Introduction
Colorectal cancer (CRC) is one of the most common
types of cancer and the fourth leading cause of cancer-related
mortality worldwide (1). CRC may
be associated with pain and compromised quality of life (2,3).
Although this type of cancer is mainly treated by surgery,
radiotherapy is often used in the neoadjuvant setting to shrink the
size of large tumours prior to surgery (4). When surgery is not an option,
radiotherapy can be used in combination with chemotherapy as a
radical treatment approach (5).
External beam radiotherapy has been used for >120
years (6) and is currently
considered as a high-precision cancer treatment type that relies on
inverse treatment planning and a magnitude of instrumental
solutions that allow highly conformal dose delivery (7-10).
Despite high dose conformity and intensity, some tumours exhibit a
high degree of intrinsic radioresistance, leading to poor therapy
outcome (11). Factors responsible
for a high intrinsic radioresistance include tumour hypoxia and
poor reoxygenation during treatment, repopulation between
radiotherapy fractions, efficient repair of DNA damage and a high
degree of heterochromatin (11-13).
With respect to CRC, there is considerable interest
in devising biomarkers to predict pathological complete response
(pCR) of the tumour to a combination of chemoradiotherapy (CRT) and
surgery (14). Responsiveness of
the tumour to preoperative CRT is crucial, as it may predict
clinical outcome and even help decide whether post-CRT surgery is
advisable. Han et al (14)
analysed several clinical factors to determine their suitability as
pCR biomarkers, and found that tumour location and post-CRT levels
of the carcinoembryonic antigen (CEA) were promising predictive
factors. However, the specificity and sensitivity of these as
biomarkers were fairly low (area under the receiver operating
characteristic curve comparing sensitivity vs. specificity =
0.638); therefore, additional biomarkers are needed.
Cell response to radiation can be estimated by
analysing the release of microRNAs (miRNAs/miRs) to the surrounding
medium. miRNAs are small, single-stranded non-coding RNAs that
serve an important role in the regulation of gene expression.
miRNAs bind to the 3'-untranslated regions of target mRNAs and
control protein synthesis by increasing mRNA degradation or
inhibiting translation (15).
miRNAs released extracellularly are contained in extracellular
vesicles and circulate in the body (16). miRNA levels and serum composition
are promising biomarkers of cancer detection and progression in
various types of cancer, including CRC (17-20).
Selected miRNA levels in the serum of patients who
underwent radiotherapy for head and neck cancer were found to be
correlated with disease remission (21,22).
Promising results have also been reported for other types of
cancer, including breast or head and neck cancer (23,24).
We became interested in identifying miRNAs that
could be used to predict the response of colorectal tumours to
radiotherapy. Since miRNAs isolated from the serum of patients
undergoing treatment reflect the response of both cancer and normal
cells to radiation, it was decided to start the analyses with an
in vitro study using a CRC cell line. This approach aims to
identify cancer cell-specific changes in the extracellular miRNA
content, which, at a later step, can be validated in patient serum.
In the present study, the expression of extracellular miRNAs
secreted from irradiated HCT116 CRC cells was quantified by
microarray and correlated with the frequency of cytogenetic damage.
Cells were exposed to single-fraction X-irradiation to assess the
basal response of the cell; however, fractionated irradiation is
performed in clinical radiotherapy (e.g., 2 Gy/day x 25 fractions).
Delivering a small fraction of the total radiation dose allows time
for normal cells to repair themselves between treatments, thereby
reducing side effects.
Materials and methods
Cell preparation and culture
The human colorectal cell line HCT116 was purchased
from RIKEN BioResource Center. HCT116 cells were maintained in
RPMI-1640 medium (Thermo Fisher Scientific, Inc.) supplemented with
10% heat-inactivated foetal bovine serum (Japan Bioserum, Co.,
Ltd.) and 1% penicillin/streptomycin (Thermo Fisher Scientific,
Inc.) in a humidified atmosphere at 37˚C and 5% CO2.
Irradiation
X-ray irradiation (150 kVp, 20 mA with 0.5-mm
aluminium and 0.3-mm copper filters) was performed using an X-ray
generator (MBR-1520R-3; Hitachi Medical Co., Ltd.), with a distance
of 45 cm between the focus and target. The dose was monitored with
a thimble ionisation chamber placed next to the sample during
irradiation. The dose rate was 1 Gy/min.
Apoptosis assay
The apoptotic cells were examined via direct
immunofluorescence flow cytometry (Cell Lab QuantaTM Sc
MPL; Beckman Coulter Immunotech). Cells harvested using a single
pipette were washed twice using Annexin V Binding Buffer (cat. no.
422201) and immunostained following the manufacturer's instructions
(BioLegend, Inc.). Following addition of fluorochrome-labelled
protein, Annexin V-FITC (cat. no. 640906; BioLegend, Inc.), and
propidium iodide (PI; MilliporeSigma), fluorescence intensity was
quantified using flow cytometry (Cell Lab QuantaTM Sc MPL) and
Kaluza Analysis Software (ver.2.0; Beckman Coulter, Inc.).
Cytokinesis-block micronucleus (CBMN)
assay
The CBMN assay was conducted based of the criteria
described by Fenech (25). After
exposure of cells to X-irradiation, 15 µg/ml cytochalasin B (Cyt-B;
FujiFilm Wako Pure Chemical Corporation) was added to RPMI-1640
medium, and then the cells were harvested after 3 days, which is
when the maximum population of binucleated cells (BNCs) could be
observed. Harvesting was performed by fixing the cells with 99%
methanol at -20˚C for 3 min and staining with 1 µg/ml Hoechst 33342
(MilliporeSigma) for 30 min at room temperature. The evaluation was
conducted using a fluorescence/bright-field microscope at x400
magnification (IX71; Olympus Corporation). The BNC fraction was
scored per slide for at least 103 BNCs. At the same
time, the nuclear division index (NDI) was determined from the
frequency of >500 viable cells (‘N’ indicates the number of
viable cells counted per slide) with 1, 2, 3 or 4 nuclei (M1, M2,
M3 or M4, respectively) and calculated using the formula, NDI = [M1
+ (2 x M2) + (3 x M3) + (4 x M4)]/N by fluorescence microscopic
observation (25).
