Introduction
Glioma, an intracranial tumour originating from
glial cells, is the most common primary intracranial tumour. There
are nearly 100,000 newly diagnosed patients worldwide each year
(1). Its high disability rate and
mortality rate seriously affect people's quality of life and
threaten people's lives (2). At
present, the diagnosis of gliomas mainly relies on imaging
technology. Comprehensive treatment includes extensive surgical
resection, adjuvant chemoradiotherapy, immunotherapy and
photodynamic therapy (3,4). Although effective interventions have
been developed for the occurrence and development of gliomas, the
survival rate of glioma patients has not been significantly
improved (5). The poor prognosis is
mainly due to the stage and the degree of malignancy of the tumour.
The 5-year survival time of early cancer patients is relatively
long (6). Therefore, it is
necessary to look for reliable and sensitive predictors associated
with tumour staging and prognosis to evaluate the occurrence and
progression of tumours early, further improve the prognosis of
patients and prolong the survival time of patients.
Through a large number of literature searches, we
have gained a preliminary understanding of the pathological
mechanism of gliomas and have discovered some biological targets
for the diagnosis and treatment of gliomas (7). For example, there are reports that
TERT gene mutations can affect the survival time of glioma patients
by controlling the interaction between the TERT promoter mutation
and IDH mutation in glioma (8).
Furthermore, inhibition of the TP53 GOF mutation can inhibit
inflammation in glioblastoma multiforme (GBM). Bioinformatics
analysis also shows that inducing TP53 GOF mutation upregulation
can significantly shorten the survival time of glioma patients
(9). EGFR, CCDC26, PHLDB1 and other
genes are also involved in the growth and development of glioma
(10). These biological targets
have been used for the diagnosis and treatment of gliomas. This is
mainly because there are many molecular subtypes of gliomas, and
the pathological mechanism is more complicated and is not
determined by a single factor. Therefore, an in-depth understanding
of the complex pathogenesis of gliomas and the discovery of new
molecular predictors for gliomas are essential.
Ras homology family member C (RHOC), a vital member
of the Rho GTPase family, has various biological functions, such as
regulating cytoskeletal reorganization and affecting cell adhesion
and migration (11). Some
biological functions of RHOC in malignant tumour cells have been
explored by many scholars. RHOC plays a potential subtle role in
invasive melanoma, gastric cancer, oesophageal squamous cell
carcinoma and other malignant tumours by influencing the biological
behaviours of malignant tumour cells, such as proliferation and
apoptosis (11-13).
It has been reported that RHOC knockout can inhibit the invasion
and migration of cholangiocarcinoma cells by inhibiting MMP2, MMP3,
MMP9 and epithelial-mesenchymal transition (EMT) (14). Similar to this finding, the
metastasis and invasion of ovarian cancer were also found to be
regulated by MMP9, which was closely related to the significant
inhibition of MMP9 expression by RHOC (15). In addition, in oesophageal squamous
cell carcinoma (ESCC), the expression of RHOC was positively
correlated with the depth of tumour invasion and the degree of
lymph node metastasis (16).
Although the biological function of RHOC has been explored in many
tumour cells, there are few studies on the relationship between
RHOC and glioma.
To the best of our knowledge, the expression
patterns and molecular functions of RHOC have not been entirely
characterized in gliomas. To uncover this information, we used RNA
sequencing (RNA-Seq) data from the Chinese Glioma Genome Atlas
(CGGA) data set to comprehensively describe the molecular patterns
and clinical relevance of RHOC in glioma. At the same time, to make
our research results more reliable, we also used the RNA-Seq data
of glioma samples from The Cancer Genome Atlas (TCGA) data set as a
validation cohort. This comprehensive study is the first to reveal
the expression patterns and molecular functions of RHOC in
glioma.
Materials and methods
Data collection
We downloaded 749 RNA-Seq data points of glioma
samples from the CGGA database. Additionally, we obtained another
666 glioma tissue RNA-Seq data points from the TCGA database and
obtained the clinical information of the corresponding patients. To
further verify the credibility of our trial, we collected 5 glioma
tissue samples and 5 non-tumour brain tissue samples from daily
surgery, stored them in a liquid nitrogen environment, and then
transferred them to a -80˚C freezer. These 10 samples were used to
determine the RHOC expression level by reverse
transcription-polymerase chain reaction (RT-PCR). For the sample
trial, we obtained written informed consent from the corresponding
patients.
Gene set enrichment analysis (GSEA) of
RHOC
GSEA is a genetic probe based on the evaluation of
data from microarrays. It can be used to determine whether a
predetermined gene shows a statistically significant difference
between two biological states. We used GSEA to predefine the level
of RHOC expression. The genes were clearly divided into the RHOC
high expression group and the RHOC low expression group and
analysed for statistical significance. The genome was permuted 1000
times per analysis. We considered genes with P<0.05 and false
discovery rate (FDR) <0.25 to be statistically significant.
Finally, we also used the Kyoto Encyclopedia of Genes and Genomes
(KEGG) database for enrichment analysis of the data set. After
exploring the signal pathway through GSEA and KEGG enrichment
analysis, we further explored the coexpression genes related to
RHOC gene expression through coexpression analysis, and analyzed
the coexpression genes by Pearson's correlation coefficient method.
The analysis and mapping were based on R language software
(v.3.6.1).
RNA isolation and RT-qPCR
analysis
RT-PCR is a technique that combines RNA RT with cDNA
PCR. The RNA strand was reverse transcribed into cDNA and then
amplified into a DNA template by PCR technology. In our study,
total RNA was extracted from gliomas and normal brain tissues, and
the total absorbance at 260 nm was determined by a
spectrophotometer. The cDNA was then reverse transcribed from the
total RNA using the First Strand cDNA Synthesis Kit (Roche). RT
qPCR was performed according to the guidelines of Faststart
Universal SYBR Green Master (Rox) (Roche, Germany). The results
were quantified using quantum Studio software (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
internal reference was GAPDH, and the primer sequences were
5'-CAAGGTCATCCATGACAACTTTG-3' (F) and 5'-GTCCACCACCCTGTTGCTGTAG-3'
(R). The primer sequences for RHOC were 5'-CCTGAGCCTTGACTTCATCTC-3'
(F) and 5'-CCACCTCAATGTCCGCAATA-3' (R). RT-PCR data were analysed
by the ΔCT method. The paired t-test was used to analyse the two
groups of data, and P<0.05 was considered to be statistically
significant.
