Introduction
Among the different gynecologic cancers, ovarian
cancer (OC) is the leading cause of death (1). In comparison to the breast cancer, OC
mortality rate is approximately three times higher despite its
lower incidence rate (2). This is
attributed to its asymptomatic initial stages and late diagnosis at
advanced stages (3,4). The incidence of OC in Saudi Arabia has
increased 4-fold between 1990 and 2016(5). It affects more than 3% of Saudi women
in their lifetime (6,7). There are different types of ovarian
malignancy; however, the epithelial ovarian cancer (EOC) is the
most common malignancy (90%) (4,8-10).
This EOC is a heterogeneous disease with several histologic
subtypes that exhibited distinct cytogenetic features, molecular
signatures and onco-pathological signaling pathways. It is a highly
invasive with five distinct histologic subtypes: High grade serous
which accounts for 70% of EOC, endometrioid constitutes 10%, clear
cell (10%), low grade serous 5% and mucinous 3% of EOC (5).
With huge milestones being achieved in the genomic
analysis, some inherited mutations of specific genes, such as BRCA1
and BRCA2, have been shown to increase OC risk. In fact, genomic
events such as amplification (HER2/KRAS) and mutations (KRAS, PIK,
RAD, BRAD) have been reported in OC (11-13).
In contrast, oral contraceptive pills, plurality, hysterectomy, and
breastfeeding have lower OC incidence (14,15).
Studies have shown that the 5-year survival rate of
patients with OC depends mainly on the disease stage at diagnosis.
In general, patients diagnosed at an early stage (I/II) have higher
5 year survivals than those at late stage (III/IV). Furthermore, a
noticeable increase in OC post-relapse survival was reported over
the years during the last decades (16). With the advent of high-throughput
sequencing technologies, the overall aim of precision medicine for
patients with OC is to predict, prevent, and treat the disease
while improving the clinical outcomes and reducing the adverse
effects associated with invasive therapeutic approaches (17). Therefore, there is a need for
devising new effective strategies based on genomic and epigenomic
signatures to identify relevant molecular biomarkers and establish
personalized therapeutics (18,19).
Genomic instability is one of the main causes of
cancer onset attributed to the interactions among a patient's
susceptible genome, environment, and lifestyle; hence, some DNA
alterations occur at the epigenomic level. Epigenetic alterations
encompass histone acetylation, noncoding RNAs, and CpG island
methylation, are among the most frequent epigenetic alterations
observed in OC (20-22).
Currently, there is an increasing interest in examining the
specific patterns of hypermethylation of CpG islands in OC. Both
targeted and whole-genome analyses for DNA methylation patterns
have shown potential in identifying methylation biomarkers and
their prognostic value in OC (23-26).
Moreover, epigenetic silencing of some genes, including hMLH1,
RASSF1A, and FANCF due to methylation is associated with ovarian
tumor drug resistance (27).
Consequently, some key epigenetic events are likely to provide
cancer cells an advantage to proliferate and invade, thereby
promoting metastasis (28,29). Thus, further studies are required to
analyze the methylation profiles of several genes to gain deeper
insights into the epigenetic alterations in OC, especially in the
Arabian Peninsula, where only a few studies have been reported
(25,26,30-32).
In this context, klotho (KL) gene is an interesting candidate aging
marker gene that could be associated with aging-related diseases
e.g. cancer. In fact, KL gene was reported with an aberrantly
methylated promoter reported in several malignancies (33). Initially discovered as an anti-aging
factor (34), KL is located in
chromosome 13q12. It is 50 kb in length and is composed of five
exons and four introns (35). It is
highly expressed in the kidneys and brain but can also be expressed
in the placenta, breast, ovaries, uterus, and fallopian tube
tissues (36). In the female
reproductive system, particularly in the ovary, the level of
abnormal expression of KL in granulosa cells is tightly linked to
the severity of ovary-related diseases (37). KL expression levels decrease in the
brain with advancing age in mammals. This decrease could be due to
hypermethylation of the promoter region of KL (38,39).
Furthermore, aberrant methylation in the promoter region of KL
decreases gene transcription (40).
Recently, KL has been reported as a potential tumor suppressor gene
that inhibits the IGF-1 pathway and is a novel target gene for
epigenetic silencing in cancers of breast (41), cervix (34), stomach (42), bladder, and ovaries (43). However, low expression levels of KL
are associated with weakness in skeletal muscle, difficulty in
executing daily activities, and increased mortality in the elderly.
Deficiency in IGF-1 expression level is a reported feature of
aging, and is likely affected by methylated KL (40). The 1439-bp long promoter region of
KL lacks a TATA box and instead, contains four potential Sp1
binding sites (35,44). Moreover, KL expression can be
restored using demethylation reagents (35). So far, only few studies have
assessed the methylation profiles of KL in OC and showed potential
prognosis value. In that, higher KL promoter methylation profiles
were associated with cancer progression (45,46).
Therefore, additional studies are required to further understand
the prognosis value of KL methylation status in OC, particularly,
in the Arabian Peninsula population. In this study, we analyzed KL
gene promoter methylation profiles in OC and assessed its
prognostic value.
Patients and methods
Patients and samples
This retrospective study comprised 63 formalin-fixed
paraffin-embedded (FFPE) tissues that were surgically resected
after obtaining written consent from female patients who were
diagnosed with OC and were selected based on the availability of
both tissues and patient clinicopathological follow-up data. The
FFPE samples were collected between 1995 and 2014 at the
Departments of Pathology & Gynecology, King Abdulaziz
University Hospital (KAUH).
