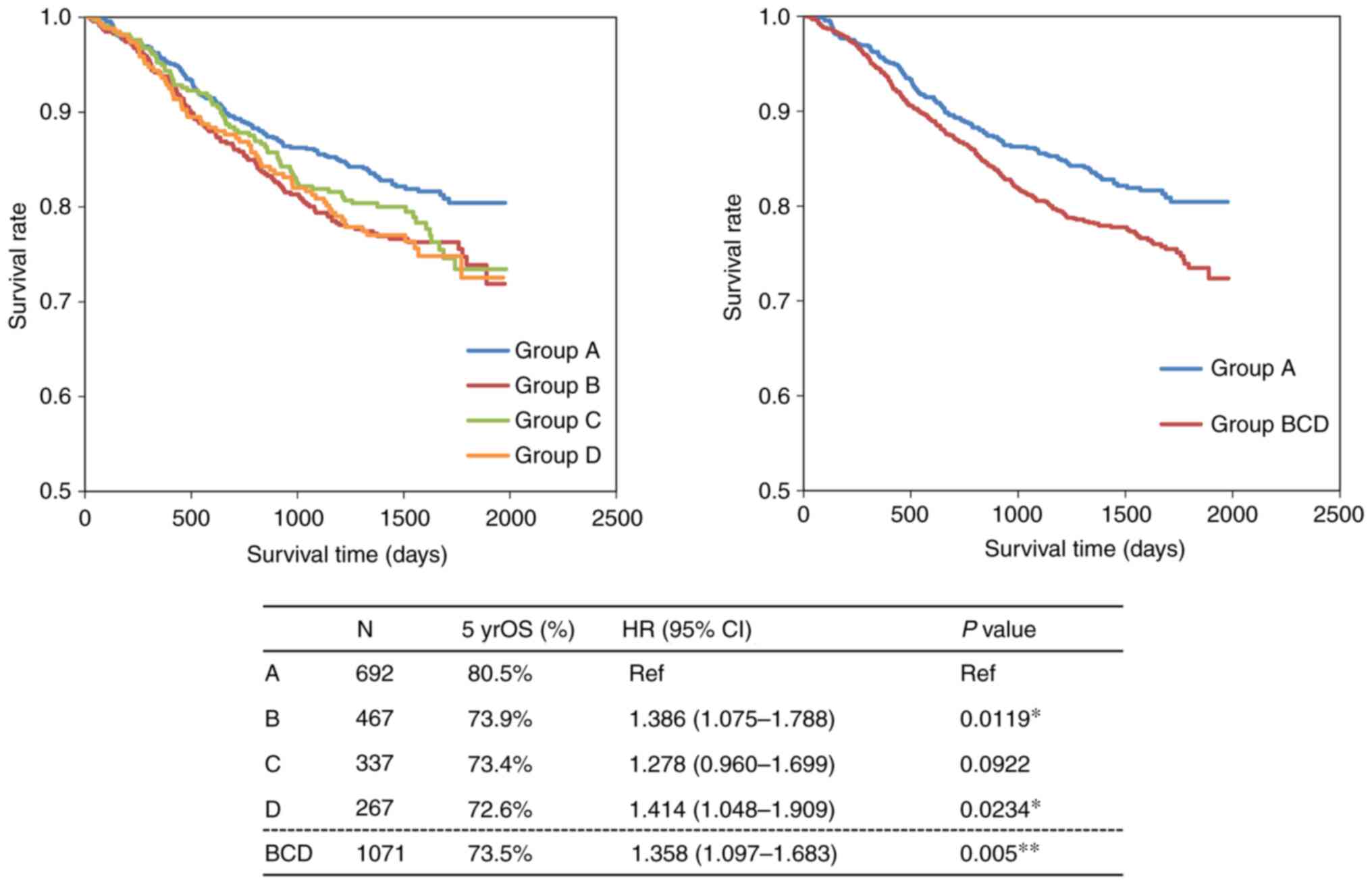
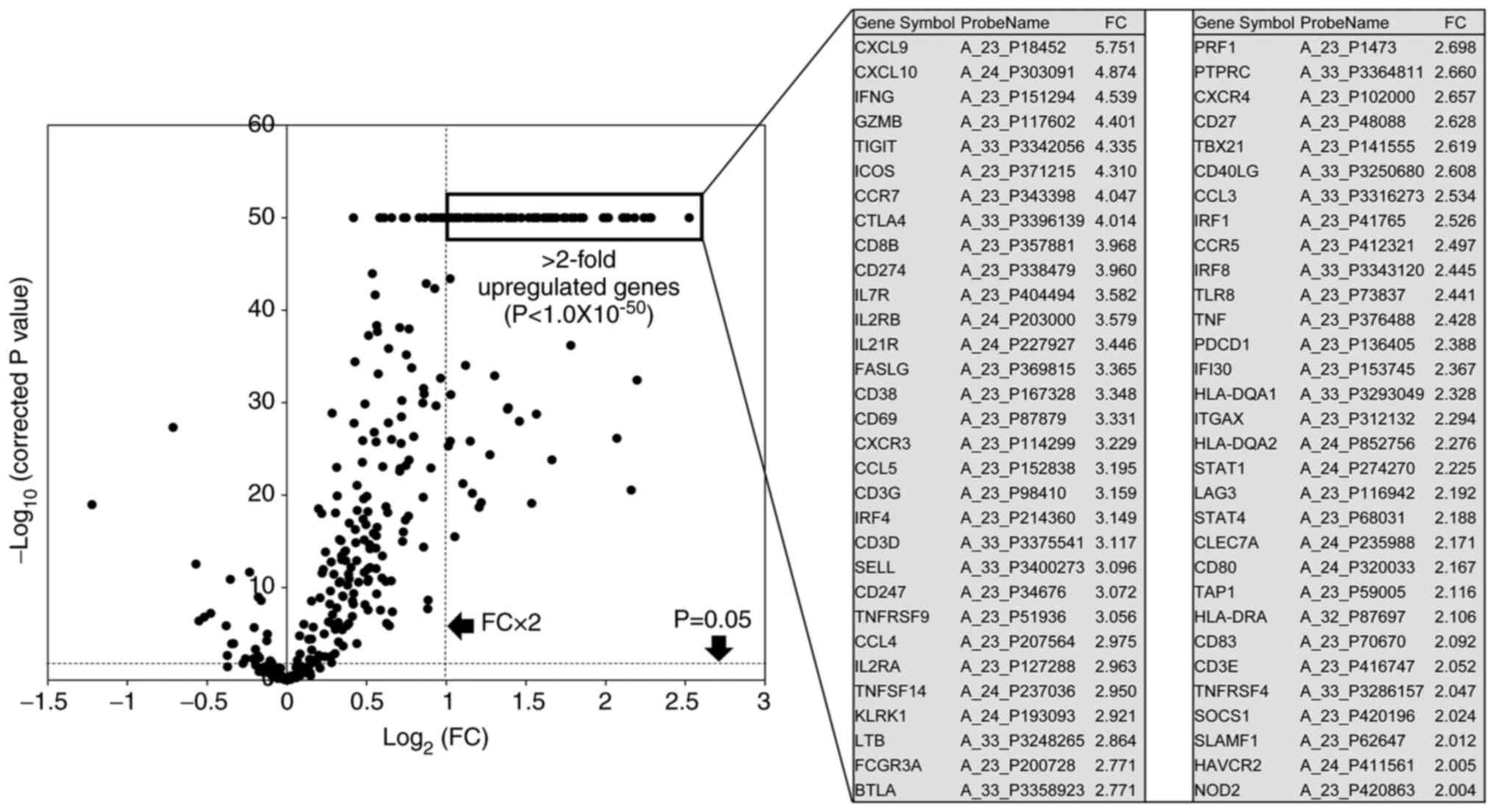
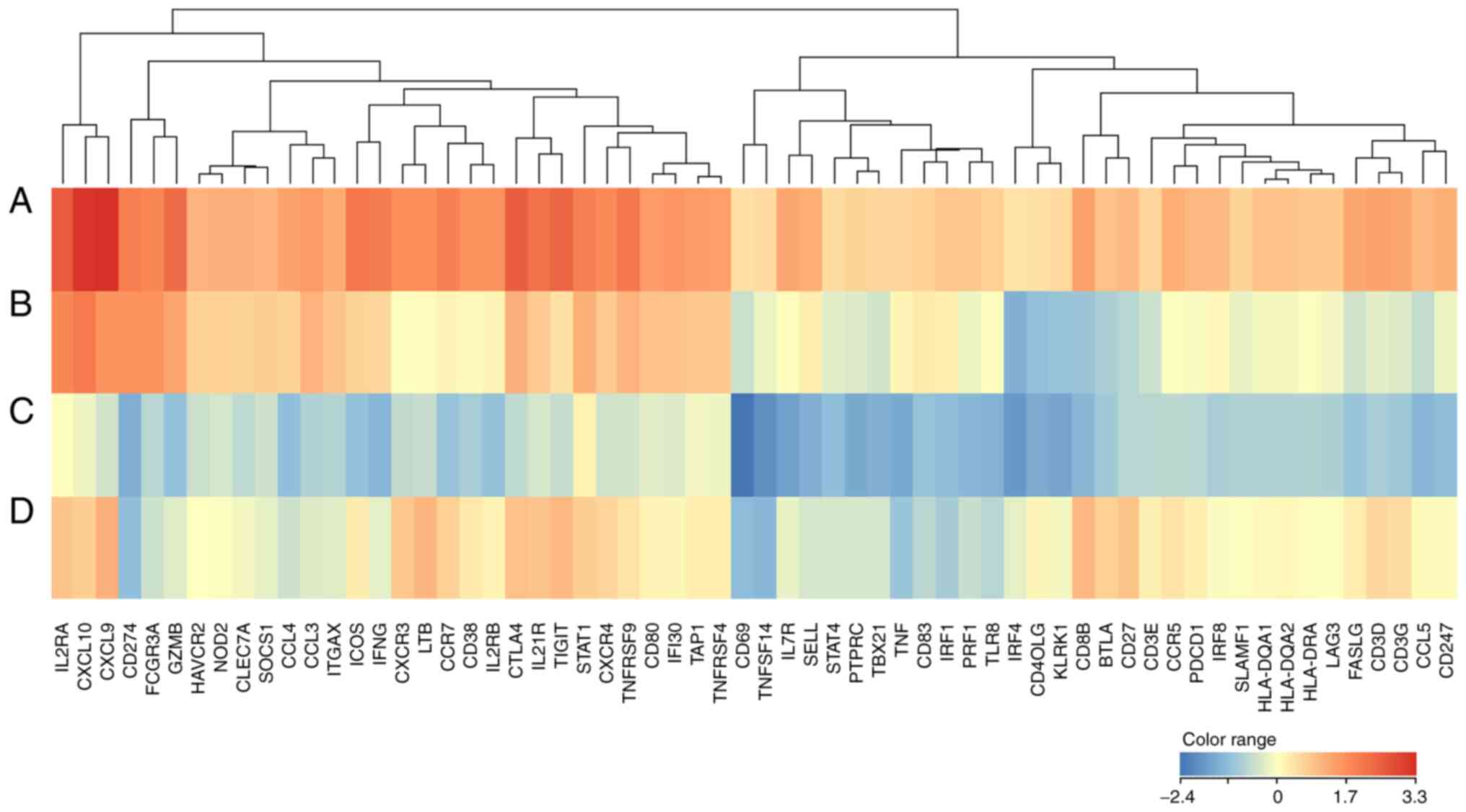
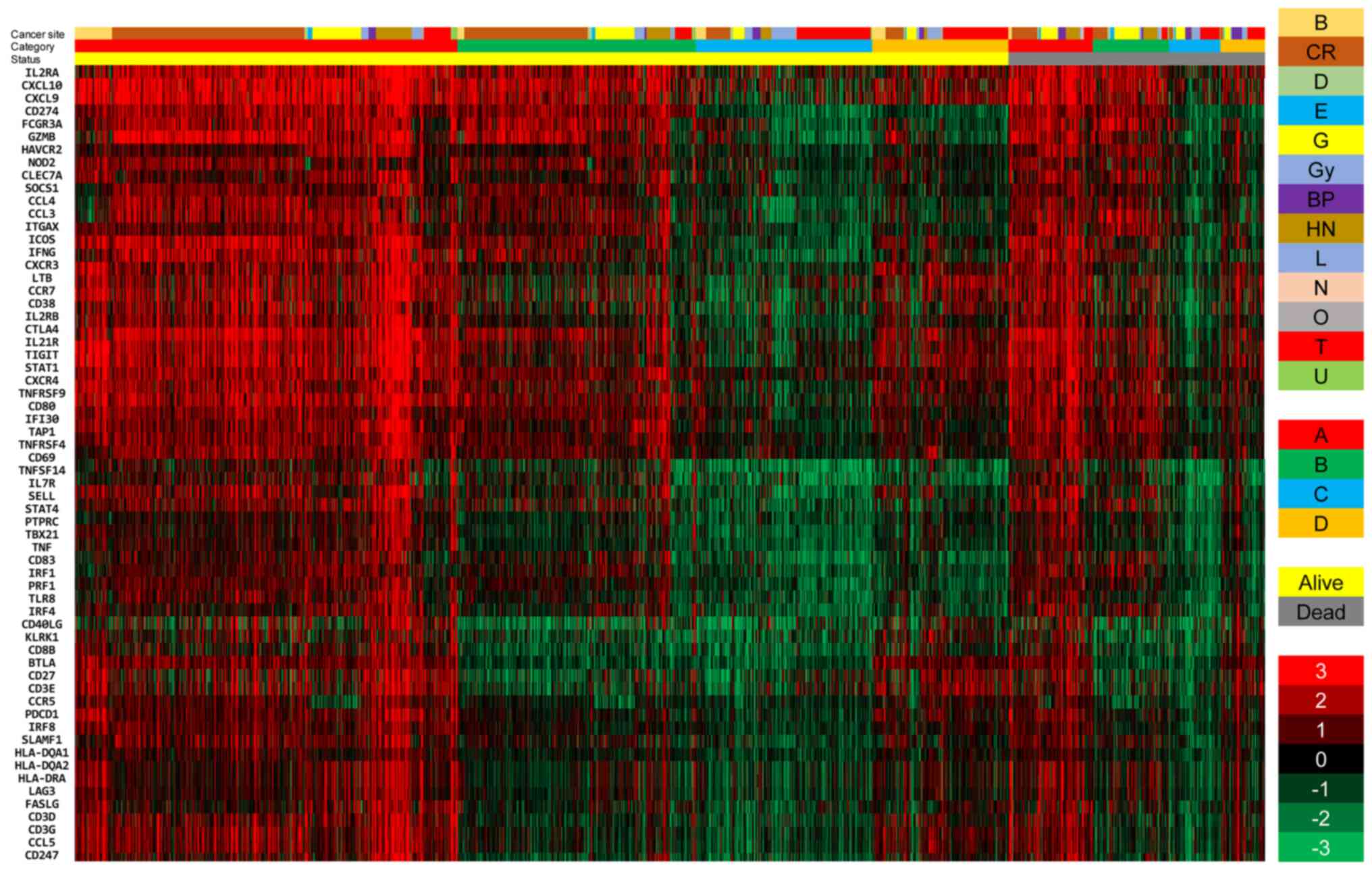
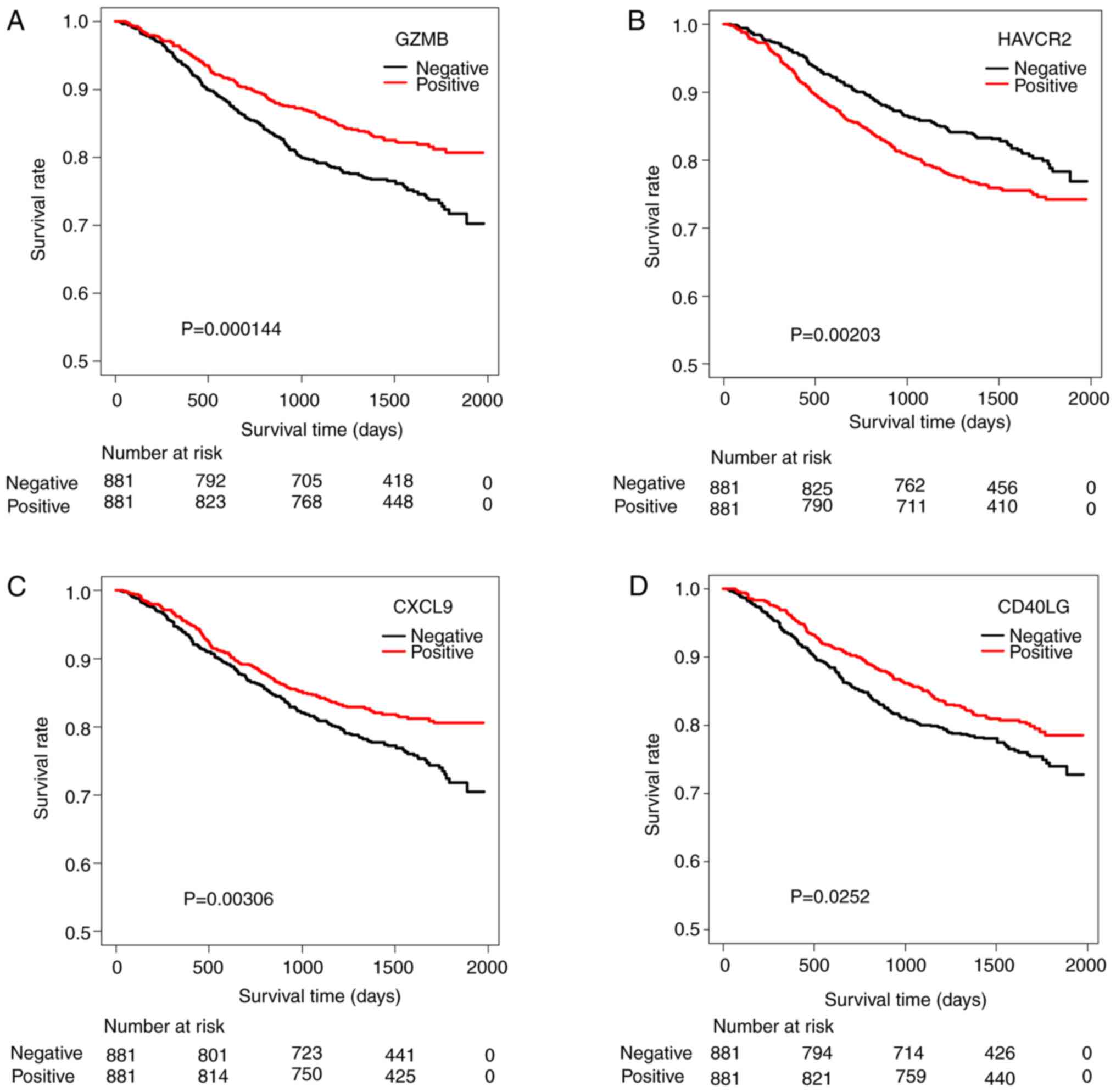
|
1
|
Weber JS, O'Day S, Urba W, Powderly J,
Nichol G, Yellin M, Snively J and Hersh E: Phase I/II study of
ipilimumab for patients with metastatic melanoma. J Clin Oncol.
26:5950–5956. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Topalian SL, Hodi FS, Brahmer JR,
Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD,
Sosman JA, Atkins MB, et al: Safety, activity, and immune
correlates of anti-PD-1 antibody in cancer. N Engl J Med.
366:2443–2454. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ,
Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunski K, et al:
Safety and activity of anti-PD-L1 antibody in patients with
advanced cancer. N Engl J Med. 366:2455–2465. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ascierto RA, Capone M, Urba WJ, Bifuco CB,
Botti G, Lugli A, Marincola FM, Ciliberto G, Galon J and Fox BA:
The additional facet of immunoscore: Immunoprofiling as a possible
predictive tool for cancer treatment. J Transl Med.
