Introduction
Cervical cancer (CC), one of the most prevalent
malignancies of the female reproductive system worldwide, is
primarily driven by persistent infection with high-risk human
papillomavirus (HPV) (1). It is
noteworthy that increasing evidence indicates that a subset of CC
progresses through HPV-independent mechanisms, a process
intricately linked to dynamic changes in the tumor microenvironment
(TME) and complex intercellular communication (2,3).
Currently, CC continues to pose important challenges in terms of
drug resistance, precise diagnosis and effective treatment.
Previous studies have shown that tumor heterogeneity and genomic
instability can lead to resistance to chemotherapy and radiotherapy
(4,5). Meanwhile, the infiltration of
immunosuppressive cells, impaired T-cell function and upregulation
of immune checkpoint molecules within the CC microenvironment
collectively promote tumor immune evasion, thereby limiting the
efficacy of immunotherapy (6,7).
Furthermore, beyond anti-angiogenic agents, there remains a lack of
effective targeted treatment strategies for metastatic or recurrent
CC in clinical practice (8).
However, current screening strategies and biomarker research for CC
remain predominantly focused on HPV testing and cytological
abnormalities. Although these methods can, to certain extent,
predict disease progression and monitor treatment response, they
exhibit notable limitations in accurately identifying precancerous
lesions, addressing HPV-independent carcinogenic mechanisms, and
monitoring treatment response and recurrence in patients with
advanced-stage disease. Therefore, developing reliable biomarkers
that can reflect TME alterations and disease evolution, and
building more effective treatment strategies based on such
biomarkers, has become a critical issue that urgently needs to be
addressed in the clinical management of CC.
Extracellular vesicles (EV) have emerged as
promising tools in liquid biopsy and tumor mechanism research due
to their ability to carry various bioactive molecules such as
nucleic acids, proteins and metabolites (9). As a class of endogenous
nanoparticles, EVs possess inherent characteristics, including low
immunogenicity, high stability, and an exceptional ability to cross
biological barriers, that make them ideal, universal platforms for
cancer research and clinical application, far beyond a single
cancer type (10). This broad
utility is particularly evidenced in the field of biomarker
discovery, where EV-based analyses have demonstrated significant
diagnostic and prognostic value across a spectrum of malignancies,
including breast, pancreatic and lung cancers (11,12).
EVs are widely present in multiple biological fluids such as blood,
saliva and urine (13-15).
Among these, plasma holds particular value in EV research due to
its easy accessibility, abundant availability, and capacity to
rapidly and systematically reflect the body's pathophysiological
status. EVs in plasma originate from tissues and organs throughout
the body, including tumor tissues from different anatomical sites,
enabling them to comprehensively reflect the body's tumor burden
and heterogeneity (16,17). Building upon this universal
potential of plasma EVs, their role in CC was investigated. During
the development and progression of CC, plasma EVs are specifically
regulated by the pathological state of CC cells, and their
molecular composition carries disease-specific biological
information (18). Therefore,
analyzing the specific molecular characteristics of plasma EVs in
the context of CC not only provides new perspectives for
understanding the biological mechanisms of the disease, but also
offers powerful tools for developing non-invasive diagnostic
strategies.
At the molecular mechanism level, transcriptomics
microRNA (miRNA or miR) sequencing has become a widely used
approach for investigating cellular phenotypes and functions by
deciphering upstream regulatory networks of gene expression
(19,20). Specifically, miRNAs carried by EVs
are not only protected from degradation but also functionally
contribute to investigating the cause of disease, thus classifying
cancer subtypes or acting as carriers targeting different target
cells (12). They play important
roles in a variety of cancer types and inflammatory diseases,
including CC (21). However,
transcriptomic analyses alone often fail to capture the
multidimensional regulatory complexity of the disease due to
inherent limitations in resolution and functional annotation. To
overcome these limitations, proteomics offers a complementary and
indispensable layer of information. As a downstream executor of
gene expression, proteomic analysis provides detailed insights into
post-translational modifications and functional protein interaction
networks (22). This downstream
perspective is critical for revealing the functional molecular
mechanisms of diseases, identifying new biomarkers, and advancing
personalized medicine. Building on the strengths of both fields, an
integrated multi-omics strategy has emerged as a powerful paradigm.
Indeed, previous studies have shown that integrating
transcriptomics and corresponding proteomics data from EV samples
cannot only reveal the association between genes and proteins, and
explore the potential mechanisms of molecular networks, but also
significantly enhance the identification of potential biomarkers
for disease diagnosis (23,24).
Consequently, monitoring circulating EV-derived miRNAs and proteins
in body fluids offers a promising strategy for accurate,
non-invasive and rapid disease diagnosis. Therefore, it was
hypothesized that concurrently monitoring EV-derived miRNAs and
proteins in plasma could provide a more accurate, non-invasive and
comprehensive strategy for understanding CC pathogenesis and
identifying diagnostic signatures.
In the present study, the aforementioned integrated
multi-omics approach was employed to systematically reveal the
molecular features and regulatory networks of plasma EV in CC
progression. Combined analysis of EV miRNA transcriptomics and
proteomics identified 22 differentially expressed miRNAs (DEMs) and
49 differentially expressed proteins (DEPs). These molecules were
co-enriched in core pathways implicated in CC, including HPV
infection, p53 signaling, PI3K-Akt pathway and
complement/coagulation cascades. Notably, through multi-omics
integration, the Homo sapiens (hsa)-miR-1-3p-LRP1 axis was
identified and experimentally validated as a potential key
regulatory target in CC. The present findings not only provide
biological evidence supporting the use of plasma EVs as disease
biomarkers, but also reveal novel signature molecules involved in
CC onset and progression, thereby outlining a viable pathway for
EV-mediated liquid biopsy and the development of targeted
therapeutic strategies.
Materials and methods
Clinical samples
All participants were recruited from the Department
of Gynecology, Qinghai University Affiliated Hospital (Qinghai,
China). The cohort included 3 female patients with CC and 3 female
healthy controls (HCs). The ages of the patients with CC were 44,
47 and 69 years (median: 53 years; range: 44-69 years), while the
HCs were 58, 61, and 61 years old (median: 61 years; range: 58-61
years). This sample size aligns with standard practices for
exploratory analysis using omics technologies and meets the
requirements for methodological reproducibility (25,26).
The present study was approved by the Institutional Review Board of
Qinghai University Affiliated Hospital (approval no. P-SL-202157)
in compliance with ethical standards, and written informed consent
was obtained from all participants prior to enrollment. CC
diagnoses were pathologically confirmed according to the
International Federation of Gynecology and Obstetrics (FIGO)
staging system (2018) (27).
Venous blood samples were collected immediately after admission.
Plasma was separated by centrifugation at 4˚C and 1,000 x g for 15
min (cat. no. HT165; Cence; https://www.xiangyilxj.com/) and stored at -80˚C until
further analysis.
EV preparation
EV were isolated from 2 ml plasma using a
standardized ultracentrifugation (UC) protocol (28). Briefly, plasma samples were diluted
10-fold with PBS (cat. no. G4202; Wuhan Servicebio Technology Co.,
Ltd.) to a final volume of 20 ml. Sequential centrifugation steps
were then performed at 2,000 x g for 10 min at 4˚C to eliminate
cellular debris and at 10,000 x g for 20 min at 4˚C to pellet
larger apoptotic bodies, and filtered through a 0.22-µm pore-size
membrane filter (cat. no. SLGP033R; MilliporeSigma) to remove large
particulate contaminants. The supernatant was subsequently
ultracentrifuged at 120,000 x g for 120 min at 4˚C (cat. no. OPTIMA
L100XP; Beckman Coulter, Inc.) to obtain purified EV pellets, which
were resuspended in 400 µl sterile PBS and stored at -80˚C until
further analysis.
Nanoparticle tracking analysis
(NTA)
NTA was performed using a NanoSight NS300 system
(Malvern Panalytical, Ltd.) equipped with a 488-nm laser and a
high-sensitivity sCMOS (scientific complementary
metal-oxide-semiconductor) camera, according to the manufacturer's
instructions. The samples were then diluted with PBS to achieve an
optimal concentration for accurate particle counting and sizing,
resulting in a final concentration of 30-50 particles per frame.
The particles per frame value was 30-60, with a camera detection
threshold of 15 and an automated injection flow rate of 30
µl/min.
