1. Introduction
The secretion associated Ras related GTPase 1B
(SAR1B) gene is a key regulator of coat protein complex II (COPII)
coat protein complex formation and plays an important role in
maintaining cellular homeostasis by mediating protein secretion,
regulating lipid metabolism, and influencing autophagosome
synthesis. It is established that SAR1B gene defects cause
chylomicron retention disease, a lipid metabolism disorder
(1). In recent years, research has
demonstrated that the SAR1B gene is involved in the regulation of
various biological behaviors of tumor cells, such as participating
in the epithelial-mesenchymal transition (EMT) of tumor cells,
regulating membrane synthesis and energy metabolism by affecting
lipid metabolism, controlling the secretion of key proteins in the
signaling pathway of tumor cells to influence signaling, and
regulating the tumor development process by affecting the synthesis
of autophagosomes under stress. It has been found that the SAR1B
gene is abnormally expressed in numerous malignant tumors, such as
colorectal cancer (CRC) and lung cancer (LC), and has the potential
to be used as a prognostic marker. The functional basis of
mediating the formation of COPII envelope protein complexes can be
used as a target for novel drug research. However, the specific
mechanism underlying the role of the SAR1B gene in tumor cell
development, migration and invasion has not been systematically
elucidated. Further research is warranted to investigate the
clinical potential of the SAR1B gene and explore the possibility of
using this gene as a therapeutic target for malignant tumors.
2. Structure, origin and physiological
function of the SAR1B gene
Structure and origin of the SAR1B
gene
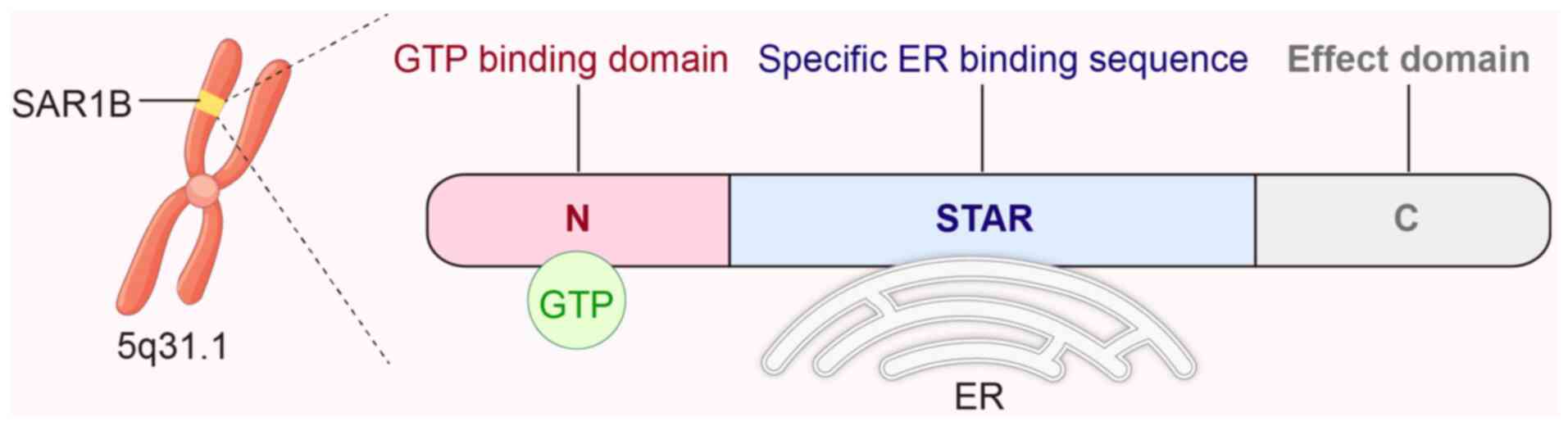
The human SAR1B gene is located on chromosome 5q31.1
and encodes a GTPase protein containing 198 amino acids, a core
component of the COPII complex. It is evolutionarily highly
conserved and widespread in eukaryotes. The SAR1B protein contains
a GTP-binding structural domain (N-terminal) and an effector
structural domain (C-terminal) that contains a unique Sar1-NH2
terminal activation recruitment (STAR) motif composed of nine amino
acids. This evolutionarily conserved cluster of bulky hydrophobic
amino acids facilitates membrane binding and contains specific
sequence information necessary for COPII complex formation
(2) (Fig. 1). SAR1B and its paralogous
homologue SAR1A are jointly involved in the formation of the COPII
complex. SAR1A and SAR1B share up to 89% amino acid sequence
identity and have a similar secondary structure of the N-terminal
amphiphilic α-helix (3). During
COPII vesicle outgrowth, they can compensate for each other's
functional defects.
SAR1B is a member of the ADP-ribosylation factor
(Arf) subfamily of the small GTPase family. The small GTPase
family, also known as the small guanosine triphosphatase (GTPase)
superfamily, is a family of proteins that can act as signaling
commanders, play a regulatory role in a variety of physiological
functions of cells, and affect the development, invasion and
migration of tumor cells through various pathways. The rat sarcoma
virus (RAS) oncogene was discovered in 1980; this gene can
stimulate the proliferation and differentiation of cells in human
cancer cells and was classified as a small GTPase (4). Thereafter, the large family of small
G proteins was discovered through continuous and in-depth
exploration and research. According to their structure and
function, these proteins have been subdivided into five subfamilies
(that is, Ras, Rho, Rab, Arf/Sar and Ran) (5). The gene encoding the Ras superfamily
of small GTPases is one of the most frequently mutated or
dysregulated genes in human cancers.
Although these subfamilies have different
physiological functions, they have a common mechanism of action,
namely they can act as ‘molecular switches’. Under the binding
effect of the regulatory factors guanine nucleotide exchange factor
(GEF) and GTPase activating protein (GAP), they can realize the
cycle of activation and inactivation states through
phosphorylation/dephosphorylation cycles, and transmit the upstream
information to the downstream effectors through conformational
changes (5). For example, members
of the Ras family can regulate cell proliferation-related pathways
through the activation of plasma membrane-localized GEFs, while
members of the Arf family rely on endoplasmic reticulum
(ER)-localized GEFs (for example, SEC12, an activator of SAR1) to
initiate vesicular transport programs. The mechanism by which SAR1B
of the Arf family initiates protein vesicular transport is
discussed below.
Physiological mechanisms of
SAR1B-mediated COPII complex formation
After synthesis, the protein undergoes preliminary
processing and modification in the rough ER and is correctly folded
and assembled to form a protein with a certain spatial structure.
