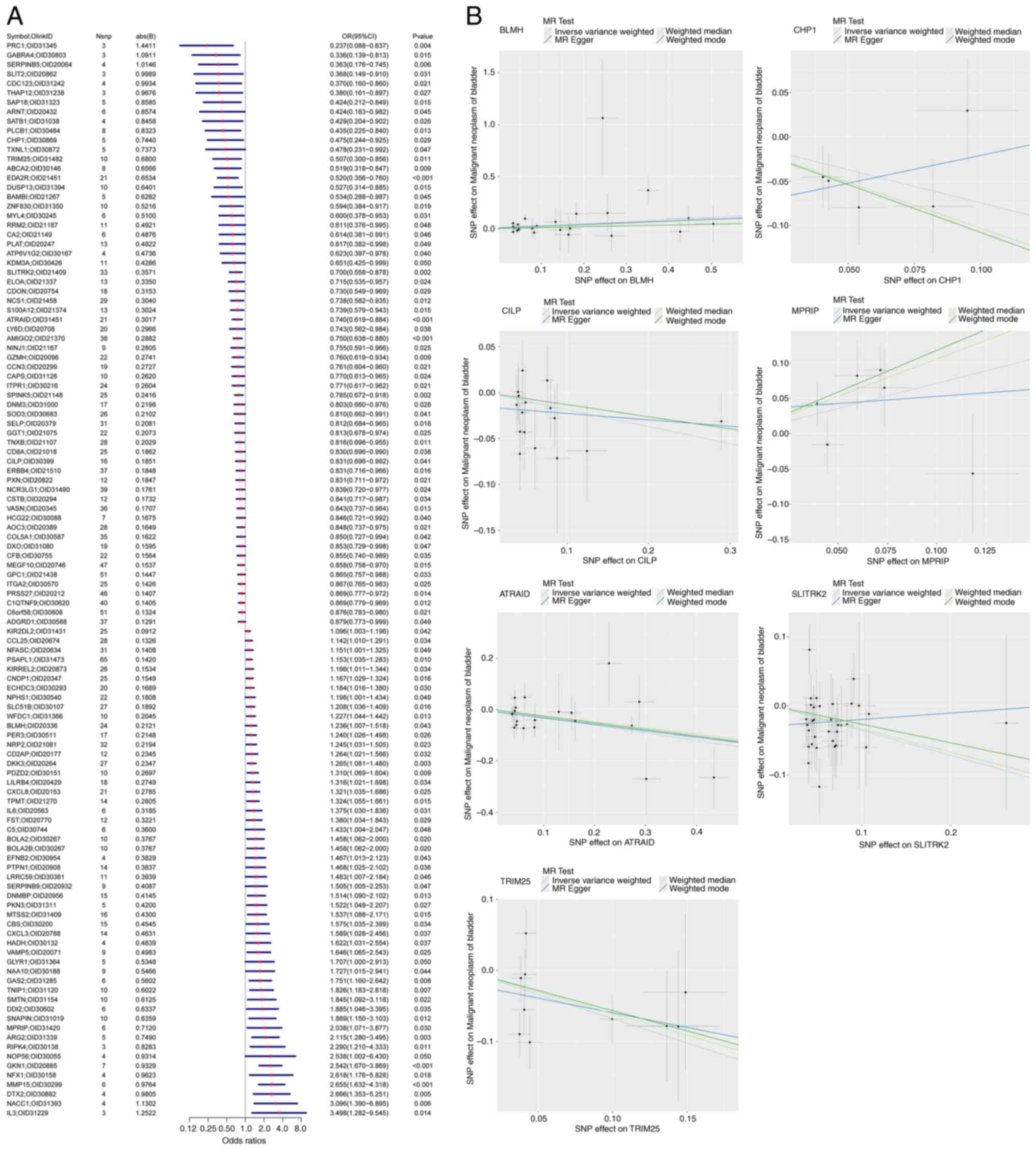
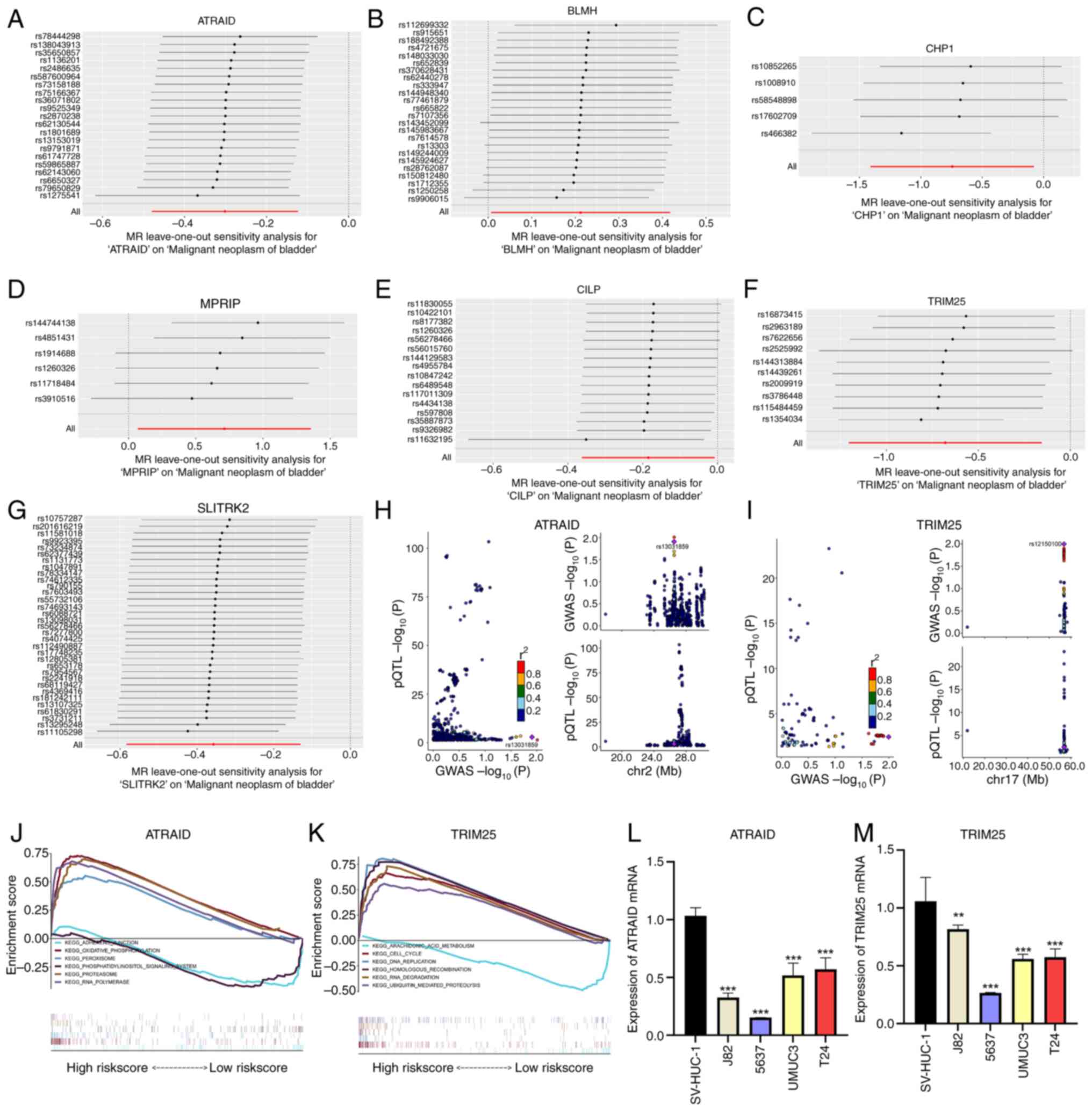
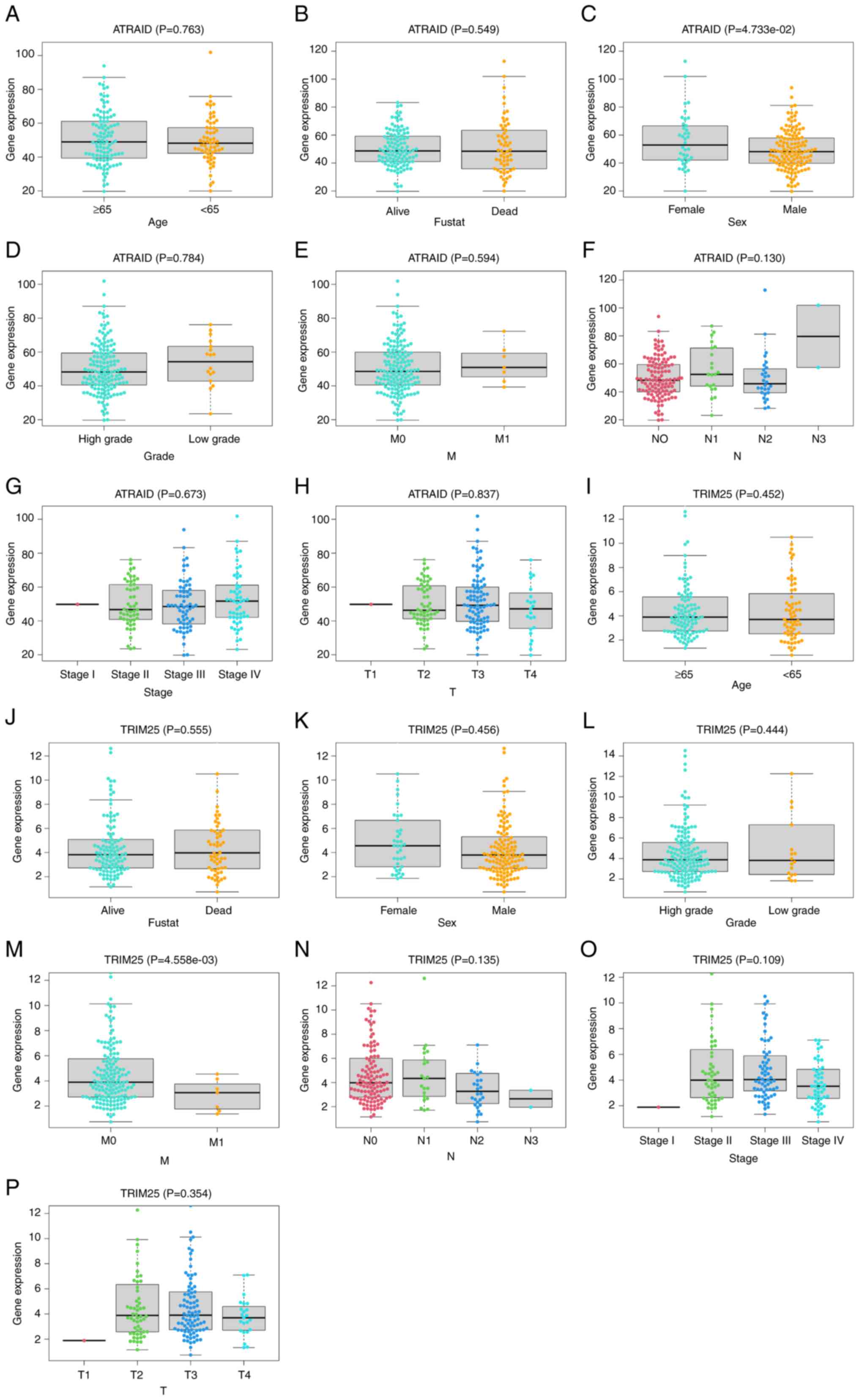