The cohort of patients was selected from a total of
100 accessible samples based on the availability of patients'
annotated clinicopathological follow-up data, and the
quality/quantity of extracted DNA. Patients' cohort was classified
based on histopathological features using the tumor node metastasis
(TNM) classification system.
The main clinicopathological features (such as age,
menopausal status, stage, grade, and lymph node status) and
follow-up and survival data are summarized in Table I. The samples and data were
retrieved from the patients after obtaining all the relevant
ethical approvals according to the guidelines of the Ethical
Committee of King Abdulaziz University Hospital (Ref. number:
KAU-189-14).
 | Table IAssociation between KL methylation
profiles and clinicopathological features of patients with ovarian
cancer. |
Table I
Association between KL methylation
profiles and clinicopathological features of patients with ovarian
cancer.
| | KL methylation
profile | |
|---|
| Clinicopathological
features | N (%) | 0 unmethylated, n
(%) | 1 hypermethylated,
n (%) |
P-valuea |
|---|
| Age, years | | | | |
|
<50 | 40 (63.5) | 19 (79.2) | 21 (55.3) | 0.064 |
|
>50 | 22 (34.9) | 5 (20.8) | 17 (44.7) | |
|
NA | 1 (1.6) | | | |
| Diagnosis
method | | | | |
|
Surgery | 58 (92.1) | 22 (91.7) | 36 (94.7) | 0.637 |
|
Biopsy | 4 (6.3) | 2 (8.3) | 2 (5.3) | |
|
NA | 1 (1.6) | | | |
| Tumor site | | | | |
|
Right | 11 (17.5) | 4 (16.7) | 7 (18.4) | 0.039b |
|
Left | 11 (17.5) | 8 (33.3) | 3 (7.9) | |
|
Bilateral | 40 (63.5) | 12 (50.0) | 28 (73.7) | |
|
NA | 1 (1.6) | | | |
| Tumor size, cm | | | | |
|
1-5 | 18 (28.6) | 9 (39.1) | 9 (23.7) | 0.370 |
|
6-10 | 16 (25.4) | 6 (26.1) | 10 (26.3) | |
|
>10 | 27 (42.9) | 8 (34.8) | 19 (50.0) | |
|
NA | 2 (3.2) | | | |
| Tumor subtype | | | | |
|
Serous | 30 (47.6) | 12 (50.0) | 18 (47.4) |
<0.001b |
|
Mucinous | 17 (27.0) | 1 (4.2) | 16 (42.1) | |
|
Other | 14 (22.2) | 11 (45.8) | 3 (7.9) | |
|
NA | 2 (3.2) | 0 (0.0) | 1 (2.6) | |
| Tumor grade | | | | |
|
Undetermined
grade (GX) | 10 (15.9) | 5 (23.8) | 5 (16.1) | 0.825 |
|
Low
malignant potential (GB) | 5 (7.9) | 2 (9.5) | 3 (9.7) | |
|
Well-differentiated
(G1) | 3 (4.8) | 2 (9.5) | 1 (3.2) | |
|
Moderately
differentiated (G2) | 15 (23.8) | 5 (23.8) | 10 (32.3) | |
|
Undifferentiated
(G3 to G4) | 19 (30.2) | 7 (33.3) | 12 (38.7) | |
|
NA | 11 (17.5) | | | |
| Lymph node
status | | | | |
|
Positive | 11 (17.5) | 2 (22.2) | 9 (56.3) | 0.208 |
|
Negative | 14 (22.2) | 7 (77.8) | 7 (43.8) | |
|
NA | 38 (60.3) | | | |
| Body mass
index | | | | |
|
<23 | 2 (3.2) | 0 (0.0) | 2 (9.5) | 0.118 |
|
23-26 | 12 (19.0) | 8 (47.1) | 4 (19.0) | |
|
>26 | 24 (38.1) | 9 (52.9) | 15 (71.4) | |
|
NA | 25 (39.7) | | | |
| Parity | | | | |
|
Parous | 29 (46.0) | 10 (62.5) | 19 (73.1) | 0.510 |
|
Nulliparous | 13 (20.6) | 6 (37.5) | 7 (26.9) | |
|
NA | 21 (33.3) | | | |
| Menopausal
status | | | | |
|
Premenopausal | 41 (65.1) | 19 (79.2) | 22 (57.9) | 0.104 |
|
Postmenopausal | 21 (33.3) | 5 (20.8) | 16 (42.1) | |
|
NA | 1 (1.6) | | | |
| Tumor stage | | | | |
|
Stage I | 16 (25.4) | 8 (38.1) | 8 (22.9) | 0.660 |
|
Stage
II | 4 (6.3) | 1 (4.8) | 3 (8.6) | |
|
Stage
III | 31 (49.2) | 10 (47.6) | 21 (60.0) | |
|
Stage
IV | 5 (7.9) | 2 (9.5) | 3 (8.6) | |
|
NA | 7 (11.1) | | | |
| Endpoint
status | | | | |
|
Living | 24 (38.1) | 11 (73.3) | 13 (41.9) | 0.063 |
|
Deceased | 22 (34.9) | 4 (26.7) | 18 (58.1) | |
|
NA | 17 (27.0) | | | |
| Recurrence
status | | | | |
|
No | 20 (31.7) | 7 (41.2) | 13 (48.1) | 0.760 |
|
Yes | 24 (83.1) | 10 (58.8) | 14 (51.9) | |
|
NA | 19 (30.2) | | | |
DNA extraction
Before DNA extraction, 10 µm thin FFPE slices were
deparaffinized using xylol (Sigma-Aldrich) and then washed with
ethanol 99-100% (Sigma-Aldrich). DNA extraction was performed using
the QIAamp DNA FFPE Tissue Kit (Qiagen) according to the
manufacturer's protocol. The quality and concentration of all DNA
samples were assessed using NanoDrop 2000 Spectrophotometer (Thermo
Fisher Scientific, Inc.).
