Introduction
Signet ring cell carcinoma (SRCC) is a rare
histological type in colorectal cancer (CRC) and it dominates only
1% of them (1). The prognosis of
SRCC patients is extremely poor and it is difficult to improve it
in spite of the multimodal treatment including surgery (2). Thus, to fight against this type of
deadly disease, several analyses were attempted to find early
clinicopathological signs and genetic features that specifically
reflect the nature of SRCC in the colorectal cancer field (1-6).
The commonly speculated clinical factors of
colorectal SRCC are relatively younger patients, female-dominated,
advanced disease stage, right-sided, and treatment-resistant
(7-14).
Additionally, CpG island methylator phenotype- high (CIMP-high),
microsatellite instability-high (MSI-high), and BRAF mutation were
commonly reported as genetic features of SRCC (15-18).
According to these analyses, the specific character of colorectal
SRCC was gradually being unveiled. However, there still be a long
way to go to establish a more efficient treatment strategy for SRCC
because it is not fully characterized yet.
Consensus Molecular Subtype (CMS) analysis was
proposed by Guinney et al (19). They tried to characterize colorectal
cancer into 4 subtypes by utilizing gene expression profile data
set being aggregated from different gene expression analysis
platforms. Several clinical trials were performed and the benefit
of CMS analysis was reported (20-22).
The morbidity of SRCC in colorectal cancer is so rare that it is
not clearly known about where this histological type of cancer is
designated in CMS analysis.
The purpose of this study is to re-confirm the
clinicopathological specificity of SRCC in colorectal cancer and
try to elucidate the SRCC's gene expression profile and the type of
classification in CMS analysis. Of course, there would be some
limitations while utilizing the CMS analysis because the
classification is not widely clinical used yet and still needs to
be fine-tuned because 20% of the case does not fall within the four
subtypes, however through this study, some clues of biological
insight can be dug out about this extremely rare histological type
of colon cancer.
Materials and methods
Study patients and diagnosis
A total of 1,350 patients who had been diagnosed
with colorectal cancer and had undergone complete resection of the
tumor from 1997 to 2011 in our department were enrolled in this
study. Of these patients, 14 were pathologically diagnosed with
SRCC, and the remaining 1,336 were diagnosed with well to
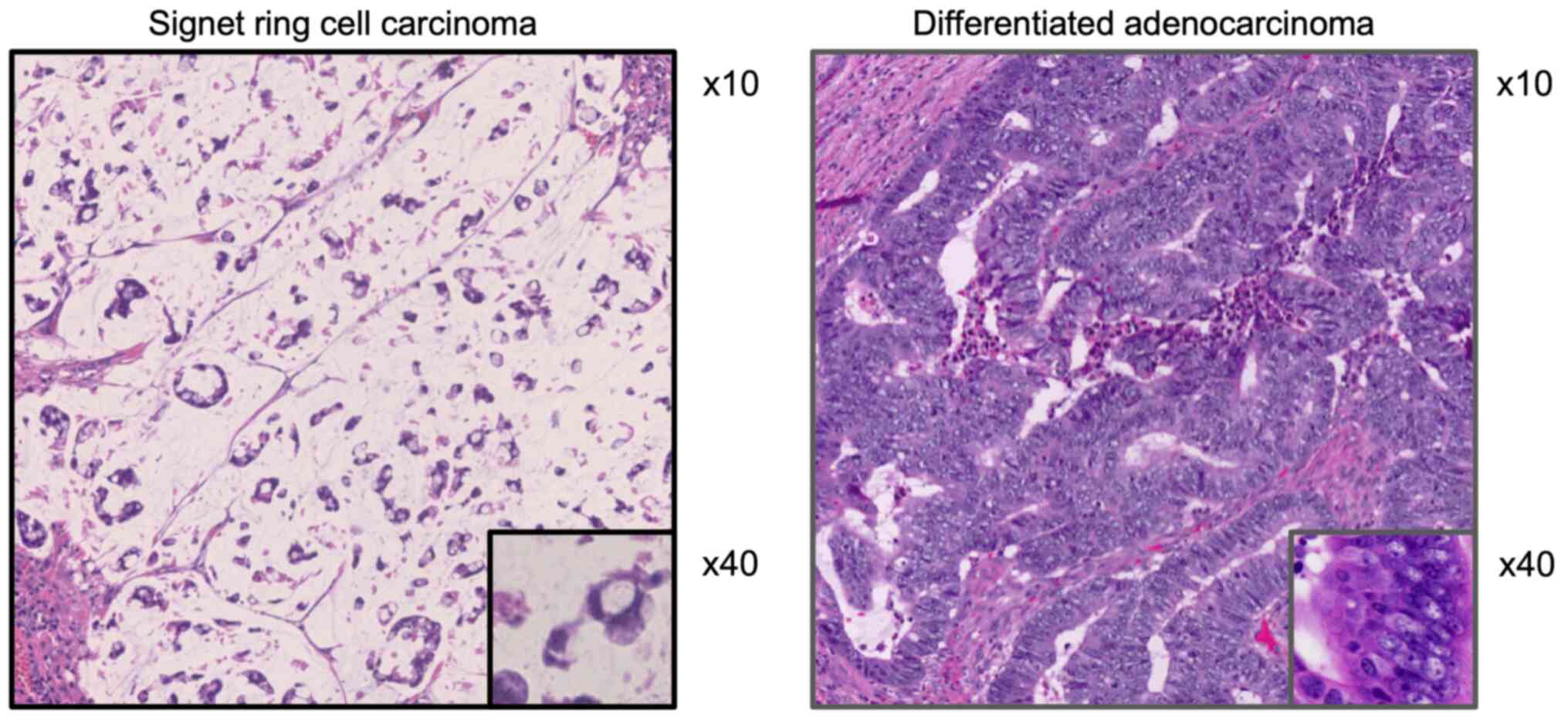
moderately differentiated adenocarcinoma (well-mode DAC) (Fig. 1). Postoperative pathological staging
was determined according to the seventh edition of the UICC TNM
classification of malignant tumors. None of the enrolled patients
underwent any chemotherapeutic treatment prior to surgery.
RNA extraction
Total RNA was extracted from frozen tumor sections
(two SRCC, two differentiated AC, and five normal tissue samples)
using the RNeasy Mini kit (Qiagen) according to the manufacturer's
instructions, and the samples were stored at -80˚C.
RNA amplification and labeling
cDNA was amplified from 20 ng total RNA using the
Ovation® Pico WTA System V2 (NuGEN Technologies)
according to the manufacturer's instructions. The amplified cDNA
yield was checked using the NanoDrop 2000 spectrophotometer.
