Introduction
Gastric cancer (GC) is the fifth most common
malignancy and the fifth leading cause of cancer-related mortality
worldwide (1). Several factors
contribute to GC initiation and progression, including
Helicobacter pylori (HP) infection, chronic gastritis and
genetic alterations (2,3). Despite its high incidence, GC is
often diagnosed at an advanced stage due to the non-specific nature
of its clinical symptoms. Consequently, late detection typically
leads to poor prognosis, with the 5-year survival rate remaining
<30% (4,5). This scenario underscores the urgent
need to identify novel therapeutic targets to enhance treatment
efficacy for GC.
The Eph receptor family, first identified in a human
hepatocellular carcinoma cell line known for erythropoietin
production, has been implicated in multiple types of cancer due to
its frequent upregulation in receptor tyrosine kinase (RTK)
screenings (6). For example,
ephrin-A1 is upregulated in hepatocellular carcinoma and drives its
progression by promoting proliferation and invasion (7). Ephrin-B3 expression is upregulated
in glioma biopsies and promotes invasion of glioma cells (8). This discovery emerged during
research aimed at identifying novel tyrosine kinase receptors,
highlighting the significance of Eph receptors in oncogenic
processes (9). Ephrin-B2 (EFNB2),
a membrane-anchored ligand of the Eph family of RTKs (10), interacts with the EphB4 receptor
as part of the largest RTK subfamily. The EFNB2-EphB4 signaling
axis serves a key role in regulating cellular adhesion and
repulsion in diverse biological contexts (11,12). Furthermore, EFNB2 can function in
a cell-autonomous manner, independent of Eph receptor interactions,
and participates in complex signaling pathways, including the
AKT/mTOR and ERK pathways (13-15), thereby modulating various
physiological events. Notably, EFNB2 has been demonstrated to
regulate malignant progression in various tumor cells (15-18). For instance, EFNB2 is highly
expressed in pancreatic cancer, and facilitates proliferation,
migration and invasion of pancreatic cancer via activation of the
p53/p21 signaling axis (16).
Conversely, EFNB2 expression has been associated with good
prognosis in breast cancer, and is known to inhibit cell
proliferation and migration (17). However, to the best of our
knowledge, the role of EFNB2 in GC remains unclear and requires
further investigation.
Integrated genomic analysis has identified
dysregulation of the Wnt/β-catenin pathway in ~46% of GC cases
(19). This evolutionarily
conserved signaling pathway, frequently disrupted by component
mutations, regulates key cellular processes, including fate
determination, proliferation, differentiation, migration, apoptosis
and stemness maintenance (20-22). Under basal conditions, β-catenin
levels are tightly controlled by a destruction complex containing
GSK3β, which targets β-catenin for proteasomal degradation through
ubiquitination-dependent mechanisms (21). Upon pathway activation, stabilized
β-catenin is translocated into the nucleus, where it forms
transcriptional complexes with co-activators to modulate gene
expression programs, including c-myc controlling cell proliferation
and differentiation (21,22). Considering the potential roles of
EFNB2 and the Wnt/β-catenin pathway in tumors, we hypothesized that
EFNB2 may regulate the malignant progression of GC through the
Wnt/β-catenin signaling cascade.
The present study investigated the expression levels
of EFNB2 and its prognostic significance in patients with GC
through a comprehensive analysis of public databases and tissue
microarray (TMA)-based immunohistochemical (IHC) assessment. To
further elucidate the functional roles of EFNB2 in GC pathogenesis,
a series of cellular assays were carried out and murine models were
established to examine its regulatory effects on cellular
proliferation, apoptotic processes and cell cycle progression both
in vitro and in vivo. Mechanistically, CHIR99021 and
differentiation-inducing factor-3 (DIF-3) were employed to assess
whether the effects of EFNB2 were primarily mediated through the
Wnt/β-catenin signaling cascade.
Materials and methods
Bioinformatics analysis
Various public online databases, including Gene
Expression Profiling Interactive Analysis (GEPIA) (23), LinkedOmics (24), Kaplan-Meier Plotter (25), Tumor Immune Estimation Resource
(TIMER) (26) and Gene Expression
Omnibus (GEO; https://www.ncbi.nlm.nih.gov/gds), were used to
explore EFNB2 expression and prognosis, and perform associated
analyses. In GEPIA, the option 'Match TCGA normal and GTEx data'
was selected for EFNB2 expression analysis; the cutoff setting
'Cutoff-High versus Cutoff-Low: 30 versus 30' was employed for
survival analysis; and Spearman correlation analysis was performed
to assess the associations of EFNB2 with GSK3β, catenin β1 and MYC.
In Kaplan-Meier Plotter, the option 'Exclude GSE62254' was selected
for survival analysis (27).
Therefore, the analysis included datasets GSE14210, GSE15459,
GSE22377, GSE29272 and GSE51105 (28-32), with the cutoff set to 'Auto select
best cutoff'. Other parameters were left at the default.
Additionally, two GEO datasets (GSE13195 and GSE184336) were used
to compare the expression levels of EFNB2 and perform correlation
analyses (33,34).
Tissue samples
A commercial GC tissue microarray (cat. no.
HStmA180Su3) was purchased from Shanghai Outdo Biotech Co., Ltd.,
consisting of 96 GC tissues and 84 adjacent normal tissues (ANTs)
collected between December 2013 and September 2015. The mean age of
the patients was 59 years, with a range of 40-83 years. The
inclusion criteria were as follows: i) Age ≥18 years, ii)
pathologically diagnosed with gastric adenocarcinoma; and iii)
underwent radical gastrectomy. The exclusion criteria included the
following: i) Underwent preoperative radiotherapy or chemotherapy;
ii) pathological specimens unavailable; iii) mortality within 1
month following operation; and iv) refusal to participate in
follow-up. Levels of the tumor markers AFP, CEA, CA50, CA125, CA153
and CA199 were assessed in patients with GC 1-3 days prior to
surgery. Comprehensive clinical, pathological and follow-up data
were available for all participants, with the exception of 24
patients lacking HP infection data and 2 patients with missing HER2
expression data. The diagnosis and staging were examined based on
the 8th edition of the American Joint Committee on Cancer
guidelines (35). Patients
underwent follow-up every 3-6 months, with a maximum follow-up of
60 months. Among the 96 cases, 67 patients died between June 2014
and March 2020, while the remaining 29 patients survived until the
end of the follow-up period. The clinicopathological
characteristics of the patients with GC are presented in Table I. Ethical approval for the present
study was obtained from the Ethics Committee of the First
Affiliated Hospital of Anhui Medical University (approval no.
2024137; Hefei, China).
 | Table IRelationship between EFNB2 expression
and clinicopathological characteristics of patients with gastric
cancer (n=96). |
Table I
Relationship between EFNB2 expression
and clinicopathological characteristics of patients with gastric
cancer (n=96).
| Parameters | Total, n | EFNB2
expression | P-value |
|---|
| Low, n (n=48) | High, n (n=48) |
|---|
| Age (years) | | | | 0.307 |
| <60 | 47 | 26 | 21 | |
| ≥60 | 49 | 22 | 27 | |
| Sex | | | | 0.811 |
| Female | 23 | 12 | 11 | |
| Male | 73 | 36 | 37 | |
| Helicobacter
pylori infection status | | | | 0.479 |
| Negative | 36 | 16 | 20 | |
| Positive | 36 | 19 | 17 | |
| Missing value | 24 | | | |
| Tumor location | | | | 0.128 |
| Cardia | 15 | 4 | 11 | |
| Gastric body | 15 | 9 | 6 | |
| Antrum | 66 | 35 | 31 | |
| Histological
grade | | | | 0.128 |
| G1 | 10 | 8 | 2 | |
| G2 | 38 | 17 | 21 | |
| G3 | 48 | 23 | 25 | |
| Tumor size
(cm) | | | | <0.001a |
| <4 | 48 | 35 | 13 | |
| ≥4 | 48 | 13 | 35 | |
| T stage | | | | 0.011a |
| T1-T2 | 25 | 18 | 7 | |
| T3-T4 | 71 | 30 | 41 | |
| Lymph node
metastasis | | | | 0.369 |
| Negative | 28 | 16 | 12 | |
| Positive | 68 | 32 | 36 | |
| M stage | | | | 0.037a |
| M0 | 83 | 45 | 38 | |
| M1 | 13 | 3 | 10 | |
| TNM stage | | | | 0.004a |
| Ⅰ-Ⅱ | 46 | 30 | 16 | |
| Ⅲ-Ⅳ | 50 | 18 | 32 | |
| HER2 | | | | 0.130b |
| Negative | 87 | 45 | 42 | |
| Positive | 7 | 1 | 6 | |
| Missing value | 2 | | | |
H&E, IHC and TUNEL staining
H&E and IHC staining of human and mouse GC
tissues was conducted according to the standard procedures
previously described (36,37).
The sections were incubated with primary antibodies at 4°C
overnight. The antibodies used were as follows: EFNB2 antibody
(1:500; cat. no. sc-398735; Santa Cruz Biotechnology, Inc.),
β-catenin antibody (1:500; cat. no. 51067-2-AP; Proteintech Group,
Inc.) and Ki67 antibody (1:500; cat. no. ab15580; Abcam). The One
Step TUNEL Apoptosis Assay Kit (cat. no. C1088; Beyotime
Biotechnology) was used according to the manufacturer's
instructions. Briefly, tumor sections were fixed in 4%
paraformaldehyde at 25°C for 60 min and then washed three times
with PBS. The TUNEL reaction solution was prepared by mixing 5
μl TdT enzyme with 45 μl fluorescent labeling
solution. Each section was treated with 50 μl of the TUNEL
solution at 37°C for 60 min in the dark, and then washed three
times with PBS. Finally, the sections were mounted with one drop of
Antifade Mounting Medium with DAPI (1:1,000; cat. no. P0131;
Beyotime Biotechnology) at 37°C for 3 min and examined under an
inverted fluorescence microscope (Olympus Corporation). Three
random fields were captured for analysis. The percentage of Ki67 or
TUNEL-positive cells was calculated as the number of positive
cells/total number of cells per field.
TMA construction and IHC evaluation
A TMA of GC tissues and ANTs was constructed using
formalin-fixed and paraffin-embedded blocks (cat. no. HStmA180Su32;
Shanghai Outdo Biotech Co., Ltd.) to evaluate the expression levels
of EFNB2. After IHC staining as described previously (36,37), TMA sections were scanned and
digitalized using the Panoramic MIDI system (3D Histech; 3DHISTECH,
Ltd.). IHC staining findings were independently assessed by two
senior pathologists who were blinded to the clinical outcomes of
the patients with GC. EFNB2 or β-catenin expression was quantified
using an integrated IHC score, which combined both staining
intensity and proportion (38,39). The samples were categorized into
two groups according to the median IHC score: The low and high
EFNB2 expression groups.
