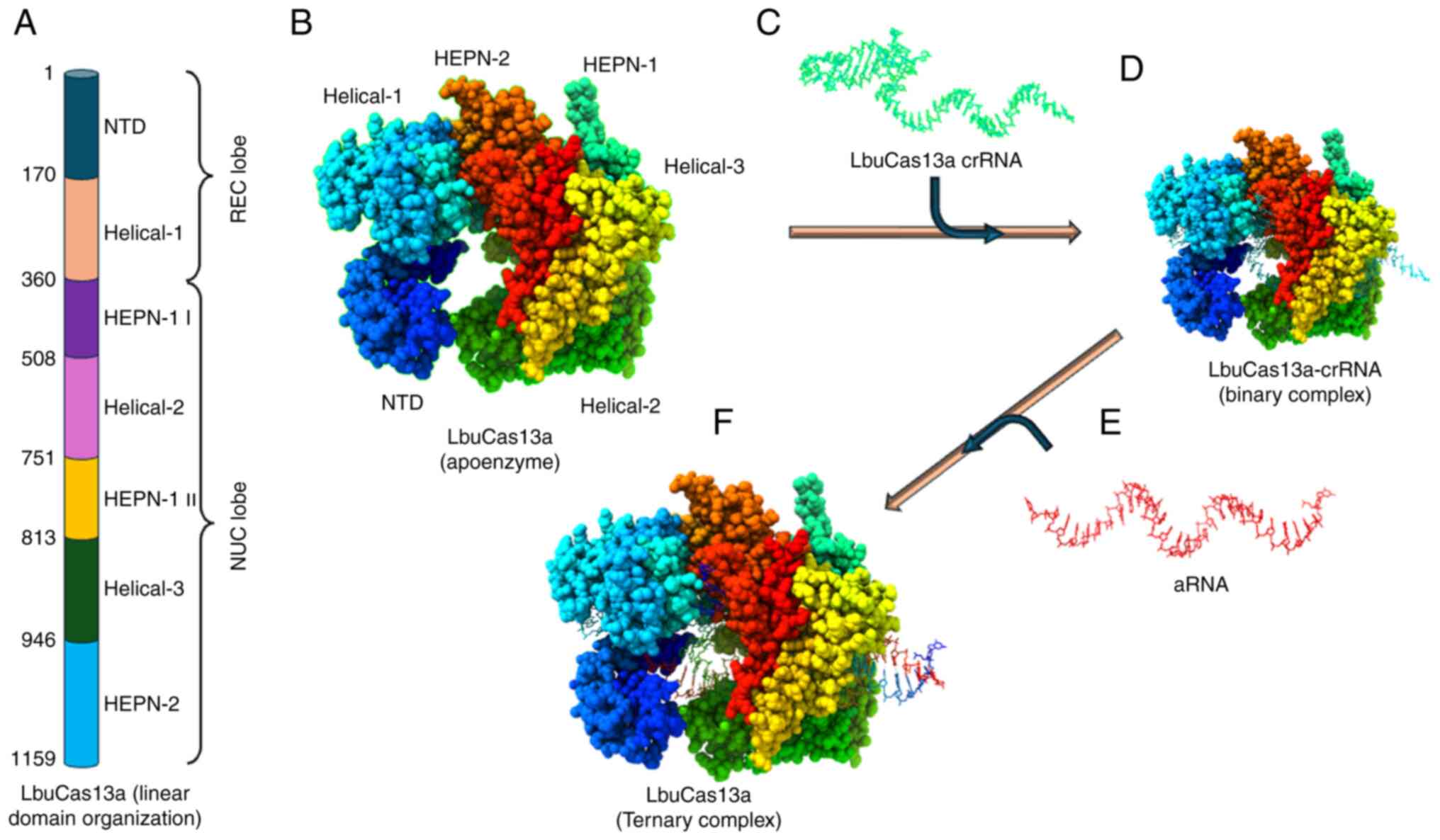 | Figure 4Basic structural features of type
VI-A CRISPR/Cas effectors. (A) Linear domain organization of
LbuCas13a, presenting different domains (REC and NUC lobes)
annotated with the corresponding amino acid number. (B) Cartoon
representation of the LbuCas13a apoenzyme. (C) Secondary structure
of LbuCas13a crRNA and its nucleotide sequence. (D) Binary
structure of the LbuCas13a-crRNA complex. (E) Secondary structure
of aRNA. (F) LbuCas13a-crRNA-aRNA ternary complex. The crystal
structure of LbuCas13a-crRNA-target RNA ternary complex was
downloaded from the PDB website https://www.rcsb.org/ (PDB ID: 5XWP) and was edited
using UCSF ChimeraX, version 1.8 (https://www.rbvi.ucsf.edu/chimerax). LbuCas13a,
Leptotrichia buccalis Cas13a; CRISPR, Clustered Regularly
Interspaced Short Palindromic Repeats; Cas, CRISPR-associated
sequence; HEPN, higher eukaryote and prokaryote nucleotide-binding;
REC, recognition; NUC, nuclease; crRNA, CRISPR RNA; aRNA, activator
RNA; PDB, protein data bank; NTD, N-terminal domain. |
Cas13a uses spacers with different sequence
identities and lengths (20-28 nucleotides) to successfully utilize
crRNAs (64,65). The 5′ region of the spacer is
hidden in the NUC lobe cavity in binary complex structures
(Fig. 4D) that are currently
known, and it adopts a deformed conformation stabilized by
significant interactions with the sugar-phosphate backbone
(51,65,66). The central portion of the spacer
surfaces out from the NUC lobe and passes through a groove created
between the NTD and Helical-2 domains (51,65,66). In Cas13a-crRNA binary complexes,
the center and 3′ region maintain an almost A-form helical
conformation; however, this is not apparent in all orthologs
(51,53).
The Cas13a functional aspects, including aRNA
binding, HEPN nuclease site activation and RNA interference, have
been well-studied. Sequence interrogation and propagation of the
crRNA spacer-aRNA duplex begins when a possible aRNA binds to the
central seed region of the crRNA (57,65). To further stabilize the duplex,
base pairing between aRNA and the remaining spacer is necessary
(57). Sequence interrogation and
propagation of the crRNA spacer-aRNA duplex eventually leads to
synergistic conformational changes in both crRNA and Cas13a, thus
forming a ternary complex with a fully active HEPN nuclease
position that is prepared to interfere with RNA (Fig. 4F). Exposure of the hidden
R-X4-H domain of the HEPN-1 region towards the exterior
shifts it closer to the R-X4-H domain in the HEPN-2
region, which activates the HEPN nuclease active site, permitting
interactions within the NUC duplex lobe and widening the binding
channel to accommodate the crRNA-aRNA duplex (51,57,65,66). The LbuCas13a binary complex and
its ternary complex provide the most pertinent insights into these
conformational changes (51).
The aRNA-bound spacer of crRNA in the ternary
complex of LbuCas13a approached a normal A-form helix (51,67). The duplex base pairs were mainly
in contact with the HEPN-1, Helical-2 and Helical-3 domains, while
they were bound inside the positively charged central channel of
the NUC lobe. LbuCas13a primarily interacts with nucleotides 7-15
(which often correspond to the seed area and portions of the HEPN
region), some nucleotides of spacer crRNAs and aRNA nucleotides
(51,53). A multitude of residues for spacer
interaction in LbuCas13a are important for ssRNA cleavage (51). The reason that Cas13a effectors
can employ spacers of >20 nucleotides with comparable efficacy
is because the duplex beyond the 24th base pair is situated outside
of the protein (51,68). Cas13a effectors cleave ssRNA
sequences in either cis or trans mode during RNA interference
(65,69,70). A groove on the protein surface
houses the HEPN active site, which is located away from the central
channel where the crRNA can bind with aRNA to form a crRNA-aRNA
complex. However, the catalytic centers of type II and V Cas
proteins are embedded within the proteins (51,65,66). For cleavage in trans, any RNA in
solution can readily approach the catalytic center; however, for
cleavage in cis, the effector-bound aRNA must be sufficiently
long.
The structure of the LbuCas13a ternary complex
demonstrated how RNA interacts with HEPN nuclease residues during
interference (Fig. 4F and G).
This interaction is performed by the nucleotide at the 5′ terminal
of aRNA swinging away from the crRNA-activator RNA duplex and being
inserted into the HEPN active center of the adjacent LbuCas13a
(65,71). An HEPN-1 β-hairpin stretches into
the main groove of the crRNA-aRNA duplex and contacts it through
some weak interactions and catches RNA in the immediate region of
the catalytic site (51,72). Its importance in capturing ssRNA
is demonstrated by the reduction in both trans and cis RNA
intrusion that result from interaction with the 5′ nucleotide of
aRNA (51).
CRISPR/Cas13b
In 2016, new computational procedures such as
position-specific iterative basic local alignment search tool and
hidden Markov models prediction, in addition to the ideas from
previous views and no firm requirement for cas1 and cas2
identification in this type, led to the discovery of Cas13b. Cas13b
consists of 1,100-1,200 amino acid effectors of type VI-B systems
(62,73). This subtype was found in bacteria
supported by other CRISPR/Cas systems, getting support from cas1
and cas2 genes. This suggests that the VI-B platform acquire
spacers in trans (73).
Type VI-B systems have several unique features: i)
The orientation of the processed mature crRNA is different from
that of the Cas13a and Cas13d crRNAs (73); ii) the size (36 nucleotides) of
crRNA direct repeat sequences (DRs) are all conserved; iii) the 5′
non-C and the 3′ NNA or NAN protospacer flanking sequences (PFSs)
limit the interference caused by Cas13b; iv) a small accessory
protein (~200 amino acids), either Csx27 or Csx28, controls the
interference of Cas13b; and v) ssRNA cleaves at the pyrimidine
base, preferring to cleave uracil. Csx28 boosts Cas13b-mediated
cleavage, which is routinely observed in subtype VI-B2 systems,
whereas Csx27 represses Cas13b-mediated cleavage. Csx27 is
infrequently present in subtype VI-B1 systems. Both accessory
proteins possess the potential to regulate orthogonal type VI-B
systems, thereby increasing their potential applications (73,74).
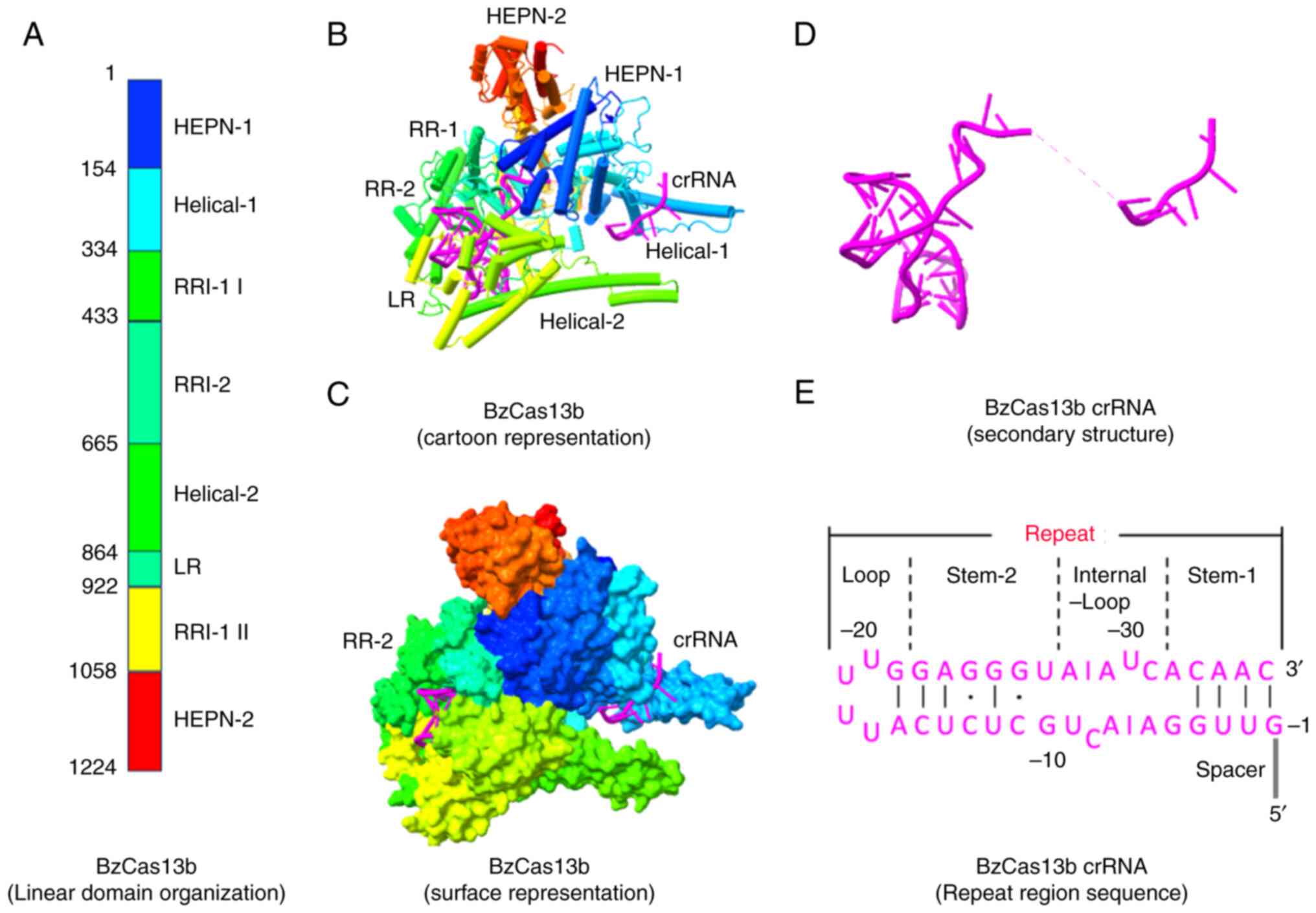
The general domain organization and mechanistic
action of Cas13b have recently been identified by determining the
structures of subtype VI-B1 Bergeyella zoohelcum (Bz)
(Fig. 5) and subtype VI-B2
Prevotella buccae (Pbu) Cas13b-crRNA binary complexes
(75-77). Cas13b adopts a distinct pyramidal
shape in both configurations with a positively charged central
cavity that holds the crRNA. The binary complex of Cas13b-crRNA
lacks a bilobed architecture, and its domain organization is
notably different from that of other type VI effectors (75,76). The two termini of the protein
consist of two domains, HEPN-1 and HEPN-2, placing the two
R-X4-H motifs relatively close to each other (Fig. 5B and C) (75,76). A sizable positively charged
channel connecting the solvent and the internal cavity was found at
the base of the pyramidal structures of both PbuCas13b and BzCas13b
(75,76). The Helical-1 and Helical-2 regions
surround this channel in BzCas13b and extend to resemble pincers
(Fig. 5B and C) (75). However, the purpose of these
pincers, which are specific to PbuCas13b, is not fully understood.