Cyanine-3-labeled cDNA was prepared from 2.0 µg cDNA using the
SureTag Complete DNA Labeling Kit (Agilent Technologies) according
to the manufacturer's instructions, and then purified and
concentrated using Amicon Ultra-0.5 ml Centrifugal Filters (Merck
Millipore). Dye incorporation and cDNA yield were checked using the
NanoDrop 2000 spectrophotometer.
cDNA microarray analysis
The cyanine-3-labeled cDNA was mixed with 1x
blocking agent and 1X hybridization buffer (50 µl total volume),
and the solution was hybridized to SurePrint G3 Human GE 8x60K
Microarray v2.0 slides for 17 h at 65˚C in a rotating hybridization
oven. After hybridization, the microarrays were washed with GE Wash
Buffer 1 for 1 min at room temperature followed by GE Wash Buffer 2
for 1 min at 37˚C and then dried immediately by brief
centrifugation. The microarrays were scanned using the
High-Resolution Microarray Scanner (Agilent Technologies) to
determine fluorescence intensity.
CMS classification
To perform consensus molecular subtype (CMS)
classification, statistical computational software R (ver3.6.1: R
Core Team (2019). R: A language and environment for statistical
computing. R Foundation for Statistical Computing, Vienna, Austria.
URL https://www.R-project.org/) was
utilized. The data for the CMS analysis was prepared according to
the report of Guinney et al (19) and the actual CMS classification was
done following the instruction of R-package CMScaller (ver0.99.1)
(23). Prior to the analysis,
acquired cDNA microarray raw data were normalized and standardized.
According to the subtypes of CMS1, CMS2, CMS3, and CMS4
classification, two of each subtype data (total 8 data) were chosen
from the crc TCGA subset (colorectal TCGA gene expression data with
subtype annotation) included in the CMScaller and utilized as a
reference data. SurePrint G3 microarray chip used in this SRCC
study mounted more than 60,000 genes thus 5,000 gene expression
data that is matched with the genes utilized in CMScaller were
extracted. In the SurePrint G3 chip the same genes were analyzed
multiply so that those genes data were applied to the CMS analysis
after calculating normalization values according to the average of
each expression data by global scaling. After the data adjustment,
the emat data sheet was developed and CMS caller analysis was
performed.
Statistical analysis
Analyses of the clinicopathological variables and
histological status of the patients were performed using JMP Pro
14.0.0 statistical software (SAS Institute, Inc.).
Clinicopathological factor correlations were compared between the
SRCC and case-control data set using Student's t-test and the
chi-square test. Progression-free survival (PFS) and
cancer-specific survival (CSS) rates were calculated using the
Kaplan-Meier method and compared using the log-rank test and
Wilcoxon test. Correlations between clinicopathological factors and
cancer-related death were estimated using the Cox proportional
hazards model. A P-value <0.05 was considered to indicate
significance.
Results
Clinicopathological characteristics of
patients and outline of treatment with SRCC patients
Patients who had been treated for colorectal cancer
from 1997 to 2011 at the Department of Surgery of Kurume University
Hospital were enrolled in this study. Of 1,350 total patients, 14
were pathologically diagnosed with signet ring cell carcinoma
(SRCC). The background and clinicopathological summary variables of
SRCC the enrolled patients are summarized in Table I. There were 12 of 14 cases were
male patients and the percentage of female patients was 14.2% in
SRCC. The median value of SRCC patients' age was 62.57±10.68. There
were only two cases of T1-T2 in tumor depth (14.2%) on the contrary
remaining 14 cases were all T4a or T4b. Nine cases showed N2 or
more lymph node metastasis and 5 cases were N0. There were 4 out of
5 cases presented peritoneal dissemination in distant metastasis.
As a result of that pathological stage was as follows: Stage I 2
cases (14.2%), stage II 1 case (7.1%), stage III 6 cases (42.9%)
and stage IV 4 cases (28.6%).
 | Table IClinicopathological summary of
fourteen cases of signet ring cell carcinoma. |
Table I
Clinicopathological summary of
fourteen cases of signet ring cell carcinoma.
| Patient no. | Sex | Age | Tumor depth | Lymph node
metastasis | Distant metastasis
(location) | Pathological
stage | Tumor location
right/left (location) | Surgical
procedure | Adjuvant
therapy | Recurrence | Familial
history |
|---|
| 1 | M | 55 | T3 | N0 | M0 | IIA | R(A) | RHC+D3 | no | no | no |
| 2 | M | 69 | T4a | N2b | M0 | IIIC | L(Rb) | APR+D3 | no | lung | no |
| 3 | M | 53 | T4a | N2a | M0 | IIIC | R(A) | RHC+D3 | CDDP+5FU | no | no |
| 4 | M | 65 | T1 | N0 | M0 | I | R(C) | ICR+D3 | no | no | yes |
| 5 | M | 52 | T4a | N2a | M0 | IIIC | L(Rb) | APR+D3 | UFT | local | yes |
| 6 | M | 66 | T4a | N2a | M1a (liver) | IVA | R(A) | RHC+D3 | no | no | no |
| 7 | M | 76 | T4b | N2 | M0 | IIIC | L(Rb) | APR+D3 | UFT+radiation | local | yes |
| 8 | F | 55 | T4a | N0 | M1b
(peritoneum) | IVB | R(C) | ICR+D3 |
IRIS->mFOLFOX-> FOLFIRI | peritoneum | no |
| 9 | M | 58 | T4a | N0 | M1b
(peritoneum) | IVB | R(A) | ICR+D3 | FOLFIRI+Bv | no | yes |
| 10 | F | 82 | T4a | N2a | M0 | IIIC | R(A) | RHC+D3 | S-1 | no | no |
| 11 | M | 80 | T4b | N2 | M0 | IIIC | L(Rb) | APR+D3 | no | local | no |
| 12 | M | 58 | T1 | N0 | M0 | I | L(Rb) | LAR+D3 | no | no | no |
| 13 | M | 49 | T4a | N3 | M1b
(peritoneum) | IVB | R(C) | RHC+D3 | FOLFOX | peritoneum | no |
| 14 | M | 58 | T4a | N3 | M1b
(peritoneum) | IVB | L(Rb) | LAR+D3 | FOLFOX | no | yes |
The locations of the primary tumors were right-sided
in 8 (57.1%) cases and left-sided in 6 cases (42.9%). All of the
SRCC patients had taken a primary tumor resection and D3
lymphadenectomy. Adjuvant chemotherapy or chemotherapy plus
radiotherapy was performed in 8 out of 14 cases. There were 6 out
of 8 cases who underwent adjuvant therapy experienced tumor
recurrence and metastasis. Lung metastasis was verified in one case
and the remaining 5 cases were all peritoneal dissemination
recurrence.