Subsequently, this protein is transported to the Golgi apparatus
via vesicle transport at the exit site of the ER and then
transferred to other organelles or secreted out of the cell from
the Golgi apparatus. Whereas the process of transport from the ER
to the Golgi requires that proteins be concentrated and packaged
into vesicles, SAR1B can initiate vesicle assembly. The vesicles
that package cargo proteins are known as the COPII shell protein
complex and have the functions of selecting, binding, and
concentrating the transported cargo proteins. The COPII shell
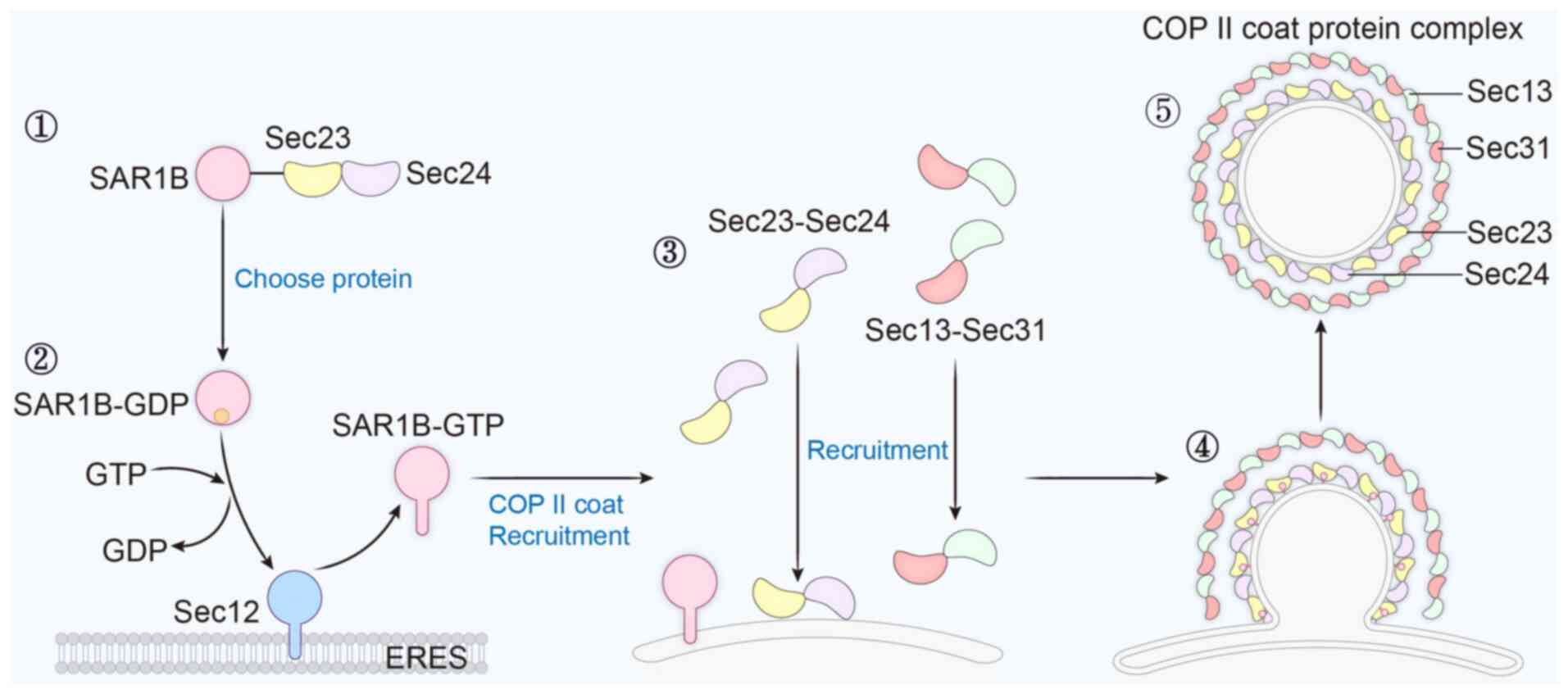
protein complex consists of five core proteins (that is, SAR1B and
the shell proteins SEC23, SEC24, SEC13 and SEC31). Among them,
SAR1B is the most important component of the COPII shell protein
complex (6). Initially, the
selection of cargoes transported in the ER is performed by the
SAR1B-SEC23-SEC24 complex. Subsequently, SAR1B is recruited to the
ER exit site by SEC12, which is activated by SEC12 and binds to the
ER membrane to initiate COPII shell assembly. SEC12 is a unique
activator of SAR1B, an integral membrane glycoprotein localized to
the ER that serves as a GEF, and a fragment of which was previously
named as the transcription factor prolactin regulatory element
binding protein. It has been shown that knockdown of SEC12 using
small interfering RNA results in diminished SAR1B activation
signaling at the ER exit (7).
Activated SAR1B sequentially recruits the SEC23-SEC24 complex and
the SEC13-SEC31 complex to complete the assembly of the inner and
outer layers of the COPII coat, respectively (8). SEC23 and SEC31 acting as a GAP to
dephosphorylate SAR1B. After cargo recruitment and concentration in
the ER lumen mediated by SEC24, SAR1B recruits SEC13 and SEC31 to
form the outer coating. Continuous formation of the outer coating
leads to the deformation of the ER membrane, which is highly curved
and subsequently shrinks and breaks off at the neck, ultimately
forming the intact vesicle. Finally, the COPII shell protein
complex carries cargo proteins for transport to the Golgi apparatus
(Fig. 2). Although the process by
which SAR1B initiates the assembly of COPII vesicles is well
understood, the mechanism underlying its role in the process of
membrane outgrowth and rupture remains poorly understood and needs
to be explored (9).
This precise regulation guarantees the accurate
transport of proteins in normal physiological activities and plays
an important role in the transport of proteins and other nutrients
in tumor cells. SAR1B is also able to play a dual role in basal
metabolism and response to stress by receiving different signals
from the cell and regulating the formation of COPII vesicles.
3. SAR1B regulates lipid metabolism in
malignant tumor cells
SAR1B has a higher affinity for the shell protein
SEC23 than SAR1A, leading to its unique ability of lipoprotein
secretion (10). It was found that
SAR1B is typically able to produce larger vesicles than SAR1A and
highly associated with impaired lipid metabolism (3). Lipid metabolism is critical in cancer
cell proliferation, survival and other biochemical processes. Lipid
metabolism disorders have become a hallmark of malignant tumors.
When nutrients (for example, lipids, proteins and nucleic acids)
are enriched, the oncogenic signaling pathway directly enhances
nutrient acquisition and promotes tumor cell proliferation, which
is inextricably linked to the regulatory role of SAR1B in lipid
transport.
Experimental results have shown that SAR1B is
overexpressed in CRC cells. Knocking out SAR1B can inhibit the
proliferation of CRC cells and induce apoptosis in RKO cells. This
suggests that SAR1B may promote the proliferation of CRC cells and
has a certain tumor-inducing effect (11). This may be related to the fact that
SAR1B serves as a hub for lipid metabolism in tumor cells.
