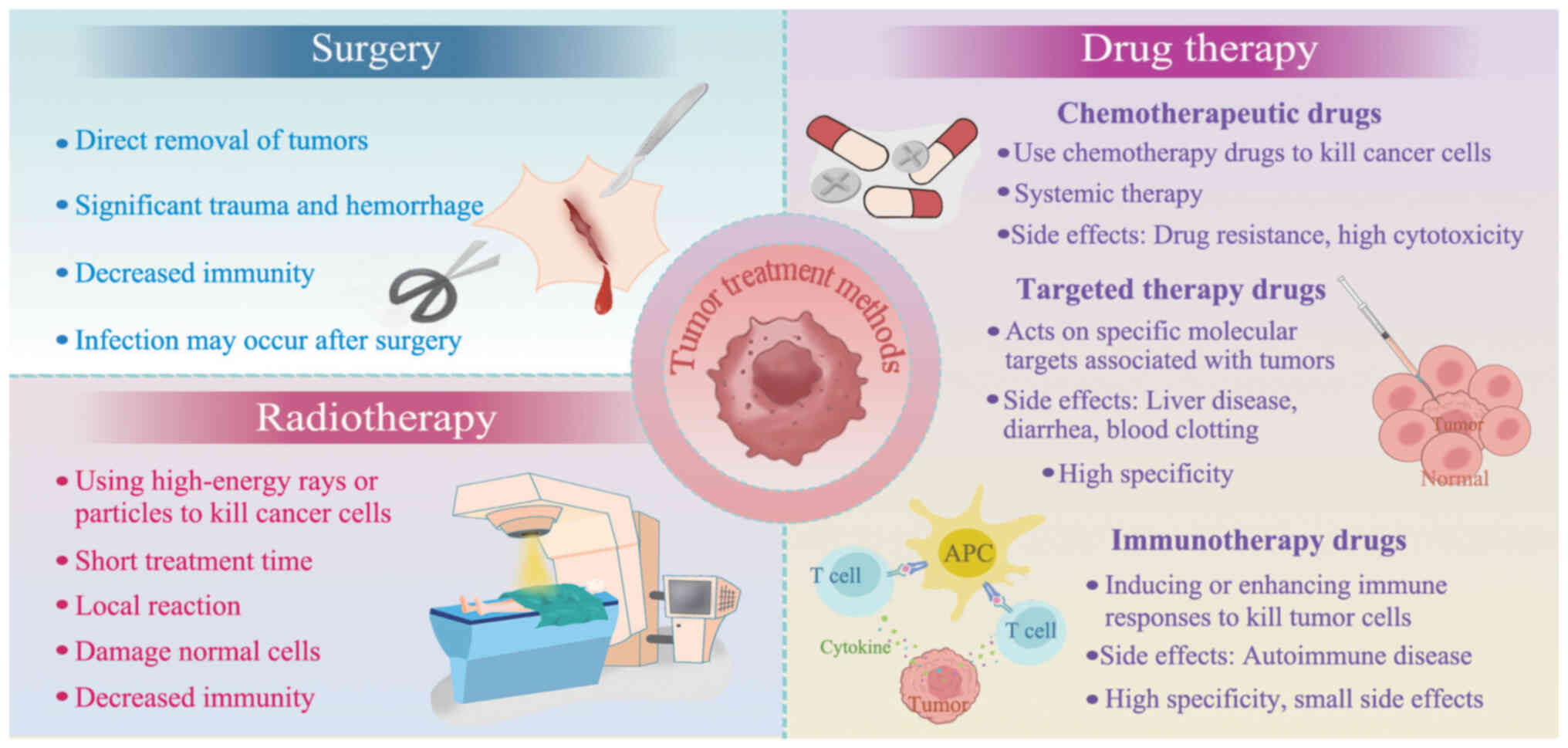
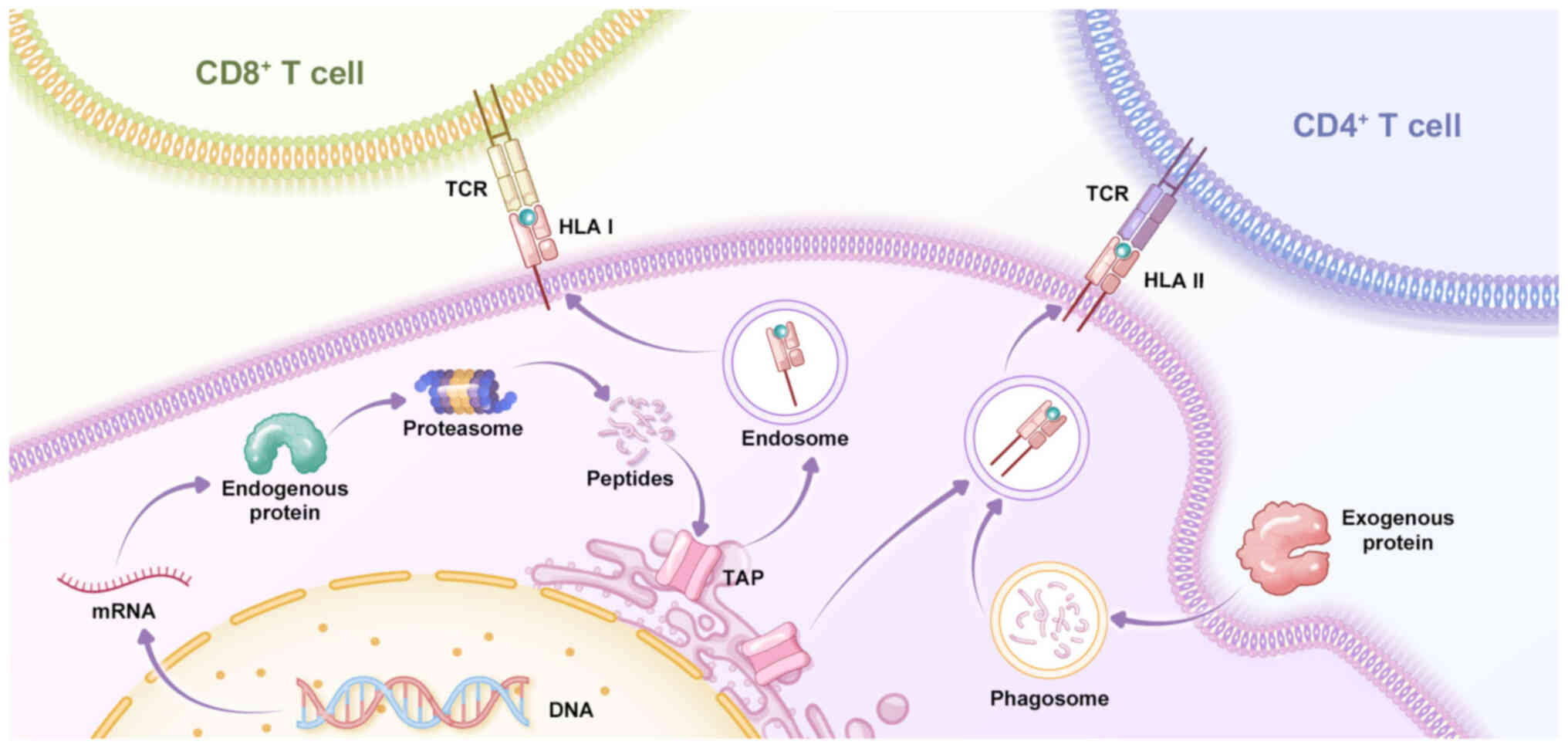
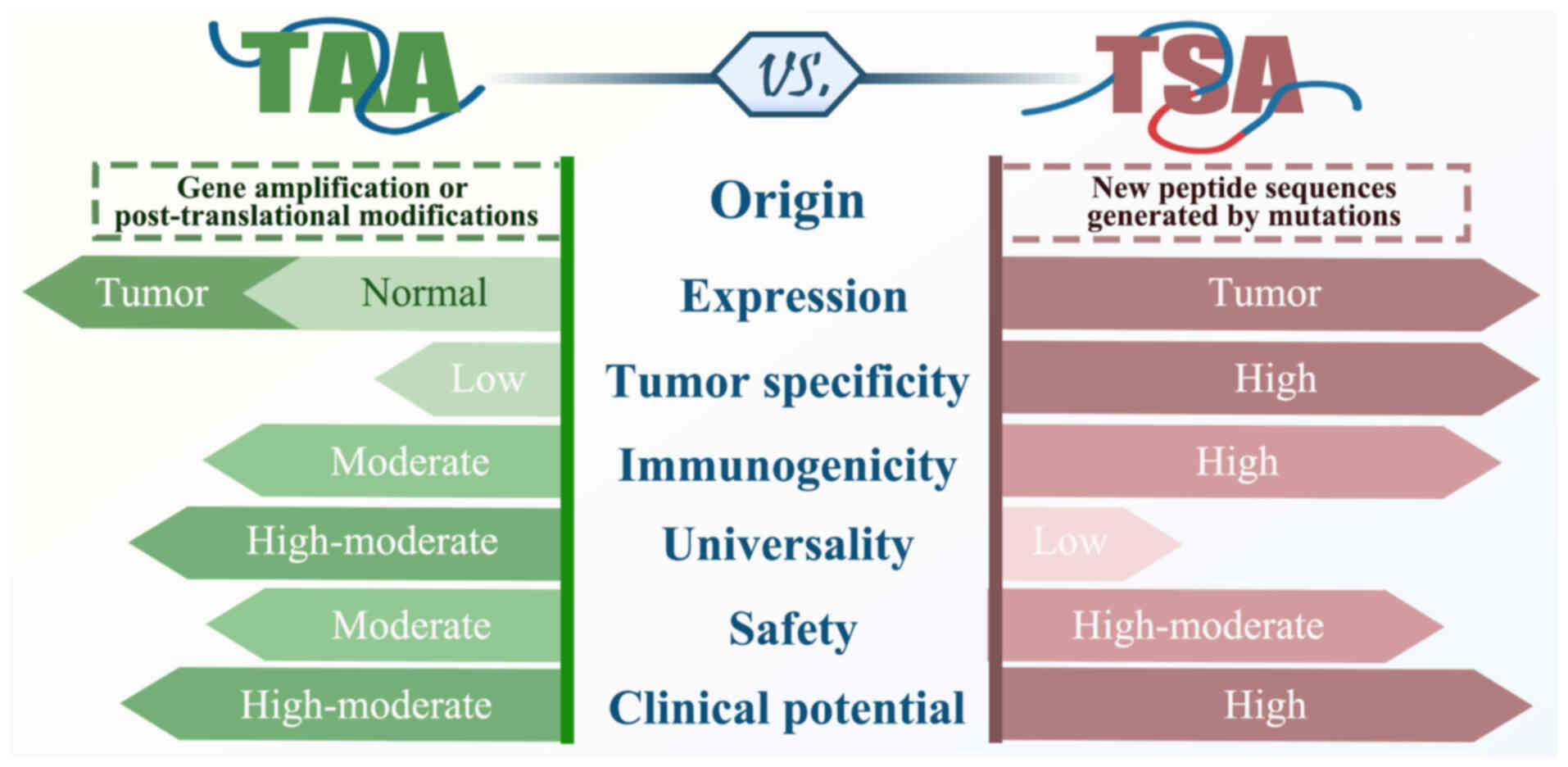
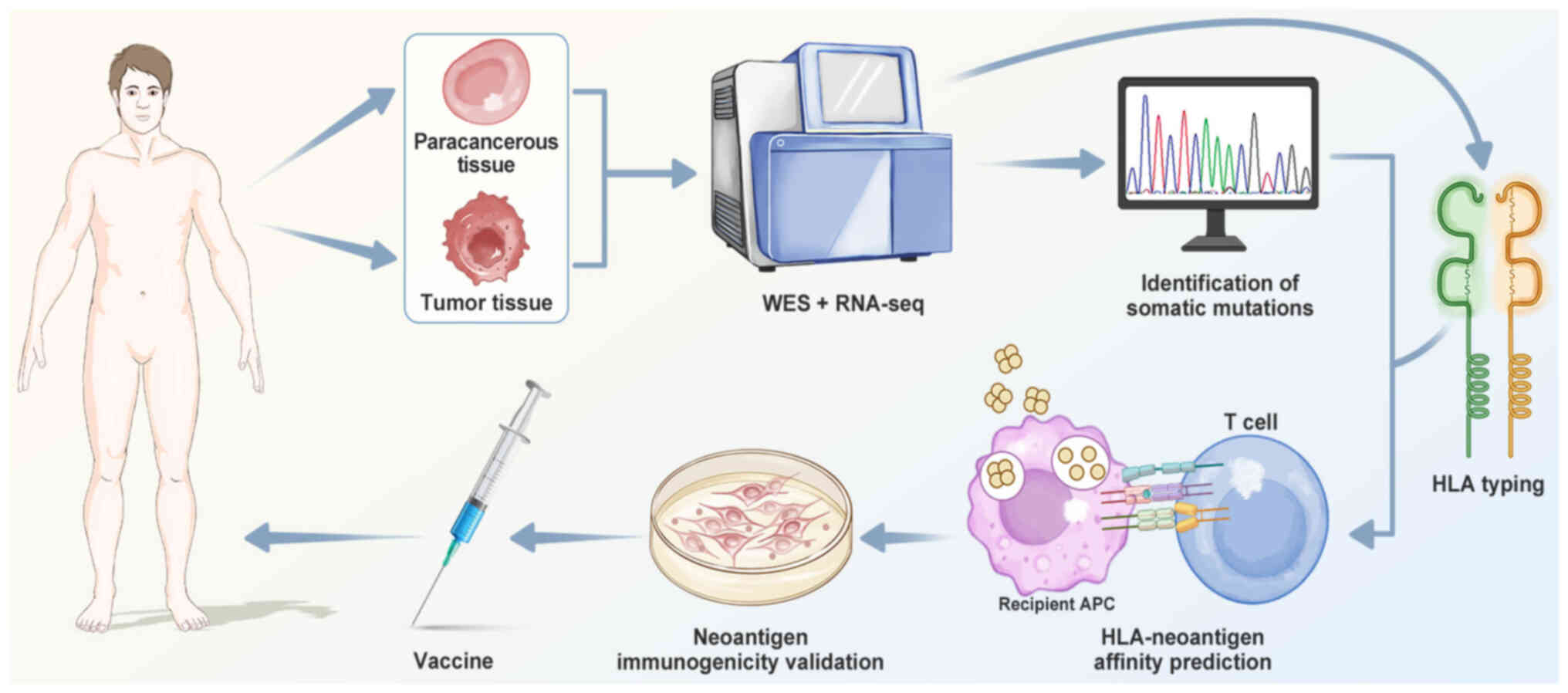
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Health Organization: Global cancer
burden growing, amidst mounting need for services. 2024.
