Introduction
Asthma is a one of the most common pediatric
diseases and is characterized by eosinophilic airway inflammation,
reversible airway obstruction, hyperresponsiveness and airway
remodeling (1,2). The prevalence of asthma is increasing
in most countries (3,4). Asthma causes substantial social
impact and costs to public and private healthcare systems (3,4).
Pediatric asthma is different from adult asthma in terms of
severity, natural history, response to treatment and mechanisms
(5,6). Early diagnosis of pediatric asthma,
although challenging, is highly important for effective treatment.
To date, the available treatment modalities have not been
sufficient to satisfactorily control asthma in children (7). Therefore, a new method for treating
pediatric asthma is urgently required.
microRNAs (miRNAs) are a class of small, noncoding,
single-stranded RNAs that regulate gene expression by binding to
their target mRNA and triggering either repression of protein
translation or RNA degradation (8,9).
miRNAs regulate development, differentiation, stem-cell
differentiation, growth control, apoptosis and immune functions
(8,9). Several miRNAs are involved in the
pathogenesis of asthma (10,11).
An miRNA array analysis in murine models of acute
and chronic asthma revealed that miRNA expression in the lungs
changed following exposure to allergens, and suggested that several
miRNAs may regulate the biological processes during the course of
asthma development (12). However,
these findings were in contrast to those of an miRNA array analysis
in asthmatic adults in another study, which had shown that changes
in miRNA expression do not appear to be involved in the development
of the asthmatic phenotype or the anti-inflammatory action of the
corticosteroid budesonide in adult asthmatic patients (13). To date, very little is known about
miRNA expression profiles in pediatric asthma.
In the present study, miRNA array analysis was
performed to determine miRNA expression profiles in asthmatic
children. Sprouty-related EVH1 domain-containing protein (Spred)
negatively regulates allergen-induced airway inflammation and
hyperresponsiveness in murine asthma models by modulating
interleukin (IL)-5 signaling (14). Among the upregulated miRNAs in
pediatric asthma, miRNA-221 and miRNA-485-3p were predicted to bind
to Spred-2 mRNA by bioinformatic analysis. Upregulation of
miRNA-221 and miRNA-485-3p among pediatric asthmatics and murine
asthma models were verified by real-time PCR. Spred-2 protein level
was downregulated in murine asthma models.
Materials and methods
Study population
The study population consisted of 12 children (age,
4–6 years) admitted to Nanjing Children’s Hospital, China. Six of
these children were diagnosed as having allergic asthma. The other
6 were considered as the control group. Asthma was diagnosed on the
basis of the recommendations of the Global Initiative for Asthma
(GINA); according to GINA, pediatric patients are defined as having
asthma if they have visited the hospital within the past 12 months
due to wheezing without evidence of common cold, and if their
forced expiratory volume in 1 second (FEV1) after inhalation of a
β2 agonist increased by 12% compared to prior to the
inhalation. The results of the skin-prick tests were positive for
all the asthmatic children. The children were first diagnosed as
having allergic asthma and did not undergo any treatment. Venous
blood samples were obtained from all the children. The study was
approved by the Medical Ethics Committee of Nanjing Children’s
Hospital. Written consent was obtained from all parents.
miRNA microarray assay
Lymphocytes were collected from the blood by using a
lymphocyte separation medium. Total RNA was isolated using TRIzol
(Invitrogen, Carlsbad, CA, USA) and an miRNeasy Mini kit (Qiagen,
Hilden, Germany). The sixth generation of miRCURY™ LNA Array
(v.16.0) (Exiqon) contains more than 1,891 capture probes, covering
all human, mouse and rat miRNAs annotated in miRBase 16.0, as well
as all viral miRNAs associated with these species. In addition,
this array contains capture probes for 66 new miRPlus™ human
miRNAs.
