Introduction
Breast cancer is the most common malignancy
worldwide and it is estimated that one out of eight women will
develop breast cancer in their life time (1,2).
Unfortunately, in spite of the enormous efforts invested to treat
breast cancer there has been limited success because many patients
still diagnosed too late and several types of breast tumours
develop resistance to the current chemotherapies (3,4). There
is therefore a need to develop more effective diagnostic and
therapeutic approaches to treat this devastating disease. One
important strategy to fight breast cancer is to identify more
effective diagnostic targets for this disease.
The T-box family of transcription factors are
important developmental regulators and have been shown to
contribute to several human syndromes (5,6). In
addition to their key role in development, extensive investigations
suggested that overexpression of some T-box factors, including TBX2
and TBX3 may drive cancer (6–12). Both
transcription factors are upregulated in a number of cancers
including melanoma (13,14) and breast cancer (15,16)
where it was shown to be required for tumour formation and cell
migration (6,17–19).
Importantly, knocking down TBX2 and TBX3 was shown to reverse key
features of the melanoma and breast cancer phenotype suggesting
that it might be a useful target in the development of novel
anti-cancer drugs to treat these cancers (12,20).
Previous study suggested TBX3 as a potential biomarker for early
stages of breast cancer (21). The
same study showed that malignant cells of primary breast cancer
tissue express higher TBX3 levels than normal breast epithelial
cells in both nucleus and cytoplasm. The elevated cytoplasmic TBX3
protein level suggests that TBX3 might leak into the cytoplasm and
subsequently enter the blood stream from the tumor tissue. This was
further supported by another interested study which showed that
TBX3 can be used as component of multiparameter monitoring of
ovarian and breast cancer at the early stages (22). All together these data highlighted
the importance of TBX3 as a potential biomarker for breast cancer.
In this regard, the current study aimed to test whether TBX3 is
overexpressed in different stages of breast cancer tissues and to
test its significance as a diagnostic biomarker to identify tumor
cells from normal cells in breast tissue. Our study demonstrated
that TBX3 is overexpressed in different stages of breast cancer
tissues and it might be reliable marker to distinguish between
breast cancer cells and normal cells in the same tissue. This is
significant to facilitate and to improve breast cancer
diagnosis.
Materials and methods
Breast cancer samples
This study has a retrospective design and the
protocol was approved by the Helsinki Committee of the Palestinian
Health Research Council under approval number (PHRC/HC/93/16). All
pathologically verified breast cancer cases admitted to the
European Gaza Hospital and subjected to mastectomy during 2016
(n=51) were included in this study. For each patient, two
paraffin-embedded tissue blocks containing cancer and non-cancer
tissues were included. Important to note that after the
histopathological examinations, 7 samples of the non-cancer tissues
were found to be invaded by cancer cells and they were excluded
from the non-cancer tissues. Therefore, the total number of samples
in this study was 51 cancer tissues and 44 non-cancer tissues of
the same patients and no samples of healthy volunteers were
included.
None of the patients received neo-adjuvant
chemotherapy. The clinical and pathological features of the
population-based breast cancer tissue samples are shown in Table I.
 | Table I.Clinical and pathological
characteristics of patients. |
Table I.
Clinical and pathological
characteristics of patients.
| Characteristic | No. of patients, n
(%) |
|---|
| Total | 51 (100) |
| Age (year) |
|
| ≤50 | 17 (33.3) |
|
>50 | 34 (66.7) |
| Tumour size
(cm) |
|
| ≤2 | 4 (7.8) |
|
2–5 | 23 (45.1) |
|
>5 | 23 (45.1) |
|
Unknown | 1 (2) |
| Lymph node
metastasis |
|
| 0 | 19 (37.3) |
|
1–3 | 13 (25.5) |
|
4–9 | 8 (15.7) |
|
≥10 | 11 (21.6) |
| Cancer
metastasis |
|
| M0 | 41 (80) |
| M1 | 9 (18) |
|
Unknown | 1 (2) |
| Histological
type |
|
|
IDC | 38 (74.5) |
|
ILC | 12 (23.5) |
|
DCIS | 1 (2.0) |
| ER scoring |
|
| 0 | 10 (19.6) |
| 1 | 9 (17.6) |
| 2 | 7 (13.7) |
| 3 | 5 (9.8) |
|
Unknown | 20 (39.2) |
| PR scoring |
|
| 0 | 10 (19.6) |
| 1 | 10 (19.6) |
| 2 | 4 (7.8) |
| 3 | 7 (13.7) |
|
Unknown | 20 (39.2) |
| HER2 scoring |
|
| 0 | 15 (29.4) |
| 1 | 2 (4.0) |
| 2 | 3 (5.9) |
| 3 | 11 (21.6) |
|
Unknown | 20 (39.2) |
| CA15-3 antigen
(U/ml) |
|
|
≤30 | 12 (23.5) |
|
>30 | 4 (7.8) |
|
Unknown | 35 (68.6) |
Immunohistochemistry
Tissues embedded in a paraffin blocks were cut into
4 µm thick sections. The sections were deparaffinized with a xylene
substitute and rehydrated using decreasing concentrations of ethyl
alcohol. For retrieval, hydration and washing triology (Cell
Marque™; Merck KGaA, Darmstadt, Germany) was used and for blocking
hydrogen peroxide was used. The sections were incubated with a
1:150 dilution of rabbit anti-human TBX3 (ab89220, Abcam,
Cambridge, UK). Primary antibody detected by HRP HiDef 2-Step
Polymer Detection™ kit (954D, Cell Marque™). For all reactions, the
DAB chromogen kit (957D, Cell Marque™) was used. Finally all
sections were counterstained with haematoxylin, dehydrated and
mounted. Negative controls were treated identically, except of the
primary antibody. The slides were examined using an iScan Coreo
Digital Microscope (Version 3.3, Ventana Medical Systems, Inc.,
Tucson, AZ, USA) (21). Notably, due
the heterogeneity of the tumour tissues it's not possible to
separately extract proteins from cancer cells and normal cells.