|
|
3
|
Siegel RL, Kratzer TB, Giaquinto AN, Sung
H and Jemal A: Cancer statistics, 2025. CA Cancer J Clin. 75:10–45.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de Visser KE and Joyce JA: The evolving
tumor microenvironment: From cancer initiation to metastatic
outgrowth. Cancer Cell. 41:374–403. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cai Z, Yin Y, Shen C, Wang J, Yin X, Chen
Z, Zhou Y and Zhang B: Comparative effectiveness of preoperative,
postoperative and perioperative treatments for resectable gastric
cancer: A network meta-analysis of the literature from the past 20
years. Surg Oncol. 27:563–574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krall JA, Reinhardt F, Mercury OA,
Pattabiraman DR, Brooks MW, Dougan M, Lambert AW, Bierie B, Ploegh
HL, Dougan SK and Weinberg RA: The systemic response to surgery
triggers the outgrowth of distant immune-controlled tumors in mouse
models of dormancy. Sci Transl Med. 10:eaan34642018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu WD, Sun G, Li J, Xu J and Wang X:
Mechanisms and therapeutic potentials of cancer immunotherapy in
combination with radiotherapy and/or chemotherapy. Cancer Lett.
452:66–70. 2019. View Article : Google Scholar
|
|
8
|
Pich O, Muiños F, Lolkema MP, Steeghs N,
Gonzalez-Perez A and Lopez-Bigas N: The mutational footprints of
cancer therapies. Nat Genet. 51:1732–1740. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sonis ST: Mucositis: The impact, biology
and therapeutic opportunities of oral mucositis. Oral Oncol.
45:1015–1020. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao H, Xiong L, Song X, Jin P, Chen L,
Chen X, Yao H, Wang Y and Wang L: Angelica sinensis polysaccharides
ameliorate stress-induced premature senescence of hematopoietic
cell via protecting bone marrow stromal cells from oxidative
injuries caused by 5-fluorouracil. Int J Mol Sci. 18:22652017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Probin V and Zhou DH: Cancer
therapy-induced residual bone marrow injury-mechanisms of induction
and implication for therapy. Curr Cancer Ther Rev. 2:271–279. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ryan JL: Ionizing radiation: The good, the
bad, and the ugly. J Invest Dermatol. 132:985–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dracham CB, Shankar A and Madan R:
Radiation induced secondary malignancies: A review article. Radiat
Oncol J. 36:85–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gupta K, Walton R and Kataria SP:
Chemotherapy-induced nausea and vomiting: Pathogenesis,
recommendations, and new trends. Cancer Treat Res Commun.
26:1002782021. View Article : Google Scholar
|
|
15
|
Spears N, Lopes F, Stefansdottir A, Rossi
V, De Felici M, Anderson RA and Klinger FG: Ovarian damage from
chemotherapy and current approaches to its protection. Hum Reprod
Update. 25:673–693. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Urruticoechea A, Alemany R, Balart J,
Villanueva A, Viñals F and Capellá G: Recent advances in cancer
therapy: An overview. Curr Pharm Des. 16:3–10. 2010. View Article : Google Scholar
|
|
17
|
Cavalcanti IDL and Soares JCS: Advances in
cancer treatment: From systemic chemotherapy to targeted therapy.
Springer Nature. 1–109. 2021.
|
|
18
|
Baudino TA: Targeted cancer therapy: The
next generation of cancer treatment. Curr Drug Discov Technol.
12:3–20. 2015. View Article : Google Scholar
|
|
19
|
Lollini PL, Cavallo F, Nanni P and Forni
G: Vaccines for tumour prevention. Nat Rev Cancer. 6:204–216. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pinho MP, Sundarasetty BS, Bergami-Santos
PC, Steponavicius-Cruz K, Ferreira AK, Stripecke R and Barbuto JAM:
Dendritic-tumor cell hybrids induce tumor-specific immune responses
more effectively than the simple mixture of dendritic and tumor
cells. Cytotherapy. 18:570–580. 2016. View Article : Google Scholar
|
|
21
|
Guzhova IV and Margulis BA: HSP70-based
anti-cancer immunotherapy. Hum Vaccin Immunother. 12:2529–2535.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Howell LM and Forbes NS: Bacteria-based
immune therapies for cancer treatment. Semin Cancer Biol.
86:1163–1178. 2022. View Article : Google Scholar
|
|
23
|
Eshhar Z: The T-body approach: Redirecting
T cells with antibody specificity. Handb Exp Pharmacol.
181:329–342. 2008. View Article : Google Scholar
|
|
24
|
Dai R, Liu M, Nik Nabil WN, Xi Z and Xu H:
Mycomedicine: A unique class of natural products with potent
anti-tumour bioactivities. Molecules. 26:11132021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lau TTY, Sefid Dashti ZJ, Titmuss E,
Pender A, Topham JT, Bridgers J, Loree JM, Feng X, Pleasance ED,
Renouf DJ, et al: The neoantigen landscape of the coding and
noncoding cancer genome space. J Mol Diagn. 24:609–618. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lybaert L, Lefever S, Fant B, Smits E, De
Geest B, Breckpot K, Dirix L, Feldman SA, van Criekinge W,
Thielemans K, et al: Challenges in neoantigen-directed
therapeutics. Cancer Cell. 41:15–40. 2023. View Article : Google Scholar
|
|
27
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761. 2017.
View Article : Google Scholar
|
|
28
|
Sánchez-Danés A and Blanpain C:
Deciphering the cells of origin of squamous cell carcinomas. Nat
Rev Cancer. 18:549–561. 2018. View Article : Google Scholar
|
|
29
|
Kim J, Lee BJ, Moon S, Lee H, Lee J, Kim
BS, Jung K, Seo H and Chung Y: Strategies to overcome hurdles in
cancer immunotherapy. Biomater Res. 28:00802024. View Article : Google Scholar
|
|
30
|
Farhadi Rad H, Tahmasebi H, Javani S,
Hemati M, Zakerhamidi D, Hosseini M, Alibabaei F, Banihashemian SZ,
Oksenych V and Eslami M: Microbiota and cytokine modulation:
Innovations in enhancing anticancer immunity and personalized
cancer therapies. Biomedicines. 12:27762024. View Article : Google Scholar
|
|
31
|
Ritu, Chandra P and Das A: Immune
checkpoint targeting antibodies hold promise for combinatorial
cancer therapeutics. Clin Exp Med. 23:4297–4322. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luo Q, Zhang L, Luo C and Jiang M:
Emerging strategies in cancer therapy combining chemotherapy with
immunotherapy. Cancer Lett. 454:191–203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Weiner GJ: Building better monoclonal
antibody-based therapeutics. Nat Rev Cancer. 15:361–370. 2015.
View Article : Google Scholar :
|
|
34
|
Wei G, Zhang H, Zhao H, Wang J, Wu N, Li
L, Wu J and Zhang D: Emerging immune checkpoints in the tumor
microenvironment: Implications for cancer immunotherapy. Cancer
Lett. 511:68–76. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Oka Y, Tsuboi A, Oji Y, Kawase I and
Sugiyama H: WT1 peptide vaccine for the treatment of cancer. Curr
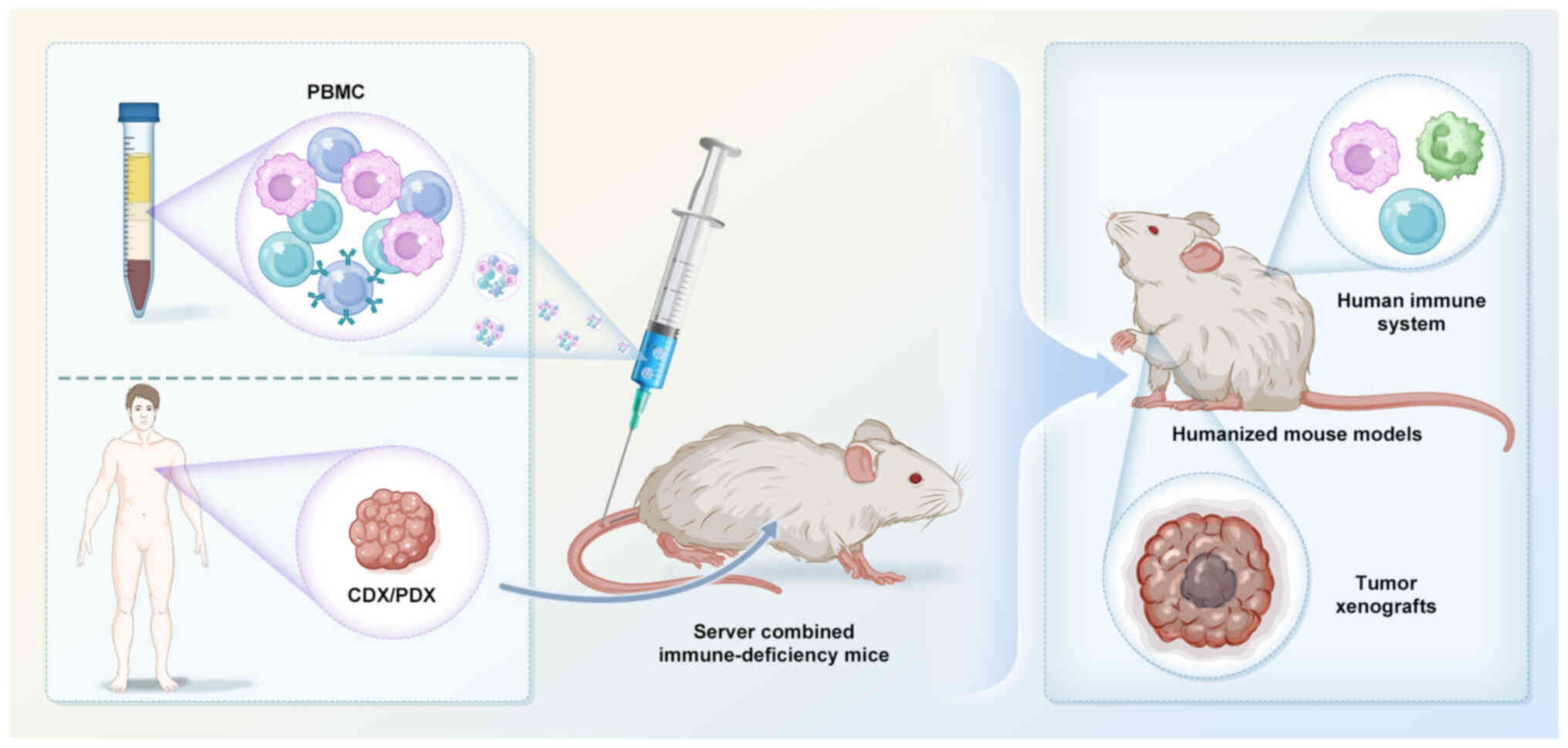
Opin Immunol. 20:211–220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Castle JC, Kreiter S, Diekmann J, Löwer M,
van de Roemer N, de Graaf J, Selmi A, Diken M, Boegel S, Paret C,
et al: Exploiting the mutanome for tumor vaccination. Cancer Res.
