1. Introduction
Cancer remains one of the leading causes of
morbidity and mortality worldwide, despite notable advances in
diagnostic technologies and therapeutic interventions. Traditional
treatments such as chemotherapy and radiotherapy often cause
damaging side effects, exhibit limited specificity and frequently
lead to therapeutic resistance, thus collectively undermining the
quality of life of patients. For instance, ~40% of patients
undergoing chemotherapy experience severe adverse effects,
including gastrointestinal toxicity and myelosuppression (1,2),
while ~50% of those with advanced solid tumors develop drug
resistance (3,4). These limitations underscore an urgent
need for more precise and effective therapeutic strategies.
Recent advances in molecular biology and
computational analysis have revealed that RNA regulatory networks
play pivotal roles in cancer progression by orchestrating key
biological processes such as proliferation, apoptosis, angiogenesis
and drug resistance (5,6). Non-coding RNAs (ncRNAs), including
microRNAs (miRNAs or miRs), long non-coding RNAs (lncRNAs) and
their variants, have emerged as central players in these networks
(7,8). Notably, novel isoforms such as
isomiRs and mechanisms such as the competitive endogenous RNA
(ceRNA) network have introduced new layers of regulatory
complexity, with notable implications for cancer diagnostics and
treatment.
The present review provides a comprehensive and
integrative perspective on RNA regulatory networks in oncology,
with a particular focus on isomiRs and ceRNA interactions. Unlike
previous reviews, this article highlights how multi-omics
profiling, single-cell RNA sequencing (scRNA-seq), and CRISPR-based
functional screening enable high-resolution mapping of RNA
interactions and support their translation into clinical
applications. These tools offer the potential to address persistent
challenges in the field, such as dynamic heterogeneity,
spatiotemporal regulation and the inference of causality within
complex networks (9-11).
By synthesizing recent findings, the present article
highlights emerging molecular mechanisms, clinical relevance and
the translational potential of RNA networks in cancer. In addition,
barriers to their application were also discussed, particularly in
multi-omics integration, and promising technological directions
that may redefine precision oncology in the future were
described.
Recent clinical experiences have underscored the
need to align early clinical signals with mechanistic annotation in
RNA oncology. Durable benefit appears to depend as much on delivery
engineering and proactive mitigation of innate-immune sensing as on
the RNA payload itself. Representative programs using optimized
carriers and tissue-directed delivery (for example, lipid
nanoparticles and ligand conjugates) illustrate how organ/lesion
exposure, endosomal escape, and immune quieting can determine
translational success (12,13).
Specific trial details are summarized in ‘Strategies and
prospects’, and failure modes with practical lessons are discussed
in ‘Existing problems’.
2. Basic composition and mechanism of action
of RNA regulatory networks
Understanding the regulatory interplay among
different RNA species is fundamental to deciphering cellular
homeostasis and disease pathogenesis. In the following section, the
basic functions of mRNA, miRNA and lncRNA were reviewed, the
theoretical basis of the competitive ceRNA network was discussed
and the generation of isomiRs and their unique regulatory roles
were described.
Introduction to the basic functions of
mRNA, miRNA and lncRNA
mRNAs are the key intermediates between DNA and
proteins. They carry genetic information from the nucleus to the
cytoplasm, where they serve as templates for protein synthesis. The
translation of mRNAs into proteins is tightly regulated at multiple
levels, ensuring that proteins are produced in response to the
developmental stage of the cell and environmental cues. Previous
studies have shown that alterations in mRNA expression patterns are
not only reflective of cellular states, but also actively
contribute to the progression of diseases, including cancer
(5,7,11).
In contrast to mRNAs, miRNAs are short,
~22-nucleotide ncRNAs that function predominantly in
post-transcriptional gene regulation. By binding to complementary
sequences in the 3' untranslated regions (3' UTRs) of target mRNAs,
miRNAs mediate mRNA degradation or inhibit translation. This mode
of regulation is highly conserved and enables a single miRNA to
target multiple mRNAs simultaneously, thus orchestrating complex
gene expression networks (14,15).
lncRNAs are a diverse group of transcripts that are
>200 nucleotides in length, and lack protein-coding potential.
Despite their inability to code for proteins, lncRNAs are emerging
as critical regulators of gene expression. They modulate chromatin
structure, influence transcriptional activity and participate in
post-transcriptional regulatory events. Notably, numerous lncRNAs
function as molecular sponges for miRNAs by sequestering them away
from their target mRNAs and thereby modulating downstream signaling
pathways (16,17). Recent advances in single-cell
sequencing have elucidated the cell type-specific expression and
function of lncRNAs, reinforcing their importance in both
physiological and pathological contexts (18).
Collectively, the interplay between mRNAs, miRNAs
and lncRNAs forms the backbone of the RNA regulatory network,
ensuring precise control over gene expression in both normal
cellular processes and in diseases such as cancer.
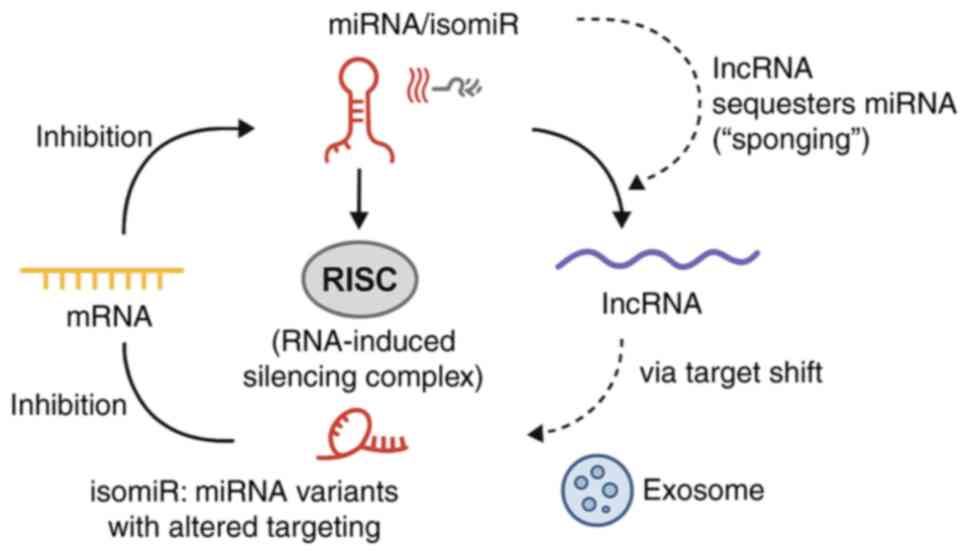
Theoretical basis of the ceRNA
network
The ceRNA hypothesis, first proposed in 2011, has
fundamentally reshaped the understanding of post-transcriptional
gene regulation. This model posits that RNA molecules containing
shared miRNA response elements (MREs) can compete for binding to
miRNAs, thereby influencing the stability and translational
efficiency of target mRNAs (16).
In essence, RNA transcripts, including mRNAs, lncRNAs and
pseudogene-derived RNAs, can function as ‘molecular sponges’,
sequestering miRNAs and reducing their availability to other
targets, thus indirectly modulating gene expression across a
regulatory network.
Several factors determine the efficacy of ceRNA
interactions. The foremost among these is the relative abundance of
the competing RNAs, whereby highly expressed ceRNAs are more likely
to effectively bind and sequester miRNAs. This concept is
formalized in the ‘miRNA sponge threshold hypothesis’, which posits
that only ceRNAs expressed above a certain quantitative threshold
can exert functional competition. Mathematical models have
substantiated this hypothesis, demonstrating non-linear regulatory
outcomes in response to ceRNA concentration (19).
In addition to transcript abundance, the number and
binding affinity of MREs within each RNA molecule shape ceRNA
regulatory capacity. For instance, transcripts harboring multiple
high-affinity sites for a given miRNA may be more susceptible to
ceRNA-mediated modulation than those with fewer or weaker binding
motifs (17,20). Furthermore, subcellular
localization plays a critical role, as ceRNA interactions are
spatially restricted to cellular compartments where miRNAs and
their targets are co-localized.
Emerging evidence has reinforced the biological
relevance of ceRNA networks, particularly in the context of cancer.
These interactions have been linked to tumor proliferation,
metastasis, immune evasion and therapeutic resistance.
Context-specific ceRNA interactions have been identified in breast,
liver, lung, pancreatic and brain cancers, demonstrating diverse
regulatory roles across tumor types (17,21,22).
A comprehensive overview of these lncRNA-miRNA interactions in
various cancers further underscores their central regulatory role
and translational potential (23).
CeRNA axes discovered in discovery sets have
reproduced prognostic separation in independent patient cohorts,
including proliferation-linked networks in lung adenocarcinoma
(LUAD) and colon cancer (24,25).
Nevertheless, the generalizability of the ceRNA
model across all cancer types remains under debate. Increasing
evidence suggests that ceRNA network robustness is highly
context-dependent, varying substantially between tumor types.
Factors such as tissue-specific RNA expression, tumor subtype
heterogeneity and the tumor microenvironment (TME) can constrain or
enhance ceRNA-mediated regulation. These foundational interactions
within the RNA regulatory landscape are illustrated in Fig. 1.
