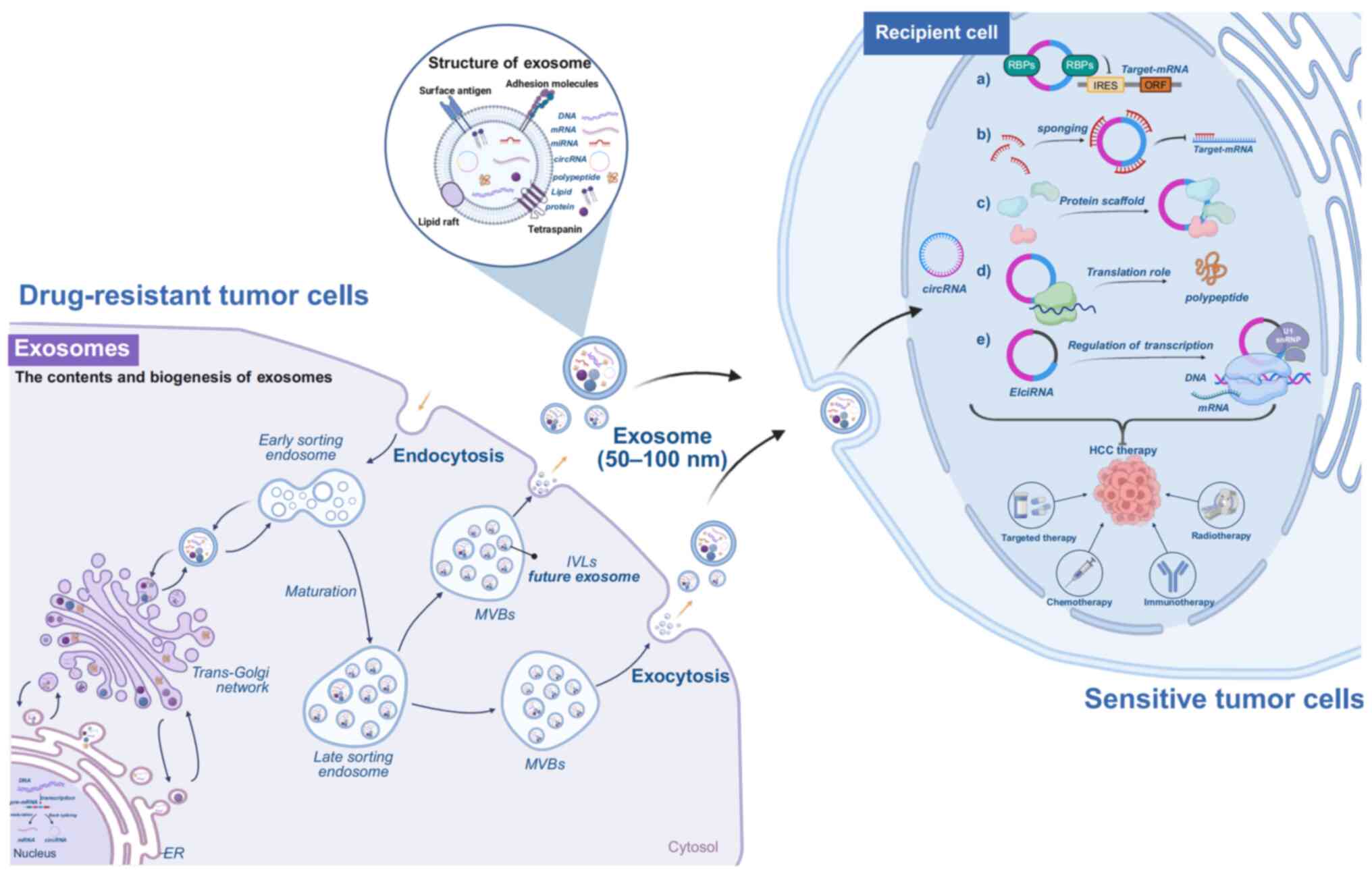
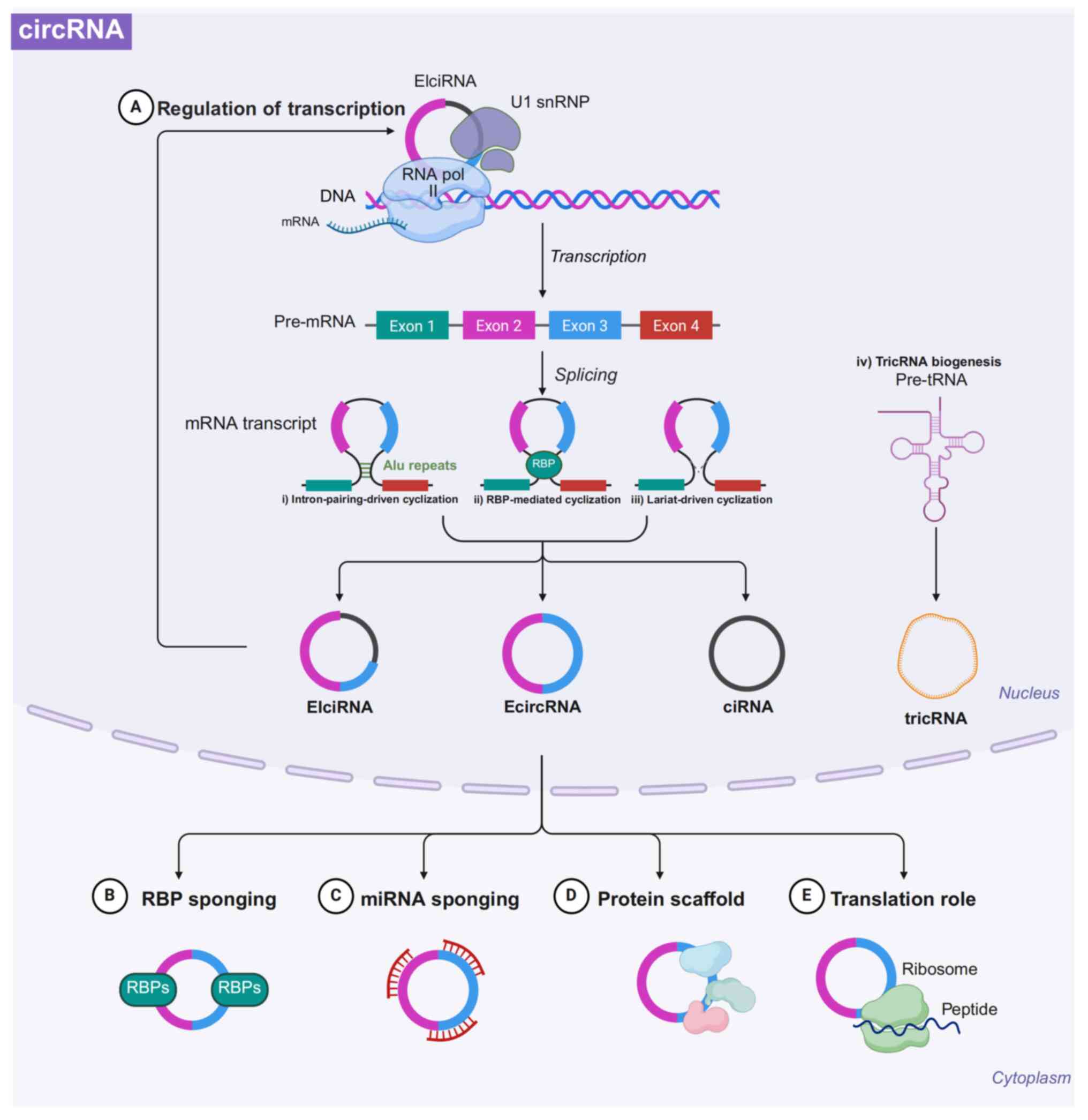
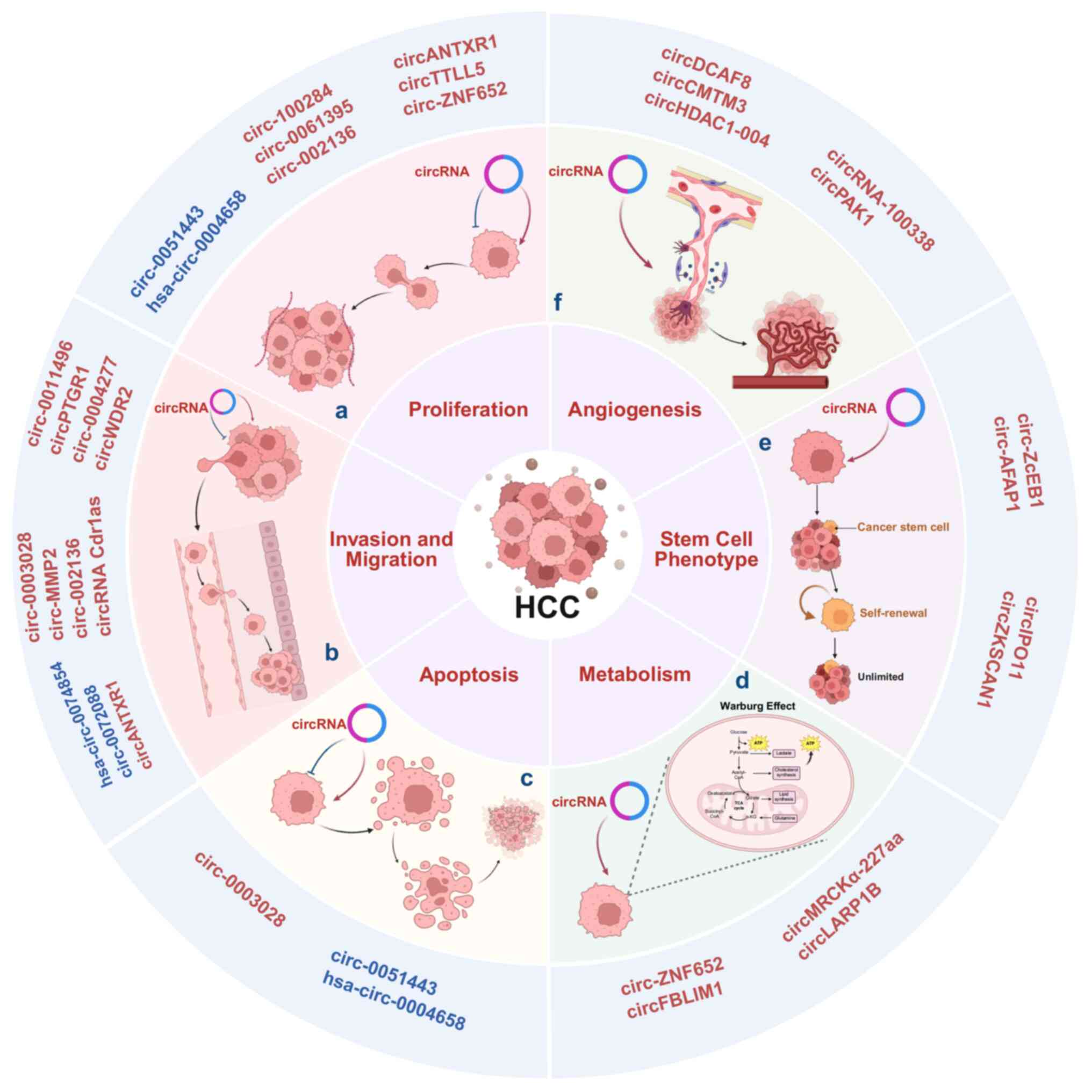
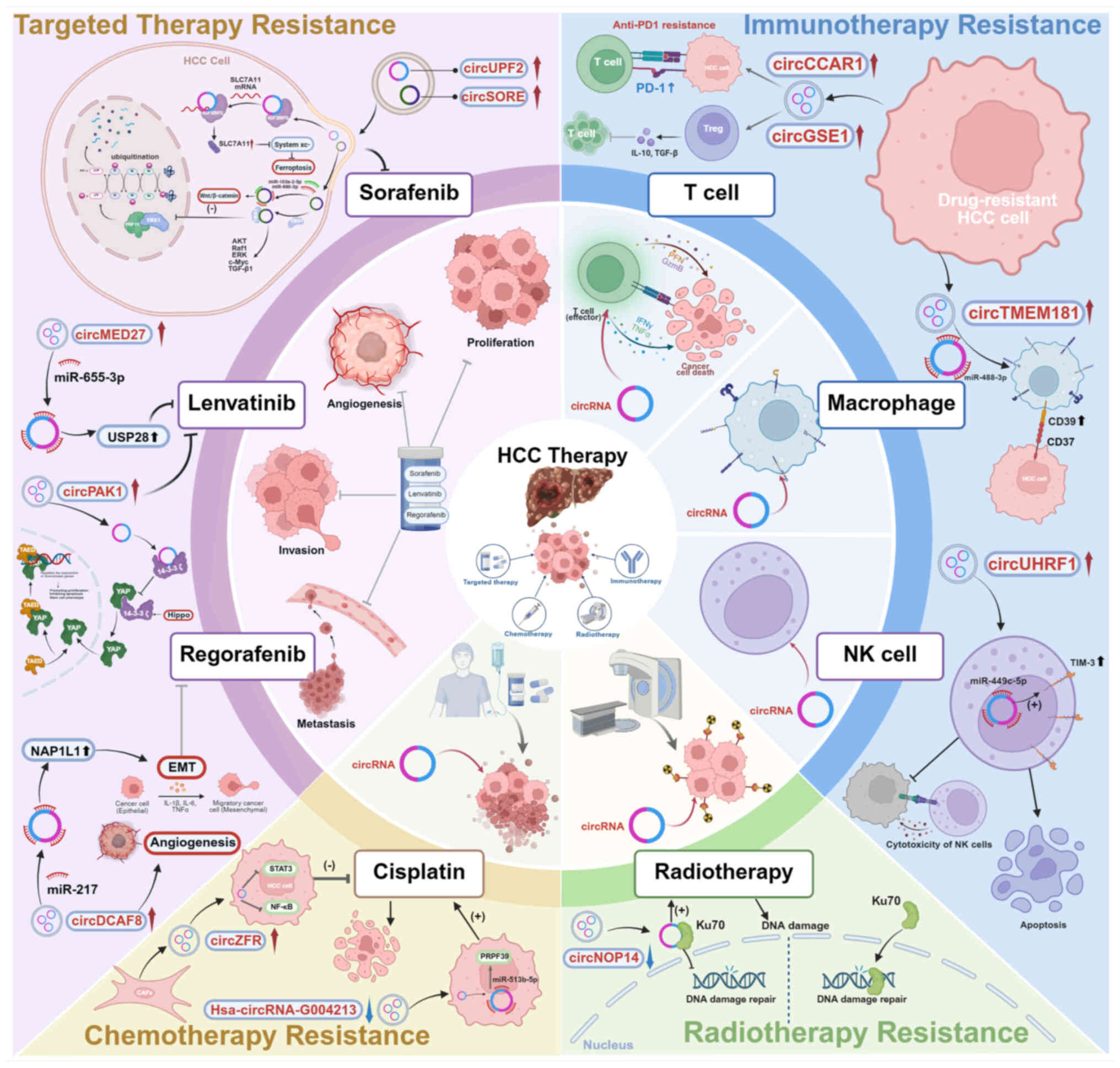
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Siegel RL, Kratzer TB, Giaquinto AN, Sung
H and Jemal A: Cancer statistics, 2025. CA Cancer J Clin. 75:10–45.
2025.PubMed/NCBI
|
|
3
|
Si J, Zou Y, Gao Y, Chen J, Jiang W, Shen
X, Zhu C and Yao Q: tRF-3a-Pro: A transfer RNA-derived small RNA as
a novel biomarker for diagnosis of Hepatitis B Virus-related
hepatocellular carcinoma. Cell Prolif. 58:e700062025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang CH, Cheng Y, Zhang S, Fan J and Gao
Q: Changing epidemiology of hepatocellular carcinoma in Asia. Liver
Int. 42:2029–2041. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jemal A, Ward EM, Johnson CJ, Cronin KA,
Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, et al:
Annual report to the nation on the status of cancer, 1975-2014,
featuring survival. J Natl Cancer Inst. 109:djx0302017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ladd AD, Duarte S, Sahin I and Zarrinpar
A: Mechanisms of drug resistance in HCC. Hepatology. 79:926–940.