Transmission electron microscope (TEM)
analysis
After fixation with an equal volume of 4%
paraformaldehyde (cat. no. BL539A; Biosharp Life Sciences) at 4˚C
for 30 min, the EV suspension was deposited onto
Formvar/carbon-coated copper grids (cat. no. BZ11032b; Henan
Zhongjingkeyi Technology Co., Ltd.) and allowed to adsorb for 30
min at room temperature (RT). The grids were then washed twice with
PBS, followed by post-fixation with 1% glutaraldehyde for 5 min.
After two washes with ultrapure water, the samples were negatively
stained with 2% uranyl acetate (cat. no. GZ02625, Tianjin Ruixin
Technology Co., Ltd.) at RT for 40 sec. Morphological observation
was ultimately performed using a Talos L120C G2 TEM (Thermo Fisher
Scientific, Inc.) operated at 120 kV.
Western blot (WB) analysis
Purified EVs were quantified using a the
Bicinchoninic Acid Assay Kit (BCA; cat. no. P0011; Beyotime
Institute of Biotechnology) according to the manufacturer's
instructions. Deformed EV proteins (loaded with 20 µg of protein
per lane) were separated by 10% SDS-PAGE using the One-Step PAGE
Gel Rapid Preparation Kit (cat. no. PG212; Shanghai Epizyme
Biopharmaceutical Technology Co., Ltd.; Ipsen Pharma) and
transferred to a PVDF membrane (cat. no. ISEQ00010;
MilliporeSigma). Membranes were blocked with 5% (w/v) non-fat milk
(cat. no. 232100; Sangon Biotech Co., Ltd.) in PBS containing 0.1%
Tween-20 (cat. no. 30189380; Shanghai HUSHI, Ltd.) for 1 h at RT
and subsequently incubated overnight at 4˚C with the following
primary antibodies diluted in blocking buffer: Anti-Alix (1:1,000;
cat. no. 18269; Cell Signaling Technology, Inc.), anti-Flotillin-1
(1:1,000; cat. no. 610820; BD Biosciences) and anti-CD63 (1:1,000;
cat. no. ab68418; Abcam). After five washes with PBS, membranes
were incubated with a horseradish peroxidase conjugated goat
anti-rabbit IgG secondary antibody (1:3,000; cat. no. 7074; Cell
Signaling Technology, Inc.) for 1 h at RT. Protein bands were
visualized using a ChemiDoc Imaging System (Clinx Science
Instruments, Co., Ltd.) with an enhanced chemiluminescence
substrate (cat. no. WBKLS0500; MilliporeSigma).
miRNA sequencing and bioinformatic
analysis
Total RNA was extracted from purified EV using
RNAiso for miRNA reagent (cat. no. 9753A, Takara Bio, Inc.), a
specialized reagent designed for the simultaneous isolation of both
large and small RNAs. RNA concentration and purity were assessed
using a NanoDrop One spectrophotometer (cat. no. 840-317400;
NanoDrop Technologies; Thermo Fisher Scientific, Inc.). Briefly, EV
samples were thoroughly mixed with 140 µl chloroform (cat. no.
C2432; MilliporeSigma) and centrifuged at 12,000 x g for 15 min at
4˚C. The aqueous phase was collected and combined with 1.5 vol
absolute ethanol. The mixture was transferred to RNeasy
purification columns (cat. no. 74104; Qiagen GmbH) for RNA
purification. Small RNA libraries were constructed from 1 µg of
total RNA per sample. Subsequently, 4 µl miRNA was subjected to 18
cycles of microamplification using the SMARTer® Starfish Total
RNA-seq Kit (cat. no. 634413; Takara Bio, Inc.), and the amplified
products were purified with AMPure XP beads (cat. no. A63881;
Beckman Coulter, Inc.). Libraries were then constructed from 1 ng
complementary DNA (cDNA) using the Transpose DNA Library Prep Kit
for Illumina, Inc. (cat. no. K0012; LifeInt) with 14 amplification
cycles. The final libraries were quality-controlled on an Agilent
2100 Bioanalyzer (cat. no. G2939A; Agilent Technologies, Inc.) to
ensure a peak distribution between 140-160 bp, corresponding to
miRNA inserts, and sequenced on an Illumina NovaSeq 6000 platform
(Illumina, Inc.), generating paired-end 150-bp reads from six
samples.
For bioinformatic analysis, raw sequencing data were
processed with Trim Galore (v0.6.7; The Babraham Institute) to
remove adapter sequences and low-quality reads (Phred quality score
<30). High-quality reads were aligned to the human reference
genome GRCh38/hg38 using Bowtie2 (v2.4.5). miRNA expression
quantification was performed with miRDeep2 (v2.0.1.3) to calculate
read counts, and miRNAs with null expression in ≥50% of samples
were filtered out. DEMs were identified using DESeq2 (v1.32.0) with
thresholds of fold-change (FC) >1.2 and adjusted P<0.05.
miRNA target genes were predicted based on the miRTarBase database
(v9.0; https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment analyses were conducted using
ClusterProfiler (v4.2.2) (29),
which is available from Bioconductor (https://bioconductor.org/packages/clusterProfiler/). A
significance threshold of P<0.05 was applied. The results were
visualized with ggplot2 (v3.3.5).
Proteomic profiling and bioinformatic
analysis
EV samples were thawed at -80˚C and lysed on ice
with 1% (v/v) protease inhibitor cocktail using ultrasonication.
The lysate was centrifuged at 12,000 x g for 10 min at 4˚C to
remove cellular debris, and the supernatant was transferred to
fresh microcentrifuge tubes. Protein concentration was quantified
using a BCA assay kit (cat. no. P0011; Beyotime Institute of
Biotechnology). For each sample, 50 µg EV protein was adjusted to
100 µl with lysis buffer. Proteins were reduced with 5 mM
dithiothreitol (cat. no. 43815; MilliporeSigma) at 56˚C for 30 min,
followed by alkylation with 11 mM iodoacetamide (cat. no. I1149;
MilliporeSigma) in the dark at RT for 15 min. Urea concentration
was diluted to <2 M using tetraethylammonium bromide. Sequential
tryptic digestion was performed with trypsin. Peptides were
reconstituted in 0.1% formic acid (FA: cat. no. 94318;
MilliporeSigma) and separated on an EASY-nLC 1200 UHPLC system
(Thermo Fisher Scientific, Inc.). The mobile phases consisted of
0.1% FA + 2% acetonitrile (ACN; phase A) and 0.1% FA + 90% ACN
(phase B). The gradient program was: 0-68 min (6-23% B), 68-82 min
(23-32% B), 82-86 min (32-80% B) and 86-90 min (80% B) at a flow
rate of 500 nl/min. Peptides were ionized via a nano-electrospray
ionization source and analyzed on an Orbitrap Exploris™ 480 mass
spectrometer. The ion source voltage was set to 2.3 kV. FAIMS
compensation voltages were configured at 45 and -65 V.
High-resolution Orbitrap detection was applied for both precursor
(400-1,200 m/z; 60,000 resolution) and fragment ions (fixed start
at 110 m/z; 15,000 resolution), with TurboTMT disabled.
Data-dependent acquisition mode was used to select the top 25 most
intense precursors per cycle for fragmentation (AGC target 100%,
signal threshold 5x104 ions/sec). Raw data were
processed through Proteome Discoverer (v2.4.1.15) against the
UniProt human proteome database (Homo_SP_20201214.fasta; 20,395
sequences). Screening thresholds for differential proteins were as
follows: FC>1.5 and P<0.05. Protein-protein interaction (PPI)
networks were analyzed using the STRING database (v11.5; https://cn.string-db.org/; confidence level >0.9)
and visualized using Cytoscape (v3.10.3). Functional enrichment
analysis was conducted using ClusterProfiler (v4.2.2), and the
results were visualized with ggplot2 (v3.3.5). Tissue localization
of EV proteins was inferred by mapping expression profiles to
immunohistochemical data from the Human Protein Atlas (https://www.proteinatlas.org).
Integrative analysis of DEMs and
proteomics
The co-expression associations between DEMs and DEPs
were evaluated using Spearman's correlation coefficient, and were
visualized via heatmaps (v1.0.12). By integrating KEGG pathway
annotations of DEM target genes and DEPs, pathways significantly
enriched in both omics datasets were identified. Overlapping
pathways were illustrated using Venn diagrams (v1.6.20), available
on CRAN (https://cran.r-project.org/package=VennDiagram)
(30). Functional prioritization
of shared pathways was performed based on enrichment factor and
gene/protein coverage. From the miRNA-protein co-expression
network, highly dense functional modules were extracted using the
MCODE plugin (Cytoscape, v2.0.0; https://apps.cytoscape.org/apps/mcode). Core
regulatory hubs were identified via the Degree algorithm in the
CytoHubba plugin (Cytoscape, v0.1), which were defined as molecules
with the highest topological centrality.
