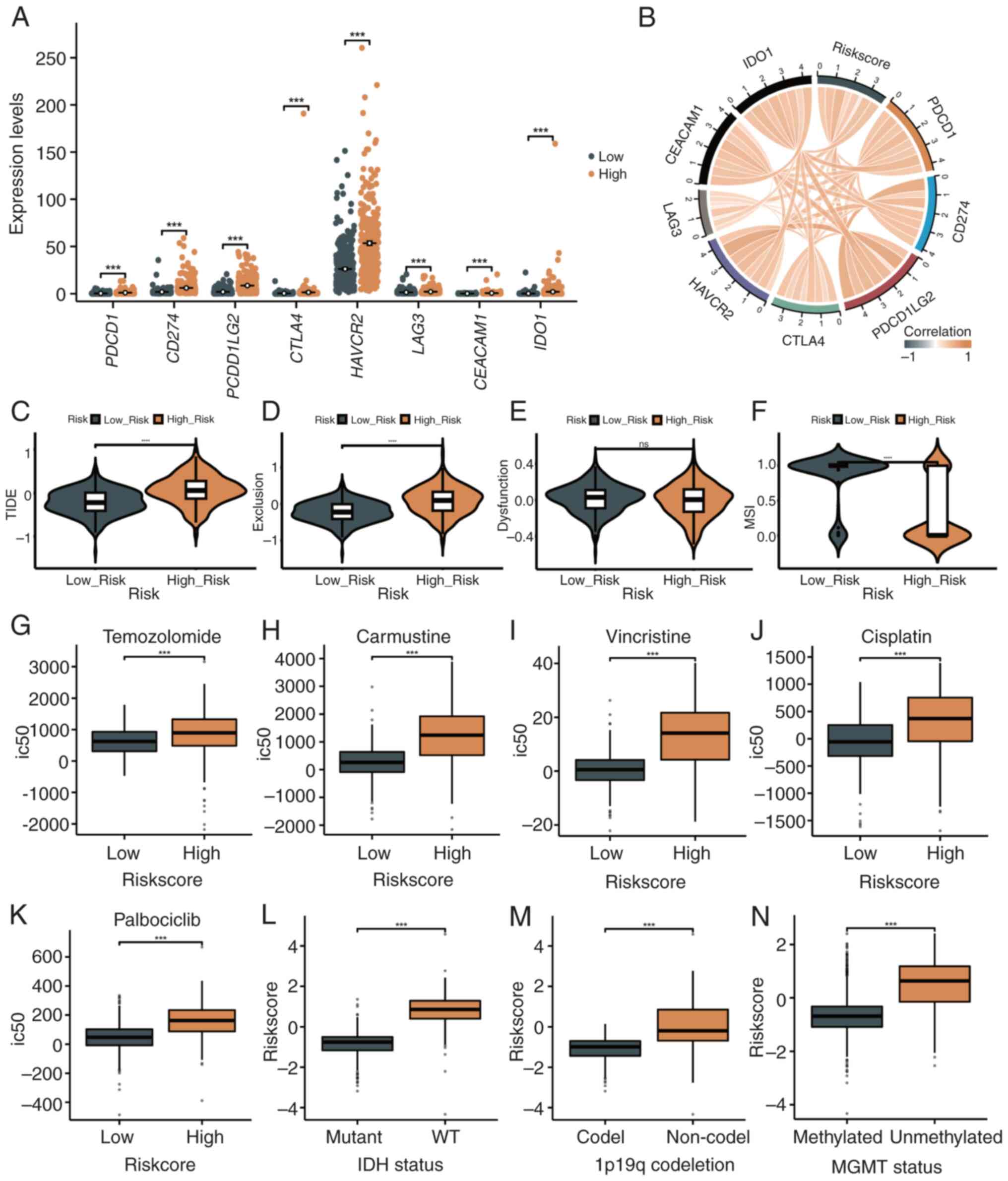
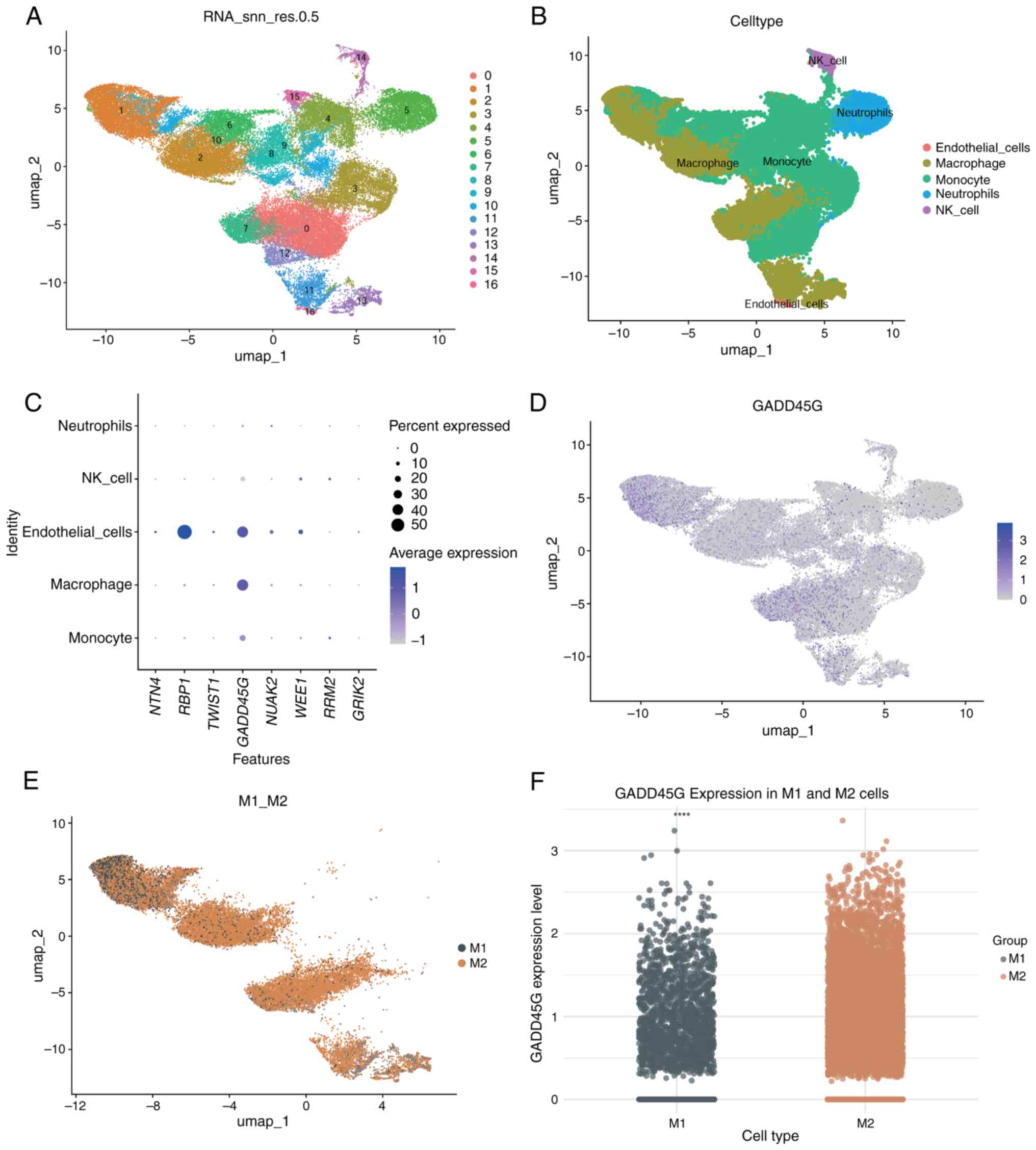
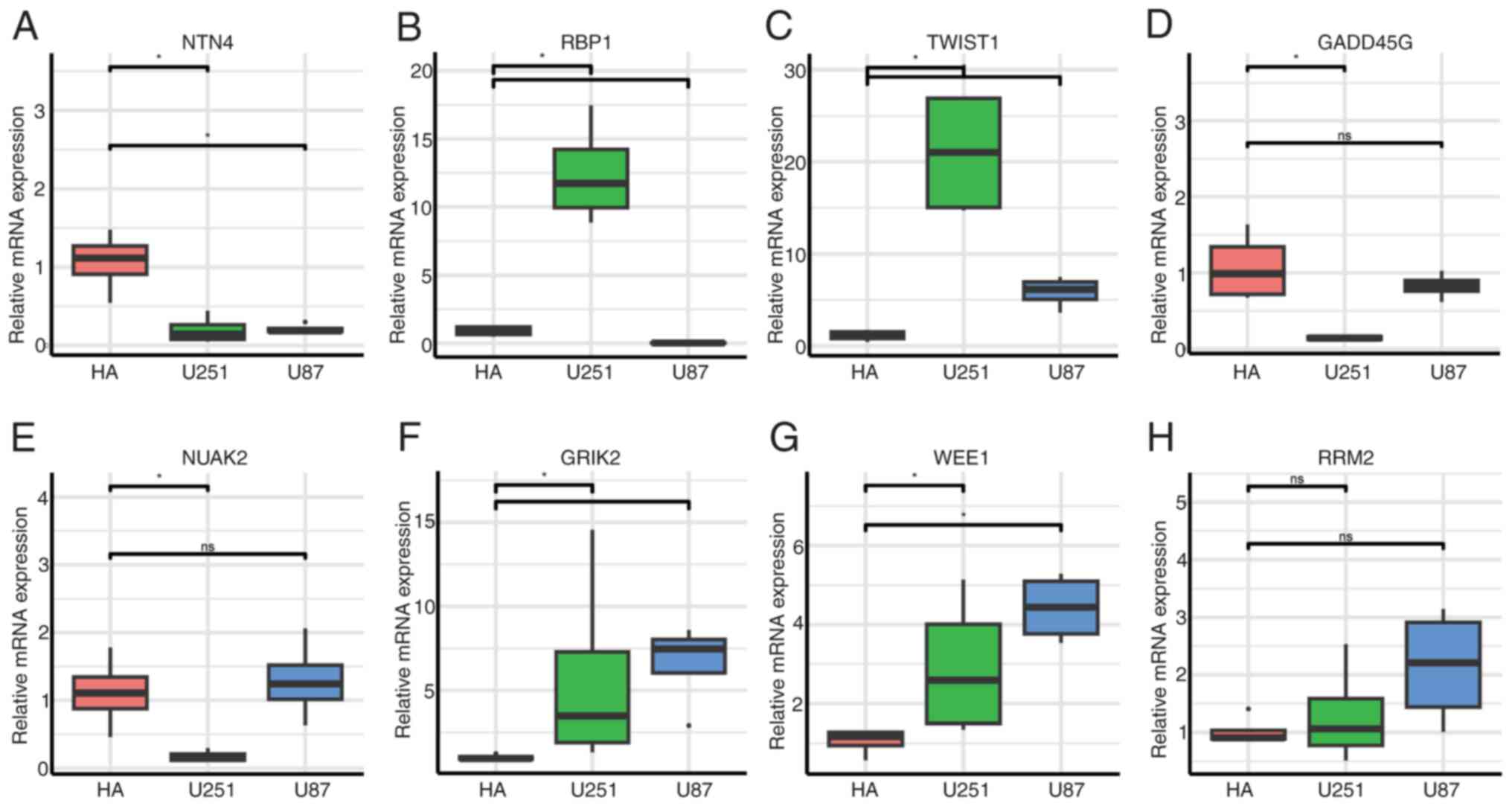
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ostrom QT, Gittleman H, Truitt G, Boscia
A, Kruchko C and Barnholtz-Sloan JS: CBTRUS Statistical report:
Primary brain and other central nervous system tumors diagnosed in
the United States in 2011-2015. Neuro Oncol. 20 (Suppl 4):iv1–iv86.
2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Campisi J: Aging, cellular senescence, and
cancer. Annu Rev Physiol. 75:685–705. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lopez-Otin C, Blasco MA, Partridge L,
Serrano M and Kroemer G: The hallmarks of aging. Cell.
153:1194–1217. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
De Carvalho DD, You JS and Jones PA: DNA
methylation and cellular reprogramming. Trends Cell Biol.
20:609–617. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hinshaw DC and Shevde LA: The tumor
microenvironment innately modulates cancer progression. Cancer Res.
79:4557–4566. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hoxhaj G and Manning BD: The PI3K-AKT
network at the interface of oncogenic signalling and cancer
metabolism. Nat Rev Cancer. 20:74–88. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Weller M, van den Bent M, Preusser M, Le
Rhun E, Tonn JC, Minniti G, Bendszus M, Balana C, Chinot O, Dirven
L, et al: EANO guidelines on the diagnosis and treatment of diffuse
gliomas of adulthood. Nat Rev Clin Oncol. 18:170–186.