Cas13b recognizes its specific crRNA, resembling an L-shaped
architecture, with the 3′ segment of the spacer region and the bulk
of the repeat region protected inside the protein (75). The twisted hairpin loop (36
nucleotide repeat region) is almost perpendicular to the spacer
direction. The repeat region has four distinct subregions: Stem-1,
stem-2, internal loop and U-rich loop. Watson-Crick base pairing
and multiple H-bonds are responsible for maintaining intramolecular
contacts (75,76) Additionally, crRNAs have several
flipped nucleotides in their repeat regions. Except for HEPN-2,
crRNA interacts extensively with all Cas13b domains; the majority
of these interactions occur with the repeat region interacting
domain-2, which includes the lid and Helical-1 III (78).
The interactions between the repeat region of crRNA
and Cas13b stabilize the phosphate backbone of this RNA type, and
this interaction is based more on the crRNA structure than its
nucleotide sequence; however, this collaboration is
ortholog-specific (75,76). The spacer region of the crRNA in
BzCas13b is surrounded by different regions, such as the HEPN-1,
Helical-1 and repeat region domains on one side, and the Helical-2
domain on the other (75). Due to
the extreme spacer flexibility, the central region (nucleotides
9-15) is invisible (75). The
nucleotide sequence between 16-22 is in the surface groove near the
surface of the protein. The spacer orientation was modified in both
PbuCas13b and BzCas13b in relation to RR by preventing the initial
spacer nucleotide from moving, although this was achieved through
different sets of contacts (75).
This shows that for both Cas13b orthologs, effective activator RNA
binding and spacer trajectory determination depend on the proper
placement of the first spacer nucleotide. The lid protein domain
processes the pre-crRNA towards the downstream using tightly
conserved arginine and lysine residues (75,76). Nucleotide A(-37) was not
recognized by catalytic residues in a base-specific way (75).
A Cas13b-based computational model was analyzed by
studying the crystal structure of PbuCas13b complexed with
36-nucleotide direct repeat sequence and a 5-nucletide spacer at
1.6 Å and illustrated aRNA binding. According to this model, Cas13b
binds to crRNA and probes ssRNA at the spacer repeat
region-proximal (3′-end) (76).
If complementarity occurs, the HEPN-1 and Helical-2 domains open in
response to the initial binding of the potential aRNA to the crRNA
spacer, allowing the RNA to enter the positively charged core
cavity. Prior to full conformational activation of the bipartite
HEPN site, the remaining RNA sequence is scanned to complete
complementarity using the spacer (76). Nevertheless, the channel that has
been suggested as a pathway for aRNA to enter the central cavity is
absent from the structure of the BzCas13b-crRNA binary complex
(75,76). Furthermore, the narrower PbuCas13b
channel might not be open enough to maintain aRNA since the
interdomain linker connects the HEPN-1 and Helical-1 domains, which
probably prevents the HEPN-1 domain from moving as much.
Additionally, in both PbuCas13b and BzCas13b, the spacer 3′ region
is buried within the polypeptide chain. However, in the binary
molecule BzCas13b-crRNA, the 9-15 nucleotides traverse wide
solvent-reachable channel-like type VI-A systems (75). Accordingly, while tandem
mismatches are intolerant across the spacer, only one mismatch is
sufficient to eliminate PbuCas13a HEPN enzymatic activity (76). As a result, the spacer middle
region most likely probes aRNA first, with aRNA binding then moving
towards the spacer 5′ and 3′ ends.
CRISPR/Cas13c
The Cas13c subtype has not been well studied, mostly
due to its lower efficiency in RNA targeting and interference
compared with the other Cas13 subtypes (53). The Cas13c Fusobacterium
perfoetens ortholog is mostly used in the limited
investigations that have been conducted (79,80). Cas13c was initially discovered in
Fusobacterium and Clostridium using bioinformatics;
however, compared with the other Cas13 types, its functional
characterization is far less comprehensive. With ~1,120 amino
acids, this protein is identical to Cas13a in terms of locus and
crRNA structure. However, like Cas13b, it lacks an adaptation
module (Cas1 and Cas2). On the 5′ end of the crRNA, this subtype
displays a DR with a spacer size of 28-30 nucleotides (80).
CRISPR/Cas13d
To date, Cas13d is the smallest type VI effector
with ~930 amino acids (10,11,81). Extending the search to smaller
effectors and refining the search tactics of earlier computational
pipelines led to the discovery of Cas13d (10,11,82). The core structure of Cas13d has
only a distant relationship with Cas13a or other type-VI effectors
outside the two HEPN domains (Fig.
3) (10,11,13). However, its ternary structure is
similar to that of Cas13a effectors (Fig. 6).
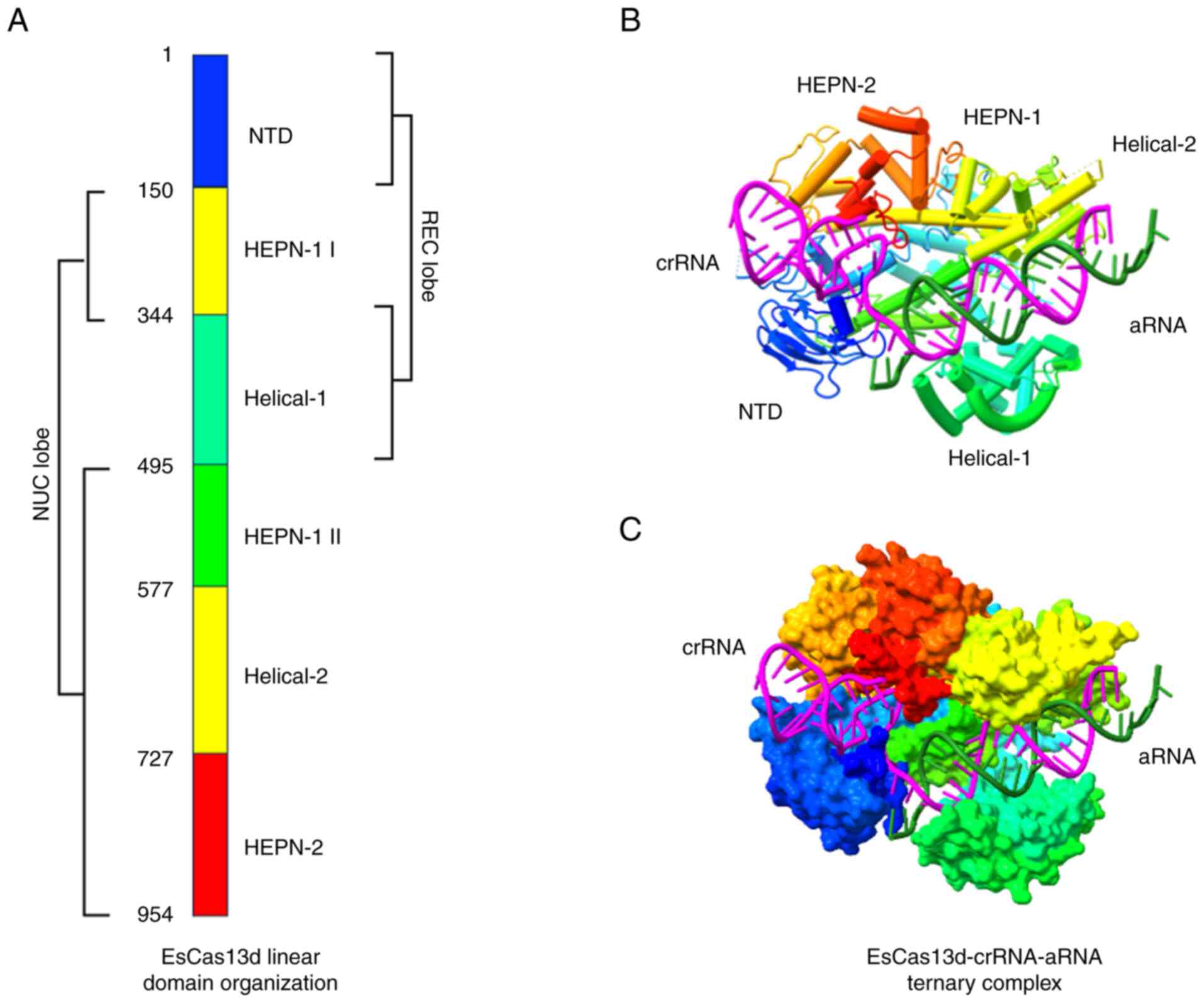 | Figure 6Diagrammatic representation of type
VI-D CRISPR/Cas effectors employing the EsCas13d-crRNA-aRNA ternary
complex. (A) Linear organization of EsCas13d showing the REC and
NUC lobes. (B) Cartoon representation of EsCas13d presenting the
position of crRNA (magenta) and aRNA (mountain green). (C) Surface
representation of EsCas13d. The crystal structure of EsCas13d
ternary complex was downloaded from the PDB website https://www.rcsb.org/ (PDB ID: 6E9F) and edited using
UCSF ChimeraX, version 1.8 (https://www.rbvi.ucsf.edu/chimerax). EsCas13d,
Escherichia coli Cas13d; CRISPR, Clustered Regularly
Interspaced Short Palindromic Repeats; Cas, CRISPR-associated
sequence; HEPN, higher eukaryote and prokaryote nucleotide-binding;
REC, recognition; NUC, nuclease; crRNA, CRISPR RNA; aRNA, activator
RNA; PDB, protein data bank; NTD, N-terminal domain. |
Ruminococcus and Eubacterium are two
benign gram-positive gut bacteria that are the source of the
majority of type VI-D CRISPR/Cas loci (10,11,83,84). Type VI-D loci are characterized by
notable divergence in locus organization and, in most cases, by the
absence of the Cas1 or Cas2 adaptation system in their immediate
proximity (Fig. 3) (10,11,85). For this effector, the repeat
section in the crRNA is highly conserved, with a stem containing an
A/U-rich loop (10,86). It has been observed that Cas13d
targets phage DNA rather than phage RNA, as assumed earlier, which
is consistent with other type VI systems (11,58).
The CRISPR array is processed by Cas13d and forms
mature crRNAs with a 5′-30 nucleotide repeat region (10,13). pre-crRNA processing tests
conducted in vitro have suggested that Cas13d does not
completely process the 3′ direct-repeat region. This results in
truncation, producing mature crRNAs with varying spacer lengths
(11). Overall, it is
hypothesized that Cas13d uses a base-catalyzed method to process
pre-crRNA in which the highly conserved basic residues of the
HEPN-2 domain are essential (13). Metal ions are not necessary;
however, their presence improves processing by increasing Cas13d
binding affinity for crRNA at lower Cas13d-crRNA ratios (13,75). Similar to other effectors of
Cas13, the Cas13d RNase potential is dependent on the availability
of Mg2+ ions and spacer-matching aRNA (10,11). With little secondary structure,
preference for uracil bases and no restrictions imposed by PFS,
Cas13d cleaves ssRNA sequences (10,11,77). Although strong collateral
non-specific RNase activity of Cas13d has been described in
vitro, no similar activity has been observed in mammalian cells
(10).
The cryo-electron microscopy structure of EsCas13d
in an apo-to-ternary state exhibits a bilobed presentation
(Fig. 6) (13,87,88). The Cas13d protein is composed of
five domains that are separated into two lobes: The NUC lobe is
composed of HEPN-1, Helical-2, Helical-1 and NTD domains, whereas
the REC lobe is composed of Helical-1 and NTD domains. The HEPN-1
domain functions as a hinge and structural scaffold that joins the
two lobes (13,75). Except for the Helical-1 domain of
Cas13a, all Cas13a domains have equivalents in the basic sequence
Cas13d (11). However, all five
protein segments are necessary for the enzymatic activity of Cas13d
due to their compact size (75).
Binding of crRNA leads to the stabilization of the REC segment and
portions of the HEPN-2 domain. This phenomenon also causes the
development of a positively charged area connecting the REC and NUC
domains (75). The spacer is
enclosed in an area composed of all protein segments other than the
N-terminus, and the repeat region of the crRNA is fastened within
the HEPN-1, HEPN-2 and NTD domains of this channel (13,75). Due to the small size of Cas13d, a
portion of the repeat region stem-loop protrudes into the solvent,
enabling truncation of the superfluous crRNA region in this area
(13). Cas13d forms extensive
contact with the crRNA repeat region, sugar-phosphate backbone and
nucleobases (13,75). Conserved base-specific contacts
are crucial for preserving the appropriate crRNA binding and
placement (13,75).
For ssRNA cleavage, the repeat region nucleotide
sequence of crRNA and its structure are important (13). The sugar-phosphate backbone plays
a major role in the interaction between Cas13d and the spacer
region, stabilizing its configuration and setting it up for aRNA
attachment (13,75). The spacer is reordered to create
an A-form RNA helix with aRNA upon engagement, eliminating most of
its prior interactions with Cas13d (75). Simultaneously, new contacts are
created between the crRNA spacer, aRNA and different domains of
Cas13d. Cas13d undergoes multiple conformational changes from
binary to ternary states, repositioning HEPN for ssRNA breakdown
and placing the target RNA in the channel (75). Strong associations exist between
aRNA binding and HEPN active-site activation. Partial
conformational rearrangement of Cas13d requires a minimum of 18
nucleotides with strict complementarity, half-maximum cleavage
activity requires 18-20 nucleotides and optimal cleavage activity
requires >21 nucleotides with complementarity (34,75). It was first considered that the
Cas13d crRNA spacer lacks a distinct seed region (75). Based on the available data, it
appears that a mismatch tolerance exists that is likely
ortholog-dependent, and a very mismatch-intolerant area inside the
central 3′ spacer nucleotides exists.
CRISPR/Cas13x and CRISPR/Cas13y
Cas13x (also known as Cas13bt3) and Cas13y have
been identified as subtypes of Cas13b. Using metagenomic data, a
recent study suggested additional categories of Cas13b exist, based
on clades of the derived phylogenetic tree (54,89). Undoubtedly, the effectiveness of
each subtype differs, depending on several factors. Being the
smallest Cas proteins, Cas13x and Cas13y have lengths between 775
and 800 amino acids, whereas the other Cas proteins have lengths
between 1,000 and 1,200 amino acids (52,85) (Fig.