Reverse transcription-quantitative PCR
(RT-qPCR)
Following plasma EV extraction from the HC and CC
groups, total RNA was isolated according to the manufacturer's
protocol of TRIzol™ reagent (cat. no. 15596026; Invitrogen; Thermo
Fisher Scientific, Inc.). The key steps included lysing the sample
in TRIzol™, followed by 5-min incubation at RT, addition of
chloroform (cat. no. C2432; MilliporeSigma), centrifugation to
separate the phases, collection of the aqueous phase, precipitation
of RNA with isopropanol (cat. no. 34863; MilliporeSigma), two
washes with 75% ethanol, air-drying and resuspension of the RNA
pellet in DEPC-treated water. RNA concentration and purity were
assessed using a NanoDrop spectrophotometer. For RT, LRP1 mRNA was
transcribed using the PrimeScript™ RT Kit (cat. no. RR047; Takara
Bio, Inc.) with 1 µg total RNA as a template, and the product was
diluted 5-fold for later use. To ensure specific detection of
mature hsa-miR-1-3p, a targeted stem-loop RT primer was used for
RT. qPCR was then performed using the SYBR Green Premix (cat. no.
AG11701; Hunan Accurate Bio-Medical Technology Co., Ltd.), which
was selected for its cost-effectiveness and compatibility with
simultaneous detection of both miRNA and mRNA targets, with a
reaction volume of 20 µl, including 2 µl diluted cDNA, 0.8 µl
forward and reverse primers (10 µM), 10 µl pre-mixed SYBR Green
Premix, and 6.4 µl ddH2O. The program was set to 95˚C
pre-denaturation for 1 min, followed by 45 cycles of 94˚C for 20
sec and 58˚C for 30 sec. Melting curve analysis was systematically
performed to verify amplification specificity (LineGene 9600 Plus;
Hangzhou Bioer Co., Ltd.). All samples were analyzed in triplicate
wells with blank controls, and relative expression levels were
calculated using the 2-ΔΔCq method (31). The internal references were GAPDH
(for LRP1) and U6 (for miR-1-3p). The primer sequences are shown in
Table I.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Primer | Sequence
(5'-3') | Note |
|---|
| hsa-miR-1-3p | F:
TGGAATGTAAAGAAGTATGTAT | Targets the mature
miR-1-3p sequence |
| | R:
GTGCAGGGTCCGAGGT | Universal reverse
primer |
| LRP1 | F:
CTATCGACGCCCCTAAGACTT | |
| | R:
CATCGCTGGGCCTTACTCT | |
| GAPDH |
TGTGGGCATCAATGGATTTGG | |
| |
ACACCATGTATTCCGGGTCAAT | |
Statistical analysis
Graphical and statistical analyses were performed
using the ggplot2 package (v3.3.5), GraphPad Prism (v8.0.12;
GraphPad; Dotmatics) and Cytoscape software (v3.10.2). Data from
three independent experiments are presented as the mean ± standard
deviation. Statistical comparisons between the HC and CC groups
were performed using an unpaired, two-tailed Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results and Discussion
EV isolation and characterization from
plasma samples
The present study included a total of 6 plasma
samples, comprising 3 patients with CC and 3 HCs. CC diagnosis was
confirmed according to the 2018 FIGO staging criteria (32), supplemented by imaging, molecular
pathology, and histopathological biopsy results. The overall
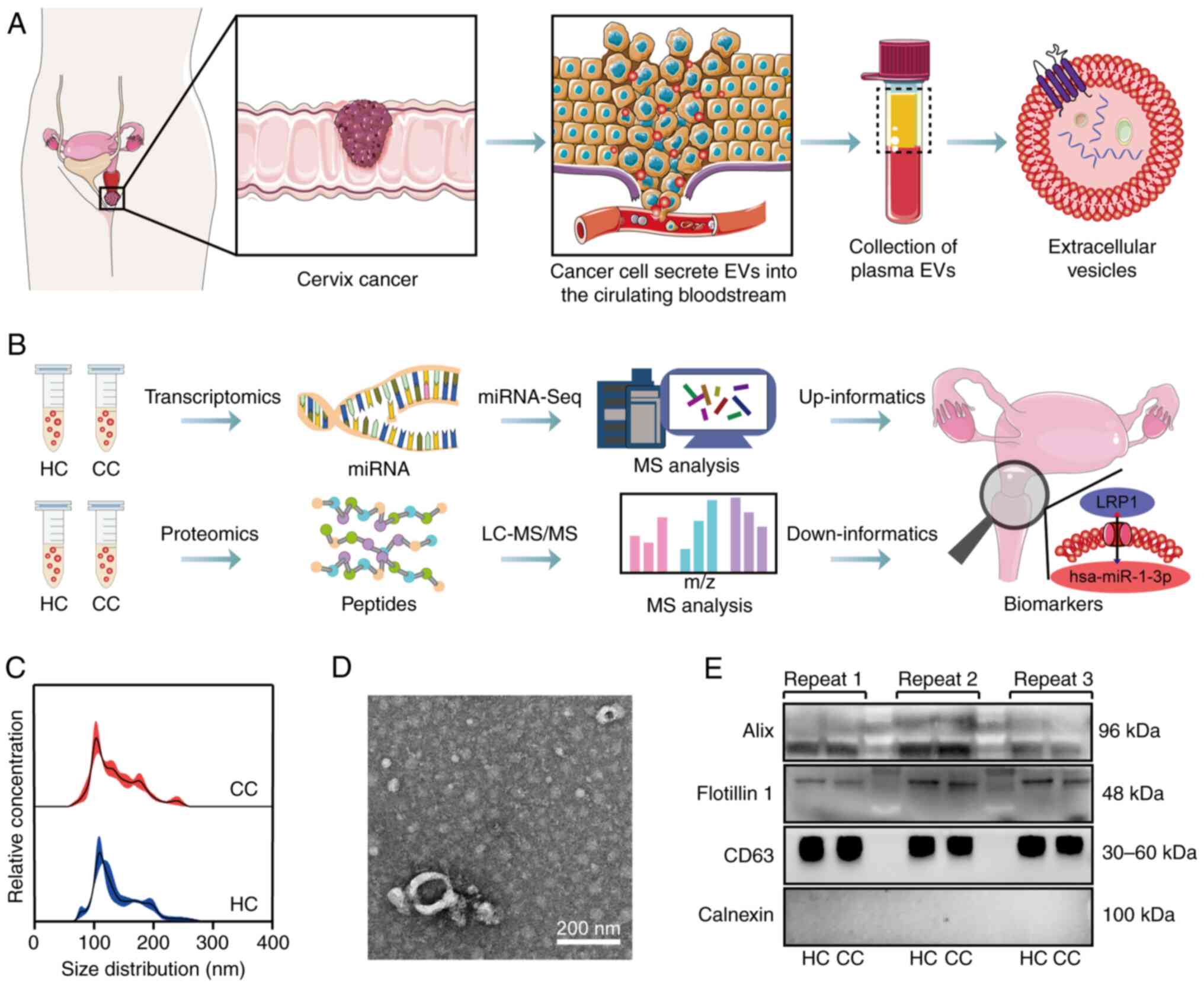
experimental workflow is revealed in Fig. 1A, while the analytical pipeline for
miRNA transcriptomics, proteomics and multi-omics integration is
outlined in Fig. 1B. EVs were
isolated from plasma using UC. NTA showed EV sizes predominantly
ranging from 50-200 nm, with peak sizes of 105 nm (CC) and 111 nm
(HC) (Fig. 1C). No significant
intergroup differences were observed in particle size distribution
or concentration (~1.5x1010 particles/ml). TEM imaging
demonstrated EV exhibiting typical cup-shaped or biconcave discoid
morphology (Fig. 1D). Following
the MISEV2018 guidelines (33), WB
analysis confirmed EV' purity, demonstrating consistent expression
of positive markers (Alix, Flotillin-1 and CD63) and absence of
calnexin contamination (20 µg protein load; Fig. 1E). Semi-quantitative analysis
showed stable marker expression without significant differences
between the HC and CC groups (Fig.
S1A-C).
