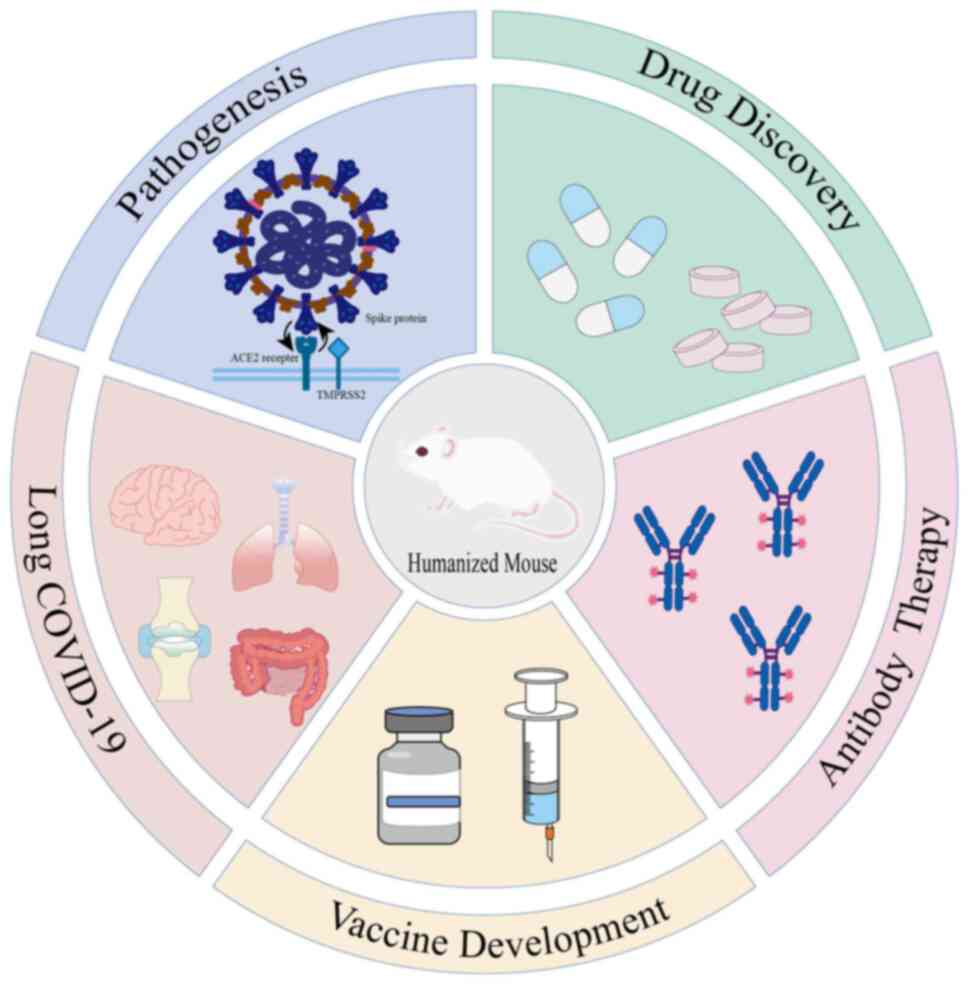
At the end of 2019, a novel coronavirus disease
(COVID-19) caused by severe acute respiratory syndrome coronavirus
2 (SARS-CoV-2) spread rapidly around the globe, causing an
unprecedented impact on the public health. SARS-CoV-2 belongs to
the genus β-coronavirus, a single-stranded positive RNA virus with
a genome length of ~30 kb (1). Among
the structural proteins encoded by its genome, the spike protein
consists of the S1 (S protein 1) and S2 (S protein 2) subunits, and
binds to angiotensin-converting enzyme 2 (ACE2) on the surface of
the host cells via a receptor-binding domain (RBD), mediating viral
invasion and triggering infection. The process relies on the
activation of S protein cleavage by host proteases, namely
transmembrane serine protease 2 (TMPRSS2) (2). The highly transmissible nature of the
virus allows for widespread dissemination through droplets, contact
and aerosolization, resulting in progression from asymptomatic
infection to severe pneumonia, acute respiratory distress syndrome
and multi-organ failure with heterogeneous clinical presentations
(3,4). Elderly patients with comorbid diseases
have significantly higher rates of severe illness and mortality,
highlighting the key role of host genetic background and viral
interactions in the process of the disease (5). The long-term consequences of viral
infections are gradually becoming apparent, with ~10-30% of
recovered patients experiencing persistent fatigue, cognitive
impairment and multi-system functional abnormalities; this
phenomenon has been defined as long COVID (6).
In order to reveal the viral pathogenesis and
develop intervention strategies, it has become imperative to
establish animal models that can mimic human pathological
characteristics. The mouse, as the most convenient laboratory
mammal, is an ideal candidate due to its well-characterized genetic
background and short reproductive cycle. While studies have
identified alternative transmembrane proteins, such as CD147 and
CD26 that may facilitate viral entry, ACE2 remains the primary
gateway for SARS-CoV-2 to enter host cells (7). However, a natural species barrier
exists: The mouse ACE2 (mACE2) receptor differs from human ACE2
(hACE2) at key amino acid residues, rendering mice naturally
insensitive to SARS-CoV-2(8). This
biological bottleneck has driven researchers to develop humanized
mouse models through genetic engineering and immune reconstitution
techniques. These models introduce human genes, cells, or tissues
into immunodeficient mice to overcome species restrictions and
replicate human infection characteristics. Numerous studies
(9-13)
have demonstrated the utility of humanized mouse models in COVID-19
research, including applications in viral pathogenesis, antiviral
drug discovery and vaccine development. The present review aimed to
systematically document the construction strategies of receptor
humanization, immune system humanization and composite humanization
models. We highlight their translational value in elucidating
virus-host interactions, developing broad-spectrum antiviral
therapies, optimizing antibody-based treatments and investigating
the pathophysiology of long COVID (Fig.
1).
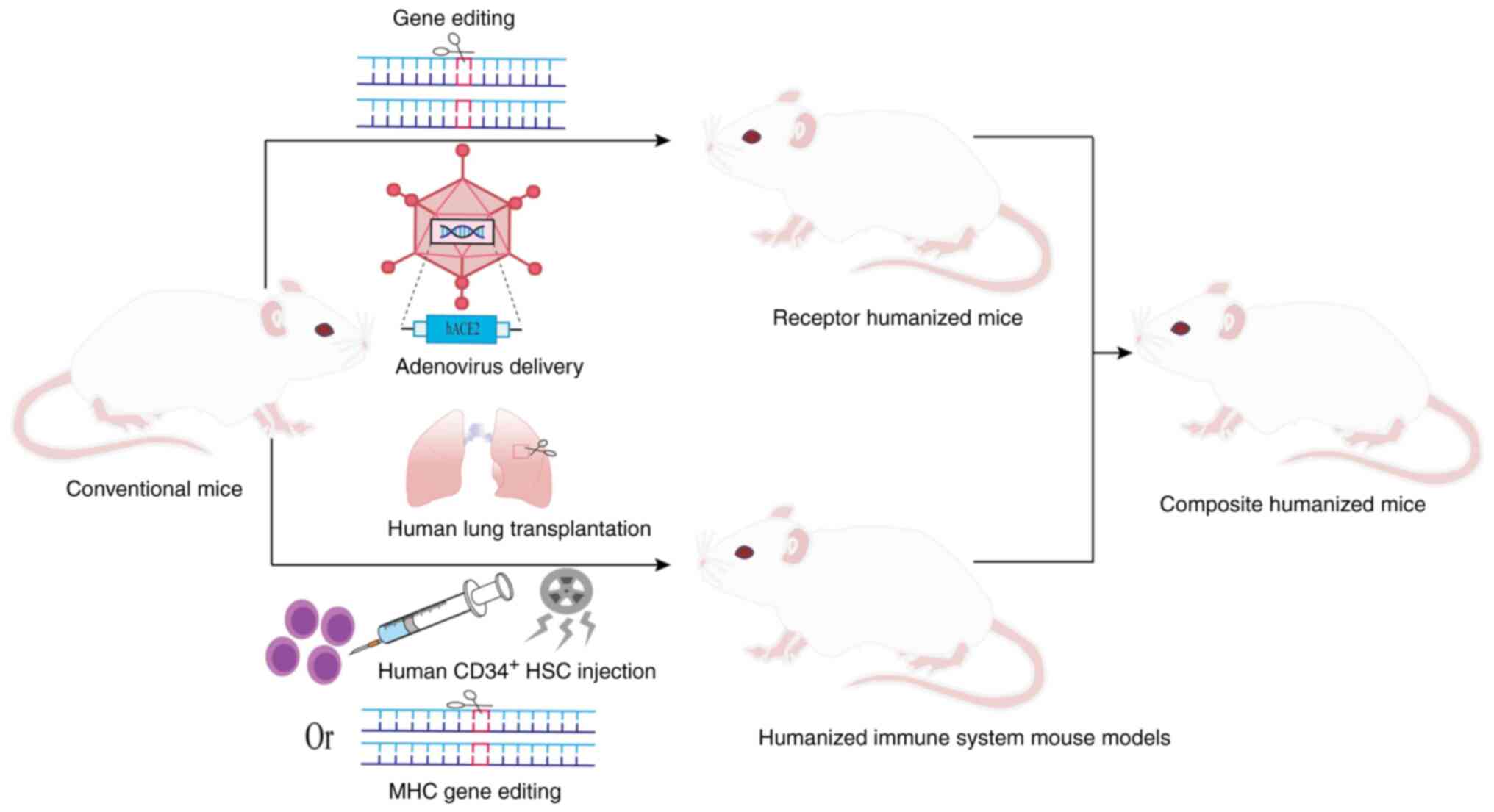
To overcome the species barrier and simulate human
SARS-CoV-2 infection, a variety of humanization strategies have
been employed (Fig. 2), as described
below.
