Introduction
More people die of cancer than any other disease in
the world today (1). Accordingly,
both physicians and researchers strive to provide reliable
monitoring for disease, diagnosis and prognosis as well as
resistance prediction. The primary goal is to provide patients with
adequate treatment and restore their well-being. Lung cancer is the
most common malignancy and contributes to the greatest number of
cancer deaths (2). Colorectal
cancer is the third-most frequently contracted malignant disease
worldwide (3). Among women, breast
cancer is the most common type of cancer (4).
In previous years, liquid biopsy techniques have
been used to treat a number of different types of cancer (5-7).
This less invasive testing method (compared with traditional
biopsy) offers potential for a satisfactory outcome, higher
recovery rate and more accurate results (5,6).
Changes in circulating tumor DNA (ctDNA) are used for cancer
screening in asymptomatic people, detecting mutations for
theranostic consideration and monitoring tumor dynamics and genetic
evolution (5). Repeated analysis
and quantitation of ctDNA may provide information on changes in
clonal composition over time, allowing for modification of
treatment regime (8). KRAS,
BRAF and EGFR mutations may be identified via liquid
biopsy in patients with colon cancer, melanoma and lung cancer
(5,6).
Numerous types of tumor are dependent on oncogenes:
Oncogene addiction has been noted in numerous types of neoplasm,
such as lung cancer (9). Among
patients with lung adenocarcinoma, ~50% have at least one driver
mutation representing a potential target for clinical intervention.
For example, activating EGFR mutations are predictive of
susceptibility to EGFR inhibitors, such as erlotinib and
gefitinib. ctDNA analysis has been shown to diagnose EGFR
exon 19 deletions and L858R mutations with a sensitivity of 82-87%
and a precision of 97-98% and may thus be an alternative to tissue
genetic analysis. Most patients taking EGFR-inhibiting
medication experience disease development after 2 years of therapy
and 60% of resistance to treatment is due to a disease clone
containing a secondary EGFR T790M mutation, which inhibits
drug access to the target kinase. T790M is more common in exon 19
deletion than in L858R among patients with acquired resistance to
EGFR tyrosine kinase inhibitors (TKIs) (7,10).
Among patients with non-small cell lung cancer
(NSCLC), 1% have chromosome translocations in the RET gene.
The College of American Pathologists, the International Association
for the Study of Lung Cancer and the Association for Molecular
Pathology published recommendations including ROS1 testing
for all patients with adenocarcinoma, the use of additional genes
(ERBB2, MET, BRAF, KRAS and RET)
for laboratories performing next-generation sequencing (NGS) panels
and immunohistochemistry as an alternative to fluorescence in
situ hybridization. Acquired resistance mutation C797S may
occur in tumors that have progressed following osimertinib
treatment with T790M mutation (7).
It is estimated that 30-40% of estrogen and/or
progesterone receptor-positive breast cancer cases have
PIK3CA mutations (11).
Increased activation of the PI3K/AKT/mTOR pathway contributes to
various aspects of cancer, including acquired growth signals,
inhibition of apoptosis, vessel generation and insensitivity to
anti-growth signals. The PI3K/AKT/mTOR pathway is associated tumor
development and progression in lung cancer. Therefore, this pathway
represents a novel target for anticancer treatment (12). The most validated anti-oncogenic
effect of PTEN is inhibition of the PI3K/AKT/mTOR oncogenic
signaling system; other documented effects include chromosomal
integrity and DNA repair (13).
Cancer-associated gene variants may be found in
apparently healthy people, arising in part from clonal
hematopoiesis (14).
Age-associated clonal hematopoiesis, often referred to as
indeterminate potential clonal hematopoiesis, is distinguished by
recurrent somatic variants that are associated with peripheral
blood hematological cancer. The most commonly involved genes are
DNMT3A, TET2 and ASXL1; other genes that are
often mutated include TP53, JAK2, SF3B1,
GNB1, PPM1D, GNAS and BCORL1. Due to
limited data, caution is required when interpreting ctDNA variants
in these genes and more research is needed to understand how to
interpret and report ctDNA variants in these genes (14,15).
In the present study, a comprehensive liquid biopsy
panel was performed on 474 patients to assess the importance and
spectrum of recurrent cancer somatic mutations.
Materials and methods
Patients
The present study was approved by the Ethics
Committee at the University of Health Sciences, Dr. Abdurrahman
Yurtaslan Ankara Oncology Training and Research Hospital (Ankara,
Turkey). Written informed consent was obtained from all patients.
Most patients visited the Medical Genetics Clinic at the Department
of Medical Genetics, University of Health Sciences, Dışkapı
Yıldırım Beyazıt Training and Research Hospital and the Department
of Medical Genetics, University of Health Sciences, Dr. Abdurrahman
Yurtaslan Ankara Oncology Training and Research Hospital (Ankara,
Turkey) with a diagnosis of advanced resistant cancer. Clinical
histories and molecular results were reviewed for 474 patients
examined at the Department of Medical Oncology, Hacettepe
University, Faculty of Medicine, Department of Medical Genetics,
University of Health Sciences, Dışkapı Yıldırım Beyazıt Training
and Research Hospital and the Department of Medical Genetics,
University of Health Sciences, Dr. Abdurrahman Yurtaslan Ankara
Oncology Training and Research Hospital (Ankara, Turkey). The
patients underwent a comprehensive liquid biopsy panel between
January 2018 and December 2020 at the Ankara Central Genetic
Laboratory (Turkey). They were evaluated according to the National
Comprehensive Cancer Network guidelines for breast-ovarian cancer
and Lynch syndrome (16,17). Patients with a strong family
history of cancer underwent a familial cancer panel. Patients with
missing data were excluded.