Differential expression and functional
implications of EV-derived miRNAs in CC
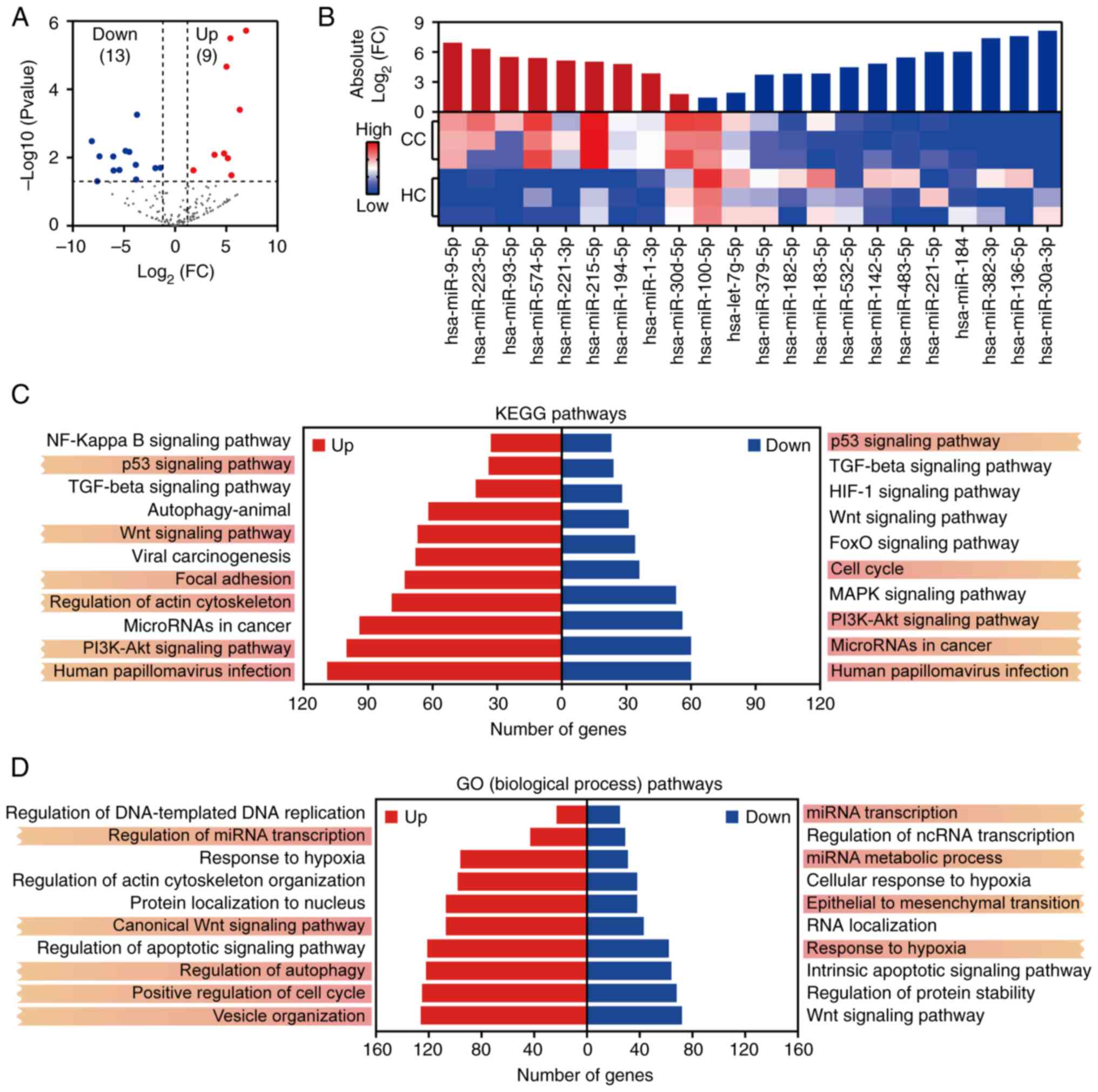
miRNA sequencing identified 2,656 miRNAs, with 22
DEMs (9 upregulated and 13 downregulated in CC) using DESeq2 (FC
>1.2, P<0.05; Fig. 2A).
Hierarchical clustering showed distinct disease-specific expression
profiles (Fig. 2B). To further
investigate the biological functions of these DEMs, their target
genes were predicted using the miRTarBase database. These predicted
targets were then subjected to KEGG and GO enrichment analyses.
KEGG analysis showed that DEM targets were significantly enriched
in several key cancer-related pathways, including the p53 signaling
pathway, PI3K-Akt signaling pathway, Wnt signaling pathway, focal
adhesion and HPV infection (Fig.
2C). These pathways are critically involved in HPV-associated
oncogenesis, immune evasion mechanisms, chemoresistance and TME
signaling (34,35). GO enrichment analysis further
revealed that these miRNAs were involved in biological processes
such as regulation of miRNA transcription, canonical Wnt signaling
pathway, vesicle organization, regulation of autophagy and positive
regulation of the cell cycle (Fig.
2D). Collectively, these findings suggest that EV-derived
miRNAs may contribute to CC progression through multiple functional
modules, including HPV-induced cell cycle dysregulation,
EV-mediated tumor-stroma communication and stress response
regulation (36,37).
Specific miRNA-pathway interactions were further
investigated. The target genes of miR-221-3p and miR-182-5p were
significantly enriched in HPV infection and PI3K-Akt signaling,
while hsa-miR-30a-3p and hsa-miR-194-5p were enriched in p53
signaling. This suggests that EV-derived miRNAs may synergize with
viral oncoproteins to remodel oncogenic signaling networks and
promote chemoresistance. For example, miR-30a-3p may enhance
chemoresistance by inhibiting downstream targets of p53(38). This indicates that EV miRNAs may
act as amplifiers of HPV oncogenesis and promote tumor
heterogeneity through intercellular communication; thus, targeting
these miRNAs may reverse chemoresistance. Furthermore, the
enrichment of DEM target genes in positive regulation of the cell
cycle, vesicle organization, miRNA transcription and regulation of
actin cytoskeleton organization pathways likely contributes to CC
proliferation and invasion. Specifically, hsa-miR-221-3p and
hsa-miR-30d-5p both target CASP3, a key regulator of apoptosis and
tumor cytotoxicity (39), which
may influence apoptotic processes. Additionally, hsa-miR-221-3p may
affect transcription factor activity (40). hsa-miR-9-5p and hsa-miR-93-5p were
associated with miRNA transcription, suggesting their potential
involvement in epigenetic feedback loops that drive tumor
heterogeneity (41,42).