2024. View Article : Google Scholar
|
|
7
|
Liu CX and Chen LL: Circular RNAs:
Characterization, cellular roles, and applications. Cell.
185:2016–2034. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L,
Gorshkov K, Mao Q, Xia S, Cen D, et al:
N6-methyladenosine-modified CircRNA-SORE sustains
sorafenib resistance in hepatocellular carcinoma by regulating
β-catenin signaling. Mol Cancer. 19:1632020. View Article : Google Scholar
|
|
9
|
Zhang XY, Li SS, Gu YR, Xiao LX, Ma XY,
Chen XR, Wang JL, Liao CH, Lin BL, Huang YH, et al: CircPIAS1
promotes hepatocellular carcinoma progression by inhibiting
ferroptosis via the miR-455-3p/NUPR1/FTH1 axis. Mol Cancer.
23:1132024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fei D, Wang F, Wang Y, Chen J, Chen S, Fan
L, Yang L, Ren Q, Duangmano S, Du F, et al: Circular RNA ACVR2A
promotes the progression of hepatocellular carcinoma through
mir-511-5p targeting PI3K-Akt signaling pathway. Mol Cancer.
23:1592024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji Y, Ni C, Shen Y, Xu Z, Tang L, Yu F,
Zhu L, Lu H, Zhang C, Yang S and Wang X: ESRP1-mediated biogenesis
of circPTPN12 inhibits hepatocellular carcinoma progression by
PDLIM2/NF-κB pathway. Mol Cancer. 23:1432024. View Article : Google Scholar
|
|
12
|
Liu Y, Song J, Zhang H, Liao Z, Liu F, Su
C, Wang W, Han M, Zhang L, Zhu H, et al: EIF4A3-induced circTOLLIP
promotes the progression of hepatocellular carcinoma via the
miR-516a-5p/PBX3/EMT pathway. J Exp Clin Cancer Res. 41:1642022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peng R, Cao J, Su BB, Bai XS, Jin X, Wang
AQ, Wang Q, Liu RJ, Jiang GQ, Jin SJ, et al: Down-regulation of
circPTTG1IP induces hepatocellular carcinoma development via
miR-16-5p/RNF125/JAK1 axis. Cancer Lett. 543:2157782022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meldolesi J: Exosomes and ectosomes in
intercellular communication. Curr Biol. 28:R435–R444. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang F, Jiang J, Qian H, Yan Y and Xu W:
Exosomal circRNA: Emerging insights into cancer progression and
clinical application potential. J Hematol Oncol. 16:672023.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dong FL, Xu ZZ, Wang YQ, Li T, Wang X and
Li J: Exosome-derived circUPF2 enhances resistance to targeted
therapy by redeploying ferroptosis sensitivity in hepatocellular
carcinoma. J Nanobiotechnology. 22:2982024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song
X, Gorshkov K, Sun Q, Lin H, Zheng X, et al: CircRNA-SORE mediates
sorafenib resistance in hepatocellular carcinoma by stabilizing
YBX1. Signal Transduct Target Ther. 5:2982020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gong J, Han G, Chen Z, Zhang Y, Xu B, Xu
C, Gao W and Wu J: CircDCAF8 promotes the progression of
hepatocellular carcinoma through miR-217/NAP1L1 Axis, and induces
angiogenesis and regorafenib resistance via exosome-mediated
transfer. J Transl Med. 22:5172024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harding C, Heuser J and Stahl P:
Receptor-mediated endocytosis of transferrin and recycling of the
transferrin receptor in rat reticulocytes. J Cell Biol. 97:329–339.
1983. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Johnstone RM, Adam M, Hammond JR, Orr L
and Turbide C: Vesicle formation during reticulocyte maturation.
Association of plasma membrane activities with released vesicles
(exosomes). J Biol Chem. 262:9412–9420. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
van Niel G, D'Angelo G and Raposo G:
Shedding light on the cell biology of extracellular vesicles. Nat
Rev Mol Cell Biol. 19:213–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Camussi G, Deregibus MC, Bruno S,
Cantaluppi V and Biancone L: Exosomes/microvesicles as a mechanism
of cell-to-cell communication. Kidney Int. 78:838–848. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Panda SS, Sahoo RK, Patra SK, Biswal S and
Biswal BK: Molecular insights to therapeutic in cancer: Role of
exosomes in tumor microenvironment, metastatic progression and drug
resistance. Drug Discov Today. 29:1040612024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Q, Zhang Y, Jin P, Chen Y, Zhang C,
Geng X, Mun KS and Phang KC: New insights into the potential of
exosomal circular RNAs in mediating cancer chemotherapy resistance
and their clinical applications. Biomed Pharmacother.
177:1170272024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang T and Zhang H: Exploring the roles
and molecular mechanisms of RNA binding proteins in the sorting of
noncoding RNAs into exosomes during tumor progression. J Adv Res.
65:105–123. 2024. View Article : Google Scholar :
|
|
27
|
Jiang Z, Liu G and Li J: Recent progress
on the isolation and detection methods of exosomes. Chem Asian J.
15:3973–3982. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan H, Li Y, Cheng S and Zeng Y: Advances
in analytical technologies for extracellular vesicles. Anal Chem.
93:4739–4774. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and
Chen M: Circular RNA: An emerging key player in RNA world. Brief
Bioinform. 18:547–557. 2017.
|
|
31
|
Conn VM, Hugouvieux V, Nayak A, Conos SA,
Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta
C and Conn SJ: A circRNA from SEPALLATA3 regulates splicing of its
cognate mRNA through R-loop formation. Nat Plants. 3:170532017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen
W, Yuan X, Yin W, Xu J, Chen K, et al: CircRNA inhibits DNA damage
repair by interacting with host gene. Mol Cancer. 19:1282020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zheng J, He T, Chen J, Zhao J, Zhang S and
Yang Z: Decoding the functional network of circular RNAs encoding
proteins in hepatocellular carcinoma: From carcinogenesis to
clinical transformation. J Adv Res. Sep 11–2025. View Article : Google Scholar : Epub ahead of
print.
|
|
34
|
Liu Z, Wang Q, Wang X, Xu Z, Wei X and Li
J: Circular RNA cIARS regulates ferroptosis in HCC cells through
interacting with RNA binding protein ALKBH5. Cell Death Discov.
6:722020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Panda AC: Circular RNAs Act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Su Y, Xu C, Liu Y, Hu Y and Wu H: Circular
RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression
via multiple miRNAs sponge. Aging (Albany NY). 11:3362–3375. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-Protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R,
Thorne RF, Zhang XD, Hu W and Wu M: CircACC1 regulates assembly and
activation of AMPK complex under metabolic stress. Cell Metab.
30:157–173.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Margvelani G, Maquera KAA, Welden JR,
Rodgers DW and Stamm S: Translation of circular RNAs. Nucleic Acids
Res. 53:gkae11672025. View Article : Google Scholar :
|
|
41
|
Lan T, Gao F, Cai Y, Lv Y, Zhu J, Liu H,
Xie S, Wan H, He H, Xie K, et al: The protein circPETH-147aa
regulates metabolic reprogramming in hepatocellular carcinoma cells
to remodel immunosuppressive microenvironment. Nat Commun.
16:3332025. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Song R, Ma S, Xu J, Ren X, Guo P, Liu H,
Li P, Yin F, Liu M, Wang Q, et al: A novel polypeptide encoded by
the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR.