7A). Furthermore, the corresponding single guide RNAs (sgRNAs)
differ fundamentally from one another in terms of sequence and
structure (for example, the sgRNA mounted by Cas13b has a spacer at
its 5′ end, whereas the sgRNA mounted by Cas13a has a spacer at its
3′ end) (55). Such CRISPR/Cas13
systems have been used for a number of purposes other than their
intended use in relatively recent times (71,90).
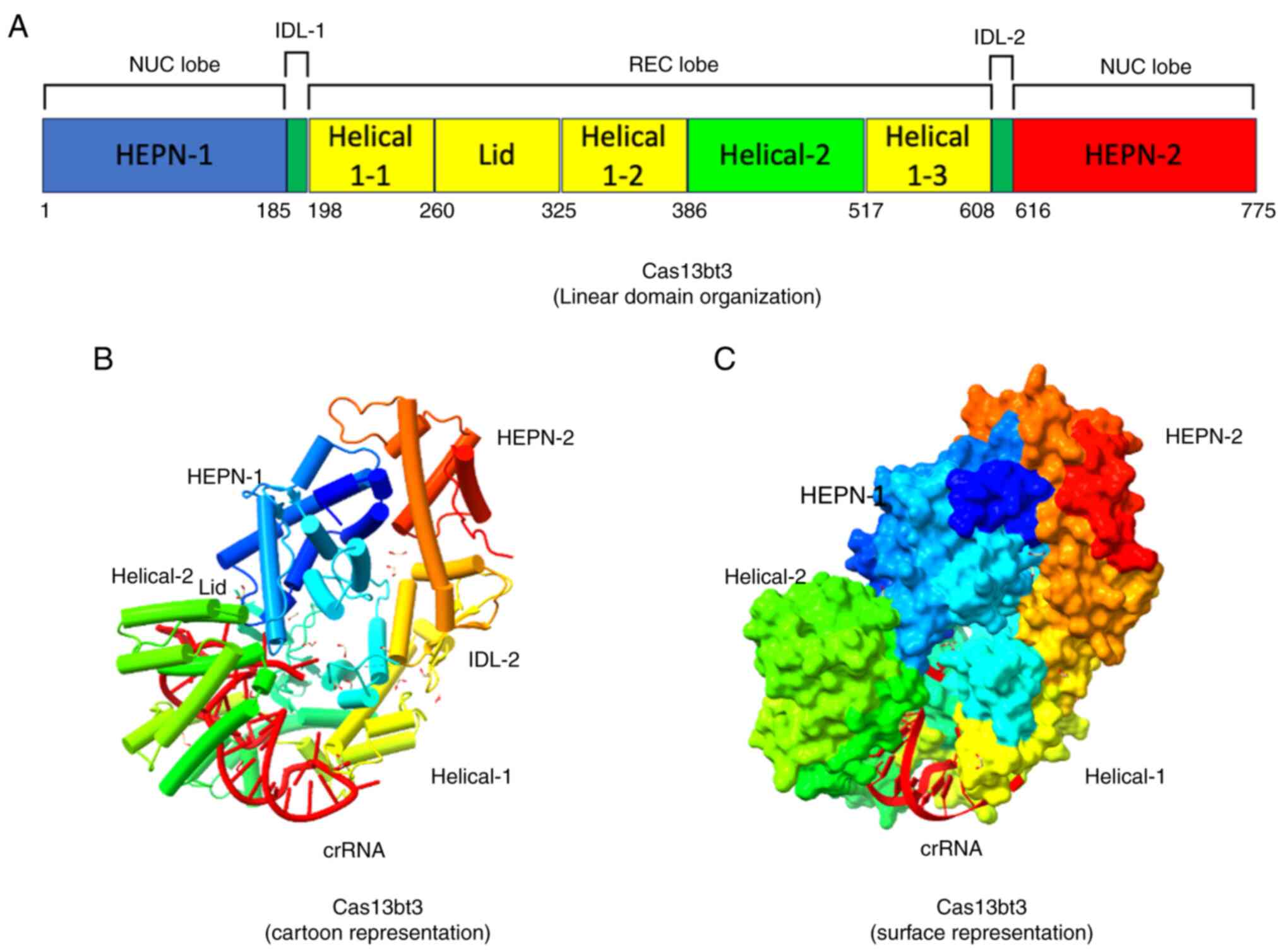 | Figure 7Structural features of the VI-X
CRISPR/Cas effector binary complex. (A) Linear domain organization
of Cas13bt3, presenting different domains (REC and NUC lobes)
annotated with corresponding amino acid numbers. (B) Cartoon
representation presenting the position of crRNA (red). (C) Surface
representation of Cas13bt3. The protein structure was downloaded
from the PDB website https://www.rcsb.org/ (PDB ID: 7VTI) and the binary
complex was edited using UCSF ChimeraX, version 1.8 (https://www.rbvi.ucsf.edu/chimerax). Cas13bt3,
Planctomycetota bacterium Cas13x; CRISPR, Clustered
Regularly Interspaced Short Palindromic Repeats; Cas,
CRISPR-associated sequence; HEPN, higher eukaryote and prokaryote
nucleotide-binding; REC, recognition; NUC, nuclease; crRNA, CRISPR
RNA; PDB, protein data bank. |
The VI-X and VI-Y families of bacteria produce all
known Cas13 orthologs. These orthologs originate from hypersaline
habitats, which lowers the possibility of preexisting immunity, as
observed in Cas9 and Cas12 orthologs that were found in pathogenic
bacterial samples that are closely related to humans (91). Additionally, to circumvent the
difficulty of delivering several large Cas13-based base editors
in vivo, Cas13x.1 has been truncated from amino acids 775 to
445 via structure-guided engineering. This allowed for the creation
of a minimum RNA base editor for C-to-U or A-to-I editing of a
variety of RNA loci in mammalian cells (52). Furthermore, it is anticipated that
in vivo RNA-editing-based research and therapeutic
applications will benefit from the use of these RNA editors with a
compact Cas13x.1 system (92-94). The summarized differences between
the various subtypes of the type VI CRISPR/Cas system are presented
in Table I.
 | Table IComparison between different subtypes
of type VI CRISPR/Cas systems. |
Table I
Comparison between different subtypes
of type VI CRISPR/Cas systems.
|
Characteristics | VI-A | VI-B | VI-C | VI-D | VI-X | VI-Y |
|---|
| Cas effector | Cas13a | Cas13b | Cas13c | Cas13d | Cas13x | Cas13y |
| Size (no. of amino
acids) | ~1,250 | ~1,150 | ~1,121 | ~930 | ~775 | ~790 |
| Cas
architecture | REC and NUC
lobe | Pyramidal (binary
complex) | ND | REC and NUC
lobe | REC and NUC
lobe | ND |
| Location of
pre-crRNA processing | Helical-1 and
HEPN-2 domains | RRI-2 domain | ND | HEPN-2 domain | Not process | ND |
| Site of RNase
activity |
Surface-exposed |
Surface-exposed | ND |
Surface-exposed | ND | ND |
| Preference for
ssRNA cleavage | A- and U-cleaving
effectors | Pyrimidine base
(mostly U) | ND | U | ND | ND |
| Specificity | High | High | Unclear | High | High | High |
| Efficiency (for
KRAS), % | 57.5 | 62.9 | Low | >90.0 | >90.0 | >90.0 |
| Represented by
PFS | LwaCas13a | PspCas13b | FpCas13c | CasRx | Cas13x.1 | Cas13y.1 |
| 5′ non-G | 5′ non-C, 3′NAN or
NNA | ND | No
restrictions | No constraints | ND |
| Requirement of
small accessory proteins | None | Csx27 and
Csx28 | ND | WYL
domain-containing proteins | ND | ND |
| crRNA
orientation | 5′-3′ | 3′-5′ | ND | 5′-3′ | 3′-5′ | ND |
| crRNA repeat
length, nucleotides | Varies, 27-32 | 36 (short), 88
(long) | ND | 30 | 36 | ND |
| Repeat
architecture | Stem-loop | Distorted
stem-loop | ND | Stem-loop | ND | ND |
| Applications | RNA knockdown and
nucleic acid detection | RNA knockdown,
imaging and epigenetic engineering | Unclear | RNA knockdown,
imaging, epigenetic engineering and circular RNA screening | RNA knockdown and
epigenetic engineering | RNA knockdown |
Innovative applications of CRISPR/Cas13
platforms for the detection of tumor-related nucleic acids and
proteins
With great efficiency and precision, the
CRISPR/Cas13 system can be employed for targeted cancer therapy by
destroying and modifying the transcripts associated with
malignancy. Furthermore, CRISPR/Cas13-based RNA editing is a
valuable choice for studying the biology of drug resistance
mechanisms, cancer research and the identification of new
therapeutic targets (22,23,95). Recently, the identification of
cancer-based RNAs, proteins and circulating tumor DNA (ctDNA) from
liquid biopsy tests has been successfully performed using
Cas13-powered biosensors (96).
In vitro and in vivo Cas13 has also been utilized to
efficiently degrade a variety of cancer-associated RNAs.
Furthermore, a plethora of RNA-manipulation platforms based on the
CRISPR/Cas13 system have been engineered, providing a vital toolkit
for modeling, researching drug-resistance biology and identifying
new therapeutic targets in cancer (97-99). The adaptability of the Cas13
systems and their applications in tumor research, therapy and
diagnosis are discussed below. In addition, the benefits and
drawbacks of recent research and the promise of Cas13-based
techniques in the emerging fields of cancer treatment and diagnosis
are discussed.
Identification of different circulating
RNAs (cRNAs)
Compared with single RNA marker assessment, the
identification of numerous cRNAs in different biological samples
provides improved specificity and sensitivity for detecting the
initial stages of cancer (100-102). In this context, early-stage
non-small cell lung cancer (NSCLC) detection has been performed by
identifying the expression levels of two mRNAs, epidermal growth
factor receptor (EGFR) and thyroid transcription factor-1, and four
micro (mi)RNAs (miR-17, miR-19b, miR-155 and miR-210), based on an
electrochemical biosensor provided by the CRISPR/Cas13 system
(23,103).
In one biosensor, a catalytic hairpin assay was
initiated by the collateral cleavage activity of Cas13, which
ultimately produced a current signal. The biosensor achieved high
sensitivity with a limit of detection (LOD) of 50 aM for RNA
detection, thus increasing the overall signal owing to the combined
effect of both the catalytic hairpin assay and collateral RNA
breakdown. The turnaround time was 36 min, and it was possible to
use a single biosensor chip 37 times continuously without any
sensitivity loss. Serum samples were obtained from four cohorts of
patients with NSCLC: 30 healthy individuals, 12 patients with
benign lung disease, 20 patients in the early stage (stages I-II)
of the disease and 55 patients in the late stage (stages III-IV) of
the disease. In total, six different RNA types from serum samples
were selected for disease evaluation. Of the 117 samples, 114 were
classified using this biosensor. Additionally, this platform
demonstrated 97.3% sensitivity and 95.2% specificity in
differentiating patients with NSCLC (stages I-IV) from non-cancer
populations, with 90% sensitivity and 95.2% specificity in
differentiating patients with initial-phase NSCLC (stages I and
II). Therefore, compared with reverse transcription-quantitative
PCR (RT-qPCR), the proposed biosensor offers more notable serum
sample discrimination. In this study, the detection cost was
estimated as $1.62 for each of the six RNAs; thus, multiple marker
analysis with this biosensor is reasonably priced (22). However, this method requires two
steps, which makes it less user friendly. Moreover, although the
biosensor can be reused, it cannot identify multiple RNAs
simultaneously. This lengthens the duration of multiple RNA
analysis. However, by improving the Cas13 platform for RNA
detection within the chip, a single-step technique could be
developed. This platform can also be coupled with a multichannel
biosensor. This research demonstrates that various RNAs in biopsy
samples can be analyzed by employing Cas13-based electrochemical
sensors to help diagnose cancer at the early stages. These precise
and sensitive biosensors can also be utilized to track clonal
evolution, therapeutic responses and tumor heterogeneity (22).
Identification of ctDNA
A promising marker for managing and diagnosing
cancer is the detection of ctDNA (104,105). The concept of CRISPR-based
diagnostics employing Cas13 was first presented by Zhang et
al (106). This approach can
be used in several applications, such as genotyping, the
identification of cancer-based alterations in DNA samples from
cellular extracts and the identification of harmful bacteria and
viruses (56).