Statistical analysis
R software (v.3.6.1) was used to perform statistical
analysis, and the Wilcoxon signed-rank test was used to examine the
expression of RHOC in glioma and non-tumour brain tissues. The
potential relationship between the expression level of RHOC and the
overall survival of glioma patients was analysed using the Cox
regression and Kaplan-Meier methods, and the receiver operating
characteristic (ROC) curve also confirmed the potential value of
RHOC in glioma diagnosis. Univariate and multivariate analyses were
used to explore the relationship between the clinical features and
overall survival time of glioma patients. Welch's t-test was used
to analyze the differences between groups. The Wilcoxon rank sum
test and Kruskal-Wallis test were used to analyse the association
between RHOC expression and clinical features.
Results
Patient characteristics
We obtained a total of 749 glioma samples from the
CGGA database, which contains a variety of complete clinical data,
such as polygenic risk score (PRS) type, age, sex, histological
type, chemotherapy status, 1p19 codeletion status and IDH mutation
status. More detailed clinical feature information is shown in
Table I.
 | Table ICharacteristics of patients with
glioma based on the Chinese glioma genome atlas. |
Table I
Characteristics of patients with
glioma based on the Chinese glioma genome atlas.
| Characteristics | Number of cases | Percentages, % |
|---|
| PRS type | | |
|
Primary | 502 | 67.02 |
|
Recurrent | 222 | 29.64 |
|
Secondary | 25 | 3.34 |
| Histology | | |
|
A | 55 | 7.34 |
|
AA | 39 | 5.21 |
|
AO | 22 | 2.94 |
|
AOA | 80 | 10.68 |
|
GBM | 176 | 23.50 |
|
O | 35 | 4.67 |
|
OA | 95 | 12.68 |
|
rA | 20 | 2.67 |
|
rAA | 36 | 4.81 |
|
rAO | 15 | 2.00 |
|
rAOA | 48 | 6.41 |
|
rGBM | 90 | 12.02 |
|
rO | 4 | 0.53 |
|
rOA | 9 | 1.20 |
|
sGBM | 25 | 3.34 |
| Grade | | |
|
WHO II | 218 | 29.11 |
|
WHO III | 240 | 32.04 |
|
WHO IV | 291 | 38.85 |
| Sex | | |
|
Male | 307 | 40.99 |
|
Female | 442 | 59.01 |
| Age, years | | |
|
≤41 | 343 | 45.79 |
|
>41 | 406 | 54.21 |
| Radiotherapy
status | | |
|
Yes | 625 | 83.44 |
|
No | 124 | 16.56 |
| Chemotherapy
status | | |
|
Yes | 520 | 69.43 |
|
No | 229 | 30.57 |
| IDH mutation
status | | |
|
Mutant | 410 | 54.74 |
|
Wild-type | 339 | 45.26 |
| 1p19q codeletion