72:1081–1091. 2012. View Article : Google Scholar
|
|
37
|
Rosenberg SA and Restifo NP: Adoptive cell
transfer as personalized immunotherapy for human cancer. Science.
348:62–68. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang J and Wang L: The emerging world of
TCR-T cell trials against cancer: A systematic review. Technol
Cancer Res Treat. 18:15330338198310682019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao QJ, Jiang Y, Xiang SX, Kaboli PJ,
Shen J, Zhao YS, Wu X, Du FK, Li MX, Cho CH, et al: Engineered
TCR-T cell immunotherapy in anticancer precision medicine: Pros and
cons. Front Immunol. 12:6587532021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sterner RC and Sterner RM: CAR-T cell
therapy: Current limitations and potential strategies. Blood Cancer
J. 11:692021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Daher M, Melo Garcia L, Li Y and Rezvani
K: CAR-NK cells: The next wave of cellular therapy for cancer. Clin
Transl Immunology. 10:e12742021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen XT, Yang J, Wang LF and Liu BR:
Personalized neoantigen vaccination with synthetic long peptides:
Recent advances and future perspectives. Theranostics.
10:6011–6023. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li L, Goedegebuure SP and Gillanders WE:
Preclinical and clinical development of neoantigen vaccines. Ann
Oncol. 28(Suppl 12): xii11–xii17. 2017. View Article : Google Scholar
|
|
44
|
Lybaert L, Thielemans K, Feldman SA, van
der Burg SH, Bogaert C and Ott PA: Neoantigen-directed therapeutics
in the clinic: Where are we? Trends Cancer. 9:503–519. 2023.
View Article : Google Scholar
|
|
45
|
Luo Y, Zhou H, Mizutani M, Mizutani N, Liu
C, Xiang R and Reisfeld RA: A DNA vaccine targeting Fos-related
antigen 1 enhanced by IL-18 induces long-lived T-cell memory
against tumor recurrence. Cancer Res. 65:3419–3427. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ribatti D: The concept of immune
surveillance against tumors: The first theories. Oncotarget.
8:7175–7180. 2017. View Article : Google Scholar
|
|
47
|
Gross L: Intradermal immunization of C3H
mice against a sarcoma that originated in an animal of the same
line. Cancer Res. 3:326–333. 1943.
|
|
48
|
Baldwin RW: Tumour-specific immunity
against spontaneous rat tumours. Int J Cancer. 1:257–264. 1966.
View Article : Google Scholar
|
|
49
|
Dietrich CH, Allen JM, Lemmon AR, Lemmon
EM, Takiya DM, Evangelista O, Walden KKO, Grady PGS and Johnson KP:
Anchored hybrid enrichment-based phylogenomics of leafhoppers and
treehoppers (Hemiptera: Cicadomorpha: Membracoidea). Insect Syst
Divers. 1:57–72. 2017. View Article : Google Scholar
|
|
50
|
Richters MM, Xia H, Campbell KM,
Gillanders WE, Griffith OL and Griffith M: Best practices for
bioinformatic characterization of neoantigens for clinical utility.
Genome Med. 11:562019. View Article : Google Scholar
|
|
51
|
Aljabali AAA, Hamzat Y, Alqudah A and
Alzoubi L: Neoantigen vaccines: Advancing personalized cancer
immunotherapy. Explor Immunol. 5:10031902025. View Article : Google Scholar
|
|
52
|
Fennemann FL, de Vries IJM, Figdor CG and
Verdoes M: Attacking tumors from all sides: Personalized multiplex
vaccines to tackle intratumor heterogeneity. Front Immunol.
10:8242019. View Article : Google Scholar
|
|
53
|
Peng M, Mo Y, Wang Y, Wu P, Zhang Y, Xiong
F, Guo C, Wu X, Li Y, Li X, et al: Neoantigen vaccine: An emerging
tumor immunotherapy. Mol Cancer. 18:1282019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rojas LA, Sethna Z, Soares KC, Olcese C,
Pang N, Patterson E, Lihm J, Ceglia N, Guasp P, Chu A, et al:
Personalized RNA neoantigen vaccines stimulate T cells in
pancreatic cancer. Nature. 618:144–150. 2023. View Article : Google Scholar
|
|
55
|
Ehx G and Perreault C: Discovery and
characterization of actionable tumor antigens. Genome Med.
11:292019. View Article : Google Scholar
|
|
56
|
Finn OJ: Human tumor antigens yesterday,
today, and tomorrow. Cancer Immunol Res. 5:347–354. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sotirov S and Dimitrov I: Tumor-derived
antigenic peptides as potential cancer vaccines. Int J Mol Sci.
25:49342024. View Article : Google Scholar
|
|
58
|
Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J,
Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, et al: An
immunogenic personal neoantigen vaccine for patients with melanoma.
Nature. 547:217–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hu Z, Leet DE, Allesøe RL, Oliveira G, Li
S, Luoma AM, Liu J, Forman J, Huang T, Iorgulescu JB, et al:
Personal neoantigen vaccines induce persistent memory T cell
responses and epitope spreading in patients with melanoma. Nat Med.
27:515–525. 2021. View Article : Google Scholar :
|
|
60
|
Keskin DB, Anandappa AJ, Sun J, Tirosh I,
Mathewson ND, Li S, Oliveira G, Giobbie-Hurder A, Felt K, Gjini E,
et al: Neoantigen vaccine generates intratumoral T cell responses
in phase Ib glioblastoma trial. Nature. 565:234–239. 2019.
View Article : Google Scholar :
|
|
61
|
Ali OA, Lewin SA, Dranoff G and Mooney DJ:
Vaccines combined with immune checkpoint antibodies promote
cytotoxic T-cell activity and tumor eradication. Cancer Immunol
Res. 4:95–100. 2016. View Article : Google Scholar
|
|
62
|
Karyampudi L, Lamichhane P, Scheid AD,
Kalli KR, Shreeder B, Krempski JW, Behrens MD and Knutson KL:
Accumulation of memory precursor CD8 T cells in regressing tumors
following combination therapy with vaccine and anti-PD-1 antibody.
Cancer Res. 74:2974–2985. 2014. View Article : Google Scholar
|
|
63
|
Robert C, Schachter J, Long GV, Arance A,
Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al:
Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med.
372:2521–2532. 2015. View Article : Google Scholar
|
|
64
|
Qiu Z, Chen Z, Zhang C and Zhong W:
Achievements and futures of immune checkpoint inhibitors in
non-small cell lung cancer. Exp Hematol Oncol. 8:192019. View Article : Google Scholar :
|
|
65
|
Zou W, Wolchok JD and Chen L: PD-L1
(B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms,
response biomarkers, and combinations. Sci Transl Med.
8:328rv42016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kantoff PW, Higano CS, Shore ND, Berger
ER, Small EJ, Penson DF, Redfern CH, Ferrari AC, Dreicer R, Sims
RB, et al: Sipuleucel-T immunotherapy for castration-resistant
prostate cancer. N Engl J Med. 363:411–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Higano CS, Armstrong AJ, Sartor AO,
Vogelzang NJ, Kantoff PW, McLeod DG, Pieczonka CM, Penson DF, Shore
ND, Vacirca J, et al: Real-world outcomes of sipuleucel-T treatment
in PROCEED, a prospective registry of men with metastatic
castration-resistant prostate cancer. Cancer. 125:4172–4180. 2019.
View Article : Google Scholar
|
|
68
|
Lin G, Elkashif A, Saha C, Coulter JA,
Dunne NJ and McCarthy HO: Key considerations for a prostate cancer
mRNA vaccine. Crit Rev Oncol Hematol. 208:1046432025. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Blum JS, Wearsch PA and Cresswell P:
Pathways of antigen processing. Annu Rev Immunol. 31:443–473. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Babbitt BP, Allen PM, Matsueda G, Haber E
and Unanue ER: Binding of immunogenic peptides to Ia
histocompatibility molecules. Nature. 317:359–361. 1985. View Article : Google Scholar
|
|
71
|
Abdel-Aal ABM, Lakshminarayanan V,
Thompson P, Supekar N, Bradley JM, Wolfert MA, Cohen PA, Gendler SJ
and Boons GJ: Immune and anticancer responses elicited by fully
synthetic aberrantly glycosylated MUC1 tripartite vaccines modified
by a TLR2 or TLR9 agonist. Chembiochem. 15:1508–1513. 2014.
View Article : Google Scholar
|
|
72
|
Vlad AM, Kettel JC, Alajez NM, Carlos CA
and Finn OJ: MUC1 immunobiology: From discovery to clinical
applications. Adv Immunol. 82:249–293. 2004. View Article : Google Scholar
|
|
73
|
Arab A, Yazdian-Robati R and Behravan J:
HER2-positive breast cancer immunotherapy: A focus on vaccine
development. Arch Immunol Ther Exp (Warsz). 68:22020. View Article : Google Scholar
|
|
74
|
Baxevanis CN, Sotiriadou NN, Gritzapis AD,
Sotiropoulou PA, Perez SA, Cacoullos NT and Papamichail M:
Immunogenic HER-2/neu peptides as tumor vaccines. Cancer Immunol
Immunother. 55:85–95. 2006. View Article : Google Scholar
|
|
75
|
Zanetti M: A second chance for telomerase
reverse transcriptase in anticancer immunotherapy. Nat Rev Clin
Oncol. 14:115–128. 2017. View Article : Google Scholar
|
|
76
|
Slingluff CL Jr, Chianese-Bullock KA,
Bullock TNJ, Grosh WW, Mullins DW, Nichols L, Olson W, Petroni G,
Smolkin M and Engelhard VH: Immunity to melanoma antigens: From
self-tolerance to immunotherapy. Adv Immunol. 90:243–295. 2006.
View Article : Google Scholar
|
|
77
|
Guo Q, Wang J, Xiao J, Wang L, Hu X, Yu W,
Song G, Lou J and Chen JF: Heterogeneous mutation pattern in tumor
tissue and circulating tumor DNA warrants parallel NGS panel
testing. Mol Cancer. 17:1312018. View Article : Google Scholar :
|
|
78
|
Hollingsworth RE and Jansen K: Turning the
corner on therapeutic cancer vaccines. NPJ Vaccines. 4:72019.
View Article : Google Scholar
|
|
79
|
Malacopol AT and Holst PJ: Cancer
vaccines: Recent insights and future directions. Int J Mol Sci.
25:112562024. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Leisegang M, Engels B, Schreiber K, Yew
PY, Kiyotani K, Idel C, Arina A, Duraiswamy J, Weichselbaum RR,
Uckert W, et al: Eradication of large solid tumors by gene therapy
with a T-cell receptor targeting a single cancer-specific point
mutation. Clin Cancer Res. 22:2734–2743. 2016. View Article : Google Scholar
|
|
81
|
Chen P, Fang QX, Chen DB and Chen HS:
Neoantigen vaccine: An emerging immunotherapy for hepatocellular
carcinoma. World J Gastrointest Oncol. 13:673–683. 2021. View Article : Google Scholar
|
|
82
|
Pan RY, Chung WH, Chu MT, Chen SJ, Chen
HC, Zheng L and Hung SI: Recent development and clinical
application of cancer vaccine: Targeting neoantigens. J Immunol
Res. 2018:43258742018. View Article : Google Scholar
|
|
83
|
Vormehr M, Türeci Ö and Sahin U:
Harnessing tumor mutations for truly individualized cancer
vaccines. Annu Rev Med. 70:395–407. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A,
Børresen-Dale AL, et al: Signatures of mutational processes in
human cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Turajlic S, Litchfield K, Xu H, Rosenthal
R, McGranahan N, Reading JL, Wong YNS, Rowan A, Kanu N, Al Bakir M,
et al: Insertion-and-deletion-derived tumour-specific neoantigens
and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol.