Receptor humanized mouse models were constructed to
systematically mimic the functional properties of human receptors.
K18 (cytokeratin 18)-hACE2 transgenic mice expressing hACE2 under
the K18 promoter are susceptible to SARS-CoV-2(14). In the hACE2 mouse model created by
Snouwaert et al (15) using
homologous substitution, mouse promoter-driven ACE2 was highly
expressed in airway club cells, whereas human promoter-driven ACE2
was expressed in alveolar type II cells. Ge et al (16) developed a fully humanized ACE2 mouse
model that achieves tissue distribution and expression levels
closer to those of human ACE2, thereby avoiding the expression
abnormalities caused by heterologous promoters. Using clustered
regularly interspaced short palindromic repeats (CRISPR)-Cas9 to
introduce a single amino acid substitution (H353K) in the
mouse ACE2 gene, the mACE2H353K mouse model was
successfully constructed (17,18). The
application of complementary embryonic stem cell and tetraploid
techniques has greatly accelerated model construction (19). hACE2-knock-in (KI) mice constructed
by knocking hACE2 into mice via CRISPR-Cas technology have been
used to study the mechanisms of pulmonary-intestinal dual
infections and intestinal flora disruption and the pathology of tau
protein in long COVID (20,21). Similarly, the generation of
hACE2-KI-NOD/SCID gamma (NSG) mice by replacing mACE2 with hACE2 in
NSG-immunodeficient mice significantly enhances their viral
susceptibility (22). Furthermore,
using hACE2 and/or hTMPRSS2 KI mice, researchers have found that
Delta strains utilize membrane fusion for host entry, whereas
Omicron strains employ endocytic pathways (23).
The use of conditional regulation systems enables
the spatiotemporal control of receptor expression. Beyond
hACE2-floxed (flanked by loxP sites) mice constructed by Cre
recombinase-mediated knockout (24),
an inducible bi-gene system was developed to tandemly connect hACE2
with green fluorescent protein downstream of the floxed STOP
cassette, achieve spatiotemporal specific expression by
tamoxifen-induced Ubi-Cre (ubiquitin promoter-driven Cre
recombinase) (25). This research
was extended to non-ACE2 receptors and synergistic mechanisms
through the humanized dendritic cell (hDC)-specific intercellular
adhesion molecule-3-grabbing non-integrin (SIGN) mouse model
constructed by replacing mouse CD209a with human DC-SIGN (26), as well as through a humanized
transferrin receptor (hTfR) mouse model (27) and a humanized model of the CD147
receptor (28). Among them, the
severity of pulmonary fibrosis in the hCD147 mouse model exceeds
that of the conventional hACE2 model. A summary of the general
information on receptor-based construction of humanized mouse
models is presented in Table I.
The major histocompatibility complex (MHC) is known
as the human leukocyte antigen (HLA) system in humans and is
crucial for adaptive immunity. In the humanized mouse models of the
immune system, a DRAGA mouse model was constructed by the
post-irradiation infusion of HLA-matched umbilical cord
blood-derived CD34+ hematopoietic stem cells (HSCs) and
combined with transgenic technology to stably express HLA in the
thymus (29,30). This design not only avoids
graft-vs.-host disease but also promotes immunoglobulin class
switching in human B cells, enabling long-term stable immune system
reconstitution. Humanized HLA transgenic mice mimic human T-cell
epitope presentation and post-vaccination immune responses, which
avoid the severe pathological manifestations caused by the high
susceptibility of traditional hACE2 transgenic mice (31,32).
Furthermore, the humanized mouse model of the localized lung was
constructed by transplanting human lung tissues into
immunodeficient mice. In the SCID-hu model, transplanted human
fetal lung tissues developed mature bronchioles, alveolar sacs, and
vascular systems, and expressed the key receptor ACE2, providing
target cells for SARS-CoV-2 infection (33). Similarly, the NSG-L model
demonstrated efficient viral replication in grafts through the
subcutaneous implantation of human lung tissue (34).
The construction of composite humanized mouse models
can more comprehensively and accurately simulate human infection
and immune response to SARS-CoV-2 than a single humanized mouse.
The MISTRG6 model supports the development of human myeloid and
lymphoid lineage cells by replacing the corresponding mouse genes
with human homologs. This model further achieves the dual
humanization of the immune system and receptor expression by
combining adeno-associated virus delivery of hACE2 to the lung
(35-37).
Similarly, the respiratory tract and immune system humanization in
immunodeficient mice is achieved by the adenoviral delivery of the
hACE2 gene to the lung of NIKO mice, combined with CD34+
HSCs transplantation (38). The
HHD-DR1 model combines immune system and receptor humanization by
knocking out mouse MHC molecules, introducing human MHC alleles,
and driving hACE2 expression in the lung and brain (39). Additionally, the Alpha variant was
found to be more infectious than the earlier virus 614D in the
TKO-BLT-L mouse model constructed by transplanting bone marrow,
liver, thymus and autologous lung tissue grafts (40). The HNFL mouse model constructed by
combining human lung tissues with myeloid-enhanced immune system
revealed the central role of myeloid immune cells in protecting
lung tissues from SARS-CoV-2 infection (41). It is recommended that constructing
single-, dual- and triple-gene composite models through precise
knock-in of human ACE2, TMPRSS2 and Fcγ genes can meet the
requirements for co-expression of multiple receptors (42).
In summary, the humanized mouse models constructed
by various strategies lay an important foundation for the study of
the pathogenic mechanism of SARS-CoV-2, the discovery of
anti-SARS-CoV-2 drugs and the development of vaccines.
The humanized mouse model has greatly enhanced the
understanding of the pathogenesis of SARS-CoV-2 and provides a key
basis for targeted therapy and for predicting the progression of
the disease. Ye et al (43)
demonstrated that, in hACE2 mice, that the virus targets
non-neuronal olfactory epithelial cells, causing structural damage,
immune cell infiltration and the downregulation of olfactory
receptor genes, mirroring the transient anosmia of patients with
COVID-19. A previous study using the K18-hACE2 model revealed that
SARS-CoV-2 induces non-classical NLR family pyrin domain containing
3 (NLRP3) inflammasome activation via caspase-11, promoting the
release of IL-1β/IL-18 and aggravating lung inflammation and
mortality (9). In hACE2 mice, the
RBD binding of the SARS-CoV-2 spike protein to ACE2 on mast cells
triggers aggregation, degranulation, histamine and protease
release, causing tracheobronchial and lung inflammation (44,45).
Sefik et al (37) further
demonstrated that, in MISTRG6-hACE2 mice, infected macrophages
activated the NLRP3 inflammasome following viral uptake via CD16
and ACE2, driving chronic lung fibrosis. Additionally, neutrophils
combat infection via phagocytosis, cytotoxic mediators and
neutrophil extracellular traps (NETs). In K18-hACE2 mice, NETs trap
SARS-CoV-2 particles; however, their excessive release damages
tissue, highlighting the dual role of NETs (46). Notably, host factors play a key role
in viral pathogenesis. Chuang et al (47) found that, in hACE2-KI mice, spike
protein enhances viral susceptibility by activating ACE2
phosphorylation, inhibiting its degradation and promoting exosomal
ACE2 propagation. The SARS-CoV-2 virulence factor, open reading
frame (ORF)8 enhances pathogenicity by modulating host immune
dynamics, including early interferon enhancement and late
inflammatory suppression. Its deletion has been shown to reduce the
mortality of K18-hACE2 mice by 40% (48). These models not only clarify the
pathogenic mechanisms of COVID-19, but also provide a basis for
immunomodulation strategies, such as inflammasome inhibition.
The interplay between the age, sex and genetic
background of the host, and viral virulence significantly
influences the disease course of COVID-19. For example, aged hACE2
mice infected with SARS-CoV-2 exhibit high mortality rates due to
suppressed early inflammatory responses in lung endothelial cells.