Mol Cancer. 22:162023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li S, Gao Y, Tang Y, Dong A, Hu S, Wang R,
Gong X and Peng Z: Circular SNX25 encoded radioresistance augmenter
facilitates DNA damage repair in hepatocellular carcinoma by
targeting BAG6-GET4 interaction. Cell Death Dis. 16:7342025.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Conn VM, Chinnaiyan AM and Conn SJ:
Circular RNA in cancer. Nat Rev Cancer. 24:597–613. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li J, Zhou W, Wang H, Huang M and Deng H:
Exosomal circular RNAs in tumor microenvironment: An emphasis on
signaling pathways and clinical opportunities. MedComm (2020).
5:e700192024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Seimiya T, Otsuka M, Iwata T, Shibata C,
Tanaka E, Suzuki T and Koike K: Emerging roles of exosomal circular
RNAs in cancer. Front Cell Dev Biol. 8:5683662020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen W, Quan Y, Fan S, Wang H, Liang J,
Huang L, Chen L, Liu Q, He P and Ye Y: Exosome-transmitted circular
RNA hsa_circ_0051443 suppresses hepatocellular carcinoma
progression. Cancer Lett. 475:119–128. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang L, Zhang J, Li P, Li T, Zhou Z and
Wu H: Exosomal hsa_circ_0004658 derived from RBPJ
overexpressed-macrophages inhibits hepatocellular carcinoma
progression via miR-499b-5p/JAM3. Cell Death Dis. 13:322022.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dai X, Chen C, Yang Q, Xue J, Chen X, Sun
B, Luo F, Liu X, Xiao T, Xu H, et al: Exosomal circRNA_100284 from
arsenite-transformed cells, via microRNA-217 regulation of EZH2, is
involved in the malignant transformation of human hepatic cells by
accelerating the cell cycle and promoting cell proliferation. Cell
Death Dis. 9:4542018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu Y, Bian L, Liu R, Wang Y and Xiao X:
Circular RNA hsa_circ_0061395 accelerates hepatocellular carcinoma
progression via regulation of the miR-877-5p/PIK3R3 axis. Cancer
Cell Int. 21:102021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yuan P, Song J, Wang F and Chen B:
Exosome-transmitted circ_002136 promotes hepatocellular carcinoma
progression by miR-19a-3p/RAB1A pathway. BMC Cancer. 22:12842022.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Huang C, Yu W, Wang Q, Huang T and Ding Y:
CircANTXR1 contributes to the malignant progression of
hepatocellular carcinoma by promoting proliferation and metastasis.
J Hepatocell Carcinoma. 8:1339–1353. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu C, Ren C, Guo L, Yang C and Yu Q:
Exosome-mediated circTTLL5 transfer promotes hepatocellular
carcinoma malignant progression through miR-136-5p/KIAA1522 axis.
Pathol Res Pract. 241:1542762023. View Article : Google Scholar
|
|
55
|
Li Y, Zang H, Zhang X and Huang G:
Exosomal Circ-ZNF652 promotes cell proliferation, migration,
invasion and glycolysis in hepatocellular carcinoma via
miR-29a-3p/GUCD1 axis. Cancer Manag Res. 12:7739–7751. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang L, Li B, Yi X, Xiao X, Zheng Q and Ma
L: Circ_0036412 affects the proliferation and cell cycle of
hepatocellular carcinoma via hedgehog signaling pathway. J Transl
Med. 20:1542022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Du A, Li S, Zhou Y, Disoma C, Liao Y,
Zhang Y, Chen Z, Yang Q, Liu P, Liu S, et al: M6A-mediated
upregulation of circMDK promotes tumorigenesis and acts as a
nanotherapeutic target in hepatocellular carcinoma. Mol Cancer.
21:1092022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang N, Zheng L, Zhan Y and Zhang Y: A
novel taspine derivative suppresses human liver tumor growth and
invasion in vitro and in vivo. Oncol Lett. 6:855–859. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hunter KW: Host genetics and tumour
metastasis. Br J Cancer. 90:752–755. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mu W, Gu P, Li H, Zhou J, Jian Y, Jia W
and Ge Y: Exposure of benzo[a]pyrene induces HCC exosome-circular
RNA to activate lung fibroblasts and trigger organotropic
metastasis. Cancer Commun (Lond). 44:718–738. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo
J, Zhang Y, Li H, Zhang Q, Yang Y and Chen G: Three isoforms of
exosomal circPTGR1 promote hepatocellular carcinoma metastasis via
the miR449a-MET pathway. EBioMedicine. 40:432–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Allgayer H, Mahapatra S, Mishra B, Swain
B, Saha S, Khanra S, Kumari K, Panda VK, Malhotra D, Patil NS, et
al: Epithelial-to-mesenchymal transition (EMT) and cancer
metastasis: The status quo of methods and experimental models 2025.
Mol Cancer. 24:1672025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Amicone L, Marchetti A and Cicchini C:
Exosome-associated circRNAs as key regulators of EMT in cancer.
Cells. 11:17162022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hu M, Li X, Jiang Z, Xia Q, Hu Y, Guo J
and Fu L: Exosomes and circular RNAs: Promising partners in
hepatocellular carcinoma from bench to bedside. Discov Oncol.
14:602023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Huang JL, Fu YP, Gan W, Liu G, Zhou PY,
Zhou C, Sun BY, Guan RY, Zhou J, Fan J, et al: Hepatic stellate
cells promote the progression of hepatocellular carcinoma through
microRNA-1246-RORα-Wnt/β-catenin axis. Cancer Lett. 476:140–151.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Luo Y, Fu Y, Huang R, Gao M, Liu F, Gui R
and Nie X: CircRNA_101505 sensitizes hepatocellular carcinoma cells
to cisplatin by sponging miR-103 and promotes oxidored-nitro
domain-containing protein 1 expression. Cell Death Discov.
5:1212019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhu C, Su Y, Liu L, Wang S, Liu Y and Wu
J: Circular RNA hsa_circ_0004277 stimulates malignant phenotype of
hepatocellular carcinoma and Epithelial-mesenchymal transition of
peripheral cells. Front Cell Dev Biol. 8:5855652020. View Article : Google Scholar
|
|
68
|
Wang Y, Gao R, Li J, Tang S and Li S, Tong
Q and Li S: Downregulation of hsa_circ_0074854 suppresses the
migration and invasion in hepatocellular carcinoma via interacting
with HuR and via suppressing exosomes-mediated macrophage M2
polarization. Int J Nanomedicine. 16:2803–2818. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Liu L, Liao R, Wu Z, Du C, You Y, Que K,
Duan Y, Yin K and Ye W: Hepatic stellate cell exosome-derived
circWDR25 promotes the progression of hepatocellular carcinoma via
the miRNA-4474-3P-ALOX-15 and EMT axes. Biosci Trends. 16:267–281.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang T, Sun Q, Shen C and Qian Y:
Circular RNA circ_0003028 regulates cell development through
modulating miR-498/ornithine decarboxylase 1 axis in hepatocellular
carcinoma. Anticancer Drugs. 34:507–518. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu D, Kang H, Gao M, Jin L, Zhang F, Chen
D, Li M and Xiao L: Exosome-transmitted circ_MMP2 promotes
hepatocellular carcinoma metastasis by upregulating MMP2. Mol
Oncol. 14:1365–1380. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A
and Qi F: CircRNA Cdr1as functions as a competitive endogenous RNA
to promote hepatocellular carcinoma progression. Aging Albany (NY).
11:8183–8203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lin Y, Zheng ZH, Wang JX, Zhao Z and Peng
TY: Tumor Cell-derived exosomal Circ-0072088 suppresses migration
and invasion of hepatic carcinoma cells through regulating MMP-16.
Front Cell Dev Biol. 9:7263232021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Prager GW, Poettler M, Unseld M and
Zielinski CC: Angiogenesis in cancer: Anti-VEGF escape mechanisms.