18:1009–1021. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yang W, Lee KW, Srivastava RM, Kuo F,
Krishna C, Chowell D, Makarov V, Hoen D, Dalin MG, Wexler L, et al:
Immunogenic neoantigens derived from gene fusions stimulate T cell
responses. Nat Med. 25:767–775. 2019. View Article : Google Scholar :
|
|
87
|
Zhang J, White NM, Schmidt HK, Fulton RS,
Tomlinson C, Warren WC, Wilson RK and Maher CA: INTEGRATE: Gene
fusion discovery using whole genome and transcriptome data. Genome
Res. 26:108–118. 2016. View Article : Google Scholar :
|
|
88
|
Snyder A, Makarov V, Merghoub T, Yuan J,
Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et
al: Genetic basis for clinical response to CTLA-4 blockade in
melanoma. N Engl J Med. 371:2189–2199. 2014. View Article : Google Scholar
|
|
89
|
Chen DS and Mellman I: Elements of cancer
immunity and the cancer-immune set point. Nature. 541:321–330.
2017. View Article : Google Scholar
|
|
90
|
Borden ES, Buetow KH, Wilson MA and
Hastings KT: Cancer neoantigens: Challenges and future directions
for prediction, prioritization, and validation. Front Oncol.
12:8368212022. View Article : Google Scholar
|
|
91
|
Anczuków O and Krainer AR: Splicing-factor
alterations in cancers. RNA. 22:1285–1301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kahles A, Lehmann KV, Toussaint NC, Hüser
M, Stark SG, Sachsenberg T, Stegle O, Kohlbacher O and Sander C;
Cancer Genome Atlas Research Network, Gunnar Rätsch: Comprehensive
analysis of alternative splicing across tumors from 8,705 patients.
Cancer Cell. 34:211–224 e6. 2018. View Article : Google Scholar
|
|
93
|
Cheng R, Xu Z, Luo M, Wang P, Cao H, Jin
X, Zhou W, Xiao L and Jiang Q: Identification of alternative
splicing-derived cancer neoantigens for mRNA vaccine development.
Brief Bioinform. 23:bbab5532022. View Article : Google Scholar
|
|
94
|
Weller C, Bartok O, McGinnis CS, Palashati
H, Chang TG, Malko D, Shmueli MD, Nagao A, Hayoun D, Murayama A, et
al: Translation dysregulation in cancer as a source for targetable
antigens. Cancer Cell. S1535-6108(25)00082-02025.Epub ahead of
print.
|
|
95
|
Carreno BM, Magrini V, Becker-Hapak M,
Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie WR, Hildebrand WH,
Mardis ER and Linette GP: Cancer immunotherapy. A dendritic cell
vaccine increases the breadth and diversity of melanoma
neoantigen-specific T cells. Science. 348:803–808. 2015. View Article : Google Scholar :
|
|
96
|
Sahin U, Derhovanessian E, Miller M, Kloke
BP, Simon P, Löwer M, Bukur V, Tadmor AD, Luxemburger U, Schrörs B,
et al: Personalized RNA mutanome vaccines mobilize poly-specific
therapeutic immunity against cancer. Nature. 547:222–226. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hacohen N, Fritsch EF, Carter TA, Lander
ES and Wu CJ: Getting personal with neoantigen-based therapeutic
cancer vaccines. Cancer Immunol Res. 1:11152013. View Article : Google Scholar
|
|
98
|
Capietto AH, Hoshyar R and Delamarre L:
Sources of cancer neoantigens beyond single-nucleotide variants.
Int J Mol Sci. 23:101312022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Schumacher T, Bunse L, Pusch S, Sahm F,
Wiestler B, Quandt J, Menn O, Osswald M, Oezen I, Ott M, et al: A
vaccine targeting mutant IDH1 induces antitumour immunity. Nature.
512:324–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wang QJ, Yu Z, Griffith K, Hanada KI,
Restifo NP and Yang JC: Identification of T-cell receptors
targeting KRAS-mutated human tumors. Cancer Immunol Res. 4:204–214.
2016. View Article : Google Scholar
|
|
101
|
Chheda ZS, Kohanbash G, Okada K, Jahan N,
Sidney J, Pecoraro M, Yang X, Carrera DA, Downey KM, Shrivastav S,
et al: Novel and shared neoantigen derived from histone 3 variant
H3.3K27M mutation for glioma T cell therapy. J Exp Med.
215:141–157. 2018. View Article : Google Scholar :
|
|
102
|
Wang Y, Shi T, Song X, Liu B and Wei J:
Gene fusion neoantigens: Emerging targets for cancer immunotherapy.
Cancer Lett. 506:45–54. 2021. View Article : Google Scholar
|
|
103
|
Wei Z, Zhou C, Zhang Z, Guan M, Zhang C,
Liu Z and Liu Q: The landscape of tumor fusion neoantigens: A
pan-cancer analysis. iScience. 21:249–260. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Dai X, Theobard R, Cheng H, Xing M and
Zhang J: Fusion genes: A promising tool combating against cancer.
Biochim Biophys Acta Rev Cancer. 1869:149–160. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Smith CC, Selitsky SR, Chai S, Armistead
PM, Vincent BG and Serody JS: Alternative tumour-specific antigens.
Nat Rev Cancer. 19:465–478. 2019. View Article : Google Scholar :
|
|
106
|
Diederichs S, Bartsch L, Berkmann JC,
Fröse K, Heitmann J, Hoppe C, Iggena D, Jazmati D, Karschnia P,
Linsenmeier M, et al: The dark matter of the cancer genome:
Aberrations in regulatory elements, untranslated regions, splice
sites, non-coding RNA and synonymous mutations. EMBO Mol Med.
8:442–457. 2016. View Article : Google Scholar :
|
|
107
|
Laumont CM, Vincent K, Hesnard L, Audemard
É, Bonneil É, Laverdure JP, Gendron P, Courcelles M, Hardy MP, Côté
C, et al: Noncoding regions are the main source of targetable
tumor-specific antigens. Sci Transl Med. 10:eaau55162018.
View Article : Google Scholar
|
|
108
|
Huang D, Zhu X, Ye S, Zhang J, Liao J,
Zhang N, Zeng X, Wang J, Yang B, Zhang Y, et al: Tumour circular
RNAs elicit anti-tumour immunity by encoding cryptic peptides.
Nature. 625:593–602. 2024. View Article : Google Scholar
|
|
109
|
Robbins PF, Lu YC, El-Gamil M, Li YF,
Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, et al:
Mining exomic sequencing data to identify mutated antigens
recognized by adoptively transferred tumor-reactive T cells. Nat
Med. 19:747–752. 2013. View Article : Google Scholar
|
|
110
|
van Buuren MM, Calis JJ and Schumacher TN:
High sensitivity of cancer exome-based CD8 T cell neo-antigen
identification. Oncoimmunology. 3:e288362014. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Biswas N, Chakrabarti S, Padul V, Jones LD
and Ashili S: Designing neoantigen cancer vaccines, trials, and
outcomes. Front Immunol. 14:11054202023. View Article : Google Scholar :
|
|
112
|
Roudko V, Greenbaum B and Bhardwaj N:
Computational prediction and validation of tumor-associated
neoantigens. Front Immunol. 11:272020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Yadav M, Jhunjhunwala S, Phung QT,
Lupardus P, Tanguay J, Bumbaca S, Franci C, Cheung TK, Fritsche J,
Weinschenk T, et al: Predicting immunogenic tumour mutations by
combining mass spectrometry and exome sequencing. Nature.
515:572–576. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Chen S, Zhou Y, Chen Y and Gu J: fastp: An
ultra-fast all-in-one FASTQ preprocessor. Bioinformatics.
34:i884–i890. 2018. View Article : Google Scholar :
|
|
115
|
Chen S: Ultrafast one-pass FASTQ data
preprocessing, quality control, and deduplication using fastp.
Imeta. 2:e1072023. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Dobin A, Davis CA, Schlesinger F, Drenkow
J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR: STAR:
Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21.
2013. View Article : Google Scholar
|
|
119
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar :
|
|
120
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Kim D, Paggi JM, Park C, Bennett C and
Salzberg SL: Graph-based genome alignment and genotyping with
HISAT2 and HISAT-genotype. Nat Biotechnol. 37:907–915. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R; 1000 Genome
Project Data Processing Subgroup: The sequence alignment/map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Shumate A, Wong B, Pertea G and Pertea M:
Improved transcriptome assembly using a hybrid of long and short
reads with StringTie. PLoS Comput Biol. 18:e10097302022. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
van der Auwera G and O'Connor BD: Genomics
in the cloud: Using docker, GATK, and WDL in Terra. O'Reilly Media,
Incorporated; 2020
|
|
126
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar
|
|
127
|
Lai Z, Markovets A, Ahdesmaki M, Chapman
B, Hofmann O, McEwen R, Johnson J, Dougherty B, Barrett JC and Dry
JR: VarDict: A novel and versatile variant caller for
next-generation sequencing in cancer research. Nucleic Acids Res.
44:e1082016. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Kim S, Scheffler K, Halpern AL, Bekritsky
MA, Noh E, Källberg M, Chen X, Kim Y, Beyter D, Krusche P and
Saunders CT: Strelka2: Fast and accurate calling of germline and
somatic variants. Nat Methods. 15:591–594. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Haas BJ, Dobin A, Stransky N, Li B, Yang
X, Tickle T, Bankapur A, Ganote C, Doak TG, Pochet N, et al:
STAR-fusion: Fast and accurate fusion transcript detection from
RNA-Seq. bioRxiv: 120295. 2017.
|
|
130
|
Yang H and Wang K: Genomic variant
annotation and prioritization with ANNOVAR and wANNOVAR. Nat
Protoc. 10:1556–1566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Mayakonda A, Lin DC, Assenov Y, Plass C
and Koeffler HP: Maftools: Efficient and comprehensive analysis of
somatic variants in cancer. Genome Res. 28:1747–1756. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Gubin MM, Artyomov MN, Mardis ER and
Schreiber RD: Tumor neoantigens: Building a framework for
personalized cancer immunotherapy. J Clin Invest. 125:3413–3421.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Shiina T, Hosomichi K, Inoko H and Kulski
JK: The HLA genomic loci map: expression, interaction, diversity
and disease. J Hum Genet. 54:15–39. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Klein J and Sato A: The HLA system. First
of two parts. N Engl J Med. 343:702–709. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Neefjes J, Jongsma M, Paul P and Bakke O:
Towards a systems understanding of MHC class I and MHC class II
antigen presentation. Nat Rev Immunol. 11:823–836. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Aurisicchio L, Pallocca M, Ciliberto G and
Palombo F: The perfect personalized cancer therapy: Cancer vaccines
against neoantigens. J Exp Clin Cancer Res. 37:862018. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Bontadini A: HLA techniques: Typing and
antibody detection in the laboratory of immunogenetics. Methods.
56:471–476. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
De Mattos-Arruda L, Vazquez M, Finotello
F, Lepore R, Porta E, Hundal J, Amengual-Rigo P, Ng CKY, Valencia
A, Carrillo J, et al: Neoantigen prediction and computational
perspectives towards clinical benefit: Recommendations from the
ESMO precision medicine working group. Ann Oncol. 31:978–990. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Hosomichi K, Shiina T, Tajima A and Inoue
I: The impact of next-generation sequencing technologies on HLA
research. J Hum Genet. 60:665–673. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Danzer M, Niklas N, Stabentheiner S, Hofer
K, Pröll J, Stückler C, Raml E, Polin H and Gabriel C: Rapid,
scalable and highly automated HLA genotyping using next-generation
sequencing: A transition from research to diagnostics. BMC
Genomics. 14:2212013. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Smith AG, Pereira S, Jaramillo A, Stoll
ST, Khan FM, Berka N, Mostafa AA, Pando MJ, Usenko CY, Bettinotti
MP, et al: Comparison of sequence-specific oligonucleotide probe vs
next generation sequencing for HLA-A, B, C, DRB1, DRB3/B4/B5, DQA1,
DQB1, DPA1, and DPB1 typing: Toward single-pass high-resolution HLA
typing in support of solid organ and hematopoietic cell transplant
programs: Toward single-pass high-resolution HLA typing in support
of solid organ and hematopoietic cell transplant programs. HLA.
