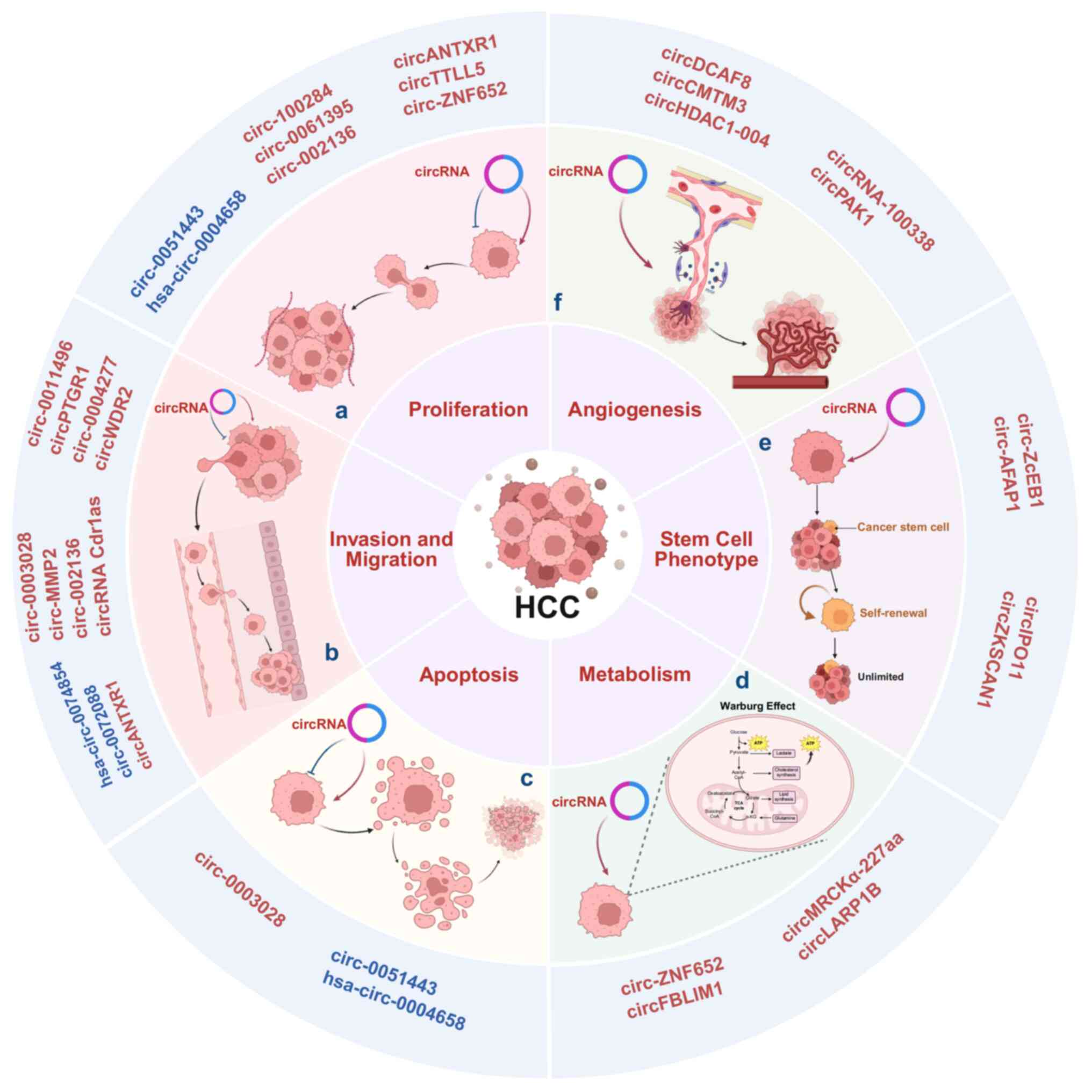
Introduction
Liver cancer is the sixth most common cancer and the
third leading cause of cancer-specific mortality worldwide. In
2022, liver cancer accounted for ~866,136 cases and 758,725 deaths
globally, with an estimated 42,240 new cases and 30,090 related
deaths expected in the United States in 2025 (1,2).
Although the relative survival rate for liver cancer has shown
notable improvement over the past 5 years, its prognosis remains
the least favorable compared with lung, esophageal and pancreatic
cancer. Hepatocellular carcinoma (HCC) is the most common type of
primary liver cancer, accounting for 75-85% of primary liver cancer
cases (1). However, the
pathogenesis of HCC is very insidious and there are often no
specific clinical manifestations in the early stages of the
disease, which poses a considerable obstacle and difficulty in
diagnosis. Therefore, the majority of patients are typically
diagnosed only when the disease has progressed to an advanced stage
and apparent symptoms such as severe jaundice and ascites have
appeared (3). In the later stages
of HCC, due to the severity of the condition, the widespread
proliferation or metastasis of tumor cells and the limitations of
local treatment, conventional treatments such as surgical
resection, liver transplantation or local percutaneous tumor
ablation are unable to effectively treat the disease, resulting in
a high mortality rate (4).
Despite significant progress and breakthroughs in liver cancer
treatment in recent years, encompassing interventional therapies,
local ablation, chemotherapy, targeted treatments and
immunotherapy, the prognosis for patients with HCC remains
unsatisfactory, with a 5-year survival rate of no more than 18%
(5). This is largely attributed
to the propensity of the tumor to metastasize and its resistance to
various therapeutic approaches. Malignant HCC can exhibit drug
resistance that is classified into two categories: Primary and
acquired resistance. The development of acquired resistance in HCC
is influenced by a variety of factors, including alterations in the
tumor microenvironment (TME), modifications in cellular signaling
pathways, dysregulated apoptosis, the presence of cancer stem cells
(CSCs), the involvement of microRNAs (miRNAs), mechanisms related
to DNA repair, variations in the immunophenotype, shifts in drug
metabolism and drug efflux and uptake as well as conditions of
tumor hypoxia (6). Since acquired
resistance to HCC can significantly reduce the therapeutic effect
of anti-cancer drugs, resulting in poor patient prognosis, reducing
HCC drug resistance has become a critical task in the field of
liver cancer research and treatment and is also one of the key
issues that need to be urgently addressed.
The advancement of high-throughput and
second-generation sequencing technologies has significantly
enhanced the capacity to identify aberrant non-coding RNAs
(ncRNAs), leading to the discovery of an increasing number of these
transcripts. Circular RNAs (circRNAs) are distinguished by their
unique covalently closed configuration, setting them apart from the
plethora of ncRNAs. circRNAs play a pivotal role in modulating gene
expression both during and after transcription through various
mechanisms, such as serving as sponges for miRNAs, coding for
polypeptides, functioning as scaffolds for proteins and
establishing stable complexes with RNA and proteins to influence
subsequent biological activities (7). Numerous studies indicate that
circRNAs are fundamentally associated with HCC progression and
treatment resistance (8-13).
Exosomes are small extracellular vesicles
originating from endocytosis, released by various cells, measuring
50-150 nm in diameter; they contain a rich and diverse array of
biologically active substances such as proteins, nucleic acids,
lipids and metabolites (14).
These bioactive compounds are pivotal in critical tumor processes,
including proliferation, invasion, metastasis, metabolism and
resistance to treatment, all of which significantly influence tumor
initiation and progression (15).
In 2015, a study first revealed that circRNAs are present within
exosomes. This groundbreaking finding suggested that serum exosomal
circRNAs may serve as promising circulating biomarkers for cancer
diagnosis (16). Furthermore,
studies have shown that exosomal circRNAs can be transferred
between HCC and non-HCC cells. In this process, they can promote or
inhibit the progression of HCC and mediate the therapeutic
resistance of HCC cells (17-19). It can therefore be seen that
exosomal circRNAs are a highly promising cancer diagnostic marker
and therapeutic target, and an in-depth exploration of the role of
exosomal circRNAs in the occurrence and development of HCC and drug
resistance is of significant clinical value.
The present review provides a thorough overview of
the various roles and intricate mechanisms of exosomal circRNAs in
the progression of HCC. In particular, how these molecules
influence HCC resistance to a range of treatment strategies,
including chemotherapy, radiotherapy, targeted therapies and
immunotherapy, is discussed. Through a comprehensive analysis of
current evidence, exosomal circRNA-mediated mechanisms of HCC
resistance, which involve alterations in cell signaling pathways,
modulation of the TME and metabolic reprogramming of cancer cells,
are described. These findings provide a solid theoretical
foundation for overcoming HCC resistance and highlight innovative
strategies to advance HCC treatment. Furthermore, targeting
exosomal circRNAs critical to treatment resistance has emerged as a
highly promising research direction in HCC therapeutics, garnering
notable attention and investment from the scientific community.
Biogenesis of exosomes and drug resistance
transfer
Exosomes, small extracellular vesicles surrounded by
a lipid bilayer, were first identified in 1983 in sheep
reticulocytes and are released by a wide range of cells under
physiological and pathophysiological conditions (20,21). Their generation involves the
inward folding of localized membranes within late endosomes,
leading to the creation of intraluminal vesicles (ILVs). These ILVs
accumulate, resulting in the formation of multivesicular bodies
(MVBs). Subsequently, an MVB merges with the cell membrane, which
releases the ILVs into the extracellular environment, transforming
them into exosomes (22).
Exosomes are packed with a diverse array of biologically active
components, including DNA, mRNA, miRNAs, long ncRNAs (lncRNAs),
circRNAs, proteins and lipids. These biologically active substances
can be transmitted between different cells, thus mediating
transcellular communication and regulating biological processes
such as tumor proliferation, metastasis, drug resistance, stemness
and metabolism (Fig. 1) (23). Exosomes are pivotal in mediating
the transfer of molecules that imbue drug resistance between tumor
cells. Drug-resistant tumor cells transfer biologically active
substances such as proteins and ncRNAs encapsulated in exosomes to
sensitive tumor cells, which can interfere with tumor cell
signaling pathways or promote drug efflux, resulting in a decrease
in the concentration of the drug in the tumor cells, which in turn
affects the cell cycle, invasion and metastasis, apoptosis,
angiogenesis and metabolic reprogramming of the cells (24-26). In addition, exosomes can mediate
tumor drug resistance by acting on tumor stem cell phenotypes,
epithelial-mesenchymal transition (EMT) and DNA repair damage,
among other mechanisms. For examination, exosomes are typically
obtained from blood or cell culture fluids, and their detection
hinges on the extraction of exosomes of high purity. Of the various
methods available, ultracentrifugation stands out as the most
prevalent technique and is often considered the 'gold standard' for
isolating exosomes (27). In
addition to ultracentrifugation, size exclusion chromatography,
ultrafiltration, immunoaffinity chromatography, microfluidics and
other new exosome isolation techniques have also emerged (28).
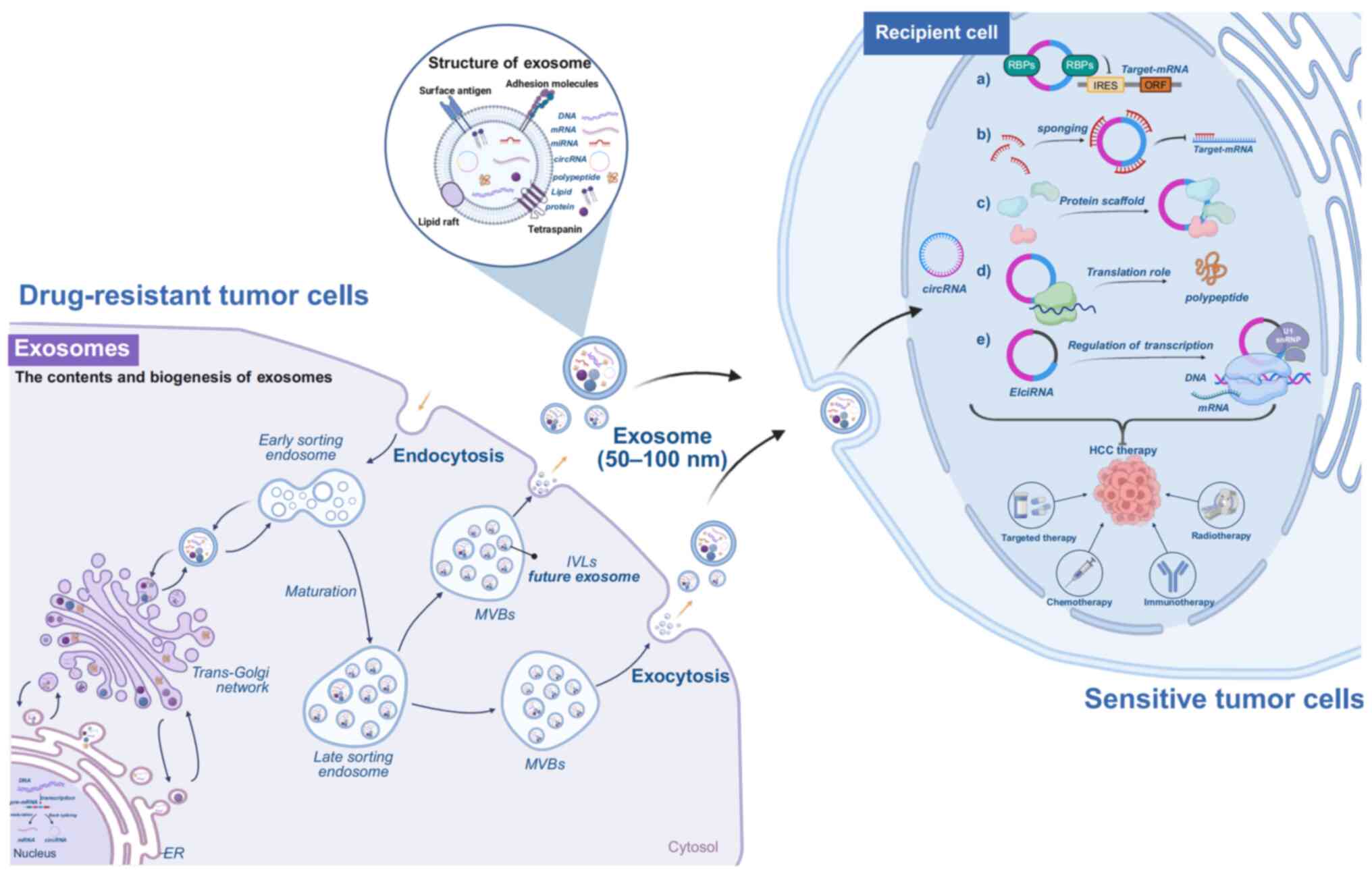 | Figure 1Schematic illustrating the formation
and secretion process of exosomes as well as their structure.
Secretory cells form early endosomes through inward budding, which
gradually evolve into mature MVBs. These MVBs fuse with the plasma
membrane to release exosomes. Exosomes contain bioactive substances
such as DNA, mRNA, miRNA, circRNA, proteins and lipids. These
components are delivered to recipient cells via endocytosis,
enabling them to exert biological effects. The primary biological
functions of circRNAs include. (A) Interacting with RBPs to
regulate gene expression (B) acting as competing sponges for miRNAs
to modulate target gene expression (C) serving as protein scaffolds
to facilitate interactions between different proteins (D) some
circRNAs can be translated by ribosomes into protein polypeptides
to perform regulatory functions; and (E) binding to U1 snRNP, which
forms complexes with RNA polymerase II to regulate gene
transcription or splicing. circRNA, circular RNA; ER, endoplasmic
reticulum; RBPs, RNA-binding protein; MVBs, multivesicular bodies;
IVLs, intraluminal vesicle; IRES, internal ribosome entry site;
ORF, open reading frame; U1 snRNP, U1 small nuclear
ribonucleoprotein particles. Figure was created with biorender.com. |
circRNAs in exosomes
Among the numerous biologically active substances in
exosomes, circRNAs and ncRNAs are of paramount importance and have
been extensively studied. circRNAs and ncRNAs are present in all
eukaryotic cells. Primarily, they are generated through the
'aberrant splicing' (also termed reverse splicing) of precursor
mRNAs (pre-mRNAs) (7). circRNAs
can be classified into various subtypes based on their distinct
splicing patterns. These include exon circRNAs (ecircRNAs),
exon-intron circRNAs (EIciRNAs), circular intron RNAs and tRNA
intron circRNAs (tricRNAs) (29).
In contrast to traditional linear RNAs that feature distinct 5' and
3' ends, circRNA molecules possess a unique closed-loop structure.
This design not only shields them from RNA exonuclease activity but
also enhances their stability during expression. As a result,
circRNAs are less susceptible to degradation and exhibit
specificity to particular tissues and cells (30). circRNAs serve several key
biological roles that can be grouped into five main categories
(Fig. 2): i) Influencing gene
transcription and splicing. circRNAs can engage in RNA-RNA
interactions, forming R loops at gene loci, which may modify the
splicing of parent transcripts or linear mRNAs. Additionally, they
can collaborate with transcription factors to enhance
transcriptional activity. CircSEP3, which originates from exon 6 of
the SEPALLATA3 gene, facilitates the accumulation of its homologous
exon 6-skipped splice variant. This is achieved by binding to the
parental genomic locus, leading to the formation of an RNA-DNA
hybrid (R-loop), which subsequently induces transcriptional pausing
and recruits splicing factors (31). Similarly, circSMARCA5 promotes the
formation of an R-loop at exon 15 of its host gene, SNF2 related
chromatin remodeling ATPase 5 (SMARCA5), which results in premature
transcriptional termination and consequently elevates the level of
a truncated, non-functional isoform of SMARCA5 mRNA (32). ii) Functioning as sponges for
RNA-binding proteins (RBPs). RBPs play vital roles in RNA cleavage,
stability and the translation of mRNAs. circRNAs, through their
unique sequences with specific RBP binding sites, can bind these
proteins, forming RNA-protein complexes that inhibit the activity
of RBPs and affect the expression of their associated genes
(33). For example, the circRNA
CIARS (hsa_circ_0008367) interacts with ALKBH5 to modulate
ferroptosis in HCC cells, thereby underscoring the significant role
of circRNAs in the regulation of cell death pathways (34). iii) Serving as miRNA sponges.
circRNAs possess numerous binding sites for miRNAs, effectively
soaking up these molecules and preventing them from interacting
with mRNAs, which in turn modulates the expression of target genes
and influences cellular processes (35). The circ-0001649 acts as a
molecular sponge for miR-127-5p, miR-612 and miR-4688, thereby
derepressing their target gene SHPRH and functioning as a tumor
suppressor in HCC (36). iv)
Acting as protein scaffolds. circRNAs can serve as scaffolding
agents, facilitating interactions and assembly among various
proteins. circRNAs facilitate protein-protein interactions and
promote the spatial colocalization of associated proteins by
engaging target proteins at specific subcellular compartments. This
is mediated through their intrinsic binding sites, which may
include domains for enzyme or substrate association (37). An illustration of this mechanism
is the formation of a ternary complex involving circACC1 and the
regulatory β and γ subunits of AMP-activated protein kinase (AMPK).