Patients were divided based on cancer type into the
following groups: Lung (n=379, 79.9%), breast (n=72, 15.2%),
gastrointestinal (n=11, 2.3%) and other (n=12, 2.5%). The other
group included patients with rare or unspecific cancer (including
carcinoma of unknown primary, melanoma and bladder, gallbladder,
liver, laryngeal and endometrial cancer).
DNA panels and NGS
From blood samples (10 ml) collected in EDTA tubes,
genomic DNA was extracted according to the manufacturer's procedure
using a QIAamp DNA Blood Midi kit and QIAcube (both Qiagen, Inc.).
Paired-end sequencing was performed with a loading concentration of
1.6 pM. The concentration was measured with an Invitrogen Qubit 3
Fluorometer (Thermo Fisher Scientific, Inc.). Amplicon lengths were
265 bp for Sophia Genetics 56 G Oncology Solution (Sophia Genetics)
and 250 bp for the ArcherDx Reveal ctDNA 28 kit (ArcherDx,
Inc.).
Two different multigene panels were used: ArcherDx
Reveal ctDNA 28 kit (AKT1, CTNNB1, ESR1,
IDH2, MAP2K2, NTRK1, RET, ALK,
DDR2, FGFR1, KIT, MET, NTRK3,
ROS1, AR, EGFR, HRAS, KRAS,
MTOR, PDGFRA, SMAD4, BRAF,
ERBB2, IDH1, MAP2K1, NRAS,
PIK3CA, TP53) and Sophia Genetics 56 G Oncology
Solution (ABL1, AKT1, ALK, APC,
ATM, BRAF, CDH1, CDKN2A, CSF-1R,
CTNNB1, DDR2, DNMT3A, EGFR,
ERBB2, ERBB4, EZH2, FBXW7,
FGFR1, FGFR2, FGFR3, FLT3, FOXL2,
GNA11, GNAQ, GNAS, HNF1A, HRAS,
IDH1, IDH2, JAK2, JAK3, KDR,
KIT, KRAS, MAP2K1, MET, MLH1,
MPL, MSH6, NOTCH1, NPM1, NRAS,
PDGFRA, PIK3CA, PTEN, PTPN11,
RB1, RET, STK11, SMAD4, SMARCB1,
SMO, SRC, TP53, TSC1, VHL). The
Sophia Genetics 56G Oncology Solution was used between January 2018
and November 2020 and ArcherDx Reveal ctDNA 28 kit has been used
since January 2020. The sequencing was performed on an Illumina
MiSeq system (Illumina, Inc.). The data were analyzed using the
Archer Analysis Platform (ArcherDx, Inc.) for the ArcherDx Reveal
ctDNA 28 kit and Sophia DDM software v4 (Sophia Genetics) for the
Sophia Genetics 56G Oncology Solution. Visualization of the data
was performed with Integrative Genomics Viewer 2.7.2 (Broad
Institute) software.
Statistical analysis
The available evidence (population frequency
information, case notes, case/control and functional tests,
internal co-occurrence and co-segregation data, evolutionary
conservation data and in silico predictions) for all
variants, except previously characterized benign alterations, was
thoroughly evaluated and analyzed. Data are presented as the mean.
In compliance with the recommendations issued by the Association
for Molecular Pathology, the American Society of Clinical Oncology
and the College of American Pathologists, variants were categorized
into tiers as follows: I, variants with strong clinical
significance; II, variants with potential clinical significance;
III, variants with unknown clinical significance and IV, variants
that are benign or likely benign (18). Tier I, II and III variations were
included in the study. Fig. 1,
Fig. 2 and Fig. 3 were prepared using Python (version
3.9.2, https://docs.python.org); Fig. 4 was prepared using
Lollipops-v1.3.5(19).