status | | |
|
Non-codel | 155 | 20.69 |
|
Codel | 594 | 79.31 |
Correlation between RHOC expression
and clinical characteristics in glioma patients
Our study found that the expression level of RHOC is
indeed significantly correlated with age, tumour grade,
chemotherapy status, 1p19 codeletion status, IDH mutation status
and histological type. The expression of RHOC increases as the
tumour grade increases (Fig. 1).
The RHOC expression level of patients <41 years was
significantly lower than that of patients >41 years. The
expression of RHOC in the 1p19 codeletion and IDH mutant groups was
significantly lower than that in the non-1p19 codeletion
(P<0.001) and IDH wild-type (P<0.001) groups. In addition, we
found that the expression levels of RHOC in the GBM and recurrent
GBM groups increased significantly.
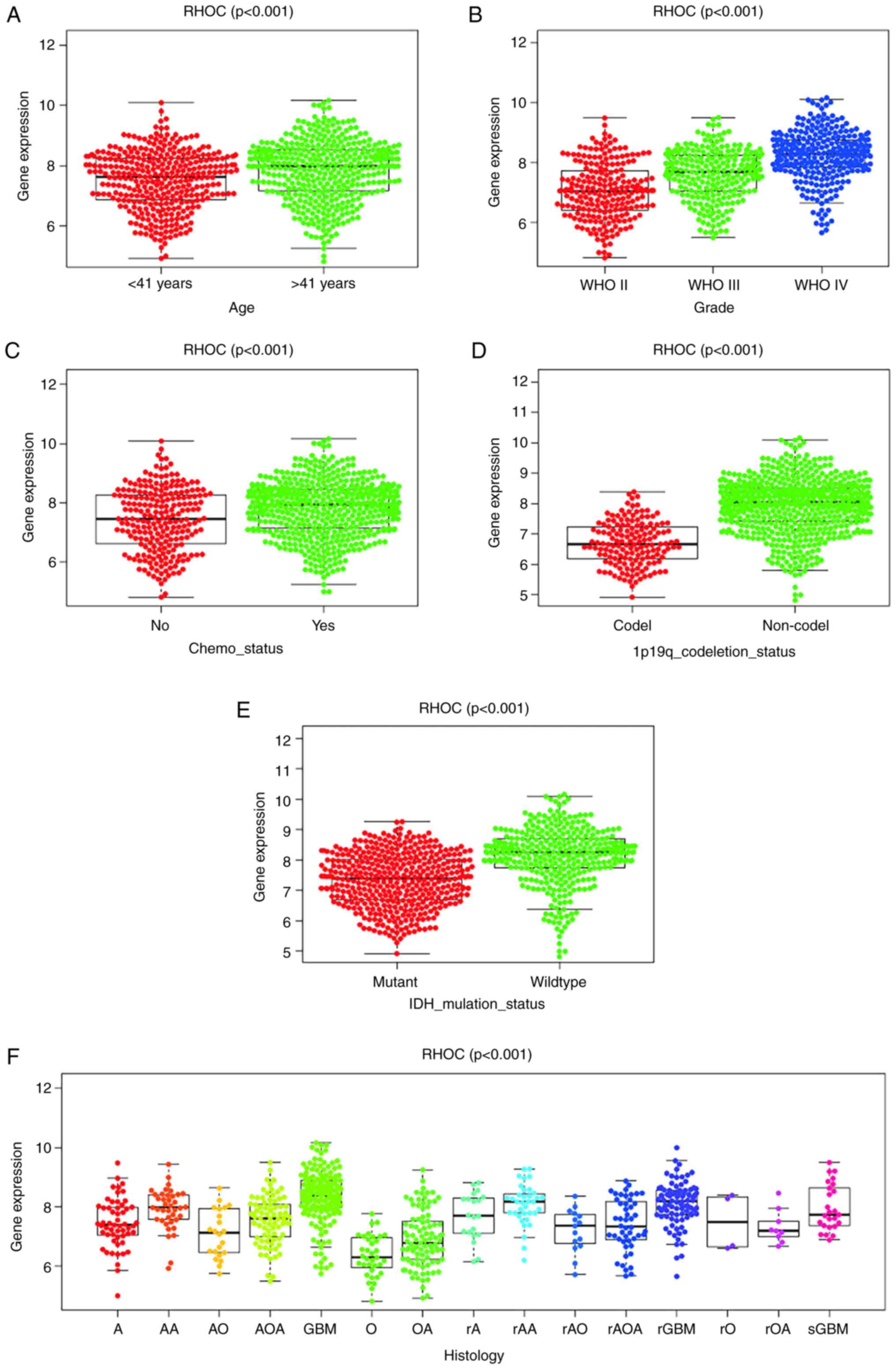 | Figure 1RHOC expression in patients with
glioma from the Chinese Glioma Genome Atlas. Association between
RHOC expression and clinicopathological characteristics, including
(A) age, (B) grade, (C) chemotherapy status, (D) 1p19q codeletion
status, (E) IDH mutation status and (F) histology. A, astrocytoma;
AA, anaplastic astrocytoma; AO, anaplastic oligodendroglioma; AOA,
anaplastic oligoastrocytoma; GBM, glioblastoma multiforme; O,
oligodendroglioma; OA, oligoastrocytoma; r, recurrence; sGBM,
secondary GBM; RHOC, Ras homology family member C; WHO, World
Health Organization; IDH, isocitrate dehydrogenase. |
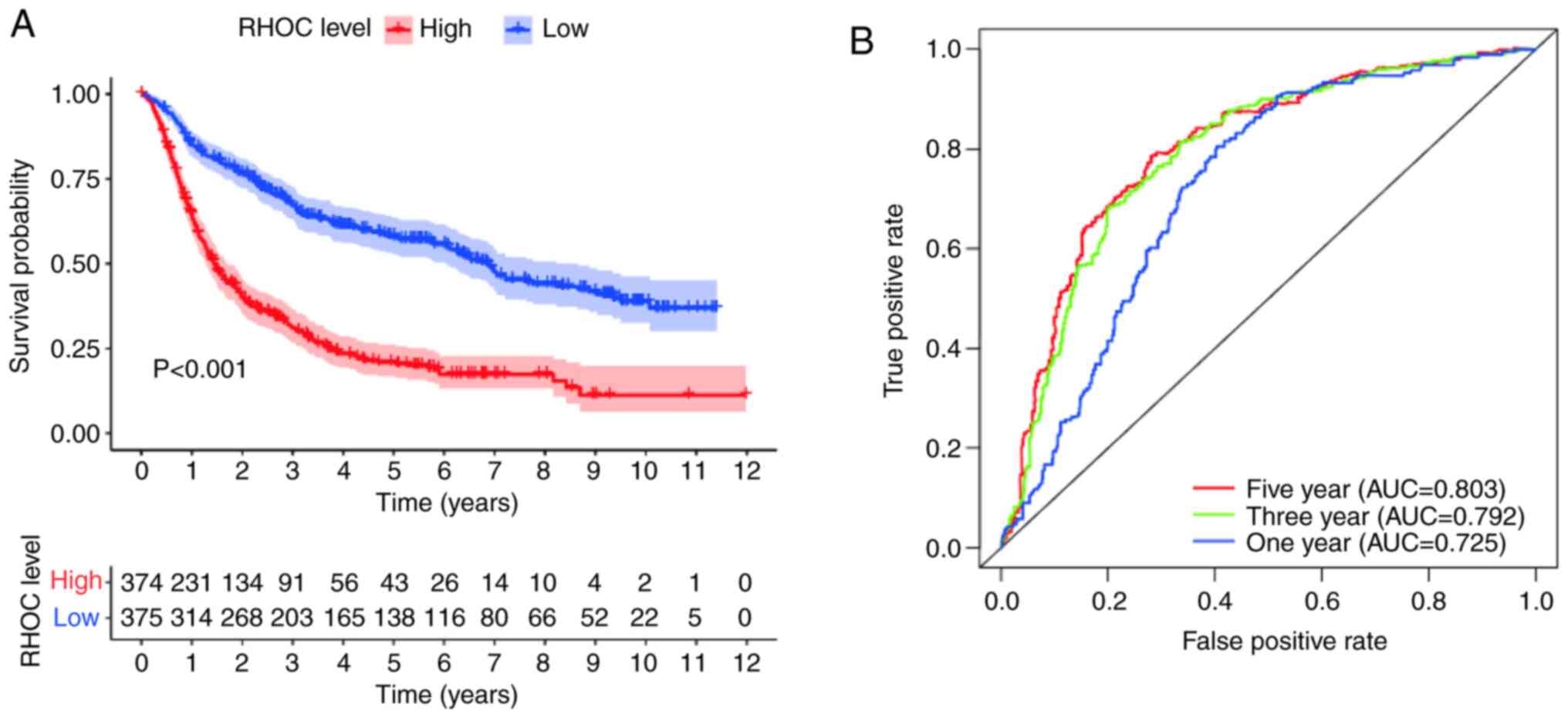
Survival outcomes and clinical
diagnostic value of RHOC in patients with glioma
The Kaplan-Meier survival analysis method explored
the potential link between overall survival and RHOC expression
levels in glioma patients. As shown in Fig. 2, the analysis of the CGGA database
showed that there were obvious abnormalities between the RHOC high
expression group and the RHOC low expression group. In terms of
survival time, glioma patients with high RHOC expression had
significantly worse survival times than those with low RHOC
expression. Moreover, the area under the curve (AUC) values for
survival in the first year, the third year and the fifth year were
0.725, 0.792 and 0.803, respectively, which were all >0.7. It
was further confirmed that RHOC had moderate diagnostic value.