This association confers stabilization and potentiates the
catalytic activity of the complete AMPK holoenzyme (38). v) The ability of circRNA to
undergo translation was originally discovered by Pamudurti et
al (39) in 2017. Comprising
translatable circRNAs and their resulting products, recent studies
have highlighted that a minority of circRNAs, which have open
reading frames and internal ribosome entry sites, or those modified
by m6A in their 5' untranslated region, possess the capability for
translation into polypeptides that are crucial for cellular
functions (33,40). circRNA-encoded proteins constitute
a newly recognized regulatory layer in HCC. Acting as a key
effector from tumor-associated macrophages, circPETH drives HCC
metastasis and immune evasion via its encoded peptide (41). These proteins not only modulate
oncogenic signaling cascades and serve as critical functional
effectors in tumor biology and clinically relevant biomarkers, but
also significantly contribute to the regulation of chemotherapy
resistance in cancer cells (33).
circRNA-encoded proteins are instrumental in dictating HCC drug
resistance. Mechanistically, these proteins influence pivotal
signaling cascades, modulate key drug efflux mechanisms and engage
with the TME, thereby conferring resistance to diverse therapies
(42,43).
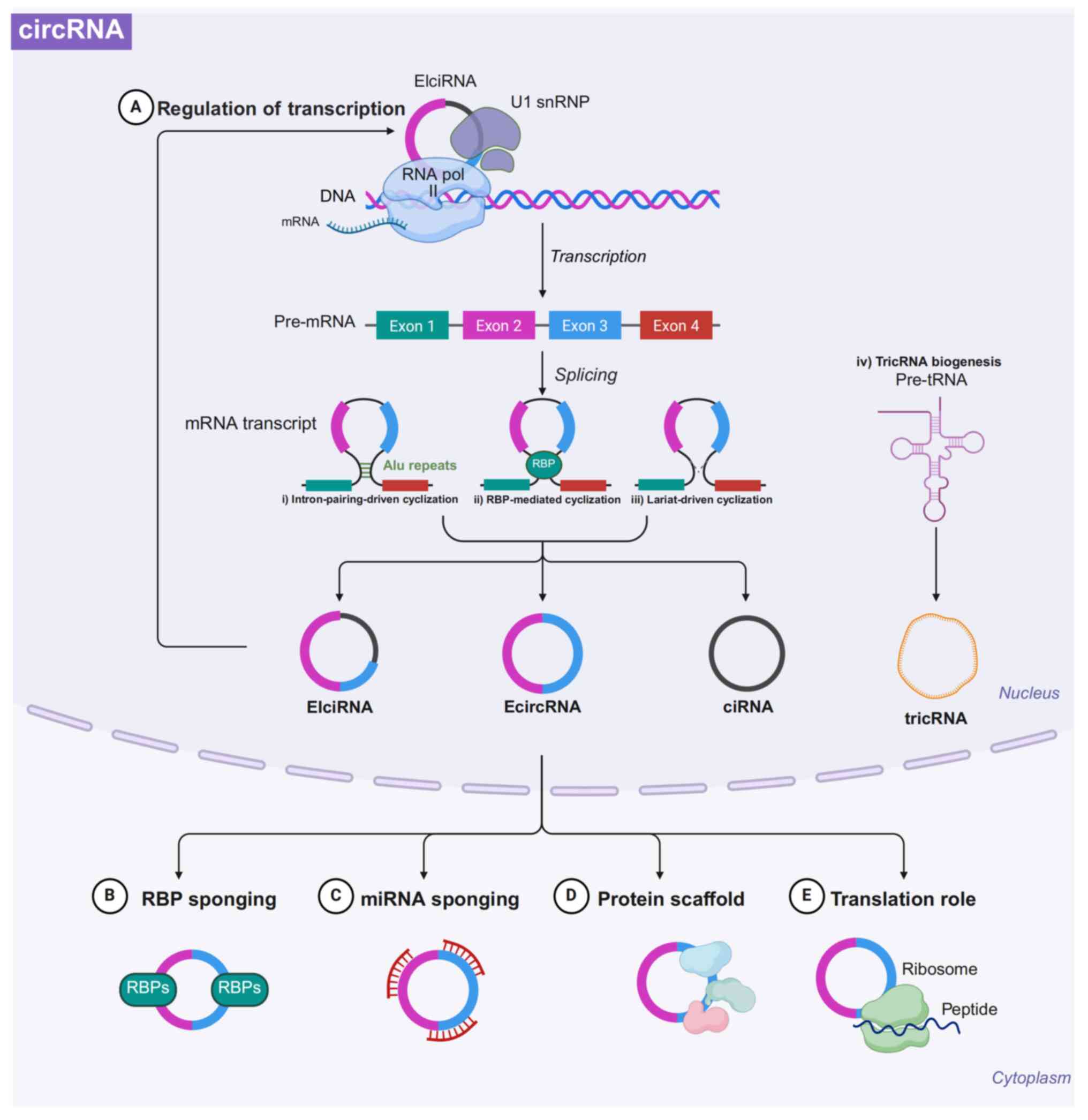 | Figure 2Biogenesis and function of circRNAs.
circRNAs are primarily generated from pre-mRNA through
back-splicing, which encompasses three unique mechanisms: i) Intron
pairing-driven cyclization, which depends on base-pairing
interactions between reverse complementary sequences, such as ALU
repeats, found in the introns adjacent to exons; ii) RBP-mediated
cyclization utilizes the involvement of RBPs to bring splicing
sites closer together, thereby facilitating the creation of
circRNAs; iii) lariat-driven cyclization aids in forming an
exon-containing lariat structure during exon skipping events,
leading to the production of EIciRNA or ecircRNA; and iv) the
generation of tricRNAs occurs through the enzymatic cleavage of
pre-tRNA, yielding tricRNAs and the remaining fragment which is
further processed into mature tRNAs. The functions of circRNAs can
be categorized into the following five classes. (A) Regulation of
RNA transcription (B) acting as sponges for RBPs (C) acting as
sponges for miRNAs (D) functioning as protein scaffolds; and (E)
translation of proteins. circRNA, circular RNA; EIciRNA,
exon-intron circRNA; EcircRNA, exonic circRNA; ciRNA, intronic
circRNA; TricRNA, tRNA intronic circular RNA; U1 snRNP, U1 small
nuclear ribonucleoprotein particles. Figure was created with
biorender.com. |
Based on the aforementioned biological functions,
circRNAs play a crucial role in key biological processes such as
cancer proliferation, invasion, metastasis and drug resistance,
highlighting them as potential targets for the early diagnosis and
treatment of various disease states (44).
Function of exosomal circRNAs in HCC
progression
Cancer markers are typically seen as a collection of
traits that human cells develop as they transition from a normal
state to one of tumor growth, and these traits are crucial for the
emergence of malignant tumors. As our understanding of the TME
evolves, the discussion surrounding cancer markers is becoming
increasingly refined. Recently, several new potential cancer
markers have emerged, including aspects such as the unlocking of
phenotypic plasticity, mutation-free epigenetic reprogramming, a
diverse microbiome and the presence of senescent cells, among
others (45). Exosomes play a
pivotal role in facilitating the transfer of circRNA, thereby
enabling signal transduction among cells. This process not only
fosters communication between nearby or distant recipient cells but
also influences several vital biological functions within those
cells. Such functions include cell proliferation, invasion and
metastasis, angiogenesis, apoptosis, metabolic reprogramming in
tumor cells and the attainment of stem cell characteristics
(Fig. 3). Recently, research on
exosomal circRNAs has shown that they play an essential role in the
development and progression of cancer (46,47). This section examines the role of
exosomal circRNAs in the development and progression of HCC,
highlighting relevant markers that are currently under extensive
investigation.
Exosomal circRNA and the proliferative
capacity of HCC cells
Sustained cell proliferation, a well-known cancer
hallmark, and the proliferative ability of HCC are crucial for its
malignant progression. In recent years, the role played by exosomal
circRNAs in regulating HCC cell proliferation has become
increasingly prominent. Certain exosomal circRNAs can exert an
inhibitory effect on the proliferative capacity of HCC, thereby
helping to impede disease progression. For example, plasma exosomes
and tissues from patients with HCC exhibit significantly lower
levels of exosomal circ-0051443 compared with healthy controls
(48). Research focusing on
molecular mechanisms has revealed that circ-0051443 is capable of
transferring from normal cells to HCC cells through exosomes; it
enhances the expression of Bcl-2 homologous antagonist/killer 1
(BAK1) in HCC cells by competing with miR-331-3p for binding sites.
Additionally, exosomal circ-0051443 reduces proliferation through
pro-apoptotic proteins and halts the cell cycle (48). A related study revealed that
exosomal hsa_circ_0004658, originating from macrophages that have
upregulated recombination signal binding protein for immunoglobulin
κJ region (RBPJ), a key transcription factor which functions as the
central effector of the Notch signaling pathway, is transferred to
HCC cells. Subsequently, it acts as a competing endogenous RNA
(ceRNA) to sponge miR-499b-5p, resulting in the inhibition of
junctional adhesion molecule 3 (JAM3) and suppression of HCC cell
growth (49).
Conversely, certain exosomal circRNAs promote the
malignancy of HCC cells, thereby facilitating disease advancement
toward a more malignant state. For example, exosomal circ-100284,
released by human L-02 liver epithelial cells that have undergone
malignant transformation due to prolonged arsenite exposure, can be
transmitted to healthy hepatocytes, enhancing cell cycle
progression and increasing proliferation via interactions with
miR-217 (50). This illustrates
that exosomal circRNAs serve as communicators for transcellular
interaction, facilitating malignant advancement in healthy
hepatocytes. Circ_0061395, an exosomal circRNA significantly
upregulated in the serum of patients with HCC, promotes tumor
progression by enhancing cell proliferation; it functions by
regulating the miR-877-5p/phosphoinositide-3-kinase regulatory
subunit 3 (PIK3R3) axis, where PIK3R3 (a key regulatory subunit of
the oncogenic PI3K signaling pathway) is upregulated. Knockdown of
circ_0061395 suppresses tumor growth in vivo and induces
cell cycle arrest, apoptosis, and inhibition of proliferation,
invasion and migration in vitro (51). Moreover, the expression of
circ_002136 is elevated in HCC tissues and cells and it can be
transmitted from one cell to another via exosomes secreted by HCC
cells (52). Functionally,
circ_002136 acts as a molecular sponge for miR-19a-3p, which leads
to the upregulation of RAB1A (an isoform of the Rab1 protein) and
consequently enhances HCC cell proliferation, migration and
invasion (52). Similarly, the
exosomal circANTXR1 released by HCC cells can promote the
proliferation and metastatic capacity of HCC by sponging miR-532-5p
to inhibit the expression of X-ray repair cross-complementary
protein 5 (XRCC5) (53); exosomal
circTTLL5 can bind to miR-136-5p, relieving its suppression on
KIAA1522 expression, thereby enhancing the proliferative ability of
HCC cells (54); and circ-ZNF652
enhances HCC cell growth via the miR-29a-3p/guanosine cyclase
domain-containing protein 1 (GUCD1) pathway (55).
In other studies, circRNAs have been shown to play
essential roles in HCC cell cycle regulation. For example,
circ_0036412 enhances the expression of GLI family zinc finger
protein 2, which is a target gene within the Hedgehog signaling
pathway. This process occurs as circ_0036412 competes with
miR-579-3p for binding, ultimately influencing the Hedgehog
signaling pathway and driving the proliferation of HCC cells as
well as their progression through the cell cycle (56). In another study, circMDK was shown
to promote HCC progression by sponging miR-346/874-3p to upregulate
autophagy related 16 like 1, a key autophagy-related protein
essential for autophagosome formation. Notably, this study was the
first to show that the poly(β-amino ester)-small interfering RNA
(siRNA) complex, a nanoparticle-based delivery system for siRNA,
targeting circMDK offers a direct and effective way to suppress
HCC, presenting a promising nanotherapeutic strategy (57).
In summary, exosomal circRNAs play the role of
signaling molecules for transcellular communication. Through this
mechanism, exosomal circRNAs can promote the proliferation of HCC
cells, ultimately contributing to the malignant progression of
HCC.
Exosomal circRNA and the invasive
metastatic ability of HCC cells
The metastatic stage represents the most critical
and life-threatening phase of tumor progression, as the majority of
cancer-related mortalities are attributed to the proliferation of
metastatic lesions at secondary sites, rather than the primary
tumor itself (58,59). Given this, significant attention
should be given to the mechanisms of tumor cell invasion and
metastasis, and targeting these mechanisms is a key means of
preventing the development of malignant tumors. Considerable
research has studied the impact of exosomal circRNAs on the
invasion and metastasis of HCC. One notable study revealed that
benzo[a]pyrene, a carcinogenic byproduct of combustion,
significantly elevates the levels of exosomal circ_0011496 secreted
from HCC cells. This particular exosomal circRNA is transferred to
lung fibroblasts, where it competitively binds to miR-486-5p. This
interaction facilitates the transformation of fibroblasts into
cancer-associated fibroblasts (CAFs) by influencing the downstream
mechanisms of Twinfilin-1 (TWF1) and matrix metalloproteinase-9
(MMP-9). Furthermore, the upregulation of TWF1 enhances the
communication between cancer cells and the surrounding matrix by
boosting the angiogenic potential of vascular endothelial growth
factor. Through both these regulatory mechanisms, exosomal
circ_0011496 promotes the metastatic ability of HCC by regulating
the TME and thus promoting metastasis (60). circPTGR1 exists in three different
isoforms and is specifically expressed in the exosomes of
metastatic HCC cells, both in low-metastatic (97L) and
high-metastatic (LM3) varieties; its expression is elevated in the
serum of patients with HCC and is linked to clinical stage and
overall prognosis. Further mechanistic investigations revealed that
circPTGR1 enhances the expression of MET proto-oncogene, receptor
tyrosine kinase (MET), encoding the hepatocyte growth factor
receptor, a tyrosine kinase known to promote cell motility and
invasion. This upregulation, mediated via the sponging of miR-449a,
consequently increases the invasive and metastatic potential of HCC
cells (61).
EMT is a key process in cancer cell metastasis,
which enables cancer cells to break through the basement membrane,
dismantle intercellular junctions, downregulate E-calmodulin and
break free from the primary site restrictions, ultimately allowing
the cancer cells to migrate and invade and obtain mesenchymal cell
properties, which further facilitates their mobility in tissues
(62). This helps these cancerous
cells to enter the circulatory system, where they are carried to
distant sites through the bloodstream or lymphatic flow. Exosomal
circRNAs serve as key regulators in EMT. Numerous reports have
revealed that exosomal circRNAs can enhance the invasive and
metastatic capabilities of HCC by influencing EMT (63-66). Previous findings indicate that
circ-0004277 is markedly elevated in HCC cells, tissues and plasma
exosomes. Upregulated expression of circ-0004277 promotes the
proliferation, migration and EMT of HCC cells both in vivo
and in vitro. Mechanistically, circ-0004277 contributes to
the aggressive traits of HCC cells by competing for binding to Hu
antigen R (HuR) proteins, leading to the inhibition of tight
junction protein 1 and promoting EMT progression. Notably, exosomal
circ-0004277 can be secreted by HCC and serves as a transcellular
communication molecule to enhance the EMT process in peripheral
cells, further escalating the invasive and metastatic potential of
HCC (67). In a separate study,
the suppression of hsa_circ_0074854 similarly inhibited the
migration and invasion of HCC cells by interacting with HuR.
Moreover, the exosomes secreted by HCC cells following the
downregulation of hsa_circ_0074854 impeded M2 macrophage
polarization, thereby hampering the migration and invasion of HCC
cells both in vitro and in vivo (68). Similarly, exosome-derived circWDR2
from hepatic stellate cells advanced HCC development via regulation
of the circWDR25/miR-4474-3p/arachidonate 15-lipoxygenase axis and
EMT pathways (69). Additionally,
circ_0003028 was shown to exert tumorigenic effects through the
miR-498/ornithine decarboxylase 1 (ODC1) signaling axis and induced
EMT of HCC cells via an exosomal pathway (70). Together, these studies show that
exosomal circRNAs can influence EMT in HCC cells, affecting the
invasion and metastasis of these cells. Exosomal circRNAs may serve
as a biomarker for HCC invasion, metastasis and malignant
advancement.
In other studies, exosomal circRNAs have been shown
to affect the invasive and metastatic ability of HCC cells. For
instance, highly metastatic HCC cell lines, particularly the 97H
and LM3 variants, release exosomal circ_MMP2, which targets L02 and
HepG2 cells. This process enhances the metastatic potential of HCC
cells by increasing the expression of MMP2 by sponging miR-136-5p
(71). It has been reported that
high-abundance exosomal circ_002136 targets the miR-19a-3p/RAB1A
pathway in cancer cells, disrupts the stable microenvironment
inside tumors, exacerbates the malignant progression of HCC tumors
and can promote the higher invasive capacity of recipient cells
through exosome delivery (52).
circRNA Cdr1as has been found to be significantly upregulated in
both HCC cell lines and tissues, and its upregulation enhances the
proliferation and migration of HCC cells. Mechanistically, Cdr1as
facilitates the upregulation of human α-fetoprotein by sponging
miR-1270 and it promotes the proliferation and migratory capacity
of normal cells nearby via exosomal transport (72). Similarly, circANTXR1 is stably and
highly expressed in HCC, where it positively regulates XRCC5 to
promote the proliferation and metastasis of HCC cells by sponging
miR-532-5p and mediates transcellular communication via exosomes
(53). Notably, exosomal circRNAs
can also hamper the invasive and migratory capacity of HCC cells.
For example, exosomal circ-0072088 secreted by HCC cells can impede
HCC metastasis by facilitating the degradation of miR-375 and
increasing the expression of MMP16 (73).
As outlined above, exosomal circRNA may serve as
diagnostic and/or prognostic indicators of HCC invasion and
metastasis and may serve as promising therapeutic targets for the
management of HCC.