In addition, SAR1B can also affects lipid metabolism
in tumor cells by regulating the transport of lipoproteins. SAR1B
regulates lipoprotein delivery in concert with surfeit locus
protein 4 (SURF4), a cargo receptor that is localized in the ER
membrane. Several lipoproteins, including chymotrypsin, very
low-density lipoprotein, and its transformation product low-density
lipoprotein (LDL), transport insoluble lipids within the
apolipoprotein envelope. There is evidence that SURF4 is involved
in the transport of lipoproteins, and knockdown of SURF4 in
hepatocytes drastically reduces plasma total cholesterol and
triglyceride levels (12). LDL is
the main carrier of cholesterol, cholesterol is an important
structural component of cell membranes, malignant tumor cells
proliferate rapidly and require a large amount of cholesterol to
form new cell membranes, and the binding of LDL to its LDL receptor
can deliver cholesterol to tumor cells and provide raw materials
for cell membrane synthesis in rapidly proliferating tumor
cells.
Cholesterol plays a crucial role in maintaining the
integrity and function of cell membranes on the one hand. On the
other hand, when cholesterol is in excess, it can lead to
cytotoxicity. Cholesterol is an amphiphilic sterol molecule;
SREBF1/sterol regulatory element-binding transcription factor 1
(SREBP-1) is a key lipogenic transcription factor and plays a
critical role in maintaining cholesterol homeostasis in tumor cells
(13). The activation of the
PI3K/Akt/mTOR signaling pathway can induce the transcription of
SREBPs and promote cholesterol uptake and synthesis to meet the
needs of tumor cells (14). It has
been confirmed that the deficiency of SAR1B can lead to a decrease
in the level of SREBP-1, resulting in reduced lipogenesis (15). Therefore, SAR1B can indirectly
regulate cholesterol homeostasis in tumor cells and play different
roles at different stages of tumor progression.
The small intestine is an important site for
transporting dietary fat in the form of lipoproteins. The lipolytic
products generated by the digestion of food are absorbed by small
intestinal epithelial cells. Chylomicrons, triglyceride-rich
lipoproteins, and dietary lipid carriers produced by the processing
of enterocytes will transfer dietary fat into the bloodstream
through the lymphatic system for the body to utilize (15). Genetic defects in SAR1B will
inhibit the transport of chylomicrons within enterocytes from the
ER to the Golgi apparatus. As a result, the lipids and fat-soluble
vitamins absorbed in the form of chylomicrons cannot be released
into the bloodstream, leading to a large amount of lipid
accumulation in enterocytes. It has been shown that knocking out
SAR1B in the human colorectal adenocarcinoma cell line Caco-2/15,
which secretes chylomicrons, disrupts lipid homeostasis and leads
to lipid accumulation (10). The
deficiency of SAR1B not only interferes with lipid homeostasis in
enterocytes, but lipid accumulation has also been observed in
SAR1B-deficient livers (12).
Cells with lipid metabolism disorders typically exhibit enhanced
fatty acid synthesis, increased lipid uptake, and inhibited
oxidation. This then gives rise to lipotoxicity, which causes
oxidative stress. Oxidative stress promotes DNA damage in cancer,
further leading to malignancy and carcinogenesis (14), and can trigger a series of
intracellular signaling cascades, exacerbating tumor progression.
Lipid accumulation can also enhance the downstream pro-cancer
signal transduction by forming lipid rafts to enrich growth factor
receptors (such as EGFR and HER2). It can be observed that SAR1B
has a certain anticancer effect in some cases, although the
specific mechanism remains to be explored.
SAR1B also regulates the formation of lipid
droplets. Lipid droplets are intracellular organelles that are
present in numerous types of cells and are generated after budding
from the ER via the COPII vesicle pathway. Lipids stored in lipid
droplets in tumor cells will provide energy support to tumor cells
and are involved in the regulation of cell membrane production and
signaling. In addition, lipid droplets can sequester
chemotherapeutic drugs, reducing the effective concentration of
drugs in tumor cells and making tumor cells resistant to
chemotherapeutic drugs. The core of lipid droplets consists of
triacylglycerol surrounded by phospholipid monolayers and surface
proteins such as perilipin 2 (PLIN2). It has been shown that SAR1B
mutations will result in reduced PLIN2 expression and can lead to
mis-localization of PLIN2, which affects the morphology and
integrity of lipid droplets and their ability to uptake and store
lipids (16).
4. SAR1B responds to stress and regulates
autophagy levels in cells
Unlike the physiological mechanism of SAR1B-mediated
COPII vesicle formation, SAR1B can also regulate the level of
cellular autophagy to maintain cellular homeostasis by forming the
precursor components of autophagosome membranes under stress.
Autophagy is a type of self-protection function during cellular
stress, which is like a ‘cleaner’ of the cell, capable of removing
damaged organelles (for example, mitochondria and ER), misfolded or
aggregated proteins, as well as invasive pathogens. In this way,
SAR1B can ensure the cell's stability in the presence of nutrients.
Consequently, it guarantees cell survival under unfavorable
conditions, such as nutrient deficiency and oxidative stress.
Moreover, autophagy plays a key role in the development of
organisms, immunity, aging, and the onset and progression of a
variety of diseases (for example, neurodegenerative diseases and
cancer) (17). The core of
autophagy is the formation of autophagosomes. Autophagosomes are
double-membrane vesicles produced during autophagy. They
encapsulate aged or damaged organelles, proteins and other
substances, and transport them to lysosomes for degradation. The
resulting macromolecules are then recycled and reused.
Autophagosome membranes are mainly obtained from the ER-Golgi
transport system, and membrane precursors of autophagosomes are
generated by membrane reorganization.
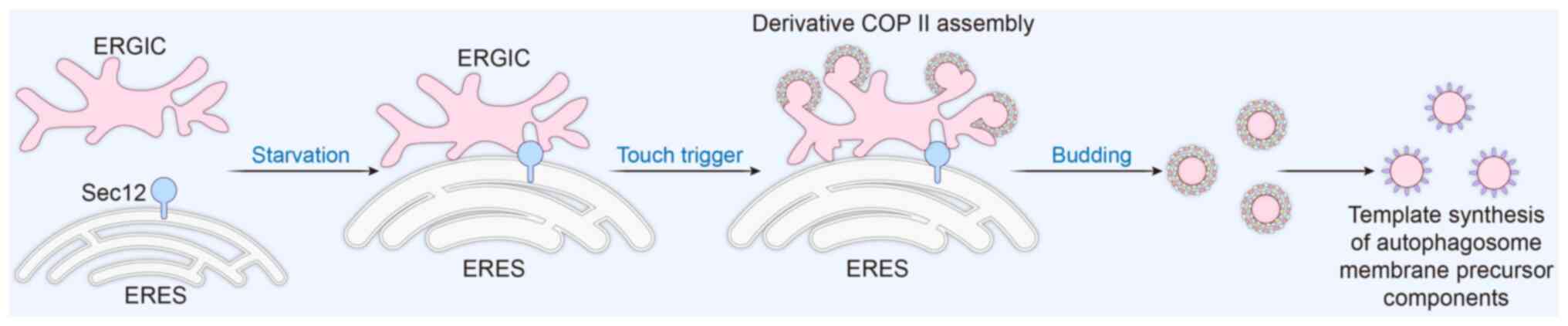
Under stress conditions such as starvation, the ER
will generate two important sites: ER-Golgi intermediate
compartment (ERGIC) and ER exit sites (ERES). Under normal
homeostasis, ERGIC and ERES accomplish vesicular transport of
proteins through SAR1-mediated generation of the COPII shell
protein complex. Sec12 is an activator of COPII vesicle formation.
