1. Introduction
The development of the bacterial artificial
chromosome (BAC) system was partly developed through the Human
Genome Project with a view to construct genomic DNA libraries and
physical maps for genomic sequencing (1). The presence of BAC clones has become
a valuable tool for identifying genomic imbalances in pregnancies
to detect chromosomal abnormalities in at-risk fetuses.
The use of this technology has increased successful
detection of risk-related abnormalities and provided an alternative
for an enhanced level of screening for chromosomal abnormalities in
high-risk pregnancies (2).
Microarray-detected chromosomal abnormality rates are estimated to
range between 5 and 17% in prenatal diagnosis, compared to normal
results obtained from previous routine cytogenetic testing
(3).
The implementation of array comparative genomic
hybridization (CGH) in postnatal diagnosis has been thoroughly
evaluated in the adolescent and adult population, and is now
recommended as the first-line diagnostic test for clinically
suspected genetic disorders (4,5).
However, there are no available concise guidelines establishing the
chromosomal microarray analysis (CMA) applications and platforms
for a prenatal setting. The controversial question concerns whether
or not CMA technology is likely to or should replace the standard
karyotype in prenatal diagnostic practice and whether karyotyping
and fluorescent in situ hybridization (FISH) remain
essential.
In this article, we reviewed the current literature
regarding array genomic hybridization in prenatal diagnosis and
discussed the benefits and issues regarding the use of microarrays
for the prenatal diagnosis of genetic diseases.
2. Conventional cytogenetic analysis
Cytogenetic analysis has provided fundamental
insight into the molecular pathogenesis of prenatal diagnosis and
has been a useful diagnostic tool for the identification of
chromosomal abnormalities in at-risk pregnancies (3). Cytogenetic methods including
karyotyping, FISH, CGH and multiplex-FISH or spectral karyotyping
(SKY) have previously provided valuable diagnostic and prognostic
information for the detection of genomic defects in prenatal
diagnosis (6,7). Since the development of chromosome
banding techniques in the late 1960s, microscopic analysis has been
used as the gold standard for prenatal diagnosis, while in
situ hybridization methods have proven to be a useful and
reliable technique for identifying and characterizing genomic
imbalances. However, these conventional methods have technical
limitations, thus resulting in the underestimation of the degree of
chromosomal changes. In addition, these methods are also limited by
their ability to detect individual DNA screening targets only
rather than the entire genome. Furthermore, hidden mosaics,
patients with uniparental disomy and complex patterns of meiotic
crossing over led to chromosomal aberrations, none of which could