Ovalbumin (OVA)-induced murine asthma
models
The BALB/c mice were randomly distributed into two
groups (n=6). The mice were kept for one week prior to the
experiment. On day 0 and day 14, mice were sensitized with 20 μg of
OVA and 20 mg Al(OH)3 in 0.2 ml PBS. Following
sensitization, mice were exposed to either aerosolized 1% OVA/PBS
or PBS only for 20 min once a day on days 27, 28, 29 and 30. On day
31, mice were analyzed for cell numbers in the bronchoalveolar
lavage fluid (BALF) and histological study.
Histological study
Lungs were removed from mice, fixed in 10% formalin
for 24 h, dehydrated, mounted in paraffin, sectioned to a thickness
of 4 μm, and stained with hematoxylin and eosin (HE).
Real-time polymerase chain reaction
(PCR)
Real-time PCR was performed as described previously
(15). Briefly, the miRNA
first-strand synthesis kit was used to perform the first-strand
synthesis. SYBR-Green PCR was then performed. U6 spliceosomal RNA
(snRNA) was used as the endogenous control.
Bioinformatic analysis of miRNAs
Bioinformatic analysis of miRNAs was performed by a
method described in the literature (12,13).
miRNA targets were analyzed using the public database TargetScan
6.0 (http://www.targetscan.org).
Western blotting
The removed lung was lysed in protein lysis buffer
(50 mM Tris, 150 mM NaCl, 10 mM EDTA, 1% Triton X-100, 200 mM
sodium fluoride and 4 mM sodium orthovanadate-containing protease
inhibitors; pH 7.5). Protein concentration was measured by the
Bradford method. Proteins were separated by 10% sodium dodecyl
sulfate-polyacrylamide electrophoresis (SDS-PAGE) and transferred
to nitrocellulose membranes.
Membranes were blocked with 5% bovine serum albumin
(BSA) in TBST (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween-20).
The membrane was incubated in 5% BSA in TBST containing Spred-2
antibody (1:1000). Membranes were then washed extensively with TBST
and then incubated with an appropriate secondary horseradish
peroxidase-labeled antibody (Santa Cruz Biotechnology, Santa Cruz,
CA, USA) at a 1:4,000 dilution. Bands were visualized by enhanced
chemiluminescence (Amersham Biosciences, Piscataway, NJ, USA).
Statistical analysis
Statistical analysis was performed using the t-test.
Differences between the groups were considered statistically
significant when the p-value was <0.05.
Results
Differences in miRNA expression among the
asthmatic and control-group children
The 6 asthmatic children had visited the hospital in
the past 12 months due to wheezing without evidence of common cold,
and their FEV1 values after inhalation of the β2 agonist were 12%
more than those pre-inhalation. The results of skin-prick tests
were positive for all of the asthmatic children. The control-group
children did not have a history of wheezing or chest tightness.
The microarray was used to detect the differences in
the miRNA expression levels between these 2 groups. The expression
levels of 36 miRNAs were significantly higher (more than two-fold)
in the asthmatic children (n=6) than in the control-group children
(p<0.05). In addition, the expression levels of 47 miRNAs were
significantly lower (more than two-fold) in the asthmatic children
than in the control-group children (p<0.05; Table I).
 | Table IDifferences in miRNA expression among
the asthmatic and control-group children. |
Table I
Differences in miRNA expression among
the asthmatic and control-group children.