KL methylation analysis
Bisulfite treatment of the KL promoter region was
performed using the Qiagen EpiTect® Bisulfite Conversion
kit as detailed in the manufacturer's handbook and reported by
Dallol et al (47). Briefly,
0.5 µg extracted DNA was incubated with
NaH2SO4 (3.12 M). Only the unmethylated
cytosine (C) residues were converted into uracil (U) residues. The
MethyLight assay for the candidate gene was performed using the
EpiTect® MethyLight PCR Kit (Qiagen, Inc.) and
EpiTect® Control DNA Set (Qiagen, Inc.) according to the
protocol guidelines, fluorescent dual-labeled TaqMan Probe
6FAM-CGGTTGGGTTAATCGCGTTTT-BHQ1, and specifically designed primers:
Forward primer 5'-AGCGTTTGTAGGACGTTTAC-3', and reverse primer
5'-TAACGAAAACAAAACTCCGC-3') (48);
qPCR was performed using the StepOnePlus™ real-time PCR
system (Applied Biosystems).
Statistical analysis
Statistical analyses were performed using
SPSS® version 19 (IBM Corp.). Differentially
hypo/hypermethylated genes' profiles were assessed compared to the
EpiTect® Control DNA Set using a t-test. The association
between KL methylation profile and clinicopathological features of
patients was analyzed using Fisher-Freeman-Halton Exact test.
Furthermore, disease-specific survival (DSS) and disease-free
survival (DFS) based on KL methylation profiles were calculated by
univariate Kaplan-Meier analysis along with hazard ration
calculation at 95% CI, and equality of the survival functions were
determined by log-rank (Mantel-Cox) test. Additionally,
Cox-regression multivariate analysis was performed to assess
possible independent prognostic impact. Results with P<0.05 were
considered statistically significant.
Results
Patient cohort overview
As shown in Table I,
most of our patients diagnosed with OC (64.5%) were <50 years of
age. Additionally, most patients (66.1%) were premenopausal women.
Furthermore, a higher number of patients in our cohort were parous
women. Notably, a high proportion of patients were diagnosed with
OC at advanced stages (64.3%) and a high grade (36.5%), where the
size of the eradicated tumors was >10 cm3. Moreover,
a high percentage of our cohort was diagnosed with the serous
histological subtype; incidentally, among these, many women were
obese with BMI >26 (Table
I).
KL methylation profiles and
clinicopathological features
Our results revealed highly frequent methylation of
KL among OC patients (n=39, 61.9%). Further, correlation analysis
between KL methylation profile and clinicopathol ogical features of
our cohort using Fisher-Freeman-Halton Exact test revealed
significant associations between KL methylation pattern and tumor
subtype (P<0.0001), tumor site (P=0.036), and borderline
significance for endpoint status (P=0.063), and age (P=0.064)
(Table I). In fact, KL promoter
methylation was observed in the majority (77%) of old OC patients
(>50 years). Particularly, this KL hypermethylation was more
prevalent (70%) in bilateral OC patients. However, no significant
associations with grade, BMI, age, and stage (P>0.05) were
observed.
KL methylation profiles and survival
outcomes
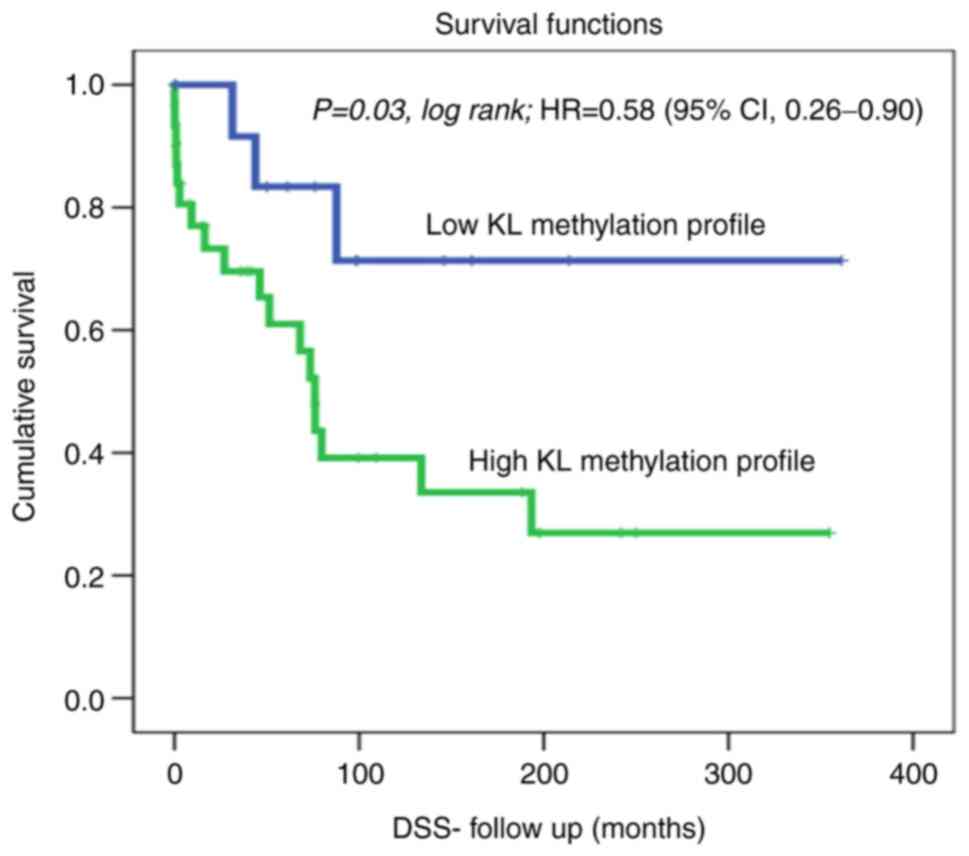
Kaplan-Meier analysis of DSS revealed a significant
difference between patients with OC showing high KL methylation,
who lived for a significantly shorter duration (shorter DSS) than
those with non-methylated KL using the cut-off [unmethylated (0; 15
cases) vs. hypermethylated (1; 31 cases)] as a determinant of DSS
in univariate (Kaplan-Meier) analysis (Fig. 1; P=0.03, log-rank). For example, in
a 1-year follow-up period, all patients with non-methylated KL
(100%) were alive compared to only 75% of those with methylated KL.