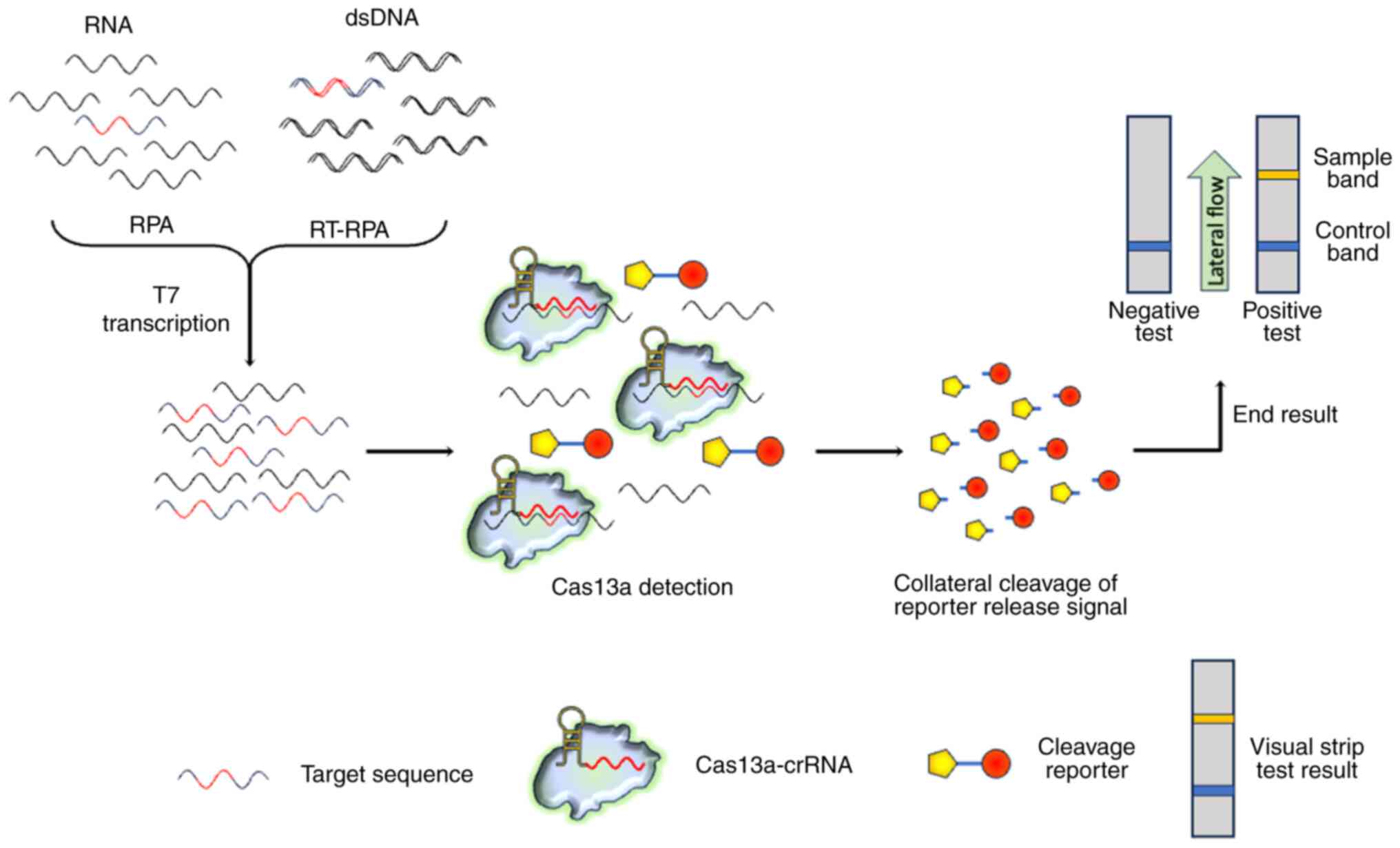
For the rapid identification of RNA or DNA
sequences, the Specific High-Sensitivity Enzymatic Reporter
UnLOCKing (SHERLOCK) platform was engineered using a recombinase
polymerase amplification (RPA) system and collateral RNA breakdown
potential provided by Cas13 (107). SHERLOCK possesses a high
sensitivity and specificity for RNA and DNA identification
(Fig. 8). Cancer-related
BRAFV600E and EGFR-L858R mutations have been identified using the
SHERLOCK approach, and as few as 0.1% of mutant alleles were
identified in mock cell-free DNA samples (Table II) (56). The sensitivity of SHERLOCK is
comparable to those of qPCR and droplet digital PCR. In another
study on patients with NSCLC, liquid biopsy was used to identify
EGFR exon 19, EGFR-T790M and EGFR-L858R deletion changes in DNA
samples using fluorescence and readout techniques (108). The SHERLOCK approach was able to
significantly identify the mutant allele, even though in one
patient with the EGFR-T790M mutation, a 0.6% ratio of mutant
alleles in cell-free DNA was observed (108). SHERLOCK provides attomolar
sensitivity and single-base specificity for the detection of DNA
and RNA in clinical samples (97,109). This approach works well with
quick and easy techniques for the isolation of nucleic acids
(56). Additionally, each
SHERLOCK reagent costs less than $0.60 per test and can be utilized
in one-step reactions after freeze-drying (56). Moreover, orthogonal Cas13 enzymes
can identify several tumor-related nucleotide changes in a single
tube (108). A thorough
procedure that encompasses the LwCas13a enzyme purification step
has been previously suggested for the SHERLOCK assay (97). In conclusion, extremely sensitive
and specific ctDNA identification can be achieved at cheaper rates
without the need for complex instrumentation with Cas13-mediated
detection.
 | Table IISummary of the different markers
present in various cancer types and their detection by the
CRISPR/Cas13-based approach. |
Table II
Summary of the different markers
present in various cancer types and their detection by the
CRISPR/Cas13-based approach.
| First author/s,
year | Cancer type | Sample used | Marker type and
target used | Method of
preamplification | Readout test,
sensitivity and assay time | (Refs.) |
|---|
| Tian et al,
2021 | Breast cancer | Cancer cell extract
and patient serum | miRNA (miR-17 and
miR-21) | Free of
amplification | Fluorescence, 100
aM and 60 min | (156) |
| Shan et al,
2019 | Breast cancer | Mock serum, cancer
cell extract and tumor tissue | miRNA (miR-17) |
Amplification-free | Fluorescence, 450
fM and 30 min | (14) |
| Zhou et al,
2021 | Breast cancer | Cancer cell extract
and mock serum | miRNA
(miR-10b) | Rolling circle
amplification | Colorimetric, 1 fM
and 120 min | (157) |
| Sha et al,
2021 | Breast cancer | Cancer cell
extract, mock serum and patient serum | miRNA (miR-17) | Amplification
free | Fluorescence, 1.33
fM and 125 min | (158) |
| Zhou et al,
2020 | Breast cancer | Cancer cell
extract | miRNA (miR-17) | Isothermal
exponential amplification | Electrochemilumi
nescence, 1 fM and 90 min | (112) |
| Cui et al,
2021 | Breast cancer | Mock serum | miRNA (miR-21) | Catalytic hairpin
assembly | Electrochemical,
2.6 fM and 150 min | (159) |
| Huang et al,
2020 | Breast cancer | Cancer cell extract
and mock serum | miRNA (miR-17) | Hyper-branching
rolling circle amplification | Fluorescence, 200
aM and 170 min | (160) |
| Abudayyeh et
al, 2016 | NSCLC | Mock serum | ctDNA (EGFR, L858R
and BRAF-V600E) | RPA and T7
transcription | Fluorescence, 0.1%
of mutant allele and 150 min | (25) |
| He et al,
2020 | NSCLC | Patient serum | Protein (serum
exosomal PD-L1) | T7 and RPA
transcription | Fluorescence, 10
particles/ml and 60 min | (120) |
| Gootenberg et
al, 2018 | NSCLC | Patient serum | ctDNA (EGFR-T790M,
EGFR L858R and EGFR exon 19 deletion) | T7 and RPA
transcription | Fluorescence
lateral-flow, 0.6% of mutant allele and 60-90 min | (108) |
| Sheng et al,
2021 | NSCLC | Patient serum | miRNA and mRNA
(miR-17, miR-19b, miR-155, miR-210 and TTF-1 and EGFR mRNAs) | Catalytic hairpin
assembly | Electrochemical, 50
aM and 36 min | (103) |
| Bruch et al,
2021 |
Medulloblastoma | - | miRNA (miR-19b and
miR-20a) |
Amplification-free | Electrochemical 10
nM (on-chip), 210 min | (114) |
| Bruch et al
2019 |
Medulloblastoma | Patient serum | miRNA (miR-19b and
miR-20a) |
Amplification-free | Electrochemical, 10
pM (off-chip), 2.2 nM (on-chip) and 210 min | (113) |
Identification of circulating miRNA
The identification of tumors at early stages by the
detection of circulating miRNAs represents an innovative strategy
for cancer diagnosis (110,111). The development of fast,
affordable, sensitive and targeted miRNA detection based on CRISPR
systems may help make cancer screening more common and enable early
detection. Compared with other CRISPR-associated enzymes utilized
in nucleic acid detection, Cas13 targets ssRNA directly and, when
activated, it demonstrates collateral ssRNA cleavage (15). Cas13 can be employed for RNA
detection without the need for reverse transcription or nucleic
acid amplification, as shown in Fig.
8. In this context, a one-step Cas13a-based miRNA detection
technique was designed to allow for amplification-free measurement
of miRNAs with a sensitivity of 450 fM and single-base specificity.
In this procedure, miR-17 was measured in the total RNA from mock
serum samples, cancer cell lines and tumor tissues from patients
with mammary cancer to highlight the potential of this
fluorescent-mediated technique (14). The Cas13-based approach showed a
potential increase in miR-17 levels in 18 out of 20 cancer tests,
and the results were comparable to those of RT-qPCR. According to
this study, tissue and liquid biopsy samples can be used to test
for cancer using the Cas13-based miRNA detection approach. To
improve the sensitivity of the original technique, Zhou et
al (112) developed several
novel biosensors based on this innovative approach (Table II).
A Cas13-mediated biosensor has been designed to
identify miR-19b and miR-20, which are elevated in patients with
medulloblastoma (113). In this
assay, the binding of miRNA-activates the Cas13 endonuclease. The
FAM-tagged reporter ssRNA is cleaved by active Cas13, leading to
the unbound washing out of glucose oxidase labeled with an anti-FAM
antibody. Glucose oxidase generates H2O2 in
electrochemical cells when the target miRNA is absent from the
sample. Consequently, the presence of the sample led to a decrease
in the signal. The instrument allowed the testing of 10 pM miR-19b
and miR-20a when the Cas13-based miRNA detection phase was carried
out outside the chip. Patients with medulloblastoma in both a full
remission state and a progressive state could be identified by the
biosensor. Furthermore, for the additional optimization of this
technique to achieve multiway detection, a new biosensor was
designed to simultaneously detect up to eight miRNAs (114). This led to the successful
detection of miR-20a and miR-19b of up to a10 nM range. These
studies demonstrated the role of Cas13-based electrochemical miRNA
sensors as potential tools for cancer diagnosis.
Detection of liquid biopsy proteins
Extracellular vesicles known as Exosomes are
released by various cell types including cancer cells (115). These vesicles are present in
bodily fluids such as saliva, blood and urine, and are composed of
lipids, proteins and nucleic acids. Exosomes are intriguing
candidates for liquid biopsy investigations in cancer diagnosis
(116). In particular, several
exosomal proteins have been identified for the identification of
cancer in its early stages, tracking the disease course and
estimating the prognosis (117,118).
Traditional protein analysis techniques, such as
mass spectrometry, ELISA and immunoblotting, require considerable
processing time, labor and expensive equipment. These techniques
also require several purification steps. New protein analysis
techniques have been developed to identify exosomal proteins,
including nanoplasmonic exosome sensors and flow cytometry.
However, these techniques are costly and require specialized
equipment (119). To overcome
these complications, RPA- and Cas13-mediated signaling has been
performed using DNA aptamer-based protein identification to
engineer an exosomal protein detection biosensor (120). This biosensor was devised to
identify exosomal programmed death ligand 1 (PD-L1), an important
biomarker for monitoring immune-based therapies. Exosomes were
extracted from the blood of patients using a mixture of antibodies.
The exosomes that were bound were mixed with DNA-based aptamers
that exclusively attached to PD-L1. After the unbound aptamers were
removed by washing, aptamers bound to PD-L1 were removed, and their
amplification was performed using RPA and T7 to produce RNAs. This
triggered Cas13a to break the reporter ssRNAs based on fluorescence
detection. This approach resulted in an LOD of 10 particles/ml, and
the detection range was notably more precise than that of ELISA for
identifying minute quantities of exosomal PD-L1. The researchers
followed a treatment plan with anti-PD-1 therapy for
non-progressive and progressive NSCLC. The PD-L1 levels were
examined in two groups of patients, and the biosensor verified that
only the progressive disease patient group showed a statistically
significant increase in exosomal PD-L1 expression.
In a different study, Chen et al (121) designed an immunosorbent assay
driven by Cas13 that could detect proteins at a femtomolar level
and was at least 100 times more sensitive than regular ELISA
testing. This technique is based on the idea that when a target
antigen is present in the sample, it attaches to a dsDNA sample and
adheres to the well via a sensing antibody. The dsDNA template is
transcriptionally transcribed by T7 to produce RNA copies, which
trigger Cas13 to break the ssRNA, a fluorescence-based reporter.
This procedure is called the CRISPR/Cas13a signal amplification
linked immunosorbent assay (CLISA). The detection range of CLISA is
very high, and the detection range for VEGF and IL-6 was shown to
be 0.81 and 2.29 fM, respectively (121) (Table II). According to these
experiments, tumor marker protein detection techniques with
Cas13-enhanced protein detection can be carried out quickly,
quantitatively, sensibly and specifically, without the requirement
for sophisticated equipment. By facilitating biopsy analysis for
tumor detection and therapeutic strategies, these techniques can
improve treatment outcomes and enable the initial stage of tumor
diagnosis.
Although, the CRISPR/Cas13 system shows great
promise for cancer diagnostics, a fully integrated Cas13-based
biosensor has not yet been engineered. A multiplex hypothetical
biosensor for the diagnosis of NSCLC-based nucleic acid detection
has been proposed for future cancer detection (22) (Fig.
9). This biosensor can extract nucleic acids from whole blood,
which are then divided into multiple channels for detection by
Cas13 present in the detection panel of the biosensor. This
biosensor model was proposed to facilitate the initial detection of
cancer and to provide an overview of the response to treatment
therapy (22).
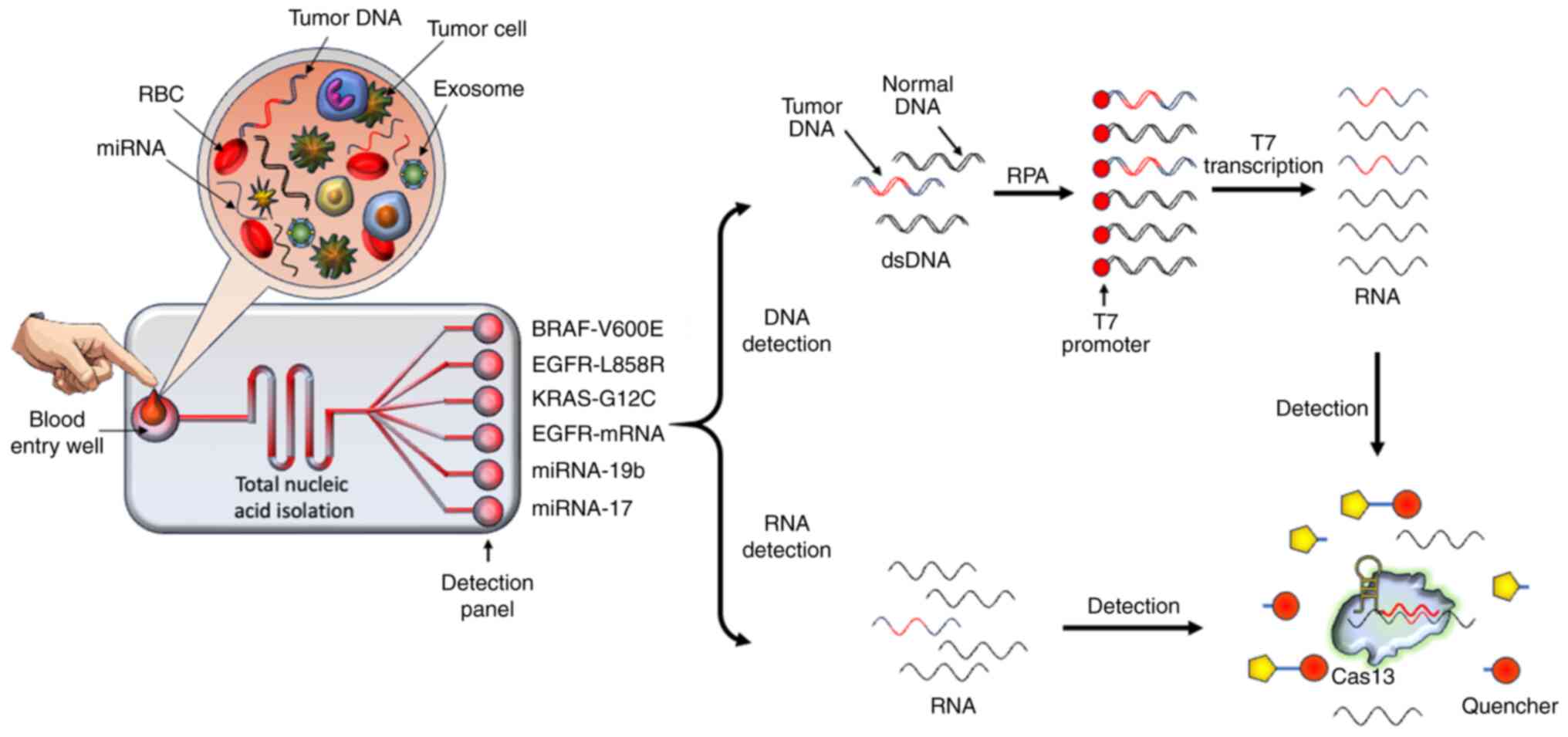 | Figure 9A proposed all-in-one Cas13-based
biosensor devise for future cancer diagnosis. As a microfluidic
sensor, the sample preparation and Cas13-based RNA and DNA
detection steps are integrated for initial-stage NSCLC diagnosis.