Background matched case-control
study
A case-controlled cohort that matched with the SRCC
cohort in the same treatment period in well and moderately
differentiated adenocarcinoma cases was constructed to re-confirm
the clinicopathological and prognostic features of SRCC. The
cancer-specific survival (CSS) and the progression-free survival
(PFS) were compared between the SRCC group and the case-control
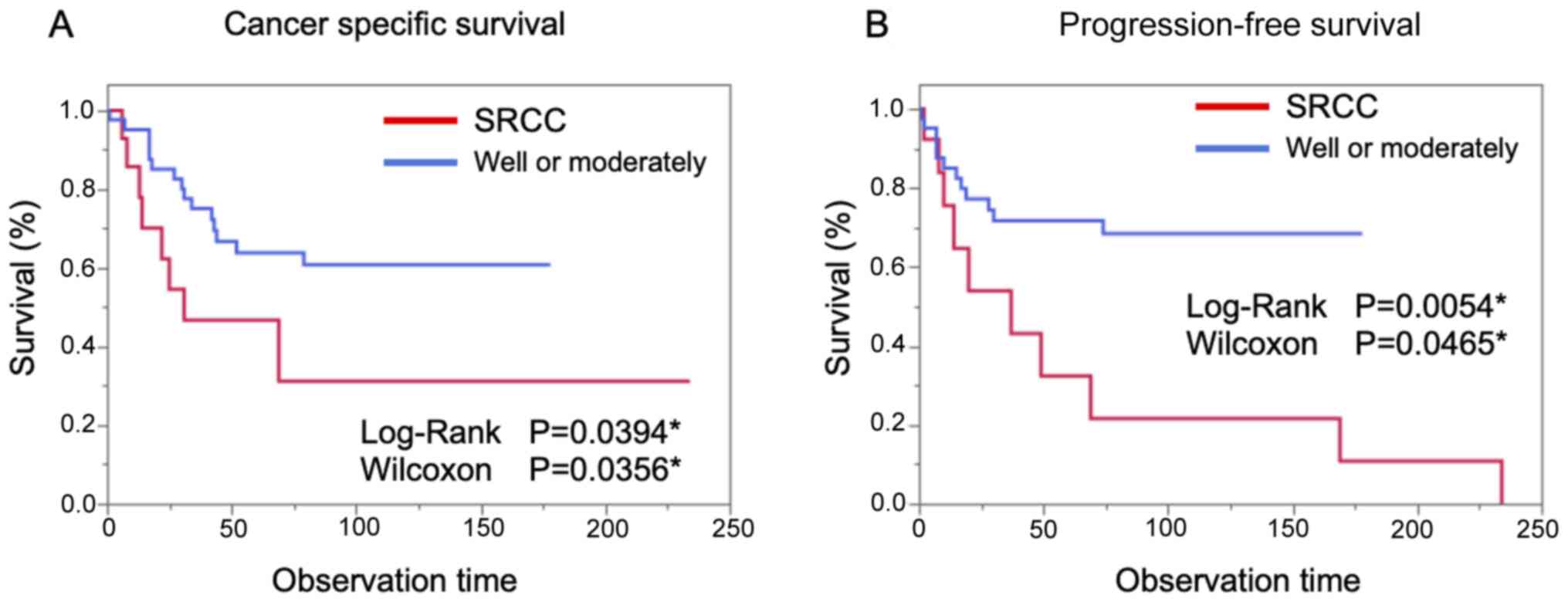
cohort group. As shown in Fig. 2
the prognosis of SRCC in CSS and in PFS was significantly worse
than that of the case-control group.
Clinicopathological variables were compared between
the SRCC group and the case-control group. There was no significant
difference in age, gender, tumor depth, lymph node metastasis,
distant metastasis, and pathological stage between the two groups
(Table II). The primary location
of the tumor is more frequently right-sided in the SRCC group than
that in the case-control group. There was a more frequent tendency
of lymphatic invasion in the SRCC group than that of in a
case-control group. On the other hand, there was no significant
difference in vascular invasion between the two groups. There was
no significant difference between liver and lung metastasis in both
groups but peritoneal dissemination more frequently occurred in the
SRCC group. There was no significant difference in the status of
familial history between the two groups Table II.
 | Table IIComparison of clinicopathological
characteristics of signet ring cell carcinoma group with well to
moderately differentiated adenocarcinoma group in the case-control
cohort. |
Table II
Comparison of clinicopathological
characteristics of signet ring cell carcinoma group with well to
moderately differentiated adenocarcinoma group in the case-control
cohort.
| Clinicopathological
variables | Signet ring cell
group (n=14) | Well to moderately
differentiated group (n=42) | P-values |
|---|
| Age mean ± SD | 62.57±10.68 | 62.57±10.42 | 1.0 |
| Sex (%) | | | 0.22 |
|
M | 12 (85.71) | 29 (69.05) | |
|
F | 2 (14.29) | 13 (30.95) | |
| Tumor depth
(%) | | | 0.29 |
|
T1-T2 | 2 (14.29) | 12 (28.57) | |
|
T3-T4 | 12 (85.07) | 30 (71.43) | 0.29 |
| Lymph node
metastasis (%) | | | 0.53 |
|
N- | 5 (35.71) | 19 (45.24)/ | |
|
N+ | 9 (64.29) | 23 (54.76) | |
| Distant metastasis
(%) | | | 0.39 |
|
M- | 9 (64.29) | 32 (76.19) | |
|
M+ | 5 (35.71) | 10 (23.81) | |
| TNM stage (%) | | | 0.11 |
|
0-II | 3 (21.43) | 19 (45.25) | |
|
II-IV | 11 (78.57) | 23 (54.76) | |
| Tumor location
(%) | | | 0.0063a |
|
Right | 8 (57.14) | 8 (19.05) | |
|
Left | 6 (42.86) | 34 (80.95) | |
| Lymphatic invasion
(%) | | | 0.053 |
|
Ly- | 2 (14.29) | 18 (42.58) | |
|
Ly+ | 12 (85.71) | 24 (57.18) | |
| Vascular invasion
(%) | | | 0.22 |
|
V- | 2 (14.29) | 13 (30.95) | |
|
V+ | 12 (85.71) | 29 (69.05) | |
| Liver Distant
Metastasis (%) | | | 0.29 |
|
- | 13 (92.86) | 34 (80.95) | |
|
+ | 1 (7.14) | 8 (19.05) | |
| Lung Distant
Metastasis (%) | | | 0.56 |
|
- | 14(100) | 41 (97.62) | |
|
+ | 0 (0) | 1 (2.38) | |
| Peritoneum (%) | | | 0.0358a |
|
- | 10 (71.43) | 39 (92.86) | |
|
+ | 4 (28.53) | 3 (7.14) | |
| Familial history of
lynch syndrome related cancer (%) | | | 0.92 |
|
Yes | 6 (42.86) | 17 (41.46) | |
|
No | 8 (57.16) | 24 (58.58) | |
Microarray analysis of altered genes
in SRCC
Microarray analysis was performed on mRNA samples
extracted from frozen tumor tissues of SRCC and well-differentiated
adenocarcinoma. Of the 58,717 differentially expressed genes
extracted from the analysis, 1,445 showed significant differences
between the SRCC and well-differentiated adenocarcinoma cases. The
top 50 most significantly altered genes are shown in Tables III and IV.