Exosomal circRNAs and HCC
angiogenesis
Tumor angiogenesis plays a crucial role throughout
the life cycle of a tumor, from initial occurrence to subsequent
gradual development to infiltration of surrounding tissues and
metastasis to distant sites (74). Exosomal circRNAs originating from
HCC cells can foster the development of vascular endothelial cells
and stimulate the creation of new vasculature, which in turn aids
in the distant metastasis of tumor cells (75). An earlier study has indicated that
circRNA-100338 derived from HCC cells influences the proliferation
of cells, angiogenesis, permeability and the ability to form new
vasculature in human umbilical vein endothelial cells (HUVECs)
through both in vitro and in vivo analyses.
Mechanistic experiments have shown that circRNA-100338, which is
delivered via exosomes, can be transferred to HUVECs and bind to
its target receptors to promote tumor neoangiogenesis through the
mammalian target of rapamycin (mTOR) signaling pathway, thereby
promoting HCC progression (76).
circPAK1 is a newly discovered circRNA significantly expressed in
HCC tissues and cell lines and is correlated with poor prognosis in
patients with HCC. In vitro experiments have demonstrated
that circPAK1 is capable of promoting angiogenesis in HCC in a
controlled environment; it achieves this by competing with
Yes-associated protein (YAP) to bind to 14-3-3ζ, facilitating the
nuclear localization of YAP. This process ultimately contributes to
the advancement of HCC by inhibiting the Hippo signaling pathway
(77). Moreover, circDCAF8 is
transferred from HCC cells to HUVECs through exosomes, which
stimulates angiogenesis in HCC (19). Exosomal circCMTM3 directly binds
to miR-3619-5p, targeting the downstream sex-determining region Y
(SRY)-box transcription factor 9 (SOX9). This interaction acts as a
catalyst for the proliferation, migration, invasion and
angiogenesis of HUVECs; it also promotes their viability and
induces tumor growth (78). A
recent study showed that circHDAC1_004 expression is upregulated in
HUVECs, where it promotes HCC angiogenesis through exosomes
(79).
These studies show that exosomal circRNAs produced
by HCC can be transferred to HUVECs using exosomes as a vector and
then bind to the corresponding target receptors to induce HUVEC
proliferation and promote the formation of new blood vessels. In
turn, the new blood vessels enhance the proliferation, invasion and
metastasis of HCC cells, ultimately leading to the malignant
progression of HCC. Consequently, focusing on exosomal circRNAs
could be an effective approach for reducing tumor metastasis and
improving sensitivity to therapy.
Exosomal circRNAs and HCC cell
apoptosis
Apoptosis results in the removal of damaged cells
and can prevent the proliferation of abnormal cells. Apoptosis is
an important mechanism for preventing tumorigenesis; however, a
hallmark of cancer is overcoming apoptosis. The levels of
circ-0051443 were found to be downregulated in HCC tissues and the
plasma exosomes of patients with HCC. Conversely, increasing its
expression resulted in cell cycle arrest during the G0/G1 phase and
facilitated apoptosis in HCC cells (48). BAK1 plays a pivotal role in
regulating cell death by triggering mitochondria-dependent
apoptosis through various protein interactions (80). In a study, exosomes that carried
circ-0051443 secreted by normal cells, were taken up by HCC cells.
Within these cells, circ-0051443 acted as a sponge for miR-331-3p,
thereby modulating BAK1 and promoting apoptosis in HCC cells, which
in turn inhibited the advancement of HCC (48). In another study, exosomal circRNA
derived from macrophages with upregulated RBPJ expression,
specifically hsa_circ_0004658, hampered the proliferation of HCC
cells and promoted apoptosis. hsa_circ_0004658 sponged miR-499b-5p,
thereby enhancing the expression of JAM3 (49). In addition, circ_0003028, which is
upregulated in HCC tissues and cells, has been shown to promote the
expression of ODC1 by targeting miR-498, thus playing a role in
tumorigenesis. Research has indicated that silencing circ_0003028
can rein in cell proliferation and metastasis, trigger apoptosis
and offer promising biomarkers and therapeutic targets for HCC
(70).
In summary, numerous studies have reported the
regulation of HCC cell apoptosis by circRNAs. However, the
mechanism by which exosomal circRNAs mediate HCC cell apoptosis is
incompletely understood and likely involves the interplay of
several circRNAs and the regulation of several signaling
pathways.
Exosomal circRNAs and HCC metabolism
Considerable research has focused on the association
of alterations in the metabolism of malignant cells. During the
development and progression of a tumor, the metabolic state of its
cells undergoes a series of adaptive changes known as tumor cell
metabolic reprogramming. These adaptations are of vital
significance in the emergence and advancement of tumors, allowing
cells to respond and adapt to changing conditions. Exosomal
circRNAs primarily influence the glycolysis of HCC cells, a
phenomenon also known as the Warburg effect. This effect is
characterized by a metabolic shift in cancer cells towards aerobic
glycolysis, leading to lactate production (81). For example, exosomal circ-ZNF652
is upregulated in the serum of patients with HCC and can be
delivered to HCC cells via exosomes. Mechanistic investigations
have revealed that exosomal circ-ZNF652 enhances the glycolytic
levels of HCC cells by acting as a sponge for miR-29a-3p to target
GUCD1 (55). Exosomal circFBLIM1
has the same effect. circFBLIM1 is upregulated in serum-derived
exosomes and HCC cells. CircFBLIM1 promotes glycolysis in HCC cells
by sponging miR-338, resulting in the upregulation of low-density
lipoprotein receptor-related protein 6 and a consequent enhancement
of glucose utilization. This previous study further established a
xenograft mouse model to verify that the depletion of circFBLIM1
inhibited the advancement of HCC cells and tumor development in
vivo (82).
A recent study showed that peptides encoded by
circRNAs can also affect metabolic processes in HCC cells. The
study revealed that tumor-associated macrophages promote glycolysis
and the advancement of tumors by increasing the expression of
circMRCKα in HCC cells. This particular circRNA can encode a novel
functional peptide consisting of 227 amino acids, referred to as
circMRCKα-227aa. On a mechanistic level, circMRCKα-227aa interacts
with ubiquitin-specific peptidase 22, elevating its protein
expression levels. This interaction prevents the degradation of
hypoxia-inducible factor 1 α, a key transcription factor that
mediates cellular responses to low oxygen, via the
ubiquitin-proteasome pathway, ultimately boosting glycolysis and
tumor progression in HCC (83).
Previous circRNA research has focused on their functioning as miRNA
sponges. However, research on the role of the peptides encoded by
circRNAs is now gaining traction.
In addition to affecting glucose metabolism, certain
circRNAs can affect lipid metabolism in HCC cells. For example,
circLARP1B promotes cell metastasis and lipid accumulation by
promoting fatty acid synthesis in HCC. Mechanistically, circLARP1B
interacts with heterogeneous nuclear ribonucleoprotein D (HNRNPD)
in the cytoplasm. This interaction results in the binding of HNRNPD
to liver kinase B1 (LKB1) mRNA, ultimately destabilizing it and
lowering LKB1 protein levels. Consequently, this process influences
the AMPK pathway, facilitating the metastasis of HCC and impacting
lipid metabolism (84). Of note,
in another study, an effective traditional Chinese medicine, FZXZP,
significantly inhibited HCC growth by improving lipid and glucose
metabolism in HCC cells and regulating a circRNA-miRNA-mRNA network
to restore metabolic homeostasis (85). This highlights a potential
treatment for targeting metabolic reprogramming in HCC cells.
The idea of targeting the circRNAs associated with
the metabolic reprogramming of HCC cells for therapeutic benefits
remains in the exploratory phase and requires in-depth
investigation. Moreover, the role of exosomal circRNAs in the
metabolic processes of HCC cells, apart from the recognized
metabolic pathways, remains unclear. Additionally, in-depth
analysis and interpretations are required.
Exosomal circRNAs and HCC stem cells
CSCs represent a distinctive subset within the
broader category of cancer cells. These cells possess the
remarkable ability to self-renew and play a crucial role in the
onset and progression of cancer. CSCs are instrumental in driving
the metastatic spread of tumors and contribute to the challenges of
treatment resistance and recurrence. Recent research indicates that
circRNAs from exosomes released by liver cancer CSCs are crucial in
HCC progression. For instance, exosomal circ-ZEB1 and circ-AFAP1,
which are primarily secreted by liver cancer CSCs, are markedly
upregulated in HCC tissues compared with adjacent non-cancerous
tissues. Moreover, there is a positive correlation between their
expression levels and that of the stemness marker CD133 in HCC
cells, indicating their potential role in processes related to
stemness within these cancerous cells. Subsequent experiments have
demonstrated that upregulation of circ-AFAP1 not only promotes
tumor growth but also increases the stemness of HCC cells and EMT.
This shows that circ-ZEB1 and circ-AFAP1, present in the exosomes
of liver CSCs, may facilitate the malignant transfer between CSCs
and non-CSCs. Thus, they may mediate the increase in malignancy of
non-CSCs and the poor prognosis of patients with HCC by enhancing
the stemness and EMT process of HCC (86).
circIPO11, which is highly expressed in HCC tumor
tissues and liver CSCs, is necessary for the self-renewal of liver
cancer CSCs; it functions by recruiting DNA topoisomerase I to the
promoter region of the glioma-associated oncogene homolog 1 (GLI1)
gene. This recruitment consequently triggers GLI1 transcription,
leading to the activation of the Hedgehog signaling pathway, which
in turn drives both the self-renewal of liver CSCs and HCC
progression (87). Certain highly
expressed circRNAs can enhance the stemness of HCC cells, while the
downregulation of other circRNAs has the same effect. circZKSCAN1,
which is downregulated by the quaking protein in HCC cells, serves
as a promising regulator of stemness in HCC and can inhibit
multiple malignant behaviors by suppressing stemness. circZKSCAN1
functions as a sponge for RBPs, competing with fragile X mental
retardation protein to inhibit the Wnt/β-catenin signaling pathway,
ultimately leading to its inactivation (88). The Hedgehog and Wnt signaling
pathways are critically involved in tumorigenesis, progression and
therapeutic response (89). These
pathways participate in crucial processes, including tumor cell
proliferation, differentiation, invasion, metastasis and the
maintenance of CSC properties (90). Following aberrant activation of
these pathways, CSCs promote cancer progression via their stem cell
characteristics, such as continuous self-renewal and drug
resistance, hampering the effectiveness of cancer treatments. These
pathways also make CSCs the main targets of anti-CSC therapy
(91).
Taken together, circRNA can affect the stem cell
phenotype of HCC cells by influencing key effector molecules in
signaling pathways, indicating that targeting circRNAs may be an
essential means of addressing naturally drug-resistant CSCs,
reducing cancer recurrence rates and increasing the effectiveness
of tumor therapies. Further studies are necessary to understand the
means by which exosomal circRNAs facilitate communication between
HCC CSCs and their non-stem cell counterparts, ultimately fostering
the increased stemness of HCC cells.
Effects of exosomal circRNAs on drug
resistance
The current treatment options for HCC primarily
consist of chemotherapy, radiotherapy, targeted therapy,
immunotherapy and novel cell and gene therapies. In clinical
treatment, the commonly used chemotherapeutic drugs are cisplatin,
oxaliplatin and doxorubicin. In terms of targeted therapy,
sorafenib, lenvatinib and regorafenib are widely employed molecular
targeted drugs. Immunotherapy, as a relatively novel treatment
method, primarily involves the selection of immune checkpoint
inhibitors/monoclonal antibodies such as programmed cell death
protein 1 (PD-1), PD-1 ligand (PD-L1) and receptor cytotoxic T
lymphocyte antigen-4 (92).
Furthermore, while studies on cell and gene therapy of the tumor
immune microenvironment exist, the research is relatively limited.
However, HCC can develop drug resistance, resulting in the failure
of treatments. In this section, the effects of exosomal circRNAs on
the drug resistance of HCC (Fig.
4 and Table SI) are
discussed.
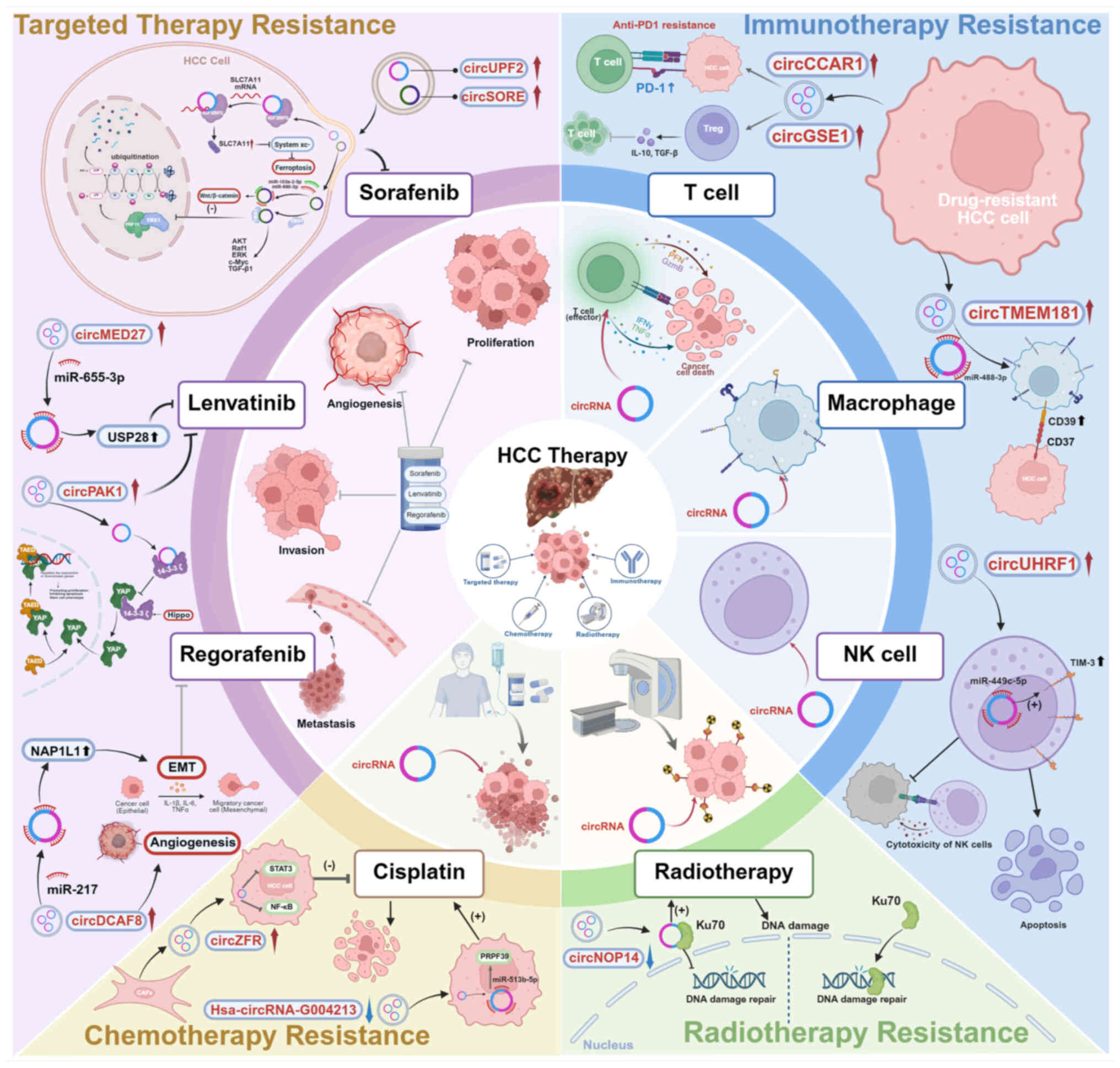 | Figure 4Exosomal circRNAs related to
therapeutic resistance in HCC. The mechanism of representative
exosomal circRNAs that show regulatory effects in chemotherapy,
radiotherapy, targeted therapy and immunotherapy resistance of HCC.
circRNA, circular RNA; SLC7A11, solute carrier family 7 member 11;
IGF2BP2, insulin-like growth factor 2 mRNA-binding protein 2;
USP28, ubiquitin specific peptidase 28; YAP, Yes-associated
protein; NAP1L1, nucleosome assembly protein 1 like 1; EMT,
epithelial-mesenchymal transition; Ku70, Ku autoantigen, 70kD.
Figure was created with biorender.com. |
Exosomal circRNAs in HCC chemotherapy
treatment resistance
Chemotherapy is a cornerstone treatment in the
management of malignant tumors; however, the challenge of acquired
drug resistance undermines its effectiveness, resulting in the
setback of treatment for several patients. Research has highlighted
that exosomal circRNAs play a crucial role in the development of
chemotherapy resistance in a range of cancer types (93).
Cisplatin remains the primary chemotherapeutic agent
in the clinical management of HCC, yet resistance poses a
significant challenge to its effectiveness. Research has
established that exosomal circ-G004213 is positively associated
with the prognosis of patients with HCC following transarterial
chemoembolization. Specifically, it enhances cisplatin sensitivity
by sponging miR-513b-5p to upregulate pre-mRNA processing factor 39
(PRPF39), a critical splicing factor, thereby modulating the
miR-513b-5p/PRPF39 axis (94).
circZFR is upregulated in CAFs and CAF exosomes. Mechanistically,
it can be delivered by exosomes and it suppresses the STAT3/NF-κB
pathway in HCC cells, in turn facilitating HCC progression and
cisplatin resistance (95). The
STAT3/NF-κB pathway is a key pathway in HCC, and the crosstalk
between the two in the development of HCC is an essential factor in
HCC progression (96). circARNT2,
which is upregulated in HCC, can regulate cisplatin resistance in
HCC cells through the miR-155-5p/pyruvate dehydrogenase kinase 1
(PDK1) axis, where PDK1 is a central metabolic kinase that shifts
energy production toward glycolysis and confers survival advantages
to cancer cells (97). circRNAs
can also promote cisplatin sensitivity in HCC cells. For example,
when circRNA_101505 is upregulated in HCC cells, it sponges
miR-103, resulting in the upregulated expression of oxidored-nitro
domain-containing protein 1, increasing the sensitivity of HCC
cells to cisplatin (66).
circ_0003418, an antitumor circRNA, is downregulated in HCC tissues
and cell lines; it can enhance the cisplatin sensitivity of HCC
cells by activating the Wnt/β-catenin pathway within these cells
(98).