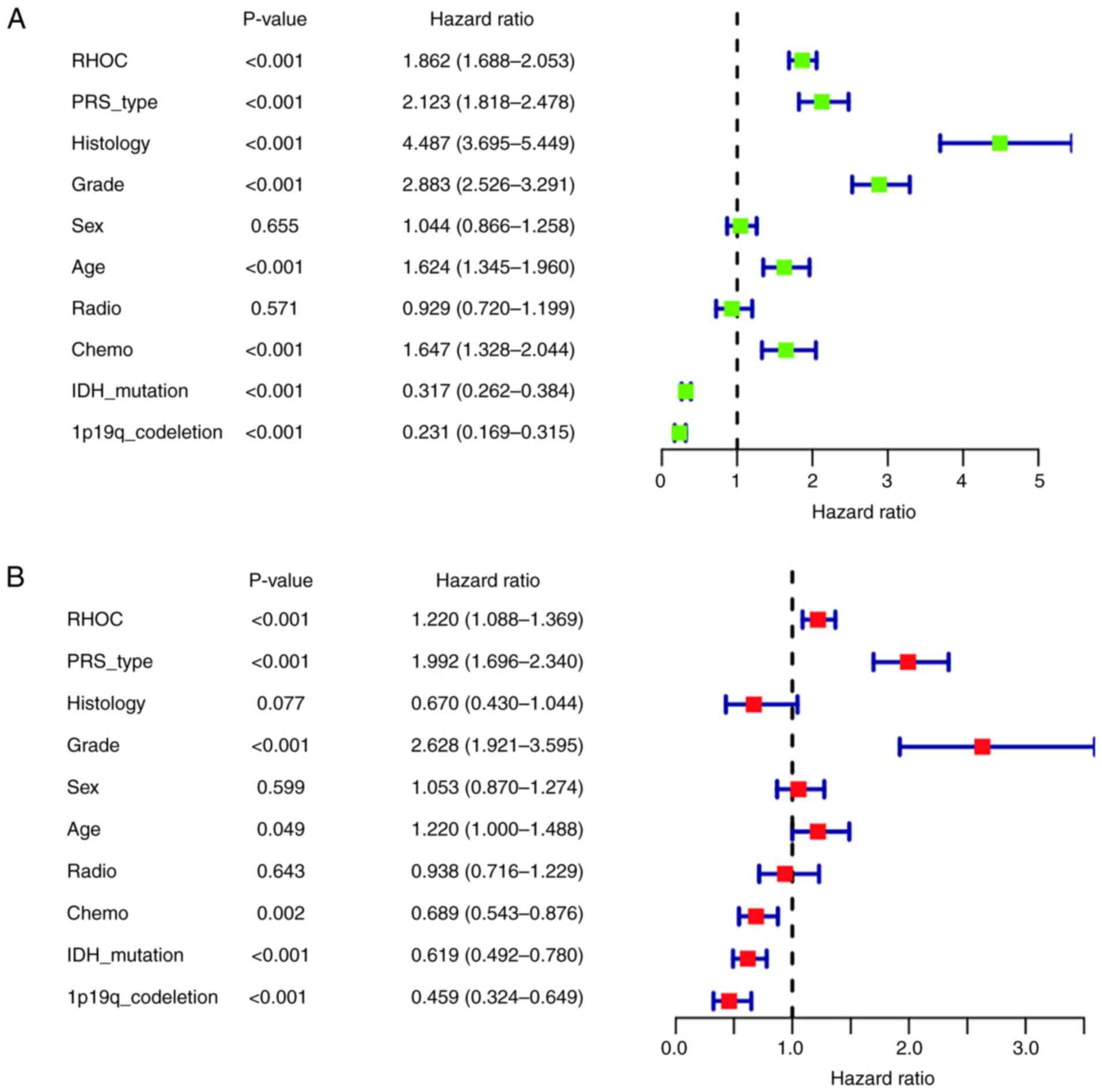
Univariate and multivariate
analyses
After univariate analysis by the Cox regression
model (shown in Fig. 3A), high RHOC
expression, PRS type, histological type, tumour grade, age and
receiving chemotherapy were closely associated with the poor
prognosis of glioma patients (P<0.001) [(hazard ratio
(HR)=1.862; 95% confidence interval (CI), 1.688-2.053), (HR=2.123;
95% CI, 1.818-2.478), (HR=4.487; 95% CI, 3.695-5.449), (HR=2.883;
95% CI, 2.526-3.291), (HR=1.624; 95% CI, 1.345-1.960), and
(HR=1.647; 95% CI, 1.328-2.044)]. In addition, IDH mutation status
and 1p19q codeletion status were strongly associated with good
prognosis (P<0.001) [(HR=0.317; 95% CI, 0.262-0.384) and
(HR=0.231; 95% CI, 0.169-0.315)].
Multivariate analysis by the Cox regression model
(Fig. 3B) showed that high RHOC
expression (P<0.001), PRS type (P<0.001) and high tumour
grade (P<0.001) were significantly associated with poor
prognosis [(HR=1.220; 95% CI, 1.088-1.369), (HR=1.992; 95% CI,
1.696-2.340), and (HR=2.628; 95% CI, 1.921-3.595)]. However, we
also found that glioma patients with IDH mutation status were
significantly associated with good prognosis. Moreover, patients
with 1p19q codeletion status were significantly associated with
good prognosis (P<0.001). Taken together, the univariate and
multivariate analyses suggested that the high expression of RHOC
might be a poor prognostic factor.
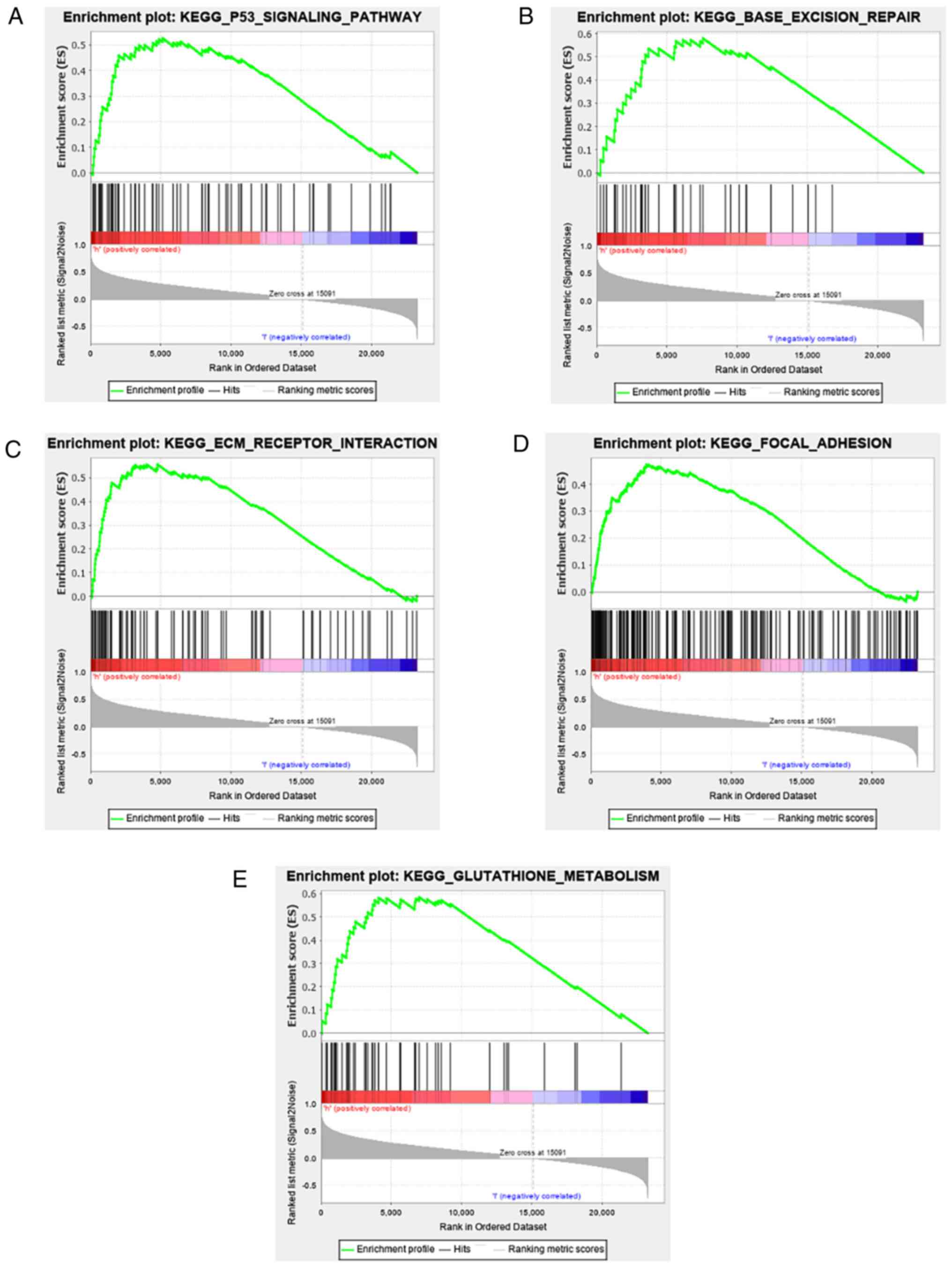
RHOC-related signalling pathways
identified by GSEA
As a bioinformatics analysis tool, GSEA analysed the
cancer signalling pathways between the RHOC high expression group
and the RHOC low expression group. As shown in Fig. 4 and Table II, among all the factors analysed,
p53, BASE_EXCISION_REPAIR, ECM receptor, focal adhesion and
glutathione metabolism showed significant differential enrichment
