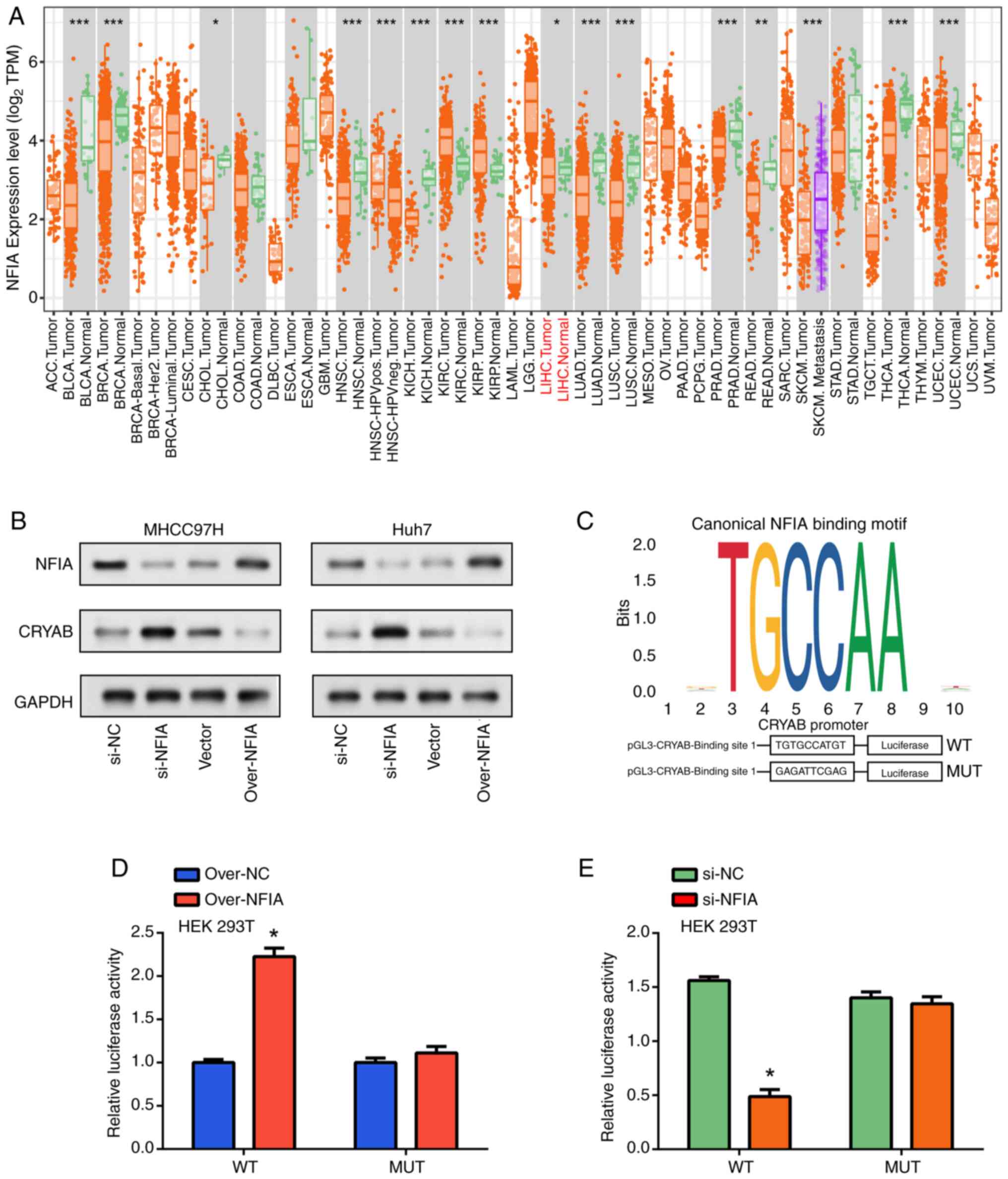
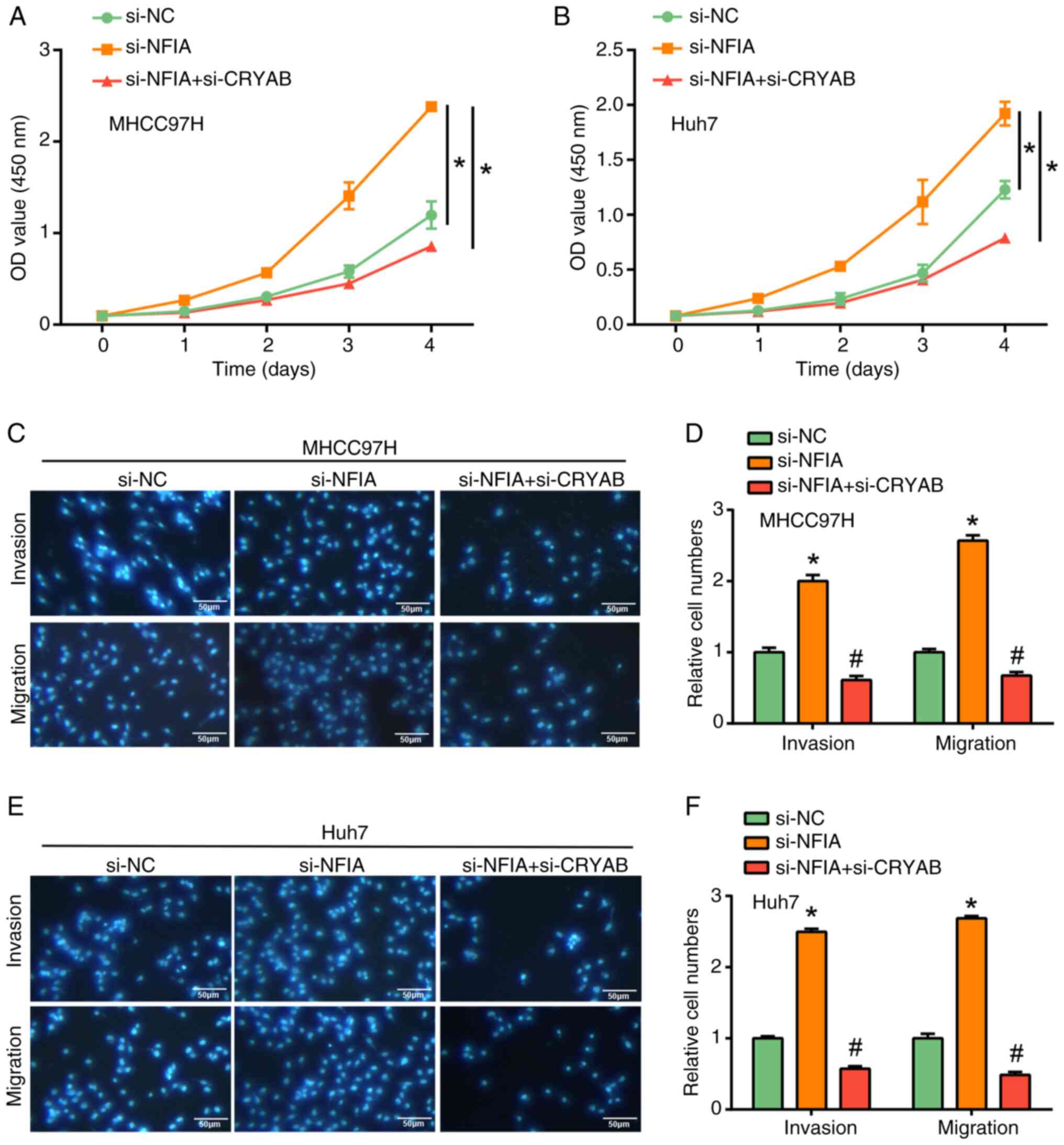
|
1
|
Qiu S, Cai J, Yang Z, He X, Xing Z, Zu J,
Xie E, Henry L, Chong CR, John EM, et al: Trends in Hepatocellular
Carcinoma Mortality Rates in the US and Projections Through 2040.
JAMA Netw Open. 7(e2445525)2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Giri S and Singh A: Epidemiology of
Hepatocellular Carcinoma in India-An Updated Review for 2024. J
Clin Exp Hepatol. 14(101447)2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hahn JW, Woo S, Park J, Lee H, Kim HJ, Ko
JS, Moon JS, Rahmati M, Smith L, Jang J, et al: Global, Regional,
and National Trends in Liver Disease-Related Mortality Across 112
Countries From 1990 to. 2021, With Projections to 2050:
Comprehensive Analysis of the WHO Mortality Database. J Korean Med
Sci. 39(e292)2024.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sagnelli E, Macera M, Russo A, Coppola N
and Sagnelli C: Epidemiological and etiological variations in
hepatocellular carcinoma. Infection. 48:7–17. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
McGlynn KA, Petrick JL and El-Serag HB:
Epidemiology of hepatocellular carcinoma. Hepatology. 73:4–13.
2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Pandyarajan V, Govalan R and Yang JD: Risk
factors and biomarkers for chronic hepatitis B associated
hepatocellular carcinoma. Int J Mol Sci. 22(479)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Li W and Ni CF: Current status of the
combination therapy of transarterial chemoembolization and local
ablation for hepatocellular carcinoma. Abdom Radiol (NY).