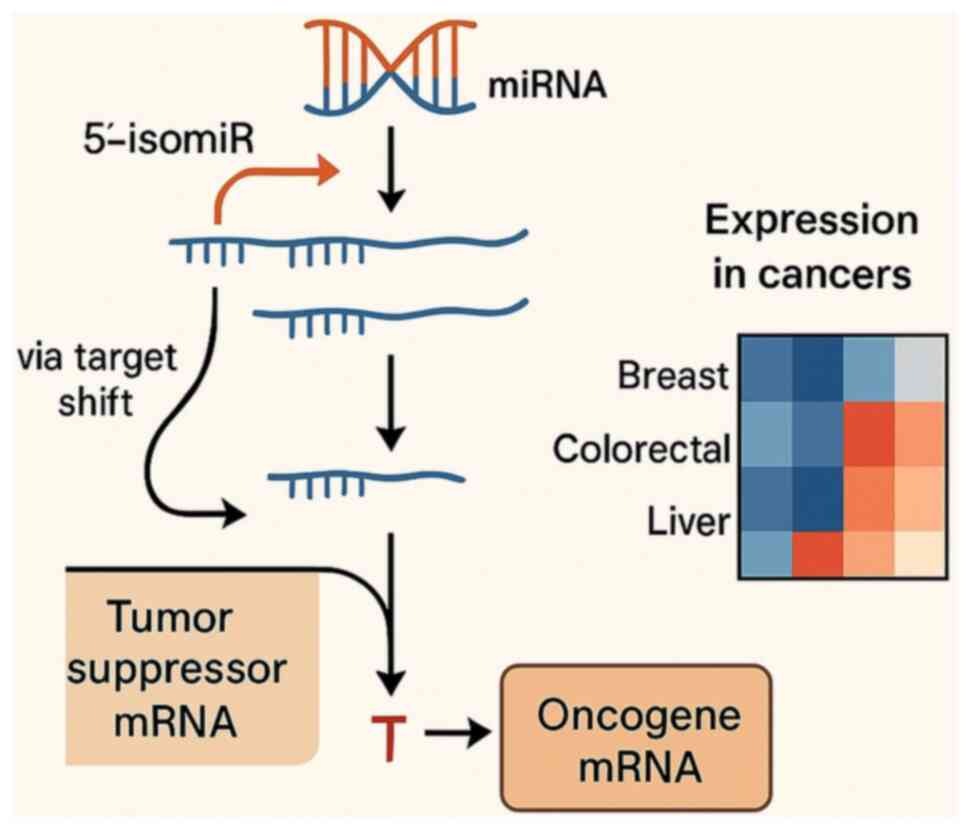
Generation of isomiRs and their
regulatory importance
Canonical miRNA loci produce multiple sequence
variants, termed isomiRs, through imprecise cleavage by
Drosha/Dicer and post-transcriptional tailing or trimming (such as
adenylation/uridylation) (17,20,23).
These edits alter the length and sequence at the 5' or 3' ends.
Alterations at the 5' end are particularly consequential because
they shift the seed (nts 2-8), thereby redefining the target
repertoire and potentially rewiring regulatory programs. By
contrast, numerous 3' variants primarily influence Argonaute
loading, stability or binding affinity without altering seed
identity (20).
In cancer, isomiRs show context-specific expression
relative to canonical miRNAs and to matched normal tissues,
implicating them in tumor-type-dependent pathway control (17,20).
They may also reshape ceRNA competition by engaging distinct MREs,
modifying post-transcriptional regulation in a manner that depends
on both sequence and tissue context. This principle is illustrated
in Fig. 2, using 5'-isomiRs with
notable tumor-normal differences in three TCGA cohorts, including
breast (BRCA), colorectal (COAD/READ) and liver (LIHC), displayed
as z-scored log2(RPM+1) heatmaps. The cancer-type-specific patterns
oppose a pan-cancer stereotype and instead support selective
deployment of 5'-isomiRs across tumor contexts.
Despite their promise, the functional independence
of numerous isomiRs remains debated. Some 5'-shifted variants
exhibit distinct targetomes and phenotypes, whereas others appear
largely redundant with their canonical counterparts (20,26).
Most evidence to date is correlative or prediction-based. Rigorous
validation such as isoform-resolved CLIP/CLASH, seed-swap
reporters, rescue assays and locus-specific perturbations are still
needed to establish causal mechanisms and to assess biomarker or
therapeutic value at isoform resolution.
3. Role of the RNA regulatory network in
cancer occurrence and progression
RNA regulatory networks play a pivotal role in
tumorigenesis by modulating important cellular processes such as
cell cycle control, proliferation, apoptosis and the TME. The
interplay among various RNA species, including mRNAs, miRNAs,
lncRNAs and other ncRNAs, provides a complex regulatory framework
that influences cancer initiation, progression and therapeutic
response. A comprehensive understanding of these networks not only
elucidates the molecular mechanisms driving cancer but also
highlights potential targets for intervention.
RNA regulatory networks in cell cycle,
proliferation and apoptosis
Dysregulation of the cell cycle and proliferation
pathways is a hallmark of cancer. Under physiological conditions,
cyclins, cyclin-dependent kinases (CDKs) and their inhibitors
coordinate orderly progression through the cell cycle. Disruptions
in these circuits can lead to uncontrolled cell division. RNA
regulatory networks, particularly those involving ceRNA
interactions, modulate the expression of these critical regulators.
For example, miRNAs targeting cyclin genes (such as CCNA2 and
CCND1) or CDK inhibitors (such as p21 and p27) can alter cell cycle
dynamics, thereby promoting or inhibiting tumor growth (27).
In parallel, these RNA networks also regulate
apoptosis by balancing the expression of pro- and anti-apoptotic
factors such as BCL2, BAX and CASP9. Dysregulation of RNA-mediated
controls may tip this balance toward survival, enabling cancer
cells to evade programmed cell death and develop therapeutic
resistance. Certain lncRNAs have been shown to stabilize mRNAs
encoding anti-apoptotic proteins, further contributing to tumor
progression (28).
MiRNA signatures that regulate these circuits (for
example, miR-21 and miR-145) correlate with recurrence and survival
in retrospective/prospective cohorts and in liquid-biopsy studies,
underscoring translational relevance (29,30-33).
Moreover, emerging evidence suggests that isomiRs,
which are functionally diverse miRNA isoforms, can fine-tune RNA
regulatory effects by altering target specificity, thereby
influencing processes such as proliferation and apoptosis in a
context-dependent manner (29).
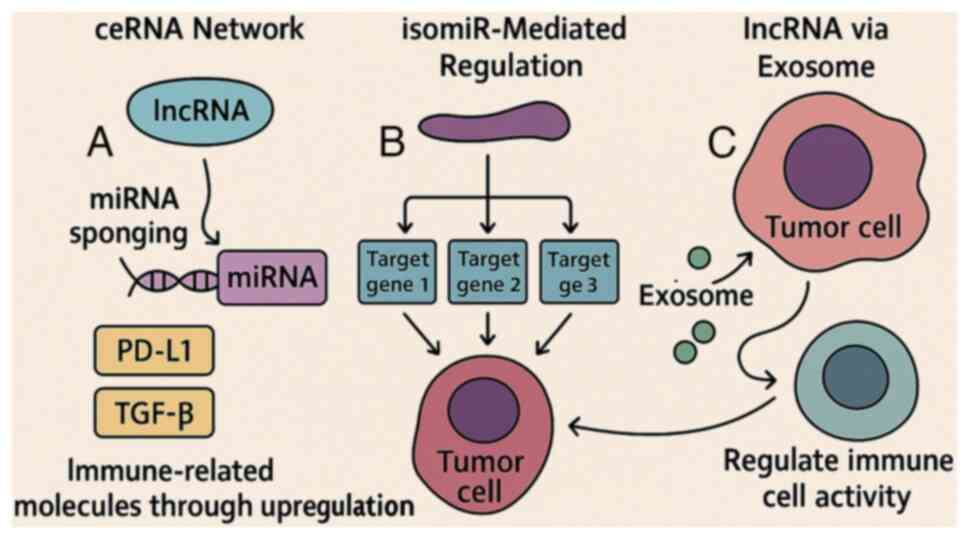
RNA interactions and regulation of the
TME
Beyond their roles in intrinsic tumor cell
functions, RNA regulatory networks profoundly influence the TME.
ncRNAs, including miRNAs and lncRNAs, are not only active
intracellularly, but can also be secreted via extracellular
vesicles (such as exosomes) to mediate intercellular communication
with immune cells, fibroblasts and endothelial cells (34,35).
These interactions affect immune infiltration, cytokine production,
angiogenesis and extracellular matrix remodeling, creating a
microenvironment conducive to tumor progression (36).
For instance, some miRNAs suppress T-cell activation
or promote the polarization of macrophages toward an
immunosuppressive M2 phenotype, facilitating immune escape
(28). Similarly, lncRNAs can act
as decoys or scaffolds that sequester transcription factors and
chromatin remodelers involved in immune regulation. This
multilayered regulatory network underscores the potential of
targeting RNA interactions to enhance immunotherapy and overcome
resistance. Ongoing clinical studies are exploring strategies to
disrupt oncogenic RNA circuits or restore tumor-suppressive RNAs to
therapeutically reprogram the TME (37).
These RNA-mediated mechanisms of immune modulation
are schematically summarized in Fig.
3.