In stress environments such as starvation, it can trigger the
formation of a special type of COPII vesicle, known as ERGIC-COPII
vesicles or derivative COPII vesicles. The formation and assembly
of derivative COPII vesicles are also regulated by SAR1B. A key
step in autophagosome biogenesis is the lipidation of
microtubule-associated protein 1 light chain 3 (LC3) to generate
the autophagosome membrane, and these derivative vesicles will
serve as the membrane template for LC3 lipidation. The formation of
derivative COPII vesicles is triggered by membrane contact, ER can
form different membrane contacts with different organelles, such as
mitochondria, to realize intercellular information transfer
(18). Derived COPII vesicles are
localized in ERGIC, and SEC12 is localized in ERES.
Starvation-induced ERES enlarges and becomes tightly apposed to
ERGIC and undergoes a membrane contact, which is typically <2 nm
(19). Close membrane contact
enables Sec12 located at the ERES to trans-activate the assembly of
COPII on the ERGIC, triggers the generation of derivative COPII
vesicles here, and further regulates the formation of
autophagosomes (19).
Subsequently, derivative vesicles generated at the ERGIC site can
assume normal protein transport functions or continue to be used to
generate autophagosome membrane precursors. It has been
experimentally demonstrated that both the size and SEC12 of ERGIC
are affected by starvation (20)
(Fig. 3). Meanwhile, experiments
have indicated that the overexpression of the Sar1 mutant protein
leads to the inhibition of autophagosome formation, and knocking
down Sar1 can reduce autophagosome formation (21). In an experiment searching for drugs
against liver cancer drug resistance, knocking down SAR1B in liver
tumor-initiating stem-like cells inhibited the formation of the
autophagy-related gene LC3 II. When SAR1B was re-expressed, the
inhibition of LC3 II formation was relieved, which supports the
involvement of SAR1B in the formation of autophagosomes in tumor
cells (22). Other families of
small G proteins are also involved in this process. Following the
formation of membrane precursors, the lipid composition of
autophagosome membranes and the modification of membrane proteins
are regulated by Rab family small GTPases (23,24).
In addition, after their formation, intact autophagosomes approach
lysosomes through microtubule networks and fuse with them to form
autophagic lysosomes, which begin to degrade autophagosome contents
and remove damaged organelles and aged proteins. The stability of
the fusion process is regulated by Rho family small GTPases through
the regulation of the cytoskeleton and affects the transport of
autophagosomes within the cell, while Ras family small GTPases
promote the expression of autophagy-related genes (25-28).
In states such as viral infection or oxidation, the
intracellular protein structure is damaged. When a large number of
proteins are misfolded or unfolded, they accumulate in the ER and
cannot be transported out normally, causing a dysregulation of the
ER homeostasis, which is termed ER stress (29). The deletion of SAR1B expression
also induces an accumulation of proteins in the ER, which may
result in the saturation of the ER's folding capacity (30). At this point, the body triggers an
adaptive mechanism, that is, unfolded protein response, which
restores ER homeostasis and exerts cytoprotective effects by
attenuating protein translation, increasing the ER folding
capacity, and increasing the ER protein degradation capacity.
Unfolded protein response induces SAR1B expression, thus
accelerating protein transport. However, in case of excessive or
prolonged ER stress, autophagy is induced. Autophagy helps the
cells to remove unfolded proteins and damaged ER, thereby relieving
the ER stress, protecting the cell function, and preventing cell
death (29).
Autophagy has contradictory effects on tumor cells
and tumor progression. Autophagy can inhibit tumors in the early
stages of tumorigenesis and maintain the genomic stability of cells
by removing carcinogens and damaged components from the cells.
Moreover, in the late stage of tumor development, autophagy can
help tumor cells survive in harsh environments such as nutrient
deficiency and hypoxia, provide tumor cells with necessary
nutrients and energy, and promote tumor growth and metastasis
(31). The application of SAR1B to
regulate the autophagy level of tumor cells in different periods is
particularly important and deserves further investigation.
5. SAR1B gene is involved in tumor cell
signaling
SAR1B is involved in regulating the
tumor Wnt/β-catenin signaling pathway and affects tumor
migration
SAR1B is associated with the transduction of several
tumor cell signaling pathways. Among them, SAR1B is involved in Wnt
protein secretion of the Wnt/β-catenin signaling pathway. Aberrant
activation of this signaling pathway is closely associated with the
development of numerous tumors, and 90% of CRCs show β-catenin
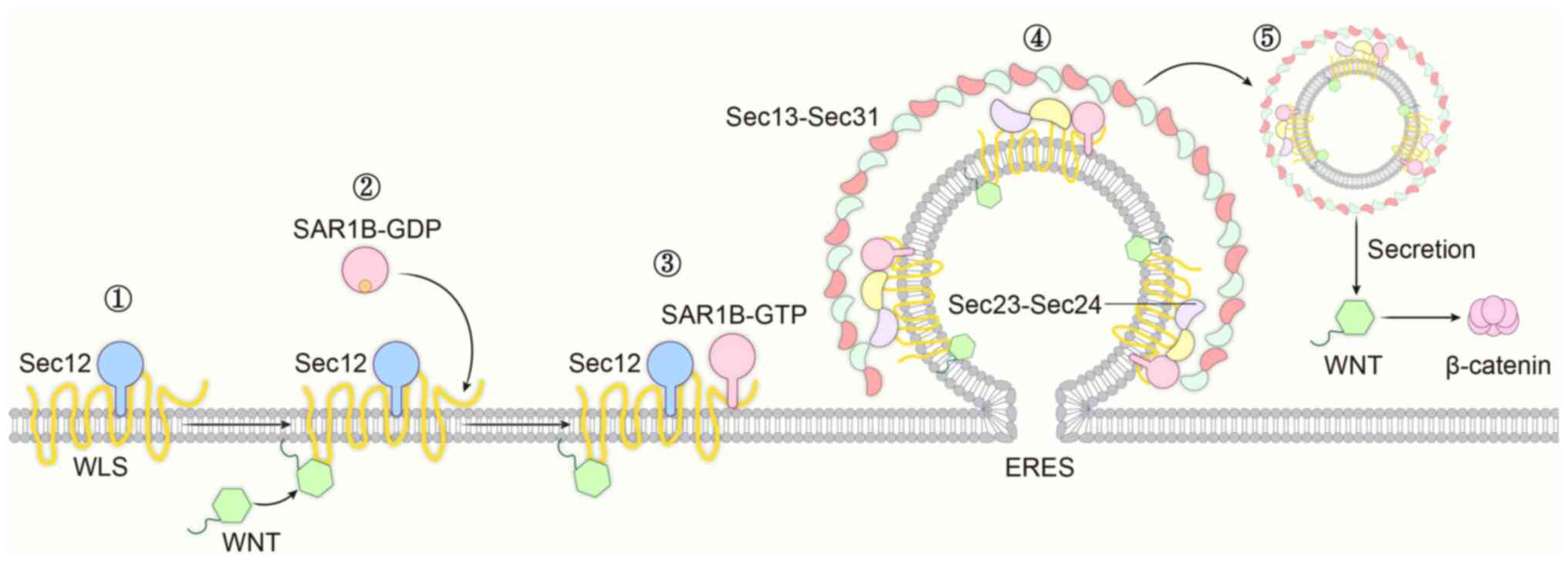
overexpression. Wnt is a secreted glycoprotein that is transported
extracellularly via COPII vesicles to play its role, and it is
essential for embryonic development and morphogenesis of adult
tissues (32). Wnt is secreted
into vesicles to control the output of functional ligands through
the ER mechanism, and Wntless is an important intermediate in this
pathway and a key regulator of Wnt protein secretion; its aberrant
expression level affects Wnt signaling. Wnt ligand secretion
mediator (Wls) enters the ER and binds to SEC12 and SAR1B, thereby
synergistically initiating the assembly of the COPII complex. Wls
is also able to bind to Wnt, facilitating its own interaction with
SEC12 and SAR1B in the ER, while regulating the movement of mature
Wnt vesicles out of the ER to the cell surface to perform their
functions. Although the underlying mechanism remains unclear,
Wnt-Wls binding in the ER may affect Wls-SEC12 binding through a
change in Wls conformation that stabilizes Wls-SEC12 binding or
exposes more SEC12 binding sites (33). When the Wnt molecule is defective
in palmitoylation, this process is compromised, and the output of
functional ligands is limited, potentially leading to the
development of certain diseases (33). Abnormalities of SAR1B in this
process lead to impaired Wnt secretion, while the downstream
β-catenin continues to accumulate and activate downstream target
genes. As a result, the cells gain the ability to proliferate
indefinitely, which is also an important basis for tumorigenesis
(Fig. 4).