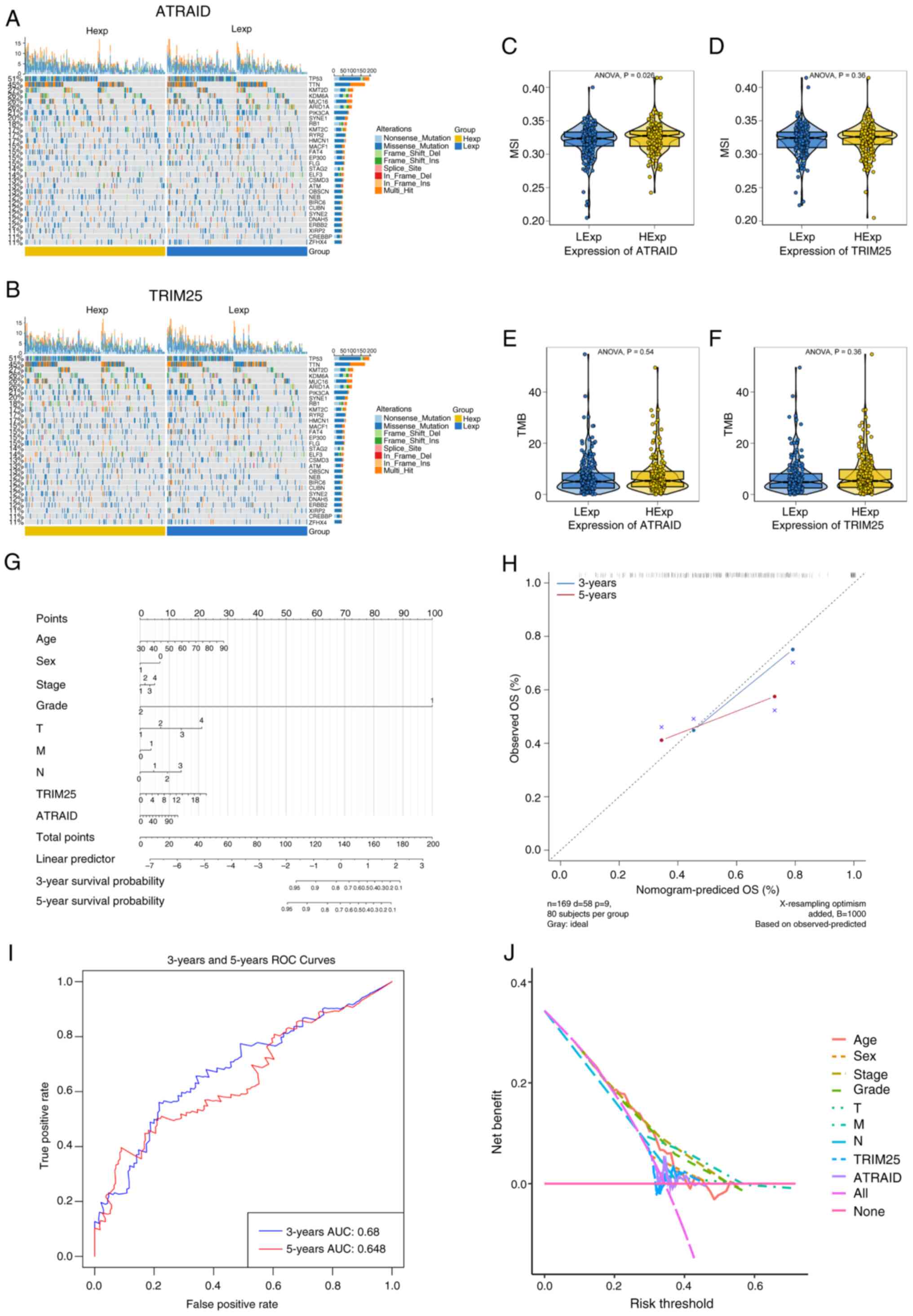
|
1
|
Zhang Y, Rumgay H, Li M, Yu H, Pan H and
Ni J: The global landscape of bladder cancer incidence and
mortality in 2020 and projections to 2040. J Glob Health.
13(04109)2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Peng M, Chu X, Peng Y, Li D, Zhang Z, Wang
W, Zhou X, Xiao D and Yang X: Targeted therapies in bladder cancer:
Signaling pathways, applications, and challenges. MedComm (2020).