Case studies in various cancer
types
LUAD: ceRNA networks in cell cycle regulation and
prognosis. In LUAD, ceRNA networks have been implicated in
regulating cell cycle-related gene expression and tumor
proliferation, with prognostic importance. A representative ceRNA
axis involved the lncRNA VPS9D1-AS1, miR-30a-5p and downstream
targets such as CCNA2, KIF11 and MKI67(24). VPS9D1-AS1 acts as a molecular
sponge for miR-30a-5p, thereby upregulating key mitotic regulators
and proliferation markers. This dysregulation contributes to
unchecked tumor growth. Notably, large-scale bioinformatics
analyses have highlighted these ceRNA circuits as potential
biomarkers for patient stratification and therapeutic
targeting.
Cholangiocarcinoma: SNHG6/miR-101-3p/E2F8
axis. In cholangiocarcinoma, the lncRNA SNHG6 serves as
a ceRNA that sequesters miR-101-3p, leading to derepression of the
transcription factor E2F8, which promotes cell cycle
progression. This ceRNA axis enhances proliferation, migration and
angiogenesis in cholangiocarcinoma cells, supporting its role in
aggressive tumor phenotypes (38).
The SNHG6/miR-101-3p/E2F8 pathway exemplifies the mechanisms
by which ceRNA dysregulation facilitates oncogenic processes
through post-transcriptional control.
Other cancer types: Breast, colorectal and liver
cancer. RNA regulatory networks are also perturbed in other
cancer types. In breast cancer, lncRNAs and miRNAs jointly modulate
the PI3K/AKT/mTOR signaling pathway, contributing to metastasis and
therapy resistance (39-41).
In colorectal cancer, the LCMT1-AS2/miR-454-3p/RPS6KA5 ceRNA
axis functions as a prognostic indicator of poor survival (25). In hepatocellular carcinoma, ncRNAs
orchestrate PI3K/AKT signaling cascades to influence angiogenesis
and immune evasion (42,43).
These case studies underscore the context-specific
architecture and functional diversity of ceRNA networks across
different malignancies (44).
Despite shared mechanisms, such as lncRNA sponging and
miRNA-mediated repression, the biological outcomes are highly
dependent on tumor type, target gene context and network topology.
Understanding these disease-specific ceRNA circuits is important
for developing targeted RNA-based interventions in precision
oncology. These differences in ceRNA network strength, biological
relevance and experimental validation across cancer types are
systematically summarized in Table
I.
 | Table IComparative effectiveness of ceRNA
networks across cancer types. |
Table I
Comparative effectiveness of ceRNA
networks across cancer types.
| First author/s,
year | Cancer type | Regulatory
strength | Key influencing
factors | Representative
ceRNA axis | Functional
validation method | (Refs.) |
|---|
| Zhang et al,
2014 | Breast cancer | High | High lncRNA and
miRNA abundance |
HOTAIR/miR-331-3p/HER2 | In vitro
knockdown and luciferase reporter assays | (89) |
| Poliseno et
al, 2010 | Glioblastoma | Low | Low miRNA levels,
high heterogeneity |
PTENP1/miR-19b/PTEN | Bioinformatics
analysis, low experimental confirmation | (90) |
| Chen et al,
2021 | Hepatocellular
carcinoma | Medium-High | High lncRNA
expression, immune complexity |
LINC00160/miR-132/AKT1 | Animal models and
expression rescue experiments | (91) |
| Xie et al,
2020 | Pancreatic
cancer | Low | Immunosuppressive
tumor microenvironment, scarce lncRNAs |
MALAT1/miR-200c/ZEB1 | Limited functional
studies | (92) |
4. Application of RNA regulatory networks in
cancer diagnosis and prognosis
As the understanding of RNA regulatory networks
improves, their clinical implications in oncology have become
increasingly evident. From identifying prognostic markers to
guiding molecular classification and precision medicine, RNA-based
tools hold promise for improving cancer diagnosis, treatment and
patient outcomes.
Screening of prognostic markers based
on RNA networks
RNA molecules, including mRNAs, miRNAs, lncRNAs and
circRNAs, have gained traction as diagnostic and prognostic
biomarkers due to their tissue-specific expression patterns,
stability in biofluids and critical regulatory roles in
tumorigenesis (45). In
particular, panels of differentially expressed RNAs have been
correlated with disease stages and patient survival across various
cancer types (30,31). For instance, specific lncRNAs have
been linked to metastasis and recurrence in breast and colorectal
cancers, while unique miRNA signatures have been proposed as early
detection markers for lung and pancreatic cancers (32). Multi-center studies of circulating
RNAs report diagnostic AUCs in the ~0.80-0.90 range for several
malignancies, and tissue lncRNA/miRNA panels track stage,
recurrence and survival (30,31,33,45-47).
Moreover, circulating RNAs, those detectable in
plasma, serum and exosomes, offer a minimally invasive means of
cancer detection and monitoring (33). By quantifying changes in RNA
expression, clinicians can potentially identify high-risk patients,
track tumor progression and evaluate therapeutic response. Despite
these advances, larger, multicenter clinical studies are needed to
validate the robustness, reproducibility and clinical utility of
RNA-based prognostic panels before they can be widely implemented
in routine practice (48).
Role of RNA molecules in molecular
typing and precision medicine
RNA signatures have proven invaluable for the
molecular classification of tumors, which is a key step in
precision medicine. For example, gene expression profiling has led
to the identification of distinct subtypes in breast cancer (such
as luminal A, luminal B, HER2-enriched and basal-like) and the
stratification of diffuse large B-cell lymphoma into molecularly
defined categories (49). These
classifications guide therapeutic decisions, such as the use of
targeted therapies (such as HER2 inhibitors in HER2-enriched breast
cancer) and immunotherapies for tumors harboring specific
expression profiles.
Additionally, RNA-based molecular typing extends
beyond mRNA to include ncRNAs. LncRNAs and miRNAs have emerged as
critical regulators of oncogenic pathways and have been associated
with tumor subtypes, therapeutic resistance and disease outcomes
(46). Integrating RNA signatures
into diagnostic frameworks enables clinicians to tailor treatment
regimens to the unique molecular features of tumors of individual
patients, improving efficacy and minimizing toxicity (47).
Clinical detection technology and its
limitations
Various technologies have been developed to harness
RNA molecules for cancer diagnosis and prognosis. High-throughput
sequencing (RNA-seq) enables comprehensive transcriptome profiling,
allowing detection of novel transcripts and splicing variants
(50). Microarray-based platforms
and reverse transcription-quantitative PCR are widely used for
validating candidate RNA biomarkers due to their ease of use and
cost-effectiveness (51). More
recently, digital droplet PCR (ddPCR) has gained popularity for its
high sensitivity in detecting low-abundance circulating RNAs
(52).
Despite these advances, challenges persist.
Variability in sample collection, RNA extraction and data analysis
can affect the sensitivity and reproducibility of RNA-based
diagnostics. Standardization of protocols and optimization across
pre-analytical steps are important (53). Furthermore, tumor heterogeneity and
the dynamic nature of RNA expression require repeated sampling to
capture the evolving transcriptome. Large, multicenter clinical
studies and robust bioinformatics pipelines are crucial to ensure
the clinical utility of RNA-based tests.
Clinical applications and
translational progress
Despite the growing body of evidence supporting
RNA-based biomarkers and regulatory networks in cancer biology,
their clinical translation remains limited. However, several
RNA-targeted therapeutics have made notable strides. For instance,
Givosiran and Inclisiran, two small interfering RNA (siRNA) drugs
approved by the FDA, have demonstrated the feasibility of RNA-based
therapy in clinical settings, although outside oncology. In the
cancer field, MRX34, a liposomal mimic of the tumor-suppressive
miR-34a, was the first miRNA therapeutic to enter clinical trials,
marking a milestone despite its early termination (54).
In parallel, RNA expression profiling has become a
valuable tool in molecular subtyping and precision medicine. For
example, the PAM50 RNA panel informs hormone and HER2-targeted
therapies in breast cancer based on intrinsic molecular subtypes
(49). However, several
technological barriers still hinder the routine clinical adoption
of RNA-based diagnostics. Although ddPCR offers high sensitivity
for detecting low-abundance RNAs in liquid biopsies, its limited
multiplexing capability and cost pose challenges for large-scale
implementation (52). Similarly,
while RNA sequencing provides comprehensive transcriptomic data,
issues such as batch effects, variable library preparation and
complex data analysis reduce inter-laboratory reproducibility
(51).
To address these challenges, emerging strategies
focus on standardizing workflows, minimizing RNA input requirements
and developing FDA-aligned diagnostic guidelines. These
developments represent a critical transition from bench to bedside,
underscoring the translational promise of RNA regulatory networks
in precision oncology.
5. Strategies and prospects of RNA
regulatory networks in cancer treatment
RNA regulatory networks have become attractive
therapeutic targets in oncology. By modulating miRNAs, lncRNAs and
their downstream effectors, it may be possible to slow tumor
progression, overcome drug resistance and improve outcomes
(55,56).
Therapeutic strategies targeting RNA
networks
miRNA/lncRNA mimics and inhibitors restore
tumor-suppressive functions or block oncogenic ncRNAs, using
synthetic oligonucleotides such as mimics, antagomiRs and ASOs
(57-59).
Modulating ceRNA interactions represents another avenue:
Downregulating oncogenic lncRNAs or other competing RNAs can
liberate tumor-suppressive miRNAs to repress their targets
(60).