Transl Lung Cancer Res. 1:14–25. 2012.PubMed/NCBI
|
|
75
|
Zhu Y, He Q and Qi M: Exosomal circPTPRK
promotes angiogenesis after radiofrequency ablation in
hepatocellular carcinoma. Exp Biol Med (Maywood). 249:100842024.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Huang XY, Huang ZL, Huang J, Xu B, Huang
XY, Xu YH, Zhou J and Tang ZY: Exosomal circRNA-100338 promotes
hepatocellular carcinoma metastasis via enhancing invasiveness and
angiogenesis. J Exp Clin Cancer Res. 39:202020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Hao X, Zhang Y, Shi X, Liu H, Zheng Z, Han
G, Rong D, Zhang C, Tang W and Wang X: CircPAK1 promotes the
progression of hepatocellular carcinoma via modulation of YAP
nucleus localization by interacting with 14-3-3ζ. J Exp Clin Cancer
Res. 41:2812022. View Article : Google Scholar
|
|
78
|
Hu K, Li NF, Li JR, Chen ZG, Wang JH and
Sheng LQ: Exosome circCMTM3 promotes angiogenesis and tumorigenesis
of hepatocellular carcinoma through miR-3619-5p/SOX9. Hepatol Res.
51:1139–1152. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xu B, Jia W, Feng Y, Wang J, Wang J, Zhu
D, Xu C, Liang L, Ding W, Zhou Y and Kong L: Exosome-transported
circHDAC1_004 promotes proliferation, migration, and angiogenesis
of hepatocellular carcinoma by the miR-361-3p/NACC1 axis. J Clin
Transl Hepatol. 11:1079–1093. 2023.PubMed/NCBI
|
|
80
|
Deng M, Sun S, Zhao R, Guan R, Zhang Z, Li
S, Wei W and Guo R: The pyroptosis-related gene signature predicts
prognosis and indicates immune activity in hepatocellular
carcinoma. Mol Med. 28:162022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lai Z, Wei T, Li Q, Wang X, Zhang Y and
Zhang S: Exosomal circFBLIM1 promotes hepatocellular carcinoma
progression and glycolysis by regulating the miR-338/LRP6 axis.
Cancer Biother Radiopharm. 38:674–683. 2023.
|
|
83
|
Yu S, Su S, Wang P, Li J, Chen C, Xin H,
Gong Y, Wang H, Ye X, Mao L, et al: Tumor-associated
macrophage-induced circMRCKα encodes a peptide to promote
glycolysis and progression in hepatocellular carcinoma. Cancer
Lett. 591:2168722024. View Article : Google Scholar
|
|
84
|
Li J, Wang X, Shi L, Liu B, Sheng Z, Chang
S, Cai X and Shan G: A mammalian conserved circular RNA CircLARP1B
regulates hepatocellular carcinoma metastasis and lipid metabolism.
Adv Sci (Weinh). 11:e23059022024. View Article : Google Scholar
|
|
85
|
Liu C, Huang R, Yu H, Gong Y, Wu P, Feng Q
and Li X: Fuzheng Xiaozheng prescription exerts anti-hepatocellular
carcinoma effects by improving lipid and glucose metabolisms via
regulating circRNA-miRNA-mRNA networks. Phytomedicine.
103:1542262022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Han T, Chen L and Li K, Hu Q, Zhang Y, You
X, Han L, Chen T and Li K: Significant CircRNAs in liver cancer
stem cell exosomes: Mediator of malignant propagation in liver
cancer? Mol Cancer. 22:1972023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Gu Y, Wang Y, He L, Zhang J, Zhu X, Liu N,
Wang J, Lu T, He L, Tian Y and Fan Z: Circular RNA circIPO11 drives
self-renewal of liver cancer initiating cells via Hedgehog
signaling. Mol Cancer. 20:1322021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin
XM, Yang S, Zhao Q, Wu T, Li ZX, et al: Circular RNAs negatively
regulate cancer stem cells by physically binding FMRP against CCAR1
complex in hepatocellular carcinoma. Theranostics. 9:3526–3540.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sarkar FH, Li Y, Wang Z and Kong D: The
role of nutraceuticals in the regulation of Wnt and Hedgehog
signaling in cancer. Cancer Metastasis Rev. 29:383–394. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Takebe N, Harris PJ, Warren RQ and Ivy SP:
Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog
pathways. Nat Rev Clin Oncol. 8:97–106. 2011. View Article : Google Scholar
|
|
91
|
Chatterjee S and Sil PC: Targeting the
crosstalks of Wnt pathway with Hedgehog and Notch for cancer
therapy. Pharmacol Res. 142:251–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Lei YR, He XL, Li J and Mo CF: Drug
resistance in hepatocellular carcinoma: Theoretical basis and
therapeutic aspects. Front Biosci (Landmark Ed). 29:522024.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Guo X, Gao CY, Yang DH and Li SL: Exosomal
circular RNAs: A chief culprit in cancer chemotherapy resistance.
Drug Resist Updat. 67:1009372023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Qin L, Zhan Z, Wei C, Li X, Zhang T and Li
J: Hsa-circRNA-G004213 promotes cisplatin sensitivity by regulating
miR-513b-5p/PRPF39 in liver cancer. Mol Med Rep. 23:4212021.
View Article : Google Scholar :
|
|
95
|
Zhou Y, Tang W, Zhuo H, Zhu D, Rong D, Sun
J and Song J: Cancer-associated fibroblast exosomes promote
chemoresistance to cisplatin in hepatocellular carcinoma through
circZFR targeting signal transducers and activators of
transcription (STAT3)/nuclear factor-kappa B (NF-κB) pathway.
Bioengineered. 13:4786–4797. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
He G and Karin M: NF-κB and STAT3-key
players in liver inflammation and cancer. Cell Res. 21:159–168.
2011. View Article : Google Scholar
|
|
97
|
Li Y, Zhang Y, Zhang S, Huang D, Li B,
Liang G, Wu Y, Jiang Q, Li L, Lin C, et al: circRNA circARNT2
suppressed the sensitivity of hepatocellular carcinoma cells to
cisplatin by targeting the miR-155-5p/PDK1 axis. Mol Ther Nucleic
Acids. 23:244–254. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Chen H, Liu S, Li M, Huang P and Li X:
circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance
through Wnt/β-catenin pathway in hepatocellular carcinoma. Onco
Targets Ther. 12:9539–9549. 2019. View Article : Google Scholar :
|
|
99
|
Xie H, Yao J, Wang Y and Ni B:
Exosome-transmitted circVMP1 facilitates the progression and
cisplatin resistance of non-small cell lung cancer by targeting
miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 29:1257–1271. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Shao N, Song L and Sun X: Exosomal
circ_PIP5K1A regulates the progression of non-small cell lung
cancer and cisplatin sensitivity by miR-101/ABCC1 axis. Mol Cell
Biochem. 476:2253–2267. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Hanssen KM, Wheatley MS, Yu DMT, Conseil
G, Norris MD, Haber M, Cole SPC and Fletcher JI: GSH facilitates
the binding and inhibitory activity of novel multidrug resistance
protein 1 (MRP1) modulators. FEBS J. 289:3854–3875. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zang R, Qiu X, Song Y and Wang Y: Exosomes
mediated transfer of Circ_0000337 contributes to cisplatin (CDDP)
resistance of esophageal cancer by regulating JAK2 via miR-377-3p.
Front Cell Dev Biol. 9:6732372021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li Y, Wu A, Chen L, Cai A, Hu Y, Zhou Z,
Qi Q, Wu Y, Xia D, Dong P, et al: Hsa_circ_0000098 is a novel
therapeutic target that promotes hepatocellular carcinoma
development and resistance to doxorubicin. J Exp Clin Cancer Res.
41:2672022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang Z, Wang L, Yin G, Li H, Zhang R, Feng
Y and Chang W: Hsa_circ_0088036 promotes tumorigenesis and
chemotherapy resistance in hepatocellular carcinoma via the
miR-140-3p/KIF2A axis. Histol Histopathol. 40:1239–1251. 2025.
|
|
105
|
Li J, Qin X, Wu R, Wan L, Zhang L and Liu
R: Circular RNA circFBXO11 modulates hepatocellular carcinoma
progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1
axis. J Cell Mol Med. 24:5152–5161. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Ren Y, Dong X, Chen L, Sun T, Alwalid O,
Kan X, Su Y, Xiong B, Liang H, Zheng C and Han P: Combined
ultrasound and CT-Guided Iodine-125 seeds implantation for
treatment of residual hepatocellular carcinoma located at complex
sites after transcatheter arterial chemoembolization. Front Oncol.