Similarly, in the 5-year follow-up period, 40% of the patients with
OC who had methylated KL, had died, whereas, 85% of those with
non-methylated KL were still alive. Our DSS results showed a hazard
ratio=0.58; (95% CI, 0.26-0.90); This means that at any follow
time, the OC patients with methylated KL group has 0.58 times as
likely more than the control (unmethylated) KL group. Which means
that patients with unmethylated KL has 0.42 times to die less than
the KL methylated group. Therefore, OC with methylated KL is more
aggressive and a predictive indicator of poor prognosis in OC
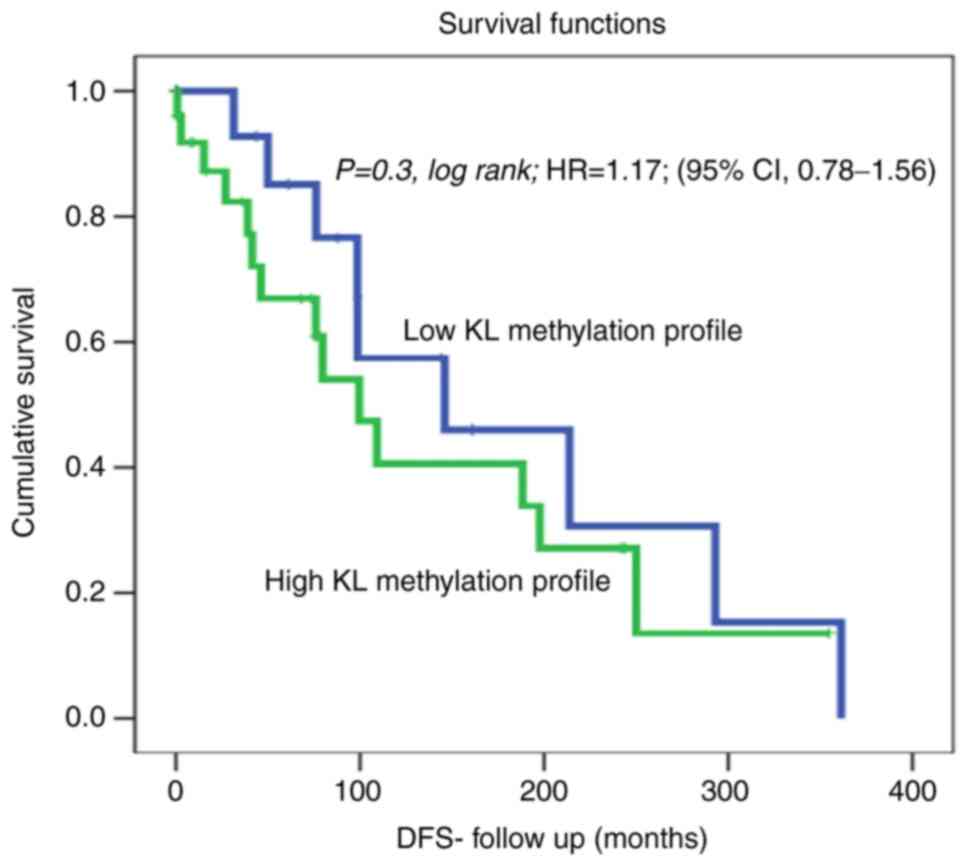
patients. Furthermore, a slight trend of reduced recurrence was
observed in OC patients with non-methylated KL. In fact, DFS
analysis using the cut-off [unmethylated (0; 17 cases) vs.
hypermethylated (1; 27 cases)] as a determinant of DFS in
univariate (Kaplan-Meier) analysis showed no significant
differences between patients with non-methylated KL, and those with
hypermethylated KL and living with no recurrence [Fig. 2; P=0.3, log-rank, hazard ratio=1.17;
(95% CI, 0.78-1.56)].