Clinical status and delivery
lessons
The liposomal miR-34a mimic MRX34 reached
first-in-human testing but was terminated for immune-related
toxicities, highlighting the need to minimize innate immune
activation and to optimize delivery and preclinical toxicology
(61). By contrast, siRNA agents
have demonstrated that careful carrier design enables
tissue-specific delivery with manageable safety, as illustrated by
patisiran in non-oncology indications (12) and by RNA nanoparticles with
favorable biodistribution and conditional endosomal escape
(13). Oncology-focused programs
span locoregional depots such as siG12D-LODER for KRAS^G12D
pancreatic cancer (62,63), systemically delivered LNP siRNAs
such as DCR-MYC (64,65), and newer platforms exemplified by
the GSTP-targeting NBF-006 evaluated in KRAS-mutant NSCLC (66,67).
Collectively, these data emphasize the centrality of delivery
engineering, target and context selection, and early immune-risk
mitigation, which are elaborated below.
6. Existing problems and limitations
While RNA regulatory networks present promise in
cancer research and treatment, several challenges remain in
understanding their full potential. Data integration, technical
limitations and the need for more refined experimental approaches
hinder the progress in this field. Addressing these issues will be
crucial for optimizing RNA-based therapeutic strategies and
improving the clinical application of RNA regulatory networks in
oncology.
Challenges of data integration and
network construction in existing studies
Integrating high-throughput RNA sequencing data
remains a major challenge due to the complexity of transcriptomic
interactions. RNA molecules, including mRNAs, miRNAs, lncRNAs and
isomiRs, form intricate, context-dependent networks that vary
across tissue types, disease stages and microenvironments. A single
RNA species may regulate or interact with multiple targets in
diverse ways. However, most current reconstruction methods rely on
statistical correlations rather than causal or directional
relationships, resulting in oversimplified or incomplete network
models (68). While numerous
computational tools have been developed to predict miRNA-lncRNA
interactions, such as correlation-based and
sequence-complementarity methods, these tools often produce
inconsistent outputs and require further experimental validation
(69).
The lack of standardized computational pipelines and
network inference frameworks further impedes data integration and
cross-platform comparability. Although numerous tools have been
developed, few can integrate multi-omics datasets (such as RNA-seq
with ATAC-seq) at the single-cell level to construct cell
type-specific regulatory maps (70). Emerging tools such as IReNA and
pySCENIC attempt to address these issues by incorporating
transcription factor motifs, chromatin accessibility and regulatory
logic into network inference (71). However, robust integration of
large-scale omics data and reproducibility across computational
platforms remain unresolved challenges.
Moreover, scRNA-seq faces limitations in detecting
low-abundance RNAs such as miRNAs and lncRNAs. These datasets are
also prone to batch effects and technical noise, affecting
reproducibility and interpretation. Although multi-omics
integration tools are advancing, fully capturing dynamic RNA
regulatory networks at single-cell resolution remains an ongoing
area of research (10).
Technical and biological problems in
isomiR research
IsomiRs, the variant forms of miRNAs, have garnered
increasing attention for their ability to expand the miRNA
regulatory landscape. However, several technical and biological
challenges hinder their accurate identification and functional
characterization. A major limitation lies in the resolution of
current sequencing technologies, which may fail to detect
low-abundance isomiRs or differentiate between closely related
isoforms, particularly those with 5' seed shifts, which can alter
target specificity (26).
Furthermore, incomplete or inconsistent annotation in public
databases hampers reliable mapping and interpretation (20).
Biologically, the roles of isomiRs in cancer remain
only partially understood. Variants that differ at the 5' end may
possess novel seed sequences, leading to distinct mRNA targeting
and potentially divergent effects on oncogenic signaling pathways
(17). Functional validation of
such isomiRs is still limited, and systematic studies across
different tissues and tumor types are required to determine their
contribution to tumorigenesis, metastasis and therapy resistance.
Advances such as dual-index library construction for reproducible
isomiR sequencing (26), along
with isomiR-specific databases [such as isomiRTar (72) and TIE], are beginning to address
some of these limitations and offer promising platforms for future
isomiR discovery and clinical application.
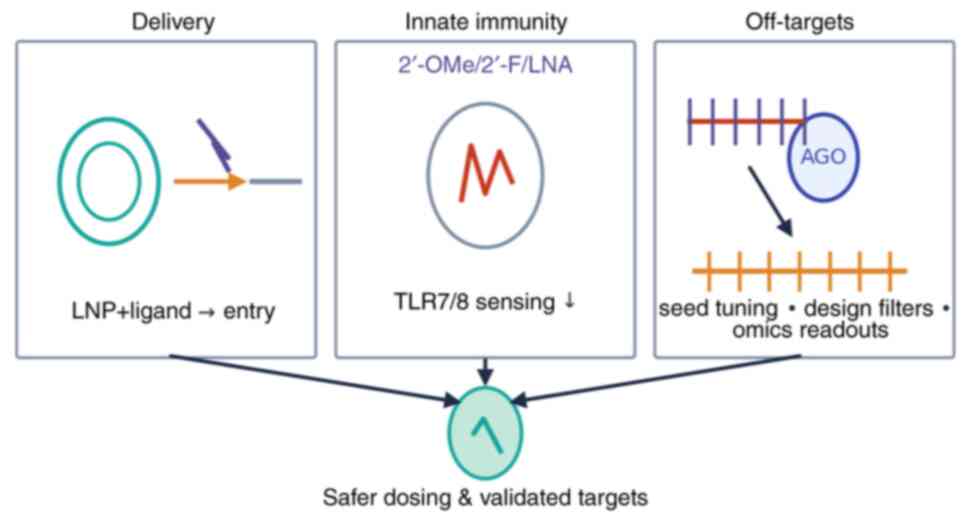
In the present review, three recurrent challenges
that have hampered the clinical success of RNA therapeutics in
oncology (delivery efficiency, innate immune activation, and
off-target effects) were highlighted, and each paired with
evidence-informed mitigation strategies.
Delivery. Effective delivery to tumor tissue
remains a major obstacle. For systemically administered agents, the
composition of lipid nanoparticles (LNPs), including the
pKa of ionizable lipids, helper lipids, and PEG-lipids,
critically influences stability, cellular uptake and endosomal
escape (73-75).
Clinical experience demonstrates that rational LNP design can
successfully translate to humans, as exemplified by patisiran,
which achieved robust hepatic gene silencing using an MC3-based LNP
and exhibited a manageable safety profile (12). Tumor selectivity can be further
enhanced through ligand-mediated targeting (for example, GalNAc,
antibodies, or aptamers) or via locoregional depot systems.
Oncology case studies (for example, siG12D-LODER and DCR-MYC) have
been described in Section 5, illustrating how delivery mode, namely
local depot versus systemic LNP, critically shapes tumor exposure
and clinical feasibility (62-65).
Practical strategies to improve delivery include refining ionizable
lipid structures, modulating PEG content, incorporating validated
targeting ligands, utilizing localized administration where
possible, and prioritizing the assessment of tumor exposure and
endosomal escape during early development (68-78).
Innate immune activation. The case of MRX34,
which was discontinued despite evidence of target engagement,
highlights the clinical consequences of uncontrolled innate immune
stimulation, often mediated through endosomal Toll-like receptors
(for example, TLR7/8) (61).
Mitigation approaches incorporate sequence ‘de-immunization’ using
chemically modified nucleotides (such as 2'-OMe, 2'-F, or LNA),
optimized PEGylation strategies, and early in vitro
screening using human whole-blood assays or PBMC cultures to
quantify cytokine release (79,80).
These measures reduce TLR-dependent immune recognition, dampen
excessive cytokine production, and help establish safer starting
doses and premedication regimens for first-in-human trials
(63,79,80).
Off-target effects. siRNA candidates can
induce seed region-mediated off-target silencing via interactions
with 3' UTRs, leading to unintended transcriptome-wide effects
(81). Comprehensive off-target
profiling involves coupling AGO2-centered methods (for example,
HITS-CLIP or PAR-CLIP) with dose-responsive transcriptomics
(RNA-seq) and proteomics. Computational filtering and CRISPR-based
validation help establish causal relationships between observed
effects and off-target binding (82,83).
Design-based solutions include modifying the seed sequence,
introducing chemical modifications that reduce miRNA-like activity,
and selecting guide strands with minimal predicted off-target
potential in relevant cellular models (81-83).
Functional verification of
isomiRs
Demonstrating function requires evidence of seed
dependence, direct target engagement and a measurable cellular
consequence in the relevant context. To establish robust evidence
for isomiR-mediated targeting, a multi-tier framework can be
applied. Initial screening with dual-luciferase reporter assays
comparing wild-type versus seed-mutant 3'UTRs, including canonical
and 5'-shifted isomiRs, provides seed-specific activity, while
seed-reversion rescues confirm specificity (61). Genetic perturbation at the
pri-miRNA locus (CRISPRi/a or base editing) modulates isomiR
production and can be linked to phenotypic outcomes using pooled
single-cell transcriptomics such as Perturb-seq or CROP-seq
(79,80). Direct binding is then established
by AGO-CLIP (HITS-CLIP or PAR-CLIP) (61), and functional repression verified
by dose-responsive RNA-seq and quantitative proteomics (61). At higher resolution, these steps
can be integrated with single-cell multi-omics to assign effects to
specific cell states (68,69,73).