in the RHOC high expression group. The above results show that RHOC
can produce relevant pathological changes in the occurrence and
development of glioma through these pathways.
 | Table IIResults from gene set enrichment
analysis. |
Table II
Results from gene set enrichment
analysis.
| Gene set name | NES | NOM P-value |
|---|
|
KEGG_P53_SIGNALING_PATHWAY | 1.6437907 | 0.028 |
|
KEGG_BASE_EXCISION_REPAIR | 1.6104667 | 0.028 |
|
KEGG_ECM_RECEPTOR_INTERACTIN | 1.6973425 | 0.039 |
|
KEGG_FOCAL_ADHESION | 1.7078017 | 0.033 |
|
KEGG_GLUTATHIONE_METABOLISM | 1.7351005 | 0.009 |
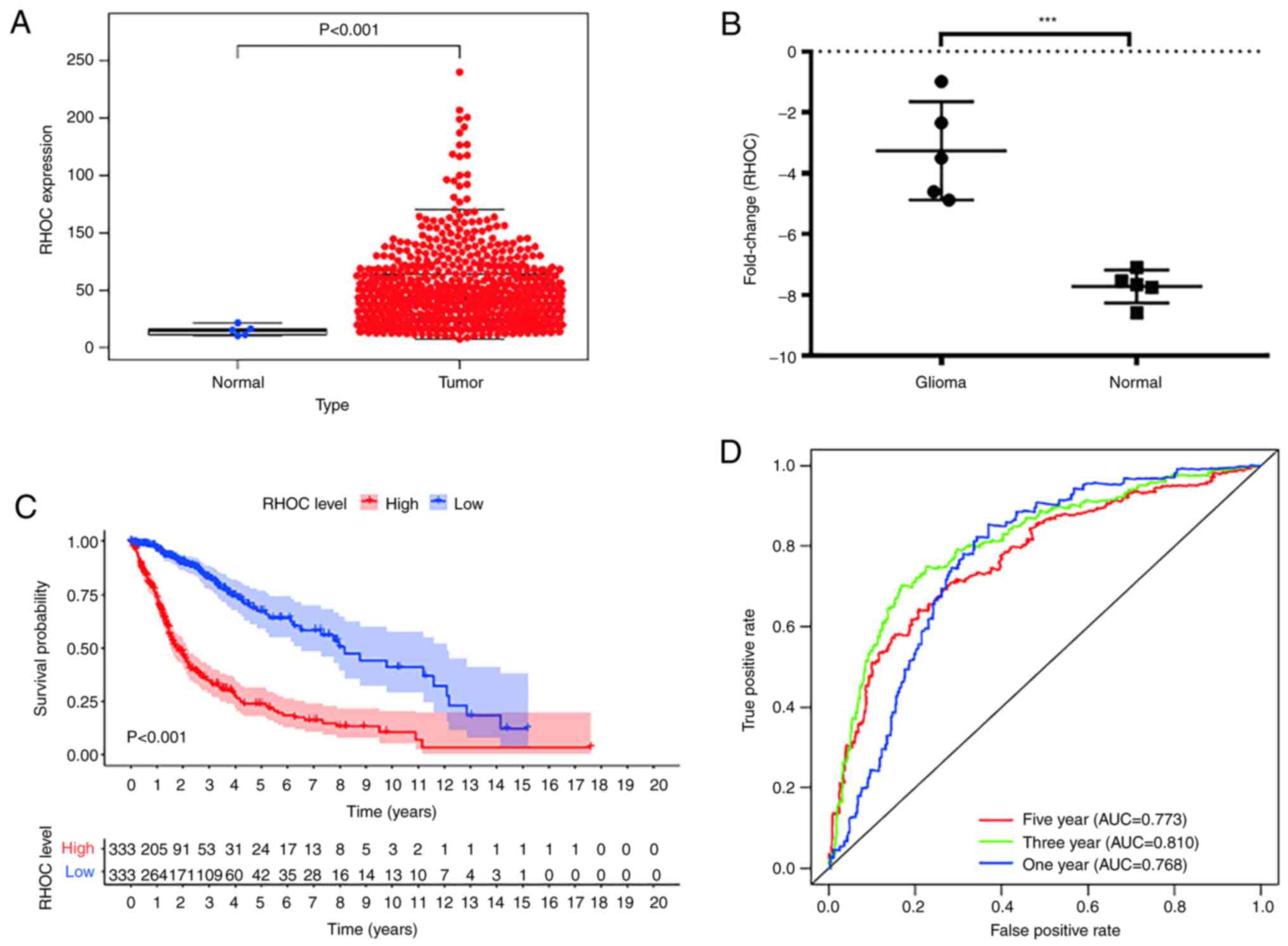
Verifying the RHOC bioinformatic
analysis results
To make our findings more credible, the data of a
total of 666 glioma patients with complete clinical information and
gene expression profile data were retrieved from the TCGA database
as shown in Table III. The rnaseq
data of 666 gliomas and 5 normal brain tissues were downloaded from
TCGA database. Wilcox method was used to detect the expression
level of RHOC in gliomas and normal brain tissues. Welch t-test was
used to compare the differences between the two groups. The results
showed that RHOC was significantly overexpressed in gliomas
(Fig. 5A). In addition, we
collected 5 glioma tissues and 5 non-tumour tissues from the clinic
and detected the expression level of RHOC by RT-PCR technology. The
results showed that compared with non-tumour tissues, RHOC was
highly expressed in glioma tissues (Fig. 5B). Finally, we derived the overall
survival time of glioma patients from the TCGA database and further
analysed the data using the Kaplan-Meier method (as shown in
Fig. 5C). We found that high RHOC
expression indeed leads to poor prognosis in glioma patients. The
ROC curve also confirmed the results of previous studies that RHOC
has a moderate diagnostic value in glioma (Fig. 5D).