In studies of other cancer types, exosomal circRNAs
can also affect the cisplatin resistance of tumor cells. For
example, exosomal circVMP1 promotes the progression and cisplatin
resistance of non-small cell lung cancer (NSCLC) by targeting the
miR-524-5p/methyltransferase-like 3/SOX2 axis (99); exosomal circ_PIP5K1A modulates the
miR-101/ATP binding cassette subfamily C member 1 (ABCC1) pathway
to enhance the advancement and cisplatin resistance of NSCLC
(99,100). ABCC1 is a multidrug resistance
protein which exports chemotherapeutic agents such as cisplatin out
of cancer cells, thereby conferring treatment resistance (101). In esophageal cancer,
exosome-mediated circ_0000337 transfer regulates Janus kinase 2
(JAK2; a non-receptor tyrosine kinase and pivotal component of the
JAK-STAT signaling pathway) via miR-377-3p, promoting resistance to
cisplatin in esophageal cancer cells (102).
It has been shown that circ_0000098 can increase
doxorubicin (DOX) resistance by sponging miR-383 to increase the
expression of P-glycoprotein and intracellular ATP levels in HCC
cells. More notably, it was demonstrated that DOX/sh-1@PLT,
produced by encapsulating DOX and short hairpin RNA (sh-1)
targeting circ_0000098 using platelets (PLTs) as drug carriers,
showed favorable therapeutic effects in mouse models (103), and thus provides guiding
significance for the use of targeted circRNAs in the management of
HCC. As for oxaliplatin (OXA) resistance, a study has shown that
hsa_circ_0088036 promotes HCC tumorigenesis and OXA resistance by
activating the PI3K/Akt and Notch pathways through regulation of
the miR-140-3p/KIF2A signaling cascade (104); circFBXO11 promotes OXA
resistance in HCC via the circFBXO11/miR-605/FOXO3/ABCB1 axis
(105).
Based on the current research progress, exosomal
circRNAs have been confirmed to play a crucial regulatory role by
mediating resistance to chemotherapy in HCC and are hypothesized to
serve as potential interventional targets for reversing resistance
to chemotherapy. However, the specific molecular mechanism by which
drug-resistant phenotypes mediated by exosomal circRNA are
transmitted between HCC cells remains relatively understudied. This
direction urgently requires more in-depth studies. Understanding
the mechanisms will not only help to clarify the drug-resistant
transmission networks at play in the TME but may also highlight a
theoretical basis for the clinical development of novel diagnostic
markers or treatment strategies, which have scientific significance
and clinical applications.
Exosomal circRNAs in HCC radiotherapy
treatment resistance
Radiotherapy is a widely used treatment for solid
tumors, which suppresses tumor progression primarily by inducing
DNA damage in tumor cells. Research has indicated that
125I seed brachytherapy, as a burgeoning adjuvant
therapy, holds great promise for treating HCC (106). Recent research has revealed that
elevated serum levels of exosomal circTMEM56 are positively
correlated with improved radiotherapy response and favorable
clinical outcomes in patients with HCC. Mechanistically, elevated
exosomal circTMEM56 specifically targets dendritic cells, thereby
enhancing radiotherapy-induced antitumor immune responses in HCC.
This occurs through the promotion of cyclic GMP-AMP synthase
(cGAS)-stimulator of interferon genes (STING) pathway activation
via the miR-136-5p/STING axis, effectively remodeling TME (107). cGAS is characterized as a key
DNA sensor that initiates innate immune responses by producing the
secondary messenger cyclic GMP-AMP, which subsequently binds to and
activates the adaptor protein STING. The cGAS-STING pathway
represents an attractive therapeutic target, as it functions as an
intrinsic barrier to tumorigenesis by linking DNA damage to
multiple antitumor mechanisms, including immune surveillance,
cellular senescence and cell death (108). By focusing on the interplay
between exosomal circTMEM56 and the cGAS-STING pathway, the
aforementioned study introduces a novel approach to augment the
efficacy of radiotherapy in HCC treatment (107). Another significant novel
discovery is circSNX25 (has_circ_0004874), a circRNA identified
with coding potential that is strongly associated with HCC
radioresistance. The novel protein SNX25-215, encoded by circSNX25,
promotes HCC cell resistance to radiotherapy both in vitro
and in vivo. The underlying mechanism of circSNX25 involves
the interaction of the amino acid residues H207 and E214 of
SNX25-215 with the Golgi-associated endoplasmic reticulum
transporter 4 homolog (GET4). This interaction inhibits the binding
of BCL2-associated anaplastic gene 6 (BAG6) to GET4. Crucially,
this disruption exposes the nuclear localization sequence of BAG6,
which subsequently facilitates its nuclear translocation. Nuclear
BAG6 then enhances DNA damage repair, ultimately resulting in an
increased resistance of HCC cells to ionizing radiation and thereby
augmenting overall HCC radioresistance (43). A study revealed that exosomal
circNOP14 is strongly associated with the radiosensitivity of HCC
cells; it can enhance the radiosensitivity of HCC cells both in
vitro and in vivo (109). Ku70 is abundantly expressed and
plays a role in the DNA damage response (DDR). Specifically, it is
involved in the C-NHEJ pathway, which is the most prevalent DNA
double-strand break repair pathway, to fix the DNA damage caused by
radiotherapy (110). circNOP14,
conversely, increases the sensitivity to radiotherapy by
interacting with Ku70 and preventing its nuclear translocation,
thereby increasing irradiation-induced DNA damage (109). Other research has indicated that
cZNF292 activates the Wnt/β-catenin pathway by interacting with
SOX9 protein to inhibit it's nuclear translocation, enhancing DDR
capacity and radiotherapy resistance in HCC cells (111); circ-LARP1B silencing suppresses
HCC tumorigenesis and increases radiosensitivity by modulating the
miR-578/insulin-like growth factor 1 receptor axis (112); circSMARCA5 is markedly
upregulated in HCC cells and has been shown to promote resistance
to radiotherapy (113);
circ_0071662 downregulation in HCC cells may be associated with the
development of radiotherapy resistance (114); and high expression of circROBO1
in HCC can promote HCC radiotherapy resistance by targeting
miR-136-5p to regulate a novel adhesion protein core subunit RAD21
(115).
Exosomal circRNAs also play a crucial role in
inducing radiotherapy resistance in other types of cancer. For
example, in endometrial cancer (EC), M2-polarized tumor-associated
macrophages can markedly reduce the radiosensitivity of EC cells by
releasing exosomes rich in hsa_circ_0001610. Among these exosomes,
hsa_circ_0001610 attenuates the radiosensitivity of EC cells by
binding miR-139-5p to allow cyclin B1 expression (116). In glioblastoma (GBM), low-dose
radiation-induced exosomal circ-METRN can promote GBM progression
and radiation resistance via the miR-4709-3p/growth factor receptor
bound protein 14 (GRB14)/platelet derived growth factor receptor α
(PDGFRα) pathway (117), where
GRB14 is a signaling adapter protein and PDGFRα is a key tyrosine
kinase receptor for cell proliferation.
Radiotherapy can inhibit tumor progression by
inducing DNA damage, and exosomal circRNA can significantly affect
tumor radiosensitivity by regulating the DDR. The aforementioned
findings suggest that exosomal circRNAs are key regulators of the
response of a tumor to radiotherapy, and analysis of their
mechanisms may highlight novel targets for developing resistance
reversal strategies or increasing radiosensitization.
Exosomal circRNAs in targeted treatment
resistance in HCC Sorafenib
Sorafenib was the first molecular-targeted drug
approved by the FDA for clinical use as first-line therapy for
advanced HCC. However, after a period of sorafenib therapy, drug
resistance may emerge (118).
Resistance limits the therapeutic effectiveness of sorafenib and
severely impacts the prognosis of patients with HCC. Recently,
numerous studies have indicated that exosomal circRNAs affect the
emergence of sorafenib resistance in HCC (Table SI). For example, circUPF2 is
significantly enriched in exosomes produced by sorafenib-resistant
HCC cell lines and can transmit resistance to non-resistant HCC
cells via exosomes. circUPF2-enriched exosomes increase resistance
to sorafenib by upregulating solute carrier family 7 member 11
(SLC7A11, also known as xCT) expression and preventing ferroptosis
in HCC cells. Subsequent research on the resistance mechanism
showed that circUPF2 serves as a scaffolding structure to promote
the formation of the circUPF2-insulin-like growth factor 2
mRNA-binding protein 2-SLC7A11 ternary complex. This complex, in
turn, increases SLC7A11 expression (a cystine/glutamate reverse
transporter protein) and enhances the functionality of System Xc-
in HCC cells. As a result, the cells become less prone to
ferroptosis and more resistant to sorafenib (17). The increased activity of System
Xc-, which is closely associated with the advancement of HCC,
comprises a transport component (SLC7A11) alongside a heavy chain
element referred to as 4F2hc (SLC3A2). This mechanism plays a
crucial role in exchanging extracellular L-cystine for
intracellular L-glutamate, with SLC7A11 primarily driving the
functional role of the transporter (119-122). A recent study identified
circTTC13, which is upregulated in HCC tissue and is associated
with tumor malignancy and the ferroptosis of tumor cells. Further
investigation of the mechanism revealed that circTTC13 directly
targets and reduces the expression of miR-513a-5p, leading to the
upregulation of SLC7A11 to inhibit the ferroptosis process in HCC
cells. In addition, silencing circTTC13 induces tumor ferroptosis
and reverses sorafenib resistance in HCC (123). Another study showed that
circRNA-SORE, upregulated in sorafenib-resistant HCC, is loaded
into exosomes, facilitating the transfer of sorafenib resistance to
HCC cells. On a mechanistic level, circRNA-SORE interacts with the
key oncogenic protein Y-box binding protein-1 (YBX1) within the
cytoplasm (18). YBX1, commonly
referred to as YB-1, is strongly associated with tumor
proliferation, resistance to drugs, cancer advancement and overall
prognosis across multiple cancer types (124). Notably, it is circRNA-SORE that
stabilizes YBX1 by impeding its PRP19-mediated ubiquitination and
degradation. The stabilized YBX1 then promotes the expression of
its downstream oncogenic targets, such as AKT, Raf1, ERK, c-Myc and
TGF-β1 (125,126), thereby driving sorafenib
resistance. It was previously shown that sorafenib resistance could
be effectively tackled and the therapeutic effectiveness could be
increased in various HCC mouse models by locally injecting siRNA to
silence circRNA-SORE (18). In
another study, it was demonstrated that circRNA-SORE can
competitively trigger the Wnt/β-catenin pathway and induce
sorafenib resistance by functioning as an miRNA sponge that
specifically binds to miR-103a-2-5p and miR-660-3p (8). New research indicates that CSF3R-AS,
an antisense circRNA, is significantly upregulated in HCC and is
strongly correlated with a poor patient prognosis. Mechanistically,
CSF3R-AS functions by directly binding to its parental mRNA, CSF3R,
through base-complementary pairing sites. Crucially, CSF3R-AS acts
as an intermediate scaffolding molecule to recruit the RBP RBMS3, a
key post-transcriptional regulator, which subsequently leads to the
stabilization of CSF3R mRNA. This stabilization further promotes
the activation of the downstream JAK2/STAT3 signaling pathway,
thereby enhancing acquired resistance to sorafenib in HCC cells
(127).
Additionally, certain circRNAs can boost the
sensitivity of HCC cells to sorafenib. circMEMO1 is significantly
downregulated and its expression levels are an independent
prognostic factor for patients with HCC. From a mechanistic
perspective, circMEMO1 inhibits HCC metastasis and stemness by
regulating the miR-106b-5p/ten-eleven translocation methylcytosine
dioxygenase 1 (TET1)/5-hydroxymethylcytosine (5hmC) axis and
suppressing the EMT process. Specifically, it upregulates TET1, an
enzyme that catalyzes the oxidation of 5-methylcytosine to form
5hmC, thereby initiating active DNA demethylation and altering the
transcriptional program (128).
Moreover, it also modulates the sensitivity of HCC cells to
sorafenib treatment (129).
circRNA-encoded secreted peptides can similarly influence the
progression of sorafenib resistance. A recent study found that
circZKSaa, a peptide secreted by circZKSCAN1, plays a crucial role
in reducing HCC progression while also increasing the sensitivity
of HCC cells to sorafenib. Through mechanistic investigations, it
was found that the overexpression of circZKSaa increased the
interaction between F-box and WD repeat domain-containing 7, a
critical tumor-suppressing E3 ubiquitin ligase, and mTOR. This
interaction facilitated the ubiquitination of mTOR, leading to the
inhibition of the PI3K/AKT/mTOR signaling pathway, which ultimately
influenced how HCC cells responded to sorafenib treatment (42).
Lenvatinib
Lenvatinib is the second first-line agent approved
for advanced HCC after sorafenib. Lenvatinib demonstrates
non-inferior overall survival compared with sorafenib in patients
with previously untreated advanced HCC (130). Furthermore, it represents a
valuable therapeutic option for patients with HCC recurrence
following liver transplantation (131). Although the initial
effectiveness of lenvatinib in sorafenib-resistant HCC cells seems
promising, based on incomplete data, >60% of patients with HCC
become tolerant to lenvatinib within a year and only a small
proportion of patients show long-term advantages (132). Exosomal circRNAs are an
essential factor in the development of resistance to levatinib.
According to a recent study, circPAK1 expression is upregulated in
HCC tumor tissues and cell lines, correlating with a poor prognosis
in patients with HCC. The overexpression of circPAK1 accelerates
HCC progression, while reducing its levels results in a decrease in
cell proliferation, migration, invasion and angiogenesis.
Mechanistically, circPAK1 facilitates YAP nuclear localization by
competitively binding 14-3-3ζ of YAP, ultimately disrupting the
Hippo signaling pathway. Furthermore, circPAK1 is transferred by
exosomes from levatinib-resistant cells to those sensitive to the
drug, thereby promoting drug resistance in the recipient cells.
More notably, this study found that applying chitosan (a
biocompatible polysaccharide nanomaterial)/si-circPAK1 nano
complexes showed favorable therapeutic effects on tumor growth and
metastasis. This provides a good guideline for developing novel
therapeutic targets for HCC (77). Expression of exosomal circMED27 is
markedly increased in the serum of patients with HCC, and higher
circMED27 levels are correlated with unfavorable clinical features
and prognosis in these patients. Upregulated circMED27 functions as
a ceRNA for miR-655-3p to upregulate the expression of
ubiquitin-specific peptidase 28 and promotes lenvatinib resistance
in HCC via this mechanism (133). An investigation highlighted the
clinical and functional significance of circRNA-mTOR, a circular
RNA highly expressed in HCC and strongly correlated with adverse
patient prognosis. Mechanistically, circRNA-mTOR exerts its
function by specifically binding to pro-RBPs PC4 and SRSF1
interacting protein 1 (PSIP1), thereby modulating the nuclear
translocation of PSIP1. This molecular event enhances HCC cell
stemness through the subsequent activation of the PSIP1/c-Myc
signaling axis. Consequently, this pathway ultimately drives HCC
progression and confers resistance to lenvatinib, establishing
circRNA-mTOR as a critical regulatory factor in the malignancy of
HCC (134). A recent study
revealed the molecular mechanism by which circCCNY enhances the
therapeutic effect of HCC through a dual mechanism. First, circCCNY
promotes the ubiquitin-mediated degradation of HSP60 by recruiting
SMURF1, a pivotal E3 ubiquitin ligase that targets specific
substrates for proteasomal degradation, leading to the release of
Raf kinase-inhibitory protein, which in turn inhibits the MAPK
signaling pathway and significantly enhances the antitumor effect
of lenvatinib. Second, circCCNY promotes the infiltration of
CD8+ T cells in the TME by blocking the MAPK/c-Myc/PD-L1
signaling axis, effectively inhibiting immune escape. These
findings suggest that circCCNY not only enhances the sensitivity of
HCC to targeted therapy but also regulates the tumor immune
microenvironment and could potentially serve as a novel target for
the combination therapy of HCC (135). Significant upregulation of
hsa_circ_0007132 was observed in serum exosomes from patients with
HCC with post-lenvatinib progression. Mechanistically,
hsa_circ_0007132 binding to the non-POU domain containing octamer
binding (NONO) protein inhibits its ubiquitination, resulting in
increased NONO stability and expression. NONO is involved in
various steps of RNA metabolism and its stabilization here
facilitates the nuclear export of zinc finger e-box binding
homeobox 1 (ZEB1) mRNA (136).
The transcription factor ZEB1 then acts as a master regulator of
EMT, a process strongly linked to drug resistance (137), thereby elevating ZEB1 levels and
ultimately mediating lenvatinib resistance (138). circPIK3C3 exhibits significantly
reduced expression in HCC and is correlated with patient prognosis,
functioning to attenuate lenvatinib resistance. Both in
vitro and in vivo investigations established that
circPIK3C3 acts as a ceRNA to sequester miR-452-5p, resulting in
the enhanced expression of SOX15. Concomitantly, this regulatory
mechanism effectively inhibits Wnt/β-catenin-related signaling
pathways. The combined effect of these actions mitigates HCC
progression and reverses acquired resistance to lenvatinib,
highlighting circPIK3C3 as a crucial suppressor of malignancy in
HCC (139). As a potential
frontline target in HCC therapeutics, the impact of exosomal
circRNA on the development of levatinib resistance in HCC still
requires further investigation.
Regorafenib
Regorafenib serves as a second-line treatment option
for individuals with advanced HCC who have already undergone
therapy with sorafenib. Regorafenib stands out as the sole systemic
treatment that has demonstrated a survival advantage for patients
whose condition has worsened despite sorafenib treatment (140,141). However, despite the richer
target profile of regorafenib, resistance remains an issue in
clinical use. Research indicates that exosomal circRNAs can affect
the development of regorafenib resistance. For instance, circDCAF8
plays a notable role in facilitating the proliferation, migration,
invasion and EMT of HCC cells. Of note, exosomal circDCAF8 enhances
angiogenesis in HCC and enables the transfer of drug resistance
from regorafenib-resistant HCC cells to those that are sensitive,
thereby imparting a resistant phenotype. This process operates
through the sponging of miR-217, which in turn increases the
expression of nucleolus assembly protein 1-like protein 1,
influencing both HCC progression and resistance to regorafenib
(19). Moreover, another study
indicated that berberine could increase the therapeutic effect of
regorafenib on HCC via the upregulation of hsa_circ_0032029 and
hsa_circ_0008928 in HCC cells (142).
Given the increasingly prominent problem of drug
resistance in targeted drug therapy for HCC, a number of recent
research efforts have validated that exosomal circRNAs are pivotal
in mediating the mechanism behind multidrug resistance. This
implies that the targeted regulation of exosomal circRNAs may serve
as potential approaches for overcoming therapeutic resistance in
HCC.
Exosomal circRNAs in immunotherapy
resistance in HCC
More recently, as in-depth studies of the immune
microenvironment during tumorigenesis and development have
increased, immunotherapy has become an essential strategy for
treating HCC (143).