Seed specificity can be assessed by comparing
canonical and 5'-shifted variants in dual-luciferase assays and by
reversing the altered seed (81).
Causality in living cells can be tested via CRISPR-based
perturbation of isomiR production or processing, with single-cell
transcriptomic readouts when cell-state resolution is required.
Physical binding to predicted sites is evaluated with AGO-centered
CLIP (HITS-CLIP or PAR-CLIP), and downstream impact is quantified
under therapeutically relevant exposures using dose-ranged RNA-seq
and quantitative proteomics (82,83).
Implications and new directions for
future cancer treatment strategies
Despite ongoing challenges, RNA-based therapeutics
are advancing rapidly, driven by innovations in molecular design,
delivery systems and target specificity. An increasing number of
therapeutic agents, including miRNA mimics, lncRNA inhibitors and
small molecules that modulate RNA-RNA or RNA-protein interactions,
are under active development. These approaches offer fine-tuned
control over key pathways such as cell proliferation, apoptosis and
immune regulation, particularly in tumors resistant to traditional
treatments (60).
At the same time, progress in RNA drug delivery is
expanding the clinical applicability of these strategies. LNP
formulations, ligand-targeted conjugates and chemically modified
oligonucleotides are being optimized to enhance stability, reduce
off-target effects and achieve tumor-specific accumulation. The
success of RNA-based drugs in non-cancer diseases, such as
Patisiran for transthyretin-mediated amyloidosis, provides valuable
insights for oncology applications.
In the future, RNA therapeutics are hypothesized to
integrate more deeply into combination regimens alongside
immunotherapies, targeted therapies and radiotherapy. Such
synergistic approaches may improve treatment efficacy while
minimizing systemic toxicity. Moreover, advances in biomarker
discovery, particularly RNA-based expression signatures, will
facilitate patient stratification and response monitoring,
supporting a shift toward more personalized cancer care.
Ultimately, the continued refinement of RNA
therapeutics, along with robust clinical validation and regulatory
frameworks, will be important to fully realize their potential in
precision oncology.
Emerging frontiers: Hot topics in RNA
regulatory network research
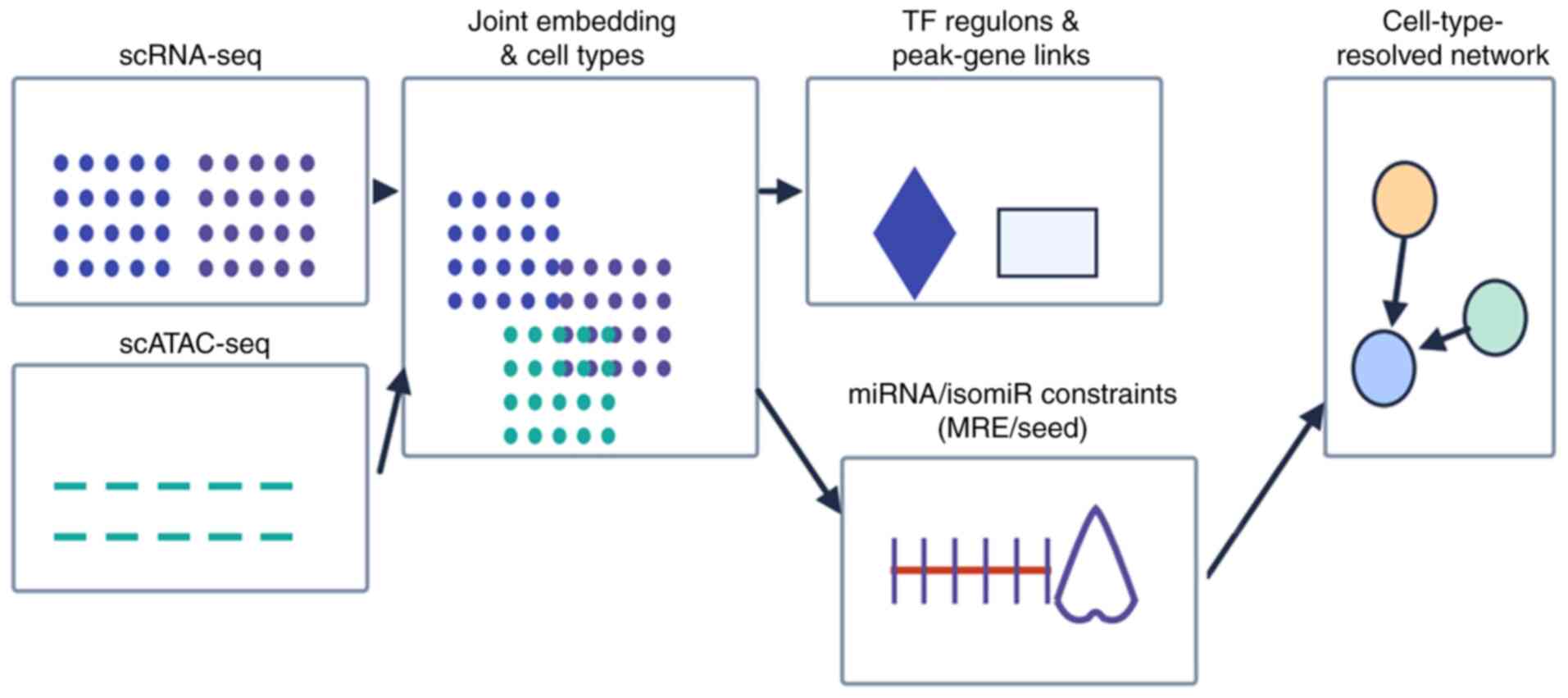
Single-cell multi-omics for high-resolution
regulatory mapping. A compact workflow for single-cell
multi-omics integration [scRNA-seq + single-cell assay for
transposase-accessible chromatin (scATAC-seq) ± spatial] is
provided in Fig. 4, from joint
embedding and TF/peak-gene linking to MRE-constrained edge
inference and cell type-resolved networks.
Concise description of single-cell multi-omics
integration. ScRNA-seq and scATAC-seq datasets were
quality-controlled and batch-corrected to obtain a joint embedding
and a reproducible cell-type map (68,69).
Chromatin peaks are linked to genes by correlating accessibility
with expression within cell types and by motif/footprint analysis
to define transcription-factor regulons with per-cell activity
scores (71). Integrated
regulatory networks are inferred with methods that couple scRNA-seq
and chromatin-accessibility constraints (such as IReNA) (70). Candidate ceRNA/isomiR relationships
are then filtered by the presence of microRNA-response elements
and, where available, orthogonal support from isomiR-target
resources (72). When spatial
transcriptomics are available, edges that co-localize within the
TME are prioritized. Robustness is assessed by stability across
donors/batches and held-out performance (68,73).
Single-cell sequencing technologies, particularly
when integrated with transcriptomic, epigenomic and proteomic
modalities (such as scRNA-seq, scATAC-seq and CITE-seq), are
transforming the current understanding of RNA regulatory networks
in cancer. These tools enable the resolution of cell type-specific
expression profiles and uncover spatial and temporal heterogeneity
in ceRNA and isomiR interactions, especially within the TME
(10,84). Such fine-grained insights are
crucial for identifying rare subpopulations that drive therapy
resistance, metastasis or immune escape.
Previous efforts have also applied single-cell
spatial transcriptomics to validate ceRNA networks in situ,
revealing topological constraints and microenvironmental influences
on RNA-based regulation (16,19,23).
These approaches provide unprecedented opportunities to reconstruct
RNA interaction maps at a cellular resolution.
Emerging multi-omics integration strategies are
moving beyond single-cell approaches to provide robust, pan-cancer
insights into post-transcriptional regulation. Representative cases
illustrate this evolution:
In cholangiocarcinoma, integrated transcriptomic and
proteomic profiling, combining RNA-seq with quantitative mass
spectrometry, has uncovered functional isomiR-mediated networks.
Predictions of isomiR-target interactions were corroborated by
corresponding protein-level changes, highlighting cross-regulatory
RNA crosstalk and prioritizing mechanistically relevant edges for
experimental validation (21).
Pan-cancer integrative frameworks have further
enabled the identification of consensus ceRNA networks across large
cohorts. By aggregating miRNA-lncRNA-mRNA data from multiple tumor
types, these approaches derive robust cross-cancer regulatory
edges, which are subsequently evaluated through dataset stability
tests and survival analysis, offering a reproducible template for
bulk multi-omics synthesis (23,68,69).
Curated multi-omics databases now systematically
consolidate single-cell, spatial and bulk genomic data to enhance
ceRNA inference. These resources support cross-modality validation,
such as confirming co-expression in bulk datasets, spatial
co-localization in tissue maps, and recurrence of edges across
cancer types, enabling more reliable triage of candidate
interactions (10).
Additionally, constraint-guided methods that
incorporate chromatin features, such as motif accessibility,
foot-printing data, and open chromatin states, are increasing the
specificity of regulatory network inference. Although often applied
in single-cell analyses, these priors are equally valuable in bulk
multi-omics integration, where they help reduce false-positive
predictions and refine edges based on functional genomic
constraints (70,71).