94:296–306. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Wittig M, Anmarkrud JA, Kässens JC, Koch
S, Forster M, Ellinghaus E, Hov JR, Sauer S, Schimmler M, Ziemann
M, et al: Development of a high-resolution NGS-based HLA-typing and
analysis pipeline. Nucleic Acids Res. 43:e702015. View Article : Google Scholar :
|
|
144
|
Boegel S, Löwer M, Schäfer M, Bukur T, de
Graaf J, Boisguérin V, Türeci O, Diken M, Castle JC and Sahin U:
HLA typing from RNA-seq sequence reads. Genome Med. 4:1022012.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Warren RL, Choe G, Freeman DJ, Castellarin
M, Munro S, Moore R and Holt RA: Derivation of HLA types from
shotgun sequence datasets. Genome Med. 4:952012. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Warren RL: HLA predictions from long
sequence read alignments, streamed directly into HLAminer. arXiv:
2209.09155. 2022.
|
|
147
|
Kim HJ and Pourmand N: HLA typing from
RNA-seq data using hierarchical read weighting [corrected]. PLoS
One. 8:e678852013. View Article : Google Scholar
|
|
148
|
Liu C, Yang X, Duffy B, Mohanakumar T,
Mitra RD, Zody MC and Pfeifer JD: ATHLATES: Accurate typing of
human leukocyte antigen through exome sequencing. Nucleic Acids
Res. 41:e1422013. View Article : Google Scholar :
|
|
149
|
Szolek A, Schubert B, Mohr C, Sturm M,
Feldhahn M and Kohlbacher O: OptiType: Precision HLA typing from
next-generation sequencing data. Bioinformatics. 30:3310–3316.
2014. View Article : Google Scholar
|
|
150
|
Shukla SA, Rooney MS, Rajasagi M, Tiao G,
Dixon PM, Lawrence MS, Stevens J, Lane WJ, Dellagatta JL, Steelman
S, et al: Comprehensive analysis of cancer-associated somatic
mutations in class I HLA genes. Nat Biotechnol. 33:1152–1158. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Huang Y, Yang J, Ying D, Zhang Y,
Shotelersuk V, Hirankarn N, Sham PC, Lau YL and Yang W:
HLAreporter: A tool for HLA typing from next generation sequencing
data. Genome Med. 7:252015. View Article : Google Scholar :
|
|
152
|
Nariai N, Kojima K, Saito S, Mimori T,
Sato Y, Kawai Y, Yamaguchi-Kabata Y, Yasuda J and Nagasaki M:
HLA-VBSeq: Accurate HLA typing at full resolution from whole-genome
sequencing data. BMC Genomics. 16(Suppl 2): S72015. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Xie C, Yeo ZX, Wong M, Piper J, Long T,
Kirkness EF, Biggs WH, Bloom K, Spellman S, Vierra-Green C, et al:
Fast and accurate HLA typing from short-read next-generation
sequence data with xHLA. Proc Natl Acad Sci USA. 114:8059–8064.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Kawaguchi S, Higasa K, Shimizu M, Yamada R
and Matsuda F: HLA-HD: An accurate HLA typing algorithm for
nextgeneration sequencing data. Hum Mutat. 38:788–797. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Kawaguchi S, Higasa K, Yamada R and
Matsuda F: Comprehensive HLA typing from a current allele database
using next-generation sequencing data. Methods Mol Biol.
1802:225–233. 2018. View Article : Google Scholar
|
|
156
|
Kawaguchi S and Matsuda F: High-definition
genomic analysis of HLA genes via comprehensive HLA allele
genotyping. Methods Mol Biol. 2131:31–38. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Ka S, Lee S, Hong J, Cho Y, Sung J, Kim
HN, Kim HL and Jung J: HLAscan: Genotyping of the HLA region using
next-generation sequencing data. BMC Bioinformatics. 18:2582017.
View Article : Google Scholar :
|
|
158
|
Hayashi S, Yamaguchi R, Mizuno S, Komura
M, Miyano S, Nakagawa H and Imoto S: ALPHLARD: A Bayesian method
for analyzing HLA genes from whole genome sequence data. BMC
Genomics. 19:7902018. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Bai Y, Wang D and Fury W: PHLAT: Inference
of high-resolution HLA types from RNA and whole exome sequencing.
Methods Mol Biol. 1802:193–201. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Orenbuch R, Filip I, Comito D, Shaman J,
Pe'er I and Rabadan R: arcasHLA: High-resolution HLA typing from
RNAseq. Bioinformatics. 36:33–40. 2020. View Article : Google Scholar :
|
|
161
|
Bauer DC, Zadoorian A, Wilson LOW;
Melbourne Genomics Health Alliance; Thorne NP: Evaluation of
computational programs to predict HLA genotypes from genomic
sequencing data. Brief Bioinform. 19:179–187. 2018.
|
|
162
|
Kiyotani K, Mai TH and Nakamura Y:
Comparison of exome-based HLA class I genotyping tools:
Identification of platform-specific genotyping errors. J Hum Genet.
62:397–405. 2017. View Article : Google Scholar
|
|
163
|
Schumacher TN and Schreiber RD:
Neoantigens in cancer immunotherapy. Science. 348:69–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Ferrari G, Kostyu DD, Cox J, Dawson DV,
Flores J, Weinhold KJ and Osmanov S: Identification of highly
conserved and broadly cross-reactive HIV type 1 cytotoxic T
lymphocyte epitopes as candidate immunogens for inclusion in
Mycobacterium bovis BCG-vectored HIV vaccines. AIDS Res Hum
Retroviruses. 16:1433–1443. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Kessler JH, Benckhuijsen WE, Mutis T,
Melief CJM, van der Burg SH and Drijfhout JW: Competition-based
cellular peptide binding assay for HLA class I. Curr Protoc
Immunol. Chapter 18: Unit 18.12. 2004. View Article : Google Scholar
|
|
166
|
Wulf M, Hoehn P and Trinder P:
Identification and validation of T-cell epitopes using the
IFN-gamma ELISPOT assay. Methods Mol Biol. 524:439–446. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Yang B, Jeang J, Yang A, Wu TC and Hung
CF: DNA vaccine for cancer immunotherapy. Hum Vaccin Immunother.
10:3153–3164. 2014. View Article : Google Scholar
|
|
168
|
Vita R, Mahajan S, Overton JA, Dhanda SK,
Martini S, Cantrell JR, Wheeler DK, Sette A and Peters B: The
immune epitope database (IEDB): 2018 Update. Nucleic Acids Res.
47(D1): D339–D343. 2019. View Article : Google Scholar :
|
|
169
|
Rammensee H, Bachmann J, Emmerich NP,
Bachor OA and Stevanović S: SYFPEITHI: Database for MHC ligands and
peptide motifs. Immunogenetics. 50:213–219. 1999. View Article : Google Scholar
|
|
170
|
Bhasin M, Singh H and Raghava GPS: MHCBN:
A comprehensive database of MHC binding and non-binding peptides.
Bioinformatics. 19:665–666. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Blass E and Ott PA: Advances in the
development of personalized neoantigen-based therapeutic cancer
vaccines. Nat Rev Clin Oncol. 18:215–229. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Garcia-Garijo A, Fajardo CA and Gros A:
Determinants for neoantigen identification. Front Immunol.
10:13922019. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Falk K, Rötzschke O, Stevanović S, Jung G
and Rammensee HG: Allele-specific motifs revealed by sequencing of
self-peptides eluted from MHC molecules. Nature. 351:290–296. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Rötzschke O, Falk K, Stevanović S, Jung G,
Walden P and Rammensee HG: Exact prediction of a natural T cell
epitope. Eur J Immunol. 21:2891–2894. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Nielsen M, Lundegaard C, Worning P,
Lauemøller SL, Lamberth K, Buus S, Brunak S and Lund O: Reliable
prediction of T-cell epitopes using neural networks with novel
sequence representations. Protein Sci. 12:1007–1017. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Nielsen M and Andreatta M: NetMHCpan-3.0;
Improved prediction of binding to MHC class I molecules integrating
information from multiple receptor and peptide length datasets.
Genome Med. 8:332016. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Peters B and Sette A: Generating
quantitative models describing the sequence specificity of
biological processes with the stabilized matrix method. BMC
Bioinformatics. 6:1322005. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Kim Y, Sidney J, Pinilla C, Sette A and
Peters B: Derivation of an amino acid similarity matrix for
peptide: MHC binding and its application as a Bayesian prior. BMC
Bioinformatics. 10:3942009. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Lundegaard C, Lund O and Nielsen M:
Prediction of epitopes using neural network based methods. J
Immunol Methods. 374:26–34. 2011. View Article : Google Scholar :
|
|
180
|
Schueler-Furman O, Altuvia Y, Sette A and
Margalit H: Structure-based prediction of binding peptides to MHC
class I molecules: Application to a broad range of MHC alleles.
Protein Sci. 9:1838–1846. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Kumar N and Mohanty D: MODPROPEP: A
program for knowledge-based modeling of protein-peptide complexes.
Nucleic Acids Res. 35(Web Server Issue): W549–W555. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Hattotuwagama CK, Doytchinova IA and
Flower DR: Toward the prediction of class I and II mouse major
histocompatibility complex-peptide-binding affinity: In silico
bioinformatic step-by-step guide using quantitative
structure-activity relationships. Methods Mol Biol. 409:227–245.
2007. View Article : Google Scholar
|
|
183
|
Luo H, Ye H, Ng HW, Shi L, Tong W,
Mendrick DL and Hong H: Machine learning methods for predicting
HLA-peptide binding activity. Bioinform Biol Insights. 9(Suppl 3):
S21–S29. 2015.
|
|
184
|
Zhang H, Lund O and Nielsen M: The
PickPocket method for predicting binding specificities for
receptors based on receptor pocket similarities: Application to
MHC-peptide binding. Bioinformatics. 25:1293–1299. 2009. View Article : Google Scholar :
|
|
185
|
Wang M, Lei C, Wang J, Li Y and Li M:
TripHLApan: Predicting HLA molecules binding peptides based on
triple coding matrix and transfer learning. Brief Bioinform.
25:bbae1542024. View Article : Google Scholar
|
|
186
|
Boehm KM, Bhinder B, Raja VJ, Dephoure N
and Elemento O: Predicting peptide presentation by major
histocompatibility complex class I: An improved machine learning
approach to the immunopeptidome. BMC Bioinformatics. 20:72019.
View Article : Google Scholar
|
|
187
|
Mei S, Li F, Xiang D, Ayala R, Faridi P,
Webb GI, Illing PT, Rossjohn J, Akutsu T, Croft NP, et al: Anthem:
A user customised tool for fast and accurate prediction of binding
between peptides and HLA class I molecules. Brief Bioinform.
22:bbaa4152021. View Article : Google Scholar
|
|
188
|
Reche PA, Glutting JP and Reinherz EL:
Prediction of MHC class I binding peptides using profile motifs.
Hum Immunol. 63:701–709. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Reche PA, Glutting JP, Zhang H and
Reinherz EL: Enhancement to the RANKPEP resource for the prediction
of peptide binding to MHC molecules using profiles. Immunogenetics.
56:405–419. 2004. View Article : Google Scholar
|
|
190
|
Reche PA and Reinherz EL: Prediction of
peptide-MHC binding using profiles. Methods Mol Biol. 409:185–200.
2007. View Article : Google Scholar
|
|
191
|
Buus S, Lauemøller SL, Worning P, Kesmir
C, Frimurer T, Corbet S, Fomsgaard A, Hilden J, Holm A and Brunak
S: Sensitive quantitative predictions of peptide-MHC binding by a
'Query by Committee' artificial neural network approach. Tissue
Antigens. 62:378–384. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Lundegaard C, Lamberth K, Harndahl M, Buus
S, Lund O and Nielsen M: NetMHC-3.0: Accurate web accessible
predictions of human, mouse and monkey MHC class I affinities for
peptides of length 8-11. Nucleic Acids Res. 36(Web Server Issue):
W509–W512. 2008. View Article : Google Scholar
|
|
193
|
Andreatta M and Nielsen M: Gapped sequence
alignment using artificial neural networks: Application to the MHC
class I system. Bioinformatics. 32:511–517. 2016. View Article : Google Scholar
|
|
194
|
Sidney J, Assarsson E, Moore C, Ngo S,
Pinilla C, Sette A and Peters B: Quantitative peptide binding
motifs for 19 human and mouse MHC class I molecules derived using
positional scanning combinatorial peptide libraries. Immunome Res.
4:22008. View Article : Google Scholar :
|
|
195
|
Jurtz V, Paul S, Andreatta M, Marcatili P,
Peters B and Nielsen M: NetMHCpan-4.0: Improved peptide-MHC class I
interaction predictions integrating eluted ligand and peptide
binding affinity data. J Immunol. 199:3360–3368. 2017. View Article : Google Scholar
|
|
196
|
Hoof I, Peters B, Sidney J, Pedersen LE,
Sette A, Lund O, Buus S and Nielsen M: NetMHCpan, a method for MHC
class I binding prediction beyond humans. Immunogenetics. 61:1–13.