Results
Patients
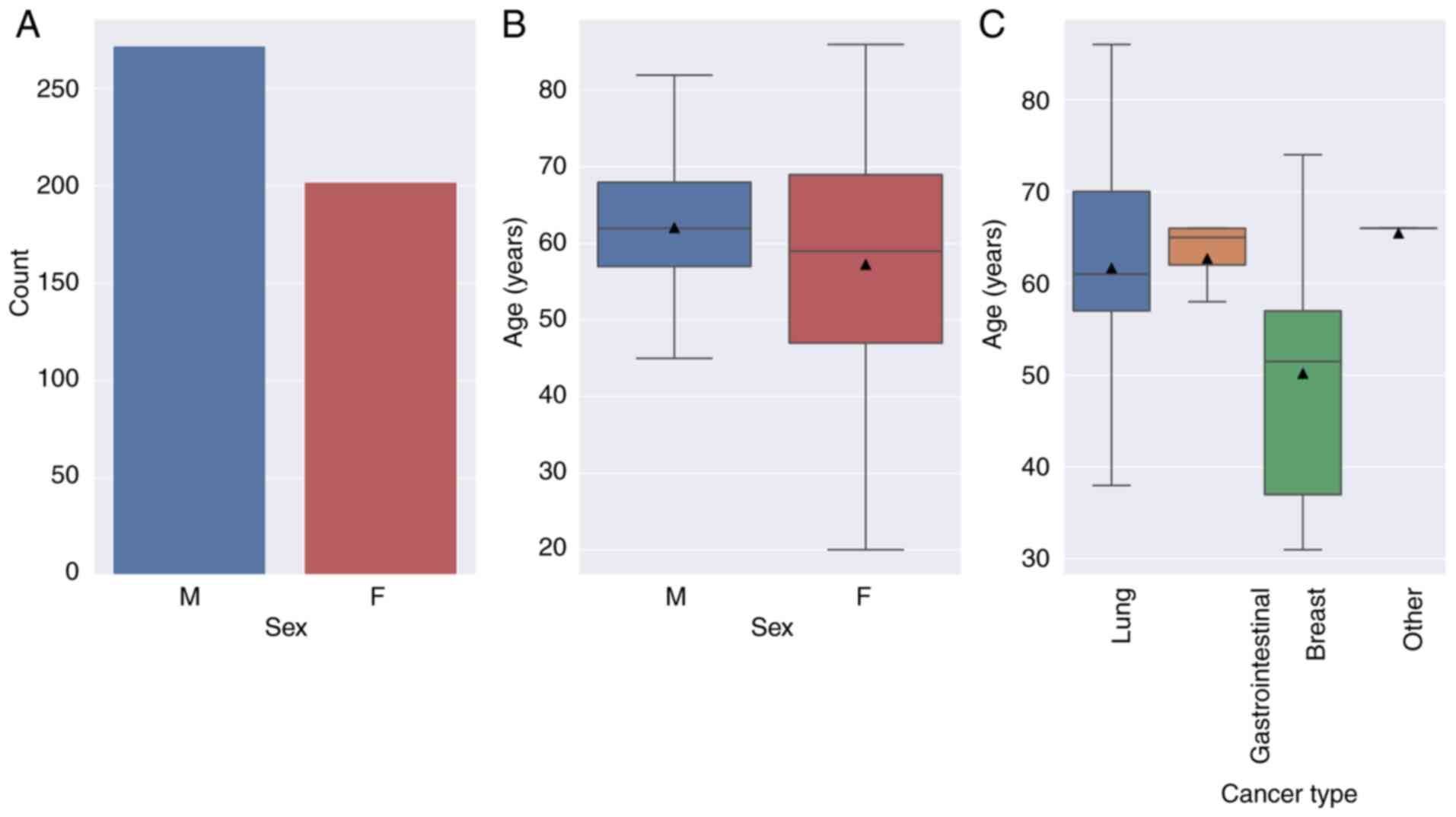
The mean age of the participants was 60 years, with
a range of 20-86 years (Fig. 1).
Most patients were between the ages of 50 and 70 years. The mean
age in each group was as follows: breast, 50.2; gastrointestinal,
62.7; lung, 61.6 years and other, 65.5 years. Patients with
advanced resistant breast cancer were referred to the clinic at an
earlier age when compared with other groups. There were notably
more male patients (n=272, 57.4%) than females (n=202, 42.6%;
Fig. 1).
Mutations
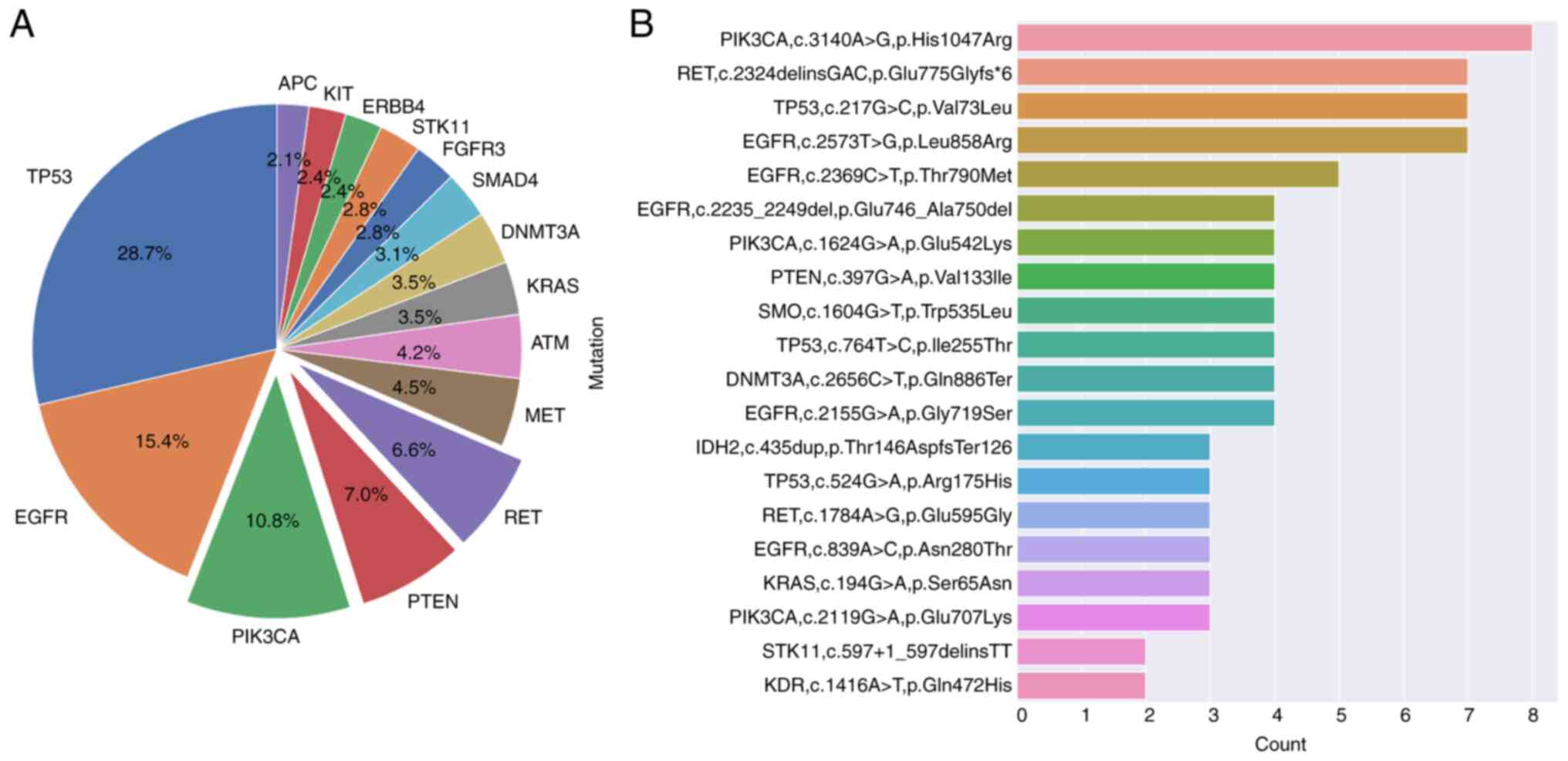
Mutations were detected in 357 patients and the
majority of variant fractions were 0.1-10.0% (data not shown). A
total of 131 mutations were nonsense or frameshifts. The most
commonly mutated genes detected in patients were TP53,
EGFR, PIK3CA, RET, PTEN, MET,
ATM and KRAS. The most common mutations detected were
‘PIK3CA, c.3140A>G, p.His1047Arg’, ‘RET,
c.2324delinsGAC, p.Glu775Glyfs*6’, ‘TP53, c.217G>C,