11:5825442021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yuan L, Wang Y, Cheng J, Lin S, Ma A, Li
K, Zheng Y, Zeng Z, Ke A, Gao C and Du S: Cancer-derived exosomal
circTMEM56 enhances the efficacy of HCC radiotherapy through the
miR-136-5p/STING axis. Cancer Biol Med. 22:396–411. 2025.PubMed/NCBI
|
|
108
|
Li T and Chen ZJ: The cGAS-cGAMP-STING
pathway connects DNA damage to inflammation, senescence, and
cancer. J Exp Med. 215:1287–1299. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Lin L, Hu P, Luo M, Chen X, Xiao M, Zhong
Z, Peng S, Chen G, Yang G, Zhang F and Zhang Y: CircNOP14 increases
the radiosensitivity of hepatocellular carcinoma via inhibition of
Ku70-dependent DNA damage repair. Int J Biol Macromol.
264:1305412024. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhao B, Rothenberg E, Ramsden DA and
Lieber MR: The molecular basis and disease relevance of
non-homologous DNA end joining. Nat Rev Mol Cell Biol. 21:765–781.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yang W, Liu Y, Gao R, Xiu Z and Sun T:
Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721
cell proliferation, vasculogenic mimicry, and radioresistance. Cell
Signal. 60:122–135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhu S, Chen Y, Ye H, Wang B, Lan X, Wang
H, Ding S and He X: Circ-LARP1B knockdown restrains the
tumorigenicity and enhances radiosensitivity by regulating
miR-578/IGF1R axis in hepatocellular carcinoma. Ann Hepatol.
27:1006782022. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Chang Z, Song Y, Luo F, Yang X, Cai Y and
Guo H: Circular RNA SMARCA5 promotes a poor prognosis and
radiotherapy resistance for patients with hepatocellular carcinoma.
Ann Clin Lab Sci. 53:573–577. 2023.PubMed/NCBI
|
|
114
|
Wang X, Zhang J, Luo F and Shen Y:
Application of circular RNA Circ_0071662 in the diagnosis and
prognosis of hepatocellular carcinoma and its response to
radiotherapy. Dig Dis. 41:431–438. 2023. View Article : Google Scholar
|
|
115
|
Yang K, Ding Y, Han J and He R: CircROBO1
knockdown improves the radiosensitivity of hepatocellular carcinoma
by regulating RAD21. Ann Hepatol. 29:1015362024. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Gu X, Shi Y, Dong M, Jiang L, Yang J and
Liu Z: Exosomal transfer of tumor-associated macrophage-derived
hsa_circ_0001610 reduces radiosensitivity in endometrial cancer.
Cell Death Dis. 12:8182021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Wang X, Cao Q, Shi Y, Wu X, Mi Y, Liu K,
Kan Q, Fan R, Liu Z and Zhang M: Identification of Low-dose
radiation-induced exosomal circ-METRN and miR-4709-3p/GRB14/PDGFRα
pathway as a key regulatory mechanism in Glioblastoma progression
and radioresistance: Functional validation and clinical theranostic
significance. Int J Biol Sci. 17:1061–1078. 2021. View Article : Google Scholar :
|
|
118
|
Gusani NJ, Jiang Y, Kimchi ET,
Staveley-O'Carroll KF, Cheng H and Ajani JA: New pharmacological
developments in the treatment of hepatocellular cancer. Drugs.
69:2533–2540. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
He J, Wang X, Chen K, Zhang M and Wang J:
The amino acid transporter SLC7A11-mediated crosstalk implicated in
cancer therapy and the tumor microenvironment. Biochem Pharmacol.
205:1152412022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Koppula P, Zhang Y, Zhuang L and Gan B:
Amino acid transporter SLC7A11/xCT at the crossroads of regulating
redox homeostasis and nutrient dependency of cancer. Cancer Commun
(Lond). 38:122018.PubMed/NCBI
|
|
121
|
Lin W, Wang C, Liu G, Bi C, Wang X, Zhou Q
and Jin H: SLC7A11/xCT in cancer: Biological functions and
therapeutic implications. Am J Cancer Res. 10:3106–3126.
2020.PubMed/NCBI
|
|
122
|
Bassi MT, Gasol E, Manzoni M, Pineda M,
Riboni M, Martín R, Zorzano A, Borsani G and Palacín M:
Identification and characterisation of human xCT that co-expresses,
with 4F2 heavy chain, the amino acid transport activity system xc.
Pflugers Arch. 442:286–296. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhang Y, Yao R, Li M, Fang C, Feng K, Chen
X, Wang J, Luo R, Shi H, Chen X, et al: CircTTC13 promotes
sorafenib resistance in hepatocellular carcinoma through the
inhibition of ferroptosis by targeting the miR-513a-5p/SLC7A11
axis. Mol Cancer. 24:322025. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kuwano M, Oda Y, Izumi H, Yang SJ, Uchiumi
T, Iwamoto Y, Toi M, Fujii T, Yamana H, Kinoshita H, et al: The
role of nuclear Y-box binding protein 1 as a global marker in drug
resistance. Mol Cancer Ther. 3:1485–1492. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Kwabiah D, Nagati V and Tripathi MK:
Transcription factor YBX1 orchestrates drug resistance and tumor
progression in HCC. Drug Discov Today. 30:1044392025. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Prabhu L, Hartley AV, Martin M, Warsame F,
Sun E and Lu T: Role of post-translational modification of the Y
box binding protein 1 in human cancers. Genes Dis. 2:240–246. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Feng Z, Gao Y, Cai C, Tan J, Liu P, Chen
Y, Deng G, Ouyang Y, Liu X, Cao K, et al: CSF3R-AS promotes
hepatocellular carcinoma progression and sorafenib resistance
through the CSF3R/JAK2/STAT3 positive feedback loop. Cell Death
Dis. 16:2172025. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Lan Y, Banks KM, Pan H, Verma N, Dixon GR,
Zhou T, Ding B, Elemento O, Chen S, Huangfu D and Evans T:
Stage-specific regulation of DNA methylation by TET enzymes during
human cardiac differentiation. Cell Rep. 37:1100952021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Dong ZR, Ke AW, Li T, Cai JB, Yang YF,
Zhou W, Shi GM and Fan J: CircMEMO1 modulates the promoter
methylation and expression of TCF21 to regulate hepatocellular
carcinoma progression and sorafenib treatment sensitivity. Mol
Cancer. 20:752021. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Kudo M, Finn RS, Qin S, Han KH, Ikeda K,
Piscaglia F, Baron A, Park JW, Han G, Jassem J, et al: Lenvatinib
versus sorafenib in first-line treatment of patients with unresec\
hepatocellular carcinoma: A randomised phase 3 non-inferiority
trial. Lancet. 391:1163–1173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Magyar CTJ, Perera S, Rajendran L, Li Z,
Almugbel FA, Feng S, Choi WJ, Aceituno L, Vogel A, Grant RC, et al:
Comparative outcome analysis of lenvatinib versus sorafenib for
recurrence of hepatocellular carcinoma after liver transplantation.