Additionally, the mortality rate in middle-aged male mice following
high-dose infection (80%) far exceeds that in females (40%),
reflecting the age and sex-related risk profiles observed in human
cases of COVID-19 (49,50). Conversely, Haoyu et al
(51) demonstrated that hACE2 mice
with premature aging exhibit only mild pathology post-infection,
indicating that progeria itself is not a direct risk factor for
severe COVID-19. Furthermore, Snouwaert et al (15) identified tissue-specific and
sex-specific differences in ACE2 expression in hACE2 mice.
García-Ayllón et al (52)
observed that the reduction in full-length ACE2 and the increase in
truncated forms following SARS-CoV-2 infection in K18-hACE2 mice
closely resembled the ACE2 dynamics during the acute phase of
infection in humans. The expression levels and isoform dynamics of
ACE2 provide further insight into the molecular basis of host
susceptibility.
Inhibitors targeting key enzymes of viral
replication exhibit significant promise. The small-molecule
compound 172 can target the 3-chymotrypsin-like protease variant of
SARS-CoV-2 and inhibit its dimerization to block viral replication.
Chan et al (60) found that
compound 172 significantly reduced the viral load in K18-hACE2 mice
and demonstrated broad-spectrum antiviral activity against various
SARS-CoV-2 variants and human coronaviruses. Additionally,
SARS-CoV-2 infection upregulates the host cathepsin L (CTSL), which
facilitates viral entry by cleaving S protein. Zhao et al
(10) observed that amantadine, a
CTSL inhibitor, significantly reduced viral load and alleviated
pathological lung damage in hACE2 mice, highlighting the potential
of CTSL as a therapeutic target. Moreover, the SARS-CoV-2 spike
protein induces metabolic reprogramming in hACE2 mouse hepatocytes.
Mercado-Gómez et al (61)
found that metformin reduced the risk of SARS-CoV-2 infection and
attenuated metabolic disorders by modulating ACE2 expression and
metabolic pathways, providing a novel therapeutic approach for
patients with metabolic liver disease and COVID-19. Given the high
mutation rate of SARS-CoV-2, developing drugs targeting host
factors is essential to prevent resistance. Frasson et al
(62) identified multiple SARS-CoV-2
variants co-dependent on host genes involved in oxidative stress
and mitochondrial function-related pathways. The antioxidant
N-acetylcysteine significantly reduced variant infection in
hACE2 mice by inhibiting reactive oxygen species production
required for early viral replication (62). Notably, Deshpande et al
(63) found that SARS-CoV-2
infection may alter the pharmacokinetics of metabolic enzymes and
transport proteins in hACE2 mice. This underscores the need for a
systematic assessment of drug-host interactions and optimization of
clinical drug safety.
The interaction of RBD with host cell ACE2 receptors
is critical for SARS-CoV-2 invasion. Clinical antibodies mostly
neutralize the virus by blocking this interaction, and some of
these also enhance the antiviral capacity through Fc-mediated
effector functions, such as the clearance of infected cells and the
activation of immune responses (72). For example, monoclonal antibody (mAb)
ch2H2 and h11B11 both target the host ACE2 receptor to block viral
binding. These antibodies exhibit broad-spectrum neutralization in
the K18-hACE2 mouse model and are effective against various
mutants, including the Omicron variant (11,73).
Conversely, the 17T2 mAb targets a conserved region within the
receptor-binding motif of the spike protein, maintaining
pan-neutralizing activity against Omicron subvariants (BA.5, XBB)
and demonstrating preventive and therapeutic efficacy in K18-hACE2
mice (74). Another notable mAb,
NT-193, exhibits broad neutralizing activity against
SARS-associated coronaviruses by targeting a conserved RBD site in
the heavy chain and blocking ACE2 binding in the light chain
(75). However, the efficacy of mAbs
may be compromised by viral variation. Polyclonal antibodies
(PAbs), which target multiple epitopes, may provide greater
efficacy against mutant strains compared to mAbs. Vanhove et
al (76) demonstrated that the
polyclonal antibody, XAV-19, targeted multiple epitopes without
inducing drug-resistant mutations. This antibody significantly
reduced viral load in hACE2 mouse lung tissues, highlighting the
potential advantages of PAbs in mitigating the impact of viral
evolution (76).
Antibodies that simultaneously target multiple
antigens with a single molecule provide advantages over traditional
mAb cocktails. For example, the SARS-CoV-2 spike-targeting
bispecific T-cell engager (S-BiTE) blocks viral entry and activates
T-cell-mediated clearance of infected cells. Li et al
(80) demonstrated that, in hACE2
mice, the viral load was significantly lower in the S-BiTE
treatment group compared to the neutralizing antibody-only group,
with comparable effects on the original strain and the Delta
variant. Additionally, it was previously demonstrated that both
bispecific humanized heavy chain antibodies and the IgG-VHH
bispecific antibody, SYZJ001, exhibited protective efficacy in
prophylactic and therapeutic experiments in hACE2 mice (81,82).
Furthermore, Titong et al (83) found that intranasal prophylaxis with
the tri-specific antibody, ABS-VIR-001, prevented infection and
mortality in hACE2 mice, with a 50-fold reduction in viral load
post-treatment. These findings highlight the potential of
multi-specific antibodies in enhancing therapeutic efficacy and
reducing the risk of viral escape.
Humanized mice mimic the human immune response and
are used in vaccine development to assess immunogenicity, safety
and to explore new formulations and vaccination strategies. Turan
et al (12) found that
K18-hACE2 mice vaccinated with the inactivated OZG-38.61.3 vaccine
had significantly lower mean viral loads in the high-dose group
compared to unvaccinated infected controls, without observing
significant toxicity, providing crucial evidence for clinical
translation. Tai et al (87)
demonstrated that an mRNA vaccine encapsulated in lipid
nanoparticles induced a potent CD8+ T-cell response in
humanized HLA transgenic mice. Freitag et al (88) investigated adenoviral vector 5
vaccines (Ad5-RBD and Ad5-S) and found that intranasal vaccination
in humanized HLA mice elicited mucosal IgA/IgG-neutralizing
antibodies and cytotoxic T-cell responses, providing effective
protection against the Beta variant. Compared to intramuscular
injection, intranasal administration avoids systemic viral vector
dissemination, enhancing safety and presenting a viable alternative
or supplement to existing vaccines. Gu et al (89) validated the Ad5 vector-based vaccine
in the hACE2 mouse model, demonstrating no adverse effects on
myocardial function even at high doses. This confirms its cardiac
safety and provides an experimental basis for vaccinating high-risk
cardiovascular patients (89).
Additionally, García-Arriaza et al (90) demonstrated that the modified vaccinia
virus Ankara (MVA) vector vaccine, MVA-S, exhibited potent
immunogenicity and complete protective efficacy in hACE2 mice. A
single dose prevented lethal infection, while two doses cleared the
pulmonary virus, supporting its clinical translation (90). These findings highlight the potential
of various vaccine platforms in combating SARS-CoV-2 and its
variants.
While mRNA and viral vector vaccines have
significantly advanced prevention strategies for COVID-19, their
long-term safety, particularly in vulnerable populations such as
children, pregnant women and immunocompromised individuals, and
their cross-protective efficacy against emerging mutants, remain
areas for improvement. Recombinant subunit vaccines provide a
promising alternative due to their established safety and
tolerability. For instance, Baiya-Vax-2, a recombinant plant-based
SARS-CoV-2 RBD vaccine, induced high levels of neutralizing
antibodies in K18-hACE2 mice, and two doses of the vaccine
significantly lowered viral loads and prevented severe disease
(91). The immunogenicity of the
vaccine can be further enhanced by targeting strategies. Marlin
et al (92) found that
subunit vaccines targeting the viral antigen CD40-expressing
antigen-presenting cells (αCD40.RBD) induced specific T-cell and
B-cell responses in immune-system-humanized mice and established a
persistent immune memory.
Vaccines have been effective in preventing COVID-19;
however, concerns regarding vaccine-associated enhanced respiratory
disease (VAERD) persist. Although the vaccine reduces viral load
and mortality in hACE2 mice, it may trigger VAERD due to Th2/Th17
immune bias, manifested by pulmonary eosinophilic infiltration and
IL-17-mediated systemic inflammation, which underscores the need
for balanced immune responses and avoidance of the risk of Th2/Th17
pathology in vaccine development (93). T-cell epitopes are essential in
immunity, with CD4+ T-cells regulating immune responses
and CD8+ T-cells eliminating infected cells. Humanized
MHC transgenic mouse models can generate specific cellular immune
responses after inoculation with inactivated viruses, enabling
rapid screening of T cell epitopes and accelerating vaccine design
(94). Beyond conventional vaccine
targets, regions of the virus not typically covered by existing
vaccines are gaining attention as potential breakthroughs. The
study by Weingarten-Gabbay et al (95) found that nonclassical ORF epitopes
induced a more potent IFN-γ+ T-cell response associated
with early viral protein expression and were more immunogenic than
classical epitopes in immune-system-humanized mice. This suggests
that non-classical epitopes may provide novel targets for vaccine
design (95). Targeting highly
conserved B-cell epitopes in spike protein or conserved T-cell
epitopes in multiple coronaviruses induces potent neutralizing
antibodies and cross-reactive T-cell responses in humanized mice.