The Arf subfamily proteins to which SAR1B belongs,
as well as their GEF and GAP, are aberrantly expressed in different
cancer cell types and human cancers through their involvement in
regulating actin remodeling. In sequencing data from The Cancer
Genome Atlas, the most common Arf genetic alterations are gene
amplification in prostate, breast, squamous lung, esophageal and
ovarian cancers, as well as invasive metastatic potential in
glioblastomas, uveal melanomas, breast, colon, prostate and
laryngeal squamous cell cancers, among others. Deletions of the Arf
GTPase genome in cancer also occur, albeit more rarely (34).
SAR1B and the other members of the Arf subfamily are
also capable of influencing the migration process of tumor cells
through EMT. β-catenin is a key regulator of EMT, a biological
process that causes epithelial cells to lose polarity and
intercellular junctions and acquire mesenchymal cell properties. It
confers the ability to metastasize and invade, and plays an
important role in embryonic development, tissue repair and tumor
metastasis. Most β-catenin is located on the cell membrane of
epithelial cells. Under normal physiological conditions,
intracellular β-catenin is at low levels and binds to E-cadherin
(CDH1) to form a complex that anchors it to the actin cytoskeleton,
enhancing intercellular adhesion and maintaining the integrity and
stability of epithelial tissue. Once this binding is disrupted,
intercellular adhesion is weakened, the polarity and organization
of epithelial cells are affected, and cells are more likely to
detach from epithelial tissues, which is important in the process
of tumor cell invasion and metastasis. In a variety of tumors, such
as breast, colorectal and gastric cancers, the reduced expression
of CDH1 is accompanied by abnormal activation and nuclear
accumulation of β-catenin, which is closely related to tumor
aggressiveness and poor prognosis (35). Experiments have shown that
knockdown of SAR1B in CRC cells enhances the migration and invasion
abilities of CRC cells and simultaneously leads to a significant
reduction in the levels of the epithelial marker CDH1 and an
increase in the mRNA expression of the mesenchymal marker vimentin.
These findings suggested that the cells shifted towards a more
mesenchymal phenotype. Therefore, SAR1B may inhibit the motility
and metastasis of CRC cells by regulating EMT. Meanwhile, the
deficiency of SAR1B can also stimulate EMT and promote cell
motility and Transwell invasion (36). Lower levels of SAR1B mRNA
expression were significantly associated with reduced survival and
advanced disease stage in patients with CRC, patients with high
expression of SAR1B have an improved prognosis, revealing a
potential role for SAR1B in inhibiting CRC progression. SAR1B may
serve as a prognostic biomarker and a potential inhibitor of
metastasis in CRC tumor cells (36). Moreover, experiments have shown
that SAR1A expression levels are elevated in patients with
osteosarcoma, and SAR1A levels increase osteosarcoma lung
metastasis. Furthermore, knocking down SAR1A can reduce the
formation of platelet-like pseudopods in osteosarcoma cells,
inhibit the EMT of osteosarcoma cells, and reduce the ability of
osteosarcoma cells to invade and migrate (37). The function of SAR1A as a
paralogous homolog of SAR1B, to some extent, also reveals the
potential function of SAR1B.
As aforementioned, SAR1B can either promote or
inhibit the proliferation of tumor cells by regulating lipid
metabolism. Meanwhile, experiments have confirmed that SAR1B can
promote the proliferation and inhibit the apoptosis of CRC cells.
Additionally, SAR1B can suppress the migration and invasion of CRC
by regulating EMT. It was found that SAR1B plays different roles at
different stages of CRC development, bringing either positive or
negative impacts on tumor cell activities. It is worth noting that
numerous genes play different regulatory roles in the biological
behavior of tumor cells within the same type of tumor. For example,
Patched 1, a gene encoding a transmembrane protein, inhibits the
proliferation of non-small cell LC (NSCLC) through its encoded
protein. However, it also promotes the migration, invasion and
adhesion of NSCLC cells through its 3'-untranslated region
(38). A similar situation exists
for the SAR1B gene in CRC cells. This interesting phenomenon is
worthy of in-depth consideration and exploration.
SAR1B is involved in the regulation of
the mTOR pathway. SAR1B as a leucine sensor for the mTOR signaling
pathway
SAR1B is involved in the mTOR pathway through two
pathways. The mTOR pathway is a key signaling pathway in cells.
mTOR is a serine/threonine protein kinase that, together with a
variety of regulatory-related proteins, form the mTOR complex 1
(mTORC1), which senses intracellular nutrients (for example, amino
acids and glucose), energy levels, growth factors, and stress
signals and promotes cell proliferation by regulating various
aspects of protein synthesis, ribosome biogenesis and cell cycle
progression (39). The small
GTPase SAR1B plays a key role in the activation, signaling and
functional regulation of the mTOR pathway.