Cell culture and reagents
GC cell lines (MKN-45, AGS, MGC-803, BGC-823, and
HGC-27) and the normal gastric mucosa epithelial cell line (GES-1)
were obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. lAGS cells were cultured in F12 medium
(HyClone; Cytiva). HGC-27 cells were cultured in DMEM (HyClone;
Cytiva). MKN-45, MGC-803, BGC-823 and GES-1 cells were cultured in
RPMI-1640 medium (HyClone; Cytiva). All media were supplemented
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (MilliporeSigma). The cells were cultured
in a humidified incubator at 37°C with 5% CO2. For the
rescue experiments, cells in designated groups were treated at 37°C
for 24 h with either CHIR99021 (5 μM; MedChemExpress) to
activate the Wnt/β-catenin signaling cascade or DIF-3 (30
μM; MedChemExpress) to inhibit the Wnt/β-catenin signaling
cascade.
Short hairpin RNA (shRNA/sh) and plasmid
transfection
Transfection of GC cells was conducted using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
shRNAs, the negative control (NC) shRNA, overexpression plasmids
and empty vector were synthesized by Shanghai GeneChem Co., Ltd.
hU6-MCS-CBh-gcGFP-IRES-puromycin was used as the vector for
sh-EFNB2 and NC. The target sequence of EFNB2 shRNAs for AGS cells
was as follows: sh1-EFNB2, 5'-CGATTTCCAAATCGATAGTTT-3'; and
sh2-EFNB2, 5'-AGCAGACAGATGCACTATTAA-3'. A random sequence control
shRNA (5'-TTCTCCGAACGTGTCACGT-3') was used as NC. For EFNB2
overexpression, HGC-27 cells were transfected with the
pcDNA3.1-EGFP-P2A-EFNB2-3FLAG plasmid utilizing
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Empty vector was used as a control for
overexpression experiments. The transfection protocol involved the
introduction of 10 μg of the designated plasmids into the
AGS or HGC-27 cells. GC cells were infected with the lentiviral
vector at 37°C for 48 h (MOI, 10). Following a 2-week selection
with 4 μg/ml puromycin (Wuhan Servicebio Technology Co.,
Ltd.), the stable clones were kept in culture with a maintenance
dose of 2 μg/ml of puromycin (Wuhan Servicebio Technology
Co., Ltd.). After the 48-h transfection and subsequent 2-week
selection, the efficiency of EFNB2 knockdown or overexpression was
verified at the protein level.
Western blotting
Western blotting was conducted as previously
described (25-27). GES-1, MKN-45, AGS, MGC-803,
BGC-823 and HGC-27 cells were lysed with RIPA lysis buffer
(Beyotime Biotechnology). This buffer is formulated to prevent
protein degradation and preserve protein phosphorylation. The
details of the primary antibodies used were as follows: EFNB2
(1:1,000; cat. no. sc-398735; Santa Cruz Biotechnology, Inc.),
Bcl-2 (1:2,000; cat. no. ab182858; Abcam), Bax (1:2,000; cat. no.
ab32503; Abcam), CyclinD1 (1:1,000, cat. no. 2978; Cell Signaling
Technology, Inc.), CDK4 (1:1,000; cat. no. 12790; Cell Signaling
Technology, Inc.), GSK3β (1:5,000; cat. no. 82061-1-RR; Proteintech
Group, Inc.), phosphorylated (p-)GSK3β (Ser389; 1:1,000; cat. no.
14850-1-AP; Proteintech Group, Inc.), β-catenin (1:5,000; cat. no.
51067-2-AP; Proteintech Group, Inc.), c-myc (1:5,000; cat. no.
808451-RR; Proteintech Group, Inc.) and β-actin (1:1,000; cat. no.
4970; Cell Signaling Technology, Inc.). After incubation with the
primary antibody at 4°C overnight, the membranes were incubated
with an HRP-labeled secondary antibody (1:5,000; cat. nos. ZB-5301
and ZB-5305; Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) and
visualized using a Tanon 5200 Multi-Imaging System (Tanon Science
and Technology Co., Ltd.).
Cell Counting Kit-8 (CCK-8) and colony
formation assays
The protocols for the CCK-8 and colony formation
assays have been described in detail previously (16,26,27). CCK-8 (Dojindo Laboratories, Inc.)
was used to assess the proliferation of GC cells according to
manufacturer's protocol. Briefly, AGS or HGC-27 cells were seeded
into a 96-well plate at a density of ~4×103 cells/well.
The absorbance was evaluated at 450 nm using a microplate reader
(Tecan Group, Ltd.). For the colony formation assay, equal numbers
(500 cells/well) of GC cells were seeded into 6-well plates and
cultured in complete growth medium at 37°C. After 14 days of
incubation, colonies were fixed with 4% paraformaldehyde for 20 min
and stained with 0.1% crystal violet solution (Sangon Biotech Co.,
Ltd.) for 30 min, both at room temperature. Images of the visible
colonies in each well were subsequently captured, and colonies were
quantified using ImageJ software (version 1.54k; National
Institutes of Health).
Cell migration and invasion assays
A Transwell chamber (8 μm pore size; Corning,
Inc.) precoated with Matrigel (1:8 dilution; BD Biosciences) at
37°C for 1 h was used for invasion assays, while a Transwell
chamber without Matrigel was employed for migration assays. For the
AGS cells (2×105 cells/well), serum-free F12 medium was
plated in the upper chamber, and F12 medium containing 20% FBS was
added to the lower chamber. For the HGC-27 cells (1×105
cells/well), serum-free DMEM was used in the upper chamber, and
DMEM with 20% FBS was used in the lower chamber. After 48 h of
incubation at 37°C, non-migratory cells were removed with a cotton
swab, while migratory cells were fixed and stained with 0.1%
crystal violet (Sangon Biotech Co., Ltd.) for 30 min at room
temperature. The wells were examined under an Olympus IX70 light
microscope (Olympus Corporation), and cells were counted in at
least three randomly selected fields per well.
Flow cytometry
AGS and HGC-27 cells were cultured and harvested.
Apoptosis was detected using the APC Annexin V Apoptosis Detection
Kit with 7-AAD (cat. no. 640930; BioLegend, Inc.). Briefly, GC
cells were resuspended in 1X binding buffer at a density of
1×106 cells/ml and then incubated with 5 μl
annexin V-APC and 5 μl 7-AAD for 15 min at 25°C in the dark.
Samples were analyzed using a FACS Canto II flow cytometer (BD
Biosciences) and FlowJo v10.7.1 (BD Biosciences).
The cell cycle distribution was assessed by flow
cytometry after PI staining (cat. no. C1052; Beyotime
Biotechnology). Briefly, GC cells were fixed with 70% cold ethanol
on ice for 60 min. The cells were washed with PBS three times and
stained with PI solution in the dark for 30 min at 37°C. Finally,
the cell cycle distribution was analyzed using a FACS Canto II flow
cytometer (BD Biosciences). The percentages of cells in the
G0/G1, S and G2/M phases were
analyzed using ModFit LT 4.1 software (Verity Software House,
Inc.).
Nude mouse xenograft model
The Animal Ethics Committee of the First Affiliated
Hospital of Anhui Medical University granted approval for this
protocol (approval no. LLSC20240221; Hefei, China). Female BALB/c
nude mice (5-6 weeks old; weight, 18-22 g) were obtained from
Beijing Vital River Laboratory Animal Technology Co., Ltd. A total
of 16 mice were housed under specific pathogen-free conditions in
an air-conditioned room (22±2°C) with a 12/12 h light/dark cycle
and 60-65% relative humidity. The mice were provided with ad
libitum access to food and water. First, the encoder carried
out randomization using a random number table to assign mice into
four groups (n=4 per group). Each mouse was marked with an ear tag
for identification, and the correspondence between coded groups and
experimental groups was established. Second, without knowledge of
the tumor suspension assignments or mouse group identities, the
experimenters established the cell-derived xenograft (CDX) model.
HGC-27 cells stably transfected with either the empty vector or the
EFNB2 vector were inoculated subcutaneously into the groin of the
mice at a dose of 1×107 cells per mouse, suspended in
100 μl PBS. DIF-3 was administered via peritumoral
subcutaneous injection twice a week at a concentration of 50
μM and a dose of 100 μl per mouse. The control group
received PBS at the same frequency and dose. Animal welfare was
maximized, and pain and distress were minimized, while achieving
scientific research objectives. The humane endpoints used were as
follows: i) Body weight loss ≥20% of initial body weight; ii) tumor
volume exceeding 2,000 mm3; iii) tumor ulceration area
≥10 mm2; iv) detection of tumor metastasis to other
organs, such as the lungs; v) persistent vocalization, trembling or
self-mutilation behavior; and vi) inability to access food or water
autonomously, accompanied by signs of severe dehydration or
malnutrition. All mice in the present study were subjected to daily
health monitoring. Following the establishment of the subcutaneous
tumor-bearing models, this frequency was ensured to be no less than
twice daily to facilitate timely assessment of their condition.
Tumor size was measured with a vernier caliper every 3-4 days, and
the tumor volume was calculated using the following formula:
(Length × width2) × 0.5. A total of 28 days after
injection, mice were humanely euthanized via CO2
asphyxiation using a gradual-fill approach, with the gas introduced
at a rate of 30-50% of the chamber volume/min. Cervical dislocation
was subsequently performed to confirm death. Tumors were harvested
and weighed. All data were recorded according to the pre-assigned
identification numbers. The encoder and researchers jointly
unblinded the study by verifying the correspondence between the
codes and the actual group assignments. The harvested tumor tissues
were subsequently processed for histological staining.
Statistical analysis
Statistical analyses were carried out using SPSS
Statistics 22.0 (IBM Corp.), while graphical representations were
generated with GraphPad Prism 7.0 (Dotmatics). Each experiment was
performed in at least triplicate, with data presented as the mean ±
SD. Q-Q plots and the Shapiro-Wilk test were used to assess the
normality of data. For the comparison of two groups, paired or
unpaired Student's t-tests were used when the continuous variable
satisfied the assumptions of normal distribution and homogeneity of
variance; the Mann-Whitney U test was used when the variable was
non-continuous or when these assumptions (normality and/or
homogeneity of variance) were violated. For comparisons among
multiple groups, one-way ANOVA followed by Tukey's post hoc test
was applied when the continuous variable satisfied the assumptions
of normal distribution and homogeneity of variance; otherwise, the
Kruskal-Wallis H test followed by Dunn's post hoc test was used.
The relationship between EFNB2 expression and clinicopathological
characteristics was examined using Pearson's χ2 test.
When the total sample size was ≥40 and at least one expected
frequency was between 1 and 5, Yates' continuity correction was
applied. Survival analysis was conducted using the Kaplan-Meier
approach, with significance determined by the log-rank test.
Multivariate analysis was conducted using the Cox proportional
hazards model. P<0.05 (two-tailed) was considered to indicate a
statistically significant difference.