This approach markedly reduces viral load and attenuates lung
inflammation (31,96). These findings highlight the central
value of conserved epitopes in overcoming viral mutations and
cross-species transmission, laying the groundwork for the
development of a pan-coronavirus vaccine.
In summary, humanized mouse models not only provide
an essential preclinical platform for refining current vaccines,
but also represent a critical technology for addressing future
coronavirus outbreaks.
Although the COVID-19 pandemic has transitioned into
a phase of normalized prevention and control, the potential risk of
SARS-CoV-2-induced long COVID remains a key concern. Long COVID is
defined as a multisystem syndrome with symptoms persisting for ≥12
weeks post-infection, characterized by pulmonary fibrosis,
cognitive impairment, and multiorgan dysfunction (13). The heterogeneous clinical
presentation of long COVID is linked to multifactorial pathogenic
mechanisms, including persistent viral residues, autoimmune
abnormalities, and chronic inflammatory cascades (97). Lung fibrosis is a hallmark of long
COVID. Cui et al (98)
simulated long COVID lung fibrosis in a humanized mouse model and
found that chronic immune activation drove fibroblast
differentiation and extracellular matrix deposition, leading to
irreversible lung injury. Additionally, Heath et al
(99) discovered that the SARS-CoV-2
spike protein disrupts the renin-angiotensin-aldosterone system
(RAAS), exacerbating coagulation abnormalities and inhibiting
fibrinolysis in hACE2-KI mice. This disruption leads to
thromboembolic cerebrovascular complications and cognitive
impairment (99). Similarly, the
SARS-CoV-2 spike protein was found to exacerbate cerebrovascular
oxidative stress and inflammation through the activation of
RAAS-damaging pathways, resulting in vascular thinning and
deteriorated cognitive function in diabetic hACE2 mice (100). These studies, leveraging humanized
mouse models, have unveiled the pathological mechanisms of long
COVID and provided a foundation for clinical interventions
targeting immunomodulation and RAAS pathways.
Humanized mouse models have revealed that long-term
residual SARS-CoV-2 RNA is associated with skeletal system
abnormalities. Using K18-hACE2 mice, Haudenschild et al
(101) found that the spike protein
increased the risk of fractures by triggering osteoclast
activation, bone loss and the thinning of the growth plate. This
occurs either through direct binding to skeletal cell ACE2
receptors or indirectly through inflammatory hypoxia (101). Notably, SARS-CoV-2 can also infect
the gastrointestinal tract and disrupt intestinal flora-host
interactions. Research using the hACE2 mouse model has demonstrated
that viral infection triggers intestinal barrier damage and flora
disruption, characterized by a reduction in the amounts of
beneficial bacteria and the proliferation of opportunistic
pathogens. This leads to abnormally high flora diversity even in
mild infections. Among these changes, the persistent reduction of
the mucosal immune-critical bacterium Akkermansia
muciniphila has been shown to be significantly associated with
fatigue and gastrointestinal symptoms in long COVID (20,102).
The study by Edwinson et al (103) using colony-humanized mice, further
demonstrated that the gut flora reduces the risk of viral
infections by inhibiting ACE2 expression. The regulation of ACE2 by
healthy flora is stable, providing a theoretical basis for
colony-targeted intervention strategies such as probiotic
modulation (103). These findings
indicate that the sequelae of COVID-19 not only involve the
respiratory and cardiovascular systems, but that interactions
between the skeletal, intestinal and immune systems may also lead
to long-term health issues, underscoring the need for comprehensive
management of the long-term effects of the virus.
Although the present review systematically
summarizes the key applications of humanized mouse models using in
the research into SARS-CoV-2, certain limitations remain. Firstly,
due to the uncertainty of ongoing virus evolution, the strategies
for model construction (e.g., reliance on hACE2) and validation
(e.g., a particular antibody or vaccine) are often based on ‘then
known’ viral characteristics. The predictive value of the original
highly specialized model may be reduced or even invalidated when a
mutant strain with an altered invasion pathway emerges. Secondly,
as regards the models themselves, even with the most advanced
construction strategies, humanized mice struggle to fully replicate
the complexity of the human immune system (38). On the one hand, these models are
typically established on immunodeficient backgrounds, making it
difficult to reproduce the complete immune network and interactions
within the tissue microenvironment. On the other hand, the majority
of models focus on reconstructing specific receptors or
single-organ functions, failing to accurately reflect the systemic
dynamic responses and multi-organ coordination mechanisms triggered
by the virus (17,40). Simultaneously, fundamental
differences exist between humans and mice in basic physiological
structures, metabolic rates, and lifespan, resulting in significant
shortcomings, particularly in simulating chronic pathological
processes, such as ‘long COVID’. Additionally, the present review
primarily relied on published literature and may not incorporate
the latest preprints or ongoing research findings in a timely
manner. In summary, while humanized mice provide a crucial platform
for SARS-CoV-2 research, careful evaluation of their
representativeness and applicability is essential when translating
findings to clinical settings.
Humanized mouse models have been pivotal in
uncovering SARS-CoV-2 pathogenicity and driving intervention
strategies since the epidemic's onset. Through gene editing,
receptor optimization and other technologies, researchers have
constructed receptor humanized, immune humanized and composite
humanized models to systematically simulate the infection
characteristics and pathological process of COVID-19. Receptor
humanization models have not only revealed the ACE2-dependent
mechanism of viral invasion, but have also led to the discovery of
the roles of novel co-receptors, such as CD147, TfR and DC-SIGN,
which laid the foundation for multi-target drug design. In addition
to breaking the species barrier by modifying viral receptors, the
reconstruction of the human immune lineage can more comprehensively
mimic the host immune response to COVID-19. Immune humanization
models recapitulated the human-specific T/B cell response and the
abnormal activation of myeloid lineage cells, and elucidated the
roles of immune imbalance in long-term sequelae. By integrating
human receptors and the immune system, the composite humanization
model recapitulates the complex phenotypes of long-term viral
retention, pulmonary fibrosis and neurodegeneration at the animal
level, which provides important clues to analyze the mechanism of
long COVID. These models validate the effectiveness of
broad-spectrum antiviral strategies targeting ACE2, and they also
accelerate the clinical translation of antibody cocktails,
bispecific antibodies and mucosal vaccines.
Although significant progress has been made in the
development of humanized mouse models, numerous challenges persist.
Discrepancies in the spatial and temporal expression patterns of
receptors compared to real human tissues can impact mechanistic
accuracy. A novel live-virus-free mouse model has been developed,
which obviates the need for viral adaptation or humanization. By
administering GU-enriched ribooligonucleotides (mimicking the
Delta/Omicron variant) via an oropharyngeal drip in combination
with low-dose bleomycin-induced lung injury, this model
successfully replicates key COVID-19 pathological features
(104). Existing models
predominantly focus on the acute infection phase, providing limited
insight into long-term COVID-19 mechanisms, such as viral latency
and reactivation or multi-organ interactions. Moreover, the low
replication of the Omicron variant in some models reveals
insufficient simulation of receptor-protease co-evolution. Further
research is required to focus on developing modular rapid-response
systems to keep pace with viral mutations.
The future development direction can break through
from the following aspects, for model technology innovation, the
integration of organoid and bioengineering models significantly
improves the accuracy of pathology simulation. The humanized lung
organoid-immunity chimeric model breaks through the bottleneck in
the study of latent viral infections and trans-organ transmission
by synchronously reconstructing the lung tissues and immune
microenvironment (105). The
two-dimensional air-liquid interface system constructed based on
fetal lung bud-tip organoids accurately reproduced the specific
infection of alveolar type II epithelium by SARS-CoV-2 and the
interferon response mechanism (106). Bioengineered lung models
dynamically simulate early COVID-19 infection characteristics by
incorporating pathogen stimulation modules, with standardized
construction systems supporting personalized medicine and
large-scale drug development (107). Dynamic monitoring technologies
combining in vivo imaging and single-cell sequencing enable
the real-time analysis of infection processes. The
68Ga-NOTA-PEP4 PET imaging agent dynamically tracks
changes in hACE2 expression (108),
while radiolabeled pseudoviruses combined with SPECT/CT and PET
imaging allow for the visualization of viral dynamics from invasion
to clearance (109). For model
optimization, the construction of a lung-intestinal-brain
multi-tissue chimera model combined with an inducible viral latent
system can systematically simulate the trans-organ damage mechanism
of long COVID. Notably, the novel finding that the intestinal flora
influences ACE2 expression through metabolic regulation suggests
that the flora-immunity co-humanization model may become a critical
breakthrough for revealing individualized susceptibility
differences.
