1. Introduction
Epithelial ovarian cancer (EOC) is the 5th leading
cause of cancer-associated death in females worldwide. In >70%
of cases, it is diagnosed at an advanced stage [International
Federation of Gynecology and Obstetrics (FIGO) stage III or IV], as
the symptoms are frequently nonspecific (1,2).
Standard initial treatment is cytoreductive surgery followed by
carboplatin and paclitaxel with bevacizumab as the first-line
treatment of ‘high-risk’ patients to improve progression-free
survival (PFS). Although 80% respond well to initial treatment,
>70% develop recurrent disease within the first two years and
eventually become chemoresistant (2).
Treatment with bevacizumab [a monoclonal antibody
that binds to vascular endothelial growth factor (VEGF inhibitor)]
has improved the PFS (currently 3.3-4.0 months) in EOC, whereas no
significant difference in overall survival was reported (3-11).
Unfortunately, no biomarker predictive of response to bevacizumab
is currently available.
Poly(adenosine diphosphate ribose) polymerase (PARP)
inhibitors have changed the clinical management of EOC by targeting
the homologous recombination repair pathway. PARP inhibitors are
successfully implemented in treating high-grade serous ovarian
carcinoma (HGSC) by leveraging inherent defects in DNA repair
mechanisms presented in ~50% of HGSC (12). Several trials have confirmed the
positive prognostic impact of maintenance treatment in
platinum-sensitive females harboring BRCA1/2 pathogenic mutations
(13-19).
Despite these novel treatment strategies, the leading cause of
death in HGSC remains chemoresistance. Predicting response to
platinum-based chemotherapy in the primary and the recurrent
setting is not possible yet (20,21).
In this regard, organoids, a 3-dimensional (3D) cell
culture derived from stem cells, provide a novel in vitro
platform to predict drug response. Organoids are in vitro 3D
cultures grown from stem cells and consisting of organ-specific
cell types (22-24).
Following this definition, the first organoids were described in
2009 in murine intestinal cells, further developed for other
organs, and eventually translated into human cells (25). Patients' tumor biopsies are
dissociated into fragments and cells, embedded in a 3D
extracellular matrix scaffold (such as Matrigel) and cultured in a
cocktail of growth and signaling factors, which must be defined and
optimized for each cancer type (26). Organoids have evolved through the
last decade and now closely reflect primary tissue's biology and
pathology, enabling their use in a broad range of applications,
such as drug development and drug screening (22,27-29).
Furthermore, organoids may be cultured from tumor
tissue and expanded within one month. Multiple organoids may be
generated from different tumor areas to mimic tumor heterogeneity
(30,31). Of note, tumor organoids capture
inter- and intratumor heterogeneity (29). The establishment of tumor-derived
organoids from various cancer types such as colon, pancreatic,
gastric, prostate, breast, esophageal, bladder and endometrial
cancers has been reported (30-37).
The rate of successful growth of organoid cultures varies by cancer
type and is highest in colon cancer, reaching 65-70% (28,38-42).
Furthermore, organoids may be used for drug
screening. For instance, drug screening of a library containing
>50 drugs was performed with 19 colorectal cancer (CRC)
organoids, demonstrating a correlation between drug sensitivity and
genetic aberrations in patient-derived organoids (PDOs) (43). Yan et al (32) screened 37 anticancer drugs in nine
gastric cancer organoids, identifying possible responses toward
novel targets. Recent studies have indicated that in-vitro
drug screening using PDOs predict patient response to chemotherapy
and targeted drugs in patients with CRC (28,38).
In EOC research, organoid cultures have been steadily emerging
throughout the last years. Regular fallopian tube and ovarian
surface epithelium-derived organoids that capture the genomic
features of the respective tissues are established. They may offer
a platform for studying cancer initiation from these potential
origins (44-46).
In addition to the healthy organoid lines, both short- and
long-term organoid cultures from EOC have been established from
several histologic subtypes (Table
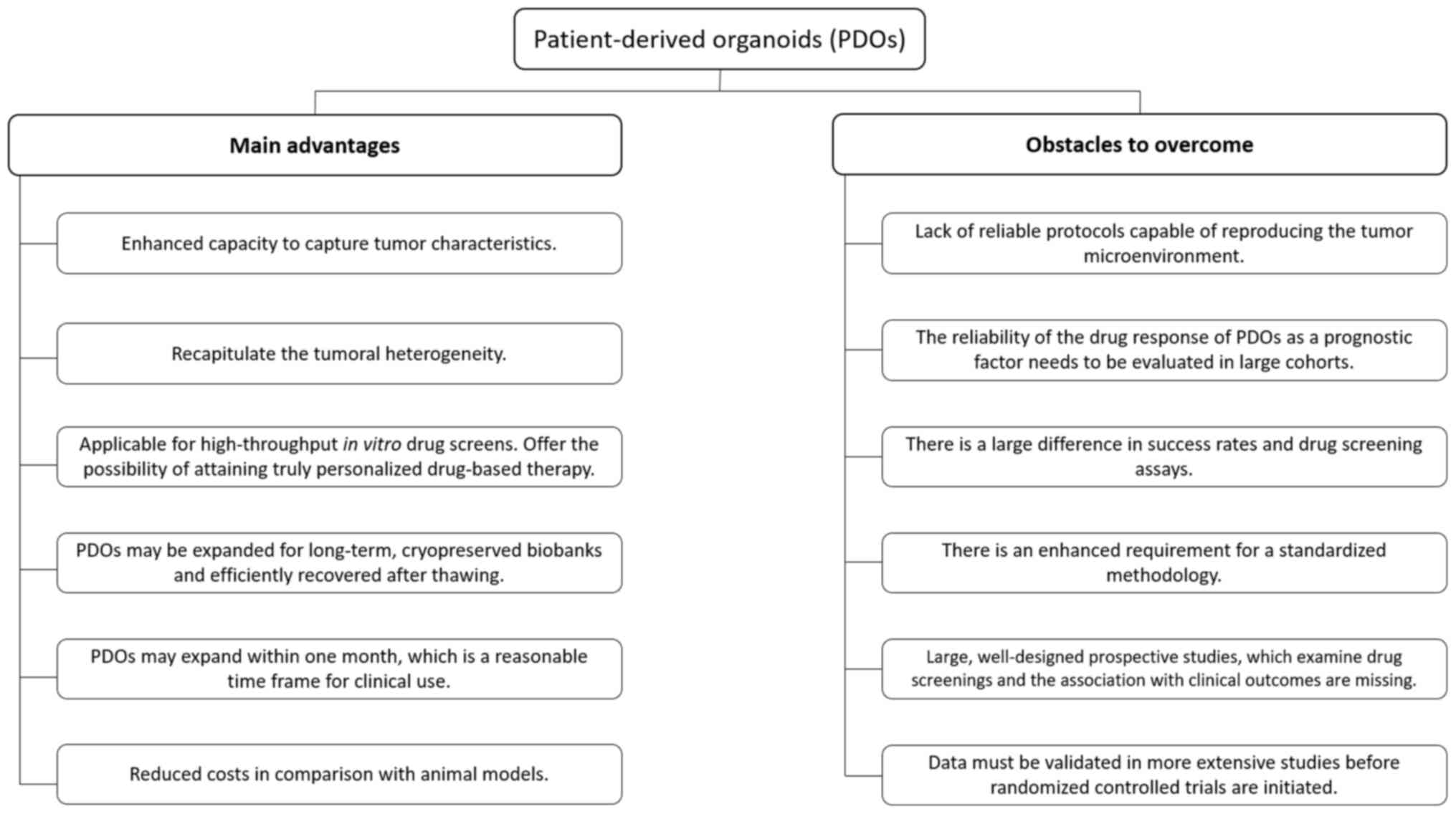
I). However, there are also obstacles to address in the use of
PDOs. Reliable protocols are required to reproduce the tumor
microenvironment, and there is currently a lack of large,
well-designed prospective studies examining drug screenings
regarding clinical outcomes. This may be the reason why PDOs are
still on an investigational level and not routinely used in the
clinic for EOC and other cancer types (Fig. 1).