Proteomic profiling of EV reveals
immune, metabolic and coagulation signatures in CC
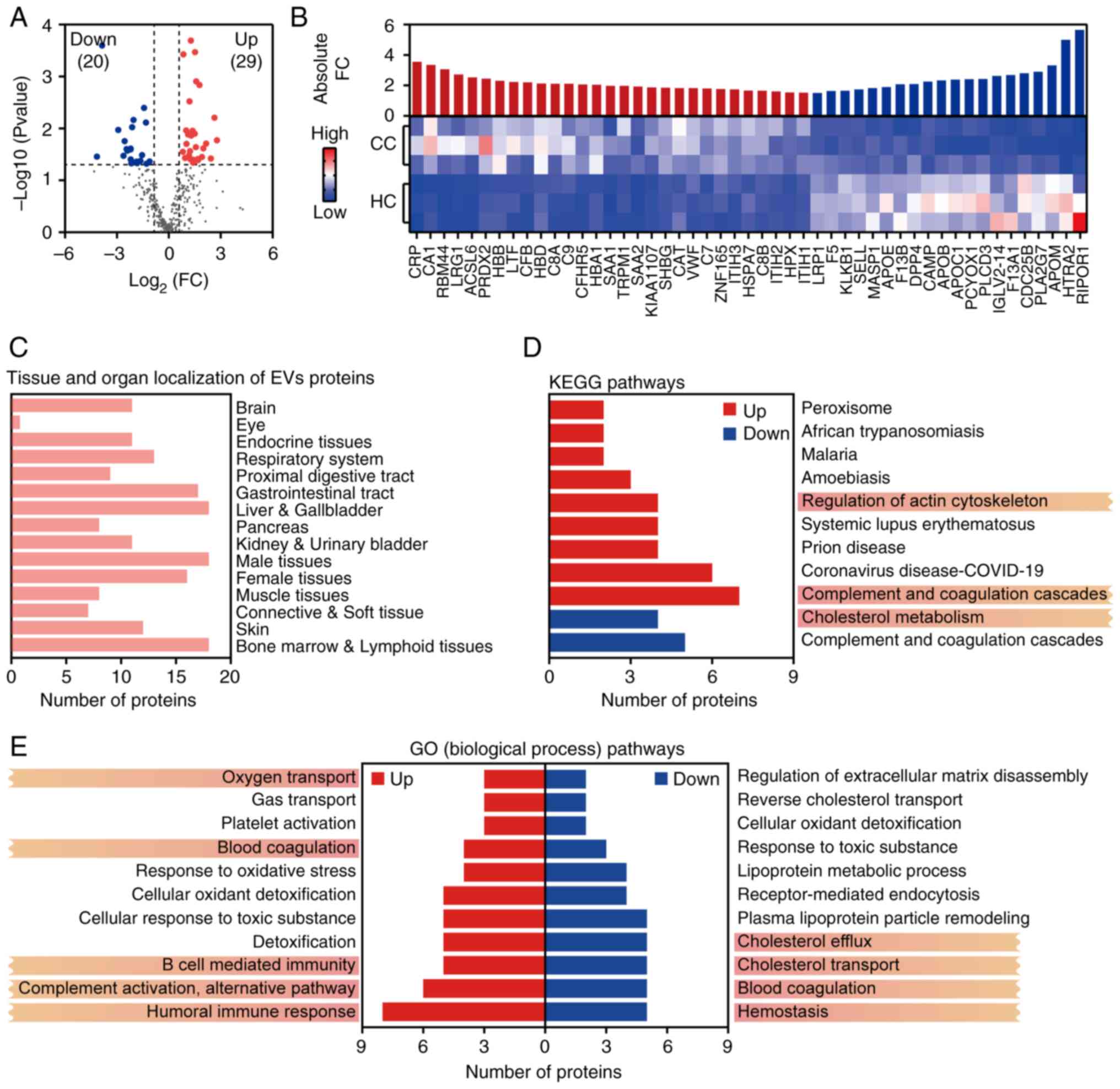
Proteomic analysis identified 363 EV-associated
proteins, with 49 DEPs (29 upregulated and 20 downregulated) using
limma (FC >1.5, P<0.05; Fig.
3A). Hierarchical clustering revealed disease-specific protein
signatures (Fig. 3B). Tissue
origin analysis indicated that DEPs were primarily derived from the
liver, gallbladder, female reproductive organs and gastrointestinal
tract (Fig. 3C). This
tissue-specific distribution highlights EVs as promising biomarkers
for reflecting pathophysiological alterations, particularly in
reproductive and digestive system-related malignancies. To explore
the functional roles of these DEPs, GO and KEGG enrichment analyses
were conducted. KEGG pathway analysis showed significant enrichment
of DEPs in the complement and coagulation cascades, regulation of
actin cytoskeleton, and cholesterol metabolism pathways (Fig. 3D). GO enrichment further identified
DEP involvement in complement activation, the alternative
complement pathway, blood coagulation and lipopolysaccharide
(LPS)-mediated signaling (Fig.
3E). These pathways may collectively drive CC progression by
modulating immunosuppressive TMEs, thus enhancing cell motility and
invasion, and facilitating HPV genome integration.
Previous studies have shown that HPV-infection tumor
cells can escape immune escape by inhibiting the activity of
natural killer and T cells (43).
The present findings further support this mechanism, as multiple
complement-related proteins (including C5, C7, C8A/B, C9 and CFB)
were significantly upregulated in CC-EV and enriched in the
alternative complement activation pathway (44,45).
These observations align with previous research suggesting that EV
contribute to immunosuppressive environments via complement system
modulation (46). Additionally,
inflammatory mediators such as CRP and SAA1/2 were highly expressed
and mapped to pathways related to LPS signaling and acute
inflammatory responses (47,48).
These proteins may contribute to an inflammatory microenvironment
that supports persistent HPV infection and tumor progression. EVs
also carry elevated levels of antioxidant enzymes, including
catalase and peroxiredoxin-2, which are known to neutralize
reactive oxygen species and maintain redox balance, thereby
preserving tumor cell viability under oxidative stress (49,50).
Lipid metabolism-related proteins, such as APOE, APOB and APOC1,
were significantly enriched in the cholesterol metabolism pathway,
while pattern recognition receptors such as TLR4 were involved in
activating the NF-κB pathway. For example, overexpression of APOE
in EVs may enhance CC cell invasion via the PI3K-Akt signaling
pathway (51). This suggests that
EV act as molecular hubs integrating metabolic reprogramming and
inflammatory signaling to support a tumor-permissive
microenvironment and sustain malignant transformation. In the
current study, aberrant expression of F5, F13A1 and F13B was
observed. These coagulation factors, which are involved in pathways
such as blood coagulation and platelet activation, may be
associated with CC angiogenesis and metastasis. Collectively, these
specific protein signatures offer promising targets for EV-based
liquid biopsy, and may provide new avenues for mechanistic studies
and therapeutic intervention in CC.
Integrated multi-omics analysis
reveals a key miRNA-protein regulatory axis in CC-EV
To systematically dissect the molecular regulatory
network mediated by EV in CC, an integrative analysis combining
EV-derived miRNA expression profiles and proteomic data was
performed. This multidimensional approach aimed to identify key
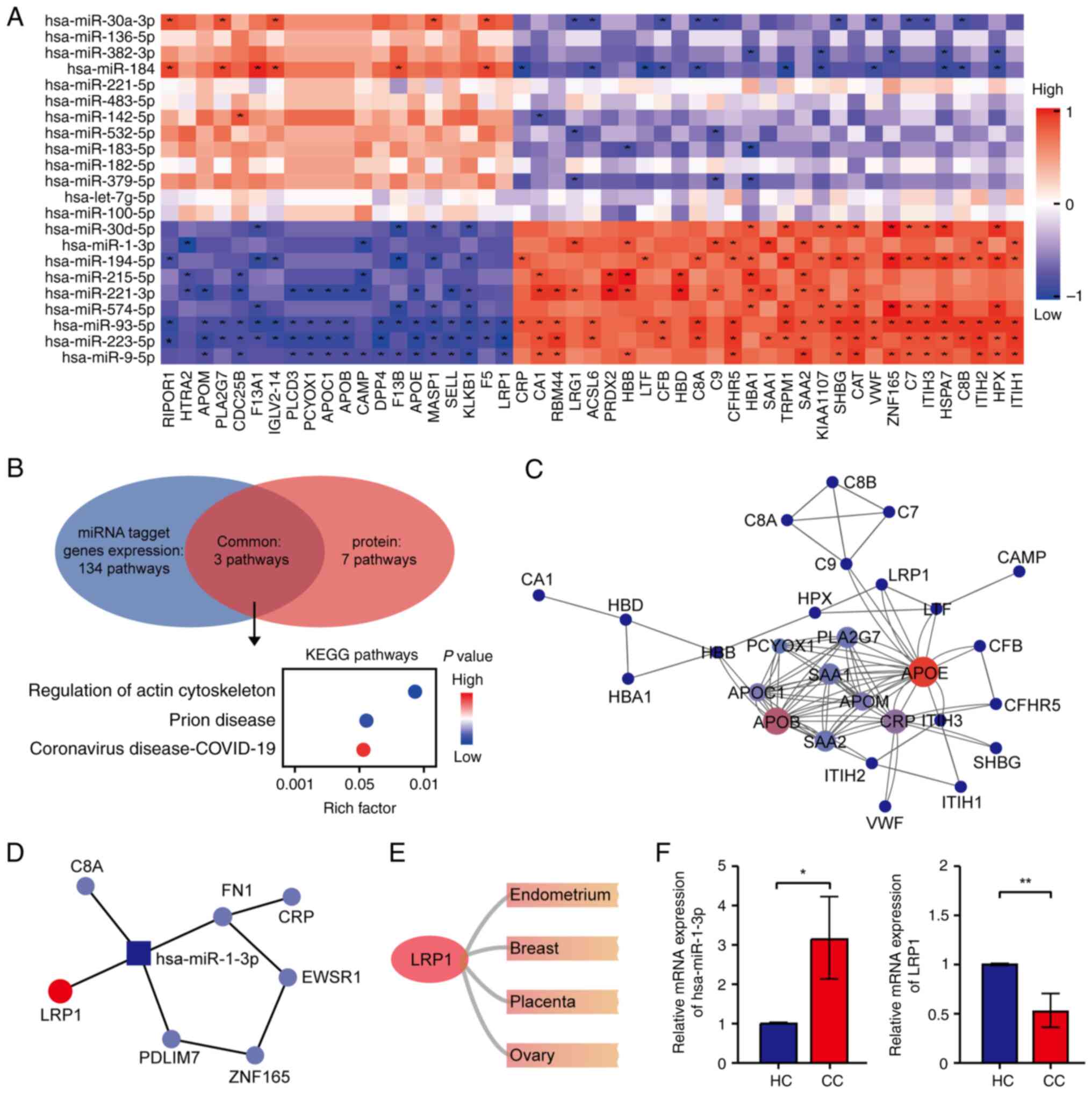
regulatory axes and diagnostic biomarkers. Spearman correlation
analysis revealed significant positive and negative associations
between DEMs and DEPs, suggesting that EV-contained miRNAs may
regulate downstream protein expression via direct targeting
(Fig. 4A). Pathway enrichment
analysis of co-expressed DEMs and DEPs demonstrated convergence in
three major biological pathways, namely regulation of the actin
cytoskeleton, prion disease and coronavirus disease-19 (Fig. 4B). These results suggested that
dysregulated EV-derived miRNAs may modulate cytoskeletal
remodeling, influence viral or misfolded protein handling, and
shape inflammatory responses within the TME. Notably, targeting of
the actin cytoskeleton regulator LRP1 by EV miRNAs suggests a
mechanistic link to enhanced CC cell motility and invasiveness
(52,53).