Multivariate analysis
A multivariable analysis was carried out to
determine if the survival association is independent of other
factors. In fact, multivariate Cox's regression analysis suggests
that KL methylation is an independent poor survival marker
(P=0.029) in relation to patient's age, tumor grade and most
importantly independent of the histological subtype of the tumor
(Table II).
 | Table IICox regression analysis of the
prognostic values of KL methylation, age at diagnosis, grade and
histological subtypes. |
Table II
Cox regression analysis of the
prognostic values of KL methylation, age at diagnosis, grade and
histological subtypes.
| Feature | P-value | Standard error
value | Relative risk | 95% CI |
|---|
| KL methylation (low
vs. high methylation) | 0.029 | 0.716 | 4.758 | 1.169-19.366 |
| Age at diagnosis
(<50 vs. >50 years) | 0.843 | 0.540 | 1.113 | 0.386-3.205 |
| Histological
subtypes (serous vs. mucinous) | 0.470 | 0.296 | 1.238 | 0.693-2.213 |
| Tumor grade (low
vs. high grade) | 0.140 | 0.186 | 1.317 | 0.914-1.898 |
Discussion
OC is one of the deadliest gynecological cancers
mainly because of delayed diagnosis and asymptomatic early-stage
progression. Moreover, the deleterious effects of OC can be
attributed to the anatomical position of the ovaries within the
deep pelvic cavity which may hide the disease expansion and delay
the diagnosis (49-51).
Scientists are seeking to improve the detection, prognosis,
management, and treatment approaches toward precision oncology to
alleviate the burden of this disease on patients, their families,
and the healthcare system (52).
With the development of high-throughput sequencing technologies, it
is possible to assess changes in the genetic and epigenetic
profiles of the target genes/genomes and identify potential
molecular biomarkers for OC (21,53-56).
KL is located at chromosome 13q12, with a length of 50 kb including
a promoter region of 1,439 bp. It is a known tumor suppressor gene
that has been reported to undergo aberrant promoter methylation in
many tumors (35). The level of
abnormal expression of KL in granulosa cells of the ovary is
tightly linked with the acuteness of ovary-related diseases
(37). Thus, in this study, the
methylation pattern of KL were determined to evaluate its potential
as a promising molecular biomarker in OC and assess its prognostic
value. To the best of our knowledge, this is the first study
conducted in the Arabian Peninsula to assess this biomarker in
patients with OC and particular clinicopathological features.
Unlike previous studies, most of our cohort was diagnosed with OC
before menopause (66.1%) and was younger than 50 years of age
(64.5%). In contrast, other studies reported OC as an age-related
disease, and as mainly postmenopausal (57). Moreover, the incidence increased
more noticeably in women over 65 years of age (58), and the median age at diagnosis was
between 50 and 79 years (59,60).
According to the data released by Cancer Research UK, 53% of
British women diagnosed with OC were 65 years and older. Moreover,
according to the American Cancer Society report, 50% of the
diagnosed OC patients were aged 63 years or above (61). These findings highlight an
interesting 15-year shift in the onset of OC in the Arabian
Peninsula, and therefore, warrant additional studies to demystify
such an early onset. Parity has been reported to reduce the risk of
OC in contrast to nulliparous women (62). However, the majority of our patient
cohort was parous women (69%), which may be attributed to the
genomic blueprint, environmental factors, and/or lifestyle choices;
however, this needs further investigation.
Till date, only Wiley et al have assessed the
KL methylation profiles in 215 epithelial OC patients. They found
that KL was hypermethylated in 14% of the OC patients compared with
the controls and the hypermethylation status was associated with a
significant reduction in DFS (46).
Strikingly, approximately 62% of our cohort had hypermethylated KL,
which is 4-fold higher than the results reported by Wiley et
al (46). Our results showed
significant associations between KL methylation profile and tumor
subtype (P<0.0001). Different OC subtypes have been established
with distinct molecular signatures (63,64).
Our data showed KL promoter hypermethylation mainly in mucinous and
serous subtypes. KL methylation showed borderline significance with
age (P=0.064) (Table I). In
contrast to the findings of Wiley et al (46), analysis of KL methylation profiles
of OC patients did not reveal a significant correlation with DFS in
our cohort (Fig. 2); however, a
trend of faster recurrence in patients with KL promoter methylation
was observed (Fig. 2). Moreover,
DSS was significantly associated with KL methylation (P=0.03;
log-rank test; Fig. 1). Our data
showed that patients with higher KL promoter methylation were more
likely to die earlier than those with non-methylated KL. In fact,
in a 1-year follow-up, 75% of the patients with methylated KL were
alive compared to those with non-methylated KL (100%). These
results are in line with those of other tumors, mainly gastric,
hepatocellular, and cervical carcinoma, where KL methylation
profiles are associated with poor DFS (33,34,42).
However, despite the association between downregulation of KL
expression in follicular cells and ovarian diseases (37), the molecular mechanism of KL action
and its relationship with aging and other diseases remains poorly
understood and requires further investigation.
We acknowledge that our current OC study is a pilot
study that gives clues about KL methylation prognosis value, and
represents only the cohort of participants with possible inherent
and unavoidable bias due to the size of the cohort and the
available biological material and associated follow-up data. A
large-scale multi-centric validation study is required to confirm
these findings.