Results
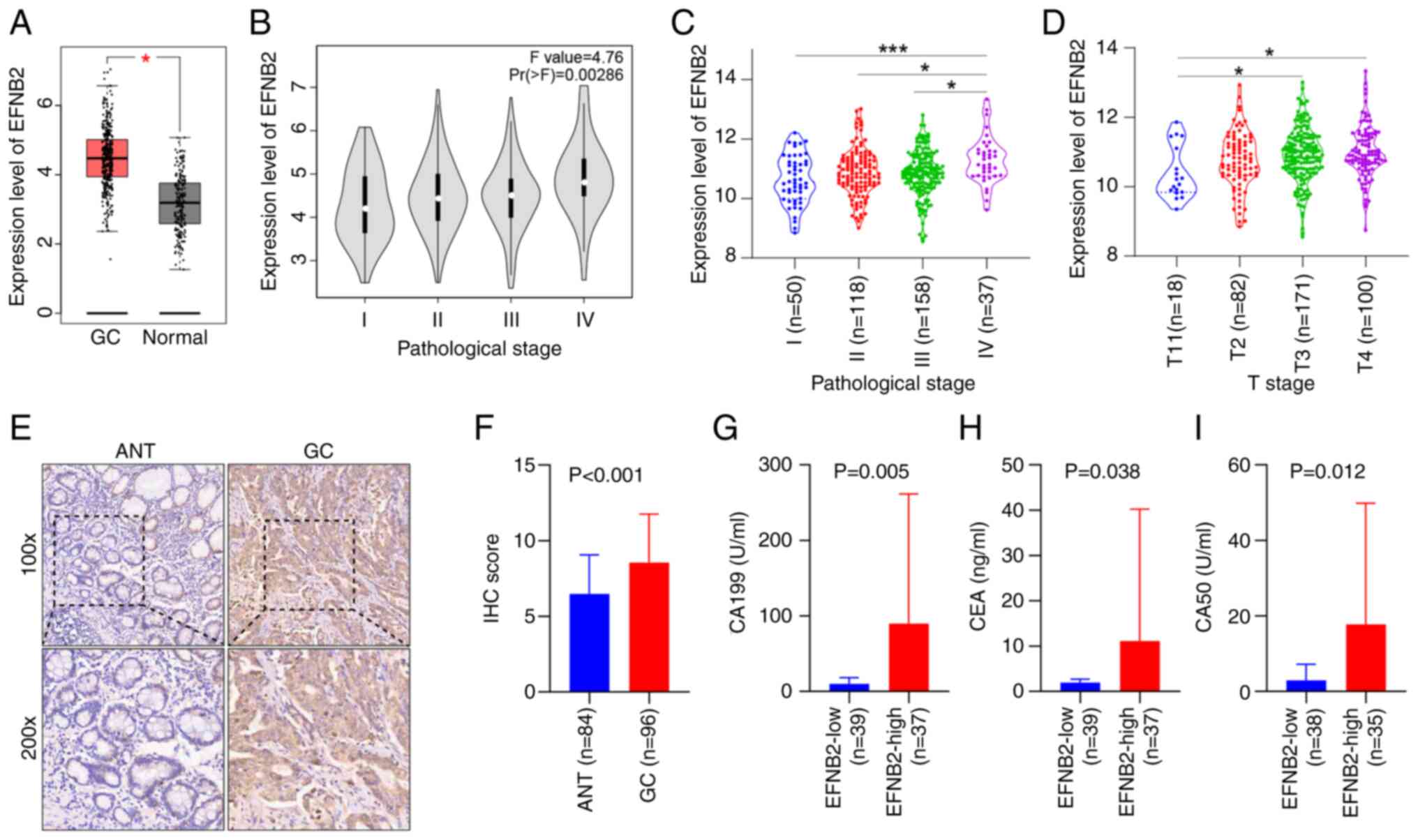
EFNB2 is highly expressed in GC tissues
and is associated with clinicopathologic characteristics
An analysis of EFNB2 expression across 33 cancer
types was performed using GEPIA, comparing tumor samples to
corresponding normal tissues from healthy individuals. Analysis
revealed that EFNB2 expression levels were increased in GC when
comparing 408 GC tissues and 211 normal tissues (Fig. 1A). EFNB2 exhibited abnormal
expression patterns in several solid malignancies, including colon
adenocarcinoma, esophageal carcinoma and pancreatic adenocarcinoma
(Fig. S1A). Furthermore, the
independent GEO database analysis confirmed that EFNB2 was highly
expressed in GC tissues compared with ANTs or normal gastric mucosa
tissues (Fig. S1B and C).
Analysis of data from GEPIA and Linkedomics databases demonstrated
that the expression levels of EFNB2 increased with the progression
of GC staging (Fig. 1B and C).
EFNB2 expression levels were significantly increased in GC with
advanced T stages (T3 or T4) compared with earlier T stages (T1)
(Fig. 1D).
To further validate these observations, a TMA was
constructed for IHC analysis utilizing a total of 180 samples,
comprising 96 GC tissues and 84 ANTs. Analysis revealed elevated
expression levels of EFNB2 in GC tissues (Fig. 1E and F). Furthermore, EFNB2
expression levels were significantly associated with tumor size, T
stage, M stage and TNM stage. However, non-significant associations
were identified between EFNB2 expression and other clinical
parameters of patients with GC, including age, sex, HP infection,
tumor location, histological grade, lymph node metastasis and HER2
status (Table I). Additionally,
the high EFNB2 expression group exhibited increased levels of
specific tumor markers, including CA199, CEA and CA50 (Fig. 1G-I). However, no significant
differences in AFP, CA125 or CA153 levels were observed (Fig. S1D-F).
Upregulated EFNB2 expression is
associated with poor prognosis of patients with GC
Subsequently, public databases were used to
investigate the association between EFNB2 and the prognosis of
patients with GC. Analysis of data from the Kaplan-Meier plotter
demonstrated that EFNB2 expression was negatively associated with
overall survival (OS; n=592) and disease-free survival (DFS; n=221)
in patients with GC (Fig. 2A and
B). The EFNB2-high expression group exhibited a 1.25-fold
elevated risk of GC-related mortality compared with the low
expression group. Accordingly, the median OS was shorter in the
high-expression group (19.0 months) than in the low-expression
group (24.5 months) (Fig. 2A and
B). The EFNB2-high expression group exhibited a 1.47-fold
higher risk of relapse compared with the low expression group. The
upper quartile DFS times were 5.3 and 3.5 months for the low and
high expression groups, respectively (Fig. 2A and B). Similarly, analysis using
the GEPIA database (n=383) demonstrated that high EFNB2 expression
was significantly associated with worse overall prognosis and
shorter time to recurrence in patients with GC (Fig. 2C and D). Notably, analysis of the
GC cohort in the present study whose samples were included in the
TMA (n=96) verified that high EFNB2 expression was an adverse
factor for the overall prognosis of patients with GC (Fig. 2E). The median OS times of the low
and high expression groups were 42 and 30 months, respectively.
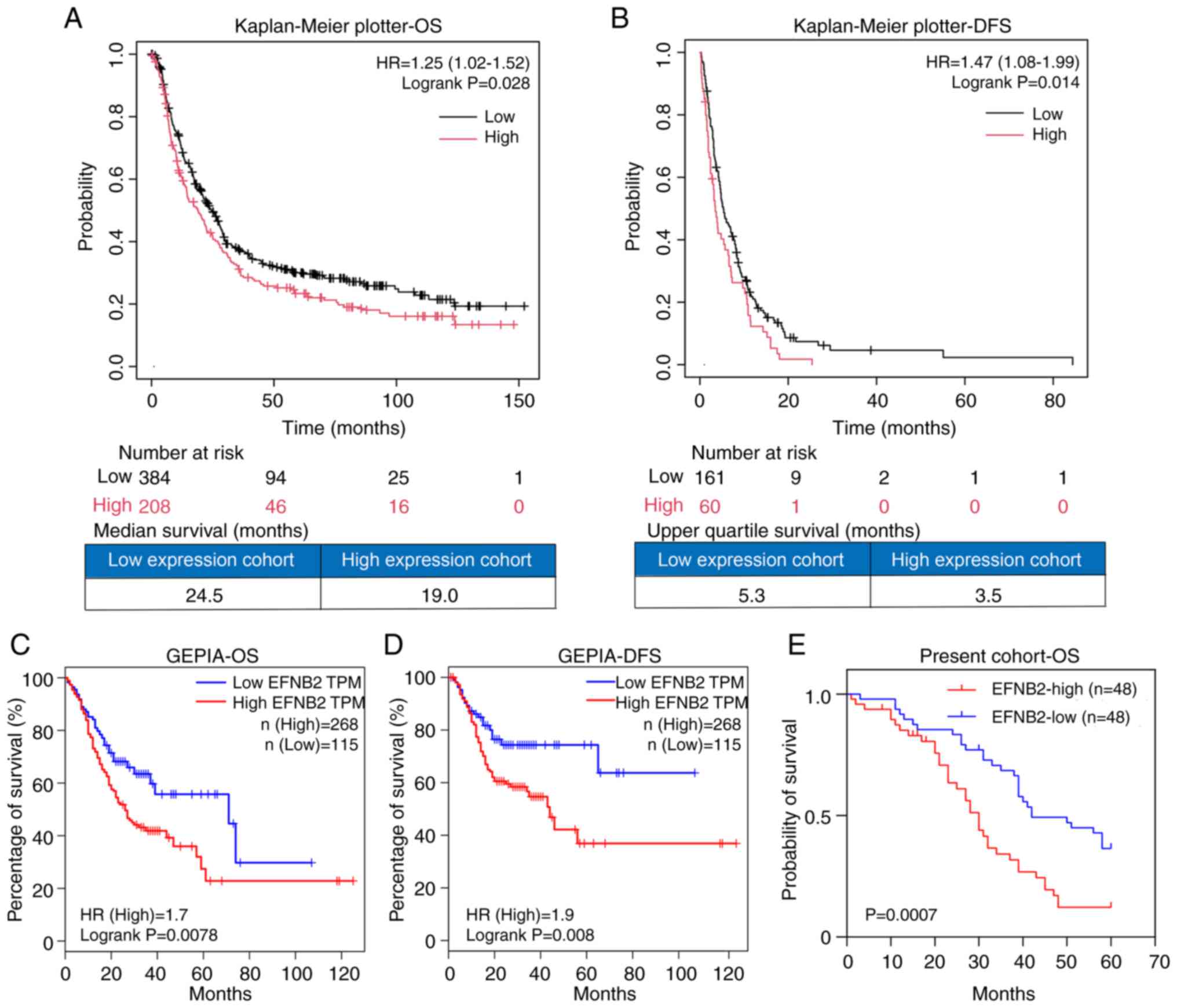 | Figure 2Upregulated EFNB2 expression is
associated with poor prognosis of patients with GC. The
Kaplan-Meier plotter demonstrated that EFNB2 was not only
associated with (A) OS (P=0.028), but also served as a negative
factor for (B) DFS in GC (P=0.014), when excluding the dataset
GSE62254. In the GEPIA database, the EFNB2-high expression group
exhibited shorter (C) OS and (D) DFS compared with the EFNB2-low
expression group, with the conditions set as 'Cutoff-High vs.