To further investigate protein-level regulatory
hubs, PPI network analysis identified APOE, CRP and APOB, which are
lipid metabolism-related proteins, as central nodes among DEPs
(Fig. 4C), thus highlighting lipid
metabolic reprogramming as a major driver of CC-associated EV
signaling. Cross-referencing this network with miRNA-protein
interactions revealed that hsa-miR-1-3p was the only miRNA directly
connected to multiple hub proteins, including LRP1, FN1, C8A and
PDLIM7 (Fig. 4D). Notably,
tissue-specific expression analysis showed that LRP1 was highly
expressed in female reproductive tissues, including the
endometrium, ovary and breast (Fig.
4E), thus underscoring its relevance in gynecological
malignancies. Previous research has shown that hsa-miR-1-3p
promotes non-small cell lung cancer progression by participating in
mechanosensitive mitotic processes (54), while activation of LRP1
fucosylation suppresses epithelial-mesenchymal transition (EMT) and
metastasis in CC (55). In the
present study, hsa-miR-1-3p was significantly upregulated in EV
from patients with CC, whereas LRP1 was significantly downregulated
(Fig. 4F), suggesting that this
miRNA may inhibit LRP1 expression to drive CC pathogenesis. These
findings indicate that elevated hsa-miR-1-3p in EV may inhibit
LRP1-mediated inflammatory chemotaxis, increase oxidative stress
within the TME, and activate Wnt/β-catenin signaling to promote
EMT. Such a mechanism aligns with previously described roles of
EV-carried miRNAs in tumor metastasis, and highlights the
hsa-miR-1-3p-LRP1 axis as a potential diagnostic and therapeutic
target (56). In summary, this
integrative multi-omics analysis uncovered a key EV-derived
miRNA-protein regulatory circuit in CC, providing novel insights
into disease progression and supporting the clinical utility of
hsa-miR-1-3p and LRP1 as candidate biomarkers for EV-based liquid
biopsy in CC.
Through integrated analysis of miRNA transcriptomics
and proteomics from plasma EV of HCs and patients with CC, the
present study unveiled a complex molecular regulatory network
during CC progression, which not only confirmed the potential of EV
as disease biomarkers, but, more importantly, revealed that
upregulated hsa-miR-1-3p in CC may participate in disease
progression by suppressing LRP1 expression.
The current study has demonstrated that
plasma-derived EV from patients with CC undergo marked alterations
in both miRNA and protein composition. The identified DEMs and
their predicted target genes were significantly enriched in
classical CC-related pathways such as p53 and PI3K-Akt (57,58).
Notably, through multi-omics integration analysis of plasma EV, it
was identified that activity changes in these pathways could be
carried and transmitted by EV. For instance, miR-221-3p and
miR-182-5p may simultaneously affect both HPV infection and the
PI3K-Akt signaling pathway, indicating that EV miRNAs may serve as
‘regulatory nodes’ coordinating the cross-talk between different
carcinogenic pathways, thereby amplifying the oncogenic effects of
HPV viral oncoproteins (59-62).
At the protein level, the present study revealed significant
enrichment of complement and coagulation cascades, suggesting that
tumor-derived EV may shape an immunosuppressive TME by modulating
the innate immune and coagulation systems, thus promoting immune
escape and angiogenesis (63,64).
Meanwhile, the dysregulation of cholesterol metabolism-related
proteins (such as APOE and APOB) coexists with the upregulation of
inflammatory mediators (such as CRP and SAA1/2), revealing a
vicious cycle where metabolic reprogramming and chronic
inflammation mutually promote each other in CC, with EVs serving as
important carriers for transmitting these signals (65,66).
Importantly, the current study found that EV-derived hsa-miR-1-3p
may suppress LRP1 expression, disrupt cytoskeletal stability and
subsequently activate pro-invasive signaling pathways such as
Wnt/β-catenin, ultimately accelerating CC metastasis. The present
research strategy connecting upstream regulatory molecules with
downstream functional proteins was able to construct a complete
signaling cascade from cause to effect, providing a new explanatory
framework for understanding the molecular mechanisms of CC.
In summary, the present study has systematically
delineated the molecular landscape of plasma EVs in CC through an
integrated multi-omics strategy and has proposed for the first time
that hsa-miR-1-3p-LRP1 is a key regulatory axis involved in disease
progression. This finding not only deepens the current
understanding of CC molecular mechanisms, but also lays a solid
theoretical foundation for developing EV-based non-invasive
diagnostic biomarkers and targeted therapeutic strategies.
Despite the aforementioned findings, the present
study has several limitations. First, the relatively small sample
size makes it difficult to accurately evaluate the dynamic changes
of the hsa-miR-1-3p-LRP1 axis during CC initiation and progression,
requiring validation through longitudinal studies in larger
cohorts. Second, the current study primarily provides correlative
evidence at the bioinformatics level; whether hsa-miR-1-3p directly
targets, and regulates LRP1 and its specific biological functions
need verification through functional experiments such as luciferase
reporter assays or gene knockdown/overexpression. Furthermore, the
mechanism of this regulatory axis in HPV-independent carcinogenesis
remains to be further explored through future in vitro and
in vivo models, which would be crucial for elucidating its
specific value in different CC subtypes.
Supplementary Material
Western blot analysis of EVs marker
proteins. (A-C) Expression of (A) Alix (~96 kDa), (B) Flotillin-1
(~48 kDa) and (C) CD63 (~30-60 kDa) in plasma-derived EVs from HC
and CC. Left panels show representative western blot bands; right
panels display semi-quantitative analysis results based on band
intensity (n=3; ns: no significant difference). EVs, extracellular
vesicles; CC, cervical cancer; HC, healthy controls.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the 2024 Qinghai
Provincial Clinical Key Specialty Construction Project Qinghui
Health Office [grant no. (2024) 23].
Availability of data and materials
The data generated in the present study may be found
in the Gene Expression Omnibus under accession number GSE310070 or
at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE310070;
and in the PRIDE Archive under accession number PXD071100 or at the
following URL: https://www.ebi.ac.uk/pride/archive/projects/PXD071100.
Authors' contributions
LQ and RY designed the present study and contributed
to the writing of the manuscript. TB, WL and XL collected clinical
samples performed, clinical analysis and contributed to
interpretation of data. YY and WZ were responsible for data
analysis and interpretation, manuscript revision, response to
reviewers' comments, and finalization of the manuscript. All
authors read and approved the final version of the manuscript. LQ
and RY confirm the authenticity of all raw data.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki. Ethical approval (approval no.
P-SL-202157) was obtained from the Ethics Committee of Qinghai
University Hospital (Xining, China). All participants received
written and verbal information about the study, including the right
to voluntary participation and withdrawal. Written and verbal
informed consent was obtained by all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Perkins RB, Wentzensen N, Guido RS and
Schiffman M: Cervical cancer screening: A review. JAMA.
330:547–558. 2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yuan Y, Cai X, Shen F and Ma F: HPV
post-infection microenvironment and cervical cancer. Cancer Lett.