 | Table IIITop 50 upregulated genes in SRCC. |
Table III
Top 50 upregulated genes in SRCC.
| Gene symbol | Description |
|---|
| DMGDH | Dimethylglycine
dehydrogenase |
| CART | Cocaine- and
amphetamine-regulated transcript protein |
| ASB5 | Ankyrin repeat and
SOCS box-containing 5 |
| SEPT4 | Septin 4 |
| SNTG2 | Syntrophin, gamma
2 |
| RYR2 | Ryanodine receptor
2 |
| CACNA2D1 | Calcium channel,
voltage-dependent, alpha 2/delta subunit 1 |
| LRRTM1 | Leucine rich repeat
transmembrane neuronal 1 |
| TNFSF18 | Tumor necrosis
factor (ligand) super family, member 18 |
| CNGA3 | Cyclic nucleotide
gated channel alpha 3 |
| EFHD1 | EF-hand domain
family, member D1 |
| ARHGEF4 | Rho guanine
nucleotide exchange factor (GEF) 4 |
| HSPB3 | Heat shock 27 kDa
protein 3 |
| PEG3 | Paternally
expressed 3 |
| INFG | Interferon,
gamma |
| SMPX | Small muscle
protein, X-linked |
| NPTX1 | Neuronal pentraxin
I |
| KCNJ8 | Potassium
inwardly-rectifying channel, subfamily J, member 8 |
| TMOD2 | Tropomodulin 2
(neuronal) |
| FAM49A | Family with
sequence similarity 49, member A |
| DPP6 |
Dipeptidyl-peptidase 6 |
| IL17B | Interleukin
17B |
| PRG-3 | Proteoglycan 3 |
| CDH19 | Cadherin 19, type
2 |
| MPDZ | Multiple PDZ domain
protein |
| GPM6A | Glycoprotein
M6A |
| KRTAP9-4 | Keratin associated
protein 9-4 |
| MN1 | Meningioma
(disrupted in balanced translocation)1 |
| ARHGAP6 | Rho GTPase
activating protein 6 |
| NPC1L1 | NPC1 (Niemann-Pick
disease, type C1, gene)-like 1 |
| BNC2 | Basonuclin 2 |
| ROR1 | Receptor tyrosine
kinase-like orphan receptor 1 |
| PRDM8 | PR domain
containing 8 |
| PCDHB5 | Protocadherin beta
5 |
| ZNF221 | Zinc finger protein
221 |
| CDH9 | Cadherin 9, type 2
(T1-cadherin) |
| GRID2 | Glutamate receptor,
ionotropic, delta 2 |
| GRIK4 | Glutamate receptor,
ionotropic, kainate 4 |
| SLC4A3 | Solute carrier
family 4, anion exchanger, member 3 |
| CD38 | CD38 molecule |
| LRRC21 | Leucine rich repeat
containing 21 |
| KIAA1102 | LIM and calponin
homology domain 1 |
| SERPINB11 | Serpin peptidase
inhibitor, clade B (ovalbumin), member 11 |
| LAMP5 | Lysosome associated
membrane protein |
| GPIHBP1 |
Glycosylphosphatidylinositol anchored high
density lipoprotein binding |
| GIT1 | G protein-coupled
receptor kinase interactor 1 |
| LRCH2 | Leucine-rich
repeats and calponin homology (CH) domain containing 2 |
| DNAJB5 | DnaJ (Hsp40)
homolog, subfamily B, member 5 |
| STK33 | Serine/threonine
kinase 33 |
| ZNF659 | Zinc finger protein
659 |
 | Table IVTop 50 downregulated genes in
SRCC. |
Table IV
Top 50 downregulated genes in
SRCC.
| Gene symbol | Description |
|---|
| FAM3D | Family with
sequence similarity 3, member D |
| CDCA7 | Cell division cycle
associated 7 |
| AREG | Amphiregulin
(schwannoma-derived growth factor) |
| DSG2 | Desmoglein 2 |
| CEACAM5 | Carcinoembryonic
antigen-related cell adhesion molecule 5 |
| TPRT | Decaprenyl
Diphosphate Synthase Subunit 1 |
| B3GNT3 | UDP-GlcNAc:betaGal
beta-1,3-N Acetylglucosaminyltransferase 3 |
| PHLDA2 | Pleckstrin
homology-like domain, family A, member 2 |
| ESRRA | Estrogen-related
receptor alpha |
| GCSH | Glycine cleavage
system protein H (aminomethyl carrier) |
| SLC22A18 | Solute carrier
family 22 (organic cation transporter), member 18 |
| TFRC | Transferrin
receptor (p90, CD71) |
| C20ORF42 | Chromosome 20 open
reading frame 42 |
| TPP2 | Tripeptidyl
peptidase II |
| FLJ20272 | Tetratricopeptide
Repeat Domain 27 |
| APPBP1 | Amyloid beta
precursor protein binding protein 1 |
| ST14 | Suppression of
tumorigenicity 14 (colon carcinoma) |
| SMPD3 | Sphingomyelin
phosphodiesterase 3, neutral membrane (neutral sphingomyelinase
II) |
| SMP3 |
Phosphatidylinositol glycan Anchor
biosynthesis class Z |
| TM2D1 | TM2 domain
containing 1 |
| MRPL45 | Mitochondrial
ribosomal protein L45 |
| PRKDC | Protein kinase,
DNA-activated, catalytic polypeptide |
| ABCC3 | ATP-binding
cassette, sub-family C (CFTR/MRP), member 3 |
| FBP1 |
Fructose-1,6-bisphosphatase 1 |
| MGC3265 | Prenylcysteine
Oxidase 1 Like |
| ZNF217 | Zinc finger protein
217 |
| OCIAD2 | OCIA domain
containing 2 |
| FLJ22662 | Phospholipase B
Domain Containing 1 |
| RFC3 | Replication factor
C (activator 1) 3, 38 kDa |
| SORL1 | Sortilin-related
receptor, L(DLR class) A repeats-containing |
| CDH1 | Cadherin 1, type 1,
E-cadherin (epithelial) |
| RARS | Arginyl-tRNA
synthetase |
| LEFTY1 | Left-right
determination factor 1 |
| EED | Embryonic ectoderm
development |
| SCCPDH | Saccharopine
dehydrogenase (putative) |
| C20ORF24 | Chromosome 20 open
reading frame 24 |
| TMEM45B | Transmembrane
protein 45B |
| SDC1 | Syndecan 1 |
| MTRF1 | Mitochondrial
translational release factor 1 |
| ATP1B1 | ATPase,
NA+/K+ transporting, beta 1 polypeptide |
| EPB41L1 | Erythrocyte
membrane protein band 4,1-like 1 |
| TBL2 | Transducin
(beta)-like 2 |
| MSI2 | Musashi homolog 2
(Drosophila) |
| FANK1 | Fibronectin type
III and ankyrin repeat domains 1 |
| LOC283537 | Not annotated |
| NOC3L | Nucleolar complex
associated 3 homolog (S, cerevisiae) |
| ALDH18A1 | Aldehyde
dehydrogenase 18 family, member A1 |
| GLB1 | Galactosidase, beta
1 |
| CELSR1 | Cadherin, EGF LAG
seven-pass G-type receptor 1(flamingo homolog,
Drosophila) |
| MGC17299 | Transmembrane
protein 125 |
In those tables, inflammation-related genes such as
Tumor Necrosis Factor (TNF), heat shock protein (HSP),
interferon-gamma (IF-γ), and interleukin-17 (IL-17) were shown as a
commonly up-regulated genes in the SRCC group. Conversely, such as
cadherin 1 (CDH1) and cadherin EGF Lag seven-pass G-type receptor
(CELSR1) both genes related to epithelial cells were identified as
a commonly down-regulated gene in SRCC.