RNA extraction
Total extracellular RNAs released from irradiated
cells (four replicates performed in parallel) were extracted from
the cell culture supernatant using ISOGEN II (Nippon Gene Co.,
Ltd.), according to the manufacturer's instructions. The collection
of RNAs in the cell culture supernatant was performed on day 3
after cell irradiation. This time point was decided as maximum RNA
concentration in the cell culture supernatant and maximum
percentage of apoptotic cells (26). The concentration of extracted RNAs
was examined using Quant-iT RiboGreen RNA Reagent and Kit (Thermo
Fisher Scientific, Inc.) and Fluoroskan Ascent (Thermo Fisher
Scientific, Inc.). In addition, the peaks of small RNAs were
confirmed using Agilent 2100 Bioanalyzer (Agilent Technologies,
Inc.). The purity and concentration of extracellular RNAs were
assessed using the NanoDrop spectrophotometer (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). The RNA samples had
260/280 nm absorbance ratios of 1.8-2.0.
Microarray analysis
Cy3-labelled miRNA was synthesised from 100 ng total
RNA of X-irradiated and non-irradiated cell culture supernatant
using miRNA Complete Labelling Reagent and Hyb kit (Agilent
Technologies, Inc.). A SurePrint G3 human miRNA microarray slide
(8x60 K, Ver. 21.0) was hybridised with the Cy3-labelled miRNA in a
hybridisation solution prepared with a Gene Expression
Hybridisation kit (Agilent Technologies, Inc.), following the
manufacturer's instructions. Cy3 fluorescence signal images on the
slide were obtained by using a microarray scanner (Agilent SureScan
Microarray Scanner G 2600 D; Agilent Technologies, Inc.) and
processed using the Feature Extraction version 10.7 software
(Agilent Technologies, Inc.) based on the manufacturer's
instructions. In the present study, miRNA samples were collected
from cell culture supernatants and the extracellular miRNA content
was examined. The expression data were processed using GeneSpring
GX14.5 software (Agilent Technologies, Inc.) for normalisation to
percentile shift 90% of all values on the respective microarrays,
followed by normalisation of the median expression level of all
samples, since using an internal control, such as U6, is not
appropriate. The lower raw data signals (lower cut-off of 50) were
removed. In addition, the coefficient of variation was also set to
<50.0% to remove miRNAs with large variations between
experiments in advance. The average expression of each miRNA from
cells exposed to X-irradiation at 2-10 Gy was compared to control
within this normalised dataset (Fig.
S1). The results were visualised with the help of heat maps.
The microarray low data in the present study were uploaded onto the
Gene Expression Omnibus database (GSE184174; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE184174).
Statistical analysis
Statistical analysis was performed using OriginLab
software version 9.1 (OriginLab) and Excel 2013 (Microsoft
Corporation) with the add-in software Statcel 3 (OMS Publishing,
Inc.). The significant differences in miRNA expression analysis and
cell damage analysis were determined by Pearson's correlation test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Analysis of HCT116 cell damage from
X-irradiation
We previously reported that the clonogenic potential
of HCT116 cells exposed to high-dose-rate X-irradiation (1 Gy/min)
is reduced in a dose-dependent manner, and the number of apoptotic
cells also increases in a dose-dependent manner (26). The percentage of late apoptotic
cells (Annexin V+/PI+) and early apoptotic
cells (Annexin V+/PI-) by exposure to 2-10 Gy
X-irradiation was found to be 19-50% and 5-17%, respectively, and
it increased in a dose-dependent manner (Fig. 1A). Based on these responses, a CBMN
assay was performed using fluorescence microscopy to elucidate
whether these cells exhibit dose-dependent nuclear damage by
ionising radiation (Fig. 1B and
C). The NDI, which is a marker of
cell proliferation in cultures and is considered as a measure of
general cytotoxicity (Fig. 1B),
was significantly decreased in HCT116 cells between days 1 and 4
after exposure to 6 and 10 Gy compared with non-irradiated controls
(Fig. 1D). As regards the
frequency of micronuclei (MN), an increase (76.0±6.81 MN/103 BNCs)
was observed for doses up to 6 Gy; however, a decreased frequency
was observed at doses >6 Gy (Fig.
1E).
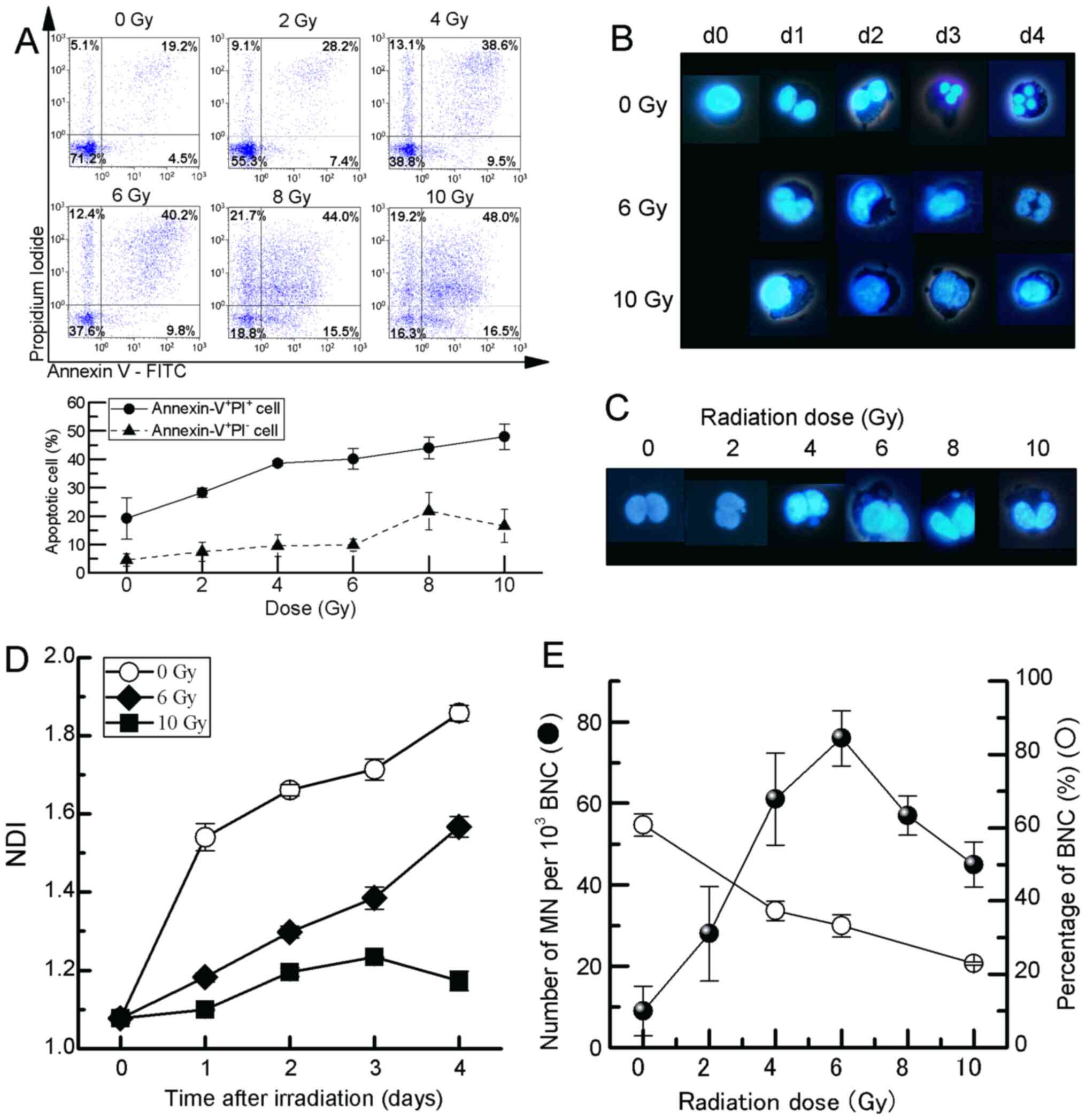 | Figure 1Analysis of CBMN assay in HCT116
cells exposed to X-irradiation. (A) Representative flow cytometry
images and percentage of apoptotic cells, (B) representative images
of nuclear division after irradiation, (C) BNCs with MN, (D)
response of NDI and (E) MN frequency. The cell behaviour in HCT116
cells exposed to 2-10 Gy X-irradiation was observed until day 4.