Therefore immunohistochemistry was used to evaluate TBX3 level in
both types of cells and not western blot analysis.
Scoring method for TBX3 level
The immunostained slides were scored for TBX3 level
by two independent pathologists, both of them are not involved in
any step of immunohistochemistry protocol preparations and without
any information of clinical patient's reports. The tissue slides
were scored using Leake et al (23) scoring system with some modifications.
Briefly, 20 normal cells (in the normal sections) or 20 tumor cells
(in the tumor sections) were randomly chosen and scored. Both
nuclear and cytoplasmic staining were considered. Two scoring
systems were used: i) Staining intensity based system; and ii)
proportion of stained cells based system. For proportion staining
score, both nuclear and cytoplasmic proportion were respectively
classified into six grades (0–5). The sum of these two scores 0 to
8 was given to describe the nuclear and cytoplasmic staining
separately (23,24). By combining these two cytoplasmic and
nuclear scores, the total tbx3 level score was 0 to 16. Suggested
scoring system is summarized in Table
II.
 | Table II.Scoring system for nuclear staining,
cytoplasmic staining and total score. |
Table II.
Scoring system for nuclear staining,
cytoplasmic staining and total score.
| A, Scores for
nuclear staining |
|---|
|
|---|
| Proportion
score | Intensity
score |
|---|
| 0=No staining | 0=No staining |
| 1=<1%
staining | 1=Weak
staining |
| 2=1–10%
staining | 2=Moderate
staining |
| 3=11–33%
staining | 3=Strong
staining |
| 4=34–66%
staining | – |
| 5=67–100%
staining | – |
| Rules | Adding the two
scores together gives a nuclear score of (0–8). |
|
| B, Scores for
cytoplasmic staining |
|
| Proportion
score | Intensity
score |
|
| 0=No staining | 0=No staining |
| 1=<1%
staining | 1=Weak
staining |
| 2=1–10%
staining | 2=Moderate
staining |
| 3=11–33%
staining | 3=Strong
staining |
| 4=34–66%
staining | – |
| 5=67–100%
staining | – |
| Rules | Adding the two
scores together gives a cytoplasmic score (0–8). |
|
| C, Total
score |
|
| Total
score | Staining
levels |
|
| 0–1 | Negative
staining |
| 2–6 | Weak positive
staining |
| 7–11 | Moderate positive
staining |
| 12–16 | Strong positive
staining |
| Rules | Adding the nuclear
score (0–8) and cytoplasmic score (0–8) together gives the total
protein level of cell (0–16). |
Our modifications include combining the cytoplasmic
and nuclear grades to measurer the total TBX3 level in the cells.
The TBX3 level scores obtained ranged from 0 to 16, and divided
into five groups: (0–1) Negative staining, (2–6) weak
positive staining, (7–11) moderate positive staining and
(12–16) strong positive staining. Applied
scoring system is summarized in (Table
II).
Statistical analysis
TBX3 expression in normal and breast cancer tissue
was analyzed using standard statistical methods. All statistical
analyses were performed using SPSS software package version 22.0
(IBM Corp., Armonk, NY, USA). Percentages and frequencies were used
to represent most personal information variables. Then the mean,
standard deviation, Median, Range, Maximum and Minimum were
calculated for variables; t-test was used to compare between TBX3
expression in tumor and normal tissues.
Correlations were performed by Spearman's rank
correlations test to determine the relation between TBX3 level and
clinicopathological features including: Age, tumor size, lymph node
size, lymph node metastasis and common breast cancer biomarkers
including: CA15-3, ER, PR and HER2. This test is based on the study
of the relationship between two variables.
Mann-Whitney U-test (MW) was used to study the
relationships between TBX3 expression and clinicopathological
features including: IDC and ILC breast cancer types, and also
non-metastatic and metastatic breast cancer. For example, this
analysis was used to compare between the total TBX3 level in
invasive lobular carcinoma (ILC) and in invasive ductal breast
cancer (IDC). Also to compare between total TBX3 level of M0 and M1
for tumor metastasis. Kruskal-Wallis H-test (KW) was applied for
statically comparison between three study groups or more. This
assay was used to study the relationships between TBX3 expression
and clinicopathological features include breast cancer stages (I,
II, III and IV) and histological grades (1,2 and 3). P<0.05 was
considered to indicate a statistically significant difference.
Diagnostic accuracy of TBX3 levels were detected by
receiver operating characteristic curve (ROC). Cut-off value and
area under a ROC curve (AUC) were used to determine the
significance of nuclear, cytoplasmic and total TBX3 levels as tumor
marker. The values of the maximum sum of sensitivity and
specificity were considered as the optimal cut off values. In
addition, sensitivity, specificity, accuracy, positive predictive
value (PPV) and negative predictive value (NPV) were
calculated.