Cutoff-Low: 30 vs. 30' (P=0.0078 and 0.008, respectively). (E)
Analysis of the cohort of patients whose samples were included in
the tissue microarray (n=96) demonstrated that patients with GC
with high EFNB2 expression had a worse clinical outcome compared
with those with low EFNB2 expression (P=0.0007). OS, overall
survival; DFS, disease-free survival; HR, hazard ratio; EFNB2,
ephrin-B2; GC, gastric cancer; GEPIA, Gene Expression Profiling
Interactive Analysis; TPM, transcript per million. |
The univariate analysis revealed that EFNB2
expression, tumor size, T stage, M stage and TNM stage were
associated with the prognosis of patients with GC (Table II). These variables were
subsequently included in a Cox proportional hazards regression
model. Multivariate analysis demonstrated that only T stage
remained an independent adverse prognostic factor (Table II). Furthermore, subgroup
analysis of patients with GC demonstrated that high EFNB2
expression was associated with a worse prognosis in subgroups of
patients with the following characteristics: Age ≥60 years, male
sex, HP infection-positive, tumor location antrum, histological
G2-3 grade, T stage T3-T4, lymph node metastasis-positive, M stage
M0 and HER2-negative (Table
III; Fig. S2).
 | Table IIUnivariate and multivariate analyses
for prognostic factors of gastric cancer. |
Table II
Univariate and multivariate analyses
for prognostic factors of gastric cancer.
| Parameters | Univariate
analysis | Multivariate
analysis |
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (≥60 vs. <60
years) | 1.453
(0.896-2.355) | 0.130 | - | - |
| Sex (male vs.
female) | 1.113
(0.642-1.932) | 0.702 | - | - |
| Helicobacter
pylori infection (positive vs. negative) | 0.713
(0.413-1.229) | 0.223 | - | - |
| Tumor location
(cardia + gastric body vs. antrum) | 0.927
(0.549-1.565) | 0.776 | - | - |
| Histological grade
(G1 vs. G2-G3) | 1.151
(0.549-2.414) | 0.710 | - | - |
| Tumor size (≥4 vs.
<4 cm) | 2.315
(1.411-3.797) | 0.001a | 1.368
(0.687-2.726) | 0.373 |
| T stage (T1-T2 vs.
T3-T4) | 3.348
(1.695-6.614) | 0.001a | 2.385
(1.112-5.113) | 0.026a |
| Lymph node
metastasis (positive vs. negative) | 1.709
(0.971-3.008) | 0.063 | - | - |
| M stage (M0 vs.
M1) | 3.149
(1.687-5.880) | <0.001a | 1.731
(0.870-3.444) | 0.118 |
| TNM stage (I-II vs.
III-IV) | 2.813
(1.689-4.684) | <0.001a | 1.428
(0.741-2.751) | 0.287 |
| HER2 (positive vs.
negative) | 1.990
(0.851-4.651) | 0.112 | - | - |
| EFNB2 expression
(high vs. low) | 2.257
(1.381-3.689) | 0.001a | 1.300
(0.688-2.456) | 0.419 |
 | Table IIIPrognostic relevance of EFNB2
expression in subgroups of patients with GC in which the worse
overall survival of patients with GC is significantly associated
with high EFNB2 expression. |
Table III
Prognostic relevance of EFNB2
expression in subgroups of patients with GC in which the worse
overall survival of patients with GC is significantly associated
with high EFNB2 expression.
| Subgroups | HR | 95% CI | P-value |
|---|
| Age ≥60 years | 2.846 | 1.421-5.700 | 0.0032 |
| Male | 2.367 | 1.308-4.282 | 0.0044 |
| HP infection,
positive | 3.028 | 1.312-6.986 | 0.0094 |
| Tumor location,
antrum | 3.385 | 1.867-6.135 | <0.0001 |
| Histological grade,
G2-G3 | 2.825 | 1.639-4.868 | 0.0002 |
| T stage, T3-T4 | 2.166 | 1.261-3.719 | 0.0051 |
| Lymph node
metastasis, positive | 2.727 | 1.522-4.889 | 0.0008 |
| M stage, M0 | 2.493 | 1.381-4.500 | 0.0024 |
| HER2, negative | 2.548 | 1.465-4.433 | 0.0009 |
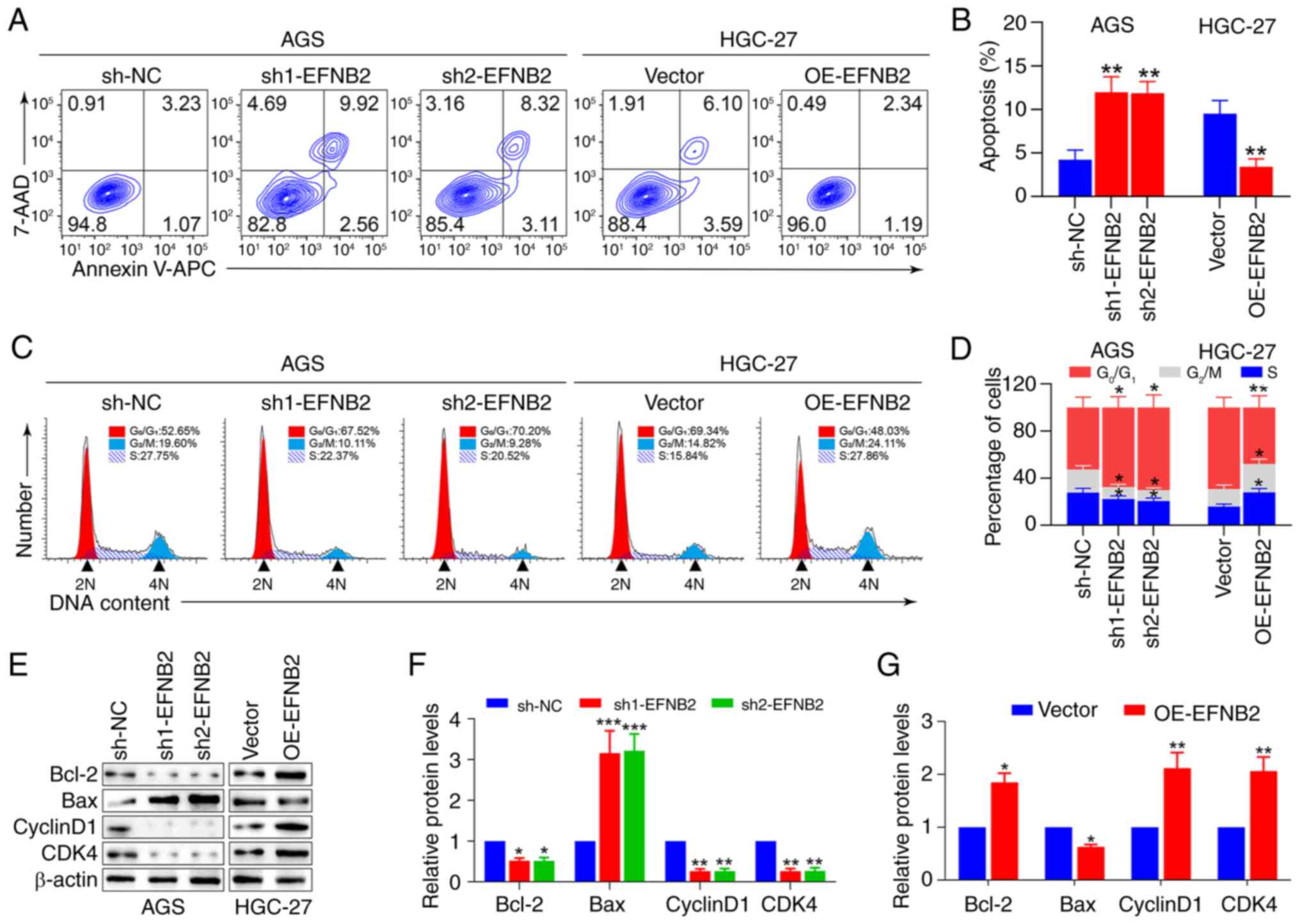
EFNB2 promotes proliferation, inhibits
apoptosis and alters the cell cycle distribution of GC cells in
vitro
To explore the roles of EFNB2 in GC cells, EFNB2
levels in various GC cell lines were measured. Compared with those
in human normal GES-1 cells, the expression levels of EFNB2 were
higher in multiple GC cell lines (Fig. 3A and B). For the subsequent
experiments, two cell lines were selected based on intrinsic EFNB2
expression levels: AGS cells (relatively high) and HGC27 cells
(relatively low). EFNB2 was then knocked down in the AGS cells and
overexpressed in the HGC27 cells using lentiviral transfection
(40). The successful stable
knockdown and overexpression of EFNB2 were verified at the protein
level (Fig. 3C and D). Analysis
of CCK-8 and colony formation assay data indicated that EFNB2
knockdown inhibited the proliferation and cell viability compared
with those in the NC group in AGS cells. Reciprocally, EFNB2
overexpression promoted these cellular properties in HGC-27 cells
(Fig. 3E-G). However, the
alteration of EFNB2 levels had a non-significant effect on cell
invasion and migration in GC cells (Fig. S3A-C). Additionally, analysis of
data from the TIMER database indicated that the infiltration of
CD8+ T cells and neutrophils in GC tissues was
negatively associated with EFNB2 expression levels, while a
non-significant relationship was found with the infiltration of
macrophages or dendritic cells (Fig.
S3D). These data indicated that EFNB2 serves a key role in the
malignant progression of GC.