 | Table ISummary of studies included. |
Table I
Summary of studies included.
| First author
(year) | Population | Origin of
biological material | Number of organoids
(success rate of growth in %) | Recapture of
molecular features | Recapture of
heterogeneity | Number of patients
with drug screening ability | Comparison of drug
screening results with clinical outcomes | (Refs.) |
|---|
| Kopper (2019) | N=32 Histology: All
main subtypes. FIGO: IA-IVB | Ovary,
uterus,peritoneum, omentum, pleural effusion, lymph node,
diaphragm, bowel, abdominal wall and ascites | 56 (65%) | + | + | N=25 | None | (46) |
| Phan (2019) | N=4 Histology:
HGSC, ovariancarcinosarcoma, high-grade peritoneal carcinoma,
high-grade mixed type. FIGO: IIIC-IVB | Ovary and
peritoneum | 4 (no details) | + | + | N=4 | Organoids from two
patients with persistent disease were platinum-resistant | (47) |
| Maru (2019) | N=15 Histology:
BBT, EMC, HGSC, MB, MC, SBT. FIGO: IA-IIIA1 | Ovary | 9 (60%) | + | + | N=9 | None | (48) |
| Jabs (2017) | N=9 Histology:
HGSC. FIGO: IIIC-IV | Ovary, ascites and
pleural effusion | 9 (no details) | + | + | N=9 | None | (49) |
| Hill (2018) | N=22 Histology:
HGSC, LGSC. FIGO: No details | Ovary, omentum,
pleural effusion, mesentery and diaphragm | 33 (80-90%) | + | + | N=33 | None | (50) |
| Hoffmann
(2020) | N=13 Histology:
HGSC. FIGO: IIIC-IVB | Omentum and
peritoneum | 15 (29%) | + | + | N=3 | None | (51) |
| Maenhoudt
(2020) | N=27 Histology:
HGSC, CCC, MC. FIGO: IIB-IVB | Ovary, omentum and
rectum. Some of the samples are not specified | 12 (44%) | + | No details | N=4 | None | (26) |
| Nanki (2020) | N=7 Histology:
HGSC, EMC, CCC. FIGO: IA-IIIC | Ovary | 7(80) | + | No details | N=7 | Concordance between
drug screening results and time to recurrence in two patients | (44) |
| de Witte
(2020) | N=23 Histology:
HGSC, LGSC, MC, CCC, EMC. FIGO: IA-IVB | Ovary, omentum,
ascites, lymph nodes, adnexae, peritoneum, uterus, ligamentum
latum, abdominal wall and bladder | 36 (no
details) | + | + | N=36 | Seven PDOs were
exposed to carboplatin and paclitaxel. A significant correlation
with clinical response was found | (27) |
| Sun (2020) | N=10 Histology:
Serous. FIGO: IA-IVB | No details | 10 (no
details) | No details | No details | N=10 | A total of 10 PDOs
were established from cisplatin-sensitive (n=4) and
cisplatin-resistant (n=6) ovarian cancer tissue. Chemosensitivity
to cisplatin was verified with drug screenings | (53) |
The present systematic review reported on the
current status of PDOs and their use in EOC to predict treatment
response.