p.Val73Leu’, ‘EGFR, c.2155G>A, p.Gly719Ser’,
‘PIK3CA, c.1624G>A, p.Glu542Lys’, ‘PTEN,
c.397G>A, p.Val133Ile’, ‘DNMT3A, c.2656C>T,
p.Gln886Ter’, ‘EGFR, c.2235_2249del, p.Glu746_Ala750del’,
‘SMO, c.1604G>T, p.Trp535Leu’ and ‘TP53,
c.764T>C, p.Ile255Thr’. The ‘PIK3CA, c.3140A>G,
p.His1047Arg’ mutation was observed eight times (2.24%; Fig. 2). The total number of different
EGFR exon 19 deletions exceeded other mutations (n=9).
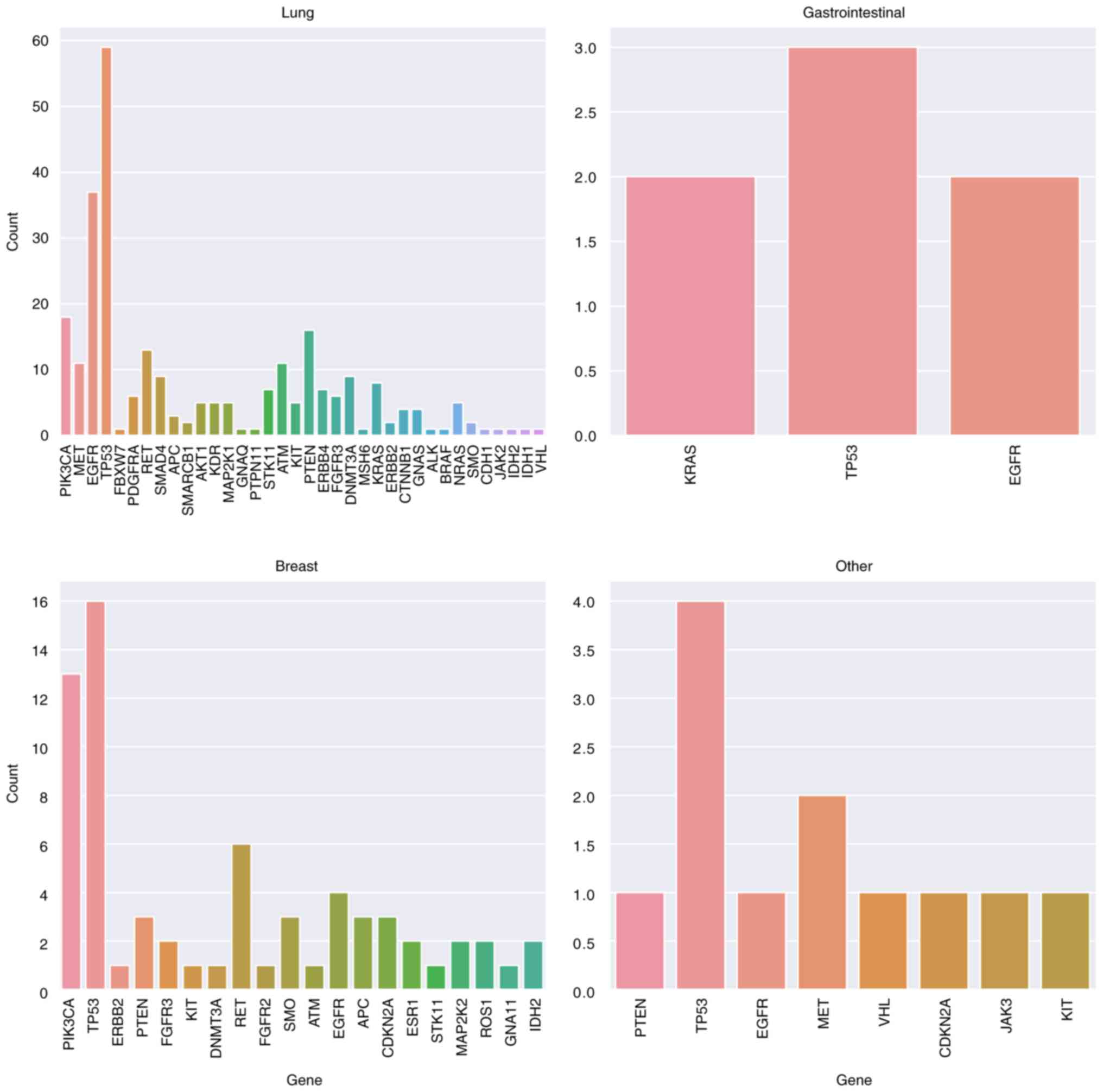
The most commonly mutated genes in each group were
as follows: Breast, TP53 (n=16, 23.5%), PIK3CA (n=13,
19.1%), RET (n=6, 8.8%) and EGFR (n=4, 5.9%);
gastrointestinal, TP53 (n=3, 42.8%), EGFR (n=2,
28.6%) and KRAS (n=2, 28.6%); lung, TP53 (n =59,
21.8%), EGFR (n=37, 13.7%), PIK3CA (n=18, 6.6%),
PTEN (n=6, 5.9%), RET (n=13, 4.8%), ATM (n=11,
4%) and MET (n=11, 4%) and other, TP53 (n=4, 33.3%)
and MET (n=2, 16.6%; Fig.
3).
The most common mutations in each group were as