be detected by standard cytogenetics or comparative CGH methods. In
order to detect such abnormalities, a high-resolution technique is
required.
3. Chromosomal microarray analysis
CMA circumvents the limitations of conventional
cytogenetic techniques. It simultaneously evaluates regions across
the entire genome with a higher resolution and an excellent
throughput in patients with suspected genome imbalance. This method
accurately identifies novel genomic aberrations of possibly
uncertain clinical significance not described previously and
provides readily interpretable results, suitable for clinical risk
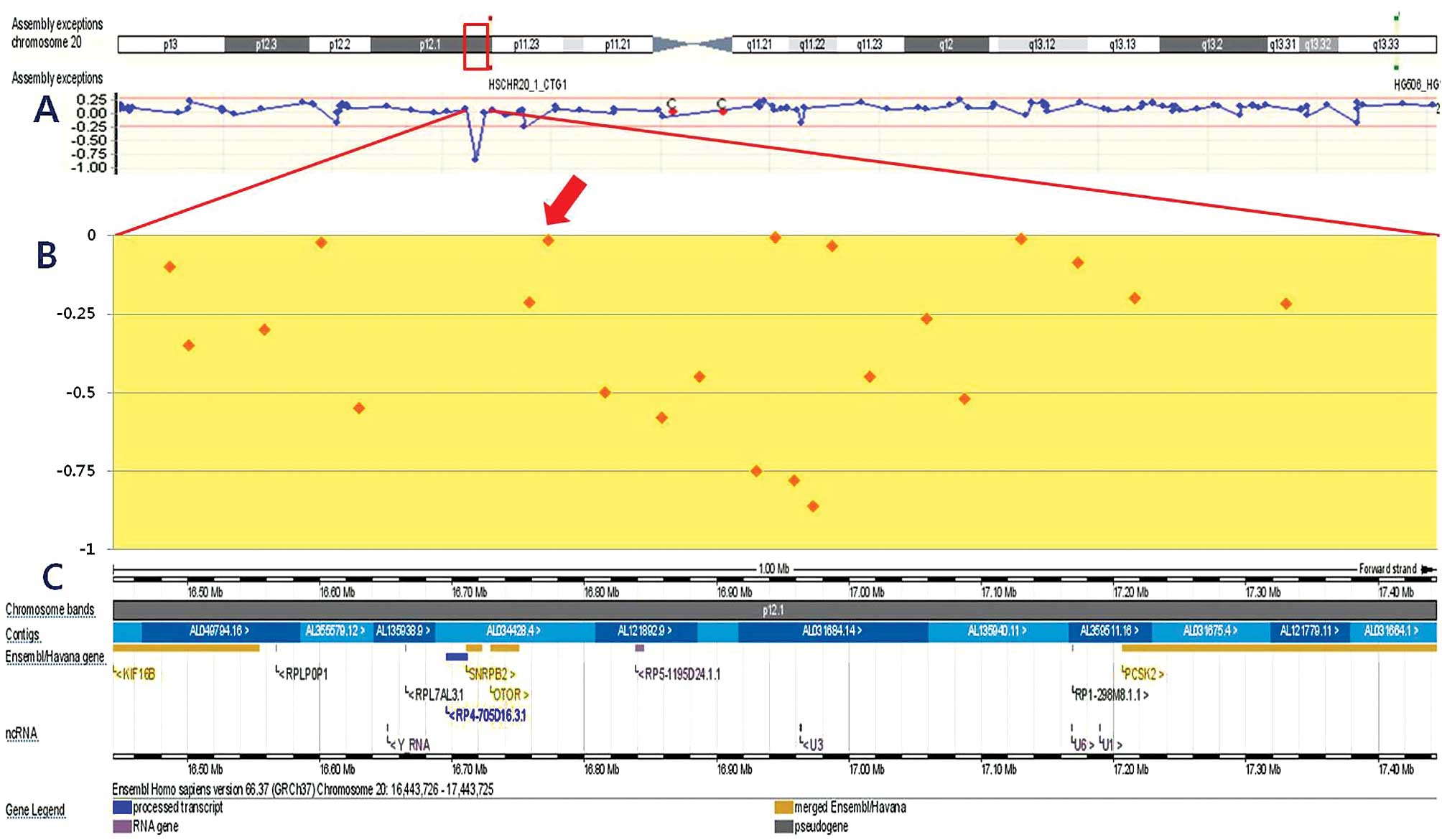
stratification and treatment planning (8) (Fig.
1).
The principle behind the array CGH technology is the
detection of chromosomal deletions and duplications by comparing
equal amounts of genomic DNA from a patient and a healthy control
(9). In the array CGH, the two
genomes (patient and control) are labeled and co-hybridized onto a
glass microscope slide, on which cloned DNA segments have been
immobilized (10). The analytic
principle involves competition between a differentially-labeled
fragmented test and a control diploid DNA, with imbalances due to
copy number changes in the test DNA resulting in a shift in the
fluorescence spectra (11).
The evaluation is performed by a scanner and the
information is then computer-integrated to determine any
quantitative deviations in the DNA of the test sample. The primary
advantage of CMA is the enhanced detection of copy number
anomalies: the deviations that are measurable by molecular means
are orders of magnitude smaller than those detectable by light
microscopy (12). Common protocols
for the application and interpretation of genomic arrays in
prenatal diagnosis are capable of decreasing the risk of unexpected
findings.
4. Clinical utility of CMA in prenatal
diagnosis
Array technology is rapidly taking over cytogenetic
laboratories, resulting in ability for greatly improved
visualization and validation. The increased diagnostic potential of
the microarray has naturally led to the need for its use in the
prenatal setting. In recent years, the application of
microarray-based genomic copy number analysis has proven to be
beneficial, allowing for proper counseling and providing the
parents with all the tools for a conscious decision regarding their
pregnancy.
There are several studies available aiming to assess
the diagnostic ability of array CGH in the screening of hidden
chromosomal aberrations in prenatal genetic diseases with an
apparently normal karyotype (13–15).
Depending on the ascertainment criteria and the level of resolution
achieved in the cytogenetic assessment, the microarray prevails in
the detection of copy number anomalies, by identifying pathogenic
abnormalities in up to 16% of fetuses with an abnormal ultrasound
and normal karyotype (13).