| Upregulation | Downregulation |
|---|
|
|
|---|
| miRNA | Fold | miRNA | Fold | miRNA | Fold |
|---|
| hsa-miR-22 | 2 | hsa-miR-126 | 0.43 | hsa-let-7g | 0.48 |
| hsa-miR-106b | 2.2 | hsa-miR-140-5p | 0.12 | hsa-miR-3614-3p | 0.17 |
| hsa-miR-320a | 2.4 | hsa-let-7i | 0.47 | hsa-miR-23b | 0.38 |
| hsa-miR-615-3p | 2.4 | hsa-miR-142-3p | 0.27 | hsa-miR-3926 | 0.19 |
| hsa-miR-891a | 2 | hsa-miR-148a | 0.28 | hsa-miR-20a | 0.14 |
| hsa-miR-877 | 2.7 | hsa-miR-182 | 0.37 | hsa-miR-33a | 0.2 |
| hsa-miR-937 | 4.4 | hsa-miR-193a-3p | 0.44 | hsa-let-7d | 0.46 |
| hsa-miR-196a | 3 | hsa-miR-29b | 0.29 | hsa-miR-30a | 0.36 |
| hsa-miR-492 | 2.9 | hsa-miR-3607-5p | 0.23 | hsa-let-7a | 0.23 |
| hsa-miR-485-3p | 2.2 | hsa-miR-335 | 0.11 | hsa-miR-27b | 0.22 |
| hsa-miR-640 | 2.9 | hsa-miR-98 | 0.23 | hsa-miR-4284 | 0.13 |
| hsa-miR-675 | 2.6 | hsa-miR-96 | 0.45 | | |
| hsa-miR-554 | 5.4 | hsa-miR-195 | 0.1 | | |
| hsa-let-7b | 2.7 | hsa-miR-143 | 0.37 | | |
| hsa-miR-551b | 2.9 | hsa-miR-660 | 0.47 | | |
| hsa-miR-320c | 2.5 | hsa-miR-532-5p | 0.45 | | |
| hsa-miR-513b | 3.2 | hsa-miR-192 | 0.33 | | |
| hsa-miR-320d | 2.2 | hsa-miR-4301 | 0.12 | | |
| hsa-miR-605 | 3.7 | hsa-miR-362-3p | 0.21 | | |