The patient's blood possesses cancer-derived cells, miRNAs,
circulating mRNAs, tumor DNAs and exosomes. In this proposed
device, the patient's blood sample is introduced into the entry
well directly. Following the quick nucleic acid isolation step, the
RNAs and DNAs flow within the channel and proceed to the detection
panel. In this panel, each well for detection contains reagents
needed to check for the NSCLC-based DNA mutation or RNA
quantification. Specific RNAs are targeted directly by Cas13, which
is activated in the presence of the CRISPR RNA spacer sequence. The
fluorescent reporter as well as target RNA are cleaved by the
activated Cas13. Additionally, cancer-associated DNAs are amplified
by RPA. After the addition of T7 promoter, the transcription of RPA
amplicons is performed. The mutation-based transcripts after
binding with Cas13 subsequently activate it, which concomitantly
cleaves the fluorescent reporter RNAs, thus providing a detectable
fluorescent signal. Cas13, CRISPR-associated sequence 13; NSCLC,
non-small cell lung cancer; miRNA, microRNA; RPA, recombinase
polymerase amplification; RBC, red blood cell; dsDNA,
double-stranded DNA. |
Delivery and expressional approaches of
CRISPR/Cas13 in cancer cells
The targeting of Cas13 expression in cancer cells
holds great promise; however, careful consideration of certain
factors such as off-target effects, immune response, tumor
heterogeneity and delivery challenges is crucial to maximize the
therapeutic efficacy. Therefore, Cas13-based cancer targeting can
be made safer by using delivery techniques that are specific to
tumor tissue or expression that is particular to cancer cells. Due
to the high specificity of Cas13, tumor-specific delivery
techniques and cancer cell-specific expression, studies employing
CRISPR/Cas13 for cancer cell targeting have been conducted without
causing harm to normal cells in vitro (122-124) or to normal tissues in
vivo (125,126). The safety of CRISPR-Cas13-based
RNA-targeting may be further enhanced using newly identified
anti-CRISPR inhibitors of Cas13a (127,128) for the optogenetic regulation of
Cas13 activity (129) or the
cancer tissue-specific activation of Cas13a enzymes (130). For the selective Cas13a-mediated
degradation of oncogenic mRNAs in cancer cells, Fan et al
(131) integrated tumor-specific
delivery, tumor-specific expression and laser-controlled release
techniques in a well-designed study. They coupled vascular
endothelial growth factor receptor 2 (VEGFR2) monoclonal antibodies
with the lipid component of nanoparticles for tumor-specific
delivery since invasive bladder cancer has upregulated VEGFR2
expression (132). Cas13a was
expressed from human telomerase reverse transcriptase (hTERT), a
synthetic promoter unique to bladder cancer (133). Using a photothermal agent,
liposomes were released under laser control. By combining these
strategies, intelligent liposomes were installed in the bladder and
orthotopic bladder cancer tumors were effectively targeted
(131).
Using promoters unique to cancer cells is another
approach of cancer cell targeting. Gao et al (99) employed a decoy minimum promoter,
which is unique to nuclear factor κB (NF-κB), to express Cas13a
exclusively in cancer cells. It has been demonstrated that this
promoter only permits effective transcription when NF-κB, which is
upregulated in a number of malignancies, is present in high
concentrations (134-136). Gao et al (99) demonstrated that only the tumor
tissue expressed the Cas13a enzyme and that the target transcripts
were exclusively broken down in the tumor tissue, even though
adeno-associated viruses (AAVs) carrying Cas13a and crRNAs were
administered intravenously to numerous tissues (99).
Zhang et al (124) employed chemically-locked
nanoparticles containing plasmids encoding PD-L1-targeting crRNA
and Cas13a. Due to the low pH and high H2O2
concentration found in the tumor microenvironment, these
nanoparticles were engineered to only release the plasmids in tumor
tissues, and the Cas13a and crRNA-encoding plasmids were primarily
absorbed by cancer cells. Similarly, Jiang et al (125) ensured targeted delivery of
Cas13d/crRNA expression cassettes to the pancreas of orthotopic
tumor-bearing mice using a capsid-optimized AAV8. A study that used
the CRISPR-Cas13d technology to treat age-related macular
degeneration also found no in vivo harm (93). Together, these findings
demonstrate that the RNA-targeting CRISPR/Cas13 system is highly
precise and that both tumor tissue-specific viral and non-viral
delivery strategies, as well as expression unique to cancer cells,
can guarantee safety.
Pre-clinical and clinical applications of
CRISPR/Cas13 for the management of cancer
In addition to the detection of different tumor
related markers such as cRNAs, ctDNAs, circulating miRNAs and
proteins, the CRISPR/Cas13a system has also been used in real-world
clinical application for cancer therapy (22). By precisely identifying and
lowering the mRNA expression of oncogenes in cancer cells, the
CRISPR/Cas13a system has been used to stop tumor growth. This has
resulted in regulated incidental breakage and the induction of
programmed cell death in cancer cells (95). However, more research is necessary
to determine whether CRISPR/Cas13a, a potent RNA knockdown
technique, can be used in all types of cancer cells. The
pre-clinical and clinical applications of CRISPR/Cas13 system for
the management of different cancers is elaborated on below.
Pancreatic cancer
Due to its quick onset, low incidence of early
diagnosis, high surgical mortality rate and low cure rate,
pancreatic cancer has surpassed breast cancer; it is a very
challenging task to identify and treat pancreatic cancer and this
disease is expected to be the third leading cause of cancer-related
death in the United States (137,138). Due to the high mortality rate of
pancreatic cancer, effective novel therapeutic approaches are
desperately needed (122). A
mutant oncogene, known as Kirsten rat sarcoma viral oncogene
homolog (KRAS), is the main cause of pancreatic cancer (139). One successful antitumor method
is to suppress the expression of the KRAS mutant at the mRNA level.
It is reported that the expression of mutant KRAS mRNA is markedly
reduced by the bacterial Cas13a protein and crRNA (25), demonstrating that the
CRISPR/Cas13a system can achieve up to 94% expressional knockdown.
In several KRAS-driven pancreatic cancer models, the Cas13a-crRNA
complex efficiently inhibits the KRAS-G12D mutation signaling
pathway, resulting in apoptosis and tumor growth inhibition both
in vitro and in vivo (140). This suggests that the CRISPR/
Cas13a system can be employed as a targeted therapy for patients
with pancreatic cancer harboring mutant KRAS (141). More notably, CRISPR/Cas13a
knockdown of KRAS-G12D can stop the growth of pancreatic cancer
cells, and CRISPR/Cas13a-mediated mRNA downregulation can cause
tumor cells to undergo apoptosis and cause marked tumor shrinkage
in vitro. Therefore, it can be concluded that the CRISPR/
Cas13a system is the most effective method for influencing the
efficient and focused suppression of carcinogenic mRNAs since it is
a flexible targeted therapeutic tool (22,142).
In a recent study, researchers employed an
innovative approach known as Cas13a Allele-Specific PCR Enzyme
Recognition (CASPER), an allele-specific PCR preamplification
technique, to increase the sensitivity and specificity of detecting
KRAS G12D with minimal DNA input (140). CASPER identified KRAS mutations
in DNA samples obtained from patients with pancreatic cancer by
ultrasound-guided fine-needle aspiration fluid; thus, implementing
this approach may be a versatile and reliable method for the
detection of point mutations in pancreatic cancer DNA samples.
Bladder cancer
Globally, bladder cancer is one of the most
prevalent cancer types, with a high incidence and recurrence rate
(143). This type of cancer had
a 5-year frequency of >1.7 million cases in 2020, making it the
sixth most common cancer worldwide (144). To the best of our knowledge, the
use of CRISPR/Cas13a-based therapeutic approaches for intravesical
instillation in bladder cancer is still unexplored, although
several experimental procedures have shown effectiveness in
enabling execution of CRISPR/Cas13a-based cellular functions. In
one study, the researchers used a CRISPR/Cas13a nanoplatform that
significantly reduced the expression of programmed death ligand 1
(PD-L1) after its intravesical instillation. The transmembrane
peptide, trans-activator of transcription and CRISPR/Cas13 were
genetically fused to generate the fusion protein, calpastatin
(CAST). Fluorinated chitosan (FCS), a transepithelial delivery
vehicle was assembled with CAST, which acted as a powerful
transmembrane RNA editor. The CAST-crRNAa/ FCS nanoparticles had
notable transepithelial properties and markedly reduced PD-L1
expression in tumor tissues when administered intravesically into
the bladder (145). The
researchers used a fenbendazole (FBZ) intravesical system to
increase immune activation in the tumor microenvironment. This
system was constructed by bovine serum albumin encapsulation and
FCS, which establishes FBZ as a potent chemo-immunological agent.
This nanoformulation method reorganized the immunological
microenvironment, synergistically decreased PD-L1 expression and
showed marked tumor cell death in a bladder cancer model. The study
suggested a possible synergistic treatment approach and revealed a
unique RNA editor nano-agent formulation. This strategy greatly
increased therapeutic efficacy and showed potential for the
clinical application of cancer perfusion therapies based on
CRISPR/Cas13 (145).
Cervical cancer
Cervical cancer is a common cancer that is a major
threat to women's health and is a major global public health issue.
Globally, there were 348,189 cervical cancer-related deaths and
661,021 new cases in 2022 (146). Multiple types of human
papillomavirus (HPV) have been linked to cervical cancer; however,
the primary causes are the high-risk HPVs, HPV16 and HPV18
(147). Cervical cancer is
largely caused by the E6 and E7 oncoproteins, which are encoded by
the HPV genome and lower the expression levels of the tumor
suppressor proteins, p53 and retinoblastoma. This results in
increased cell proliferation and decreased apoptosis, which
furthers the development of cervical cancer (148). Therefore, it is anticipated that
antiviral drugs that suppress the production of E6/E7 oncoproteins
will be used to treat cervical cancer. According to previous
research, the CRISPR/Cas13a system can efficiently and precisely
knockdown HPV16/18 E6/E7 mRNA (123). The HPV16 knockdown by the
CRISPR/Cas13a system markedly decreased tumor weight and volume in
a subcutaneous xenograft tumor growth model. According to the
aforementioned study, the ability of the CRISPR/Cas13a system to
target HPV E6/E7 mRNA may make it a viable treatment option for
cervical cancer linked to HPV.
Glioma
The most prevalent type of malignant brain cancer
is glioblastoma multiform (GBM), with an incidence rate of 3.23 per
100,000 population globally (149). Since there are several forms of
GBM and its prognosis is typically poor, research on its
therapeutic approach is especially crucial (150). The CRISPR/Cas13a system can kill
glioma cells that have upregulated EGFR VIII expression, a distinct
mutant subtype of EGFR observed in glioma. The CRISPR/Cas13a system
has been characterized by non-specific RNA cleavage, according to a
single-cell RNA sequencing investigation conducted on
U87-Cas13a-EGFR VIII cells (151). In addition, CRISPR/Cas13a has
been shown to inhibit glioma formation in intracranial tumors. In a
mouse tumor model, U87-F3-T3 cells expressing Cas13a prevented
glioma cells from proliferating and growing, demonstrating
CRISPR/Cas13a-based suppression (151).
Challenges and prospects
Understanding the scope and limitations of RNA
editing by the Cas13 system and avoiding potential errors are
crucial, although this system has numerous uses in clinical
diagnostics, scientific research and possible therapeutic
applications. For targeted RNA knockdown, high-fidelity Cas13
variants reduce the risk of unintentional collateral RNA
degradation. However, recent research has revealed unexpected
effects of expressing either Cas13 or gRNA alone. These unexpected
effects include off-target effects, intrinsic targeting of host RNA
and cellular toxicity (70). The
creation of an innovative Cas13 platform for a range of uses will
be made easier with an improved comprehension of these findings and
the underlying mechanisms.
The prospects to enhance the specificity and
minimize unintended RNA cleavage by the CRISPR/Cas13 system in the
near future may be challenging, which include different novel
approaches, such as i) optimizing the gRNA design: The new designs
of the gRNAs can significantly improve the targeting specificity
within cancer cells. This approach includes the chemical
modification of nucleotides and other structural alterations to
enhance the gRNA stability and reduce off-target effects. The
chemical modifications into gRNAs can enhance their binding
affinity and specificity for target RNA sequence (78); ii) use of high-fidelity Cas13
variants: The off-target effects of Cas13 can be further reduced by
using high-fidelity Cas13 variants. These variants are engineered
to have high specificity for their target RNA sequence (152); and iii) employment of
computational tools: The prediction and minimization of Cas13
off-target effects for potential gRNA-target interactions can be
advanced by computational tools and algorithms (153).
Additional challenges regarding the use of the
Cas13 system within the cells of interest include its delivery
options. The Cas13 system may have difficulty in penetrating
certain tissues or cell types (such as HeLa and 293T cells), which
limits its effectiveness. Off-target effects or unintended
consequences can raise ethical concerns, particularly in clinical
applications. Specifically, germline editing using this RNA editing
system raises ethical questions regarding the modification of
future generations. Despite these limitations, ongoing research is
addressing these challenges and exploring innovative approaches to
improve the specificity of the Cas13 RNA-editing strategy, enhance
its specificity and improve delivery approaches for potential
therapeutic applications.