11(54)2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gnjatic S, Bronte V, Brunet LR, Butler MO,
Disis ML, Galon J, Hakansson LG, Hanks BA, Karanikas V, Khleif SN,
et al: Identifying baseline immune-related biomarkers to predict
clinical outcome of immunotherapy. J Immunother Cancer.
5(44)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Johnson DB, Frampton GM, Rioth MJ, Yusko
E, Xu Y, Guo X, Ennis RC, Fabrizio D, Chalmers ZR, Greenbowe J, et
al: Targeted next generation sequencing identifies markers of
response to PD-1 blockade. Cancer Immunol Res. 4:959–967.
2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dudley JC, Lin MT, Le DT and Eshleman JR:
Microsatellite instability as a biomarker for PD-1 blockade. Clin
Cancer Res. 22:813–820. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yarchoan M, Johnson BA III, Lutz ER,
Laheru DA and Jaffee EM: Targeting neoantigen to augment antitumor
immunity. Nat Rev Cancer. 17:209–222. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lin Y, Xu J and Lan H: Tumor-associated
macrophages in tumor metastasis: Biological roles and clinical
therapeutic applications. J Hematol Oncol. 12(76)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Fleming B, Hu X, Weber R, Nagibin V, Groth
C, Artevogt P, Utical J and Umansky V: Targeting myeloid-derived
suppressor cells to bypass tumor-induced immunosuppression. Front
Immunol. 9(398)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Rooney MS, Shukla SA, Wu CJ, Getz G and
Hacohen N: Molecular and genetic properties of tumors associated
with local immune cytolytic activity. Cell. 160:48–61.
2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ock CY, Keam B, Kim S, Lee JS, Kim M, Kim
TM, Jeon YK, Kim DW, Chung DH and Heo DS: Pan-cancer immunogenic
perspective on the tumor microenvironment based on PD-L1 and CD8
T-cell infiltration. Clin Cancer Res. 22:2261–2270. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Teng MW, Ngiow SF, Ribas A and Smyth MJ:
Classifying cancers based on T-cell infiltration and PD-L1. Cancer
Res. 75:2139–2145. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yarchoan M, Hopkins A and Jaffee EM: Tumor
mutational burden and response rate to PD-1 inhibition. N Engl J
Med. 377:2500–2501. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bruni D, Angell HK and Galon J: The immune
contexture and immunoscore in cancer prognosis and therapeutic
efficacy. Nat Rev Cancer. 20:662–680. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Snyder A, Makarov V, Merghoub T, Yuan J,
Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et
al: Genetic basis for clinical response to CTLA-4 blockade in
melanoma. N Engl J Med. 371:2189–2199. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Iwai Y, Ishida M, Tanaka Y, Okazaki T,
Honjo T and Minato N: Involvement of PD-L1 on tumor cells in the
escape from host immune system and tumor immunotherapy by PD-L1
blockade. Proc Natl Acad Sci USA. 99:12293–12297. 2002.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Petty AJ, Dai R, Lapalombella R, Baiocchi
RA, Benson DM, Li Z, Huang X and Yang Y: Hedgehog-induced PD-L1 on
tumor-associated macrophages is critical for suppression of
tumor-infiltrating CD8+ T cell function. JCI Insight.
6(e146707)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kondou R, Iizuka A, Nonomura C, Miyata H,
Ashizawa T, Nagashima T, Ohshima K, Urakami K, Kusuhara M,
Yamaguchi K and Akiyama Y: Classification of tumor microenvironment
immune types based on immune response-associated gene expression.