CMS analysis of SRCC cases
According to the subtypes of CMS1, CMS2, CMS3, and
CMS4 classification, two each data were chosen from the CRC TCGA
subset (colorectal TCGA gene expression data with subtype
annotation) included in the CMScaller and utilized as a reference
data. SRCC microarray data were processed with the reference
data.
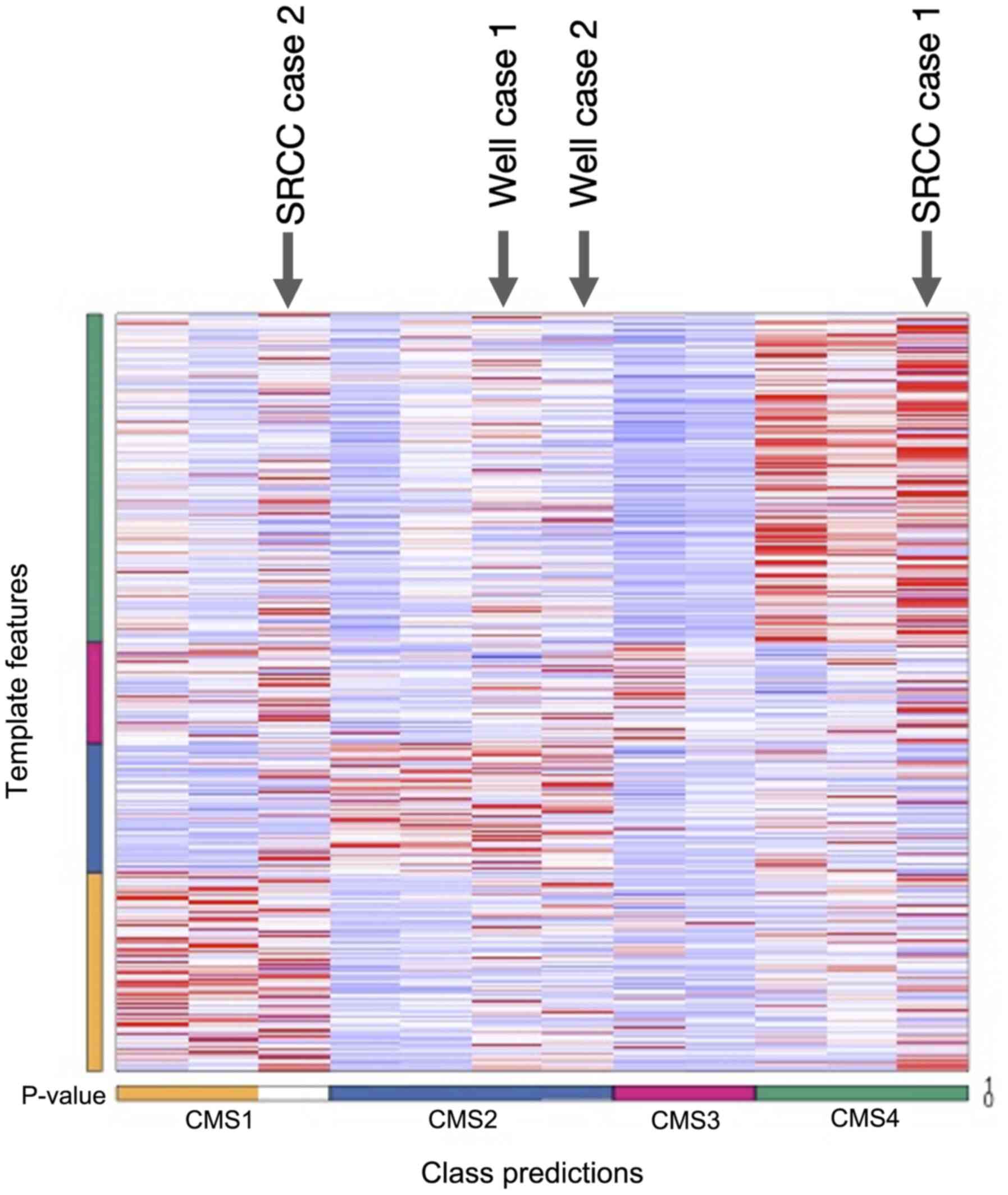
One of the SRCC data was assigned into the CMS1
however it was not statistically significant and the other data
were assigned into CMS4 with a statistically significant
correlation (Fig. 3). Both two
cases of well-differentiated adenocarcinoma were assigned to CMS2
(Table V).
 | Table VStatistical analysis of CMS
classification of signet ring cell carcinoma and well
differentiated adenocarcinoma cases. |
Table V
Statistical analysis of CMS
classification of signet ring cell carcinoma and well
differentiated adenocarcinoma cases.