Apoptotic analysis and MN frequency in cells exposed to 2-10 Gy
X-irradiation was estimated using flow cytometry (Annexin
V+/PI- early apoptosis and Annexin
V+/PI+ late apoptosis) and the CBMN assay,
respectively. The cells were cultured with Cyt-B, and MN and NDI
were scored at 72 h after exposure to X-irradiation. The values are
the mean ± SE of four separate experiments. CBMN, cytokinesis-block
micronucleus; MN, micronuclei; NDI, nuclear division index; BNC,
binucleated cell; Cyt-B, cytochalasin B. |
Extracellular miRNA expression
following exposure to X-irradiation
To determine the miRNAs released from HCT116 cells
X-irradiated with 2-10 Gy, the cell culture supernatants were
collected, and miRNAs were isolated at day 3 when the maximum ratio
of apoptotic cells and MN frequency were reached (26,27).
These samples were applied to 8x60 K format miRNA microarrays, and
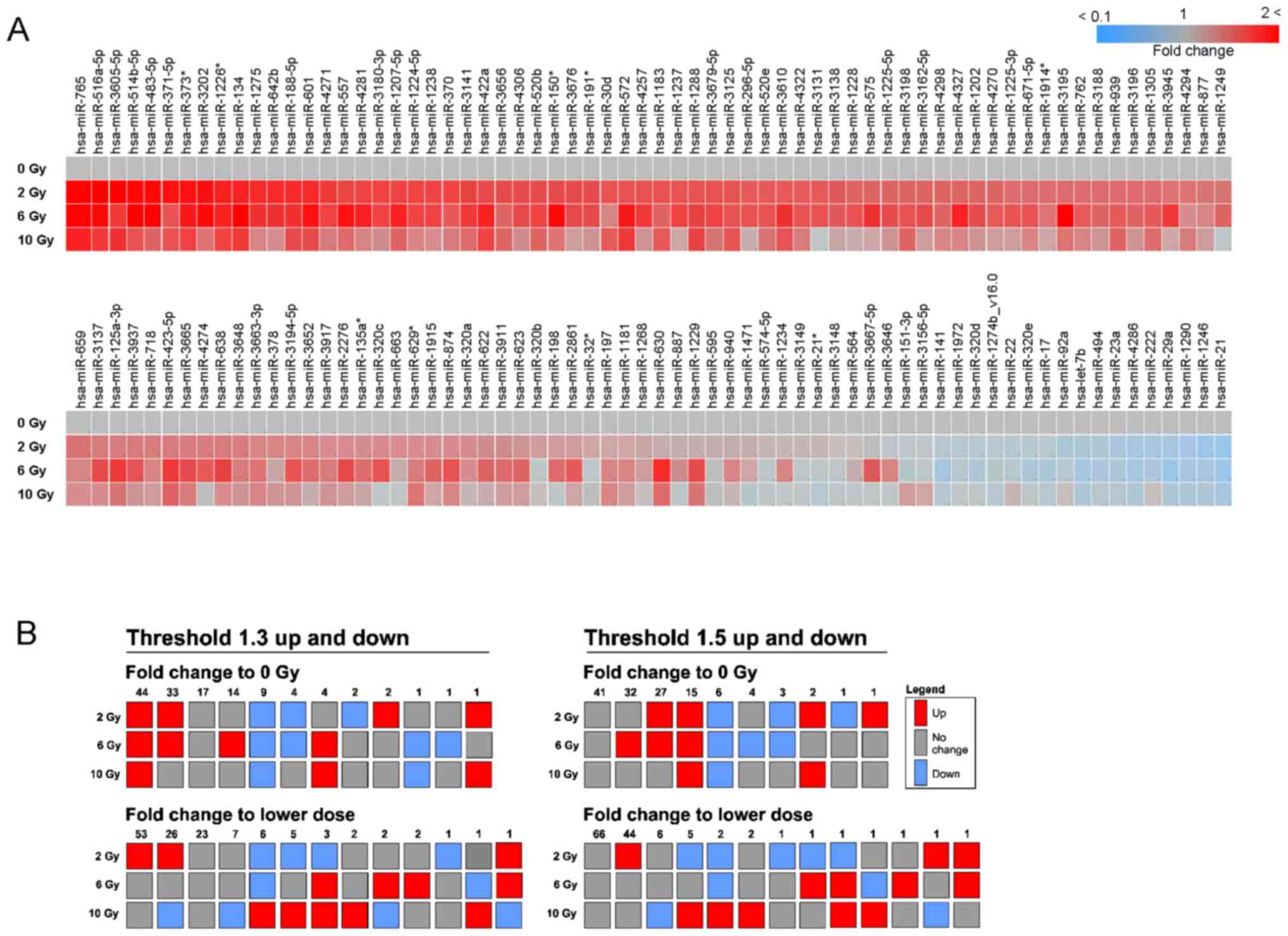
132 human miRbase-annotated miRNAs were detected (Table SI, Fig. 2A). To identify miRNAs responding in
a radiation dose-dependent manner, the miRNAs up- and downregulated
by 1.3- or 1.5-fold were considered, as the number of detectable
miRNAs was too small (1 or 0) at an up-/downregulation threshold of
2-fold. Following exposure to 2-10 Gy, 44 and 15 miRNAs were
upregulated by 1.3- and 1.5-fold, respectively, vs. the 0 Gy
control group. In addition, 9 and 6 miRNAs were downregulated vs.