44:2268–2275. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Viveiros P, Riaz A, Lewandowski RJ and
Mahalingam D: Current state of liver-directed therapies and
combinatory approaches with systemic therapy in hepatocellular
carcinoma (HCC). Cancers (Basel). 11(1085)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nazzal M, Gadani S, Said A, Rice M, Okoye
O, Taha A and Lentine KL: Liver targeted therapies for
hepatocellular carcinoma prior to transplant: Contemporary
management strategies. Glob Surg Feb. 15:2018.PubMed/NCBI View Article : Google Scholar : (Epub ahead of
print).
|
|
10
|
Fuster-Anglada C, Mauro E, Ferrer-Fàbrega
J, Caballol B, Sanduzzi-Zamparelli M, Bruix J, Fuster J, Reig M,
Díaz A and Forner A: Histological predictors of aggressive
recurrence of hepatocellular carcinoma after liver resection. J
Hepatol. 81:995–1004. 2024.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Glavas D, Bao QR, Scarpa M, Ruffolo C,
Brown ZJ, Pawlik TM and Spolverato G: Treatment and prognosis of
fibrolamellar hepatocellular carcinoma: A systematic review of the
recent literature and meta-analysis. J Gastrointest Surg.
27:705–715. 2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xu Y, Zhang X, Zhang R, Sun Y, Liu J, Luo
C, Yang J, Fang W, Guo Q and Wei L: AFP deletion leads to
anti-tumorigenic but pro-metastatic roles in liver cancers with
concomitant CTNNB1 mutations. Cancer Lett.
566(216240)2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zheng Y, Zhu M and Li M: Effects of
alpha-fetoprotein on the occurrence and progression of
hepatocellular carcinoma. J Cancer Res Clin Oncol. 146:2439–2446.
2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xu W, Guo Y, Huang Z, Zhao H, Zhou M,
Huang Y, Wen D, Song J, Zhu Z, Sun M, et al: Small heat shock
protein CRYAB inhibits intestinal mucosal inflammatory responses
and protects barrier integrity through suppressing IKKβ activity.
Mucosal Immunol. 12:1291–1303. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yin B, Tang S, Xu J, Sun J, Zhang X, Li Y
and Bao E: CRYAB protects cardiomyocytes against heat stress by
preventing caspase-mediated apoptosis and reducing F-actin
aggregation. Cell Stress Chaperones. 24:59–68. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Huang Z, Mou T, Luo Y, Pu X, Pu J, Wan L,
Gong J, Yang H, Liu Y, Li Z, et al: Inhibition of miR-450b-5p
ameliorates hepatic ischemia/reperfusion injury via targeting
CRYAB. Cell Death Dis. 11(455)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang J, Liu J, Wu J, Li W, Chen Z and
Yang L: Progression of the role of CRYAB in signaling pathways and
cancers. Onco Targets Ther. 12:4129–4139. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Fane M, Harris L, Smith AG and Piper M:
Nuclear factor one transcription factors as epigenetic regulators
in cancer. Int J Cancer. 140:2634–2641. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wu YZ, Chan KYY, Leung KT, Lam HS, Tam YH,
Lee KH, Li K and Ng PC: The miR-223/nuclear factor I-A axis
regulates inflammation and cellular functions in intestinal tissues
with necrotizing enterocolitis. FEBS Open Bio. 11:1907–1920.
2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lee J, Hoxha E and Song HR: A novel
NFIA-NFκB feed-forward loop contributes to glioblastoma cell
survival. Neuro Oncol. 19:524–534. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu C, Duan P, Li B, Huang C, Jing Y and
Yan W: miR-29a activates Hes1 by targeting Nfia in esophageal
carcinoma cell line TE-1. Oncol Lett. 9:96–102. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kaczorowski M, Matysiak J, Kielb P,
Malkiewicz B and Halon A: Nuclear Factor IA Is Down-regulated in
Muscle-invasive and High-grade Bladder Cancers. Anticancer Res.