follows: Breast, ‘PIK3CA, c.3140A>G, p.His1047Arg’,
‘RET, c.2324delinsGAC, p.Glu775Glyfs*6’ and ‘TP53,
c.217G>C, p.Val73Leu’; gastrointestinal, ‘KRAS,
c.194G>A, p.Ser65Asn’, ‘TP53, c.217G>C, p.Val73Leu’
and ‘EGFR, c.2155G>A, p.Gly719Ser’; lung, ‘PIK3CA,
c.1624G>A, p.Glu542Lys’, ‘EGFR, c.2235_2249del,
p.Glu746_Ala750del’, ‘DNMT3A, c.2656C>T, p.Gln886Ter’,
‘TP53, c.764T>C, p.Ile255Thr’, ‘RET,
c.2324delinsGAC, p.Glu775Glyfs*6’, ‘EGFR, c.2573T>G,
p.L858R’, ‘TP53, c.524G>A, p.Arg175His’, ‘RET,
c.1784A>G, p.Glu595Gly’, ‘EGFR, c.2369C>T,
p.Thr790Met’ and ‘PIK3CA, c.3140A>G, p.His1047Arg’ and
other, ‘MET, c.3380T>C, p.Val1127Ala’ and ‘TP53,
c.403T>C, p.Cys135Arg’ (Fig.
3).
In 117 (24.7%) patients, no responsible mutation was
identified. TP53, PIK3CA and RET gene
mutations were predominant in patients aged <50 years; in those
aged ≥50, TP53, EGFR, PIK3CA and PTEN
gene mutations were predominant. PIK3CA, PTEN and
RET variants showed a higher incidence in the breast and
lung groups (Fig. 3).