Not applicable.
Funding: The present study was supported by grants from the
National Key Research and Development Project (grant no.
2023YFC2605603), the Program for Innovative Research Team (in
Science and Technology) in University of Henan Province (grant no.
25IRTSTHN038) and the National Natural Science Foundation of China
(grant no. 82273696).
Not applicable.
HY conceptualized the study. XF, YW, YL, JL and FL
performed the literature search. XF wrote the manuscript. All
authors have read and approved the final manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H,
Wang W, Song H, Huang B, Zhu N, et al: Genomic characterisation and
epidemiology of 2019 novel coronavirus: Implications for virus
origins and receptor binding. Lancet. 395:565–574. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hoffmann M, Kleine-Weber H, Schroeder S,
Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH,
Nitsche A, et al: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2
and is blocked by a clinically proven protease inhibitor. Cell.
181:271–280.e8. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu
Y, Zhang L, Fan G, Xu J, Gu X, et al: Clinical features of patients
infected with 2019 novel coronavirus in Wuhan, China. Lancet.
395:497–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Rai P, Kumar BK, Deekshit VK and
Karunasagar I and Karunasagar I: Detection technologies and recent
developments in the diagnosis of COVID-19 infection. Appl Microbiol
Biotechnol. 105:441–455. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sursal T, Gandhi CD, Clare K, Feldstein E,
Frid I, Kefina M, Galluzzo D, Kamal H, Nuoman R, Amuluru K, et al:
Significant mortality associated with COVID-19 and comorbid
cerebrovascular disease: A quantitative systematic review. Cardiol
Rev. 31:199–206. 2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Scholkmann F and May CA: COVID-19,
post-acute COVID-19 syndrome (PACS, ‘long COVID’) and post-COVID-19
vaccination syndrome (PCVS, ‘post-COVIDvac-syndrome’): Similarities
and differences. Pathol Res Pract. 246(154497)2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Radzikowska U, Ding M, Tan G, Zhakparov D,
Peng Y, Wawrzyniak P, Wang M, Li S, Morita H, Altunbulakli C, et
al: Distribution of ACE2, CD147, CD26, and other SARS-CoV-2
associated molecules in tissues and immune cells in health and in
asthma, COPD, obesity, hypertension, and COVID-19 risk factors.
Allergy. 75:2829–2845. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Adams LE, Dinnon III KH, Hou YJ, Sheahan
TP, Heise MT and Baric RS: Critical ACE2 determinants of SARS-CoV-2
and group 2B coronavirus infection and replication. mBio.
12:e03149–20. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Rodrigues TS, Caetano CCS, de Sá KSG,
Almeida L, Becerra A, Gonçalves AV, Lopes LS, Oliveira S,
Mascarenhas DPA, Batah SS, et al: CASP4/11 contributes to NLRP3
activation and COVID-19 exacerbation. J Infect Dis. 227:1364–1375.
2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhao MM, Yang WL, Yang FY, Zhang L, Huang
WJ, Hou W, Fan CF, Jin RH, Feng YM, Wang YC and Yang JK: Cathepsin
L plays a key role in SARS-CoV-2 infection in humans and humanized
mice and is a promising target for new drug development. Signal
Transduct Target Ther. 6(134)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Du Y, Shi R, Zhang Y, Duan X, Li L, Zhang
J, Wang F, Zhang R, Shen H, Wang Y, et al: A broadly neutralizing
humanized ACE2-targeting antibody against SARS-CoV-2 variants. Nat
Commun. 12(5000)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Turan RD, Tastan C, Dilek Kancagi D,
Yurtsever B, Sir Karakus G, Ozer S, Abanuz S, Cakirsoy D,
Tumentemur G, Demir S, et al: Gamma-irradiated SARS-CoV-2 vaccine
candidate, OZG-38.61.3, confers protection from SARS-CoV-2
challenge in human ACEII-transgenic mice. Sci Rep.
11(15799)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Perumal R, Shunmugam L, Naidoo K, Abdool
Karim SS, Wilkins D, Garzino-Demo A, Brechot C, Parthasarathy S,
Vahlne A and Nikolich JŽ: Long COVID: A review and proposed
visualization of the complexity of long COVID. Front Immunol.
14(1117464)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yinda CK, Port JR, Bushmaker T, Offei
Owusu I, Purushotham JN, Avanzato VA, Fischer RJ, Schulz JE,
Holbrook MG, Hebner MJ, et al: K18-hACE2 mice develop respiratory
disease resembling severe COVID-19. PLoS Pathog.
17(e1009195)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Snouwaert JN, Jania LA, Nguyen T, Martinez
DR, Schäfer A, Catanzaro NJ, Gully KL, Baric RS, Heise M, Ferris
MT, et al: Human ACE2 expression, a major tropism determinant for
SARS-CoV-2, is regulated by upstream and intragenic elements. PLoS
Pathog. 19(e1011168)2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ge C, Salem AR, Elsharkawy A, Natekar J,
Guglani A, Doja J, Ogala O, Wang G, Griffin SH, Slivano OJ, et al:
Development and characterization of a fully humanized ACE2 mouse
model. BMC Biol. 23(194)2025.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Song IW, Washington M, Leynes C, Hsu J,
Rayavara K, Bae Y, Haelterman N, Chen Y, Jiang MM, Drelich A, et
al: Generation of a humanized mAce2 and a conditional hACE2 mouse
models permissive to SARS-COV-2 infection. Mamm Genome. 35:113–121.
2024.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li K, Verma A, Li P, Ortiz ME, Hawkins GM,
Schnicker NJ, Szachowicz PJ, Pezzulo AA, Wohlford-Lenane CL, Kicmal
T, et al: Adaptation of SARS-CoV-2 to ACE2H353K mice
reveals new spike residues that drive mouse infection. J Virol.
98(e0151023)2024.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu FL, Wu K, Sun J, Duan Z, Quan X, Kuang
J, Chu S, Pang W, Gao H, Xu L, et al: Rapid generation of ACE2
humanized inbred mouse model for COVID-19 with tetraploid
complementation. Natl Sci Rev. 8(nwaa285)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang Y, Ma Y, Sun W, Zhou X, Wang R, Xie
P, Dai L, Gao Y and Li J: Exploring gut-lung axis crosstalk in
SARS-CoV-2 infection: Insights from a hACE2 mouse model. J Med
Virol. 96(e29336)2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Choi CY, Gadhave K, Villano J, Pekosz A,
Mao X and Jia H: Generation and characterization of a humanized
ACE2 mouse model to study long-term impacts of SARS-CoV-2
infection. J Med Virol. 96(e29349)2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ma MT, Jiang Q, Chen CH, Badeti S, Wang X,
Zeng C, Evans D, Bodnar B, Marras SAE, Tyagi S, et al: S309-CAR-NK
cells bind the Omicron variants in vitro and reduce SARS-CoV-2
viral loads in humanized ACE2-NSG mice. J Virol.
98(e0003824)2024.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Verma SK, Ana-Sosa-Batiz F, Timis J,
Shafee N, Maule E, Pinto PBA, Conner C, Valentine KM, Cowley DO,
Miller R, et al: Influence of Th1 versus Th2 immune bias on viral,
pathological, and immunological dynamics in SARS-CoV-2
variant-infected human ACE2 knock-in mice. EBioMedicine.
108(105361)2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tang AT, Buchholz DW, Szigety KM, Imbiakha
B, Gao S, Frankfurter M, Wang M, Yang J, Hewins P, Mericko-Ishizuka
P, et al: Cell-autonomous requirement for ACE2 across organs in
lethal mouse SARS-CoV-2 infection. PLoS Biol.
21(e3001989)2023.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bruter AV, Korshunova DS, Kubekina MV,
Sergiev PV, Kalinina AA, Ilchuk LA, Silaeva YY, Korshunov EN,
Soldatov VO and Deykin AV: Novel transgenic mice with Cre-dependent
co-expression of GFP and human ACE2: A safe tool for study of
COVID-19 pathogenesis. Transgenic Res. 30:289–301. 2021.PubMed/NCBI View Article : Google Scholar : (Epub ahead of
print).
|
|
26
|
Tu Y, Fang Y, Zheng R, Lu D, Yang X, Zhang
L, Li D, Sun Y, Yu W, Luo D and Wang H: A murine model of DC-SIGN
humanization exhibits increased susceptibility against SARS-CoV-2.
Microbes Infect. 26(105344)2024.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liao Z, Wang C, Tang X, Yang M, Duan Z,
Liu L, Lu S, Ma L, Cheng R, Wang G, et al: Human transferrin
receptor can mediate SARS-CoV-2 infection. Proc Natl Acad Sci USA.