2009. View Article : Google Scholar
|
|
197
|
Reynisson B, Alvarez B, Paul S, Peters B
and Nielsen M: NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved
predictions of MHC antigen presentation by concurrent motif
deconvolution and integration of MS MHC eluted ligand data. Nucleic
Acids Res. 48(W1): W449–W454. 2020. View Article : Google Scholar
|
|
198
|
Bassani-Sternberg M, Chong C, Guillaume P,
Solleder M, Pak H, Gannon PO, Kandalaft LE, Coukos G and Gfeller D:
Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen
predictions and identifies allostery regulating HLA specificity.
PLoS Comput Biol. 13:e10057252017. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Gfeller D, Schmidt J, Croce G, Guillaume
P, Bobisse S, Genolet R, Queiroz L, Cesbron J, Racle J and Harari
A: Improved predictions of antigen presentation and TCR recognition
with MixMHCpred2.2 and PRIME2.0 reveal potent SARS-CoV-2
CD8+ T-cell epitopes. Cell Syst. 14:72–83.e5. 2023.
View Article : Google Scholar
|
|
200
|
Liu G, Li D, Li Z, Qiu S, Li W, Chao CC,
Yang N, Li H, Cheng Z, Song X, et al: PSSMHCpan: A novel PSSM-based
software for predicting class I peptide-HLA binding affinity.
Gigascience. 6:1–11. 2017. View Article : Google Scholar
|
|
201
|
O'Donnell TJ, Rubinsteyn A and Laserson U:
MHCflurry 2.0: Improved pan-allele prediction of MHC class
I-presented peptides by incorporating antigen processing. Cell
Syst. 11:42–48.e7. 2020. View Article : Google Scholar
|
|
202
|
O'Donnell TJ, Rubinsteyn A, Bonsack M,
Riemer AB, Laserson U and Hammerbacher J: MHCflurry: Open-source
class I MHC binding affinity prediction. Cell Syst. 7:129–132.e4.
2018. View Article : Google Scholar
|
|
203
|
Phloyphisut P, Pornputtapong N, Sriswasdi
S and Chuangsuwanich E: MHCSeqNet: A deep neural network model for
universal MHC binding prediction. BMC Bioinformatics. 20:2702019.
View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Hu Y, Wang Z, Hu H, Wan F, Chen L, Xiong
Y, Wang X, Zhao D, Huang W and Zeng J: ACME: Pan-specific
peptide-MHC class I binding prediction through attention-based deep
neural networks. Bioinformatics. 35:4946–4954. 2019. View Article : Google Scholar
|
|
205
|
Wu J, Wang W, Zhang J, Zhou B, Zhao W, Su
Z, Gu X, Wu J, Zhou Z and Chen S: DeepHLApan: A deep learning
approach for neoantigen prediction considering both HLA-peptide
binding and immunogenicity. Front Immunol. 10:25592019. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Sarkizova S, Klaeger S, Le PM, Li LW,
Oliveira G, Keshishian H, Hartigan CR, Zhang W, Braun DA, Ligon KL,
et al: A large peptidome dataset improves HLA class I epitope
prediction across most of the human population. Nat Biotechnol.
38:199–209. 2020. View Article : Google Scholar
|
|
207
|
Shao XM, Bhattacharya R, Huang J,
Sivakumar IKA, Tokheim C, Zheng L, Hirsch D, Kaminow B, Omdahl A,
Bonsack M, et al: High-throughput prediction of MHC class I and II
neoantigens with MHCnuggets. Cancer Immunol Res. 8:396–408. 2020.
View Article : Google Scholar
|
|
208
|
Yang X, Zhao L, Wei F and Li J:
DeepNetBim: Deep learning model for predicting HLA-epitope
interactions based on network analysis by harnessing binding and
immunogenicity information. BMC Bioinformatics. 22:2312021.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Albert BA, Yang Y, Shao XM, Singh D, Smit
KN, Anagnostou V and Karchin R: Deep neural networks predict class
I major histocompatibility complex epitope presentation and
transfer learn neoepitope immunogenicity. Nat Mach Intell.
5:861–872. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Sturniolo T, Bono E, Ding J, Raddrizzani
L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP,
Sinigaglia F and Hammer J: Generation of tissue-specific and
promiscuous HLA ligand databases using DNA microarrays and virtual
HLA class II matrices. Nat Biotechnol. 17:555–561. 1999. View Article : Google Scholar
|
|
211
|
Singh H and Raghava GP: ProPred:
Prediction of HLA-DR binding sites. Bioinformatics. 17:1236–1237.
2001. View Article : Google Scholar
|
|
212
|
Wan J, Liu W, Xu Q, Ren Y, Flower DR and
Li T: SVRMHC prediction server for MHC-binding peptides. BMC
Bioinformatics. 7:4632006. View Article : Google Scholar
|
|
213
|
Nielsen M, Lundegaard C and Lund O:
Prediction of MHC class II binding affinity using SMM-align, a
novel stabilization matrix alignment method. BMC Bioinformatics.
8:2382007. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Nielsen M and Lund O: NN-align. An
artificial neural network-based alignment algorithm for MHC class
II peptide binding prediction. BMC Bioinformatics. 10:2962009.
View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Jensen KK, Andreatta M, Marcatili P, Buus
S, Greenbaum JA, Yan Z, Sette A, Peters B and Nielsen M: Improved
methods for predicting peptide binding affinity to MHC class II
molecules. Immunology. 154:394–406. 2018. View Article : Google Scholar
|
|
216
|
Nielsen M, Lundegaard C, Blicher T, Peters
B, Sette A, Justesen S, Buus S and Lund O: Quantitative predictions
of peptide binding to any HLA-DR molecule of known sequence:
NetMHCIIpan. PLoS Comput Biol. 4:e10001072008. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Andreatta M, Karosiene E, Rasmussen M,
Stryhn A, Buus S and Nielsen M: Accurate pan-specific prediction of
peptide-MHC class II binding affinity with improved binding core
identification. Immunogenetics. 67:641–650. 2015. View Article : Google Scholar :
|
|
218
|
Kaabinejadian S, Barra C, Alvarez B, Yari
H, Hildebrand WH and Nielsen M: Accurate MHC motif deconvolution of
immunopeptidomics data reveals a significant contribution of DRB3,
4 and 5 to the total DR immunopeptidome. Front Immunol.
13:8354542022. View Article : Google Scholar :
|
|
219
|
Pfeifer N and Kohlbacher O: Multiple
instance learning allows MHC class II epitope predictions across
alleles. Lect Notes Comput Sci. 5251:210–221. 2008. View Article : Google Scholar
|
|
220
|
Bordner AJ and Mittelmann HD: MultiRTA: A
simple yet reliable method for predicting peptide binding
affinities for multiple class II MHC allotypes. BMC Bioinformatics.
11:4822010. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Zhang L, Chen Y, Wong HS, Zhou S,
Mamitsuka H and Zhu S: TEPITOPEpan: Extending TEPITOPE for peptide
binding prediction covering over 700 HLA-DR molecules. PLoS One.
7:e304832012. View Article : Google Scholar
|
|
222
|
Chen B, Khodadoust MS, Olsson N, Wagar LE,
Fast E, Liu CL, Muftuoglu Y, Sworder BJ, Diehn M, Levy R, et al:
Predicting HLA class II antigen presentation through integrated
deep learning. Nat Biotechnol. 37:1332–1343. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Racle J, Michaux J, Rockinger GA, Arnaud
M, Bobisse S, Chong C, Guillaume P, Coukos G, Harari A, Jandus C,
et al: Robust prediction of HLA class II epitopes by deep motif
deconvolution of immunopeptidomes. Nat Biotechnol. 37:1283–1286.
2019. View Article : Google Scholar
|
|
224
|
Racle J, Guillaume P, Schmidt J, Michaux
J, Larabi A, Lau K, Perez MAS, Croce G, Genolet R, Coukos G, et al:
Machine learning predictions of MHC-II specificities reveal
alternative binding mode of class II epitopes. Immunity.
56:1359–1375.e13. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Abelin JG, Harjanto D, Malloy M, Suri P,
Colson T, Goulding SP, Creech AL, Serrano LR, Nasir G, Nasrullah Y,
et al: Defining HLA-II ligand processing and binding rules with
mass spectrometry enhances cancer epitope prediction. Immunity.
51:766–779.e17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Liu Z, Jin J, Cui Y, Xiong Z, Nasiri A,
Zhao Y and Hu J: DeepSeqPanII: An interpretable recurrent neural
network model with attention mechanism for peptide-HLA class II
binding prediction. IEEE/ACM Trans Comput Biol Bioinform.
19:2188–2196. 2022. View Article : Google Scholar
|
|
227
|
Rammensee HG, Friede T and Stevanoviíc S:
MHC ligands and peptide motifs: First listing. Immunogenetics.
41:178–228. 1995. View Article : Google Scholar
|
|
228
|
Chicz RM, Urban RG, Lane WS, Gorga JC,
Stern LJ, Vignali DA and Strominger JL: Predominant naturally
processed peptides bound to HLA-DR1 are derived from MHC-related
molecules and are heterogeneous in size. Nature. 358:764–768. 1992.
View Article : Google Scholar
|
|
229
|
Wang P, Sidney J, Dow C, Mothé B, Sette A
and Peters B: A systematic assessment of MHC class II peptide
binding predictions and evaluation of a consensus approach. PLoS
Comput Biol. 4:e10000482008. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Yu K, Petrovsky N, Schönbach C, Koh JYL
and Brusic V: Methods for prediction of peptide binding to MHC
molecules: A comparative study. Mol Med. 8:137–148. 2002.
View Article : Google Scholar
|
|
231
|
Peters B, Bui HH, Frankild S, Nielson M,
Lundegaard C, Kostem E, Basch D, Lamberth K, Harndahl M, Fleri W,
et al: A community resource benchmarking predictions of peptide
binding to MHC-I molecules. PLoS Comput Biol. 2:e652006. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Gowthaman U, Chodisetti SB, Parihar P and
Agrewala JN: Evaluation of different generic in silico methods for
predicting HLA class I binding peptide vaccine candidates using a
reverse approach. Amino Acids. 39:1333–1342. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Bonsack M, Hoppe S, Winter J, Tichy D,
Zeller C, Küpper MD, Schitter EC, Blatnik R and Riemer AB:
Performance evaluation of MHC class-I binding prediction tools
based on an experimentally validated MHC-peptide binding data set.
Cancer Immunol Res. 7:719–736. 2019. View Article : Google Scholar
|
|
234
|
Trolle T, Metushi IG, Greenbaum JA, Kim Y,
Sidney J, Lund O, Sette A, Peters B and Nielsen M: Automated
benchmarking of peptide-MHC class I binding predictions.
Bioinformatics. 31:2174–2181. 2015. View Article : Google Scholar :
|
|
235
|
Engels B, Engelhard VH, Sidney J, Sette A,
Binder DC, Liu RB, Kranz DM, Meredith SC, Rowley DA and Schreiber
H: Relapse or eradication of cancer is predicted by peptide-major
histocompatibility complex affinity. Cancer Cell. 23:516–526. 2013.
View Article : Google Scholar :
|
|
236
|
Nogueira C, Kaufmann JK, Lam H and
Flechtner JB: Improving cancer immunotherapies through empirical
neoantigen selection. Trends Cancer. 4:97–100. 2018. View Article : Google Scholar
|
|
237
|
Liao WWP and Arthur JW: Predicting peptide
binding to major histocompatibility complex molecules. Autoimmun
Rev. 10:469–473. 2011. View Article : Google Scholar
|
|
238
|
Backert L and Kohlbacher O:
Immunoinformatics and epitope prediction in the age of genomic
medicine. Genome Med. 7:1192015. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Türeci Ö, Löwer M, Schrörs B, Lang M,
Tadmor A and Sahin U: Challenges towards the realization of
individualized cancer vaccines. Nat Biomed Eng. 2:566–569. 2018.