2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xie Y, He L, Lugano R, Zhang Y, Cao H, He
Q, Chao M, Liu B, Cao Q, Wang J, et al: Key molecular alterations
in endothelial cells in human glioblastoma uncovered through
single-cell RNA sequencing. JCI Insight. 6(e150861)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Macosko EZ, Basu A, Satija R, Nemesh J,
Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck
EM, et al: Highly parallel Genome-wide expression profiling of
individual cells using nanoliter droplets. Cell. 161:1202–1214.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Habicht J, Mooneyham A, Shetty M, Zhang X,
Shridhar V, Winterhoff B, Zhang Y, Cepela J, Starr T, Lou E and
Bazzaro M: UNC-45A is preferentially expressed in epithelial cells
and binds to and co-localizes with interphase MTs. Cancer Biol
Ther. 20:1304–1313. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Becht E, McInnes L, Healy J, Dutertre CA,
Kwok IWH, Ng LG, Ginhoux F and Newell EW: Dimensionality reduction
for visualizing single-cell data using UMAP. Nat Biotechnol: Dec 3,
2018 (Epub ahead of print) doi: 10.1038/nbt.4314.
|
|
15
|
Aran D, Looney AP, Liu L, Wu E, Fong V,
Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al:
Reference-based analysis of lung single-cell sequencing reveals a
transitional profibrotic macrophage. Nat Immunol. 20:163–172.
2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Childs BG, Durik M, Baker DJ and van
Deursen JM: Cellular senescence in aging and age-related disease:
From mechanisms to therapy. Nat Med. 21:1424–1435. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li L, Huang Y, Gao Y, Shi T, Xu Y, Li H,
Hyytiäinen M, Keski-Oja J, Jiang Q, Hu Y and Du Z: EGF/EGFR
upregulates and cooperates with Netrin-4 to protect glioblastoma
cells from DNA damage-induced senescence. BMC Cancer.
18(1215)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Guo Z, Zhao Y, Wu Y, Zhang Y, Wang R, Liu
W, Zhang C and Yang X: Cellular retinol-binding protein 1: A
therapeutic and diagnostic tumor marker. Mol Biol Rep.
50:1885–1894. 2023.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939.
2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yu X, He T, Tong Z, Liao L, Huang S,
Fakhouri WD, Edwards DP and Xu J: Molecular mechanisms of
TWIST1-regulated transcription in EMT and cancer metastasis. EMBO
Rep. 24(e56902)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hollander MC, Sheikh MS, Bulavin DV,
Lundgren K, Augeri-Henmueller L, Shehee R, Molinaro TA, Kim KE,
Tolosa E, Ashwell JD, et al: Genomic instability in
Gadd45a-deficient mice. Nat Genet. 23:176–184. 1999.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Qiu W, David D, Zhou B, Chu PG, Zhang B,
Wu M, Xiao J, Han T, Zhu Z, Wang T, et al: Down-regulation of
growth arrest DNA damage-inducible gene 45beta expression is
associated with human hepatocellular carcinoma. Am J Pathol.
162:1961–1974. 2003.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang W, Huper G, Guo Y, Murphy SK, Olson
JA Jr and Marks JR: Analysis of methylation-sensitive transcriptome
identifies GADD45a as a frequently methylated gene in breast
cancer. Oncogene. 24:2705–2714. 2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li Y, Song X, Liu L and Yue L: NUAK2
silencing inhibits the proliferation, migration and
epithelial-to-mesenchymal transition of cervical cancer cells via
upregulating CYFIP2. Mol Med Rep. 24(817)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Namiki T, Yaguchi T, Nakamura K, Valencia
JC, Coelho SG, Yin L, Kawaguchi M, Vieira WD, Kaneko Y, Tanemura A,
et al: NUAK2 amplification coupled with PTEN deficiency promotes
melanoma development via CDK activation. Cancer Res. 75:2708–2715.
2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tang L, Tong SJ, Zhan Z, Wang Q, Tian Y
and Chen F: Expression of NUAK2 in gastric cancer tissue and its
effects on the proliferation of gastric cancer cells. Exp Ther Med.