CRISPR-based functional validation and network
causality. Traditional studies of RNA networks rely heavily on
correlational analyses, which limit causal inference. CRISPR-based
screening technologies, particularly CRISPR interference and
activation (CRISPRa), now offer a powerful means of interrogating
RNA function. When combined with single-cell transcriptomics (such
as Perturb-seq and CRISPRa-Perturb-seq), these tools allow
high-throughput, context-specific validation of lncRNAs, miRNAs and
ceRNA nodes (9,85). These screens have successfully
identified key regulatory elements that contribute to drug
resistance and tumor progression, enhancing the biological
credibility of proposed networks. As RNA-targeted therapies
continue to develop, such functional genomics approaches will play
a pivotal role in bridging predictive models and therapeutic
translation.
RNA structural biology and non-canonical
regulatory layers. Beyond sequence-level regulation, emerging
evidence points to the importance of RNA secondary and tertiary
structures, especially G-quadruplexes (G4s), in shaping RNA
regulatory landscapes. G4 motifs are highly stable, guanine-rich
structures found in both coding and non-coding regions, where they
influence miRNA binding, splicing and translation (17,86).
These structures can modulate ceRNA stability or alter the
seed-region recognition capacity of isomiRs, introducing novel
dimensions of regulatory control. Structure-based targeting of RNA
is gaining traction as a therapeutic strategy. Molecules that
selectively bind or stabilize RNA G4s have shown antiproliferative
effects in cancer models, offering new possibilities for
intervention (87,88). Future work may focus on
programmable modulation of such structures using CRISPR-guided
systems or synthetic ligands.
These emerging technologies, including single-cell
multi-omics, CRISPR-based functional validation and RNA
structure-focused regulation, are redefining the landscape of RNA
network research. Their integration will unlock novel therapeutic
avenues and deepen the mechanistic understanding of cancer biology
at single-cell and molecular resolutions.
Dynamic heterogeneity in RNA
regulatory networks and its regulatory mechanisms
Recent studies have revealed that cellular
heterogeneity within tumors and their microenvironments influence
the construction and function of RNA regulatory networks (10,20,33).
First, distinct cell types, such as immune cells versus tumor
cells, exhibit fundamentally different RNA network regulatory
mechanisms. Immune cells, which play key roles in antigen
presentation, cytokine secretion and immune modulation, tend to
have ceRNA networks tailored to regulating immune responses. By
contrast, tumor cells often display ceRNA networks that primarily
promote proliferation, evade immune surveillance and drive
metabolic reprogramming. For example, immune cell ceRNA
interactions are closely linked to immune response regulation,
whereas in tumor cells these networks are more frequently involved
in cell cycle and metabolic pathway regulation (37).
Furthermore, there is mounting evidence that ceRNA
networks undergo dynamic remodeling during tumor progression. In
early-stage tumors, ceRNA networks may serve to maintain a balance
between proliferation and apoptosis. However, as tumors advance,
these networks can be restructured to support metastasis, drug
resistance and immune evasion. In LUAD, key ceRNA components have
been observed to undergo notable changes in both expression and
interaction patterns as the disease progresses, indicating a
dynamic adjustment of the network architecture over time (24).
Lastly, integrating time- and space-resolved
expression data is crucial for reconstructing accurate RNA
regulatory maps. Single-cell multi-omics and spatial
transcriptomics technologies now enable researchers to capture RNA
expression dynamics across different tumor regions and time points.
For instance, the LnCeCell 2.0 database leverages such data to
reveal the spatiotemporal specificity of ceRNA interactions among
various cell types, providing powerful tools for dissecting the
complex heterogeneity of tumor tissues (10).
Collectively, these insights underscore the
importance of considering both cellular and temporal heterogeneity
in RNA regulatory network analyses. A deeper understanding of these
dynamic changes not only enriches the knowledge of tumor biology
but also informs the development of personalized therapeutic
strategies based on the spatiotemporal modulation of gene
expression. These considerations are consolidated in Fig. 5, which links the major constraints
to practical mitigation levers and validation readouts.
7. Future prospects and emerging
directions
The rapid evolution of RNA regulatory research is
opening new frontiers in cancer biology and therapy. While marked
progress has been made in identifying the components and functions
of RNA networks, several challenges remain. These include the need
for robust, context-specific functional validation, improved
delivery of RNA-targeted therapeutics and standardization in data
integration across multi-omics platforms.
In the future, a key direction may be the
convergence of mechanistic insight and clinical application.
High-resolution technologies such as single-cell multi-omics and
CRISPR-based screening may continue to play a vital role in
decoding network dynamics and identifying cell-specific
vulnerabilities. However, their full translational potential
depends on the development of clinically viable tools for
manipulating RNA interactions in vivo, including
programmable RNA editors, structure-targeted ligands and
combination therapies that modulate multiple layers of regulation
simultaneously.
Another promising avenue lies in integrating
RNA-based approaches with other therapeutic modalities, such as
immunotherapy and metabolism-targeting drugs. RNA biomarkers and
regulatory signatures may enable more refined patient
stratification and dynamic monitoring of treatment response,
supporting the vision of adaptive, personalized oncology.
Ultimately, the future of RNA regulatory network
research hinges on the ability of researchers to move from
descriptive models to predictive, actionable frameworks. This may
require not only technological innovation but also
interdisciplinary collaboration across molecular biology,
bioinformatics, structural biology and clinical oncology. With
continued progress, RNA networks may shift from being viewed as
passive readouts of gene activity to becoming active therapeutic
blueprints that guide next-generation cancer treatment.
Design implications begin with a biology-driven
approach: Prioritizing clinical evaluation in adjuvant or minimal
residual disease settings, or other immune-permissive contexts, to
maximize therapeutic responsiveness (12,13,79,80).
When monotherapy cytotoxicity is insufficient, intentional
combination strategies, such as pairing RNA-based agents with
checkpoint inhibitors or pathway-sensitizing therapeutics, should
be employed (66,67,79,80).
Emphasis should be placed on target quality and dependency rather
than tumor mutational burden alone, favoring high-quality
neoantigens or functionally validated targets (79,80).
Biomarker plans must be pre-specified, integrating
diagnostic-enrichment-pharmacodynamic (D-E-P) chains and systematic
tissue or ctDNA sampling into the trial's statistical design
(12,13). Furthermore, innate immune sensing
should be de-risked prior to first-in-human studies through
optimized sequence chemistry, formulation, and delivery routes, as
underscored by the clinical experience with MRX34, where
immune-related toxicity led to discontinuation despite evidence of
target engagement (61). By
contrast, previous systemic siRNA programs demonstrate that careful
carrier tuning and chemical modification can markedly improve
uptake and endosomal escape while mitigating immunostimulatory
risks (64,67).
8. Conclusion
RNA regulatory networks offer compelling
opportunities to improve cancer diagnosis, prognosis and treatment,
yet translation is constrained by challenges in data integration,
context-specific network inference, delivery and the isoform-level
biology of regulators such as isomiRs. New modalities including
single-cell multi-omics and CRISPR-based functional genomics are
enabling causal mapping and more reliable biomarkers and
targets.
Real-world experience points to clear design rules
for RNA therapeutics: Optimize delivery, proactively mitigate
innate immune sensing, and build rigorous, isoform-aware
validation. If these hurdles are addressed through integrated
multi-omics and improved delivery platforms, RNA networks can
realize their potential to transform precision oncology.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Key
Research and Development Program of China (grant no.
2022YFF1301602) and the National Natural Science Foundation of
China (grant nos. 32570497, 32270442, 31872219 and 31370401).
Availability of data and materials
Not applicable.
Authors' contributions
WR conceived the topic area, conducted literature
searches, evaluated all relevant literature, designed the research
framework, wrote the manuscript, and critically revised the
intellectual content. The author read and approved the final
version of the manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The author declares that he has no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, artificial
intelligence tools were used to improve the readability and
language of the manuscript, and subsequently, the authors revised
and edited the content produced by the artificial intelligence
tools as necessary, taking full responsibility for the ultimate
content of the present manuscript.
References
|
1
|
Nurgali K, Jagoe RT and Abalo R: Adverse
effects of cancer chemotherapy: Anything new to improve tolerance
and reduce sequelae? Front Pharmacol. 9(245)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pearce A, Haas M, Viney R, Pearson SA,
Haywood P, Brown C and Ward R: Incidence and severity of
self-reported chemotherapy side effects in routine care: A
prospective cohort study. PLoS One. 12(e0184360)2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Vasan N, Baselga J and Hyman DM: A view on
drug resistance in cancer. Nature. 575:299–309. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang X, Zhang H and Chen X: Drug
resistance and combating drug resistance in cancer. Cancer Drug
Resist. 2:141–160. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang Y, Tang J, Wang C, Zhang Q, Zeng A
and Song L: Autophagy-related lncRNAs in tumor progression and drug
resistance: A double-edged sword. Genes Dis. 11:367–381.
2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18.
2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yan H and Bu P: Non-coding RNA in cancer.
Essays Biochem. 65:625–639. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mitra R, Adams C and Eischen C: Decoding
the lncRNAome across diverse cellular stresses reveals core
p53-effector pan-cancer suppressive lncRNAs. Cancer Res Commun.