View Article : Google Scholar
|
|
240
|
Schumacher TN, Scheper W and Kvistborg P:
Cancer neoantigens. Annu Rev Immunol. 37:173–200. 2019. View Article : Google Scholar
|
|
241
|
Xie N, Shen G, Gao W, Huang Z, Huang C and
Fu L: Neoantigens: Promising targets for cancer therapy. Signal
Transduct Target Ther. 8:92023. View Article : Google Scholar :
|
|
242
|
Bassani-Sternberg M, Bräunlein E, Klar R,
Engleitner T, Sinitcyn P, Audehm S, Straub M, Weber J,
Slotta-Huspenina J, Specht K, et al: Direct identification of
clinically relevant neoepitopes presented on native human melanoma
tissue by mass spectrometry. Nat Commun. 7:134042016. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Sette A, Vitiello A, Reherman B, Fowler P,
Nayersina R, Kast WM, Melief CJ, Oseroff C, Yuan L, Ruppert J, et
al: The relationship between class I binding affinity and
immunogenicity of potential cytotoxic T cell epitopes. J Immunol.
153:5586–5592. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Bjerregaard AM, Nielsen M, Jurtz V, Barra
CM, Hadrup SR, Szallasi Z and Eklund AC: An analysis of natural T
cell responses to predicted tumor neoepitopes. Front Immunol.
8:15662017. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Shah K, Al-Haidari A, Sun J and Kazi JU: T
cell receptor (TCR) signaling in health and disease. Signal
Transduct Target Ther. 6:4122021. View Article : Google Scholar
|
|
246
|
Larsen MV, Lundegaard C, Lamberth K, Buus
S, Brunak S, Lund O and Nielsen M: An integrative approach to CTL
epitope prediction: A combined algorithm integrating MHC class I
binding, TAP transport efficiency, and proteasomal cleavage
predictions. Eur J Immunol. 35:2295–2303. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Stranzl T, Larsen MV, Lundegaard C and
Nielsen M: NetCTLpan: Pan-specific MHC class I pathway epitope
predictions. Immunogenetics. 62:357–368. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Zhou C, Zhu C and Liu Q: Toward in silico
identification of tumor neoantigens in immunotherapy. Trends Mol
Med. 25:980–992. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Chen P, Chen D, Bu D, Gao J, Qin W, Deng
K, Ren L, She S, Xu W, Yang Y, et al: Dominant neoantigen
verification in hepatocellular carcinoma by a single-plasmid system
coexpressing patient HLA and antigen. J Immunother Cancer.
11:e0063342023. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Chudley L, McCann KJ, Coleman A, Cazaly
AM, Bidmon N, Britten CM, van der Burg SH, Gouttefangeas C, Jandus
C, Laske K, et al: Harmonisation of short-term in vitro culture for
the expansion of antigen-specific CD8(+) T cells with detection by
ELISPOT and HLA-multimer staining. Cancer Immunol Immunother.
63:1199–1211. 2014. View Article : Google Scholar :
|
|
251
|
Slota M, Lim JB, Dang Y and Disis ML:
ELISpot for measuring human immune responses to vaccines. Expert
Rev Vaccines. 10:299–306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
252
|
Czerkinsky CC, Nilsson LA, Nygren H,
Ouchterlony O and Tarkowski A: A solid-phase enzyme-linked
immunospot (ELISPOT) assay for enumeration of specific
antibody-secreting cells. J Immunol Methods. 65:109–121. 1983.
View Article : Google Scholar
|
|
253
|
Taguchi T, McGhee JR, Coffman RL, Beagley
KW, Eldridge JH, Takatsu K and Kiyono H: Detection of individual
mouse splenic T cells producing IFN-gamma and IL-5 using the
enzyme-linked immunospot (ELISPOT) assay. J Immunol Methods.
128:65–73. 1990. View Article : Google Scholar
|
|
254
|
Miyahira Y, Murata K, Rodriguez D,
Rodriguez JR, Esteban M, Rodrigues MM and Zavala F: Quantification
of antigen specific CD8+ T cells using an ELISPOT assay. J Immunol
Methods. 181:45–54. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
255
|
Verneris MR, Karimi M, Baker J, Jayaswal A
and Negrin RS: Role of NKG2D signaling in the cytotoxicity of
activated and expanded CD8+ T cells. Blood. 103:3065–3072. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Wonderlich J, Shearer G, Livingstone A,
Brooks A, Soloski MJ and Presby MM: Induction and measurement of
cytotoxic T lymphocyte activity. Curr Protoc Immunol.
120:3.11.1–3.11.29. 2018.
|
|
257
|
Chan JK, Hamilton CA, Cheung MK, Karimi M,
Baker J, Gall JM, Schulz S, Thorne SH, Teng NN, Contag CH, et al:
Enhanced killing of primary ovarian cancer by retargeting
autologous cytokine-induced killer cells with bispecific
antibodies: A preclinical study. Clin Cancer Res. 12:1859–1867.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Clevers H and Tuveson DA: Organoid models
for cancer research. Annu Rev Cancer Biol. 3:223–234. 2019.
View Article : Google Scholar
|
|
259
|
Jacob F, Salinas RD, Zhang DY, Nguyen PTT,
Schnoll JG, Wong SZH, Thokala R, Sheikh S, Saxena D, Prokop S, et
al: A patient-derived glioblastoma organoid model and biobank
recapitulates inter- and intra-tumoral heterogeneity. Cell.
180:188–204.e22. 2020. View Article : Google Scholar
|
|
260
|
Lee SH, Hu W, Matulay JT, Silva MV,
Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, Williams AB, et
al: Tumor evolution and drug response in patient-derived organoid
models of bladder cancer. Cell. 173:515–528.e17. 2018. View Article : Google Scholar
|
|
261
|
Cattaneo CM, Dijkstra KK, Fanchi LF,
Kelderman S, Kaing S, van Rooij N, van den Brink S, Schumacher TN
and Voest EE: Tumor organoid-T-cell coculture systems. Nat Protoc.
15:15–39. 2020. View Article : Google Scholar
|
|
262
|
Weng G, Tao J, Liu Y, Qiu J, Su D, Wang R,
Luo W and Zhang T: Organoid: Bridging the gap between basic
research and clinical practice. Cancer Lett. 572:2163532023.
View Article : Google Scholar : PubMed/NCBI
|
|
263
|
Shultz LD, Ishikawa F and Greiner DL:
Humanized mice in translational biomedical research. Nat Rev
Immunol. 7:118–130. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Rongvaux A, Takizawa H, Strowig T,
Willinger T, Eynon EE, Flavell RA and Manz MG: Human
hemato-lymphoid system mice: Current use and future potential for
medicine. Annu Rev Immunol. 31:635–674. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Morillon YM II, Sabzevari A, Schlom J and
Greiner JW: The development of next-generation PBMC humanized mice
for preclinical investigation of cancer immunotherapeutic agents.
Anticancer Res. 40:5329–5341. 2020. View Article : Google Scholar
|
|
266
|
Ehx G, Ritacco C and Baron F:
Pathophysiology and preclinical relevance of experimental
graft-versus-host disease in humanized mice. Biomark Res.
12:1392024. View Article : Google Scholar : PubMed/NCBI
|
|
267
|
Chuprin J, Buettner H, Seedhom MO, Greiner
DL, Keck JG, Ishikawa F, Shultz LD and Brehm MA: Humanized mouse
models for immuno-oncology research. Nat Rev Clin Oncol.
20:192–206. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
268
|
Guil-Luna S, Sedlik C and Piaggio E:
Humanized mouse models to evaluate cancer immunotherapeutics. Annu
Rev Cancer Biol. 5:119–136. 2021. View Article : Google Scholar
|
|
269
|
De La Rochere P, Guil-Luna S, Decaudin D,
Azar G, Sidhu SS and Piaggio E: Humanized mice for the study of
immuno-oncology. Trends Immunol. 39:748–763. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
270
|
Camacho RE, Wnek R, Fischer P, Shah K,
Zaller DM, Woods A, La Monica N, Aurisicchio L, Fitzgerald-Bocarsly
P and Koo GC: Characterization of the NOD/scid-[Tg]DR1 mouse
expressing HLA-DRB1*01 transgene: A model of SCID-hu mouse for
vaccine development. Exp Hematol. 35:1219–1230. 2007. View Article : Google Scholar
|
|
271
|
Spranger S, Frankenberger B and Schendel
DJ: NOD/scid IL-2Rg(null) mice: A preclinical model system to
evaluate human dendritic cell-based vaccine strategies in vivo. J
Transl Med. 10:302012. View Article : Google Scholar
|
|
272
|
Li R, Han DS, Shi JP, Han YX, Tan P, Zhang
R and Li JM: Choosing tumor mutational burden wisely for
immunotherapy: A hard road to explore. Biochim Biophys Acta Rev
Cancer. 1874:1884202020. View Article : Google Scholar
|
|
273
|
Ahmed J, Das B, Shin S and Chen A:
Challenges and future directions in the management of tumor
mutational burden-high (TMB-H) advanced solid malignancies. Cancers
(Basel). 15:58412023. View Article : Google Scholar : PubMed/NCBI
|
|
274
|
Teixido C, Castillo P, Martinez-Vila C,
Arance A and Alos L: Molecular markers and targets in melanoma.
Cells. 10:23202021. View Article : Google Scholar : PubMed/NCBI
|
|
275
|
Yarchoan M, Hopkins A and Jaffee EM: Tumor
mutational burden and response rate to PD-1 inhibition. N Engl J
Med. 377:2500–2501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
276
|
Ott PA, Hu-Lieskovan S, Chmielowski B,
Govindan R, Naing A, Bhardwaj N, Margolin K, Awad MM, Hellmann MD,
Lin JJ, et al: A phase Ib trial of personalized neoantigen therapy
plus anti-PD-1 in patients with advanced melanoma, non-small cell
lung cancer, or bladder cancer. Cell. 183:347–362.e24. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
277
|
Mørk SK, Kadivar M, Bol KF, Draghi A,
Wulff Westergaard MC, Skadborg SK, Overgaard N, Sørensen AB,
Rasmussen IS, Andreasen LV, et al: Personalized therapy with
peptide-based neoantigen vaccine (EVX-01) including a novel
adjuvant, CAF®09b, in patients with metastatic melanoma.
Oncoimmunology. 11:20232552022. View Article : Google Scholar
|
|
278
|
Weber JS, Carlino MS, Khattak A, Meniawy
T, Ansstas G, Taylor MH, Kim KB, McKean M, Long GV, Sullivan RJ, et
al: Individualised neoantigen therapy mRNA-4157 (V940) plus
pembrolizumab versus pembrolizumab monotherapy in resected melanoma
(KEYNOTE-942): A randomised, phase 2b study. Lancet. 403:632–644.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
279
|
Ding Z, Li Q, Zhang R, Xie L, Shu Y, Gao
S, Wang P, Su X, Qin Y, Wang Y, et al: Personalized neoantigen
pulsed dendritic cell vaccine for advanced lung cancer. Signal
Transduct Target Ther. 6:262021. View Article : Google Scholar :
|
|
280
|
Gainor JF, Patel MR, Weber JS, Gutierrez
M, Bauman JE, Clarke JM, Julian R, Scott AJ, Geiger JL, Kirtane K,
et al: T-cell responses to individualized neoantigen therapy
mRNA-4157 (V940) alone or in combination with pembrolizumab in the
phase 1 KEYNOTE-603 study. Cancer Discov. 14:2209–2223. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
281
|
Lee JM, Spicer J, Nair S, Khattak A, Brown
M, Meehan RS, Shariati NM, Deng X, Samkari A and Chaft JE: The
phase 3 INTerpath-002 study design: Individualized neoantigen
therapy (INT) V940 (mRNA-4157) plus pembrolizumab vs placebo plus
pembrolizumab for resected early-stage non-small-cell lung cancer
(NSCLC). J Clin Oncol. 42(16 Suppl): TPS81162024. View Article : Google Scholar
|
|
282
|
Li F, Deng L, Jackson KR, Talukder AH,
Katailiha AS, Bradley SD, Zou Q, Chen C, Huo C, Chiu Y, et al:
Neoantigen vaccination induces clinical and immunologic responses
in non-small cell lung cancer patients harboring EGFR mutations. J
Immunother Cancer. 9:e0025312021. View Article : Google Scholar : PubMed/NCBI
|
|
283
|
Gao S, Wang J, Zhu Z, Fang J, Zhao Y, Liu
Z, Qin H, Wei Y, Xu H, Dan X, et al: Effective personalized
neoantigen vaccine plus anti-PD-1 in a PD-1 blockade-resistant lung
cancer patient. Immunotherapy. 15:57–69. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
284
|
Awad MM, Govindan R, Balogh KN, Spigel DR,
Garon EB, Bushway ME, Poran A, Sheen JH, Kohler V, Esaulova E, et
al: Personalized neoantigen vaccine NEO-PV-01 with chemotherapy and
anti-PD-1 as first-line treatment for non-squamous non-small cell
lung cancer. Cancer Cell. 40:1010–1026.e11. 2022. View Article : Google Scholar
|
|
285
|
McCann K, von Witzleben A, Thomas J, Wang
C, Wood O, Singh D, Boukas K, Bendjama K, Silvestre N, Nielsen FC,
et al: Targeting the tumor mutanome for personalized vaccination in
a TMB low non-small cell lung cancer. J Immunother Cancer.