In the study by Vialard et al (14), array CGH diagnosed two de
novo unbalanced karyotypes and four additional abnormalities
that could not be identified with conventional cytogenetic methods
in classic microdeletion syndromes and subtelomeric rearrangements
in 39 fetuses with multiple congenital abnormalities after the
pregnancy was terminated. A previous analysis of eight prenatal
studies using the array technology from various platforms also
concluded that array CGH detected a 3.6% increase in genomic
imbalances when conventional karyotyping was normal, regardless of
the indication for referral. When the referral indication was
abnormal in the ultrasound, this percentage increased to 5.2%
(13).
More recently, a Canadian study using array CGH, has
demonstrated the identification of an additional 8.2% of positive
diagnosis in 49 fetuses with major malformations that were not
visible with karyotyping (12). In
the experiment of Le Caignec et al, (16) the array platform detected all
cytogenetic abnormalities previously analyzed by G banding and
revealed new rearrangements in 7–10% of the cases from chorionic
villus culture in 41 products of conception. Emerging data from
D’Amours et al (17) also
reported a relatively high percentage of findings of unclear
clinical significance in 12.2% of the tested fetuses. These
observations demonstrated that the potential of the array CGH to
reveal the cryptic and/or complex nature of chromosome arrangements
otherwise undetectable by chromosome analysis markedly increases
the elucidation of prenatal genetic diseases.
Additional cases have also emphasized the importance
of further investigation on microarray technology since other
imbalances underlying more serious consequences may be present.
Maitz et al (18) reported
a characteristic case concerning a 21-week gestation fetus with a
complex congenital heart defect and no other ultrasound
abnormalities. The karyotype carried out by conventional
cytogenetic analysis was normal. FISH analysis by the Di
George/VCFS probe, combined with a control probe mapping to the
22q13.3 region (ARSA) was performed, excluding the 22q11.2 deletion
and showing only one signal from the ARSA locus. Microarray
analysis demonstrated that a 6.5 Mb interstitial deletion was in
fact present at 22q13.3, leading to hemizygosity in several genes
(19). Findings of a similar study
by Wat et al (20) also
demonstrated that the high frequency of cardiac and diaphragmatic
defects associated with 8p23.1 interstitial deletions that were
detected by microarray analysis were not identified by conventional
chromosome analysis. These findings prove that in isolated
ultrasound heart abnormalities and a normal karyotype, FISH
analysis is not adequate, and should therefore be substituted by
microarray analysis.
Considering the advantages and the lack of
additional risk of array CGH for the patients, it is reasonable to
suggest that this test be offered to all women already undergoing
invasive testing (21). In the
study conducted by Van den Veyver et al (2), only 4 (22%) of 18 abnormal prenatal
array CGH cases had abnormal ultrasound findings as the sole
indication for testing, suggesting that testing should not be
limited to pregnancies with known abnormal ultrasound findings. Wat
et al (20) also suggested
that array CGH be performed on all prenatal cases with congenital
cardiac and/or diaphragm defects. When offered to choose between
karyotyping and array CGH, 74% of the parents chose the latter
method (21).
Another crucial instance requiring the application
of microarray is the presence of a de novo reciprocal
translocation or a de novo supernumerary marker chromosome
in the fetal karyotype (22).
Previous studies (23–25) demonstrated that cryptic deletions
are present either at the translocation breakpoints or elsewhere in
the genome in approximately 40% of the de novo reciprocal
translocations detected in patients with phenotypic abnormalities,
explaining the phenotype-genotype correlation. Since the
breakpoints of the great majority of reciprocal translocations are
non-recurrent, it is obvious that only the array platforms covering
most of the genome have the potential to detect deletions
associated with reciprocal translocations.
A high-resolution array platform covering the entire
genome would therefore provide much more informative results than
one containing only low coverage limited to prenatal
disease-associated regions. Although microarray technology does not
have the potential to detect polyploidy and balanced chromosomal
rearrangements, these are relatively infrequent causes of abnormal
phenotypes in a typical referral population. The frequency of
pathogenic de novo reciprocal translocations due to the
breakage of a dosage-sensitive gene or its long-range controlling
region is extremely low (22), and
polyploidy is almost always lethal during fetal life and is
generally detected on ultrasound investigation (26). In case of such a suspicion,
conventional karyotyping detects balanced chromosomal
rearrangements. The majority of truly balanced translocations
generate no phenotypic abnormality (27) and their identification leads to
difficult clinical decisions during pregnancy. The American College
of Obstetricians and Gynecologists (ACOG) (28) suggested that conventional
karyotyping remains the principal tool for prenatal diagnosis and
targeted arrays be offered as an adjunct in cases with abnormal
prenatal anatomical findings and a normal conventional karyotype
(21).