Results
TBX3 is overexpressed in different
stages breast cancer tissues
Previous studies showed that TBX3 is overexpressed
in a subset of breast cancer cell lines (25) and primary breast cancer tissues
(21). In the current study, we
tested whether TBX3 is overexpressed in different stages and types
of breast cancer tissues. TBX3 protein level could be evaluated in
breast cancer tissues by different techniques such as
immunohistochemistry and western blot analysis. However, tumor
tissues usually include mixtures of both normal and malignant
cells, and therefore, only a subpopulation of tumor cells represent
the nature of cancer cells. To overcome this problem, we performed
immunohistochemistry analysis to evaluate TBX3 protein level in
breast cancer cells included in the tumor tissues in comparison to
normal cells out of the tumor tissues. Tumor and normal sections
were stained and scored as described in the materials and methods
section (21,23,26).
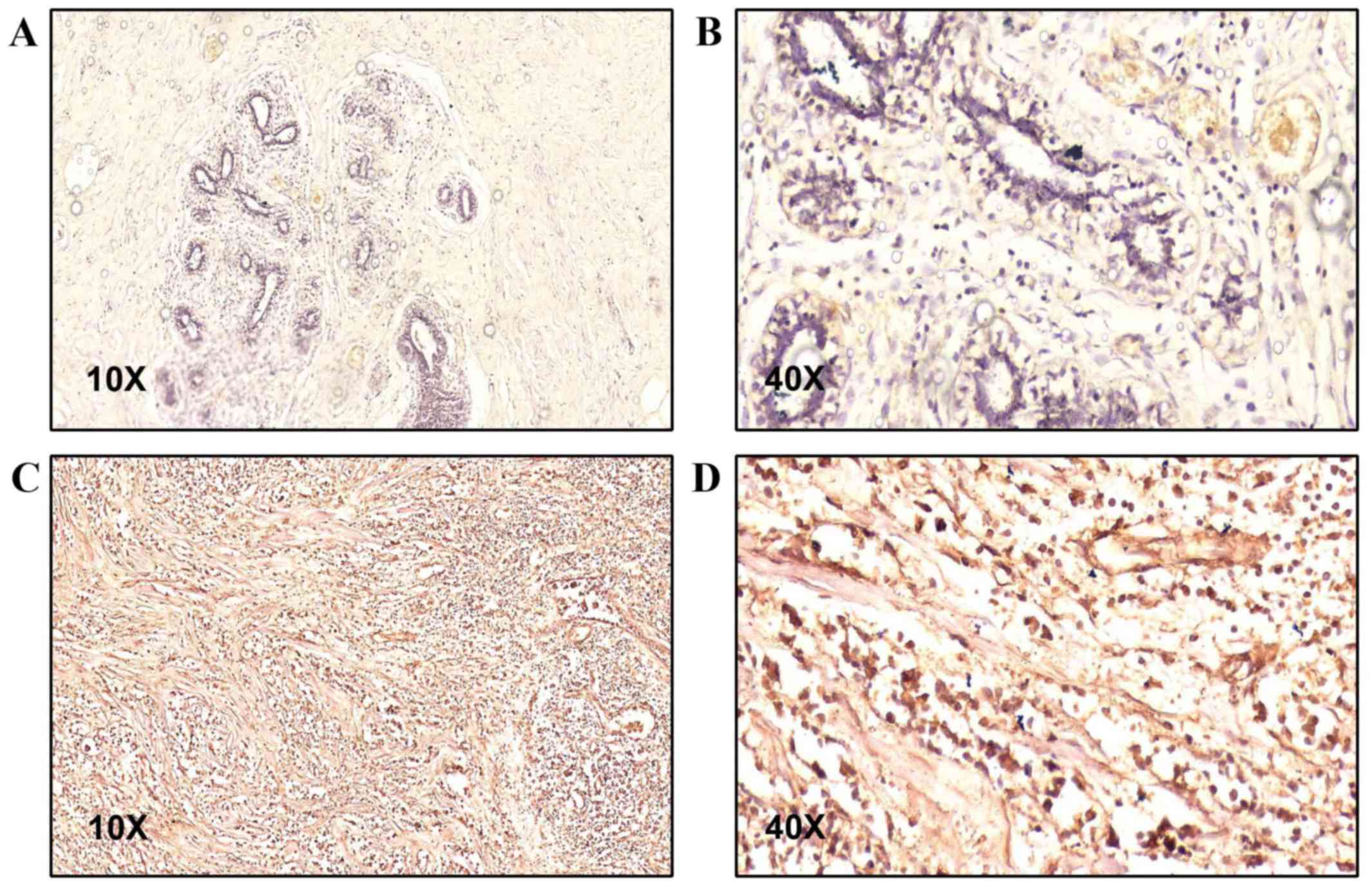
Results showed that TBX3 protein is overexpressed in tumour tissues
compared with normal tissues among breast cancer patients (Fig. 1). The mean of TBX3 level in tumour
tissues is 10.9±2.5 and in normal tissues is 6.7±3.8 (P<0.001).