Int J Oncol. 54:219–228. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ohshima K, Hatakeyama K, Nagashima T,
Watanabe Y, Kanto K, Doi Y, Ide T, Shimoda Y, Tanabe T, Ohnami S,
et al: Integrated analysis of gene expression and copy number
identified potential cancer driver genes with
amplification-dependent overexpression in 1,454 solid tumors. Sci
Rep. 7(641)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Doaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Akiyama Y, Kondou R, Iizuka A, Ohshima K,
Urakami K, Nagashima T, Shimoda Y, Tanabe T, Ohnami S, Ohnami S, et
al: Immune response-associated gene analysis of 1,000 cancer
patients using whole-exome sequencing and gene expression
profiling-Project HOPE. Biomed Res. 37:233–242. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EGR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gubin MM, Esaulova E, Ward JP, Malkova ON,
Runci D, Wong P, Noguchi T, Arthur CD, Meng W, Alspach E, et al:
High-dimensional analysis delineates myeloid and lymphoid
compartment remodeling during successful immune-checkpoint cancer
therapy. Cell. 175:1014–1030. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Guo X, Zhang Y, Zheng L, Zheng C, Song J,
Zhang Q, Kang B, Liu Z, Jin L, Xing R, et al: Global
characterization of T cells in non-small-cell lung cancer by
single-cell sequencing. Nat Med. 24:978–985. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sathe A, Grimes SM, Lau BT, Chen J, Suarez
C, Huang RJ, Poultsides G and Ji HP: Single-cell genomic
characterization reveals the cellular reprogramming of the gastric
tumor microenvironment. Clin Cancer Res. 26:2640–2653.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hossain MA, Liu G, Dai B, Si Y, Yang Q,
Wazir J, Birnbaumer L and Yang Y: Reinvigorating exhausted CD8+
cytotoxic T lymphocytes in the tumor microenvironment and current
strategies in cancer immunotherapy. Med Res Rev. 41:156–201.
2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Xiao Z, Locasale JW and Dai Z: Metabolism
in the tumor microenvironment: Insights from single-cell analysis.
Oncoimmunology. 9(1726556)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kumagai S, Togashi Y, Kamada T, Sugiyama
E, Nishinakamura H, Takeuchi Y, Vitaly K, Itahashi K, Maeda Y,
Matsui S, et al: The PD-1 expression balance between effector and
regulatory T cells predicts the clinical efficacy of PD-1 blockade
therapies. Nat Immunol. 21:1346–1358. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Opzoomer JW, Sosnowska D, Anstee J, Spicer
JF and Arnold JN: Cytotoxic chemotherapy as an immune stimulus: A
molecular perspective on turning up the immunological heat on
cancer. Front Immunol. 10(1654)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yu WD, Sun G, Li J, Xu J and Wang X:
Mechanisms and therapeutic potentials of cancer immunotherapy in
combination with radiotherapy and/or chemotherapy. Cancer Lett.
452:66–70. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Turgeon GA, Weickhardt A, Azad AA, Solomon
B and Siva S: Radiotherapy and immunotherapy: A synergistic effect
in cancer care. Med J Aust. 210:47–53. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Rodriguez-Ruiz ME, Vanpouille-Box C,
Melero I, Formenti SC and Demaria S: Immunological mechanisms
responsible for radiation-induced abscopal effect. Trends Immunol.
39:644–655. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Mitsuya K, Akiyama Y, Iizuka A, Miyata H,
Deguchi S, Hayashi N, Maeda C, Kondou R, Kanematsu A, Watanabe K,
et al: Alpha-type-1 polarized dendritic cell-based vaccination in
newly diagnosed high-grade glioma: A phase II clinical trial.
Anticancer Res. 40:6473–6484. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Simon S, Voillet V, Vignard V, Wu Z,
Dabrowski C, Jouand N, Beauvais T, Khammari A, Braudeau C, Josien
R, et al: PD-1 and TIGIT coexpression identifies a circulating CD8
T cell subset predictive of response to anti-PD-1 therapy. J
Immunother Cancer. 8(e001631)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Bleharski JR, Niazi KR, Sieling PA, Cheng
G and Modlin RL: Signaling lymphocytic activation molecule is
expressed on CD40 ligand-activated dendritic cells and
directly augments production of inflammatory cytokines. J Immunol.
167:3174–3181. 2001.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Schetters ST, Rodriguez E, Kruijssen LJ,
Crommentuijn MHW, Boon L, Van den Bossche J, Den Haan JMM and Van
Kooyk Y: Monocyte-derived APCs are central to the response of PD1
checkpoint blockade and provide a therapeutic target for
combination therapy. J Immunother Cancer. 8(e000588)2020.PubMed/NCBI View Article : Google Scholar
|