Discussion
TP53, which encodes a tumor suppressor
protein, was the most commonly mutated gene. According to the
present data, the most commonly mutated genes were TP53,
EGFR, PIK3CA, PTEN, RET, MET,
ATM, KRAS and DNMT3A. The most common mutation was
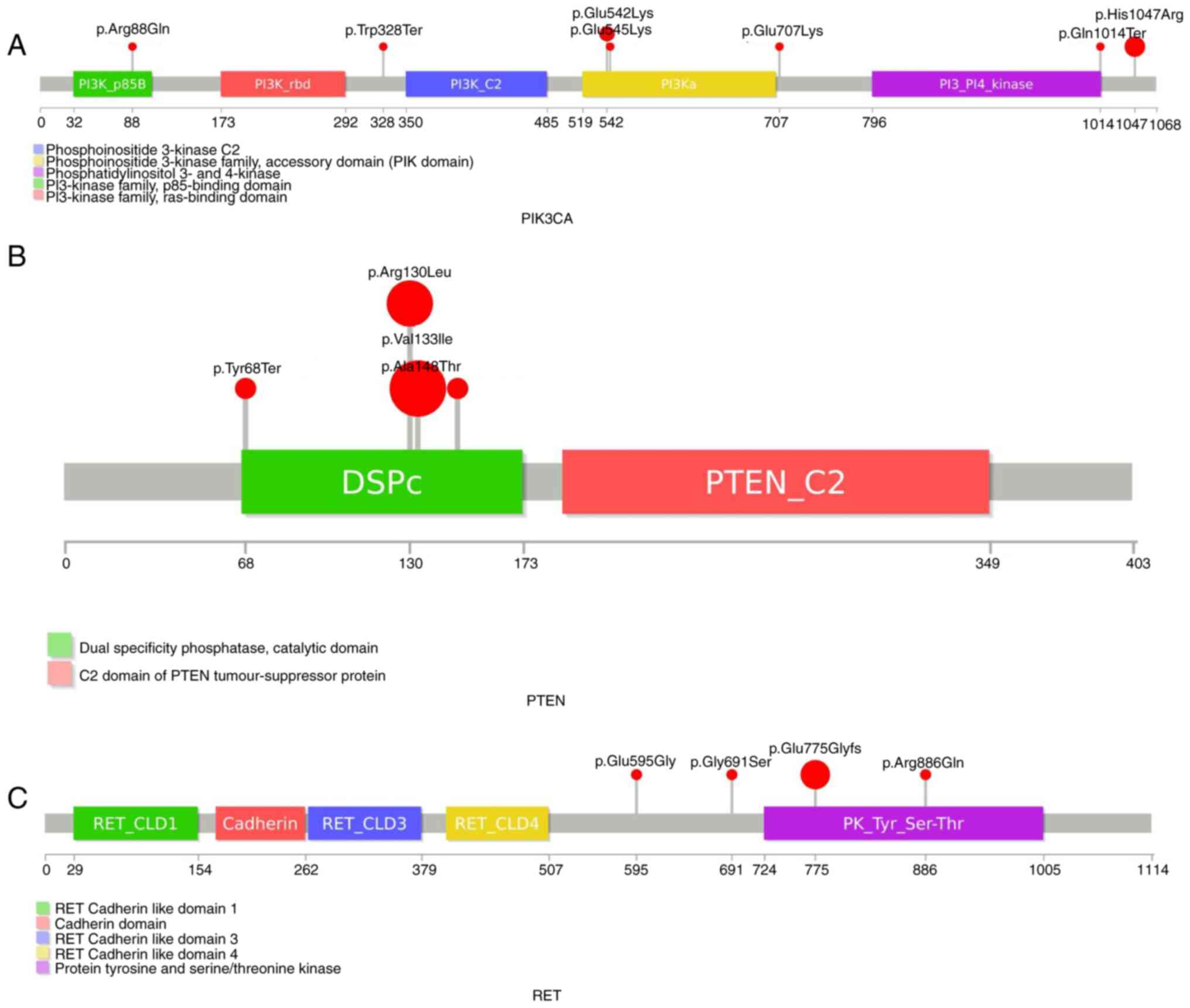
‘PIK3CA, c.3140A>G, p.His1047Arg.’ PIK3CA
mutations were primarily detected in domains associated with the
PI3K/AKT/mTOR pathway (Fig. 4).
Most patients were diagnosed with lung cancer and this distribution
varies in comparison with prior research (11,12).
‘PIK3CA, c.1624G>A, p.Glu542Lys’ and ‘PIK3CA,
c.1633G>A, p.Glu545Lys’ have been reported together, but in the
present study these mutations were detected separately and
‘PIK3CA, c.1624G>A, p.Glu542Lys’ was primarily
predominant in the lung group. ‘PIK3CA, c.3140A>G,
p.His1047Arg’ was primarily detected in the breast group. In one
patient, ‘PIK3CA, c.1624G>A, p.Glu542Lys’ was observed
with EGFR exon 19 deletion and T790M mutations. Nonsense
mutations were infrequent in PIK3CA. PIK3CA signaling
pathways serve a crucial role in replication, differentiation and
apoptosis. EGFR activation promotes tumor growth, invasion
and migration via the PIK3CA and mTOR pathways.
PIK3CA mutations may cause resistance to
EGFR-targeting therapies in patients with lung cancer
(20,21).
RET mutations were most frequent in the
breast (8.8%) and lung groups (4.8%). The ‘RET,
c.2324delinsGAC, p.Glu775Glyfs*6’ mutation was most common and most
of RET mutations were around the kinase domain (Fig. 4). PTEN is one of the most
commonly inactivated tumor suppressor genes in a wide variety of
cancer types. PTEN loss and the activation of PI3K/mTOR
signaling are associated with EGFR TKIs resistance in
patients with NSCLC (20,21). PTEN mutations were observed
in the breast (4.4%) and lung (6%) groups. ‘PTEN,
c.397G>A, p.Val133Ile’ was the most common mutation. Half of the
PTEN mutations were nonsense or frameshift and most
mutations were around the catalytic domain. Similarly, most
APC mutations were nonsense or frameshift. While most
germline APC mutations were missense, the majority of the
somatic APC mutations were nonsense or frameshift. This may
help to distinguish between germline and somatic mutations.
It has been reported that patients with EGFR
exon 19 deletions who receive long-term EGFR-TKI therapy show a
high prevalence of T790M mutations (22). In the present study, T790M mutation
was detected in two patients who also exhibited the exon 19
deletion. In one patient, the ‘ALK, c.3626delG,
p.Arg1209Glnfs*49’ mutation was detected and no ROS
mutations were detected. Another patient exhibited an ALK
L1198F mutation in addition to the C1156Y mutation. L1198F
substitution confers resistance to lorlatinib via steric
interference with drug binding. However, L1198F paradoxically
enhances binding to crizotinib, negating the effect of C1156Y and
re-sensitizing resistant cancer to crizotinib. The patient received
crizotinib, following which cancer-associated symptoms and liver
failure resolved (23). Further
studies are needed to detect and clarify the effects of these
neutralizing mutations.