3:842–859. 2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Guo Q, Liu Q, He D, Xin M, Dai Y, Sun R,
Li H, Zhang Y, Li J, Kong C, et al: LnCeCell 2.0: An updated
resource for lncRNA-associated ceRNA networks and web tools based
on single-cell and spatial transcriptomics sequencing data. Nucleic
Acids Res. 53:D107–D115. 2025.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Slack FJ and Chinnaiyan AM: The role of
non-coding RNAs in oncology. Cell. 179:1033–1055. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Adams D, Gonzalez-Duarte A, O'Riordan WD,
Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk
JL, et al: Patisiran, an RNAi therapeutic, for hereditary
transthyretin amyloidosis. N Engl J Med. 379:11–21. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xu C, Haque F, Jasinski DL, Binzel DW, Shu
D and Guo P: Favorable biodistribution, specific targeting and
conditional endosomal escape of RNA nanoparticles in cancer
therapy. Cancer Lett. 414:57–70. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Naeli P, Winter T, Hackett AP, Alboushi L
and Jafarnejad SM: The intricate balance between microRNA-induced
mRNA decay and translational repression. FEBS J. 290:2508–2524.
2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Nejad C, Stunden HJ and Gantier MP: A
guide to miRNAs in inflammation and innate immune responses. FEBS
J. 285:3695–3716. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zelli V, Compagnoni C, Capelli R, Corrente
A, Cornice J, Vecchiotti D, Di Padova M, Zazzeroni F, Alesse E and
Tessitore A: Emerging role of isomiRs in cancer: State of the art
and recent advances. Genes (Basel). 12(1447)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kim GD, Shin SI, Jung SW, An H, Choi SY,
Eun M, Jun CD, Lee S and Park J: Cell type- and age-specific
expression of lncRNAs across kidney cell types. J Am Soc Nephrol.
35:870–885. 2024.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bosia C, Pagnani A and Zecchina R:
Modelling competing endogenous RNA networks. PLoS One.
8(e66609)2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wagner V, Meese E and Keller A: The
intricacies of isomiRs: From classification to clinical relevance.
Trends Genet. 40:784–796. 2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Guo L, Dou Y, Yang Y, Zhang S, Kang Y,
Shen L, Tang L, Zhang Y, Li C, Wang J, et al: Protein profiling
reveals potential isomiR-associated cross-talks among RNAs in
cholangiocarcinoma. Comput Struct Biotechnol J. 19:5722–5734.
2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Drillis G, Goulielmaki M, Spandidos DA,
Aggelaki S and Zoumpourlis V: Non-coding RNAs (miRNAs and lncRNAs)
and their roles in lymphogenesis in all types of lymphomas and
lymphoid malignancies. Oncol Lett. 21(393)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sun B, Liu C, Li H, Zhang L, Luo G, Liang
S and Lü M: Research progress on the interactions between long
non-coding RNAs and microRNAs in human cancer. Oncol Lett.
19:595–605. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yang Y, Zhang S and Guo L:
Characterization of cell cycle-related competing endogenous RNAs
using robust rank aggregation as prognostic biomarker in lung
adenocarcinoma. Front Oncol. 12(807367)2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liang F, Xu Y, Zheng H and Tang W:
Establishing a carcinoembryonic antigen-associated competitive
endogenous RNA network and forecasting an important regulatory axis
in colon adenocarcinoma patients. J Gastrointest Oncol. 15:220–236.
2024.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang J, Zhang W, Zhang Y, Zhou J, Li J,
Zhang M, Wen S, Gao X, Zhou N, Li H, et al: Reproducible and high
sample throughput isomiR next-generation sequencing for cancer
diagnosis. J Clin Oncol. 42(e15013)2024.
|
|
27
|
Rahmani F, Zandigohar M, Safavi P, Behzadi
M, Ghorbani Z, Payazdan M, Ferns G, Hassanian SM and Avan A: The
interplay between noncoding RNAs and p21 signaling in
gastrointestinal cancer: From tumorigenesis to metastasis. Curr
Pharm Des. 29:766–776. 2023.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yu Z, Rong Z, Sheng J, Luo Z, Zhang J, Li
T, Zhu Z, Fu Z, Qiu Z and Huang C: Aberrant non-coding RNA
expressed in gastric cancer and its diagnostic value. Front Oncol.
11(606764)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ruan Z, Chi D, Wang Q, Jiang J, Quan Q,
Bei J and Peng R: Development and validation of a prognostic model
and gene co-expression networks for breast carcinoma based on
scRNA-seq and bulk-seq data. Ann Transl Med.
10(1333)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Rahman MS, Ghorai S, Panda K, Santiago MJ,
Aggarwal S, Wang T, Rahman I, Chinnapaiyan S and Unwalla HJ: Dr.
Jekyll or Mr. Hyde: The multifaceted roles of miR-145-5p in human
health and disease. Noncoding RNA Res. 11:22–37. 2024.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yuan X, Mao Y and Ou S: Diagnostic
accuracy of circulating exosomal circRNAs in malignancies: A
meta-analysis and systematic review. Medicine (Baltimore).
102(e33872)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang D, Zhang W, Zhang C, Wang L, Chen H
and Xu J: Exosomal non-coding RNAs have a significant effect on
tumor metastasis. Mol Ther Nucleic Acids. 29:16–35. 2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Natarelli N, Boby A, Aflatooni S, Tran JT,
Diaz MJ, Taneja K and Forouzandeh M: Regulatory miRNAs and lncRNAs
in skin cancer: A narrative review. Life (Basel).
13(1696)2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Benedetti A, Turco C, Fontemaggi G and
Fazi F: Non-coding RNAs in the crosstalk between breast cancer
cells and tumor-associated macrophages. Noncoding RNA.
8(16)2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhan DT and Xian HC: Exploring the
regulatory role of lncRNA in cancer immunity. Front Oncol.
13(1191913)2023.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang H, Wang L, Tang L, Luo J, Ji H, Zhang
W, Zhou J, Li Q and Miao L: Long noncoding RNA SNHG6 promotes
proliferation and angiogenesis of cholangiocarcinoma cells through
sponging miR-101-3p and activation of E2F8. J Cancer. 11:3002–3012.
2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Abu-Alghayth MH, Khan FR, Belali TM,
Abalkhail A, Alshaghdali K, Nassar SA, Almoammar NE, Almasoudi HH,
Hessien KBG, Aldossari MS and Binshaya AS: The emerging role of
noncoding RNAs in the PI3K/AKT/mTOR signalling pathway in breast
cancer. Pathol Res Pract. 255(155180)2024.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Golhani V, Ray SK and Mukherjee S: Role of
microRNAs and long non-coding RNAs in regulating angiogenesis in
human breast cancer: A molecular medicine perspective. Curr Mol
Med. 22:882–893. 2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zheng M, Wu L, Xiao RC, Zhou Y, Cai J and
Shen SR: Integrated analysis of coexpression and a tumor-specific
ceRNA network revealed a potential prognostic biomarker in breast
cancer. Transl Cancer Res. 12:949–964. 2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hussain MS, Moglad E, Afzal M, Gupta G,
Almalki WH, Kazmi I, Alzarea SI, Kukreti N, Gupta S, Kumar D, et
al: Non-coding RNA mediated regulation of PI3K/Akt pathway in
hepatocellular carcinoma: therapeutic perspectives. Pathol Res
Pract. 258(155303)2024.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Liu J, Xiao S and Chen J: Development of
an inflammation-related lncRNA-miRNA-mRNA network based on
competing endogenous RNA in breast cancer at single-cell
resolution. Front Cell Dev Biol. 10(839876)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhan H, Tu S, Zhang F, Shao A and Lin J:
MicroRNAs and long non-coding RNAs in c-Met-regulated cancers.
Front Cell Dev Biol. 8(145)2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Cui J, Chen M, Zhang L, Huang S, Xiao F
and Zou L: Circular RNAs: Biomarkers of cancer. Cancer Innov.
1:197–206. 2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Panoutsopoulou K, Avgeris M and Scorilas
A: miRNA and long non-coding RNA: molecular function and clinical
value in breast and ovarian cancers. Expert Rev Mol Diagn.
18:963–979. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhong X, Li J, Wu X, Wu X, Hu L, Ding B
and Qian L: Identification of N6-methyladenosine-related lncRNAs
for predicting overall survival and clustering of a potentially
novel molecular subtype of breast cancer. Front Oncol.
11(742944)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Helsmoortel H, Everaert C, Lumen N, Ost P
and Vandesompele J: Detecting long non-coding RNA biomarkers in
prostate cancer liquid biopsies: Hype or hope? Noncoding RNA Res.
3:64–74. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Islakoglu YO, Noyan S, Aydos A and
Dedeoglu BG: Meta-microRNA biomarker signatures to classify breast
cancer subtypes. OMICS. 22:709–716. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lee BI, Oades K, Vo L, Lee J, Landers M,
Wang Y and Monforte J: NGS-based targeted RNA sequencing for
expression analysis of patients with triple-negative breast cancer
using a modulized, 96-gene biomarker panel. J Clin Oncol.
30(56)2012.
|
|
51
|
Narrandes S and Xu W: Gene expression
detection assay for cancer clinical use. J Cancer. 9:2249–2265.