Our results showed that KL promoter hypermethylation
is a poor prognosticator in OC patients. Patients with higher KL
promoter methylation are more likely to die earlier, and therefore,
more vigilant follow-up and integrative therapeutic approaches are
required. Due to the increase in OC related mortality and the
absence of early diagnosis or effective therapeutics, KL
methylation could be used as an OC prognosticator to achieve better
and personalized clinical management of this gynecological
malignancy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Deanship of Scientific
Research in KAU (grant no. G:205-247-1440).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MHAZ and AB contributed to study design, statistical
analysis and contributed in both drafting and critical revision of
the manuscript. FMY and MA contributed to data collection,
performing the experiments and manuscript drafting. AD performed
part of the experiments and contributed to data analysis. AB and MA
have checked and confirmed the authenticity of the data. All
authors critically reviewed and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The samples and data were retrieved from the
patients after obtaining all the relevant ethical approvals
according to the guidelines of the Ethical Committee of King
Abdulaziz University Hospital (Jeddah, Saudi Arabia; ref. no.
KAU-189-14). Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian Cancer Statistics, 2018. CA Cancer J Clin. 68:284–296.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yoneda A, Lendorm ME, Couchman JR and
Multhaupt HA: Breast and ovarian cancers: A survey and possible
roles for the cell surface heparan sulfate proteoglycans. J
Histochem Cytochem. 60:9–21. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lage H and Denkert C: Resistance to
chemotherapy in ovarian carcinoma. In: Targeted Therapies in
Cancer. Dietel M (ed). Springer Berlin Heidelberg, Berlin,
Heidelberg, pp51-60, 2007.
|
|
4
|
Taylor SE and Kirwan JM: Ovarian
cancer-current management and future directions. Obstet Gynaecol
Reprod Med. 19:130–135. 2009.
|
|
5
|
Althubiti MA and Nour Eldein MM: Trends in
the incidence and mortality of cancer in Saudi Arabia. Saudi Med J.
39:1259–1262. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Almutairi AA, Edali AM, Khan SA, Aldihan
WA and Alkhenizan AH: Yield of prostate cancer screening at a
community based clinic in Saudi Arabia. Saudi Med J,. 40:681–686.
2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Karst AM and Drapkin R: Ovarian cancer
pathogenesis: A model in evolution. J Oncol.
2010(932371)2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Prat J: New insights into ovarian cancer
pathology. Ann Oncol. 23 (Suppl 10):x111–x117. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bower M and Waxman J: Ovarian cancer. In:
Lecture Notes: Oncology. 3rd edition. Wiley and Sons, London,
pp.186-192, 2016.
|
|
11
|
Manna PR, Molehin D and Ahmed AU:
Dysregulation of aromatase in breast, endometrial, and ovarian
cancers: An overview of therapeutic strategies. In: Molecular and
Cellular Changes in the Cancer Cell. Vol. 144. Pruitt K (ed).
Elsevier Academic Press Inc., San Diego, CA pp487-537, 2016.
|
|
12
|
Hesketh PJ, Kris MG, Basch E, Bohlke K,
Barbour SY, Clark-Snow RA, Danso MA, Dennis K, Dupuis LL, Dusetzina
SB, et al: Antiemetics: American Society of Clinical Oncology
clinical practice guideline update. J Clin Oncol. 35:3240–3261.
2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Norquist BM, Brady MF, Harrell MI, Walsh
T, Lee MK, Gulsuner S, Bernards SS, Casadei S, Burger RA, Tewari
KS, et al: Mutations in homologous recombination genes and outcomes
in ovarian carcinoma patients in GOG 218: An NRG
oncology/gynecologic oncology group study. Clin Cancer Res.
24:777–783. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Maradeo ME and Cairns P: Translational
application of epigenetic alterations: Ovarian cancer as a model.
FEBS Lett. 585:2112–2120. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Salehi F, Dunfield L, Phillips KP, Krewski
D and Vanderhyden BC: Risk factors for ovarian cancer: An overview
with emphasis on hormonal factors. J Toxicol Environ Health B Crit
Rev. 11:301–321. 2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Irodi A, Rye T, Herbert K, Churchman M,
Bartos C, Mackean M, Nussey F, Herrington CS, Gourley C and Hollis
RL: Patterns of clinicopathological features and outcome in
epithelial ovarian cancer patients: 35 years of prospectively
collected data. BJOG. 127:1409–1420. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sapiezynski J, Taratula O,
Rodriguez-Rodriguez L and Minko T: Precision targeted therapy of
ovarian cancer. J Control Release. 243:250–268. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Currie G and Delles C: Precision medicine
and personalized medicine in cardiovascular disease. In:
Sex-Specific Analysis of Cardiovascular Function. Vol. 1065.
Kerkhof PLM and Miller VM (eds). Springer International Publishing
AG, Cham, pp589-605, 2018.
|
|
19
|
Schmid BC and Oehler MK: New perspectives
in ovarian cancer treatment. Maturitas. 77:128–136. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Keita M, Wang ZQ, Pelletier JF, Bachvarova
M, Plante M, Gregoire J, Renaud MC, Mes-Masson AM, Paquet ÉR and
Bachvarov D: Global methylation profiling in serous ovarian cancer
is indicative for distinct aberrant DNA methylation signatures
associated with tumor aggressiveness and disease progression.
Gynecol Oncol. 128:356–363. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ganzfried BF, Riester M, Haibe-Kains B,
Risch T, Tyekucheva S, Jazic I, Wang XV, Ahmadifar M, Birrer MJ,
Parmigiani G, et al: curatedOvarianData: Clinically annotated data
for the ovarian cancer transcriptome. Database (Oxford).
2013(bat013)2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Rajendram R, Rajendram R, Patel VB and
Preedy VR: Biomarkers in health and disease: Cancer further
knowledge. In: Biomarkers in Cancer. Biomarkers in Disease:
Methods, Discoveries and Applications. Preedy V, Patel V (eds).