2. Method
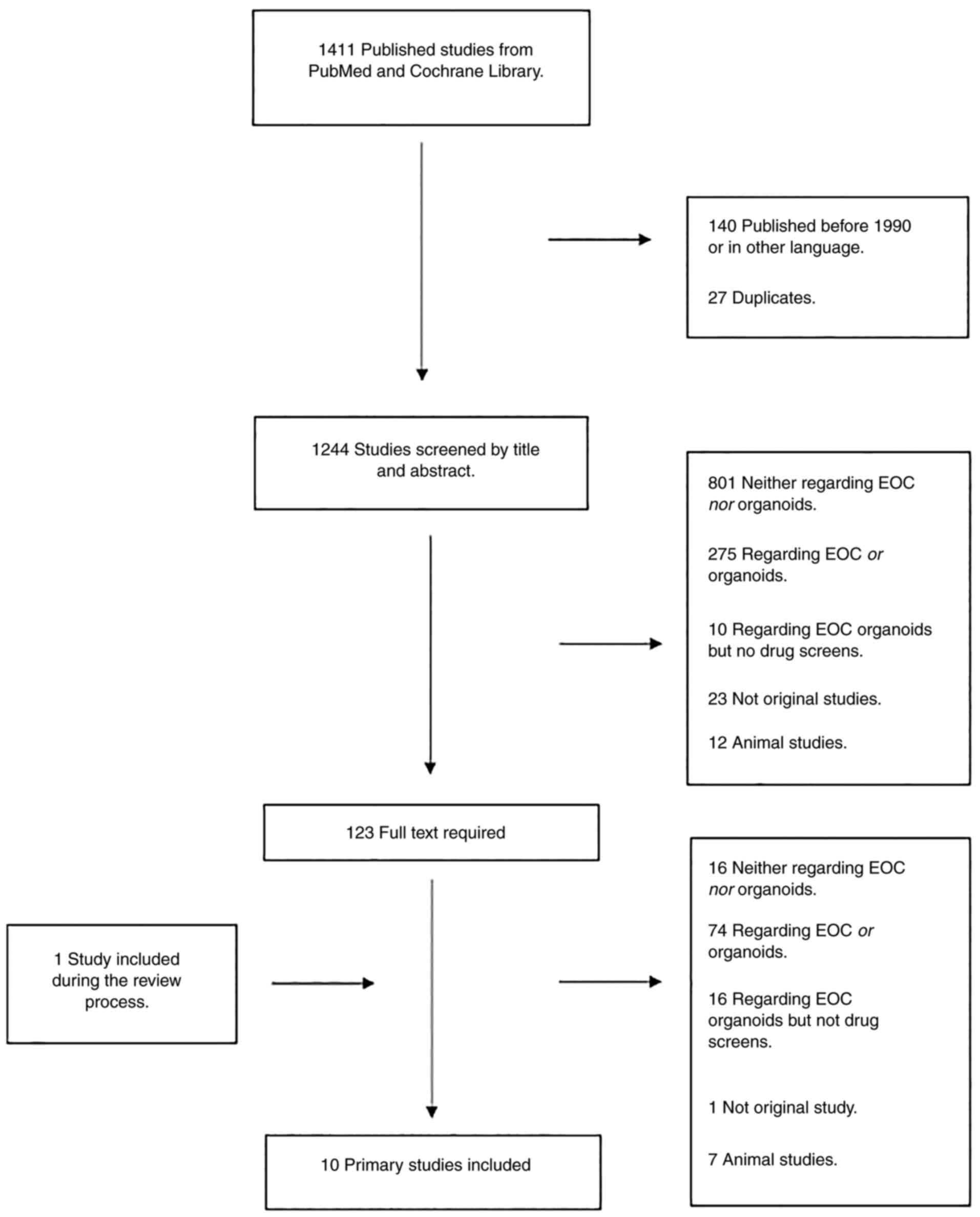
The search in PubMed and the Cochrane Library was
performed on July 10, 2020 (Fig.
2). The following search criteria were used: ‘Ovarian Cancer’,
‘Organoids’ and ‘Cell culture techniques’ (Fig. S1, Fig. S2, Fig. S3, Fig. S4 and Fig. S5). The literature search was
combined with a screening of the reference sections of relevant
studies, which did not add any new studies. The first author
performed a screening of all identified articles by title and
abstract. In total, 1,244 studies were screened and the full text
of 123 studies was examined. Only original studies with successful
EOC organoid growth and drug screening ability were included.
Studies were excluded if they had included other cancer types than
EOC, were not original studies or were performed on animals.
Furthermore, studies in languages other than English, Danish,
Swedish or Norwegian were excluded. One of the studies was included
during the review process. Finally, a total of 10 studies were
included in the present review (Fig.
2).
3. Overview of the publications
A recent study by Kopper et al (46) from 2019 published a breakthrough in
EOC organoids (Table I). They
presented a protocol capable of cultivating long-term organoids of
all major subtypes of EOC and established 56 organoids (success
rate of growth was 65%) from 32 different patients. The organoids
maintained the genomic landscape, histological aspects and tumor
heterogeneity of the original tumor. Finally, they demonstrated
that EOC organoids may be used for drug screening assays and
capture different tumor subtype responses to standard
platinum-based chemotherapy. Furthermore, organoids were tested for
homologous recombination (HR) deficiency (HRD) using the
recombination capacity test and it was determined that cells with
HRD were sensitive to PARP inhibitors.
In another recent publication by Phan et al
(47), a method of cultivating
organoids derived from four patients was performed, including one
patient with high-grade mixed type carcinoma with a component of
HGSC (40%) and a clear cell carcinoma (60%), one patient with HGSC,
one patient with ovarian carcinosarcoma and one patient with
high-grade peritoneal carcinoma. Drug screening was achievable
within one week from harvesting the original tumor. Tumor
heterogeneity was preserved and multiple drugs were tested during
the same period. Drug screening results reflected clinical outcomes
and revealed a positive correlation; one patient had persistent
disease despite aggressive debulking surgery and treatment with
carboplatin and paclitaxel. Resistance to carboplatin was also
observed in the high-throughput assay. Another patient was
diagnosed with progressive, platinum-resistant HGSC and was heavily
pretreated prior to sample procurement. The organoids from this
patient were also platinum-resistant, with no reduction of
viability observed upon treating the cells with carboplatin.
Maru et al (48) reported on the propagation of nine
ovarian organoid cultures derived from HGSC, mucinous and
endometrioid ovarian carcinoma with an overall growth success rate
of 60%. These organoids were long-term cultured. However, the exact
duration of the cultivation was not specified. Intratumor
heterogeneity was maintained and represented the original
histological and genetic features. Maru et al (48) also established a drug sensitivity
assay using spheroids derived from the ovarian organoids.
Jabs et al (49) harvested organoids for drug testing
from 2D cell cultures of HGSC. However, these organoids were grown
within 10 days. They screened cells from nine patients with HGSC
with clinically relevant drugs and determined that homologous
recombination deficiency scores correlated with drug effects in
organoids. The HRD score in this study was defined as the number of
loss of heterozygosity regions observed in a tumor sample. HRD
scores in this study varied between 3 and 22. They correlated with
cytotoxic responses to carboplatin and all its combinations and
paclitaxel, azacytidine and decitabine responses. However, these
drugs do not directly affect DNA structure or repair. Furthermore,
positive HRD scores (≥10) determined for tumor tissues co-occurred
with high drug-induced cytotoxicity and fast organoid growth.
Hill et al (50) reported on short-lived (2 weeks)
HGSC organoids for drug testing. The organoids maintained their
parent heterogeneity and did not develop any novel mutations. This
group used organoids to assess DNA damage repair defects and their
influence on immune-oncologic agents, apparently providing a
possible tool to predict patient response to therapy. By testing
the HR defects of 33 organoids (growth success rate 80-90%) in 22
patients, the study confirmed that HRD in the organoids is related
to sensitivity to PARP inhibitors regardless of the mutation status
of DNA repair genes.