Given the potential described in this review, we
anticipate the array CGH to be the initial prenatal diagnostic
approach for the identification of chromosomal abnormalities in the
near future. Although clinical application of array CGH as a
universal routine test for genetic diagnosis is premature, further
investigation may allow for an evaluation of the overall diagnostic
yield of microarray technology over routine prenatal testing with
conventional karyotype, as well as cost effectiveness (29).
5. Conclusions and final considerations
In this review, we presented the potential utility
of array CGH for the detection of chromosomal abnormalities in
prenatal diagnosis. This new platform, with its potential to
decrease the risk of unexpected findings, is likely to be the
first-line test for detecting chromosomal abnormalities in prenatal
diagnosis.
In order to reach a consensus regarding the optimum
configuration of an array, additional investigations carried out on
large-scale populations that have undergone both karyotyping and a
commercially reproducible array are required.
Acknowledgements
This study was financed by the research fund of the
Korea Nazarene University in 2012.
References
|
1
|
De Braekeleer E, Douet-Guilbert N, Basinko
A, et al: Using bacterial artificial chromosomes in leukemia
research: the experience at the university cytogenetics laboratory
in Brest, France. J Biomed Biotechnol. 3294712011.
|
|
2
|
Van den Veyver IB, Patel A, Shaw CA, et
al: Clinical use of array comparative genomic hybridization (aCGH)
for prenatal diagnosis in 300 cases. Prenat Diagn. 29:29–39.
2009.PubMed/NCBI
|
|
3
|
Lee CN, Lin SY, Lin CH, Shih JC, Lin TH
and Su YN: Clinical utility of array comparative genomic
hybridisation for prenatal diagnosis: a cohort study of 3171
pregnancies. BJOG. Feb 07–2012.(E-pub ahead of print).
|
|
4
|
Shen Y, Dies KA, Holm IA, et al: Clinical
genetic testing for patients with autism spectrum disorders.
Pediatrics. 125:727–735. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miller DT, Adam MP, Aradhya S, et al:
Consensus statement: Chromosomal microarray is a first-tier
clinical diagnostic test for individuals with developmental
disabilities or congenital anomalies. Am J Hum Genet. 86:749–764.
2010. View Article : Google Scholar
|
|
6
|
Kang JU, Koo SH, Kwon KC, Park JW, Shin
SY, Kim JM and Jung SS: High frequency of genetic alterations in
non-small cell lung cancer detected by multi-target fluorescence in
situ hybridization. J Korean Med Sci. 22:S47–S51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kang JU, Koo SH, Jeong TE, Kwon KC, Park
JW and Jeon CH: Multitarget fluorescence in situ hybridization and
melanoma antigen genes analysis in primary bladder carcinoma.
Cancer Genet Cytogenet. 164:32–38. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang JU, Koo SH, Kwon KC and Park JW:
Frequent silence of chromosome 9p, homozygous DOCK8,
DMRT1 and DMRT3 deletion at 9p24.3 in squamous cell
carcinoma of the lung. Int J Oncol. 37:327–335. 2010.PubMed/NCBI
|
|
9
|
Kang JU, Koo SH, Kwon KC, Park JW and Kim
JM: Identification of novel candidate target genes, including
EPHB3, MASP1 and SST at 3q26.2-q29 in squamous cell carcinoma of
the lung. BMC Cancer. 9:2372009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shaffer LG and Bejjani BA: Medical
applications of array CGH and the transformation of clinical
cytogenetics. Cytogenet Genome Res. 115:303–309. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maciejewski JP and Mufti GJ: Whole genome
scanning as a cytogenetic tool in hematologic malignancies. Blood.
15:965–974. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Duncan A and Langlois S: SOGC Genetics
Committee: CCMG Prenatal Diagnosis Committee. Use of array genomic
hybridization technology in prenatal diagnosis in Canada. J Obstet
Gynaecol Can. 33:1256–1259. 2011.PubMed/NCBI
|
|
13
|
de Ravel TJ, Devriendt K, Fryns JP and
Vermeesch JR: What’s new in karyotyping? The move towards array
comparative genomic hybridisation (CGH). Eur J Pediatr.