121(e2317026121)2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wu J, Chen L, Qin C, Huo F, Liang X, Yang
X, Zhang K, Lin P, Liu J, Feng Z, et al: CD147 contributes to
SARS-CoV-2-induced pulmonary fibrosis. Signal Transduct Target
Ther. 7(382)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Brumeanu TD, Vir P, Karim AF, Kar S,
Benetiene D, Lok M, Greenhouse J, Putmon-Taylor T, Kitajewski C,
Chung KK, et al: Human-immune-system (HIS) humanized mouse model
(DRAGA: HLA-A2.HLA-DR4.Rag1KO.IL-2RγcKO.NOD) for COVID-19. Hum
Vaccin Immunother. 18(2048622)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Brumeanu TD, Vir P, Karim AF, Kar S,
Benetiene D, Lok M, Greenhouse J, Putmon-Taylor T, Kitajewski C,
Chung KK, et al: A Human-Immune-System (HIS) humanized mouse model
(DRAGA: HLA-A2. HLA-DR4. Rag1 KO.IL-2Rγc KO. NOD) for COVID-19.
bioRxiv [Preprint]: 2020.08.19.251249, 2021.
|
|
31
|
Prakash S, Srivastava R, Coulon PG,
Dhanushkodi NR, Chentoufi AA, Tifrea DF, Edwards RA, Figueroa CJ,
Schubl SD, Hsieh L, et al: Genome-wide asymptomatic B-Cell, CD4
+ and CD8 + T-cell epitopes, that are highly
conserved between human and animal coronaviruses, identified from
SARS-CoV-2 as immune targets for pre-emptive pan-coronavirus
vaccines. bioRxiv [Preprint]: 2020.09.27.316018, 2020.
|
|
32
|
Li S, Han X, Hu R, Sun K, Li M, Wang Y,
Zhao G, Li M, Fan H and Yin Q: Transcriptomic profiling reveals
SARS-CoV-2-infected humanized MHC mice recapitulate human post
vaccination immune responses. Front Cell Infect Microbiol.
15(1634577)2025.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Fu W, Wang W, Yuan L, Lin Y, Huang X, Chen
R, Cai M, Liu C, Chen L, Zhou M, et al: A SCID mouse-human lung
xenograft model of SARS-CoV-2 infection. Theranostics.
11:6607–6615. 2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sun R, Zhao Z, Fu C, Wang Y, Guo Z, Zhang
C, Liu L, Zhang C, Shu C, He J, et al: Humanized mice for
investigating SARS-CoV-2 lung infection and associated human immune
responses. Eur J Immunol. 52:1640–1647. 2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sefik E, Israelow B, Zhao J, Qu R, Song E,
Mirza H, Kaffe E, Halene S, Meffre E, Kluger Y, et al: A humanized
mouse model of chronic COVID-19 to evaluate disease mechanisms and
treatment options. Res Sq [Preprint]: rs.3.rs-279341, 2021.
|
|
36
|
Sefik E, Israelow B, Mirza H, Zhao J, Qu
R, Kaffe E, Song E, Halene S, Meffre E, Kluger Y, et al: A
humanized mouse model of chronic COVID-19. Nat Biotechnol.
40:906–920. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sefik E, Qu R, Junqueira C, Kaffe E, Mirza
H, Zhao J, Brewer JR, Han A, Steach HR, Israelow B, et al:
Inflammasome activation in infected macrophages drives COVID-19
pathology. Nature. 606:585–593. 2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yong KSM, Anderson DE, Zheng AKE, Liu M,
Tan SY, Tan WWS, Chen Q and Wang LF: Comparison of infection and
human immune responses of two SARS-CoV-2 strains in a humanized
hACE2 NIKO mouse model. Sci Rep. 13(12484)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Le Chevalier F, Authié P, Chardenoux S,
Bourgine M, Vesin B, Cussigh D, Sassier Y, Fert I, Noirat A,
Nemirov K, et al: Mice humanized for MHC and hACE2 with high
permissiveness to SARS-CoV-2 omicron replication. Microbes Infect.
25(105142)2023.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Di Y, Lew J, Goncin U, Radomska A, Rout
SS, Gray BET, Machtaler S, Falzarano D and Lavender KJ: SARS-CoV-2
variant-specific infectivity and immune profiles Are detectable in
a humanized lung mouse model. Viruses. 14(2272)2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kenney DJ, O'Connell AK, Turcinovic J,
Montanaro P, Hekman RM, Tamura T, Berneshawi AR, Cafiero TR, Al
Abdullatif S, Blum B, et al: Humanized mice reveal a
macrophage-enriched gene signature defining human lung tissue
protection during SARS-CoV-2 infection. Cell Rep.
39(110714)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jarnagin K, Alvarez O, Shresta S and Webb
DR: Animal models for SARS-Cov2/Covid19 research-A commentary.
Biochem Pharmacol. 188(114543)2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Ye Q, Zhou J, He Q, Li RT, Yang G, Zhang
Y, Wu SJ, Chen Q, Shi JH, Zhang RR, et al: SARS-CoV-2 infection in
the mouse olfactory system. Cell Discov. 7(49)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Cao JB, Zhu ST, Huang XS, Wang XY, Wu ML,
Li X, Liu FL, Chen L, Zheng YT and Wang JH: Mast cell
degranulation-triggered by SARS-CoV-2 induces tracheal-bronchial
epithelial inflammation and injury. Virol Sin. 39:309–318.
2024.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wu ML, Liu FL, Sun J, Li X, He XY, Zheng
HY, Zhou YH, Yan Q, Chen L, Yu GY, et al: SARS-CoV-2-triggered mast
cell rapid degranulation induces alveolar epithelial inflammation
and lung injury. Signal Transduct Target Ther.
6(428)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Dos Ramos Almeida CJL, Veras FP, Paiva IM,
Schneider AH, da Costa Silva J, Gomes GF, Costa VF, Silva BMS,
Caetite DB, Silva CMS, et al: Neutrophil virucidal activity against
SARS-CoV-2 is mediated by neutrophil extracellular traps. J Infect
Dis. 229:1352–1365. 2024.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Chuang HC, Hsueh CH, Hsu PM, Huang RH,
Tsai CY, Chung NH, Chow YH and Tan TH: SARS-CoV-2 spike protein
enhances MAP4K3/GLK-induced ACE2 stability in COVID-19. EMBO Mol
Med. 14(e15904)2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Bello-Perez M, Hurtado-Tamayo J, Mykytyn
AZ, Lamers MM, Requena-Platek R, Schipper D, Muñoz-Santos D,
Ripoll-Gómez J, Esteban A, Sánchez-Cordón PJ, et al: SARS-CoV-2
ORF8 accessory protein is a virulence factor. mBio.
14(e0045123)2023.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Park U, Lee JH, Kim U, Jeon K, Kim Y, Kim
H, Kang JI, Park MY, Park SH, Cha JS, et al: A humanized ACE2 mouse
model recapitulating age- and sex-dependent immunopathogenesis of
COVID-19. J Med Virol. 96(e29915)2024.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Subramaniam S, Kenney D, Jayaraman A,
O'Connell AK, Walachowski S, Montanaro P, Reinhardt C, Colucci G,
Crossland NA, Douam F and Bosmann M: Aging is associated with an
insufficient early inflammatory response of lung endothelial cells
in SARS-CoV-2 infection. Front Immunol. 15(1397990)2024.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Haoyu W, Meiqin L, Jiaoyang S, Guangliang
H, Haofeng L, Pan C, Xiongzhi Q, Kaixin W, Mingli H, Xuejie Y, et
al: Premature aging effects on COVID-19 pathogenesis: New insights
from mouse models. Sci Rep. 14(19703)2024.PubMed/NCBI View Article : Google Scholar
|
|
52
|
García-Ayllón MS, Moreno-Pérez O,
García-Arriaza J, Ramos-Rincón JM, Cortés-Gómez M, Brinkmalm G,
Andrés M, León-Ramírez JM, Boix V, Gil J, et al: Plasma ACE2
species are differentially altered in COVID-19 patients. FASEB J.
35(e21745)2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Lu T, Zhang C, Li Z, Wei Y, Sadewasser A,
Yan Y, Sun L, Li J, Wen Y, Lai S, et al: Human
angiotensin-converting enzyme 2-specific antisense oligonucleotides
reduce infection with SARS-CoV-2 variants. J Allergy Clin Immunol.
154:1044–1059. 2024.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Ikemura N, Taminishi S, Inaba T, Arimori
T, Motooka D, Katoh K, Kirita Y, Higuchi Y, Li S, Suzuki T, et al:
An engineered ACE2 decoy neutralizes the SARS-CoV-2 Omicron variant
and confers protection against infection in vivo. Sci Transl Med.