497:243–254. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
De la Fuente-Hernandez MA,
Alanis-Manriquez EC, Ferat-Osorio E, Rodriguez-Gonzalez A,
Arriaga-Pizano L, Vazquez-Santillan K, Melendez-Zajgla J,
Fragoso-Ontiveros V, Alvarez-Gomez RM and Maldonado Lagunas V:
Molecular changes in adipocyte-derived stem cells during their
interplay with cervical cancer cells. Cell Oncol (Dordr).
45:85–101. 2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bower JJ, Vance LD, Psioda M, Smith-Roe
SL, Simpson DA, Ibrahim JG, Hoadley KA, Perou CM and Kaufmann WK:
Patterns of cell cycle checkpoint deregulation associated with
intrinsic molecular subtypes of human breast cancer cells. NPJ
Breast Cancer. 3(9)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liu Z, Lu Q, Zhang Z, Feng Q and Wang X:
TMPRSS2 is a tumor suppressor and its downregulation promotes
antitumor immunity and immunotherapy response in lung
adenocarcinoma. Respir Res. 25(238)2024.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kandathil SA, Peter Truta I, Kadletz-Wanke
L, Heiduschka G, Stoiber S, Kenner L, Herrmann H, Huskic H and
Brkic FF: Lymphocyte-to-monocyte ratio might serve as a prognostic
marker in young patients with tongue squamous cell carcinoma. J
Pers Med. 14(159)2024.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang Y, Zhang Q, Han X, Han L, Wang T, Hu
J, Li L, Ding Z, Shi X and Qian X: SLAMF8, a potential new immune
checkpoint molecule, is associated with the prognosis of colorectal
cancer. Transl Oncol. 31(101654)2023.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Garcia J, Hurwitz HI, Sandler AB, Miles D,
Coleman RL, Deurloo R and Chinot OL: Bevacizumab (Avastin®) in
cancer treatment: A review of 15 years of clinical experience and
future outlook. Cancer Treat Rev. 86(102017)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pegtel DM and Gould SJ: Exosomes. Annu Rev
Biochem. 88:487–514. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Quan J, Liu Q, Li P, Yang Z, Zhang Y, Zhao
F and Zhu G: Mesenchymal stem cell exosome therapy: Current
research status in the treatment of neurodegenerative diseases and
the possibility of reversing normal brain aging. Stem Cell Res
Ther. 16(76)2025.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jahan S, Mukherjee S, Ali S, Bhardwaj U,
Choudhary RK, Balakrishnan S, Naseem A, Mir SA, Banawas S,
Alaidarous M, et al: Pioneer role of extracellular vesicles as
modulators of cancer initiation in progression, drug therapy, and
vaccine prospects. Cells. 11(490)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lee Y, Ni J, Beretov J, Wasinger VC,
Graham P and Li Y: Recent advances of small extracellular vesicle
biomarkers in breast cancer diagnosis and prognosis. Mol Cancer.
22(33)2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Fang SB, Zhou ZR, Peng YQ, Liu XQ, He BX,
Chen DH, Chen D and Fu QL: Plasma EVs display antigen-presenting
characteristics in patients with allergic rhinitis and promote
differentiation of Th2 cells. Front Immunol.
12(710372)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Woo HK, Park J, Ku JY, Lee CH, Sunkara V,
Ha HK and Cho YK: Urine-based liquid biopsy: Non-invasive and
sensitive AR-V7 detection in urinary EVs from patients with
prostate cancer. Lab Chip. 19:87–97. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hu L, Zhang T, Ma H, Pan Y, Wang S, Liu X,
Dai X, Zheng Y, Lee LP and Liu F: Discovering the secret of
diseases by incorporated tear exosomes analysis via rapid-isolation
system: iTEARS. ACS Nano. 16:11720–11732. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Casanova-Salas I, Aguilar D,
Cordoba-Terreros S, Agundez L, Brandariz J, Herranz N, Mas A,
Gonzalez M, Morales-Barrera R, Sierra A, et al: Circulating tumor
extracellular vesicles to monitor metastatic prostate cancer
genomics and transcriptomic evolution. Cancer Cell.
42:1301–1312.e7. 2024.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lucien F, Gustafson D, Lenassi M, Li B,
Teske JJ, Boilard E, von Hohenberg KC, Falcón-Perez JM, Gualerzi A,
Reale A, et al: MIBlood-EV: Minimal information to enhance the
quality and reproducibility of blood extracellular vesicle
research. J Extracell Vesicles. 12(e12385)2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Thippabhotla S, Zhong C and He M: 3D cell
culture stimulates the secretion of in vivo like extracellular
vesicles. Sci Rep. 9(13012)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li Y, Zhao J, Yu S, Wang Z, He X, Su Y,
Guo T, Sheng H, Chen J, Zheng Q, et al: Extracellular vesicles long
RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human
blood as potential biomarkers for cancer diagnosis. Clin Chem.
65:798–808. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Garcia-Martin R, Wang G, Brandão BB,
Zanotto TM, Shah S, Kumar Patel S, Schilling B and Kahn CR:
MicroRNA sequence codes for small extracellular vesicle release and
cellular retention. Nature. 601:446–451. 2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hasanzadeh M, Movahedi M, Rejali M, Maleki
F, Moetamani-Ahmadi M, Seifi S, Hosseini Z, Khazaei M, Amerizadeh
F, Ferns GA, et al: The potential prognostic and therapeutic
application of tissue and circulating microRNAs in cervical cancer.
J Cell Physiol. 234:1289–1294. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hoshino A, Kim HS, Bojmar L, Gyan KE,
Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H,
Heissel S, et al: extracellular vesicle and particle biomarkers
define multiple human cancers. Cell. 182:1044–1061.e18.
2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Blaser MC, Buffolo F, Halu A, Turner ME,
Schlotter F, Higashi H, Pantano L, Clift CL, Saddic LA, Atkins SK,
et al: Multiomics of tissue extracellular vesicles identifies
unique modulators of atherosclerosis and calcific aortic valve
stenosis. Circulation. 148:661–678. 2023.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yuan X, Sun L, Jeske R, Nkosi D, York SB,
Liu Y, Grant SC, Meckes DG Jr and Li Y: Engineering extracellular
vesicles by three-dimensional dynamic culture of human mesenchymal
stem cells. J Extracell Vesicles. 11(e12235)2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang H, Deng B, Jiang Y, et al: Proteomic
analysis of cerebrospinal fluid exosomes derived from cerebral
palsy children. Chinese Journal of Tissue Engineering Research.
27:903–908. 2023.(In Chinese). https://d.wanfangdata.com.cn/periodical/xdkf202306016.
|
|
26
|
Sheng J and Zhang WY: Identification of
biomarkers for cervical cancer in peripheral blood lymphocytes
using oligonucleotide microarrays. Chin Med J (Engl).
123:1000–1005. 2010.PubMed/NCBI
|
|
27
|
Saleh M, Virarkar M, Javadi S, Elsherif
SB, de Castro Faria S and Bhosale P: Cervical cancer: 2018 revised
international federation of gynecology and obstetrics staging
system and the role of imaging. AJR Am J Roentgenol. 214:1182–1195.
2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tian Y, Gong M, Hu Y, Liu H, Zhang W,
Zhang M, Hu X, Aubert D, Zhu S, Wu L and Yan X: Quality and
efficiency assessment of six extracellular vesicle isolation
methods by nano-flow cytometry. J Extracell Vesicles.
9(1697028)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2(100141)2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen H and Boutros PC: VennDiagram: A
package for the generation of highly-customizable Venn and Euler
diagrams in R. BMC Bioinformatics. 12(35)2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Salib MY, Russell JHB, Stewart VR,
Sudderuddin SA, Barwick TD, Rockall AG and Bharwani N: 2018 FIGO
staging classification for cervical cancer: Added benefits of
imaging. Radiographics. 40:1807–1822. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Théry C, Witwer KW, Aikawa E, Alcaraz MJ,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin-Smith GK, et al: Minimal information for studies of
extracellular vesicles 2018 (MISEV2018): A position statement of
the international society for extracellular vesicles and update of
the MISEV2014 guidelines. J Extracell Vesicles.
7(1535750)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Aguayo F, Perez-Dominguez F, Osorio JC,
Oliva C and Calaf GM: PI3K/AKT/mTOR signaling pathway in HPV-driven
head and neck carcinogenesis: Therapeutic implications. Biology
(Basel). 12(672)2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sacconi A, Muti P, Pulito C, Urbani G,
Allegretti M, Pellini R, Mehterov N, Ben-David U, Strano S, Bossi P
and Blandino G: Immunosignatures associated with TP53 status and
co-mutations classify prognostically head and neck cancer patients.