| hsa-miR-523 | 2.2 | hsa-miR-15b | 0.45 | | |
| hsa-miR-665 | 4.4 | hsa-miR-744 | 0.41 | | |
| hsa-miR-1260 | 2.2 | hsa-miR-15a | 0.3 | | |
| hsa-miR-3202 | 2.1 | hsa-miR-30e | 0.27 | | |
| hsa-miR-224 | 5.6 | hsa-miR-7 | 0.2 | | |
| hsa-miR-221 | 2.3 | hsa-miR-199a-5p | 0.43 | | |
| hsa-miR-4288 | 2.1 | hsa-miR-374a | 0.33 | | |
| hsa-miR-4300 | 2 | hsa-miR-125b | 0.32 | | |
| hsa-miR-491-3p | 3.4 | hsa-miR-126 | 0.45 | | |
| hsa-miR-4306 | 2 | hsa-miR-576-5p | 0.36 | | |
| hsa-miR-4268 | 3.1 | hsa-miR-324-5p | 0.47 | | |
| hsa-miR-3171 | 2.6 | hsa-miR-20b | 0.32 | | |
| hsa-miR-1246 | 2.4 | hsa-miR-20a | 0.28 | | |
| hsa-miR-620 | 2 | hsa-miR-451 | 0.16 | | |
| hsa-miR-938 | 3 | hsa-miR-138-1 | 0.35 | | |
| hsa-miR-1280 | 2.3 | hsa-miR-424 | 0.19 | | |
| hsa-miR-483-3p | 4.9 |
hsa-miRPlus-I874 | 0.41 | | |
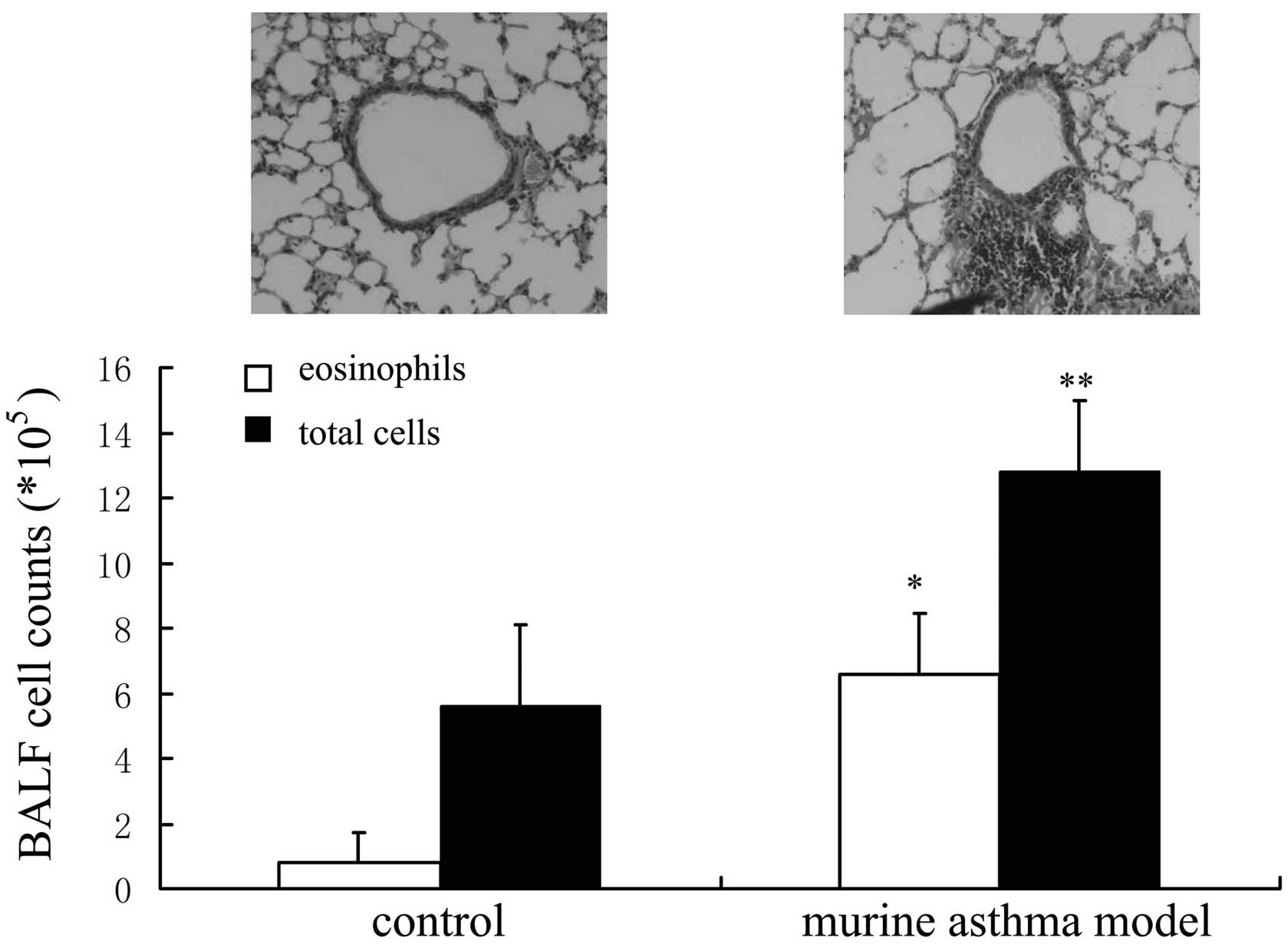
OVA-induced murine asthma models
The inflammation of airways and the infiltration of
leukocytes to lung tissue are the two major features of asthma. The
total cell and eosinophil counts were higher in OVA-induced murine
asthma models compared with controls. The infiltration of
leukocytes to lung tissue was observed in OVA-induced murine asthma
models (Fig. 1).