Immunotherapy: Immune checkpoint
inhibitors
Immune checkpoints are regulatory molecules found on
immune cells that are crucial for sustaining self-immune function
and are involved in various diseases, including NSCLC, melanoma,
colon cancer and HCC (144-147). PD-1/PD-L1 has emerged as the
predominant target in immunotherapy for HCC. Although immunotherapy
targeting immune checkpoints has shown promising efficacy in
certain patients with HCC, the problem of relapse and drug
resistance remains to be solved. Recent studies suggest that
circRNAs may play either immunostimulatory or immunosuppressive
roles within the tumor immune microenvironment. circRNAs appear to
influence the activity of immune cells and the expression of
molecules associated with immunity, highlighting the potential of
exosomal circRNAs as crucial contributors to resistance against
immunotherapy (147). A recent
investigation has elucidated the pivotal role of novel proteins
translated from circRNAs in mediating immune therapy resistance and
sustaining malignant biological behaviors in HCC cells. The study
identified circPETH as a critical component, specifically an
exosomal circRNA that is packaged within and secreted by
tumor-associated macrophages and subsequently internalized by
recipient HCC cells. The findings demonstrated that
tumor-associated macrophage-derived exosomal circPETH actively
promotes HCC cell invasion, migration and aerobic glycolysis.
Mechanistically, a key function of circPETH within the HCC cytosol
is its capacity to recruit ribosomes and initiate cap-independent
translation, resulting in the expression of the novel protein
circPETH-147aa. Further mechanistic dissection revealed that
circPETH-147aa maintains HuR phosphorylation at the S100 site by
specifically occupying surface MEG pockets. This interaction
enhances HuR-dependent SLC43A2 mRNA stability, which consequently
promotes the heightened uptake of methionine and leucine by HCC
cells. This metabolic shift compromises CD8+ T cell
antitumor cytotoxicity and impairs anti-HCC immunity. Notably, the
study also identified norathyriol, a small-molecule compound that
reverses the oncogenic effects of circPETH-147aa by competitively
occupying the MEG pocket. This targeted intervention potentiates
anti-PD-1 efficacy and restores cytotoxic CD8+ T cell
function. These results position norathyriol as a highly promising
therapeutic agent to improve treatment outcomes for patients with
HCC and mitigate resistance to immune checkpoint blockade therapy
(41). Emerging evidence
indicates that Circ-CDYL plays a critical role in mediating
immunotherapy resistance. Mechanistically, Circ-CDYL binds to and
interacts with hornerin (HRNR), thereby preventing its degradation
via the ubiquitin-proteasome pathway. This stabilization of HRNR
promotes activation of the downstream mTORC1 signaling pathway,
leading to the increased expression of PD-L1. Consequently, this
heightened PD-L1 expression drives the secretion of
PD-L1-containing exosomes, which subsequently suppress the
proliferation and cytotoxicity of CD8+ T cells within the TME. This
Circ-CDYL-mediated cascade ultimately diminishes the efficacy of
anti-PD-L1 immunotherapy in HCC and confers resistance to immune
checkpoint blockade (148).
circCCAR1 is upregulated in the exosomes derived from tumor cells,
tumor tissues and the plasma of patients with HCC. Exosomes
secreted by HCC cells can be taken up by CD8+ T cells,
which leads to the dysfunction of these T cells by stabilizing the
PD-1 protein, ultimately increasing resistance to anti-PD-1
immunotherapy. From a mechanistic analysis, a positive feedback
loop involving circCCAR1, miR-127-5p and wilms' tumor 1-associating
protein, a key regulator of m6A RNA methylation that guides the
methyltransferase complex to specific RNA targets, can enhance the
expression of circCCAR1. This RNA can be secreted from HCC cells to
CD8+ T cells in a manner dependent on heterogeneous
nuclear ribonucleoprotein A2/B1, which stabilizes the PD-1 protein
and results in T cell dysfunction, thereby increasing resistance to
anti-PD-1 treatment. Furthermore, the elevated levels CCAR1 in HCC
cells may also contribute to resistance against anti-PD-1
immunotherapy through its interaction with β-catenin, which
facilitates the transcription of PD-L1 (149). Focusing on the associated
circRNA offers a novel approach to enhance immunotherapy
effectiveness in patients with HCC. Another discovery indicated
that exosomal circTMEM181 enhances the immunosuppressive
environment in HCC and increases resistance to anti-PD-1 therapy in
HCC cells. In terms of the underlying mechanism, exosomal
circTMEM181 can act as a sponge for miR-488-3p within macrophages,
thereby upregulating the expression of CD39 in macrophages. The
expression of cell-specific CD39 in macrophages and CD73 in HCC
cells synergistically activates the eATP-adenosine pathway
increasing adenosine production. As a consequence, the function of
CD8+ T cells is hampered and the immune microenvironment
of HCC is remodeled, resulting in resistance to anti-PD-1 therapy
in HCC cells (150). It has been
reported that high circUHRF1 levels are associated with unfavorable
clinical outcomes and impairs natural killer (NK) cell function in
patients with HCC and that it can be secreted by HCC cells into the
plasma in the form of exosomes, resulting in a reduced NK cell
ratio and decreased NK cell tumor infiltration. Mechanistic
analysis revealed that circUHRF1 inhibits NK cell function by
degrading miR-449c-5p to upregulate the expression of T-cell
immunoglobulin mucin-3 (TIM-3) and impede the ability of NK cells
to generate IFN-γ and TNF-α, thereby inhibiting NK cell function
and inducing HCC progression in an NK cell-dependent manner.
Additionally, the investigation confirmed that HCC cells with
knocked down circUHRF1 expression demonstrated increased
sensitivity to anti-PD-1 therapy and exhibited improved overall
survival (OS) rates (151). This
suggests that targeting exosomal circUHRF1 may be a promising and
effective method to restore HCC sensitivity to anti-PD-1 therapy.
Moreover, in another study, elevated levels of circMET were shown
to trigger EMT in HCC cells; its overexpression induced an
immunosuppressive microenvironment in HCC via the
miR-30-5p/Snail/dipeptidyl peptidase 4 (DPP4)/C-X-C motif chemokine
ligand 10 (CXCL10) axis (152).
In this axis, DPP4 functions as a serine protease that cleaves and
inactivates the T-cell chemoattractant CXCL10. DPP4 inhibitors are
a class of drugs used to treat type 2 diabetes, but in this study,
it was shown to increase CXCL10 expression in tumor cells by
inhibiting the cleavage of the chemokine CXCL10 by DPP4, thereby
triggering effective tumor immunity and enhancing the clinical
efficacy of immunotherapy. It is worth noting that in animal
experiments, the combination of the DPP4 inhibitor sitagliptin and
anti-PD-1 therapy was significantly effective and synergistic,
suggesting that sitagliptin can improve the efficacy of PD-1
blockade immunotherapy, highlighting the potential value of
repurposing established therapeutics for other diseases in the
management of HCC (152).
circRHBDD1 exhibits increased expression in patients with HCC
responsive to anti-PD-1 therapy and facilitates glycolysis within
HCC cells. Targeting the circRHBDD1/YTH N6-methyladenosine RNA
binding protein 1/PIK3R1 axis can improve anti-PD-1 therapy in a
mouse model with normal immune function (153). circSOD2 can promote immune
escape and anti-PD-1 resistance in HCC by targeting the
miR-497-5p/Annexin A11 axis (154).
According to the aforementioned studies, exosomal
circRNAs can relay information between tumors and immune cells via
the secretion of exosomes. These circRNAs can influence immune
cells and immune-related molecules within the tumor immune
microenvironment, leading to the immunosuppression of tumor cells,
which in turn may result in resistance to immunotherapy.
Immunotherapy is an effective treatment strategy and its
relationship with exosomal circRNAs is still unclear. Exosomal
circRNAs may become an effective way to improve sensitivity to
anti-PD-1 therapy.
Immunotherapy: Adoptive cell therapy
Evidence suggests that monotherapy with immune
checkpoint inhibitors alone may not effectively tackle the
underlying issues related to immune system failure and the
suppression of immune cells responsible for targeting cancer cells
(155). The TME is another key
factor contributing to resistance to immunotherapy. In the TME, the
dysfunction of immune effector cells induces immunosuppression,
mediating tumor cell immune escape.
T cells
Immunotherapy is a practical approach for improving
the efficacy of immune checkpoint inhibitors, especially for
patients with cancer who have developed resistance to checkpoint
inhibitors, by inducing cancer-specific T cells (156). Specifically, T cells can be
genetically edited to express T cell receptors or chimeric antigen
receptors (CARs) on their cell surface that recognize tumor
antigens, thereby enhancing the specificity and reactivity of
immune cells (157).
Nonetheless, certain patients may experience a gradual relapse
following a successful response to immunotherapy, leading to
acquired drug resistance (158).
A key factor that may contribute to this phenomenon is that,
throughout treatment, cancer cells can evolve strategies to escape
immune detection or diminish the effectiveness of T cells,
ultimately resulting in suboptimal treatment outcomes (159).
Studies have shown that exosomal circRNAs are
involved in immune cell therapy resistance. For instance, recent
research has indicated that exosomal circGSE1 induces the
proliferation of regulatory T cells (Tregs) by modulating the
miR-324-5p/TGFBR1/Smad3 axis. These proliferated Tregs are capable
of regulating the immune escape of HCC by inhibiting
CD4+ T cells and stimulating CD8+ T cells,
which in turn leads to resistance to HCC immunotherapy (160). In another study, the levels of
circCCAR1 were shown to be markedly higher in patients with HCC,
and this increase promoted HCC growth and metastasis both in
vivo and in vitro. More notably, exosomes secreted by
HCC cells containing circCCAR1 were absorbed by CD8+ T
cells, causing CD8+ T cell dysfunction and thus
promoting HCC immunosuppression. From a mechanistic perspective,
circCCAR1 impairs activated CD8+ T cells by suppressing
their proliferation, accelerating their apoptosis, diminishing
their cytotoxicity and cytokine secretion as well as increasing the
expression of lymphocyte-activation gene 3, PD1, TIM-3 and T cell
immunoreceptor with immunoglobulin and ITIM domains on the surface
of CD8+ T cells (149).
Exosomal circRNAs often play similar roles in the
immunotherapy of other types of cancer. For example, in NSCLC,
exosomal circUSP7 from cancer cells triggered CD8+ T
cell dysfunction by regulating a miR-934/Src homology 2 domain
containing protein tyrosine phosphatase 2 (SHP2) axis. SHP2, a key
signaling node that transduces signals from various immune
checkpoints, contributes to immunosuppression in NSCLC (161). In bladder cancer (BCa),
exosome-derived circTRPS1 from BCa cells can induce
immunosuppression by regulating the intracellular reactive oxygen
species balance and CD8+ T cell depletion via a
circTRPS1/miR141-3p/glutaminase 1 axis, where GLS1 is a key
metabolic enzyme that catalyzes the conversion of glutamine to
glutamate (162).
NK cells
Liver NK cells are a crucial component of the
immune system, serving an indispensable function in supporting
liver immunity and protection against HCC. NK cells contribute
significantly to the liver's immune defense by employing a range of
strategies, including the direct elimination of tumor cells and the
release of cytokines (163).
CAR-NK therapy has become a promising method for treating patients
with HCC (164,165). It has been reported that
HCC-derived exosomal circRNAs target NK cells to promote the
formation of an immunosuppressive microenvironment. For instance,
circUHRF1, primarily released by HCC cells in exosomes, plays a
role in fostering immunosuppression by breaking down miR-449c-5p to
enhance TIM-3 expression. This process leads to the dysfunction and
eventual death of NK cells, ultimately impairing their activity. In
addition, exosomal circUHRF1 also hinders the production of IFN-γ
and TNF-α by NK cells, further suppressing their functionality
(151). In addition to targeting
NK cells to promote immunosuppression, certain circRNAs promote NK
cell-mediated cytotoxicity against HCC. For instance, in HCC cell
lines, the expression of hsa_circ_0007456 is significantly
decreased. This change significantly affects their susceptibility
to NK cells, which may impact the immunotherapeutic effect of HCC.
Research into the underlying mechanisms has shown that
hsa_circ_0007456 primarily influences the sensitivity of HCC cells
to NK cells by sponging miR-6852-3p and regulating the expression
of intercellular adhesion molecule-1 (ICAM-1) in HCC cells
(166). The expression of ICAM-1
by human cancer cells can affect NK cell function by binding to
lymphocyte function-associated antigen 1 on NK cells, thereby
regulating the adhesion between NK cells and target cells (167).
Macrophages
Studies have found that macrophages are among the
key target cells for exosomal circRNAs (168). Within the TME, HCC cells deliver
biologically active materials such as circRNAs to macrophages in
the form of vesicles. These circRNAs may interfere with the normal
immune function of macrophages, for example, by inhibiting their
antigen presentation ability (150) and regulating their polarization
state (68). This allows cancer
cells to evade immune surveillance, creating favorable conditions
for tumor growth and metastasis (150). Exosomes derived from HCC
containing circTMEM181, are capable of enhancing the expression of
CD39 in macrophages. This is achieved through the sequestration of
miR-488-3p. As a result, CD39 collaboratively activates the
eATP-adenosine pathway along with the expression of CD73 in HCC
cells. This joint action leads to the generation of larger
quantities of adenosine, ultimately undermining the function of
CD8+ T cells (150).
Another study revealed that exosomal hsa_circ_0074854 is elevated
in HCC tissues and cell lines, and silencing hsa_circ_0074854
reduced HCC proliferation both in vitro and in vivo.
Mechanistically, hsa_circ_0074854 is transferred from HCC cells to
macrophages via exosomes. M2 macrophages play a crucial role in
creating an immunosuppressive TME and facilitating immune evasion,
whereas the exosomes from cells with hsa_circ_0074854 knockdown
impeded the polarization of macrophages toward an M2 phenotype
(68). This shows that targeting
hsa_circ_0074854 may be a novel strategy for preventing
immunosuppression in HCC.
In summary, current research has found that one of
the critical factors in immune cell therapy resistance is the
activity of immune cells. Therapeutically, circRNAs often suppress
immune cell function, which further leads to immunosuppression of
HCC cells, making it difficult for immunotherapeutics to achieve
the desired effect. However, by targeting circRNAs, which is like a
precise 'key', the 'shackles' on the function of immune cells can
be removed, thereby lifting the immunosuppression of HCC cells and
allowing immune cells to resume their tasks. This could have
revolutionary implications for tackling immunotherapy resistance in
HCC, offering a novel approach for overcoming this complex
issue.
Potential of exosomal circRNAs in clinical
applications
Exosomal circRNAs function as clinical
biomarkers
Exosomal circRNAs derived from HCC cells are
notably abundant and stable. Secreted into the TME and subsequently
disseminated via systemic circulation, these circRNAs are
transferred to diverse target cells. This delivery mechanism
enables them to critically modulate malignant phenotypes, including
tumor proliferation, invasion, metastasis, angiogenesis, immune
evasion and drug resistance (169). Emerging evidence consistently
demonstrates that exosomes critically contribute to HCC progression
by actively remodeling the TME and inducing an immunosuppressive
state within it (170).
Consequently, exosomal circRNAs are pivotal in mediating
intercellular crosstalk within the TME, thereby either promoting or
inhibiting the invasion and metastasis of HCC. This regulatory
capacity confers upon them significant potential as robust
biomarkers for the diagnosis, therapeutic target and prognostic
stratification of HCC (171,172). Liquid biopsy represents a
non-invasive methodology that harnesses various bodily fluids,
including blood, plasma, serum, urine and gastric fluid, to
accurately monitor and reflect the overall disease status (173). Given their characteristic
dysregulation in disease and notable abundance in stable exosomal
carriers, circRNAs are readily detectable in bodily fluids (such as
blood, serum, urine, saliva and cerebrospinal fluid). These
attributes collectively position circRNAs as ideal biomarkers for
liquid biopsy (16,174).
For instance, hsa_circ_0004001, hsa_circ_0004123
and hsa_circ_0075792 have been identified as upregulated in serum
exosomes of patients with HCC, exhibiting enhanced diagnostic
sensitivity and specificity. Notably, the combined application of
these three biomarkers notably improves diagnostic accuracy,
achieving a sensitivity of 90.5% and an area under the curve (AUC)
of 0.89. This evidence strongly supports the utility of this
circRNA panel as a valuable diagnostic signature for HCC (175). circ-0072088 is primarily
secreted by HCC cells via exosomes, exhibiting significantly higher
expression in HCC tissues and cells compared with adjacent
non-cancerous tissues and healthy hepatocytes. Receiver operating
characteristic (ROC) curve analysis has revealed the diagnostic
efficacy of circ-0072088 for HCC, yielding an AUC of 0.899.
Moreover, Kaplan-Meier survival curves and Cox regression analyses
consistently established that elevated circ-0072088 expression is
associated with significantly reduced 5-year survival rates,
thereby positioning it as an independent prognostic factor for
patients with HCC. Collectively, these results highlight that
extracellular vesicle-derived circ-0072088 functions as both a
diagnostic and prognostic marker for HCC (73). The expression level of
circ_0051443 in patient plasma exosomes is significantly
downregulated compared with healthy controls, positioning it as a
potential diagnostic and prognostic biomarker. Mechanistically,
circ_0051443 can suppress malignant biological behaviors by
regulating key processes such as apoptosis, proliferation and cell
cycle arrest (48). In an
independent cohort, reverse transcription-quantitative PCR was
employed to determine the expression levels of circAKT3 in
circulating exosomes from 124 patients with HCC and 100 healthy
controls. The results demonstrated that exosomal circAKT3
expression was significantly elevated in patients with HCC compared
with the healthy subjects. More critically, high circAKT3
expression was correlated with increased tumor recurrence rates and
elevated mortality among patients with HCC, thereby establishing
circAKT3 as a crucial biomarker for predicting HCC recurrence and
unfavorable prognosis (176). A
recent study demonstrated that circYTHDC2 is significantly
upregulated across HCC tissues, serum and cell lines, and exhibits
notable stability against RNase degradation. ROC analysis confirmed
the high diagnostic accuracy of circYTHDC2 in both tissue
(AUC=0.846) and serum (AUC=0.788) samples. Furthermore, elevated
levels of circYTHDC2 significantly were correlated with adverse
clinicopathological features, including advanced Barcelona Clinic
Liver Cancer staging, larger tumor volume, intrahepatic metastasis
and portal vein invasion. Collectively, these findings establish
circYTHDC2 as a stable and clinically viable HCC biomarker with
notable diagnostic accuracy and prognostic value (177). Furthermore, another preliminary
investigation assessed the diagnostic and prognostic utility of
circulating circ_0009910 and circ_0027478 in conjunction with
miR-1236-3p in patients with HCC. The study reported a significant
upregulation of circ_0009910 and circ_0027478 in the plasma of
these patients, contrasted by the downregulation of miR-1236-3p.