Mol Cancer. 22(192)2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Khokhar M, Kartha P, Hassan S and Pandey
RK: Decoding dysregulated genes, molecular pathways and microRNAs
involved in cervical cancer. J Gene Med. 26(e3713)2024.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cao MX, Zhang WL, Yu XH, Wu JS, Qiao XW,
Huang MC, Wang K, Wu JB, Tang YJ, Jiang J, et al: Retraction Note:
Interplay between cancer cells and M2 macrophages is necessary for
miR-550a-3-5p down-regulation-mediated HPV-positive OSCC
progression. J Exp Clin Cancer Res. 40(310)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Park D, Kim H, Kim Y and Jeoung D: miR-30a
Regulates the expression of CAGE and p53 and regulates the response
to anti-cancer drugs. Mol Cells. 39:299–309. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bhat AA, Thapa R, Afzal O, Agrawal N,
Almalki WH, Kazmi I, Alzarea SI, Altamimi ASA, Prasher P, Singh SK,
et al: The pyroptotic role of Caspase-3/GSDME signalling pathway
among various cancer: A Review. Int J Biol Macromol. 242 (Pt
2)(124832)2023.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Shao X, Zheng Y, Huang Y, Li G, Zou W and
Shi L: Hsa-miR-221-3p promotes proliferation and migration in
HER2-positive breast cancer cells by targeting LASS2 and MBD2.
Histol Histopathol. 37:1099–1112. 2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kania EE, Carvajal-Moreno J, Hernandez VA,
English A, Papa JL, Shkolnikov N, Ozer HG, Yilmaz AS, Yalowich JC
and Elton TS: hsa-miR-9-3p and hsa-miR-9-5p as post-transcriptional
modulators of DNA topoisomerase IIα in human leukemia K562 cells
with acquired resistance to etoposide. Mol Pharmacol. 97:159–170.
2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Schoen C, Glennon JC, Abghari S, Bloemen
M, Aschrafi A, Carels CEL and Von den Hoff JW: Differential
microRNA expression in cultured palatal fibroblasts from infants
with cleft palate and controls. Eur J Orthod. 40:90–96.
2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li M, Song J, Wang L, Wang Q, Huang Q and
Mo D: Natural killer cell-related prognosis signature predicts
immune response in colon cancer patients. Front Pharmacol.
14(1253169)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
West EE, Kolev M and Kemper C: Complement
and the regulation of T cell responses. Annu Rev Immunol.
36:309–338. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Afshar-Kharghan V: The role of the
complement system in cancer. J Clin Invest. 127:780–789.
2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Loh JT, Zhang B, Teo JKH, Lai RC, Choo
ABH, Lam KP and Lim SK: Mechanism for the attenuation of neutrophil
and complement hyperactivity by MSC exosomes. Cytotherapy.
24:711–719. 2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Shiels MS, Katki HA, Hildesheim A,
Pfeiffer RM, Engels EA, Williams M, Kemp TJ, Caporaso NE, Pinto LA
and Chaturvedi AK: Circulating inflammation markers, risk of lung
cancer, and utility for risk stratification. J Natl Cancer Inst.
107(djv199)2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang W, Yin Y, Jiang Y, Yang Y, Wang W,
Wang X, Ge Y, Liu B and Yao L: Relationship between vaginal and
oral microbiome in patients of human papillomavirus (HPV) infection
and cervical cancer. J Transl Med. 22(396)2024.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Nadif R, Mintz M, Jedlicka A, Bertrand JP,
Kleeberger SR and Kauffmann F: Association of CAT polymorphisms
with catalase activity and exposure to environmental oxidative
stimuli. Free Radic Res. 39:1345–1350. 2005.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chen P, Zhong X, Song Y, Zhong W, Wang S,
Wang J, Huang P, Niu Y, Yang W, Ding Z, et al: Triptolide induces
apoptosis and cytoprotective autophagy by ROS accumulation via
directly targeting peroxiredoxin 2 in gastric cancer cells. Cancer
Lett. 587(216622)2024.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wan S, He QY, Yang Y, Liu F, Zhang X, Guo
X, Niu H, Wang Y, Liu YX, Ye WL, et al: SPARC Stabilizes ApoE to
Induce cholesterol-dependent invasion and sorafenib resistance in
hepatocellular carcinoma. Cancer Res. 84:1872–1888. 2024.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Garcia-Arcos I, Park SS, Mai M,
Alvarez-Buve R, Chow L, Cai H, Baumlin-Schmid N, Agudelo CW,
Martinez J, Kim MD, et al: LRP1 loss in airway epithelium
exacerbates smoke-induced oxidative damage and airway remodeling. J
Lipid Res. 63(100185)2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Shen S, Zhang S, Liu P, Wang J and Du H:
Potential role of microRNAs in the treatment and diagnosis of
cervical cancer. Cancer Genet. 248-249:25–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Song Y, Wang Z, He L, Sun F, Zhang B and
Wang F: Dysregulation of pseudogenes/lncRNA-Hsa-miR-1-3p-PAICS
pathway promotes the development of NSCLC. J Oncol.
2022(4714931)2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
He L, Guo Z, Wang W, Tian S and Lin R:
FUT2 inhibits the EMT and metastasis of colorectal cancer by
increasing LRP1 fucosylation. Cell Commun Signal.
21(63)2023.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Yang Y, Mao F, Guo L, Shi J, Wu M, Cheng S
and Guo W: Tumor cells derived-extracellular vesicles transfer
miR-3129 to promote hepatocellular carcinoma metastasis by
targeting TXNIP. Dig Liver Dis. 53:474–485. 2021.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wei W and Liu C: Prognostic and predictive
roles of microRNA-411 and its target STK17A in evaluating
radiotherapy efficacy and their effects on cell migration and
invasion via the p53 signaling pathway in cervical cancer. Mol Med
Rep. 21:267–281. 2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Fu K, Zhang L, Liu R, Shi Q, Li X and Wang
M: MiR-125 inhibited cervical cancer progression by regulating VEGF
and PI3K/AKT signaling pathway. World J Surg Oncol.
18(115)2020.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Yi Y, Liu Y, Wu W, Wu K and Zhang W: The
role of miR-106p-5p in cervical cancer: From expression to
molecular mechanism. Cell Death Discov. 4(36)2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Weiss BG, Anczykowski MZ, Ihler F,
Bertlich M, Spiegel JL, Haubner F, Canis M, Küffer S, Hess J, Unger
K, et al: MicroRNA-182-5p and microRNA-205-5p as potential
biomarkers for prognostic stratification of p16-positive
oropharyngeal squamous cell carcinoma. Cancer Biomark. 33:331–347.
2022.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Babion I, Miok V, Jaspers A, Huseinovic A,
Steenbergen RDM, van Wieringen WN and Wilting SM: Identification of
deregulated pathways, key regulators, and novel miRNA-mRNA
interactions in HPV-mediated transformation. Cancers (Basel).
12(700)2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Cui G, Wang L and Huang W: Circular RNA
HIPK3 regulates human lens epithelial cell dysfunction by targeting
the miR-221-3p/PI3K/AKT pathway in age-related cataract. Exp Eye
Res. 198(108128)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Becker A, Thakur BK, Weiss JM, Kim HS,
Peinado H and Lyden D: Extracellular vesicles in cancer:
Cell-to-cell mediators of metastasis. Cancer Cell. 30:836–848.
2016.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Dobrolecki LE, Airhart SD, Alferez DG,
Aparicio S, Behbod F, Bentires-Alj M, Brisken C, Bult CJ, Cai S,
Clarke RB, et al: Patient-derived xenograft (PDX) models in basic
and translational breast cancer research. Cancer Metastasis Rev.
35:547–573. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Vendrov AE, Lozhkin A, Hayami T, Levin J,
Silveira Fernandes Chamon J, Abdel-Latif A, Runge MS and Madamanchi
NR: Mitochondrial dysfunction and metabolic reprogramming induce
macrophage pro-inflammatory phenotype switch and atherosclerosis
progression in aging. Front Immunol. 15(1410832)2024.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Sarwar MS, Cheng D, Peter RM, Shannar A,
Chou P, Wang L, Wu R, Sargsyan D, Goedken M, Wang Y, et al:
Metabolic rewiring and epigenetic reprogramming in leptin
receptor-deficient db/db diabetic nephropathy mice. Eur J
Pharmacol. 953(175866)2023.PubMed/NCBI View Article : Google Scholar
|