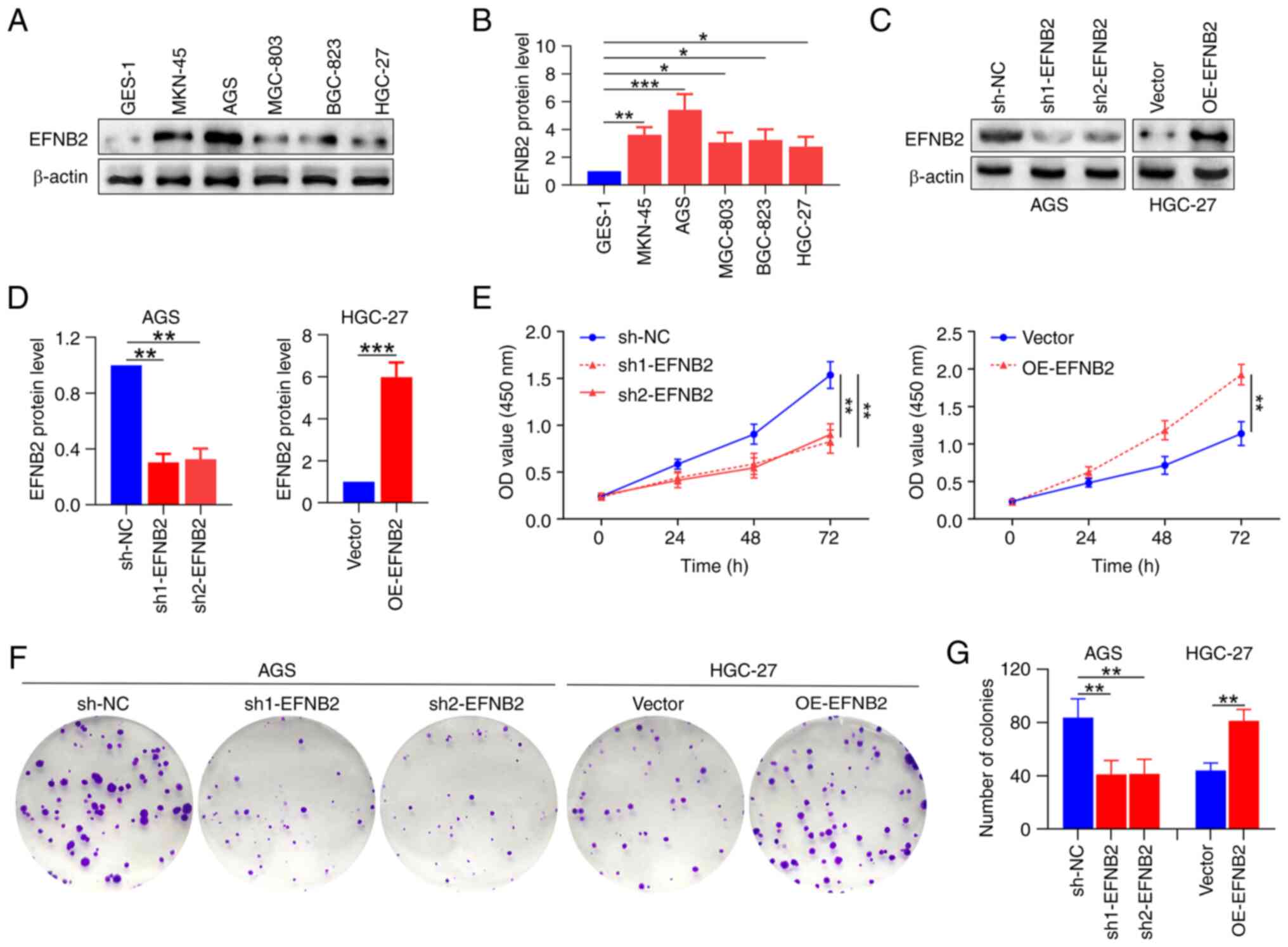 | Figure 3EFNB2 promotes the proliferation and
viability of GC cells in vitro. (A) Representative western
blot bands. (B) Expression levels of EFNB2 were detected in
multiple GC cell lines and human GES-1 normal gastric mucosal
epithelial cells using western blotting (one-way ANOVA). (C)
Representative western blot bands. (D) The effects of EFNB2
knockdown and overexpression were verified by western blotting
(one-way ANOVA for AGS cells; unpaired Student's test for HGC-27
cells). (E) Cell Counting Kit-8 assays revealed the OD values at
450 nm after 0, 24, 48 and 72 h (one-way ANOVA). (F) Representative
colony formation assay images. (G) Colony formation was assessed by
counting the number of colonies in various groups (one-way ANOVA
for AGS cells; unpaired Student's test for HGC-27 cells). Data are
presented as the mean ± SD. *P<0.05,
**P<0.01, ***P<0.001. GC, gastric
cancer; EFNB2, ephrin-B2; OD, optical density; OE, overexpression
vector; sh, short hairpin RNA; NC, negative control. |
Apoptosis and cell cycle regulation were next
examined, as both are key factors influencing cell proliferation.
Flow cytometry and cell cycle analysis revealed that EFNB2
knockdown elevated the proportion of apoptotic cells and quiescent
period G0/G1 arrest, and decreased the
proportions of cells in the G2/M and S stages (Fig. 4A-D). Similarly, EFNB2
overexpression reduced the proportion of apoptotic cells and
quiescent period G0/G1 arrest, and increased
the proportions of cells in the G2/M and S stages
(Fig. 4A-D). Furthermore,
apoptosis-related and cell cycle-associated proteins were
evaluated. Analysis revealed that the expression levels of
anti-apoptosis factor Bcl-2 were decreased, whereas those of
pro-apoptosis protein Bax were elevated, and the levels of CyclinD1
and CDK4 were decreased following EFNB2 knockdown (Fig. 4E-G). Correspondingly, the level of
Bcl-2 was upregulated, while the level of Bax was downregulated,
and the expression levels of CyclinD1 and CDK4 were elevated
following EFNB2 overexpression (Fig.
4E-G). These findings indicated that EFNB2 facilitated cell
proliferation by reducing apoptosis and influencing the cell
cycle.
Wnt/β-catenin signaling pathway is
responsible for EFNB2-mediated biological effects in GC cells
To further explore the potential mechanisms by which
EFNB2 promotes cell proliferation, the correlation modules of
public databases were explored. Data obtained from GEPIA and TIMER
databases demonstrated significant correlations between EFNB2
expression and the levels of GSK3β, catenin β1 and its downstream
effector MYC in GC (Fig. 5A and
B), suggesting EFNB2 may carry out a biological role via the
Wnt/β-catenin signaling pathway. The independent GSE184336 dataset
(n=231) also confirmed the aforementioned associations (Fig. S3E). Emerging studies have
highlighted the frequent disruption of the Wnt/β-catenin signaling
pathway in GC, which is associated with aggressive tumor cell
proliferation and unfavorable outcomes (19,21,41). Subsequently, the changes in
Wnt/β-catenin pathway-associated proteins in response to modulated
EFNB2 expression were assessed. The findings demonstrated that
knockdown of EFNB2 decreased the p-GSK3β/GSK3β ratio, as well as
the protein levels of β-catenin and its downstream target c-myc
(Fig. 5C). Reciprocally, EFNB2
overexpression increased the p-GSK3β/GSK3β ratio and the expression
of β-catenin and downstream c-myc (Fig. 5D).
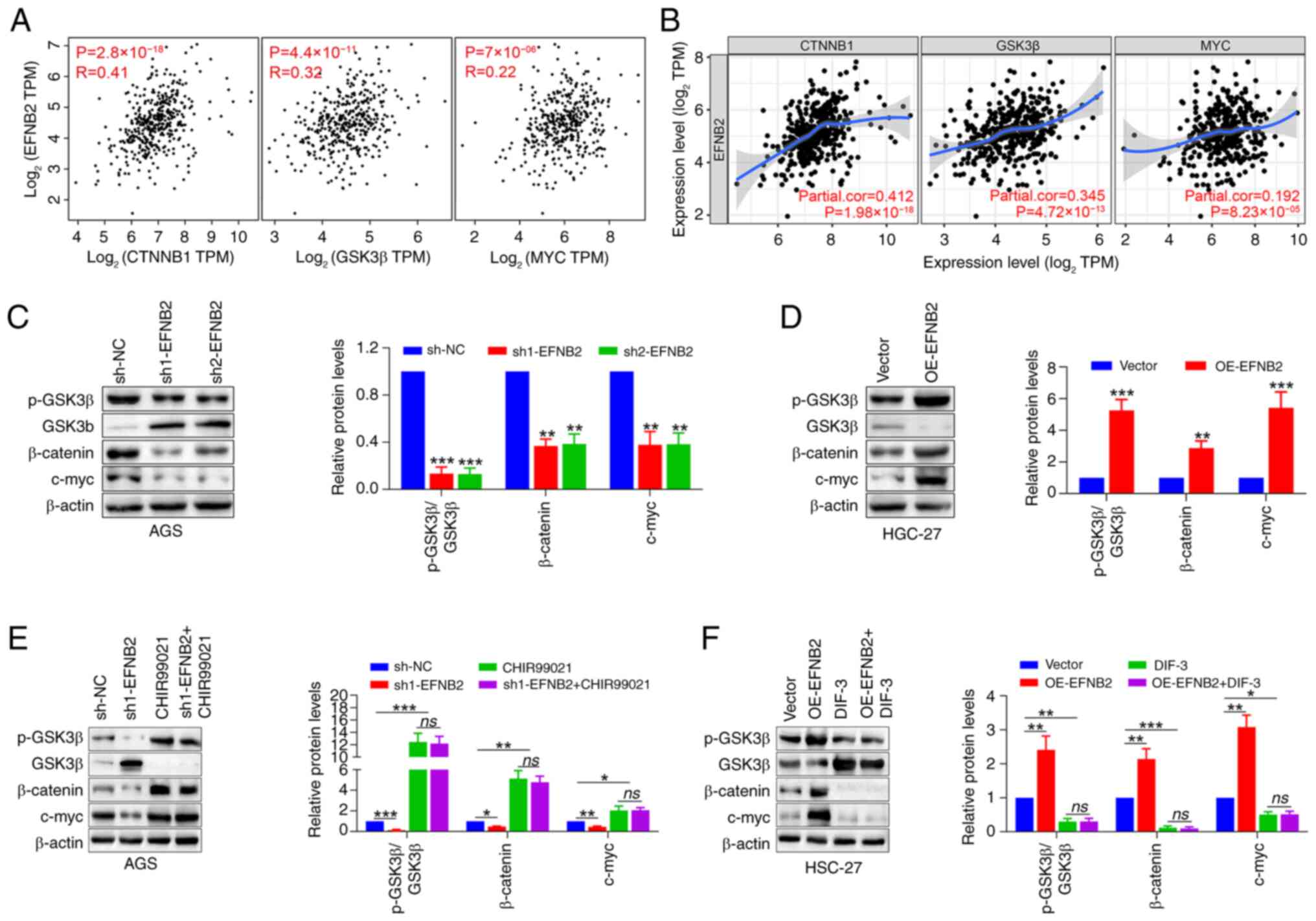 | Figure 5Wnt/β-catenin signaling pathway is
responsible for EFNB2-mediated biological effects in gastric cancer
cells. (A) Gene Expression Profiling Interactive Analysis and (B)
Tumor Immune Estimation Resource databases were employed to explore
the transcriptional correlation between EFNB2 and CTNNB1, GSK3β and
downstream MYC (Spearman). Representative western blot bands and
semi-quantification of protein expression levels of p-GSK3β, GSK3β,
β-catenin and c-myc detected under the condition of EFNB2 (C)
knockdown and (D) overexpression (one-way ANOVA for AGS cells;
unpaired Student's test for HGC-27 cells). Representative western
blot bands and semi-quantification of protein expression levels in
the presence of (E) an agonist (CHIR99021) and (F) an inhibitor
(DIF-3) of the Wnt/β-catenin signaling pathway in the EFNB2
knockdown or overexpression groups, respectively. Protein levels of
p-GSK3β, GSK3β, β-catenin and c-myc were detected by western
blotting (one-way ANOVA). Data are presented as the mean ± SD.