Hoffmann et al (51) created 15 organoids (growth rate,
29%) from 13 primary deposits from patients with advanced HGSC.
They indicated that the mutational and phenotypic profile of the
organoids closely matched those of the parental tumor. Preliminary
tests with carboplatin indicated individual differences in drug
response of organoids from three patients.
Maenhoudt et al (26) established organoid cultures of
tissue from patient-derived EOC, particularly from HGSC. A total of
27 patients were included and twelve organoids were established.
The overall growth rate was 36% for patients with HGSC and 44% for
all patients. The organoids established exhibited tumor-associated
morphology and disease characteristics and recapitulated the parent
tumors' marker expression and mutational landscape. Furthermore,
the organoids displayed tumor-specific sensitivity to clinical
chemotherapeutic drugs.
Nanki et al (44) established seven patient-derived EOC
organoids in <3 weeks with a growth rate of 80%. These organoids
captured the characteristics of histological cancer subtypes and
replicated the mutational landscape of the primary tumors. Seven
pairs of organoids (three HGSC, one clear cell, three endometrioid)
and original tumors shared 59% of the variants identified.
Furthermore, drug screening was possible and the organoid harboring
a BRCA1 mutation had a higher sensitivity to olaparib and platinum
drugs than the other organoids. They also compared the time to
recurrence after completion of the first-line platinum regimen
against drug screening results in two patients and observed
concordance between the results. Of note, one patient was sensitive
to paclitaxel, docetaxel, topotecan, SN-38, gemcitabine and
trabectedin, and her time to recurrence after completing the
first-line platinum regimen was 18 months. The time to recurrence
in another patient who exhibited resistance to all tested drugs,
except trabectedin, was nine months.
de Witte et al (27) included 36 organoids [29 of which
have been established previously by Kopper et al (46) from 23 patients with EOC. PDOs
resembled the tumors from which they were derived, with an average
overlap of 67% of single nucleotide variants and similar copy
number alteration profiles. Intra-patient tumor heterogeneity
assessment in seven patients with organoids derived from multiple
tumor locations revealed a differential response to at least one
drug for all patients. Furthermore, organoids displayed inter- and
intra-patient drug response heterogeneity to chemotherapy and
targeted drugs. A total of seven PDOs (derived from five patients)
were exposed to carboplatin and paclitaxel combination treatment
in vitro. It was possible to directly compare their response
with the patient's clinical response. These PDO drug responses had
a statistically significant correlation with the clinical response,
as measured by the chemotherapy response score, normalization of
the serum biomarker carbohydrate antigen-125 and radiological
responses [response evaluation criteria in solid tumors (RECIST)]
(52). Furthermore, they
demonstrated that organoid establishment and drug screening are
feasible within three weeks.
Sun et al (53) established organoids (n=10) from
cisplatin-sensitive and cisplatin-resistant ovarian cancer tissues.
The PDOs verified chemosensitivity to cisplatin. RNA sequencing was
employed to compare the expression of chemosensitivity-related
genes in four cisplatin-sensitive and six cisplatin-resistant PDOs.
Significantly higher expression levels of Aurora-A were observed in
PDOs from cisplatin-resistant patients.
4. Discussion
In recent years, there has been an increased
interest in EOC organoids, reflected in a large number of published
studies. The present review aimed to uncover the current status of
PDOs and their ability to perform drug screenings in EOC and
thereby predict treatment response. A total of 10 studies fulfilled
the inclusion criteria for this review. In all studies, attempts to
grow EOC-organoids were successful, which were able to recapture
the genomic and mutational profile and heterogeneity of the donor
tissue, and drug screenings were performed (26,27,44,46-52).
Other recent reviews have examined the use of PDOs
in EOC (54,55). A total of two studies focused on
EOC, also included in the present review (27,47).
Both reviews concluded that PDOs were able to predict treatment
response and may guide therapeutic decisions in the future. Yet,
they also concluded that PDOs require to be generated and expanded
efficiently to enable drug screening in a clinically meaningful
time window. In addition to these reviews, the present study
discussed how the growth rates may be improved and whether the
histological subtypes may have a role in this context. Furthermore,
the lack of clinical outcomes in numerous studies included was
addressed, which is critical for the clinical use of PDOs.
Data are still limited, which is reflected by the
small number of studies and number of patients. Furthermore, there
is a considerable variation in the reported growth rates (29-90%)
and in four studies, the growth rates were not stated (Table I). The number and success rates of
organoids used for drug screens were not reported in any of the
studies included. Furthermore, there are differences in
histological diagnoses. While the studies by Kopper et al
(46), Maru et al (48) and de Witte et al (27) comprised all major histological
subtypes of EOC, Hoffmann et al (51) and Jabs et al (49) only included patients with HGSC
(Table I). At the same time, none
of the studies examined the differences in growth between the
different histological subgroups. The number of tissue samples was
unknown and the histological subgroups were too small for a
conclusive statistical analysis (46). It is well known that EOC consists
of several distinct subtypes that differ in their clinical and
molecular profile and should be considered and treated as uniform
entities in clinical and research settings (2). Thus, optimally, organoid studies
should obtain a homogenous study population by including specific
subtypes of EOC to optimize growth conditions and growth rates by
adapting growth medium according to histologic subtypes. Comparing
these results with organoid studies from patients with
gastrointestinal cancers, it was indicated that the growth rate of
organoids was consistently 65-70% in these studies (28,38-42).
Furthermore, success rates of drug screenings (25-80%) were
mentioned and causes of failed organoid growth, such as no or few
tumor cells in the biopsies, quality control problems or bacterial
infections, were addressed. Clinical parameters such as sex, biopsy
location and prior systemic treatment were also examined and it was
determined that they did not influence the success of culture.
However, even in CRC, where the use of organoids is included in
clinical studies, there is still a substantial fraction of patients
for whom no organoid-informed decision may be made. Growth rates
may be further improved by obtaining multiple core biopsies,
together with a pathologist's direct evaluation of the biopsies to
identify samples with low cellularity, as suggested by
Vlachogiannis et al (28).
One of the explanations for the higher growth rates
observed in gastrointestinal cancers may be that they are
morphologically more homogenous than EOC (56,57).