14(eabn7737)2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Zhang L, Dutta S, Xiong S, Chan M, Chan
KK, Fan TM, Bailey KL, Lindeblad M, Cooper LM, Rong L, et al:
Engineered ACE2 decoy mitigates lung injury and death induced by
SARS-CoV-2 variants. Nat Chem Biol. 18:342–351. 2022.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Zhang L, Dutta S, Xiong S, Chan M, Chan
KK, Fan TM, Bailey KL, Lindeblad M, Cooper LM, Rong L, et al:
Engineered high-affinity ACE2 peptide mitigates ARDS and death
induced by multiple SARS-CoV-2 variants. bioRxiv [Preprint]:
2021.12.21.473668, 2021.
|
|
57
|
Hwang J, Kim BK, Moon S, Park W, Kim KW,
Yoon JH, Oh H, Jung S, Park Y, Kim S, et al: Conversion of host
cell receptor into virus destructor by immunodisc to neutralize
diverse SARS-CoV-2 variants. Adv Healthc Mater.
13(e2302803)2024.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dong W, Wang J, Tian L, Zhang J, Mead H,
Jaramillo SA, Li A, Zumwalt RE, Whelan SPJ, Settles EW, et al: FXa
cleaves the SARS-CoV-2 spike protein and blocks cell entry to
protect against infection with inferior effects in B.1.1.7 variant.
bioRxiv [Preprint]: 2021.06.07.447437, 2021.
|
|
59
|
Yu F, Liu X, Ou H, Li X, Liu R, Lv X, Xiao
S, Hu M, Liang T, Chen T, et al: The histamine receptor H1 acts as
an alternative receptor for SARS-CoV-2. mBio.
15(e0108824)2024.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Chan CC, Guo Q, Chan JF, Tang K, Cai JP,
Chik KK, Huang Y, Dai M, Qin B, Ong CP, et al: Identification of
novel small-molecule inhibitors of SARS-CoV-2 by chemical genetics.
Acta Pharm Sin B. 14:4028–4044. 2024.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Mercado-Gómez M, Prieto-Fernández E,
Goikoetxea-Usandizaga N, Vila-Vecilla L, Azkargorta M, Bravo M,
Serrano-Maciá M, Egia-Mendikute L, Rodríguez-Agudo R,
Lachiondo-Ortega S, et al: The spike of SARS-CoV-2 promotes
metabolic rewiring in hepatocytes. Commun Biol.
5(827)2022.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Frasson I, Diamante L, Zangrossi M,
Carbognin E, Pietà AD, Penna A, Rosato A, Verin R, Torrigiani F,
Salata C, et al: Identification of druggable host dependency
factors shared by multiple SARS-CoV-2 variants of concern. J Mol
Cell Biol. 16(mjae004)2024.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Deshpande K, Lange KR, Stone WB, Yohn C,
Schlesinger N, Kagan L, Auguste AJ, Firestein BL and Brunetti L:
The influence of SARS-CoV-2 infection on expression of
drug-metabolizing enzymes and transporters in a hACE2 murine model.
Pharmacol Res Perspect. 11(e01071)2023.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Mazzarella L, Santoro F, Ravasio R,
Fumagalli V, Massa PE, Rodighiero S, Gavilán E, Romanenghi M, Duso
BA, Bonetti E, et al: Inhibition of the lysine demethylase LSD1
modulates the balance between inflammatory and antiviral responses
against coronaviruses. Sci Signal. 16(eade0326)2023.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Xiong S, Zhang L, Richner JM, Class J,
Rehman J and Malik AB: Interleukin-1RA mitigates SARS-CoV-2-induced
inflammatory lung vascular leakage and mortality in humanized
K18-hACE-2 mice. Arterioscler Thromb Vasc Biol. 41:2773–2785.
2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Botella-Asunción P, Rivero-Buceta EM,
Vidaurre-Agut C, Lama R, Rey-Campos M, Moreno A, Mendoza L,
Mingo-Casas P, Escribano-Romero E, Gutierrez-Adan A, et al: AG5 is
a potent non-steroidal anti-inflammatory and immune regulator that
preserves innate immunity. Biomed Pharmacother.
169(115882)2023.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Weiss CM, Liu H, Ball EE, Hoover AR, Wong
TS, Wong CF, Lam S, Hode T, Keel MK, Levenson RM, et al:
N-dihydrogalactochitosan reduces mortality in a lethal mouse model
of SARS-CoV-2. PLoS One. 18(e0289139)2023.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Yeung ST, Premeaux TA, Du L, Niki T,
Pillai SK, Khanna KM and Ndhlovu LC: Galectin-9 protects
humanized-ACE2 immunocompetent mice from SARS-CoV-2 infection.
Front Immunol. 13(1011185)2022.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Meier M, Becker S, Levine E, DuFresne O,
Foster K, Moore J, Burnett FN, Hermanns VC, Heath SP, Abdelsaid M
and Coucha M: Timing matters in the use of renin-angiotensin system
modulators and COVID-related cognitive and cerebrovascular
dysfunction. PLoS One. 19(e0304135)2024.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Silva-Santos Y, Pagni RL, Gamon THM, de
Azevedo MSP, Bielavsky M, Darido MLG, de Oliveira DBL, de Souza EE,
Wrenger C, Durigon EL, et al: Lisinopril increases lung ACE2 levels
and SARS-CoV-2 viral load and decreases inflammation but not
disease severity in experimental COVID-19. Front Pharmacol.
15(1414406)2024.PubMed/NCBI View Article : Google Scholar
|
|
71
|
da Silva-Santos Y, Pagni RL, Gamon THM, de
Azevedo MSP, Darido MLG, de Oliveira DBL, Durigon EL, Luvizotto
MCR, Ackerman HC, Marinho CRF, et al: Angiotensin-converting enzyme
inhibition and/or angiotensin receptor blockade modulate cytokine
profiles and improve clinical outcomes in experimental COVID-19
infection. Int J Mol Sci. 26(7663)2025.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Corti D, Purcell LA, Snell G and Veesler
D: Tackling COVID-19 with neutralizing monoclonal antibodies. Cell.
184:3086–3108. 2021.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Sun CP, Chiu CW, Wu PY, Tsung SI, Lee IJ,
Hu CW, Hsu MF, Kuo TJ, Lan YH, Chen LY, et al: Development of
AAV-delivered broadly neutralizing anti-human ACE2 antibodies
against SARS-CoV-2 variants. Mol Ther. 31:3322–3336.
2023.PubMed/NCBI View Article : Google Scholar
|
|
74
|
de Campos-Mata L, Trinité B, Modrego A,
Tejedor Vaquero S, Pradenas E, Pons-Grífols A, Rodrigo Melero N,
Carlero D, Marfil S, Santiago C, et al: A monoclonal antibody
targeting a large surface of the receptor binding motif shows
pan-neutralizing SARS-CoV-2 activity. Nat Commun.
15(1051)2024.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Onodera T, Kita S, Adachi Y, Moriyama S,
Sato A, Nomura T, Sakakibara S, Inoue T, Tadokoro T, Anraku Y, et
al: A SARS-CoV-2 antibody broadly neutralizes SARS-related
coronaviruses and variants by coordinated recognition of a
virus-vulnerable site. Immunity. 54:2385–2398.e10. 2021.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Vanhove B, Marot S, So RT, Gaborit B,
Evanno G, Malet I, Lafrogne G, Mevel E, Ciron C, Royer PJ, et al:
XAV-19, a swine glyco-humanized polyclonal antibody against
SARS-CoV-2 spike receptor-binding domain, targets multiple epitopes
and broadly neutralizes variants. Front Immunol.
12(761250)2021.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Fu D, Zhang G, Wang Y, Zhang Z, Hu H, Shen
S, Wu J, Li B, Li X, Fang Y, et al: Structural basis for SARS-CoV-2
neutralizing antibodies with novel binding epitopes. PLoS Biol.
19(e3001209)2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Hansen J, Baum A, Pascal KE, Russo V,
Giordano S, Wloga E, Fulton BO, Yan Y, Koon K, Patel K, et al:
Studies in humanized mice and convalescent humans yield a
SARS-CoV-2 antibody cocktail. Science. 369:1010–1014.
2020.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Wang F, Li L, Dou Y, Shi R, Duan X, Liu H,
Zhang J, Liu D, Wu J, He Y, et al: Etesevimab in combination with
JS026 neutralizing SARS-CoV-2 and its variants. Emerg Microbes
Infect. 11:548–551. 2022.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Li F, Xu W, Zhang X, Wang W, Su S, Han P,
Wang H, Xu Y, Li M, Fan L, et al: A spike-targeting bispecific T
cell engager strategy provides dual layer protection against
SARS-CoV-2 infection in vivo. Commun Biol. 6(592)2023.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Casasnovas JM, Margolles Y, Noriega MA,
Guzmán M, Arranz R, Melero R, Casanova M, Corbera JA,
Jiménez-de-Oya N, Gastaminza P, et al: Nanobodies protecting from
lethal SARS-CoV-2 infection target receptor binding epitopes
preserved in virus variants other than omicron. Front Immunol.