42:493–500. 2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chen M, Wu GB, Xie ZW, Shi DL and Luo M: A
novel diagnostic four-gene signature for hepatocellular carcinoma
based on artificial neural network: Development, validation, and
drug screening. Front Genet. 13(942166)2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jin Y, Dai Y, Qiao O, Hu P and Han J:
miR-1972 inhibits hepatocellular carcinoma proliferation by
targeting GZMH-mediated DNA replication in the cell cycle. J Pharm
Pharmacol: Apr 18, 2024 (Epub ahead of print).
|
|
25
|
Xie Z, Huang J, Li Y, Zhu Q, Huang X, Chen
J, Wei C, Luo S, Yang S and Gao J: Single-cell RNA sequencing
revealed potential targets for immunotherapy studies in
hepatocellular carcinoma. Sci Rep. 13(18799)2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression
analyses for RNA-sequencing and microarray studies. Nucleic Acids
Res. 43(7)2015.doi: 10.1093/nar/gkv007.
|
|
27
|
Jia A, Xu L and Wang Y: Venn diagrams in
bioinformatics. Brief Bioinform. 22(bbab108)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang S, Shi YN, Gu J, He P, Ai QD, Zhou
XD, Wang W and Qin L: Mechanisms of dihydromyricetin against
hepatocellular carcinoma elucidated by network pharmacology
combined with experimental validation. Pharm Biol. 61:1108–1119.
2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang H, Cheng W, Hu P, Ling T, Hu C, Chen
Y, Zheng Y, Wang J, Zhao T and You Q: Integrative analysis
identifies oxidative stress biomarkers in non-alcoholic fatty liver
disease via machine learning and weighted gene co-expression
network analysis. Front Immunol. 15(1335112)2024.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Jin AL, Zhang CY, Zheng WJ, Xian JR, Yang
WJ, Liu T, Chen W, Li T, Wang BL, Pan BS, et al: CD155/SRC complex
promotes hepatocellular carcinoma progression via inhibiting the
p38 MAPK signalling pathway and correlates with poor prognosis.
Clin Transl Med. 12(e794)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ballout N, Etievant L and Viallon V: On
the use of cross-validation for the calibration of the adaptive
lasso. Biom J. 65(2200047)2023.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lee M, Teber ET, Holmes O, Nones K, Patch
AM, Dagg RA, Lau LMS, Lee JH, Napier CE, Arthur JW, et al: Telomere
sequence content can be used to determine ALT activity in tumours.
Nucleic Acids Res. 46:4903–4918. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yan C, Niu Y, Ma L, Tian L and Ma J:
System analysis based on the cuproptosis-related genes identifies
LIPT1 as a novel therapy target for liver hepatocellular carcinoma.
J Transl Med. 20(452)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Koch DT, Yu H, Beirith I, Schirren M,
Drefs M, Liu Y, Knoblauch M, Koliogiannis D, Sheng W, De Toni EN,
et al: Tigecycline causes loss of cell viability mediated by
mitochondrial OXPHOS and RAC1 in hepatocellular carcinoma cells. J
Transl Med. 21(876)2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhao S, Zhang Y, Lu X, Ding H, Han B, Song
X, Miao H, Cui X, Wei S, Liu W, et al: CDC20 regulates the cell
proliferation and radiosensitivity of P53 mutant HCC cells through
the Bcl-2/Bax pathway. Int J Biol Sci. 17:3608–3621.
2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kong J, Wang X, Wang J and Yu G: Silencing
of RAB42 down-regulated PD-L1 expression to inhibit the immune
escape of hepatocellular carcinoma cells through inhibiting the E2F
signaling pathway. Cell Signal. 108(110692)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Fornes O, Castro-Mondragon JA, Khan A, Van
der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M,
Baranašić D, et al: JASPAR 2020: Update of the open-access database
of transcription factor binding profiles. Nucleic Acids Res.
48(D1):D87–D92. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhong H, Shi Q, Wen Q, Chen J, Li X, Ruan
R, Zeng S, Dai X, Xiong J, Li L, et al: Pan-cancer analysis reveals
potential of FAM110A as a prognostic and immunological biomarker in
human cancer. Front Immunol. 14(1058627)2023.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hsiao CY, Chen PD and Huang KW: A
prospective assessment of the diagnostic value of contrast-enhanced
ultrasound, dynamic computed tomography and magnetic resonance
imaging for patients with small liver tumors. J Clin Med.
8(1353)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Park HJ, Jang HY, Kim SY, Lee SJ, Won HJ,
Byun JH, Choi SH, Lee SS, An J and Lim YS: Non-enhanced magnetic
resonance imaging as a surveillance tool for hepatocellular
carcinoma: Comparison with ultrasound. J Hepatol. 72:718–724.