13:676–680. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang R, Su D, Liu Y, Huang H, Qiu J, Cao
Z, Yang G, Chen H, Luo W, Tao J, et al: The NF-kappaB/NUAK2
signaling axis regulates pancreatic cancer progression by targeting
SMAD2/3. iScience. 27(109406)2024.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wu CS, Lu YJ, Li HP, Hsueh C, Lu CY, Leu
YW, Liu HP, Lin KH, Hui-Ming Huang T and Chang YS: Glutamate
receptor, ionotropic, kainate 2 silencing by DNA hypermethylation
possesses tumor suppressor function in gastric cancer. Int J
Cancer. 126:2542–2552. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sanai N, Li J, Boerner J, Stark K, Wu J,
Kim S, Derogatis A, Mehta S, Dhruv HD, Heilbrun LK, et al: Phase 0
Trial of AZD1775 in First-recurrence glioblastoma patients. Clin
Cancer Res. 24:3820–3828. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li C, Zheng J, Chen S, Huang B, Li G, Feng
Z, Wang J and Xu S: RRM2 promotes the progression of human
glioblastoma. J Cell Physiol. 233:6759–6767. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Aarts M, Sharpe R, Garcia-Murillas I,
Gevensleben H, Hurd MS, Shumway SD, Toniatti C, Ashworth A and
Turner NC: Forced mitotic entry of S-phase cells as a therapeutic
strategy induced by inhibition of WEE1. Cancer Discov. 2:524–539.
2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xiao Y, Li M, Ma T, Ning H and Liu L:
AMG232 inhibits angiogenesis in glioma through the p53-RBM4-VEGFR2
pathway. J Cell Sci. 136(jcs260270)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhao Y, Chen Y, Wei L, Ran J, Wang K, Zhu
S and Liu Q: p53 inhibits the Urea cycle and represses polyamine
biosynthesis in glioma cell lines. Metab Brain Dis. 38:1143–1153.
2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Herranz N and Gil J: Mechanisms and
functions of cellular senescence. J Clin Invest. 128:1238–1246.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ryan MB and Corcoran RB: Therapeutic
strategies to target RAS-mutant cancers. Nat Rev Clin Oncol.
15:709–720. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zuchegna C, Leone S, Romano A, Porcellini
A and Messina S: KRAS is a molecular determinant of platinum
responsiveness in glioblastoma. BMC Cancer. 24(77)2024.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen Z, Yan X, Miao C, Liu L, Liu S, Xia
Y, Fang W, Zheng D and Luo Q: Targeting MYH9 represses
USP14-mediated NAP1L1 deubiquitination and cell proliferation in
glioma. Cancer Cell Int. 23(220)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Stine ZE, Walton ZE, Altman BJ, Hsieh AL
and Dang CV: MYC, Metabolism, and Cancer. Cancer Discov.
5:1024–1039. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Qiu X, Li Y and Zhang Z: Crosstalk between
oxidative phosphorylation and immune escape in cancer: A new
concept of therapeutic targets selection. Cell Oncol (Dordr).
46:847–865. 2023.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ashton TM, McKenna WG, Kunz-Schughart LA
and Higgins GS: Oxidative phosphorylation as an emerging target in
cancer therapy. Clin Cancer Res. 24:2482–2490. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kuo CL, Ponneri Babuharisankar A, Lin YC,
Lien HW, Lo YK, Chou HY, Tangeda V, Cheng LC, Cheng AN and Lee AY:
Mitochondrial oxidative stress in the tumor microenvironment and
cancer immunoescape: Foe or friend? J Biomed Sci.
29(74)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Mun JY, Leem SH, Lee JH and Kim HS: Dual
relationship between stromal cells and immune cells in the tumor
microenvironment. Front Immunol. 13(864739)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Deng Y, Chen Q, Wan C, Sun Y, Huang F, Hu
Y and Yang K: Microglia and macrophage metabolism: A regulator of
cerebral gliomas. Cell Biosci. 14(49)2024.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Li C, Jiang P, Wei S, Xu X and Wang J:
Regulatory T cells in tumor microenvironment: New mechanisms,
potential therapeutic strategies and future prospects. Mol Cancer.
19(116)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Feng B, Wu J, Shen B, Jiang F and Feng J:
Cancer-associated fibroblasts and resistance to anticancer
therapies: Status, mechanisms, and countermeasures. Cancer Cell
Int. 22(166)2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Borst R, Meyaard L and Pascoal Ramos MI:
Understanding the matrix: Collagen modifications in tumors and
their implications for immunotherapy. J Transl Med.
22(382)2024.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wei R, Zhou J, Bui B and Liu X: Glioma
actively Orchestrate a self-advantageous extracellular matrix to
promote recurrence and progression. BMC Cancer.