| Case | Prediction | d.CMS1 | d.CMS2 | d.CMS3 | d.CMS4 | P-value | FDR |
|---|
| SRCC case 1 | CMS4 | 0.6885880805 | 0.7941236723 | 0.7319281894 | 0.5330759305 | 0.001a | 0.001 |
| SRCC case 2 | NA | 0.6179980022 | 0.7074984863 | 0.6460235957 | 0.713021697 | 0.569 | 0.569 |
| Well case 1 | CMS2 | 0.7670768658 | 0.6185472389 | 0.7328083907 | 0.7341745467 | 0.001a | 0.001 |
| Well case 2 | CMS2 | 0.7690166154 | 0.6682347108 | 0.7220604844 | 0.8455248645 | 0.001a | 0.001 |
Gene Set Analysis packaged in CMS caller was
performed to investigate the comprehensively altered genes in the
CMS4 assigned SRCC case, then the up-regulation of epithelial
Mesenchymal transition (EMT) related genes and the down-regulation
of fatty acid, glycolysis, differentiation, MYC, and DNA
repair-related genes were identified as mainly changed gene
expression elements in SRCC (Table
VI).
 | Table VIGene set analysis of SRCC case 1,
utilizing CMS caller. |
Table VI
Gene set analysis of SRCC case 1,
utilizing CMS caller.
| Gene set name | N Genes | Direction | P-values | FDR |
|---|
| EMT | 199 | Up | 0.079 | 0.122 |
| TGF-β | 60 | Up | 0.380 | 0.443 |
| LGR5
stem-cells | 62 | Down | 0.622 | 0.663 |
| CDX2 | 35 | Down | 0.124 | 0.173 |
| Fatty acids | 157 | Down |
<0.001a | <0.001 |
| Glycolysis | 200 | Down |
<0.001a | <0.001 |
|
Differentiation | 612 | Down | 0.001a | 0.017 |
| Cell cycle | 200 | Down | 0.000 | 0.000 |
| WNT | 13 | Down | 0.245 | 0.312 |
| MYC | 58 | Down |
<0.001a | <0.001 |
| MSS | 80 | Down | 0.000 | 0.000 |
| HNF4α | 58 | Down |
<0.001a | <0.001 |
| DNA repair | 148 | Down |
<0.001a | <0.001 |
| MSI | 29 | Down | 0.991 | 0.991 |
Discussion
Through this study, we re-confirmed that the
morbidity of SRCC in colorectal cancer is quite low and the
prognosis of it was significantly worse compared with the
conventional histological type as previously reported. Comparison
of clinicopathological factors also verified that the primary
location of the tumor was right-sided and the peritoneal
dissemination was frequently observed in SRCC. Microarray and
following CMS analyses revealed that there was a specific gene
expression pattern in colorectal SRCC.
There were only 14 SRCC cases (1.04%) out of 1,336
colorectal cancer cases who had been treated in our department from
1997 to 2011. There were several papers about the prevalence of
SRCC in colorectal cancer (24-28)
and each paper reported it around 1-2%. Our result re-confirmed
that the prevalence of SRCC in colorectal cancer is quite rare. In
this study, the primary location of SRCC was significantly more
than on the right side compared with the case-control cohort group.
The same fact was reported by Kim et al and they stated that
the tumor histologically containing SRCC element tended to be on
the right side of the colon (29).
So it can be mentioned that there must be a common
clinicopathological nature in SRCC characterizing to be in specific
locations.
There was no significant difference among lymph node
metastasis between the SRCC group and case-control cohort group
however, the lymphatic invasion was relatively higher and
peritoneal dissemination was significantly higher in the SRCC
group. The invasive character of SRCC is also clearly shown in
accordance with a previous report on the pathological
characteristics of SRCC (1,17,30).
The prognosis between the SRCC group and
case-control cohort group was compared and both CSS and PFS were
significantly worse in the SRCC group than the case-control cohort
group. The reference cohort group contains relatively old cases
however a whole of cases was case-controlled and stage IVB cases
were assigned in both groups. Now, multimodal treatment was
introduced in colorectal cancer therapeutic strategy and as shown
in Table I FOLFOX or FOLFIRI were
performed even in the SRCC group so that both two groups do not
have any difference in clinical background. This result is implying
that SRCC has chemotherapy resistance and it leads to a worse
prognosis.
In the microarray analysis, the top 50 up- and
down-regulated genes were selected (Tables III and IV). As shown in the tables, numerous
genes that have not been reported in SRCC studies were identified.
Notably, NPC1L1 and GPIHBP1 are involved in transporting
cholesterol and absorbing lipoprotein respectively. NPC1L1 is an
intracellular cholesterol transporter gene and the gene was
reported to be connected with colitis-associated tumorigenesis
(31). GP1HBP1 was reported to be
associated with lipoprotein nutrient utilization in glioma
(32). As shown in Table V, fatty acid and glycolysis
associated genes were down-regulated in SRCC thus, up-regulation of
these genes may imply that both energy transporting molecules are
necessary for the survival of SRCC and would be a key biological
target for the treatment of SRCC.
One of the SRCC cases was assigned into CMS4 and the
other case was assigned into CMS1 (Fig.
3; Tables V and VI) in the CMS analysis. In the CMS
analysis, colorectal cancer was classified into CMS1 to
CMS4(19), and, in brief, CMS1 is
MSI immune type, CMS2 is canonical type, CMS3 is a metabolic type
and CMS4 is mesenchymal type. In this study, SRCC case 1 was
classified into CMS4 so case 1 was revealed to have a genetic
background mainly related to epithelial-mesenchymal transition
(EMT).
There was a report about a bio-physiological change
in colorectal cancer cell lines after the cell lines acquired chemo
agent resistance (33). And after
that, a couple of clinical documents presented the association of
chemoresistance and EMT related gene alteration (34-38).
EMT is a gene alteration that can explain the both phenomenon of
invasive and chemoresistant characteristics of SRCC so that the
result is very compatible.
Another SRCC case was assigned into CMS1 but it was
not statistically significant. CMS1 is an MSI immune type and it
has a character of MSI-high, CIMP high, hypermutation, BRAF
mutation, and worse survival after relapse. In a study of the
genetic background of colorectal cancer, RAS mutations and
especially BRAF mutations were found to be particularly common in
SRCC (39,40). Furthermore, to take into account the
result of the SRCC was significantly frequent in the right side
that it reminded us that the SRCC case 2 bore CMS1 specific gene
alteration on it.
The element of SRCC is considered to be the one of a
poor prognostic factor however, our study revealed that SRCC cases
contain a mixture of MSI-high and EMT enriched cases on it. As a
result, consideration of the primary therapeutic strategy could be
possible utilizing CMS analysis according to this study. One
limitation of this study is the lack of information regarding the
MSI and CIMP statuses of the study patients. Because MSI and CIMP
affect gene expression, this is critical information. However, in
this study, we could not acquire both of the information from these
SRCC cases. If the information was available, it would ensure a
result of this study furthermore.