Importantly, 21 patients (47.73%) showed strong positive TBX3
staining in tumour tissue and only 5 (11.36%) of normal tissues
showed similar levels of TBX3. While only two tumour samples
(4.54%) showed weak TBX3 level, 11 samples (25%) of normal tissues
showed weak TBX3 level. Moreover, no tumour tissues showed
completely negative TBX3 level but 2 (4.54%) normal tissues showed
negative TBX3 stain. The expression level of TBX3 in normal and
tumour tissue are summarized in Table
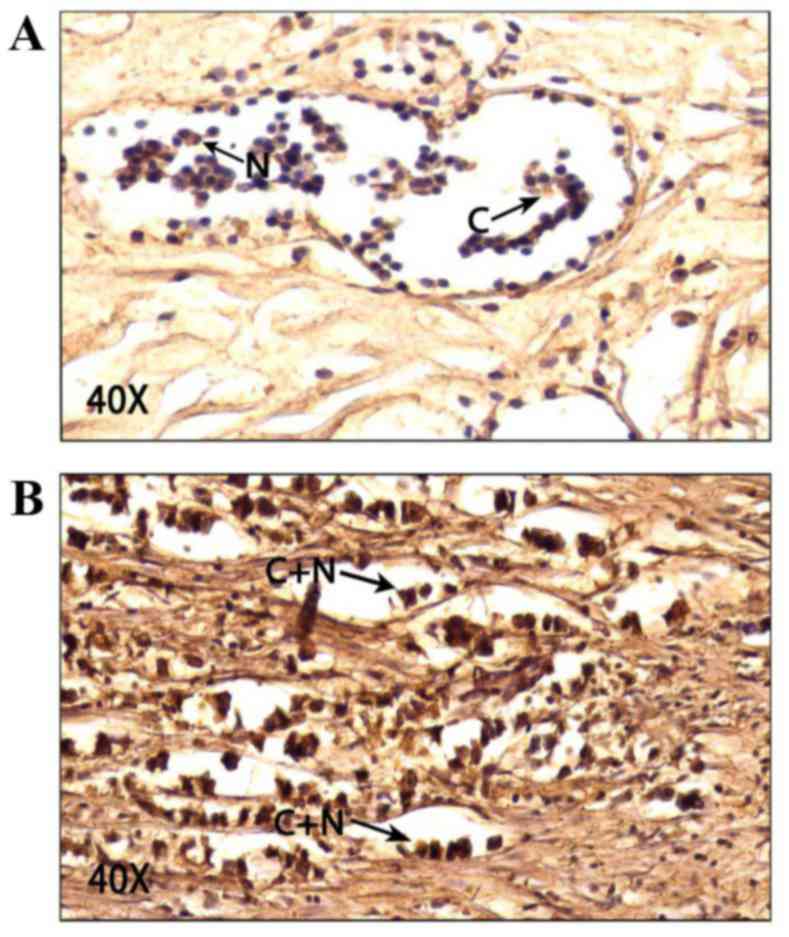
III. Importantly, our results revealed that TBX3 is mainly
cytoplasmic in both normal and breast cancer tissues (Fig. 2). According to our scoring system,
the mean of cytoplasmic TBX3 level in normal cells is 5.3±1.6
compared with 2.6±2 in the nucleus and this difference is
significant with P<0.001 (Table
IV). Similarly, the mean of cytoplasmic TBX3 level in tumour
cells is 6.7±1.4 compared with 4.2±2.2 in the nucleus and this
difference is also significant (P<0.001). These results show
that TBX3 is significantly overexpressed in breast cancer
cells.
 | Table III.Expression level of TBX3 in normal
and tumour tissues. |
Table III.
Expression level of TBX3 in normal
and tumour tissues.
|
|
| TBX3 expression
(n) |
|
|---|
|
|
|
|
|
|---|
|
| Total no. | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | P-value |
|---|
| Normal | 44 | 2 (4.54) | 11 (25) | 26 (59.1) | 5 (11.36) | <0.001 |
| Tumour | 44 | 0 (0) | 2 (4.54) | 21 (47.73) | 21 (47.73) | <0.001 |
 | Table IV.Cytoplasmic and nuclear Tbx3 levels
in normal and tumour tissues. |
Table IV.
Cytoplasmic and nuclear Tbx3 levels
in normal and tumour tissues.
| Tbx3 proteins | Normal cells
(n=44) | Tumour cells
(n=51) | t | P-value |
|---|
| Cytoplasmic level,
Mean ± SD (Min-Max) | 5.3±1.6 (0–8) | 6.7±1.4 (3–8) | 4.851 | <0.001 |
| Nuclear level, Mean
± SD (Min-Max) | 2.6±2 (0–6) | 4.2±2.2 (0–8) | 3.731 | <0.001 |
TBX3 is a promising diagnostic marker
for breast cancer
To explore the significance of TBX3 as a diagnostic
marker of breast cancer, we analysed the obtained data using the
ROC curve and AUC. Youden's index was used to determine the cut-off
value for TBX3 level in tumour tissues compared to normal tissues.
These methods were used by several studies to show the diagnostic
value of tumour makers including carcinoembryonic antigen (CEA),
cancer antigen 19-9 (CA19-9), cancer antigen 125 (CA125), CA15-3
and tissue polypeptide-specific antigen (TPS) in metastatic breast
cancer (27–30).