4(e455)2023.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Thomas J and Sonpavde G: Molecularly
targeted therapy towards genetic alterations in advanced bladder
cancer. Cancers (Basel). 14(1795)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Anderson NL and Anderson NG: The human
plasma proteome: History, character, and diagnostic prospects. Mol
Cell Proteomics. 1:845–867. 2002.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Suhre K, McCarthy MI and Schwenk JM:
Genetics meets proteomics: Perspectives for large population-based
studies. Nat Rev Genet. 22:19–37. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Roslan A, Sulaiman N, Mohd Ghani KA and
Nurdin A: Cancer-associated membrane protein as targeted therapy
for bladder cancer. Pharmaceutics. 14(2218)2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sarafidis M, Lambrou GI, Zoumpourlis V and
Koutsouris D: An integrated bioinformatics analysis towards the
identification of diagnostic, prognostic, and predictive key
biomarkers for urinary bladder cancer. Cancers (Basel).
14(3358)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kojima T, Kawai K, Miyazaki J and
Nishiyama H: Biomarkers for precision medicine in bladder cancer.
Int J Clin Oncol. 22:207–213. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pietzner M, Wheeler E, Carrasco-Zanini J,
Cortes A, Koprulu M, Wörheide MA, Oerton E, Cook J, Stewart ID,
Kerrison ND, et al: Mapping the proteo-genomic convergence of human
diseases. Science. 374(eabj1541)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ferkingstad E, Sulem P, Atlason BA,
Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, Gunnarsdottir K,
Helgason A, Oddsson A, Halldorsson BV, et al: Large-scale
integration of the plasma proteome with genetics and disease. Nat
Genet. 53:1712–1721. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang J, Dutta D, Köttgen A, Tin A,
Schlosser P, Grams ME and Harvey B: CKDGen Consortium. Yu B,
Boerwinkle E, et al: Plasma proteome analyses in individuals of
European and African ancestry identify cis-pQTLs and models for
proteome-wide association studies. Nat Genet. 54:593–602.
2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sun BB, Maranville JC, Peters JE, Stacey
D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran
P, et al: Genomic atlas of the human plasma proteome. Nature.
558:73–79. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Suhre K, Arnold M, Bhagwat AM, Cotton RJ,
Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, DeLisle RK, et
al: Connecting genetic risk to disease end points through the human
blood plasma proteome. Nat Commun. 8(14357)2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Davies NM, Holmes MV and Davey Smith G:
Reading mendelian randomisation studies: A guide, glossary, and
checklist for clinicians. BMJ. 362(k601)2018.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Xin J, Gu D, Chen S, Ben S, Li H, Zhang Z,
Du M and Wang M: SUMMER: A Mendelian randomization interactive
server to systematically evaluate the causal effects of risk
factors and circulating biomarkers on pan-cancer survival. Nucleic
Acids Res. 51 (D1):D1160–D1167. 2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sun BB, Chiou J, Traylor M, Benner C, Hsu
YH, Richardson TG, Surendran P, Mahajan A, Robins C,
Vasquez-Grinnell SG, et al: Plasma proteomic associations with
genetics and health in the UK Biobank. Nature. 622:329–338.
2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yu Z, Liao J, Chen Y, Zou C, Zhang H,
Cheng J, Liu D, Li T, Zhang Q, Li J, et al: Single-cell
transcriptomic map of the human and mouse bladders. J Am Soc
Nephrol. 30:2159–2176. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Burgess S, Butterworth A and Thompson SG:
Mendelian randomization analysis with multiple genetic variants
using summarized data. Genet Epidemiol. 37:658–665. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bowden J, Davey Smith G and Burgess S:
Mendelian randomization with invalid instruments: Effect estimation
and bias detection through Egger regression. Int J Epidemiol.
44:512–525. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bowden J, Davey Smith G, Haycock PC and
Burgess S: Consistent estimation in mendelian randomization with
some invalid instruments using a weighted median estimator. Genet
Epidemiol. 40:304–314. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hartwig FP, Davey Smith G and Bowden J:
Robust inference in summary data Mendelian randomization via the
zero modal pleiotropy assumption. Int J Epidemiol. 46:1985–1998.