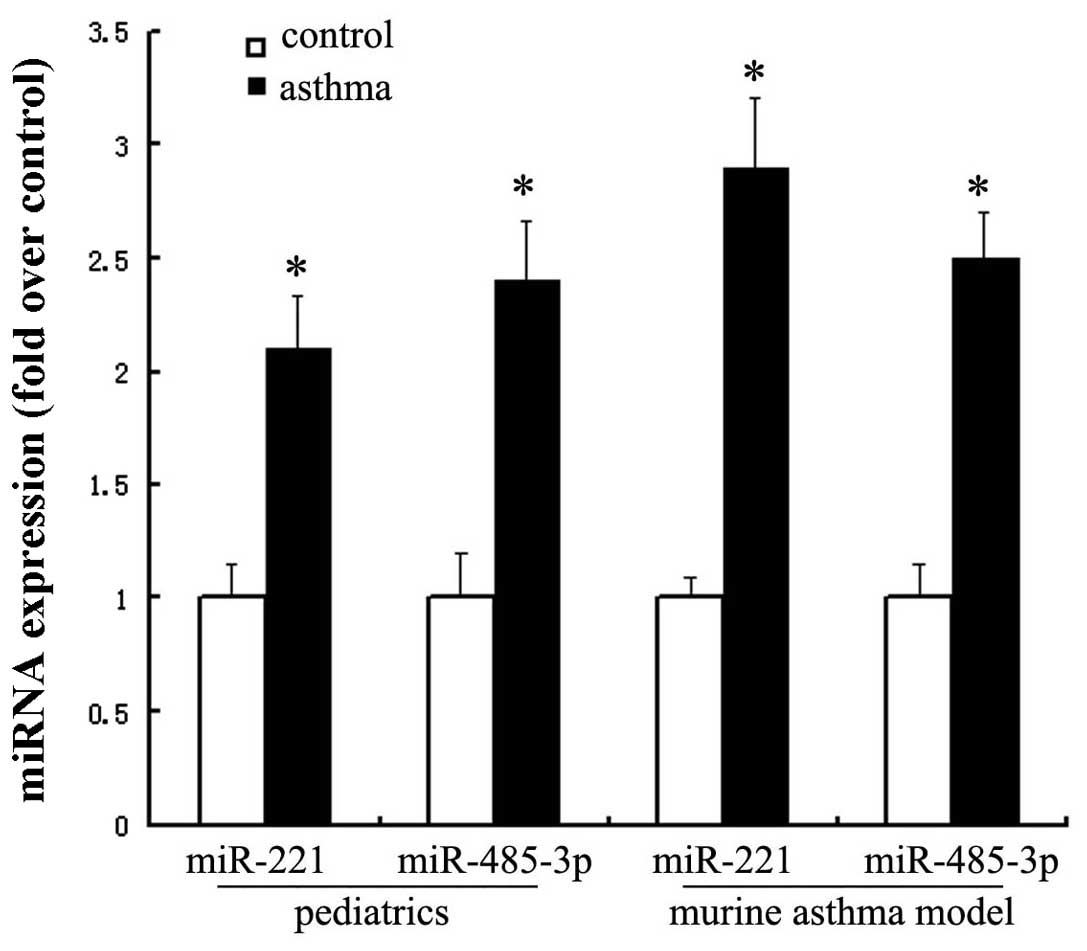
Upregulation of miRNA-221 and
miRNA-485-3p in pediatric asthmatics and murine asthma models
Real-time PCR was used to confirm the differential
expression of miRNA-221 and miRNA-485-3p in pediatric asthmatics
and murine asthma models. miRNA-221 was upregulated approximately
two-fold in pediatric asthmatics and approximately three-fold in
murine asthma models. miRNA-485-3p was upregulated approximately
2.5-fold in pediatric asthmatics and murine asthma models (Fig. 2).