Optimizing the delivery of CRISPR/Cas13 to cancer
cells is challenging but highly significant for enhancing its
therapeutic efficacy. Some novel strategies to improve the delivery
of CRISPR/Cas13 in target cells include: i) Viral vectors:
CRISPR/Cas13 components can be easily delivered by AAVs and
lentiviruses into target cells. These vectors can be modified to
target cancer cells specifically, ensuring efficient delivery and
expression of the CRISPR system (153); ii) nanoparticle-based delivery:
CRISPR/Cas13 components can also be delivered efficiently in target
cancer cells by utilizing nanoparticles, such as lipid
nanoparticles or polymer-based nanoparticles. These nanoparticles
can be engineered to target specific tumor cells, enhancing
specificity and reducing off-target effects (154); iii) cell-penetrating peptides
(CPPs): CPPs can facilitate the delivery of CRISPR/Cas13 components
into cancer cells by enhancing cellular uptake. These peptides can
be conjugated with CRISPR components to improve their intracellular
delivery (152); iv)
electroporation: This method involves applying an electric field to
cells to increase the permeability of the cell membrane, allowing
CRISPR/Cas13 components to enter the cells more effectively. It is
particularly useful for ex vivo applications, such as
modifying immune cells before reintroducing them into the patient
(155); and v) targeted delivery
systems: Developing targeted delivery systems that use ligands or
antibodies to recognize and bind to specific receptors on cancer
cells can enhance the precision of CRISPR/Cas13 delivery. This
approach minimizes off-target effects and maximizes therapeutic
impact (154). All these
strategies collectively contribute to a more effective and precise
delivery of CRISPR/Cas13 in cancer therapy, potentially leading to
improved treatment outcomes.
Furthermore, the prospects of Cas13 RNA-targeting
and editing approaches in the near future include the correction of
genetic defects, such as in cystic fibrosis, Huntington's disease
and sickle cell anemia, for proper disease management. In addition,
this system could be employed to target and destroy viral RNA to
combat viral infections, such as HIV and influenza, and emerging
viruses, such as SARS-CoV-2. Cas13-based diagnostic tools can be
developed for rapid and sensitive detection of genetic mutations,
pathogens and other biomarkers.
Conclusions
Since its discovery in 2016, the CRISPR/Cas13
platform has revolutionized the art of RNA-based diagnostics and
therapeutics, in addition to the fundamental study of RNA biology.
Cas13 is a potent gene targeting tool due to its ability to
precisely and efficiently target nearly any nuclear or cytoplasmic
RNA sequence. The potential of CRISPR/ Cas13 as a therapeutic
platform is further enhanced by its small size and inherent
flexibility. With its single-nucleotide accuracy and specificity,
the constantly growing CRISPR/ Cas13 toolset enables a wide variety
of RNA modifications. Safer treatments for a variety of illnesses
are made possible by the capacity to accurately modify RNAs,
including substitutions or nucleotide additions, without changing
DNA. However, the structural and biochemical aspects of the gene
editing strategies of the CRISPR/Cas13 system are still
insufficient to comprehend its thorough use in the clinical field.
The prospects to enhance the specificity and minimize unintended
RNA cleavage by the CRISPR/Cas13 system is highly significant but
challenging. These challenges can be circumvented by using new gRNA
designs, high-fidelity Cas13 variants and the employment of new
computational tools. Furthermore, optimizing the delivery
approaches of the CRISPR/Cas13 system into cancer cells may be
utilized by engineered nanoparticles, viral vectors, CPPs,
electroporation and other targeted delivery systems. We anticipate
that Cas13 technologies will continue to advance, opening
fascinating possibilities for RNA-based diagnostics, RNA-targeting
treatments and biological research.
Availability of data and material
Not applicable.
Authors' contributions
Conceptualization was conducted by KSA and AAK;
writing the original draft was conducted by KSA, AHR and AAK;
reviewing and editing the manuscript was conducted by KSA, AHR,
NMA, FMA, GhadahMA, GhadeerMA and AAK; data curation was conducted
by KSA, AHR, NMA, FMA, GhadahMA, GhadeerMA and AAK. All authors
read and approved the final version of the manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patent consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The researchers would like to thank the Deanship of
Graduate Studies and Scientific Research at Qassim University for
financial support (QU-APC-2025).
Funding
This study was supported by the Deanship of Graduate Studies and
Scientific Research at Qassim University (QU-APC-2025).
References
|
1
|
Marraffini LA: CRISPR-Cas immunity in
prokaryotes. Nature. 526:55–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allemailem KS, Alsahli MA, Almatroudi A,
Alrumaihi F, Alkhaleefah FK, Rahmani AH and Khan AA: Current
updates of CRISPR/Cas9-mediated genome editing and targeting within
tumor cells: An innovative strategy of cancer management. Cancer
Commun (Lond). 42:1257–1287. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Piergentili R, Del Rio A, Signore F, Umani
Ronchi F, Marinelli E and Zaami S: CRISPR-Cas and its wide-ranging
applications: From human genome editing to environmental
implications, technical limitations, hazards and bioethical issues.
Cells. 10:9692021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Allemailem KS, Almatroudi A, Rahmani AH,
Alrumaihi F, Alradhi AE, Alsubaiyel AM, Algahtani M, Almousa RM,
Mahzari A, Sindi AAA, et al: Recent updates of the CRISPR/ Cas9
genome editing system: Novel approaches to regulate its
spatiotemporal control by genetic and physicochemical strategies.
Int J Nanomedicine. 31:5335–5363. 2024. View Article : Google Scholar
|
|
5
|
Meng X, Wu TG, Lou QY, Niu KY, Jiang L,
Xiao QZ, Xu T and Zhang L: Optimization of CRISPR-Cas system for
clinical cancer therapy. Bioeng Transl Med. 8:e104742022.
View Article : Google Scholar
|
|
6
|
Heler R, Marraffini LA and Bikard D:
Adapting to new threats: The generation of memory by CRISPR-Cas
immune systems. Mol Microbial. 93:1–9. 2014. View Article : Google Scholar
|
|
7
|
Faure G, Shmakov SA, Yan WX, Cheng DR,
Scott DA, Peters JE, Makarova KS and Koonin EV: CRISPR-Cas in
mobile genetic elements: Counter-defence and beyond. Nat Rev
Microbiol. 17:513–525. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hille F, Richter H, Wong SP, Bratovič M,
Ressel S and Charpentier E: The biology of CRISPR-Cas: Backward and
forward. Cell. 172:1239–1259. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Van Der Oost J, Westra ER, Jackson RN and
Wiedenheft B: Unravelling the structural and mechanistic basis of
CRISPR-Cas systems. Nat Rev Microbiol. 12:479–492. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Konermann S, Lotfy P, Brideau NJ, Oki J,
Shokhirev MN and Hsu PD: Transcriptome engineering with
RNA-targeting type VI-D CRISPR effectors. Cell. 173:665–676.e14.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan WX, Chong S, Zhang H, Makarova KS,
Koonin EV, Cheng DR and Scott DA: Cas13d is a compact RNA-targeting
type VI CRISPR effector positively modulated by a
WYL-domain-containing accessory protein. Mol Cell. 70:327–339.e5.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao F, Zhang T, Sun X, Zhang X, Chen L,
Wang H, Li J, Fan P, Lai L, Sui T and Li Z: A strategy for Cas13
miniaturization based on the structure and AlphaFold. Nat Commun.
14:55452023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Ye Y, Ye W, Perčulija V, Jiang H,
Chen Y, Li Y, Chen J, Lin J, Wang S, et al: Two HEPN domains
dictate CRISPR RNA maturation and target cleavage in Cas13d. Nat
Commun. 10:25442019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shan Y, Zhou X, Huang R and Xing D:
High-fidelity and rapid quantification of miRNA combining crRNA
programmability and CRISPR/Cas13a trans-cleavage activity. Anal
Chem. 91:5278–5285. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang H and Patel DJ: Structures,
mechanisms and applications of RNA-centric CRISPR-Cas13. Nat Chem
Biol. 20:673–688. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhong L, Li Y, Xiong L, Wang W, Wu M, Yuan
T, Yang W, Tian C, Miao Z, Wang T and Yang S: Small molecules in
targeted cancer therapy: Advances, challenges, and future
perspectives. Signal Transduct Target Ther. 6:2012021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Allemailem KS, Almatroodi SA, Almatroudi
A, Alrumaihi F, Al Abdulmonem W, Al-Megrin WAI, Aljamaan AN,
Rahmani AH and Khan AA: Recent advances in genome-editing
technology with CRISPR/Cas9 variants and stimuli-responsive
targeting approaches within tumor cells: A future perspective of
cancer management. Int J Mol Sci. 24:70522023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pulumati A, Pulumati A, Dwarakanath BS,
Verma A and Papineni RVL: Technological advancements in cancer
diagnostics: Improvements and limitations. Cancer Rep (Hoboken).
6:e17642023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Dai C, Jiang L, Tong G, Xiong Y,
Khan K, Tang Z, Chen X and Zeng H: Advanced devices for tumor
diagnosis and therapy. Small. 17:e21000032021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Allemailem KS, Alsahli MA, Almatroudi A,
Alrumaihi F, Al Abdulmonem W, Moawad AA, Alwanian WM, Almansour NM,
Rahmani AH and Khan AA: Innovative strategies of reprogramming
immune system cells by targeting CRISPR/Cas9-based genome-editing
tools: A new era of cancer management. Int J Nanomedicine.
18:5531–5559. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Allemailem KS, Almatroudi A, Alrumaihi F,
Alradhi AE, Theyab A, Algahtani M, Alhawas MO, Dobie G, Moawad AA,
Rahmani AH and Khan AA: Current updates of CRISPR/Cas system and
anti-CRISPR proteins: Innovative applications to improve the genome
editing strategies. Int J Nanomedicine. 19:10185–10212. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Palaz F, Kalkan AK, Can Ö, Demir AN,
Tozluyurt A, Özcan A and Ozsoz M: CRISPR-Cas13 system as a
promising and versatile tool for cancer diagnosis, therapy, and
research. ACS Synth Biol. 10:1245–1267. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Durán-Vinet B, Araya-Castro K, Calderón J,
Vergara L, Weber H, Retamales J, Araya-Castro P and Leal-Rojas P:
CRISPR/ Cas13-based platforms for a potential next-generation
diagnosis of colorectal cancer through exosomes micro-RNA
detection: A review. Cancers (Basel). 13:46402021. View Article : Google Scholar
|
|
24
|
Xu D, Cai Y, Tang L, Han X, Gao F, Cao H,
Qi F and Kapranov P: A CRISPR/Cas13-based approach demonstrates
biological relevance of vlinc class of long non-coding RNAs in
anticancer drug response. Sci Rep. 10:17942020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abudayyeh OO, Gootenberg JS, Konermann S,
Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E,
Minakhin L, et al: C2c2 is a single-component programmable
RNA-guided RNA-targeting CRISPR effector. Science. 353:aaf55732016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Núñez-Álvarez Y, Espie-Caullet T, Buhagiar
G, Rubio-Zulaika A, Alonso-Marañón J, Luna-Pérez E, Blazquez L and
Luco RF: A CRISPR-dCas13 RNA-editing tool to study alternative
splicing. Nucleic Acids Res. 52:11926–11939. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Apostolopoulos A, Kawamoto N, Chow SYA,
Tsuiji H, Ikeuchi Y, Shichino Y and Iwasaki S: dCas13-mediated
translational repression for accurate gene silencing in mammalian
cells. Nat Commun. 15:22052024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deltcheva E, Chylinski K, Sharma CM,
Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J and Charpentier
E: CRISPR RNA maturation by trans-encoded small RNA and host factor
RNase III. Nature. 471:602–607. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barrangou R, Fremaux C, Deveau H, Richards
M, Boyaval P, Moineau S, Romero DA and Horvath P: CRISPR provides
acquired resistance against viruses in prokaryotes. Science.
315:1709–1712. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rananaware SR, Vesco EK, Shoemaker GM,
Anekar SS, Sandoval LSW, Meister KS, Macaluso NC, Nguyen LT and
Jain PK: Programmable RNA detection with CRISPR-Cas12a. Nat Commun.
14:54092023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Allemailem KS: Recent advances in
understanding the molecular mechanisms of multidrug resistance and
novel approaches of CRISPR/Cas9-based genome-editing to combat this
health emergency. Int J Nanomedicine. 19:1125–1143. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
McGinn J and Marraffini LA: Molecular
mechanisms of CRISPR-Cas spacer acquisition. Nat Rev Microbiol.
17:7–12. 2019. View Article : Google Scholar
|
|
33
|
Makarova KS, Wolf YI and Koonin EV:
Evolutionary classification of CRISPR-Cas systems. Crispr: Biology
and Applications. 13–38. 2022. View Article : Google Scholar
|
|
34
|
Koonin EV, Makarova KS and Zhang F:
Diversity, classification and evolution of CRISPR-Cas systems. Curr
Opin Microbiol. 37:67–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yan F, Wang W and Zhang J: CRISPR-Cas12
and Cas13: The lesser known siblings of CRISPR-Cas9. Cell Biol
Toxicol. 35:489–492. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Makarova KS, Haft DH, Barrangou R, Brouns
SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI,
Yakunin AF, et al: Evolution and classification of the CRISPR-Cas
systems. Nat Rev Microbiol. 9:467–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Steens JA, Bravo JPK, Salazar CR, Yildiz
C, Amieiro AM, Köstlbacher S, Prinsen SHP, Andres AS, Patinios C,
Bardis A, et al: Type III-B CRISPR-Cas cascade of proteolytic
cleavages. Science. 383:512–519. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shabalina SA and Koonin EV: Origins and
evolution of eukaryotic RNA interference. Trends Ecol Evol.
23:578–587. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Makarova KS, Wolf YI, Iranzo J, Shmakov
SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH,
Horvath P, et al: Evolutionary classification of CRISPR-Cas
systems: A burst of class 2 and derived variants. Nat Rev
Microbiol. 18:67–83. 2020. View Article : Google Scholar
|
|
40
|
Brouns SJ, Jore MM, Lundgren M, Westra ER,
Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV and
Van Der Oost J: Small CRISPR RNAs guide antiviral defense in
prokaryotes. Science. 321:960–964. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen XD, Chen Z, Wythes G, Zhang Y, Orr
BC, Sun G, Chao YK, Navarro Torres A, Thao K, Vallurupalli M, et
al: Helicase-assisted continuous editing for programmable
mutagenesis of endogenous genomes. Science. 386:eadn58762024.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jinek M, Chylinski K, Fonfara I, Hauer M,
Doudna JA and Charpentier E: A programmable dual-RNA-guided DNA
endonuclease in adaptive bacterial immunity. Science. 337:816–821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chylinski K, Makarova KS, Charpentier E
and Koonin EV: Classification and evolution of type II CRISPR-Cas
systems. Nucleic Acids Res. 42:6091–6105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Marraffini LA and Sontheimer EJ: CRISPR
interference limits horizontal gene transfer in staphylococci by
targeting DNA. Science. 322:1843–1845. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stella G and Marraffini L: Type III
CRISPR-Cas: Beyond the Cas10 effector complex. Trends Biochem Sci.