2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Giambartolomei C, Vukcevic D, Schadt EE,
Franke L, Hingorani AD, Wallace C and Plagnol V: Bayesian test for
colocalisation between pairs of genetic association studies using
summary statistics. PLoS Genet. 10(e1004383)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bonneville R, Krook MA, Kautto EA, Miya J,
Wing MR, Chen HZ, Reeser JW, Yu L and Roychowdhury S: Landscape of
microsatellite instability across 39 cancer types. JCO Precis
Oncol. 2017(PO.17.00073)2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Geeleher P, Cox N and Huang RS:
pRRophetic: An R package for prediction of clinical
chemotherapeutic response from tumor gene expression levels. PLoS
One. 9(e107468)2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hao Y, Hao S, Andersen-Nissen E, Mauck WM
III, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al:
Integrated analysis of multimodal single-cell data. Cell.
184:3573–3587.e29. 2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Cieslak MC, Castelfranco AM, Roncalli V,
Lenz PH and Hartline DK: t-Distributed stochastic neighbor
embedding (t-SNE): A tool for eco-physiological transcriptomic
analysis. Mar Genomics. 51(100723)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang Q, Yu B, Zhang Y, Tian Y, Yang S,
Chen Y and Wu H: Combination of single-cell and bulk RNA seq
reveals the immune infiltration landscape and targeted therapeutic
drugs in spinal cord injury. Front Immunol.
14(1068359)2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Aran D, Looney AP, Liu L, Wu E, Fong V,
Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al:
Reference-based analysis of lung single-cell sequencing reveals a
transitional profibrotic macrophage. Nat. Immunol. 20:163–172.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang W, Lu Z, Wang M, Liu Z, Wu B, Yang C,
Huan H and Gong P: The cuproptosis-related signature associated
with the tumor environment and prognosis of patients with glioma.
Front Immunol. 13(998236)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li Y, Zou X, Gao J, Cao K, Feng Z and Liu
J: APR3 modulates oxidative stress and mitochondrial function in
ARPE-19 cells. FASEB J: fj201800001RR, 2018 (Epub ahead of
print).
|
|
34
|
Zhu F, Yan W, Zhao ZL, Chai YB, Lu F, Wang
Q, Peng WD, Yang AG and Wang CJ: Improved PCR-based subtractive
hybridization strategy for cloning differentially expressed genes.
Biotechniques. 29:310–313. 2000.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang Y, Li Q, Huang W, Zhang J, Han Z,
Wei H, Cui J, Wang Y and Yan W: Increased expression of
apoptosis-related protein 3 is highly associated with tumorigenesis
and progression of cervical squamous cell carcinoma. Hum Pathol.
44:388–393. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang P, Zhou C, Jing Q, Gao Y, Yang L, Li
Y, Du J, Tong X and Wang Y: Role of APR3 in cancer: Apoptosis,
autophagy, oxidative stress, and cancer therapy. Apoptosis.
28:1520–1533. 2023.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cao HL, Gu MQ, Sun Z and Chen ZJ:
miR-144-3p contributes to the development of thyroid tumors through
the PTEN/PI3K/AKT pathway. Cancer Manag Res. 12:9845–9855.
2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kwon SC, Yi H, Eichelbaum K, Föhr S,
Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW and Kim VN:
The RNA-binding protein repertoire of embryonic stem cells. Nat
Struct Mol Biol. 20:1122–1130. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Choudhury NR, Heikel G, Trubitsyna M,
Kubik P, Nowak JS, Webb S, Granneman S, Spanos C, Rappsilber J,
Castello A and Michlewski G: RNA-binding activity of TRIM25 is
mediated by its PRY/SPRY domain and is required for ubiquitination.
BMC Biol. 15(105)2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Heikel G, Choudhury NR and Michlewski G:
The role of Trim25 in development, disease and RNA metabolism.
Biochem Soc Trans. 44:1045–1050. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liu Y, Tao S, Liao L, Li Y, Li H, Li Z,
Lin L, Wan X, Yang X and Chen L: TRIM25 promotes the cell survival
and growth of hepatocellular carcinoma through targeting Keap1-Nrf2
pathway. Nat Commun. 11(348)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Rahimi-Tesiye M, Zaersabet M, Salehiyeh S
and Jafari SZ: The role of TRIM25 in the occurrence and development
of cancers and inflammatory diseases. Biochim Biophys Acta Rev
Cancer. 1878(188954)2023.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Shu XS, Zhao Y, Sun Y, Zhong L, Cheng Y,
Zhang Y, Ning K, Tao Q, Wang Y and Ying Y: The epigenetic modifier
PBRM1 restricts the basal activity of the innate immune system by
repressing retinoic acid-inducible gene-I-like receptor signalling
and is a potential prognostic biomarker for colon cancer. J Pathol.