Transplantation. 109:681–690. 2025. View Article : Google Scholar
|
|
132
|
Hu B, Zou T, Qin W, Shen X, Su Y, Li J,
Chen Y, Zhang Z, Sun H, Zheng Y, et al: Inhibition of EGFR
overcomes acquired lenvatinib resistance driven by STAT3-ABCB1
signaling in hepatocellular carcinoma. Cancer Res. 82:3845–3857.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Zhang P, Sun H, Wen P, Wang Y, Cui Y and
Wu J: circRNA circMED27 acts as a prognostic factor and mediator to
promote lenvatinib resistance of hepatocellular carcinoma. Mol Ther
Nucleic Acids. 27:293–303. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Tang Y, Yuan F, Cao M, Ren Y, Li Y, Yang
G, Zhong Z, Liang H, Xiong Z, He Z, et al: CircRNA-mTOR promotes
hepatocellular carcinoma progression and lenvatinib resistance
through the PSIP1/c-Myc axis. Adv Sci (Weinh). 12:e24105912025.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Yang L, Tan W, Wang M, Wei Y, Xie Z, Wang
Q, Zhang Z, Zhuang H, Ma X, Wang B, et al: circCCNY enhances
lenvatinib sensitivity and suppresses immune evasion in
hepatocellular carcinoma by serving as a scaffold for SMURF1
mediated HSP60 degradation. Cancer Lett. 612:2174702025. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Hou YR, Diao LT, Hu YX, Zhang QQ, Lv G,
Tao S, Xu WY, Xie SJ, Zhang Q and Xiao ZD: The conserved LncRNA
DIO3OS restricts hepatocellular carcinoma stemness by interfering
with NONO-mediated nuclear export of ZEB1 mRNA. Adv Sci (Weinh).
10:e23019832023. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Orellana-Serradell O, Herrera D, Castellón
EA and Contreras HR: The transcription factor ZEB1 promotes
chemoresistance in prostate cancer cell lines. Asian J Androl.
21:460–467. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Cao M, Li Y, Su X, Tang Y, Yuan F, Ren Y,
Deng M and Yao Z: Exosome-derived hsa_circ_0007132 promotes
lenvatinib resistance by inhibiting the ubiquitin-mediated
degradation of NONO. Noncoding RNA Res. 14:1–13. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Yuan F, Tang Y, Liang H, Cao M, Ren Y, Li
Y, Yang G, Zhong Z, Xiong Z, He Z, et al: CircPIK3C3 inhibits
hepatocellular carcinoma progression and lenvatinib resistance by
suppressing the Wnt/β-catenin pathway via the miR-452-5p/SOX15
axis. Genomics. 117:1109992025. View Article : Google Scholar
|
|
140
|
Bruix J, Qin S, Merle P, Granito A, Huang
YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al:
Regorafenib for patients with hepatocellular carcinoma who
progressed on sorafenib treatment (RESORCE): A randomised,
double-blind, placebo-controlled, phase 3 trial. Lancet. 389:56–66.
2017. View Article : Google Scholar
|
|
141
|
Ueshima K, Nishida N and Kudo M:
Sorafenib-regorafenib sequential therapy in advanced hepatocellular
carcinoma: A Single-institute experience. Dig Dis. 35:611–617.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Wang K, Yu G, Lin J, Wang Z, Lu Q, Gu C,
Yang T, Liu S and Yang H: Berberine sensitizes human hepatoma cells
to regorafenib via modulating expression of circular RNAs. Front
Pharmacol. 12:6322012021. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Li M, Bhoori S, Mehta N and Mazzaferro V:
Immunotherapy for hepatocellular carcinoma: The next evolution in
expanding access to liver transplantation. J Hepatol. 81:743–755.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Raskova Kafkova L, Mierzwicka JM,
Chakraborty P, Jakubec P, Fischer O, Skarda J, Maly P and Raska M:
NSCLC: From tumorigenesis, immune checkpoint misuse to current and
future targeted therapy. Front Immunol. 15:13420862024. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Klobuch S, Seijkens TTP, Schumacher TN and
Haanen J: Tumour-infiltrating lymphocyte therapy for patients with
advanced-stage melanoma. Nat Rev Clin Oncol. 21:173–184. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu
W, Wang Z, Wu Q, Peng H, Wei H, et al: Blockade of the checkpoint
receptor TIGIT prevents NK cell exhaustion and elicits potent
anti-tumor immunity. Nat Immunol. 19:723–732. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Jiang W, Pan S, Chen X, Wang ZW and Zhu X:
The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in
cancer immunotherapy. Mol Cancer. 20:1162021. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Fu J, Liu F, Bai S, Jiang X, Song H, Zhang
M, Zhao R, Ouyang T, Yu M, Qian H, et al: Circular RNA CDYL
facilitates hepatocellular carcinoma stemness and PD-L1+
exosomes-mediated immunotherapy resistance via stabilizing hornerin
protein by blocking synoviolin 1-mediated ubiquitination. Int J
Biol Macromol. 310:1432462025. View Article : Google Scholar
|
|
149
|
Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y,
Jiang J, Wang L, Mang Y and Gao Y: Exosome-derived circCCAR1
promotes CD8 + T-cell dysfunction and anti-PD1 resistance in
hepatocellular carcinoma. Mol Cancer. 22:552023. View Article : Google Scholar
|
|
150
|
Lu JC, Zhang PF, Huang XY, Guo XJ, Gao C,
Zeng HY, Zheng YM, Wang SW, Cai JB, Sun QM, et al: Amplification of
spatially isolated adenosine pathway by tumor-macrophage
interaction induces anti-PD1 resistance in hepatocellular
carcinoma. J Hematol Oncol. 14:2002021. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ,
Shi GM, Cai JB and Ke AW: Cancer cell-derived exosomal circUHRF1
induces natural killer cell exhaustion and may cause resistance to
anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer.
19:1102020. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Huang XY, Zhang PF, Wei CY, Peng R, Lu JC,
Gao C, Cai JB, Yang X, Fan J and Ke AW: Circular RNA circMET drives
immunosuppression and anti-PD1 therapy resistance in hepatocellular
carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 19:922020.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Cai J, Chen Z, Zhang Y, Wang J, Zhang Z,
Wu J, Mao J and Zuo X: CircRHBDD1 augments metabolic rewiring and
restricts immunotherapy efficacy via m6A modification in
hepatocellular carcinoma. Mol Ther Oncolytics. 24:755–771. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Ye R, Lu X, Liu J, Duan Q, Xiao J, Duan X,
Yue Z and Liu F: CircSOD2 contributes to tumor progression, immune
evasion and Anti-PD-1 resistance in hepatocellular carcinoma by
targeting miR-497-5p/ANXA11 axis. Biochem Genet. 61:597–614. 2023.
View Article : Google Scholar
|
|
155
|
Hao L, Li S, Ye F, Wang H, Zhong Y, Zhang
X, Hu X and Huang X: The current status and future of
targeted-immune combination for hepatocellular carcinoma. Front
Immunol. 15:14189652024. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Mizukoshi E and Kaneko S: Immune cell
therapy for hepatocellular carcinoma. J Hematol Oncol. 12:522019.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Lu F, Ma XJ, Jin WL, Luo Y and Li X:
Neoantigen specific T cells derived from T Cell-derived induced
pluripotent stem cells for the treatment of hepatocellular
carcinoma: Potential and challenges. Front Immunol. 12:6905652021.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Gutwillig A, Santana-Magal N,
Farhat-Younis L, Rasoulouniriana D, Madi A, Luxenburg C, Cohen J,
Padmanabhan K, Shomron N, Shapira G, et al: Transient cell-in-cell
formation underlies tumor relapse and resistance to immunotherapy.
Elife. 11:e803152022. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
O'Donnell JS, Teng MWL and Smyth MJ:
Cancer immunoediting and resistance to T cell-based immunotherapy.
Nat Rev Clin Oncol. 16:151–167. 2019. View Article : Google Scholar
|
|
160
|
Huang M, Huang X and Huang N: Exosomal
circGSE1 promotes immune escape of hepatocellular carcinoma by
inducing the expansion of regulatory T cells. Cancer Sci.
113:1968–1983. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D,
Long X, Lin K, Lu F, Xu JJ and Wu YB: Cancer cell-derived exosomal
circUSP7 induces CD8+ T cell dysfunction and anti-PD1 resistance by
regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer. 20:1442021.