24(974)2024.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ellert-Miklaszewska A, Poleszak K,
Pasierbinska M and Kaminska B: Integrin signaling in glioma
pathogenesis: From biology to therapy. Int J Mol Sci.
21(888)2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wu M, Huang Q, Xie Y, Wu X, Ma H, Zhang Y
and Xia Y: Improvement of the anticancer efficacy of PD-1/PD-L1
blockade via combination therapy and PD-L1 regulation. J Hematol
Oncol. 15(24)2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Pulanco MC, Madsen AT, Tanwar A, Corrigan
DT and Zang X: Recent advancements in the B7/CD28 immune checkpoint
families: New biology and clinical therapeutic strategies. Cell Mol
Immunol. 20:694–713. 2023.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Vidyarthi A, Agnihotri T, Khan N, Singh S,
Tewari MK, Radotra BD, Chatterjee D and Agrewala JN: Predominance
of M2 Macrophages in gliomas leads to the suppression of local and
systemic immunity. Cancer Immunol Immunother. 68:1995–2004.
2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ravi VM, Neidert N, Will P, Joseph K,
Maier JP, Kückelhaus J, Vollmer L, Goeldner JM, Behringer SP,
Scherer F, et al: T-cell dysfunction in the glioblastoma
microenvironment is mediated by myeloid cells releasing
interleukin-10. Nat Commun. 13(925)2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Puviindran BJ, Wallace S, Ayasoufi K,
Lerner E and Fecci PE: Within and beyond the tumor: Mechanisms of
glioblastoma-induced immunosuppression. Neurooncol Adv. 7 (Suppl
4):iv4–iv18. 2025.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Zajac A, Sumorek-Wiadro J, Langner E,
Wertel I, Maciejczyk A, Pawlikowska-Pawlęga B, Pawelec J, Wasiak M,
Hułas-Stasiak M, Bądziul D, et al: Involvement of PI3K pathway in
glioma cell resistance to temozolomide treatment. Int J Mol Sci.
22(5155)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Meng J, Qian W, Yang Z, Gong L, Xu D,
Huang H, Jiang X, Pu Z, Yin Y and Zou J: p53/E2F7 axis promotes
temozolomide chemoresistance in glioblastoma multiforme. BMC
Cancer. 24(317)2024.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Perrault EN, Shireman JM, Ali ES, Lin P,
Preddy I, Park C, Budhiraja S, Baisiwala S, Dixit K, James CD, et
al: Ribonucleotide reductase regulatory subunit M2 drives
glioblastoma TMZ resistance through modulation of dNTP production.
Sci Adv. 9(eade7236)2023.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Pokorny JL, Calligaris D, Gupta SK,
Iyekegbe DO Jr, Mueller D, Bakken KK, Carlson BL, Schroeder MA,
Evans DL, Lou Z, et al: The efficacy of the wee1 inhibitor MK-1775
combined with temozolomide is limited by heterogeneous distribution
across the Blood-brain barrier in glioblastoma. Clin Cancer Res.
21:1916–1924. 2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Hussain SF, Yang D, Suki D, Aldape K,
Grimm E and Heimberger AB: The role of human glioma-infiltrating
microglia/macrophages in mediating antitumor immune responses.
Neuro Oncol. 8:261–279. 2006.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Quail DF and Joyce JA: The
microenvironmental landscape of brain tumors. Cancer Cell.
31:326–341. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Deng R, Qin J, Wang L, Li H, Wen N, Qin K,
Dong J, Wu J, Zhu D and Sun X: Energy metabolism-related GLUD1
contributes to favorable clinical outcomes of IDH-mutant glioma.
BMC Neurol. 24(344)2024.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Familiari P, Lapolla P, Picotti V,
Palmieri M, Pesce A, Carosi G, Relucenti M, Nottola S, Gianno F,
Minasi S, et al: Role of 1p/19q codeletion in diffuse Low-grade
glioma tumour prognosis. Anticancer Res. 43:2659–2670.
2023.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Mansouri A, Hachem LD, Mansouri S, Nassiri
F, Laperriere NJ, Xia D, Lindeman NI, Wen PY, Chakravarti A, Mehta
MP, et al: MGMT promoter methylation status testing to guide
therapy for glioblastoma: Refining the approach based on emerging
evidence and current challenges. Neuro Oncol. 21:167–178.
2019.PubMed/NCBI View Article : Google Scholar
|