the 0 Gy control group. On the other hand, no miRNAs were stepwise
up- or downregulated by 1.3- and 1.5-fold, respectively, compared
with the adjacent lower dose in any of the combinations (2 vs. 0, 6
vs. 2 or 10 vs. 6 Gy). There were still some miRNAs responding in
accordance with the 6-Gy peak seen for the MN frequency. When
comparing to the adjacent lower dose, patterns of
up-up-down/up-up-no change and down-down-up/down-down-no change
were found for 1 (miR-3195: 1.3- and 1.5-fold upregulation), 6
(miR-17, miR-29a, miR-98, miR-320d, miR-320e and miR-4286: 1.3-fold
downregulation) and 2 (miR-29a and miR-98: 1.5-fold downregulation)
miRNAs, respectively. The absolute level of miRNAs may also be a
relevant factor allowing for detection in the serum, and miR-572,
miR-939 and miR-3610 were the most highly expressed miRNAs on
average (Table SI).
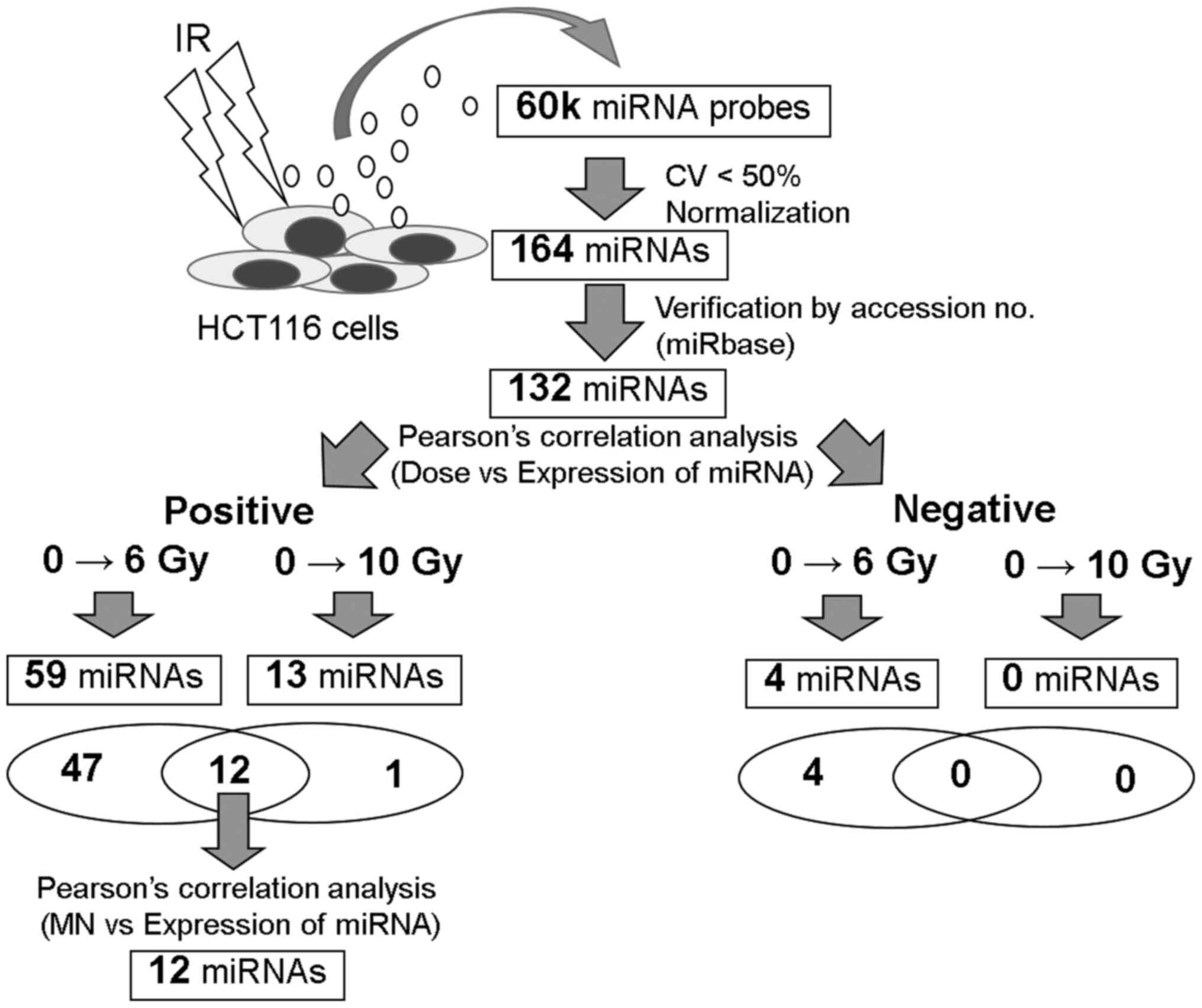
To elucidate the statistical correlation between
radiation doses and miRNA expression, the 132 miRNA dataset was
used, which was created based on the criteria described in
Materials and methods. Using samples between 0 and 6 Gy, 59 miRNAs
exhibiting a positive correlation and 4 miRNAs exhibiting a
negative correlation were identified (Fig. 3, Table SII). In addition, for samples
between 0 and 10 Gy, 13 miRNAs exhibiting a positive correlation
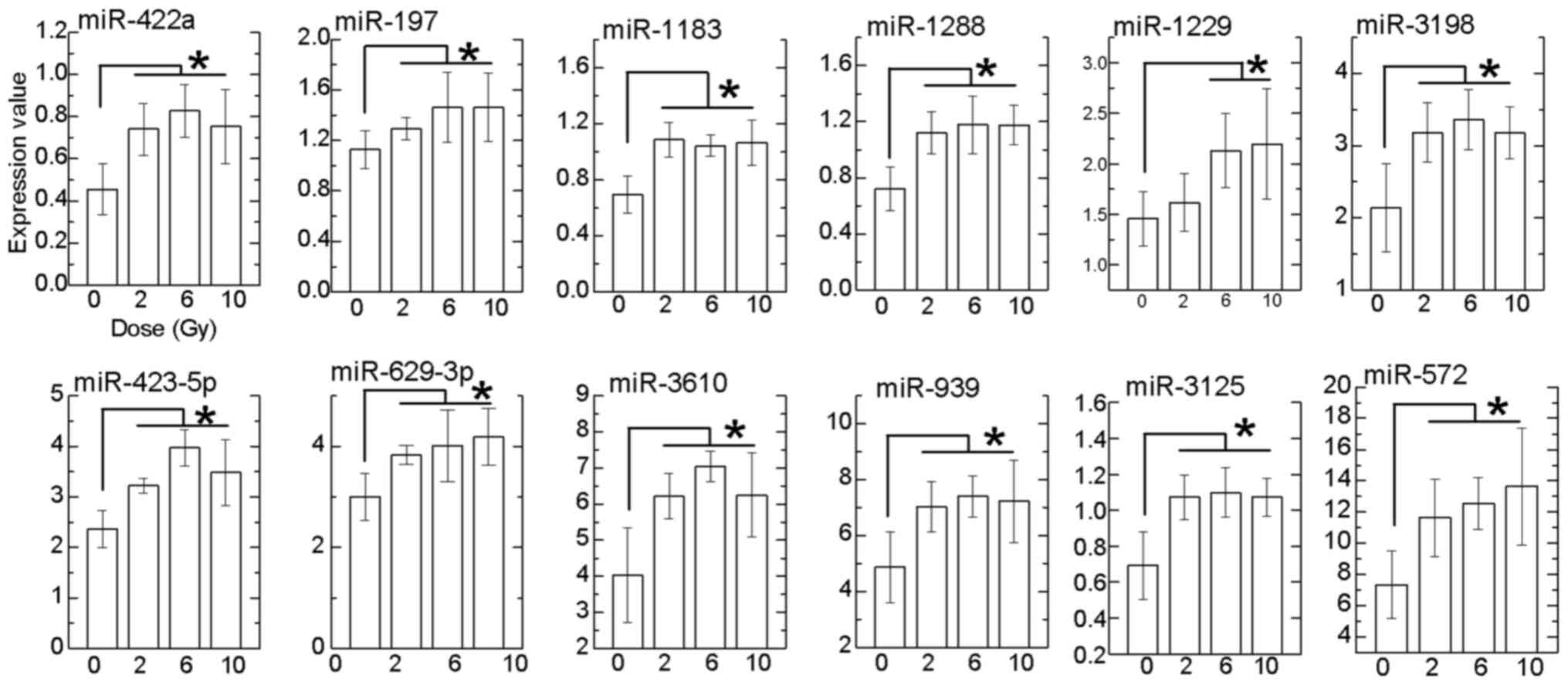
were identified. Of these, 12 miRNAs exhibited significantly
upregulated expression at doses between 6 and 10 Gy compared with 0
Gy (Fig. 4). It is known that the
maximum limiting dose of MN frequency in blood cells is 3-4 Gy
(25). In the present study on HCT
116 cells, the maximum limiting dose was up to 6 Gy (Fig. 1E). To predict the cellular damage
and induction of apoptosis by the expression of extracellular
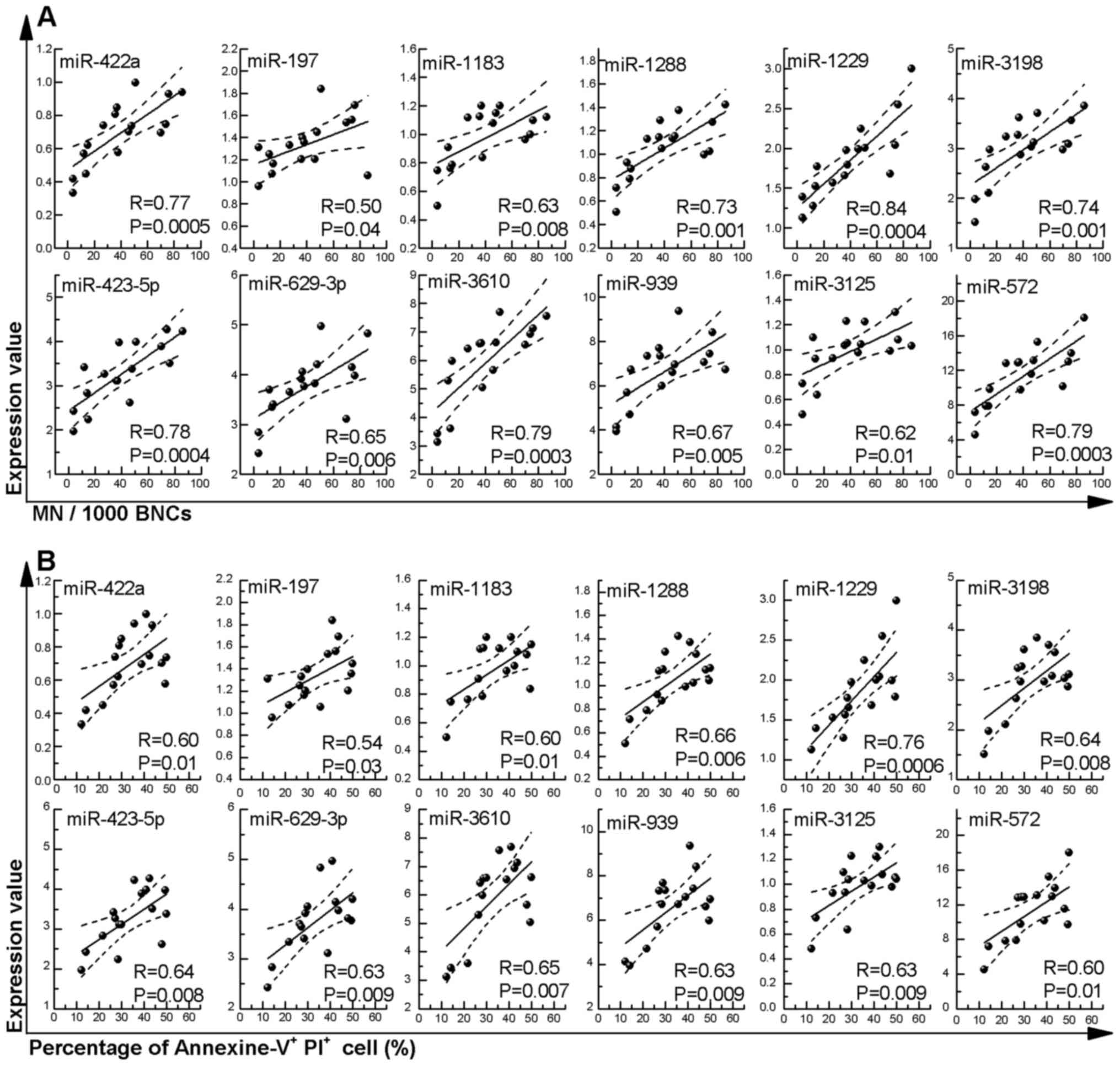
miRNAs, we focused on the relationship between the 12
dose-dependently upregulated miRNAs and MN frequency (Fig. 5A), and the percentage of Annexin
V+/PI+ cells (Fig. 5B). A significantly positive
correlation of all miRNAs (miR-422a, R=0.77; miR-197, R=0.50;
miR-1183, R=0.63; miR-1288, R=0.73; miR-1229, R=0.84; miR-3198,
R=0.74; miR-423-5p, R=0.78; miR-629-3p, R=0.65; miR-3610, R=0.79;
miR-939, R=0.67; miR-3125, R=0.62; and miR-572, R=0.79) was
observed between the expression level and MN frequency (Fig. 5A). In addition, a significantly