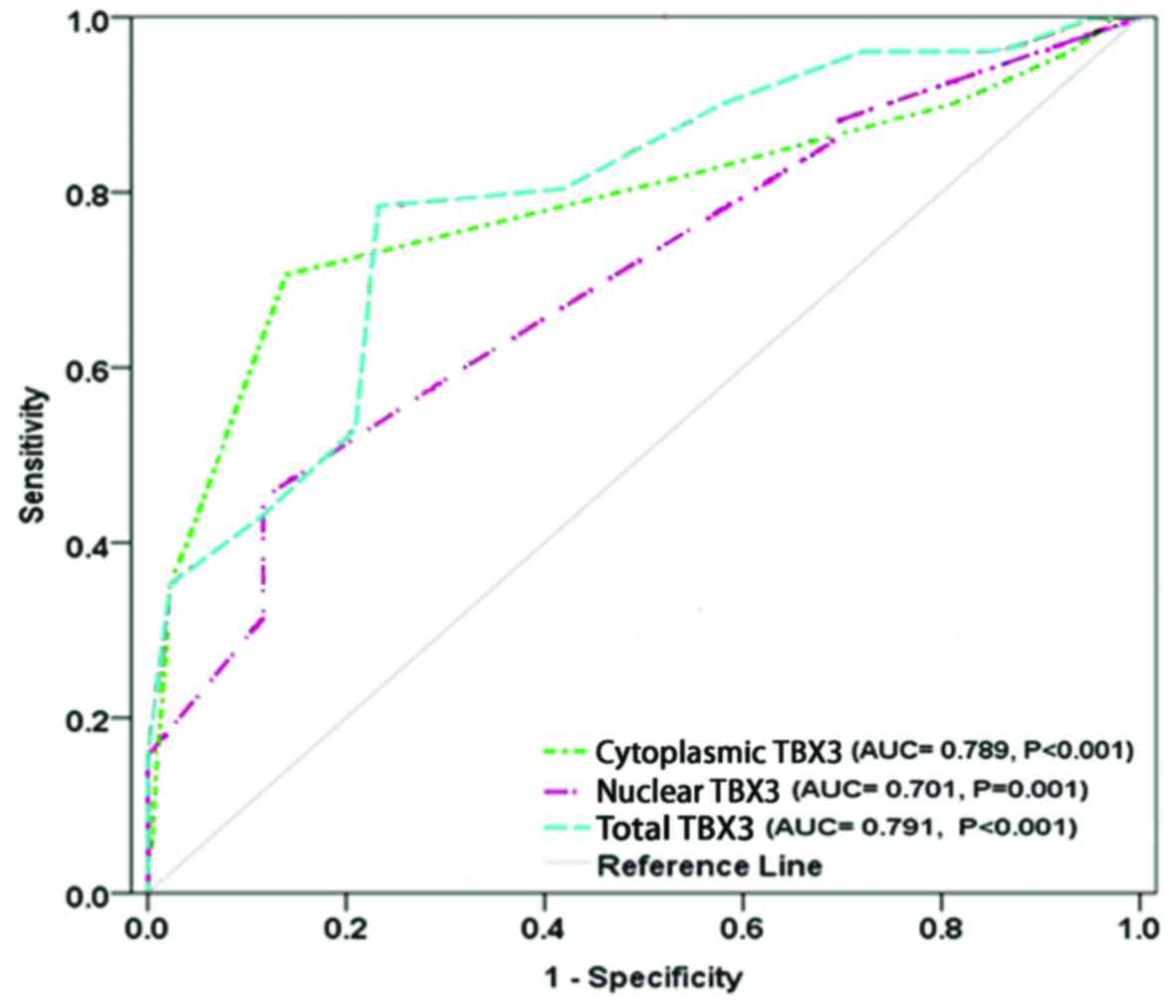
The results showed that TBX3 protein is
statistically significant diagnostic marker for breast cancer
(P<0.001). The cut-off value for TBX3 level was 9 of a total
score 16. The AUC was 0.791 (P<0.001), sensitivity and
specificity were 78.4 and 79.6% respectively. PPV was 80%, NPV was
78%, and accuracy of TBX3 as diagnostic breast cancer marker was
79%. The cut-off value for cytoplasmic TBX3 was 6 of a total score
8. The AUC was 0.798 (P<0.001), sensitivity was 70.6%,
specificity was 86.4%, PPV was 85.7, NPV was 71.7, and accuracy was
77.9%. In addition to the cytoplasmic TBX3, the cut-off value of
nuclear TBX3 was 4 of a total score 8, the AUC was 0.701 (P=0.001),
sensitivity was 45.1%, specificity was 88.6%, PPV was 82.1, NPV was
58.2, and accuracy was 65.3% (Table
V and Fig. 3). Furthermore,
cytoplasmic TBX3 has higher accuracy than nuclear TBX3 whereas
nuclear TBX3 has higher specificity than cytoplasmic TBX3. However,
in comparison to the cytoplasmic and nuclear TBX3, total TBX3 level
showed the best diagnosability for breast cancer with the highest
sensitivity of 78.4%, highest AUC of 0.791 (0.700–0.882) at cut-off
value=9. Which means that TBX3 staining assay has the ability to
distinguish between normal and tumour tissues. Together, these
findings suggest total TBX3 level as a potential diagnostic marker
for breast cancer.
 | Table V.Youden index cut-off values,
sensitivity, specificity, PPV, NPV and AUC of TBX3 biomarker for
breast cancer diagnoses. |
Table V.
Youden index cut-off values,
sensitivity, specificity, PPV, NPV and AUC of TBX3 biomarker for
breast cancer diagnoses.