*P<0.05, **P<0.01,
***P<0.001 vs. sh-NC or vector group. ns, not
significant; NC, negative control; DIF-3, differentiation-inducing
factor-3; p-, phosphorylated; OE, overexpression vector; sh, short
hairpin RNA; CTNNB1, catenin β1; TPM, transcript per million;
EFNB2, ephrin-B2. |
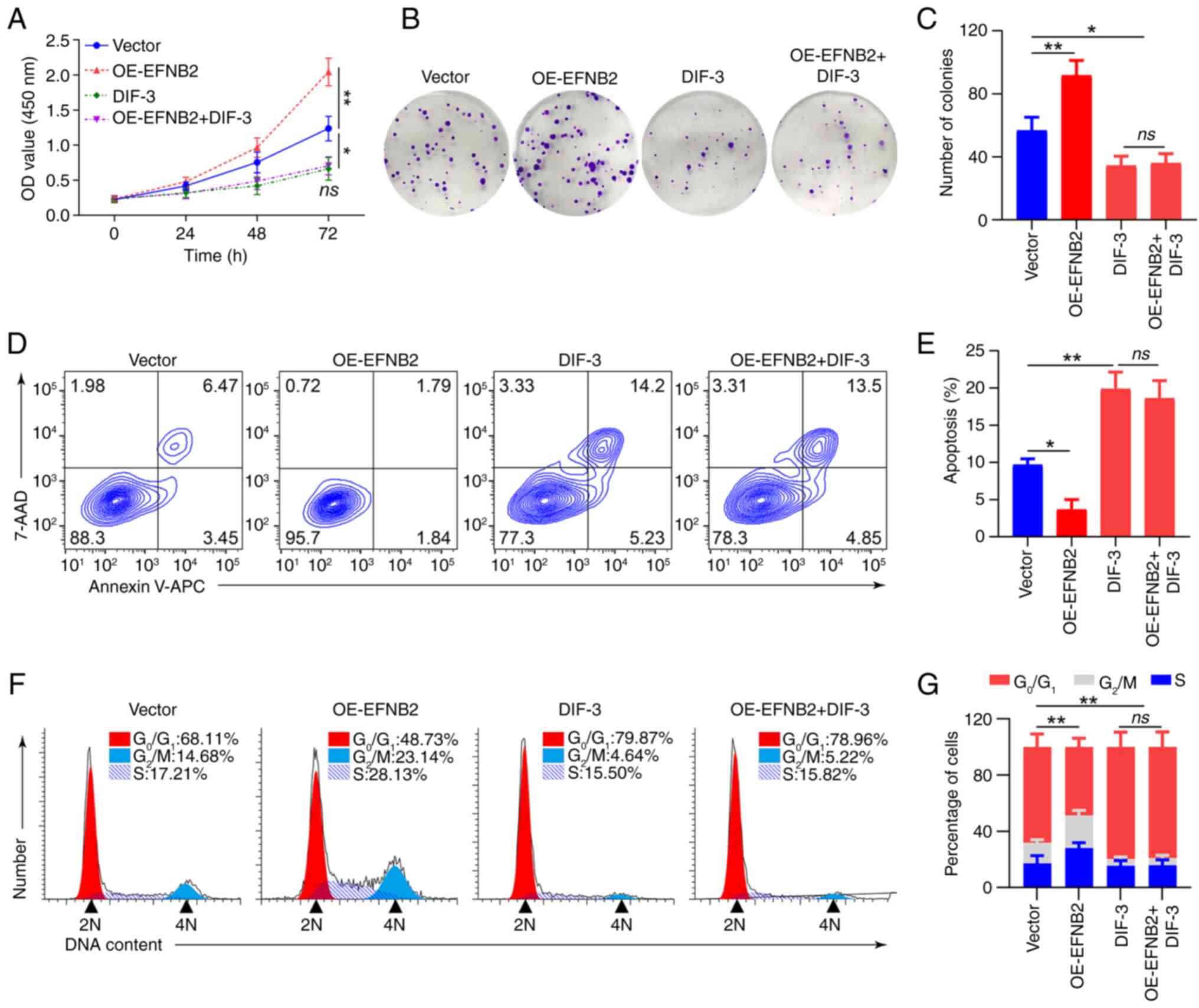
Rescue experiments were conducted utilizing the
Wnt/β-catenin signaling pathway agonist CHIR99021 and inhibitor
DIF-3. Western blotting revealed that CHIR99021 abolished the
inhibitory effect of EFNB2 knockdown on the Wnt/β-catenin pathway,
whereas DIF-3 abrogated the promoting effect of EFNB2
overexpression on this pathway (Fig.
5E and F). When the Wnt/β-catenin pathway was activated by
CHIR99021 or inhibited by DIF-3, EFNB2 lost its regulatory effect
on the pathway. Notably, DIF-3 eliminated the effects of EFNB2 on
proliferation, viability, apoptosis and cell cycle regulation in GC
cells (Fig. 6). These results
suggested that the biological functions of EFNB2 were dependent on
the activation of the Wnt/β-catenin signaling pathway.
EFNB2 promotes the tumor growth of GC
through the Wnt/β-catenin signaling pathway in vivo
To further clarify whether elevated EFNB2 expression
could promote tumor growth in vivo, a CDX mouse model was
established by subcutaneously inoculating HGC-27 cell suspensions
into immunodeficient nude mice and tumor growth was monitored. At 4
weeks post-injection, the xenograft tumors of the EFNB2
overexpression group had a higher volume and weight compared with
those of the control group (Fig.
7A-C), suggesting that EFNB2 could effectively promote tumor
growth in vivo. Additionally, Wnt/β-catenin pathway
inhibitor DIF-3 successfully dampened GC progression and
counteracted the promoting effects of EFNB2 overexpression on tumor
growth in vivo (Fig.
7A-C). Subsequently, H&E and IHC staining were carried out
(Fig. 7D). The expression levels
of EFNB2 in xenograft tumors from CDX models in the EFNB2
overexpression group and the control group were assessed by IHC
staining (Fig. 7D and E). The
findings also demonstrated that DIF-3 abrogated the promoting
effect of EFNB2 overexpression on β-catenin (Fig. 7D and E). Furthermore, Ki67
staining was used to assess the proliferative state of tumor in
vivo. Compared with the control group, the proportion of
Ki67-positive cells in EFNB2 overexpression group was increased,
while DIF-3 eliminated the promoting effect of EFNB2 on
proliferation in CDX models (Fig. 7D
and E). TUNEL staining was used to evaluate the apoptotic
condition of tumors. The proportion of apoptosis was lower in the
EFNB2 overexpression group compared with the control group, while
DIF-3 reversed the inhibitory effect of EFNB2 on apoptosis in
vivo (Fig. 7D and E). These
data suggested that EFNB2 accelerated the tumor growth of GC via
the Wnt/β-catenin signaling pathway in vivo.
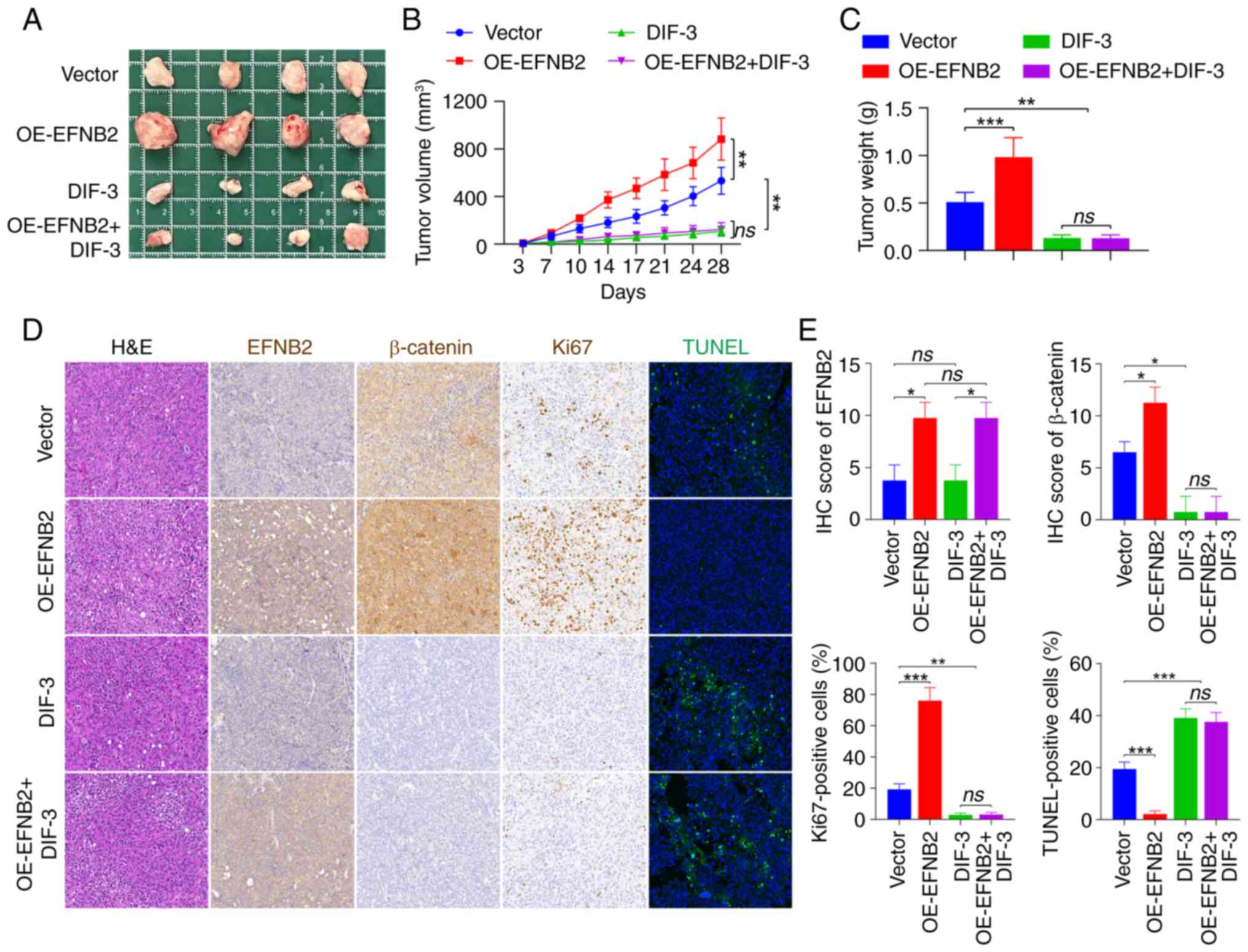 | Figure 7EFNB2 promotes the tumor growth of
gastric cancer via the Wnt/β-catenin signaling pathway in
vivo. (A) Tumors from the immunodeficient nude mice in the four
experimental groups were excised and imaged. (B) Subcutaneous tumor
growth (mm3) was measured every 3-4 days following
various treatments (one-way ANOVA). (C) Excised tumor masses (g)
were weighed and compared across groups (one-way ANOVA). (D)
Histopathological evaluation of xenograft tumors included H&E
staining to examine tissue morphology, IHC analysis of EFNB2,
β-catenin and Ki67 expression, and TUNEL staining for apoptotic
cell evaluation. All images were captured at a magnification of
×200. (E) Quantification of IHC scores for EFNB2 and β-catenin, and
the percentage of positive cells for Ki67 and TUNEL staining
(Kruskal-Wallis H test for EFNB2 and β-catenin; one-way ANOVA for
Ki67 and TUNEL staining). Data are presented as the mean ± SD.