 | Table IIICharacteristics of patients with
glioma based on the cancer genome atlas. |
Table III
Characteristics of patients with
glioma based on the cancer genome atlas.
|
Characteristics | Number of
cases | Percentages, % |
|---|
| Sex | | |
|
Male | 385 | 57.81 |
|
Female | 281 | 42.19 |
| Age, years | | |
|
≤51 | 399 | 59.91 |
|
>51 | 267 | 40.09 |
| Grade | | |
|
WHO II | 243 | 36.49 |
|
WHO III | 260 | 39.04 |
|
WHO IV | 163 | 24.47 |
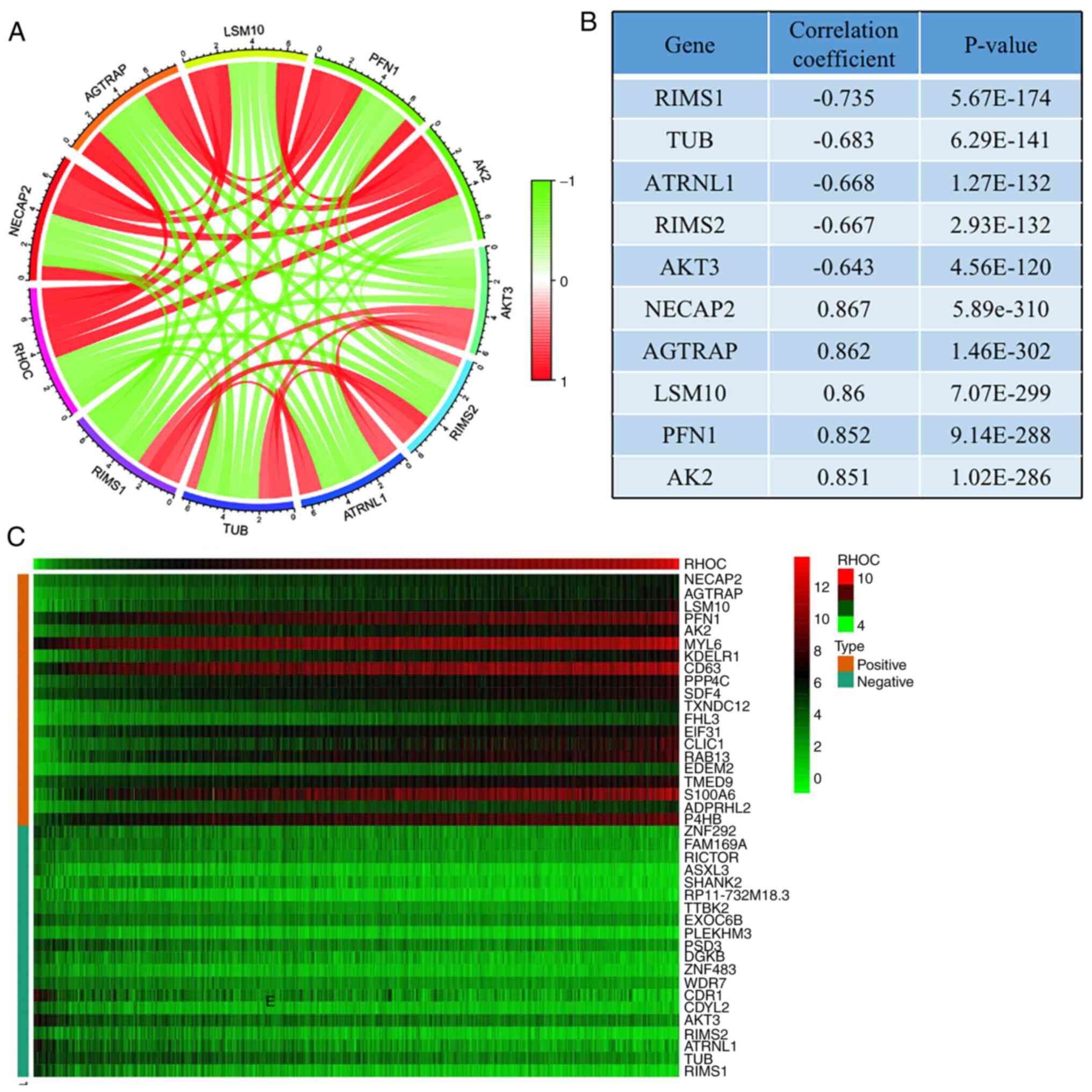
Co-expression network to predict genes
that are critical to glioma pathogenesis
We used a heat map to represent the top 20 genes
that were most positively and negatively correlated with RHOC
expression (Fig. 6C). Co-expression
networks consist of gene expression data relative to normal
expression levels. In a co-expression network, each node represents
one gene in the network, and the gap represents two related genes.
String concatenation is a close association between these genes,
indicating a possible regulatory relationship. Among them, genes
such as NECAP2, AGTRAP, LSM10, PFN1, and AK2 were significantly
positively related to RHOC, while genes such as AKT3, RIMS2,
ATRNL1, TUB, and RIMS1 were significantly negatively related to
RHOC (Fig. 6A). We also tabulated
their connection values, as shown in Fig. 6B.
Discussion
Recently, the role of RHOC has attracted much
attention from academia. Numerous studies have shown that RHOC has
unique potential value in malignant tumour cells (17). RHOC has been found to be highly
expressed in lung cancer, breast cancer, liver cancer and many
other tumour tissues and seriously affects the prognosis of
patients (18-20).
However, few studies have explored the relationship between RHOC
and glioma. Therefore, our report will be a pioneering experimental
study to clarify the potential diagnostic and prognostic value of
RHOC expression in glioma.