| Biomarker | Cut-off point | Tumor tissues
(n=51) | Normal Tissues
(n=44) | Sensitivity
(%) | Specificity
(%) | Accuracy (%) | PPV (%) | NPV (%) | AUC (95% CI) | P-value |
|---|
| Cytoplasmic | ≤6 | 15 | 38 | 70.6 | 86.4 | 77.9 | 85.7 | 71.7 | 0.789 | <0.001 |
| TBX3 (0–8) | >6 | 36 | 6 |
|
|
|
|
| (0.695–0.882) |
|
| Nuclear | ≤4 | 28 | 39 | 45.1 | 88.6 | 65.3 | 82.1 | 58.2 | 0.701 | 0.001 |
| TBX3 (0–8) | >4 | 23 | 5 |
|
|
|
|
| (0.597–0.805) |
|
| Total TBX3 | ≤9 | 11 | 34 | 78.4 | 79.6 | 79.0 | 80.0 | 78.0 | 0.791 | <0.001 |
| level (0–16) | >9 | 40 | 10 |
|
|
|
|
| (0.700–0.882) |
|
Associations between
clinicopathological parameters and TBX3 expression
Our results indicate that there are no significant
correlations between TBX3 protein level and clinicopathological
parameters in breast cancer at confidence level 0.05. These
clinicopathological parameters include age, anatomic stage, cancer
type, histopathological grade, tumour size, lymph node size, lymph
node metastasis and tumour metastasis. Similarly, no significant
correlation between TBX3 level and ER, PR, HER2 and CA15-3 levels
as tested by Spearman rank correlation, KW(H) test and Mann-Whitney
(U) test (Tables VI and VII). Therefore, these findings suggest
TBX3 overexpression as biomarker for human breast cancer regardless
of age, stage, grade, metastasis, non-metastasis, and breast cancer
type.
 | Table VI.Correlations between TBX3 level and
clinicopathological parameters among study population. |
Table VI.
Correlations between TBX3 level and
clinicopathological parameters among study population.
|
| Correlation with
TBX3 level in tumor cells |
|---|
|
|
|
|---|
| Parameters | R | P-value |
|---|
| Age (years) | −0.130 | 0.364 |
| Tumour size
(cm) | −0.038 | 0.791 |
| LN metastasis
(%) | 0.160 | 0.263 |
| LN size (cm) | −0.072 | 0.638 |
| ER | −0.004 | 0.982 |
| PR | 0.016 | 0.930 |
| HER2 | 0.092 | 0.622 |
| CA15-3 | −0.115 | 0.670 |
 | Table VII.Differences of TBX3 levels among
tumour stages in breast cancer patients. |
Table VII.
Differences of TBX3 levels among
tumour stages in breast cancer patients.
|
|
| Total TBX3
(0–16) | Statistical
analysis |
|---|
|
|
|
|
|
|---|
| Variable | Total n | Median | IQR
(25th-75th) | MW/KW | P-value |
|---|
| Type of tumour |
|
|
| 203.017 | 0.567 |
|
ILC | 12 | 7.5 | 0–13 |
|
|
|
IDC | 38 | 7 | 0–12 |
|
|
| Tumour
metastasis |
|
|
| 178.525 | 0.882 |
| M0 | 41 | 11 | 10–13 |
|
|
| M1 | 9 | 11 | 9–13 |
|
|
| Histopathological
grade |
|
|
| 0.215 | 0.898 |
| Grade
1 | 3 | 9 | 0–11 |
|
|
| Grade
2 | 31 | 7 | 0–12 |
|
|
| Grade
3 | 2 | 6 | 0–12 |
|
|
| Stage of
cancer |
|
|
| 7.011 | 0.072 |
| I | 2 | 14 | 14–14 |
|
|
| II | 19 | 10 | 8–12 |
|
|
|
III | 20 | 12 | 10–13 |
|
|
| IV | 9 | 11 | 9–13 |
|
|
Discussion
TBX3 is a member of T-box transcription factors
family that plays critical roles in development and oncogenic
process (31). Many evidence point
out a functional linkage between T-box genes, cellular
differentiation/proliferation, and tumorigenesis, especially breast
cancers (32). Also, it has been
involved in a widespread range of carcinomas like, ovarian,
cervical, pancreatic, bladder, liver and melanoma.
The present study is a cross-sectional and
hospital-based study included 51 breast cancer patient samples.
Tissues were collected by mastectomy from breast cancer patients
and embedded in paraffin blocks. Usually, in breast cancer samples
there are cancer cells and normal cells and generally the normal
cells are located in the edge of the sample, therefore the level of
TBX3 was estimated by immunohistochemistry analysis in both normal
and cancer cells. Histologically normal tissue adjacent to the
tumour (NAT) is commonly used as a control in cancer studies.
However, little is known about the transcriptomic profile of NAT,
how it is influenced by the tumour, and how the profile compares
with non-tumour-bearing tissues (33). Still, the study of tumour biology,
irrespective of approach, requires controls. Using normal adjacent
tissue as this control has many advantages, such as the relative
ease of access and the control for variability between individuals
and anatomic sites. Our results showed that TBX3 is significantly
overexpressed in breast cancer tissues. This is in agreement with
several previous in vitro and in vivo studies. For
example, an early study tested several breast cancer cell lines
showed that TBX3 is overexpressed in many breast cancer cell lines,
where it plays a role in inhibition of the senescence (25). A study by Yarosh et al
(21) suggested that TBX3 might be
overexpressed in patients breast cancer tissues. This study was
carried out in the United States and showed that TBX3 is
significantly overexpressed in 42 breast cancer tissues in
comparison to normal tissues. Importantly, Yarosh et al
tested breast cancer samples of early stages but the current study
included all stages of breast cancer patients. Additionally, while
the previous study showed that TBX3 is mainly cytoplasmic in breast
cancer cells only, we showed that TBX3 is mainly cytoplasmic
protein in both normal and breast cancer tissues. Significantly,
the current study provides more statistical analysis to show the
importance of TBX3 as a marker of breast cancer cells. Another
recent study found that TBX3 protein has different levels and
localizations in the different stages of cell cycle (1). The study showed that TBX3 levels
increases during G1 and peaks in S-phase. While the protein is both
nuclear and cytoplasmic in G1 and G2, it is predominantly nuclear
in S-phase cells. These results provide evidence that TBX3 levels
and localization is regulated during the cell cycle and that it may
have a more important role in S-phase. Other studies showed that
TBX3 stabilization and subcellular localization depend on its
phosphorylation at specific sites (14). All together, these studies and our
current findings show that TBX3 is overexpressed in breast cancer
tissues where it could be used as diagnostic or prognostic marker.