View Article : Google Scholar :
|
|
162
|
Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y,
Chen X, Chen Y, Xu C, Hu Y, et al: Exosome-derived circTRPS1
promotes malignant phenotype and CD8+ T cell exhaustion in bladder
cancer microenvironments. Mol Ther. 30:1054–1070. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Peng H, Wisse E and Tian Z: Liver natural
killer cells: Aubsets and roles in liver immunity. Cell Mol
Immunol. 13:328–336. 2016. View Article : Google Scholar
|
|
164
|
Balkhi S, Zuccolotto G, Di Spirito A,
Rosato A and Mortara L: CAR-NK cell therapy: Promise and challenges
in solid tumors. Front Immunol. 16:15747422025. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Li S, Jing J, Chen Y, Chi E, Wang B, Xie
Z, Yang W, Shen H and Pan J: Precision sniper for solid tumors:
CAR-NK cell therapy. Cancer Immunol Immunother. 74:2752025.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH,
Wang YG and Zhang YQ: Hsa_circ_0007456 regulates the natural killer
cell-mediated cytotoxicity toward hepatocellular carcinoma via the
miR-6852-3p/ICAM-1 axis. Cell Death Dis. 12:942021. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Jeong JU, Uong TNT, Chung WK, Nam TK, Ahn
SJ, Song JY, Kim SK, Shin DJ, Cho E, Kim KW, et al: Effect of
irradiation-induced intercellular adhesion molecule-1 expression on
natural killer cell-mediated cytotoxicity toward human cancer
cells. Cytotherapy. 20:715–727. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Liu G, Liu Q, Jia L, Chai Z, Jing L, Xu F
and Fan Y: Exosomal circRNAs: Key modulators in breast cancer
progression. Cell Death Discov. 11:1962025. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Shen H, Liu B, Xu J, Zhang B, Wang Y, Shi
L and Cai X: Circular RNAs: Characteristics, biogenesis, mechanisms
and functions in liver cancer. J Hematol Oncol. 14:1342021.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Xu HZ, Lin XY, Xu YX, Xue HB, Lin S and Xu
TW: An emerging research: The role of hepatocellular
carcinoma-derived exosomal circRNAs in the immune microenvironment.
Front Immunol. 14:12271502023. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Zhang Q, Li H, Liu Y, Li J, Wu C and Tang
H: Exosomal Non-coding RNAs: New insights into the biology of
hepatocellular carcinoma. Curr Oncol. 29:5383–5406. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Tonon F, Grassi C, Tierno D, Biasin A,
Grassi M, Grassi G and Dapas B: Non-coding RNAs as potential
diagnostic/prognostic markers for hepatocellular carcinoma. Int J
Mol Sci. 25:122352024. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Plebani M, Scott S, Simundic AM, Cornes M,
Padoan A, Cadamuro J, Vermeersch P, Çubukçu HC, González Á, Nybo M,
et al: New insights in preanalytical quality. Clin Chem Lab Med.
63:1682–1692. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu
H, Xu X, Liang Q, Christiani DC, Wang M, et al: Circular RNAs in
body fluids as cancer biomarkers: The new frontier of liquid
biopsies. Mol Cancer. 20:132021. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Sun XH, Wang YT, Li GF, Zhang N and Fan L:
Serum-derived three-circRNA signature as a diagnostic biomarker for
hepatocellular carcinoma. Cancer Cell Int. 20:2262020. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Luo Y, Liu F and Gui R: High expression of
circulating exosomal circAKT3 is associated with higher recurrence
in HCC patients undergoing surgical treatment. Surg Oncol.
33:276–281. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Gong Y, Li X and Wang M: Elevated
CircYthdc2 expression is correlated with aggressive features and
poor progression-free survival in hepatocellular carcinoma. BMC
Gastroenterol. 25:6582025. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Awed MS, Ibrahim A, Ezzat O, Fawzy A,
Sabir DK and Radwan AF: Preliminary evaluation of plasma
circ_0009910, circ_0027478, and miR-1236-3p as diagnostic and
prognostic biomarkers in hepatocellular carcinoma. Int J Mol Sci.
26:48422025. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang
L, Deng Q and Zheng F: Using circular RNA SMARCA5 as a potential
novel biomarker for hepatocellular carcinoma. Clin Chim Acta.
492:37–44. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Li C, Wei Y, Li X, Jiang P, Dong X, Guo Q,
Xu Q, Cao X, Xia J and Wang Z: An Ultrasensitive and robust CircRNA
nanobiosensor via magnetic control depuration and catalytic hairpin
assembly cascade amplification for clinically accessible liquid
biopsy. Anal Chem. 97:23939–23949. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Weng Q, Chen M, Li M, Zheng YF, Shao G,
Fan W, Xu XM and Ji J: Global microarray profiling identified
hsa_circ_0064428 as a potential immune-associated prognosis
biomarker for hepatocellular carcinoma. J Med Genet. 56:32–38.
2019. View Article : Google Scholar
|
|
182
|
Zhang L, Xu T, Li Y, Pang Q and Ding X:
Serum hsa_circ_0000615 is a prognostic biomarker of sorafenib
resistance in hepatocellular carcinoma. J Clin Lab Anal.
36:e247412022. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Qiao GL, Chen L, Jiang WH, Yang C, Yang
CM, Song LN, Chen Y, Yan HL and Ma LJ: Hsa_circ_0003998 may be used
as a new biomarker for the diagnosis and prognosis of
hepatocellular carcinoma. Onco Targets Ther. 12:5849–5860. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Lyu L, Yang W, Yao J, Wang H, Zhu J, Jin
A, Liu T, Wang B, Zhou J, Fan J, et al: The diagnostic value of
plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma.
Biomark Med. 15:359–371. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Wang Y, Pei L, Yue Z, Jia M, Wang H and
Cao LL: The potential of serum exosomal hsa_circ_0028861 as the
novel diagnostic biomarker of HBV-derived hepatocellular cancer.
Front Genet. 12:7032052021. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Huang S, Qin H, Dai B, Liu M and Shen J:
Establishment and evaluation of a circAdpgk-0001 knockdown method
using CRISPR-Cas13d RNA-targeting technology. PeerJ. 13:e201232025.
View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Wang L, Wang L, Dong C, Liu J, Cui G, Gao
S and Liu Z: Exploring the potential and advancements of circular
RNA therapeutics. Exploration (Beijing). 5:e202400442025.
View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Cai J, Qiu Z, Chi-Shing Cho W, Liu Z, Chen
S, Li H, Chen K, Li Y, Zuo C and Qiu M: Synthetic circRNA
therapeutics: Innovations, strategies, and future horizons. MedComm
(2020). 5:e7202024. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Ruan YL, Chen TY, Zheng LB, Cai JW, Zhao
H, Wang YL, Tao LY, Xu JJ, Ji L and Cai XJ: cDCBLD2 mediates
sorafenib resistance in hepatocellular carcinoma by sponging
miR-345-5p binding to the TOP2A coding sequence. Int J Biol Sci.
19:4608–4626. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Yang Q and Wu G: CircRNA-001241 mediates
sorafenib resistance of hepatocellular carcinoma cells by sponging
miR-21-5p and regulating TIMP3 expression. Gastroenterol Hepatol.
45:742–752. 2022. View Article : Google Scholar
|
|
191
|
Qiu R and Zeng Z: Hsa_circ_0006988
promotes sorafenib resistance of hepatocellular carcinoma by
modulating IGF1 using miR-15a-5p. Can J Gastroenterol Hepatol.
2022:12061342022.
|
|
192
|
Li X, Yin X, Bao H and Liu C: Circular RNA
ITCH increases sorafenib-sensitivity in hepatocellular carcinoma
via sequestering miR-20b-5p and modulating the downstream
PTEN-PI3K/Akt pathway. Mol Cell Probes. 67:1018772023. View Article : Google Scholar
|
|
193
|
Zhang X, Wang W, Mo S and Sun X: DEAD-box
helicase 17 circRNA (circDDX17) reduces sorafenib resistance and
tumorigenesis in hepatocellular carcinoma. Dig Dis Sci.
69:2096–2108. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Wang S, Liu D, Wei H, Hua Y, Shi G and
Qiao J: The hsa_circRNA_102049 mediates the sorafenib sensitivity
of hepatocellular carcinoma cells by regulating Reelin gene
expression. Bioengineered. 13:2272–2284. 2022. View Article : Google Scholar : PubMed/NCBI
|