*P<0.05, **P<0.01,
***P<0.001. ns, not significant; EFNB2, ephrin-B2;
DIF-3, differentiation-inducing factor-3; OE, overexpression
vector; IHC, immunohistochemical. |
Discussion
The findings of the present study revealed a
significant upregulation of EFNB2 expression in GC tissues compared
with ANTs, which was inversely associated with patient prognosis,
as evidenced by analysis of multiple public databases and IHC
analysis of a TMA. Functional experiments indicated that EFNB2
knockdown suppressed cellular proliferation, induced apoptosis and
arrested cell cycle progression in vitro. By contrast,
overexpression of EFNB2 exhibited the opposite effect. Notably, the
Wnt/β-catenin signaling pathway was identified as a key mediator of
EFNB2-driven oncogenic processes in GC progression.
Accumulating clinical research supports EFNB2 as a
prognostic biomarker in multiple solid tumors (42-44). In head and neck squamous cell
carcinoma (HNSCC), increased EFNB2 expression has been identified
as an indicator of unfavorable clinical outcomes and reduced
therapeutic responsiveness (42).
Similarly, in esophageal squamous cell carcinoma (ESCC), EFNB2
upregulation is inversely associated with patient survival
(43). Consistent with these
reports, the results of the present study demonstrated that
elevated EFNB2 expression was associated with a worse prognosis in
patients with GC. Notably, the consistent prognostic performance of
EFNB2 across different GC subgroups underscores its potential
clinical value in predicting the prognosis of patients with GC.
EFNB2 has emerged as a clinically relevant biomarker
across multiple malignancies, demonstrating distinct pathological
associations in various cancer types (42,43,45). In ESCC, transcriptional
upregulation of EFNB2 is associated with a family history of
esophageal cancer, advanced tumor stage and metastatic progression
(43). In HNSCC, elevated EFNB2
mRNA levels are largely found in human papillomavirus-negative
tumors arising from the oral cavity, especially in cases with
alcohol intake, TP53 mutations and EGFR amplification (42). In endometrial carcinoma, EFNB2
expression is associated with deep myometrial invasion >50%
(45). The present study
established a marked association between EFNB2 expression and
several GC parameters, including tumor size, T/M staging, TNM
classification and serum markers (CA199, CEA and CA50). However, no
statistically significant association with patient demographics
(age and sex), HP infection, tumor localization, histological
grade, nodal involvement and HER2 expression levels was observed.
Additionally, multivariate analysis demonstrated that only T stage
was an independent prognostic factor, while EFNB2 expression levels
did not reach statistical significance. These findings may be
attributed to the excessive number of variables included in the
analysis and the limited sample size, potentially leading to biased
estimates in the clinicopathological and prognostic assessments.
Future studies with larger sample sizes are warranted to validate
these preliminary findings and obtain more reliable results.
Krusche et al (44) revealed that EFNB2 enhanced
perivascular invasion and proliferation in glioblastoma stem-like
cells by facilitating RhoA-mediated anchorage-independent
cytokinesis. In colorectal cancer with mutant p53, Alam et
al (15) reported that DNA
damage-induced EFNB2 reverse signaling contributed to
chemoresistance and promoted epithelial-mesenchymal transition.
Conversely, EFNB2 could function as a tumor suppressor in specific
types of cancer. For instance, Magic et al (17) found that EFNB2 suppressed cell
proliferation and motility in vitro, and upregulated EFNB2
expression was associated with prolonged metastasis-free survival
in breast cancer. Similarly, Wang et al (46) observed that
EFNB2 decreased the migration and invasion of lung cancer cells.
The role of EFNB2 varies considerably with in different tumor
types, highlighting the need for further studies to fully
understand its complex functions.
The results of the present study indicated that
EFNB2 expression was significantly higher in GC cells and tissues,
and EFNB2 could increase cell survival and proliferation by
inhibiting apoptosis and accelerating the cell cycle. EFNB2
knockdown elevated Bax levels, while reducing Bcl-2 levels, whereas
EFNB2 overexpression exhibited the opposite effect. Bax and Bcl-2
are key regulators of the intrinsic apoptotic pathway (39,47), functioning by accumulating in the
mitochondrial outer membrane to modulate its permeability (47). The balance between Bax-mediated
pro-apoptotic signals and Bcl-2-mediated anti-apoptotic signals is
important for maintaining cellular apoptosis homeostasis (40,47). Dysregulation of EFNB2 disturbed
the Bcl-2/Bax equilibrium, resulting in altered apoptotic activity.
EFNB2 downregulation decreased the levels of cell cycle-related
proteins CDK4 and CyclinD1, while EFNB2 overexpression increased
their expression. CyclinD1 and CDK4 serve a key role in the
G1-to-S-phase transition by forming a complex that
drives cell cycle progression, thereby fueling cellular
proliferation and tumor growth (48,49). Collectively, these alterations in
protein levels point to an underlying change in the proliferative
capacity of GC cells following EFNB2 dysregulation.
Previous studies have identified the Wnt/β-catenin
signaling pathway as a key regulator in the progression of GC
(50-52) The nuclear translocation of
β-catenin activates multiple transcription factors, initiating
downstream signaling networks and driving the expression of
multiple proteins, including c-myc (51). Conversely, GSK3β serves a
counteracting role by phosphorylating β-catenin, which accelerates
its degradation and inhibits the transcriptional activity of
β-catenin target genes within the nucleus (52). Additionally, the oncogenic
transcription factor c-myc is dysregulated in >70% of different
types of human cancer, where it drives tumorigenesis by modulating
key cellular functions (53). As
a transcriptional regulator, it directly controls ~15% of genomic
targets, including key cell cycle components such as CyclinD1 and
CDK4, thereby orchestrating cellular proliferation, differentiation
and apoptosis (53). The present
study demonstrated that EFNB2 knockdown decreased the
phosphorylation of GSK3β, reduced β-catenin protein levels and
inhibited β-catenin-mediated c-myc expression, ultimately impairing
the proliferative capacity of GC cells. Conversely, EFNB2
overexpression exerted the opposite effects. EFNB2 could not only
regulate p-GSK3β levels but also directly influenced the levels of
total GSK3β. Although the opposite trends observed in GSK3β and
p-GSK3β levels further support the conclusions, the exact molecular
mechanism underlying these effects remains to be elucidated.
Rescue experiments offered additional evidence that
EFNB2 regulated GC cell proliferation, apoptosis and the cell cycle
via the Wnt/β-catenin signaling pathway. These in vitro
results were further validated using CDX murine models, which
substantiated the involvement of the Wnt/β-catenin pathway in
EFNB2-mediated effects. Notably, although previous studies have
suggested that Wnt/β-catenin signaling is involved in GC metastasis
(54,55), the data in the present did not
reveal a notable effect of EFNB2 expression on the migration and
invasion of GC cells. This may be attributed to the involvement of
other signaling pathways that counteract the effects of the
Wnt/β-catenin pathway on migration and invasion. This observation
further underscores the complexity of the downstream signaling
networks of EFNB2, and the specific mechanisms involved require
further investigation.
In summary, the expression levels of EFNB2 were
higher in GC tissues compared with ANTs, and were associated with
an unfavorable clinical prognosis in patients with GC. EFNB2
effectively regulated cell proliferation, apoptosis and the cell
cycle in GC in vitro. Furthermore, EFNB2 promoted the tumor
growth and malignant progression of GC in vivo.
Mechanistically, EFNB2 exerted its oncogenic effects by activating
the Wnt/β-catenin signaling pathway. Consequently, EFNB2 may be a
potential molecular target for the treatment of GC.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
DD contributed to writing the original draft, data
visualization, experimental design and implementation. XW, RX, RL
and YZ contributed to experimental design and implementation, data
visualization, and writing, review and editing. ZW contributed to
supervision, project administration, and analysis and
interpretation of data. DD and ZW confirmed the authenticity of all
the raw data. All authors have read and approved the final version
of the manuscript.
Ethics approval and consent to
participate
All animal procedures were carried out according to
the protocol approved by the Animal Ethics Committee of the First
Affiliated Hospital of Anhui Medical University (approval no.
LLSC20240221; Hefei, China). The human tissue microarray was
purchased from Shanghai Outdo Biotech Co., Ltd. (HStmA180Su32)
after the approval of the Ethics Committee of the First Affiliated
Hospital of Anhui Medical University (approval no. 2024137; Hefei,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Scientific Research Fund
Project of Anhui Medical University (grant no. 2023xkj225) and Key
Natural Science Project of Anhui Provincial Universities (grant no.
2023AH053303).
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Chen YC, Malfertheiner P, Yu HT, Kuo CL,
Chang YY, Meng FT, Wu YX, Hsiao JL, Chen MJ, Lin KP, et al: Global
prevalence of helicobacter pylori infection and incidence of
gastric cancer between 1980 and 2022. Gastroenterology.
166:605–619. 2024. View Article : Google Scholar
|
|
3
|
Onoyama T, Ishikawa S and Isomoto H:
Gastric cancer and genomics: Review of literature. J Gastroenterol.
57:505–516. 2022. View Article : Google Scholar
|
|
4
|
Qiu H, Cao S and Xu R: Cancer incidence,
mortality, and burden in China: A time-trend analysis and
comparison with the United States and United Kingdom based on the
global epidemiological data released in 2020. Cancer Commun (Lond).
41:1037–1048. 2021. View Article : Google Scholar
|
|
5
|
Guan WL, He Y and Xu RH: Gastric cancer
treatment: Recent progress and future perspectives. J Hematol
Oncol. 16:572023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pasquale EB: Eph receptors and ephrins in
cancer: Bidirectional signalling and beyond. Nat Rev Cancer.
10:165–180. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iida H, Honda M, Kawai HF, Yamashita T,
Shirota Y, Wang BC, Miao H and Kaneko S: Ephrin-A1 expression
contributes to the malignant characteristics of {alpha}-fetoprotein
producing hepatocellular carcinoma. Gut. 54:843–851. 2005.
View Article : Google Scholar
|
|
8
|
Nakada M, Drake KL, Nakada S, Niska JA and
Berens ME: Ephrin-B3 ligand promotes glioma invasion through
activation of Rac1. Cancer Res. 66:8492–8500. 2006. View Article : Google Scholar
|
|
9
|
Hirai H, Maru Y, Hagiwara K, Nishida J and
Takaku F: A novel putative tyrosine kinase receptor encoded by the
eph gene. Science. 238:1717–1720. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Piffko A, Uhl C, Vajkoczy P, Czabanka M
and Broggini T: EphrinB2-EphB4 signaling in neurooncological
disease. Int J Mol Sci. 23:16792022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rutkowski R, Mertens-Walker I, Lisle JE,
Herington AC and Stephenson SA: Evidence for a dual function of
EphB4 as tumor promoter and suppressor regulated by the absence or
presence of the ephrin-B2 ligand. Int J Cancer. 131:E614–E624.