Normally, mTOR helps cells proliferate, as well as
and maintain their function and structure. However, abnormally high
activity of mTOR may lead to excessive cell proliferation, which in
turn may lead to the development of malignant solid tumors (for
example, breast cancer, LC and ovarian cancer) and hematologic
malignancies (for example, leukemia and lymphoma) (40). In addition, abnormalities of mTOR
have been associated with the development of certain neurological
diseases, such as cognitive impairment and Parkinson's disease
(41). Sustained activation of the
mTOR signaling pathway promotes cell proliferation and survival and
inhibits autophagy. In neurological diseases, it leads to abnormal
changes in neurons, interferes with the normal transmission and
processing of neural signals, and thus affects the brain's
information integration and cognitive functions. For example, it
may cause increased neuronal excitability or excessive synaptic
growth. Alternatively, it may result in the failure to clear
unhelpful proteins in the brain through autophagy, leading to their
deposition in the brain and damage to neuronal functions. As an
illustration, the deposition of amyloid-β (Aβ) in the brain to form
amyloid plaques causes Alzheimer's disease (42). In tumors, the continuous activation
of mTOR is manifested as promoting the proliferation and metastasis
of tumor cells. For instance, the abnormal activation of the
PI3K-Akt-mTOR signaling pathway can promote tumor cell
proliferation, survival, metabolic reprogramming and angiogenesis
(43).
Leucine can act as an upstream regulator of the mTOR
pathway, entering the cell via amino acid transporter carriers on
the cell membrane and regulating the mTORC1 signaling pathway and
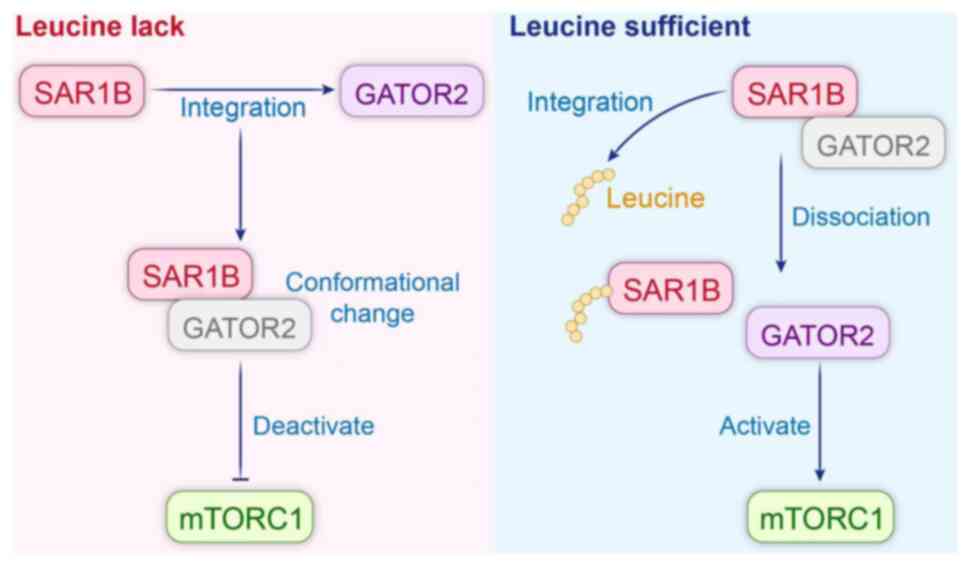
subsequent protein synthesis processes (44). SAR1B can act as a leucine sensor to
regulate mTORC1 signaling based on intracellular leucine levels.
GAP activity towards Rags 2 (GATOR2) is an activator of mTORC1;
SAR1B senses the intracellular leucine level through GATOR2 to
regulate mTORC1 signaling. In the absence of leucine, SAR1B binds
to GATOR2 and undergoes a conformational change, losing its
activating function for mTORC1 and, thus, inhibiting mTORC1. Under
leucine-sufficient conditions, SAR1B binds to leucine, undergoes a
conformational change, and dissociates from GATOR2, which acts to
activate mTORC1. Binding status does not affect their function in
mTORC1 regulation (Fig. 5)
(45). Knockdown of SAR1B renders
mTORC1 insensitive to amino acid depletion and specifically affects
the regulation of mTORC1 by leucine, without influencing other
amino acids. Re-expression of SAR1B can restore the sensitivity of
mTORC1 to leucine. The SAR1B mutant fails to bind to GATOR2 and
loses its inhibitory effect on mTORC1 under leucine-deficient
conditions. Therefore, it can be observed that SAR1B is essential
for the inhibition of mTORC1 under leucine-deficient conditions,
and the leucine-sensitive interaction between SAR1B and GATOR2 is
crucial for the regulatory effect of SAR1B on mTORC1(45).
Deficiency of the small GTPase SAR1B is associated
with lung carcinogenesis. In a mouse lung in situ xenograft
tumor model, deficiency of SAR1B and its homologue SAR1A eliminated
leucine sensitivity and failed to inhibit mTORC1 and significantly
promoted the mTORC1-dependent tumor growth. Immunohistochemical
staining revealed higher levels of the anabolic marker pS6 and the
proliferation marker Ki-67 and lower levels of the catabolic marker
light chain 3B in SAR1A- and SAR1B-deficient tumors. Silencing of
SAR1A and SAR1B in cells resulted in sustained activation of
mTORC1, which in turn led to tumorigenesis. Bioinformatics analysis
showed that SAR1B is frequently absent in human lung squamous cell
carcinoma and lung adenocarcinoma, and that patients with cancer
with lower levels of SAR1B expression have a poorer prognosis.
Therefore, SAR1B is most likely a tumor suppressor in LC (45). The mechanism of cancer inhibition
may be related to the modulation of leucine levels by SAR1B, which
affects the mTOR signaling pathway. SAR1B can be used as a
prognostic marker for LC, and compounds selectively targeting
SAR1B-dependent mTORC1 signaling may have potential for use in the
treatment of LC.
SAR1B regulates the formation of autophagosomal
membrane precursors associated with the mTOR pathway. In
addition to the core molecular mechanisms involved in the formation
of autophagosomes and autophagosomal membranes, the complex
signaling cascades that control autophagy are also particularly
important. Among them, the mTOR pathway is the core but not the
only one. mTORC1 regulates anabolism and catabolism to meet the
needs of cell proliferation. In proliferating cells, highly active
mTORC1 promotes the synthesis of biomolecules while inhibiting
autophagy. When mTORC1 is activated, it can phosphorylate and
inhibit autophagy-associated proteins, thereby inhibiting the
formation of autophagosomes and the autophagic process involving
the small GTPase SAR1B. By contrast, when the activity of mTORC1 is
inhibited under nutrient-deficient or stress conditions, the
autophagic process is activated, and cells can degrade and recycle
intracellular proteins and organelles through the formation of
autophagosomes to maintain cell survival and energy (46).
Dysregulation of mTORC1 is associated with
autophagy-deficient diseases. Autophagy requires the involvement of
COPII-derived vesicle transport, and vesicle generation is
regulated by SAR1B. The mTOR inhibitor rapamycin can treat cancer
by upregulating autophagy levels. Drugs have been successfully
developed based on the GTP-binding ability of SAR1 and its ability
to mediate vesicle formation to regulate autophagy levels.