Several studies have evaluated the concordance
between KRAS, BRAF and NRAS point mutations
detected by ctDNA and tumor-tissue analysis (24,25).
More mutations were detected by ctDNA analysis than by tumor
biopsy, reflecting the ability of liquid biopsy to reflect tumor
heterogeneity. Moreover, the shorter turnaround time required to
perform ctDNA analysis compared with tumor tissue analysis makes it
possible to start treatment earlier (24).
Familial cancer syndromes account for 5-10% of all
malignancies caused by inherited mutations; they raise the risk of
tumor development and are typically characterized by early-onset
cancer (26). In the present
study, patients with high variant fraction mutations in APC,
ATM, MLH1, MSH6, PTEN, PTPN11,
RB1, RET, STK11, TP53, TSC1 and
VHL underwent testing with a familial cancer panel. Even
though the variant fractions of the mutations were between 1 and
10%, there may be some risk associated with germline inheritance.
Family screening and genetic counseling are important for suspected
germline mutations, primarily when the variant fraction is between
50 and 100%.
The majority of patients presented with advanced
metastatic tumors from different cities around the country.
Treatment and survival information could not be collected for all
the patients and follow-up tests could not be performed. Only
mutations that can be targeted with treatment were reported and
discussed here. Despite these limitations, the liquid biopsy test
helped to identify the resistance mechanism in most cases (Figs. 2, 3 and 4).
The treatment of patients with lung and breast
cancer with PIK3CA, PTEN and RET mutations has
not yet been defined. In the present study, PIK3CA mutations
occurred in ~6.6% of patients with lung adenocarcinoma and 19% of
patients with breast cancer. It has been reported that
PIK3CA-positive patients have a worse prognosis (2). PI3K/AKT/mTOR pathway inhibitors may
be considered for patients with PIK3CA and PTEN
mutations. To the best of our knowledge, the present study is the
first to concentrate on PIK3CA, PTEN and RET
mutations in the context of breast and lung adenocarcinoma and
evaluate the genetic variability. The findings support the
potential of using gene therapy to target mutant PIK3CA,
PTEN and RET genes.
In conclusion, the findings of the present study
suggested that patients with solid tumors, particularly lung and
breast cancer, should undergo PIK3CA, PTEN and
RET sequencing to assess clinical characteristics and
prognosis. Greater understanding of the structure and mechanisms of
PIK3CA, PTEN and RET may help to inform more
clinically meaningful therapeutic approaches for patients with
cancer. Moreover, developments in assessing and researching novel
variants of known cancer genes will serve a key role in improving
individual cancer risk prediction, therapy and prognosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to restrictions of the
Ministry of Health of Turkey but are available from the
corresponding author on reasonable request.
Authors' contributions
IS designed the study, analyzed and interpreted the
results and wrote the manuscript. HS collected the data, analyzed
and interpreted the results and reviewed the manuscript. SA and OD
evaluated the patients and collected the clinical data. HBE and TB
collected the data and analyzed and interpreted the results. All
authors read and approved the final version of the manuscript. IS,
HS, HBE and TB confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved (approval no.
03/1072) by the Ethics Committee at the University of Health
Sciences, Dr. Abdurrahman Yurtaslan Ankara Oncology Training and
Research Hospital. Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
HS, ORCID no. 0000-0002-6087-5947; IS, ORCID no.
0000-0002-6050-816X.
References
|
1
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Song Z, Yu X and Zhang Y: Mutation and
prognostic analyses of PIK3CA in patients with completely
resected lung adenocarcinoma. Cancer Med. 5:2694–2700.
2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mattiuzzi C, Sanchis-Gomar F and Lippi G:
Concise update on colorectal cancer epidemiology. Ann Transl Med.
7(609)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Dafni U, Tsourti Z and Alatsathianos I:
Breast cancer statistics in the european union: Incidence and
survival across european countries. Breast Care. 14:344–353.
2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Franczak C, Filhine-Tressarieu P, Broséus
J, Gilson P, Merlin JL and Harlé A: Clinical interest of
circulating tumor DNA in oncology. Arch Med Res. 49:297–305.
2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Arneth B: Update on the types and usage of
liquid biopsies in the clinical setting: A systematic review. BMC
Cancer. 18(527)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lindeman NI, Cagle PT, Aisner DL, Arcila
ME, Beasley MB, Bernicker EH, Colasacco C, Dacic S, Hirsch FR, Kerr
K, et al: Updated molecular testing guideline for the selection of
lung cancer patients for treatment with targeted tyrosine kinase
inhibitors: Guideline from the college of American pathologists,
the international association for the study of lung cancer, and the
association for molecular pathology. Arch Pathol Lab Med.