Our objective was to investigate the relationship
between RHOC and the clinical features of gliomas. We obtained the
clinical features of glioma patients from the CGGA database and
found that RHOC was highly expressed in glioma. As shown in
Fig. 1, the expression level of
RHOC was positively correlated with the age of glioma patients and
World Health Organization (WHO) grade and had a potential
relationship with chemotherapy status, histological type, 1p19q
codeletion status and IDH mutation status. The expression level of
RHOC was also significantly higher in colorectal cancer tissue
cells than in normal and adjacent tissues, and in particular, local
intestinal damage in colorectal cancer tissue was also closely
associated with the biological function of RHOC (21). Moreover, the invasion and metastasis
of hepatocellular carcinoma (HCC) are closely related to the
expression and activity of the RHOC protein (22). Some studies have found that RHOC is
a key factor in the invasion and metastasis of head and neck
squamous cell carcinoma (23). Many
studies have shown that the expression level of RHOC is related to
the clinical characteristics of malignant tumours. We have reason
to believe that the expression level of RHOC also has some
potential correlation with the clinical characteristics of glioma,
which was further confirmed by our experimental results.
However, the specific effect of RHOC on glioma cells
is still unclear, and we further used the Kaplan-Meier method to
analyse the correlation between the RHOC expression level and
overall survival of patients. As shown in Fig. 2A, in terms of overall survival, we
found that glioma patients with significantly high RHOC expression
had better overall survival than those with low RHOC expression. In
addition, as shown in Fig. 2B, the
ROC curve further confirmed our finding that high RHOC expression
leads to poor prognosis in glioma patients, with moderate
diagnostic reference value. Studies have shown that overexpression
of RHOC may lead to poor prognosis in liver cancer by promoting
vasculogenic mimicry (VM) induced by the EMT mechanism (24). Kleer CG found that the expression of
RHOC increased with the progression of breast cancer tissue and
that high RHOC expression was significantly associated with
decreased patient survival (25).
To confirm the role of RHOC expression in glioma cells and to
further verify whether our findings are inevitable or contingent
factors, we performed univariate and multivariate analyses
(Fig. 3) and carefully observed
that RHOC can indeed serve as an independent risk factor and has
moderate diagnostic value.
From the above results, we did not identify the
mechanism of RHOC in glioma patients with adverse prognoses. We
used GSEA to explore the specific role of RHOC in cell signalling
pathways. As shown in Fig. 4, RHOC
was significantly enriched in cell signalling pathways (e.g., the
p53 signalling pathway, base excision and repair, ECM receptor
interaction, focal adhesion, glutathione metabolism). Among them,
the ECM receptor interaction pathway, which may be associated with
breast cancer, was also reported in previous studies. It has also
been found that Twist2 promotes the proliferation and invasion of
renal cell carcinoma (RCC) cells by regulating ECM receptor
interactions (26). Most
surprisingly, the role of focal adhesion in glioma was also
elucidated. HOXA2 was found to be highly expressed in gliomas and
could affect the proliferation of gliomas by regulating the focal
adhesion pathway (27). CD155/PVR,
as a regulatory factor of adhesion signalling, promotes the
proliferation of glioma cells by regulating adhesion signalling and
local adhesion kinetics (28). Our
results also show that RHOC is significantly enriched in the focal
adhesion pathway, and it is reasonable to believe that RHOC can
affect the occurrence and development of glioma by regulating focal
adhesion.
The occurrence and development of glioma itself is
very complex, involving many biological processes not only related
to RHOC. Thus, in order to predict the pathogenesis of glioma more
accurately, we further conducted co-expression analysis and the
results showed that NECAP2, AGTRAP, LSM10, PFN1 and AK2 are
conducive to the occurrence and development of glioma. AKT3, RIMS2,
ATRNL1, TUB and RIMS1 contribute to prognosis and survival.
Previous studies have shown that PFN1 phosphorylation can obviously
promote the development of glioma (29), and targeting Akt3 may become an
effective method for treating glioma patients (30). Although the remaining eight genes
have not currently been found to be significant in glioma by
researchers, we strongly believe that they will be studied by
scholars in the near future to discover their potential value.
Although we performed an extensive analysis using
public databases to gain more insight into the relationship between
RHOC and glioma, there are still some limitations. Our sample came
from a public database so it has inherent shortcomings, such as
differences in the inclusion criteria, geographical differences,
differences in the extent of surgical resection, and differences in
the dose of chemoradiotherapy. However, public databases have
unique advantages, such as multicentre design, large sample sizes,
and ethnic diversity. Second, we used bioinformatics methods to
deeply understand the potential link between RHOC and glioma.
Because of the complexity of glioma pathogenesis and the wide range
of gene functions, this topic still needs to be explored by future
scholars to further understand the relationship between RHOC and
glioma more comprehensively. Our study provides a basis for future
scholars to guide progress in glioma research.
Through a series of analysis methods, this study
concluded that the abnormally high expression of RHOC could serve
as a novel oncogene independently leading to poor prognosis in
glioma. In addition, we identified possible important carcinogenic
pathways. We firmly believe that RHOC will become a new biomarker
for the diagnosis and prognosis of glioma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Central Plains Thousand
Talents Plan of Henan Province (grant no. ZYQR201912111).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
BZ, JY and XL conceived and designed the experiment.
BL provided reagents/analytical tools, performed data analysis and
participated in drafting the manuscript. YW and HW have also made
contributions to the design of this experiment and part of the data
acquisition. JW and MZ have made contributions to data acquisition,
data analysis and interpretation. YaZ, YoZ and RL analyzed the data
and prepared charts. YG provided the final approval of the released
version, and made a contribution to the data analysis and
interpretation. BZ, JY and XL confirm the authenticity of all the
raw data to ensure its validity. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All procedures performed in the present study were
in accordance with the ethical standards of the Ethical Committee
of the Henan Provincial People's Hospital. The experimental scheme
was also approved by the Ethics Committee of Henan Provincial
People's Hospital [approval no. 2020 lunshenzi (08)]. All patients
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
R, Torre L and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang H, Xu T, Huang Q, Jin W and Chen J:
Immunotherapy for malignant glioma: Current status and future
directions. Trends Pharmacol Sci. 41:123–138. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kong Z, Yan C, Zhu R, Wang J, Wang Y, Wang
Y, Wang R, Feng F and Ma W: Imaging biomarkers guided
anti-angiogenic therapy for malignant gliomas. Neuroimage Clin.
20:51–60. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Delgado-López P, Corrales-García E,
Martino J, Lastra-Aras E and Dueñas-Polo M: Diffuse low-grade
glioma: A review on the new molecular classification, natural
history and current management strategies. Clin Transl Oncol.