CD133+ liver tumor-initiating stem cell-like cells
isolated from mouse and human liver tumors drive early tumor
initiation and recurrence and are chemo-resistant to mTOR
inhibitors, which may lead to drug-resistant epilepsy and
hepatocellular carcinoma recurrence. The chemo-sensitizing drug
baicalein (BC), which was developed based on the resistance of
hepatocellular carcinoma cells to mTOR inhibitors, can block mTOR
inhibitor-induced autophagy through competitive inhibition of the
GTP-binding ability of SAR1B GTPase, and thus has a
chemo-sensitizing effect. BC binds and inhibits small GTPases
involved in vesicle transport, and the binding of SAR1B to GTP is
blocked. Without the binding of SAR1B to GTP, vesicles cannot be
generated, the autophagosome membrane cannot be synthesized
normally, and autophagosome generation is reduced, thus inhibiting
the autophagy process. BC-treated tumor tissues show a significant
reduction of active SAR1B. Nanobead pull-down and mass
spectrometry, biochemical binding assay and three-dimensional
computational simulation, as well as activity analysis of BC
binding site showed that the main binding site of BC is the NH
2-terminal motif of SAR1, which is responsible for the activation
of SAR1 as well as involved in its interactions with GEF SEC12,
including GEF-binding and GAP-binding. The BC-mediated
SAR1B-dependent autophagy inhibition was attributed to its
chemo-sensitizing effect; inhibition is the basis of its
chemo-sensitizing effect. Therefore, SAR1B could be a novel target
for anticancer effects in liver tumor-initiating stem cell-like
cells and HCC cells that are resistant to mTORC1 inhibition
(22).
6. Discussion and future perspectives
As a core regulator of COPII vesicle trafficking,
SAR1B plays a role in tumor cell genesis, development,
proliferation, migration, substance metabolism, autophagy level,
signal transduction, and therapeutic resistance by mediating
protein secretion. SAR1B has the potential to be used as a
prognostic marker in lung and CRCs, and it has become a therapeutic
target for drug resistance in hepatocellular carcinoma. SAR1B has
been shown to promote cell proliferation and inhibit tumor cell
metastasis in CRC cells, maintain cellular homeostasis by
regulating autophagy and removing intracellular carcinogens under
normal cellular stress, and help tumor cells regulate autophagy to
ensure their survival under nutrient-deficient conditions and
provide energy support to tumor cells by affecting their lipid
metabolism. Therefore, SAR1B has opposite functions in various
biological behaviors of malignant tumors. Of course, there are
still some thorny issues to be resolved. The mechanism by which
SAR1B promotes the proliferation of tumor cells in CRC, the
mechanism through which SAR1B inhibits the proliferation of LC
cells, the reason why it has completely opposite effects in
different types of tumors, whether SAR1B inhibits mTORC1 and
suppresses cancer proliferation regardless of the presence of
leucine, and if the effect of SAR1B on the mTOR pathway can be
observed in tumor types other than LC, remain unexplored. The
application of the two sides of SAR1B, rational utilization of its
cancer inhibitory function, and identification of therapeutic
targets against its cancer-promoting mechanism have become major
challenges in this field. In addition, whether SAR1B also mediates
the transport of some proteins that are highly relevant to various
biological behaviors of tumor cells remains unknown. Thus, further
investigation is warranted to address these challenges.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant nos. 81372931 and 82003238) and
Bethune's Advanced Medical Research Fund in China (grant no.
2023-YJ-152-J-005).
Availability of data and materials
Not applicable.
Authors' contributions
HL was responsible for establishing the framework of
the manuscript. QY undertook the task of literature retrieval,
composed the manuscript, and crafted the accompanying
illustrations. GH, CL and ZL offered professional guidance and were
actively involved in the meticulous revision and refinement of the
article. All authors read and approved the final version of the
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shoulders CC, Stephens DJ and Jones B: The
intracellular transport of chylomicrons requires the small GTPase,
Sar1b. Curr Opin Lipidol. 15:191–197. 2004.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Huang M, Weissman JT, Beraud-Dufour S,
Luan P, Wang C, Chen W, Aridor M, Wilson IA and Balch WE: Crystal
structure of Sar1-GDP at 1.7 A resolution and the role of the NH2
terminus in ER export. J Cell Biol. 155:937–948. 2001.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Loftus AF, Hsieh VL and Parthasarathy R:
Modulation of membrane rigidity by the human vesicle trafficking
proteins Sar1A and Sar1B. Biochem Biophys Res Commun. 426:585–589.
2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Reiner DJ and Lundquist EA: Small GTPases.
WormBook. 2018:1–65. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Takai Y, Sasaki T and Matozaki T: Small
GTP-binding proteins. Physiol Rev. 81:153–208. 2001.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Melville D, Gorur A and Schekman R:
Fatty-acid binding protein 5 modulates the SAR1 GTPase cycle and
enhances budding of large COPII cargoes. Mol Biol Cell. 30:387–399.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Maeda M, Arakawa M, Komatsu Y and Saito K:
Small GTPase ActIvitY ANalyzing (SAIYAN) system: A method to detect
GTPase activation in living cells. J Cell Biol.
223(e202403179)2024.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Van der Verren SE and Zanetti G: The small
GTPase Sar1, control centre of COPII trafficking. FEBS Lett.
597:865–882. 2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tang VT, Xiang J, Chen Z, McCormick J,
Abbineni PS, Chen XW, Hoenerhoff M, Emmer BT, Khoriaty R, Lin JD
and Ginsburg D: Functional overlap between the mammalian Sar1a and
Sar1b paralogs in vivo. Proc Natl Acad Sci USA.
121(e2322164121)2024.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Melville DB, Studer S and Schekman R:
Small sequence variations between two mammalian paralogs of the
small GTPase SAR1 underlie functional differences in coat protein
complex II assembly. J Biol Chem. 295:8401–8412. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lu Y, Zhou SK, Chen R, Jiang LX, Yang LL
and Bi TN: Knockdown of SAR1B suppresses proliferation and induces
apoptosis of RKO colorectal cancer cells. Oncol Lett.
20(186)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang X, Wang H, Xu B, Huang D, Nie C, Pu
L, Zajac GJM, Yan H, Zhao J, Shi F, et al: Receptor-mediated ER
export of lipoproteins controls lipid homeostasis in mice and
humans. Cell Metab. 33:350–366.e7. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Geng F and Guo D: SREBF1/SREBP-1
concurrently regulates lipid synthesis and lipophagy to maintain
lipid homeostasis and tumor growth. Autophagy. 20:1183–1185.
2024.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Deng CF, Zhu N, Zhao TJ, Li HF, Gu J, Liao
DF and Qin L: Involvement of LDL and ox-LDL in cancer development
and its therapeutical potential. Front Oncol.
12(803473)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sané A, Ahmarani L, Delvin E, Auclair N,
Spahis S and Levy E: SAR1B GTPase is necessary to protect
intestinal cells from disorders of lipid homeostasis, oxidative
stress, and inflammation. J Lipid Res. 60:1755–1764.