Notably, circ_0009910 exhibited excellent diagnostic performance
(AUC=0.90), and its elevated expression was positively correlated
with adverse clinicopathological features, including tumor size,
metastasis and α-fetoprotein levels. These findings underscore the
potential of circ_0009910 as an independent diagnostic and
prognostic biomarker for HCC (178). Although this study primarily
utilized total plasma samples for detection, its findings offer
compelling evidence that circulating circRNAs accurately reflect
the pathological characteristics and malignancy of HCC.
Furthermore, in a separate analysis, circSMARCA5 expression levels
in the plasma from patients with HCC were found to be significantly
lower than those in healthy controls. This downregulation conferred
high discriminatory accuracy (AUC=0.938), demonstrating the
potential of circSMARCA5 as a robust biomarker for distinguishing
patients with HCC from healthy subjects (179). A recent study has demonstrated
significant progress, clinically validating a nanobiosensor that
successfully detects circSMARCA5 expression levels in both liver
tissue and whole blood samples, thus enabling the accurate
discrimination of patients with HCC from healthy individuals. This
breakthrough establishes a highly promising circulating RNA-based
liquid biopsy tool with clinical application potential (180).
Exosomal circRNAs are also capable of reflecting
the presence of tumor-infiltrating lymphocytes (TILs) within HCC
tissues, thereby serving as a robust prognostic biomarker. Research
indicates that patients with HCC presenting with high TIL levels
typically exhibit significantly improved OS outcomes (181). Specifically, the expression of
hsa_circ_0064428 was found to be significantly reduced in high-TIL
patients but conversely increased in those with low-TIL
infiltration. Furthermore, hsa_circ_0064428 expression demonstrated
a significant negative correlation with patient survival rates,
tumor size and metastasis. These findings collectively establish
hsa_circ_0064428 as a potent prognostic biomarker for HCC,
potentially reflecting the antitumor immune microenvironment
(181). Most notably, exosomal
circRNA also serves as a prognostic biomarker for treatment
resistance in HCC. A study has indicated that hsa_circ_0000615 is
significantly upregulated in patients with sorafenib-resistant HCC
compared with those with chemotherapy-sensitive HCC, with its
expression level positively correlated with advanced disease stage.
Furthermore, hsa_circ_0000615 exhibits moderately good AUC values,
and elevated expression strongly correlates with shorter OS in
patients with chemotherapy-resistant HCC. These findings suggest
that hsa_circ_0000615 holds promising value as a novel prognostic
biomarker for sorafenib resistance, thus offering diagnostic
utility for detecting chemoresistance in HCC (182). Additionally, specific exosomal
circRNAs exhibit potential as markers for the early identification
and differential diagnosis of HCC from high-risk populations, such
as individuals with chronic hepatitis B virus infection and
cirrhosis. Examples include serum exosomal hsa_circ_0028861,
exosomal circ_0070396 and hsa_circ_0003998, which demonstrate
utility in discriminating HCC from these background liver diseases
(183-185).
Given the established potential of exosomal
circRNAs as emerging clinical biomarkers for HCC diagnosis and
prognosis, targeted clinical validation trials remain essential.
Supported by the accumulation of robust future research data,
exosomal circRNA is highly anticipated to evolve into a more
reliable and clinically actionable tumor biomarker, ultimately
facilitating its successful translation into routine clinical
applications.
Therapeutic strategies based on exosomal
circRNA
Exosomal circRNAs have emerged not only as
biomarkers but also as a promising therapeutic strategy.
Specifically, the utilization of exosomal RNA delivery systems for
the treatment of malignant tumors, including HCC, has demonstrated
considerable potential as a novel and robust approach in modern
cancer therapy.
Strategies targeting oncogenic circRNAs can be
employed to inhibit HCC progression or drug resistance.
Specifically, technologies such as RNA interference, antisense
oligonucleotides and CRISPR-Cas13a have been engineered for the
effective targeting of these oncogenic circRNAs (186). For example, a prior study
reported that the oncogenic circRNA, circ_0000098, drives HCC
progression via the miR-383/mitochondrial calcium uniporter
regulator 1 axis. Notably, this research engineered PLTs to
encapsulate both DOX and a short hairpin RNA targeting
circ_0000098, termed DOX/sh-1@ PLT. In both in vitro and
in vivo models, DOX/sh-1@PLT demonstrated effective delivery
to HCC cells, promoting intracellular DOX accumulation and
successfully reversing drug resistance in HCC (103). The superior stability, minute
size and low toxicity of exosomes establish them as compelling
candidates for drug delivery systems. Therefore, strategies
employing exosome-mediated transfer of anticancer circRNAs hold
great promise for the inhibition of HCC progression and the
reversal of acquired drug resistance (42,94,107,129,135).
Beyond the aforementioned studies, research
exploring the potential utility of circRNAs in cancer vaccines,
gene editing therapies and adoptive cell therapies continues to
rapidly expand (187,188). However, further research is
essential to fully delineate the potential of circRNAs as a
therapeutic intervention for HCC. Collectively, exosomal circRNA is
emerging as a novel category of RNA therapeutics. While challenges
persist, its distinctive benefits poise it to become a significant
asset in the future of HCC therapy.
Conclusions and future perspectives
In conclusion, the regulatory roles of exosomal
circRNAs in HCC pathogenesis, including their impact on
proliferation, invasion-metastasis capabilities, angiogenesis,
metabolic reprogramming and stem cell-like phenotypes, have been
discussed. The present comprehensive review highlights recent
advances in understanding how exosomal circRNAs mediate therapeutic
resistance in HCC. Moreover, the promising role of exosomal
circRNAs as prospective therapeutic targets is highlighted,
shedding light on their clinical significance and presenting
innovative approaches to address treatment resistance and enhance
patient prognosis. However, compared with lncRNAs, studies on the
precise functional mechanisms and clinical applications of exosomal
circRNAs remain limited.
Significant knowledge gaps exist. Current research
primarily emphasizes the effect of miRNA sponging by circRNAs,
leaving other regulatory mechanisms insufficiently investigated.
Notably, certain circRNAs encode functional peptides with specific
biological activities, suggesting that protein-coding circRNAs may
represent a novel frontier in HCC therapeutics. Additionally, key
inquiries concerning the biogenesis of exosomal circRNAs continue
to be unanswered, especially the intricate molecular regulatory
networks that dictate their selective packaging, directional
sorting and eventual degradation. Elucidating these mechanisms
holds significant scientific value for developing innovative
technologies to regulate exosomal circRNA secretion and uptake
within TMEs.
Breakthroughs in this field will improve our
understanding of tumor evolution and advance translational
research, ultimately enabling targeted modulation of exosomal
circRNAs for precision oncology. With continuous advancements in
detection technologies and bioinformatics algorithms, the
diagnostic and therapeutic significance of exosomal circRNAs in
malignancies is becoming increasingly apparent. Further
investigations into the functional mechanisms of exosomal circRNAs
in HCC may critically inform the optimization of clinical
management strategies and accelerate their practical implementation
in personalized cancer therapy.
Supplementary Data
Availability of data and materials
Not applicable.
Authors' contributions
ZL and YuW collected the literature and wrote the
manuscript. YaW and YZ wrote, conceived and reviewed the manuscript
critically. Data authentication is not applicable. All authors have
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
We thank Miss Mohan Liu (China-Japan Union Hospital
of Jilin University, Changchun, Jilin, 130031, P.R. China) for the
related discussions.
Funding
This review was funded by Science and Technology Development
Project of Jilin Province of Jilin Province (grant no.
YDZJ202401252ZYTS).
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Siegel RL, Kratzer TB, Giaquinto AN, Sung
H and Jemal A: Cancer statistics, 2025. CA Cancer J Clin. 75:10–45.
2025.PubMed/NCBI
|
|
3
|
Si J, Zou Y, Gao Y, Chen J, Jiang W, Shen
X, Zhu C and Yao Q: tRF-3a-Pro: A transfer RNA-derived small RNA as
a novel biomarker for diagnosis of Hepatitis B Virus-related
hepatocellular carcinoma. Cell Prolif. 58:e700062025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang CH, Cheng Y, Zhang S, Fan J and Gao
Q: Changing epidemiology of hepatocellular carcinoma in Asia. Liver
Int. 42:2029–2041. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jemal A, Ward EM, Johnson CJ, Cronin KA,
Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, et al:
Annual report to the nation on the status of cancer, 1975-2014,
featuring survival. J Natl Cancer Inst. 109:djx0302017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ladd AD, Duarte S, Sahin I and Zarrinpar
A: Mechanisms of drug resistance in HCC. Hepatology. 79:926–940.
2024. View Article : Google Scholar
|
|
7
|
Liu CX and Chen LL: Circular RNAs:
Characterization, cellular roles, and applications. Cell.
185:2016–2034. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L,
Gorshkov K, Mao Q, Xia S, Cen D, et al:
N6-methyladenosine-modified CircRNA-SORE sustains
sorafenib resistance in hepatocellular carcinoma by regulating
β-catenin signaling. Mol Cancer. 19:1632020. View Article : Google Scholar
|
|
9
|
Zhang XY, Li SS, Gu YR, Xiao LX, Ma XY,
Chen XR, Wang JL, Liao CH, Lin BL, Huang YH, et al: CircPIAS1
promotes hepatocellular carcinoma progression by inhibiting
ferroptosis via the miR-455-3p/NUPR1/FTH1 axis. Mol Cancer.
23:1132024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fei D, Wang F, Wang Y, Chen J, Chen S, Fan
L, Yang L, Ren Q, Duangmano S, Du F, et al: Circular RNA ACVR2A
promotes the progression of hepatocellular carcinoma through
mir-511-5p targeting PI3K-Akt signaling pathway. Mol Cancer.
23:1592024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji Y, Ni C, Shen Y, Xu Z, Tang L, Yu F,
Zhu L, Lu H, Zhang C, Yang S and Wang X: ESRP1-mediated biogenesis
of circPTPN12 inhibits hepatocellular carcinoma progression by
PDLIM2/NF-κB pathway. Mol Cancer. 23:1432024. View Article : Google Scholar
|
|
12
|
Liu Y, Song J, Zhang H, Liao Z, Liu F, Su
C, Wang W, Han M, Zhang L, Zhu H, et al: EIF4A3-induced circTOLLIP
promotes the progression of hepatocellular carcinoma via the
miR-516a-5p/PBX3/EMT pathway. J Exp Clin Cancer Res. 41:1642022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peng R, Cao J, Su BB, Bai XS, Jin X, Wang
AQ, Wang Q, Liu RJ, Jiang GQ, Jin SJ, et al: Down-regulation of
circPTTG1IP induces hepatocellular carcinoma development via
miR-16-5p/RNF125/JAK1 axis. Cancer Lett. 543:2157782022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meldolesi J: Exosomes and ectosomes in
intercellular communication. Curr Biol. 28:R435–R444. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang F, Jiang J, Qian H, Yan Y and Xu W:
Exosomal circRNA: Emerging insights into cancer progression and
clinical application potential. J Hematol Oncol. 16:672023.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dong FL, Xu ZZ, Wang YQ, Li T, Wang X and
Li J: Exosome-derived circUPF2 enhances resistance to targeted
therapy by redeploying ferroptosis sensitivity in hepatocellular
carcinoma. J Nanobiotechnology. 22:2982024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song
X, Gorshkov K, Sun Q, Lin H, Zheng X, et al: CircRNA-SORE mediates
sorafenib resistance in hepatocellular carcinoma by stabilizing
YBX1. Signal Transduct Target Ther. 5:2982020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gong J, Han G, Chen Z, Zhang Y, Xu B, Xu
C, Gao W and Wu J: CircDCAF8 promotes the progression of
hepatocellular carcinoma through miR-217/NAP1L1 Axis, and induces
angiogenesis and regorafenib resistance via exosome-mediated
transfer. J Transl Med. 22:5172024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harding C, Heuser J and Stahl P:
Receptor-mediated endocytosis of transferrin and recycling of the
transferrin receptor in rat reticulocytes. J Cell Biol. 97:329–339.
1983. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Johnstone RM, Adam M, Hammond JR, Orr L
and Turbide C: Vesicle formation during reticulocyte maturation.
Association of plasma membrane activities with released vesicles
(exosomes). J Biol Chem. 262:9412–9420. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
van Niel G, D'Angelo G and Raposo G:
Shedding light on the cell biology of extracellular vesicles. Nat
Rev Mol Cell Biol. 19:213–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Camussi G, Deregibus MC, Bruno S,
Cantaluppi V and Biancone L: Exosomes/microvesicles as a mechanism
of cell-to-cell communication. Kidney Int. 78:838–848. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Panda SS, Sahoo RK, Patra SK, Biswal S and
Biswal BK: Molecular insights to therapeutic in cancer: Role of
exosomes in tumor microenvironment, metastatic progression and drug
resistance. Drug Discov Today. 29:1040612024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Q, Zhang Y, Jin P, Chen Y, Zhang C,
Geng X, Mun KS and Phang KC: New insights into the potential of
exosomal circular RNAs in mediating cancer chemotherapy resistance
and their clinical applications. Biomed Pharmacother.
177:1170272024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang T and Zhang H: Exploring the roles
and molecular mechanisms of RNA binding proteins in the sorting of
noncoding RNAs into exosomes during tumor progression. J Adv Res.
65:105–123. 2024. View Article : Google Scholar :
|
|
27
|
Jiang Z, Liu G and Li J: Recent progress
on the isolation and detection methods of exosomes. Chem Asian J.
15:3973–3982. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan H, Li Y, Cheng S and Zeng Y: Advances
in analytical technologies for extracellular vesicles. Anal Chem.
93:4739–4774. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and
Chen M: Circular RNA: An emerging key player in RNA world. Brief
Bioinform. 18:547–557. 2017.
|
|
31
|
Conn VM, Hugouvieux V, Nayak A, Conos SA,
Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta
C and Conn SJ: A circRNA from SEPALLATA3 regulates splicing of its
cognate mRNA through R-loop formation. Nat Plants. 3:170532017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen
W, Yuan X, Yin W, Xu J, Chen K, et al: CircRNA inhibits DNA damage
repair by interacting with host gene. Mol Cancer. 19:1282020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zheng J, He T, Chen J, Zhao J, Zhang S and
Yang Z: Decoding the functional network of circular RNAs encoding
proteins in hepatocellular carcinoma: From carcinogenesis to
clinical transformation. J Adv Res. Sep 11–2025. View Article : Google Scholar : Epub ahead of
print.
|
|
34
|
Liu Z, Wang Q, Wang X, Xu Z, Wei X and Li
J: Circular RNA cIARS regulates ferroptosis in HCC cells through
interacting with RNA binding protein ALKBH5. Cell Death Discov.
6:722020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Panda AC: Circular RNAs Act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Su Y, Xu C, Liu Y, Hu Y and Wu H: Circular
RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression
via multiple miRNAs sponge. Aging (Albany NY). 11:3362–3375. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-Protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R,
Thorne RF, Zhang XD, Hu W and Wu M: CircACC1 regulates assembly and
activation of AMPK complex under metabolic stress. Cell Metab.
30:157–173.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Margvelani G, Maquera KAA, Welden JR,
Rodgers DW and Stamm S: Translation of circular RNAs. Nucleic Acids
Res. 53:gkae11672025. View Article : Google Scholar :
|
|
41
|
Lan T, Gao F, Cai Y, Lv Y, Zhu J, Liu H,
Xie S, Wan H, He H, Xie K, et al: The protein circPETH-147aa
regulates metabolic reprogramming in hepatocellular carcinoma cells
to remodel immunosuppressive microenvironment. Nat Commun.
16:3332025. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Song R, Ma S, Xu J, Ren X, Guo P, Liu H,
Li P, Yin F, Liu M, Wang Q, et al: A novel polypeptide encoded by
the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR.
Mol Cancer. 22:162023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li S, Gao Y, Tang Y, Dong A, Hu S, Wang R,
Gong X and Peng Z: Circular SNX25 encoded radioresistance augmenter
facilitates DNA damage repair in hepatocellular carcinoma by
targeting BAG6-GET4 interaction. Cell Death Dis. 16:7342025.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Conn VM, Chinnaiyan AM and Conn SJ:
Circular RNA in cancer. Nat Rev Cancer. 24:597–613. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li J, Zhou W, Wang H, Huang M and Deng H:
Exosomal circular RNAs in tumor microenvironment: An emphasis on
signaling pathways and clinical opportunities. MedComm (2020).
5:e700192024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Seimiya T, Otsuka M, Iwata T, Shibata C,
Tanaka E, Suzuki T and Koike K: Emerging roles of exosomal circular
RNAs in cancer. Front Cell Dev Biol. 8:5683662020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen W, Quan Y, Fan S, Wang H, Liang J,
Huang L, Chen L, Liu Q, He P and Ye Y: Exosome-transmitted circular
RNA hsa_circ_0051443 suppresses hepatocellular carcinoma
progression. Cancer Lett. 475:119–128. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang L, Zhang J, Li P, Li T, Zhou Z and
Wu H: Exosomal hsa_circ_0004658 derived from RBPJ
overexpressed-macrophages inhibits hepatocellular carcinoma
progression via miR-499b-5p/JAM3. Cell Death Dis. 13:322022.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dai X, Chen C, Yang Q, Xue J, Chen X, Sun
B, Luo F, Liu X, Xiao T, Xu H, et al: Exosomal circRNA_100284 from
arsenite-transformed cells, via microRNA-217 regulation of EZH2, is
involved in the malignant transformation of human hepatic cells by
accelerating the cell cycle and promoting cell proliferation. Cell
Death Dis. 9:4542018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu Y, Bian L, Liu R, Wang Y and Xiao X:
Circular RNA hsa_circ_0061395 accelerates hepatocellular carcinoma
progression via regulation of the miR-877-5p/PIK3R3 axis. Cancer
Cell Int. 21:102021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yuan P, Song J, Wang F and Chen B:
Exosome-transmitted circ_002136 promotes hepatocellular carcinoma
progression by miR-19a-3p/RAB1A pathway. BMC Cancer. 22:12842022.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Huang C, Yu W, Wang Q, Huang T and Ding Y:
CircANTXR1 contributes to the malignant progression of
hepatocellular carcinoma by promoting proliferation and metastasis.