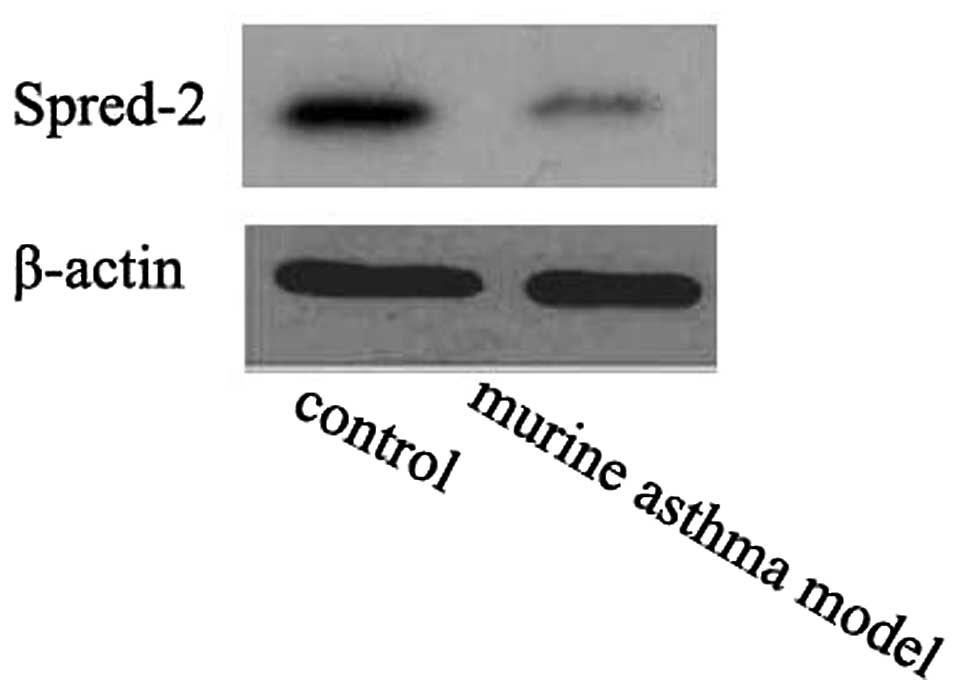
Spred-2 protein level was downregulated
in murine asthma models
The miRNA-221 and miRNA-485-3p targets were analyzed
using the public database TargetScan 6.0 (http://www.targetscan.org). Spred-2 was the predicted
target of miRNA-221 and miRNA-485-3p. The Spred-2 protein level was
downregulated in murine asthma models compared to the controls
(Fig. 3).
Discussion
In this study, 36 miRNAs were significantly
upregulated and 47 significantly downregulated two-fold in the
asthmatic group, compared to the respective miRNAs in the control
group. A previous miRNA array analysis showed that miRNAs are not
involved in the development of allergic asthma in adults (13). However, an miRNA array analysis in
murine models of acute and chronic asthma revealed that miRNA
expression in the lung changes on exposure to allergens and that
several miRNAs regulate biological processes during the course of
asthma development (12).
Functional studies of miRNA in asthma also proved
the critical role of miRNAs in asthma. Mice deficient in miRNA-155
show increased airway remodeling (10). Downregulation of miRNA-133a in a
mouse model of allergic bronchial asthma has been shown to
upregulate RhoA, resulting in increased airway contraction
(11). In vivo inhibition
of let-7 miRNAs inhibits the production of allergic cytokines and
the development of the asthmatic phenotype (16). miRNA-26a is capable of regulating
the hypertrophy of human airway smooth muscle cells by modulating
the levels of glycogen synthase kinase-3β (17). Selective in vivo blockade of
miRNA-126 suppresses the asthmatic phenotype (18). All these observations from previous
studies prove that miRNAs play a critical role in the development
of allergic asthma.
Spred-2 was downregulated in murine asthma models.
Spred has been identified as a negative regulator of growth
factor-mediated, Ras-dependent ERK activation (14). Spred negatively regulates
allergen-induced airway inflammation and hyperresponsiveness in
murine asthma models by modulating IL-5 signaling (14). Among the upregulated miRNAs in
pediatric asthma, miRNA-221 and miRNA-485-3p were predicted to bind
to Spred-2 mRNA by bioinformatic analysis.
Upregulation of miRNA-221 and miRNA-485-3p in
pediatric asthmatics and murine asthma models were verified by
real-time PCR. miRNA-221 favored mast cell adhesion and migration
towards stem cell factor or antigen in Transwell migration assays,
as well as cytokine production and degranulation in response to
IgE-antigen complexes (19).
miRNA-221 regulated cell cycle checkpoints in mast cells in
response to acute activation stimuli (20). These results indicated that
miRNA-221 may contribute to mast cell-related pathological
conditions, such as asthma.