Springer, Dordrecht, pp981-986, 2015.
|
|
23
|
Strathdee G, Appleton K, Illand M, Millan
DW, Sargent J, Paul J and Brown R: Primary ovarian carcinomas
display multiple methylator phenotypes involving known tumor
suppressor genes. Am J Pathol. 158:1121–1127. 2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu JH, Liang XA, Wu YM, Li FS and Dai YM:
Identification of DNA methylation of SOX9 in cervical cancer using
methylated-CpG island recovery assay. Oncol Rep. 29:125–132.
2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lee MP and Dunn BK: Influence of genetic
inheritance on global epigenetic states and cancer risk prediction
with DNA methylation signature: Challenges in technology and data
analysis. Nutr Rev. 66 (Suppl 1):S69–S72. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shen SC, Liao CH, Lo YF, Tsai HP, Kuo WL,
Yu CC, Chao TC, Chen MF, Chang HK, Lin YC, et al: Favorable outcome
of secondary axillary dissection in breast cancer patients with
axillary nodal relapse. Ann Surg Oncol. 19:1122–1128.
2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Balch C, Huang TH, Brown R and Nephew KP:
The epigenetics of ovarian cancer drug resistance and
resensitization. Am J Obstet Gynecol. 191:1552–1572.
2004.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ahluwalia A, Yan P, Hurteau JA, Bigsby RM,
Jung SH, Huang TH and Nephew KP: DNA methylation and ovarian
cancer. I. Analysis of CpG island hypermethylation in human ovarian
cancer using differential methylation hybridization. Gynecol Oncol.
82:261–268. 2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zeller C, Dai W, Curry E, Siddiq A, Walley
A, Masrour N, Kitsou-Mylona I, Anderson G, Ghaem-Maghami S, Brown R
and El-Bahrawy M: The DNA methylomes of serous borderline tumors
reveal subgroups with malignant- or benign-like profiles. Am J
Pathol. 182:668–677. 2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Shi H, Wei SH, Leu YW, Rahmatpanah F, Liu
JC, Yan PS, Nephew KP and Huang TH: Triple analysis of the cancer
epigenome: An integrated microarray system for assessing gene
expression, DNA methylation, and histone acetylation. Cancer Res.
63:2164–2171. 2003.PubMed/NCBI
|
|
33
|
Xie B, Zhou J, Yuan L, Ren F, Liu DC, Li Q
and Shu G: Epigenetic silencing of Klotho expression correlates
with poor prognosis of human hepatocellular carcinoma. Hum Pathol.
44:795–801. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lee J, Jeong DJ, Kim J, Lee S, Park JH,
Chang B, Jung SI, Yi L, Han Y, Yang Y, et al: The anti-aging gene
Klotho is a novel target for epigenetic silencing in human cervical
carcinoma. Mol Cancer. 9(109)2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Rubinek T and Wolf I: Klotho tumor
suppressor. In: Encyclopedia of Cancer. Schwab M (ed). Springer,
Berlin, Heidelberg, pp1-5, 2016.
|
|
36
|
Fan CF, Tan C and Wang SQ: α-Klotho: A
novel regulator in female reproductive outcomes and hormone-related
cancer. Int J Clin Exp Med. 10:8511–8521. 2017.
|
|
37
|
Xie T, Ye W, Liu J, Zhou L and Song Y: The
emerging key role of Klotho in the hypothalamus-pituitary-ovarian
axis. Reprod Sci. 28:322–331. 2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Duce JA, Podvin S, Hollander W, Kipling D,
Rosene DL and Abraham CR: Gene profile analysis implicates Klotho
as an important contributor to aging changes in brain white matter
of the rhesus monkey. Glia. 56:106–117. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
King GD, Rosene DL and Abraham CR:
Promoter methylation and age-related downregulation of Klotho in
rhesus monkey. Age. 34:1405–1419. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Semba RD, Moghekar AR, Hu J, Sun K, Turner
R, Ferrucci L and O'Brien R: Klotho in the cerebrospinal fluid of
adults with and without Alzheimer's disease. Neurosci Lett.
558:37–40. 2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Rubinek T, Shulman M, Israeli S, Bose S,
Avraham A, Zundelevich A, Evron E, Gal-Yam EN, Kaufman B and Wolf
I: Epigenetic silencing of the tumor suppressor Klotho in human
breast cancer. Breast Cancer Res Treat. 133:649–657.
2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang LJ, Wang X, Wang XJ, Jie P, Lu H,
Zhang S, Lin X, Lam EK, Cui Y, Yu J and Jin H: Klotho is silenced
through promoter hypermethylation in gastric cancer. Am J Cancer
Res. 1:111–119. 2011.PubMed/NCBI
|
|
43
|
Yan Y, Wang Y, Xiong Y, Lin X, Zhou P and
Chen Z: Reduced Klotho expression contributes to poor survival
rates in human patients with ovarian cancer, and overexpression of
Klotho inhibits the progression of ovarian cancer partly via the
inhibition of systemic inflammation in nude mice. Mol Med Rep.
15:1777–1785. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Matsumura Y, Aizawa H, Shiraki-Iida T,
Nagai R, Kuro-o M and Nabeshima Y: Identification of the human
klotho gene and its two transcripts encoding membrane and secreted
klotho protein. Biochem Biophys Res Commun. 242:626–630.