166:637–643. 2007.
|
|
14
|
Vialard F, Molina Gomes D, Leroy B, et al:
Array comparative genomic hybridization in prenatal diagnosis:
another experience. Fetal Diagn Ther. 25:277–284. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hillman SC, Pretlove S, Coomarasamy A, et
al: Additional information from array comparative genomic
hybridization technology over conventional karyotyping in prenatal
diagnosis: a systematic review and meta-analysis. Ultrasound Obstet
Gynecol. 37:6–14. 2011. View
Article : Google Scholar
|
|
16
|
Le Caignec C, Boceno M, Saugier-Veber P,
Jacquemont S, Joubert M, David A, Frebourg T and Rival JM:
Detection of genomic imbalances by array based comparative genomic
hybridisation in fetuses with multiple malformations. J Med Genet.
42:121–128. 2005.PubMed/NCBI
|
|
17
|
D’Amours G, Kibar Z, Mathonnet G, Fetni R,
Tihy F, Désilets V, Nizard S, Michaud JL and Lemyre E: Whole-genome
array CGH identifies pathogenic copy number variations in fetuses
with major malformations and a normal karyotype. Clin Genet.
81:128–141. 2012.PubMed/NCBI
|
|
18
|
Maitz S, Gentilin B, Colli AM, et al:
Expanding the phenotype of 22q13.3 deletion: report of a case
detected prenatally. Prenat Diagn. 28:978–980. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zuffardi O, Vetro A, Brady P and Vermeesch
J: Array technology in prenatal diagnosis. Semin Fetal Neonatal
Med. 16:94–98. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wat MJ, Shchelochkov OA, Holder AM, et al:
Chromosome 8p23.1 deletions as a cause of complex congenital heart
defects and diaphragmatic hernia. Am J Med Genet A. 149A:1661–1677.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lichtenbelt KD, Knoers NV and
Schuring-Blom GH: From karyotyping to array-CGH in prenatal
diagnosis. Cytogenet Genome Res. 135:241–250. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Breman AM, Bi WM and Cheung SW: Prenatal
diagnosis by array-based comparative genomic hybridization in the
clinical laboratory setting. Beijing Da Xue Xue Bao. 18:500–504.
2009.PubMed/NCBI
|
|
23
|
De Gregori M, Ciccone R, Magini P, et al:
Cryptic deletions are a common finding in ‘balanced’ reciprocal and
complex chromosome rearrangements: a study of 59 patients. J Med
Genet. 44:750–762. 2007.
|
|
24
|
Baptista J, Mercer C, Prigmore E, et al:
Breakpoint mapping and array CGH in translocations: comparison of a
phenotypically normal and an abnormal cohort. Am J Hum Genet.
82:927–936. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schluth-Bolard C, Delobel B, Sanlaville D,
et al: Cryptic genomic imbalances in de novo and inherited
apparently balanced chromosomal rearrangements: array CGH study of
47 unrelated cases. Eur J Med Genet. 52:291–296. 2009. View Article : Google Scholar
|
|
26
|
Cain CC, Saul DO, Oehler E, Blakemore K
and Stetten G: Prenatal detection of a subtle unbalanced chromosome
rearrangement by karyotyping, FISH and array comparative genomic
hybridization. Fetal Diagn Ther. 24:286–290. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Warburton D: De novo balanced chromosome
rearrangements and extra marker chromosomes identified at prenatal
diagnosis: clinical significance and distribution of breakpoints.
Am J Hum Genet. 49:995–1013. 1991.
|
|
28
|
ACOG Committee No. 446. Array comparative
genomic hybridization in prenatal diagnosis. Obstet Gynecol.
114:1161–1163. 2009. View Article : Google Scholar
|
|
29
|
Evangelidou P, Sismani C, Ioannides M,
Christodoulou C, Koumbaris G, Kallikas I, Georgiou I, Velissariou V
and Patsalis PC: Clinical application of whole-genome array CGH
during prenatal diagnosis: Study of 25 selected pregnancies with
abnormal ultrasound findings or apparently balanced structural
aberrations. Mol Cytogenet. 3:242010. View Article : Google Scholar
|