Considering these limitations, further analyses
involving additional SRCC cases are needed utilizing CMS analysis
to identify the gene expression alterations that accurately reflect
the biological features of SRCC.
Through this study, the clinicopathological severity
of SRCC could be recognized. Microarray and subsequent integrative
computational analyses were useful tools to comprehend the gene
expression signature and to infer specific groups of genes hidden
in the large information data.
Acknowledgements
The authors would like to thank Ms. Matsuo
(Department of Surgery, Kurume University School of Medicine,
Japan) and Ms. Kawaguchi (Research Center for Innovative Cancer
Therapy, Kurume University School of Medicine, Japan) for
performing the RNA extractions from the tissue samples and Ms. Otsu
(Department of Surgery, Kurume University School of Medicine,
Japan) for preparation of the slides for Hematoxylin and Eosin
staining and immunohistochemistry staining.
Funding
No funding was received.
Availability of data and materials
The datasets analyzed during the current study are
available in the Array Express repository, on the web page
https://www.ebi.ac.uk/arrayexpress/
ArrayExpress accession E-MTAB-9149.
Authors' contributions
KT conceived of the study and methodology, curated
the data and wrote and drafted the original manuscript. TS
conceived of the study, curated the data, analyzed the data and
wrote, reviewed and edited the manuscript. KI ran the software, was
involved in data validation, performed formal analysis and
visualized the data. AK performed the IHC experiment. SN, SS, KY,
MK, TY, KF, KK, TO, TY and TM were involved in data curation. FF
and JA reviewed, edited and drafted the manuscript, suggested
important key points and interpreted the data in the current study.
YA conceptualized the study, supervised the study and was involved
in project administration. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The current study was conducted in accordance with
the provision of the Declaration of Helsinki and was approved by
the Institutional Review Board of Kurume University Hospital
(approval no. 245).
Patient consent for publication
The current study acquired written informed consent
for publication from all of the patients enrolled in this
study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arifi S, Elmesbahi O and Amarti Riffi A:
Primary signet ring cell carcinoma of the colon and rectum. Bull
Cancer. 102:880–888. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fu J, Wu L, Jiang M, Tan Y, Li D, Chen F,
Jiang T and Du J: Signet ring cell carcinoma of resectable
metastatic colorectal cancer has rare surgical value. J Surg Oncol.
114:1004–1008. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cabibi D, Calascibetta A, Aragona F,
Martorana A, Campione M and Sanguedolce R: Differing expression of
metalloprotease and of adhesion molecules in signet-ring cell and
intestinal colorectal carcinoma. Anticancer Res. 29:4417–4422.
2009.PubMed/NCBI
|
|
4
|
Bellan A, Cappellesso R, Lo Mele M, Peraro
L, Balsamo L, Lanza C, Fassan M and Rugge M: Early signet ring cell
carcinoma arising from colonic adenoma: The molecular profiling
supports the adenoma-carcinoma sequence. Hum Pathol. 50:183–186.
2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wistuba II, Behrens C, Albores-Saavedra J,
Delgado R, Lopez F and Gazdar AF: Distinct K-ras mutation pattern
characterizes signet ring cell colorectal carcinoma. Clin Cancer
Res. 9:3615–3619. 2003.PubMed/NCBI
|
|
6
|
Korphaisarn K, Morris V, Davis JS, Overman
MJ, Fogelman DR, Kee BK, Dasari A, Raghav KPS, Shureiqi I, Trupti
M, et al: Signet ring cell colorectal cancer: Genomic insights into
a rare subpopulation of colorectal adenocarcinoma. Br J Cancer.
121:505–510. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liang Z, Yan D, Li G and Cheng H: Clinical
analysis of primary colorectal signet-ring cell carcinoma. Clin
Colorectal Cancer. 17:e39–e44. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Nitsche U, Friess H, Agha A, Angele M,
Eckel R, Heitland W, Jauch KW, Krenz D, Nüssler NC, Rau HG, et al:
Prognosis of mucinous and signet-ring cell colorectal cancer in a
population-based cohort. J Cancer Res Clin Oncol. 142:2357–2366.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Börger ME, Gosens MJ, Jeuken JW, van
Kempen LC, van de Velde CJ, van Krieken JH and Nagtegaal ID: Signet
ring cell differentiation in mucinous colorectal carcinoma. J
Pathol. 212:278–286. 2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen JS, Hsieh PS, Chiang JM, Yeh CY, Tsai
WS, Tang R, Changchien CR and Wu RC: Clinical outcome of signet
ring cell carcinoma and mucinous adenocarcinoma of the colon. Chang
Gung Med J. 33:51–57. 2010.PubMed/NCBI
|
|
11
|
Hyngstrom JR, Hu CY, Xing Y, You YN, Feig
BW, Skibber JM, Rodriguez-Bigas MA, Cormier JN and Chang GJ:
Clinicopathology and outcomes for mucinous and signet ring
colorectal adenocarcinoma: Analysis from the national cancer data
base. Ann Surg Oncol. 19:2814–2821. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Song IH, Hong SM, Yu E, Yoon YS, Park IJ,
Lim SB, Kim JC, Yu CS and Kim J: Signet ring cell component
predicts aggressive behaviour in colorectal mucinous
adenocarcinoma. Pathology. 51:384–391. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Fukata K, Yuasa N, Takeuchi E, Miyake H,
Nagai H, Yoshioka Y and Miyata K: Clinical and prognostic
differences between surgically resected right-sided and left-sided
colorectal cancer. Surg Today. 50:267–274. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Anthony T, George R, Rodriguez-Bigas M and
Petrelli NJ: Primary signet-ring cell carcinoma of the colon and
rectum. Ann Surg Oncol. 3:344–348. 1996.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bae JM, Kim MJ, Kim JH, Koh JM, Cho NY,
Kim TY and Kang GH: Differential clinicopathological features in
microsatellite instability-positive colorectal cancers depending on
CIMP status. Virchows Arch. 459:55–63. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Alvi MA, Loughrey MB, Dunne P, McQuaid S,
Turkington R, Fuchs MA, McGready C, Bingham V, Pang B, Moore W, et
al: Molecular profiling of signet ring cell colorectal cancer
provides a strong rationale for genomic targeted and immune
checkpoint inhibitor therapies. Br J Cancer. 117:203–209.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tajiri K, Sudou T, Fujita F, Hisaka T,
Kinugasa T and Akagi Y: Clinicopathological and corresponding
genetic features of colorectal signet ring cell carcinoma.