Only one study on pancreatic cancer stated the histopathological
diagnoses as adenocarcinomas, accounting for 98% of cases (42). The growth media used in all EOC
studies consisted mainly of the same components such as noggin,
R-spondin, EGF, Wnt, antibiotics and ROCK inhibitor (Table SI). However, minor differences in
components, such as insulin, hydrocortisone and β-estradiol, exist.
The study by Maru et al (48) is the only one that has attempted to
optimize the growth medium. Their initial growth rate was 45%,
which rose to 83% after introducing a seven-minute digestion step
with Accumax, a potent proteolytic and collagenolytic enzyme with
DNAse activity, following the routine digestion of tumors with
dispase and collagenase. This growth rate is comparable to the
growth rate of 80% obtained by Nanki et al (44). Compared with the growth medium used
by Kopper et al (46),
heregulin β-1, nicotinamide, forskolin, hydrocortisone and
estradiol were replaced by gastrin and insulin-like growth factor.
These examples highlight that growth rates may be improved with
growth media modifications and emphasize the requirement for medium
optimization in EOC.
Another major limitation of the reviewed studies is
that only four included data on clinical outcomes (27,44,47,53).
All studies indicated a positive correlation between clinical
response and drug screenings. de Witte et al (27) exposed seven PDOs to carboplatin and
paclitaxel and determined a significant correlation with clinical
response. Nanki et al (44)
found concordance between drug screening results and time to
recurrence in two patients tested for drugs, including paclitaxel,
docetaxel and olaparib. Phan et al (47), using PDOs from two patients with
persistent disease, indicated that the cancers were
platinum-resistant, in concordance with Sun et al (53). PDOs were established from
platin-sensitive and platin-resistant patients and their
chemosensitivity was verified (44,47).
However, none of the studies mention the number of organoids, which
failed to perform drug screens.
Furthermore, the number of drug-screened organoids
is minimal and thus, the results are only exploratory and require
to be validated in larger studies. In gastrointestinal cancers,
various studies have compared PDO drug screenings with clinical
outcomes in primary and recurrent settings (28,38,42,58,59).
Results from these studies suggested that PDOs mainly recapitulate
patient response to treatment. Only in one study, the results of
drug screens did not correlate for one out of three
chemotherapeutics (38). A
multicenter cohort study on metastatic breast, colon and non-small
cell lung cancers is being conducted by the Foundation Hubrecht
Organoid Technology (TUMOROID trial) in the Netherlands. This study
is ongoing and has so far indicated a positive correlation between
drug response in organoids and the clinical response of patients
according to the RECIST criteria (60-62).
The small number of studies and cases of EOC and the fact that the
studies did not compare drug responses with clinical outcomes in a
standardized manner are prone to type two errors, yielding
insignificant and inconclusive results. Furthermore, it raises
concerns about publication bias, as studies with no correlation
between drug screens and clinical outcomes may not be published.
These parameters and the lack of information may give a misleading
picture of the organoids' ability to predict the treatment effect
of chemotherapy and targeted therapy and thus overestimate their
success in a clinical context.
A high degree of inter- and intratumor heterogeneity
has been described even within specific subtypes of EOC, which
means that the molecular profile may be different, depending on the
location of the tumor sample (61). Thus, molecular analysis of a single
biopsy from a tumor may not represent other tumor parts. Therefore,
treatment based on analysis from a single biopsy may have limited
benefit as molecular pathways active in other tumor parts will lead
to tumor growth. Inter- and intratumor heterogeneity was examined
in seven studies (Table I). Only
one study performed drug screenings on organoids obtained from
different tumor sites and metastasis from the same patients. They
derived two to four PDOs from distinct cancer lesions in seven
patients and revealed a differential response to at least one drug
for all patients (27). In CRC and
pancreatic cancer, patients' organoids were established from
multiple locations from tumor and metastasis. It was reported that
responses to individual agents differed substantially between
lesions in a single patient, which further complicates the use of
PDOs to predict treatment response (38,58).
Another limitation of the studies included is the
lack of uniform statistical analyses. A total of four studies
lacked statistical analyses (44,47,48,50),
while three studies use one-way ANOVA to calculate significant
variation between two experimental groups (44,49,53).
However, none of these three studies reported the F-values, which
is critical in an ANOVA test, as it tells whether the difference in
the two groups is significant.
Other factors such as biopsy localization and size
may also have an impact on organoid growth. In addition, the time
from tissue sampling to arrival and handling in the laboratory,
including the time stored refrigerated, may also impact the growth
rates. None of the studies mentioned these circumstances.
In addition, large, well-designed prospective
studies examining standardized organoid-based drug screenings
regarding relevant clinical outcomes are currently lacking but
urgently required prior to the implementation of drug screenings
based on resistance patterns of organoids in a clinical setting
(52). Indeed, standardization of
drug screenings and transparency in all stages from growth to drug
screenings is needed. This method must meet the quality criteria
for a good test, such as reproducibility, consistency, validity,
cost-effectiveness, non-invasiveness, and high sensitivity and
specificity. So far, the most robust empirical demonstration of the
utility of a medical test is a properly designed randomized
controlled trial. Therefore, the already published data must be
validated more extensively in future studies before randomized
controlled trials with power are initiated in a clinical setting
(Fig. 1). Another factor of
importance in future studies is the time frame from tissue sampling
to drug screenings. These results should be available prior to
relevant oncological treatment in a clinical setting (62). At present, sequencing results and
molecular profiles are available within few days. Therefore,
PDO-based drug screens performed within a few weeks to supplement
molecular analyses should be mandatory.
In addition to drug screens, organoids may also
provide valuable insight into the pathogenesis of EOC. The reported
high genetic stability of healthy organoid cultures over a long
period enables the study of mutagenic processes in detail (39,63).
Particularly the role of fallopian tube epithelium and ovarian
surface epithelial cells in the development of EOC is yet to be
fully elucidated (64). In the
future, organoid biobanks developed from cancer patients' tissues
may be a powerful tool to enforce organoids and improve the
possibility of validating drug screening results. A combined effort
of the US National Cancer Institute, Cancer Research UK, the UK
Wellcome Trust Sanger Institute and the foundation of Hubrecht
Organoid Technology, Netherlands-known as the Human Cancer Models
Initiative-is ongoing to generate a large, globally accessible bank
of novel cancer cell culture models, including organoids, available
for the research community (39).