positive correlation of all miRNAs (miR-422a, R=0.60; miR-197,
R=0.54; miR-1183, R=0.60; miR-1288, R=0.66; miR-1229, R=0.76;
miR-3198, R=0.76; miR-423-5p, R=0.64; miR-629-3p, R=0.63; miR-3610,
R=0.65; miR-939, R=0.63; miR-3125, R=0.63; and miR-572, R=0.60) was
also observed with the percentage of Annexin
V+/PI+ cells (Fig. 5B).
Discussion
The aim of the present study was to quantify the
expression of extracellular miRNAs released from HCT116 CRC cells
exposed to high-dose single-fraction ionising radiation to identify
predictive extracellular markers of radiotherapeutic CRC cell
damage.
The dose-dependent radiation response at the level
of cell nuclear damage, MN (Fig.
1), was in line with the previously reported reduced clonogenic
potency for doses up to 6 Gy (26). This increased MN frequency
indicates increased number of apoptotic cells. At doses >6 Gy,
increased cell death or cell division arrest most likely
contributed to the lower frequency of MN, since a larger proportion
of cells with severe damage may have undergone apoptosis at the
72-h time point following exposure to X-irradiation.
In radiation biology, the CBMN assay is a radiation
biodosimetry tool that uses peripheral blood lymphocytes and has an
excellent dose dependency (28),
contributing to dose estimation within the range of 2-6 Gy in cases
of accidental radiation exposure (29). Clinically, 50 Gy/25 fractions or 25
Gy/5 fractions (30), which
corresponds to 2-5 Gy/fraction, are delivered to the target region
for CRC. Therefore, the exposure conditions of the present study
are in the clinical range for a single fraction.
It is extremely difficult to analyse the target
tissue cells located in deep regions using the CBMN assay in
patients undergoing radiotherapy. However, it is possible to use
body fluids to detect radiation response markers. A total of 60
specific miRNAs that were released extracellularly and their
increased or decreased expression after exposure to X-radiation
correlated with dose were identified (Fig. 4). The 12 selected miRNAs included
those positively correlated to dose when analysing both 0-6 and
0-10 Gy, which shows that there was a lower expression and MN
frequency at 10 Gy compared with those at 6 Gy. According to the
American Society for Radiation Oncology and the Japanese Society