244:36–48. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Li YH, Zhong M, Zang HL and Tian XF: The
E3 ligase for metastasis associated 1 protein, TRIM25, is targeted
by microRNA-873 in hepatocellular carcinoma. Exp Cell Res.
368:37–41. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang J, Yin G, Bian H, Yang J, Zhou P, Yan
K, Liu C, Chen P, Zhu J, Li Z and Xue T: LncRNA XIST upregulates
TRIM25 via negatively regulating miR-192 in hepatitis B
virus-related hepatocellular carcinoma. Mol Med.
27(41)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Dai H, Zhao S, Xu L, Chen A and Dai S:
Expression of Efp, VEGF and bFGF in normal, hyperplastic and
malignant endometrial tissue. Oncol Rep. 23:795–799.
2010.PubMed/NCBI
|
|
47
|
Dai H, Zhang P, Zhao S, Zhang J and Wang
B: Regulation of the vascular endothelial growth factor and growth
by estrogen and antiestrogens through Efp in Ishikawa endometrial
carcinoma cells. Oncol Rep. 21:395–401. 2009.PubMed/NCBI
|
|
48
|
Yang KS, Xu CQ and Lv J: Identification
and validation of the prognostic value of cyclic GMP-AMP
synthase-stimulator of interferon (cGAS-STING) related genes in
gastric cancer. Bioengineered. 12:1238–1250. 2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wang S, Kollipara RK, Humphries CG, Ma SH,
Hutchinson R, Li R, Siddiqui J, Tomlins SA, Raj GV and Kittler R:
The ubiquitin ligase TRIM25 targets ERG for degradation in prostate
cancer. Oncotarget. 7:64921–64931. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Qin X, Chen S, Qiu Z, Zhang Y and Qiu F:
Proteomic analysis of ubiquitination-associated proteins in a
cisplatin-resistant human lung adenocarcinoma cell line. Int J Mol
Med. 29:791–800. 2012.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Walsh LA, Alvarez MJ, Sabio EY, Reyngold
M, Makarov V, Mukherjee S, Lee KW, Desrichard A, Turcan Ş, Dalin
MG, et al: An integrated systems biology approach identifies TRIM25
as a key determinant of breast cancer metastasis. Cell Rep.
20:1623–1640. 2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Li F, Sun Q, Liu K, Zhang L, Lin N, You K,
Liu M, Kon N, Tian F, Mao Z, et al: OTUD5 cooperates with TRIM25 in
transcriptional regulation and tumor progression via
deubiquitination activity. Nat Commun. 11(4184)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhenilo S, Deyev I, Litvinova E, Zhigalova
N, Kaplun D, Sokolov A, Mazur A and Prokhortchouk E: DeSUMOylation
switches Kaiso from activator to repressor upon hyperosmotic
stress. Cell Death Differ. 25:1938–1951. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Tang H, Li X, Jiang L and Liu Z, Chen L,
Chen J, Deng M, Zhou F, Zheng X and Liu Z: RITA1 drives the growth
of bladder cancer cells by recruiting TRIM25 to facilitate the
proteasomal degradation of RBPJ. Cancer Sci. 113:3071–3084.
2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Li W, Shen Y, Yang C, Ye F, Liang Y, Cheng
Z, Ou Y, Chen W, Chen Z, Zou L, et al: Identification of a novel
ferroptosis-inducing micropeptide in bladder cancer. Cancer Lett.
582(216515)2024.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Eberhardt W, Nasrullah U and Pfeilschifter
J: TRIM25: A global player of cell death pathways and promising
target of tumor-sensitizing therapies. Cells. 14(65)2025.PubMed/NCBI View Article : Google Scholar
|