This library of cultures and corresponding clinical data is created
to aid basic research, find leads for new compounds and help
explore novel therapeutic strategies (26). Organoids may be an attractive tool
for developing precise treatment strategies. They are already used
to select optimal therapy in a metastatic setting for
gastrointestinal cancers and may be used as a future platform for
EOC drug testing as well. The major advantages of organoids are
clonality, the possibility for high-throughput screening and
reduced cost compared to animal models (22) (Fig.
1).
Currently, clinical diagnostic molecular approaches
such as next-generation sequencing are used to guide treatment
decisions based on mutations and fusions that may be biologically
targeted. While it remains challenging to predict the response of
treatment, organoids may add essential knowledge regarding
treatment response and eventually help determine optimal
treatments, particularly in the setting of a recurrent disease
course. However, drug screening results from organoids cannot stand
alone and there is still a requirement for molecular analyses such
as sequencing upfront to identify druggable targets.
5. Conclusion
Organoid growth in EOC is possible, although growth
rates vary. The grown organoids appear to recapitulate the
molecular features and heterogeneity of the original tumor. They
are suitable for drug screening assays but the success rate of drug
screening is still low and must be improved before implementation
in the clinic may be considered. Furthermore, drug screening
results must be held against clinical outcomes and examined in
adequately designed randomized clinical trials. Thus,
standardization and transparency in all stages, from growth to drug
screenings, are required. Using organoids for diagnostic insight or
biobanking may be the next step forward. Combining this technology
with molecular approaches may add to the current knowledge
regarding the treatment of EOC in the future.
Supplementary Material
PubMed search: Ovarian cancer and
organoids (July 10, 2020). EOC, epithelial ovarian cancer.
PubMed search: Ovarian cancer and cell
culture techniques (July 10, 2020). EOC, epithelial ovarian
cancer.
Cochrane Library search: Ovarian
cancer and cell culture techniques (July 10, 2020). EOC, epithelial
ovarian cancer.
Cochrane Library search: Organoids
(July 10, 2020). EOC, epithelial ovarian cancer.
Cochrane Library search: Cell culture
techniques (July 10, 2020). EOC, epithelial ovarian cancer.
List of drugs screened for in the
studies.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
YS contributed to all stages of the review, from
literature search to manuscript writing and submission. EH, CH and
TS contributed as supervisors, providing guidance and evaluation of
the academic work. All authors made substantial contributions to
the design, interpretation of data, drafting the manuscript and
revising it critically for important intellectual content. YS and
TS assessed the authenticity of all raw data to ensure their
legitimacy. All authors read and approved the final manuscript.
Ethics approval of consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Dyba T, Randi G, Bettio M, Gavin A, Visser O and Bray F: Cancer
incidence and mortality patterns in Europe: Estimates for 40
countries and 25 major cancers in 2018. Eur J Cancer. 103:356–387.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lheureux S, Gourley C, Vergote I and Oza
AM: Epithelial ovarian cancer. Lancet. 393:1240–1253.
2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Colombo N, Sessa C, du Bois A, Ledermann
J, McCluggage WG, McNeish I, Morice P, Pignata S, Ray-Coquard I,
Vergote I, et al: ESMO-ESGO consensus conference recommendations on
ovarian cancer: Pathology and molecular biology, early and advanced
stages, borderline tumours and recurrent disease. Ann Oncol.
30:672–705. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chase DM, Chaplin DJ and Monk BJ: The
development and use of vascular targeted therapy in ovarian cancer.
Gynecol Oncol. 145:393–406. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liu FW, Cripe J and Tewari KS:
Anti-angiogenesis therapy in gynecologic malignancies. Oncology
(Williston Park). 29:350–360. 2015.PubMed/NCBI
|
|
6
|
Perren TJ, Swart AM, Pfisterer J,
Ledermann JA, Pujade-Lauraine E, Kristensen G, Carey MS, Beale P,
Cervantes A, Kurzeder C, et al: A phase 3 trial of bevacizumab in
ovarian cancer. N Engl J Med. 365:2484–2496. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Burger RA, Brady MF, Bookman MA, Fleming
GF, Monk BJ, Huang H, Mannel RS, Homesley HD, Fowler J, Greer BE,
et al: Incorporation of bevacizumab in the primary treatment of
ovarian cancer. N Engl J Med. 365:2473–2483. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Aghajanian C, Blank SV, Goff BA, Judson
PL, Teneriello MG, Husain A, Sovak MA, Yi J and Nycum LR: OCEANS: A
randomized, double-blind, placebo-controlled phase III trial of
chemotherapy with or without bevacizumab in patients with
platinum-sensitive recurrent epithelial ovarian, primary
peritoneal, or fallopian tube cancer. J Clin Oncol. 30:2039–2045.
2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Aghajanian C, Goff B, Nycum LR, Wang YV,
Husain A and Blank SV: Final overall survival and safety analysis
of OCEANS, a phase 3 trial of chemotherapy with or without
bevacizumab in patients with platinum-sensitive recurrent ovarian
cancer. Gynecol Oncol. 139:10–16. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Coleman RL, Brady MF, Herzog TJ, Sabbatini
P, Armstrong DK, Walker JL, Kim BG, Fujiwara K, Tewari KS, O'Malley
DM, et al: Bevacizumab and paclitaxel-carboplatin chemotherapy and
secondary cytoreduction in recurrent, platinum-sensitive ovarian
cancer (NRG Oncology/Gynecologic Oncology Group study GOG-0213): A
multicentre, open-label, randomised, phase 3 trial. Lancet Oncol.
18:779–791. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pujade-Lauraine E, Hilpert F, Weber B,
Reuss A, Poveda A, Kristensen G, Sorio R, Vergote I, Witteveen P,
Bamias A, et al: Bevacizumab combined with chemotherapy for
platinum-resistant recurrent ovarian cancer: The AURELIA open-label
randomized phase III trial. J Clin Oncol. 32:1302–1308.
2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cancer Genome Atlas Research Network.
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ledermann J, Harter P, Gourley C,
Friedlander M, Vergote I, Rustin G, Scott C, Meier W,
Shapira-Frommer R, Safra T, et al: Olaparib maintenance therapy in
platinum-sensitive relapsed ovarian cancer. N Engl J Med.