49:28–37. 2024. View Article : Google Scholar :
|
|
46
|
Zhou Y, Bravo JPK, Taylor HN, Steens JA,
Jackson RN, Staals RHJ and Taylor DW: Structure of a type IV
CRISPR-Cas ribonucleoprotein complex. iScience. 24:1022012021.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zetsche B, Gootenberg JS, Abudayyeh OO,
Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van
Der Oost J, Regev A, et al: Cpf1 is a single RNA-guided
endonuclease of a class 2 CRISPR-Cas system. Cell. 163:759–771.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gao P, Yang H, Rajashankar KR, Huang Z and
Patel DJ: Type V CRISPR-Cas Cpf1 endonuclease employs a unique
mechanism for crRNA-mediated target DNA recognition. Cell Res.
26:901–913. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Abudayyeh OO, Gootenberg JS,
Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DB,
Kellner MJ, Regev A, et al: RNA targeting with CRISPR-Cas13.
Nature. 550:280–284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
van Beljouw SPB, Sanders J,
Rodríguez-Molina A and Brouns SJJ: RNA-targeting CRISPR-Cas
systems. Nat Rev Microbiol. 21:21–34. 2023. View Article : Google Scholar
|
|
51
|
Liu L, Li X, Ma J, Li Z, You L, Wang J,
Wang M, Zhang X and Wang Y: The molecular architecture for
RNA-guided RNA cleavage by Cas13a. Cell. 170:714–726.e10. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu C, Zhou Y, Xiao Q, He B, Geng G, Wang
Z, Cao B, Dong X, Bai W, Wang Y, et al: Programmable RNA editing
with compact CRISPR-Cas13 systems from uncultivated microbes. Nat
Methods. 18:499–506. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Perčulija V, Lin J, Zhang B and Ouyang S:
Functional features and current applications of the RNA-targeting
type VI CRISPR-Cas systems. Adv Sci (Weinh). 8:20046852021.
View Article : Google Scholar
|
|
54
|
Hu Y, Chen Y, Xu J, Wang X, Luo S, Mao B,
Zhou Q and Li W: Metagenomic discovery of novel CRISPR-Cas13
systems. Cell Discov. 8:1072022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
O'Connell MR: Molecular mechanisms of RNA
targeting by Cas13-containing type VI CRISPR-Cas systems. J Mol
Biol. 431:66–87. 2019. View Article : Google Scholar
|
|
56
|
Gootenberg JS, Abudayyeh OO, Lee JW,
Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer
NM, Freije CA, et al: Nucleic acid detection with
CRISPR-Cas13a/C2c2. Science. 356:438–442. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tambe A, East-Seletsky A, Knott GJ, Doudna
JA and O'Connell MR: RNA binding and HEPN-nuclease activation are
decoupled in CRISPR-Cas13a. Cell Rep. 24:1025–1036. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Meeske AJ, Nakandakari-Higa S and
Marraffini LA: Cas13-induced cellular dormancy prevents the rise of
CRISPR-resistant bacteriophage. Nature. 570:241–245. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tong H, Huang J, Xiao Q, He B, Dong X, Liu
Y, Yang X, Han D, Wang Z, Wang X, et al: High-fidelity Cas13
variants for targeted RNA degradation with minimal collateral
effects. Nat Biotechnol. 41:108–119. 2023. View Article : Google Scholar
|
|
60
|
VanderWal AR, Park JU, Polevoda B, Kellogg
EH and O'Connell MR: CRISPR-Csx28 forms a Cas13b-activated membrane
pore required for robust CRISPR-Cas adaptive immunity. Science.
380:66432023.
|
|
61
|
Guan J, Oromí-Bosch A, Mendoza SD,
Karambelkar S, Berry J and Bondy-Denomy J: RNA targeting with
CRISPR-Cas13a facilitates bacteriophage genome engineering.
BioRxiv. View Article : Google Scholar
|
|
62
|
Shmakov S, Abudayyeh OO, Makarova KS, Wolf
YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S,
Severinov K, et al: Discovery and functional characterization of
diverse class 2 CRISPR-Cas systems. Mol Cell. 60:385–397. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Deng X, Osikpa E, Yang J, Oladeji SJ,
Smith J, Gao X and Gao Y: Structural basis for the activation of a
compact CRISPR-Cas13 nuclease. Nat Commun. 14:58452023. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
East-Seletsky A, O'Connell MR, Burstein D,
Knott GJ and Doudna JA: RNA targeting by functionally orthogonal
type VI-A CRISPR-Cas enzymes. Mol Cell. 66:373–383.e3. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Knott GJ, East-Seletsky A, Cofsky JC,
Holton JM, Charles E, O'Connell MR and Doudna JA: Guide-bound
structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat
Struct Mol Biol. 24:825–833. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu L, Li X, Wang J, Wang M, Chen P, Yin
M, Li J, Sheng G and Wang Y: Two distant catalytic sites are
responsible for C2c2 RNase activities. Cell. 168:121–134.e12. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Charles EJ, Kim SE, Knott GJ, Smock D,
Doudna J and Savage DF: Engineering improved Cas13 effectors for
targeted post-transcriptional regulation of gene expression.
BioRxiv. View Article : Google Scholar
|
|
68
|
Molina Vargas AM, Sinha S, Osborn R,
Arantes PR, Patel A, Dewhurst S, Hardy DJ, Cameron A, Palermo G and
O'Connell MR: New design strategies for ultra-specific
CRISPR-Cas13a-based RNA detection with single-nucleotide mismatch
sensitivity. Nucleic Acids Res. 52:921–939. 2024. View Article : Google Scholar :
|
|
69
|
East-Seletsky A, O'Connell MR, Knight SC,
Burstein D, Cate JHD, Tjian R and Doudna JA: Two distinct RNase
activities of CRISPR-C2c2 enable guide-RNA processing and RNA
detection. Nature. 538:270–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ai Y, Liang D and Wilusz JE: CRISPR/Cas13
effectors have differing extents of off-target effects that limit
their utility in eukaryotic cells. Nucleic Acids Res. 50:e652022.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang T, Zhao Y, Ye J, Cao X, Xu C, Chen
B, An H, Jiao Y, Zhang F, Yang X and Zhou G: Establishing
CRISPR/Cas13a immune system conferring RNA virus resistance in both
dicot and monocot plants. Plant Biotechnol J. 17:1185–1187. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kick LM, Von Wrisberg MK, Runtsch LS and
Schneider S: Structure and mechanism of the RNA dependent RNase
Cas13a from Rhodobacter capsulatus. Commun Biol. 5:712022.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Smargon AA, Cox DBT, Pyzocha NK, Zheng K,
Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P,
Shmakov S, Makarova KS, et al: Cas13b is a type VI-B
CRISPR-associated RNA-guided RNase differentially regulated by
accessory proteins Csx27 and Csx28. Mol Cell. 65:618–630.e7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
VanderWal AR, Park JU, Polevoda B, Nicosia
JK, Molina Vargas AM, Kellogg EH and O'Connell MR: Csx28 is a
membrane pore that enhances CRISPR-Cas13b-dependent antiphage
defense. Science. 380:410–415. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang B, Ye W, Ye Y, Zhou H, Saeed AFUH,
Chen J, Lin J, Perčulija V, Chen Q, Chen CJ, et al: Structural
insights into Cas13b-guided CRISPR RNA maturation and recognition.
Cell Res. 28:1198–1201. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Slaymaker IM, Mesa P, Kellner MJ, Kannan
S, Brignole E, Koob J, Feliciano PR, Stella S, Abudayyeh OO,
Gootenberg JS, et al: High-resolution structure of Cas13b and
biochemical characterization of RNA targeting and cleavage. Cell
Rep. 26:3741–3751.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bhoobalan-Chitty Y, Stouf M and De Paepe
M: Genetic manipulation of Bacteriophage T4 utilizing the
CRISPR-Cas13b system. bioRxiv. View Article : Google Scholar
|
|
78
|
Liu L and Pei DS: Insights gained from RNA
editing targeted by the CRISPR-Cas13 family. Int J Mol Sci.
23:114002022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kavuri NR, Ramasamy M, Qi Y and Mandadi K:
Applications of CRISPR/Cas13-based RNA editing in plants. Cells.
11:26652022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Umaña A and Slade DJ: CRISPR-Cas systems
in Fusobacterium: An untapped genetic frontier. bioRxiv. View Article : Google Scholar
|
|
81
|
Hussein M, Liu Y, Vink M, Kroon PZ, Das
AT, Berkhout B and Herrera-Carrillo E: Evaluation of the effect of
RNA secondary structure on Cas13d-mediated target RNA cleavage. Mol
Ther Nucleic Acids. 35:1022782024. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wessels HH, Stirn A, Méndez-Mancilla A,
Kim EJ, Hart SK, Knowles DA and Sanjana NE: Prediction of on-target
and off-target activity of CRISPR-Cas13d guide RNAs using deep
learning. Nat Biotechnol. 42:628–637. 2024. View Article : Google Scholar
|
|
83
|
Gupta R, Ghosh A, Chakravarti R, Singh R,
Ravichandiran V, Swarnakar S and Ghosh D: Cas13d: A new molecular
scissor for transcriptome engineering. Front Cell Dev Biol.
10:8668002022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tang XZ, Tan SX, Hoon S and Yeo GW:
Pre-existing adaptive immunity to the RNA-editing enzyme Cas13d in
humans. Nat Med. 28:1372–1376. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kordyś M, Sen R and Warkocki Z:
Applications of the versatile CRISPR-Cas13 RNA targeting system.
Wiley Interdiscip Rev RNA. 13:e16942022. View Article : Google Scholar
|
|
86
|
Liu Y, Jing P, Zhou Y, Zhang J, Shi J,
Zhang M, Yang H and Fei J: The effects of length and sequence of
gRNA on Cas13b and Cas13d activity in vitro and in vivo. Biotechnol
J. 18:e23000022023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang C, Konermann S, Brideau NJ, Lotfy P,
Wu X, Novick SJ, Strutzenberg T, Griffin PR, Hsu PD and Lyumkis D:
Structural basis for the RNA-guided ribonuclease activity of
CRISPR-Cas13d. Cell. 175:212–223.e17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang K, Zhang Z, Kang J, Chen J, Liu J,
Gao N, Fan L, Zheng P, Wang Y and Sun J: CRISPR/Cas13d-mediated
microbial RNA knockdown. Front Bioeng Biotechnol. 8:8562020.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Nakagawa R, Kannan S, Altae-Tran H, Takeda
SN, Tomita A, Hirano H, Kusakizako T, Nishizawa T, Yamashita K,
Zhang F, et al: Structure and engineering of the minimal type VI
CRISPR-Cas13bt3. Mol Cell. 82:3178–3192.e5. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Mahas A, Wang Q, Marsic T and Mahfouz MM:
A novel miniature CRISPR-Cas13 system for SARS-CoV-2 diagnostics.
ACS Synth Biol. 10:2541–2551. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Mehta A and Merkel OM: Immunogenicity of
Cas9 protein. J Pharm Sci. 109:62–67. 2020. View Article : Google Scholar :
|
|
92
|
Kushawah G, Hernandez-Huertas L,
Abugattas-Nuñez Del Prado J, Martinez-Morales JR, DeVore ML, Hassan
H, Moreno-Sanchez I, Tomas-Gallardo L, Diaz-Moscoso A, Monges DE,
et al: CRISPR-Cas13d induces efficient mRNA knockdown in animal
embryos. Dev Cell. 54:805–817.e7. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhou C, Hu X, Tang C, Liu W, Wang S, Zhou
Y, Zhao Q, Bo Q, Shi L, Sun X, et al: CasRx-mediated RNA targeting
prevents choroidal neovascularization in a mouse model of
age-related macular degeneration. Natl Sci Rev. 7:835–837. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhou H, Su J, Hu X, Zhou C, Li H, Chen Z,
Xiao Q, Wang BO, Wu W, Sun Y, et al: Glia-to-neuron conversion by
CRISPR-CasRx alleviates symptoms of neurological disease in mice.
Cell. 181:590–603.e16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Granados-Riveron JT and Aquino-Jarquin G:
CRISPR-Cas13 precision transcriptome engineering in cancer. Cancer
Res. 78:4107–4113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Yazdi ZF, Roshannezhad S, Sharif S and
Abbaszadegan MR: Recent progress in prompt molecular detection of
liquid biopsy using Cas enzymes: Innovative approaches for cancer
diagnosis and analysis. J Transl Med. 22:11732024. View Article : Google Scholar
|
|
97
|
Kellner MJ, Koob JG, Gootenberg JS,
Abudayyeh OO and Zhang F: SHERLOCK: Nucleic acid detection with
CRISPR nucleases. Nat Protoc. 14:2986–3012. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Patchsung M, Jantarug K, Pattama A,
Aphicho K, Suraritdechachai S, Meesawat P, Sappakhaw K, Leelahakorn
N, Ruenkam T, Wongsatit T, et al: Clinical validation of a
Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed
Eng. 4:1140–1149. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Gao J, Luo T, Lin N, Zhang S and Wang J: A
new tool for CRISPR-Cas13a-based cancer gene therapy. Mol Ther
Oncolytics. 19:79–92. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Usuba W, Urabe F, Yamamoto Y, Matsuzaki J,
Sasaki H, Ichikawa M, Takizawa S, Aoki Y, Niida S, Kato K, et al:
Circulating miRNA panels for specific and early detection in
bladder cancer. Cancer Sci. 110:408–419. 2019. View Article : Google Scholar :
|
|
101
|
Liu R, Chen X, Du Y, Yao W, Shen L, Wang
C, Hu Z, Zhuang R, Ning G, Zhang C, et al: Serum microRNA
expression profile as a biomarker in the diagnosis and prognosis of
pancreatic cancer. Clin Chem. 58:610–618. 2012. View Article : Google Scholar
|
|
102
|
Shi J, Li X, Zhang F, Zhang C, Guan Q, Cao
X, Zhu W, Zhang X, Cheng Y, Ou K, et al: Circulating lncRNAs
associated with occurrence of colorectal cancer progression. Am J
Cancer Res. 5:2258–2265. 2015.PubMed/NCBI
|
|
103
|
Sheng Y, Zhang T, Zhang S, Johnston M,
Zheng X, Shan Y, Liu T, Huang Z, Qian F, Xie Z, et al: A
CRISPR/Cas13a-powered catalytic electrochemical biosensor for
successive and highly sensitive RNA diagnostics. Biosens
Bioelectron. 178:1130272021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Cescon DW, Bratman SV, Chan SM and Siu LL:
Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer.