In conclusion, we applied miRNA microarray technique
for screening the differential expression of miRNAs in asthmatic
children. miRNA-221 and miRNA-485-3p were upregulated in pediatric
asthmatics and murine asthma models. Spred-2, the predicted target
of miRNA-221 and miRNA-485-3p, was downregulated in murine asthma
models. miRNA-221 and miRNA-485-3p may regulate the pathogenesis of
asthma.
Acknowledgements
This project was supported by grants (QYK09163,
ZKX11012) from the Key Project supported by the Medical Science and
Technology Development Foundation, Nanjing Department of
Health.
References
|
1
|
Galli SJ, Tsai M and Piliponsky AM: The
development of allergic inflammation. Nature. 454:445–454. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tang ML and Powell CV: Childhood asthma as
an allergic disease: rationale for the development of future
treatment. Eur J Pediatr. 160:696–704. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Masoli M, Fabian D, Holt S and Beasley R:
The global burden of asthma: executive summary of the GINA
Dissemination Committee report. Allergy. 59:469–478. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Braman SS: The global burden of asthma.
Chest. 130:4S–12S. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Phelan PD, Robertson CF and Olinsky A: The
Melbourne Asthma Study: 1964–1999. J Allergy Clin Immunol.
109:189–194. 2002.
|
|
6
|
Szefler SJ: Facing the challenges of
childhood asthma: what changes are necessary? J Allergy Clin
Immunol. 115:685–688. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
MacDonald C, Sternberg A and Hunter PR: A
systematic review and meta-analysis of interventions used to reduce
exposure to house dust and their effect on the development and
severity of asthma. Environ Health Perspect. 115:1691–1695. 2007.
View Article : Google Scholar
|
|
8
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rodriguez A, Vigorito E, Clare S, et al:
Requirement of bic/microRNA-155 for normal immune function.
Science. 316:608–611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chiba Y and Misawa M: MicroRNAs and their
therapeutic potential for human diseases: MiR-133a and bronchial
smooth muscle hyperresponsiveness in asthma. J Pharmacol Sci.
114:264–268. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garbacki N, Di Valentin E, Huynh-Thu VA,
et al: MicroRNAs profiling in murine models of acute and chronic
asthma: a relationship with mRNAs targets. PLoS One. 6:e165092011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Williams AE, Larner-Svensson H, Perry MM,
et al: MicroRNA expression profiling in mild asthmatic human
airways and effect of corticosteroid therapy. PLoS One.
4:e58892009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Inoue H, Kato R, Fukuyama S, et al:
Spred-1 negatively regulates allergen-induced airway eosinophilia
and hyperresponsiveness. J Exp Med. 201:73–82. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mayoral RJ, Pipkin ME, Pachkov M, van
Nimwegen E, Rao A and Monticelli S: MicroRNA-221–222 regulate the
cell cycle in mast cells. J Immunol. 182:433–445. 2009.
|
|
16
|
Polikepahad S, Knight JM, Naghavi AO, et
al: Proinflammatory role for let-7 microRNAS in experimental
asthma. J Biol Chem. 285:30139–30149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mohamed JS, Lopez MA and Boriek AM:
Mechanical stretch up-regulates microRNA-26a and induces human
airway smooth muscle hypertrophy by suppressing glycogen synthase
kinase-3beta. J Biol Chem. 285:29336–29347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mattes J, Collison A, Plank M, Phipps S
and Foster PS: Antagonism of microRNA-126 suppresses the effector
function of TH2 cells and the development of allergic airways
disease. Proc Natl Acad Sci USA. 106:18704–18709. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mayoral RJ, Deho L, Rusca N, et al:
MiR-221 influences effector functions and actin cytoskeleton in
mast cells. PLoS One. 6:e261332011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chun-Zhi Z, Lei H, An-Ling Z, et al:
MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|