13(863831)2022.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Chi H, Wang L, Liu C, Cheng X, Zheng H, Lv
L, Tan Y, Zhang N, Zhao S, Wu M, et al: An engineered IgG-VHH
bispecific antibody against SARS-CoV-2 and its variants. Small
Methods. 6(e2200932)2022.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Titong A, Gallolu Kankanamalage S, Dong J,
Huang B, Spadoni N, Wang B, Wright M, Pham KLJ, Le AH and Liu Y:
First-in-class trispecific VHH-Fc based antibody with potent
prophylactic and therapeutic efficacy against SARS-CoV-2 and
variants. Sci Rep. 12(4163)2022.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Yang Z, Wang Y, Jin Y, Zhu Y, Wu Y, Li C,
Kong Y, Song W, Tian X, Zhan W, et al: A non-ACE2 competing human
single-domain antibody confers broad neutralization against
SARS-CoV-2 and circulating variants. Signal Transduct Target Ther.
6(378)2021.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Luo S, Zhang J, Kreutzberger AJB, Eaton A,
Edwards RJ, Jing C, Dai HQ, Sempowski GD, Cronin K, Parks R, et al:
An antibody from single human VH-rearranging mouse
neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting
membrane fusion. Sci Immunol. 7(eadd5446)2022.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Geng J, Chen L, Yuan Y, Wang K, Wang Y,
Qin C, Wu G, Chen R, Zhang Z, Wei D, et al: CD147 antibody
specifically and effectively inhibits infection and cytokine storm
of SARS-CoV-2 and its variants delta, alpha, beta, and gamma.
Signal Transduct Target Ther. 6(347)2021.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Tai W, Feng S, Chai B, Lu S, Zhao G, Chen
D, Yu W, Ren L, Shi H, Lu J, et al: An mRNA-based T-cell-inducing
antigen strengthens COVID-19 vaccine against SARS-CoV-2 variants.
Nat Commun. 14(2962)2023.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Freitag TL, Fagerlund R, Karam NL,
Leppänen VM, Ugurlu H, Kant R, Mäkinen P, Tawfek A, Jha SK,
Strandin T, et al: Intranasal administration of adenoviral vaccines
expressing SARS-CoV-2 spike protein improves vaccine immunity in
mouse models. Vaccine. 41:3233–3246. 2023.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Gu S, Chen Z, Meng X, Liu G, Xu H, Huang
L, Wu L, Gong J, Chen D, Xue B, et al: Spike-based adenovirus
vectored COVID-19 vaccine does not aggravate heart damage after
ischemic injury in mice. Commun Biol. 5(902)2022.PubMed/NCBI View Article : Google Scholar
|
|
90
|
García-Arriaza J, Garaigorta U, Pérez P,
Lázaro-Frías A, Zamora C, Gastaminza P, Del Fresno C, Casasnovas
JM, Sorzano CÓ S, Sancho D and Esteban M: COVID-19 vaccine
candidates based on modified vaccinia virus Ankara expressing the
SARS-CoV-2 spike induce robust T- and B-cell immune responses and
full efficacy in mice. J Virol. 95:e02260–20. 2021.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Phoolcharoen W, Shanmugaraj B,
Khorattanakulchai N, Sunyakumthorn P, Pichyangkul S, Taepavarapruk
P, Praserthsee W, Malaivijitnond S, Manopwisedjaroen S,
Thitithanyanont A, et al: Preclinical evaluation of immunogenicity,
efficacy and safety of a recombinant plant-based SARS-CoV-2 RBD
vaccine formulated with 3M-052-Alum adjuvant. Vaccine.
41:2781–2792. 2023.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Marlin R, Godot V, Cardinaud S, Galhaut M,
Coleon S, Zurawski S, Dereuddre-Bosquet N, Cavarelli M, Gallouët
AS, Maisonnasse P, et al: Targeting SARS-CoV-2 receptor-binding
domain to cells expressing CD40 improves protection to infection in
convalescent macaques. Nat Commun. 12(5215)2021.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Zhang T, Magazine N, McGee MC, Carossino
M, Veggiani G, Kousoulas KG, August A and Huang W: Th2 and
Th17-associated immunopathology following SARS-CoV-2 breakthrough
infection in Spike-vaccinated ACE2-humanized mice. J Med Virol.
96(e29408)2024.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Zhang J, Fang F, Zhang Y, Han X, Wang Y,
Yin Q, Sun K, Zhou H, Qin H, Zhao D, et al: Humanized major
histocompatibility complex transgenic mouse model can play a potent
role in SARS-CoV-2 human leukocyte antigen-restricted T cell
epitope screening. Vaccines (Basel). 13(416)2025.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Weingarten-Gabbay S, Klaeger S, Sarkizova
S, Pearlman LR, Chen DY, Gallagher KME, Bauer MR, Taylor HB, Dunn
WA, Tarr C, et al: Profiling SARS-CoV-2 HLA-I peptidome reveals T
cell epitopes from out-of-frame ORFs. Cell. 184:3962–3980.e17.
2021.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Zayou L, Prakash S, Vahed H, Dhanushkodi
NR, Quadiri A, Belmouden A, Lemkhente Z, Chentoufi A, Gil D, Ulmer
JB and BenMohamed L: Dynamics of spike-specific neutralizing
antibodies across five-year emerging SARS-CoV-2 variants of concern
reveal conserved epitopes that protect against severe COVID-19.
Front Immunol. 16(1503954)2025.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Klein J, Wood J, Jaycox JR, Dhodapkar RM,
Lu P, Gehlhausen JR, Tabachnikova A, Greene K, Tabacof L, Malik AA,
et al: Distinguishing features of long COVID identified through
immune profiling. Nature. 623:139–148. 2023.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Cui L, Fang Z, De Souza CM, Lerbs T, Guan
Y, Li I, Charu V, Chen SY, Weissman I and Wernig G: Innate immune
cell activation causes lung fibrosis in a humanized model of long
COVID. Proc Natl Acad Sci USA. 120(e2217199120)2023.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Heath SP, Hermanns VC, Coucha M and
Abdelsaid M: SARS-CoV-2 spike protein exacerbates thromboembolic
cerebrovascular complications in humanized ACE2 mouse model. Transl
Stroke Res. 16:1214–1228. 2025.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Burnett FN, Coucha M, Bolduc DR, Hermanns
VC, Heath SP, Abdelghani M, Macias-Moriarity LZ and Abdelsaid M:
SARS-CoV-2 spike protein intensifies cerebrovascular complications
in diabetic hACE2 Mice through RAAS and TLR signaling activation.
Int J Mol Sci. 24(16394)2023.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Haudenschild AK, Christiansen BA, Orr S,
Ball EE, Weiss CM, Liu H, Fyhrie DP, Yik JHN, Coffey LL and
Haudenschild DR: Acute bone loss following SARS-CoV-2 infection in
mice. J Orthop Res. 41:1945–1952. 2023.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Upadhyay V, Suryawanshi RK, Tasoff P,
McCavitt-Malvido M, Kumar RG, Murray VW, Noecker C, Bisanz JE,
Hswen Y, Ha CWY, et al: Mild SARS-CoV-2 infection results in
long-lasting microbiota instability. mBio.
14(e0088923)2023.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Edwinson A, Yang L, Chen J and Grover M:
Colonic expression of Ace2, the SARS-CoV-2 entry receptor, is
suppressed by commensal human microbiota. Gut Microbes.
13(1984105)2021.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Zhu H, Sharma AK, Aguilar K, Boghani F,
Sarcan S, George M, Ramesh J, Van Der Eerden J, Panda CS, Lopez A,
et al: Simple virus-free mouse models of COVID-19 pathologies and
oral therapeutic intervention. iScience. 27(109191)2024.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Pujhari S and Rasgon JL: Mice with
humanized-lungs and immune system-an idealized model for COVID-19
and other respiratory illness. Virulence. 11:486–488.
2020.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Lamers MM, van der Vaart J, Knoops K,
Riesebosch S, Breugem TI, Mykytyn AZ, Beumer J, Schipper D,
Bezstarosti K, Koopman CD, et al: An organoid-derived
bronchioalveolar model for SARS-CoV-2 infection of human alveolar
type II-like cells. EMBO J. 40(e105912)2021.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Ahmadipour M, Prado JC, Hakak-Zargar B,
Mahmood MQ and Rogers IM: Using ex vivo bioengineered lungs to
model pathologies and screening therapeutics: A proof-of-concept
study. Biotechnol Bioeng. 121:3020–3033. 2024.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Parker MFL, Blecha J, Rosenberg O, Ohliger
M, Flavell RR and Wilson DM: Cyclic 68Ga-labeled
peptides for specific detection of human angiotensin-converting
enzyme 2. J Nucl Med. 62:1631–1637. 2021.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Li D, Xiong L, Pan G, Wang T, Li R, Zhu L,
Tong Q, Yang Q, Peng Y, Zuo C, et al: Molecular imaging on
ACE2-dependent transocular infection of coronavirus. J Med Virol.
94:4878–4889. 2022.PubMed/NCBI View Article : Google Scholar
|