142:321–346. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Von Bubnoff N: Liquid biopsy: Approaches
to dynamic genotyping in cancer. Oncol Res Treat. 40:409–416.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bronte G, Ulivi P, Verlicchi A, Cravero P,
Delmonte A and Crinò L: Targeting RET-rearranged non-small-cell
lung cancer: Future prospects. Lung Cancer (Auckl). 10:27–36.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Oxnard GR, Thress KS, Alden RS, Lawrance
R, Paweletz CP, Cantarini M, Yang JC, Barrett JC and Jänne PA:
Association between plasma genotyping and outcomes of treatment
with osimertinib (AZD9291) in advanced non-small-cell lung cancer.
J Clin Oncol. 34:3375–3382. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Liu S, Li S, Wang B, Liu W, Gagea M, Chen
H, Sohn J, Parinyanitikul N, Primeau T, Do KA, et al: Cooperative
effect of oncogenic MET and PIK3CA in an HGF-dominant
environment in breast cancer. Mol Cancer Ther. 18:399–412.
2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tan AC: Targeting the PI3K/Akt/mTOR
pathway in non-small cell lung cancer (NSCLC). Thorac Cancer.
11:511–518. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gkountakos A, Sartori G, Falcone I, Piro
G, Ciuffreda L, Carbone C, Tortora G, Scarpa A, Bria E, Milella M,
et al: PTEN in lung cancer: Dealing with the problem,
building on new knowledge and turning the game around. Cancers
(Basel). 11(1141)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jaiswal S, Fontanillas P, Flannick J,
Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N,
Chavez A, et al: Age-related clonal hematopoiesis associated with
adverse outcomes. N Engl J Med. 371:2488–2498. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Snyder MW, Kircher M, Hill AJ, Daza RM and
Shendure J: Cell-free DNA comprises an in vivo nucleosome footprint
that informs its tissues-of-origin. Cell. 164:57–68.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gupta S, Provenzale D, Llor X, Halverson
AL, Grady W, Chung DC, Haraldsdottir S, Markowitz AJ, Slavin TP Jr,
Hampel H, et al: NCCN guidelines insights: Genetic/familial
high-risk assessment: Colorectal, version 2.2019. J Natl Compr
Cancer Netw. 17:1032–1041. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Daly MB, Pilarski R, Yurgelun MB, Berry
MP, Buys SS, Dickson P, Domchek SM, Elkhanany A, Friedman S, Garber
JE, et al: NCCN guidelines insights: Genetic/familial high-risk
assessment: Breast, ovarian, and pancreatic, version 1.2020. J Natl
Compr Cancer Netw. 18:380–391. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li MM, Datto M, Duncavage EJ, Kulkarni S,
Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ,
Younes A and Nikiforova MN: Standards and guidelines for the
interpretation and reporting of sequence variants in cancer: A
joint consensus recommendation of the association for molecular
pathology, american society of clinical oncology, and college of
american pathologists. J Mol Diagnostics. 19:4–23. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jay JJ and Brouwer C: Lollipops in the
clinic: Information dense mutation plots for precision medicine.
PLoS One. 11(e0160519)2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Guo Y, Song J, Wang Y, Huang L, Sun L,
Zhao J, Zhang S, Jing W, Ma J and Han C: Concurrent genetic
alterations and other biomarkers predict treatment efficacy of
egfr-tkis in EGFR-mutant non-small cell lung cancer: A Review.
Front Oncol. 10(610923)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chang WJ, Sung JS, Lee SY, Kang EJ, Kwon
NJ, Kim HM, Shin SW, Choi JY, Choi YJ, Kim JW, et al: The clinical
significance of RAS, PIK3CA, and PTEN mutations in
non-small cell lung cancer using cell-free DNA. J Clin Med.
14(2642)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Matsuo N, Azuma K, Sakai K, Hattori S,
Kawahara A, Ishii H, Tokito T, Kinoshita T, Yamada K, Nishio K and
Hoshino T: Association of EGFR Exon 19 deletion and EGFR-TKI
treatment duration with frequency of T790M mutation in EGFR-mutant
lung cancer patients. Sci Rep. 6(36458)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shaw AT, Friboulet L, Leshchiner I, Gainor
JF, Bergqvist S, Brooun A, Burke BJ, Deng YL, Liu W, Dardaei L, et
al: Resensitization to crizotinib by the lorlatinib ALK resistance
mutation L1198F . N Engl J Med. 374:54–61. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Thierry AR, El Messaoudi S, Mollevi C,
Raoul JL, Guimbaud R, Pezet D, Artru P, Assenat E, Borg C,
Mathonnet M, et al: Clinical utility of circulating DNA analysis
for rapid detection of actionable mutations to select metastatic
colorectal patients for anti-EGFR treatment. Ann Oncol.
28:2149–2159. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Formica V, Lucchetti J, Doldo E, Riondino
S, Morelli C, Argirò R, Renzi N, Nitti D, Nardecchia A, Dell'Aquila
E, et al: Clinical utility of plasma KRAS, NRAS and
BRAF mutational analysis with real time PCR in metastatic
colorectal cancer patients-the importance of tissue/plasma
discordant cases. J Clin Med. 10(87)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Howlader N, Noone AM, Krapcho M, Garshell
J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z,
Mariotto A, Lewis DR, Chen HS, Feuer EJ and Cronin KA (eds): SEER
Cancer Statistics Review, 1975-2012, National Cancer Institute.
Bethesda, MD, based on November 2014 SEER data submission, posted
to the SEER web site, 2015. https://seer.cancer.gov/archive/csr/1975_2012.
Accessed November 18, 2015.
|