10:e0038212022. View Article : Google Scholar :
|
|
286
|
Moen MD: Bevacizumab: In previously
treated glioblastoma. Drugs. 70:181–189. 2010. View Article : Google Scholar
|
|
287
|
Engelhardt B: The blood-central nervous
system barriers actively control immune cell entry into the central
nervous system. Curr Pharm Des. 14:1555–1565. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
288
|
Johanns TM, Bowman-Kirigin JA, Liu C and
Dunn GP: Targeting neoantigens in glioblastoma: An overview of
cancer immunogenomics and translational implications. Neurosurgery.
64(CN Suppl 1): S165–S176. 2017. View Article : Google Scholar
|
|
289
|
Dang L, Yen K and Attar EC: IDH mutations
in cancer and progress toward development of targeted therapeutics.
Ann Oncol. 27:599–608. 2016. View Article : Google Scholar
|
|
290
|
Ni YQ, Shen PB, Wang XC, Liu HY, Luo HY
and Han XZ: The roles of IDH1 in tumor metabolism and immunity.
Future Oncol. 18:3941–3953. 2022. View Article : Google Scholar
|
|
291
|
de la Fuente MI: Targeting IDH1/IDH2
mutations in gliomas. Curr Opin Neurol. 35:787–793. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
292
|
Pellegatta S, Valletta L, Corbetta C,
Patanè M, Zucca I, Riccardi Sirtori F, Bruzzone MG, Fogliatto G,
Isacchi A, Pollo B and Finocchiaro G: Effective immuno-targeting of
the IDH1 mutation R132H in a murine model of intracranial glioma.
Acta Neuropathol Commun. 3:42015. View Article : Google Scholar : PubMed/NCBI
|
|
293
|
Platten M, Bunse L, Wick A, Bunse T, Le
Cornet L, Harting I, Sahm F, Sanghvi K, Tan CL, Poschke I, et al: A
vaccine targeting mutant IDH1 in newly diagnosed glioma. Nature.
592:463–468. 2021. View Article : Google Scholar :
|
|
294
|
Platten M, Schilling D, Bunse L, Wick A,
Bunse T, Riehl D, Green E, Sanghvi K, Karapanagiotou-Schenkel I,
Harting I, et al: Atim-33. Noa-16: A first-in-man multicenter phase
I clinical trial of the German neurooncology working group
evaluating a mutation-specific peptide vaccine targeting idh1r132h
in patients with newly diagnosed malignant astrocytomas. Neuro
Oncol. 20(Suppl 6): vi8–vi9. 2018. View Article : Google Scholar
|
|
295
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009. View Article : Google Scholar
|
|
296
|
Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur
V, Stevanović S, Gouttefangeas C, Platten M, Tabatabai G, Dutoit V,
van der Burg SH, et al: Actively personalized vaccination trial for
newly diagnosed glioblastoma. Nature. 565:240–245. 2019. View Article : Google Scholar
|
|
297
|
Narita Y, Arakawa Y, Yamasaki F, Nishikawa
R, Aoki T, Kanamori M, Nagane M, Kumabe T, Hirose Y, Ichikawa T, et
al: A randomized, double-blind, phase III trial of personalized
peptide vaccination for recurrent glioblastoma. Neuro Oncol.
21:348–359. 2019. View Article : Google Scholar :
|
|
298
|
Bruix J and Sherman M; Practice Guidelines
Committee American Association for the Study of Liver Diseases:
Management of hepatocellular carcinoma. Hepatology. 42:1208–1236.
2005. View Article : Google Scholar
|
|
299
|
Tampaki M, Papatheodoridis GV and
Cholongitas E: Intrahepatic recurrence of hepatocellular carcinoma
after resection: An update. Clin J Gastroenterol. 14:699–713. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
300
|
European Association for the Study of the
Liver: EASL clinical practice guidelines: Management of
hepatocellular carcinoma. J Hepatol. 69:182–236. 2018. View Article : Google Scholar
|
|
301
|
Charneau J, Suzuki T, Shimomura M,
Fujinami N and Nakatsura T: Peptide-based vaccines for
hepatocellular carcinoma: A review of recent advances. J Hepatocell
Carcinoma. 8:1035–1054. 2021. View Article : Google Scholar
|
|
302
|
Liu C, Shao J, Dong Y, Xu Q, Zou Z, Chen
F, Yan J, Liu J, Li S, Liu B and Shen J: Advanced HCC patient
benefit from neoantigen reactive T cells based immunotherapy: A
case report. Front Immunol. 12:6851262021. View Article : Google Scholar :
|
|
303
|
Cai Z, Su X, Qiu L, Li Z, Li X, Dong X,
Wei F, Zhou Y, Luo L, Chen G, et al: Personalized neoantigen
vaccine prevents postoperative recurrence in hepatocellular
carcinoma patients with vascular invasion. Mol Cancer. 20:1642021.
View Article : Google Scholar : PubMed/NCBI
|
|
304
|
Peng S, Chen S, Hu W, Mei J, Zeng X, Su T,
Wang W, Chen Z, Xiao H, Zhou Q, et al: Combination neoantigen-based
dendritic cell vaccination and adoptive T-cell transfer induces
antitumor responses against recurrence of hepatocellular carcinoma.
Cancer Immunol Res. 10:728–744. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
305
|
Shen J, Wang LF, Zou ZY, Kong WW, Yan J,
Meng FY, Chen FJ, Du J, Shao J, Xu QP, et al: Phase I clinical
study of personalized peptide vaccination combined with
radiotherapy for advanced hepatocellular carcinoma. World J
Gastroenterol. 23:5395–5404. 2017. View Article : Google Scholar
|
|
306
|
Guo Z, Yuan Y, Chen C, Lin J, Ma Q, Liu G,
Gao Y, Huang Y, Chen L, Chen LZ, et al: Durable complete response
to neoantigen-loaded dendritic-cell vaccine following anti-PD-1
therapy in metastatic gastric cancer. NPJ Precis Oncol. 6:342022.
View Article : Google Scholar :
|
|
307
|
Yu YJ, Shan N, Li LY, Zhu YS, Lin LM, Mao
CC, Hu TT, Xue XY, Su XP, Shen X and Cai ZZ: Preliminary clinical
study of personalized neoantigen vaccine therapy for microsatellite
stability (MSS)-advanced colorectal cancer. Cancer Immunol
Immunother. 72:2045–2056. 2023. View Article : Google Scholar
|
|
308
|
Sultan H, Salazar AM and Celis E:
Poly-ICLC, a multi-functional immune modulator for treating cancer.
Semin Immunol. 49:1014142020. View Article : Google Scholar : PubMed/NCBI
|
|
309
|
Zeng Y, Zhang W, Li Z, Zheng Y, Wang Y,
Chen G, Qiu L, Ke K, Su X, Cai Z, et al: Personalized
neoantigen-based immunotherapy for advanced collecting duct
carcinoma: Case report. J Immunother Cancer. 8:e0002172020.
View Article : Google Scholar :
|
|
310
|
Chen Z, Zhang S, Han N, Jiang J, Xu Y, Ma
D, Lu L, Guo X, Qiu M, Huang Q, et al: A neoantigen-based peptide
vaccine for patients with advanced pancreatic cancer refractory to
standard treatment. Front Immunol. 12:6916052021. View Article : Google Scholar
|
|
311
|
Kwok DW, Stevers NO, Etxeberria I, Nejo T,
Colton Cove M, Chen LH, Jung J, Okada K, Lakshmanachetty S, Gallus
M, et al: Tumour-wide RNA splicing aberrations generate actionable
public neoantigens. Nature. 639:463–473. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
312
|
Rosenberg-Mogilevsky A, Siegfried Z and
Karni R: Generation of tumor neoantigens by RNA splicing
perturbation. Trends Cancer. 11:12–24. 2025. View Article : Google Scholar
|
|
313
|
Huang P, Wen F, Tuerhong N, Yang Y and Li
Q: Neoantigens in cancer immunotherapy: Focusing on alternative
splicing. Front Immunol. 15:14377742024. View Article : Google Scholar :
|
|
314
|
Jayasinghe RG, Cao S, Gao Q, Wendl MC, Vo
NS, Reynolds SM, Zhao Y, Climente-González H, Chai S, Wang F, et
al: Systematic analysis of splice-site-creating mutations in
cancer. Cell Rep. 23:270–281.e3. 2018. View Article : Google Scholar
|
|
315
|
Smart AC, Margolis CA, Pimentel H, He MX,
Miao D, Adeegbe D, Fugmann T, Wong KK and Van Allen EM: Intron
retention is a source of neoepitopes in cancer. Nat Biotechnol.
36:1056–1058. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
316
|
Wang F, Cai G, Wang Y, Zhuang Q, Cai Z, Li
Y, Gao S, Li F, Zhang C, Zhao B and Liu X: Circular RNA-based
neoantigen vaccine for hepatocellular carcinoma immunotherapy.
MedComm (2020). 5:e6672024. View Article : Google Scholar : PubMed/NCBI
|
|
317
|
Hu ZX, Guo XY, Li ZT, Meng ZQ and Huang
SL: The neoantigens derived from transposable elements-A hidden
treasure for cancer immunotherapy. Biochim Biophys Acta Rev Cancer.
1879:1891262024. View Article : Google Scholar
|
|
318
|
Li M, Wang Y, Wu P, Zhang S, Gong Z, Liao
Q, Guo C, Wang F, Li Y, Zeng Z, et al: Application prospect of
circular RNA-based neoantigen vaccine in tumor immunotherapy.
Cancer Lett. 563:2161902023. View Article : Google Scholar : PubMed/NCBI
|
|
319
|
Lang F, Schrörs B, Löwer M, Türeci Ö and
Sahin U: Identification of neoantigens for individualized
therapeutic cancer vaccines. Nat Rev Drug Discov. 21:261–282. 2022.
View Article : Google Scholar
|
|
320
|
Gurung HR, Heidersbach AJ, Darwish M, Chan
PPF, Li J, Beresini M, Zill OA, Wallace A, Tong AJ, Hascall D, et
al: Systematic discovery of neoepitope-HLA pairs for neoantigens
shared among patients and tumor types. Nat Biotechnol.
42:1107–1117. 2024. View Article : Google Scholar
|
|
321
|
Shen KY, Zhu Y, Xie SZ and Qin LX:
Immunosuppressive tumor microenvironment and immunotherapy of
hepatocellular carcinoma: Current status and prospectives. J
Hematol Oncol. 17:252024. View Article : Google Scholar : PubMed/NCBI
|
|
322
|
Cherepnev G, Volk HD and Kern F: Use of
peptides and peptide libraries as T-cell stimulants in flow
cytometric studies. Methods Cell Biol. 75:453–479. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
323
|
Gascoigne NR: Do T cells need endogenous
peptides for activation? Nat Rev Immunol. 8:895–900. 2008.
View Article : Google Scholar
|
|
324
|
Shemesh CS, Hsu JC, Hosseini I, Shen BQ,
Rotte A, Twomey P, Girish S and Wu B: Personalized cancer vaccines:
Clinical landscape, challenges, and opportunities. Mol Ther.
29:555–570. 2021. View Article : Google Scholar
|
|
325
|
Gerlinger M, Rowan AJ, Horswell S, Math M,
Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N,
Stewart A, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar
|
|
326
|
Prehn RT and Main JM: Immunity to
methylcholanthrene-induced sarcomas. J Natl Cancer Inst.
18:769–778. 1957.PubMed/NCBI
|
|
327
|
Hao Q, Long YH, Yang Y, Deng YQ, Ding ZY,
Yang L, Shu Y and Xu H: Development and clinical applications of
therapeutic cancer vaccines with individualized and shared
neoantigens. Vaccines (Basel). 12:7172024. View Article : Google Scholar
|
|
328
|
D'Alise AM, Brasu N, De Intinis C, Leoni
G, Russo V, Langone F, Baev D, Micarelli E, Petiti L, Picelli S, et
al: Adenoviral-based vaccine promotes neoantigen-specific
CD8+ T cell stemness and tumor rejection. Sci Transl
Med. 14:eabo76042022. View Article : Google Scholar
|
|
329
|
Tsimberidou AM: Targeted therapy in
cancer. Cancer Chemother Pharmacol. 76:1113–1132. 2015. View Article : Google Scholar
|