366:1382–1392. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Moore K, Colombo N, Scambia G, Kim BG,
Oaknin A, Friedlander M, Lisyanskaya A, Floquet A, Leary A, Sonke
GS, et al: Maintenance olaparib in patients with newly diagnosed
advanced ovarian cancer. N Engl J Med. 379:2495–2505.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Pujade-Lauraine E, Ledermann JA, Selle F,
Gebski V, Penson RT, Oza AM, Korach J, Huzarski T, Poveda A,
Pignata S, et al: Olaparib tablets as maintenance therapy in
patients with platinum-sensitive, relapsed ovarian cancer and a
BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised,
placebo-controlled, phase 3 trial. Lancet Oncol. 18:1274–1284.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mirza MR, Monk BJ, Herrstedt J, Oza AM,
Mahner S, Redondo A, Fabbro M, Ledermann JA, Lorusso D, Vergote I,
et al: Niraparib maintenance therapy in platinum-sensitive,
recurrent ovarian cancer. N Engl J Med. 375:2154–2164.
2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
González-Martín A, Pothuri B, Vergote I,
DePont Christensen R, Graybill W, Mirza MR, McCormick C, Lorusso D,
Hoskins P, Freyer G, et al: Niraparib in patients with newly
diagnosed advanced ovarian cancer. N Engl J Med. 381:2391–2402.
2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Swisher EM, Lin KK, Oza AM, Scott CL,
Giordano H, Sun J, Konecny GE, Coleman RL, Tinker AV, O'Malley DM,
et al: Rucaparib in relapsed, platinum-sensitive high-grade ovarian
carcinoma (ARIEL2 Part 1): An International, multicentre,
open-label, phase 2 trial. Lancet Oncol. 18:75–87. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Coleman RL, Oza AM, Lorusso D, Aghajanian
C, Oaknin A, Dean A, Colombo N, Weberpals JI, Clamp A, Scambia G,
et al: Rucaparib maintenance treatment for recurrent ovarian
carcinoma after response to platinum therapy (ARIEL3): A
randomised, double-blind, placebo-controlled, phase 3 trial.
Lancet. 390:1949–1961. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tsibulak I, Zeimet AG and Marth C: Hopes
and failures in front-line ovarian cancer therapy. Crit Rev Oncol
Hematol. 143:14–19. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Joo WD, Visintin I and Mor G: Targeted
cancer therapy-Are the days of systemic chemotherapy numbered?
Maturitas. 76:308–314. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Dumont S, Jan Z, Heremans R, Van Gorp T,
Vergote I and Timmerman D: Organoids of epithelial ovarian cancer
as an emerging preclinical in vitro tool: A review. J Ovarian Res.
12(105)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Maru Y and Hippo Y: Current status of
patient-derived ovarian cancer models. Cells. 8(505)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Clevers H: Modeling development and
disease with organoids. Cell. 165:1586–1597. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sato T, Vries RG, Snippert HJ, van de
Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters
PJ and Clevers H: Single Lgr5 stem cells build crypt-villus
structures in vitro without a mesenchymal niche. Nature.
459:262–265. 2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Maenhoudt N, Defraye C, Boretto M, Jan Z,
Heremans R, Boeckx B, Hermans F, Arijs I, Cox B, Van Nieuwenhuysen
E, et al: Developing organoids from ovarian cancer as experimental
and preclinical models. Stem Cell Reports. 14:717–729.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
de Witte CJ, Espejo Valle-Inclan J, Hami
N, Lõhmussaar K, Kopper O, Vreuls CPH, Jonges GN, van Diest P,
Nguyen L, Clevers H, et al: Patient-derived ovarian cancer
organoids mimic clinical response and exhibit heterogeneous inter-
and intrapatient drug responses. Cell Rep.
31(107762)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Vlachogiannis G, Hedayat S, Vatsiou A,
Jamin Y, Fernández-Mateos J, Khan K, Lampis A, Eason K, Huntingford
I, Burke R, et al: Patient-derived organoids model treatment
response of metastatic gastrointestinal cancers. Science.
359:920–926. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lõhmussaar K, Boretto M and Clevers H:
Human-derived model systems in gynecological cancer research.
Trends Cancer. 6:1031–1043. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sato T, Stange DE, Ferrante M, Vries RG,
Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J,
Siersema PD and Clevers H: Long-term expansion of epithelial
organoids from human colon, adenoma, adenocarcinoma, and Barrett's
epithelium. Gastroenterology. 141:1762–1772. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Boj SF, Hwang CI, Baker LA, Chio II, Engle
DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, et al:
Organoid models of human and mouse ductal pancreatic cancer. Cell.
160:324–338. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yan HHN, Siu HC, Law S, Ho SL, Yue SSK,
Tsui WY, Chan D, Chan AS, Ma S, Lam KO, et al: A comprehensive
human gastric cancer organoid biobank captures tumor subtype
heterogeneity and enables therapeutic screening. Cell Stem Cell.
23:882–897.e11. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gao D, Vela I, Sboner A, Iaquinta PJ,
Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora
VK, et al: Organoid cultures derived from patients with advanced
prostate cancer. Cell. 159:176–187. 2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sachs N, de Ligt J, Kopper O, Gogola E,
Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H,
et al: A living biobank of breast cancer organoids captures disease
heterogeneity. Cell. 172:373–386.e10. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li X, Francies HE, Secrier M, Perner J,
Miremadi A, Galeano-Dalmau N, Barendt WJ, Letchford L, Leyden GM,
Goffin EK, et al: Organoid cultures recapitulate esophageal
adenocarcinoma heterogeneity providing a model for clonality
studies and precision therapeutics. Nat Commun.
9(2983)2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lee SH, Hu W, Matulay JT, Silva MV,
Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, Williams AB, et
al: Tumor evolution and drug response in patient-derived organoid
models of bladder cancer. Cell. 173:515–528.e17. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Girda E, Huang EC, Leiserowitz GS and
Smith LH: The use of endometrial cancer patient-derived organoid
culture for drug sensitivity testing is feasible. Int J Gynecol
Cancer. 27:1701–1707. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ooft SN, Weeber F, Dijkstra KK, McLean CM,
Kaing S, van Werkhoven E, Schipper L, Hoes L, Vis DJ, van de Haar
J, et al: Patient-derived organoids can predict response to
chemotherapy in metastatic colorectal cancer patients. Sci Transl
Med. 11(eaay2574)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Drost J and Clevers H: Organoids in cancer
research. Nat Rev Cancer. 18:407–418. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Weeber F, van de Wetering M, Hoogstraat M,
Dijkstra KK, Krijgsman O, Kuilman T, Gadellaa-van Hooijdonk CG, van
der Velden DL, Peeper DS, Cuppen EP, et al: Preserved genetic
diversity in organoids cultured from biopsies of human colorectal
cancer metastases. Proc Natl Acad Sci USA. 112:13308–13311.