1998.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lu L, Katsaros D, Wiley A, de la Longrais
IA, Puopolo M and Yu H: Klotho expression in epithelial ovarian
cancer and its association with insulin-like growth factors and
disease progression. Cancer Invest. 26:185–192. 2008.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wiley A, Katsaros D, Lu L, de La Longrais
IAR, Puopolo M, Si J and Yu H: DNA methylation of the human Klotho
Gene: Associations with IGF-I, IGF-II, and IGFBP-3 expression and
ovarian cancer survival. Cancer Res. 66 (Suppl 8)(S1204)2006.
|
|
47
|
Dallol A, Al-Ali W, Al-Shaibani A and
Al-Mulla F: Analysis of DNA methylation in FFPE tissues using the
MethyLight technology. In: Formalin-Fixed Paraffin-Embedded
Tissues: Methods and Protocols. Al-Mulla F (ed). Humana Press,
Totowa, NJ, pp191-204, 2011.
|
|
48
|
Dallol A, Buhmeida A, Merdad A,
Al-Maghrabi J, Gari MA, Abu-Elmagd MM, Elaimi A, Assidi M,
Chaudhary AG, Abuzenadah AM, et al: Frequent methylation of the
Klotho gene and overexpression of the FGFR4 receptor in invasive
ductal carcinoma of the breast. Tumour Biol. 36:9677–9683.
2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Losi L, Fonda S, Saponaro S, Chelbi ST,
Lancellotti C, Gozzi G, Alberti L, Fabbiani L, Botticelli L and
Benhattar J: Distinct DNA methylation profiles in ovarian tumors:
Opportunities for novel biomarkers. Int J Mol Sci.
19(1559)2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Ebell MH, Culp MB and Radke TJ: A
systematic review of symptoms for the diagnosis of ovarian cancer.
Am J Prev Med. 50:384–394. 2016.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Rooth C: Ovarian cancer: Risk factors,
treatment and management. Br J Nurs. 22:S23–S30. 2013.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ramaswami R, Bayer R and Galea S:
Precision medicine from a public health perspective. In: Annual
Review of Public Health. Vol. 39. Fielding JE, Brownson RC and
Green LW (eds). Annual Reviews, Palo Alto, CA, pp153-168, 2018.
|
|
53
|
Bachvarov D, L'Esperance S, Popa I,
Bachvarova M, Plante M and Têtu B: Gene expression patterns of
chemoresistant and chemosensitive serous epithelial ovarian tumors
with possible predictive value in response to initial chemotherapy.
Int J Oncol. 29:919–933. 2006.PubMed/NCBI
|
|
54
|
L'Espérance S, Popa I, Bachvarova M,
Plante M, Patten N, Wu L, Têtu B and Bachvarov D: Gene expression
profiling of paired ovarian tumors obtained prior to and following
adjuvant chemotherapy: Molecular signatures of chemoresistant
tumors. Int J Oncol. 29:5–24. 2006.PubMed/NCBI
|
|
55
|
Sabatier R, Finetti P, Cervera N, Birnbaum
D and Bertucci F: Gene expression profiling and prediction of
clinical outcome in ovarian cancer. Crit Rev Oncol Hematol.
72:98–109. 2009.PubMed/NCBI View Article : Google Scholar
|
|
56
|
García-Sánchez A and Marqués-García F:
Review of methods to study gene expression regulation applied to
asthma. In: Molecular Genetics of Asthma. Isidoro García M (ed).
Springer New York, NY, pp71-89, 2016.
|
|
57
|
Chan JK, Urban R, Cheung MK, Osann K, Shin
JY, Husain A, Teng NN, Kapp DS, Berek JS and Leiserowitz GS:
Ovarian cancer in younger vs older women: A population-based
analysis. Br J Cancer. 95:1314–1320. 2006.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Mohammadian M, Ghafari M, Khosravi B,
Salehiniya H, Aryaie M, Bakeshei FA and Mohammadian-Hafshejani A:
Variations in the incidence and mortality of ovarian cancer and
their relationship with the human development index in European
countries in 2012. Biomed Res Ther. 4:1541–1557. 2017.
|
|
59
|
Zheng G, Yu H, Kanerva A, Försti A,
Sundquist K and Hemminki K: Familial risks of ovarian cancer by age
at diagnosis, proband type and histology. PLoS One.
13(e0205000)2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Arora N, Talhouk A, McAlpine JN, Law MR
and Hanley GE: Long-term mortality among women with epithelial
ovarian cancer: A population-based study in British Columbia,
Canada. BMC Cancer. 18(1039)2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Smith RA, Cokkinides V and Brawley OW:
Cancer screening in the United States, 2012: A review of current
American Cancer Society guidelines and current issues in cancer
screening. CA Cancer J Clin. 62:129–142. 2012.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Kim SJ, Rosen B, Fan I, Ivanova A,
McLaughlin JR, Risch H, Narod SA and Kotsopoulos J: Epidemiologic
factors that predict long-term survival following a diagnosis of
epithelial ovarian cancer. Br J Cancer. 116:964–971.
2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Earp MA and Cunningham JM: DNA methylation
changes in epithelial ovarian cancer histotypes. Genomics.
106:311–321. 2015.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Köbel M, Bak J, Bertelsen BI, Carpen O,
Grove A, Hansen ES, Levin Jakobsen AM, Lidang M, Måsbäck A, Tolf A,
et al: Ovarian carcinoma histotype determination is highly
reproducible, and is improved through the use of
immunohistochemistry. Histopathology. 64:1004–1013. 2014.PubMed/NCBI View Article : Google Scholar
|