2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kong S, Yue X, Kong S and Ren Y:
Application of contrast-enhanced ultrasound and enhanced CT in
diagnosis of liver cancer and evaluation of radiofrequency
ablation. Oncol Lett. 16:2434–2438. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Galle PR, Foerster F, Kudo M, Chan SL,
Llovet JM, Qin S, Schelman WR, Chintharlapalli S, Abada PB, Sherman
M and Zhu AX: Biology and significance of alpha-fetoprotein in
hepatocellular carcinoma. Liver Int. 39:2214–2229. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Yang Y, Yang M, Pang H, Qiu Y, Sun T, Wang
T, Shen S and Wang W: A macrophage differentiation-mediated gene:
DDX20 as a molecular biomarker encompassing the tumor
microenvironment, disease staging, and prognoses in hepatocellular
carcinoma. Oxid Med Cell Longev. 2022(9971776)2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Krajnović M, Kožik B, Božović A and
Jovanović-Ćupić S: Multiple roles of the RUNX gene family in
hepatocellular carcinoma and their potential clinical implications.
Cells. 12(2303)2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wang T, Jin H, Hu J, Li X, Ruan H, Xu H,
Wei L, Dong W, Teng F, Gu J, et al: COL4A1 promotes the growth and
metastasis of hepatocellular carcinoma cells by activating FAK-Src
signaling. J Exp Clin Cancer Res. 39(148)2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lin TC: RUNX1 and cancer. Biochim Biophys
Acta Rev Cancer. 1877(188715)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Liu Y, Wu Z, Zhao Y, Zhen M, Wang Y and
Liu Q: Apolipoprotein H-based prognostic risk correlates with liver
lipid metabolism disorder in patients with HBV-related
hepatocellular carcinoma. Heliyon. 10(e31412)2024.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Huang C and Freter C: Lipid metabolism,
apoptosis and cancer therapy. Int J Mol Sci. 16:924–949.
2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Lv S, Wang W, Wang H, Zhu Y and Lei C:
PPARγ activation serves as therapeutic strategy against bladder
cancer via inhibiting PI3K-Akt signaling pathway. BMC Cancer.
19(204)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Cai HB, Zhao MY, Li XH, Li YQ, Yu TH, Wang
CZ, Wang LN, Xu WY, Liang B, Cai YP, et al: Single cell sequencing
revealed the mechanism of CRYAB in glioma and its diagnostic and
prognostic value. Front Immunol. 14(1336187)2024.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Dai A, Guo X, Yang X, Li M, Fu Y and Sun
Q: Effects of the CRYAB gene on stem cell-like properties of
colorectal cancer and its mechanism. J Cancer Res Ther.
18:1328–1337. 2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Deng J, Chen X, Zhan T, Chen M, Yan X and
Huang X: CRYAB predicts clinical prognosis and is associated with
immunocyte infiltration in colorectal cancer. PeerJ.
9(e12578)2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Ruan H, Li Y, Wang X, Sun B, Fang W, Jiang
S and Liang C: CRYAB inhibits migration and invasion of bladder
cancer cells through the PI3K/AKT and ERK pathways. Jpn J Clin
Oncol. 50:254–260. 2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Su CH, Liu LC, Hsieh YH, Wang HC, Tsai CW,
Chang WS, Ho CY, Wu CI, Lin CH, Lane HY and Bau DT: Association of
Alpha B-Crystallin (CRYAB) genotypes with breast cancer
susceptibility in Taiwan. Cancer Genomics Proteomics. 8:251–254.
2011.PubMed/NCBI
|
|
57
|
Lu R, Tang P, Zhang D, Lin S, Li H, Feng
X, Sun M and Zhang H: SOX9/NFIA promotes human ovarian cancer
metastasis through the Wnt/β-catenin signaling pathway. Pathol Res
Pract. 248(154602)2023.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Hu Y, Zhang Y, Ding M and Xu R: Lncrna
linc00511 acts as an oncogene in colorectal cancer via sponging
mir-29c-3p to upregulate nfia. Onco Targets Ther. 13:13413–13424.
2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Sun C, Li Y, Tan Y, Zhang H, Liang Y, Zeng
J, Yu J and Zou H: A novel role for NFIA in restoring
radiosensitivity in radioresistant NSCLC cells by downregulating
the AKT and ERK pathways. Biochem Biophys Res Commun. 515:558–564.
2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zhu Z, Luo L, Xiang Q, Wang J, Liu Y, Deng
Y and Zhao Z: MiRNA-671-5p Promotes prostate cancer development and
metastasis by targeting NFIA/CRYAB axis. Cell Death Dis.
11(949)2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Shi C, Yang X, Bu X, Hou N and Chen P:
Alpha B-crystallin promotes the invasion and metastasis of
colorectal cancer via epithelial-mesenchymal transition. Biochem
Biophys Res Commun. 489:369–374. 2017.PubMed/NCBI View Article : Google Scholar
|