for Radiation Oncology (31,32),
the clinical fractionation scheme for CRC tissue is delivering ~50
Gy as a total dose in 25 fractions (2 Gy/day). Therefore, miRNA
expression in body fluids, such as the serum, has a potential as a
monitoring tool within this dose range, whether or not cancer cells
are damaged.
In research on biomarkers from body fluids, the
peripheral blood, including the metabolites from cancer tissue, has
been evaluated for diagnosis and prognosis prediction. In CRC, CEA
and carbohydrate antigen 19-9 antigens are considered as
representative markers in cancer screening tests (33). Cancer researchers have recently
become interested in detailed screening of proteins, DNA and RNA in
body fluids for clinical applications to further improve the
accuracy of cancer detection (34-37).
In recent years, miRNAs have been reported to be highly promising
as cancer biomarkers (38). miRNAs
regulate various biological processes in cells, and their
expression fluctuates with the DNA damage response to radiation
(39). Thus, extracellular miRNAs
in patients receiving radiotherapy may reflect a radiation-specific
response and may be useful for the determination of radiation
dose.
Within the data presented herein, it was observed
that 8 of 12 miRNAs (miR-422a, miR-197, miR-1183, miR-1288,
miR-1229, miR-423-5p, miR-939 and miR-572) have a function in CRC.
Intracellular expression of miR-422a, miR-572 and miR-939 serve a
role in tumour suppression; miR-422a targets AKT1 and MAPK1;
miR-572 targets the MOP-1 pathway activated by STAT3 and miR-939
targets NF-κB (40-44).
Of those, miR-572 and miR-939 were also among the three most highly
expressed miRNAs, with the highest chance of detection in the
blood. miR-197, miR-1183 and miR-1288 are associated with CRC
response (upregulation) to chemotherapy and/or radiotherapy
(45-47).
miR-423-5p and miR-1229 were reported as diagnostic circulating
biomarkers of CRC (48,49). However, the remaining miRNAs
(miR-3198, miR-629-3p, miR-3610 and miR-3125) have no previous
reports in the field of CRC. Therefore, the phenomenon seen in this
study is that the dose-dependent intracellular response to the
ionising reaction affected the mRNA release system, including
extracellular vesicles. Therefore, these 12 miRNAs may serve as
potential biomarkers of radiation response in CRC cells.