Furthermore, it provides evidence that TBX3 localizes differently
depending on its role and the cell cycle phase (34–36).
However, due to the small number of our patients, the results of
ROC analysis need validation using an independent breast cancer
cohort. Furthermore, in our study, we didn't compare between TBX3
levels in cancer patients vs. healthy subjects but we showed that
TBX3 might be suitable to distinguish between tumor cells and
normal cells in breast cancer patients. Finally, the current study
as well as different previous studies (21,23,26)
didn't use specific markers for specific organelles and
alternatively pathologists depend on the morphological features of
the organelles to identify them, which might be considered as
another a limitation of this technique.
Acknowledgements
The authors would like to acknowledge the
Palestinian Ministry of Health for their assistance in sample
collection.
Funding
The present study was supported by the Qatar Charity
under the Ibhath project for research grants, which is funded by
the Cooperation Council for the Arab States of the Gulf through the
Islamic Development Bank.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SA designed the study, supervised the experimental
procedures such as section preparations and staining, performed the
statistical analyses and was a major contributor in writing the
manuscript. AL, HH and FR diagnosed the breast cancer sections and
evaluated TBX3 levels in the cancerous and normal sections. AS was
the oncologist who diagnosed and treated all of the patients, and
AS also assisted with writing the paper. AM performed the
statistical analyses. HT collected and prepared the samples and
contributed to writing the paper. MR and MN performed sectioning
and staining the different tissues. BA designed the study and wrote
the paper with SA.
Ethics approval and consent to
participate
The present retrospective study was approved by the
Helsinki Committee of the Palestinian Health Research Council
(approval no. PHRC/HC/93/16). Each patient provided written
informed consent, which was conducted with the kind assistance of
the Palestinian Ministry of Health.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Willmer T, Peres J, Mowla S, Abrahams A
and Prince S: The T-Box factor TBX3 is important in S-phase and is
regulated by c-Myc and cyclin A-CDK2. Cell Cycle. 14:3173–3183.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheng HD, Lui YM and Freimanis RI: A novel
approach to microcalcification detection using fuzzy logic
technique. IEEE Trans Med Imaging. 17:442–450. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
American Cancer Society: Cancer Treatment
and Survivorship Facts and Figures 2012–2013. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/cancer-treatment-and-surviorship-facts-and-figures/cancer-treatment-and-survivorship-facts-and-figures-2012-2013.pdfJanuary
1–2012
|
|
4
|
Ashkenazi A: Targeting the extrinsic
apoptosis pathway in cancer. Cytokine Growth Factor Rev.
19:325–331. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Naiche LA, Harrelson Z, Kelly RG and
Papaioannou VE: T-box genes in vertebrate development. Annu Rev
Genet. 39:219–239. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peres J and Prince S: The T-box
transcription factor, TBX3, is sufficient to promote melanoma
formation and invasion. Mol Cancer. 12:1172013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bilican B and Goding CR: Cell cycle
regulation of the T-box transcription factor tbx2. Exp Cell Res.
312:2358–2366. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Burgucu D, Guney K, Sahinturk D, Ozbudak
IH, Ozel D, Ozbilim G and Yavuzer U: Tbx3 represses PTEN and is
over-expressed in head and neck squamous cell carcinoma. BMC
Cancer. 12:4812012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Demay F, Bilican B, Rodriguez M, Carreira
S, Pontecorvi M, Ling Y and Goding CR: T-box factors: Targeting to
chromatin and interaction with the histone H3 N-terminal tail.
Pigment Cell Res. 20:279–287. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hoogaars WM, Barnett P, Rodriguez M, Clout
DE, Moorman AF, Goding CR and Christoffels VM: TBX3 and its splice
variant TBX3 + exon 2a are functionally similar. Pigment Cell
Melanoma Res. 21:379–387. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Humtsoe JO, Koya E, Pham E, Aramoto T, Zuo
J, Ishikawa T and Kramer RH: Transcriptional profiling identifies
upregulated genes following induction of epithelial-mesenchymal
transition in squamous carcinoma cells. Exp Cell Res. 318:379–390.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peres J, Davis E, Mowla S, Bennett DC, Li
JA, Wansleben S and Prince S: The highly homologous T-Box
transcription factors, TBX2 and TBX3, have distinct roles in the
oncogenic process. Genes Cancer. 1:272–282. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hoek K, Rimm DL, Williams KR, Zhao H,
Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, et
al: Expression profiling reveals novel pathways in the
transformation of melanocytes to melanomas. Cancer Res.
64:5270–5282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peres J, Mowla S and Prince S: The T-box
transcription factor, TBX3, is a key substrate of AKT3 in
melanomagenesis. Oncotarget. 6:1821–1833. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Douglas NC and Papaioannou VE: The T-box
transcription factors TBX2 and TBX3 in mammary gland development
and breast cancer. J Mammary Gland Biol Neoplasia. 18:143–147.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu G, Sinclair C, Hinson S, Ingle JN,
Roche PC and Couch FJ: Structural analysis of the 17q22-23 amplicon
identifies several independent targets of amplification in breast
cancer cell lines and tumors. Cancer Res. 61:4951–4955.