2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Lange T, Gross T, Jeney Á, Scherzinger J,
Sinkala E, Niemöller C, Zimmermann S, Koltay P, Stetten F, Zengerle
R and Jeney C: Validation of scRNA-seq by scRT-ddPCR using the
example of ErbB2 in MCF7 cells. bioRxiv, 2022.
|
|
53
|
Poel D, Voortman J, Oord R, Gall H and
Verheul H: Standardization and optimization of circulating microRNA
serum profiling in patients with cancer. Cancer Res 75: doi.org/10.1158/1538-7445.AM2015-3991,
2015.
|
|
54
|
Ratti M, Lampis A, Ghidini M, Salati M,
Mirchev MB, Valeri N and Hahne JC: MicroRNAs (miRNAs) and long
non-coding RNAs (lncRNAs) as new tools for cancer therapy: First
steps from bench to bedside. Target Oncol. 15:261–278.
2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Slaby O, Laga R and Sedlacek O:
Therapeutic targeting of non-coding RNAs in cancer. Biochem J.
474:4219–4251. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Haryana S: Long non-coding RNA (lncRNA)
and microRNA (miRNA) in cancer management. J Med Sci.
48(35)2016.
|
|
57
|
Gambari R, Brognara E, Spandidos D and
Fabbri E: Targeting oncomiRNAs and mimicking tumor suppressor
miRNAs: New trends in the development of miRNA therapeutic
strategies in oncology. Int J Oncol. 49:5–32. 2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Pierouli K, Papakonstantinou E,
Papageorgiou L, Diakou I, Mitsis T, Dragoumani K, Spandidos DA,
Bacopoulou F, Chrousos GP, Goulielmos G, et al: Long noncoding RNAs
and microRNAs as regulators of stress in cancer. Mol Med Rep.
26(361)2022.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Agami R: Abstract CN01-03: Prospects for
miRNA- and lncRNA-based cancer therapeutics. Mol Cancer Ther.
12(CN01-03-CN01-03)2013.
|
|
60
|
Fu Z, Wang L, Li S, Chen F, Au-Yeung K and
Shi C: MicroRNA as an important target for anticancer drug
development. Front Pharmacol. 12(736323)2021.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Hong D, Kang Y, Borad M, Sachdev J, Ejadi
S, Lim H, Brenner A, Park K, Lee J, Kim T, et al: Phase 1 study of
MRX34, a liposomal miR-34a mimic, in patients with advanced solid
tumours. Br J Cancer. 122:1630–1637. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Varghese AM, Ang C, Dimaio CJ, Javle MM,
Gutierrez M, Yarom N, Stemmer SM, Golan T, Geva R, Semenisty V, et
al: A phase II study of siG12D-LODER in combination with
chemotherapy in patients with locally advanced pancreatic cancer
(PROTACT). J Clin Oncol. 38 (Suppl 15)(TPS4672)2020.
|
|
63
|
ClinicalTrials.gov: A phase 2 study of siG12D LODER
in combination with chemotherapy in patients with locally advanced
pancreatic cancer. Identifier: NCT01676259. Accessed Aug 21,
2025.
|
|
64
|
Tolcher AW, Papadopoulos KP, Patnaik A,
Rasco D, Martinez D, Wood D, Fielman B, Sharma MR, Janisch L, Brown
B, et al: Safety and activity of DCR-MYC, a first-in-class
Dicer-substrate siRNA targeting MYC, in a phase I study in patients
with advanced solid tumors. J Clin Oncol. 33 (Suppl
15)(11006)2015.
|
|
65
|
ClinicalTrials.gov: Phase Ib/II, multicenter dose
escalation study of DCR-MYC in hepatocellular carcinoma.
Identifier: NCT02314052 (status history/results). Accessed Aug 21,
2025.
|
|
66
|
Bazhenova L, Mamdani H, Chiappori A, Spira
A, Iams W, Tolcher A, Barve M, Gabayan A, Vandross A, Cina C, et
al: First-in-human dose-expansion study of NBF-006, a novel
investigational siRNA targeting GSTP, in patients with KRAS-mutated
non-small cell lung cancer. Cancer Res. 84 (Suppl
7)(CT040)2024.
|
|
67
|
Cina C, Majeti B, O'Brien Z, Wang L,
Clamme JP, Adami R, Tsang KY, Harborth J, Ying W and Zabludoff S: A
novel lipid nanoparticle NBF-006 encapsulating glutathione
S-Transferase P. siRNA for the treatment of KRAS-driven non-small
cell lung cancer. Mol Cancer Ther. 24:7–17. 2025.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Cantarella S, Di Nisio E, Carnevali D,
Dieci G and Montanini B: Interpreting and integrating big data in
non-coding RNA research. Emerg Top Life Sci. 3:343–355.
2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Veneziano D, Marceca GP, Di Bella S,
Nigita G, Distefano R and Croce CM: Investigating miRNA-lncRNA
interactions: Computational tools and resources. Methods Mol Biol.
1970:251–277. 2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Jiang J, Lyu P, Li J, Huang S, Blackshaw
S, Qian J and Wang J: IReNA: Integrated regulatory network analysis
of single-cell transcriptomes and chromatin accessibility profiles.
iScience. 25(105359)2021.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Kumar N, Mishra B, Athar M and Mukhtar S:
Inference of gene regulatory network from single-cell
transcriptomic data using pySCENIC. Methods Mol Biol. 2328:171–182.
2021.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Nersisyan S, Gorbonos A, Makhonin A,
Zhiyanov A, Shkurnikov M and Tonevitsky A: isomiRTar: A
comprehensive portal of pan-cancer 5'-isomiR targeting. PeerJ.
10(e14205)2022.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Hou X, Zaks T, Langer R and Dong Y: Lipid
nanoparticles for mRNA/RNA delivery: Challenges and opportunities.
Nat Rev Mater. 6:1078–1094. 2021.
|
|
74
|
Jayaraman M, Ansell SM, Mui BL, Tam YK,
Chen J, Du X, Butler D, Eltepu L, Matsuda S, Narayanannair JK, et
al: Maximizing the potency of siRNA lipid nanoparticles for in vivo
delivery. Angew Chem Int Ed. 51:8529–8533. 2012.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Tam YYC, Chen S and Cullis PR: Advances in
lipid nanoparticles for siRNA delivery. Adv Drug Deliv Rev.
65:331–339. 2013.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Springer AD and Dowdy SF: GalNAc-siRNA
conjugates: Leading the way for delivery to hepatocytes. Nucleic
Acid Ther. 28:109–118. 2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Debacker AJ, Voutila J, Catley M, Blakey D
and Habib N: Delivery of oligonucleotides to the liver with GalNAc.
Nucleic Acid Ther. 30:364–386. 2020.PubMed/NCBI View Article : Google Scholar
|
|
78
|
McNamara JO II, Andrechek ER, Wang Y,
Viles KD, Rempel RE, Gilboa E, Sullenger BA and Giangrande PH: Cell
type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat
Biotechnol. 24:1005–1015. 2006.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Karikó K, Buckstein M, Ni H and Weissman
D: Suppression of RNA recognition by Toll-like receptors: Impact of
nucleoside modifications. Immunity. 23:165–175. 2005.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Coch C, Lück C, Schwickart A, Putschli B,
Renn M, Höller T, Barchet W, Hartmann G and Schlee M: A human in
vitro whole-blood assay to predict therapeutic
oligonucleotide-induced cytokine release. PLoS One.
8(e71057)2013.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Jackson AL, Burchard J, Schelter J, Chau
N, Cleary M, Lim L and Linsley P: Widespread siRNA off-target
transcript silencing mediated by seed region interactions. Nat
Biotechnol. 24:635–637. 2006.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Chi SW, Zang JB, Mele A and Darnell RB:
Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature.
460:479–486. 2009.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Hafner M, Landthaler M, Burger L, Khorshid
M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC,
Munschauer M, et al: Transcriptome-wide identification of
RNA-binding protein and microRNA target sites by PAR-CLIP. Cell.
141:129–141. 2010.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Dai H, Jin QQ, Li L and Chen LN:
Reconstructing gene regulatory networks in single-cell
transcriptomic data analysis. Zool Res. 41:599–604. 2020.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Schmidt R, Steinhart Z, Layeghi M, Freimer
J, Bueno R, Nguyen V, Blaeschke F, Ye C and Marson A: CRISPR
activation and interference screens decode stimulation responses in
primary human T cells. Science. 375(eabj4008)2022.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Pandey S, Agarwala P and Maiti S:
Targeting RNA G-quadruplexes for potential therapeutic
applications. Top Curr Chem (Cham). 377:177–206. 2017.
|
|
87
|
Roxo C, Zielińska K and Pasternak A:
Bispecific G-quadruplexes as inhibitors of cancer cells growth.
Biochimie. 214:91–100. 2023.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Chuya N and Hiroyuki S: G-quadruplex in
cancer biology and drug discovery. Biochem Biophys Res Commun.
533:762–773. 2020.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Zhang J, Zhang P, Wang L, Piao HL and Ma
L: Long non-coding RNA HOTAIR in carcinogenesis and metastasis.
Acta Biochim Biophys Sin (Shanghai). 46:1–5. 2014.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Chen Y, Liu Y, Wu J, Xu Y, Li H and Zhao
Y: LINC00160 promotes hepatocellular carcinoma progression via the
miR-132/AKT1 axis. BMC Cancer. 21(21)2021.
|
|
92
|
Xie Z, Dang Y, Wu H, Tian Y, Zhou Y, Li J
and Gu Z: The role of lncRNA-miRNA-mRNA ceRNA network in the
development and progression of pancreatic cancer. Cancer Cell Int.
20(273)2020.PubMed/NCBI View Article : Google Scholar
|