2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
DeHaan RK, Sarvestani SK and Huang EH:
Organoid models of colorectal pathology: Do they hold the key to
personalized medicine? A systematic review. Dis Colon Rectum.
63:1559–1569. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L,
Xia F, Fu G, Deng Y, Pan M, et al: Patient-derived organoids
predict chemoradiation responses of locally advanced rectal cancer.
Cell Stem Cell. 26:17–26.e6. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
van de Wetering M, Francies HE, Francis
JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J,
Taylor-Weiner A, Kester L, et al: Prospective derivation of a
living organoid biobank of colorectal cancer patients. Cell.
161:933–945. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Nanki Y, Chiyoda T, Hirasawa A, Ookubo A,
Itoh M, Ueno M, Akahane T, Kameyama K, Yamagami W, Kataoka F and
Aoki D: Patient-derived ovarian cancer organoids capture the
genomic profiles of primary tumours applicable for drug sensitivity
and resistance testing. Sci Rep. 10(12581)2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kessler M, Hoffmann K, Brinkmann V, Thieck
O, Jackisch S, Toelle B, Berger H, Mollenkopf HJ, Mangler M,
Sehouli J, et al: The Notch and Wnt pathways regulate stemness and
differentiation in human fallopian tube organoids. Nat Commun.
6(8989)2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kopper O, de Witte CJ, Lõhmussaar K,
Valle-Inclan JE, Hami N, Kester L, Balgobind AV, Korving J, Proost
N, Begthel H, et al: An organoid platform for ovarian cancer
captures intra- and interpatient heterogeneity. Nat Med.
25:838–849. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Phan N, Hong JJ, Tofig B, Mapua M,
Elashoff D, Moatamed NA, Huang J, Memarzadeh S, Damoiseaux R and
Soragni A: A simple high-throughput approach identifies actionable
drug sensitivities in patient-derived tumor organoids. Commun Biol.
2(78)2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Maru Y, Tanaka N, Itami M and Hippo Y:
Efficient use of patient-derived organoids as a preclinical model
for gynecologic tumors. Gynecol Oncol. 154:189–198. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Jabs J, Zickgraf FM, Park J, Wagner S,
Jiang X, Jechow K, Kleinheinz K, Toprak UH, Schneider MA, Meister
M, et al: Screening drug effects in patient-derived cancer cells
links organoid responses to genome alterations. Mol Syst Biol.
13(955)2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Hill SJ, Decker B, Roberts EA, Horowitz
NS, Muto MG, Worley MJ Jr, Feltmate CM, Nucci MR, Swisher EM,
Nguyen H, et al: Prediction of DNA repair inhibitor response in
short-term patient-derived ovarian cancer organoids. Cancer Discov.
8:1404–1421. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Hoffmann K, Berger H, Kulbe H,
Thillainadarasan S, Mollenkopf HJ, Zemojtel T, Taube E,
Darb-Esfahani S, Mangler M, Sehouli J, et al: Stable expansion of
high-grade serous ovarian cancer organoids requires a low-Wnt
environment. EMBO J. 39(e104013)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Sun H, Wang H and Wang X, Aoki Y and Wang
X, Yang Y, Cheng X, Wang Z and Wang X: Aurora-A/SOX8/FOXK1
signaling axis promotes chemoresistance via suppression of cell
senescence and induction of glucose metabolism in ovarian cancer
organoids and cells. Theranostics. 10:6928–6945. 2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Nero C, Vizzielli G, Lorusso D, Cesari E,
Daniele G, Loverro M, Scambia G and Sette C: Patient-derived
organoids and high grade serous ovarian cancer: From disease
modeling to personalized medicine. J Exp Clin Cancer Res.
40(116)2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wensink GE, Elias SG, Mullenders J,
Koopman M, Boj SF, Kranenburg OW and Roodhart JML: Patient-derived
organoids as a predictive biomarker for treatment response in
cancer patients. NPJ Precis Oncol. 5(30)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Fleming M, Ravula S, Tatishchev SF and
Wang HL: Colorectal carcinoma: Pathologic aspects. J Gastrointest
Oncol. 3:153–173. 2012.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Haeberle L and Esposito I: Pathology of
pancreatic cancer. Transl Gastroenterol Hepatol.
4(50)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Tiriac H, Belleau P, Engle DD, Plenker D,
Deschênes A, Somerville TDD, Froeling FEM, Burkhart RA, Denroche
RE, Jang GH, et al: Organoid profiling identifies common responders
to chemotherapy in pancreatic cancer. Cancer Discov. 8:1112–1129.
2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Weeber F, Ooft SN, Dijkstra KK and Voest
EE: Tumor organoids as a Pre-clinical cancer model for drug
discovery. Cell Chem Biol. 24:1092–1100. 2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Aboulkheyr Es H, Montazeri L, Aref AR,
Vosough M and Baharvand H: Personalized cancer medicine: An
organoid approach. Trends Biotechnol. 36:358–371. 2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Klement GL, Arkun K, Valik D, Roffidal T,
Hashemi A, Klement C, Carmassi P, Rietman E, Slaby O, Mazanek P, et
al: Future paradigms for precision oncology. Oncotarget.
7:46813–46831. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Matchar DB: Chapter 1: Introduction to the
methods guide for medical test reviews. J Gen Intern Med. 27 (Suppl
1):S4–S10. 2012.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Behjati S, Huch M, van Boxtel R, Karthaus
W, Wedge DC, Tamuri AU, Martincorena I, Petljak M, Alexandrov LB,
Gundem G, et al: Genome sequencing of normal cells reveals
developmental lineages and mutational processes. Nature.
513:422–425. 2014.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Drost J, van Boxtel R, Blokzijl F,
Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman
JE, van Wezel T, et al: Use of CRISPR-modified human stem cell
organoids to study the origin of mutational signatures in cancer.
Science. 358:234–238. 2017.PubMed/NCBI View Article : Google Scholar
|