J Hepatocell Carcinoma. 8:1339–1353. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu C, Ren C, Guo L, Yang C and Yu Q:
Exosome-mediated circTTLL5 transfer promotes hepatocellular
carcinoma malignant progression through miR-136-5p/KIAA1522 axis.
Pathol Res Pract. 241:1542762023. View Article : Google Scholar
|
|
55
|
Li Y, Zang H, Zhang X and Huang G:
Exosomal Circ-ZNF652 promotes cell proliferation, migration,
invasion and glycolysis in hepatocellular carcinoma via
miR-29a-3p/GUCD1 axis. Cancer Manag Res. 12:7739–7751. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang L, Li B, Yi X, Xiao X, Zheng Q and Ma
L: Circ_0036412 affects the proliferation and cell cycle of
hepatocellular carcinoma via hedgehog signaling pathway. J Transl
Med. 20:1542022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Du A, Li S, Zhou Y, Disoma C, Liao Y,
Zhang Y, Chen Z, Yang Q, Liu P, Liu S, et al: M6A-mediated
upregulation of circMDK promotes tumorigenesis and acts as a
nanotherapeutic target in hepatocellular carcinoma. Mol Cancer.
21:1092022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang N, Zheng L, Zhan Y and Zhang Y: A
novel taspine derivative suppresses human liver tumor growth and
invasion in vitro and in vivo. Oncol Lett. 6:855–859. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hunter KW: Host genetics and tumour
metastasis. Br J Cancer. 90:752–755. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mu W, Gu P, Li H, Zhou J, Jian Y, Jia W
and Ge Y: Exposure of benzo[a]pyrene induces HCC exosome-circular
RNA to activate lung fibroblasts and trigger organotropic
metastasis. Cancer Commun (Lond). 44:718–738. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo
J, Zhang Y, Li H, Zhang Q, Yang Y and Chen G: Three isoforms of
exosomal circPTGR1 promote hepatocellular carcinoma metastasis via
the miR449a-MET pathway. EBioMedicine. 40:432–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Allgayer H, Mahapatra S, Mishra B, Swain
B, Saha S, Khanra S, Kumari K, Panda VK, Malhotra D, Patil NS, et
al: Epithelial-to-mesenchymal transition (EMT) and cancer
metastasis: The status quo of methods and experimental models 2025.
Mol Cancer. 24:1672025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Amicone L, Marchetti A and Cicchini C:
Exosome-associated circRNAs as key regulators of EMT in cancer.
Cells. 11:17162022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hu M, Li X, Jiang Z, Xia Q, Hu Y, Guo J
and Fu L: Exosomes and circular RNAs: Promising partners in
hepatocellular carcinoma from bench to bedside. Discov Oncol.
14:602023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Huang JL, Fu YP, Gan W, Liu G, Zhou PY,
Zhou C, Sun BY, Guan RY, Zhou J, Fan J, et al: Hepatic stellate
cells promote the progression of hepatocellular carcinoma through
microRNA-1246-RORα-Wnt/β-catenin axis. Cancer Lett. 476:140–151.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Luo Y, Fu Y, Huang R, Gao M, Liu F, Gui R
and Nie X: CircRNA_101505 sensitizes hepatocellular carcinoma cells
to cisplatin by sponging miR-103 and promotes oxidored-nitro
domain-containing protein 1 expression. Cell Death Discov.
5:1212019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhu C, Su Y, Liu L, Wang S, Liu Y and Wu
J: Circular RNA hsa_circ_0004277 stimulates malignant phenotype of
hepatocellular carcinoma and Epithelial-mesenchymal transition of
peripheral cells. Front Cell Dev Biol. 8:5855652020. View Article : Google Scholar
|
|
68
|
Wang Y, Gao R, Li J, Tang S and Li S, Tong
Q and Li S: Downregulation of hsa_circ_0074854 suppresses the
migration and invasion in hepatocellular carcinoma via interacting
with HuR and via suppressing exosomes-mediated macrophage M2
polarization. Int J Nanomedicine. 16:2803–2818. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Liu L, Liao R, Wu Z, Du C, You Y, Que K,
Duan Y, Yin K and Ye W: Hepatic stellate cell exosome-derived
circWDR25 promotes the progression of hepatocellular carcinoma via
the miRNA-4474-3P-ALOX-15 and EMT axes. Biosci Trends. 16:267–281.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang T, Sun Q, Shen C and Qian Y:
Circular RNA circ_0003028 regulates cell development through
modulating miR-498/ornithine decarboxylase 1 axis in hepatocellular
carcinoma. Anticancer Drugs. 34:507–518. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu D, Kang H, Gao M, Jin L, Zhang F, Chen
D, Li M and Xiao L: Exosome-transmitted circ_MMP2 promotes
hepatocellular carcinoma metastasis by upregulating MMP2. Mol
Oncol. 14:1365–1380. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A
and Qi F: CircRNA Cdr1as functions as a competitive endogenous RNA
to promote hepatocellular carcinoma progression. Aging Albany (NY).
11:8183–8203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lin Y, Zheng ZH, Wang JX, Zhao Z and Peng
TY: Tumor Cell-derived exosomal Circ-0072088 suppresses migration
and invasion of hepatic carcinoma cells through regulating MMP-16.
Front Cell Dev Biol. 9:7263232021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Prager GW, Poettler M, Unseld M and
Zielinski CC: Angiogenesis in cancer: Anti-VEGF escape mechanisms.
Transl Lung Cancer Res. 1:14–25. 2012.PubMed/NCBI
|
|
75
|
Zhu Y, He Q and Qi M: Exosomal circPTPRK
promotes angiogenesis after radiofrequency ablation in
hepatocellular carcinoma. Exp Biol Med (Maywood). 249:100842024.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Huang XY, Huang ZL, Huang J, Xu B, Huang
XY, Xu YH, Zhou J and Tang ZY: Exosomal circRNA-100338 promotes
hepatocellular carcinoma metastasis via enhancing invasiveness and
angiogenesis. J Exp Clin Cancer Res. 39:202020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Hao X, Zhang Y, Shi X, Liu H, Zheng Z, Han
G, Rong D, Zhang C, Tang W and Wang X: CircPAK1 promotes the
progression of hepatocellular carcinoma via modulation of YAP
nucleus localization by interacting with 14-3-3ζ. J Exp Clin Cancer
Res. 41:2812022. View Article : Google Scholar
|
|
78
|
Hu K, Li NF, Li JR, Chen ZG, Wang JH and
Sheng LQ: Exosome circCMTM3 promotes angiogenesis and tumorigenesis
of hepatocellular carcinoma through miR-3619-5p/SOX9. Hepatol Res.
51:1139–1152. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xu B, Jia W, Feng Y, Wang J, Wang J, Zhu
D, Xu C, Liang L, Ding W, Zhou Y and Kong L: Exosome-transported
circHDAC1_004 promotes proliferation, migration, and angiogenesis
of hepatocellular carcinoma by the miR-361-3p/NACC1 axis. J Clin
Transl Hepatol. 11:1079–1093. 2023.PubMed/NCBI
|
|
80
|
Deng M, Sun S, Zhao R, Guan R, Zhang Z, Li
S, Wei W and Guo R: The pyroptosis-related gene signature predicts
prognosis and indicates immune activity in hepatocellular
carcinoma. Mol Med. 28:162022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lai Z, Wei T, Li Q, Wang X, Zhang Y and
Zhang S: Exosomal circFBLIM1 promotes hepatocellular carcinoma
progression and glycolysis by regulating the miR-338/LRP6 axis.
Cancer Biother Radiopharm. 38:674–683. 2023.
|
|
83
|
Yu S, Su S, Wang P, Li J, Chen C, Xin H,
Gong Y, Wang H, Ye X, Mao L, et al: Tumor-associated
macrophage-induced circMRCKα encodes a peptide to promote
glycolysis and progression in hepatocellular carcinoma. Cancer
Lett. 591:2168722024. View Article : Google Scholar
|
|
84
|
Li J, Wang X, Shi L, Liu B, Sheng Z, Chang
S, Cai X and Shan G: A mammalian conserved circular RNA CircLARP1B
regulates hepatocellular carcinoma metastasis and lipid metabolism.
Adv Sci (Weinh). 11:e23059022024. View Article : Google Scholar
|
|
85
|
Liu C, Huang R, Yu H, Gong Y, Wu P, Feng Q
and Li X: Fuzheng Xiaozheng prescription exerts anti-hepatocellular
carcinoma effects by improving lipid and glucose metabolisms via
regulating circRNA-miRNA-mRNA networks. Phytomedicine.
103:1542262022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Han T, Chen L and Li K, Hu Q, Zhang Y, You
X, Han L, Chen T and Li K: Significant CircRNAs in liver cancer
stem cell exosomes: Mediator of malignant propagation in liver
cancer? Mol Cancer. 22:1972023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Gu Y, Wang Y, He L, Zhang J, Zhu X, Liu N,
Wang J, Lu T, He L, Tian Y and Fan Z: Circular RNA circIPO11 drives
self-renewal of liver cancer initiating cells via Hedgehog
signaling. Mol Cancer. 20:1322021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin
XM, Yang S, Zhao Q, Wu T, Li ZX, et al: Circular RNAs negatively
regulate cancer stem cells by physically binding FMRP against CCAR1
complex in hepatocellular carcinoma. Theranostics. 9:3526–3540.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sarkar FH, Li Y, Wang Z and Kong D: The
role of nutraceuticals in the regulation of Wnt and Hedgehog
signaling in cancer. Cancer Metastasis Rev. 29:383–394. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Takebe N, Harris PJ, Warren RQ and Ivy SP:
Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog
pathways. Nat Rev Clin Oncol. 8:97–106. 2011. View Article : Google Scholar
|
|
91
|
Chatterjee S and Sil PC: Targeting the
crosstalks of Wnt pathway with Hedgehog and Notch for cancer
therapy. Pharmacol Res. 142:251–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Lei YR, He XL, Li J and Mo CF: Drug
resistance in hepatocellular carcinoma: Theoretical basis and
therapeutic aspects. Front Biosci (Landmark Ed). 29:522024.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Guo X, Gao CY, Yang DH and Li SL: Exosomal
circular RNAs: A chief culprit in cancer chemotherapy resistance.
Drug Resist Updat. 67:1009372023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Qin L, Zhan Z, Wei C, Li X, Zhang T and Li
J: Hsa-circRNA-G004213 promotes cisplatin sensitivity by regulating
miR-513b-5p/PRPF39 in liver cancer. Mol Med Rep. 23:4212021.
View Article : Google Scholar :
|
|
95
|
Zhou Y, Tang W, Zhuo H, Zhu D, Rong D, Sun
J and Song J: Cancer-associated fibroblast exosomes promote
chemoresistance to cisplatin in hepatocellular carcinoma through
circZFR targeting signal transducers and activators of
transcription (STAT3)/nuclear factor-kappa B (NF-κB) pathway.
Bioengineered. 13:4786–4797. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
He G and Karin M: NF-κB and STAT3-key
players in liver inflammation and cancer. Cell Res. 21:159–168.
2011. View Article : Google Scholar
|
|
97
|
Li Y, Zhang Y, Zhang S, Huang D, Li B,
Liang G, Wu Y, Jiang Q, Li L, Lin C, et al: circRNA circARNT2
suppressed the sensitivity of hepatocellular carcinoma cells to
cisplatin by targeting the miR-155-5p/PDK1 axis. Mol Ther Nucleic
Acids. 23:244–254. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Chen H, Liu S, Li M, Huang P and Li X:
circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance
through Wnt/β-catenin pathway in hepatocellular carcinoma. Onco
Targets Ther. 12:9539–9549. 2019. View Article : Google Scholar :
|
|
99
|
Xie H, Yao J, Wang Y and Ni B:
Exosome-transmitted circVMP1 facilitates the progression and
cisplatin resistance of non-small cell lung cancer by targeting
miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 29:1257–1271. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Shao N, Song L and Sun X: Exosomal
circ_PIP5K1A regulates the progression of non-small cell lung
cancer and cisplatin sensitivity by miR-101/ABCC1 axis. Mol Cell
Biochem. 476:2253–2267. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Hanssen KM, Wheatley MS, Yu DMT, Conseil
G, Norris MD, Haber M, Cole SPC and Fletcher JI: GSH facilitates
the binding and inhibitory activity of novel multidrug resistance
protein 1 (MRP1) modulators. FEBS J. 289:3854–3875. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zang R, Qiu X, Song Y and Wang Y: Exosomes
mediated transfer of Circ_0000337 contributes to cisplatin (CDDP)
resistance of esophageal cancer by regulating JAK2 via miR-377-3p.
Front Cell Dev Biol. 9:6732372021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li Y, Wu A, Chen L, Cai A, Hu Y, Zhou Z,
Qi Q, Wu Y, Xia D, Dong P, et al: Hsa_circ_0000098 is a novel
therapeutic target that promotes hepatocellular carcinoma
development and resistance to doxorubicin. J Exp Clin Cancer Res.
41:2672022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang Z, Wang L, Yin G, Li H, Zhang R, Feng
Y and Chang W: Hsa_circ_0088036 promotes tumorigenesis and
chemotherapy resistance in hepatocellular carcinoma via the
miR-140-3p/KIF2A axis. Histol Histopathol. 40:1239–1251. 2025.
|
|
105
|
Li J, Qin X, Wu R, Wan L, Zhang L and Liu
R: Circular RNA circFBXO11 modulates hepatocellular carcinoma
progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1
axis. J Cell Mol Med. 24:5152–5161. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Ren Y, Dong X, Chen L, Sun T, Alwalid O,
Kan X, Su Y, Xiong B, Liang H, Zheng C and Han P: Combined
ultrasound and CT-Guided Iodine-125 seeds implantation for
treatment of residual hepatocellular carcinoma located at complex
sites after transcatheter arterial chemoembolization. Front Oncol.
11:5825442021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yuan L, Wang Y, Cheng J, Lin S, Ma A, Li
K, Zheng Y, Zeng Z, Ke A, Gao C and Du S: Cancer-derived exosomal
circTMEM56 enhances the efficacy of HCC radiotherapy through the
miR-136-5p/STING axis. Cancer Biol Med. 22:396–411. 2025.PubMed/NCBI
|
|
108
|
Li T and Chen ZJ: The cGAS-cGAMP-STING
pathway connects DNA damage to inflammation, senescence, and
cancer. J Exp Med. 215:1287–1299. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Lin L, Hu P, Luo M, Chen X, Xiao M, Zhong
Z, Peng S, Chen G, Yang G, Zhang F and Zhang Y: CircNOP14 increases
the radiosensitivity of hepatocellular carcinoma via inhibition of
Ku70-dependent DNA damage repair. Int J Biol Macromol.
264:1305412024. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhao B, Rothenberg E, Ramsden DA and
Lieber MR: The molecular basis and disease relevance of
non-homologous DNA end joining. Nat Rev Mol Cell Biol. 21:765–781.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yang W, Liu Y, Gao R, Xiu Z and Sun T:
Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721
cell proliferation, vasculogenic mimicry, and radioresistance. Cell
Signal. 60:122–135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhu S, Chen Y, Ye H, Wang B, Lan X, Wang
H, Ding S and He X: Circ-LARP1B knockdown restrains the
tumorigenicity and enhances radiosensitivity by regulating
miR-578/IGF1R axis in hepatocellular carcinoma. Ann Hepatol.
27:1006782022. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Chang Z, Song Y, Luo F, Yang X, Cai Y and
Guo H: Circular RNA SMARCA5 promotes a poor prognosis and
radiotherapy resistance for patients with hepatocellular carcinoma.
Ann Clin Lab Sci. 53:573–577. 2023.PubMed/NCBI
|
|
114
|
Wang X, Zhang J, Luo F and Shen Y:
Application of circular RNA Circ_0071662 in the diagnosis and
prognosis of hepatocellular carcinoma and its response to
radiotherapy. Dig Dis. 41:431–438. 2023. View Article : Google Scholar
|
|
115
|
Yang K, Ding Y, Han J and He R: CircROBO1
knockdown improves the radiosensitivity of hepatocellular carcinoma
by regulating RAD21. Ann Hepatol. 29:1015362024. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Gu X, Shi Y, Dong M, Jiang L, Yang J and
Liu Z: Exosomal transfer of tumor-associated macrophage-derived
hsa_circ_0001610 reduces radiosensitivity in endometrial cancer.
Cell Death Dis. 12:8182021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Wang X, Cao Q, Shi Y, Wu X, Mi Y, Liu K,
Kan Q, Fan R, Liu Z and Zhang M: Identification of Low-dose
radiation-induced exosomal circ-METRN and miR-4709-3p/GRB14/PDGFRα
pathway as a key regulatory mechanism in Glioblastoma progression
and radioresistance: Functional validation and clinical theranostic
significance. Int J Biol Sci. 17:1061–1078. 2021. View Article : Google Scholar :
|
|
118
|
Gusani NJ, Jiang Y, Kimchi ET,
Staveley-O'Carroll KF, Cheng H and Ajani JA: New pharmacological
developments in the treatment of hepatocellular cancer. Drugs.
69:2533–2540. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
He J, Wang X, Chen K, Zhang M and Wang J:
The amino acid transporter SLC7A11-mediated crosstalk implicated in
cancer therapy and the tumor microenvironment. Biochem Pharmacol.
205:1152412022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Koppula P, Zhang Y, Zhuang L and Gan B:
Amino acid transporter SLC7A11/xCT at the crossroads of regulating
redox homeostasis and nutrient dependency of cancer. Cancer Commun
(Lond). 38:122018.PubMed/NCBI
|
|
121
|
Lin W, Wang C, Liu G, Bi C, Wang X, Zhou Q
and Jin H: SLC7A11/xCT in cancer: Biological functions and
therapeutic implications. Am J Cancer Res. 10:3106–3126.
2020.PubMed/NCBI
|
|
122
|
Bassi MT, Gasol E, Manzoni M, Pineda M,
Riboni M, Martín R, Zorzano A, Borsani G and Palacín M:
Identification and characterisation of human xCT that co-expresses,
with 4F2 heavy chain, the amino acid transport activity system xc.