Anticancer Res. 37:3817–3823. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yalcin S and Onguru O: BRAF mutation in
colorectal carcinomas with signet ring cell component. Cancer Biol
Med. 14:287–292. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Guinney J, Dienstmann R, Wang X, de
Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda
G, Angelino P, et al: The consensus molecular subtypes of
colorectal cancer. Nat Med. 21:1350–1356. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Lenz HJ, Ou FS, Venook AP, Hochster HS,
Niedzwiecki D, Goldberg RM, Mayer RJ, Bertagnolli MM, Blanke CD,
Zemla T, et al: Impact of consensus molecular subtype on survival
in patients with metastatic colorectal cancer: Results from
CALGB/SWOG 80405 (Alliance). J Clin Oncol. 37:1876–1885.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Stintzing S, Wirapati P, Lenz HJ,
Neureiter D, Fischer von Weikersthal L, Decker T, Kiani A, Kaiser
F, Al-Batran S, Heintges T, et al: Consensus molecular subgroups
(CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI
plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial.
Ann Oncol. 30:1796–1803. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mooi JK, Wirapati P, Asher R, Lee CK,
Savas P, Price TJ, Townsend A, Hardingham J, Buchanan D, Williams
D, et al: The prognostic impact of consensus molecular subtypes
(CMS) and its predictive effects for bevacizumab benefit in
metastatic colorectal cancer: Molecular analysis of the AGITG MAX
clinical trial. Ann Oncol. 29:2240–2246. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Eide PW, Bruun J, Lothe RA and Sveen A:
CMScaller: An R package for consensus molecular subtyping of
colorectal cancer pre-clinical models. Sci Rep.
7(16618)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Almagro UA: Primary signet-ring carcinoma
of the colon. Cancer. 52:1453–1457. 1983.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Secco GB, Fardelli R, Campora E, Lapertosa
G, Gentile R, Zoli S and Prior C: Primary mucinous adenocarcinomas
and signet-ring cell carcinomas of colon and rectum. Oncology.
51:30–34. 1994.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Belli S, Aytac HO, Karagulle E, Yabanoglu
H, Kayaselcuk F and Yildirim S: Outcomes of surgical treatment of
primary signet ring cell carcinoma of the colon and rectum: 22
cases reviewed with literature. Int Surg. 99:691–698.
2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Nitsche U, Zimmermann A, Spath C, Müller
T, Maak M, Schuster T, Slotta-Huspenina J, Käser SA, Michalski CW,
Janssen KP, et al: Mucinous and signet-ring cell colorectal cancers
differ from classical adenocarcinomas in tumor biology and
prognosis. Ann Surg. 258:775–783. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hugen N, Verhoeven RH, Lemmens VE, van
Aart CJ, Elferink MA, Radema SA, Nagtegaal ID and de Wilt JH:
Colorectal signet-ring cell carcinoma: Benefit from adjuvant
chemotherapy but a poor prognostic factor. Int J Cancer.
136:333–339. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kim D, Kim SY, Lee JS, Hong YS, Kim JE,
Kim KP, Kim J, Jang SJ, Yoon YK and Kim TW: Primary tumor location
predicts poor clinical outcome with cetuximab in RAS wild-type
metastatic colorectal cancer. BMC Gastroenterol.
17(121)2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tung YS, Wu SC and Chen CP: Primary signet
ring cell carcinoma of colorectum: An age- and sex-matched
controlled study. Am J Gastroenterol. 91:2195–2199. 1996.PubMed/NCBI
|
|
31
|
He J, Shin H, Wei X, Kadegowda AK, Chen R
and Xie SK: NPC1L1 knockout protects against colitis-associated
tumorigenesis in mice. BMC Cancer. 15(189)2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hu X, Matsumoto K, Jung RS, Weston TA,
Heizer PJ, He C, Sandoval NP, Allan CM, Tu Y, Vinters HV, et al:
GPIHBP1 expression in gliomas promotes utilization of
lipoprotein-derived nutrients. Elife. 8(e47178)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yang DA, Fan F, Camp ER, van Buren G, Liu
W, Somcio R, Gray MJ, Cheng H, Hoff PM and Ellis LM: Chronic
oxaliplatin resistance induces epithelial-to-mesenchymal transition
in colorectal cancer cell lines. Clin Cancer Res. 12:4147–4153.
2006.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hwang WL, Yang MH, Tsai ML, Lan HY, Su SH,
Chang SC, Teng HW, Yang SH, Lan YT, Chiou SH and Wang HW: SNAIL
regulates interleukin-8 expression, stem cell-like activity, and
tumorigenicity of human colorectal carcinoma cells.
Gastroenterology. 141:279–291, 291.e1-e5. 2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gasiulė S, Dreize N, Kaupinis A, Ražanskas
R, Čiupas L, Stankevičius V, Kapustina Ž, Laurinavičius A, Valius M
and Vilkaitis G: Molecular insights into miRNA-driven resistance to
5-fluorouracil and oxaliplatin chemotherapy: miR-23b modulates the
epithelial-mesenchymal transition of colorectal cancer cells. J
Clin Med Res. 8(2115)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Bhangu A, Wood G, Mirnezami A, Darzi A,
Tekkis P and Goldin R: Epithelial mesenchymal transition in
colorectal cancer: Seminal role in promoting disease progression
and resistance to neoadjuvant therapy. Surg Oncol. 21:316–323.
2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sun L, Ke J, He Z, Chen Z, Huang Q, Ai W,
Wang G, Wei Y, Zou X, Zhang S, et al: HES1 promotes colorectal
cancer cell resistance To 5-Fu by inducing Of EMT and ABC
transporter proteins. J Cancer. 8:2802–2808. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fujikawa H, Tanaka K, Toiyama Y, Saigusa
S, Inoue Y, Uchida K and Kusunoki M: High TrkB expression levels
are associated with poor prognosis and EMT induction in colorectal
cancer cells. J Gastroenterol. 47:775–784. 2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ogino S, Brahmandam M, Cantor M, Namgyal
C, Kawasaki T, Kirkner G, Meyerhardt JA, Loda M and Fuchs CS:
Distinct molecular features of colorectal carcinoma with signet
ring cell component and colorectal carcinoma with mucinous
component. Mod Pathol. 19:59–68. 2006.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kakar S, Deng G, Smyrk CT, Cun L, Sahai V
and Kim SY: Loss of heterozygosity, aberrant methylation, BRAF
mutation and KRAS mutation in colorectal signet ring cell
carcinoma. Mod Pathol. 25:1040–1047. 2012.PubMed/NCBI View Article : Google Scholar
|