2001.PubMed/NCBI
|
|
17
|
Jacobs JJ, Keblusek P, Robanus-Maandag E,
Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ,
Koh EY, Daley GQ, et al: Senescence bypass screen identifies TBX2,
which represses Cdkn2a (p19 (ARF)) and is amplified in a subset of
human breast cancers. Nat Genet. 26:291–299. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Redmond KL, Crawford NT, Farmer H, D'Costa
ZC, O'Brien GJ, Buckley NE, Kennedy RD, Johnston PG, Harkin DP and
Mullan PB: T-box 2 represses NDRG1 through an EGR1-dependent
mechanism to drive the proliferation of breast cancer cells.
Oncogene. 29:3252–3262. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu J, Ma X, Cheung KF, Li X, Tian L, Wang
S, Wu CW, Wu WK, He M, Wang M, et al: Epigenetic inactivation of
T-box transcription factor 5, a novel tumor suppressor gene, is
associated with colon cancer. Oncogene. 29:6464–6474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wansleben S, Davis E, Peres J and Prince
S: A novel role for the anti-senescence factor TBX2 in DNA repair
and cisplatin resistance. Cell Death Dis. 4:e8462013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yarosh W, Barrientos T, Esmailpour T, Lin
L, Carpenter PM, Osann K, Anton-Culver H and Huang T: TBX3 is
overexpressed in breast cancer and represses p14 ARF by interacting
with histone deacetylases. Cancer Res. 68:693–699. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lomnytska M, Dubrovska A, Hellman U,
Volodko N and Souchelnytskyi S: Increased expression of cSHMT, Tbx3
and utrophin in plasma of ovarian and breast cancer patients. Int J
Cancer. 118:412–421. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Leake R, Barnes D, Pinder S, Ellis I,
Anderson L, Anderson T, Adamson R, Rhodes T, Miller K and Walker R:
Immunohistochemical detection of steroid receptors in breast
cancer: A working protocol. UK Receptor Group, UK NEQAS, The
Scottish Breast Cancer Pathology Group, and The Receptor and
Biomarker Study Group of the EORTC. J Clin Pathol. 53:634–635.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sasaki T, Ryo A, Uemura H, Ishiguro H,
Inayama Y, Yamanaka S, Kubota Y, Nagashima Y, Harada M and Aoki I:
An immunohistochemical scoring system of prolyl isomerase Pin1 for
predicting relapse of prostate carcinoma after radical
prostatectomy. Pathol Res Pract. 202:357–364. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fan W, Huang X, Chen C, Gray J and Huang
T: TBX3 and its isoform TBX3+2a are functionally distinctive in
inhibition of senescence and are overexpressed in a subset of
breast cancer cell lines. Cancer Res. 64:5132–5139. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sasaki K, Bastacky SI, Zynger DL and
Parwani AV: Use of immunohistochemical markers to confirm the
presence of vas deferens in vasectomy specimens. Am J Clin Pathol.
132:893–898. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Gao H, Chen Z, Zhang L, Zhu X, Wang
S and Peng W: Diagnosis of breast cancer based on
microcalcifications using grating-based phase contrast CT. Eur
Radiol. 28:3742–3750. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu SG, He ZY, Zhou J, Sun JY, Li FY, Lin
Q, Guo L and Lin HX: Serum levels of CEA and CA15-3 in different
molecular subtypes and prognostic value in Chinese breast cancer.
Breast. 23:88–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Di Gioia D, Blankenburg I, Nagel D,
Heinemann V and Stieber P: Tumor markers in the early detection of
tumor recurrence in breast cancer patients: CA 125, CYFRA 21-1,
HER2 shed antigen, LDH and CRP in combination with CEA and CA 15-3.
Clin Chim Acta. 461:1–7. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang W, Xu X, Tian B, Wang Y, Du L, Sun T,
Shi Y, Zhao X and Jing J: The diagnostic value of serum tumor
markers CEA, CA19-9, CA125, CA15-3, and TPS in metastatic breast
cancer. Clin Chim Acta. 470:51–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Willmer T, Hare S, Peres J and Prince S:
The T-box transcription factor TBX3 drives proliferation by direct
repression of the p21(WAF1) cyclin-dependent kinase inhibitor. Cell
Div. 11:62016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Horton AC, Mahadevan NR, Minguillon C,
Osoegawa K, Rokhsar DS, Ruvinsky I, de Jong PJ, Logan MP and
Gibson-Brown JJ: Conservation of linkage and evolution of
developmental function within the Tbx2/3/4/5 subfamily of T-box
genes: Implications for the origin of vertebrate limbs. Dev Genes
Evol. 218:613–628. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aran D, Camarda R, Odegaard J, Paik H,
Oskotsky B, Krings G, Goga A, Sirota M and Butte AJ: Comprehensive
analysis of normal adjacent to tumor transcriptomes. Nat Commun.
8:10772017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kunasegaran K, Ho V, Chang TH, De Silva D,
Bakker ML, Christoffels VM and Pietersen AM: Transcriptional
repressor Tbx3 is required for the hormone-sensing cell lineage in
mammary epithelium. PLoS One. 9:e1101912014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sunwoo HH and Suresh MR: Cancer markers.
In The ImmunoassayHandbook. 1. 4th edition. Elsevier Ltd.; Oxford:
pp. 673–693. 2013
|
|
36
|
Willmer T, Cooper A, Peres J, Omar R and
Prince S: The T-Box transcription factor 3 in development and
cancer. Biosci Trends. 11:254–266. 2017. View Article : Google Scholar : PubMed/NCBI
|