1:276–290. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Rothwell DG, Ayub M, Cook N,
Thistlethwaite F, Carter L, Dean E, Smith N, Villa S, Dransfield J,
Clipson A, et al: Utility of ctDNA to support patient selection for
early phase clinical trials: The TARGET study. Nat Med. 25:738–743.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhang X, Tian Y, Xu L, Fan Z, Cao Y, Ma Y,
Li H and Ren F: CRISPR/Cas13-assisted hepatitis B virus covalently
closed circular DNA detection. Hepatol Int. 16:306–315. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Antropov DN and Stepanov GA: Molecular
mechanisms underlying CRISPR/Cas-based assays for nucleic acid
detection. Curr Issues Mol Biol. 45:649–662. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Gootenberg JS, Abudayyeh OO, Kellner MJ,
Joung J, Collins JJ and Zhang F: Multiplexed and portable nucleic
acid detection platform with Cas13, Cas12a, and Csm6. Science.
360:439–444. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Myhrvold C, Freije CA, Gootenberg JS,
Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM,
Parham LA, et al: Field-deployable viral diagnostics using
CRISPR-Cas13. Science. 360:444–448. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Bianchi F, Nicassio F, Marzi M, Belloni E,
Dall'Olio V, Bernard L, Pelosi G, Maisonneuve P, Veronesi G and Di
Fiore PP: A serum circulating miRNA diagnostic test to identify
asymptomatic high-risk individuals with early stage lung cancer.
EMBO Mol Med. 3:495–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Zhang L, Xu Y, Jin X, Wang Z, Wu Y, Zhao
D, Chen G, Li D, Wang X, Cao H, et al: A circulating miRNA
signature as a diagnostic biomarker for non-invasive early
detection of breast cancer. Breast Cancer Res Treat. 154:423–434.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhou T, Huang R, Huang M, Shen J, Shan Y
and Xing D: CRISPR/Cas13a powered portable electrochemiluminescence
chip for ultrasensitive and specific MiRNA detection. Adv Sci
(Weinh). 7:19036612020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Bruch R, Baaske J, Chatelle C, Meirich M,
Madlener S, Weber W, Dincer C and Urban GA: CRISPR/Cas13a-powered
electrochemical microfluidic biosensor for nucleic acid
amplification-free miRNA diagnostics. Adv Mater. 31:e19053112019.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Bruch R, Johnston M, Kling A, Mattmüller
T, Baaske J, Partel S, Madlener S, Weber W, Urban GA and Dincer C:
CRISPR-powered electrochemical microfluidic multiplexed biosensor
for target amplification-free miRNA diagnostics. Biosens
Bioelectron. 177:1128872021. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Ruivo CF, Adem B, Silva M and Melo SA: The
biology of cancer exosomes: Insights and new perspectives. Cancer
Res. 77:6480–6488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Contreras-Naranjo JC, Wu HJ and Ugaz VM:
Microfluidics for exosome isolation and analysis: Enabling liquid
biopsy for personalized medicine. Lab Chip. 17:3558–3577. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Li W, Li C, Zhou T, Liu X, Liu X, Li X and
Chen D: Role of exosomal proteins in cancer diagnosis. Mol Cancer.
16:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Li A, Zhang T, Zheng M, Liu Y and Chen Z:
Exosomal proteins as potential markers of tumor diagnosis. J
Hematol Oncol. 10:1752017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Shao H, Im H, Castro CM, Breakefield X,
Weissleder R and Lee H: New technologies for analysis of
extracellular vesicles. Chem Rev. 118:1917–1950. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
He Y, Wu Y, Wang Y, Wang X, Xing S, Li H,
Guo S, Yu X, Dai S, Zhang G, et al: Applying CRISPR/Cas13 to
construct exosomal PD-L1 ultrasensitive biosensors for dynamic
monitoring of tumor progression in immunotherapy. Adv Therap.
3:20000932020. View Article : Google Scholar
|
|
121
|
Chen Q, Tian T, Xiong E, Wang P and Zhou
X: CRISPR/Cas13a signal amplification linked immunosorbent assay
for femtomolar protein detection. Anal Chem. 92:573–577. 2020.
View Article : Google Scholar
|
|
122
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Chen Y, Jiang H, Wang T, He D, Tian R, Cui
Z, Tian X, Gao Q, Ma X, Yang J, et al: In vitro and in vivo growth
inhibition of human cervical cancer cells via human papillomavirus
E6/ E7 mRNAs' cleavage by CRISPR/Cas13a system. Antiviral Res.
178:1047942020. View Article : Google Scholar
|
|
124
|
Zhang Z, Wang Q, Liu Q, Zheng Y, Zheng C,
Yi K, Zhao Y, Gu Y, Wang Y, Wang C, et al: Dual-locking
nanoparticles disrupt the PD-1/PD-L1 pathway for efficient cancer
immunotherapy. Adv Mater. 31:e19057512019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Jiang W, Li H, Liu X, Zhang J, Li T, Liu L
and Yu X: Precise and efficient silencing of mutant
KrasG12D by CRISPR-CasRx controls pancreatic cancer
progression. Theranostics. 10:115072020. View Article : Google Scholar :
|
|
126
|
Yue H, Huang R, Shan Y and Xing D:
Delivery of Cas13a/ crRNA by self-degradable black phosphorus
nanosheets to specifically inhibit Mcl-1 for breast cancer therapy.
J Mater Chem B. 8:11096–11106. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Lin P, Qin S, Pu Q, Wang Z, Wu Q, Gao P,
Schettler J, Guo K, Li R, Li G, et al: CRISPR-Cas13 inhibitors
block RNA editing in bacteria and mammalian cells. Mol Cell.
78:850–861. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Meeske AJ, Jia N, Cassel AK, Kozlova A,
Liao J, Wiedmann M, Patel DJ and Marraffini LA: A phage-encoded
anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas
immunity. Science. 369:54–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Bubeck F, Hoffmann MD, Harteveld Z,
Aschenbrenner S, Bietz A, Waldhauer MC, Börner K, Fakhiri J,
Schmelas C, Dietz L, et al: Engineered anti-CRISPR proteins for
optogenetic control of CRISPR-Cas9. Nat Methods. 15:924–927. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Lee J, Mou H, Ibraheim R, Liang SQ, Liu P,
Xue W and Sontheimer EJ: Tissue-restricted genome editing in vivo
specified by microRNA-repressible anti-CRISPR proteins. RNA.
25:1421–1431. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Fan J, Liu Y, Liu L, Huang Y, Li X and
Huang W: A multifunction lipid-based CRISPR-Cas13a genetic circuit
delivery system for bladder cancer gene therapy. ACS Synth Biol.
9:343–355. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Kopparapu PK, Boorjian SA, Robinson BD,
Downes M, Gudas LJ, Mongan NP and Persson JL: Expression of VEGF
and its receptors VEGFR1/VEGFR2 is associated with invasiveness of
bladder cancer. Anticancer Res. 33:2381–2390. 2013.PubMed/NCBI
|
|
133
|
Liu L, Liu Y, Zhang T, Wu H, Lin M, Wang
C, Zhan Y, Zhou Q, Qiao B, Sun X, et al: Synthetic Bax-Anti Bcl2
combination module actuated by super artificial hTERT promoter
selectively inhibits malignant phenotypes of bladder cancer. J Exp
Clin Cancer Res. 35:32016. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Wang D, Tang H, Xu X, Dai W, Wu J and Wang
J: Control the intracellular NF-κB activity by a sensor consisting
of miRNA and decoy. Int J Biochem Cell Biol. 95:43–52. 2018.
View Article : Google Scholar
|
|
135
|
Dai W, Wu J, Wang D and Wang J: Cancer
gene therapy by NF-κB-activated cancer cell-specific expression of
CRISPR/ Cas9 targeting telomeres. Gene Ther. 27:266–280. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Wang D, Dai W and Wang J: A cell-specific
nuclear factor-kappa B-activating gene expression strategy for
delivering cancer immunotherapy. Hum Gene Ther. 30:471–484. 2019.
View Article : Google Scholar
|
|
137
|
Rahib L, Wehner MR, Matrisian LM and Nead
KT: Estimated projection of US cancer incidence and death to 2040.
JAMA Netw Open. 4:e2147082021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Halbrook CJ, Lyssiotis CA, di Magliano MP
and Maitra A: Pancreatic cancer: Advances and challenges. Cell.
186:1729–1754. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Linehan A, O'Reilly M, McDermott R and
O'Kane GM: Targeting KRAS mutations in pancreatic cancer:
Opportunities for future strategies. Front Med (Lausanne).
11:13691362024. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Amintas S, Cullot G, Boubaddi M, Rébillard
J, Karembe L, Turcq B, Prouzet-Mauléon V, Bedel A, Moreau-Gaudry F,
Cappellen D and Dabernat S: Integrating allele-specific PCR with
CRISPR-Cas13a for sensitive KRAS mutation detection in pancreatic
cancer. J Biol Eng. 18:532024. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Zhao X, Liu L, Lang J, Cheng K, Wang Y, Li
X, Shi J, Wang Y and Nie G: A CRISPR-Cas13a system for efficient
and specific therapeutic targeting of mutant KRAS for pancreatic
cancer treatment. Cancer Lett. 431:171–181. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Zhang Y, Li S, Li R, Qiu X, Fan T, Wang B,
Zhang B and Zhang L: Advances in application of CRISPR-Cas13a
system. Front Cell Infect Microbiol. 14:12915572024. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Kluth LA, Black PC, Bochner BH, Catto J,
Lerner SP, Stenzl A, Sylvester R, Vickers AJ, Xylinas E and Shariat
SF: Prognostic and prediction tools in bladder cancer: A
comprehensive review of the literature. Eur Urol. 68:238–253. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Wéber A, Vignat J, Shah R, Morgan E,
Laversanne M, Nagy P, Kenessey I and Znaor A: Global burden of
bladder cancer mortality in 2020 and 2040 according to GLOBOCAN
estimates. World J Urol. 42:2372024. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Liang M, Wang Y, Liu L, Deng D, Yan Z,
Feng L, Kong C, Li C, Li Y and Li G: Synergistic intravesical
instillation for bladder cancer: CRISPR-Cas13a and fenbendazole
combination therapy. J Exp Clin Cancer Res. 43:2232024. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Zhou L, Li Y, Wang H, Qin R, Han Z and Li
R: Global cervical cancer elimination: Quantifying the status,
progress, and gaps. BMC Med. 23:672025. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Matsukura T and Sugase M: Pitfalls in the
epidemiologic classification of human papillomavirus types
associated with cervical cancer using polymerase chain reaction:
Driver and passenger. Int J Gynecol Cancer. 18:1042–1050. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Goodwin EC and DiMaio D: Repression of
human papillomavirus oncogenes in HeLa cervical carcinoma cells
causes the orderly reactivation of dormant tumor suppressor
pathways. Proc Natl Acad Sci USA. 97:12513–12518. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Pellerino A, Caccese M, Padovan M,
Cerretti G and Lombardi G: Epidemiology, risk factors, and
prognostic factors of gliomas. Clin Transl Imaging. 10:467–475.
2022. View Article : Google Scholar
|
|
150
|
Verhaak RGW, Hoadley KA, Purdom E, Wang V,
Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al:
Integrated genomic analysis identifies clinically relevant subtypes
of glioblastoma characterized by abnormalities in PDGFRA, IDH1,
EGFR, and NF1. Cancer Cell. 17:98–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Wang Q, Liu X, Zhou J, Yang C, Wang G, Tan
Y, Wu Y, Zhang S, Yi K and Kang C: The CRISPR-Cas13a gene-editing
system induces collateral cleavage of RNA in glioma cells. Adv Sci
(Weinh). 6:19012992019. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Di Carlo E and Sorrentino C: State of the
art CRISPR-based strategies for cancer diagnostics and treatment.
Biomark Res. 12:1562024. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Azeez SS, Hamad RS, Hamad BK, Shekha MS
and Bergsten P: Advances in CRISPR-Cas technology and its
applications: Revolutionising precision medicine. Front Genome Ed.
6:15099242024. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Chehelgerdi M, Chehelgerdi M,
Khorramian-Ghahfarokhi M, Shafieizadeh M, Mahmoudi E, Eskandari F,
Rashidi M, Arshi A and Mokhtari-Farsani A: Comprehensive review of
CRISPR-based gene editing: mechanisms, challenges, and applications
in cancer therapy. Mol Cancer. 23:92024. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Mousavi Kahaki SA, Ebrahimzadeh N, Fahimi
H and Moshiri A: Development of an optimized protocol for
generating knockout cancer cell lines using the CRISPR/Cas9 system,
with emphasis on transient transfection. PLoS One. 19:e03103682024.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Tian T, Shu B, Jiang Y, Ye M, Liu L, Guo
Z, Han Z, Wang Z and Zhou X: An ultralocalized Cas13a assay enables
universal and nucleic acid amplification-free single-molecule RNA
diagnostics. ACS Nano. 15:1167–1178. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Zhou T, Huang M, Lin J, Huang R and Xing
D: High-fidelity CRISPR/Cas13a trans-cleavage-triggered rolling
circle amplified DNAzyme for visual profiling of microRNA. Anal
Chem. 93:2038–2044. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Sha Y, Huang R, Huang M, Yue H, Shan Y, Hu
J and Xing D: Cascade CRISPR/cas enables amplification-free
microRNA sensing with fM-sensitivity and single-base-specificity.
Chem Commun (Camb). 57:247–250. 2021. View Article : Google Scholar
|
|
159
|
Cui Y, Fan S, Yuan Z, Song M, Hu J, Qian
D, Zhen D, Li J and Zhu B: Ultrasensitive electrochemical assay for
microRNA-21 based on CRISPR/Cas13a-assisted catalytic hairpin
assembly. Talanta. 224:1218782021. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Huang M, Huang R, Yue H, Shan Y and Xing
D: Ultrasensitive and high-specific microRNA detection using
hyper-branching rolling circle amplified CRISPR/Cas13a biosensor.
Sens Actuat B Chem. 325:1287992020. View Article : Google Scholar
|