2012. View Article : Google Scholar
|
|
12
|
Piffko A, Broggini T, Harms C, Adams RH,
Vajkoczy P and Czabanka M: Ligand-dependent and ligand-independent
effects of Ephrin-B2-EphB4 signaling in melanoma metastatic spine
disease. Int J Mol Sci. 22:80282021. View Article : Google Scholar
|
|
13
|
Dai B, Shi X, Ma N, Ma W, Zhang Y, Yang T,
Zhang J and He L: HMQ-T-B10 induces human liver cell apoptosis by
competitively targeting EphrinB2 and regulating its pathway. J Cell
Mol Med. 22:5231–5243. 2018. View Article : Google Scholar
|
|
14
|
Zhong S, Pei D, Shi L, Cui Y and Hong Z:
Ephrin-B2 inhibits Aβ25-35-induced apoptosis by alleviating
endoplasmic reticulum stress and promoting autophagy in HT22 cells.
Neurosci Lett. 704:50–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alam SK, Yadav VK, Bajaj S, Datta A, Dutta
SK, Bhattacharyya M, Bhattacharya S, Debnath S, Roy S, Boardman LA,
et al: DNA damage-induced ephrin-B2 reverse signaling promotes
chemoresistance and drives EMT in colorectal carcinoma harboring
mutant p53. Cell Death Differ. 23:707–722. 2016. View Article : Google Scholar :
|
|
16
|
Zhu F, Dai SN, Xu DL, Hou CQ, Liu TT, Chen
QY, Wu JL and Miao Y: EFNB2 facilitates cell proliferation,
migration, and invasion in pancreatic ductal adenocarcinoma via the
p53/p21 pathway and EMT. Biomed Pharmacother. 125:1099722020.
View Article : Google Scholar
|
|
17
|
Magic Z, Sandström J and Perez-Tenorio G:
Ephrin-B2 inhibits cell proliferation and motility in vitro and
predicts longer metastasis-free survival in breast cancer. Int J
Oncol. 55:1275–1286. 2019.
|
|
18
|
Zhang Y, Zhang R, Ding X and Ai K: EFNB2
acts as the target of miR-557 to facilitate cell proliferation,
migration and invasion in pancreatic ductal adenocarcinoma by
bioinformatics analysis and verification. Am J Transl Res.
10:3514–3528. 2018.
|
|
19
|
Akhavanfar R, Shafagh SG, Mohammadpour B,
Farahmand Y, Lotfalizadeh MH, Kookli K, Adili A, Siri G and Eshagh
Hosseini SM: A comprehensive insight into the correlation between
ncRNAs and the Wnt/β-catenin signalling pathway in gastric cancer
pathogenesis. Cell Commun Signal. 21:1662023. View Article : Google Scholar
|
|
20
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mirabelli CK, Nusse R, Tuveson DA and
Williams BO: Perspectives on the role of Wnt biology in cancer. Sci
Signal. 12:eaay44942019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lien WH and Fuchs E: Wnt some lose some:
Transcriptional governance of stem cells by Wnt/β-catenin
signaling. Genes Dev. 28:1517–1532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar
|
|
24
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar
|
|
25
|
Győrffy B: Survival analysis across the
entire transcriptome identifies biomarkers with the highest
prognostic power in breast cancer. Comput Struct Biotechnol J.
19:4101–4109. 2021. View Article : Google Scholar
|
|
26
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar
|
|
27
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A,
Michalowski A and Green JE: A gene expression signature of acquired
chemoresistance to cisplatin and fluorouracil combination
chemotherapy in gastric cancer patients. PLoS One. 6:e166942011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ooi CH, Ivanova T, Wu J, Lee M, Tan IB,
Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al: Oncogenic
pathway combinations predict clinical prognosis in gastric cancer.
PLoS Genet. 5:e10006762009. View Article : Google Scholar :
|
|
30
|
Förster S, Gretschel S, Jöns T, Yashiro M
and Kemmner W: THBS4, a novel stromal molecule of diffuse-type
gastric adenocarcinomas, identified by transcriptome-wide
expression profiling. Mod Pathol. 24:1390–1403. 2011. View Article : Google Scholar
|
|
31
|
Wang G, Hu N, Yang HH, Wang L, Su H, Wang
C, Clifford R, Dawsey EM, Li JM, Ding T, et al: Comparison of
global gene expression of gastric cardia and noncardia cancers from
a high-risk population in china. PLoS One. 8:e638262013. View Article : Google Scholar
|
|
32
|
Busuttil RA, George J, Tothill RW,
Ioculano K, Kowalczyk A, Mitchell C, Lade S, Tan P, Haviv I and
Boussioutas A: A signature predicting poor prognosis in gastric and
ovarian cancer represents a coordinated macrophage and stromal
response. Clin Cancer Res. 20:2761–2772. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheng L, Yang S, Yang Y, Zhang W, Xiao H,
Gao H, Deng X and Zhang Q: Global gene expression and functional
network analysis of gastric cancer identify extended pathway maps
and GPRC5A as a potential biomarker. Cancer Lett. 326:105–113.
2012. View Article : Google Scholar
|
|
34
|
Lou S, Zhang J, Yin X, Zhang Y, Fang T,
Wang Y and Xue Y: Comprehensive characterization of tumor purity
and its clinical implications in gastric cancer. Front Cell Dev
Biol. 9:7825292022. View Article : Google Scholar :
|
|
35
|
Fang C, Wang W, Deng JY, Sun Z, Seeruttun
SR, Wang ZN, Xu HM, Liang H and Zhou ZW: Proposal and validation of
a modified staging system to improve the prognosis predictive
performance of the 8th AJCC/UICC pTNM staging system for gastric
adenocarcinoma: A multicenter study with external validation.
Cancer Commun (Lond). 38:672018.PubMed/NCBI
|
|
36
|
Liu Q, Li Y, Niu Z, Zong Y, Wang M, Yao L,
Lu Z, Liao Q and Zhao Y: Atorvastatin (Lipitor) attenuates the
effects of aspirin on pancreatic cancerogenesis and the
chemotherapeutic efficacy of gemcitabine on pancreatic cancer by
promoting M2 polarized tumor associated macrophages. J Exp Clin
Cancer Res. 35:332016. View Article : Google Scholar
|
|
37
|
Shen J, Cao B, Wang Y, Ma C, Zeng Z, Liu
L, Li X, Tao D, Gong J and Xie D: Hippo component YAP promotes
focal adhesion and tumour aggressiveness via transcriptionally
activating THBS1/FAK signalling in breast cancer. J Exp Clin Cancer
Res. 37:1752018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Liu Q, Liu J and Liao Q:
Upregulated CD58 is associated with clinicopathological
characteristics and poor prognosis of patients with pancreatic
ductal adenocarcinoma. Cancer Cell Int. 21:3272021. View Article : Google Scholar :
|
|
39
|
Zhang Y, Liu Q, Yang S and Liao Q:
Knockdown of LRRN1 inhibits malignant phenotypes through the
regulation of HIF-1α/notch pathway in pancreatic ductal
adenocarcinoma. Mol Ther Oncolytics. 23:51–64. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Y, Zhang R, Luo G and Ai K: Long
noncoding RNA SNHG1 promotes cell proliferation through PI3K/AKT
signaling pathway in pancreatic ductal adenocarcinoma. J Cancer.
9:2713–2722. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han R, Yang J, Zhu Y and Gan R: Wnt
signaling in gastric cancer: Current progress and future prospects.
Front Oncol. 14:14105132024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Oweida A, Bhatia S, Hirsch K, Calame D,
Griego A, Keysar S, Pitts T, Sharma J, Eckhardt G, Jimeno A, et al:
Ephrin-B2 overexpression predicts for poor prognosis and response
to therapy in solid tumors. Mol Carcinog. 56:1189–1196. 2017.
View Article : Google Scholar
|
|
43
|
Ni Q, Chen P, Zhu B, Li J, Xie D and Ma X:
Expression levels of EPHB4, EFNB2 and caspase-8 are associated with
clinicopathological features and progression of esophageal squamous
cell cancer. Oncol Lett. 19:917–929. 2020.
|
|
44
|
Krusche B, Ottone C, Clements MP,
Johnstone ER, Goetsch K, Lieven H, Mota SG, Singh P, Khadayate S,
Ashraf A, et al: EphrinB2 drives perivascular invasion and
proliferation of glioblastoma stem-like cells. Elife. 5:e148452016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Takai N, Miyazaki T, Fujisawa K, Nasu K
and Miyakawa I: Expression of receptor tyrosine kinase EphB4 and
its ligand ephrin-B2 is associated with malignant potential in
endometrial cancer. Oncol Rep. 8:567–573. 2001.
|
|
46
|
Wang L, Peng Q, Sai B, Zheng L, Xu J, Yin
N, Feng X and Xiang J: Ligand-independent EphB1 signaling mediates
TGF-β-activated CDH2 and promotes lung cancer cell invasion and
migration. J Cancer. 11:4123–4131. 2020. View Article : Google Scholar :
|
|
47
|
Peña-Blanco A and García-Sáez AJ: Bax, Bak
and beyond-mitochondrial performance in apoptosis. FEBS J.
285:416–431. 2018. View Article : Google Scholar
|
|
48
|
Tchakarska G and Sola B: The double
dealing of cyclin D1. Cell Cycle. 19:163–178. 2020. View Article : Google Scholar
|
|
49
|
Ziegler DV, Parashar K and Fajas L: Beyond
cell cycle regulation: The pleiotropic function of CDK4 in cancer.
Semin Cancer Biol. 98:51–63. 2024. View Article : Google Scholar
|
|
50
|
Koushyar S, Powell AG, Vincan E and Phesse
TJ: Targeting Wnt signaling for the treatment of gastric cancer.
Int J Mol Sci. 21:39272020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang
X, Zhou Z, Shu G and Yin G: Wnt/β-catenin signalling: Function,
biological mechanisms, and therapeutic opportunities. Signal
Transduct Target Ther. 7:32022. View Article : Google Scholar
|
|
52
|
Zhang Y and Wang X: Targeting the
Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol.
13:1652020. View Article : Google Scholar
|
|
53
|
Madden SK, de Araujo AD, Gerhardt M,
Fairlie DP and Mason JM: Taking the Myc out of cancer: Toward
therapeutic strategies to directly inhibit c-Myc. Mol Cancer.
20:32021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xue W, Yang L, Chen C, Ashrafizadeh M,
Tian Y and Sun R: Wnt/β-catenin-driven EMT regulation in human
cancers. Cell Mol Life Sci. 81:792024. View Article : Google Scholar
|
|
55
|
Wu C, Zhuang Y, Jiang S, Liu S, Zhou J, Wu
J, Teng Y, Xia B, Wang R and Zou X: Interaction between
Wnt/β-catenin pathway and microRNAs regulates
epithelial-mesenchymal transition in gastric cancer (review). Int J
Oncol. 48:2236–2246. 2016. View Article : Google Scholar
|