Pflugers Arch. 442:286–296. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhang Y, Yao R, Li M, Fang C, Feng K, Chen
X, Wang J, Luo R, Shi H, Chen X, et al: CircTTC13 promotes
sorafenib resistance in hepatocellular carcinoma through the
inhibition of ferroptosis by targeting the miR-513a-5p/SLC7A11
axis. Mol Cancer. 24:322025. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kuwano M, Oda Y, Izumi H, Yang SJ, Uchiumi
T, Iwamoto Y, Toi M, Fujii T, Yamana H, Kinoshita H, et al: The
role of nuclear Y-box binding protein 1 as a global marker in drug
resistance. Mol Cancer Ther. 3:1485–1492. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Kwabiah D, Nagati V and Tripathi MK:
Transcription factor YBX1 orchestrates drug resistance and tumor
progression in HCC. Drug Discov Today. 30:1044392025. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Prabhu L, Hartley AV, Martin M, Warsame F,
Sun E and Lu T: Role of post-translational modification of the Y
box binding protein 1 in human cancers. Genes Dis. 2:240–246. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Feng Z, Gao Y, Cai C, Tan J, Liu P, Chen
Y, Deng G, Ouyang Y, Liu X, Cao K, et al: CSF3R-AS promotes
hepatocellular carcinoma progression and sorafenib resistance
through the CSF3R/JAK2/STAT3 positive feedback loop. Cell Death
Dis. 16:2172025. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Lan Y, Banks KM, Pan H, Verma N, Dixon GR,
Zhou T, Ding B, Elemento O, Chen S, Huangfu D and Evans T:
Stage-specific regulation of DNA methylation by TET enzymes during
human cardiac differentiation. Cell Rep. 37:1100952021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Dong ZR, Ke AW, Li T, Cai JB, Yang YF,
Zhou W, Shi GM and Fan J: CircMEMO1 modulates the promoter
methylation and expression of TCF21 to regulate hepatocellular
carcinoma progression and sorafenib treatment sensitivity. Mol
Cancer. 20:752021. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Kudo M, Finn RS, Qin S, Han KH, Ikeda K,
Piscaglia F, Baron A, Park JW, Han G, Jassem J, et al: Lenvatinib
versus sorafenib in first-line treatment of patients with unresec\
hepatocellular carcinoma: A randomised phase 3 non-inferiority
trial. Lancet. 391:1163–1173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Magyar CTJ, Perera S, Rajendran L, Li Z,
Almugbel FA, Feng S, Choi WJ, Aceituno L, Vogel A, Grant RC, et al:
Comparative outcome analysis of lenvatinib versus sorafenib for
recurrence of hepatocellular carcinoma after liver transplantation.
Transplantation. 109:681–690. 2025. View Article : Google Scholar
|
|
132
|
Hu B, Zou T, Qin W, Shen X, Su Y, Li J,
Chen Y, Zhang Z, Sun H, Zheng Y, et al: Inhibition of EGFR
overcomes acquired lenvatinib resistance driven by STAT3-ABCB1
signaling in hepatocellular carcinoma. Cancer Res. 82:3845–3857.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Zhang P, Sun H, Wen P, Wang Y, Cui Y and
Wu J: circRNA circMED27 acts as a prognostic factor and mediator to
promote lenvatinib resistance of hepatocellular carcinoma. Mol Ther
Nucleic Acids. 27:293–303. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Tang Y, Yuan F, Cao M, Ren Y, Li Y, Yang
G, Zhong Z, Liang H, Xiong Z, He Z, et al: CircRNA-mTOR promotes
hepatocellular carcinoma progression and lenvatinib resistance
through the PSIP1/c-Myc axis. Adv Sci (Weinh). 12:e24105912025.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Yang L, Tan W, Wang M, Wei Y, Xie Z, Wang
Q, Zhang Z, Zhuang H, Ma X, Wang B, et al: circCCNY enhances
lenvatinib sensitivity and suppresses immune evasion in
hepatocellular carcinoma by serving as a scaffold for SMURF1
mediated HSP60 degradation. Cancer Lett. 612:2174702025. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Hou YR, Diao LT, Hu YX, Zhang QQ, Lv G,
Tao S, Xu WY, Xie SJ, Zhang Q and Xiao ZD: The conserved LncRNA
DIO3OS restricts hepatocellular carcinoma stemness by interfering
with NONO-mediated nuclear export of ZEB1 mRNA. Adv Sci (Weinh).
10:e23019832023. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Orellana-Serradell O, Herrera D, Castellón
EA and Contreras HR: The transcription factor ZEB1 promotes
chemoresistance in prostate cancer cell lines. Asian J Androl.
21:460–467. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Cao M, Li Y, Su X, Tang Y, Yuan F, Ren Y,
Deng M and Yao Z: Exosome-derived hsa_circ_0007132 promotes
lenvatinib resistance by inhibiting the ubiquitin-mediated
degradation of NONO. Noncoding RNA Res. 14:1–13. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Yuan F, Tang Y, Liang H, Cao M, Ren Y, Li
Y, Yang G, Zhong Z, Xiong Z, He Z, et al: CircPIK3C3 inhibits
hepatocellular carcinoma progression and lenvatinib resistance by
suppressing the Wnt/β-catenin pathway via the miR-452-5p/SOX15
axis. Genomics. 117:1109992025. View Article : Google Scholar
|
|
140
|
Bruix J, Qin S, Merle P, Granito A, Huang
YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al:
Regorafenib for patients with hepatocellular carcinoma who
progressed on sorafenib treatment (RESORCE): A randomised,
double-blind, placebo-controlled, phase 3 trial. Lancet. 389:56–66.
2017. View Article : Google Scholar
|
|
141
|
Ueshima K, Nishida N and Kudo M:
Sorafenib-regorafenib sequential therapy in advanced hepatocellular
carcinoma: A Single-institute experience. Dig Dis. 35:611–617.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Wang K, Yu G, Lin J, Wang Z, Lu Q, Gu C,
Yang T, Liu S and Yang H: Berberine sensitizes human hepatoma cells
to regorafenib via modulating expression of circular RNAs. Front
Pharmacol. 12:6322012021. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Li M, Bhoori S, Mehta N and Mazzaferro V:
Immunotherapy for hepatocellular carcinoma: The next evolution in
expanding access to liver transplantation. J Hepatol. 81:743–755.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Raskova Kafkova L, Mierzwicka JM,
Chakraborty P, Jakubec P, Fischer O, Skarda J, Maly P and Raska M:
NSCLC: From tumorigenesis, immune checkpoint misuse to current and
future targeted therapy. Front Immunol. 15:13420862024. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Klobuch S, Seijkens TTP, Schumacher TN and
Haanen J: Tumour-infiltrating lymphocyte therapy for patients with
advanced-stage melanoma. Nat Rev Clin Oncol. 21:173–184. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu
W, Wang Z, Wu Q, Peng H, Wei H, et al: Blockade of the checkpoint
receptor TIGIT prevents NK cell exhaustion and elicits potent
anti-tumor immunity. Nat Immunol. 19:723–732. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Jiang W, Pan S, Chen X, Wang ZW and Zhu X:
The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in
cancer immunotherapy. Mol Cancer. 20:1162021. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Fu J, Liu F, Bai S, Jiang X, Song H, Zhang
M, Zhao R, Ouyang T, Yu M, Qian H, et al: Circular RNA CDYL
facilitates hepatocellular carcinoma stemness and PD-L1+
exosomes-mediated immunotherapy resistance via stabilizing hornerin
protein by blocking synoviolin 1-mediated ubiquitination. Int J
Biol Macromol. 310:1432462025. View Article : Google Scholar
|
|
149
|
Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y,
Jiang J, Wang L, Mang Y and Gao Y: Exosome-derived circCCAR1
promotes CD8 + T-cell dysfunction and anti-PD1 resistance in
hepatocellular carcinoma. Mol Cancer. 22:552023. View Article : Google Scholar
|
|
150
|
Lu JC, Zhang PF, Huang XY, Guo XJ, Gao C,
Zeng HY, Zheng YM, Wang SW, Cai JB, Sun QM, et al: Amplification of
spatially isolated adenosine pathway by tumor-macrophage
interaction induces anti-PD1 resistance in hepatocellular
carcinoma. J Hematol Oncol. 14:2002021. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ,
Shi GM, Cai JB and Ke AW: Cancer cell-derived exosomal circUHRF1
induces natural killer cell exhaustion and may cause resistance to
anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer.
19:1102020. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Huang XY, Zhang PF, Wei CY, Peng R, Lu JC,
Gao C, Cai JB, Yang X, Fan J and Ke AW: Circular RNA circMET drives
immunosuppression and anti-PD1 therapy resistance in hepatocellular
carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 19:922020.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Cai J, Chen Z, Zhang Y, Wang J, Zhang Z,
Wu J, Mao J and Zuo X: CircRHBDD1 augments metabolic rewiring and
restricts immunotherapy efficacy via m6A modification in
hepatocellular carcinoma. Mol Ther Oncolytics. 24:755–771. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Ye R, Lu X, Liu J, Duan Q, Xiao J, Duan X,
Yue Z and Liu F: CircSOD2 contributes to tumor progression, immune
evasion and Anti-PD-1 resistance in hepatocellular carcinoma by
targeting miR-497-5p/ANXA11 axis. Biochem Genet. 61:597–614. 2023.
View Article : Google Scholar
|
|
155
|
Hao L, Li S, Ye F, Wang H, Zhong Y, Zhang
X, Hu X and Huang X: The current status and future of
targeted-immune combination for hepatocellular carcinoma. Front
Immunol. 15:14189652024. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Mizukoshi E and Kaneko S: Immune cell
therapy for hepatocellular carcinoma. J Hematol Oncol. 12:522019.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Lu F, Ma XJ, Jin WL, Luo Y and Li X:
Neoantigen specific T cells derived from T Cell-derived induced
pluripotent stem cells for the treatment of hepatocellular
carcinoma: Potential and challenges. Front Immunol. 12:6905652021.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Gutwillig A, Santana-Magal N,
Farhat-Younis L, Rasoulouniriana D, Madi A, Luxenburg C, Cohen J,
Padmanabhan K, Shomron N, Shapira G, et al: Transient cell-in-cell
formation underlies tumor relapse and resistance to immunotherapy.
Elife. 11:e803152022. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
O'Donnell JS, Teng MWL and Smyth MJ:
Cancer immunoediting and resistance to T cell-based immunotherapy.
Nat Rev Clin Oncol. 16:151–167. 2019. View Article : Google Scholar
|
|
160
|
Huang M, Huang X and Huang N: Exosomal
circGSE1 promotes immune escape of hepatocellular carcinoma by
inducing the expansion of regulatory T cells. Cancer Sci.
113:1968–1983. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D,
Long X, Lin K, Lu F, Xu JJ and Wu YB: Cancer cell-derived exosomal
circUSP7 induces CD8+ T cell dysfunction and anti-PD1 resistance by
regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer. 20:1442021.
View Article : Google Scholar :
|
|
162
|
Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y,
Chen X, Chen Y, Xu C, Hu Y, et al: Exosome-derived circTRPS1
promotes malignant phenotype and CD8+ T cell exhaustion in bladder
cancer microenvironments. Mol Ther. 30:1054–1070. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Peng H, Wisse E and Tian Z: Liver natural
killer cells: Aubsets and roles in liver immunity. Cell Mol
Immunol. 13:328–336. 2016. View Article : Google Scholar
|
|
164
|
Balkhi S, Zuccolotto G, Di Spirito A,
Rosato A and Mortara L: CAR-NK cell therapy: Promise and challenges
in solid tumors. Front Immunol. 16:15747422025. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Li S, Jing J, Chen Y, Chi E, Wang B, Xie
Z, Yang W, Shen H and Pan J: Precision sniper for solid tumors:
CAR-NK cell therapy. Cancer Immunol Immunother. 74:2752025.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH,
Wang YG and Zhang YQ: Hsa_circ_0007456 regulates the natural killer
cell-mediated cytotoxicity toward hepatocellular carcinoma via the
miR-6852-3p/ICAM-1 axis. Cell Death Dis. 12:942021. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Jeong JU, Uong TNT, Chung WK, Nam TK, Ahn
SJ, Song JY, Kim SK, Shin DJ, Cho E, Kim KW, et al: Effect of
irradiation-induced intercellular adhesion molecule-1 expression on
natural killer cell-mediated cytotoxicity toward human cancer
cells. Cytotherapy. 20:715–727. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Liu G, Liu Q, Jia L, Chai Z, Jing L, Xu F
and Fan Y: Exosomal circRNAs: Key modulators in breast cancer
progression. Cell Death Discov. 11:1962025. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Shen H, Liu B, Xu J, Zhang B, Wang Y, Shi
L and Cai X: Circular RNAs: Characteristics, biogenesis, mechanisms
and functions in liver cancer. J Hematol Oncol. 14:1342021.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Xu HZ, Lin XY, Xu YX, Xue HB, Lin S and Xu
TW: An emerging research: The role of hepatocellular
carcinoma-derived exosomal circRNAs in the immune microenvironment.
Front Immunol. 14:12271502023. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Zhang Q, Li H, Liu Y, Li J, Wu C and Tang
H: Exosomal Non-coding RNAs: New insights into the biology of
hepatocellular carcinoma. Curr Oncol. 29:5383–5406. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Tonon F, Grassi C, Tierno D, Biasin A,
Grassi M, Grassi G and Dapas B: Non-coding RNAs as potential
diagnostic/prognostic markers for hepatocellular carcinoma. Int J
Mol Sci. 25:122352024. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Plebani M, Scott S, Simundic AM, Cornes M,
Padoan A, Cadamuro J, Vermeersch P, Çubukçu HC, González Á, Nybo M,
et al: New insights in preanalytical quality. Clin Chem Lab Med.
63:1682–1692. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu
H, Xu X, Liang Q, Christiani DC, Wang M, et al: Circular RNAs in
body fluids as cancer biomarkers: The new frontier of liquid
biopsies. Mol Cancer. 20:132021. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Sun XH, Wang YT, Li GF, Zhang N and Fan L:
Serum-derived three-circRNA signature as a diagnostic biomarker for
hepatocellular carcinoma. Cancer Cell Int. 20:2262020. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Luo Y, Liu F and Gui R: High expression of
circulating exosomal circAKT3 is associated with higher recurrence
in HCC patients undergoing surgical treatment. Surg Oncol.
33:276–281. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Gong Y, Li X and Wang M: Elevated
CircYthdc2 expression is correlated with aggressive features and
poor progression-free survival in hepatocellular carcinoma. BMC
Gastroenterol. 25:6582025. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Awed MS, Ibrahim A, Ezzat O, Fawzy A,
Sabir DK and Radwan AF: Preliminary evaluation of plasma
circ_0009910, circ_0027478, and miR-1236-3p as diagnostic and
prognostic biomarkers in hepatocellular carcinoma. Int J Mol Sci.
26:48422025. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang
L, Deng Q and Zheng F: Using circular RNA SMARCA5 as a potential
novel biomarker for hepatocellular carcinoma. Clin Chim Acta.
492:37–44. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Li C, Wei Y, Li X, Jiang P, Dong X, Guo Q,
Xu Q, Cao X, Xia J and Wang Z: An Ultrasensitive and robust CircRNA
nanobiosensor via magnetic control depuration and catalytic hairpin
assembly cascade amplification for clinically accessible liquid
biopsy. Anal Chem. 97:23939–23949. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Weng Q, Chen M, Li M, Zheng YF, Shao G,
Fan W, Xu XM and Ji J: Global microarray profiling identified
hsa_circ_0064428 as a potential immune-associated prognosis
biomarker for hepatocellular carcinoma. J Med Genet. 56:32–38.
2019. View Article : Google Scholar
|
|
182
|
Zhang L, Xu T, Li Y, Pang Q and Ding X:
Serum hsa_circ_0000615 is a prognostic biomarker of sorafenib
resistance in hepatocellular carcinoma. J Clin Lab Anal.
36:e247412022. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Qiao GL, Chen L, Jiang WH, Yang C, Yang
CM, Song LN, Chen Y, Yan HL and Ma LJ: Hsa_circ_0003998 may be used
as a new biomarker for the diagnosis and prognosis of
hepatocellular carcinoma. Onco Targets Ther. 12:5849–5860. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Lyu L, Yang W, Yao J, Wang H, Zhu J, Jin
A, Liu T, Wang B, Zhou J, Fan J, et al: The diagnostic value of
plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma.
Biomark Med. 15:359–371. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Wang Y, Pei L, Yue Z, Jia M, Wang H and
Cao LL: The potential of serum exosomal hsa_circ_0028861 as the
novel diagnostic biomarker of HBV-derived hepatocellular cancer.
Front Genet. 12:7032052021. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Huang S, Qin H, Dai B, Liu M and Shen J:
Establishment and evaluation of a circAdpgk-0001 knockdown method
using CRISPR-Cas13d RNA-targeting technology. PeerJ. 13:e201232025.
View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Wang L, Wang L, Dong C, Liu J, Cui G, Gao
S and Liu Z: Exploring the potential and advancements of circular
RNA therapeutics. Exploration (Beijing). 5:e202400442025.
View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Cai J, Qiu Z, Chi-Shing Cho W, Liu Z, Chen
S, Li H, Chen K, Li Y, Zuo C and Qiu M: Synthetic circRNA
therapeutics: Innovations, strategies, and future horizons. MedComm
(2020). 5:e7202024. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Ruan YL, Chen TY, Zheng LB, Cai JW, Zhao
H, Wang YL, Tao LY, Xu JJ, Ji L and Cai XJ: cDCBLD2 mediates
sorafenib resistance in hepatocellular carcinoma by sponging
miR-345-5p binding to the TOP2A coding sequence. Int J Biol Sci.
19:4608–4626. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Yang Q and Wu G: CircRNA-001241 mediates
sorafenib resistance of hepatocellular carcinoma cells by sponging
miR-21-5p and regulating TIMP3 expression. Gastroenterol Hepatol.
45:742–752. 2022. View Article : Google Scholar
|
|
191
|
Qiu R and Zeng Z: Hsa_circ_0006988
promotes sorafenib resistance of hepatocellular carcinoma by
modulating IGF1 using miR-15a-5p. Can J Gastroenterol Hepatol.
2022:12061342022.
|
|
192
|
Li X, Yin X, Bao H and Liu C: Circular RNA
ITCH increases sorafenib-sensitivity in hepatocellular carcinoma
via sequestering miR-20b-5p and modulating the downstream
PTEN-PI3K/Akt pathway. Mol Cell Probes. 67:1018772023. View Article : Google Scholar
|
|
193
|
Zhang X, Wang W, Mo S and Sun X: DEAD-box
helicase 17 circRNA (circDDX17) reduces sorafenib resistance and
tumorigenesis in hepatocellular carcinoma. Dig Dis Sci.
69:2096–2108. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Wang S, Liu D, Wei H, Hua Y, Shi G and
Qiao J: The hsa_circRNA_102049 mediates the sorafenib sensitivity
of hepatocellular carcinoma cells by regulating Reelin gene
expression. Bioengineered. 13:2272–2284. 2022. View Article : Google Scholar : PubMed/NCBI
|