2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Makiyama T, Obama T, Watanabe Y and Itabe
H: Sar1 affects the localization of perilipin 2 to lipid droplets.
Int J Mol Sci. 23(6366)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yang Z and Klionsky DJ: Mammalian
autophagy: Core molecular machinery and signaling regulation. Curr
Opin Cell Biol. 22:124–131. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li S, Yan R, Xu J, Zhao S, Ma X, Sun Q,
Zhang M, Li Y, Liu JG, Chen L, et al: A new type of ERGIC-ERES
membrane contact mediated by TMED9 and SEC12 is required for
autophagosome biogenesis. Cell Res. 32:119–138. 2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li S, Zhang M and Ge L: A new type of
membrane contact in the ER-Golgi system regulates autophagosome
biogenesis. Autophagy. 17:4499–4501. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ge L, Zhang M, Kenny SJ, Liu D, Maeda M,
Saito K, Mathur A, Xu K and Schekman R: Remodeling of ER-exit sites
initiates a membrane supply pathway for autophagosome biogenesis.
EMBO Rep. 18:1586–1603. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zoppino FC, Militello RD, Slavin I,
Alvarez C and Colombo MI: Autophagosome formation depends on the
small GTPase Rab1 and functional ER exit sites. Traffic.
11:1246–1261. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wu R, Murali R, Kabe Y, French SW, Chiang
YM, Liu S, Sher L, Wang CC, Louie S and Tsukamoto H: Baicalein
targets GTPase-mediated autophagy to eliminate liver
tumor-initiating stem cell-like cells resistant to mTORC1
inhibition. Hepatology. 68:1726–1740. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Morgan NE, Cutrona MB and Simpson JC:
Multitasking Rab proteins in autophagy and membrane trafficking: A
focus on Rab33b. Int J Mol Sci. 20(3916)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yu L, Chen Y and Tooze SA: Autophagy
pathway: Cellular and molecular mechanisms. Autophagy. 14:207–215.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Till A, Saito R, Merkurjev D, Liu JJ, Syed
GH, Kolnik M, Siddiqui A, Glas M, Scheffler B, Ideker T, et al:
Evolutionary trends and functional anatomy of the human expanded
autophagy network. Autophagy. 11:1652–1667. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang Y and Li G: A tumor suppressor DLC1:
The functions and signal pathways. J Cell Physiol. 235:4999–5007.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cao W, Li J, Yang K and Cao D: An overview
of autophagy: Mechanism, regulation and research progress. Bull
Cancer. 108:304–322. 2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang X, Cheng Q, Yin H and Yang G:
Regulation of autophagy and EMT by the interplay between p53 and
RAS during cancer progression (Review). Int J Oncol. 51:18–24.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Qi Z and Chen L: Endoplasmic reticulum
stress and autophagy. Adv Exp Med Biol. 1206:167–177.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Caruso ME, Jenna S, Bouchecareilh M,
Baillie DL, Boismenu D, Halawani D, Latterich M and Chevet E:
GTPase-mediated regulation of the unfolded protein response in
Caenorhabditis elegans is dependent on the AAA+ ATPase CDC-48. Mol
Cell Biol. 28:4261–4274. 2008.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Debnath J, Gammoh N and Ryan KM: Autophagy
and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol.
24:560–575. 2023.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Schatoff EM, Leach BI and Dow LE: Wnt
signaling and colorectal cancer. Curr Colorectal Cancer Rep.
13:101–110. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sun J, Yu S, Zhang X, Capac C, Aligbe O,
Daudelin T, Bonder EM and Gao N: A Wntless-SEC12 complex on the ER
membrane regulates early Wnt secretory vesicle assembly and mature
ligand export. J Cell Sci. 130:2159–2171. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sztul E, Chen PW, Casanova JE, Cherfils J,
Dacks JB, Lambright DG, Lee FS, Randazzo PA, Santy LC, Schürmann A,
et al: ARF GTPases and their GEFs and GAPs: Concepts and
challenges. Mol Biol Cell. 30:1249–1271. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sun L, Xing J, Zhou X, Song X and Gao S:
Wnt/β-catenin signalling, epithelial-mesenchymal transition and
crosslink signalling in colorectal cancer cells. Biomed
Pharmacother. 175(116685)2024.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Huang M and Wang Y: Targeted quantitative
proteomic approach for probing altered protein expression of small
GTPases associated with colorectal cancer metastasis. Anal Chem.
91:6233–6241. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhan F, Deng Q, Chen Z, Xie C, Xiang S,
Qiu S, Tian L, Wu C, Ou Y, Chen J and Xu L: SAR1A regulates the
RhoA/YAP and autophagy signaling pathways to influence osteosarcoma
invasion and metastasis. Cancer Sci. 113:4104–4119. 2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wan X, Kong Z, Chu K, Yi C, Hu J, Qin R,
Zhao C, Fu F, Wu H, Li Y and Huang Y: Co-expression analysis
revealed PTCH1-3'UTR promoted cell migration and invasion by
activating miR-101-3p/SLC39A6 axis in non-small cell lung cancer:
Implicating the novel function of PTCH1. Oncotarget. 9:4798–4813.
2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Mossmann D, Park S and Hall MN: mTOR
signalling and cellular metabolism are mutual determinants in
cancer. Nat Rev Cancer. 18:744–757. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Fu W and Wu G: Targeting mTOR for
anti-aging and anti-cancer therapy. Molecules.
28(3157)2023.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhu Z, Yang C, Iyaswamy A, Krishnamoorthi
S, Sreenivasmurthy SG, Liu J, Wang Z, Tong BC, Song J, Lu J, et al:
Balancing mTOR signaling and autophagy in the treatment of
Parkinson's disease. Int J Mol Sci. 20(728)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Davoody S, Asgari Taei A, Khodabakhsh P
and Dargahi L: mTOR signaling and Alzheimer's disease: What we know
and where we are? CNS Neurosci Ther. 30(e14463)2024.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Mafi S, Mansoori B, Taeb S, Sadeghi H,
Abbasi R, Cho WC and Rostamzadeh D: mTOR-mediated regulation of
immune responses in cancer and tumor microenvironment. Front
Immunol. 12(774103)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ananieva EA, Powell JD and Hutson SM:
Leucine metabolism in T cell activation: mTOR signaling and beyond.
Adv Nutr. 7:798S–805S. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen J, Ou Y, Luo R, Wang J, Wang D, Guan
J, Li Y, Xia P, Chen PR and Liu Y: SAR1B senses leucine levels to
regulate mTORC1 signalling. Nature. 596:281–284. 2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Senapati PK, Mahapatra KK, Singh A and
Bhutia SK: mTOR inhibitors in targeting autophagy and
autophagy-associated signaling for cancer cell death and therapy.
Biochim Biophys Acta Rev Cancer. 1880(189342)2025.PubMed/NCBI View Article : Google Scholar
|