Radiotherapy causes activation of several processes
in the tumour microenvironment, such as inflammation, cycling
hypoxia, immunomodulation, revascularisation, and extracellular
matrix remodelling coordinated by cancer-associated fibroblasts and
fibrosis (50). The release of
miRNAs from tumour cells may contribute to these effects, and it is
possible that metabolite control differs among patients with
cancer. Of note, there is still a need for improved estimates of
which patients are responding well or less well to treatment;
therefore, these miRNAs may serve as markers with the purpose of
optimising target exposure to the radiation dose during
radiotherapy. Further detailed in vitro and in vivo
studies are needed to validate these data and to analyse if the
expression pattern of these specific miRNAs also contributes to the
radiosensitivity of CRC. A limitation of the present study was the
lack of investigation of miRNA expression in clinical samples
(e.g., colon cancer vs. normal adjacent non-cancerous samples). To
verify the evidence on the 12 released extracellular miRNAs, the
reproducibility in animal models and/or clinical specimens, and the
response of CRC cell exposed to fractionated irradiation, should be
further investigated in the future. In addition, we plan to test
the role of these 12 miRNAs in endocytosis by CRC cells exposed to
high-dose rate ionising radiation.
In conclusion, the findings of the present study
suggest that specific extracellular miRNAs have the potential to
serve as cell injury markers induced by single high doses of
ionising radiation in CRC cells.
Supplementary Material
Three-dimensional scatter plot showing
the association among radiation dose, micronuclei frequency and
expression of miRNAs. The 12 miRNAs of focus in the present study
were plotted. miRNA/miR, microRNA.
The 132 detected miRNAs list using
microarray analysis with the annotation of human miRbase.
The correlation analysis between
radiation dose and miRNA expression in 132 detected miRNAs.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by KAKENHI,
Grants-in-Aid for Scientific Research, Fund for the Promotion of
Joint International Research (Fostering Joint International
Research) (project no. 17KK0181) and Grant-in-Aid for challenging
Exploratory Research (project no. 19K22731). The funders had no
role in study design, data collection and analysis, decision to
publish or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SM, TU, YMa and LL designed the study, prepared the
manuscript draft and substantively participated in revising the
manuscript. SM, TU, YMo and MC contributed by analysing the
biological data. TU and AW contributed by normalizing by the
microarray data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Araghi M, Soerjomataram I, Jenkins M,
Brierley J, Morris E, Bray F and Arnold M: Global trends in
colorectal cancer mortality: Projections to the year 2035. Int J
Cancer. 144:2992–3000. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
van Gijn W, Marijnen CA, Nagtegaal ID,
Kranenbarg EM, Putter H, Wiggers T, Rutten HJ, Påhlman L, Glimelius
B and van de Velde CJ: Dutch Colorectal Cancer Group. Preoperative
radiotherapy combined with total mesorectal excision for resectable
rectal cancer: 12-year follow-up of the multicentre, randomised
controlled TME trial. Lancet Oncol. 12:575–582. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kye BH and Cho HM: Overview of radiation
therapy for treating rectal cancer. Ann Coloproctol. 30:165–174.
2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
No authors listed. News of Science.
Science. 125:18–22. 1957.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Brock KK, McShan DL, Ten Haken RK,
Hollister SJ, Dawson LA and Balter JM: Inclusion of organ
deformation in dose calculations. Med Phys. 30:290–295.
2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
De Ruysscher D, Belderbos J, Reymen B, van
Elmpt W, van Baardwijk A, Wanders R, Hoebers F, Vooijs M, Ollers M
and Lambin P: State of the art radiation therapy for lung cancer
2012: A glimpse of the future. Clin Lung Cancer. 14:89–95.
2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Emami B, Lyman J, Brown A, Coia L, Goitein
M, Munzenrider JE, Shank B, Solin LJ and Wesson M: Tolerance of
normal tissue to therapeutic irradiation. Int J Radiat Oncol Biol
Phys. 21:109–122. 1991.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Marks LB, Yorke ED, Jackson A, Ten Haken
RK, Constine LS, Eisbruch A, Bentzen SM, Nam J and Deasy JO: Use of
normal tissue complication probability models in the clinic. Int J
Radiat Oncol Biol Phys. 76 (Suppl):S10–S19. 2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Baumann M, Krause M, Overgaard J, Debus J,
Bentzen SM, Daartz J, Richter C, Zips D and Bortfeld T: Radiation
oncology in the era of precision medicine. Nat Rev Cancer.
16:234–249. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tang L, Wei F, Wu Y, He Y, Shi L, Xiong F,
Gong Z, Guo C, Li X, Deng H, et al: Role of metabolism in cancer
cell radioresistance and radiosensitization methods. J Exp Clin
Cancer Res. 37(87)2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen T, Zhang Y, Guo WH, Meng MB, Mo XM
and Lu Y: Effects of heterochromatin in colorectal cancer stem
cells on radiosensitivity. Chin J Cancer. 29:270–276.
2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Han YD, Kim WR, Park SW, Cho MS, Hur H,
Min BS, Baik SH, Lee KY and Kim NK: Predictors of pathologic
complete response in rectal cancer patients undergoing total
mesorectal excision after preoperative chemoradiation. Medicine
(Baltimore). 94(e1971)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Czochor JR and Glazer PM: microRNAs in
cancer cell response to ionizing radiation. Antioxid Redox Signal.
21:293–312. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bhat SA, Majid S and Hassan T: MicroRNAs
and its emerging role as breast cancer diagnostic marker - A
review. Adv Biomarker Sci Technol. 1:1–8. 2019.
|
|
19
|
Yu H, Guan Z, Cuk K, Zhang Y and Brenner
H: Circulating microRNA biomarkers for lung cancer detection in
East Asian populations. Cancers (Basel). 11(415)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Masuda T, Hayashi N, Kuroda Y, Ito S,
Eguchi H and Mimori K: MicroRNAs as biomarkers in colorectal
cancer. Cancers (Basel). 9(124)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen L, Wen Y, Zhang J, Sun W, Lui VWY,
Wei Y, Chen F and Wen W: Prediction of radiotherapy response with a
5-microRNA signature-based nomogram in head and neck squamous cell
carcinoma. Cancer Med. 7:726–735. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pasi F, Corbella F, Baio A, Capelli E, De
Silvestri A, Tinelli C and Nano R: Radiation-induced circulating
miRNA expression in blood of head and neck cancer patients. Radiat
Environ Biophys. 59:237–244. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Halimi M, Parsian H, Mohsen Asghari S,
Sariri R, Moslemi D and Yeganeh F: MicroRNAs: Are they indicators
for prediction of response to radiotherapy in breast cancer? Irn J
Med Hypotheses Ideas. 7:59–64. 2013.
|
|
24
|
Moertl S, Mutschelknaus L, Heider T and
Atkinson MJ: MicroRNAs as novel elements in personalized
radiotherapy. Transl Cancer Res. 5:S1262–S1269. 2016.
|
|
25
|
Fenech M: Cytokinesis-block micronucleus
cytome assay. Nat Protoc. 2:1084–1104. 2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ueno T, Monzen S, Chiba M, Morino Y and
Hosokawa Y: Screening for biological marker of dose-optimization in
cancer radiotherapy. Nippon Hoshasen Gijutsu Gakkai Zasshi.