19:931–944. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ho VK, Reijneveld JC, Enting RH, Bienfait
HP, Robe P, Baumert BG and Visser O: Dutch Society for
Neuro-Oncology (LWNO). Changing incidence and improved survival of
gliomas. Eur J Cancer. 50:2309–2318. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Okuyama A, Shibata A and Nishimoto H:
Critical points for interpreting patients' survival rate using
cancer registries: A literature review. J Epidemiol. 28:61–66.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gao W, Li H, Liu Y, Zhang Y, Zhao H and
Liu F: Long non-coding RNA FLVCR1-AS1 promotes glioma cell
proliferation and invasion by negatively regulating miR-30b-3p. Mol
Med Rep. 22:723–732. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Vuong HG, Altibi AMA, Duong UNP, Ngo HTT,
Pham TQ, Chan AK, Park CK, Fung KM and Hassell L: TERT promoter
mutation and its interaction with IDH mutations in glioma: Combined
TERT promoter and IDH mutations stratifies lower-grade glioma into
distinct survival subgroups-a meta-analysis of aggregate data. Crit
Rev Oncol Hematol. 120:1–9. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ham SW, Jeon HY, Jin X, Kim EJ, Kim JK,
Shin YJ, Lee Y, Kim SH, Lee SY, Seo S, et al: TP53 Gain-of-function
mutation promotes inflammation in glioblastoma. Cell Death Differ.
26:409–425. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Thomas P, Pranatharthi A, Ross C and
Srivastava S: RhoC: A fascinating journey from a cytoskeletal
organizer to a cancer stem cell therapeutic target. J Exp Clin
Cancer Res. 38(328)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wu Y, Chen Y, Sang J and Xu W: RhoC
protein stimulates migration of gastric cancer cells through
interaction with scaffold protein IQGAP1. Mol Med Rep. 4:697–703.
2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhao ZH, Tian Y, Yang JP, Zhou J and Chen
KS: RhoC, vascular endothelial growth factor and microvascular
density in esophageal squamous cell carcinoma. World J
Gastroenterol. 21:905–912. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yang H, Liang J, Zhou J, Mi J, Ma K, Fan
Y, Ning J, Wang C, Wei X and Li E: Knockdown of RHOC by shRNA
suppresses invasion and migration of cholangiocellular carcinoma
cells via inhibition of MMP2, MMP3, MMP9 and epithelial-mesenchymal
transition. Mol Med Rep. 13:5255–5261. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhao Y, Zong ZH and Xu HM: RhoC expression
level is correlated with the clinicopathological characteristics of
ovarian cancer and the expression levels of ROCK-I, VEGF, and MMP9.
Gynecol Oncol. 116:563–571. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang HZ, Liu JG, Wei YP, Wu C, Cao YK and
Wang M: Expression of G3BP and RhoC in esophageal squamous
carcinoma and their effect on prognosis. World J Gastroenterol.
13:4126–4130. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lang S, Busch H, Boerries M, Brummer T,
Timme S, Lassmann S, Aktories K and Schmidt G: Specific role of
RhoC in tumor invasion and metastasis. Oncotarget. 8:87364–87378.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Arisanty D, Harahap W, Khambri D, Rustam
R, Aliska G, Achyar A and Menra JP: The comparison of RhoC and PI3K
gene expression on the breast cancer tissue and benign tumour
tissue. Open Access Maced J Med Sci. 7:1911–1916. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shen Y, Bu L, Li R, Chen Z, Tian F and Ge
Q: Expression and biological interaction network of RHOC for
hepatic carcinoma with metastasis in PBMC samples. Onco Targets
Ther. 12:9117–9128. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bellovin DI, Simpson KJ, Danilov T,
Maynard E, Rimm DL, Oettgen P and Mercurio AM: Reciprocal
regulation of RhoA and RhoC characterizes the EMT and identifies
RhoC as a prognostic marker of colon carcinoma. Oncogene.
25:6959–6967. 2006.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang HB, Liu XP, Liang J, Yang K, Sui AH
and Liu YJ: Expression of RhoA and RhoC in colorectal carcinoma and
its relations with clinicopathological parameters. Clin Chem Lab
Med. 47:811–817. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Xie S, Zhu M, Lv G, Zhang Q and Wang G:
The role of RhoC in the proliferation and apoptosis of
hepatocellular carcinoma cells. Med Oncol. 29:1802–1809.
2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tumur Z, Katebzadeh S, Guerra C, Bhushan
L, Alkam T and Henson BS: RhoC mediates epidermal growth
factor-stimulated migration and invasion in head and neck squamous
cell carcinoma. Neoplasia. 17:141–151. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Guo JQ, Zheng QH, Chen H, Chen L, Xu JB,
Chen MY, Lu D, Wang ZH, Tong HF and Lin S: Ginsenoside Rg3
inhibition of vasculogenic mimicry in pancreatic cancer through
downregulation of VE-cadherin/EphA2/MMP9/MMP2 expression. Int J
Oncol. 45:1065–1072. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kleer CG, Griffith KA, Sabel MS, Gallagher
G, van Golen KL, Wu ZF and Merajver SD: RhoC-GTPase is a novel
tissue biomarker associated with biologically aggressive carcinomas
of the breast. Breast Cancer Res Treat. 93:101–110. 2005.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang HJ, Tao J, Sheng L, Hu X, Rong RM,
Xu M and Zhu TY: Twist2 promotes kidney cancer cell proliferation
and invasion by regulating ITGA6 and CD44 expression in the
ECM-receptor interaction pathway. Onco Targets Ther. 9:1801–1812.
2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu Z, Shen F, Wang H, Li A, Wang J, Du L,
Liu B, Zhang B, Lian X, Pang B, et al: Abnormally high expression
of HOXA2 as an independent factor for poor prognosis in glioma
patients. Cell Cycle. 19:1632–1640. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sloan KE, Stewart JK, Treloar AF, Matthews
RT and Jay DG: CD155/PVR enhances glioma cell dispersal by
regulating adhesion signaling and focal adhesion dynamics. Cancer
Res. 65:10930–10937. 2005.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fan Y, Potdar AA, Gong Y, Eswarappa SM,
Donnola S, Lathia JD, Hambardzumyan D, Rich JN and Fox PL:
Profilin-1 phosphorylation directs angiocrine expression and
glioblastoma progression through HIF-1α accumulation. Nat Cell
Biol. 16:445–456. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Mure H, Matsuzaki K, Kitazato KT,
Mizobuchi Y, Kuwayama K, Kageji T and Nagahiro S: Akt2 and Akt3
play a pivotal role in malignant gliomas. Neuro Oncol. 12:221–232.
2010.PubMed/NCBI View Article : Google Scholar
|