74:459–464. 2018.PubMed/NCBI View Article : Google Scholar : (In Japanese).
|
|
27
|
Ueno T, Monzen S, Chiba M and Hosokawa Y:
Basic investigation to optimize radiation dose using biological
evaluation in radiotherapy. Cytometry Res. 28:7–11. 2018.
|
|
28
|
International Atomic Energy Agency:
Cytogenetic Dosimetry: Applications in Preparedness for and
Response to Radiation Emergencies. EPR-Biodosimetry 2011. Vienna,
2011.
|
|
29
|
Liu Q, Cao J, Wang ZQ, Bai YS, Lü YM,
Huang QL, Zhao WZ, Li J, Jiang LP, Tang WS, et al: Dose estimation
by chromosome aberration analysis and micronucleus assays in
victims accidentally exposed to (60)Co radiation. Br J Radiol.
82:1027–1032. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Rodrigues G, Velker V and Best L:
Radiation Oncology Primer and Review, Essential Concepts and
Protocols. 1st edition. Demos MEDICAL, pp1-376, 2013.
|
|
31
|
Japanese Society for Radiation Oncology:
JASTRO Guidelines 2020 for Radiotherapy Treatment Planning. Fifth
edition. Kanehara & Co. Ltd., 2020. (In Japanese).
|
|
32
|
Wo JY, Anker CJ, Ashman JB, Bhadkamkar NA,
Bradfield L, Chang DT, Dorth J, Garcia-Aguilar J, Goff D, Jacqmin
D, et al: Radiation therapy for rectal cancer: Executive summary of
an ASTRO clinical practice guideline. Pract Radiat Oncol. 11:13–25.
2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lech G, Słotwiński R, Słodkowski M and
Krasnodębski IW: Colorectal cancer tumour markers and biomarkers:
Recent therapeutic advances. World J Gastroenterol. 22:1745–1755.
2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Babel I, Barderas R, Díaz-Uriarte R,
Martínez-Torrecuadrada JL, Sánchez-Carbayo M and Casal JI:
Identification of tumor-associated autoantigens for the diagnosis
of colorectal cancer in serum using high density protein
microarrays. Mol Cell Proteomics. 8:2382–2395. 2009.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Oh T, Kim N, Moon Y, Kim MS, Hoehn BD,
Park CH, Kim TS, Kim NK, Chung HC and An S: Genome-wide
identification and validation of a novel methylation biomarker,
SDC2, for blood-based detection of colorectal cancer. J Mol Diagn.
15:498–507. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Han M, Liew CT, Zhang HW, Chao S, Zheng R,
Yip KT, Song ZY, Li HM, Geng XP, Zhu LX, et al: Novel blood-based,
five-gene biomarker set for the detection of colorectal cancer.
Clin Cancer Res. 14:455–460. 2008.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X,
Yu L, Wang L, Wang J, Wu Y, et al: A plasma microRNA panel for
early detection of colorectal cancer. Int J Cancer. 136:152–161.
2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Faltejskova P, Svoboda M, Srutova K,
Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K,
Kiss I, et al: Identification and functional screening of microRNAs
highly deregulated in colorectal cancer. J Cell Mol Med.
16:2655–2666. 2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wei WT, Nian XX, Wang SY, Jiao HL, Wang
YX, Xiao ZY, Yang RW, Ding YQ, Ye YP and Liao WT: miR-422a inhibits
cell proliferation in colorectal cancer by targeting AKT1 and
MAPK1. Cancer Cell Int. 17(91)2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cui C, Yu J, Huang S, Zhu H and Huang Z:
Transcriptional regulation of gene expression by microRNAs as
endogenous decoys of transcription factors. Cell Physiol Biochem.
33:1698–1714. 2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Cui C, Zhai D, Cai L, Duan Q, Xie L and Yu
J: Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis
via counteracting miR-939-mediated transcriptional repression of
Bcl-xL. Cancer Res Treat. 50:992–1008. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wang N, He X, Zhou R, Jia G and Qiao Q:
STAT3 induces colorectal carcinoma progression through a novel
miR-572-MOAP-1 pathway. OncoTargets Ther. 11:3475–3484.
2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang M, Hu H, Wang Y, Huang Q, Huang R,
Chen Y, Ma T, Qiao T, Zhang Q, Wu H, et al: Long non-coding RNA
TUG1 mediates 5-fluorouracil resistance by acting as a ceRNA of
miR-197-3p in colorectal cancer. J Cancer. 10:4603–4613.
2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Della Vittoria Scarpati G, Falcetta F,
Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK,
D'Incalci M, De Placido S and Pepe S: A specific miRNA signature
correlates with complete pathological response to neoadjuvant
chemoradiotherapy in locally advanced rectal cancer. Int J Radiat
Oncol Biol Phys. 83:1113–1119. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Gopalan V, Pillai S, Ebrahimi F,
Salajegheh A, Lam TC, Le TK, Langsford N, Ho YH, Smith RA and Lam
AK: Regulation of microRNA-1288 in colorectal cancer: Altered
expression and its clinicopathological significance. Mol Carcinog.
53 (Suppl 1):E36–E44. 2014.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lu X and Lu J: The significance of
detection of serum miR-423-5p and miR-484 for diagnosis of
colorectal cancer. Clin Lab. 61:187–190. 2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ogata-Kawata H, Izumiya M, Kurioka D,
Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H,
et al: Circulating exosomal microRNAs as biomarkers of colon
cancer. PLOS ONE. 9(e92921)2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Barker HE, Paget JT, Khan AA and
Harrington KJ: The tumour microenvironment after radiotherapy:
mechanisms of resistance and recurrence. Nat Rev Cancer.
15:409–425. 2015.PubMed/NCBI View Article : Google Scholar
|